Abstract
Platelets are critical for hemostasis, thrombosis, and inflammatory responses1,2, yet the events leading to mature platelet production remain incompletely understood3. The bone marrow (BM) is proposed to be a major site of platelet production although indirect evidence points towards a potential pulmonary contribution to platelet biogenesis4-7. By directly imaging the lung microcirculation in mice8, we discovered that a large number of megakaryocytes (MKs) circulate through the lungs where they dynamically release platelets. MKs releasing platelets in the lung are of extrapulmonary origin, such as the BM, where we observed large MKs migrating out of the BM space. The lung contribution to platelet biogenesis is substantial with approximately 50% of total platelet production or 10 million platelets per hour. Furthermore, we identified populations of mature and immature MKs along with hematopoietic progenitors that reside in the extravascular spaces of the lung. Under conditions of thrombocytopenia and relative stem cell deficiency in the BM9, these progenitors can migrate out of the lung, repopulate the BM, completely reconstitute blood platelet counts, and contribute to multiple hematopoietic lineages. These results position the lung as a primary site of terminal platelet production and an organ with considerable hematopoietic potential.
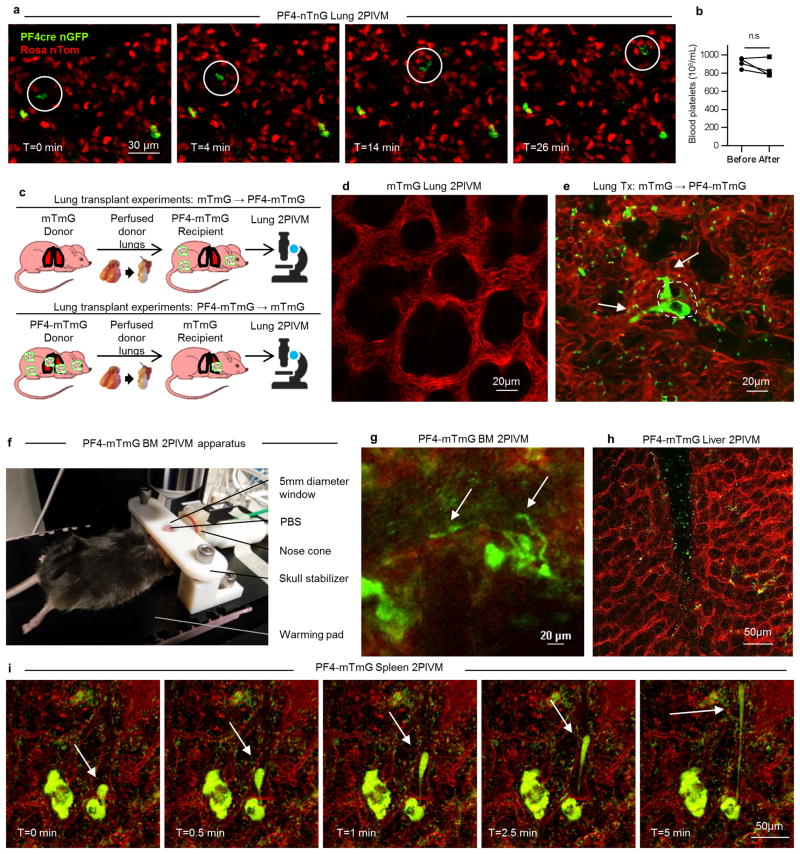
Platelets are released from MKs, but incredibly, since their discovery in the 19th century, we incompletely understand the mechanisms by which platelets are produced. Based on previous work showing the presence of MKs in the lung10 and more platelets and fewer MKs in the blood exiting than entering the lungs4,11, we hypothesized that the lung could have a major role in platelet biogenesis, and directly investigated this process using lung 2-photon intravital microscopy (2PIVM) and fluorescent mouse strains. We used PF4-Cre × mTmG (hereafter called PF4-mTmG) reporter mice, in which PF4-Cre12 drives membrane GFP expression in MKs and platelets, while all other cells are labeled with membrane tomato, and observed large circulating GFP+ cells pass through the lung microcirculation where they produce GFP+ extensions in a flow-dependent manner (Fig. 1a-b and Supplementary Video 1). These events strikingly resembled proplatelet and preplatelet formation from cultured MKs3,13,14. In the lung, the entire sequence of these events varied from approximately 20 to 60 minutes (Fig. 1a-b and Supplementary Video 1). Many of the GFP+ cells contained large nuclei (>10 μm), which appeared as unlabeled dark holes that remained intact during this process (Fig. 1b and Supplementary Video 2) and resulted in naked intravascular nuclei after platelets were released (Supplementary Video 2). We confirmed that we labeled large mobile nucleated cells by imaging the lung microcirculation of PF4-Cre × nTnG (hereafter called PF4-nTnG) reporter mice, in which a fluorescence switch allows GFP+ nuclei to be tracked (Extended Data Fig. 1a and Supplementary Video 3).
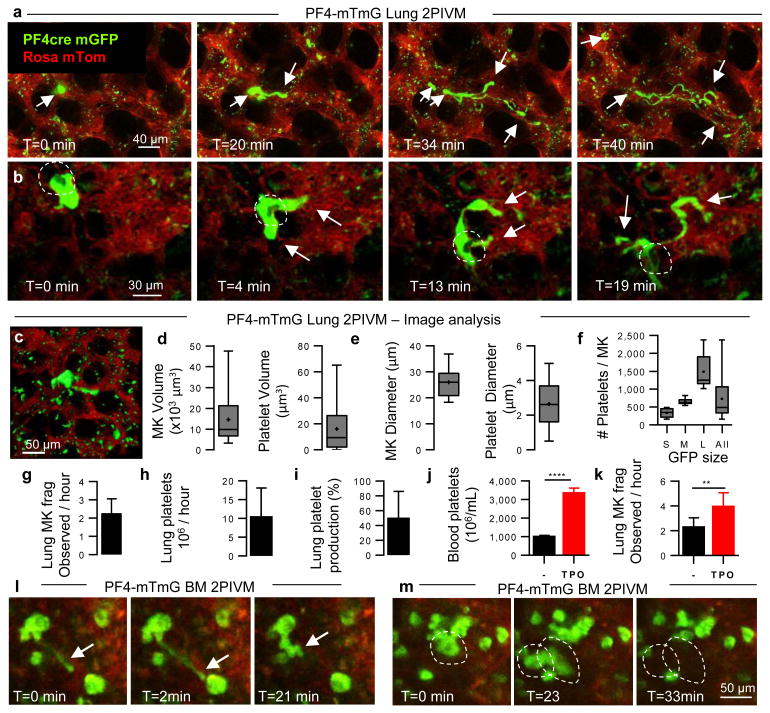
Figure 1. The lung is an important site of MK circulation and platelet production.
(a-c) Visualization of MKs and platelet production in the lung circulation by 2-photon intravital microscopy (2PIVM) in PF4-mTmG mice. (a,b) Sequential images show large-sized MK (green) in the lung capillaries (red) where it undergoes proplatelet formation (arrows). (b) Dark hole in the cytoplasm (circled) indicates the nucleus. Time elapsed is indicated. (c-f) Characterization of PF4+ events by image analysis. (c) Representative image of surface analysis of the GFP channel. (d) Volume distribution and (e) equivalent diameter of MKs and platelets. (f) Number of platelets produced by one MK according to its size: Small (<500 platelets, n=18), Medium (500-1000 platelets, n=7), Large (>1000 platelets, n=10). (d-f) Min-to-max boxplots are presented. (g-i) Quantification of lung platelet production. (g) Number of MKs releasing platelets observed per hour in imaged volume of lung (2 hour movies, n=10). (h) Estimation of the number and (i) the percentage of platelets produced by the lung. (j) Platelet counts in the blood and (k) number of MKs releasing platelets in the lung 5 days after TPO treatment. n≥5 mice per group. Unpaired t-test: ****P < 0.0001, **P < 0.005. (g-k) Mean ± S.D. are presented. (l,m) Visualization of proplatelet release (arrow) and MK migration (circled) in the BM sinusoids by 2PIVM in PF4-mTmG mice.
We next quantified the GFP+ MKs/proplatelets in the PF4-mTmG lung by assigning surface volumes (Fig. 1c and Supplementary Video 4). The putative MKs (large GFP+ cells undergoing platelet release) had median volumes of 10,000 μm3 and diameters of >25 μm (Fig. 1d, e), while the putative platelets (small circulating GFP+ events) had median volumes of <10 μm3 and diameters of 2-3 μm (Fig. 1d, e), values that are consistent with previous estimates for MKs and platelet sizes3. For each large GFP+ cell undergoing platelet release, we calculated the number of platelets that could be liberated in the lung circulation, and this ranged from <500 platelets for small-sized MKs or proplatelets to >1000 platelets for larger-sized MKs (Fig. 1f), with a median of ∼500 platelets per MK. Previous studies have estimated wide ranges for the number of platelets produced from a single MK (200-10,000 platelets)15-17. Our method uses direct measurement for each event, presumably yielding more accurate estimates. Altogether, we analyzed 20 hours of movies from 10 different mice, and observed an average of 2.2 ± 0.26 (n=10) MKs per hour in an imaged volume of lung of 0.07 mm3 (Fig. 1g and Supplementary Video 5). When extrapolated to the entire lung volume, this equals >10 million platelets produced per hour from the lungs (Fig. 1h, Methods, and Extended Data Table 1). Overall, when adjusted for platelet lifespan and splenic sequestration, we estimate that the lung is responsible for approximately 50% of total platelet production in the mouse (Fig. 1i, Methods, and Extended Data Table 1). Blood platelet counts were unchanged after 2PIVM (Extended Data Fig 1b). Lung platelet production is also biologically tunable, since the administration of the MK growth factor, thrombopoietin (TPO), increases blood platelets by 3-fold (Fig. 1j) and the number of MKs undergoing proplatelet formation observed per hour by 2-fold (3.9 ± 0.38, n=9) (Fig. 1k). We conclude from these experiments that the lung is a primary site of platelet biogenesis.
