Abstract
Aim
Lower species diversity, increased population densities and ecological niche enlargement are common characteristics of island faunas. However it remains to be determined if they extend to the parasite community. We tested if Haemosporidia parasite pressure varies between islands and the mainland with two different levels of analysis: i) at the host community level, and ii) with paired-species comparisons between islands and the mainland.
Location
Gulf of Guinea, West Africa.
Methods
We used molecular-based methods to identify avian Haemosporidian parasites (Plasmodium, Haemoproteus and Leucocytozoon) to describe their diversity, prevalence, host specificity and their phylogenetic relationships in five islands of the Gulf of Guinea and in nearby mainland areas.
Results
We found reduced Haemosporidia diversity on islands for Haemoproteus and Leucocytozoon, but not for Plasmodium. In addition, lower parasite prevalence on islands was found using a paired-species approach. Although the mean host specificity of the parasite community on islands did not differ from the mainland, we found a very distinct parasite species assemblage on the islands, which was composed of both the most generalist and the most specialist lineages.
Main conclusions
This study supports the hypothesis that parasite pressure is reduced on islands. Colonization is made by generalists with high host switching capacities, with some subsequently evolving into highly specialised parasites. This suggests that ‘taxon cycle’ dynamics may explain the assemblage of insular parasite communities.
Keywords: avian malaria, Haemoproteus, host specificity, island biogeography, Leucocytozoon, Plasmodium, taxon cycle
INTRODUCTION
Island faunas are often characterized by lower species diversity, ecological niche enlargement, and increased population densities (MacArthur & Wilson, 1967; MacArthur et al., 1972; Buckley & Jetz, 2007). To date, it remains uncertain if and how these patterns described for insular vertebrates apply to their parasite fauna (Illera et al., 2015; Jean et al., 2016). Although island biogeography of parasites and their colonization histories have received increasing attention in recent years (e.g. Cornuault et al., 2012; Sari et al., 2013; Clark & Clegg, 2015), studies thus far have investigated the effects of insularity on parasite diversity and phylogenetic relationships on islands only (Ishtiaq et al., 2012; Carlson et al., 2013; Olsson-Pons et al., 2015; Ricklefs et al., 2016), or have compared islands with the mainland but taking into consideration only one host species (Pérez-Rodríguez et al., 2013; Sari et al., 2013). It is therefore of interest to test if the core predictions of island biogeography theory apply to parasite communities on a broader scale, i.e. by comparing parasite parameters in multiple host species on islands and the mainland.
The first prediction of a depauperate parasite fauna on islands has been confirmed by several studies (Steadman et al., 1990; Fallon et al., 2005; Ricklefs et al., 2011; Spurgin et al., 2012). Nevertheless, few of them have specifically compared island hosts with their counterparts on the mainland, which is important given the geographical and phylogenetic factors that are known to have a strong influence on parasite distributions (e.g. Fallon et al., 2005; Beadell et al., 2006). If islands do exhibit low parasite diversity in relation to the nearby mainland areas it is expected that, on average, parasite species on islands will have broader niches (i.e. broader range of host species) than on the mainland. This would be driven by two distinct processes: i) community sorting: at the time of colonization, generalist parasites from the mainland should be more successful in establishing viable populations by being able to infect multiple and novel host species (taxon cycle; Wilson, 1961; Ricklefs & Bermingham, 2002); ii) island populations will experience less interspecific competition relatively to the species-rich mainland communities, which will open the way for niche enlargement (MacArthur & Wilson, 1967). This decrease of average host specificity will allow for the maintenance of large populations and hence decrease the extinction risk (Pérez-Rodríguez et al., 2013). Finally, to test the prediction of increased parasite density on islands one can use parasite prevalence (the proportion of individuals infected in a population) as a proxy. Increased density of hosts is thought to arise from a mixture of reduced interspecific competition and release from predation (MacArthur et al., 1972; Buckley & Jetz, 2007). It is less clear which mechanisms may affect parasites, but we can predict an increase of parasite prevalence in two complementary ways: i) by a direct effect, with a lower interspecific parasite competition allowing for higher parasite densities to build up; and ii) indirectly, with increased densities of host populations facilitating parasite transmission (see review in McCallum et al., 2001).
Understanding the characteristics of insular parasites is important, as it should play a major role on the evolutionary dynamics and assembly of communities on islands. First, a release from ‘natural enemies’ (e.g. pathogens and predators) in island environments is often postulated as one of the key factors promoting demographic and evolutionary change on islands (MacArthur & Wilson, 1967; Ricklefs & Bermingham, 2002; Torchin et al., 2003). Second, parasite pressure is expected to influence the hosts’ investment in immunity (Lindström et al., 2004; Matson, 2006; Beadell et al., 2007), and hence environmental variation in pathogen levels should be a main determinant of the evolution of pathogen resistance. If island hosts harbour fewer parasites, they are expected to have evolved reduced defences and, as a consequence, island hosts are often portrayed as being particularly vulnerable to introduced diseases (Wikelski et al., 2004). This is illustrated most notably by the dramatic population declines and extinction of endemic birds in the Hawaiian islands following the introduction of avian malaria and their vectors (Warner, 1968; van Riper et al., 1986; LaPointe et al., 2012).
In this study, we tested the predictions of island biogeography theory in parasites using avian haemosporidian parasites in five Gulf of Guinea islands (Africa) and nearby mainland areas. Birds are commonly infected by three closely related genera (Plasmodium, Haemoproteus and Leucocytozoon) that are transmitted by the bite of an arthropod host: Plasmodium is transmitted by mosquitoes (Culicidae), Haemoproteus by midges (Ceratopogonidae) and louse flies (Hippoboscidae), and Leucocytozoon by black flies (Simulidae; Valkiūnas, 2005; LaPointe et al., 2012).
Here, we considered two different levels and types of analysis. First, we compared how the parasite pressure differs between islands and the mainland using the whole host community. Second, at a finer scale, we compared island birds with their mainland counterpart (conspecific population or sister/closely-related species). We examined how insularity affects: 1) the diversity of Plasmodium, Haemoproteus and Leucocytozoon lineages and their phylogenetic relationships, 2) their evolutionary strategy in terms of host specificity (i.e. generalist versus specialist) and 3) their prevalence. According to the predictions of island biogeography theory, we expected lower diversity, lower host specificity of parasites and higher prevalence in island hosts.
MATERIALS AND METHODS
Study area
Sampling was carried out on five islands of the Gulf of Guinea: a land-bridge island (Bioko), three oceanic islands (Príncipe, São Tomé, Annobón) and one islet (Boné), and on the nearby mainland in Cameroon and Gabon (Fig. 1; Appendix S1, Table S1.1). The islands are the offshore part of the 1000 km Cameroon Line of Tertiary to Recent volcanoes, which extends from Annobón to the Mandara Mountains on the Nigeria-Cameroon border (Burke, 2001). Volcanic activity in the continental and oceanic sector has been contemporaneous and more or less continuous since the Cretaceous (Burke, 2001).
Figure 1.
