SUMMARY
In both prokaryotes and eukaryotes insight into gene function is typically obtained by in silico homology searches and/or phenotypic analyses of strains bearing mutations within open reading frames. However, the studies herein illustrate how mRNA function is not limited to the expression of a cognate protein. We demonstrate that a stress-induced protein-encoding mRNA (irvA) from the dental caries pathogen Streptococcus mutans directly modulates target mRNA (gbpC) stability through seed pairing interactions. The 5’ untranslated region of irvA mRNA is a trans-riboregulator of gbpC and a critical activator of the DDAG stress response, whereas IrvA functions independently in the regulation of natural competence. The irvA riboregulatory domain controls GbpC production by forming irvA-gbpC hybrid mRNA duplexes that prevent gbpC degradation by an RNase J2-mediated pathway. These studies implicate a potentially ubiquitous role for typical protein-encoding mRNAs as riboregulators, which could alter current concepts in gene regulation.
INTRODUCTION
With the recent explosion of bacterial RNA-seq studies, it is apparent that bacteria produce a surprising abundance of uncharacterized noncoding RNAs (Li et al., 2013; Romby and Charpentier, 2010; Toffano-Nioche et al., 2013). It has also recently become evident that small noncoding RNA regulators (sRNAs) are ubiquitously employed as critical nodes within bacterial genetic networks and can regulate gene expression through a highly diverse array of regulatory mechanisms (Storz et al., 2011). Consequently, RNA regulators are vital for the regulation of gene expression: perhaps of equal or greater importance to protein regulators. This function has thus far been solely attributed to sRNAs. There is also a significant pool of mRNAs in the cell containing 5’ untranslated regions (UTRs) that regulate translation in cis by folding into complex secondary structures (Gripenland et al., 2010; Romby and Charpentier, 2010). In addition, mRNAs may contain decoy sites that sequester riboregulatory RNAs (Figueroa-Bossi et al., 2009; Overgaard et al., 2009; Plumbridge et al., 2014). However, mRNAs have not been traditionally recognized as trans regulators of heterologous mRNA stability or translation.
Most trans-acting riboregulatory sRNAs indirectly control gene expression either positively or negatively through complementary base pairing interactions (seed pairing) that modulate translation initiation, since translation has an inherent stabilizing effect upon mRNAs (Frohlich and Vogel, 2009; Gottesman, 2011). In some instances, sRNA interactions may directly alter target mRNA stability by modifying target accessibility to RNases (Bandyra et al., 2012; Desnoyers et al., 2009; Papenfort et al., 2013; Pfeiffer et al., 2009; Rice et al., 2012). In addition, a small subset of riboregulatory sRNAs, referred to as dual-function sRNAs, also contains translated open reading frames (ORFs) (Vanderpool et al., 2011). The ORFs of dual-function sRNAs all encode peptides ranging in size from 26 – 53 amino acids and only a fraction have known functions (Balaban and Novick, 1995; Berghoff et al., 2009; Mangold et al., 2004; Roberts and Scott, 2007; Shimizu et al., 2002; Sonnleitner et al., 2011; Wadler and Vanderpool, 2007). The existence of such sRNAs implies that a single RNA molecule can serve as both a trans-acting riboregulator and a template for translation. It remains to be determined whether dual-function sRNAs are simply an unusual subset of otherwise noncoding riboregulatory RNAs or they are indicative of a wider regulatory role for translated RNAs.
The irvA gene of S. mutans encodes a putative transcription repressor and was originally discovered due to its induction by a variety of genetic mutations. In a wild-type background, irvA gene expression is extremely low, but its expression increases >100-fold in various mutant backgrounds (Merritt et al., 2005; Tsang et al., 2006). Besides triggering irvA expression, these mutations also share a variety of common phenotypes, such as deficiencies in lantibiotic bacteriocin production and natural genetic competence (Merritt et al., 2005; Niu et al., 2010; Niu et al., 2008). The key regulator of irvA transcription, irvR, is located directly adjacent on the chromosome and encodes a LexA-like self-cleaving transcription repressor responsible for preventing irvA expression under normal growth conditions (Niu et al., 2010; Niu et al., 2008). A mutation of irvR constitutively derepresses irvA and triggers each of the previously described irvA-dependent phenotypes in addition to the constitutive activation of the dextran-dependent aggregation (DDAG) stress response (Niu et al., 2010; Niu et al., 2008). In a wild-type background, the characteristic cellular aggregation phenotype of the DDAG stress response is only detectable in the presence of the various environmental stresses that trigger the production of a critical surface exposed lectin called GbpC (Sato et al., 2002; Sato et al., 1997). It is unknown how stress activates GbpC production, but its lectin activity normally serves as a major adhesin for biofilm development (Banas and Vickerman, 2003; Idone et al., 2003; Lynch et al., 2007). This is presumably due to its high affinity for the dextran and glucan polymers within the exopolysaccharide matrix of S. mutans biofilms. In an irvR mutant background, GbpC production is constitutively activated in an irvA-dependent manner implicating IrvA as its regulator (Niu et al., 2010; Niu et al., 2008).
Our current analysis of irvA unexpectedly revealed that the regulation of GbpC production is in fact, not due to the putative transcription regulatory function of the IrvA protein. Remarkably, the entire process is mediated solely by a trans-acting riboregulatory function encoded within the 5’ UTR of irvA mRNA. Therefore, irvA is a dual-function mRNA serving as both a template for translation and as a seed pairing posttranscriptional regulator via interactions with its 5’ UTR. These results implicate a much broader role for mRNAs as highly versatile regulators of gene expression in addition to their traditionally assigned role as templates for translation.
RESULTS
The irvA 5’ UTR is required for the Dextran-Dependent Aggregation Stress Response
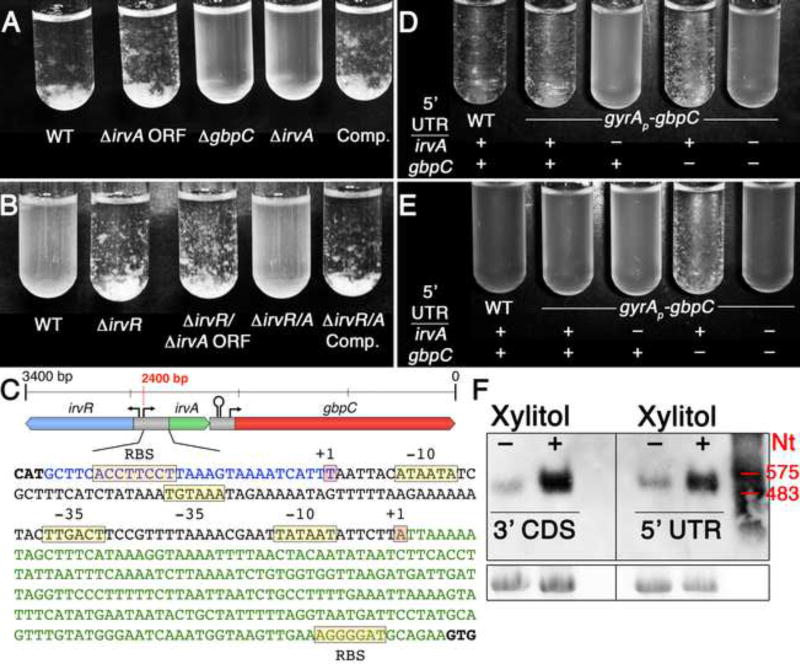
Our previous genetic studies of irvR suggested that environmental stress was likely to be the endogenous signal responsible for relieving IrvR repression upon irvA. In addition, an irvR deletion stimulated the expression of gbpC and constitutively activated the dextran-dependent aggregation (DDAG) stress response (Niu et al., 2010; Niu et al., 2008). From these results, we hypothesized that the unknown endogenous stress- induced DDAG pathway was likely functioning through irvA. Consistent with previous reports (Sato et al., 1997, 2000), we found a range of environmental stresses to be effective at triggering the characteristic cellular aggregation phenotype of the DDAG response including the commonly utilized food sweetener xylitol. However, in contrast to our expectations, the irvA mutant exhibited a wild-type DDAG response to xylitol (Fig. 1A), which prompted us to reexamine our previous genetic data. Interestingly, a double mutation of the entire irvR/A locus yielded a constitutive DDAG− phenotype, whereas separate deletions of the irvR and irvA open reading frames (ORF) resulted in a constitutive DDAG+ phenotype (Figs. 1B and C). We identified the transcription start sites for both irvR and irvA, and curiously, irvR was found to have a minimal 5’ UTR (29 nt), whereas irvA possesses a 235 nt 5’ UTR (Fig. 1C). Using this information, we assayed the DDAG phenotype of an irvA deletion mutant devoid of both the 5’ UTR and ORF. The more complete deletion of irvA rendered the strain fully incapable of engaging the DDAG response in the presence of environmental stress (Fig. 1A). This was in stark contrast to the irvA ORF deletion mutant, which behaved similarly as the wild-type (Fig. 1A).