To investigate the origin of the intravascular MKs/proplatelets in the lung, we used a novel approach to adoptively transfer lung resident cells—the orthotopic single lung transplant model in mice18. We transplanted a mTmG lung (no Cre or GFP expression) into a PF4-mTmG recipient and vice versa (Extended Data Fig 1c-e and Supplementary Video 6). Using 2PIVM, we observed proplatelet formation from GFP+ MKs in the lung vasculature following mTmG → PF4-mTmG lung transplants, but not in the reverse transplant (PF4-mTmG → mTmG). These experiments confirmed that MKs releasing platelets in the lung circulation are of extrapulmonary origin. We hypothesized that the BM19, spleen, or liver could be the source of these intravascular MKs/proplatelets. We imaged the calvarial BM (Extended Data Fig. 1f) and the spleen in PF4-mTmG mice and observed extravascular MKs releasing proplatelets into the sinusoids of the BM (Fig. 1l, Extended Data Fig. 1g, and Supplementary Video 7) and spleen (Extended Data Fig. 1i and Supplementary Video 9). In the BM, we also observed large MKs exiting the BM space (Fig. 1m and Supplementary Video 8). In contrast to observations in the lung, we did not observe any intravascular MKs undergoing proplatelet formation. We did not observe any MKs in the liver (Extended Data Fig. 1h).
In addition to intravascular MKs, we also observed large cells in the perivascular lung interstitium in PF4-Cre × ROSA26-tdTomato (hereafter called PF4-tomato) and PF4-mTmG mice (Fig. 2a,b, Extended Data Fig. 2a, and Supplementary Video 10), and in PF4-mTmG→mTmG lung transplants (Extended Data Fig. 2b). These extravascular cells were sessile during our imaging (up to 4 hours) and contained large nuclei (Fig. 2c). While comparatively large, these extravascular cells were on average one-third the volume and approximately half the diameter of the intravascular MKs (Fig. 2d) and also smaller than resident MKs in the BM and spleen (Extended Data Fig. 2c-e). Using image analysis, we detected ∼2000 PF4-tomato cells per mm3 of lung tissue or >1 million cells per lung (Fig. 2e). The nuclear diameters of these cells were significantly larger than non-GFP+ cells (Fig. 2f) and had more complex shapes (Fig. 2g). We used a method of intravascular labeling prior to lung digestion20 to determine the relative proportion of intra- vs. extravascular MKs (Fig. 2i) and found that 85% of PF4-tomato events are extravascular and 15% intravascular (Fig. 2j).
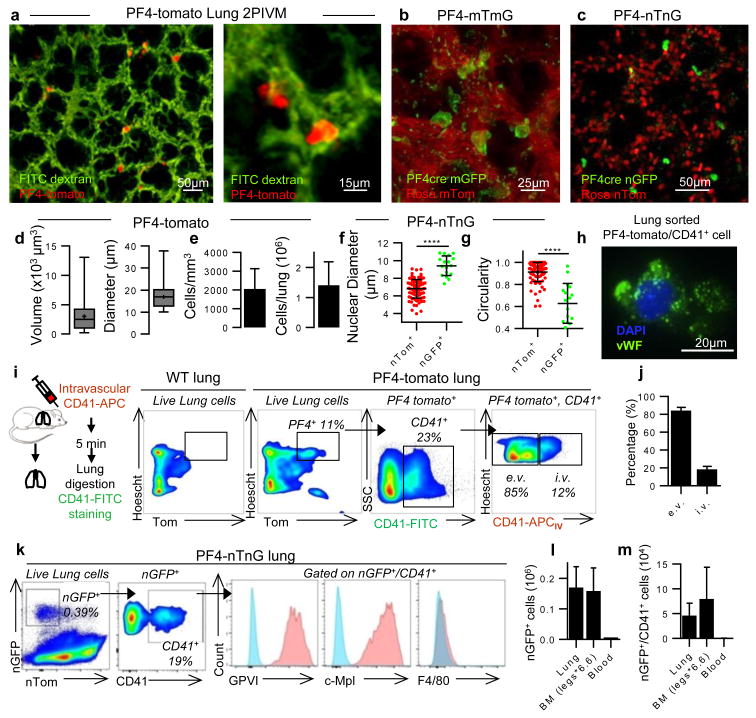
Figure 2. Resident MKs are present in the extravascular spaces of the lung.
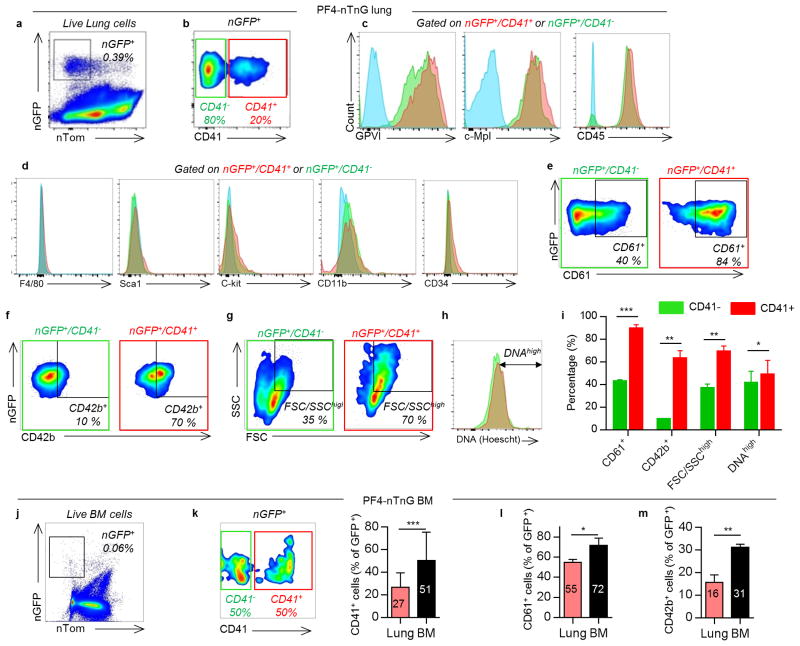
(a-c) Visualization of resident/static MKs in the lung by 2PIVM of (a) PF4-tomato, (b) PF4-mTmG, or (c) PF4-nTnG mice. (d) Size characterization of PF4+ cells (red, >10μm) by quantitative image analysis of PF4-tomato lungs. Min-to-max boxplots are presented. (e) Quantification of PF4+ cells (red, >10μm). (f) Comparison of nuclear size and (g) circularity between PF4+ cells (green) and all other lung cells (red) by quantitative image analysis of PF4-nTnG lungs. Unpaired t-test: ****P < 0.0001. (h) Representative immunofluorescence images of PF4 and CD41+ cells sorted from perfused PF4-tomato lung and stained with anti-vWF (green) and DAPI (blue). (i,j) Intravascular (i.v.) or extravascular (e.v.) localization of PF4+ and CD41-FITC+ cells: (i) experimental schema, representative FACS plots, and (j) percentage (mean of 4 experiments, n=8 mice). (k) FACS gating strategy and surface expression of nucleated PF4+ and CD41+ cells from PF4-nTnG lungs. (l) FACS quantification of nucleated PF4+ and (m) nucleated PF4+/CD41+ cells present in PF4-nTnG whole lung, BM (2 femurs, 2 tibias × 6.6) and blood (1.5mL). (e-g,l,m) Mean ± S.D. are presented.
