Summary
The isolation and characterization of mitochondrial membrane proteins is technically challenging because they natively reside within the specialized environment of the lipid bilayer, an environment that must be recapitulated to some degree during reconstitution to ensure proper folding, stability and function. Here we describe protocols for the assembly of a membrane protein into lipid bilayer nanodiscs in a series of cell-free reactions. Cell-free expression of membrane proteins circumvents problems attendant with in vivo expression such as cytotoxicity, low expression levels and the formation of inclusion bodies. Nanodiscs are artificial membrane systems comprised of discoidal lipid bilayer particles bound by annuli of amphipathic scaffold protein that shield lipid acyl chains from water. They are therefore excellent platforms for membrane protein reconstitution and downstream solution-based biochemical and biophysical analysis. This chapter details the procedures for the reconstitution of a mitochondrial membrane protein into nanodiscs using two different types of approaches: co-translational and post-translational assembly. These strategies are broadly applicable for different mitochondrial membrane proteins. They are also applicable for the use of nanodiscs with distinct lipid compositions that are biomimetic for different mitochondrial membranes and that recapitulate lipid profiles associated with pathological disorders in lipid metabolism.
Keywords: Nanodisc, mitochondria, membrane protein, cell-free translation
1. Introduction
Mitochondria are morphologically complex organelles that are bound by two membranes, each with a distinct lipid and protein composition. The outer membrane (OM) contains both α-helical and β-barrel transmembrane proteins and a membrane protein content of ~50% by mass. By contrast, the highly convoluted inner membrane (IM) contains membrane proteins that are exclusively α-helical, with a remarkable membrane protein content of ~75% by mass (Alder, 2011). Considered another way, among the mitochondrial proteome (~1500 individual proteins in mammals or ~900 individual proteins in yeast), about 25% of mitochondria-resident proteins are associated with or embedded in the OM or IM (Distler et al., 2008; Schmidt et al., 2010). The ability to express, isolate, and functionally characterize membrane proteins is therefore vital to our understanding of mitochondrial physiology. More generally, because membrane proteins constitute roughly one-half of pharmaceutical targets (Sanders and Myers, 2004), their production and functional characterization is critical for drug development. But owing to their hydrophobicity, integral membrane proteins are notoriously difficult to express and purify by traditional experimental approaches, creating a major bottleneck in their biochemical, biophysical and structural analysis. This chapter describes procedures that leverage the technical advantages of cell free biosynthetic systems with advances in the use of nanoscale lipid bilayers for the expression and functional reconstitution of a mitochondrial membrane protein.
Cell-free protein translation systems are based on biosynthetic machinery that is isolated from metabolically active cells with relatively low amounts of endogenous messenger RNA (mRNA). The most commonly used lysates include those from rabbit reticulocytes, from bacterial cells, and from wheat germ embryos. These lysates include the components essential for translation: ribosomes, transfer RNA (tRNA), aminoacyl tRNA synthetases, and translation initiation and elongation and termination factors. In cell-free translation reactions, these lysates are supplemented with mixtures that enhance protein translation, including energy generating systems and amino acids. Some systems entail programming the reaction with plasmid or linear DNA that encodes the protein of interest and from which mRNA transcripts are synthesized by endogenous RNA polymerases. In other systems, purified mRNA transcripts that are prepared during a separate in vitro transcription reaction are added to the cell-free translation.
Cell-free protein synthesis reactions confer several advantages over conventional expression systems (e.g., in live Escherichia coli, yeast, or insect cells), particularly for the expression of membrane proteins. Most notably, because living cells are not involved in the reaction, cell free systems avoid the potential cytotoxicity of expressing hydrophobic proteins, plasmid instability, and issues associated with cellular protein trafficking and membrane integration, all of which can result in low protein yield (Junge et al., 2011; Sachse et al., 2014). Moreover, because cell-free expression systems are open, they allow for unrestricted experimental access to the reaction for the inclusion of additives such as cofactors and probes for site-specific protein labeling (e.g., with fluorescent or EPR probes). Of course, the native milieu of a membrane protein is a lipid bilayer, and several strategies have been utilized that provide additives that mimic the nonpolar membrane environment within cell-free systems for proper folding and stability of translated proteins. By one approach, reactions contain mild nonionic detergents that are compatible with the biosynthetic machinery and allow for the cotranslational stabilization of membrane proteins in proteo-micelle complexes (Junge et al., 2011; Klammt et al., 2005). In addition, synthetic surfactants including organic amphipathic polymers (amphipols), fluorinated surfactants, and designed peptide surfactants have been shown to stabilize membrane proteins produced by cell-free translation (Bazzacco et al., 2012; Blesneac et al., 2012; Corin et al., 2011; Park et al., 2007; Park et al., 2011; Popot et al., 2011; Wang et al., 2011).
Recent advances in model membrane systems have provided new mechanisms for the reconstitution of membrane proteins into a lipidic environment. By comparison with the above hydrophobic additives, such model membranes allow proteins to fold into physiologically relevant lamellar lipid bilayers and can satisfy requirements of specific lipid interactions for membrane protein function (Andersen and Koeppe, 2007; Cross et al., 2013; Lee, 2004). Cell-free reconstitution strategies for membrane proteins have been reported using liposomes, spherical lipid vesicles of different diameter (Hovijitra et al., 2009; Kalmbach et al., 2007; Long et al., 2012; Moritani et al., 2010) and bicelles, discoidal assemblies of lipids with outer edges of short-chain lipid or detergent (Durr et al., 2012; Lyukmanova et al., 2012; Uhlemann et al., 2012). However, nanoscale lipid bilayers belted by rings of amphipathic protein or copolymers have emerged as excellent membrane mimetics for experimentation due to their small size, stability, monodispersity, and wide range of compatible lipids (Malhotra and Alder, 2014). Most notable for cell-free translation systems, nanodiscs – discoidal nanoscale lipid bilayers that are stabilized by annuli of amphipathic polypeptides – allow for the reconstitution of membrane proteins into lamellar bilayer systems that are amenable to solution-based biochemical, biophysical and structural analysis. Developed originally by Sligar and colleagues, nanodiscs comprise bilayers of synthetic or naturally derived lipids enclosed by engineered variants of apolipoprotein A-1, termed membrane scaffold protein (MSP) (Bayburt et al., 2002; Bayburt and Sligar, 2003; Denisov et al., 2004). Nanodiscs have most widely been used for the reconstitution of overexpressed and purified membrane proteins, as recently reviewed (Schuler et al., 2013). However, several groups have recently shown the utility of nanodiscs in the reconstitution of membrane proteins synthesized by cell-free systems (Cappuccio et al., 2008; Henrich et al., 2015; Katzen et al., 2008; Lyukmanova et al., 2012; Proverbio et al., 2013; Roos et al., 2012; Yang et al., 2011). In this regard, multiple reconstitution strategies have been reported, including the cotranslational insertion of membrane proteins into pre-assembled nanodiscs added to the translation reaction and the simultaneous formation of nanodiscs and the target membrane protein.
