Abstract
Lymphangioleiomyomatosis (LAM) is a rare, low-grade, metastasizing neoplasm that arises from an unknown source, spreads via the lymphatics, and targets the lungs. All pulmonary structures become infiltrated with benign-appearing spindle and epithelioid cells (LAM cells) that express smooth-muscle and melanocyte-lineage markers, harbor mTOR-activating mutations in tuberous sclerosis complex (TSC) genes, and recruit abundant stromal cells. Elaboration of lymphangiogenic growth factors and matrix remodeling enzymes by LAM cells enables their access to lymphatic channels and likely drives the cystic lung remodeling that often culminates in respiratory failure. Dysregulated cellular signaling results in a shift from oxidative phosphorylation to glycolysis as the preferred mode of energy generation, to allow for the accumulation of biomass required for cell growth and tolerance of nutrient-poor, anaerobic environments. Symptomatic LAM occurs almost exclusively in females after menarche, highlighting the central but as yet poorly understood role for sex-restricted anatomical structures and/or hormones in disease pathogenesis. LAM is an elegant model of malignancy because biallelic mutations at a single genetic locus confer all features that define cancer upon the LAM cell—metabolic reprogramming and proliferative signals that drive uncontrolled growth and inappropriate migration and invasion, the capacity to exploit the lymphatic circulation as a vehicle for metastasis and access to the lungs, and destruction of remote tissues. The direct benefit of the study of this rare disease has been the rapid identification of an effective FDA-approved therapy, and the collateral benefits have included elucidation of the pivotal roles of mTOR signaling in the regulation of cellular metabolism and the pathogenesis of cancer.
Keywords: lymphangiomyomatosis, tuberous sclerosis, perivascular epithelioid cell tumor (PECOMA), tumor suppressor syndrome
OVERVIEW
Lymphangioleiomyomatosis (LAM) is a rare cystic lung disease affecting young and middle-aged women that typically presents with insidiously progressive dyspnea on exertion. Pulmonary function testing often reveals an obstructive defect, but because the initial chest radiograph is usually unremarkable, the patient is most frequently assumed to have asthma or chronic obstructive lung disease and treated with inhaler therapy. The correct diagnosis may be delayed for three to five years from onset of symptoms, when computed tomography (CT) of the chest obtained for progressive dyspnea, pneumothorax, pleural effusion, or unrelated reasons (such as to rule out pulmonary embolism) reveals distinctive cystic changes in the pulmonary parenchyma. Even if the radiologist and primary care physician are unfamiliar with LAM, this unique CT pattern or the discovery of chyle on pleural tap often triggers referral to a pulmonary specialist and leads to a diagnosis. In addition to LAM, the other rare cystic lung diseases that are typically considered in the differential include Birt-Hogg-Dubé syndrome, Langerhans cell histiocytosis, light chain deposition disease, and Sjögren syndrome. The definitive diagnosis is most often made on clinical and serological grounds using defined criteria (1–4), but in approximately 20–30% of cases a biopsy may be necessary. Histological analysis of lung sections reveals atypical smooth muscle–like cells infiltrating all lung structures, including blood vessels, conducting airways, alveolar septa, lymphatics, and pleura (5). Both micronodular and cystic lesions are apparent at low power, and although the spindle and epithelioid morphology of the LAM cells on hematoxylin and eosin staining is usually sufficiently characteristic to make a diagnosis without further testing, the diagnosis can be confirmed by staining with an antibody to gp100 protein called HMB-45. Median survival in LAM likely exceeds 15–20 years, but most patients suffer from progressive respiratory failure and are dyspneic with walking on flat ground within a decade of the onset of symptoms. Recurrent pneumothoraces develop in about two-thirds of patients and chylous complications in about one-third.
Until recently, the treatment options available to the LAM patient included watchful waiting, unproven hormonal therapies, and, in advanced cases, lung transplantation. However, remarkable progress in our understanding of LAM—driven by patient advocacy, abundant clues from nature [restriction to females, recurrence in transplanted lungs, occurrence in patients with tuberous sclerosis complex (TSC), etc.], and intense scientific interest in the pathway—has yielded an effective therapy in under 20 years since efforts began in earnest. In the process, study of this rare monogenic neoplasm and the parent disease, TSC, has yielded astonishing insights into cellular metabolism that are informing our approach to more common cancers.
THE GENETIC BASIS OF LYMPHANGIOLEIOMYOMATOSIS
Prior to the 1990s, few physicians considered LAM to be a genetic disorder. The disease was classified among the fibrotic, interstitial lung diseases, and familial transmission was not apparent in patients presenting for evaluation in pulmonary clinics. However, LAM was reported in what was thought to be a few percent of patients with TSC, an autosomal dominant disorder that typically presents in childhood with benign tumors (brain, kidney, and skin), cognitive defects, and epilepsy. These reports of TSC-LAM patients led to genetic studies in patients with the sporadic form of the disease (S-LAM), which occurs in patients who do not have TSC (6, 7). The two genes associated with TSC, TSC1 and TSC2, were discovered by linkage analysis of large TSC families conducted by multinational consortia in 1997 and 1993, respectively. Before the functions of the genes were known, somatic mutations in TSC2 were found in LAM lesions in the lungs, kidneys, and lymph nodes but not in circulating leukocytes or normal tissues of S-LAM patients. The finding of identical TSC2 mutations in neoplastic cells within the lung and kidney suggested a common origin for the tumors in those organs, and a metastatic theory for LAM was validated by genetic confirmation of recipient origins of recurrent LAM lesions within the donor allografts of LAM patients who had undergone lung transplantation. The source of LAM cells that populate the lung remains obscure to this day, with favored candidates being the uterus (8), angiomyolipomas (9), the lymphatic system, and the bone marrow. The prevalence of axial lymph node involvement is highest in the lower abdomen and diminishes in a gradient fashion toward the chest, suggesting an origin in the pelvis (10).
LAM develops through the “two-hit” mechanism that was described by Knudson (11) as common to the tumor suppressor syndromes. In patients with TSC-LAM, a mutation in a TSC gene (either TSC1 or TSC2) is present in the germline, and a second mutation in the other allele of the same locus occurs in a somatic tissue, resulting in loss of heterozygosity for the normal allele (6, 7) and neoplastic transformation of cells that ultimately metastasize to the lung. S-LAM has a similar genetic pathogenesis, except that germline mutations are not detected, only TSC2 mutations have been reported, and both “hits” are thought to occur in somatic tissues post conception (12). Although not frequently considered, it is also possible that the first “hit” in S-LAM reflects very low-level germline or somatic mosaicism, in which only a small fraction of cells harbor the mutation at levels that have been undetectable with the sequencing techniques applied.