Fig. 1. The irvA 5’ UTR mediates the dextran-dependent aggregation (DDAG) stress response.
A) DDAG response assay performed in the presence of xylitol stress. The strains from left to right are: wild-type, irvA ORF deletion, gbpC deletion, irvA 5’ UTR + ORF deletion, and irvA 5’ UTR + ORF deletion complemented in trans. B) DDAG response assay performed in normal growth conditions. The strains from left to right are: wild- type, irvR deletion, separate deletions of the irvR and irvA ORFs, deletion of the entire irvR/A locus, and the irvR/A locus deletion expressing irvA in trans. C) Illustration of the irvR/A intergenic region. The irvR 5’ UTR is shown in blue and the irvA 5’ UTR in green. D) DDAG response assay performed in the presence of xylitol stress. E) DDAG response assay performed in normal growth conditions at late-log/early stationary phase. F) Northern blots of wild-type irvA mRNA in both normal and xylitol stress growth conditions. Probe target locations are listed under the samples. An RNA ladder is shown on the right and the bottom panel is a 16S rRNA loading control. See also Figure S1.
To determine whether the irvA 5’ UTR was solely responsible for triggering the DDAG response during stress, we replaced the irvA ORF with the green fluorescent protein (gfp) ORF to create a chimeric irvA 5’ UTR-gfp fusion mRNA (Fig. S1). We also replaced the gbpC promoter with that of the constitutive DNA gyrase promoter gyrAP to eliminate potentially confounding effects due to changes in gbpC transcription. Despite the constitutive expression of gbpC from the gyrA promoter fusion, the strain did not exhibit a constitutive DDAG phenotype under normal growth conditions, but still exhibited a stress-inducible DDAG response that was critically dependent upon the irvA 5’ UTR-gfp ORF fusion (Fig. 1D). We also found that the gbpC 5’ UTR was not required for DDAG, but may play a minor inhibitory role (Fig. 1D & E). This indicated that irvA activation is highly unlikely to function via the gbpC 5’ UTR. In addition, northern blot results demonstrated that the DDAG regulatory function of the irvA 5’ UTR occurs in the context of a single irvA RNA containing both the 5’ UTR and irvA coding sequence (CDS) (Figs. 1F and S1). Thus, we found no evidence suggesting that the UTR is processed into an sRNA in either normal or stress growth conditions. Though, we did note a substantial increase in irvA mRNA abundance due to xylitol stress (Fig. 1F).
The irvA 5’ UTR controls GbpC protein production by modulating gbpC mRNA stability
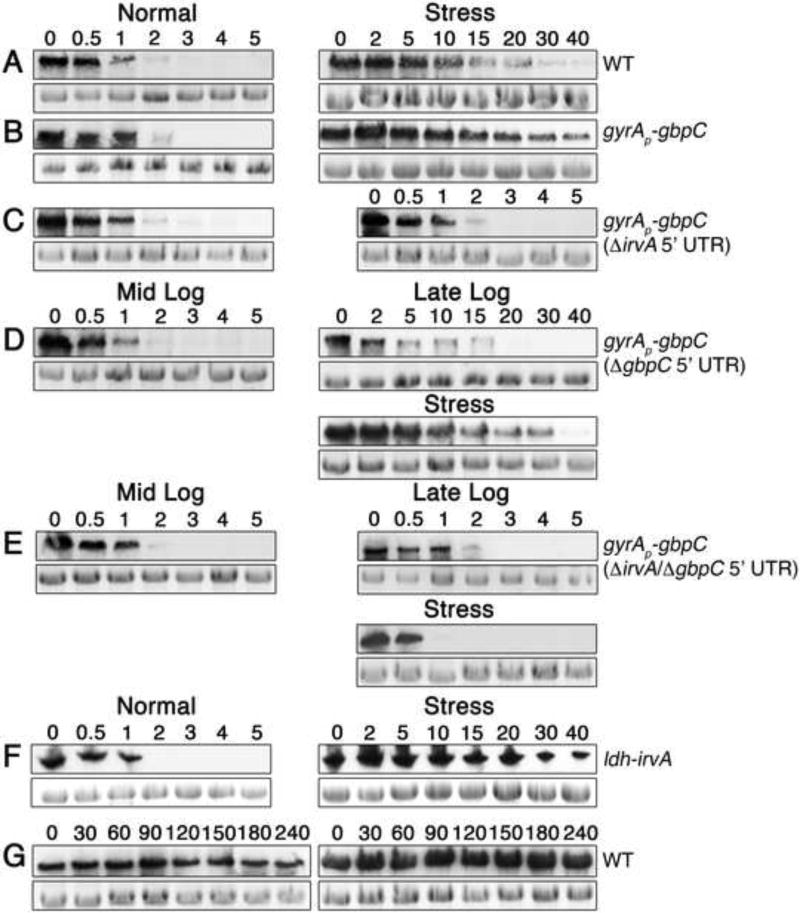
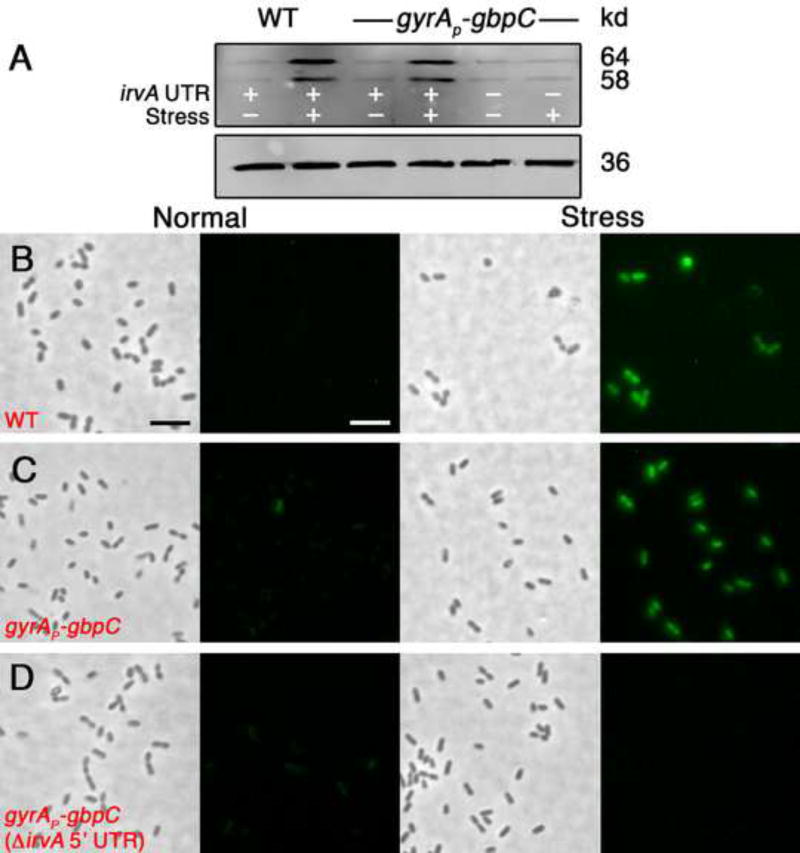
Based upon the results with the gyrAP–gbpC fusion strain, it appeared that one or more steps occurring after gbpC transcription are primarily responsible for controlling the DDAG phenotype. Since gbpC mRNA normally exhibits an extremely short half-life of <1 minute (Biswas et al., 2007), we were curious whether environmental stress might stabilize the message. As shown in figure 2, xylitol stress increased gbpC half-life by greater than an order of magnitude in both the wild-type and gyrAP–gbpC fusion strains (Figs. 2 A & B). This effect was also specifically dependent upon the irvA 5’ UTR (Fig. 2C). To further examine the correlation between irvA, gbpC mRNA stability, and the DDAG response, we also compared gbpC mRNA stability in cells grown in conditions that we previously determined to trigger three distinct DDAG phenotypes (Figs. 1D & E). Consistent with our previous results, the severity of the DDAG response was directly proportional to the stability of gbpC mRNA and was critically dependent upon the irvA 5’ UTR (Figs. 2D & E). In addition, the effect upon gbpC stability was identical using trans expressed irvA indicating that gbpC stabilization was not due to irvA cis effects (Fig. 2F). Unlike gbpC, xylitol stress did not elicit a reciprocal increase in irvA mRNA stability (Figs. 2G and S2). As expected, changes in gbpC mRNA stability directly correlated with protein abundance (Fig. 3A – D).
Fig. 2. gbpC mRNA stability is regulated by the irvA 5’ UTR.