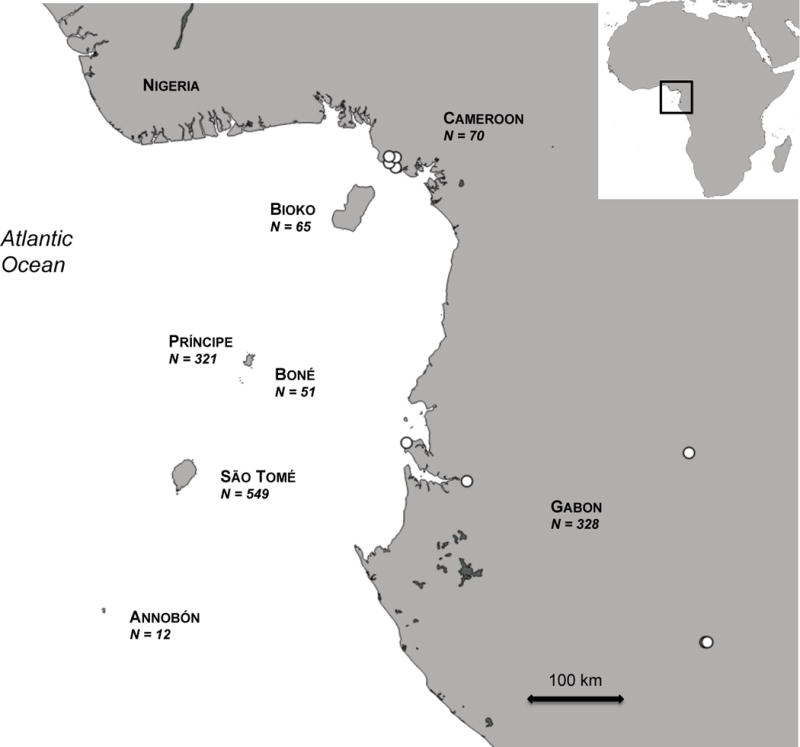
Map of the Gulf of Guinea, in central-west Africa, illustrating the seven regions sampled within the five islands and the mainland sampling areas (located in Cameroon and Gabon). White dots represent sampling sites on the mainland and the numbers of sampled individuals are given for each region.
The region has an equatorial climate with the year being divided into rainy and dry seasons. The natural habitat type of the Gulf of Guinea islands has been described as rainforest (Exell, 1944; Exell, 1973). The forest is stratified with altitude into lowland rainforest (0–800m), montane forest (800–1400), and mist forest (1400–2500), the latter being absent from Príncipe and Annobón (Exell, 1944). On the islands, most of the lowland forests and some montane forests are replaced by coffee and cocoa plantations, and only south-west and central São Tomé and the South of Príncipe remain covered by relatively undisturbed forest. Today, we can define four types of habitats in order of increasing anthropogenic influence: old-growth rainforest, secondary forest, shade plantation and non-forested habitats (de Lima et al., 2013). We sampled in old-growth rainforest, in shade plantation and in non-forested habitats. Shade plantation refers to agroforestry areas that have crops (cocoa or coffee) growing under the canopy of trees, and the non-forested category includes man-made habitats, such as oil palm monocultures and savannahs (de Lima et al., 2013). In our analyses, we grouped together the habitats “shade plantation” and “non-forested habitat” under the term “plantation” that refers to modified-human habitat in contrast to “forest” which is undisturbed (i.e. old-growth rainforest). On the mainland, we sampled birds in similar types of habitats.
Host species and data collection
Data collection took place from October to April (2002–2014) coinciding with the most important breeding periods both on the islands and the mainland (Appendix S1, Table S1.2). Birds were captured with ECOTONE (Poland) mist-nets, banded with a metal ring, measured and weighted. In addition, a small amount of blood was collected from the brachial vein and stored in ethanol for subsequent molecular analyses.
Sampling sites used for the analysis at the community level were located from sea level to 2400 m and they were either in forest or in plantations (Appendix S1, Table S1.1). For the paired-species analysis, among all the sites, we used samples from sites located only in plantations at the same altitude (below 600 m) on both islands and mainland to control for the habitat and altitude effect.
At the community level, we captured a total of 1396 birds from 13 families and 72 species, targeting both endemic and non-endemic host species, between 2002 and 2014 (Table 1; Appendix S1, Table S1.3). We also gathered additional data from the MalAvi database (Bensch et al., 2009; Accessed on 05 September 2016) from the same 13 families of three countries bordering the Gulf of Guinea: Cameroon, Gabon and Nigeria (Fig. 2; Table 1).
Table 1.
Blood parasite diversity and prevalence in birds from the Gulf of Guinea, West Africa. Numbers of lineages of each parasite genera and the prevalence in percentage are given for each of the sampled regions, as well as the additional numbers of lineages gathered online from three mainland regions (#).
| Plasmodium | Haemoproteus | Leucocytozoon | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| N individuals | N lineages | Prevalence (%) | N lineages | Prevalence (%) | N lineages | Prevalence (%) | ||
| Islands | Annobón | 12 | 0 | 0 | 1 | 30 | 1 | 25 |
| Bioko | 65 | 5 | 39.47 | 3 | 18.42 | 4 | 49.23 | |
| Boné | 51 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Príncipe | 321 | 12 | 34.68 | 4 | 4.43 | 8 | 9.87 | |
| São Tomé | 549 | 18 | 18.68 | 10 | 16.09 | 6 | 29.12 | |
| Total | 998 | 29 | 24 | 17 | 11.1 | 15 | 22.8 | |
|
| ||||||||
| Mainland | Cameroon | 70 | 8 | 22.95 | 7 | 26.23 | 7 | 17.14 |
| Gabon | 328 | 30 | 45.68 | 22 | 20.68 | 35 | 37.19 | |
| Total | 398 | 35 | 42.1 | 28 | 21.5 | 42 | 34.2 | |
| Cameroon (#) | 24 | 10 | 1 | |||||
| Gabon (#) | 12 | 19 | 0 | |||||
| Nigeria (#) | 27 | 21 | 24 | |||||
Figure 2.
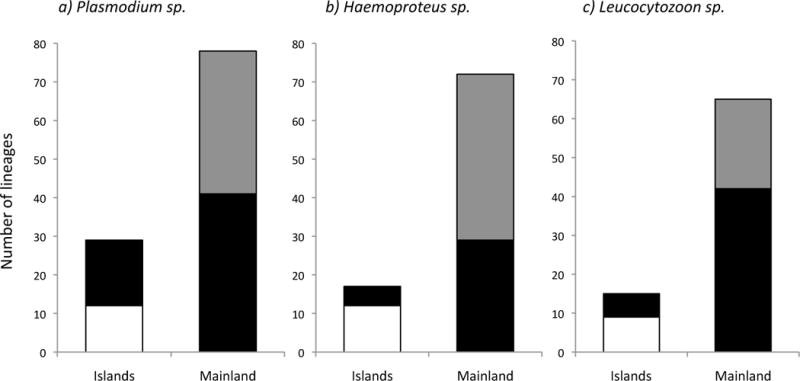
Number of parasite lineages found on birds from the Gulf of Guinea for a) Plasmodium sp., b) Haemoproteus sp., and c) Leucocytozoon sp. Islands: numbers of lineages found on the islands, with the white part representing lineages found exclusively on the islands; Mainland: number of lineages found on the mainland, with the grey part representing lineages from the mainland retrieved from the MalAvi database.
For the paired-species analysis, we restricted the samples to those collected in 2013 and 2014 (N=580 individuals) from six of the 13 families, for a total of 21 species corresponding to 11 groups of two or three species (Table 2). Species present in both island and mainland locations were paired together; this was the case of Ploceus cucullatus, Euplectes hordeaceus, Lonchura cucullata and Cyanomitra olivacea. Otherwise we paired species according to their phylogenetic proximity (Table 2), based on molecular phylogenies or on current taxonomy (e.g. Turdus olivaceofuscus, Melo et al., 2010). If the closest relative did not occur in the sampling site on the mainland, we used the congeneric species present (e.g. Estrilda spp; see also Lobato et al., 2017). In addition, Ploceus cucullatus is the closest mainland relative of both the São Tomé endemic P. grandis and the Príncipe endemic P. princeps (Staffan Andersson, Göteborg University, pers. comm.) and hence, in addition to being paired to a conspecific population present on São Tomé, it was compared to the two island endemics in another paired comparison (pairs 8 and 9; Table 2).
