Abstract
Bile acids play a critical role in the regulation of glucose, lipid, and energy metabolism through activation of the nuclear bile acid receptor farnesoid X receptor (FXR) and membrane G protein-coupled bile acid receptor-1 (Gpbar-1, aka TGR5). Agonist activation of FXR and TGR5 improves insulin and glucose sensitivity and stimulates energy metabolism to prevent diabetes, obesity, and non-alcoholic fatty liver disease (NAFLD). Bile acids have both pro- and anti-inflammatory actions through FXR and TGR5 in the intestine and liver. In the intestine, bile acids activate FXR and TGR5 to stimulate stimulate fibroblast growth factor 15 and glucagon-like peptide-1 secretion. FXR and TGR5 agonists may have therapeutic potential for treating liver-related metabolic diseases, such as diabetes and NAFLD.
Keywords: Bile acid metabolism, Cholestatic liver, diseases Metabolic diseases
1. Introduction
Bile acids are the end-products of cholesterol catabolism in the liver. Bile acids are physiological detergents important for emulsification of dietary fats, drugs, and lipid-soluble vitamins in the intestine and subsequent absorption and transport to the liver for metabolism and distribution to other tissues and organs. More recent studies have demonstrated that bile acids are signaling molecules that activate the nuclear receptor farnesoid X receptor (FXR) and G protein-coupled bile acid receptor-1 (Gpbar-1, aka TGR5) to regulate glucose, lipid, and energy metabolism. This review will discuss detailed bile acid synthesis and metabolism, regulation of bile acid synthesis, the roles of bile acid-activated nuclear receptors FXR and TGR5 in metabolic regulation, and bile acids as therapeutic drugs to treat liver diseases, diabetes, and obesity.
2. Bile acid metabolism
2.1. Bile acid synthesis in the liver
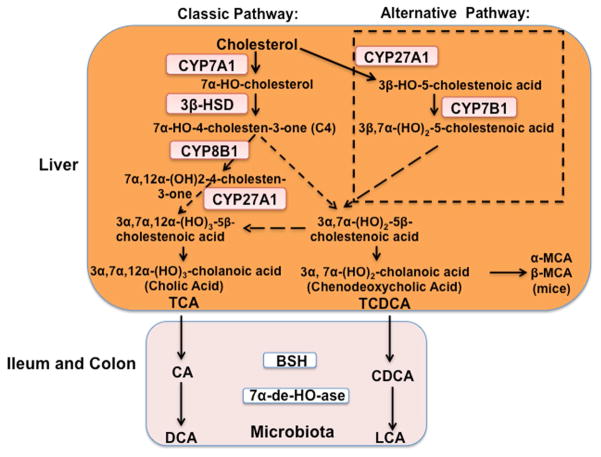
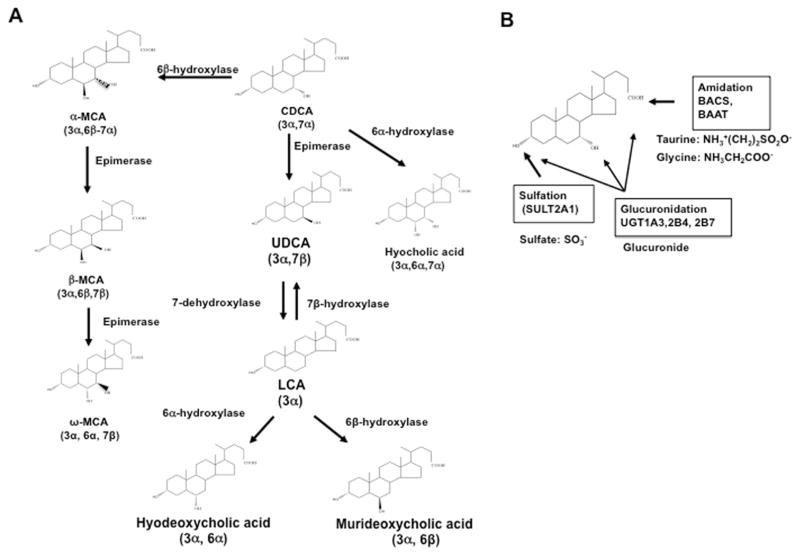
In human liver, two primary bile acids, cholic acid (CA) and chenodeoxycholic (CDCA), are synthesized from cholesterol through two pathways (Fig. 1) [1]. The classic pathway is initiated by the rate-limiting enzyme cholesterol 7α-hydroxylase (CYP7A1). For CA (3α,7α,12α-trihydroxy-cholan-24-oic acid) synthesis, sterol 12α-hydroxylase (CYP8B1) is required for 12α-hydroxylation of 7α-hydroxy-4-cholesten-3-one, an intermediate and marker for the rate of bile acid synthesis. Mitochondrial steroid 27-hydroxylase (CYP27A1) catalyzes steroid side-chain oxidation, which is followed by oxidative cleavage of a 3-carbon-side-chain to form C24-bile acids, CA and CDCA. In the alternative pathway, CYP27A1 initiates bile acid synthesis by hydroxylation and oxidation of cholesterol to 3β-hydroxy-5-cholestenoic acid, which is then 7α-hydroxylated by oxysterol 7α-hydroxylase (CYP7B1) to form 3β,7α-dihydroxy-5-cholestenoic acid. These reactions also occur in the macrophages and steroidogenic tissues. It has been suggested that conversion of oxysterols formed in the macrophages to bile acids in the liver is a reverse cholesterol transport pathway for protection against atherosclerosis [2]. Bile acids synthesized in the liver are primary bile acids. Different primary bile acids are synthesized by various hydroxylases and epimerases, depending on species. In mouse liver, CDCA (3α,7α-dihydroxy-cholan-24-oic acid) is converted to α-muricholic acid (α-MCA, 3α, 6β, 7α), which is then epimerized to β-MCA (3α, 6β, 7β) and ω-MCA (3α, 6α, 7β) (Fig. 2A) [3]. These MCAs are highly soluble. Mouse liver is capable of converting the secondary bile acid lithocholic acid (LCA, 3α) formed in the intestine to CDCA by 7α-hydroxylation. In humans and mice, some CDCA can be epimerized to ursodeoxycholic acid (UDCA, 3α,7β), a highly soluble bile acid. CDCA can be converted to hyocholic acid (3α, 6α, 7α) by 6α-hydroxylation. In mice, LCA can be hydroxylated to hyodeoxycholic acid (3α, 6α) and murideoxycholic acid (3α, 6β). Hyodeoxycholic acid can also be derived from MCA and hyocholic acid [4]. Bile acids form Na2+ salts and are conjugated to glycine or taurine by bile acid:Coenzyme A synthase and bile acid aminotransferase for secretion into bile via canalicular bile salt export peptide (Fig. 2B and Fig. 3). Glucuronidation of bile acids by UDP-glucuronosyltransferase also