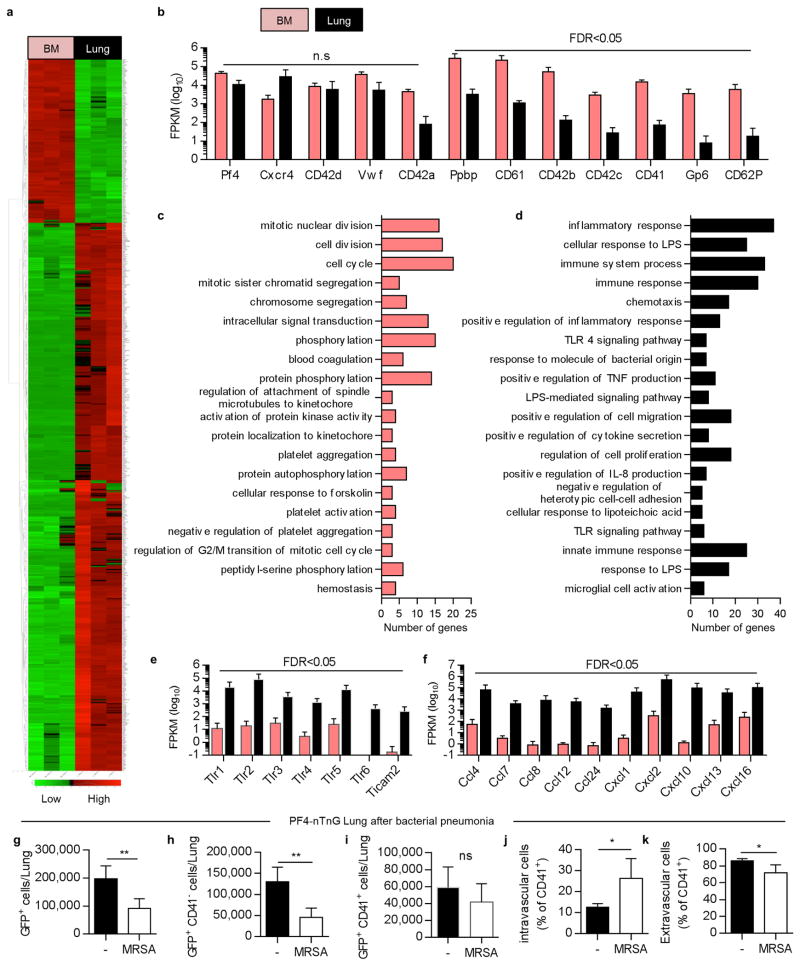
To further characterize lung MKs, we sorted PF4-tomato+ and CD41+ cells from perfused and digested lungs and stained with the MK/platelet marker, von Willebrand factor, (vWF). The large, PF4-tomato+/CD41+ cells with complex nuclei also stained positive for vWF in a granular pattern, which is consistent with MKs (Fig. 2h). To avoid cell aggregation with PF4-tomato+ platelets during flow cytometry staining, we prepared digested lungs from PF4-nTnG mice where GFP is targeted to the cell nucleus and thus does not stain anucleate platelets. We gated on nGFP+/CD41+ events as the putative lung MK pool (Fig. 2k and Extended Data Fig. 3a) and this population co-stained for the MK/platelet-specific markers, glycoprotein VI (GP VI) and the TPO receptor c-Mpl (Fig. 2k), but did not stain for markers of other lung resident cells, such as F4/80 for macrophages (Fig. 2k and Extended Data Fig. 3a-d) confirming that these lung-derived cells are MKs. The majority of the nGFP+ cells in the lung are CD41- (Extended Data Fig. 3b), but both CD41+ and negative cells co-stain for GPVI and c-Mpl confirming their MK lineage (Extended Data Fig. 3c). The nGFP+/CD41- cells have a more immature profile with lower CD61 and CD42b expression, smaller size and lower DNA content than the nGFP+/CD41+ cells (Extended Data Fig.3e-i). Overall, lung MKs are more immature than BM MKs (Extended Data Fig 3j-m), but the total number of MKs in the lung is comparable to the BM (Fig. 2l-m). We also used RNA-sequencing to characterize lung and BM MKs in PF4-nTnG mice (nGFP+/CD41+ cells). We discovered over 700 differentially-expressed genes (Extended Data Fig. 4a, Supplementary Table 1) with many MK/platelet pathways represented in both groups, but with less expression of mature MK markers in the lung group (Extended Data Fig. 4b,c) consistent with our profiling by immunostaining.
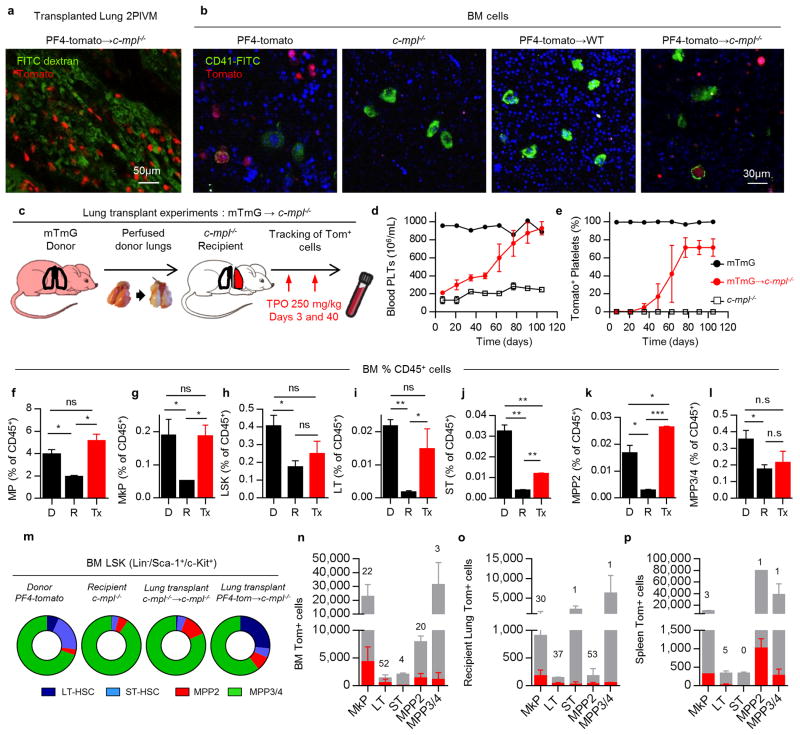
Lung 2PIVM indicated that only the intravascular MKs were active in the release of platelets. To test the function of extravascular MKs in the lung, we used the orthotopic single lung transplant model to adoptively transfer lung resident cells. PF4-tomato donor lungs were extensively perfused and the left lung was immediately transplanted into wild-type (WT) or c-mpl-/- thrombocytopenic recipients (Fig. 3a). These transplanted mice were injected with TPO at 3 and 40 days post-transplantation and bled weekly to track the numbers of platelets (Fig. 3a, c). Peripheral blood tomato+ events after lung transplantation are by definition of donor lung origin. In WT recipients, we detected low-level (1-2% of total CD41+ events) and transient production of tomato+ events (Fig. 3d, Extended Data 5i). However, in the majority (70%) of c-mpl-/- recipients, we detected a large and sustained (90 days) production of tomato+ events (Fig. 3c, d and Extended Data Fig. 5c, d) that fully reconstituted platelet counts (Fig. 3b and Extended Data Fig. 5g, h). We observed the same response in 2 out of 5 c-mpl-/- recipients not treated with TPO post-transplantation (Extended Data Fig. 5a, b, e, f). The tomato+/CD41+ events were also CD42+, GPVI+, and c-Mpl+ (Extended Data Fig. 6a, b) and expressed CD62P when stimulated with thrombin (Extended Data Fig. 6c-e), confirming their platelet nature.
Figure 3. Lung-derived progenitors reconstitute platelet counts and hematopoietic stem cell deficiency in thrombocytopenic mice.
(a-f) PF4-tomato to c-mpl-/- lung transplants. (a) Experimental schema. (b) Blood platelet counts. (c,d) %donor-derived platelets was analyzed by counting tomato+ events in the CD41+/FSClow gate. (b,d) Mean ± S.E.M. are presented. (e,f) BM cells from donor (PF4-tom), recipient (c-mpl-/-), or transplanted mice with 10 months sustained donor-derived platelet production (Lung Tx) were analyzed. (e) FACS analysis of BM cells reveals Tomato+ cells (CD41+ and CD41- populations). Percentage of lineage negative BM cells positive for Tomato and CD41. (f) Representative immunofluorescence image of Tomato+ cell (red) in BM of a transplanted mouse stained with anti-CD41 (green) and Syto60 nucleic acid stain (blue). (g-n) mTmG to c-mpl-/- lung transplants. BM cells from donor (D, mTmG), recipient (R, c-mpl-/-), or transplanted (Tx) mice with sustained donor-derived platelet production (3 months) were analyzed. (g) Representative FACS analysis of the myeloerythroid progenitor compartment (MP) and the MK progenitor population (MkP). (h) Percentage of the MkP population within the MP compartment. (i,j) Percentage of donor origin Tomato+ cells in the MkP population. (k) Representative FACS analysis of the LSK compartment (LSK). Multipotent Progenitors 2 (MPP2), Multipotent Progenitors 3 and 4 (MPP3/4), Short-term HSC (ST) and Long-term HSC (LT) population frequencies within the LSK compartment. (l) Percentage of the LT HSC population within the LSK compartment. (m,n) Percentage of donor origin Tomato+ cells in the LT population. (h,j,l,n) Mean ± S.D. are presented (n=2-4 mice per group). Unpaired t-test: **P < 0.01, *P < 0.05
In selected experiments, c-mpl-/- lung transplant recipients were followed for up to 10 months post-transplantation and we observed sustained production of tomato+ platelets and sustained reconstitution of platelet counts (Extended Data Fig. 5j, k). Since the platelet lifespan is 3-5 days in mice21, the persistence of donor-origin platelets for >3 months suggested the presence in the transplanted lung of a progenitor population capable of long-term reconstitution of mature MKs/platelets. Indeed, the smaller size of the extravascular MKs compared to the intravascular MKs in the lung and to the extravascular MKs in the BM and spleen could point to the presence of MK progenitors.
We imaged lungs at 3 months post-transplantation and confirmed the persistence of PF4-tomato cells (Extended Data Fig. 7a). We also detected the presence of tomato+/CD41+ cells in the BM of PF4-tomato→c-mpl-/- lung transplant recipients using flow cytometry (Fig. 3e) and immunofluorescence (Fig. 3f and Extended Data Fig. 7b). To test for lung MK progenitors and to track donor cells in recipient mice, we performed mTmG→c-mpl-/- lung transplants where all cells and platelets are tomato+ (Extended Data Fig.7c-e). We next quantified the numbers of MkP in the BM of mTmG→c-mpl-/- lung transplant recipients by staining myeloid progenitors (MP; Lin-/Sca-1-/c-Kit+) for CD41 and CD150 (Fig. 3g)22. We found increased MP and MkP in the BM of the lung transplant recipients relative to c-mpl-/- BM, which was similar to the frequency of MP and MkP normally found in the BM of WT (mTmG) mice (Fig. 3h and Extended Data Fig. 7f,g). One third of the MkP in the BM of mTmG→c-mpl-/- BM were tomato+ (Fig. 3i, j).