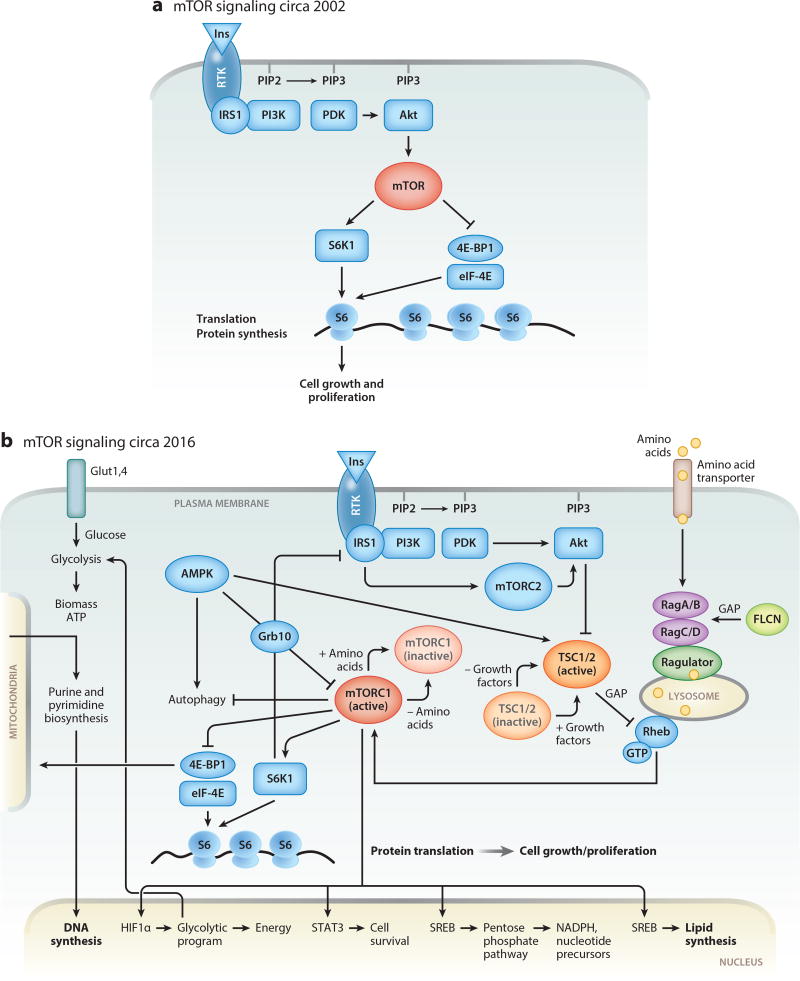
The TSC1 locus on chr 9q34 encodes hamartin, a 130-kDa protein with 1,163 amino acids and no obvious informative homologies in the National Center for Biotechnology Information database when first cloned (13). The TSC2 locus on chr 16p13.3 encodes tuberin, a 200-kDa protein with 1,804 amino acids and a C-terminal GTPase activating protein (GAP) domain (14). The function of tuberin as a regulator of cell size, cell cycle, and cell growth was revealed by several Drosophila laboratories about 15 years ago (15–19). In a series of epistatic experiments, tuberin was positioned within the PI3K (phosphatidylinositol-4,5-bisphosphate 3 kinase) pathway downstream of Akt and upstream of Rheb, mTOR (mechanistic target of rapamycin), and S6 kinase (Figure 1a). Hamartin and tuberin were shown to form a complex, with hamartin functioning as a stabilizing scaffold, to suppress the activity of Rheb through the GAP domain of tuberin. Mutations in TSC1 or TSC2 result in ubiquitination and degradation of the complex, which in turn lead to derepression of Rheb and constitutive activation of downstream effectors such as S6 and 4EBP1 that drive protein translation and cell growth (Figure 1b).
Figure 1.
(a) Schematic representation of the mTOR signaling pathway before discovery of TSC function. (b) Simplified schematic representation of our current understanding of the mTOR signaling pathway. In normal cells, growth factors or insulin (Ins) activate their cognate receptor tyrosine kinases (RTKs), leading to autophosphorylation. Activated RTKs recruit PI3K, a key enzyme that transduces extracellular signaling into cellular responses by activating serine-threonine kinase Akt. Akt phosphorylates TSC (a complex of two proteins, TSC1 and TSC2), releasing the small GTPase Rheb from TSC suppression, which results in activation of mTORC1. In lymphangioleiomyomatosis (LAM), the TSC1/2 complex is mutationally inactivated and does not suppress Rheb. Activated Rheb and amino acids, through the complex composed of Ragulator, RagA/B, and RagC/D, recruit mTORC1 to the lysosomal surface. Constitutively activated mTORC1 induces metabolic reprogramming by (1) upregulating transcription of HIF1α, thereby modulating the expression of glycolytic enzymes, SREBP transcription factors mediating synthesis of fatty acids and cholesterol, and enzymes of the pentose phosphate pathway (PPP); (2) increasing translation of ribosomal and mitochondrial proteins, thereby inducing ribosomal and mitochondrial biogenesis; and (3) inducing de novo synthesis of proteins, lipids, and nucleotides (purines and pyrimidines). STAT3 is required for LAM cell survival, consistent with a role for the mTORC1-STAT3 axis in regulation of prosurvival genes in TSC1/2-deficient cells.
EVADING GROWTH SUPPRESSION
The TSC tumor suppressor complex is an upstream negative regulator of mTOR (20). Together, TSC1 and TSC2 constitute a critical node that integrates growth factor, nutrient, energy, and stress signaling (21, 22). mTOR forms the catalytic core of two functionally distinct complexes, mTORC1 and mTORC2, which are distinguished by their differential sensitivity to the bacterial macrolide rapamycin (23). mTORC1 is exquisitely sensitive to rapamycin and controls protein translation, lipid metabolism, glycolysis, glucose uptake, and purine and pyrimidine synthesis. In contrast, mTORC2 is relatively insensitive to rapamycin, is activated by insulin, and regulates key aspects of cell proliferation, including assembly of the actin cytoskeleton and cellular survival (24–26). Rapamycin inhibits mTORC1 by binding to FKBP12 (FK506-binding protein of 12 kDa) and bridging the interaction of the peptide–macrolide complex with the FKBP12-binding domain of mTOR (27).
A fundamental hallmark of cancer cells is sustained proliferative capacity (28). Growth of normal cells is regulated by extracellular signals that activate or repress intracellular signaling cascades such as the PI3K–mTOR axis (29). Gain-of-function or loss-of-function mutations in the genes encoding signaling proteins within these intracellular pathways can result in constitutive signals for growth. The cellular function of the tumor suppressor complex TSC1/2 is to regulate cell growth via the mTOR signaling axis. In LAM, mutations of TSC genes uncouple the mTOR signaling cascade from obligate regulation by PI3K-dependent growth-promoting factors, release the cell from context-specific growth regulation, and facilitate uncontrolled growth.