increases solubility for secretion of bile acids to bile via canalicular bile acid transporter multidrug resistance protein-related protein 2. Bile acids can also be sulfated by sulfotransferases to reduce toxicity and promote secretion into urine. Bile acids are stored in the gallbladder as mixed micelles with cholesterol and phosphatidylcholine. After a meal, bile acids are secreted into the intestinal tract to emulsify dietary fats, steroids, and lipid-soluble vitamins. In the intestine (colon), gut microbial bile salt hydrolases deconjugate conjugated-bile acids, and bacterial 7α-dehydroxylases convert the primary bile acids CA and CDCA to deoxycholic acid (3α, 7α) and LCA, respectively (Fig. 1). Most bile acids are re-conjugated to glycine or taurine, reabsorbed in the ileum, and transported back to the liver via the portal vein. The enterohepatic circulation (EHC) of bile acids recovers approximately 95% of bile acids (Fig 3). In humans, the classic bile acid synthesis pathway is the predominant pathway (82%) for synthesis of a highly hydrophilic bile acid pool consisting of approximately 30% each of CA, CDCA, and deoxycholic acid. In contrast, in mice, the two pathways contribute equally to generate a highly hydrophilic bile acid pool containing approximately 50% CA and 50% α- and β-MCAs. In humans, bile acids are glycine- and taurine-conjugated at a ratio of 3 to 1, whereas in mice most bile acids (95%) are taurine-conjugated. The relative contribution of the classic and alternative bile acid synthesis pathways to bile acid synthesis determines the bile acid pool size and composition.
Fig. 1. Bile acid synthesis pathways.
Two bile acid synthesis pathways are involved in the conversion of cholesterol to bile acids in the liver. The classic pathway is initiated by cholesterol 7α-hydroxylase (CYP7A1), and the alternative pathway is initiated by steroid 27-hydroxylase (CYP27A1). 3β-hydroxysteroid dehydrogenase (3β-HSD) converts 7α-hydroxycholesterol to 7α-hydroxy-4-cholesten-3-one (C4). Serum C4 level has been used as a marker for the rate of bile acid synthesis. Sterol 12-hydroxylase (CYP8B1) is a branch enzyme that synthesizes cholic acid (CA). Without 12α-hydroxylation, chenodeoxycholic acid (CDCA) is synthesized. Mitochondrial CYP27A1 catalyzes oxidation of the steroid side chain, and the peroxisomal β-oxidation reaction cleaves a 3C unit to form C24 cholestenoic acid, the backbone of most bile acids. CA and CDCA are the two primary bile acids synthesized in human liver. In mice, CDCA is converted to α- and β-muricholic acids (α-MCA and β-MCA, respectively). Bile acids are immediately conjugated to the amino acids taurine or glycine (TCA or TCDCA, respectively) for secretion into bile. In the ileum, TCA and TCDCA are deconjugated by bacterial bile salt hydrolase (BSH) activity, and the 7α-hydroxyl group is removed by bacterial 7α-dehydroxylase activity to form deoxycholic acid (DCA) and lithocholic acid (LCA), respectively. Bile acids (TCA, TDCA, TCDCA, Tα-MCA, and Tβ-MCA) are re-conjugated and circulated back to the liver. LCA is secreted into feces, and a small amount is circulated to the liver, conjugated to sulfite, and secreted into urine.
Fig. 2. Bile acid metabolism.
A. Primary bile acids are metabolized to other bile acids in the liver and intestine. In mice, CDCA is converted to α-MCA, β-MCA, and ω-MCA. In humans and mice, CDCA can be converted to ursodeoxycholic acid (UDCA) or hyocholic acid. UDCA can be 7-dehydroxylated to LCA, which can be hydroxylated to hyodeoxycholic acid and muricholic acid. B. Bile acids can be conjugated to the amino acids taurine or glycine at the C24OOH group by bile acid Co-A synthase (BACS) and bile acid amino transferase (BAAT). Sulfotransferase (SULT2A1) transfers a sulfate to the 3β-HO position. UDP-glucuronosyltransferase (UGT) transfers a glucuronide group to the C3-OH, C7-OH, and C24-OOH groups. In the intestine, FXR induces fibroblast growth factor (FGF) 15, which is secreted into portal circulation to activate the FGF receptor 4/β-klotho complex, which activates ERK1/2 and JNK of the MAPK pathway to inhibit bile acid synthesis.
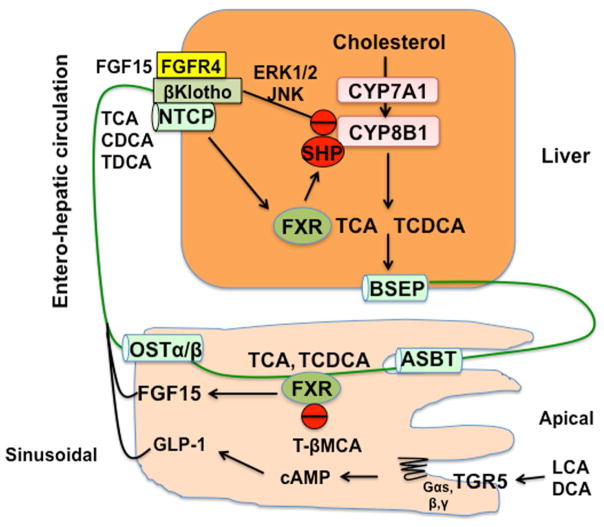
Fig. 3. Mechanisms of bile acid feedback inhibition of bile acid synthesis.
