Abstract
Consumption of human breast milk (HBM) attenuates the incidence of necrotizing enterocolitis (NEC), which remains a leading and intractable cause of mortality in preterm infants. Here, we report that this diminution correlates with alterations in the gut microbiota, particularly enrichment of Propionibacterium species. Transfaunation of microbiota from HBM-fed preterm infants or a newly identified and cultured Propionibacterium strain, P. UF1, to germfree mice conferred protection against pathogen infection and correlated with profound increases in intestinal Th17 cells. The induction of Th17 cells was dependent on bacterial dihydrolipoamide acetyltransferase (DlaT), a major protein expressed on the P. UF1 surface layer (S-layer). Binding of P. UF1 to its cognate receptor, SIGNR1, on dendritic cells resulted in the regulation of intestinal phagocytes. Importantly, transfer of P. UF1 profoundly mitigated induced NEC-like injury in neonatal mice. Together, these results mechanistically elucidate the protective effects of HBM and P. UF1–induced immunoregulation, which safeguard against proinflammatory diseases, including NEC.
Keywords: Immunology, Inflammation
Keywords: Adaptive immunity
Introduction
Despite major advancements in medicine, the rate of preterm birth has not decreased and remains one of the leading global causes of infant mortality (1). Maternal pathogen-induced inflammation potentially contributes to both preterm birth and fetal inflammatory responses (2). This detrimental inflammation results in significant deterioration of the microbial consortium of the neonatal gut (3) that skews immunity toward proinflammatory responses, possibly resulting in necrotizing enterocolitis (NEC), a potentially fatal inflammatory bowel necrosis primarily affecting preterm infants with risk factors (e.g., intestinal injury) (4). While the multifactorial mechanisms involved in NEC and potential therapeutic approaches remain elusive (5), the need for novel therapeutic interventions that redirect inflammation and reduce disease severity, leading to its regression, is urgent (6). Maintaining gut homeostasis to swiftly resolve gastrointestinal infections, which otherwise induce deleterious signals, is critically important and highly dependent on the fitness of host regulatory mechanisms. These mechanisms are dictated by critical nutrients in human breast milk (HBM) and the mother’s and infant’s microbiomes (7), which together make up a pivotal triad intricately involved in infant intestinal immunity (8). Errors in these mechanisms may increase pathologic signals, resulting in dysfunctional gut homeostasis, potentially leading to NEC.
Thus far, it has been postulated that HBM not only increases beneficial bacteria within balanced gut microbiota, but also promotes host immune regulation and reduces the risk of NEC in preterm infants (9–12). However, further mechanistic insights are required to better understand the immune regulation orchestrated by HBM, the beneficial bacteria within, and critical induced metabolites, all of which may control T cell responses (e.g., Th17 cells) intricately involved in mucosal protection and the pathogenesis of proinflammatory diseases (13). This includes NEC, in which the recruitment of inflammatory cells is robustly increased, while functional Tregs are significantly decreased (14). Thus, if severe induced inflammation leads to NEC, its redirection by novel therapeutic approaches involving beneficial commensals, including Propionibacterium (P.) University of Florida 1 (UF1) (P. UF1), may enhance proinflammatory NEC regression.
Results
Characterization of P. UF1 bacterium.
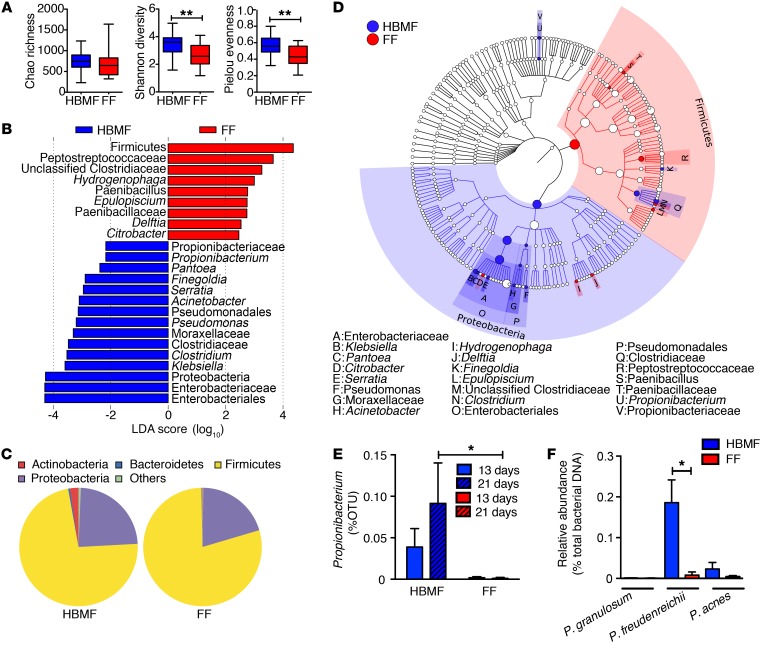
Focusing on the status of the microbiota of 2 discrete cohorts, we demonstrate that the composition and diversity of the gut microbiota from human breast milk–fed (HBMF) preterm infants (n = 20) were distinct from those of formula-fed (FF) preterm infants (n = 20). Microbiota analyses of these 2 cohorts revealed major differences in the Shannon diversity, which estimates the total number of operational taxonomic units existing in the given bacterial community, and the Pielou’s evenness index, which demonstrates how evenly the individuals in the community are distributed over different operational taxonomic units (P < 0.01) (Figure 1A). Together, the microbial evenness and diversity were found to be significantly increased in the HBMF cohort’s microbiota, while a trend toward an increase in bacterial richness was also observed (Figure 1A). Moreover, microbiota compositions between the HBMF and FF preterm infants were principally different in the Actinobacteria phylum (e.g., Propionibacteriaceae), among other bacteria, including Bacteriodetes, Firmicutes, and Proteobacteria (Figure 1, B–D). Thus, the focus was drawn to key species of this Actinobacteria phylum, particularly Propionibacterium, which contributed heavily to these differences (Figure 1, B–D). Importantly, the proportion of Propionibacterium, including P. freudenreichii, in the microbiota of the HBMF preterm infants was significantly increased throughout days 13 to 21 after birth, while these bacteria were poorly detected in the FF preterm infants’ fecal samples (Figure 1, E and F). Subsequently, Propionibacterium was isolated from HBMF preterm infants’ fecal samples using carbohydrate separation and cultured optimally in de Man, Rogosa, and Sharpe (MRS) broths and lactate.