In this chapter, we describe methods for the cell-free synthesis and nanodisc reconstitution of Tim23, the central subunit of the TIM23 protein transport complex of the mitochondrial inner membrane. The workflow (Figure 1) includes all necessary steps for the setup of the cell-free system, which is based on wheat germ lysates and programmed with purified mRNA transcripts that encode Tim23. We provide procedures for co-translational assembly of Tim23-containing nanodiscs (wherein Tim23 is synthesized in the presence of preformed nanodiscs) as well as for post-translational assembly (wherein pre-synthesized Tim23 is added to nanodisc assembly reactions). These two distinct approaches are provided because different target proteins may be more amenable for reconstitution by one approach over the other.
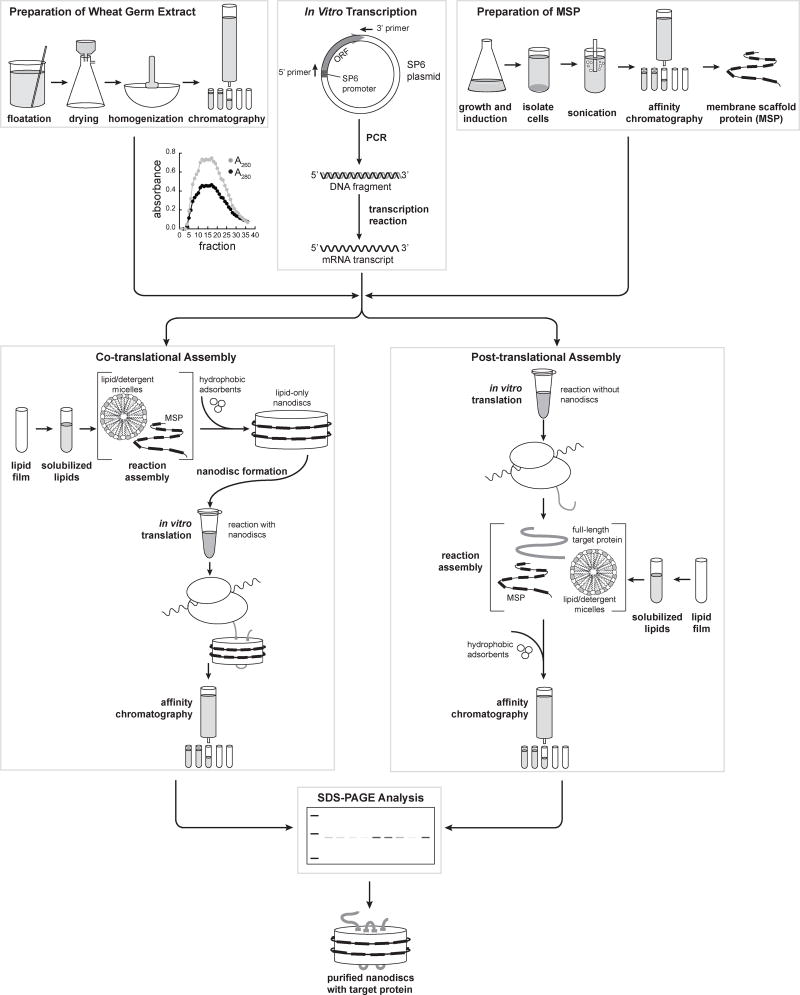
Fig. 1.
Protocol flowchart, including: Preparation of wheat germ extract (Sections 2.1 and 3.1), In vitro transcription (Sections 2.2 and 3.2), Preparation of MSP (Sections 2.3 and 3.3), Co- and post-translational nanodisc assembly (Sections 2.4 and 3.4), and SDS-PAGE analysis (Sections 2.5 and 3.5).
2. Materials
2.1 Preparation of Wheat Germ Extract (see Note 1)
Wheat germ, untreated (Sigma W-0125) (see Note 2).
Mortar and pestle, six inch.
Floatation Solvent: carbon tetrachloride and cyclohexane (600:200, v/v).
Homogenization Buffer: 40 mM HEPES-KOH, pH 7.5, 100 mM KOAc, 1 mM Mg(OAc)2, 2 mM CaCl2, and 4 mM DTT (see Note 3).
Column Buffer: 40 mM HEPES-KOH, pH 7.5, 100 mM KOAc, 5 mM Mg(OAc)2, and 4 mM DTT (see Note 3).
Buchner funnel and Whatman #1 filter paper.
Centrifuge and 50 ml centrifuge tubes.
Column (approximately 2.5 cm I.D. × 28 cm length; bed volume 140 ml) packed with gel filtration medium (Sephadex G-25, fine) and equilibrated with Column Buffer.
Fraction collector
1% (v/v) SDS.
Absorbance spectrophotometer and 1 ml quartz cuvette.
2.2 In Vitro Transcription (see Note 4)
Transcription vector containing the gene of interest with an SP6 promoter (see Note 5).
Primers for DNA amplification of the ORF. The upstream (5′) oligonucleotide is complementary to the plasmid SP6 promoter and the downstream (3′) oligonucleotide is complementary to the gene sequence downstream of the stop codon (see Note 6).
DNA polymerase (Bio-Rad #170-8870) and dNTP mix (Bio-Rad #170-8874).
PCR Cleanup Kit (QIAquick PCR Purification Kit, Qiagen #28106).
PCR thermal cycler and thin-walled PCR tubes (see Note 7).
Refrigerated microcentrifuge.
Transcription Buffer (10× Master Mix): 1M HEPES-KOH, pH 7.5, 200 mM MgCl2, and 25 mM spermidine.
100 mM DTT
Ribonucleotide (rNTP) solution mixture: 100 mM each of ATP, GTP, UTP and CTP in 20 mM Tris-HCl, pH 7.5.
0.1 U/µl diguanosine triphosphate [G(5′)ppp(5′)G] (New England Biolabs #S1407).
20 U/µl Ribonuclease Inhibitor (RNasin).
0.05 U/µl pyrophosphatase
SP6 RNA Polymerase (see Note 8).
3M NaOAc, pH 5.2.
100% Ethanol
TE Buffer (10 mM Tris, pH 7.5, 1 mM EDTA, pH 7.5)
Rotary evaporator
2.3 Preparation of MSP
YT Culture Media: 0.5% (w/v) yeast extract, 0.8% (w/v) tryptone, 80 mM NaCl.
YT-Agar Selective Media: 0.5% (w/v) yeast extract, 0.8% (w/v) tryptone, 80 mM NaCl, 0.75% (w/v) agar, 25 µg/ml kanamycin (see Note 9).
SOC Media: 0.5% (w/v) yeast extract, 2% (w/v) tryptone, 8.5 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 20 mM D-glucose (see Note 10).
20% (w/v) D-glucose, filter sterilized with 0.22 µm filter.
BL21 (DE3) Star competent E. coli cells (Invitrogen).
pET28a:MSP1E3D1 plasmid DNA.
50 mg/ml kanamycin.