Two mechanisms have been proposed for bile acid feedback inhibition of CYP7A1, CYP8B1, and bile acid synthesis. In the liver, FXR induces small heterodimer partner (SHP), a negative nuclear receptor, to inhibit CYP7A1 and CYP8B1 gene transcription. Bile acids are excreted into bile via canalicular bile salt export pump (BSEP). In the ileum, apical sodium-dependent bile salt transporter (ASBT) reabsorbs bile acids into enterocytes, and the bile acids are then secreted into portal circulation via sinusoidal organic solute transporter α/β (OSTαβ). Bile acids are transported into hepatocytes via Na2+-dependent taurocholate co-transport peptide (NTCP) located on the sinusoidal membrane. This EHC of bile acids from the intestine to the liver inhibits bile acid synthesis and maintains bile acid homeostasis. In the intestine, bile acid-activated FXR induces fibroblast growth factor (FGF) 15 to activate FGF receptor (FGFR4)/β-Klotho signaling, which inhibits CYP7A1 gene transcription via JNK and ERK1/2 MAPK pathways. Antagonism of FXR activity by T-βMCA reduces FGF15, thus increasing CYP7A1 expression and bile acid synthesis. The secondary bile acids produced in the colon, LCA and DCA, activate intestinal TGR5 to activate cAMP signaling, which stimulates glucagon-like peptide-1 (GLP-1) secretion from enteroendocrine L cells.
2.2. Regulation of bile acid synthesis
Regulation of bile acid synthesis is extremely complicated and not completely understood. The rate of bile acid synthesis, pool size, and composition varies by species and gender and is influenced by diet, circadian rhythms, hormones, drugs, the gut microbiota, pathological states, and genetic background. Bile acids maintain liver metabolic homeostasis and have anti-inflammatory properties under normal physiological conditions. Accumulation of high levels of hydrophobic bile acids in cholestasis causes liver inflammation and injury. Thus, bile acid concentrations have to be tightly regulated to maintain very low levels in the liver and blood circulation.
The EHC of bile acids from the intestine to the liver inhibits bile acid synthesis mainly by transcriptional repression of the rate-limiting enzyme CYP7A1 and the branch enzyme for cholic acid synthesis CYP8B1 (Fig. 3). The EHC of bile acids is highly efficient, occurs seven to eight times a day, and recycles approximately 95% of the bile acids in the pool. Two mechanisms of bile acid feedback regulation of bile acid synthesis have been suggested based on animal model studies (Fig. 3). In the liver, bile acids activate the nuclear receptor FXR to induce a negative nuclear receptor called small heterodimer partner (SHP). SHP inhibits trans-activating activity of hepatocyte nuclear factor 4 and liver-related homologue-1, which bind to the Cyp7a1 and Cyp8b1 gene promoters [5]. In FXR deficient mice, bile acid synthesis is upregulated and bile acids pool size is increased [6]. In SHP deficient mice, bile acid synthesis is still inhibited, suggesting other pathways may be involved in bile acid feedback regulation [7]. SHP is a transcriptional repressor that plays a critical role in regulation of bile acid homeostasis and prevention against cholestatic injury [8]. Recent studies have identified several long non-coding RNAs involved in the regulation of bile acid metabolism. SHP regulates H19, which is involved in Bcl1-induced cholestatic injury [9]. Maternally expressed 3 gene induces cholestatic injury by interaction with polypyrimidine tract binding protein 1 to facilitate SHP mRNA decay [9, 10]. A long noncoding RNA interacts with heterogeneous nuclear ribonuceoprotein family A2/B1 to repress Cyp7a1 gene transcription [11]. SHP may play a role in protection against liver fibrosis and liver injury by regulating the transcription factor elongation factor 2F1 and the fibrogenic factor early growth response factor 1 [12]. In the intestine, bile acids activate FXR to induce fibroblast growth factor (FGF) 15, which is secreted into portal circulation to activate the hepatic FGF receptor 4 (FGFR4)/β-Klotho complex. This then leads to activation of extracellular signal-regulated kinase 1/2 and c-Jun N-terminal kinase of the mitogen-activated protein kinase pathways and subsequent inhibition of Cyp7a1 and Cyp8b1 gene transcription [13]. Recent studies have revealed that T-αMCA and T-βMCAs are potent antagonists that inhibit FXR induction of FGF15 in the ileum [14]. In germ-free mice, all bile acids are conjugated, and T-MCAs accumulate. Therefore, bile acid synthesis and pool size are increased because of T-MCA antagonism of intestinal FXR signaling. The gut-to-liver axis plays a critical role in regulating bile acid synthesis and metabolism. Bile acids control gut bacterial overgrowth [15], and the gut microbiota controls bile acid synthesis and composition [16, 17]. Thus, gut-liver signaling cross-talk plays a critical role in regulation of host metabolism [18].
3. Bile acid receptors in metabolic regulation
Bile acids regulate hepatic metabolism via activation of FXR and TGR5. These two bile acid-activated receptors are highly expressed in the gastrointestinal tract and play critical roles in the regulation of liver metabolism and homeostasis. The gut microbiota controls the release of gut hormones/peptides, such as peptide YY and glucagon-like peptide-1 (GLP-1) [19]. These hormones regulate insulin secretion from pancreatic β-cells and, in turn, glucose homeostasis. The gut microbiota also causes low-grade inflammation and initiates obesity and insulin resistance [20]. Dysbiosis has been associated with inflammatory bowel diseases, obesity, and type 2 diabetes as well as non-alcoholic fatty liver disease, cirrhosis, and liver cancer [21–24].
3.1. FXR in metabolic regulation
The role of FXR in the regulation of hepatic lipid and glucose metabolism has been studied extensively. Activation of FXR by bile acids and a potent FXR agonist GW4064 reduces serum triglycerides and improves glucose tolerance and insulin resistance in diabetic mice [25, 26]. In diabetes, serum bile acid levels, especially 12α-hydroxylated bile acids, are increased. Hyperglycemia may stimulate acetylation of Cyp7a1 via epigenetic mechanisms to increase basal levels of bile acids in diabetes [27]. Increasing 12α-hydroxylated bile acids increases dietary absorption of fats and cholesterol. Glucagon and cAMP suppress, whereas insulin stimulates, CYP7A1 expression in human hepatocytes [28]. Activation of FXR induces SHP to inhibit bile acid and fatty acid synthesis by inhibiting oxysterol-regulated steroid response element binding protein-1c and gluconeogenesis in hepatocytes [25].