Figure 1. Abundance of Propionibacterium in the fecal samples of HBMF preterm infants.
Fecal samples were collected from HBMF (n = 20) and FF preterm infants (n = 20), and microbiota composition was analyzed by 16S rDNA sequencing. (A) Summary box plots of Chao richness, Shannon diversity, and Pielou’s evenness indices derived from analyses of fecal samples of HBMF (blue) and FF (red) preterm infants on day 13 ± 2 to 3. (B) Linear discriminant analysis (LDA) of taxons between HBMF and FF preterm infants’ microbiota on day 13 ± 2 to 3. Taxa enriched in HBMF preterm infants’ microbiota have a negative score (blue), and taxa enriched in FF preterm infants’ microbiota have a positive score (red). Only taxa with an absolute value of LDA score of more than 2 are shown. (C) Phylum structure of the abundant bacteria in fecal samples of HBMF and FF preterm infants on days 21 ± 3. (D) Taxonomic cladogram of HBMF versus FF preterm infants’ bacterial fecal samples on day 21 ± 3 (blue, HBMF-enriched taxa; red, FF-enriched taxa). (E) Percentage of operational taxonomic units (OTUs) of Propionibacteria in the fecal samples of HBMF and FF preterm infants by day 13 ± 2 to 3 and day 21 ± 3. (F) Relative abundance of different Propionibacteria (e.g., P. freudenreichii) in fecal samples of HBMF and FF preterm infants. Error bars indicate mean ± SEM. *P < 0.05; **P < 0.01, 2-tailed unpaired t test (A, E, and F). Kruskal-Wallis test (B and D).
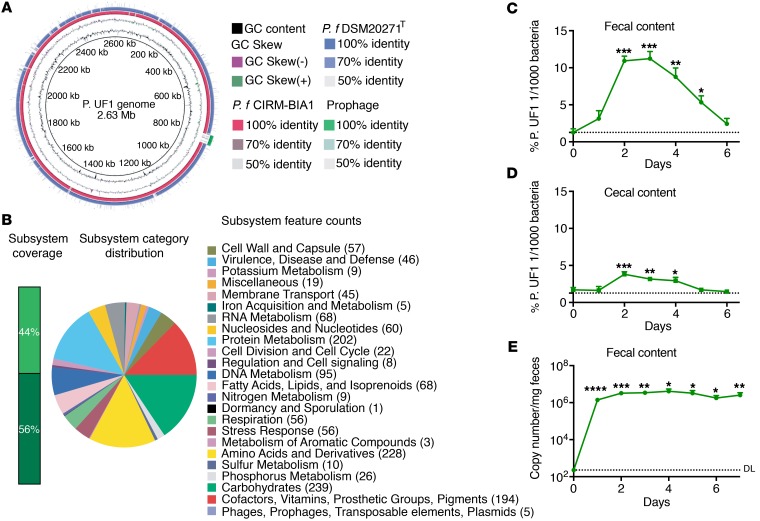
The first draft genome-sequence analyses of one of these newly identified Propionibacterium strains, tentatively designated P. UF1, exhibited 90% identity to known Propionibacterium species, including P. freudenreichii subsp. freudenreichii DSM20271T and subsp. shermanii CIRM-BIA1 (Figure 2A). The genome of P. UF1 (2.63 Mb) encodes for critical enzymes involved in fermentation, catabolism, and biosynthetic pathways for all amino acids (Figure 2B). Furthermore, introducing this bacterium to conventional C57BL/6 mice via a single oral gavage resulted in transient gut colonization, potentially due to bacterial competition for space and nutritional resources (15, 16), whereby this bacterium was no longer detectable in the fecal or cecal contents of C57BL/6 mice after 5 to 6 days (Figure 2, C and D). In contrast, P. UF1 permanently colonized the gut of germfree (GF) mice, as detected in the fecal contents of these mice until day 7 (Figure 2E). Such colonization in GF mice was expected, as P. UF1, like other beneficial (e.g., Bifidobacterium bifidum) (17) or pathogenic bacteria (e.g., Citrobacter rodentium) (18), demonstrates the same trend due to the lack of competitive commensal bacteria.
Figure 2. Characterization of P. UF1 bacterium.