1M isopropyl-β-D-thiogalactopyranoside (IPTG) in H2O.
500 mM phenylmethanesulfonylfluoride (PMSF) in 100% ethanol.
10% (v/v) TritonX-100.
Ni-NTA agarose.
Resuspension Buffer: 20 mM Na-phosphate, pH 7.5
Column Wash 1 Buffer: 40 mM Tris-HCl, pH 8.0, 300 mM NaCl, 1% (v/v) TritonX-100.
Column Wash 2 Buffer: 40 mM Tris-HCl, pH 8.0, 300 mM NaCl, 20 mM imidazole, 50 mM Na-cholate.
Column Wash 3 Buffer: 40 mM Tris-HCl, pH 8.0, 300 mM NaCl, 50 mM imidazole.
Column Elution Buffer: 40 mM Tris-HCl, pH 8.0, 300 mM NaCl, 400 mM imidazole.
Dialysis Buffer: 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.5 mM EDTA.
2.4 Assembly of Nanodiscs with Target Protein
Pyrex 50 ml round-bottom centrifuge tube.
Rotary evaporator.
Source of compressed nitrogen gas.
Stocks of synthetic lipids in chloroform, including: a) 1-hexadecanoyl-2-(9Z-octacecenoyl)-sn-glycero-3-phosphocholine (POPC; Avanti #850457C); b) 1-hexadecanoyl-2-(9Z-octacecenoyl)-sn-glycero-3-phosphocholine (POPE; Avanti #850757C); c) 1,1′,2,2′-tetra-(9Z-octadecenoyl) cardiolipin (TOCL; Avanti #710335C) (see Note 11).
ND Buffer A: 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.5 mM EDTA.
ND Buffer B: 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.5 mM EDTA, 59 mM Na-cholate.
ND Column Buffer 1: 40 mM Tris-HCl, pH 8.0, 300 mM NaCl.
ND Column Buffer 2: 40 mM Tris-HCl, pH 8.0, 300 mM NaCl, 400 mM imidazole
ND Storage Buffer: 20 mM HEPES-KOH, pH 7.5, 200 mM NaCl, 50 mM KOAc, 5 mM Mg(OAc)2, 5% (v/v) glycerol.
Laboratory vortex mixer.
Bath sonicator.
15 ml conical tubes
Laboratory rotisserie
BioBeads SM-2 (Bio-Rad) hydrophobic adsorbents.
Column with Ni-NTA agarose matrix.
Amicon Ultra Centrifigalfilter Units (Millipore), 3,000 MWCO, 5 ml capacity.
Translation Buffer (10×): 200 mM HEPES-KOH, pH 7.5, 1 M KOAc, pH 7.5, 25 mM Mg (OAc)2, 2 mM spermidine, 0.08 mM S-adenosylmethionine.
Protease Inhibitor Mixture (200×): 50 µg/ml each of Antipain, Chymostatin, Leupeptin, Pepstatin A and 5% (v/v) of Aprotinin.
Energy Generating System and Amino Acid Mixture (EGS): 90 mM HEPES-KOH, pH 7.5, 15 mM ATP, pH 7.5, 15 mM GTP, pH 7.5, 120 mM creatine phosphate, 0.12 U/µl creatine phosphokinase, 0.38 mM each of 20 common L-amino acids (see Note 12).
Wheat germ extract (prepared as in Step 3.1)
L-[35S]-methionine.
2.5 Gel-Based Analysis of Assembly
Mini-PROTEAN Tetra Cell (Bio-Rad 165-8004).
PowerPac Universal Power Supply (Bio-Rad 164-5070).
Mini-Protean TGX Precast Gel (Bio-Rad 456-1044S)
Electrophoresis running buffer (25 mM Sigma 7–9 buffer, 0.2 M glycine, 0.1 (w/v) sodium dodecyl sulfate, 0.045% (w/v) bromophenol blue, 0.1 M DTT).
SDS-PAGE Sample Buffer (125 mM Tris-Base, 18%(v/v) glycerol, 3.6%(w/v) sodium dodecyl sulfate, 0.045%(w/v) bromophenol blue, 0.1M DTT).
Destain solution (50%(v/v) methanol, 10%(v/v) acetic acid)
Coomassie stain solution (50%(v/v) methanol, 10%(v/v) acetic acid, 0.5%(w/v) Coomassie G-250).
Gel imaging system capable of imaging radiolabeled samples (see Note 13).
Gel drying apparatus (Bio-Rad 165-1789 Hydrotech system including Model 583 gel dryer and vacuum pump).
Imaging screen-K, 20 × 25 cm phosphor imaging screen (Bio-Rad 170-7843) and exposure cassette-K (Bio-Rad 170-7861).
14C-methylated protein molecular weight markers (Perkin-Elmer #NEC81100).
Precision Plus All Blue Prestained Standards (Bio-Rad 161-0373).
3. Methods
3.1 Preparation of Wheat Germ Extract
These procedures are based on protocols modified from Erickson and Blobel (Erickson and Blobel, 1983). Floatation enriches the wheat germ for viable, intact embryos. The extract is then prepared in three steps: homogenization of the floated germ, homogenate centrifugation, and gel filtration of the supernatant.
3.1.1. Floatation of Wheat Germ
Prepare Floatation Solvent in a 1 L beaker in a well-ventilated hood. Stir with a glass rod until no schlieren lines are visible.
Carefully pour 50 g of wheat germ to the top of the solution and gently stir. During the 2–3 minute separation process, damaged embryos will settle and intact, viable embryos will float.
Decant the floating embryos into a Buchner funnel fitted with Whatman #1 filter paper and dry the wheat germ embryos by pulling air through the funnel for 30 min (see Note 14).
Spread the dried wheat germ onto sterile foil to continue drying in fume hood.
Repeat steps 1–4 with another 50 g of wheat germ (see Note 15).
Combine and weigh the dried embryos. The target yield of the floated wheat germ is 30–40% (w/w) (see Note 16).
3.1.2. Preparation of Wheat Germ Extract (see Note 17)
Place dried wheat germ in a pre-chilled 6 in. mortar, cover with liquid N2, and grind wheat germ to a fine powder with a pestle. The total grinding time should be about two minutes (see Note 18). Add the dried powder to a sterile sheet of foil.
Add Homogenization Buffer to the mortar at a sufficient volume to saturate the powder (see Note 19), slowly add powdered wheat germ, and grind until a thick paste is obtained.
Transfer the homogenate into two 50 ml pre-chilled centrifuge tubes using a sterile rubber policemen. Centrifuge at 23,000 × g at 4°C for 10 min.
Transfer the supernatant to fresh 50 ml centrifuge tubes using a sterile Pasteur pipette (see Note 20).
Repeat centrifugation step (23,000 × g at 4°C for 10 min) and transfer the supernatants to a fresh tube. Measure the extract volume.
Perform gel filtration of the extract by adding supernatant to the G-25 column and elute with Column Buffer under gravity flow. Collect eluate in 1 ml fractions (see Note 21).