Transgenic overexpression of Cyp7A1 in mice increases bile acid synthesis and insulin and glucose tolerance, reduces inflammation, and protects against high fat-diet induced obesity and steatosis [29]. In these mice, bile acid pool size is doubled with reduced CA but increased CDCA, which may activate FXR/SHP mechanisms to reduce hepatic steatosis and increase glucose and insulin sensitivity. Surprisingly, Cyp7a1 deficient mice are also protected from high-fat-high-cholesterol diet-induced metabolic disorder [30]. In Cyp7a1−/− mice, the bile acid pool was reduced by 40%, and CA was reduced, while MCAs were increased. This was likely due to activation of the alternative bile acid synthesis pathway to compensate for inactivation of the classic bile acid synthesis pathway. CA is highly efficient at absorbing dietary cholesterol and fats. Reduction of CA in the bile acid pool may improve glucose and lipid metabolism. However, adenovirus-mediated overexpression of Cyp7a1 ameliorated lipopolysaccharide-induced hepatic inflammation through FXR activation, which inhibited nuclear factor-kB activity [31]. Cyp7a1−/− mice displayed more severe hepatic steatosis, oxidative stress, apoptosis, and fibrosis than wild type mice when fed a methionine-choline deficient diet [31]. Remarkably, adenovirus-mediated overexpression of Cyp7a1 in Cyp7a1−/− mice reversed methionine-choline deficient diet-induced hepatic fibrosis and injury. Thus, maintaining bile acid and cholesterol homeostasis is important for protection against liver injury and hepatic steatosis.
Germ-free and antibiotic-treated mice have increased bile acid pools because of increased T-MCAs, which antagonize FXR activity to increase bile acid synthesis in hepatocytes [14]. Recent studies have demonstrated that intestinal Fxr gene deficiency or intestinal FXR antagonism by the antioxidant tempol increased T-MCAs and protected against diet-induced obesity and diabetes [32, 33]. Consistent with this, the intestine-selective FXR inhibitor Gly-MCA, which is not absorbed into the circulation, increases T-MCAs and prevents obesity and NAFLD in mice [34]. In contrast, another recent study reported that activation of intestinal Fxr by the selective intestinal FXR agonist fexaramine improved diabetes and obesity in mice [35]. It is likely that antibiotic treatment or ablation of intestinal FXR alters the gut microbiota and bile acid metabolism to protect against nonalcoholic steatohepatitis and diabetes. In contrast, activation of intestinal FXR by fexaramine increases FGF15 to stimulate energy metabolism and improve insulin sensitivity and glucose tolerance. Fexaramine markedly increased LCA, which may activate TGR5 to stimulate GLP-1 secretion and improve insulin sensitivity. In general, activation of FXR signaling is beneficial for reducing liver inflammation and lipogenesis and protecting against cholestatic liver diseases and NAFLD. A recent study revealed that UDCA treatment (20 mg/kg/day) for 3 weeks increased bile acid synthesis by reducing FGF19 and liver and serum low density lipoprotein-cholesterol but increased triglyceride content in 40 morbidly obese patients [36]. It was suggested that UDCA may antagonize intestinal FXR to stimulate bile acid synthesis.
3.2. TGR5 in metabolic regulation
The role of TGR5 in the regulation of hepatic metabolism has not been studied in depth. TGR5 is widely expressed in many tissues, including the intestine, gallbladder, liver, and brain [37–39]. In the liver, TGR5 is expressed in Kupffer and sinusoidal endothelial cells but not in hepatocytes [39, 40]. In the gastrointestinal tract, activation of TGR5 by bile acids and agonists protects intestinal barrier function, reduces inflammation, and stimulates gallbladder refilling and GLP-1 secretion from enteroendocrine L cells [41]. GLP-1 secretion then increases postprandial insulin secretion from pancreatic β-cells to improve insulin resistance [42]. GLP-1 is an intestinal incretin produced in L cells and is released in response to meal intake [43]. GLP-1 secretion is stimulated by nutrients, such as carbohydrates, fats, and proteins, in the intestinal lumen. The synthetic GLP-1 analog exendin-4 reduces hepatic steatosis by decreasing lipogenesis and inducing fatty acid oxidation [44]. It has been reported that activation of TGR5 induces the thyroid hormone deiodinase 2 to stimulate energy metabolism in brown adipose tissues [45]. Activation of TGR5 stimulates adenylyl cyclase to convert ATP to cAMP, which activates protein kinase A and subsequently cAMP response element binding protein, resulting in alleviation of obesity and hepatic steatosis in diet-induced obese mice [42].
4. Bile acids as therapeutic agents
4.1. Cholestatic liver diseases
Cholestasis is a pathological condition in which normal bile flow out of the liver is reduced or disrupted, leading to intrahepatic accumulation of bile acids. Accumulation of cytotoxic bile acids causes liver inflammation by activating NF-kB-mediated pro-inflammatory cytokine production. High levels of toxic bile acids damage the bile duct epithelium and elevate biliary pressure to rupture the bile duct and expose hepatocytes to high concentrations of bile acids and inflammatory infiltration. This then leads to hepatocyte cell death. Cholestasis can result from genetic defects in canalicular transporters (e.g. bile salt export peptide), mechanical obstruction of the bile duct by gallstones or tumors, factors associated with pregnancy (intrahepatic cholestasis of pregnancy), autoimmune destruction of the bile ducts in and outside of the liver, or drug-induced liver toxicity [46, 47]. Bile acid composition and pool size play a critical role in control of metabolism and inflammation in the gastrointestinal tract in various cholestatic conditions. Hydrophobic bile acids are especially cytotoxic and cause hepatocyte cell death via various direct and indirect mechanisms. Chronic cholestasis causes liver fibrosis, cirrhosis, liver failure, hepatocellular carcinoma, and cholangiocarcinoma. Primary biliary cholangitis (PBC) and primary sclerosing cholangitis (PSC) are two common types of progressive chronic cholestasis diagnosed in humans [48, 49]. The etiologies of PBC and PSC are not fully clear but involve genetic, immune, and environmental factors. PBC is characterized by autoimmune destruction of the small intrahepatic bile duct, leading to inflammation and impaired hepatic bile acid secretion and accumulation. PSC is an idiopathic liver disease characterized by chronic liver inflammation and progressive destruction of intrahepatic and extrahepatic bile ducts. PBC is predominantly diagnosed in females, while PSC is more prevalent in men.