The Lysosome Is the Signaling Hub for Growth Regulation by Amino Acids and Growth Factors
One of the landmark discoveries that resulted from the study of TSC1/2-deficient cells was the exquisite spatial regulation of cellular signaling and metabolism that occurs on the surface of the lysosome. The TSC1/2 tumor suppressor complex regulates mTOR by directly controlling the activity of the GAP domain of the small GTPase, Rheb (30). Under normal conditions in nonproliferating cells, TSC1/2 has strong affinity for Rheb in its GDP-bound inactive form on the lysosomal surface (31), maintaining Rheb in the quiescent, or “off,” state. In the presence of growth factors and nutrients, the GAP activity of the TSC1/2 complex is turned off by Akt-mediated phosphorylation, allowing the GTP-bound active form of Rheb to accumulate and activate mTORC1. Mutations of TSC1/2 proteins that prevent association of the TSC complex with the lysosome impedeTSC1/2-dependent hydrolysis of GTP into GDP by the Rheb GTPase, thereby facilitating constitutive signals for cell growth (32–34).
In normal cells, maximal activation of mTORC1 requires dual input from growth factors and amino acids (24) (Figure 1). In LAM, uncoupling of mTOR from regulation by growth factors and the TSC1/2 complex facilitates unilateral amino acid–mediated activation of mTOR by another class of small GTPases (in addition to Rheb), the Rag GTPases (35). Amino acids activate multi-protein complexes on lysosomes, which contain Rag GTPases and recruit mTORC1 to the membrane, in position to be phosphorylated by Rheb (24, 35). Amino acid–induced mTOR activation is also regulated by the tumor suppressor protein folliculin (FLCN), which modulates Rag GTPase activity (36). Interestingly, FLCN mutations cause the Birt-Hogg-Dubé syndrome, a rare disease that, like LAM, results in formation of lung cysts as well as kidney tumors (37, 38). Signaling pathway intersections between ultrarare diseases that share unusual disease manifestations provide unique insights into disease pathogenesis and mechanisms of organ development, neoplasia, and lung remodeling that are difficult to obtain in any other way.
Feedback Loops Restrain Neoplastic Growth in TSC1/2-Deficient Cells
The study of TSC-deficient cell models has revealed multiple intracellular and extracellular feedback loops that modulate the anabolic effects of activated mTOR. When complexed with Raptor, Rheb regulates mTOR-mediated phosphorylation of p70 S6 kinase (S6K1) and 4E-BP1 (22). In turn, S6K1 activation suppresses PI3K signaling through an intracellular negative feedback loop in which pS6K1 phosphorylates insulin receptor substrate 1 (IRS1) and targets it for degradation through ubiquitin-mediated mechanisms, extinguishing its PI3K-activating actions (39, 40). Grb10 is a cytoplasmic protein that suppresses signaling by insulin in a manner that complements the S6K1–IRS1 feedback loop (41–43). Phosphorylation of Grb10 by activated mTORC1 potentiates its ability to suppress growth-promoting signals from insulin (41–43). TSC1/2-deficient cells also secrete modulators of mTOR actions. A recent study employing a quantitative proteomics approach revealed that an IGF-1-binding protein, IGFBP5, acts as an extracellular negative regulator of mTORC signaling by inhibiting growth induced by IGF-1-mediated pathways (44). It is possible that IGFBP5 secreted by TSC1/2-deficient cells may restrain growth of adjacent cells, thus providing a competitive advantage in nutrient availability for TSC-deficient tumor growth (44). The presence of multiple feedback mechanisms that restrain the cellular actions of activated mTOR may explain the rarity of malignant transformation in TSC and LAM (45) and has important implications for the development of therapeutic strategies for LAM and other neoplasms.
SUSTAINING PROLIFERATION BY METABOLIC REPROGRAMMING
mTOR Activation Alone Is Sufficient to Activate Warburg Metabolism
Continuous LAM cell proliferation driven by mTOR activation requires major adjustments in energy metabolism (46). When oxygen is available, normal cells use glucose to produce ATP in two ways: via aerobic glycolysis in the cytosol, forming pyruvate and two molecules of ATP, and via the NADP-driven enzymatic process of oxidative phosphorylation in the mitochondria, which produces 36 molecules of ATP and carbon dioxide. In cancer, energy production is predominantly limited to aerobic glycolysis through a metabolic shift called the Warburg effect. This cellular choice seems counterintuitive, given the relatively poor efficiency of ATP generation by glycolysis alone (47). The advantage it confers to cancer cells, however, is the diversion of glycolytic intermediates into key biosynthetic and metabolic pathways to generate macromolecules that constitute the substrate for assembly of new cells and organelles (47). The transcription factor hypoxia-inducible factor 1α (HIF1α) (48–51), a key regulator of tumor initiation, is upregulated by low oxygen levels and serves to prevent damaging effects of hypoxia on cell function by promoting glycolysis, glucose transport, and angiogenesis (52, 53). HIF1α upregulates glycolytic enzymes to maintain cellular ATP levels in “glucose-addicted” cancer cells (46) and induces vascular endothelial growth factor (VEGF) expression, resulting in the neovasculogenesis required for adequate nutrient delivery to sustain growth. In LAM, even under aerobic conditions, inappropriately activated mTORC1 increases HIF1α gene transcription and translation (51). Thus, the study of LAM and TSC cell models has revealed that this exquisitely simple monogenic neoplasm exhibits the classic Warburg metabolism that is a signature of neoplastic growth, and that mTOR activation is sufficient to activate the glycolytic pathway.
mTOR Links mRNA Translation to Lipid, Protein, and Ribonucleotide Metabolism
mTORC1 regulates protein synthesis predominantly by regulating the transcription of genes encoding rRNAs and tRNAs (54). Activated mTOR stimulates RNA translation by regulating the assembly of the eIF4F complex and by phosphorylating downstream targets, including 4E-BPs (eukaryotic-translation-initiation-factor-binding proteins) and S6K1. Importantly, mTORC1 controls mitochondrial biogenesis and activity by selectively promoting translation of mitochondria-related mRNAs through 4E-BPs (55). In LAM, mTORC1 induces constitutive activation of S6K1 (56) and phosphorylation of 4E-BP1 (57), driving the accumulation of cellular biomass and the heightened cellular metabolism required to support it. Neoplastic cells also support their anabolic activities by biosynthesis of purines and pyrimidines, which are building blocks not only for ATP but also for RNA and DNA synthesis (58). The study of TSC-deficient cells has recently revealed that mTORC1 is a master regulator of purine (59) and pyrimidine (60) biosynthesis (58, 61). Purine synthesis takes place on specific enzyme complexes named purinosomes (61), which colocalize with mitochondria and require mTORC1 activation (58). Activated mTORC1 drives the expression of genes regulating the mitochondrial tetrahydrofolate cycle to enhance production of purine nucleotides (59), thus mobilizing the mitochondrial machinery to translate growth signals into cell proliferation.