(A) Genome sequence comparison of isolated P. UF1 with known P. freudenreichii spp. Shermanii CIRM-BIA1 (red) and P. freudenreichii spp. freudenreichii DSM20271T (blue). Prophage was identified by PHAST (green). Sequence identity between P. freudenreichii strains and P. UF1 is shown. (B) Correlation of the annotated genome sequence on the RAST platform with different metabolic pathways of P. UF1. Numbers in parentheses indicate numbers of annotated genes belonging to subcategories of pathways. (C and D) C57BL/6 mice (n = 3) were gavaged with P. UF1 (109 CFU/mouse) 1 time, and mice were sacrificed every day to detect P. UF1 in the fecal (C) and cecal (D) contents. (E) GF mice (n = 5) were gavaged with P. UF1 (109 CFU/mouse) 1 time, and fecal samples were collected every day to detect P. UF1. DL indicates the qPCR detection limit. Asterisks indicate statistical significance compared with day 0. Data are representative of 1 (E) or 3 (C and D) independent experiments. Error bars indicate mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, 2-tailed unpaired t test (C–E).
Regulation of intestinal proinflammation by P. UF1.
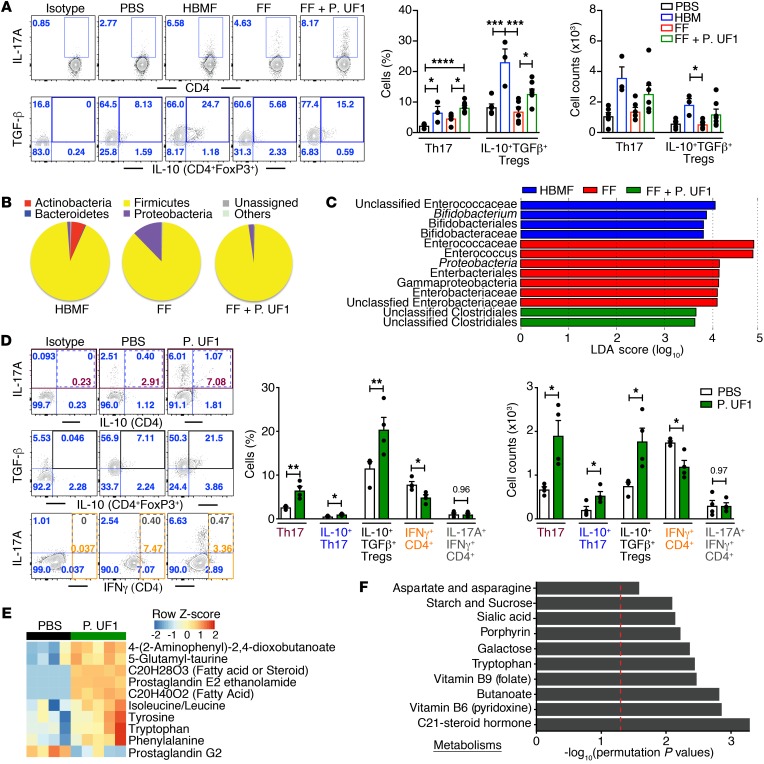
To explore the potential immunologic differences caused by the microbiota of the aforementioned cohorts, fecal microbes derived from HBMF and FF preterm infants were transfaunated into GF mice (19). GF mice receiving HBMF preterm infants’ microbiota did not exhibit the enhanced proinflammation (e.g., IL-1β) in colonic DCs that was observed in the GF mice transfaunated with FF preterm infants’ microbiota (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI95376DS1). Moreover, transfaunating GF mice with HBMF preterm infants’ microbiota increased Th17 cells and sustained IL-10+ Tregs, a trend that was less evident in GF mice transfaunated with FF preterm infants’ microbiota (Figure 3A). Not only were proinflammatory DC responses attenuated upon P. UF1 gavage of GF mice transfaunated with FF preterm infants’ microbiota, but Th17 cells and IL-10+ Tregs were also significantly increased to the levels seen in GF mice transfaunated with HBMF preterm infants’ microbiota (Supplemental Figure 1A and Figure 3A). Additionally, the composition of the gut microbiota in GF mice transfaunated with FF preterm infants’ microbiota and subsequently gavaged with P. UF1 showed changes in bacterial organization, including decreased levels of γ-Proteobacteria, when compared with the FF preterm infants’ microbiota, indicating the potential influence of P. UF1 on the FF preterm infants’ microbiota (Figure 3, B and C). The same trend was also observed in GF mice that were transfaunated with HBMF preterm infants’ microbiota, wherein γ-Proteobacteria levels were also decreased, possibly by the elevated growth of Actinobacteria, since beneficial bacteria may compete with indigenous pathobionts (e.g., γ-Proteobacteria) (20–22) in the given environment of GF mice (Figure 3B). To specifically investigate P. UF1–induced Th17 cells, GF mice were monoassociated with P. UF1 (4 oral gavages with 109 CFU/mouse, every 3 days), and DC and T cell responses were analyzed. Once again, proinflammatory cytokines (e.g., IL-1β) promoting intestinal pathology (23) were decreased in colonic DCs (Supplemental Figure 1B and Supplemental Figure 11). Additionally, cytokine transcripts (e.g., IL-1β) were not only diminished in colonic tissues of GF mice monoassociated with P. UF1, but also in sorted colonic DCs (Supplemental Figure 2, B and C). The generation of Th17 cells, particularly IL-10+IFN-γ– Th17 cells, and IL-10+ Tregs was also substantially increased in GF mice monoassociated with P. UF1, while the IFN-γ+CD4+ T cell response (Th1 cells) was decreased compared with that in the control groups (Figure 3D and Supplemental Figure 12), suggesting that P. UF1 not only critically regulates DCs, but also specifically augments the formation of Th17 cells. Such differential dependency of the Th17 cells on P. UF1 was then highlighted in GF mice monoassociated with P. UF1 or E. coli JM109 (24). Here, E. coli JM109, when compared with P. UF1, did not induce Th17 cell differentiation (Supplemental Figure 1C). Furthermore, tryptophan, tyrosine, and phenylalanine were increased in the fecal samples of the GF mice monoassociated with P. UF1 (Figure 3E), potentially regulating Th17 cell homeostasis (25, 26). Accordingly, pathway analyses (27) revealed that these enriched gut metabolites (Figure 3F) are highly involved in C21-steroid hormone biosynthesis (28), tryptophan metabolism, multiple vitamin biosynthetic pathways (e.g., vitamin B12) (29), and porphyrin biosynthesis (30).