4.2. Bile acids as therapeutic agents for liver diseases
Bile acid derivatives with FXR and TGR5 agonistic activity have recently been developed to treat metabolic diseases [50–53]. UDCA (Ursodiol™) has been used as a bile acid replacement therapy for patients with bile acid synthesis deficiency, gallstone dissolution, and digestive diseases for many years [54]. UDCA has been approved by the FDA for treatment of PBC and has been shown to significantly improve liver function and prolong the time before liver transplantation is required [55]. UDCA can activate intracellular signaling pathways, such as protein kinase C and mitogen-activated protein kinase, and alleviate liver injury in cholestasis. It increases hepatobiliary secretion and promotes biliary HCO3− secretion, which protects hepatocytes and cholangiocytes from hydrophobic bile acid insult [56–58]. In addition, UDCA exhibits anti-inflammatory and pro-survival effects in cholestasis [58, 59]. Approximately 40% of patients with PBC do not respond adequately to UDCA [60]. UDCA is not effective in treating patients with PSC, and the efficacy of UDCA in other forms of cholestasis has not been demonstrated. Nor-ursodeoxycholic acid (norUDCA) is a side chain-shortened C23 homologue of UDCA [61, 62]. It has been shown that norUDCA is passively absorbed by cholangiocytes, undergoes cholehepatic shunting, and increases HCO3− secretion [58]. NorUDCA improves sclerosing cholangitis in the Mdr2−/− model of cholangiopathy [63] and in clinical trials for PSC.
Fibrates activate peroxisome proliferator-activated receptor α and have been used to treat patients with hypertriglyceridemia. Fibrates have been used as monotherapy for patients with PBC and in combination therapy with UDCA for patients with PBC who do not adequately respond to UDCA [64]. Activation of peroxisome proliferator-activated receptor α inhibits CYP7A1, CYP8B1, and CYP27A1 to reduce bile acid synthesis and induces multidrug resistant protein 3, UDP-glucuronosyl transferase family 2B4, sulfur transferase family 2A1, and detoxification of bile acids. In clinical studies, bezafibrate reduced serum alkaline phosphatase and γ-glutamyltransferase in patients with PBC who did not adequately respond to UDCA [65]. Fibrate drugs are used as adjunct therapy for cholestasis [64].
4.3. FXR agonists for treatment of metabolic liver diseases
Obeticholic acid (OCA, 6α-ethyl CDCA) is a selective FXR agonist (EC50 = 0.099 μM) developed by Intercept Pharmaceuticals, Inc. [66, 67]. Based on an understanding of FXR regulation of bile acid metabolism and inflammation, the potential benefit of OCA in treating cholestasis has been extensively investigated in both experimental animal models and in humans [68]. OCA treatment displayed effective protection in experimental cholestasis models [66, 69]. Recent clinical trials reported that OCA significantly improved liver function [70, 71] and reduced serum alkaline phosphatase, γ-glutamyl transferase, and alanine aminotransferase activities in patients with PBC [70]. OCA was recently approved by the FDA for treatment of patients with PBC. In addition, OCA improved NASH scores in clinical trials and is a promising therapy for NASH [72, 73]. Pruritus is commonly associated with cholestasis and treatments utilizing bile acid derivatives, including both FXR and TGR5 agonists [74].
4.4. TGR5 agonists for treatment of metabolic liver diseases
Activation of TGR5 has been shown to protect against inflammation in the intestine and liver [50], stimulate GLP-1 secretion from enteroendocrine L cells and insulin synthesis and secretion from pancreatic β-cells [41, 75], and protect cholangiocytes from bile acid toxicity in cholestasis. Thus, it is a potential therapy for cholestasis [50]. However, TGR5 agonists may also promote proliferation, apoptosis, and progression of cholangiocarcinoma [76].
Several potent TGR5 selective agonists are under development for treating metabolic diseases [77, 78]. INT-777 (6α-ethyl-23(S)-methyl-CA, EC50 = 0.820 μM) is a selective TGR5 agonist developed by Intercept Pharmaceuticals, Inc. that has been shown to protect intestinal barrier function and immune responses to experimental colitis [79, 80]. Additionally, INT-777 reduces macrophage inflammation by activating cAMP signaling, which inhibits NF-kB activity and inflammatory cytokine production. Furthermore, it inhibits plaque formation and atherosclerosis in Ldlr−/− mice [81]. A previous study demonstrated that TGR5 agonists induced NO production and reduced monocyte adhesion in vascular endothelial cells [82]. A non-bile acid and potent TGR5 agonist has been shown to stimulate GLP-1 secretion and lower glucose levels [83, 84].
4.5. FXR and TGR5 dual agonist
A dual FXR and TGR5 agonist, INT-767 (6α-ethyl-3α,7α,23- trihydroxy-24-nor-5β-cholan-23-sulfate sodium salt, EC50 = 0.03 for FXR and EC50 = 0.63 for TGR5), has been developed by Intercept Pharmaceuticals, Inc. It improves diabetes in mice [85, 86] and reduces liver injury in an Mdr2−/− cholangiopathy mouse model [87]. Furthermore, one study revealed that INT-767 treatment in db/db mice improved hepatic steatosis and decreased inflammation [88]. Clinical trials of INT-767 for the treatment of NASH are expected to start soon.
5. Conclusion
Bile acids are important physiological agents for nutrient absorption and integrators of glucose, lipid, and energy metabolism control. Bile acid synthesis is tightly regulated to maintain metabolic homeostasis and prevent accumulation of highly toxic bile acids. Dysregulation of bile acid synthesis causes metabolic diseases, including cholestatic liver diseases, diabetes, and obesity. Activation of FXR and TGR5 by bile acids or their specific agonists improves insulin and glucose tolerance. These two bile acid receptors are critical for improving insulin sensitivity after vertical sleeve gastrectomy [89, 90]. Gastric bypass surgery in obese patients with diabetes rapidly improves insulin sensitivity and glycemic control before weight reduction and is associated with increased serum bile acids and GLP-1 levels [91, 92]. Activation of both FXR and TGR5 in the intestine may coordinately stimulate GLP-1 secretion to improve hepatic glucose and insulin sensitivity in diabetes. It is expected that FXR and TGR5 dual agonists will be developed as therapeutic agents for treatment of NAFLD and diabetes [85, 87, 88].