In addition to polynucleotide and protein synthesis, actively growing cells require lipids for biosynthesis of newly formed membranes. In TSC1/2-null cells, activated mTORC1 induces expression of many lipogenic genes by regulating their transcription and protein levels through activation of the sterol-regulatory-element-binding proteins (SREBPs) (51, 62), transcription factors that mediate fatty acid synthesis and cholesterol biosynthesis (51).
In summary, the study of TSC1/2-deficient cell models led to basic discoveries of key molecular mechanisms of amino acid, lipid metabolism, purine, and pyrimidine signaling. Loss of TSC tumor suppressor function uncouples mTOR from regulation by growth factors (24, 29, 35, 56) and induces metabolic reprogramming to sustain continuous LAM cell growth and proliferation by influencing diverse pathways that control cell biomass, ranging from glycolysis and mitochondrial metabolism to lipid biosynthesis (24, 35, 58).
RESISTING CELL DEATH
With changes in nutrient availability, cells modulate their metabolic state to sustain their viability. Under growth factor–controlled conditions, healthy cells maintain homeostasis by regulating the uptake of sufficient glucose and amino acid nutrients to sustain context-appropriate ATP production and macromolecular synthesis. In states of nutrient or energy depletion, mammalian cells deploy a cellular program of bulk degradation called autophagy, in which cytoplasmic macromolecules and organelles are autodigested to sustain survival. Decreases in cellular ATP activate AMP-dependent protein kinase (AMPK), which induces autophagy by phosphorylating TSC1/2 and inhibiting mTORC1 signaling (24, 63, 64). Autophagy is only a temporizing measure, however, and under extreme conditions of prolonged nutrient deprivation, programmed cell death by apoptosis ensues.
In LAM, loss of TSC1/2 disengages mTORC1 from upstream inputs, resulting in insensitivity to the prosurvival signaling mechanisms that induce formation of autophagosomes (i.e., double-membrane-bordered cellular compartments created during the process of autophagy) in response to energy stress (65). Under in vitro conditions, glucose depletion promotes TSC1/2-deficient cell apoptosis because hyperactive mTORC1 also induces endoplasmic reticulum (ER) stress, activates the unfolded protein response, and promotes accumulation of the tumor suppressor p53 (66). Importantly, pharmacological targeting of mTORC1 with rapamycin modulates the metabolic state of TSC-null cells, leading to increased survival, which has implications for balancing the therapeutic objectives of reducing tumor burden and dampening destructive cellular actions (such as cystic lung remodeling in LAM).
TSC-deficient cells can become resistant to apoptosis by deregulating metabolic and catabolic processes. They activate macroautophagy and nutrient- and stress-responsive transcription factors such as GNL3, which binds to p53 to regulate the cell cycle and is required for cellular access to secondary nitrogen sources (24, 67, 68). Loss of TSC1/2 in LAM strongly activates signal transducer and activator of transcription 3 (STAT3) (69, 70), a well established regulator of genes promoting tumorigenesis and cancer cell survival (62). Activation of Rho GTPase via mTORC2 also facilitates TSC2-deficient cell survival by downregulation of antiapoptotic Bcl2 and upregulation of proapoptotic Bim, Bok, and Puma (26). Rapamycin-induced inhibition of mTORC1 signaling is context specific and can have opposing effects on cell proliferation depending on nutrient availability (71). In vascularized regions of solid tumors, rapamycin inhibits mTORC1 signaling and cell proliferation. However, in the inner regions of nutrient-deprived tumors, mTOR inhibition may increase the use of extracellular proteins as an alternative source of nutrients and enhance growth (71). In states of nutrient starvation (or inhibition of mTOR with rapamycin), TSC-deficient cells also become dependent on glutamate metabolism (72) and the pentose phosphate pathway for survival (73–75).
The rapalogs sirolimus and everolimus have been shown in randomized clinical trials of TSC and LAM (4, 5, 76, 77) to stabilize pulmonary function decline (76), induce regression of astrocytomas (77), and reduce the size of angiomyolipomas (78), leading to FDA approvals for each of these indications. However, upon cessation of mTOR inhibitor therapy, lung function decline resumes and both subependymal giant cell astrocytomas (77) and angiomyolipomas regrow, often approaching their pretreatment volumes (76, 78). These clinical observations highlight the importance of better understanding the compensatory pathways that support TSC and LAM cell survival, since the therapeutic strategy of inhibiting mTORC1 signaling alone is not sufficient to cause cell death or induce remission in these diseases.
ACTIVATING INVASION AND METASTASIS
DNA mutations in tumor cells equip them with metastatic capacity by upregulating extracellular proteases, downregulating adhesion molecules, and activating small Rho GTPases that drive actin cytoskeleton remodeling and cellular motility.
In culture, TSC2-deficient cells demonstrate loss of contact inhibition and ability to proliferate in a nonadherent state (79). Nonadherent TSC2-null cells exhibit deregulation of β-catenin, suggesting potential involvement of Wnt signaling, and expression of MMP-7, which is known to facilitate cell invasiveness (79). Upregulation of MCP-1 (80, 81), which recruits monocytes, memory T cells, and dendritic cells and has been implicated in tumor metastasis, may also play a role in LAM cell invasion and stromal conditioning.
TSC1 regulates the activity of Rho GTPase and associates with the cytoskeletal proteins ezrin, radixin, and moesin, which serve as molecular bridges between the plasma membrane and cortical actin filaments (82). The TSC1-binding domain located in the C-terminus of TSC2 (83–85) overlaps with the Rho-activating domain of TSC1 (82), suggesting that the TSC1/2 complex is involved in the regulation of Rho GTPase activity, actin remodeling, and cell migration. TSC2 modulates actin cytoskeletal rearrangements (86) by blocking TSC1-mediated inhibition of Rac1, leading to its activation and inhibition of Rho, which together promote stress fiber disassembly and focal adhesion remodeling (86). Loss of function of either TSC1 or TSC2 due to inactivating mutations promotes the degradation of the TSC1–TSC2 complex and deregulation of Rac1 and Rho activities. The abnormal cell motility that results is central to disease pathogenesis in LAM. Consistent with this notion, primary cultures of explanted LAM cells exhibit increased invasiveness, rates of migration, and Rho activation, all of which are rescued by overexpression of TSC2 (87). Interestingly, TSC1 and TSC2 differentially regulate actin stress fiber formation and cell migration, and only TSC2 loss promotes an mTORC2-dependent promigratory cell phenotype (88).