Table 2.
Blood parasite diversity and prevalence in the Gulf of Guinea across the bird species used in a paired-species comparison between islands and the mainland. Nind: number of individuals tested; N: number of parasite lineages. Ploceus cucullatus from Gabon was used in two paired comparisons (pairs 8 and 9; see text).
| Plasmodium | Haemoproteus | Leucocytozoon | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pair | Family | Region | Species | Nind | N | Prevalence | N | Prevalence | N | Prevalence |
| 1 | Columbidae | Gabon | Turtur afer | 10 | 0 | 0 (0/10) | 1 | 10.00 (1/10) | 0 | 10.00 (1/10) |
| 1 | Príncipe | Aplopelia larvata | 13 | 0 | 0 (0/12) | 1 | 8.33 (1/12) | 0 | 0 (0/13) | |
| 1 | São Tomé | Columba malherbii | 8 | 0 | 0 (0/6) | 2 | 33.33 (2/6) | 0 | 37.50 (3/8) | |
|
| ||||||||||
| 2 | Estrildidae | Gabon | Estrilda melpoda | 14 | 1 | 7.14 (1/14) | 2 | 92.85 (13/14) | 0 | 78.57 (11/14) |
| 2 | Príncipe | Estrilda astrild | 17 | 0 | 0 (0/17) | 0 | 0 (0/17) | 0 | 0 (0/17) | |
| 2 | São Tomé | Estrilda astrild | 38 | 0 | 0 (0/38) | 0 | 0 (0/38) | 1 | 2.63 (1/38) | |
|
| ||||||||||
| 3 | Estrildidae | Gabon | Lonchura cucullata | 19 | 1 | 5.26 (1/19) | 2 | 68.42 (13/19) | 0 | 0 (0/19) |
| 3 | Príncipe | Lonchura cucullata | 19 | 0 | 0 (0/19) | 1 | 5.26 (1/19) | 0 | 0 (0/19) | |
|
| ||||||||||
| 4 | Fringillidae | Gabon | Crithagra capistrata | 21 | 2 | 28.57 (6/21) | 0 | 0 (0/21) | 1 | 14.28 (3/21) |
| 4 | São Tomé | Crithagra mozambica | 14 | 3 | 50.00 (7/14) | 0 | 0 (0/14) | 0 | 7.14 (1/14) | |
| 4 | São Tomé | Crithagra rufobrunnea | 21 | 2 | 9.52(2/21) | 1 | 75.00 (15/20) | 2 | 66.66 (14/21) | |
|
| ||||||||||
| 5 | Nectariniidae | Gabon | Cyanomitra verticalis | 15 | 7 | 53.33(8/15) | 0 | 0 (0/15) | 4 | 26.66 (4/15) |
| 5 | Príncipe | Nectarinia hartlaubii | 10 | 3 | 100 (10/10) | 0 | 0 (0/10) | 2 | 80.00 (8/10) | |
| 5 | São Tomé | Nectarinia newtonii | 29 | 6 | 24.14 (7/29) | 1 | 3.45 (1/29) | 2 | 20.69 (6/29) | |
|
| ||||||||||
| 6 | Nectariniidae | Gabon | Cyanomitra olivacea | 12 | 4 | 66.66 (6/9) | 2 | 25.00 (2/8) | 5 | 58.33 (7/12) |
| 6 | Príncipe | Cyanomitra olivacea | 17 | 5 | 64.70 (11/17) | 0 | 0 (0/17) | 0 | 17.64 (3/17) | |
|
| ||||||||||
| 7 | Ploceidae | Gabon | Euplectes hordeaceus | 27 | 5 | 59.26 (16/27) | 2 | 11.11(3/27) | 6 | 51.85 (14/27) |
| 7 | São Tomé | Euplectes hordeaceus | 23 | 1 | 4.35 (1/23) | 0 | 0 (0/23) | 0 | 0 (0/23) | |
|
| ||||||||||
| 8 | Ploceidae | Gabon | Ploceus cucullatus | 46 | 4 | 58.53(24/41) | 2 | 11.63 (5/43) | 3 | 57.14 (24/42) |
| 8 | São Tomé | Ploceus cucullatus | 33 | 4 | 70.00 (21/30) | 0 | 0 (0/30) | 0 | 0 (0/30) | |
|
| ||||||||||
| 9 | Ploceidae | Gabon | Ploceus cucullatus | 46 | 4 | 58.53(24/41) | 2 | 11.63(5/43) | 3 | 57.14(24/42) |
| 9 | Príncipe | Ploceus princeps | 53 | 8 | 52.08(25/48) | 0 | 0 (0/48) | 0 | 0 (0/48) | |
| 9 | São Tomé | Ploceus grandis | 16 | 1 | 6.25(1/16) | 0 | 0 (0/16) | 2 | 43.75 (7/16) | |
|
| ||||||||||
| 10 | Ploceidae | Gabon | Ploceus nigricollis | 30 | 3 | 51.72 (15/29) | 3 | 27.58 (8/29) | 11 | 73.33 (22/30) |
| 10 | São Tomé | Ploceus sanctithomae | 18 | 3 | 22.22 (4/18) | 0 | 0 (0/18) | 4 | 77.77 (14/18) | |
|
| ||||||||||
| 11 | Turdidae | Gabon | Turdus pelios | 24 | 4 | 95.65 (22/23) | 0 | 0 (0/23) | 1 | 66.66 (16/24) |
| 11 | São Tomé | Turdus olivaceofuscus | 33 | 4 | 45.45 (15/33) | 0 | 0 (0/33) | 1 | 18.18 (6/33) | |
Molecular analyses
Haemosporidian parasites screening
To test for the presence of Plasmodium, Haemoproteus and Leucocytozoon we used the protocol and the cycling profile conditions described in Hellgren et al. (2004). In all samples from Ploceus cucullatus from Gabon we obtained an unusual band profile. In this species, two bands of very strong intensity of approximate sizes of 480 and 250 bp were systematically obtained when amplifying the Haemoproteus and Plasmodium fragment. Because modifying cycling conditions did not help, we used the protocol of Drovetski et al. (2014), in which three primer pairs are used, to obtain the prevalence of malaria parasites for this species. Because in our comparative approach we paired P. cucullatus from Gabon with P. cucullatus and P. grandis from São Tomé, and P. princeps from Príncipe, samples from these species were also screened using the protocol of Drovetski et al. (2014) in order to be comparable with P. cucullatus from the mainland.
Sequencing and lineage identification
All samples were screened twice in independent PCRs to confirm positive or negative scores. When the two independent PCRs gave different results, a third PCR was performed to confirm positive or negative scores. All PCR reactions included at least a positive sample from previous assays and a negative control. PCR products from confirmed positives were purified for cycle sequencing reactions using ExoSAP-IT (USB Corporation) following the manufacturer’s instructions. Bi-directional sequencing was performed with dye-terminator fluorescent labeling in a 3130xl Genetic Analyzer (Applied Biosystems). The sequences were edited and aligned using the program GENEIOUS 7 (Kearse et al., 2012). Haplotypes obtained were compared with sequences available in GenBank and MalAvi (Bensch et al., 2009) databases. Parasite sequences that differed by one or more base pairs were treated as distinct lineages (Bensch et al., 2009; Appendix S2, Table S2.1 and S2.2 for a complete list of lineages and distribution). Multiple infections were observed in a number of samples. Double infections were resolved when only one double peak was observed, otherwise we gave a score of ‘multiple infection’ to the sample but we did not obtain the separate lineages (n=106).
