Abstract
Bioprocess optimization has yielded powerful clones for biotherapeutic production. However, new genomic technologies allow more targeted approaches to cell line development. Here we review efforts to enhance protein production in mammalian cells through metabolic engineering. Most efforts aimed to reduce toxic byproducts accumulation to enhance protein productivity. However, recent work highlights the possibility of regulating other desirable traits (e.g., apoptosis and glycosylation) by targeting central metabolism since these processes are interconnected. Therefore, as we further detail the pathways underlying cell growth and protein production and deploy diverse algorithms for their analysis, opportunities will arise to move beyond simple cell line designs and facilitate cell engineering strategies with complex combinations of genes that together underlie a phenotype of interest.
Graphical Abstract
Introduction
Over the past few decades, protein-based products have emerged as important biopharmaceuticals that treat complex human diseases (e.g., inflammatory disorders, cancer and infectious diseases). These drugs are predominantly synthesized in mammalian cells lines since the cells often produce high quantities of therapeutic proteins with appropriate critical quality attributes (CQAs) that impact potency and immunogenicity (e.g., glycosylation) [1]. The market for mammalian-produced therapeutic proteins has gradually grown and is projected to further increase to ~20% of the pharmaceutical market in 2017 [2].
High volumetric productivity and product titer are important to obtain more affordable protein therapeutics [3]. A cell line may achieve these goals from a combination of changes that collectively make the host system a protein “superproducer” [4]. Thus, these attributes are selected by manufacturers and include high translation efficiency, secretory capacity, growth capacity, duration of viability at maximum cell density, and human-like post-translational modifications [4, 5].
To date, most improvements in protein production have been achieved by media and bioprocess optimization (e.g., feeding strategies and process parameter control) [6]. Random mutagenesis is also been used to find cell factories with desired phenotypes. However, the availability of high-throughput omics data (genomic, transcriptomic, proteomic and metabolomic) [7] and the emergence of genome editing tools [3] provide novel opportunities for targeted genome engineering of the host cell. Indeed, they have enabled the overexpression or down-regulation of specific gene candidates to increase yield during culture and control product quality [3, 4, 8–10]. Several prominent strategies have targeted cell metabolism, cell cycle regulatory machinery, the protein secretion pathway, apoptosis, and protein glycosylation [9, 11].
Core metabolic engineering for an effective host cell alteration
The engineering approaches developed to control protein production attributes have resulted in mixed levels of success. Most successful studies targeted energy metabolism to reduce the accumulation of toxic by-products (i.e., lactate and ammonia) and/or increase metabolic efficiency (Table 1).
Table 1.
Metabolic engineering strategies used to reduce accumulation of lactate and ammonia in the culture medium
| Target gene | Effect | References |
|---|---|---|
| glucose transporter (GLUT1) | Decreased the transporter affinity to reduce flux through glycolysis and lactate production. | [12] |
| fructose transporter (GLUT5) | Transfected GLUT5 to allows cell to grow on fructose as sole carbon source which results in a reduction of sugar consumption and lactate production. | [13, 14] |
| lactate dehydrogenase (LDH) | Reduced the conversion of lactate from pyruvate and the regeneration of NAD+. | [15, 16] |
| pyruvate carboxylase (PC) | Increased the conversion of pyruvate to oxaloacetate and its entry into the TCA cycle. The overexpression of PC is often associated with a reduction in specific glucose, glutamine consumption rates and lactate to glucose yield. | [17–20] |
| Co-overexpression of alanine aminotransferase (ALT1) and taurine transporter (TAUT) | Increased transamination between 2-oxoglutarate and alanine, which accumulates early in the culture period due to the TAUT introduction. Pyruvate and glutamate were formed, thus increasing the flux through TCA cycle and reducing lactate formation. | [22] |
| carbamoyl phosphate synthetase I (CPS I) and ornithine transcarbamoylase (OTC). | Improved the first and the second steps of urea cycle, leading to decreased ammonia secretion. | [23] |
| glutamine synthetase (GS) | Improved the synthesis of glutamine from glutamate and eliminated the need of exogenous supplied glutamine and reduced ammonia accumulation | [24–26] |
| malate dehydrogenase (MDH) | Improved the conversion of oxaloacetate to malate, forced flow into TCA cycle, increased ATP and NADH intracellular levels, and improved growth. | [27] |
Several studies have successfully decreased glucose uptake by up to 50%, leading to reduced lactate production. This has been accomplished by knocking down the glucose transporter GLUT1 [12] or by transfecting the fructose transporter GLUT5 [13, 14]. An 80% reduction in lactate production was also achieved by knocking down lactate dehydrogenase [15, 16] leading to an improved product titer from 2–3-fold. Other strategies have overexpressed pyruvate carboxylase to improve the connection between glycolysis and the TCA cycle [17–20]. Interestingly, the overexpression of pyruvate carboxylase decreased glutamine consumption and extended cell viability. Zhou et al. [21] attenuated the expression of both lactate dehydrogenase and pyruvate dehydrogenase kinase using siRNAs, thus forcing the conversion of pyruvate to acetyl-coA instead of lactate. This strategy successfully reduced lactate production by almost 90% and increased specific and volumetric productivity by 75% and 68%, respectively, without decreasing cell growth. Another double target strategy to reduce lactate production was performed by [22] by overexpressing alanine aminotransferase and the taurine transporter. The overexpression of the taurine transporter in CHO cells leads to an accumulation of alanine in the early period of the culture, but an overexpression of alanine aminotransferase converts the alanine to pyruvate, which is subsequently metabolized in the TCA cycle.