The neoplastic cells that infiltrate the lung in LAM arise from an unknown source. They migrate through blood and lymphatic channels and form nodular and cystic lesions in the lung interstitium, enveloping and invading all lung structures, including lymphatics, airways, and blood vessels. LAM lesions are composed of haphazardly arranged epithelioid and spindle-shaped smooth muscle cells that stain intensely with antibodies against smooth muscle actin and more variably for estrogen receptors, progesterone receptors, gp100 (HMB-45), and other melanocytic proteins (89). Lymphangiogenic growth factors VEGF-C and VEGF-D are also expressed in the LAM lesion (90), most likely through mTOR-driven HIF1α pathway activation (48, 49). VEGF-D is elevated in the serum of most LAM patients and has proven to be a useful clinical and predictive biomarker (2, 3, 91). VEGF-C and -D are ligands for VEGF receptor 3 (VEGFR-3), which exhibits a highly restricted expression pattern in lymphatic endothelial cells and fenestrated blood vessels of endocrine organs such as the pancreas, thyroid, and adrenal glands (92–94). Other lymphatic markers that are present in the LAM lesion are podoplanin and LYVE-1 (90). Cleft-like spaces that are lined with VEGFR-3-expressing lymphatic endothelial cells are often found within both pulmonary and extrapulmonary LAM lesions (90). The “frustrated lymphangiogenesis” theory of lung destruction in LAM holds that VEGF-D-expressing LAM cells that invade the pulmonary parenchyma induce a matrix-modifying lymphangiogenic program that is “confusing” to the mature organ and culminates in cyst formation (5).
EVADING IMMUNE DETECTION
Clusters of LAM cells are found in chylous pleural fluid of patients with LAM and in the lumen of the thoracic duct (10). They are composed of alpha smooth muscle actin and VEGF-D-expressing spindle cells enveloped by a single layer of endothelial cells (90). In this Trojan horse–like manner, mutant LAM cells may cloak themselves with nonmutant endothelial cells and evade immune detection as they course through lymphatic channels to ultimately become imbedded in the lung microvasculature, in position to infiltrate the lung interstitium.
The capacity of epithelial cancer cells to metastasize and invade is primarily attributed to activating mutations of multiple oncogenes (95). It is remarkable that mutational inactivation of a single tumor suppressor gene in LAM confers all of the features that define cancer, including uncontrolled proliferation, metastatic dissemination, and destruction of remote tissues. Thus, LAM may serve as a elegant model to dissect the minimum cellular elements required for destructive cancer behaviors.
ROLE OF SEX STEROIDS IN LYMPHANGIOLEIOMYOMATOSIS PATHOGENESIS
Several lines of clinical evidence suggest that LAM development and progression are influenced by female hormones: Nearly all reported symptomatic cases are in women, LAM is exacerbated by pregnancy and supplemental estrogen therapy, and LAM is most rapidly progressive in premenopausal women (96). Unfortunately, the small retrospective case series of antihormonal therapy in the literature have yielded conflicting results, and the only randomized trial of an antiestrogen therapy (i.e., letrozole in postmenopausal women) in LAM to date underenrolled and was not adequately powered to address questions of efficacy (NCT01353209).
In vitro studies also support a role for estrogen in LAM progression. LAM cells isolated from the lungs of LAM patients express high levels of estrogen receptor alpha (ERα) (96, 97), which is known to be a potent mediator of estrogen-induced proliferation in cancers of the breast and uterus. Estradiol treatment of isolated human LAM cells promotes proliferation through nuclear and extranuclear actions of ERα, as well as activation of Erk1 (98, 99). In fact, mitogen-activated protein kinase kinase (MEK) inhibition prevents estradiol-induced proliferation, suggesting that extranuclear (nongenomic) estradiol actions are required for estradiol-mediated proliferation of LAM cells in culture (97). Similarly, estradiol promotes proliferation and blocks apoptosis in cultured TSC2-null ELT3 leiomyoma cells (100), which are derived from uterine leiomyomas of the Eker rat with spontaneous loss of TSC2 in one allele. Estrogen-promoted metastasis of ELT3 cells that have been subcutaneously injected into nude mice (100) can be blocked using inhibitors of MEK. Inactivation of the Tsc2 gene in the mouse uterus induces growth of metastasizing myometrial tumors (101), further suggesting that LAM tumors might originate from the uterine myometrium. Taken together, these data suggest that estrogen promotes LAM progression and support the need to conduct trials of antiestrogen therapy in the future (4; C Lu, HS Lee, G Pappas, et al., manuscript in preparation).
Like ERα, progesterone receptors are expressed in human LAM cells and in ELT3 cells derived from Eker rats (102). It is not clear, however, that progesterone significantly alters proliferation of LAM and leiomyoma cells in vitro. In fact, in the clinic, progesterone is considered to be antiproliferative for uterine leiomyomas, which has led to the hypothesis that progesterone might actually be useful in preventing progression of disease in LAM patients. Indeed, progesterone had been used as a treatment for LAM for nearly two decades following a sensational case report that suggested dramatic benefit. However, a careful retrospective analysis of LAM patients treated with progestins suggested no benefit (103), and there have been no prospective trials. Thus, exogenous progestin therapy is currently not recommended for patients with LAM (4; C Lu, HS Lee, G Pappas, et al., manuscript in preparation).
CONCLUSIONS
From the study of LAM and TSC, we have learned that mTOR is a master regulator that integrates cellular inputs of nutrient availability. We have learned that oxygen and energy status link LAM and TSC to outputs of protein translation; generation of macromolecular precursors, such as purines and pyrimidines; processes of cell metabolism, such as glycolysis and the accumulation of cellular biomass and macromolecules (including lipids, proteins, and polynucleotides); and cell survival, migration, and invasiveness. We have discovered that mTOR exerts these actions from its perch on the lysosomal surface, where it is ideally positioned to sample the status of nutrient availability, and through effects on transcription in the nucleus and mitochondrion. The translation of scientific discoveries in mTOR signaling from genetic dissection to astute observations in the eyes of flies to effective therapies for TSC and LAM occurred with astounding speed and ranks among the triumphs of twenty-first-century medicine. Perhaps even more remarkable, that it was driven by patient advocacy is a testament to the awesome power of that motivating force (76, 104).