Statistical analyses
Statistical analyses were performed for each parasite genus separately, both at the community level and in the paired-species design. To determine whether there were differences in parasite levels between the islands and the mainland at the community level, we used generalized linear mixed models (SAS, 1999) to investigate variation in the following variables: 1) parasite diversity, i.e., number of lineages found per region and per bird family, 2) parasite prevalence, i.e., percentage of infected individuals, and 3) host-specificity. We used and combined data from available online databases and our own sequences to estimate parasite diversity per region (five islands and three mainland regions) and to calculate host specificity indices (Hellgren et al., 2009).
To investigate differences in parasite diversity between the islands and the mainland, we gathered information on the number of lineages of Plasmodium, Haemoproteus and Leucocytozoon found per bird family with i) our sampling in each of the seven regions described above (five islands and two mainland regions; n=145 lineages) and ii) additional information from online databases in three mainland regions (Cameroon, Gabon and Nigeria, n=103; Fig. 2). We used ‘insularity’ (mainland versus island) and the status of birds (endemic or non-endemic) as fixed factors, and we accounted for host phylogeny by including a random factor ‘family’ and a random factor ‘region’ in the models. In addition, estimates of lineage diversity per region were also conducted through rarefaction and extrapolation curves (with 95% confidence intervals) using ESTIMATES 9.1.0 (Colwell et al., 2013; Appendix S2, Fig. S2.1).
For prevalence, we used a binomial distribution (infection = 0 or 1). Patterns of prevalence variation were inferred from our data only because prevalence data could not be extracted from online databases. We included a fixed factor ‘insularity’ and controlled for several other factors: habitat (forest versus plantation), altitude (continuous variable), year of sampling (2002 or 2014), as well as the random factors ‘family’ and ‘region’.
Host specificity of each parasite lineage from our survey and from the MalAvi database was estimated using the modified version of the host specificity index (Poulin & Mouillot, 2003; Hellgren et al., 2009) that accounts both for the number of hosts species of the parasite and the taxonomic distance among them, and for the variance of the taxonomic distance among host species. First, in our statistical model for the host specificity index (log-transformed), we tested the effect of insularity using the presence/absence of the lineage on islands, on the mainland or on both. We compared the host specificity indices of all lineages found on the islands versus all the lineages detected on the mainland. Then, with our data only, we compared the prevalence of specialist and generalist lineages on islands versus mainland (Fig. S2.2). We designated a lineage as specialist when it infected one, two or three species of the same genus; a lineage infecting two species of two different genera or families was considered as generalist. We used as fixed factors the ‘insularity’ effect (island versus mainland), as well as other factors (i.e. habitat, altitude, year) and the random effect ‘region’.
For the paired-species design, we used the package ‘lme4’ (Bates et al., 2015) in R (R Core Team, 2015). We tested the insularity effect using generalized linear mixed models on parasite diversity and prevalence but this time with the random effect ‘species’ nested in ‘pair’ which was in turn nested in ‘family’. Since we formed two pairs including species with different genera (pairs 1 and 5; Table 2), we performed the statistical analyses both including and excluding them (Appendix S2).
Phylogenetic analyses
For the three haemosporidia genera we recovered all sequences available on MalAvi for the Gulf of Guinea. Relationships among lineages were inferred with Bayesian methods, both as implemented in MRBAYES 3.2.6 (Huelsenbeck & Ronquist, 2001; Ronquist & Huelsenbeck, 2003) and in BEAST 1.8.2 (Drummond et al., 2012). We used PARTITIONFINDER 1.1.1 (Lanfear et al., 2012) to determine the best-fit partitioning scheme for the cytochrome b dataset (486 bp) and the respective substitution models for each partition, using the Bayesian information criterion (detailed methods in Appendix S3).
RESULTS
Parasite diversity
In this study, we described 81 new Haemosporidian lineages and recorded 64 previously described ones (Appendix S2; Table S2.1 and S2.2) from the Gulf of Guinea but also from other locations of the world. Plasmodium was represented by the highest number of lineages on the islands (n=29), followed by Haemoproteus and Leucocytozoon (17 and 15 respectively; Table 1, Fig. 2). In addition, the number of lineages found exclusively on islands (i.e. endemic lineages) was high, ranging from 65% for Haemoproteus, 53% for Leucocytozoon, and 41% for Plasmodium (Fig. 2; Appendix S2). On the mainland, we found respectively 136, 107 and 109 of Plasmodium, Haemoproteus and Leucocytozoon lineages (Fig. 2), including those retrieved from the online database MalAvi (Table 1, Fig. 2).
Parasite diversity was significantly reduced on the islands for Haemoproteus (community level: P = 0.04; paired-species design: P = 0.005) and Leucocytozoon (community level: P = 0.001; paired-species design: P = 0.0003), but not for Plasmodium (community level: P = 0.34; paired-species design: P = 0.42). These results were confirmed by rarefaction and extrapolation curves (Appendix S2, Fig. S2.1).
Prevalence
With the community approach we failed to detect any insularity effect on the prevalence of the three Haemosporidia genera (Plasmodium F1,1081 = 2.85, P = 0.091; Haemoproteus F1,1081 = 1.04, P = 0.307; Leucocytozoon F1,1327 = 0.02, P = 0.885). However, the paired-species design found a lower prevalence on the islands for all genera (Plasmodium P = 0.011; Haemoproteus P < 0.0001; Leucocytozoon P < 0.0001).
Although insularity had no effect on prevalence at the community level, other factors explained the variation in prevalence, such as altitude (Leucocytozoon F1,1327 = 76.81, P < 0.0001; higher prevalence above 300 m) and habitat (Plasmodium F1,1081 = 10.54, P = 0.001; Leucocytozoon F1,1327 = 5.51, P = 0.019). We found an opposite effect of habitat for these two genera, with a higher Plasmodium prevalence in plantation (forest: 15.62%, plantation: 33.08%) and a higher Leucocytozoon prevalence in forest (forest: 38.43%, plantation: 23.25%).
Host specificity and phylogenetic relationships
Using the online database MalAvi we calculated the host specificity indices for each lineage recovered in the Gulf of Guinea. Indices ranged from 0 (specialist parasite found in only one host species) to 90.33 (Plasmodium sp. SGS1, a generalist found in 106 species) and are given in Appendix S2 (Table S2.1). Overall we did not find an insularity effect on the mean host specificity index for any of the haemosporidian genera (Plasmodium: F1,2 = 0.03, P = 0.871; Haemoproteus: F1,2 = 0.10, P = 0.748; Leucocytozoon: F1,2 = 0.95, P = 0.332; Fig. 3). However, when looking closely at the assemblage of lineages, we found that on the islands, the community of parasites was composed of i) lineages exclusively found on islands (endemic lineages) with a low host specificity index (i.e. specialist parasites) and ii) lineages found at both islands and mainland sites with a high host specificity index (i.e. generalist parasites; Fig. 3; Appendix S2, Tables S2.1 and S2.2); whereas the lineages exclusively found on the mainland had a host specificity index two to ten times higher than on the islands (Fig. 3).
Figure 3.