In addition to lactate, efforts have been made to control other toxic byproducts that impact cell viability and product quality. For example, ammonia production has been reduced by overexpressing the first two steps of the urea cycle by up to 35% [23] and by overexpressing glutamine synthetase. Such efforts have improved the synthesis of glutamine from glutamate [24–26] and thereby improved cell phenotypes.
Beyond the aim to eliminate toxic byproducts, Chong et al. [27] engineered metabolism to improve integral cell number. They had observed that malate accumulated in the medium of CHO cultures. This suggested that a bottleneck existed at the step of the malate dehydrogenase reaction in the TCA cycle. To overcome this, they overexpressed malate dehydrogenase II to reduce NAD+ concentration and increase NADH, which was further used by oxidative phosphorylation to produce ATP, thus increasing cell viability by 1.9-fold.
Thus, numerous efforts have been made to engineer the central metabolic pathways in mammalian cell culture to reduce toxic byproducts and increase cell viability, usually by targeting just one or two genes. Further efforts will continue, especially since recent work has identified many more endogenous metabolites that inhibit growth [28] and so metabolic engineering efforts will aim to control their concentration in mammalian cell culture.
Controlling other metabolic pathways while engineering central metabolism
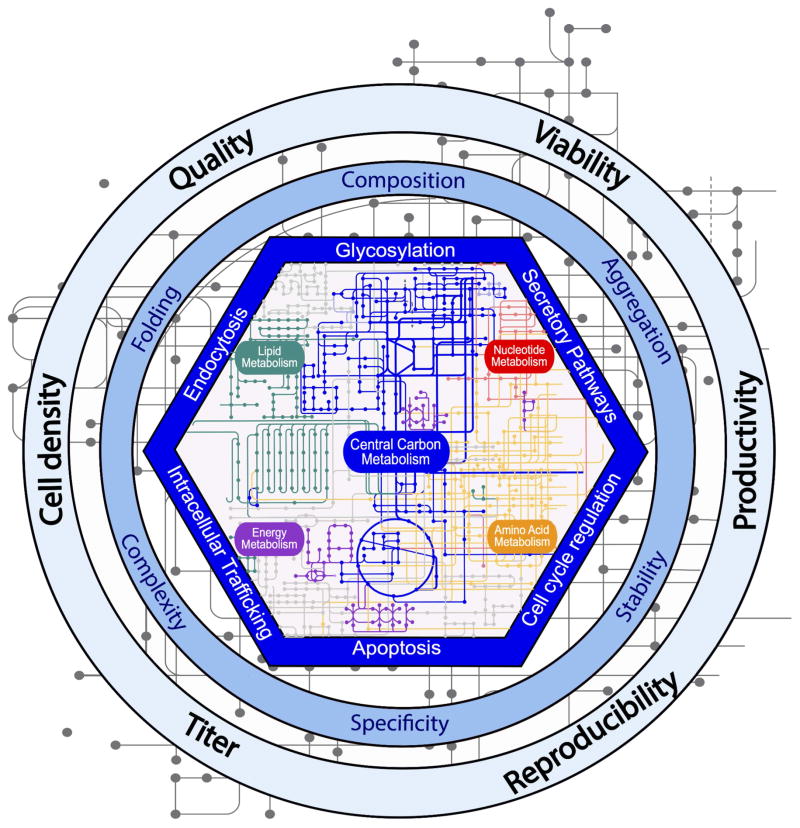
Most of the metabolic engineering efforts in the aforementioned studies aimed to reduce toxic by-product accumulation by targeting single genes related to carbohydrate metabolism (Table 1). However, studies are now tracking the influence of central carbon metabolism on other pathways, including the biosynthesis and metabolism of amino acids, nucleic acids, lipids, and ultimately protein synthesis and protein quality. The knowledge of these pathway connections can be used, for example, to track the cellular switch occurring between exponential growth and protein production, since this event changes the balance of different central metabolic pathways (e.g., pentose phosphate pathway flux and oxidative TCA cycle) [29]. For example, the ratio of flux between the pentose phosphate pathway (PPP) and glycolysis flux has been suspected to significantly impact protein production. Indeed, the PPP activity in a human cell line was much lower compared to CHO K1 cell line that produced higher amounts of protein [30, 31]. Furthermore, Mulukutla et al. [32] recently investigated the regulation of glucose metabolism, including the connections between glycolysis, the pentose phosphate pathway, nucleotide synthesis, glycerol-3 phosphate metabolism and serine/glycine/threonine biosynthesis. The interconnection between these pathways reiterates the central role of carbon metabolism of regulating the global metabolic state of a cell and other cellular processes (e.g., apoptosis, glycosylation) influencing protein quality attributes (Figure 1).
Figure 1. Central role of carbon metabolism to cell physiology, product quality, and bioprocess optimization.
