Abstract
Gene therapy is a prospective strategy to modulate gene expression level in specific cells to treat human inherited diseases, cancers, and acquired disorders. A subset of noncoding RNAs, microRNAs (miRNAs) and small interference RNAs (siRNAs), compose an important class of widely used effectors for gene therapy, especially in cancer treatment. Functioning through the RNA interference (RNAi) mechanism, miRNA and siRNA show potent ability in silencing oncogenic factors for cancer gene therapy. For a better understanding of this field, we reviewed the mechanism and biological function, the principles of design and synthesis, and the delivery strategies of noncoding RNAs with clinical potentials in cancer gene therapy.
Keywords: Noncoding RNA, MicroRNA, Small interference RNA, RNA interference, Cancer, Gene therapy
1 Introduction
Gene therapy is a method of modulating gene expression level by introducing exogenous genetic materials into specific cells to treat human diseases including cancers. The most frequently used genetic material in gene therapy is DNA and RNA. DNA molecules used in gene therapy are usually disease-related genes (or gene fragments) to achieve gain-of-function effects, as well as antisense oligonucleotides for loss-of-function results (Zamecnik and Stephenson 1978). In the case of RNA, a subset of noncoding RNAs, microRNA (miRNA) and small interference RNA (siRNA), are emerging as popular effectors for gene therapy (Table 1). They are RNAs of small molecular weight, without protein-coding potentials. They have been shown to have potent biological functions accomplished by the mechanism of RNA interference (RNAi), which is a process triggered by double-stranded RNA molecules (Fire et al. 1998; Elbashir et al. 2001) and has been proved to be a powerful approach for reducing expression of mRNAs encoding pathogenic factors.
Table 1.
Characteristics of noncoding RNAs for cancer gene therapy
| microRNA | siRNA | shRNA | |
|---|---|---|---|
| Length | Primary (various), precursor (~70 nt), mature (18–25 nt) | ~21 nt | 25–27 nt |
| RNA structure | Primary (contains a hairpin), precursor (hairpin), mature (double-stranded) | Double-stranded | Hairpin |
| Targeting specificity | Intrinsically determined by seed sequence | Artificially designed | Artificially designed |
| Production | Chemical synthesis, viral expression, transgene expression | Chemical synthesis | Viral expression |
| Delivery method | Viral, non-viral and transgene | Non-viral vectors | Viral vectors |
1.1 Mechanism and Function of Noncoding RNAs
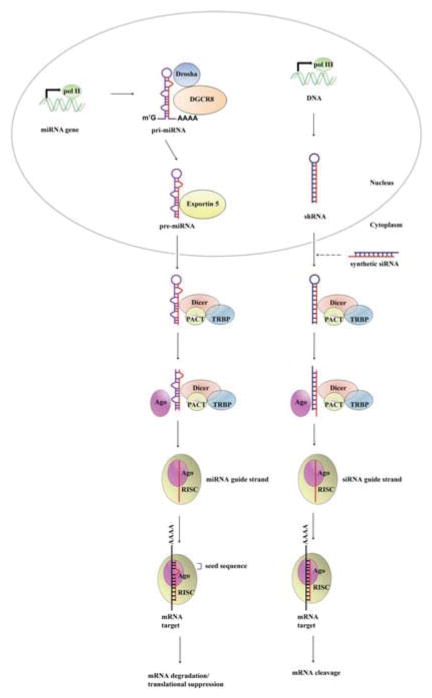
miRNAs are endogenous small noncoding RNAs of ~18–25 nucleotides in length that regulate gene expression in a sequence-specific manner via the degradation of target mRNAs or the inhibition of protein translation. Most miRNA genes are transcribed by RNA Pol II to produce primary miRNA transcripts (pri-miRNAs) that contain a 5′ cap and a 3′ poly(A) tail (Lee et al. 2004; Cai et al. 2004). Pri-miRNAs harbor local hairpin structures and flanking sequences, which are subsequently cleaved within the nucleus by Drosha and DGCR8/Pasha (Denli et al. 2004; Gregory et al. 2004; Lee et al. 2003), to generate ~70-nt hairpin precursors known as pre-miRNAs. Next, the pre-miRNA is exported into the cytoplasm by Exportin-5 and further cleaved into a mature ~22-nt miRNA:miRNA* duplex by an RNase III enzyme Dicer, and its partners TRBP (TAR-RNA binding protein)/Loquacious and PACT (protein activator of PKR) in human cells (Hutvagner et al. 2001; Ketting et al. 2001). Subsequently, an RNA-induced silencing complex called RISC is assembled with the protein Argonaute (Ago) 2 (Gregory et al. 2005; Maniataki and Mourelatos 2005). The miRNA strand is selectively incorporated into the RISC complex (Schwarz et al. 2003; Du and Zamore 2005) and guides the complex specifically to its mRNA targets through complementary base-pairing interactions between the seed sequence (base 2–8 in the 5′ end of the mature miRNA) and the binding site within target mRNAs. Through this mechanism, they exert their silencing functions by either mRNA degradation or translation inhibition (Fig. 1).