Measure the absorbance of each fraction at 260 and 280 nm by adding 5 µl fraction to 1 ml of 1% (v/v) SDS in a quartz cuvette with a spectrophotometer blanked with 5 µl of column buffer and 1 ml of 1% (v/v) SDS. The profile should be similar to that shown in Figure 1.
Pool the most turbid fractions (typically 25–30 fractions, depending on the starting amount of wheat germ) into a sterile 50 ml centrifuge tube, aliquot into microfuge tubes, flash freeze in liquid N2, and store at −80°C (see Note 22).
3.2 In Vitro Transcription
The procedure outlined here uses PCR to generate DNA fragments for the translation reaction. Alternatively, one could use plasmid linearized with appropriate restriction endonuclease(s).
3.2.1 PCR Amplification of DNA Fragments
Assemble the PCR reaction mixture (Table 1) in a PCR tube (see Note 23).
Add the reaction to a PCR thermal cycler programmed as follows: first denature (94°C, 4 min); 30 cycles of amplification (denature 94°C, 20 s; anneal 54°C, 20 s; extension 72°C, 30 s); final extension (72°C, 4 min). Optimal temperatures and time lengths will vary for different constructs.
Purify the amplified DNA fragment using the QIAquick PCR Purification Kit and elute with 50 µl of RNase-free H2O (see Note 24).
Table 1.
PCR Protocol
| Component | Volume (µl) | Final Concentration |
|---|---|---|
| Nuclease-free H2O | 80.5 | n/a |
|
| ||
| 10× PCR buffer | 10.0 | 1× |
|
| ||
| 10 mM dNTP mix | 2.0 | 0.2 mM, each dNTP |
|
| ||
| 50 mM MgCl2 | 4.0 | 2 mM |
|
| ||
| 50 µM forward primer | 1.0 | 0.5 µM |
|
| ||
| 50 µM reverse primer | 1.0 | 0.5 µM |
|
| ||
| 100 ng/µl DNA template | 1.0 | 1 ng/µl |
|
| ||
| DNA polymerase (5U/µl) | 0.5 | 0.013 U/µl |
|
| ||
| Total | 100.0 | |
3.2.2. In Vitro Transcription Reaction
Assemble the in vitro transcription mixture (Table 2) in a 1.5 ml microfuge tube and incubate the reaction at 37°C for 1.5 h (see Note 25).
Isolate and purify RNA by adding reaction to 340 µl of 100% ethanol and 13.3 µl of 3M NaOAc (pH 5.2) and precipitating mRNA on ice for 2 h; pelleting samples at maximum speed in a microfuge (4°C) and carefully aspirating supernatant; washing pellet with 1 ml 70% (v/v) ethanol, re-centrifuging and aspirating supernatant; drying pellet in a rotary evaporator for 5 min; and resuspending mRNA pellet in 100 ml TE (pH 7.5) (see Note 26).
Aliquot mRNA in volumes of 25 µl each, flash-freeze in liquid N2 and store at −80°C (see Note 27).
Table 2.
In Vitro Transcription Protocol
| Component | Volume (µl) | Final Concentration |
|---|---|---|
| Nuclease-free H2O | 43.3 | n/a |
|
| ||
| 10× Transcription Buffer | 10.0 | 100 mM HEPES, pH 7.5 |
| - | 20 mM MgCl2 | |
| - | 2.5 mM spermidine | |
|
| ||
| 100 mM DTT | 10.0 | 10 mM |
|
| ||
| 100 mM rNTP mixture | 4.0 | 4 mM, each rNTP |
|
| ||
| 0.1 U/µl (5′)ppp(5′)G | 13.0 | 0.013 U/µl |
|
| ||
| 20 U/µl RNasin | 2.0 | 0.4 U/µl |
|
| ||
| PCR product DNA | 13.0 | * |
|
| ||
| SP6 RNA polymerase | 3.5 | ** |
|
| ||
| 0.5 U/µl Pyrophosphatase | 1.2 | 0.006 U/µl |
|
| ||
| Total | 100.0 | |
variable, depending on efficiency of PCR reaction and purification
variable, depending on source of polymerase
3.3 Preparation of MSP
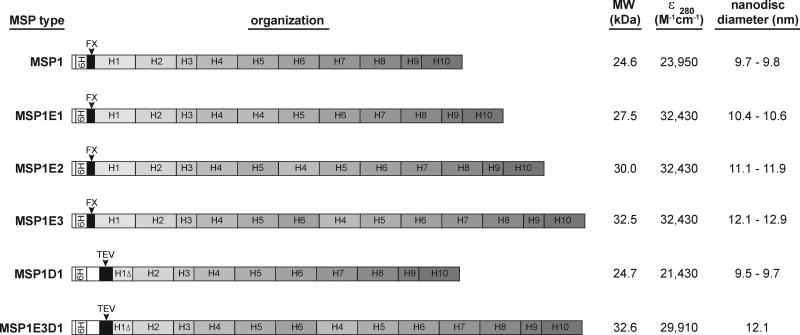
Several variants of MSP have been described (refs). Among them, plasmids comprised of the pET28a vector with the MSP coding sequence are available in the Addgene repository. These contain an N-terminal 6XHis tag followed by specific cleavage sites just upstream of the MSP (Figure 2). This protocol describes the preparation of MSP1E3D1 construct; the expression and purification of other variants is similar.
Fig. 2.
MSP constructs available through the Addgene plasmid repository. The composition of each construct includes N-terminal hexahistidine affinity tags (6H), cleavage sites including Factor X (FX) and tobacco etch virus (TEV) proteolysis sequences, and apo-I derived helices comprising 11 residues (H1Δ, H3 and H9) and 22 residues (H1, H2, H4, H5, H6, H7, H8 and H10). The molecular weights indicated are for the intact, uncleaved polypeptide. The disc diameters are published values based on gel filtration chromatography and small angle x-ray scattering (Denisov et al., 2004).
3.3.1. MSP1E3D1 Expression
Transform pre-chilled BL21 (DE3) Star cells with 25 ng pET28a:MSP1E3D1 plasmid DNA by incubation on ice for 30 min, heat shock at 42°C for 45 s, and recovery at 37°C in 0.95 ml SOC Media for 1 h.
Plate 100 µl transformed cells on kanamycin-selective plates and incubate overnight at 37°C.
Prepare 100 ml growth medium (100 ml YT culture medium, 0.2% (w/v) filter-sterilized glucose, 25 µg/ml kanamycin), divide into several 5 ml aliquots in culture tubes, inoculate each with individual colonies from transformation plate and grow overnight (37°C, 250 rpm).
Prepare 4× 500 ml growth medium (500 ml YT culture medium, 0.2% (w/v) filter-sterilized glucose, 25 µg/ml kanamycin), inoculate each with 1 ml overnight culture and grow (37°C, 250 rpm) to OD600 = 0.8 to 1.0.
Induce expression by reducing growth temperature to 26°C, adding 1 mM IPTG, and continuing incubation for 3 h.