Figure 3. Modulation of colonic immune responses by HBMF preterm infants’ microbiota.
(A–C) GF mice were transfaunated with HBMF preterm infants’ microbiota (blue), FF preterm infants’ microbiota (red), or FF preterm infants’ microbiota plus 4 treatments with P. UF1 (green) or were left untransfaunated (black). Colonic immune responses and microbiota were analyzed 2 weeks later. Representative data of flow plots, percentages, and total cell counts of Th17 cells and IL-10+TGF-β+ Tregs (A). Microbial phyla structure (B) and LDA analysis (C) of fecal samples from indicated group. (D) GF mice were monoassociated with P. UF1 (green) or gavaged with PBS (white), and induced colonic immune responses were analyzed. Representative data of flow plots, percentages, and total cell counts of Th17 cells, IL-10+TGF-β+ Tregs, and IFN-γ+CD4+ cells (Th1 cells). (E and F) Heatmap of selected metabolites differentiating fecal samples from PBS- and P. UF1–gavaged GF mice (E), and significant metabolic pathways of fecal samples from PBS- versus P. UF1–gavaged GF mice (F). Red dashed line shows permutation of P = 0.05. Data are pooled from 2 independent experiments (n = 3–7 mice/group, A–C) or representative of 3 (n = 4–5 mice/group, E and F) or 6 (n = 4 mice/group, D) independent experiments. Error bars indicate mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, ANOVA plus Tukey’s post test (A) or 2-tailed unpaired t test (D).
Bacterial dihydrolipoamide acetyltransferase drives Th17 cells.
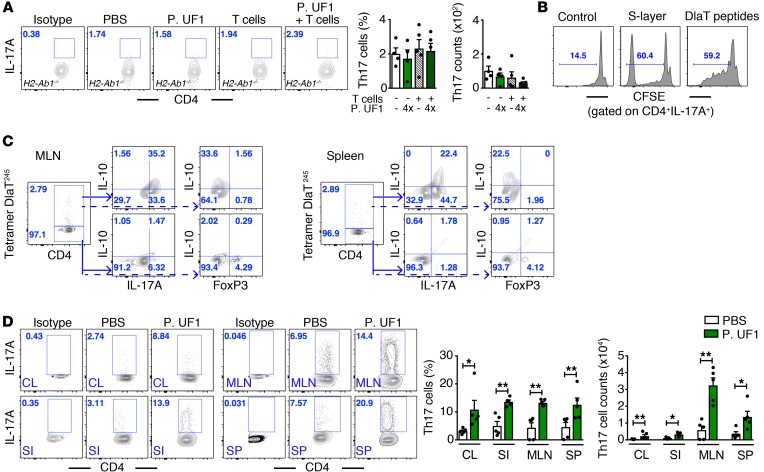
Sensing receptor interactions with the bacterial surface layer (S-layer) stimulate intestinal DCs, leading to T cell activation (31–34). Activated T cells, in general, and antigen-specific Th17 cells, in particular, are tightly controlled by gut commensals, including segmented filamentous bacteria (SFB) (35, 36) along with regulated cytokines (e.g., IL-6, TGF-β, IL-1β) (37). To elucidate the role of the P. UF1 S-layer in Th17 cell differentiation, the S-layer was isolated by guanidine hydrochloride (GdnHCl) and characterized by mass spectrometry. Dihydrolipoamide acetyltransferase (DlaT) was found to be one of the major S-layer proteins, from which three 15-mer peptides were deduced exhibiting high affinity for MHC II. Interestingly, DlaT is critically involved in pyruvate decarboxylation, which links glycolysis to the citric acid cycle (38); however, this protein could also be expressed by other, yet to be identified mechanism(s) on the S-layer to exert immune activation. Importantly, we demonstrate the dependency of P. UF1 S-layer–induced Th17 cells in H2-Ab1+/+ sufficient mice (MHC II+/+) (Supplemental Figure 3A). However, this was not observed in H2-Ab1–/– deficient mice gavaged with P. UF1 alone or with P. UF1 plus adaptively transferred CD4+ T cells (Figure 4A). Thus, these data once again illuminate the critical role of MHC II in antigen-specific CD4+ T cell differentiation, as also previously demonstrated for SFB-dependent Th17 cells (39).
Figure 4. Induction of Th17 cell differentiation by P. UF1.