Fig. 1.
The biogenesis process and RNAi mechanism of miRNA and siRNA/shRNA. Most miRNA genes are transcribed by RNA polymerase II (pol II) to produce primary miRNA transcripts (pri-miRNAs) that contain a 5′ cap and a 3′ poly(A) tail. Pri-miRNAs are subsequently cleaved within the nucleus by Drosha and DGCR8/Pasha to generate ~70-nt hairpin precursors known as pre-miRNAs. The pre-miRNA is exported into the cytoplasm by Exportin-5 and further cleaved into a mature ~22-nt miRNA:miRNA* duplex by Dicer, and its partners TRBP and PACT. Subsequently, an RNA-induced silencing complex called RISC is assembled with the protein Argonaute (Ago). The miRNA strand (guide strand) is selectively incorporated into the RISC complex and guides the complex to its mRNA targets through complementary base-pairing interactions between the seed sequence (base 2–8 in the 5′ end of the mature miRNA) and the binding site within target mRNAs. The target mRNA is silenced by either mRNA degradation or translation inhibition. Similarly, shRNA with stem-loop structure is transcribed by RNA polymerase III (pol III). In the cytoplasm, shRNA and synthetic siRNA are subject to the processing by Dicer and its partners TRBP and PACT to give rise to double-stranded ~21-nt siRNA. The guide strand of siRNA is assembled into RISC for target cleavage and gene silencing by RNAi mechanism. miRNA, microRNA; siRNA, small interference RNA; shRNA, short hairpin RNA; RISC, RNA-induced silencing complex
miRNAs exhibit a wide range of physiological functions, especially in cancer biology. Some miRNAs act as oncogenes or tumor suppressors. For example, oncogene miR-21 is upregulated in cancer cells, promotes cell growth, and suppresses apoptosis (Chan et al. 2005; Krichevsky and Gabriely 2009). Tumor suppressor let-7 family is usually downregulated or deleted in multiple cancer types, and restoration of let-7 expression leads to regression of tumors (Kumar et al. 2008; Johnson et al. 2007; Esquela-Kerscher et al. 2008; Takamizawa et al. 2004; Yang et al. 2008). miR-200 family is well known to be associated with cancer cell metastasis and apoptosis (Park et al. 2008; Gregory et al. 2008; Schickel et al. 2010). Based on the knowledge, efforts to overexpress tumor suppressor miRNAs and inhibit oncogene miRNAs to treat cancers have achieved positive results. Slack et al. delivered exogenous let-7 to established mouse models of non-small cell lung cancer and significantly reduces tumor burden (Trang et al. 2010). Naldini et al. functionally knocked down miR-223 expression by introducing decoy miRNA targets into mouse models (Gentner et al. 2009). Techniques for successful administration of miRNAs in vivo make it possible for miRNAs to act as good candidates for cancer therapy.
Much like miRNA, siRNA and shRNA are also potent mediators of sequence-specific gene silencing by RNAi mechanism. siRNA is a double-stranded small RNA of ~21 nt in length. It is exogenously synthesized as oligonucleotides (hereafter named as siRNA), or generated by 25–27-nt short hairpin RNA (shRNA) expressed from a DNA vector. The transcript of shRNA forms a stem-loop structure which can be further processed by Dicer to give rise to double-stranded ~21-nt siRNA. By either way, the functional guide strand of siRNA is assembled into RISC for target silencing by RNAi mechanism (Fig. 1). The interactions of siRNA and their targets are based on full complementarity of base pairing, which is different from miRNAs. The biological functions of siRNAs are mainly dependent on their target genes, because siRNAs can be flexibly designed and synthesized to target and modulate the function of any transcript theoretically. To date, siRNA has been extensively used in cancer gene therapy by targeting oncogenes such as BCL-2 (in chronic myeloid leukemia), tyrosine kinase receptor EphA2 gene (in ovarian cancer cells) (Landen et al. 2005), and Ews–Fli1 gene fusion (in Ewing sarcoma cells) (Hu-Lieskovan et al. 2005).