We next tested if the hematopoietic stem cell (HSC) deficiency characteristic of the c-mpl-/- BM9 could be restored by lung transplantation. We gated on the BM LSK (Lin-/Sca-1+/c-Kit+) population and probed for the following subsets: long-term (LT) HSCs (CD48-/CD150+), short-term (ST) HSCs (CD48-/CD150-), multipotent progenitor (MPP) 2 (CD48+/CD150+), and MPP3/4 (CD48+/CD150-) (Fig. 3k)23. Lung transplantation from TPO-competent donors (mTmG) restored the LSK population deficiencies (Fig. 3l and Extended Data Fig. 7h-l). Lung origin MkP, LT-HSC, ST-HSC, MPP2 and MPP3/4 were also found in the BM, spleen and recipient lung (Fig. 3m,n and Extended Data Fig. 7n-p). A non-specific post-lung transplantation response was ruled out as c-mpl-/-→c-mpl-/- lung transplants did not produce increased platelet counts (Fig. 3b) or alter the BM composition (Extended Data Fig. 7m). Taken together, these results suggest that a hematopoietic progenitor population residing in the lung is capable of migrating to the BM and reconstituting HSC defects and associated cytopenias.
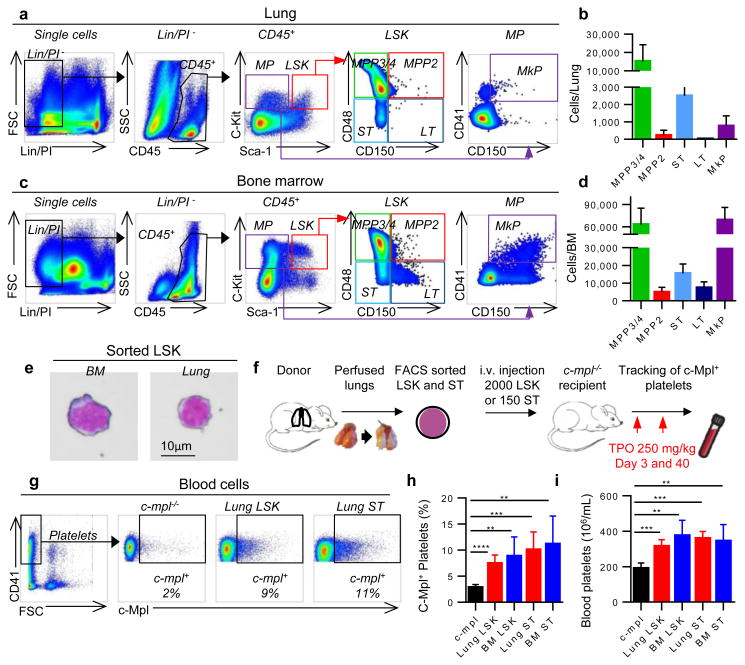
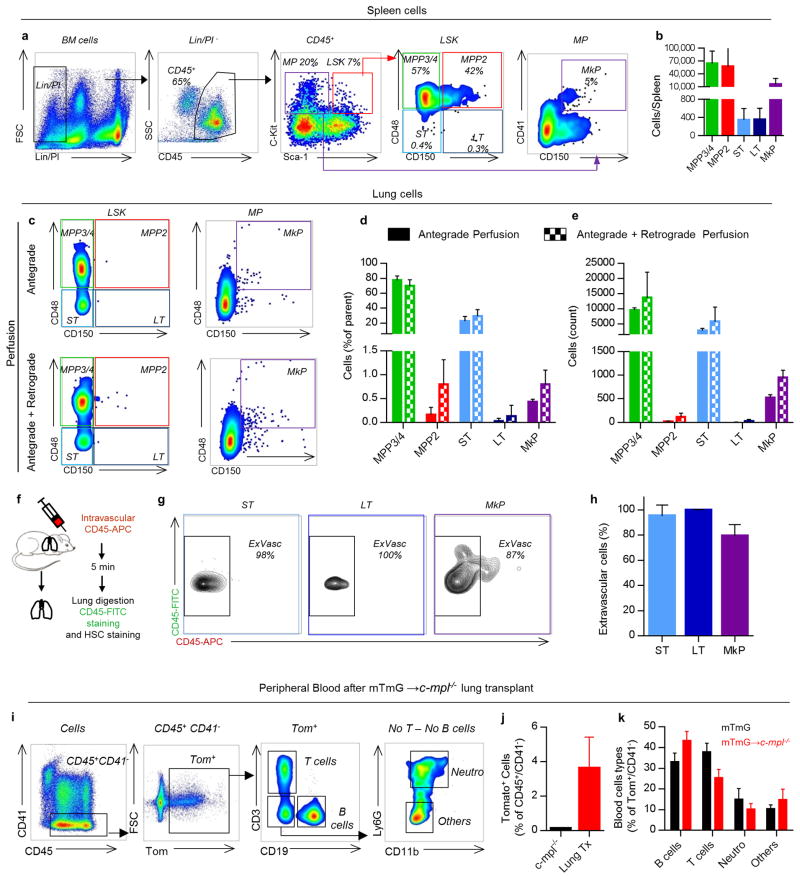
We tested for hematopoietic progenitors in the dispersed WT lung using similar gating on live CD45+/Lin-/Sca-1+/c-Kit+ cells as in the BM. We discovered that the lung contains an array of hematopoietic progenitors including ST-HSCs, MPP2, MPP3/4, and MP/MkP populations (Fig. 4a, b), which were morphologically indistinguishable from BM LSK (Fig. 4e). These cells were present at lower numbers versus the BM (Fig. 4c, d) and spleen (Extended Data Fig. 8a, b), except for larger ST-HSCs in the lung versus spleen. These cells were extravascular as they were not removed by perfusion and were not stained by intravascular CD45-APC antibody (Extended Data Fig. 8c-h). To our knowledge, this is the first description of hematopoietic progenitor cells in the adult lung, and we reasoned that they could be the source of the reconstituting effects observed with lung transplantation. To test this hypothesis, we isolated LSK and ST-HSCs from perfused WT lungs (and the BM for comparison) and injected these cells intravenously (i.v.) into c-mpl-/- recipients and tested for the presence of c-Mpl+ platelets in the peripheral blood (Fig. 4f). Transplantation of lung LSK cells and ST-HSCs increased c-Mpl+ platelets and partially restored platelet counts similar to the effect observed with BM populations (Fig. 4g-i). These results establish that the adult lung contains functional hematopoietic precursors capable of migration, BM engraftment, and reconstitution of hematopoietic defects.
Figure 4. The lung contains hematopoietic progenitors including MK progenitors.
Representative (a) lung and (c) BM FACS plots of hematopoietic progenitors within the LSK compartment (LSK) and the myeloerythroid progenitors compartment (MP). (b,d) Cell counts of hematopoietic progenitor populations in the whole lung and the BM (legs), respectively. MPP2, MPP3/4, ST-HSC, LT HSC, and MkP. (e) Representative image of Wright-Giemsa staining of LSK cells sorted from BM or lung. (f) Experimental schema. (g,h) Percentage of donor-derived platelets was analyzed by FACS counting of c-Mpl+ events in the CD41+/FSCsmall gate. (i) Blood platelet counts. (h,i) Mean ± S.D. are presented (n=4-5 mice per group). Unpaired t-test: **P < 0.01, ***P < 0.001, ****P < 0.0001
Finally, we tested whether lung hematopoietic progenitors are capable of multi-lineage BM reconstitution. Using mTmG donors to track mature lineages in the peripheral blood and BM (Extended Data Fig. 7c), we detected sustained production of donor-derived (tomato+/CD41-) cells in the peripheral blood of the c-mpl-/- recipient mice (Extended Data Fig. 8i, j), which included platelets (Extended Data Fig. 7d, e) neutrophils, and B and T cells, (Extended Data Fig. 8k). Considering there are no neutrophil, B or T cell defects in c-mpl-/- mice and therefore no impetus for donor-derived reconstitution, these results demonstrate a remarkable contribution of the lung to overall hematopoiesis.
Our results provide direct evidence that the lung is a major site of platelet biogenesis using a distinct mechanism of proplatelet release from intravascular MKs (of extrapulmonary origin) in the lung microcirculation (Extended Data Fig. 9a). These results open new lines of investigation to improve our approach to treat thrombocytopenia, which affects millions of patients worldwide and causes substantial morbidity and mortality. We propose that the lung is an ideal bioreactor for the production of mature platelets from MKs, which could advance studies of treating thrombocytopenia with cell-based therapies16. Beyond the mechanical forces that promote proplatelet formation and extension, the lung may contain unique signaling partners for MKs that promote platelet release. Interactions between MKs and endothelial cells through GPIb-vWF signaling has been shown to promote proplatelet formation in vitro24 and considering that vWF levels are particularly high in the pulmonary arteries25, this pathway could finely regulate MKs for platelet production.
The lung is a reservoir for resident MKs and hematopoietic progenitor cells (Extended Data Fig. 9b), which raises questions about the factors responsible for homing of these cells into and out of the lung, the function of these cells in the lung niche, and their roles in host defense26. Provocatively, MKs are a rich source of cytokines and growth factors that could potentially influence inflammatory/fibrotic lung diseases. Our RNA-Seq analysis revealed a skewing towards an innate immunity function of the lung MKs (Extended Data Fig. 4d-f and Supplementary Table 1) that may reflect the unique environment of the exposed lung versus the BM. Indeed, we detected changes in the resident and circulating lung MK populations in bacterial pneumonia (Extended Data Fig. 4g-k). Our findings may also find application to the field of lung transplantation where post-transplantation chimerism could affect acute and chronic allograft rejection. Our results add to the growing literature that the lung is a sophisticated organ capable of regeneration after major injury, a major site of platelet production, and with untapped potential as a contributor to hematopoiesis.