The numbers above the figures indicate the time (min.) after the addition of rifampicin for mRNA stability assays. For each figure, the top panels show target mRNA northern blots, while the bottom panels are 16S rRNA loading controls. Panels A – F probe gbpC mRNA. A) Wild-type strain in normal growth conditions (half-life < 1 min.) vs. xylitol stress (half-life > 10 min.). B) gyrAP-gbpC strain in normal growth conditions vs. xylitol stress. C) The same experiment is performed as in 2B, but the irvA 5’ UTR has been deleted. D) gyrAP-gbpC strain (without the gbpC 5’ UTR) in mid-log phase, late-log phase, and mid-log phase with added xylitol stress. E) The same experiment performed as in 2D, but both the gbpC and irvA 5’ UTRs have been deleted from the gyrAP-gbpC strain. F) irvA was expressed in trans from the ldh gene locus. Using this strain, gbpC mRNA stability was measured both in the absence and presence of xylitol stress. G) The stability of irvA mRNA in the wild-type was assayed in the absence or presence of xylitol stress at mid-log phase. See also Figures S1 and S2.
Fig. 3. gbpC mRNA stability determines GbpC protein abundance.
A) Western blot of GbpC in the wild-type and gyrAP-gbpC strains. Two bands are present because the antibody recognizes both the immature cytoplasmic and mature cell wall anchored forms of the GbpC protein. In the bottom panel, an HA-epitope tagged lactate dehydrogenase protein was used as a loading control for each condition. In panels B-D, GbpC was engineered to express a tetracysteine FlAsH tag to facilitate the in situ detection of GbpC with the FlAsH reagent. Strains were grown to mid-log phase in the absence and presence of xylitol stress and imaged using identical exposure settings. B) Wild-type C) gyrAP-gbpC strain D) gyrAP-gbpC strain with a deletion of the irvA 5’ UTR. The scalebars indicate 2 µm.
The irvA 5’ UTR interacts directly with gbpC mRNA
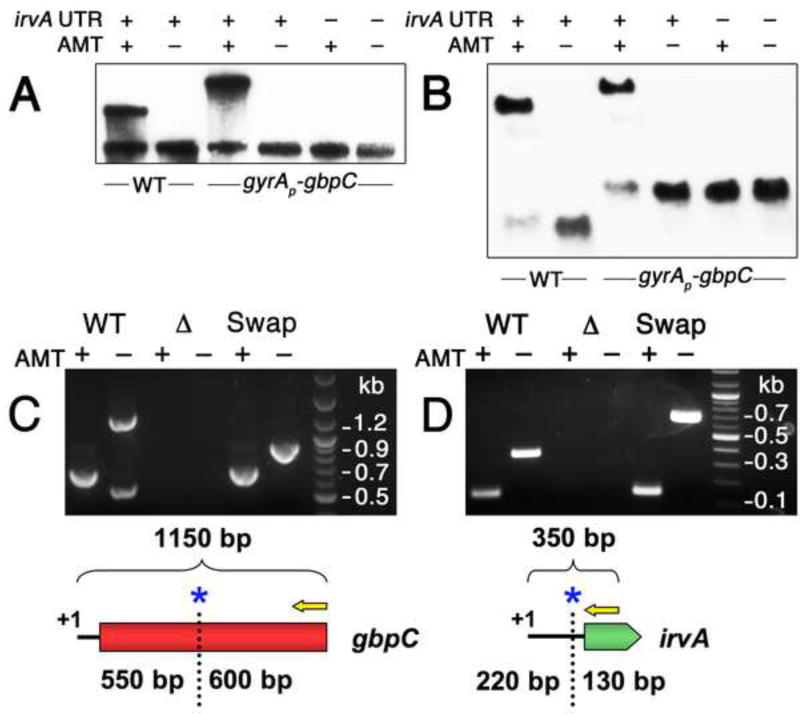
Due to the dominant effect of posttranscriptional control over gbpC expression, we hypothesized that gbpC mRNA stability was likely to be controlled through sRNA-like interactions with irvA. We began by testing an RNA mobility shift assay using in vitro transcribed irvA and gbpC. While we were able to detect an interaction, the hybrid duplex formed slowly (30 min.) and only constituted a small percentage of the total RNA (data not shown). To further examine the relevance this interaction, we developed an in vivo approach for the RNA mobility shift assay using the psoralen crosslinker 4′-aminomethyl-trioxsalen (AMT). By performing northern blots on AMT crosslinked cultures, it was possible to detect distinct mobility shifts specific to the AMT-treated strains expressing both the irvA 5’ UTR and gbpC (Figs. 4A & B). In the gyrAP-gbpC strain, a larger mobility shift was observed relative to the wild-type because the irvA 5’ UTR–gfp ORF fusion resulted in a larger mRNA than the wild-type irvA. Similarly, this was also reflected in the larger size of the uncrosslinked mRNAs in the irvA northern blots (Fig. 4B). To detect irvA in the gyrAP-gbpC strain, it was necessary to hybridize with a gfp CDS probe because the irvA 5’ UTR probe performed poorly with the crosslinked samples (data not shown), presumably due to the presence of crosslinks within the irvA 5’ UTR. This was not an issue in the wild-type strain because the irvA probe targeted the 3’ of the irvA CDS (Fig. 4B). The mobility shift observed with this probe also provided additional evidence that the full-length irvA mRNA is responsible for mediating the interaction with gbpC, rather than a separate processed form of the irvA 5’ UTR.
Fig. 4. irvA and gbpC RNAs form a complex during stress.
A) Samples were grown in the presence or absence of AMT RNA crosslinking agent and then detected with anti-gbpC probes via northern blot. The role of the irvA 5’ UTR was assessed by engineering a small deletion within a critical portion of the 5’ UTR required for DDAG. B) The same experiment is performed as in 4A, except an irvA CDS probe is used for the wild-type samples and a gfp CDS probe is used for the gyrAP-gbpC samples. Both probes were mixed in a single hybridization reaction. In panels C and D, the RACE walk procedure was used to probe the interaction region between irvA and gbpC. In both experiments, the entire procedure was repeated using mutant strains containing swapped irvA and gbpC interaction domains. The RACE walk results are also illustrated below each set of reactions. Yellow arrows represent 5’ RACE primers and blue asterisks mark the locations of the first crosslink sites. C) Anti-irvA oligonucleotides are used to precipitate RNA complexes with gbpC. 5’ RACE is performed on gbpC. D) Anti-gbpC oligonucleotides are used to precipitate RNA complexes with irvA. 5’ RACE is performed on irvA. See also Figures S1 and S3.
To further validate the RNA mobility shift experiments as well as identify the interaction region between irvA and gbpC, we modified an in vivo RNA-RNA mapping assay referred to as “RNA walk” (Lustig et al., 2010). The RNA walk assay takes advantage of the fact that reverse transcription of crosslinked RNA will result in cDNAs that are truncated at crosslinked sites. Thus, S. mutans was grown in the presence of xylitol stress, crosslinked in vivo using AMT, and then irvA-gbpC RNA complexes were purified by affinity chromatography. RT-PCR amplicons were generated using a nested adaptor primer and a gene-specific primer similarly as in 5’ RACE protocols (Fig. 4C & D). Thus, our modified version of the RNA walk procedure would be more appropriately referred to as “RACE walk”. After sequencing the PCR amplicons, we determined that the terminal 3’ boundary of the gbpC crosslinked region occurred 515 nt into the gbpC CDS, whereas the irvA crosslinked region terminated at the putative Shine-Dalgarno sequence in its 5’ UTR. Due to the fewer affinity purifications that could be performed on the samples receiving no AMT treatment, we frequently observed an extra PCR band arising in the uncrosslinked gbpC RT-PCR reactions (Fig. 4C). We sequenced this band and confirmed it to be 23S rRNA contamination. Based upon the RACE walk results, we performed an interaction domain swap experiment and were still able to detect identical 3’ terminal crosslinked sites in both molecules, whereas the untreated samples yielded full-length cDNAs of the expected sizes for the chimeric mRNAs (Fig. 4C & D). From the RNA mobility shift and RACE walk experiments, it was clear that irvA and gbpC mRNAs form a stable complex in which all of the elements required to facilitate their interaction are fully contained within the irvA 5’ UTR and the first 550 nt of gbpC.