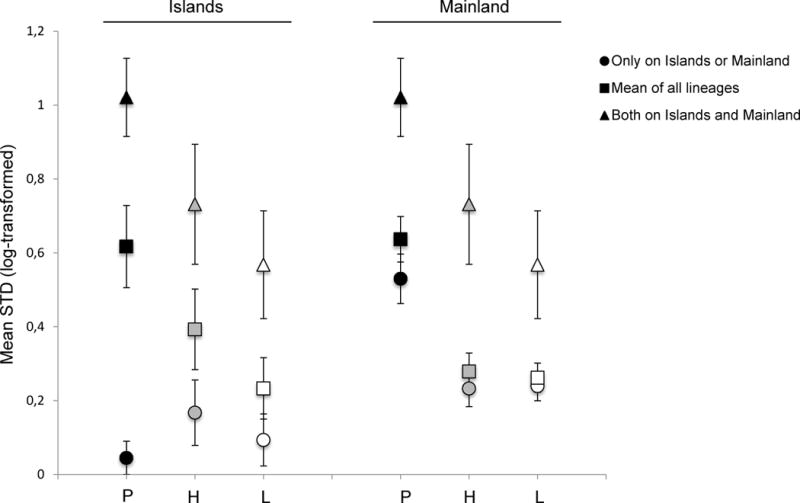
Host specificity across the three Haemosporida genera found on birds on the Gulf of Guinea islands and mainland. Host specificity estimated with Poulin & Mouillot’s (2005) STD* index (mean STD log transformed ± SE). P=Plasmodium, H=Haemoproteus, L=Leucocytozoon. Circles: lineages restricted either to islands or to the mainland; triangles: lineages found simultaneously on islands and the mainland; squares: all lineages. High values represent generalist lineages, and low values represent specialist lineages.
On the islands, we found that generalist lineages were more prevalent than the specialists for the three genera (Plasmodium: 15.4% of specialists, 84.6% of generalists; Haemoproteus: 46.25% of specialists, 53.75% of generalists; Leucocytozoon: 23.7% of specialists, 76.3% of generalists). On the mainland, Plasmodium had a similar behaviour (24.1% of specialists, 75.9% of generalists), whereas specialist lineages were more prevalent for both Haemoproteus (62.3% of specialists, 37.7% of generalists) and Leucocytozoon (78.1% of specialists, 21.9%). These differences were only significant for Leucocytozoon (F1,160 = 3.69, P = 0.05; Appendix S2, Fig. S2.2).
Finally, phylogenetic inference showed that most parasite lineages on the islands are the result of independent colonizations (Appendix S3, Fig. S3.1 and S3.2). Many of these went on to differentiate in situ leading to the origin of putatively endemic lineages through anagenesis. In only a few cases endemic lineages underwent cladogenesis within the archipelago (e.g. Haemoproteus lineages in the white-eyes, Zosteropidae) or even within-islands (e.g. Plasmodium lineages in the São Tomé thrush, Turdus olivaceofuscus). Whilst absolute divergence dates cannot be estimated due to the uncertainty regarding the rates of evolution of the cytochrome b in the three Haemosporidian genera analysed here, the data nevertheless suggest a wide range of ages for endemic lineages.
DISCUSSION
This study provides strong support for the prediction of reduced parasite diversity on islands, although this was not significant for one of the three genera of Haemosporidia analysed here (Plasmodium). Parasite prevalence (an indicator of parasite ‘population density’ and infection success) was affected by insularity but in the opposite direction to the density compensation hypothesis, i.e. it decreased instead of increasing on the islands. We also showed that the parasite community on islands was composed of both very specialized, endemic, parasites and of highly generalist lineages also present on the mainland. The arrival of generalist lineages that, in some cases, evolved in situ to become endemic specialists, is a pattern expected from the taxon cycle hypothesis (Wilson, 1961) and was supported by our phylogenetic analyses that confirmed the importance of multiple colonizations for the build-up of the parasite communities of the Gulf of Guinea islands.
Parasite diversity
Estimating true parasite species diversity is greatly affected by sampling effort (Walther et al., 1995). Additional sampling on the islands could reveal additional rare Haemosporidian lineages but we believe that additional sampling on the mainland – a highly biodiverse and poorly explored region – would lead to the discovery of a greater number of lineages than for the islands. Given the large differences found here and considering that the sampling effort was greater on the islands, our main conclusion of overall higher diversity of lineages on the mainland is, if anything, more likely to be strengthened with further sampling. One could also argue that what we considered an endemic lineage could reflect a bias in sampling effort and that these are simply lineages not found on the continent yet. This could be of concern if we relied on our sample collection only. However, we used the large database MalAvi (Bensch et al., 2009) that comprises more than 1800 records in sub-Saharan Africa including 648 described lineages. The use of this database brings a greater confidence in the interpretation of our results.
For Plasmodium, we found a relatively high diversity of lineages on islands and this could be due to two non-exclusive phenomena: 1) Plasmodium parasites may have a higher colonization rate as indicated by their lower endemicity level and 2) Plasmodium lineages may be, in general, more successful colonisers than Haemoproteus and Leucocytozoon because of their potential higher adaptability and/or a better within-host competitive success (van Rooyen et al., 2013). In relation to Plasmodium, the insular Haemoproteus and Leucocytozoon community was characterised by lower diversity and higher endemism. The co-existence of a high proportion of specialised lineages found exclusively on the islands with a few very generalist lineages, shared with the mainland, could indicate that the genera Haemoproteus and Leucocytozoon may be in the contracting phase of a taxon cycle (Wilson, 1961) – where diversity decreases as specialised lineages go extinct at a higher rate than the establishment of new colonisers from the mainland.
Prevalence
At the community level, parasite prevalence on islands did not differ from the mainland. However, when using the paired-species comparison, we found that the prevalence of parasite infections significantly decreased on islands. Hence, although we did not find evidence for density compensation in Haemosporidia arising from reduced species diversity (as predicted by the island biogeography theory), this result does further support the hypothesis that parasite pressure is lower on islands. In our study, the generalist lineages were the most prevalent on islands. This generalist trend should lead to a higher probability of co-infections. As generalist parasites are considered poor competitors (Hellgren et al., 2009), an increase of co-infections on islands could play a role in limiting parasite densities and prevalence. Previous studies on the prevalence of blood parasites on islands were inconclusive, with either higher (Illera et al., 2015) or lower prevalence on islands (Pérez-Rodríguez et al., 2013). These conflicting results may arise from the fact that parasite prevalence is also likely to vary in relation to environmental and local factors. For example, the effect of habitat on Plasmodium and Leucocytozoon prevalence is probably associated with the ecology of their distinct vectors. Plasmodium prevalence was higher in disturbed habitats, which is in accordance with several studies showing that habitat disturbance is associated with an increase of Plasmodium prevalence (Chasar et al., 2009; Sehgal, 2010). The mosquitoes that transmit Plasmodium may favour the open areas created in human-modified habitats (Patz et al., 2000; Yasuoka & Levins, 2007; Vittor et al., 2009). On the contrary, Simulium black flies, vectors of Leucocytozoon, need lotic microhabitats for laying their eggs and are particularly sensitive to physicochemical characteristics of streams (Stangler et al., 2013). This could contribute to a preference of the vectors for pristine forest where flowing water is not disturbed or polluted (Docile et al., 2015). This could also contribute to explain why Leucocytozoon prevalence was found to increase at higher altitudes, as habitat disturbance decreases with altitude (de Lima et al., 2013). Additional research will be necessary to understand the complex web of factors affecting parasite prevalence in general, and on islands in particular. This must necessarily involve comprehensive studies on the distribution and ecology of the vectors (Bataille et al., 2012; Santiago-Alarcon et al., 2012). To date, almost nothing is known about vectors in the Gulf of Guinea islands (Ribeiro et al., 1998, Mustapha et al., 2004, 2006); further study and sampling of mosquitoes, midges and black flies is therefore the next step in our ongoing research in the area.