The engineering of central carbon metabolism enables the regulation of key cellular processes (i.e., glycosylation, secretory pathways, cell cycle regulation, apoptosis, intracellular trafficking, and endocytosis), involved in the acquisition of protein quality attributes (i.e., protein composition, aggregation, stability, specificity, complexity, and folding). Moreover, it helps in efforts to control the global metabolic state of a cell to ensure the achievement of optimal culture process goals (i.e., cell density, viability, productivity, product titer, quality, and reproducibility).
With carbon metabolism connected to other cell processes, it is possible to control desired cellular attributes by only manipulating genes of central metabolism. For example, Majors et al. [33] inhibited apoptosis by modifying GLUT1 and/or hexokinase expression, since a high glycolytic activity can slow the onset of apoptosis by improving energy efficiency [34]. Moreover, recent studies have altered glycosylation patterns by maintaining efficient glucose metabolism and avoiding accumulation of toxic byproducts, since central metabolism controls the supply of nucleotide sugar precursors [35, 36]. Finally, Mulukutla et al. [32] highlighted several potential engineering targets within central metabolism (e.g. PFK, F26BP, PKM2, PGAM, 3PG, 3PGDH and 6PGD) that globally impact cell growth and protein production. Thus, knowledge of how different pathways are connected will guide future cell engineering efforts and enable the use of metabolic engineering to regulate the many pathways related to protein production.
Conclusion - A more holistic picture of mammalian cell physiology can guide more complex engineering strategies
The ideal CHO cell line for protein production does not have any exact, defined phenotypic traits in particular. However, it typically includes attributes of high cell viability, cell density, and titer. It also would exhibit robust growth, high stability of expression and control over desired post-translational modifications. To achieve these attributes, multiple modifications are needed in CHO cells, and such changes would target diverse cell pathways and physiological functions.
To begin identifying genes whose expression correlate with desirable attributes of protein synthesis and secretion, several studies have use diverse experimental data types to compare mammalian factories that produce high or low titers of recombinant protein. These have included data types such as transcriptomics [5, 37, 38], metabolomics [39], proteomic profiling [40, 41], and miRNA expression patterns [42]. These studies highlight the importance of intracellular trafficking, endocytosis, cytoskeletal elements, lipid metabolism, the unfolded protein response, protein-processing constituents in the Golgi, and cell cycle related functions (e.g., apoptosis resistance and proliferation) [3, 5]. However, we are still missing fundamental knowledge about how these cellular mechanisms are organized and link together. Furthermore, it is often not clear how they are connected to process conditions, and how these factors all impact protein production [43]. Thus, efforts to further enhance drug production will be facilitated as the molecular basis of these processes are studied and linked to protein production. Pathway maps and interaction networks [44–46] are starting points that link the processes, and can help identify new process conditions and cell engineering strategies that control product quantity and quality.