Harvest cells by centrifugation (4,000 × g for 20 min), decant medium, flash freeze pellets in liquid N2 and store at −80°C.
3.3.2. MSP1E3D1 Purification
Thaw pellets from each 500 ml culture on ice and resuspend each pellet in 20 ml Resuspension Buffer with 1 mM PMSF. Add TritonX-100 to a final concentration of 1% (v/v).
Disrupt cells by ultrasonication in an ice bath using a microtip (50% amplitude, 60 s, total, five times). Clarify the lysate by centrifugation (30,000 × g for 30 min) and combine supernatants.
Equilibrate Ni-NTA agarose column (matrix bed 8 ml) with several column volumes (CV) of Column Wash 1 Buffer; load clarified lysate supernatant onto column and wash the column with 5 CV Column Wash 1 Buffer, 5 CV Column Wash 2 Buffer, 5 CV Column Wash 3 Buffer.
Elute MSP1E3D1 with Column Elution Buffer (10× 1 ml fractions). Pool protein-containing fractions, add to dialysis cassette (MWCO 2,000) and dialyze overnight at 4°C in 3 l Dialysis Buffer with one buffer exchange.
Measure MSP concentration by A280 and known molar extinction coefficients (Figure 2). Aliquot stocks into 250 µl, flash freeze in liquid N2 and store in −80°C.
3.4. Assembly of Nanodiscs with Target Protein
3.4.1. Preparation of Solubilized Lipids
Assemble the desired lipid blend by combining chloroform stocks of lipids in a 50 ml round-bottom tube using a Hamilton syringe. To assemble a mitochondrial inner membrane biomimetic lipid combination (40 mol% POPC, 40 mol% POPE, 20 mol% TOCL), add 0.2 ml of POPC, 0.19 ml of POPE, and 0.2 ml of TOCL.
Evaporate the chloroform under a gentle nitrogen stream in a fume hood (see Note 28).
Cover the tube with parafilm with small hole on top and place in vacuum dessicator for a minimum of two hours to remove all traces of organic solvent.
Hydrate the lipid film by adding 0.8 ml of ND Buffer B (the concentration of cholate should be at least twice the concentration of lipid), vortexing lipids into solution, and placing in a bath sonicator for a total of four × one minute cycles until suspension is clear (see Note 29).
3.4.2. Assembly of Lipid-Only Nanodiscs (for Co-Translational Assembly Only)
Prepare a 2 ml lipid-only nanodisc assembly reaction in a 15 ml conical tube by assembling cholate-solubilized lipids (from Section 3.4.1) and MSP (from Section 3.3) at optimized molar ratios and adjusting the volume with ND Buffer A as shown in Table 3. Incubate assembly for 30 min.
Prepare BioBeads by adding 1.5 g of dry beads to a 15 ml conical tube and hydrating with 1.0 ml of ND Buffer A on laboratory rotisserie for 30 min. Carefully remove buffer by pipetting.
Initiate the self-assembly reaction by adding the nanodisc assembly mixture to the conical vial containing pre-hydrated BioBeads. Incubate reaction on laboratory rotisserie for 2 h (see Note 30).
Affinity purify assembled nanodiscs via batch purification with Ni-NTA agarose by adding 1 ml of Ni-NTA beads (equilibrated with ND-Buffer A) to the assembly reaction and incubating for 45 min; adding slurry to chromatography column (approx. 0.8 cm id × 4 cm length); preparing wash and elution buffers (Table 4) and adding: 2 CV of Wash 1 Buffer, 2 CV of Wash 2 Buffer, 2 CV of Wash 3 Buffer, and eluting with Elution Buffer in four-one ml fractions (see Note 31).
Perform nanodisc concentration and buffer exchange by adding elution fractions to 5 ml Millipore Centrifugal unit (3,000 MWCO), pre-equilibrated with ND Storage Buffer. Perform the following four times: centrifuge sample at 3,500 × g for 15–20 min at 4°C, discard eluate, add 3.0 ml ND Storage Buffer to nanodisc sample in filter device. Finally, concentrate the sample to approximately 0.5 ml (see Note 32).
Purified nanodiscs can be used immediately in translation reactions. Alternatively, they may be stored long-term by flash freezing in liquid N2 and storage at −80°C.
Table 3.
Lipid-Only Biomimetic Nanodisc Assembly Reaction
| Component | Volume (µl) | Final Concentration |
|---|---|---|
| 20.5 mM Lipid Blend | 590 | 2.4 mM POPC |
| 2.4 mM POPE | ||
| 1.2 mM TOCL | ||
|
| ||
| 200 µM MSP1E3D1 | 500 | 50 µM |
|
| ||
| ND-Buffer A | 910 | |
|
| ||
| Total | 2000 | |
Table 4.
Nanodisc Affinity Purification Buffers
| Buffer | ND Column Buffer 1 Volume (ml) |
ND Column Buffer 2 Volume (ml) |
|---|---|---|
| Wash 1 (no imidazone) | 25.00 | 0 |
|
| ||
| Wash 2 (20 mM imidazole) | 23.75 | 1.25 |
|
| ||
| Wash 3 (50 mM imidazole) | 21.87 | 3.13 |
|
| ||
| Elution (400 mM imidazole) | 0 | 25.00 |
|
| ||
3.4.3. Co-translational Assembly of Target Protein in Nanodiscs
Assemble Translation Reaction with Nanodiscs (Table 5) in a 1.5 ml microfuge tube and incubate the reaction at 26°C for 40 min (see Note 33).
Centrifuge the reaction mixture at 20,000 × g for 10 min to pellet precipitated protein, and transfer clarified supernatant containing nanodiscs with target protein to a new microfuge tube.
To re-purify and concentrate the [35S]Tim23-containing nanodiscs, the sample can be subjected to a second round of Ni-NTA based chromatography, exactly as detailed in Section 3.4.2, steps 4 and 5 (see Note 34).
Table 5.
Translation Reaction
| With Nanodiscs | No Nanodiscs | ||
|---|---|---|---|
| Component | Volume (µl) | Volume (µl) | Final Concentration |
| Nuclease-free H2O | 146.4 | 146.4 | n/a |
|
| |||
| 10× Translation Buffer | 50 | 50 | 20 mM HEPES, pH 7.5 |
| 100 mM KOAc, pH 7.5 | |||
| 2.5 mM Mg(OAc)2 | |||
| 200 µM spermidine | |||
| 8 µM S-adenosylmethionine | |||
|
| |||
| 100 mM DTT | 5 | 5 | 1 mM |
|
| |||
| 200× Protease Inhibitors | 2.6 | 2.6 | 1× |
|
| |||
| 20 U/µl RNasin | 2.6 | 2.6 | 0.1 U/µl |
|
| |||
| EGS | 40 | 40 | 7.2 mM HEPES, pH 7.5 |
| 1.2 mM ATP, pH 7.5 | |||
| 1.2 mM GTP, pH 7.5 | |||
| 9.6 mM creatine phosphate | |||
| 9.6 U/ml creatine phosphokinase | |||
| 47.5 µM each amino acid | |||
|
| |||
| Wheat Germ Extract | 100 | 100 | 20% (v/v) |
|
| |||
| 10µCi/µl [35S]methionine | 13.4 | 13.4 | 0.25 µCi/µl |
|
| |||
| Tim23 mRNA | 40 | 40 | 8% (v/v) |
|
| |||
| Purified Nanodiscs | 100 | n/a | 20% (v/v) nanodiscs |
|
| |||
| ND Storage Buffer | n/a | 100 | 20% (v/v) buffer |
|
| |||
| Total | 500 | 500 | |
3.4.4. Post-translational Assembly of Target Protein in Nanodiscs
Assemble Translation Reaction without Nanodiscs (Table 5) in a 1.5 ml centrifuge tube and incubate the reaction at 26°C for 40 min (see Note 33).