(A) CD4+ splenic cells derived from C57BL/6 mice gavaged with P. UF1 were transferred into H2-Ab1–/– recipient mice. H2-Ab1–/– mice were gavaged 4 times (4×) with P. UF1 (109 CFU/mouse) or with PBS (n = 4–5 mice/group). Representative data of flow plots, percentages, and total cell counts of colonic Th17 cells. (B) Splenic CD4+ T cells were sorted and labeled with CFSE. T cells were cocultured with pulsed BMDCs with S-layer (10 μg/ml) or DlaT peptides (20 μg/mg) for 5 days. Cells were stained and analyzed by flow cytometry. Representative histogram plots of CFSE-labeled IL-17A+ CD4+ T cells. (C) C57BL/6 mice were gavaged 4 times with P. UF1. Splenic and MLN tetramer DlaT245+ cells were enriched with phycoerythrin (PE) beads and analyzed. Representative plots depicting the percentage of DlaT245+ tetramer CD4+ T cells with a majority of IL-17A+IL-10+ T cells, but negative for FoxP3+ T cells. (D) Th17 cell differentiation from naive splenic Thy1.1+CD4+CD45RBhi cells in Thy1.2+ Rag1–/– mice gavaged with P. UF1 or PBS. Th17 cell differentiation was analyzed in the colon (CL), small intestine (SI), MLN, and spleen (SP) (n = 5 mice/group). Data are representative of 2 (D) or 3 (A–C) independent experiments. Error bars indicate mean ± SEM. *P < 0.05; **P < 0.01, ANOVA plus Tukey’s post test (A) or Mann-Whitney U test (D).
Additionally, it has recently been reported that bacterial-induced metabolites support the differentiation of T cells, including Th17 cells (40). To demonstrate whether P. UF1–induced metabolites can prime Th17 cell differentiation, metabolomic analyses of P. UF1–gavaged H2-Ab1–/– mice compared with H2-Ab1–/– mice plus CD4+ T cells and PBS-gavaged H2-Ab1–/– mice were performed. Data revealed different metabolic pathways, particularly in tryptophan and butanoate metabolism; however, no Th17 cells were promoted in the H2-Ab1–/– mice lacking MHC II (Supplemental Figure 3, B and C), once again highlighting the critical role of MHC II in differential T cell activation (41). Furthermore, cytokines (e.g., IL-1β, IL-6) alone were not sufficient to induce Th17 cells in mice that were gavaged with the S-layer P. UF1, denoting that the S-layer and its deduced peptides are essential in Th17 cell differentiation (Supplemental Figure 3A).
To further evaluate S-layer–specific Th17 cells, C57BL/6 mice were gavaged with P. UF1. Subsequently, splenic CD4+ T cells were sorted and cocultured with bone marrow–derived DCs pulsed with the S-layer or with the 3 DlaT-peptides. Data demonstrate the differential DlaT positivity for only splenic and mesenteric lymph node (MLN) Th17 cells, but not IL-10+ Tregs, using the DlaT-specific tetramer245 (Figure 4, B and C). To specifically determine the differentiation of Th17 cells by P. UF1, sorted naive splenic Thy1.1+CD4+CD45RBhi cells were transferred into P. UF1– or PBS-gavaged Thy1.2+ Rag1–/– mice (42) lacking mature T and B cells (43). Data document the differentiation of Th17 cells from Thy1.1+CD4+CD45RBhi T cells by P. UF1 in the intestines, MLNs, and spleens of Thy1.2+ Rag1–/– mice (Figure 4D).
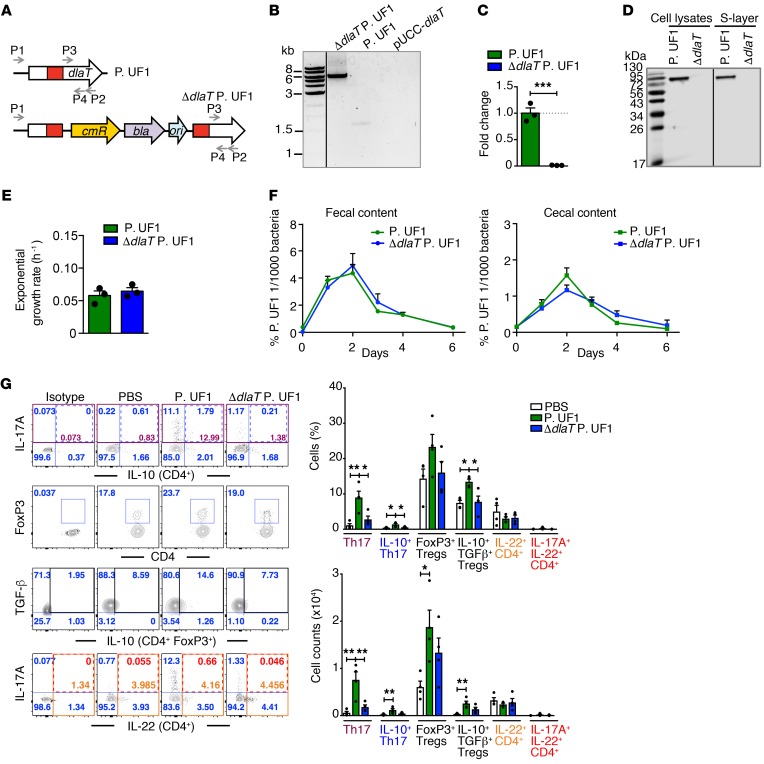
To investigate the mechanistic paradigm associated with DlaT-specific Th17 cell differentiation, the dlaT gene was deleted from the bacterial chromosome by homologous recombination with a single crossover event (44), resulting in the ΔdlaT P. UF1 (Figure 5, A–D). Importantly, the bacterial growth kinetics and detection of ΔdlaT P. UF1 in conventional C57BL/6 mouse fecal and cecal contents were comparable to P. UF1 (Figure 5, E and F). When compared with P. UF1, GF mice monoassociated with ΔdlaT P. UF1 exhibited significantly increased expression of IL-1β, IL-6, and IL-12/23p40 in colonic DCs and colonic tissue (Supplemental Figure 2, A–C). ΔdlaT P. UF1 also substantially abrogated Th17 cells, while IL-10+ Tregs were generally comparable to those of PBS-gavaged mice, suggesting a crucial role for DlaT in IL-10+ Th17 cell differentiation. Such P. UF1–induced Th17 cells did not significantly express IL-22 (45) (Figure 5G).