irvA and gbpC mRNAs interact directly through seed pairing
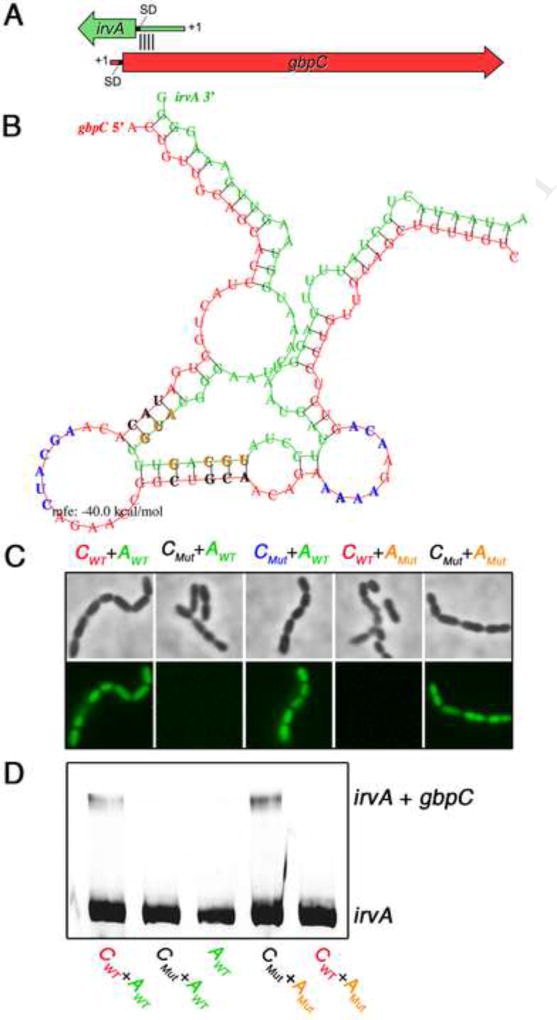
To distinguish whether irvA-gbpC mRNA complexes form via direct or indirect interactions, we were interested to determine whether there was evidence to implicate seed pairing between the RNAs. Due to the inefficient duplex formed using in vitro transcribed irvA and gbpC, in vitro interaction mapping approaches were deemed impractical. Thus, we began by designing a strategy for the high-throughput mutagenesis of irvA to first identify the irvA nucleotides critical for the DDAG phenotype (see Supplemental Methods). Consistent with our previous results, no critical residues were detected within the irvA ORF, whereas all of the critical bases were found in the latter half of the UTR with the final cluster of critical bases terminating at the identical nucleotide previously identified by RACE walk (Fig. S3). With this information, we used the program RNAhybrid (Rehmsmeier et al., 2004) to model a potential interaction between the RACE walk interaction domain of gbpC with only the DDAG-mediating portion of the irvA 5’ UTR. The predicted duplex occurred well within the gbpC CDS starting 109 nt downstream of the gbpC translation initiation codon (Fig. 5A). This model was consistent with our previous DDAG phenotypic data, which demonstrated that the gbpC 5’ UTR is not a target of irvA regulation (Fig. 1D & E). Using the irvA mutagenesis data and RNAhybrid model as guides, we created a series of mutant gbpC-gfp reporter strains containing point mutations within key irvA residues that were both required for DDAG and predicted to seed pair with gbpC. The corresponding seed pair mutations were also engineered into gbpC. When point mutations were introduced into predicted seed paired bases of either gbpC or irvA, the reporter strain lost the ability to respond to stress (i.e. constitutively dark) (Fig. 5C). In contrast, it was possible to rescue the responsiveness of the reporter by combining both sets of complementary gbpC and irvA mutations into the same strain (Fig. 5C). Mutations occurring within the predicted unpaired regions of gbpC had no impact upon the reporter (Fig. 5C). We also tested these seed pair mutations using in vitro transcribed irvA and gbpC. Mirroring the reporter results, irvA-gbpC duplexes were detectable using either wild-type or compensatory double mutant mRNAs, while no interactions occurred with either combination of wild-type + mutant mRNAs (Fig. 5D). These data strongly supported a role for seed pairing as the primary mechanism mediating the interaction between irvA and gbpC.
Fig. 5. irvA and gbpC mRNA seed pairing.
A) Illustration of the irvA-gbpC hybrid duplex. Block arrows indicate the CDS of the mRNAs, while the thinner lines represent 5’ UTR’s. The approximate location of the seed region is indicated by black lines between the RNAs. B) The predicted irvA-gbpC hybrid mRNA structure is shown with irvA RNA in green and gbpC RNA in red. Seed pairing bases colored in black (gbpC) or orange (irvA) as well as unpaired bases colored in blue (gbpC) were each targeted for mutagenesis to their complementary bases. C) The effect of these point mutations was tested in vivo in the presence of xylitol. GFP fluorescence is indicative of stable gbpC mRNA. Strains from left to right: wild-type gbpC + irvA; mutant gbpC (complement of bases shown in black) + wild-type irvA; mutant gbpC (complement of bases shown in blue) + wild-type irvA; wild-type gbpC + mutant irvA (complement of orange bases); and mutant gbpC (complement of black bases) + mutant irvA (complement of orange bases). D) The same mutations were introduced into in vitro transcribed irvA and gbpC mRNA and analyzed by RNA mobility shift assay. The reactions were visualized using an anti-irvA probe. See also Figure S3.
The interaction of irvA and gbpC mRNA protects gbpC from ribonuclease-mediated degradation
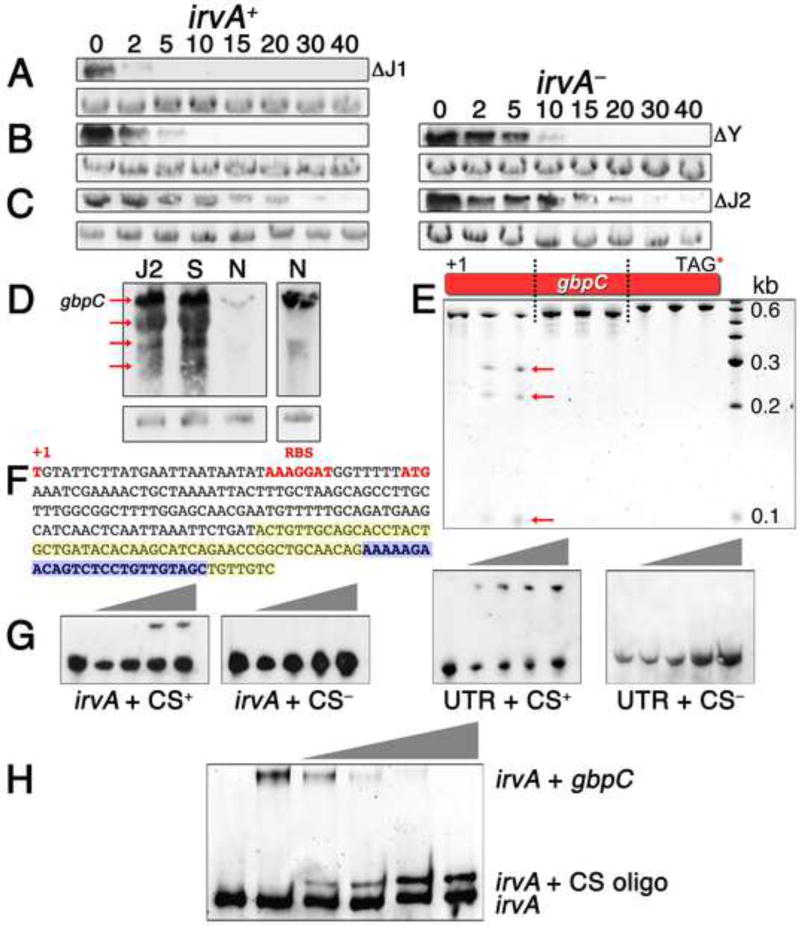
Typically, when sRNA-target interactions increase mRNA stability, the interaction changes the secondary structure of the target mRNA to facilitate the loading of ribosomes and ultimately enhance translation initiation (Frohlich and Vogel, 2009; Gottesman, 2011). However, translation did not appear to play an obvious role in regulating gbpC stability (Fig. S4). Thus, we hypothesized that irvA must directly protect gbpC mRNA from ribonuclease attack. We mutated a variety of predicted exo- and endoribonucleases and assayed for a constitutive DDAG+ phenotype. All were dispensable for DDAG except for RNase J2 and to a much lesser extent RNase Y (Fig. S4). In many Gram positive bacteria, the major endoribonuclease and 5’–3’ exoribonuclease activities in the cell are catalyzed by RNase Y, RNase J1, and RNase J2 (Condon, 2010; Lehnik-Habrink et al., 2012). To further confirm these results, we measured gbpC mRNA stability in the RNase Y, J1, and J2 backgrounds and found that all correlated strongly with the observed DDAG phenotypes (Fig. 6A – C). Furthermore, stress or an RNase J2 mutation triggers a substantial increase in the total abundance of full-length gbpC mRNA along with identical gbpC degradation intermediates (Fig. 6D). These results all implicated RNase J2 as the principal source of gbpC instability in normal growth conditions. To test this further, we purified RNase J2 and digested gbpC in vitro. RNase J2 introduced two endonuclease cleavages located within the first 575 nt of gbpC (Fig. 6E). Further cleavage analysis of this region localized the first cut site to within with the predicted irvA seed region, whereas the second site is approximately 100 nt further downstream (Figs. 6F and S5). Similarly, the addition of irvA RNA to the cleavage reaction only inhibited cleavage at the first site (Fig. S5). The region encompassing the first cleavage site was also required for an in vitro interaction between irvA and gbpC (Figs. 6G and 6H). Therefore, it is highly likely that this RNase J2 cleavage site would be sequestered within the seed region of the irvA-gbpC hybrid duplex following the onset of environmental stress.