Acknowledgments
The authors thank Dr. Kun-Lian Guan at the University of California, San Diego, for helpful discussion of the manuscript, as well as Sakshum Chadha for help with the figure. Citations were limited in number, and the authors apologize in advance for omissions of relevant articles.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Johnson SR, Cordier JF, Lazor R, et al. European Respiratory Society guidelines for the diagnosis and management of lymphangioleiomyomatosis. Eur. Respir. J. 2010;35:14–26. doi: 10.1183/09031936.00076209. [DOI] [PubMed] [Google Scholar]
- 2.Young LR, Lee HS, Inoue Y, et al. Serum VEGF-D concentration as a biomarker of lymphangioleiomyomatosis severity and treatment response: a prospective analysis of the Multicenter International Lymphangioleiomyomatosis Efficacy of Sirolimus (MILES) trial. Lancet. 2013;1:445–52. doi: 10.1016/S2213-2600(13)70090-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young LR, VanDyke R, Gulleman PM, et al. Serum vascular endothelial growth factor-D prospectively distinguishes lymphangioleiomyomatosis from other diseases. Chest. 2010;138:674–81. doi: 10.1378/chest.10-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCormack FX, Gupta N, Finlay GR, et al. Official American Thoracic Society/Japanese Respiratory Society clinical practice guidelines: lymphangioleiomyomatosis diagnosis and management. Am. J. Respir. Crit. Care Med. 2016;194:748–61. doi: 10.1164/rccm.201607-1384ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henske EP, McCormack FX. Lymphangioleiomyomatosis—a wolf in sheep’s clothing. J. Clin. Investig. 2012;122:3807–16. doi: 10.1172/JCI58709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smolarek TA, Wessner LL, McCormack FX, et al. Evidence that lymphangiomyomatosis is caused by TSC2 mutations: chromosome 16p13 loss of heterozygosity in angiolipomas and lymph nodes from women with lymphangiomyomatosis. Am. J. Hum. Genet. 1998;62:810–15. doi: 10.1086/301804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carsillo T, Astrinidis A, Henske EP. Mutations in the tuberous sclerosis complex gene TSC2 are a cause of sporadic pulmonary lymphangioleiomyomatosis. PNAS. 2000;97:6085–90. doi: 10.1073/pnas.97.11.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammes SR, Krymskaya VP. Targeted approaches toward understanding and treating pulmonary lymphangioleiomyomatosis (LAM) Horm. Cancer. 2013;4:70–77. doi: 10.1007/s12672-012-0128-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J, Astrinidis A, Henske EP. Chromosome 16 loss of heterozygosity in tuberous sclerosis and sporadic lymphangiomyomatosis. Am. J. Respir. Crit. Care Med. 2001;164:1537–40. doi: 10.1164/ajrccm.164.8.2104095. [DOI] [PubMed] [Google Scholar]
- 10.Seyama K, Kumasaka T, Kurihara M, et al. Lymphangioleiomyomatosis: a disease involving the lymphatic system. Lymphatic Res. Biol. 2010;8:21–31. doi: 10.1089/lrb.2009.0018. [DOI] [PubMed] [Google Scholar]
- 11.Knudson AG. Two genetic hits (more or less) to cancer. Nat. Rev. Cancer. 2001;1:157–62. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- 12.Juvet SC, McCormack FX, Kwiatkowski DJ, Downey GP. Molecular pathogenesis of lymphangioleiomyomatosis: lessons learned from orphans. Am. J. Respir. Cell Mol. Biol. 2006;36:398–408. doi: 10.1165/rcmb.2006-0372TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Slegtenhorst M, Hoogt RD, Hermans C, et al. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277:805–8. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- 14.European Chromosome 16 Tuberous Sclerosis Consortium. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell. 1993;75:1305–15. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- 15.Ito M, Rubin GM. gigas, a Drosophila homolog of tuberous sclerosis complex gene product-2, regulates the cell cycle. Cell. 1999;96:529–39. doi: 10.1016/s0092-8674(00)80657-1. [DOI] [PubMed] [Google Scholar]
- 16.Gao X, Pan D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 2001;15:1383–92. doi: 10.1101/gad.901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montagne J, Radimerski T, Thomas G. Insulin signaling: lessons from the Drosophila tuberous sclerosis complex, a tumor suppressor. Science’s STKE. 2001;105:PE36. doi: 10.1126/stke.2001.105.pe36. [DOI] [PubMed] [Google Scholar]
- 18.Potter CJ, Huang H, Xu T. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell. 2001;105:357–68. doi: 10.1016/s0092-8674(01)00333-6. [DOI] [PubMed] [Google Scholar]
- 19.Tapon N, Ito N, Dickson BJ, et al. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001;105:345–55. doi: 10.1016/s0092-8674(01)00332-4. [DOI] [PubMed] [Google Scholar]
- 20.McManus EJ, Alessi DR. TSC1–TSC2: a complex tale of PKB-mediated S6K regulation. Nat. Cell Biol. 2002;4:E214–E216. doi: 10.1038/ncb0902-e214. [DOI] [PubMed] [Google Scholar]
- 21.Inoki K, Ouyang H, Li Y, Guan K-L. Signaling by target of rapamycin proteins in cell growth control. Microbiol. Mol. Biol. Rev. 2005;69:79–100. doi: 10.1128/MMBR.69.1.79-100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci. Signal. 2009;2:pe24. doi: 10.1126/scisignal.267pe24. [DOI] [PubMed] [Google Scholar]
- 24.Chantranupong L, Wolfson RL, Sabatini DM. Nutrient-sensing mechanisms across evolution. Cell. 2015;161:67–83. doi: 10.1016/j.cell.2015.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1–TSC2 complex is required for proper activation of mTOR complex 2. Mol. Cell Biol. 2008;28:4104–15. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goncharova EA, Goncharov DA, Li H, et al. mTORC2 is required for proliferation and survival of TSC2-null cells. Mol. Cell Biol. 2011;31:2484–98. doi: 10.1128/MCB.01061-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Canc. Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 28.Hanahan D, Weinberg Robert A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Dibble CC, Cantley LC. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015;25:545–55. doi: 10.1016/j.tcb.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoki K, Corradetti MN, Guan K-L. Dysregulation of the TSC-mTOR pathway in human disease. Nat. Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- 31.Menon S, Dibble CC, Talbott G, et al. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156:771–85. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Gao X, Saucedo LJ, et al. Rheb is a direct target of the tuberous sclerosis tumor suppressor proteins. Nat. Cell Biol. 2003;5:578–81. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 33.Tee AR, Manning BD, Roux PP, et al. Tuberous sclerosis complex gene products, tuberin and hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 2003;13:1259–68. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 34.Tabancay AP, Jr, Gau C-L, Machado IMP, et al. Identification of dominant negative mutants of Rheb GTPase and their use to implicate the involvement of human Rheb in the activation of p70S6K. J. Biol. Chem. 2003;278:39921–30. doi: 10.1074/jbc.M306553200. [DOI] [PubMed] [Google Scholar]