Methods
Mice
Mice were housed and bred under specific pathogen-free conditions at the University of California, San Francisco (UCSF) Laboratory Animal Research Center and all experiments conformed to ethical principles and guidelines approved by the UCSF Institutional Animal Care and Use Committee. Male and female mice between 8–12 weeks of age were used for experiments. C57BL/6, PF4-Cre, Rosa26-LSL-tdTomato, mTmG, nTnG, and BoyJ mice were purchased from the Jackson Laboratories. To track platelets and/or MKs, PF4-Cre expressing mice were crossed with Rosa26-LSL-tdTomato, mTmG or nTnG reporter strains. The fluorophore of PF4 expressing cells (tdTomato or GFP) is localized to the cytoplasm, the cell membrane, or the nucleus, respectively. c-mpl-/- mice (C57BL/6 background) were obtained from a Material Transfer Agreement from Genentech.
Lung intravital imaging
We used 2PIVM to observe MK and platelet production in real-time in the mouse lung. A modified version27 of the previously published method of stabilized lung imaging was used28. Mice were anaesthetized with ketamine and xylazine and secured with tape to a custom heated microscope stage. A small tracheal cannula was inserted, sutured into place, and attached to a MiniVent mouse ventilator (Harvard Apparatus). Mice were ventilated with a tidal volume of 10 μl of compressed air (21% O2) per gram of mouse weight, a respiratory rate of 130–140 breaths per minute, and a positive-end expiratory pressure of 2–3 cm H2O. Isoflurane was continuously delivered to maintain anesthesia and mice were administered 300 μl of 0.9% saline solution i.p. every hour. The mice were placed in the right lateral decubitus position and a small surgical incision was made to expose the rib cage. A second incision was then made into the intercostal space between ribs 4 and 5, through the parietal pleura, to expose the surface of the left lung lobe. A flanged thoracic window with 8 mm coverslip was then inserted between the two ribs and secured to the stage using a set of two optical posts and a 90° angle post clamp (Thor Labs)27,29. 20–25 mm Hg of suction was applied (Amvex Corporation) to gently immobilize the lung. The two-photon microscope objective was then lowered into place over the thoracic window. In selected experiments, to permit identification of the lung vasculature, FITC dextran (50 μL of 25 mg/mL; Life Technologies) was injected i.v. into the tail vein prior to imaging.
Spleen and Liver intravital imaging
Mice were anesthetized and ventilated as noted above. To expose the spleen, a skin incision was made in the left flank. To expose the liver, an incision was made along the costal margin to expose and externalize the liver. The same window used for lung imaging was used to facilitate imaging of the spleen and liver. The mouse was placed on a 37 °C temperature controlled heated stage for the duration of the imaging and saline solution was administered i.p. every hour.
Bone Marrow Intravital imaging
Mice were anesthetized with an initial dose of ketamine and xylazine and anesthesia was maintained with isoflurane delivered through a nose cone. Hair and the underlying subcutaneous tissue were removed to expose the calvarium. The periosteum was removed using a microsurgical knife. To stabilize the skull, we 3D printed an apparatus that was fixed to the mouse skull with Vetbond and attached to the heated stage below. The microscope objective was then lowered into a 5 mm beveled hole filled with saline.
Two-photon microscopy
Intravital imaging was performed using a Nikon A1R Multi-photon microscope equipped with a Mai Tai DeepSee IR Laser (Spectra Physics) (UCSF BIDC). The MaiTai laser was tuned to 920 nm for simultaneous excitation of GFP or FITC and tdTomato. Emitted light was detected using a 25× water lens (Nikon) and using green (500-550nm) and red (570-620nm) filters. Images were captured with a high-resolution galvano scanner (1 fps, 512×512 px). The microscope was controlled by the NIS Element AR software (4.50). We captured a 1052 μm × 1578 μm x-y surface area (1.66 mm2) and z-stack images were acquired with z-depths of 5 μm (total of 40 μm z-depth). We captured a complete image every 1 minute for 120 minutes.
Image analysis
Images were analyzed using Imaris 7.6.1 (Bitplane) or NIS-Element (Nikon) software (UCSF BIDC). Surface analysis was performed to quantify and characterize the volume, diameter, or circularity of the resident and circulating MKs or platelets. MKs/MK fragments were defined as PF4+ events with a diameter >15 μm. Platelets were defined as PF4+ events with a diameter between 0.5-5 μm. To calculate the number of platelets released by each MK observed, the ratio of the MK/platelet volumes was calculated for each of the 35 fragmenting MKs/proplatelets observed during lung imaging. The number of platelets that can be produced by one MK and was divided into 3 groups according to size: Small (<500 platelets, n=18), Medium (500-1000 platelets, n=7), Large (>1000 platelets, n=10).
Quantifying platelet production in the lung (see variables in Table 1)
For each movie (∼2 hours), the MKs observed to be undergoing fragmentation in the lung (LungMKfrag) were quantified.
The number of platelets released by each MK undergoing fragmentation in the lung was calculated using the volume ratio of the MK volume to the average platelet volume.
The number of platelets produced in the lung was then calculated.
Where the Lung fraction is the ratio of the mouse total lung volume30 to the observed lung volume:
Finally, we estimated the contribution of the lung to overall thrombopoiesis.
The total number of platelets produced per day is calculated according to the number of circulating platelets in the mouse blood divided by the life span of platelets, and taking into account that one-third of the produced platelets are sequestered by the spleen.
In selected experiments, mice were treated with recombinant human thrombopoietin (rhTPO, Genentech) i.p. (250 mg/kg), 5 days before lung imaging.
Lung, BM, spleen and blood single-cell preparation for flow cytometry or cell sorting
Lung digestion
For lung HSC or MK cell sorting, lungs were perfused before collection and digestion. Lungs were placed in 2 ml of PBS with 5 μL/mL DnaseI (Roche) and 0.5 mg/mL LiberaseTM (Roche), minced with scissors in 15mL tubes, and digested 30 minutes at 37°C before filtration through a 100 μm cell strainer and red blood cell lysis. Samples were then filtered through a 40 μm filter and resuspended for subsequent FACS staining. For experiments where vascular localization was tested, mice were injected with i.v. CD41–APC (eBioscience) or CD45-APC (eBioscience) 5 min before lung collection.
BM isolation
Tibias and femurs from both legs were collected from mice following euthanasia. BM cells were flushed with PBS with 5mM EDTA before filtration through a 70 μm cell strainer and red blood cell lysis.
Spleen isolation
The spleen was collected and pressed with the end of a plunger from a 1 ml syringe into 1 mL of PBS with 5mM EDTA before filtration through a 70 μm cell strainer and red blood cell lysis.
Flow cytometry and cell sorting
For surface staining, cells or platelets were incubated with anti-Fc receptor antibody (clone 2.4G2) and stained with antibodies in Hanks Buffered Salt Solution (HBSS) with 2% fetal calf serum and 5mM EDTA for 30 min.
Antibody clones used in these studies
CD41-APC (MWReg30, eBioscience), CD41-FITC (MWReg30, BD), CD41-BV421 (MWReg30, BioLegend), CD41-BV570 (MWReg30, BioLegend), c-mpl-Biotin (AMM2, IBL) and Streptavidin PE-Cy7 (BD), GPVI-FITC (JAQ1, emfret), F4/80-PB (CI:A3-1, BioLegend), CD45-APC (30F11, BioLegend), CD42d-APC (1C2, eBioscience), CD62P-APC (Psel.KO2.3, eBioscience). HSCs were stained with Rat unconjugated Lin antibodies Gr-1 (RB6-8C5), Mac1 (M1/70), B220 (RA3-6B2), CD5 (53-7.3), CD4 (GK1.5), CD8 (53-6.7) (eBioscience), Ter-119 (BD), CD3 (17A2, BioLegend), goat anti-rat-PE-Cy5 (Invitrogen), c-Kit-APC-eFluor780 (2B8, eBioscience), Sca-1-PB (D7, BioLegend), CD48-APC (HM48-1, BioLegend), CD150-PE-Cy7 (TC15-12F12.2, BioLegend) and CD45.2-FITC (104, eBioscience) antibodies. HSC and progenitors populations were defined as followed: Myeloerythroid progenitor (MP: Lin-/CD45+/Sca-1-/c-Kit+), MK progenitor (MkP: MP/ CD150+,CD41+), LSK compartment (LSK: Lin-/CD45+/Sca-1+/c-Kit+), Multipotent Progenitors 2 (MPP2: LSK/CD48+/CD150+), Multipotent Progenitors 3 and 4 (MPP3/4: LSK/CD48+/CD150-), Short-term HSC (ST: LSK/CD48-/CD150-) and Long-term HSC (LT: LSK/CD48-/CD150+). Mature blood cells were stained with CD19-PB (6D5, BioLegend), Gr-1 APC-Cy7 (RB6-8C5, eBioscience), CD3-PerCP710 (17A2, eBioscience), and CD11b-APC (M1/70, BD). For sorting HSCs, a c-kit+ cell enrichment step was done before staining the cells using c-kit Ab-conjugated magnetic beads and MACS separation columns (Miltenyi Biotec). Stained cells were re-suspended for final analysis in Hanks Buffered Salt Solution (HBSS) with 2% serum (FCS) and 1 μg/ml propidium iodide (PI) for dead cell exclusion. Samples were analysed on a LSRII flow cytometer (BD Biosciences) and cell sorting was performed using a BD FACS Aria II (BD Biosciences) in the UCSF Flow Cytometry Core. Analysis of flow cytometry data was performed using Flowjo (Treestar)
Lung perfusion
Lungs were perfused before lung transplant experiments, FACS, or cell analysis. Mice were anesthetized with an i.p. injection of ketamine (50 mg/kg) and xylazine (10 mg/kg). A 20-gauge angiocath was inserted into the trachea and connected to a ventilator. Through a midline abdominal incision the diaphragm was incised circumferentially and 0.1 ml of heparin was injected directly into the inferior vena cava. A thoracotomy was performed to expose the heart and the lungs. 5 ml of cold PBS with 0.1 ml of heparin solution was perfused directly into the right ventricle using a 27 gauge needle. The trachea was then tied and the heart–lung block removed and placed in a small tube containing PBS. In selected experiments, we also performed retrograde lung perfusion by instilling 5 ml of cold PBS with 0.1 ml of heparin solution into the left atrium.