Host specificity and parasite community assembly on the islands
Using all the data available on MalAvi database, we found a similar average host-specificity on the mainland compared to the islands. A closer look at the distribution of generalists/specialists revealed an interesting pattern. Lineages present both on islands and on the mainland, i.e., the successful colonisers, were by far the most generalist lineages, with a host specificity index mean at least five times higher than the others. On the islands, the parasite community was composed of these generalist lineages and of lineages restricted to the islands with very narrow niches. On islands, the generalist lineages were the most prevalent. This suggests that, as predicted by island biogeography theory (MacArthur & Wilson, 1967), a broad ecological niche would be selected as a strategy that decreases the chances of extinction – in this case of parasites colonizing a new area where potential host populations are small (Beadell et al., 2006). The same pattern was also found for the Haemosporidian parasites of a songbird in the Madeira and the Canary islands (Pérez-Rodríguez et al., 2013).
Phylogenetic analyses showed that the current parasite diversity on the islands derives from multiple colonizations from the mainland followed, in many instances, by in situ speciation. Inter-island dispersal was negligible. These results parallel those found across distinct taxonomic groups showing that the Gulf of Guinea is a major centre of endemism where the biota of each island has evolved mostly in isolation from those of the other islands (Jones, 1994; Jones & Tye, 2006). The only case of inter-island dispersal followed by diversification (archipelago radiation) occurred in the lineages infecting the Zosteropidae clade, whose species in the Gulf of Guinea oceanic islands constitute one of the few examples of archipelago radiation in birds (Melo et al., 2011). This suggests that, in this case, the diversification pattern of the parasite lineages was driven by the dispersal history of the hosts rather than by the dispersal of vectors.
The phylogenetic and host specificity data suggest that the assembly of parasite communities in the Gulf of Guinea islands matches the taxon cycle hypothesis (Wilson, 1961): i) we found that generalist parasite lineages were the most successful island colonisers, and were the most prevalent lineages in the insular bird community; ii) with time, divergence from the mainland relatives accumulates and leads to the evolution of island endemics which are characterised by an increase in host specificity, becoming very narrow specialists, which we found in our system; iii) specialists will be more prone to extinction and will be replaced by new arrivals of generalists. This cyclic pattern of species turnover is described for macro-fauna, but its causes remain poorly understood. The possibility that the taxon cycle may extend to parasites is interesting given that parasites, through the evolutionary arms race with their hosts, have been proposed as one of the agents underlying macro-faunal taxon cycles (Ricklefs & Bermingham, 1999, 2002; Ricklefs et al., 2016).
Do island hosts experience a more benign parasite environment?
Our results, for Haemosporidian parasites from the Gulf of Guinea, support the hypothesis that parasite pressure is lower on islands. We found that islands had generally lower parasite diversity and either similar or reduced prevalence. As a result, in this region, island hosts face reduced diversity of infections and at least some of the host species on islands are less likely to become infected. Additionally, given low host diversity on the islands, it can also be argued that single-host parasites on islands have evolved to become less virulent, decreasing the risk of extinction of the few hosts that allow them to complete their life cycle. Studies on the pathogenicity of the different lineages of the island parasites studied here, as well as additional studies on other parasite groups and island systems, are needed to establish the generality of the patterns revealed in the present study. A reduced parasite pressure is thought to have direct consequences in terms of both health condition and immunological trade-offs of hosts. Understanding the effects of reduced parasite pressure on host life-history strategies and immunity is therefore essential to understand patterns of adaptation on islands as well as to attempt to mitigate the impact of recently introduced pathogens on the endemic communities of oceanic islands.
Supplementary Material
Appendix S1 Characteristics of sampling sites and host species.
Appendix S2 Host specificity indices and Genbank accession numbers.
Appendix S3 Phylogenetic analyses and trees.
Acknowledgments
We are extremely thankful to all the people who made the study possible in the field: Patrice Christy, Bikegila, Pedro Leitão, José Obiang, Francis Njie, Noélia Zafra Calvo, Gervásio, Lúcio Primo, Octávio Horta da Veiga and Alexandre Vaz. In São Tomé and Príncipe we received crucial logistic support from the Department of the Environment and ECOFAC, in Bioko and Annobón from the Universidad Nacional de Guinea Ecuatorial, in Cameroon from the Limbe Botanical and Zoological Garden, and in Gabon from the Station de Recherche de l’IRET at Ipassa-Makokou. Permits for sample collection were obtained from the respective authorities in the different countries. We thank Ricardo Lopes for lending us aliquots to test UNIV primers, and we thank the technical support provided by CTM (Centro de Testagem Molecular, CIBIO-INBIO, Portugal). We thank the associate editor S. Clegg and chief editor M. Carine for their comments, which greatly improved the manuscript.
CL, MM, EL, and RC were funded by the Portuguese Science and Technology Foundation (FCT; IF/00744/2014/CP1256/CT0001; SFRH/BPD/100614/2014; SFRH/BPD/80214/2011; IF/01411/2014/CP1256/CT0007); RC was additionally funded by a Marie Curie Fellowship. This study was funded by FEDER funds through the COMPETE program (FCOMP-01-0124-FEDER-028312) and by FCT, (PTDC/BIA-BIC/4556/2012), and by the project “Genomics and Evolutionary Biology” co-financed by North Portugal Regional Operational Programme 2007/2013 (ON.2 – O Novo Norte), under the National Strategic Reference Framework (NSRF), through FEDER to RC and MM, and by NIH GM063258 (USA) to RCF. Field work in Gabon and São Tomé Island in 2013 was funded by the National Geographic Society (NGS/Waitt Grant W251-12), the British Ecological Society (Small Ecological Projects 369/4558), and by the Languedoc Roussillon French region Program ‘Chercheur d’Avenir 2011’ to CD. This research was conducted under the scope of the International Associated Laboratory between the CNRS (France) and CIBIO (Portugal): LIA “Biodiversity and Evolution”.
Biographies
Claire Loiseau research explores the impacts of anthropogenic changes on host-parasite interactions. She particularly studies the avian blood parasites as model to understand how the ecological factors affect the distribution and the evolutionary strategies of parasites.
Martim Melo research focuses on the diversification of the bird fauna of Africa, and in particular in its oceanic and ecological islands – natural speciation centers that are amenable for the study of the complex processes of adaptation and speciation.
Footnotes
DR. CLAIRE LOISEAU (Orcid ID : 0000-0002-0407-2904)
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article:
DATA ACCESSIBILITY
Genetic mtDNA data generated for this study are available on GenBank: KT376897-KT376976 and KT595662-KT595669.
Editor: Sonya Clegg
Author contributions: C.L. wrote the paper together with M.M. and R.C.; M.M., R.C., E.L., C.D. and C.L. collected samples; M.M., J.S.B., R.C.F., S.R. and E.L. conducted the parasite screening; C.L. identified the lineages and conducted the statistical analyses; E.L. performed the paired-approach statistical analyses; M.M. conducted the phylogenetic analyses; R.C. designed the study; All authors contributed to revisions of the manuscript.