Fig. 6. Mechanism of gbpC stabilization by the irvA 5’ UTR.
In panels A – C, the RNase J1, Y, and J2 mutants were each compared for gbpC mRNA stability under normal growth conditions in a wild-type irvA background (irvA+) or irvA deletion mutant (irvA−). The numbers above the figures signify the time after rifampicin addition and the bottom panels are 16S rRNA loading controls. A) RNase J1 mutant B) RNase Y mutant C) RNase J2 mutant. D) The degradation profile of gbpC mRNA in a wild-type background was compared between normal (N) and stress (S) growth conditions as well as in an RNase J2 mutant (J2) background in normal growth conditions. The full-length gbpC transcript is indicated by an arrow along with multiple mRNA degradation intermediates. Due to the much weaker gbpC signal from the wild-type unstressed sample, we repeated this sample with a longer film exposure (right panel). E) Three fragments of gbpC mRNA indicated in the diagram were each in vitro transcribed and either left untreated (first lane for each fragment) or treated with increasing amounts of recombinant S. mutans RNase J2 (following two lanes). Arrows designate degradation products. F) The sequence of gbpC between the +1 site and the predicted seed region with irvA is shown. Bases highlighted in yellow correspond to the seed region predicted by RNAhybrid, while the bases highlighted in blue encompass the predicted RNase J2 cleavage site. G) An RNA mobility shift assay was performed using in vitro transcribed gbpC and increasing amounts of in vitro transcribed irvA or irvA UTR. Wild-type gbpC mRNA containing the illustrated RNase J2 cleavage site sequence (CS+) was compared to mutant gbpC lacking this sequence (CS−). The reactions were visualized with anti-irvA probes. H) A similar RNA mobility shift assay was performed using fixed quantities of wild-type irvA and gbpC mRNA together with increasing quantities of a competitor RNA oligo identical to the illustrated RNase J2 cleavage site sequence (CS oligo). See also Figures S4 and S5.
irvA is a dual-function mRNA
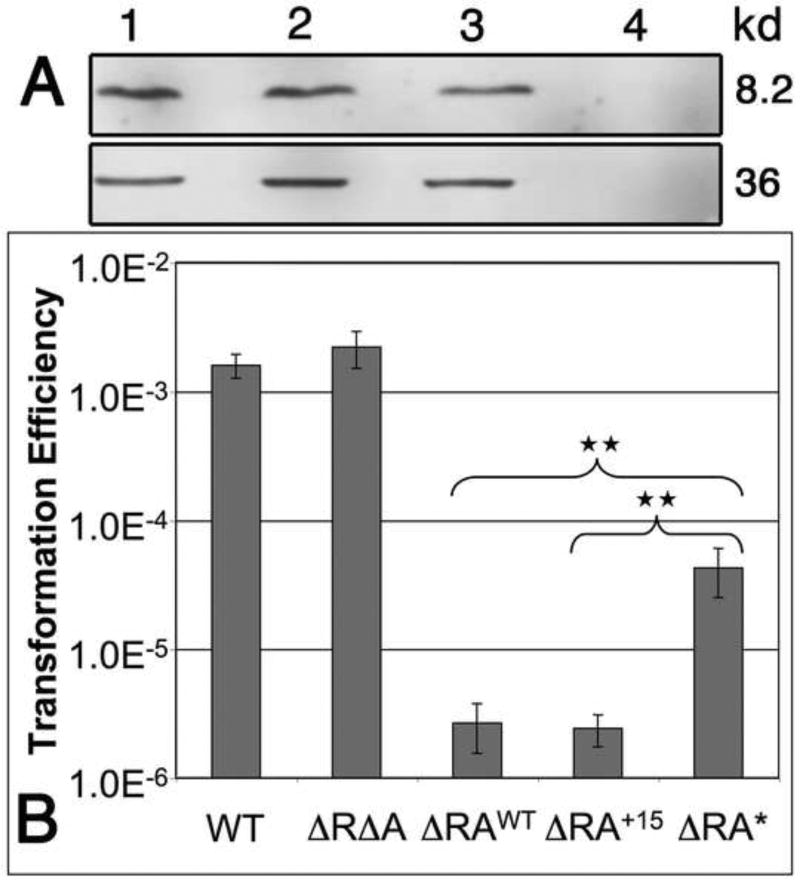
Our studies of the connection between irvA and the DDAG response established a clear role for the irvA 5’ UTR as a riboregulator of gbpC mRNA. However, it also indicated that the DDAG response functions entirely independent of the irvA CDS. This unusual result prompted us to question whether irvA truly functions as a protein-encoding mRNA or if it is simply a noncoding sRNA. Consequently, we confirmed the presence of the IrvA protein via western blot (Fig. 7A). The next question was whether the IrvA protein has a discernable physiological function in the cell, since it is fully dispensable for the DDAG response. As previously described, irvA gene expression inhibits the development of natural competence (Niu et al., 2010; Niu et al., 2008). Thus, we were curious whether the IrvA protein might play a role in this phenotype. Indeed, a deficiency in IrvA protein production did impair the ability of irvA to inhibit natural competence development (Fig. 7B). It is also apparent that both the irvA 5’ UTR and ORF are nearly identical in each of 57 recently sequenced strains of S. mutans (Cornejo et al., 2013), which further suggests that both the irvA RNA and protein are functionally conserved components of the core S. mutans genome. However, we did note that the genome reference strain UA159 used in these studies was one of only three strains in the collection encoding a slightly truncated IrvA, due to a frameshift near the 3’ end of the irvA ORF (Fig. S6). We restored the irvA reading frame in UA159 and confirmed that the frameshift had no impact upon competence regulation (Fig. 7B). From these results, we conclude that irvA mRNA serves two highly conserved and independent functions: a trans-acting riboregulator and a template for the translation of a Cro-like putative transcription regulator.
Fig. 7. irvA encodes a dual-function mRNA.
A) A FLAG tag was engineered onto the C-terminus of IrvA and visualized by western blot after growing the cultures to mid-log phase. The samples from left to right are: 1. IrvA-FLAG in xylitol stress; 2. IrvA-FLAG in ΔirvR background; 3. constitutive irvA-FLAG expression strain; and 4. true wild-type with no epitope tags in xylitol stress. The bottom panel is an HA-tagged lactate dehydrogenase loading control. B) Transformation assays of the following strains from left to right: wild-type, irvR/A double mutant, irvR mutant, irvR mutant with irvA expressing a frameshift corrected ORF, and irvR mutant with irvA expressing a translation deficient ORF. Transformation efficiency is defined as the ratio of transformants to total CFU. Values are listed as the means ± standard deviations. One-way ANOVA and a Fisher’s protected least significance difference post-hoc test were used to compare the means of the groups indicated in the graph (** P < 0.005). See also Figure S6.
DISCUSSION
The regulatory mechanism used to control the DDAG stress response in S. mutans reveals a potential role for mRNAs as postranscriptional trans–acting riboregulators. Its discovery was based upon an initially puzzling observation in which two separate deletion mutations of irvA yielded opposing DDAG phenotypes. The source of this discrepancy was a direct result of the modular two-domain architecture contained within irvA mRNA: a trans-acting riboregulatory domain in the 5’ UTR and the protein-encoding domain of the CDS. This result may have broad implications for genetic studies of other genes unknown to encode dual-function mRNAs, since mutagenesis constructs typically only target ORFs. The irvA example illustrates how such an approach could easily leave the riboregulatory function of a gene fully intact, potentially resulting in highly biased or even misleading phenotypes.
Mechanism of gbpC stabilization
Of the characterized sRNA-mRNA target interactions, most trigger the repression of gene expression. Only a handful of examples implicate trans-acting sRNAs as activators, and of these, most stabilize mRNA indirectly by remodeling ribosome binding site-occluding secondary structures (Frohlich and Vogel, 2009). Other sRNAs, such as SgrS and RydC in S. typhimurium, directly activate target gene expression by preventing access to endogenous RNase cleavage sites (Frohlich et al., 2013; Papenfort et al., 2013). This mechanism is highly analogous to the irvA-gbpC interaction, where the S. mutans RNase J2 mutation triggered a potent irvA-independent stabilization of gbpC mRNA. Since translation was not found to influence gbpC mRNA stability, the principal function of irvA is likely to directly alter gbpC accessibility to RNase J2-mediated degradation. Surprisingly, this was found to occur through a unique mechanism involving seed pairing far within the gbpC CDS. Other characterized sRNA interactions occurring within coding regions serve almost exclusively in target destabilization (De Lay et al., 2013; Romby and Charpentier, 2010; Storz et al., 2011; Vanderpool et al., 2011). To the best of our knowledge, the aforementioned SgrS sRNA is the only other example in which CDS seed pairing activates gene expression (Papenfort et al., 2013). However, unlike irvA, SgrS specifically stabilizes the downstream CDS, rather than its target. Given the location of the irvA-gbpC hybrid duplex, it might also be expected that seed pairing would simultaneously hinder GbpC translation by preventing ribosome progression. However, in vitro translation studies have demonstrated that the ribosome has intrinsic helicase activity and can read through RNA duplexes containing stretches of ≥18 paired bases (Takyar et al., 2005). The RNAhybrid model of the irvA-gbpC duplex (Fig. 5B) predicts stretches of complementarity well below this threshold.