Prepare a 2 ml target protein-nanodisc assembly reaction in a 15 ml conical tube by assembling cholate-solubilized lipids (from Section 3.4.1) and MSP (from Section 3.3) at optimized molar ratios and adjusting the volume with ND Buffer A as shown in Table 3. Incubate assembly for 30 min.
Prepare BioBeads by adding 1.5 g of dry beads to a 15 ml conical tube and hydrating with 1.0 ml of ND Buffer A on laboratory rotisserie for 30 min. Carefully remove buffer by pipetting.
Initiate the self-assembly reaction by adding the nanodisc assembly mixture to the conical vial containing pre-hydrated BioBeads. Incubate reaction on laboratory rotisserie for 2 h (see Note 30).
Affinity purify assembled nanodiscs via batch purification with Ni-NTA agarose by adding 1 ml of Ni-NTA beads (equilibrated with ND-Buffer A) to the assembly reaction and incubating for 45 min; adding slurry to chromatography column (approx. 0.8 cm id × 4 cm length); preparing wash and elution buffers (Table 4) and adding: 2 CV of Wash 1 Buffer, 2 CV of Wash 2 Buffer, 2 CV of Wash 3 Buffer, and eluting with Elution Buffer in four-one ml fractions (see Note 31).
Perform nanodisc concentration and buffer exchange by adding elution fractions to 5 ml Millipore Centrifugal unit (3,000 MWCO), pre-equilibrated with ND Storage Buffer. Perform the following four times: centrifuge sample at 3,500 × g for 15–20 min at 4°C, discard eluate, add 3.0 ml ND Storage Buffer to nanodisc sample in filter device. Finally, concentrate the sample to approximately 0.5 ml (see Note 32).
3.5 Gel-Based Analysis of Assembly
Set up the electrophoresis apparatus according to the manufacturer’s instructions.
In separate microfuge tubes, pre-mix 5 µl nanodisc purification fractions (flow through, wash steps, elution steps, and concentrated samples) with an equal volume of SDS-PAGE Sample Buffer and heat at 65°C for 10 min prior to gel loading. For gels in radioisotope scans, add 5 µl of radiolabeled molecular weight markers to an equal volume of SDS-PAGE Sample Buffer; for Coomassie gels, add 5 µl of all blue markers to an equal volume of SDS-PAGE Sample Buffer. Quantitatively load all samples onto gel. See Figure 3 for a loading schematic.
Run gels at 125 V until the bromophenol blue dye front reaches the bottom of the separating gel.
Prepare gels used for radioisotope scans by washing in 100 ml of Destain Solution for 10 min followed by two 10 min washes in water. Dry gels for 40 min at 80°C and place on phosphor imaging screen (see Note 35). Perform radioisotope scan on the molecular imager using the appropriate settings.
Prepare gels used for Coomassie stain by washing in 100 ml of Coomassie Stain Solution for 30 min followed by multiple 20 min washes in 100 ml Destain Solution. Continue destain solution until signal from protein bands are sufficiently contrasted with background.
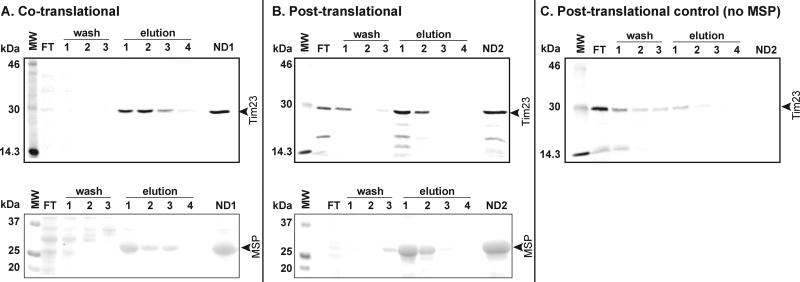
Fig. 3.
Analysis of nanodisc assembly and purification for A) co-translational assembly, B) post-translational assembly, and C) negative control (lacking MSP). Samples were resolved by SDSPAGE on 12.5% gels and visualized by radioisotope scanning (upper gels) to image [35S]Tim23 and by Coomassie staining (lower gels) to detect MSP. Individual flow through, wash, and elution fractions are indicated. ND1 and ND2 denote the final combined samples for co- and post-translational assembly reactions, respectively.
Table 6.
Target Protein-Biomimetic Nanodisc Assembly Reaction
| Component | Volume (µl) | Final Concentration |
|---|---|---|
| 20.5 mM Lipid Blend | 590 | 2.4 mM POPC |
| 2.4 mM POPE | ||
| 1.2 mM TOCL | ||
|
| ||
| 200 µM MSP1E3D1 | 500 | 50 µM |
|
| ||
| [35S]Tim23 translation | 250 | 12.5% (v/v) |
|
| ||
| 200× Protease Inhibitors | 20 | 2× |
|
| ||
| ND-Buffer A | 640 | |
|
| ||
| Total | 2000 | |
Acknowledgments
We thank members of the Alder Research Group for their useful input. This work is supported by grants to NNA from the National Science Foundation (MCB-1330695) and from the National Institutes of Health (RGM113092).
Footnotes
All glassware should be autoclaved and baked overnight to eliminate nucleases. All solutions should be prepared with Milli-Q (or double-distilled) water.
Wheat germ (typically from Triticum aestivum L.) can also be sourced from commercial mills, but note that different strains of wheat will have variable activity and should be tested. Activity is best retained if wheat germ is stored in small batches in a vacuum desiccator at 4°C.
Add DTT from prepared stocks (1M DTT) just prior to use.
As an alternative to the transcription protocol described here, one may use commercially available kits to synthesize mRNA transcripts.
We commonly use plasmids designed for in vitro transcription/translation, such as pSP65 and pGEM4z (Promega) with the open reading frame (ORF) of interest subcloned into the multicloning site. Plasmid stocks are stored at concentrations of about 100 ng/ml in TE Buffer at −20°C.
Primer stocks are stored at concentrations of 50 µM in TE Buffer at −20°C.