To completely map out the cellular pathways, it is first critical to know the genetic basis of the cells. Recent efforts to sequence the Chinese hamster genome [47–50] have enabled this for CHO cells. Furthermore, variations in the cell lines, such as mutations and epigenetic changes in individual cell lines [51], can be catalogued and analyzed to help develop engineering strategies for improved protein factories, focusing on a specific cell line.
With the genetic basis established, a holistic understanding of the cellular basis for high productivity could be achieved within the systems biology context. This will be accomplished by the development of detailed metabolic pathway and interaction maps of the major cell processes, and identifying the genes associated with the pathways [45, 52]. These metabolic networks can be converted into mathematical models that can guide engineering efforts by quantifying the connection of cellular processes to desired phenotypes and protein production using metabolic flux analysis [44, 45, 53]. Furthermore, these models will allow the analysis and integration of the avalanche of high-throughput data available at the genomic, transcriptomic, proteomic, and metabolomic levels thanks to innovations in these fields [54]. The analyses of these data in the context of cellular pathways will be particularly informative when investigated along with phenotypic differences of different cell lines, such as variations in growth media, feeding strategies, process conditions, and the type and amount of produced protein.
Finally, with the genetic basis and pathways mapped out, we can move toward the design and implementation of more complex genetic changes, using multiplex genome edits in CHO cells [55–57]. Similarly, we can harness the multiplex targeting of miRNA [58, 59]. Thus, with the guidance of predictive mathematical models of CHO cell metabolism, and emerging concepts of dynamic [60, 61] and combinatorial [8], more complex and targeted approaches can be explored to improve mammalian factories. These tools will be invaluable in the future of engineering, wherein multiplex metabolic engineering strategies account for details of cell line, culture environment and product, in the pursuit of a perfect therapeutic production factory.
Highlights.
Mammalian cell metabolism impacts therapeutic protein production
Recent sequencing efforts allow more targeted engineering approaches in CHO cells
Metabolic engineering of CHO metabolism has improved production processes
Systems biology tools can guide future engineering of metabolic phenotypes
Acknowledgments
This work was supported by generous funding from the Novo Nordisk Foundation provided to the Center for Biosustainability at the Technical University of Denmark (grant no. NNF16CC0021858), and from NIGMS (grant no. R35 GM119850).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chiang AW, Li S, Spahn PN, Richelle A, Kuo CC, Samoudi M, Lewis NE. Modulating carbohydrate-protein interactions through glycoengineering of monoclonal antibodies to impact cancer physiology. Curr Opin Struct Biol. 2016;40:104–111. doi: 10.1016/j.sbi.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh G. Biopharmaceutical Benchmarks 2014. Nat Biotechnol. 2014;32:992–1000. doi: 10.1038/nbt.3040. [DOI] [PubMed] [Google Scholar]
- 3••.Fischer S, Handrick R, Otte K. The art of CHO cell engineering: A comprehensive retrospect and future perspectives. Biotechnol Adv. 2015;33:1878–1896. doi: 10.1016/j.biotechadv.2015.10.015. A review of the main engineering techniques and the related achievements in Chinese Hamster cell lines improvements over the past three decades. [DOI] [PubMed] [Google Scholar]
- 4.Dietmair S, Nielsen LK, Timmins NE. Engineering a mammalian super producer. J of Chem Technol Biotechnol. 2011;86(7):905–914. doi: 10.1002/jctb.2576. [DOI] [Google Scholar]
- 5.Charaniya S, Karypis G, Hu WS. Mining transcriptome data for function–trait relationship of hyper productivity of recombinant antibody. Biotechnol Bioeng. 2009;102:1654–1669. doi: 10.1002/bit.22210. [DOI] [PubMed] [Google Scholar]