Model for RNase J2 control of the DDAG stress response
Most firmicutes such as S. mutans, B. subtilis, S. pyogenes, and Enterococcus faecalis encode two RNase J paralogs referred to as RNase J1 and RNase J2 (Bugrysheva and Scott, 2010; Even et al., 2005; Gao et al., 2010). In B. subtilis, RNase J1 is a pleiotropic regulator of mRNA stability, whereas the specific function of RNase J2 has remained enigmatic (Lehnik-Habrink et al., 2012). This makes RNase J2 a particularly intriguing component of the DDAG pathway. In in vitro RNase cleavage assays, the B. subtilis RNase J2 exhibits exceptionally weak 5’-3’ exoribonuclease activity (Condon, 2010; Even et al., 2005; Mathy et al., 2010) and regulates the activity of RNase J1 in heteromeric RNase J1/J2 complexes (Mader et al., 2008; Mathy et al., 2010). Consequently, it has been speculated that RNase J2 serves more of a structural and/or regulatory role for RNase J1 enzymatic activity (Figaro et al., 2013). The S. mutans DDAG response was critically dependent upon RNase J2, yet we were unable to discern an obvious role for RNase J1, which strongly argues against the possibility that the RNase J2 DDAG phenotype is a consequence of disrupted RNase J1/J2 heteromeric complexes. Consistent with this scenario, we tested D73K/D160K as well as H69A/H71A loss of function point mutations in RNase J2 (Li de la Sierra-Gallay et al., 2008) and observed constitutive DDAG phenotypes (data not shown). In addition to stabilizing gbpC mRNA, the RNase J2 mutation also triggered a distinct degradation pattern for gbpC mRNA. Likewise, gbpC is cleavable by RNase J2 in in vitro digests, making it among the first endogenous substrates known for this enzyme.
Based upon these results, we propose the following regulatory model. Under normal growth conditions, gbpC mRNA serves as a high affinity substrate for RNase J2 cleavage(s) catalyzing its rapid degradation. In response to environmental stress, irvA-gbpC hybrid duplexes form and are resistant to the initial cleavage event, thereby inhibiting the RNase J2-mediated degradation pathway (Fig. 6). However, gbpC is still subject to RNase degradation, as evidenced by the identical gbpC cleavage products produced specifically in response to environmental stress or by an RNase J2 mutation. Presumably, gbpC is a poorer substrate for this alternate RNase J2-independent degradation pathway, which would account for the observed increase in gbpC mRNA abundance and its reduced turnover rate. Whereas genetic switches are a classic feature of many regulatory circuits, the irvA regulatory mechanism is more analogous to a genetic rheostat, whereby the abundance of GbpC is ultimately adjusted by the severity of stress placed upon the cell. The equilibrium between the unstable (free gbpC) and stable (duplex gbpC) states of gbpC mRNA can be altered proportionally by the irvA transcription rate, which is itself controlled by the stress state of the cell. Given the lack of an avid in vitro interaction between irvA and gbpC, it is also possible that the equilibrium of this interaction is further influenced by an as yet unidentified component catalyzing the reaction in vivo. Presumably, this component would be activated by stress, since irvA overexpression does not increase gbpC stability under normal growth conditions [Unpublished and (Zhu et al., 2009)]. As the surface lectin activity of GbpC serves to anchor S. mutans cells to the biofilm (Lynch et al., 2007), the irvA rheostat mechanism is likely utilized to direct the appropriate cellular resources towards bolstering biofilm integrity during episodes of increased environmental stress. Concurrently, irvA expression also redirects cellular resources away from many accessory gene pathways, such as bacteriocin production and competence development (Merritt et al., 2005; Niu et al., 2010; Niu et al., 2008), since these functions are both metabolically expensive and provide little survival advantage in such conditions.
mRNA as a source of trans-acting riboregulatory molecules
While the majority of known trans-acting riboregulatory RNAs are derived from noncoding transcripts, it is clear that both eukaryotes and prokaryotes frequently utilize translated mRNAs as a source of posttranscriptional regulators. In eukaryotes, such activity is provided by the recently described class of microRNAs (miRNAs) referred to as mirtrons. While most pre-miRNAs are derived from Drosha enzyme processing of noncoding RNAs, mirtrons utilize RNA splicing mechanisms to generate pre-miRNAs derived from the introns of mRNAs (Curtis et al., 2012; Westholm and Lai, 2011). In bacteria, posttranscriptional processing of mRNA UTRs similarly provides an abundant source of riboregulatory RNAs (Caldelari et al., 2013; Chao et al., 2012; Kim et al., 2014; Loh et al., 2009; Vogel et al., 2003). In addition, the ORFs of prokaryotic genes may also contain internal promoter sequences yielding sRNA regulators (Chao et al., 2012; Guo et al., 2014). For irvA, its riboregulatory and template functions are contiguous within the same molecule (Fig. 1F), which implies that many other protein-encoding mRNAs could similarly serve as trans-acting riboregulators. Considering this possibility along with the large diversity of known mRNA-derived riboregulators, perhaps this is indicative of a much broader and more intriguing role for mRNAs as trans-acting posttranscriptional regulatory molecules, rather than simply passive templates for the production of proteins.
EXPERIMENTAL PROCEDURES
DNA manipulation and strain construction
Details of strain construction are described in Supplemental Experimental Procedures. Strains and plasmids are listed in Table S1, while primer sequences can be found Table S2. The S. mutans genome reference strain UA159 is referred to as the wild type strain throughout this study.
DDAG assay
The DDAG assay was performed similarly as previously described by Sato et al. (Sato et al., 1997). Each pair of tubes was swirled briefly and aggregation was observed as obvious clumping and cell precipitation. A robust DDAG response develops within 2 min. of dextran addition.
5’ RACE for the determination of irvR, irvA, and gyrA transcription start sites
Amplification of the 5’ ends of cDNA was performed using the FirstChoice® RLM-RACE Kit (Ambion) according the manufacturer’s instructions. The primers used for irvR, irvA, and gyrA and 5’ end determination are listed in Table S2.
mRNA stability assays
S. mutans cells were grown to mid log phase in BTR-G medium +/− 0.6% (wt/vol) xylitol. Rifampicin was added to the culture to a final concentration of 500 µg/ml. Aliquots of the culture were withdrawn at different time points after the addition of rifampicin, rapidly chilled to 4 °C by mixing with 10 ml crushed ice, and harvested by centrifugation (4000×g, 10 min, 4°C). The transcript abundance of a particular gene at each time point was determined by Northern blot analysis.
Western blot analysis
Western blot analysis was carried out using the WesternDot 625 western blot kit (Invitrogen). Following SDS- PAGE, the separated proteins were transferred to a nitrocellulose membrane (Whatman) using the mini Trans- Blot electrophoretic transfer cell (Bio-Rad) for 1 hr (200mA). The subsequent immunodetection was performed according to the manufacturer’s protocol. GbpC antiserum was kindly provided by Dr. Yutaka Sato and diluted 1:10,000 for detection. IrvA was detected used the FLAG primary antibody (Sigma) diluted 1:20,000, while LDH was detected using the HA primary antibody diluted 1:10,000 (Invitrogen).
In vivo AMT cross-linking of S. mutans cultures
Cross-linking was performed essentially as described in (Liu et al., 2003). 50 ml cultures of S. mutans cells were harvested at mid log phase and washed twice with PBS. Cells were concentrated to a density of 5×109 CFU/ml. 4'-Aminomethyl-trioxsalen hydrochloride (AMT) (Sigma) was added to the cells at a concentration of 0.2 mg ml−1. Cells treated with AMT were kept on ice for 30 min. and then irradiated using a UV lamp at 365 nm for 1 hr at 4 °C (UVP, 115V, 60Hz).
Affinity purification of gbpC-irvA mRNA coprecipitates using 5'- biotinylated oligonucleotides
50 ml mid-log phase cultures were grown in BTR-G medium + 0.6% (wt/vol) xylitol and resuspended in 1 ml hybridization buffer (20 mM pH 8.5 HEPES, 5 mM MgCl2, 300 mM KCl, 0.01% NP40, 1 mM DTT) containing 5 µl (200 U) RNaseOut ribonuclease inhibitor (Invitrogen). Cells were disrupted with glass beads using three cycles of 30 sec. homogenization with an MP FastPrep-24. Next, lysates were centrifuged at 16,000×g at 4 °C to pellet the cell debris and the supernatants were combined with 50 µL anti-gbpC or anti-irvA neutravidin beads. The neutravidin affinity matrix (Pierce) was prepared using the manufacturer’s protocol and affinity chromatography was performed as previously described (Lustig et al., 2010). Samples receiving AMT crosslinking were washed three times, whereas uncrosslinked samples received one wash. RNA was extracted from the affinity matrix using TRI reagent (Sigma).