Lung transplant experiments
Left lung transplants in mice were performed as previously described31. Lungs from PF4-tomato donor or mTmG mice were perfused and the left lung was immediately transplanted into C57BL/6 wild-type or c-mpl-/- recipients. Selected c-mpl-/- recipients received recombinant human thrombopoietin (rhTPO, Genentech) i.p. (250 mg/kg) at day 3 and day 40 after transplantation. Blood was collected from the submandibular vein every week to test for blood platelet counts, donor derived platelets (tomato+ platelets), and donor derived mature blood cells. The transplanted mice were analyzed for 3 months and then euthanized for BM, blood, and lung collection and analysis. In selected experiments, mice were aged to 10 months post-transplantation prior to analysis. For 2PIVM after lung transplantation, a mTmG perfused lung was transplanted into a PF4-mTmG recipient or vice-versa. The transplanted lung was imaged immediately after transplantation.
Blood collection and platelet counting
For whole blood analysis, blood was withdrawn from the submandibular vein for survival bleeding or by cardiac puncture in terminal experiments. Blood was collected into either acid citrate dextrose (Sigma-Aldrich) for flow cytometry analysis or EDTA tubes (BD microtainer) for platelet counting. Platelet counts in the peripheral blood were measured with a Hemavet 950 FS complete blood counting instrument (Drew Scientific) and confirmed with manual platelet counts.
Cell immunofluorescence
Lung MKs
Nucleated cells that were double positive for tomato and CD41 were sorted from digested lung and Cytospin slides were prepared. The cells were fixed with PFA 4% and permeabilized with 0.5% Triton. After saturation with PBS/BSA 3%, cells were stained overnight at 4°C with sheep anti-vWF antibody (Abcam) and 1 hour with anti-Sheep AlexaFluor488 antibody (Invitrogen). Slides were mounted with DAPI mounting medium (Molecular Probes) and analysed on a Nikon TI-E high-throughput epifluorescence microscope (UCSF Nikon Center).
BM cells after lung transplants
BM cells were isolated (see single-cell preparation methods) and resuspended in 1mL PBS-EDTA 5mM. 250μL of the suspension was stained 20 min at room temperature in the dark with 1 μM Syto60 (cell permeant nucleic acid stain) and anti CD41-FITC (1μL). Cells were washed, resuspended in PBS, and seeded in 6 wells plate for immediate imaging on a Nikon A1R Multi-photon microscope (UCSF BIDC). Cells were excited with a 920nm laser for simultaneous excitation of FITC (Green) tdTomato (Red) and Syto60 Nucleic acid stain (FarRed).
Platelet activation assay
To determine if the tomato+/CD41+ events detected in the peripheral blood after lung transplantation were bona fide platelets, we stimulated whole blood with thrombin (10 nM) to induce the expression of P-selectin (RB40.34, BD)32, which was measured by flow cytometric analysis and compared to platelets from PF4-tomato mice.
MRSA infection
We used the SF8300 strain of MRSA (Methicillin-resistant Staphylococcus aureus, obtained from Dr. Chip Chambers at UCSF), which is a minimally-passaged USA300 clinical strain representative of the epidemic clone USA300-0114. Stock solutions of SF8300 at the mid-logarithmic growth phase (1010 CFU/ml) were aliquoted and frozen at -80°C using standard techniques. On the day of the experiment, a vial of SF8300 was thawed and diluted with PBS to the concentration needed (5 × 107 CFU/mouse in 50 μL) for direct tracheal instillation into anesthetized PF4-nTnG mice. Lungs were harvested 24 hours after inoculation for single-cell preparation and flow cytometry. To test for vascular localization, mice were injected with i.v. CD41–APC (eBioscience), 5 min before lung collection and CD41-FITC staining.
RNA-Seq analysis
For RNA-sequencing experiments, we used PF4-nTnG mice and sorted PF4+/CD41+ cells from lung and BM prepared for single-cell analysis. For each experiment, 4 mice were pooled and cells were sorted directly into lysis buffer. Total mRNA was isolated from 1 × 104 – 4 × 105 purified cells using a Dynabeads mRNA DIRECT kit (Ambion-Life Technologies). Three independent replicates were used for each population. A mRNA library was prepared by the UCSF SABRE Functional Genomics Facility using low input Nugen Ovation plus Nextera kit (<100 ng RNA) and sequencing was performed using Single-end 50bp RNAseq Illumina TruSeq Stranded PolyA library kit and an Illumina HiSeq 4000 machine. Sequencing yielded ∼432 million total reads with an average read depth of 72 million reads/sample. Reads were then aligned to the mouse genome (Aligner: STAR_2.4.2a aligner, Alignment Genome: Ensembl Mouse GRCm38.78) and genes that mapped uniquely to known mRNAs were used to assess differential expression between lung and BM groups. Differential expression testing was carried out using DESeq2 v1.14.0.
Using a log ratio test and fold change cutoffs (false discovery rate FDR <0.05), 705 genes were found to be differentially expressed: 543 were upregulated by lung MKs and 162 by BM MKs. Functional pathways representative of each gene signature were analyzed for enrichment in gene categories from the Gene Ontology Biological processes (GO-BP) database (Gene Ontology Consortium) using DAVID Bioinformatics Resources. For gene signature and each GO category, the significance of the number of overlapping genes in the two sets was calculated using a Fisher's exact test performed by the DAVID software. The P-value resulting from the test reflects the probability of obtaining the observed overlap or greater by chance. GO-BP with at least 3 genes and P-value <0.001 were identified.
Statistical analysis
For surface analysis, min-to-max boxplots are represented: the line in the middle of the box is plotted at the median, the box extends from the 25th to 75th percentiles and the whiskers go down to the smallest value and up to the largest. The + indicates the mean. Other results plotted as histograms are reported as mean ± SD and were analyzed by t-test and multi-group comparisons were performed using a one-way ANOVA test and Bonferroni post-hoc test (GraphPad PRISM version 5.0; GraphPad Software Inc., La Jolla, CA). In all cases where statistical significance is provided, variance was not statistically different between groups. Sample sizes were chosen on the basis of previous experience in the lab with respect to inherent variability in intravital imaging, lung transplantation, and cellular transplantation experiments. There is a 10% surgical failure rate with the mouse orthotopic lung transplantation surgery, and it was pre-established that these mice would be excluded from the analysis. Animals within each cohort were randomly assigned to treatment groups. Blinded analysis was not performed in these studies. P values of less than or equal to 0.05 were considered to be statistically significant, indicated by ‘*’ in figures, unless otherwise noted.
Extended Data
Extended Data Figure 1. MKs and proplatelets observed in lung circulation are from an extrapulmonary source.
(a) Lung 2PIVM of a PF4-nTnG mouse (nuclear GFP). The presence of the mobile GFP+ nucleated cells (circled) indicates the presence of a nucleus in circulating MKs. (b) Platelet counts in the blood before and after imaging. (c) Experimental schema of mTmG (perfused donor lung) to PF4-mTmG (recipient mouse) and vice-versa lung transplantation followed by 2PIVM. (d) 2PIVM of a mTmG mouse lung showing no GFP signal. (e) 2PIVM of a mTmG mouse lung transplanted into a PF4-mTmG recipient mouse showing GFP+ cells from recipient origin and platelet production in the lung. (f) BM 2PIVM apparatus. (g) Representative image of proplatelet release in the BM sinusoids (arrows). (h) Liver 2PIVM of PF4-mTmG mouse. Small platelets (GFP, green) were seen in the circulation but neither resident nor circulating MKs or proplatelets were observed. (i) Spleen 2PIVM of PF4-mTmG mouse. Sequential images show resident MKs (GFP, green) releasing proplatelets (arrows) in the spleen vasculature (in red).
Extended Data Figure 2. Resident MKs in lung and other organs.
(a) Survey of PF4-tomato mouse lung visualized by 2PIVM. PF4-tomato expressing cells (in red) are found in high numbers in the lung. Lung vasculature is labeled by intravascular injection of FITC dextran (green). A total area of 2.49 mm2 (1.6 mm × 1.6 mm) was imaged. (b) Resident (static) GFP+ cells are found in PF4-mTmG (donor lung) to mTmG (recipient) transplanted mouse. (c) BM and (d) spleen 2PIVM images of PF4-mTmG mice. (c,d) Large-sized MKs (GFP, green) are found in the BM and spleen in large numbers. (e) Size characterization of resident (static) GFP+ cells by image analysis of PF4-mTmG lungs, BM, and spleen. Min-to-max boxplots are presented: the line in the middle of the box is plotted at the median, the box extends from the 25th to 75th percentiles and the whiskers range from the smallest to the largest values. The + indicates the mean.