References
- Bataille A, Fournie G, Cruz M, Cedeno V, Parker P, Cunningham AA, Goodman SJ. Host selection and parasite infection in Aedes taeniorhynchus, endemic disease vector in the Galapagos Islands. Infection Genetics and Evolution. 2012;12:1831–1841. doi: 10.1016/j.meegid.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker Ben, Walker S. Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software. 2015;67:1–48. [Google Scholar]
- Beadell JS, Ishtiaq F, Covas R, Melo M, Warren BH, Atkinson CT, Bensch S, Graves GR, Jhala YV, Peirce MA, Rahmani AR, Fonseca DM, Fleischer RC. Global phylogeographic limits of Hawaii’s avian malaria. Proceedings of the Royal Society B: Biological Sciences. 2006;273:2935–2944. doi: 10.1098/rspb.2006.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadell JS, Atkins C, Cashion E, Jonker M, Fleischer RC. Immunological change in a parasite-impoverished environment: divergent signals from four island taxa. Plos One. 2007;2:e896. doi: 10.1371/journal.pone.0000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensch S, Hellgren O, Pérez-Tris J. MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Molecular Ecology Resources. 2009;9:1353–1358. doi: 10.1111/j.1755-0998.2009.02692.x. Accessed on 05 September 2016. [DOI] [PubMed] [Google Scholar]
- Buckley LB, Walter Jetz W. Insularity and the determinants of lizard population density. Ecology Letters. 2007;10:481–489. doi: 10.1111/j.1461-0248.2007.01042.x. [DOI] [PubMed] [Google Scholar]
- Burke K. Origin of the Cameroon Line of volcano-capped swells. Journal of Geology. 2001;109:349–362. [Google Scholar]
- Carlson J, Martínez-Góme J, Valkiūnas G, Loiseau C, Bell D, Sehgal RNM. Diversity and phylogenetic relationships of haemosporidian parasites in birds of Socorro Island, México and their role in the reintroduction of the Socorro Dove Zenaida graysoni. Journal of Parasitology. 2013;99:270–276. doi: 10.1645/GE-3206.1. [DOI] [PubMed] [Google Scholar]
- Chasar A, Loiseau C, Valkiūnas G, Iezhova T, Smith TB, Sehgal RNM. Prevalence and diversity patterns of avian blood parasites in degraded African rainforest habitats. Molecular Ecology. 2009;18:4121–4133. doi: 10.1111/j.1365-294X.2009.04346.x. [DOI] [PubMed] [Google Scholar]
- Clark NJ, Clegg SM. The influence of vagrant hosts and weather patterns on the colonization and persistence of blood parasites in an island bird. Journal of Biogeography. 2015;42:641–651. [Google Scholar]
- Colwell RK. EstimateS: statistical estimation of species richness and shared species from samples. 2013 Version 9. Persistent URL < purl.oclc.org/estimates>.
- Cornuault J, Bataillard A, Warren BH, Lootvoet A, Mirleau P, Duval T, Mila B, Thebaud C, Heeb P. The role of immigration and in-situ radiation in explaining blood parasite assemblages in an island bird clade. Molecular Ecology. 2012;21:1438–1452. doi: 10.1111/j.1365-294X.2012.05483.x. [DOI] [PubMed] [Google Scholar]
- Docile TN, Figueiró R, Gil-Azevedo LH, Nessimian JL. Water pollution and distribution of the black fly (Diptera: Simuliidae) in the Atlantic Forest, Brazil. Revista de Biología Tropical. 2015;63:683–693. [PubMed] [Google Scholar]
- Drovetski SV, Aghayan SA, Mata VA, Lopes RJ, Mode NA, Harvey JA, Voelker G. Does the niche breadth or trade-off hypothesis explain the abundance-occupancy relationship in avian Haemosporidia? Molecular Ecology. 2014;23:3322–3329. doi: 10.1111/mec.12744. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exell AW. Catalogue of the vascular plants of S. Tomé (with Principe and Annobon) British Museum (Natural History); London: 1944. [Google Scholar]
- Exell AW. Angiosperms of the islands of the Gulf of Guinea (Fernando Pó, Príncipe, S. Tomé and Annobón) Bulletin of the British Museum (Natural History) Botany. 1973;4:325–411. [Google Scholar]
- Fallon SM, Bermingham E, Ricklefs RE. Host specialization and geographic localization of avian malaria parasites: A regional analysis in the Lesser Antilles. American Naturalist. 2005;165:466–480. doi: 10.1086/428430. [DOI] [PubMed] [Google Scholar]
- Hellgren O, Waldenström J, Bensch S. A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. Journal of Parasitology. 2004;90:797–802. doi: 10.1645/GE-184R1. [DOI] [PubMed] [Google Scholar]
- Hellgren O, Pérez-Tris J, Bensch S. A jack-of-all-trades and still a master of some: prevalence and host range in avian malaria and related blood parasites. Ecology. 2009;90:2840–2849. doi: 10.1890/08-1059.1. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Illera JC, Fernández-Álvarez A, Hernández-Flores CN, Foronda P. Unforeseen biogeographical patterns in a multiple parasite system in Macaronesia. Journal of Biogeography. 2015;42:1858–1870. [Google Scholar]
- Ishtiaq F, Beadell JS, Warren BH, Fleischer RC. Diversity and distribution of avian haematozoan parasites in the western Indian Ocean region: a molecular survey. Parasitology. 2012;139:221–231. doi: 10.1017/S0031182011001831. [DOI] [PubMed] [Google Scholar]
- Jean K, Burnside WR, Carlson L, Smith K, Guegan JF. An equilibrium theory signature in the island biogeography of human parasites and pathogens. Global Ecology and Biogeography. 2016;25:107–116. [Google Scholar]
- Jones PJ. Biodiversity in the Gulf of Guinea: an overview. Biodiversity and Conservation. 1994;3:772–784. [Google Scholar]
- Jones PJ, Tye A. The birds of São Tomé & Príncipe with Annobón – islands of the Gulf of Guinea. British Ornithologists’ Union and British Ornithologists’ Club; Oxford: 2006. [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- LaPointe DA, Atkinson CT, Samuel MD. Ecology and conservation biology of avian malaria. Annals of the New York Academy of Sciences. 2012;1249:211–226. doi: 10.1111/j.1749-6632.2011.06431.x. [DOI] [PubMed] [Google Scholar]
- de Lima RF, Dallimer M, Atkinson P, Barlow J. Biodiversity and land-use change: understanding the complex responses of an endemic-rich bird assemblage. Diversity and Distributions. 2013;19:411–422. [Google Scholar]
- Lindström KM, Foufopoulos J, Parn H, Wikelski M. Immunological investments reflect parasite abundance in island populations of Darwin’s finches. Proceedings of the Royal Society B: Biological Sciences. 2004;271:1513–1519. doi: 10.1098/rspb.2004.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobato E, Doutrelant C, Melo M, Reis S, Covas R. Insularity effects on bird immune parameters: a comparison between island and mainland populations in West Africa. Ecology and Evolution. 2017 doi: 10.1002/ece3.2788. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur RH, Wilson EO. The theory of island biogeography. Princeton University Press; Princeton: 1967. [Google Scholar]
- MacArthur RH, Diamond JM, Karr JR. Density compensation in island faunas. Ecology. 1972;53:330–342. [Google Scholar]