Figure 5. Differentiation of DlaT-specific Th17 cells.
(A) Genetic organization of the dlaT locus from P. UF1 and ΔdlaT P. UF1. P1, P2, P3, and P4 primers was used for identifying ΔdlaT P. UF1. cmR, chloramphenicol resistance gene (P. UF1); bla, ampicillin resistance gene (E. coli); ori, replication origin of pUC19 plasmid. (B) PCR amplification of dlaT with primers P1/P2. (C) Quantitative reverse-transcriptase PCR (qRT-PCR) analyses of dlaT mRNA expression using primers P3/P4 (n = 3 samples/group). (D) Western blot analysis of DlaT expression in the cell lysates and S-layer using polyclonal anti-DlaT antibodies. (E) Exponential growth rate of P. UF1 and ΔdlaT P. UF1 in MRS-lactate medium (n = 3 samples/group). (F) Colonization of P. UF1 and ΔdlaT P. UF1 in C57BL/6 mice (n = 2 mice/group). (G) GF mice were gavaged with P. UF1 (green), ΔdlaT P. UF1 (blue), or PBS (white). Colonic cell immune responses were analyzed 2 weeks later (n = 4 mice/group). Representative data of flow plots, percentages, and total cell counts of Th17 cells, CD4+FoxP3+ Tregs, IL-10+TGF-β+ Tregs, and IL-22+CD4+ T cells. Data are representative of 1 (F) or 3 (B–E and G) independent experiments. Error bars indicate mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, 2-tailed unpaired t test (C and E) or ANOVA plus Tukey’s post test (G).
To delineate any potential changes in ΔdlaT P. UF1 transcriptomic machinery that may affect gut homeostasis, the transcriptic and metabolic changes in ΔdlaT P. UF1 compared with P. UF1 were analyzed. Deletion of dlaT in P. UF1 resulted in significant transcriptic changes involved in the glycolytic and multiple metabolic pathways, including the metabolism of carbohydrates, nitrogen, folate, and cysteine biosynthesis (Supplemental Figure 4, A–C). More importantly, multiple tryptophan-derived metabolites (e.g., hydroxykynurenamine, formyl-acetyl-5-methoxykynurenamine) were significantly observed in P. UF1, but not in ΔdlaT P. UF1 (Supplemental Figure 4, D and E), potentially qualitatively influencing the regulation of DC and T cell responses.
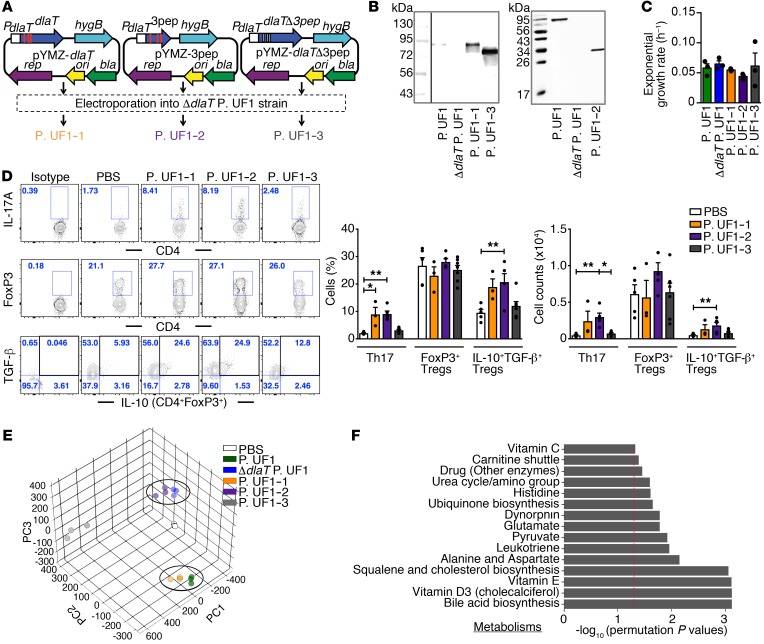
To specifically elucidate the relevance of DlaT in Th17 cell differentiation and to exclude possible ectopic effects resulting from dlaT deletion in P. UF1, we then complemented the dlaT gene (P. UF1–1), the 3 DlaT peptide sequences (P. UF1–2), or the dlaT gene without these 3 peptides (P. UF1–3) in the ΔdlaT P. UF1 (Figure 6, A and B), whereby the bacterial growth kinetics of these P. UF1 recombinants were comparable to those of P. UF1 (Figure 6C). GF mice were then monoassociated with P. UF1–1, P. UF1–2, or P. UF1–3. Data demonstrate the significance of DlaT and the 3 DlaT peptides, such that mice receiving P. UF1–1 or P. UF1–2, but not P. UF1–3, exhibited differentiation of Th17 cells and induction of IL-10+ Tregs (Figure 6D). Furthermore, metabolomic analyses of P. UF1–1 and P. UF1 separated similarly from the other groups of GF mice gavaged with ΔdlaT P. UF1, P. UF1–2, P. UF1–3, or PBS, indicating a physiologic role for DlaT in P. UF1 molecular machinery supporting bacterial metabolic homeostasis (Figure 6, E and F).
Figure 6. Restoring Th17 cell differentiation by complementation of ΔdlaT P. UF1 with dlaT.