1.2 Design and Synthesis of Noncoding RNAs for Cancer Gene Therapy
For gene therapy, the efficacy of exogenous genetic materials is largely determined by the compatibility with endogenous cellular machinery to perform their functions, as well as the delivery methods. However, multiple side effects of miRNA and siRNA in cancer gene therapy have been reported, including off-target effects, induction of immune system responses (Robbins et al. 2009), and saturation of endogenous RNAi pathway components (Khan et al. 2009; Grimm et al. 2006). The side effects sometimes can cause severe clinical outputs, thus limit the application of noncoding RNAs in gene therapy. To maximize the efficacy and minimize the side effects, it is necessary to follow some rules when designing noncoding RNAs for cancer gene therapy. Generally, it is important to consider the targeting sequences, the length, and the chemical modification of 3′ and 5′ ends of noncoding RNAs.
Synthetic miRNAs are usually present in the form of pri- or pre-miRNAs. Their targeting sequences (i.e., seed sequences) are determined by the nature of a specific miRNA. For synthetic siRNA and expressed shRNA, the targeting sequences are fully complementary to and determined by target mRNA sequences. The public TRC portal launched by the Broad Institute (The RNAi Consortium, http://www.broadinstitute.org/rnai/public/), as well as some commercial siRNA manufacturers, have developed online tools to help design specific and potent targeting sequences of siRNA based on the consideration of mRNA target sequence, secondary structures, siRNA stability, and minimizing sequence-dependent off-target effects. In addition, when designing targeting sequences for an interest gene, one should always pay attention to avoid the immunostimulatory effect of the synthetic sequences. It was reported that transfection of siRNA elicited interferon (IFN) responses (Sledz et al. 2003). A strategy to minimize this side effect is to avoid immunostimulatory sequences in siRNA design, e.g., 5′-GUCCUUCAA-3′ and 5′-UGUGU-3′ (Hornung et al. 2005; Judge et al. 2005).
In addition to the targeting sequence, the length and the modification of 3′ and 5′ ends of noncoding RNAs for cancer gene therapy also need to be paid attention to. It was reported that 27-bp double-stranded RNAs can be up to 100 times more potent than 21-mer siRNAs due to more efficient processing by Dicer, and incorporation of DNA nucleotides into siRNA also enhanced Dicer processing (Kim et al. 2004, 2005). To reduce interferon production in target cells, avoiding 5′ triphosphates of siRNA by chemical synthesis needs to be considered (Kim et al. 2004). Similarly, 2-nt 3′ overhangs alleviate interferon induction effect by resembling endogenous products processed by Dicer (e.g., mature miRNA) (Marques et al. 2006). In addition, 2′-O-methyl modification of siRNA increases the stability and retains targeting specificity, but reduces interferon production (Judge et al. 2006; Morrissey et al. 2005). Conjugating cholesterol to the sense strand of the siRNA duplex is another common modification that manifested to be a successful strategy to enhance systemic delivery efficiency by promoting liver uptake of siRNA (Soutschek et al. 2004).
1.3 Delivery of Noncoding RNAs for Cancer Gene Therapy
Besides the chemical structures (RNA sequence and end modification), in vivo delivery method is another critical determinant affecting the efficacy of noncoding RNAs for cancer gene therapy. The obstacles for in vivo delivery include protecting from endogenous nuclease digestion, evading immune detection, and promoting extravasation from blood vessels to target tissues and cells. To overcome these obstacles, a variety of in vivo delivery methods for noncoding RNAs have been developed (Table 2).
Table 2.