- 35.Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell Biol. 2013;15:555–64. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsun Z-Y, Bar-Peled L, Chantranupong L, et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt LS, Linehan WM. Clinical features, genetics and potential therapeutic approaches for Birt-Hogg-Dubé syndrome. Expert Opin. Orphan Drugs. 2015;3:15–29. doi: 10.1517/21678707.2014.987124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goncharova EA, Goncharov DA, James ML, et al. Folliculin controls lung alveolar enlargement and epithelial cell survival through E-cadherin, LKB1, and AMPK. Cell Rep. 2014;7:412–23. doi: 10.1016/j.celrep.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrington LS, Findlay GM, Gray A, et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J. Cell Biol. 2004;166:213–23. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah OJ, Wang Z, Hunter T. Inappropiate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 2004;14:1650–56. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 41.Hsu PP, Kang SA, Rameseder J, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–22. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yea SS, Fruman DA. New mTOR targets Grb attention. Science. 2011;332:1270–71. doi: 10.1126/science.1208071. [DOI] [PubMed] [Google Scholar]
- 43.Yu Y, Yoon S-O, Poulogiannis G, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–26. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding M, Bruick RK, Yu Y. Secreted IGFBP5 mediates mTORC1-dependent feedback inhibition of IGF-1 signalling. Nat. Cell Biol. 2016;18:319–27. doi: 10.1038/ncb3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manning BD, Logsdon MN, Lipovsky AI, et al. Feedback inhibition of Akt signaling limits the growth of tumors lacking Tsc2. Genes Dev. 2005;19:1773–78. doi: 10.1101/gad.1314605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeNicola GM, Cantley LC. Cancer’s fuel choice: new flavors for a picky eater. Mol. Cell. 2015;60:514–23. doi: 10.1016/j.molcel.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brugarolas J, Kaelin JWG. Dysregulation of HIF and VEGF is a unifying feature of the familial hamartoma syndromes. Cancer Cell. 2004;6:7–10. doi: 10.1016/j.ccr.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 49.Brugarolas JB, Vazquez F, Reddy A, et al. TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell. 2003;4:147–58. doi: 10.1016/s1535-6108(03)00187-9. [DOI] [PubMed] [Google Scholar]
- 50.Prabhakar NR, Semenza GL. Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Phys. Rev. 2012;92:967–1003. doi: 10.1152/physrev.00030.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Düvel K, Yecies JL, Menon S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell. 2010;39:171–83. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keith B, Johnson RS, Simon MC. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat. Rev. Cancer. 2008;8:967–75. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhat M, Robichaud N, Hulea L, et al. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015;14:261–78. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 55.Morita M, Gravel S-P, Chénard V, et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metabol. 2013;18:698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 56.Goncharova EA, Goncharov DA, Eszterhas A, et al. Tuberin regulates p70 S6 kinase activation and ribosomal protein S6 phosphorylation: a role for the TSC2 tumor suppressor gene in pulmonary lymphangioleiomyomatosis. J. Biol. Chem. 2002;277:30958–67. doi: 10.1074/jbc.M202678200. [DOI] [PubMed] [Google Scholar]
- 57.Kwiatkowski DJ, Zhang H, Bandura JL, et al. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet. 2002;11:525–34. doi: 10.1093/hmg/11.5.525. [DOI] [PubMed] [Google Scholar]
- 58.Ma EH, Jones RG. (TORC)ing up purine biosynthesis. Science. 2016;351:670–71. doi: 10.1126/science.aaf1929. [DOI] [PubMed] [Google Scholar]
- 59.Ben-Sahra I, Hoxhaj G, Ricoult SJH, et al. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–33. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–28. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.French JB, Jones SA, Deng H, et al. Spatial colocalization and functional link of purinosomes with mitochondria. Science. 2016;351:733–37. doi: 10.1126/science.aac6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J. Cell Sci. 2013;126:1713–19. doi: 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Russell RC, Yuan H-X, Guan K-L. Autophagy regulation by nutrient signaling. Cell Res. 2014;24:42–57. doi: 10.1038/cr.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011;13:1016–23. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parkhitko A, Myachina F, Morrison TA, et al. Tumorigenesis in tuberous sclerosis complex is autophagy and p62/sequestosome 1 (SQSTM1)-dependent. PNAS. 2011;108:12455–60. doi: 10.1073/pnas.1104361108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Inoki K, Kim J, Guan K-L. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu. Rev. Pharmacol. Toxicol. 2012;52:381–400. doi: 10.1146/annurev-pharmtox-010611-134537. [DOI] [PubMed] [Google Scholar]
- 67.Jacinto A, Hall MN. TOR signalling in bugs, brain and brawn. Nat. Rev. Mol. Cell Biol. 2003;4:117–26. doi: 10.1038/nrm1018. [DOI] [PubMed] [Google Scholar]
- 68.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 69.Goncharova EA, Goncharov DA, Damera G, et al. Signal transducer and activator of transcription 3 is required for abnormal proliferation and survival of TSC2-deficient cells: relevance to pulmonary lymphangioleiomyomatosis. Mol. Pharmacol. 2009;76:766–77. doi: 10.1124/mol.109.057042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.El-Hashemite N, Kwiatkowski DJ. Interferon-γ-Jak-Stat signaling in pulmonary lymphangioleiomyomatosis and renal angiomyolipoma: a potential therapeutic target. Am. J. Respir. Cell Mol. Biol. 2005;33:227–30. doi: 10.1165/rcmb.2005-0152RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Palm W, Park Y, Wright K, et al. The utilization of extracellular proteins as nutrients is suppressed by mTORC1. Cell. 2015;162:259–70. doi: 10.1016/j.cell.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Csibi A, Fendt S-M, Li C, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153:840–54. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Choo AY, Kim SG, Vander Heiden MG, et al. Glucose addiction of TSC null cells is caused by failed mTORC1-dependent balancing of metabolic demand with supply. Mol. Cell. 2010;38:487–99. doi: 10.1016/j.molcel.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parkhitko AA, Priolo C, Coloff JL, et al. Autophagy-dependent metabolic reprogramming sensitizes TSC2-deficient cells to the antimetabolite 6-aminonicotinamide. Mol. Cancer Res. 2014;12:48–57. doi: 10.1158/1541-7786.MCR-13-0258-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pusapati RV, Daemen A, Wilson C, et al. mTORC1-dependent metabolic reprogramming underlies escape from glycolysis addiction in cancer cells. Cancer Cell. 2016;29:548–62. doi: 10.1016/j.ccell.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 76.McCormack FX, Inoue Y, Moss J, et al. Efficacy and safety of sirolimus in lymphangioleiomyomatosis. N. Engl. J. Med. 2011;364:1595–606. doi: 10.1056/NEJMoa1100391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Franz DN, Leonard J, Tudor C, et al. Rapamycin causes regression of astrocytomas in tuberous sclerosis complex. Ann. Neurol. 2006;59:490–98. doi: 10.1002/ana.20784. [DOI] [PubMed] [Google Scholar]