(A) Complementation of ΔdlaT P. UF1 with the dlaT gene (P. UF1–1, orange), the 3 dlaT peptides (P. UF1–2, purple), and the dlaT gene minus the 3 peptides (P. UF1–3, gray). (B) Western blot analysis of DlaT expression and the 3 peptides in cell lysates of the complemented strains using polyclonal anti-DlaT antibodies. (C) Exponential growth rate of the complemented strains in MRS-lactate medium (n = 3 samples/group). (D) GF mice were gavaged with P. UF1–1, P. UF1–2, P. UF1–3 (109 CFU/mouse), or PBS on days 0, 3, 6, and 9. Two weeks later, induced colonic immune responses were analyzed. Representative data of flow plots, percentages, and total cell counts of Th17 cells, CD4+FoxP3+ Tregs, and IL-10+TGF-β+ Tregs. (E and F) Fecal sample metabolomic analyses of GF mice gavaged with aforementioned complemented strains or PBS. PCA of fecal sample metabolites (E) and comparison of significant pathways between P. UF1: P. UF1–1 and ΔdlaT P. UF1: P. UF1–2 (F). Red dashed line shows permutation P value of 0.05. Data are pooled from 2 independent experiments (n = 3–7 mice/group, D) or are representative of 1 (n = 3 mice/group, E and F) or 3 (B and C) independent experiments. Error bars indicate mean ± SEM. *P < 0.05; **P < 0.01, ANOVA plus Tukey’s post test (C) or Kruskal-Wallis plus Dunn’s post test (D).
Immune regulation by P. UF1 against Listeria monocytogenes infection.
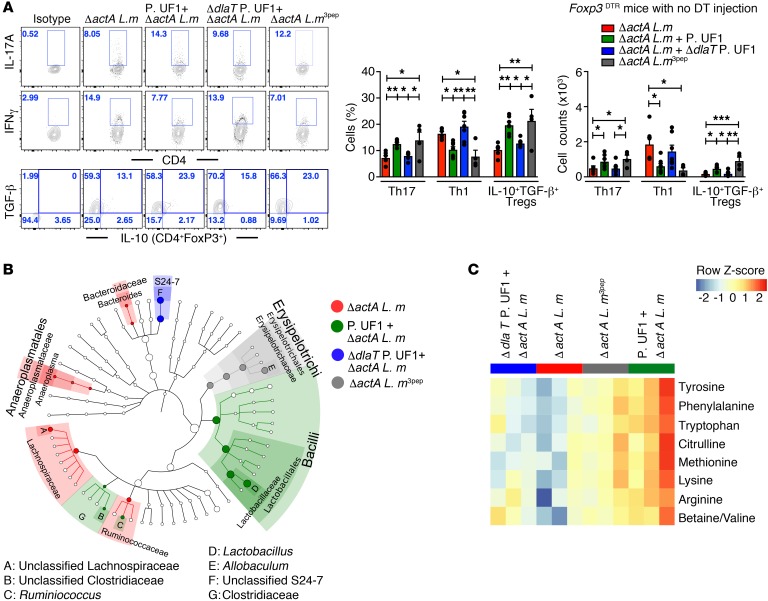
Knowing the importance of commensals in the instruction of Th17 cell differentiation (46, 47), the protective role of Th17 cells in response to intracellular pathogens, including Listeria monocytogenes (L. m), may demand consideration (48). To further evaluate the regulatory effects of P. UF1 on protective colonic DlaT-dependent Th17 cells against L. m infection, ΔactA L. m was established. ΔactA L. m lacking the ActA polypeptide cannot interact with actin filaments and is, as a consequence, nonmotile to spread in the periphery (49). Subsequently, ΔactA L. m3pep was generated by introducing pIMK2, which specifically harbors the 3 DlaT peptides into ΔactA L. m (Supplemental Figure 5, A and B). Indeed, ΔactA L. m3pep, like P. UF1, controlled DCs by attenuating proinflammatory signals (e.g., IL-1β) (Supplemental Figure 5C) and regulating IFN-γ+ Th1 cells, potentially minimizing pathogenic inflammation and induced DlaT tetramer245+IL-10+ Th17 cell responses when compared with other mouse groups infected only with ΔactA L. m or gavaged with ΔdlaT P. UF1 and infected with ΔactA L. m (Figure 7A and Supplemental Figure 6, A and B). These data highlight the critical involvement of DlaT in colonic Th17 cell differentiation, whereby its deletion significantly abated Th17 cell responses against ΔactA L. m infection (Figure 7A). As expected, these DlaT tetramer245+IL-10+ Th17 cells were completely diminished in ΔactA L. m3pep–infected Rorc(γt)GFP/GFP mice deficient for retinoid-related orphan receptor (RORγt) (50) required for the Th17 cell lineage (Supplemental Figure 6A). Importantly, IL-10+ Tregs, which contract upon L. m infection (51), were not only functionally sustained, but their frequency was markedly increased in mice orally infected with ΔactA L. m3pep or gavaged with P. UF1 and infected with ΔactA L. m, potentially regulating protective CD4+ T cell subsets, including Th1 cells and Th17 cells, all of which may synergistically contribute to the resolution of this pathogenic infection (Figure 7A). Moreover, ΔactA L. m infection was significantly reduced in mice gavaged with P. UF1 on day 4, in contrast with the other groups (Supplemental Figure 5D). When compared with that of other mouse groups, the composition of the gut microbiota of mice gavaged with P. UF1 and infected with ΔactA L. m demonstrates enrichment of Lactobacillus and Ruminococcus species (Figure 7B). This may suggest that regulation of pathogen-induced inflammation by P. UF1 not only plays a role in sustaining the microbiota upon intestinal infection, but may also critically affect the pattern of induced gut metabolites, including citrulline involved in nitric oxide production and innate signaling (52), ultimately controlling protective immune responses to ΔactA L. m infection. Such regulatory responses were not observed in mice gavaged with ΔdlaT P. UF1 or in ΔactA L. m–infected mice (Figure 7C).