Extended Data Figure 3. Surface expression of lung MKs compared to BM MKs.
(a) Flow cytometric analysis of nGFP+ cells from PF4-nTnG lungs. (b) CD41 expression defines two populations of MKs: CD41+ (red) and CD41- (green). (c) Positive surface expression was detected for the following markers: GPVI, c-Mpl and CD45 in both populations. Unstained cells are plotted in blue. (d) Negative surface expression was detected for the following markers: F4/80, CD34, CD11b, Sca-1, c-Kit. (e) The CD41+ population has a higher percentage of CD61+ cells, (f) CD42b+ cells, (g) larger cells, and (h) higher DNA content, and summarized in (i). (j) Flow cytometric analysis of nGFP+ cells from PF4-nTnG BM. Compared to the lung, the BM nGFP+ population has a higher percentage of (k) CD41+ cells, (l) CD61+ cells, and (m) CD42b+ cells. Data are representative of three or more replicates. Mean ± SD are presented. Unpaired t-test: *P < 0.05, **P < 0.01, ***P < 0.001.
Extended Data Figure 4. Gene expression analysis of lung vs BM MKs and bacterial pneumonia experiment reveal an immature profile and a potential role in immunity for lung MKs.
MKs (nGFP+/CD41+) were sorted from PF4-nTnG lungs and BM followed by mRNA isolation and sequencing. (a) Relative mRNA expression is shown on a scale from low (green) to high (red). 3 independent experiments (4 mice each) were used for statistical analysis. Heatmap of all significant differentially expressed genes (false discovery rate (FDR) <0.05). (b) Read counts for MK-related genes, which are found in both lung and BM MKs, but some are underrepresented in the lung MKs. (c,d) Gene ontology biological processes analysis related to the genes downregulated (c) or upregulated (d) in the lung MKs. Top 20 biological processes are shown. Vertical axis represents gene ontology categories, while horizontal axis indicates the number of genes in each ontology category. (e) Read counts for TLR gene pathways and (f) chemokines overexpressed in the lung MKs. FPKM: Fragments per kilobase of exon per million fragments mapped. (g-k) Lung MKs and progenitor populations are altered during infection. Flow cytometric analysis of nGFP+ cells from PF4-nTnG lungs, 24 hours after intratracheal administration of Staphylococcus aureus (MRSA, 5 × 107 cfu). CD41-APC was injected i.v before lung digestion and staining with CD41-FITC. Number of cells in normal or infected lung are shown; all nGFP+ cells (g), less mature cells (nGFP+/CD41-) (h), and mature cells (nGFP+/CD41+) (i). Percentage of intravascular MKs (j) and extravascular MKs (k) in the mature population (nGFP+/CD41+). n= 4-6 mice per group.
Extended Data Figure 5. Platelet reconstitution after lung transplantation ± TPO injection.
Blood was collected from the mandibular vein every week following lung transplant. After transplant, a group of mice represented in red and orange received TPO injection (250mg/kg, day 3 and 40). The other group was left untreated and is represented in blue and purple. A fraction of mice per group showed a sustained platelet production for more than 3 months (blue and red). In the other fraction (purple and orange), the platelet production was lower and transient (less than 3 months). The fractions of mice in each group are indicated. Data from individual mice (a,c,e,g) or group average (b,d,f,h) are plotted. (a-d,i,j) Percentage of donor-derived platelets. Percentage was analyzed by FACS, counting the tomato+ platelets (CD41+/FSCsmall gate). (e-h,k) Overall platelet counts in the peripheral blood. (j,k) Plots from mice with 10 months of sustained platelet production are shown. (i) Percentage of tomato+ platelets in control lung transplants. (l) Color code for the different lung transplant groups according to lung origin (donor), recipient mouse, treatment received (±TPO) and observed response (sustained or transient).
Extended Data Figure 6. Characterization of platelets produced after lung transplantation.
(a,b) Flow cytometric analysis of tomato+ platelets observed in the blood and stained with antibodies against CD41, CD42d, GPVI and c-Mpl. Blood from (a) PF4-tomato or (b) PF4-tomato→c-mpl-/- lung transplants. (c) Platelet activation experimental schema. (d) Flow cytometric analysis of tomato+ platelets after stimulation with thrombin (10 nM) and stained with antibody against CD62P. (e) Percentage of CD62P+ platelets before and after thrombin activation. Ctrl = PF4-tomato, Lung tx = PF4-tomato→c-mpl-/-. Mean ± SD are presented (n=2-3 mice per group). Unpaired t-test: **P < 0.01, ***P < 0.001
Extended Data Figure 7. Lung and BM analysis of transplanted mice.
Mice with sustained production of lung-derived platelets were sacrificed at least 3 months after lung transplantation. (a) Representative 2PIVM image of a lung after PF4-tomato→c-mpl-/- transplant. (b) Representative 2PIVM image of flushed BM cells from PF4-tomato and c-mpl-/- mice, and PF4-tomato→WT and PF4-tomato→c-mpl-/- lung transplants. (c) Experimental schema of mTmG→c-mpl-/- lung transplants. Blood was collected from the mandibular vein every two weeks to test for donor-derived platelets (tomato+ platelets) and overall blood platelet counts. (d) Percentage of donor-derived platelets was analyzed by FACS counting of tomato+ events in the CD41+/FSCsmall gate. (e) The platelet count in peripheral blood was determined by CBC. (d,e) Mean ± S.E.M. are presented. (f-l) BM cells from donor (mTmG, D), recipient (c-mpl-/-, R), or mTmG→c-mpl-/- lung transplants (Tx) with 3 months sustained donor-derived platelet production were analyzed. Population frequencies within the CD45+ compartment: (f) Myeloerythroid progenitors (MP: Lin-/Sca-1-/c-Kit+), (g) LSK (Lin-/Sca-1+/c-Kit+), (h) MkP, (i) LT, (j) ST, (k) MPP2, and (l) MPP3/4. (m) Population frequencies within the LSK compartment from indicated groups. (n-p) Total cell populations (grey bars), donor origin tomato+ cells (red bars), and %tomato+ cells (above bars) in (n) BM, (o) recipient native lung (right lung) and (p) spleen. Mean ± SD are presented (n=2-3 mice per group). Unpaired t-test: *P < 0.05, **P < 0.01, ***P < 0.001.
Extended Data Figure 8. Lung hematopoietic progenitors are extravascular and have multi-lineage capabilities.
(a) Representative spleen FACS plots of hematopoietic progenitors within the LSK compartment (LSK: Lin-,/Sca-1+/c-Kit+) and the myeloerythroid progenitors compartment (MP: Lin-/Sca-1-/c-Kit+). (b) Cell counts of hematopoietic progenitor populations in the spleen. (c-e) Lungs were perfused before digestion. (c) Representative FACS plot of LSK and MP compartments with antegrade perfusion +/- retrograde perfusion. (d) Frequencies and (e) cell counts of lung hematopoietic progenitor populations. (f-h) CD45-APC mAb was injected i.v. via the tail vein 5 min before lung digestion and staining with CD45-FITC mAb. (g) Hematopoietic progenitor populations were examined for labeling with injected CD45 mAb by flow cytometry. (h) Percentage of CD45-FITC+ cells positive or negative (extravascular cells) for the i.v. CD45-APC mAb. (i-k) Peripheral blood was analyzed 2-3 months after mTmG→c-mpl-/- lung transplants (i) Representative FACS plot of blood cell analysis. (j) Percentage of lung-derived cells (Tom+/CD41-) in the blood after transplant. (k) Percentage of B cells (CD19+), T cells (CD3+) or neutrophils (CD11b+, Ly6G+) in the lung-derived cells (Tom+/CD41-). Mean ± SEM are presented (n=5 mice per group).
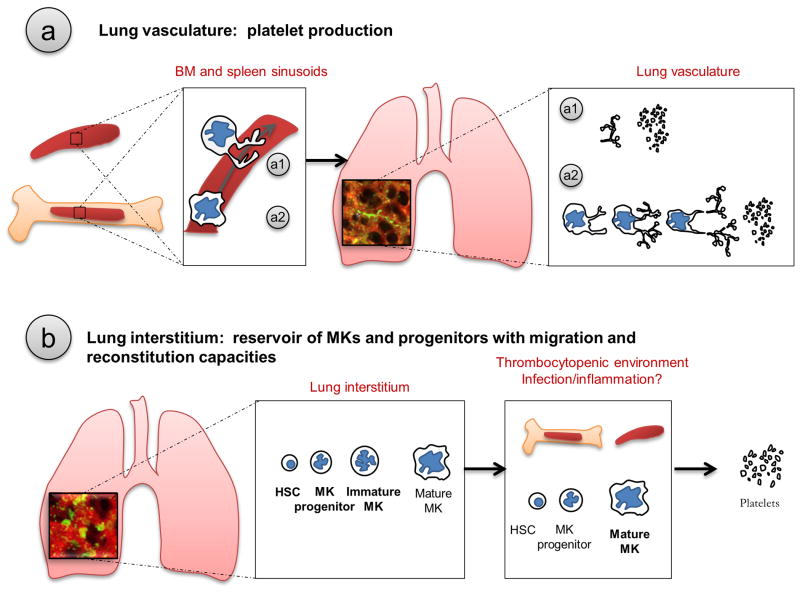
Extended Data Figure 9. Proposed schema of lung involvement in platelet biogenesis.