- 6.Dickson AJ. Enhancement of production of protein biopharmaceuticals by mammalian cell cultures: the metabolomics perspective. Curr Op in Biotechnol. 2014;30:73–79. doi: 10.1016/j.copbio.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Kildegaard HF, Baycin-Hizal D, Lewis NE, Betenbaugh MJ. The emerging CHO systems biology era: harnessing the ‘omics revolution for biotechnology. Curr Op Biotechnol. 2013;24:1102–7. doi: 10.1016/j.copbio.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Kim JY, Kim Y-G, Lee GM. CHO cells in biotechnology for production of recombinant proteins: current state and further potential. Appl Microbiol Biotechnol. 2012;93:917–930. doi: 10.1007/s00253-011-3758-5. [DOI] [PubMed] [Google Scholar]
- 9.Lim Y, Wong NSC, Lee YY, Ku SCY, Wong DCF, Yap MGS. Engineering mammalian cells in bioprocessing – current achievements and future perspectives. Biotechnol Appl Biochem. 2010;55:175–189. doi: 10.1042/BA20090363. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J. Mammalian cell protein expression for biopharmaceutical production. Biotechnol Adv. 2012;30(5):1158–1170. doi: 10.1016/j.biotechadv.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 11•.Klein T, Niklas J, Heinzle E. Engineering the supply chain for protein production/secretion in yeasts and mammalian cells. J Ind Microbiol Biotechnol. 2015;42:453. doi: 10.1007/s10295-014-1569-2. Review of the engineering strategies developed to optimize the supply chain for building blocks and energy for the production and secretion of pharmaceutical protein in yeast and mammalian cells. [DOI] [PubMed] [Google Scholar]
- 12.Paredes C, Prats E, Cairo JJ, Azorin F, Cornudella L, Godia F. Modification of glucose and glutamine metabolism in hybridoma cells through metabolic engineering. Cytotechnol. 1999;30:85–93. doi: 10.1023/A:1008012518961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue Y, Tsukamoto Y, Yamanaka M, Nakamura S, Inoue A, Nishino N, Kawahara H. Efficient production of recombinant IgG by metabolic control and co-expression with GLUT5 in a fructose-based medium. Cytotechnol. 2010;62(4):301–306. doi: 10.1007/s10616-010-9289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wlaschin KF, Hu WS. Engineering cell metabolism for high-density cell culture via manipulation of sugar transport. J Biotechnol. 2007;131:168–176. doi: 10.1016/j.jbiotec.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Chen K, Liu Q, Xie L, Sharp PA, Wang DIC. Engineering of a mammalian cell line for reduction of lactate formation and high monoclonal antibody production. Biotechnol Bioeng. 2001;72:55–61. doi: 10.1002/1097-0290(20010105)72:1<55::aid-bit8>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 16.Kim SH, Lee GM. Down-regulation of lactate dehydrogenase-A by siRNAs for reduced lactic acid formation of Chinese hamster ovary cells producing thrombopoietin. Appl Microbiol Biotechnol. 2007a;74:152–159. doi: 10.1007/s00253-006-0654-5. [DOI] [PubMed] [Google Scholar]
- 17.Elias CB, Carpentier E, Durocher Y, Bisson L, Wagner R, Kamen A. Improving Glucose and Glutamine Metabolism of Human HEK 293 and Trichoplusiani Insect Cells Engineered To Express a Cytosolic Pyruvate Carboxylase Enzyme. Biotechnol Progress. 2003;19:90–97. doi: 10.1021/bp025572x. [DOI] [PubMed] [Google Scholar]
- 18.Fogolin MB, Wagner R, Etcheverrigaray M, Kratje R. Impact of temperature reduction and expression of yeast pyruvate carboxylase on hGM-CSF-producing CHO cells. J Biotechnol. 2004;109:179–191. doi: 10.1016/j.jbiotec.2003.10.035. [DOI] [PubMed] [Google Scholar]
- 19.Irani N, Beccaría AJ, Wagner R. Expression of recombinant cytoplasmic yeast pyruvate carboxylase for the improvement of the production of human erythropoietin by recombinant BHK-21 cells. J Biotechnol. 2002;93:269–282. doi: 10.1016/s0168-1656(01)00409-6. [DOI] [PubMed] [Google Scholar]
- 20.Kim SH, Lee GM. Functional expression of human pyruvate carboxylase for reduced lactic acid formation of Chinese hamster ovary cells (DG44) Appl Microbiol Biotechnol. 2007b;76:659–665. doi: 10.1007/s00253-007-1041-6. [DOI] [PubMed] [Google Scholar]