- Matson KD. Are there differences in immune function between continental and insular birds? Proceedings of the Royal Society B: Biological Sciences. 2006;273:2267–2274. doi: 10.1098/rspb.2006.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum H, Barlow N, Hone J. How should pathogen transmission be modelled? Trends in Ecology and Evolution. 2001;16:295–300. doi: 10.1016/s0169-5347(01)02144-9. [DOI] [PubMed] [Google Scholar]
- Melo M, Bowie RCK, Voelker G, Dallimer M, Collar NJ, Jones PJ. Multiple lines of evidence support the recognition of a very rare bird species: the Príncipe thrush. Journal of Zoology. 2010;282:120–129. [Google Scholar]
- Melo M, Warren BH, Jones PJ. Rapid parallel evolution of aberrant traits in the diversification of the Gulf of Guinea white-eyes (Aves, Zosteropidae) Molecular Ecology. 2011;20:4953–4967. doi: 10.1111/j.1365-294X.2011.05099.x. [DOI] [PubMed] [Google Scholar]
- Mustapha M, Jarvis W, Post RJ. The Simuliidae (Diptera) of the Republic of São Tomé and Príncipe, including the description of a new species. African Invertebrates. 2004;45:143–155. [Google Scholar]
- Mustapha M, McCall PJ, Cheke RA, Post RJ. The blackflies (Diptera: Simuliidae) of Bioko (Republic of Equatorial Guinea) and the Gulf of Guinea with a description of the larvae of the ‘Pomeroy’ form of Simulium cervicornutum. Systematic Entomology. 2006;31:611–620. [Google Scholar]
- Olsson-Pons S, Clark NJ, Ishtiaq F, Clegg SM. Differences in host species relationships and biogeographic influences produce contrasting patterns of prevalence, community composition and genetic structure in two genera of avian malaria parasites in southern Melanesia. Journal of Animal Ecology. 2015;84:985–998. doi: 10.1111/1365-2656.12354. [DOI] [PubMed] [Google Scholar]
- Patz JA, Graczyk TK, Geller N, Vittor AY. Effects of environmental change on emerging parasitic diseases. International Journal for Parasitology. 2000;30:1395–1405. doi: 10.1016/s0020-7519(00)00141-7. [DOI] [PubMed] [Google Scholar]
- Pérez-Rodríguez A, Ramírez A, Richardson DS, Pérez-Tris J. Evolution of parasite island syndromes without long-term host population isolation: parasite dynamics in Macaronesian blackcaps Sylvia atricapilla. Global Ecology and Biogeography. 2013;22:1272–1281. [Google Scholar]
- Poulin R, Mouillot D. Parasite specialization from a phylogenetic perspective: a new index of host specificity. Parasitology. 2003;126:473–480. doi: 10.1017/s0031182003002993. [DOI] [PubMed] [Google Scholar]
- Ribeiro H, Da Cunha Ramos H, Capela RA, Alves Pires C. Os mosquitos (Diptera: Culicidae) da Ilha de São Tomé. Garcia de Orta Série Zoologica. 1998;22:1–20. [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2015. URL http://www.R-project.org/ [Google Scholar]
- Ricklefs RE, Bermingham E. Taxon cycles in the Lesser Antillean avifauna. Ostrich. 1999;70:49–59. [Google Scholar]
- Ricklefs RE, Bermingham E. The concept of the taxon cycle in biogeography. Global Ecology and Biogeography. 2002;11:353–361. [Google Scholar]
- Ricklefs RE, Gray JD, Latta SC, Svensson-Coelho M. Distribution anomalies in avian haemosporidian parasites in the southern Lesser Antilles. Journal of Avian Biology. 2011;42:570–584. [Google Scholar]
- Ricklefs RE, Soares L, Ellis VA, Latta SC. Haemosporidian parasites and avian host population abundance in the Lesser Antilles. Journal of Biogeography. 2016;43:1277–1286. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:475–481. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Santiago-Alarcon D, Palinauskas V, Schaefer HM. Diptera vectors of avian Haemosporidian parasites: untangling parasite life cycles and their taxonomy. Biological Reviews. 2012;87:928–964. doi: 10.1111/j.1469-185X.2012.00234.x. [DOI] [PubMed] [Google Scholar]
- Sari EHR, Klompen H, Parker PG. Tracking the origins of lice, haemosporidian parasites and feather mites of the Galapagos flycatcher (Myiarchus magnirostris) Journal of Biogeography. 2013;40:1082–1093. [Google Scholar]
- SAS. SAS user’s guide. SAS Institute Inc; Cary, NC, USA: 1999. [Google Scholar]
- Sehgal RNM. Deforestation and avian infectious diseases. Journal of Experimental Biology. 2010;213:955–960. doi: 10.1242/jeb.037663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurgin LG, Illera JC, Padilla DP, Richardson DS. Biogeographic patterns and co-occurrence of pathogenic infection across island populations of Berthelot’s pipit (Anthus berthelotii) Oecologia. 2012;168:691–701. doi: 10.1007/s00442-011-2149-z. [DOI] [PubMed] [Google Scholar]
- Stangler A, Halgos J, Beracko P. Balckfly (Diptera, Simuliidae) communities and species richness estimation in Carpathian montane streams. Central European Journal of Biology. 2013;8:681–692. [Google Scholar]
- Steadman DW, Greiner EC, Wood CS. Absence of blood parasites in indigenous and introduced birds from the Cook Islands, South-Pacific. Conservation Biology. 1990;4:398–404. [Google Scholar]
- Torchin ME, Lafferty KD, Dobson AP, McKenzie VJ, Kuris AM. Introduced species and their missing parasites. Nature. 2003;421:628–630. doi: 10.1038/nature01346. [DOI] [PubMed] [Google Scholar]
- Valkiūnas G. Avian malaria parasites and other Haemosporidia. CRC Press; Boca Raton, Florida: 2005. [Google Scholar]
- van Riper C, III, van Riper SG, Goff ML, Laird M. The epizootiology and ecological significance of malaria in Hawaiian land birds. Ecological Monographs. 1986;56:327–344. [Google Scholar]
- van Rooyen J, Lalubin F, Glaizot O, Christe P. Avian haemosporidian persistence and co-infection in great tits at the individuals level. Malaria Journal. 2013;12:40. doi: 10.1186/1475-2875-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittor AY, Pan W, Gilman RH, Tielsch J, Glass G, Shields T, Sanchez-Lozano W, Pinedo VV, Salas-Cobos E, Flores S, Patz JA. Linking deforestation to malaria in the Amazon: characterization of the breeding habitat of the principal malaria vector, Anopheles darlingi. American Journal of Tropical Medicine and Hygiene. 2009;81:5–12. [PMC free article] [PubMed] [Google Scholar]
- Walther BA, Cotgreave P, Price RD, Gregory RD, Clayton DH. Sampling effort and parasite species richness. Parasitology Today. 1995;14:306–310. doi: 10.1016/0169-4758(95)80047-6. [DOI] [PubMed] [Google Scholar]
- Warner RE. The role of introduced diseases in the extinction of the endemic Hawaiian avifauna. Condor. 1968;70:101–120. [Google Scholar]
- Wikelski M, Foufopoulos J, Vargas H, Snell H. Galápagos birds and diseases: invasive pathogens as threats for island species. Ecology and Society. 2004;9:5. [Google Scholar]
- Wilson EO. The nature of the taxon cycle in the Melanesian ant fauna. The American Naturalist. 1961;95:169–193. [Google Scholar]
- Yasuoka J, Levins R. Impact of deforestation and agricultural development on anopheline ecology and malaria epidemiology. American Journal of Tropical Medicine and Hygiene. 2007;76:450–460. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Characteristics of sampling sites and host species.
Appendix S2 Host specificity indices and Genbank accession numbers.
Appendix S3 Phylogenetic analyses and trees.