Delivery methods of noncoding RNAs for cancer gene therapy
| Method | RNA species delivered | Advantages | Disadvantages |
|---|---|---|---|
| Non-viral vectors | |||
| Naked delivery | miRNA, siRNA | No carriers needed | High dosage required |
| Lipid-based carriers | miRNA, siRNA | Robust, effective, and selective delivery | Sophisticated preparation needed |
| Polymersomes | siRNA | Robust, effective, and selective delivery | Sophisticated preparation needed |
| Cell-penetrating peptides | miRNA (e.g., pHLIP) | Effective and selective delivery | Expensive, sophisticated preparation |
| Inorganic nanoparticles | siRNA | Easy preparation | Limited efficiency, sometimes toxic |
| Viral vectors | miRNA, shRNA | Effective delivery, stable expression | Biosafety risk, immunogenic |
| Transgene | miRNA | Stable expression, non-immunogenic | Research purpose only |
Noncoding RNA molecules could be simply delivered in a naked form at a relatively high dosage; for example, a dose of 50 mg/kg with the inhibitor of oncomir miR-10b (antagomiR-10b) was injected via tail vein, and it successfully suppressed the metastasis of mouse breast cancer by silencing endogenous miR-10b (Ma et al. 2010). To protect noncoding RNA from degradation and enhance the delivery efficiency, a variety of synthetic vectors have been developed, such as lipid-based carriers (Li and Szoka 2007), polymersomes (Lee et al. 2005), cell-penetrating peptides (Martin and Rice 2007), and inorganic nanoparticles (Sokolova and Epple 2008). Using nanoliposomes 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC), Calin et al. demonstrated successful delivery of both miR-520d-3p and EphA2-targeting siRNA to mouse model and found that the dual therapy was more potent in antitumor efficiency than either monotherapy alone due to simultaneously targeting both EphA2 and EphB2 oncogenes (Nishimura et al. 2013). Viral vectors are also widely used to express noncoding RNAs in vivo. Commonly used viral vectors for this purpose include retrovirus, lentivirus, adenovirus, and adeno-associated virus. Using an adeno-associated virus vector, systemic administration of miR-26a in a mouse model of liver cancer resulted in retarded growth and apoptosis induction of cancer cells (Kota et al. 2009). With an adenovirus vector, Slack and colleagues successfully delivered exogenous let-7 to established mouse models of non-small cell lung cancer and significantly reduces tumor burden (Trang et al. 2010). Naldini and colleagues presented technologies to functionally knock down miRNA expression by introducing decoy miRNA targets via lentiviral vectors into mouse models (Gentner et al. 2009). In addition, novel methods for in vivo delivery of noncoding RNAs are developing very fast. Recent study by Slack et al. reported that a novel construct, attachment of peptide nucleic acid anti-miRs to a peptide with a low pH-induced transmembrane structure (pHLIP), could transport an anti-miR-155 across plasma membranes under acidic conditions and reduced tumor growth. This method could selectively target the anti-miR to the acidic tumor microenvironment, evade systemic clearance by the liver, and facilitate cell entry via a non-endocytic pathway (Cheng et al. 2015). The discovery that a small molecule enoxacin (Penetrex) could enhance the activity of the RNAi pathway may also help to increase the efficacy of in vivo delivery of miRNA and siRNA (Shan et al. 2008).
For research purpose only, the technology of transgenic animal represents a liable method that is frequently employed to study in vivo function of expressed noncoding RNAs in cancer treatment. Inducible expression of miR-21 in a conditional transgenic mouse model revealed the oncogenic role of this miRNA in inducing pre-B-cell lymphoma and supports the efforts to treat human cancers through pharmacological inactivation of miRNAs such as miR-21 (Medina et al. 2010). The transgenic method provides valuable research data and applicable experience for related clinical trials.
2 Conclusion
miRNAs and siRNAs represent an extensively used class of noncoding effectors for cancer gene therapy. They both utilize RNAi mechanism to perform their biological functions in cancer treatment. The efficiency of miRNAs and siRNAs depends on multiple factors such as targeting sequence, end modification, and systemic delivery method. The understanding of the interaction between noncoding RNAs and their targets has been applied to clinical trials. To date, the targeting siRNAs for BCL-2 (e.g., Chronic myeloid leukemia), VEGF (solid tumors), and PLK1 (e.g., liver tumor) are undergoing or have completed clinical trials (from ClinicalTrials.gov). With progress in these studies, noncoding RNAs are believed to contribute a lot more to the field of cancer gene therapy.