Acknowledgments
This work was supported by NIH grants DK44442 and DK58379.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–66. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Björkhem I, Diczfalusy U. Oxysterols: friends, foes, or just fellow passengers? Arterioscler Thromb Vasc Biol. 2002;22:734–742. doi: 10.1161/01.atv.0000013312.32196.49. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi S, Fukami T, Masuo Y, et al. Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J Lipid Res. 2016;57:2130–2137. doi: 10.1194/jlr.M071183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eyssen HJ, De Pauw G, Van Eldere J. Formation of hyodeoxycholic acid from muricholic acid and hyocholic acid by an unidentified gram-positive rod termed HDCA-1 isolated from rat intestinal microflora. Appl Environ Microbiol. 1999;65:3158–3163. doi: 10.1128/aem.65.7.3158-3163.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu TT, Makishima M, Repa JJ, et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 6.Sinal CJ, Tohkin M, Miyata M, et al. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Lee YK, Bundman D, et al. Redundant pathways for negative feedback regulation of bile Acid production. Dev Cell. 2002;2:721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 8.Park YJ, Qatanani M, Chua SS, et al. Loss of orphan receptor small heterodimer partner sensitizes mice to liver injury from obstructive cholestasis. Hepatology. 2008;47:1578–1586. doi: 10.1002/hep.22196. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Liu C, Barbier O, et al. Bcl2 is a critical regulator of bile acid homeostasis by dictating Shp and lncRNA H19 function. Scientific reports. 2016;6:20559. doi: 10.1038/srep20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Yang Z, Trottier J, et al. Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology. 2017;65:604–615. doi: 10.1002/hep.28882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan X, Yan J, Ren J, et al. A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism. Hepatology. 2016;64:58–72. doi: 10.1002/hep.28391. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Xu N, Xu J, et al. E2F1 is a novel fibrogenic gene that regulates cholestatic liver fibrosis through the Egr-1/SHP/EID1 network. Hepatology. 2014;60:919–930. doi: 10.1002/hep.27121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li T, Chiang JY. Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev. 2014;66:948–983. doi: 10.1124/pr.113.008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sayin SI, Wahlstrom A, Felin J, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell metabolism. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Inagaki T, Moschetta A, Lee YK, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Islam KB, Fukiya S, Hagio M, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 17.Jones BV, Begley M, Hill C, et al. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wahlstrom A, Sayin SI, Marschall HU, et al. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell metabolism. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Reimer RA, McBurney MI. Dietary fiber modulates intestinal proglucagon messenger ribonucleic acid and postprandial secretion of glucagon-like peptide-1 and insulin in rats. Endocrinology. 1996;137:3948–3956. doi: 10.1210/endo.137.9.8756571. [DOI] [PubMed] [Google Scholar]
- 20.Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–172. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 21.Aron-Wisnewsky J, Gaborit B, Dutour A, et al. Gut microbiota and non-alcoholic fatty liver disease: new insights. Clin Microbiol Infect. 2013;19:338–348. doi: 10.1111/1469-0691.12140. [DOI] [PubMed] [Google Scholar]
- 22.Kakiyama G, Pandak WM, Gillevet PM, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatology. 2013;58:949–955. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joyce SA, Gahan CG. The gut microbiota and the metabolic health of the host. Curr Opin Gastroenterol. 2014;30:120–127. doi: 10.1097/MOG.0000000000000039. [DOI] [PubMed] [Google Scholar]
- 24.Ridlon JM, Kang DJ, Hylemon PB, et al. Bile acids and the gut microbiome. Cur Opin Gastroenterol. 2014;30:332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe M, Houten SM, Wang L, et al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clinl Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Lee FY, Barrera G, et al. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci U S A. 2006;103:1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li T, Francl JM, Boehme S, et al. Glucose and insulin induction of bile acid synthesis: mechanisms and implication in diabetes and obesity. J Bio Chem. 2012;287:1861–1873. doi: 10.1074/jbc.M111.305789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li T, Chanda D, Zhang Y, et al. Glucose stimulates cholesterol 7alpha-hydroxylase gene transcription in human hepatocytes. J Lipid Res. 2010;51:832–842. doi: 10.1194/jlr.M002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, Owsley E, Matozel M, et al. Transgenic expression of cholesterol 7alpha-hydroxylase in the liver prevents high-fat diet-induced obesity and insulin resistance in mice. Hepatology. 2010;52:678–690. doi: 10.1002/hep.23721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrell JM, Boehme S, Li F, et al. Cholesterol 7{alpha}-hydroxylase-deficient mice are protected from high fat/high cholesterol diet-induced metabolic disorders. J Lipid Res. 2016;57:1144–1154. doi: 10.1194/jlr.M064709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong Q, Hu Z, Zhang F, et al. Fibroblast growth factor 21 improves hepatic insulin sensitivity by inhibiting mammalian target of rapamycin complex 1 in mice. Hepatology. 2016;64:425–438. doi: 10.1002/hep.28523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F, Jiang C, Krausz KW, et al. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Comun. 2013;4:2384. doi: 10.1038/ncomms3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang C, Xie C, Li F, et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J Clin Invest. 2015;125:386–402. doi: 10.1172/JCI76738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang C, Xie C, Lv Y, et al. Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction. Nat Commun. 2015;6:10166. doi: 10.1038/ncomms10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang S, Suh JM, Reilly SM, et al. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat Med. 2015;21:159–165. doi: 10.1038/nm.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller M, Thorell A, Claudel T, et al. Ursodeoxycholic acid exerts farnesoid X receptor-antagonistic effects on bile acid and lipid metabolism in morbid obesity. J Hepatology. 2015;62:1398–1404. doi: 10.1016/j.jhep.2014.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maruyama T, Miyamoto Y, Nakamura T, et al. Identification of membrane-type receptor for bile acids (M-BAR) Biochem Biophys Res Commun. 2002;298:714–719. doi: 10.1016/s0006-291x(02)02550-0. [DOI] [PubMed] [Google Scholar]