5’ RACE of affinity purified RNA (“RACE walk”)
Details of cDNA synthesis can be found in Supplemental Experimental Procedures, while RACE walk primer sequences can be found in Table S2. The protocol is slightly modified from a previously published protocol (Lustig et al., 2010). cDNA samples were treated with 2 U RNaseH for 30 min at 37 °C and then purified using QIAquick PCR purification kit (Qiagen). 5 µl of purified cDNA was ligated with 2 µl (10 mM) T3 adaptor primer in a 10 µl reaction consisting of 10 U of T4 RNA ligase I (NEB) and 1 µl buffer and then incubated at 22 °C overnight. After heat inactivation at 65 °C for 15 min., the reaction was used as a template for PCR. Two rounds of PCR were performed using two sets of nested primers. The resulting PCR products from the second round PCR reactions were subsequently cloned into the pGEM-T Easy vector (Promega) and sequenced.
In vivo RNA mobility shift assay
After UV crosslinking with AMT, total RNA was extracted from the S. mutans cells. 20 µg total RNA was treated with 5 U Turbo DNase (Ambion) at 37 °C for 30 min. to remove genomic DNA contamination. Next, 3 µl of 20 mg ml−1 proteinase K solution (Invitrogen) was added to the sample and incubated at 42 °C for 60 min. 20 µg of total RNA was separated in a 1% agarose 0.66 M formaldehyde gel and then visualized by northern blot.
In vitro RNA mobility shift assay
In vitro synthesized gbpC and irvA transcripts were purified from 6% urea polyacrylamide gels with RNA elution buffer [0.1 M sodium acetate, 0.1% (wt/vol) SDS, 10 mM EDTA]. Duplex formation was performed at 37 °C in TMN buffer (20 mM pH 7.5 Tris acetate, 10 mM magnesium acetate, 100 mM NaCl) for 30 min. 5 nM gbpC transcript (553 nt and 528 nt) was incubated with either 1000 nM irvA transcript or a range of transcripts consisting of 250 nM, 500 nM, 750 nM, and 1000 nM. Challenge experiments were performed with increasing amounts of competitor oligonucleotide (5 nM, 10 nM, 20 nM, 40 nM) in TMN buffer at 37 °C for 30 min. Duplexes were separated in 6% native polyacrylamide gels in TBE buffer and visualized using via northern blot.
In vitro RNase J2 digestion of gbpC
In vitro RNase J2 cleavage assays were modified from a previously published protocol (Condon et al., 2008). RNase J2 endonuclease activity was assayed in a 10 µl reaction volume containing 20 mM pH 8.0 Tris-HCl, 8 mM MgCl2, 100 mM NH4Cl, 0.1 mM DTT, 20 U RNasin (Promega), and 2 g RNase J2 at 37 °C. Reactions were incubated with 250 nM gbpC for 15 or 30 minutes and stopped by adding RNA loading buffer and separated on 5%, 6%, or 8% urea-polyacrylamide gels.
FLAsH labeling and fluorescence microscopy
The green fluorescent FLAsH-EDT2 labeling reagent was used according to a previously described protocol (Lei et al., 2011). Stationary phase starter cultures were diluted in BTR-G +/− 0.6% xylitol and grown to mid-log phase before fluorescence imaging using identical exposure settings with an Olympus BX61 epifluorescence microscope.
Natural competence assay
Determination of S. mutans transformation efficiency was performed using a previously described methodology (Niu et al., 2008). Data from three independent experiments were analyzed by one-way ANOVA. An F-test of the group means was followed by a post-hoc analysis using the Fisher’s protected least significant difference (PLSD) test to assess the variance between group means.
Supplementary Material
Acknowledgments
We would like to express our gratitude to Drs. Y. Sato and J. Banas for their generous gifts of GbpC antibodies. We also acknowledge the University of Oklahoma Health Sciences Center Laboratory for Molecular Biology and Cytometry Research for their efforts with the deep sequencing of the irvA mutant library. We greatly appreciate and thank Drs. D. Dubnau, R. Tweten, and J. Vogel for their critical reading and insightful comments regarding this manuscript. This work was supported by an NIH NIDCR DE018893 grant to J.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balaban N, Novick RP. Translation of RNAIII, the Staphylococcus aureus agr regulatory RNA molecule, can be activated by a 3'-end deletion. FEMS microbiology letters. 1995;133:155–161. doi: 10.1111/j.1574-6968.1995.tb07877.x. [DOI] [PubMed] [Google Scholar]
- Banas JA, Vickerman MM. Glucan-binding proteins of the oral streptococci. Crit Rev Oral Biol Med. 2003;14:89–99. doi: 10.1177/154411130301400203. [DOI] [PubMed] [Google Scholar]
- Bandyra KJ, Said N, Pfeiffer V, Gorna MW, Vogel J, Luisi BF. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Molecular cell. 2012;47:943–953. doi: 10.1016/j.molcel.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghoff BA, Glaeser J, Sharma CM, Vogel J, Klug G. Photooxidative stress-induced and abundant small RNAs in Rhodobacter sphaeroides. Molecular microbiology. 2009;74:1497–1512. doi: 10.1111/j.1365-2958.2009.06949.x. [DOI] [PubMed] [Google Scholar]
- Biswas I, Drake L, Biswas S. Regulation of gbpC expression in Streptococcus mutans. Journal of bacteriology. 2007;189:6521–6531. doi: 10.1128/JB.00825-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugrysheva JV, Scott JR. The ribonucleases J1 and J2 are essential for growth and have independent roles in mRNA decay in Streptococcus pyogenes. Molecular microbiology. 2010;75:731–743. doi: 10.1111/j.1365-2958.2009.07012.x. [DOI] [PubMed] [Google Scholar]
- Caldelari I, Chao Y, Romby P, Vogel J. RNA-Mediated Regulation in Pathogenic Bacteria. Cold Spring Harbor perspectives in medicine. 2013;3 doi: 10.1101/cshperspect.a010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3' UTRs as a genomic reservoir of regulatory small RNAs. The EMBO journal. 2012;31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C. What is the role of RNase J in mRNA turnover? RNA biology. 2010;7:316–321. doi: 10.4161/rna.7.3.11913. [DOI] [PubMed] [Google Scholar]
- Condon C, Pellegrini O, Mathy N, Benard L, Redko Y, Oussenko IA, Deikus G, Bechhofer DH. Assay of Bacillus subtilis ribonucleases in vitro. Methods in enzymology. 2008;447:277–308. doi: 10.1016/S0076-6879(08)02215-5. [DOI] [PubMed] [Google Scholar]
- Cornejo OE, Lefebure T, Bitar PD, Lang P, Richards VP, Eilertson K, Do T, Beighton D, Zeng L, Ahn SJ, et al. Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans. Molecular biology and evolution. 2013;30:881–893. doi: 10.1093/molbev/mss278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis HJ, Sibley CR, Wood MJ. Mirtrons, an emerging class of atypical miRNA. Wiley interdisciplinary reviews. 2012;3:617–632. doi: 10.1002/wrna.1122. [DOI] [PubMed] [Google Scholar]
- De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. The Journal of biological chemistry. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers G, Morissette A, Prevost K, Masse E. Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. The EMBO journal. 2009;28:1551–1561. doi: 10.1038/emboj.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B.subtilis with functional homology to E.coli RNase E. Nucleic acids research. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figaro S, Durand S, Gilet L, Cayet N, Sachse M, Condon C. Bacillus subtilis mutants with knockouts of the genes encoding ribonucleases RNase Y and RNase J1 are viable, with major defects in cell morphology, sporulation, and competence. Journal of bacteriology. 2013;195:2340–2348. doi: 10.1128/JB.00164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes & development. 2009;23:2004–2015. doi: 10.1101/gad.541609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich KS, Papenfort K, Fekete A, Vogel J. A small RNA activates CFA synthase by isoform-specific mRNA stabilization. The EMBO journal. 2013 doi: 10.1038/emboj.2013.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich KS, Vogel J. Activation of gene expression by small RNA. Current opinion in microbiology. 2009;12:674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Gao P, Pinkston KL, Nallapareddy SR, van Hoof A, Murray BE, Harvey BR. Enterococcus faecalis rnjB is required for pilin gene expression and biofilm formation. Journal of bacteriology. 2010;192:5489–5498. doi: 10.1128/JB.00725-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. Roles of mRNA stability, translational regulation, and small RNAs in stress response regulation. 2. Washington D.C.: ASM Press; 2011. [Google Scholar]
- Gripenland J, Netterling S, Loh E, Tiensuu T, Toledo-Arana A, Johansson J. RNAs: regulators of bacterial virulence. Nature reviews. 2010;8:857–866. doi: 10.1038/nrmicro2457. [DOI] [PubMed] [Google Scholar]