- 21.Zhou M, Crawford Y, Ng D, Tung J, Pynn AF, Meier A, Yuk IH, Vijayasankaran N, Leach K, Joly J, et al. Decreasing lactate level and increasing antibody production in Chinese hamster ovary cells (CHO) by reducing the expression of lactate dehydrogenase and pyruvate dehydrogenase kinases. J Biotechnol. 2011;153:27–34. doi: 10.1016/j.jbiotec.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Tabuchi H, Sugiyama T. Cooverexpression of alanine aminotransferase 1 in Chinese hamster ovary cells overexpressing taurine transporter further stimulates metabolism and enhances product yield. Biotechnol Bioeng. 2013;110:2208–2215. doi: 10.1002/bit.24881. [DOI] [PubMed] [Google Scholar]
- 23.Park H, Kim IH, Kim IY, Kim KH, Kim HJ. Expression of carbamoyl phosphate synthetase I and ornithine transcarbamoylase genes in Chinese hamster ovary dhfr- cells decreases accumulation of ammonium ion in culture media. J Biotechnol. 2000;81:129–140. doi: 10.1016/s0168-1656(00)00282-0. [DOI] [PubMed] [Google Scholar]
- 24.Bell SL, Bebbington C, Scott MF, Wardell JN, Spier RE, Bushell ME, Sanders PG. Genetic engineering of hybridoma glutamine metabolism. Enzyme Microb Technol. 1995;17:98–106. doi: 10.1016/0141-0229(94)00056-w. [DOI] [PubMed] [Google Scholar]
- 25.Cockett MI, Bebbington CR, Yarranton GT. High-level expression of tissue inhibitor of metaloproteinase in Chinese hamster ovary cells using glutamine synthetase gene amplification. Biotechnol. 1990;8:662–667. doi: 10.1038/nbt0790-662. [DOI] [PubMed] [Google Scholar]
- 26.Fan L, Kadura I, Krebs LE, Hatfield CC, Shaw MM, Frye CC. Improving the efficiency of CHO cell line generation using glutamine synthetase gene knockout cells. Biotechnol Bioeng. 2012;109:1007–1015. doi: 10.1002/bit.24365. [DOI] [PubMed] [Google Scholar]
- 27.Chong WP, Reddy SG, Yusufi FN, Lee DY, Wong NS, Heng CK, Yap MG, Ho YS. Metabolomics-driven approach for the improvement of Chinese hamster ovary cell growth: overexpression of malate dehydrogenase II. J Biotechnol. 2010;147:116–121. doi: 10.1016/j.jbiotec.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 28.Hiller GW, Mulukulta BC, Kleiman GL. Method of cell culture. Patent WO2015140708. 2015
- 29.Templeton N, Dean J, Reddy P, Young JD. Peak antibody production is associated with increased oxidative metabolism in an industrially relevant fed-batch CHO cell culture. Biotechnol Bioeng. 2013;110:2013–2024. doi: 10.1002/bit.24858. [DOI] [PubMed] [Google Scholar]
- 30.Niklas J, Sandig V, Heinzle E. Metabolite channeling and compartmentation in the human cell line AGE1. HN determined by 13C labeling experiments and 13C metabolic flux analysis. J Biosci Bioeng. 2011;112(6):616–623. doi: 10.1016/j.jbiosc.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 31.Nicolae A, Wahrheit J, Bahnemann J, Zeng A-P, Heinzle E. Non-stationary 13C metabolic flux analysis of Chinese hamster ovary cells in batch culture using extracellular labeling highlights metabolic reversibility and compartmentation. BMC Syst Biol. 2014;8:50. doi: 10.1186/1752-0509-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Mulukutla BC, Yongky A, Le T, Mashek DG, Hu Ws. Regulation of Glucose Metabolism – A Perspective From Cell Bioprocessing. Trends Biotechnol. 2016;34(8):638–51. doi: 10.1016/j.tibtech.2016.04.012. A summary of the allosteric regulation of glucose metabolism occuring in cultured cells. This review also provides a holistic view of the interplay between the signalling pathways, and transcription factors. [DOI] [PubMed] [Google Scholar]