- 38.Kawamata Y, Fujii R, Hosoya M, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 39.Keitel V, Donner M, Winandy S, et al. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem Biophys Res Commun. 2008;372:78–84. doi: 10.1016/j.bbrc.2008.04.171. [DOI] [PubMed] [Google Scholar]
- 40.Keitel V, Reinehr R, Gatsios P, et al. The G-protein coupled bile salt receptor TGR5 is expressed in liver sinusoidal endothelial cells. Hepatology. 2007;45:695–704. doi: 10.1002/hep.21458. [DOI] [PubMed] [Google Scholar]
- 41.Katsuma S, Hirasawa A, Tsujimoto G. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem Biophys Res Commun. 2005;329:386–390. doi: 10.1016/j.bbrc.2005.01.139. [DOI] [PubMed] [Google Scholar]
- 42.Thomas C, Gioiello A, Noriega L, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mojsov S, Heinrich G, Wilson IB, et al. Preproglucagon gene expression in pancreas and intestine diversifies at the level of post-translational processing. J Biol Chem. 1986;261:11880–11889. [PubMed] [Google Scholar]
- 44.Ding X, Saxena NK, Lin S, et al. Exendin-4, a glucagon-like protein-1 (GLP-1) receptor agonist, reverses hepatic steatosis in ob/ob mice. Hepatology. 2006;43:173–181. doi: 10.1002/hep.21006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe M, Houten SM, Mataki C, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 46.Srivastava A. Progressive familial intrahepatic cholestasis. J Clin Exp Hepatol. 2014;4:25–36. doi: 10.1016/j.jceh.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zollner G, Trauner M. Mechanisms of cholestasis. ClinLiver Dis. 2008;12:1–26. vii. doi: 10.1016/j.cld.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 48.Zein CO, Lindor KD. Latest and emerging therapies for primary biliary cirrhosis and primary sclerosing cholangitis. Curr Gastroenterol Rep. 2010;12:13–22. doi: 10.1007/s11894-009-0079-2. [DOI] [PubMed] [Google Scholar]
- 49.Kaplan MM, Gershwin ME. Primary biliary cirrhosis. N Engl J Med. 2005;353:1261–1273. doi: 10.1056/NEJMra043898. [DOI] [PubMed] [Google Scholar]
- 50.Pols TW, Noriega LG, Nomura M, et al. The bile acid membrane receptor TGR5: a valuable metabolic target. Dig Dis. 2011;29:37–44. doi: 10.1159/000324126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adorini L, Pruzanski M, Shapiro D. Farnesoid X receptor targeting to treat nonalcoholic steatohepatitis. Drug Discov Today. 2012;17:988–997. doi: 10.1016/j.drudis.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 52.Schaap FG, Trauner M, Jansen PL. Bile acid receptors as targets for drug development. Nat Rev Gastroenterol Hepatol. 2014;11:55–67. doi: 10.1038/nrgastro.2013.151. [DOI] [PubMed] [Google Scholar]
- 53.Arab JP, Karpen SJ, Dawson PA, et al. Bile acids and nonalcoholic fatty liver disease: Molecular insights and therapeutic perspectives. Hepatology. 2016;65:350–362. doi: 10.1002/hep.28709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lioudaki E, Ganotakis ES, Mikhailidis DP. Lipid lowering drugs and gallstones: a therapeutic option? Curr Pharm Des. 2011;17:3622–3631. doi: 10.2174/138161211798220909. [DOI] [PubMed] [Google Scholar]
- 55.Dyson JK, Hirschfield GM, Adams DH, et al. Novel therapeutic targets in primary biliary cirrhosis. Nat Rev Gastroenterol Hepatol. 2015;12:147–158. doi: 10.1038/nrgastro.2015.12. [DOI] [PubMed] [Google Scholar]
- 56.Hohenester S, Wenniger LM, Paulusma CC, et al. A biliary HCO3− umbrella constitutes a protective mechanism against bile acid-induced injury in human cholangiocytes. Hepatology. 2012;55:173–183. doi: 10.1002/hep.24691. [DOI] [PubMed] [Google Scholar]
- 57.Prieto J, Garcia N, Martí-Climent JM, et al. Assessment of biliary bicarbonate secretion in humans by positron emission tomography. Gastroenterology. 1999;117:167–172. doi: 10.1016/s0016-5085(99)70564-0. [DOI] [PubMed] [Google Scholar]
- 58.Beuers U. Drug insight: Mechanisms and sites of action of ursodeoxycholic acid in cholestasis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:318–328. doi: 10.1038/ncpgasthep0521. [DOI] [PubMed] [Google Scholar]
- 59.Poupon R. Ursodeoxycholic acid and bile-acid mimetics as therapeutic agents for cholestatic liver diseases: an overview of their mechanisms of action. Clin Res Hepatol Gastroenterol. 2012;36(Suppl 1):S3–S12. doi: 10.1016/S2210-7401(12)70015-3. [DOI] [PubMed] [Google Scholar]
- 60.Parés A, Caballería L, Rodés J. Excellent long-term survival in patients with primary biliary cirrhosis and biochemical response to ursodeoxycholic Acid. Gastroenterology. 2006;130:715–720. doi: 10.1053/j.gastro.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 61.Yoon YB, Hagey LR, Hofmann AF, et al. Effect of side-chain shortening on the physiologic properties of bile acids: hepatic transport and effect on biliary secretion of 23-nor-ursodeoxycholate in rodents. Gastroenterology. 1986;90:837–852. doi: 10.1016/0016-5085(86)90859-0. [DOI] [PubMed] [Google Scholar]
- 62.Yeh HZ, Schteingart CD, Hagey LR, et al. Effect of side chain length on biotransformation, hepatic transport, and choleretic properties of chenodeoxycholyl homologues in the rodent: studies with dinorchenodeoxycholic acid, norchenodeoxycholic acid, and chenodeoxycholic acid. Hepatology. 1997;26:374–385. doi: 10.1002/hep.510260218. [DOI] [PubMed] [Google Scholar]
- 63.Halilbasic E, Fiorotto R, Fickert P, et al. Side chain structure determines unique physiologic and therapeutic properties of norursodeoxycholic acid in Mdr2−/− mice. Hepatology. 2009;49:1972–1981. doi: 10.1002/hep.22891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghonem NS, Assis DN, Boyer JL. Fibrates and cholestasis. Hepatology. 2015;62:635–643. doi: 10.1002/hep.27744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iwasaki S, Ohira H, Nishiguchi S, et al. The efficacy of ursodeoxycholic acid and bezafibrate combination therapy for primary biliary cirrhosis: A prospective, multicenter study. Hepatol Res. 2008;38:557–564. doi: 10.1111/j.1872-034X.2007.00305.x. [DOI] [PubMed] [Google Scholar]
- 66.Pellicciari R, Fiorucci S, Camaioni E, et al. 6alpha-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J Med Chem. 2002;45:3569–3572. doi: 10.1021/jm025529g. [DOI] [PubMed] [Google Scholar]