- Guo MS, Updegrove TB, Gogol EB, Shabalina SA, Gross CA, Storz G. MicL, a new sigmaE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes & development. 2014;28:1620–1634. doi: 10.1101/gad.243485.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idone V, Brendtro S, Gillespie R, Kocaj S, Peterson E, Rendi M, Warren W, Michalek S, Krastel K, Cvitkovitch D, et al. Effect of an orphan response regulator on Streptococcus mutans sucrose-dependent adherence and cariogenesis. Infection and immunity. 2003;71:4351–4360. doi: 10.1128/IAI.71.8.4351-4360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HM, Shin JH, Cho YB, Roe JH. Inverse regulation of Fe- and Ni-containing SOD genes by a Fur family regulator Nur through small RNA processed from 3'UTR of the sodF mRNA. Nucleic acids research. 2014;42:2003–2014. doi: 10.1093/nar/gkt1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnik-Habrink M, Lewis RJ, Mader U, Stulke J. RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases. Molecular microbiology. 2012;84:1005–1017. doi: 10.1111/j.1365-2958.2012.08072.x. [DOI] [PubMed] [Google Scholar]
- Lei Y, Zhang Y, Guenther BD, Kreth J, Herzberg MC. Mechanism of adhesion maintenance by methionine sulphoxide reductase in Streptococcus gordonii. Molecular microbiology. 2011;80:726–738. doi: 10.1111/j.1365-2958.2011.07603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li de la Sierra-Gallay I, Zig L, Jamalli A, Putzer H. Structural insights into the dual activity of RNase J. Nature structural & molecular biology. 2008;15:206–212. doi: 10.1038/nsmb.1376. [DOI] [PubMed] [Google Scholar]
- Li L, Huang D, Cheung MK, Nong W, Huang Q, Kwan HS. BSRD: a repository for bacterial small regulatory RNA. Nucleic acids research. 2013;41:D233–238. doi: 10.1093/nar/gks1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Ben-Shlomo H, Xu YX, Stern MZ, Goncharov I, Zhang Y, Michaeli S. The trypanosomatid signal recognition particle consists of two RNA molecules, a 7SL RNA homologue and a novel tRNA-like molecule. The Journal of biological chemistry. 2003;278:18271–18280. doi: 10.1074/jbc.M209215200. [DOI] [PubMed] [Google Scholar]
- Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- Lustig Y, Wachtel C, Safro M, Liu L, Michaeli S. 'RNA walk' a novel approach to study RNA-RNA interactions between a small RNA and its target. Nucleic acids research. 2010;38:e5. doi: 10.1093/nar/gkp872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch DJ, Fountain TL, Mazurkiewicz JE, Banas JA. Glucan-binding proteins are essential for shaping Streptococcus mutans biofilm architecture. FEMS microbiology letters. 2007;268:158–165. doi: 10.1111/j.1574-6968.2006.00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader U, Zig L, Kretschmer J, Homuth G, Putzer H. mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Molecular microbiology. 2008;70:183–196. doi: 10.1111/j.1365-2958.2008.06400.x. [DOI] [PubMed] [Google Scholar]
- Mangold M, Siller M, Roppenser B, Vlaminckx BJ, Penfound TA, Klein R, Novak R, Novick RP, Charpentier E. Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Molecular microbiology. 2004;53:1515–1527. doi: 10.1111/j.1365-2958.2004.04222.x. [DOI] [PubMed] [Google Scholar]
- Mathy N, Hebert A, Mervelet P, Benard L, Dorleans A, Li de la Sierra-Gallay I, Noirot P, Putzer H, Condon C. Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behaviour. Molecular microbiology. 2010;75:489–498. doi: 10.1111/j.1365-2958.2009.07004.x. [DOI] [PubMed] [Google Scholar]
- Merritt J, Kreth J, Shi W, Qi F. LuxS controls bacteriocin production in Streptococcus mutans through a novel regulatory component. Molecular microbiology. 2005;57:960–969. doi: 10.1111/j.1365-2958.2005.04733.x. [DOI] [PubMed] [Google Scholar]
- Niu G, Okinaga T, Qi F, Merritt J. The Streptococcus mutans IrvR repressor is a CI-like regulator that functions through autocleavage and Clp-dependent proteolysis. Journal of bacteriology. 2010;192:1586–1595. doi: 10.1128/JB.01261-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu G, Okinaga T, Zhu L, Banas J, Qi F, Merritt J. Characterization of irvR, a novel regulator of the irvA-dependent pathway required for genetic competence and dextran-dependent aggregation in Streptococcus mutans. Journal of bacteriology. 2008;190:7268–7274. doi: 10.1128/JB.00967-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard M, Johansen J, Moller-Jensen J, Valentin-Hansen P. Switching off small RNA regulation with trap-mRNA. Molecular microbiology. 2009;73:790–800. doi: 10.1111/j.1365-2958.2009.06807.x. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell. 2013;153:426–437. doi: 10.1016/j.cell.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nature structural & molecular biology. 2009;16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- Plumbridge J, Bossi L, Oberto J, Wade JT, Figueroa-Bossi N. Interplay of transcriptional and small RNA-dependent control mechanisms regulates chitosugar uptake in Escherichia coli and Salmonella. Molecular microbiology. 2014;92:648–658. doi: 10.1111/mmi.12573. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA (New York, NY. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JB, Balasubramanian D, Vanderpool CK. Small RNA binding-site multiplicity involved in translational regulation of a polycistronic mRNA. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2691–2698. doi: 10.1073/pnas.1207927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Scott JR. RivR and the small RNA RivX: the missing links between the CovR regulatory cascade and the Mga regulon. Molecular microbiology. 2007;66:1506–1522. doi: 10.1111/j.1365-2958.2007.06015.x. [DOI] [PubMed] [Google Scholar]
- Romby P, Charpentier E. An overview of RNAs with regulatory functions in gram-positive bacteria. Cell Mol Life Sci. 2010;67:217–237. doi: 10.1007/s00018-009-0162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Senpuku H, Okamoto K, Hanada N, Kizaki H. Streptococcus mutans binding to solid phase dextran mediated by the glucan-binding protein C. COral microbiology and immunology. 2002;17:252–256. doi: 10.1034/j.1399-302x.2002.170408.x. [DOI] [PubMed] [Google Scholar]
- Sato Y, Yamamoto Y, Kizaki H. Cloning and sequence analysis of the gbpC gene encoding a novel glucan-binding protein of Streptococcus mutans. Infection and immunity. 1997;65:668–675. doi: 10.1128/iai.65.2.668-675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yamamoto Y, Kizaki H. Xylitol-induced elevated expression of the gbpC gene in a population of Streptococcus mutans cells. European journal of oral sciences. 2000;108:538–545. doi: 10.1034/j.1600-0722.2000.00928.x. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Yaguchi H, Ohtani K, Banu S, Hayashi H. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Molecular microbiology. 2002;43:257–265. doi: 10.1046/j.1365-2958.2002.02743.x. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Gonzalez N, Sorger-Domenigg T, Heeb S, Richter AS, Backofen R, Williams P, Huttenhofer A, Haas D, Blasi U. The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Molecular microbiology. 2011;80:868–885. doi: 10.1111/j.1365-2958.2011.07620.x. [DOI] [PubMed] [Google Scholar]
- Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Molecular cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Toffano-Nioche C, Luo Y, Kuchly C, Wallon C, Steinbach D, Zytnicki M, Jacq A, Gautheret D. Detection of non-coding RNA in bacteria and archaea using the DETR'PROK Galaxy pipeline. Methods (San Diego, Calif. 2013;63:60–65. doi: 10.1016/j.ymeth.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Tsang P, Merritt J, Shi W, Qi F. IrvA-dependent and IrvA-independent pathways for mutacin gene regulation in Streptococcus mutans. FEMS microbiology letters. 2006;261:231–234. doi: 10.1111/j.1574-6968.2006.00351.x. [DOI] [PubMed] [Google Scholar]
- Vanderpool CK, Balasubramanian D, Lloyd CR. Dual-function RNA regulators in bacteria. Biochimie. 2011;93:1943–1949. doi: 10.1016/j.biochi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EG. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic acids research. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm JO, Lai EC. Mirtrons: microRNA biogenesis via splicing. Biochimie. 2011;93:1897–1904. doi: 10.1016/j.biochi.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Ajdic D, Liu Y, Lynch D, Merritt J, Banas JA. Role of the Streptococcus mutans irvA gene in GbpC-independent, dextran-dependent aggregation and biofilm formation. Applied and environmental microbiology. 2009;75:7037–7043. doi: 10.1128/AEM.01015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.