Acknowledgments
This work was supported, in whole or in part, by the Recruitment Project of Hundred Person of Sun Yat-Sen University (XZ), National Natural Science Foundation 81302262 (XZ), Guangdong Province Science and Technology Project 2015A020212019 (XZ), the Basser Research Center for BRCA (LZ), the National Institutes of Health R01CA142776 (LZ), R01CA190415 (LZ), P50CA083638 (LZ), P50CA174523 (LZ), the Breast Cancer Alliance (LZ), and the Marsha Rivkin Center for Ovarian Cancer Research (LZ).
Abbreviations
- miRNA
MicroRNA
- siRNA
Small interference RNA
- RNAi
RNA interference
- Ago2
Argonaute 2
- RISC
RNA-induced silencing complex
- TRBP
TAR-RNA binding protein
- PACT
Protein activator of PKR
- shRNA
Short hairpin RNA
- TRC
The RNAi Consortium
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphatidylcholine
Contributor Information
Xiaomin Zhong, Department of Obstetrics and Gynecology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA. Center for Stem Cell Biology and Tissue Engineering, Department of Biology, Zhongshan School of Medicine, Sun Yat-Sen University, Guangzhou 510080, P.R. China.
Dongmei Zhang, Department of Obstetrics and Gynecology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA. State Key Laboratory of Biotherapy/Collaborative Innovation Center of Biotherapy, West China Hospital, Sichuan University, Chengdu 610041, P.R. China.
Minmin Xiong, Center for Stem Cell Biology and Tissue Engineering, Department of Biology, Zhongshan School of Medicine, Sun Yat-Sen University, Guangzhou 510080, P.R. China.
Lin Zhang, Department of Obstetrics and Gynecology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
References
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Cheng CJ, Bahal R, Babar IA, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518(7537):107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, et al. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132(21):4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7(6):759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gentner B, Schira G, Giustacchini A, et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods. 2009;6(1):63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, et al. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Hornung V, Guenthner-Biller M, Bourquin C, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11(3):263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- Hu-Lieskovan S, Heidel JD, Bartlett DW, et al. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 2005;65(19):8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67(16):7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Judge AD, Sood V, Shaw JR, et al. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23(4):457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Judge AD, Bola G, Lee AC, et al. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13(3):494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15(20):2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AA, Betel D, Miller ML, et al. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27(6):549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Longo M, Han Y, et al. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat Biotechnol. 2004;22(3):321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- Kim DH, Behlke MA, Rose SD, et al. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23(2):222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O’Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13(1):39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105(10):3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landen CN, Jr, Chavez-Reyes A, Bucana C, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65(15):6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, MacKay JA, Frechet JM, et al. Designing dendrimers for biological applications. Nat Biotechnol. 2005;23(12):1517–1526. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- Li W, Szoka FC., Jr Lipid-based nanoparticles for nucleic acid delivery. Pharm Res. 2007;24(3):438–449. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- Ma L, Reinhardt F, Pan E, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28(4):341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19(24):2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Devosse T, Wang D, et al. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24(5):559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- Martin ME, Rice KG. Peptide-guided gene delivery. AAPS J. 2007;9(1):E18–E29. doi: 10.1208/aapsj0901003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467(7311):86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- Morrissey DV, Lockridge JA, Shaw L, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23(8):1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Jung EJ, Shah MY, et al. Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer Discov. 2013;3(11):1302–1315. doi: 10.1158/2159-8290.CD-13-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, et al. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19(2):89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- Schickel R, Park SM, Murmann AE, et al. miR-200c regulates induction of apoptosis through CD95 by targeting FAP-1. Mol Cell. 2010;38(6):908–915. doi: 10.1016/j.molcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Shan G, Li Y, Zhang J, et al. A small molecule enhances RNA interference and promotes microRNA processing. Nat Biotechnol. 2008;26(8):933–940. doi: 10.1038/nbt.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz CA, Holko M, de Veer MJ, et al. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5(9):834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- Sokolova V, Epple M. Inorganic nanoparticles as carriers of nucleic acids into cells. Angew Chem Int Ed Engl. 2008;47(8):1382–1395. doi: 10.1002/anie.200703039. [DOI] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Trang P, Medina PP, Wiggins JF, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29(11):1580–1587. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Kaur S, Volinia S, et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008;68(24):10307–10314. doi: 10.1158/0008-5472.CAN-08-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamecnik PC, Stephenson ML. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc Natl Acad Sci U S A. 1978;75(1):280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]