We use a Bio-Rad C1000 thermal cycler and low tube strip individual PCR tubes.
SP6 polymerase is available commercially from vendors such as New England Biolabs; however, we prepare our own stocks in-house.
Media, excluding antibiotic, should be sterilized by autoclave (121°C). Kanamycin from 50 mg/ml stock can be added to medium stock once it is cooled.
Media, excluding glucose, should be sterilized by autoclave (121°C). Filter-sterilized glucose can be added to medium stock once it is cooled.
Alternatively, lipids can be purchased as powder stocks and resuspending them in detergent-containing buffer to the desired concentration.
Stocks of L-amino acids can be prepared from analytical grade kits (e.g., Sigma LAA21).
We use a Bio-Rad Pharos FX Plus Molecular Imager with external lasers. Other imaging instrumentation (e.g., the Bio-Rad PMI system) can be used for the detection of radiolabeled samples.
Floated wheat germ will turn white when dry.
The organic solvent mixture can be reused for the second set of wheat germ.
If overnight storage is necessary at this step, the wheat germ can be stored in a plastic beaker covered with parafilm and aluminum foil at −80°C.
All steps should be carried out in a cold room at 4°C.
Grinding the wheat germ in liquid N2 minimizes enzymatic degradation during cell disruption.
As a rule of thumb, the amount of Extraction Buffer should be 2× weight of dried wheat germ. Extra buffer may be added to maintain the consistency of the paste.
There will be a yellowish floating layer of lipid. Take care to avoid transferring this layer into the new tube.
The opaque, brown solution eluting just after the void volume contains the target product. The slower-running yellowish lipid-rich layer is discarded.
If desired, the combined fractions can be normalized to a specific A260 by adjusting concentration with Column Buffer. Aliquot sizes of stored wheat germ extract can vary (e.g., from 50 to 200 µl); however, after thawing lysate, any remaining stock should be discarded and not re-frozen. The activity of lysate stocks will remain high for several months when stored at −80°C.
Reagent concentrations have been optimized for the BioRad iTaq Polymerase system; optimal concentrations of reagents, particularly for MgCl2, should be tested empirically for different enzymes, plasmid templates, and primers.
Purification of PCR products is done in accordance with the manufacturer’s protocol. We typically run PCR DNA on a 1.8% agarose gel alongside DNA standards to confirm successful amplification of the desired product.
To enhance the incorporation of diguanosine triphosphate, add one-tenth the concentration of GTP initially, then supplement the reaction with 4 mM GTP after one hour.
Alternatively, the RNA can be isolated and purified using a commercially available kit such as the QIAgen RNeasy Mini Kit.
mRNA in TE (pH 7.5) can be stored in an ultracold freezer for several months and can withstand multiple freeze/thaw cycles.
Flush-dry the lipids until the first dry residue appears and continue drying for an additional 15 min. Over-drying the lipids at this stage can make resuspension difficult.
During nanodisc preparation steps, make sure that the sample temperature is above the melting temperature (Tm) of the highest melting lipid. This ensures that all lipids are in the liquid crystalline phase and is especially important for long chain, saturated lipids with high Tm.
Alternatively, the 2 ml self-assembly reaction can be done by extensive dialysis against a 2000× volume of ND-Buffer A.
Alternatively, assembled nanodiscs can be purified by size exclusion chromatography (e.g., by fractionation on a Superdex 200 10/300 GL Column equilibrated with ND-Buffer A).
Following nanodisc concentration, it is advisable to determine the final protein concentration (e.g. using a Bradford assay) and/or the final lipid concentration in order to normalize the amount of discs in subsequent reactions.
To enhance the readout of endogenous mRNA prior to addition of the [35S]methionine label, we incubate reactions at 26°C for 5 min prior to mRNA addition.
Alternatively, in order to enrich for only those nanodiscs that contain the target protein, one may perform affinity chromatography against a tag engineered onto the target protein.
Sufficient signal from 35S-labeled protein can generally be obtained after K-screen exposure for a few hours, depending on the efficiency of translation and [35S]methionine incorporation.
References
- Alder NN. Biogenesis of Lipids and Proteins within Biological Membranes. In: Yeagle PL, editor. The Structure of Biological Membranes. New York: CRC Press; 2011. pp. 315–377. [Google Scholar]
- Andersen OS, Koeppe RE., 2nd Bilayer thickness and membrane protein function: an energetic perspective. Annual review of biophysics and biomolecular structure. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- Bayburt TH, Grinkova YV, Sligar SG. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Lett. 2002;2:853–856. [Google Scholar]
- Bayburt TH, Sligar SG. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein science : a publication of the Protein Society. 2003;12:2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzacco P, Billon-Denis E, Sharma KS, Catoire LJ, Mary S, Le Bon C, Point E, Baneres JL, Durand G, Zito F, et al. Nonionic homopolymeric amphipols: application to membrane protein folding, cell-free synthesis, and solution nuclear magnetic resonance. Biochemistry. 2012;51:1416–1430. doi: 10.1021/bi201862v. [DOI] [PubMed] [Google Scholar]
- Blesneac I, Ravaud S, Juillan-Binard C, Barret LA, Zoonens M, Polidori A, Miroux B, Pucci B, Pebay-Peyroula E. Production of UCP1 a membrane protein from the inner mitochondrial membrane using the cell free expression system in the presence of a fluorinated surfactant. Biochimica et biophysica acta. 2012;1818:798–805. doi: 10.1016/j.bbamem.2011.12.016. [DOI] [PubMed] [Google Scholar]
- Cappuccio JA, Blanchette CD, Sulchek TA, Arroyo ES, Kralj JM, Hinz AK, Kuhn EA, Chromy BA, Segelke BW, Rothschild KJ, et al. Cell-free co-expression of functional membrane proteins and apolipoprotein, forming soluble nanolipoprotein particles. Molecular & cellular proteomics : MCP. 2008;7:2246–2253. doi: 10.1074/mcp.M800191-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corin K, Baaske P, Ravel DB, Song J, Brown E, Wang X, Wienken CJ, Jerabek-Willemsen M, Duhr S, Luo Y, et al. Designer lipid-like peptides: a class of detergents for studying functional olfactory receptors using commercial cell-free systems. PloS one. 2011;6:e25067. doi: 10.1371/journal.pone.0025067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross TA, Murray DT, Watts A. Helical membrane protein conformations and their environment. European biophysics journal : EBJ. 2013;42:731–755. doi: 10.1007/s00249-013-0925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. Journal of the American Chemical Society. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- Distler AM, Kerner J, Hoppel CL. Proteomics of mitochondrial inner and outer membranes. Proteomics. 2008;8:4066–4082. doi: 10.1002/pmic.200800102. [DOI] [PubMed] [Google Scholar]