- 67.Pellicciari R, Costantino G, Camaioni E, et al. Bile acid derivatives as ligands of the farnesoid X receptor. Synthesis, evaluation, and structure-activity relationship of a series of body and side chain modified analogues of chenodeoxycholic acid. J Med Chem. 2004;47:4559–4569. doi: 10.1021/jm049904b. [DOI] [PubMed] [Google Scholar]
- 68.Ali AH, Carey EJ, Lindor KD. Recent advances in the development of farnesoid X receptor agonists. Ann Transl Med. 2015;3:5. doi: 10.3978/j.issn.2305-5839.2014.12.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fiorucci S, Clerici C, Antonelli E, et al. Protective effects of 6-ethyl chenodeoxycholic acid, a farnesoid X receptor (FXR) Ligand, In estrogen induced cholestasis. J Pharmacol Exp Ther. 2005;313:604–612. doi: 10.1124/jpet.104.079665. [DOI] [PubMed] [Google Scholar]
- 70.Hirschfield GM, Mason A, Luketic V, et al. Efficacy of obeticholic acid in patients with primary biliary cirrhosis and inadequate response to ursodeoxycholic acid. Gastroenterology. 2015;148:751–61. e8. doi: 10.1053/j.gastro.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 71.Nevens F, Andreone P, Mazzella G, et al. A placebo-controlled trial of obeticholic acid in primary biliary cholangitis. N Engl J Med. 2016;375:631–643. doi: 10.1056/NEJMoa1509840. [DOI] [PubMed] [Google Scholar]
- 72.Mudaliar S, Henry RR, Sanyal AJ, et al. Efficacy and safety of the farnesoid x receptor agonist obeticholic acid in patients with type 2 diabetes and nonalcoholic Fatty liver disease. Gastroenterology. 2013;145:574–82. e1. doi: 10.1053/j.gastro.2013.05.042. [DOI] [PubMed] [Google Scholar]
- 73.Neuschwander-Tetri BA, Loomba R, Sanyal AJ, et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet. 2015;385:956–965. doi: 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alemi F, Kwon E, Poole DP, et al. The TGR5 receptor mediates bile acid-induced itch and analgesia. J Clin Invest. 2013;123:1513–1530. doi: 10.1172/JCI64551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kumar DP, Asgharpour A, Mirshahi F, et al. Activation of transmembrane bile acid receptor TGR5 modulates pancreatic islet alpha cells to promote glucose homeostasis. J Biol Chem. 2016;291:6626–6640. doi: 10.1074/jbc.M115.699504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reich M, Deutschmann K, Sommerfeld A, et al. TGR5 is essential for bile acid-dependent cholangiocyte proliferation in vivo and in vitro. Gut. 2016;65:487–501. doi: 10.1136/gutjnl-2015-309458. [DOI] [PubMed] [Google Scholar]
- 77.Sato H, Macchiarulo A, Thomas C, et al. Novel potent and selective bile acid derivatives as TGR5 agonists: biological screening, structure-activity relationships, and molecular modeling studies. J Med Chem. 2008;51:1831–1841. doi: 10.1021/jm7015864. [DOI] [PubMed] [Google Scholar]
- 78.Tiwari A, Maiti P. TGR5: an emerging bile acid G-protein-coupled receptor target for the potential treatment of metabolic disorders. Drug Discov Today. 2009;14:523–530. doi: 10.1016/j.drudis.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 79.Cipriani S, Mencarelli A, Chini MG, et al. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PloS one. 2011;6:e25637. doi: 10.1371/journal.pone.0025637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pellicciari R, Sato H, Gioiello A, et al. Nongenomic actions of bile acids. Synthesis and preliminary characterization of 23- and 6,23-alkyl-substituted bile acid derivatives as selective modulators for the G-protein coupled receptor TGR5. J Med Chem. 2007;50:4265–4268. doi: 10.1021/jm070633p. [DOI] [PubMed] [Google Scholar]
- 81.Pols TW, Nomura M, Harach T, et al. TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab. 2011;14:747–757. doi: 10.1016/j.cmet.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kida T, Tsubosaka Y, Hori M, et al. Bile acid receptor TGR5 agonism induces NO production and reduces monocyte adhesion in vascular endothelial cells. Arterioscler Thromb Vasc Biol. 2013;33:1663–1669. doi: 10.1161/ATVBAHA.113.301565. [DOI] [PubMed] [Google Scholar]
- 83.Duan H, Ning M, Chen X, et al. Design, synthesis, and antidiabetic activity of 4-phenoxynicotinamide and 4-phenoxypyrimidine-5-carboxamide derivatives as potent and orally efficacious TGR5 agonists. J Med Chem. 2012;55:10475–10489. doi: 10.1021/jm301071h. [DOI] [PubMed] [Google Scholar]
- 84.Duan H, Ning M, Zou Q, et al. Discovery of intestinal targeted TGR5 agonists for the treatment of type 2 diabetes. J Med Chem. 2015;58:3315–3328. doi: 10.1021/jm500829b. [DOI] [PubMed] [Google Scholar]
- 85.Rizzo G, Passeri D, De Franco F, et al. Functional characterization of the semisynthetic bile acid derivative INT-767, a dual farnesoid X receptor and TGR5 agonist. Mol Pharmacol. 2010;78:617–630. doi: 10.1124/mol.110.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.D’Amore C, Di Leva FS, Sepe V, et al. Design, synthesis, and biological evaluation of potent dual agonists of nuclear and membrane bile acid receptors. J Med Chem. 2014;57:937–954. doi: 10.1021/jm401873d. [DOI] [PubMed] [Google Scholar]
- 87.Baghdasaryan A, Claudel T, Gumhold J, et al. Dual farnesoid X receptor/TGR5 agonist INT-767 reduces liver injury in the Mdr2−/− (Abcb4−/−) mouse cholangiopathy model by promoting biliary HCO output. Hepatology. 2011;54:1303–1312. doi: 10.1002/hep.24537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McMahan RH, Wang XX, Cheng LL, et al. Bile acid receptor activation modulates hepatic monocyte activity and improves nonalcoholic fatty liver disease. J Biol Chem. 2013;288:11761–11770. doi: 10.1074/jbc.M112.446575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ryan KK, Tremaroli V, Clemmensen C, et al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature. 2014;509:183–188. doi: 10.1038/nature13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McGavigan AK, Garibay D, Henseler ZM, et al. TGR5 contributes to glucoregulatory improvements after vertical sleeve gastrectomy in mice. Gut. 2017;66:226–234. doi: 10.1136/gutjnl-2015-309871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Patti ME, Houten SM, Bianco AC, et al. Serum bile acids are higher in humans with prior gastric bypass: potential contribution to improved glucose and lipid metabolism. Obesity (Silver Spring) 2009;17:1671–1677. doi: 10.1038/oby.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Simonen M, Dali-Youcef N, Kaminska D, et al. Conjugated bile acids associate with altered rates of glucose and lipid oxidation after Roux-en-Y gastric bypass. Obes Surg. 2012;22:1473–1480. doi: 10.1007/s11695-012-0673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]