- 33.Majors BS, Betenbaugh MJ, Chiang GG. Links between metabolism and apoptosis in mammalian cells: Applications for anti-apoptosis engineering. Met Eng. 2007;9:317–326. doi: 10.1016/j.ymben.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Templeton N, Lewis A, Dorai H, Qian EA, Campbell MP, Smith KD, Betenbaugh MJ, Young JD. The impact of anti-apoptotic gene Bcl-2Δ expression on CHO central metabolism. Metab Eng. 2014;25:92–102. doi: 10.1016/j.ymben.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 35.Lewis AM, Croughan WD, Aranibar N, Lee AG, Warrack B, Abu-Absi NR, Patel R, Drew B, Borys MC, Reily MD, Jian Li Z. Understanding and controlling sialylation in a CHO Fc-Fusion process. Plos One. 2016;11(6):e0157111. doi: 10.1371/journal.pone.0157111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McAtee AG, Templeton N, Young JD. Role of Chinese hamster ovary central carbon metabolism in controlling the quality of secreted biotherapeutic proteins. Pharm Bioprocess. 2014;2(1):63–74. doi: 10.4155/pbp.13.65. [DOI] [Google Scholar]
- 37.Clarke C, Doolan P, Barron N, Meleady P, O’Sullivan F, Gammell P, Melville M, Leonard M, Clynes M. Large scale microarray profiling and coexpression network analysis of CHO cells identifies transcriptional modules associated with growth and productivity. J Biotechnol. 2011;155(3):350–359. doi: 10.1016/j.jbiotec.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 38.Yee JC, Gerdtzen ZP, Hu WS. Comparative transcriptome analysis to unveil genes affecting recombinant protein productivity in mammalian cells. Biotechnol Bioeng. 2009;102:246–263. doi: 10.1002/bit.22039. [DOI] [PubMed] [Google Scholar]
- 39.Ghorbaniaghdam A, Chen J, Henry O, Jolicoeur M. Analyzing clonal variation of monoclonal antibody-producing CHO cell lines using an in silico metabolomic platform. PLoS One. 2014;9(3):e90832. doi: 10.1371/journal.pone.0090832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carlage T, Hincapie M, Zang L, Lyubarskaya Y, Madden H, Mhatre R, Hancock WS. Proteomic profiling of a high-producing Chinese hamster ovary cell culture. Anal Chem. 2009;81:7357–7362. doi: 10.1021/ac900792z. [DOI] [PubMed] [Google Scholar]
- 41.Orellana CA, Marcellin E, Schulz BL, Nouwens AS, Gray PP, Nielsen LK. High-antibody-producing Chinese hamster ovary cells up-regulate intracellular protein transport and glutathione synthesis. J Proteome Res. 2015;14(2):609–618. doi: 10.1021/pr501027c. [DOI] [PubMed] [Google Scholar]
- 42.Maccani A, Hackl M, Leitner C, Steinfellner W, Graf AB, Tatto NE, Karbiener M, Scheideler M, Grillari J, Mattanovich D, et al. Identification of microRNAs specific for high producer CHO cell lines using steady-state cultivation. Appl Microbiol Biotechnol. 2014;98(17):7535–48. doi: 10.1007/s00253-014-5911-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farrell A, McLoughlin N, Milne JJ, Marison IW, Bones J. Application of Multi-Omics Techniques for Bioprocess Design and Optimization in Chinese Hamster Ovary Cells. J Proteome Res. 2014;13:3144–3159. doi: 10.1021/pr500219b. [DOI] [PubMed] [Google Scholar]
- 44.Gutierrez JM, Lewis NE. Optimizing eukaryotic cell hosts for protein production through systems biotechnology and genome-scale modeling. Biotechnol J. 2015;10:939–949. doi: 10.1002/biot.201400647. [DOI] [PubMed] [Google Scholar]
- 45••.Hefzi H, Ang KS, Hanscho M, Bordbar A, Ruckerbauer D, Lakshmanan M, Orellana CA, Baycin-Hizal D, Huang H, Ley D, et al. A consensus genome-scale reconstruction of CHO cell metabolism for improved biotherapeutic protein production. Cell Systems. 2016;3(5):434–443. doi: 10.1016/j.cels.2016.10.020. This article presents the genome-scale reconstruction of the metabolic pathways of CHO cells and the development of cell-line specific models for CHO-K1, CHO-S and CHO-DG44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46•.Kaas CS, Fan Y, Weilguny D, Kristensen C, Kildegaard HF, Andersen MR. Toward genome-scale models of the Chinese hamster ovary cells: Incentives, status and perspectives. Pharm Bioprocess. 2014;2:437–448. doi: 10.4155/pbp.14.54. Description of the possibility of using –omics data to develop computational models allowing the analysis of CHO cell metabolism and the improvement of cell line design. [DOI] [Google Scholar]
- 47.Brinkrolf K, Rupp O, Laux H, Kollin F, Ernst W, Linke B, Kofler R, Romand S, Hesse F, Budach WE, et al. Chinese hamster genome sequenced from sorted chromosomes. Nat Biotech. 2013;31(8):694–695. doi: 10.1038/nbt.2645. [DOI] [PubMed] [Google Scholar]