- Durr UH, Gildenberg M, Ramamoorthy A. The magic of bicelles lights up membrane protein structure. Chemical reviews. 2012;112:6054–6074. doi: 10.1021/cr300061w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson AH, Blobel G. Cell-free translation of messenger RNA in a wheat germ system. Methods in enzymology. 1983;96:38–50. doi: 10.1016/s0076-6879(83)96007-x. [DOI] [PubMed] [Google Scholar]
- Henrich E, Dotsch V, Bernhard F. Screening for lipid requirements of membrane proteins by combining cell-free expression with nanodiscs. Methods in enzymology. 2015;556:351–369. doi: 10.1016/bs.mie.2014.12.016. [DOI] [PubMed] [Google Scholar]
- Hovijitra NT, Wuu JJ, Peaker B, Swartz JR. Cell-free synthesis of functional aquaporin Z in synthetic liposomes. Biotechnology and bioengineering. 2009;104:40–49. doi: 10.1002/bit.22385. [DOI] [PubMed] [Google Scholar]
- Junge F, Haberstock S, Roos C, Stefer S, Proverbio D, Dotsch V, Bernhard F. Advances in cell-free protein synthesis for the functional and structural analysis of membrane proteins. New biotechnology. 2011;28:262–271. doi: 10.1016/j.nbt.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Kalmbach R, Chizhov I, Schumacher MC, Friedrich T, Bamberg E, Engelhard M. Functional cell-free synthesis of a seven helix membrane protein: in situ insertion of bacteriorhodopsin into liposomes. Journal of molecular biology. 2007;371:639–648. doi: 10.1016/j.jmb.2007.05.087. [DOI] [PubMed] [Google Scholar]
- Katzen F, Fletcher JE, Yang JP, Kang D, Peterson TC, Cappuccio JA, Blanchette CD, Sulchek T, Chromy BA, Hoeprich PD, et al. Insertion of membrane proteins into discoidal membranes using a cell-free protein expression approach. Journal of proteome research. 2008;7:3535–3542. doi: 10.1021/pr800265f. [DOI] [PubMed] [Google Scholar]
- Klammt C, Schwarz D, Fendler K, Haase W, Dotsch V, Bernhard F. Evaluation of detergents for the soluble expression of alpha-helical and beta-barrel-type integral membrane proteins by a preparative scale individual cell-free expression system. The FEBS journal. 2005;272:6024–6038. doi: 10.1111/j.1742-4658.2005.05002.x. [DOI] [PubMed] [Google Scholar]
- Lee AG. How lipids affect the activities of integral membrane proteins. Biochimica et biophysica acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Long AR, O'Brien CC, Alder NN. The cell-free integration of a polytopic mitochondrial membrane protein into liposomes occurs cotranslationally and in a lipid-dependent manner. PloS one. 2012;7:e46332. doi: 10.1371/journal.pone.0046332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyukmanova EN, Shenkarev ZO, Khabibullina NF, Kopeina GS, Shulepko MA, Paramonov AS, Mineev KS, Tikhonov RV, Shingarova LN, Petrovskaya LE, et al. Lipid-protein nanodiscs for cell-free production of integral membrane proteins in a soluble and folded state: comparison with detergent micelles, bicelles and liposomes. Biochimica et biophysica acta. 2012;1818:349–358. doi: 10.1016/j.bbamem.2011.10.020. [DOI] [PubMed] [Google Scholar]
- Malhotra K, Alder NN. Advances in the use of nanoscale bilayers to study membrane protein structure and function. Biotechnology & genetic engineering reviews. 2014;30:79–93. doi: 10.1080/02648725.2014.921502. [DOI] [PubMed] [Google Scholar]
- Moritani Y, Nomura SM, Morita I, Akiyoshi K. Direct integration of cell-free-synthesized connexin-43 into liposomes and hemichannel formation. The FEBS journal. 2010;277:3343–3352. doi: 10.1111/j.1742-4658.2010.07736.x. [DOI] [PubMed] [Google Scholar]
- Park KH, Berrier C, Lebaupain F, Pucci B, Popot JL, Ghazi A, Zito F. Fluorinated and hemifluorinated surfactants as alternatives to detergents for membrane protein cell-free synthesis. The Biochemical journal. 2007;403:183–187. doi: 10.1042/BJ20061473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KH, Billon-Denis E, Dahmane T, Lebaupain F, Pucci B, Breyton C, Zito F. In the cauldron of cell-free synthesis of membrane proteins: playing with new surfactants. New biotechnology. 2011;28:255–261. doi: 10.1016/j.nbt.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Popot JL, Althoff T, Bagnard D, Baneres JL, Bazzacco P, Billon-Denis E, Catoire LJ, Champeil P, Charvolin D, Cocco MJ, et al. Amphipols from A to Z. Annual review of biophysics. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]
- Proverbio D, Roos C, Beyermann M, Orban E, Dotsch V, Bernhard F. Functional properties of cell-free expressed human endothelin A and endothelin B receptors in artificial membrane environments. Biochimica et biophysica acta. 2013;1828:2182–2192. doi: 10.1016/j.bbamem.2013.05.031. [DOI] [PubMed] [Google Scholar]
- Roos C, Zocher M, Muller D, Munch D, Schneider T, Sahl HG, Scholz F, Wachtveitl J, Ma Y, Proverbio D, et al. Characterization of co-translationally formed nanodisc complexes with small multidrug transporters, proteorhodopsin and with the E. coli MraY translocase. Biochimica et biophysica acta. 2012;1818:3098–3106. doi: 10.1016/j.bbamem.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Sachse R, Dondapati SK, Fenz SF, Schmidt T, Kubick S. Membrane protein synthesis in cell-free systems: from bio-mimetic systems to bio-membranes. FEBS letters. 2014;588:2774–2781. doi: 10.1016/j.febslet.2014.06.007. [DOI] [PubMed] [Google Scholar]
- Sanders CR, Myers JK. Disease-related misassembly of membrane proteins. Annual review of biophysics and biomolecular structure. 2004;33:25–51. doi: 10.1146/annurev.biophys.33.110502.140348. [DOI] [PubMed] [Google Scholar]
- Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nature reviews Molecular cell biology. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- Schuler MA, Denisov IG, Sligar SG. Nanodiscs as a new tool to examine lipid-protein interactions. Methods in molecular biology. 2013;974:415–433. doi: 10.1007/978-1-62703-275-9_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlemann EM, Pierson HE, Fillingame RH, Dmitriev OY. Cell-free synthesis of membrane subunits of ATP synthase in phospholipid bicelles: NMR shows subunit a fold similar to the protein in the cell membrane. Protein science : a publication of the Protein Society. 2012;21:279–288. doi: 10.1002/pro.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Corin K, Baaske P, Wienken CJ, Jerabek-Willemsen M, Duhr S, Braun D, Zhang S. Peptide surfactants for cell-free production of functional G protein-coupled receptors. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9049–9054. doi: 10.1073/pnas.1018185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JP, Cirico T, Katzen F, Peterson TC, Kudlicki W. Cell-free synthesis of a functional G protein-coupled receptor complexed with nanometer scale bilayer discs. BMC biotechnology. 2011;11:57. doi: 10.1186/1472-6750-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]