The role of the lung in platelet biogenesis is two-fold and occurs in two different compartments. (a) Platelet production in the lung vasculature. After being released from the BM or the spleen, proplatelets (a1) and MKs (a2) are retained in the lung vasculature, the first capillary bed encountered by any cell leaving the BM, where proplatelet formation and extension and final platelet release is observed. (b) Presence of MKs and hematopoietic progenitors in the lung interstitium. Mature and immature MKs along with hematopoietic progenitors are found in the lung interstitium. In thrombocytopenic environments, hematopoietic progenitors from the lung migrate and restore BM hematopoietic deficiencies.
Extended Data Table 1. Variables used to calculate the number and percentage of platelets produced by the lung.
| Variable | Name | Method | Value |
|---|---|---|---|
| Volume of lung imaged | Volumeobserved | Calculated from x, y, z dimensions | 0.07 mm3 |
| Volume of adult mouse lung | Volumelung | Known from literature | 700 mm3 |
| # MK undergoing fragmentation imaged/hour | Lung-MKfrag/hour | Measured using Imaris | |
| #Platelets released per MK | NPlat/MK | Calculated using Imaris | |
| Platelet life-span | LifePlat | Known from literature | 4 days |
| #Platelets in adult mouse | Platblood | Known from literature | 1.5×109 |
| #Platelets sequestered in spleen | Platspleen | Known from literature | One-third |
Supplementary Material
Acknowledgments
We thank the UCSF BIDC for assistance with 2PIVM and 3D printing; A. Hérault, E. Verovskaya and S.Y. Zhang from the Passegué laboratory for assistance with hematopoietic progenitor isolation and transplantation; D. Erle and the UCSF SABRE Functional Genomics Facility for assistance with the RNA-sequencing experiments. This work was supported in part by NIH grants HL092471 to E.P., HL107386 and HL130324 to M.R.L., the UCSF Nina Ireland Program in Lung Health (M.R.L.), and the UCSF Program for Breakthrough Biomedical Research (M.R.L.).
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions: E.L. designed and conducted most of the experiments, analyzed the data, and wrote the manuscript. G.O-M. designed and conducted experiments and analyzed the data. A.C. and B.M. conducted experiments and analyzed data. F.L. performed the lung transplantation experiments. D.M.S., E.E.T., M.B.H. and T.D. assisted in designing and conducting experiments. S.R.C, M.F.K. and A.D.L. assisted in designing experiments and provided editorial support on the manuscript. E.P. assisted in designing experiments, provided technical expertise with hematopoietic progenitor analyses, and provided editorial support on the manuscript. M.R.L. designed the experiments, conducted experiments, analyzed the data, and wrote the manuscript.
The authors declare no competing financial interests.
Data Availability Statement: The RNA-sequencing data that support the findings of this study have been deposited in the NIH SRA database with the accession code “SRP097794” (https://trace.ncbi.nlm.nih.gov/Traces/sra_sub/sub.cgi?acc=SRP097794&focus=SRP097794&from=list&action=show:STUDY).
References
- 1.Semple JW, Italiano JE, Jr, Freedman J. Platelets and the immune continuum. Nat Rev Immunol. 2011;11:264–274. doi: 10.1038/nri2956. [DOI] [PubMed] [Google Scholar]
- 2.Versteeg HH, Heemskerk JW, Levi M, Reitsma PH. New fundamentals in hemostasis. Physiol Rev. 2013;93:327–358. doi: 10.1152/physrev.00016.2011. [DOI] [PubMed] [Google Scholar]
- 3.Machlus KR, Italiano JE., Jr The incredible journey: From megakaryocyte development to platelet formation. J Cell Biol. 2013;201:785–796. doi: 10.1083/jcb.201304054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howell WH, Donahue DD. The Production of Blood Platelets in the Lungs. J Exp Med. 1937;65:177–203. doi: 10.1084/jem.65.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine RF, et al. Circulating megakaryocytes: delivery of large numbers of intact, mature megakaryocytes to the lungs. European journal of haematology. 1993;51:233–246. doi: 10.1111/j.1600-0609.1993.tb00637.x. [DOI] [PubMed] [Google Scholar]
- 6.Zucker-Franklin D, Philipp CS. Platelet production in the pulmonary capillary bed: new ultrastructural evidence for an old concept. The American journal of pathology. 2000;157:69–74. doi: 10.1016/S0002-9440(10)64518-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weyrich AS, Zimmerman GA. Platelets in lung biology. Annu Rev Physiol. 2013;75:569–591. doi: 10.1146/annurev-physiol-030212-183752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Looney MR, et al. Stabilized imaging of immune surveillance in the mouse lung. Nat Methods. 2011;8:91–96. doi: 10.1038/nmeth.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexander WS, Roberts AW, Nicola NA, Li R, Metcalf D. Deficiencies in progenitor cells of multiple hematopoietic lineages and defective megakaryocytopoiesis in mice lacking the thrombopoietic receptor c-Mpl. Blood. 1996;87:2162–2170. [PubMed] [Google Scholar]
- 10.Aschoff L. Uber capillare Embolie von riesenkemhaltigen Zellen. Arch Pathol Anat Phys. 1893;134:11–14. [Google Scholar]
- 11.Kallinikos-Maniatis A. Megakaryocytes and platelets in central venous and arterial blood. Acta Haematol. 1969;42:330–335. doi: 10.1159/000208817. [DOI] [PubMed] [Google Scholar]
- 12.Tiedt R, Schomber T, Hao-Shen H, Skoda RC. Pf4-Cre transgenic mice allow the generation of lineage-restricted gene knockouts for studying megakaryocyte and platelet function in vivo. Blood. 2007;109:1503–1506. doi: 10.1182/blood-2006-04-020362. [DOI] [PubMed] [Google Scholar]
- 13.Patel SR, Hartwig JH, Italiano JE., Jr The biogenesis of platelets from megakaryocyte proplatelets. J Clin Invest. 2005;115:3348–3354. doi: 10.1172/JCI26891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thon JN, et al. Cytoskeletal mechanics of proplatelet maturation and platelet release. J Cell Biol. 2010;191:861–874. doi: 10.1083/jcb.201006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trowbridge EA, et al. The origin of platelet count and volume. Clin Phys Physiol Meas. 1984;5:145–170. doi: 10.1088/0143-0815/5/3/007. [DOI] [PubMed] [Google Scholar]
- 16.Fuentes R, et al. Infusion of mature megakaryocytes into mice yields functional platelets. J Clin Invest. 2010;120:3917–3922. doi: 10.1172/JCI43326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaufman RM, Airo R, Pollack S, Crosby WH. Circulating megakaryocytes and platelet release in the lung. Blood. 1965;26:720–731. [PubMed] [Google Scholar]
- 18.Sayah DM, et al. Neutrophil extracellular traps are pathogenic in primary graft dysfunction after lung transplantation. Am J Respir Crit Care Med. 2015;191:455–463. doi: 10.1164/rccm.201406-1086OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Junt T, et al. Dynamic visualization of thrombopoiesis within bone marrow. Science. 2007;317:1767–1770. doi: 10.1126/science.1146304. [DOI] [PubMed] [Google Scholar]
- 20.Pereira JP, An J, Xu Y, Huang Y, Cyster JG. Cannabinoid receptor 2 mediates the retention of immature B cells in bone marrow sinusoids. Nat Immunol. 2009;10:403–411. doi: 10.1038/ni.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu ZJ, et al. Expansion of the neonatal platelet mass is achieved via an extension of platelet lifespan. Blood. 2014;123:3381–3389. doi: 10.1182/blood-2013-06-508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pronk CJ, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Pietras EM, et al. Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions. Cell Stem Cell. 2015;17:35–46. doi: 10.1016/j.stem.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunois-Larde C, et al. Exposure of human megakaryocytes to high shear rates accelerates platelet production. Blood. 2009;114:1875–1883. doi: 10.1182/blood-2009-03-209205. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto K, de Waard V, Fearns C, Loskutoff DJ. Tissue distribution and regulation of murine von Willebrand factor gene expression in vivo. Blood. 1998;92:2791–2801. [PubMed] [Google Scholar]
- 26.Haas S, et al. Inflammation-Induced Emergency Megakaryopoiesis Driven by Hematopoietic Stem Cell-like Megakaryocyte Progenitors. Cell Stem Cell. 2015;17:422–434. doi: 10.1016/j.stem.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Headley MB, et al. Visualization of immediate immune responses to pioneer metastatic cells in the lung. Nature. 2016;531:513–517. doi: 10.1038/nature16985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Looney MR, et al. Stabilized imaging of immune surveillance in the mouse lung. Nat Methods. 2011;8:91–96. doi: 10.1038/nmeth.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortiz-Munoz G, et al. Aspirin-triggered 15-epi-lipoxin A4 regulates neutrophil-platelet aggregation and attenuates acute lung injury in mice. Blood. 2014;124:2625–2634. doi: 10.1182/blood-2014-03-562876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canals M, Olivares R, Rosenmann M. A radiographic method to estimate lung volume and its use in small mammals. Biol Res. 2005;38:41–47. doi: 10.4067/s0716-97602005000100006. [DOI] [PubMed] [Google Scholar]
- 31.Sayah DM, et al. Neutrophil extracellular traps are pathogenic in primary graft dysfunction after lung transplantation. Am J Respir Crit Care Med. 2015;191:455–463. doi: 10.1164/rccm.201406-1086OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cornelissen I, et al. Roles and interactions among protease-activated receptors and P2ry12 in hemostasis and thrombosis. Proc Natl Acad Sci U S A. 2010;107:18605–18610. doi: 10.1073/pnas.1013309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.