- 48.Lewis NE, Liu X, Li Y, Nagarajan H, Yerganian G, O’Brien E, Bordbar A, Roth AM, Rosenbloom J, Bian C, et al. Genomic landscapes of Chinese hamster ovary cell lines as revealed by the Cricetulus griseus draft genome. Nat Biotechnol. 2013;31:759–65. doi: 10.1038/nbt.2624. [DOI] [PubMed] [Google Scholar]
- 49•.Vishwanathan N, Bandyopadhyay AA, Fu HY, Sharma M, Johnson KC, Mudge J, Ramaraj T, Onsongo G, Siverstein KAT, Jacob NM, et al. Augmenting Chinese hamster genome assembly by identifying regions of high confidence. Biotechnol J. 2016;11:1151–1157. doi: 10.1002/biot.201500455. Identification of high confidence regions in the assembled genome of Chinese Hamster to facilitate its use for cell- and genome engineering. [DOI] [PubMed] [Google Scholar]
- 50.Xu X, Nagarajan H, Lewis NE, Pan S, Cai Z, Liu X, Chen W, Xie M, Wang W, Hammond S, et al. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat Biotechnol. 2011;29:735–741. doi: 10.1038/nbt.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feichtinger J, Hernandez I, Fischer C, Hanscho M, Auer N, Hackl M, Jadhav V, Baumann M, Krempl PM, Schmidl C, et al. Comprehensive Genome and Epigenome Characterization of CHO Cells in Response to Evolutionary Pressures and Over Time. Biotechnol Bioeng. 2016;113(10):2241–2253. doi: 10.1002/bit.25990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hefzi H, Lewis NE. From random mutagenesis to systems biology in metabolic engineering of mammalian cells. Pharm Bioprocess. 2014;2:355–358. doi: 10.4155/pbp.14.36. [DOI] [Google Scholar]
- 53•.King ZA, Lloyd CJ, Feist AM, Palsson BØ. Next-generation genome-scale models for metabolic engineering. Curr Op Biotechnol. 2015;35:23–29. doi: 10.1016/j.copbio.2014.12.016. Presentation of the use of COBRA methods for strain optimization and the development of the next-generation models for systems metabolic engineering. [DOI] [PubMed] [Google Scholar]
- 54.Hyduke DR, Lewis NE, Palsson BØ. Analysis of omics data with genome-scale models of metabolism. Mol Biosystems. 2013;9(2):167–174. doi: 10.1039/c2mb25453k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Z, Wang S, Halim A, Schulz MA, Frodin M, Rahman SH, Vester-Christensen MB, Behrens C, Kristensen C, Vakhrushev SY, et al. Engineered CHO cells for production of diverse, homogeneous glycoproteins. Nat Biotechnol. 2015;33:842–844. doi: 10.1038/nbt.3280. [DOI] [PubMed] [Google Scholar]
- 56.Grav LM, Lee JS, Gerling S, Kallehauge TB, Hansen AH, Kol S, Lee GM, Pedersen LE, Kildegaard HF. One-step generation of triple knockout CHO cell lines using CRISPR/Cas9 and fluorescent enrichment. Biotechnol J. 2015;10:1446–1456. doi: 10.1002/biot.201500027. [DOI] [PubMed] [Google Scholar]
- 57•.Lee JS, Grav LM, Lewis NE, Kildegaard HL. CRISPR/Cas9-mediated genome engineering of CHO cell factories: Applications and perspectives. Biotechnol J. 2015;10(7):979–994. doi: 10.1002/biot.201500082. Summary of the applications and the future perspectives of the CRISPR/Cas9 genome engineering techniques for CHO cell line development. [DOI] [PubMed] [Google Scholar]
- 58.Fischer S, Handrick R, Aschrafi A, Otte K. Unveiling the principle of microRNA- mediated redundancy in cellular pathway regulation. RNA Biol. 2015;12:238–247. doi: 10.1080/15476286.2015.1017238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hackl M, Borth N, Grillari J. miRNAs - pathway engineering of CHO cell factories that avoids translational burdening. Trends Biotechnol. 2012;30:405–406. doi: 10.1016/j.tibtech.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 60.Brockman IM, Prather KLJ. Dynamic metabolic engineering: New strategies for developing responsive cell factories. Biotechnol J. 2015;10:1360–1369. doi: 10.1002/biot.201400422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Le H, Vishwanathan N, Kantardjieff A, Doo I, Srienc M, Zheng X, Somia N, Hu WS. Dynamic gene expression for metabolic engineering of mammalian cells in culture. Metab Eng. 2013;20:212–220. doi: 10.1016/j.ymben.2013.09.004. [DOI] [PubMed] [Google Scholar]