- 78.Bissler JJ, McCormack FX, Young LR, et al. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N. Engl. J. Med. 2008;358:140–51. doi: 10.1056/NEJMoa063564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barnes EA, Kenerson HL, Mak BC, Yeung RS. The loss of tuberin promotes cell invasion through the β-catenin pathway. Am. J. Respir. Cell Mol. Biol. 2010;43:617–27. doi: 10.1165/rcmb.2008-0335OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pacheco-Rodriguez G, Kumaki F, Steagall WK, et al. Chemokine-enhanced chemotaxis of lymphangioleiomyomatosis cells with mutations in the tumor suppressorTSC2 gene. J. Immunol. 2009;182:1270–77. doi: 10.4049/jimmunol.182.3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goncharova EA, Goncharov DA, Fehrenbach M, et al. Prevention of alveolar destruction and airspace enlargement in a mouse model of pulmonary lymphangioleiomyomatosis (LAM) Sci. Transl. Med. 2012;4:154ra34. doi: 10.1126/scitranslmed.3003840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lamb RF, Roy C, Diefenbach TJ, et al. The TSC1 tumor suppressor hamartin regulates cell adhesion through ERM proteins and the GTPase Rho. Nat. Cell Biol. 2000;2:281–87. doi: 10.1038/35010550. [DOI] [PubMed] [Google Scholar]
- 83.Hodges AK, Li S, Maynard J, et al. Pathological mutations in TSC1 and TSC2 disrupt the interaction between hamartin and tuberin. Hum. Mol. Genet. 2001;10:2899–905. doi: 10.1093/hmg/10.25.2899. [DOI] [PubMed] [Google Scholar]
- 84.van Slegtenhorst MA, Nellist M, Nagelkerken B, et al. Interaction between hamartin and tuberin, the TSC1 and TSC2 gene products. Hum. Mol. Genet. 1998;7:1053–57. doi: 10.1093/hmg/7.6.1053. [DOI] [PubMed] [Google Scholar]
- 85.Benvenuto G, Li S, Brown SJ, et al. The tuberous sclerosis-1 (TSC-1) gene product hamartin suppresses cell growth and augments the expression of the TSC2 product tuberin by inhibiting its ubiquination. Oncogene. 2000;19:6306–16. doi: 10.1038/sj.onc.1204009. [DOI] [PubMed] [Google Scholar]
- 86.Goncharova E, Goncharov D, Noonan D, Krymskaya VP. TSC2 modulates actin cytoskeleton and focal adhesion through TSC1-binding domain and the Rac1 GTPase. J. Cell Biol. 2004;167:1171–82. doi: 10.1083/jcb.200405130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goncharova EA, Goncharov DA, Lim PN, et al. Modulation of cell migration and invasiveness by tumor suppressor TSC2 in lymphangioleiomyomatosis. Am. J. Respir. Cell Mol. Biol. 2006;34:473–80. doi: 10.1165/rcmb.2005-0374OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goncharova EA, James ML, Kudryashova TV, et al. Tumor suppressors TSC1 and TSC2 differentially modulate actin cytoskeleton and motility of mouse embryonic fibroblasts. PLoS ONE. 2014;9:e111476. doi: 10.1371/journal.pone.0111476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Matsumoto Y, Horiba K, Usuki J, et al. Markers of cell proliferation and expression of melanosomal antigen in lymphangioleiomyomatosis. Am. J. Respir. Cell Mol. Biol. 1999;21:327–36. doi: 10.1165/ajrcmb.21.3.3693. [DOI] [PubMed] [Google Scholar]
- 90.Kumasaka T, Seyama K, Mitani K, et al. Lymphangiogenesis in lymphangioleiomyomatosis: its implication in the progression of lymphangioleiomyomatosis. Am. J. Surg. Pathol. 2004;28:1007–16. doi: 10.1097/01.pas.0000126859.70814.6d. [DOI] [PubMed] [Google Scholar]
- 91.Young LR, Inoue Y, McCormack FX. Diagnostic potential of serum VEGF-D for lymphangioleiomyomatosis. N. Engl. J. Med. 2008;358:199–200. doi: 10.1056/NEJMc0707517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Baluk P, Tammela T, Ator E, et al. Pathogenesis of persistent lymphatic vessel hyperplasia in chronic airway inflammation. J. Clin. Investig. 2005;115:247–57. doi: 10.1172/JCI22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Karpanen T, Bry M, Ollila HM, et al. Overexpression of vascular endothelial growth factor-B in mouse heart alters cardiac lipid metabolism and induces myocardial hypertrophy. Circ. Res. 2008;103:1018–26. doi: 10.1161/CIRCRESAHA.108.178459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baldwin ME, Halford MM, Roufail S, et al. Vascular endothelial growth factor D is dispensable for development of the lymphatic system. Mol. Cell Biol. 2005;25:2441–49. doi: 10.1128/MCB.25.6.2441-2449.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers—a different disease. Nat. Rev. Cancer. 2007;7:778–90. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 96.Hammes SR, Krymskaya VP. Targeted approaches toward understanding and treating pulmonary lymphangioleiomyomatosis (LAM) Horm. Cancer. 2012;4:70–77. doi: 10.1007/s12672-012-0128-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu J, Astrinidis A, Howard S, Henske EP. Estradiol and tamoxifen stimulate LAM-associated angiomyolipoma cell growth and activate both genomic and nongenomic signaling pathways. Am. J. Physiol. Lung Cell Mol. Physiol. 2004;286:L694–700. doi: 10.1152/ajplung.00204.2003. [DOI] [PubMed] [Google Scholar]
- 98.Gu X, Yu JJ, Ilter D, et al. Integration of mTOR and estrogen-ERK2 signaling in lymphangioleiomyomatosis pathogenesis. PNAS. 2013;110:14960–65. doi: 10.1073/pnas.1309110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Finlay GA, York B, Karas RH, et al. Estrogen-induced smooth muscle cell growth is regulated by tuberin and associated with altered activation of platelet-derived growth factor receptor-beta and ERK-1/2. J. Biol. Chem. 2004;279:23114–22. doi: 10.1074/jbc.M401912200. [DOI] [PubMed] [Google Scholar]
- 100.Yu JJ, Robb VA, Morrison TA, et al. Estrogen promotes the survival and pulmonary metastasis of tuberin-null cells. PNAS. 2009;106:2635–40. doi: 10.1073/pnas.0810790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Prizant H, Taya M, Lerman I, et al. Estrogen maintains myometrial tumors in a lymphangioleiomyomatosis model. End Relat. Cancer. 2016;23:265–80. doi: 10.1530/ERC-15-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ohori N, Yousem SA, Sonmez-Alpan E, Colby TV. Estrogen and progesterone receptors in lymphangioleiomyomatosis, epithelial hemangioendothelioma, and sclerosing hemangioma of the lung. Am. J. Clin. Pathol. 1991;96:529–635. doi: 10.1093/ajcp/96.4.529. [DOI] [PubMed] [Google Scholar]
- 103.Taveira-DaSilva AM, Stylianou MP, Hedin CJ, et al. Decline in lung function in patients with lymphangioleiomyomatosis treated with or without progesterone. Chest. 2004;126:1867–74. doi: 10.1378/chest.126.6.1867. [DOI] [PubMed] [Google Scholar]
- 104.Ingelfinger JR, Drazen JM. Patient organizations and research on rare diseases. N. Engl. J. Med. 2011;364:1670–71. doi: 10.1056/NEJMe1102290. [DOI] [PubMed] [Google Scholar]