Figure 7. Regulation of colonic immunity by P. UF1 during ΔactA L. m infection.
(A) B6.129(Cg)-Foxp3tm3(DTR/GFP)Ayr/J (Foxp3DTR) mice with no DT injection were gavaged with PBS (red), P. UF1 (green), or ΔdlaT P. UF1 (blue) and orally infected with ΔactA L. m. One group of mice was orally infected with ΔactA L. m3pep (gray). Representative data of flow plots, percentages, and total counts of colonic Th17 cells, Th1 cells, and FoxP3+IL-10+TGF-β+ Tregs. (B) Taxonomic cladogram of fecal microbiota from experimental groups (P < 0.05, Kruskal-Wallis test) from groups of C57BL/6 mice gavaged with P. UF1 or dlaT P. UF1 and orally infected with ΔactA L. m as described in A. (C) Heatmap of selected fecal metabolites (P < 0.05, 1-way ANOVA) from mouse groups as described in A. Data are pooled from 2 independent experiments (n = 4–7 mice/group, A) or representative of 2 independent experiments (n = 3 mice/group, B and C). Error bars indicate mean ± SEM. P < 0.05; *P < 0.05; **P < 0.01; ***P < 0.001, ANOVA plus Tukey’s post test (A).
To demonstrate the role of P. UF1–induced IL-10+ Tregs in the regulation of gut homeostasis, the frequency of these cells was significantly reduced in B6.129(Cg)-Foxp3tm3(DTR/GFP)Ayr/J mice (Foxp3DTR mice) through diphtheria toxin (DT) injection (53). Conversely, regulated immunity against ΔactA L. m infection was significantly diminished when compared with functional Tregs from mice with no DT injection (Figure 7A and Supplemental Figure 7, A and B), again highlighting the regulatory role of sustained P. UF1–induced IL-10+ Tregs in controlling inflammatory responses (54) against ΔactA L. m infection.
Regulation of P. UF1–induced immunity requires SIGNR1.
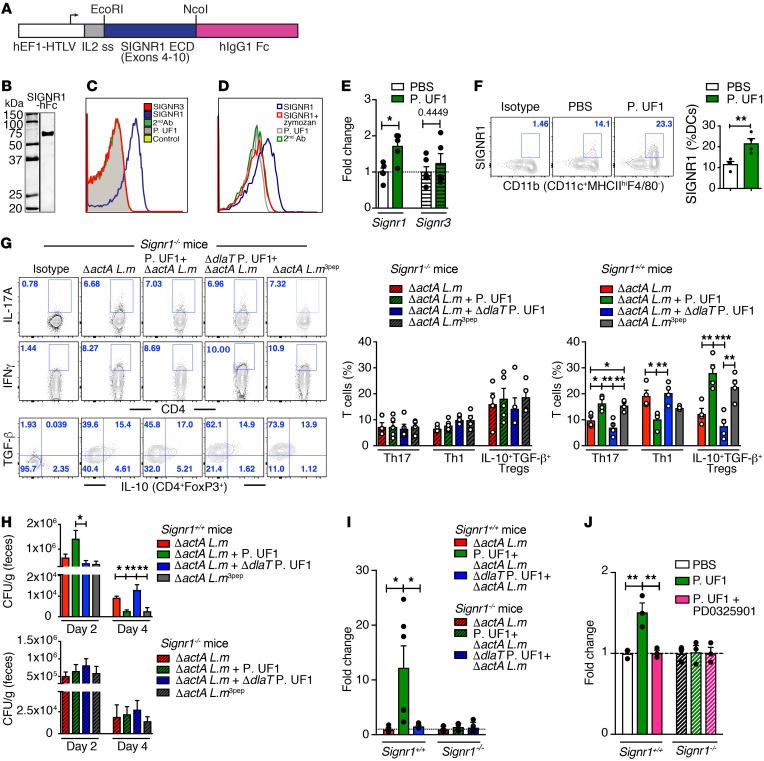
To identify the cognate receptor that binds to the P. UF1 S-layer, governing DC function and subsequent regulation of T cells, including Th17 cells, the focus was centered on C-type lectin receptors (CLRs), particularly SIGNR1 (CD209b). SIGNR1 recognizes microbes via lipoglycan moieties inducing regulatory signals controlling DCs to initiate T cell activation (55). In fact, SIGNR1, but not SIGNR3 (CD209d) (56), was identified as the binding receptor for P. UF1 when released by CHO cells in the form of a SIGNR1-hFc complex (Figure 8, A–C). Binding of P. UF1 to the SIGNR1-hFc complex was significantly reduced when SIGNR1-hFc was saturated with zymozan consisting of a protein-carbohydrate complex (Figure 8D). Additionally, gavaging mice with P. UF1 upregulated SIGNR1 in colonic tissue and in DCs (Figure 8, E and F). To assess SIGNR1 involvement in limiting pathogen-induced inflammation, Signr1+/+ and Signr1–/– mice were gavaged with P. UF1, ΔdlaT P. UF1, or PBS and then orally infected with ΔactA L. m. Data illustrate the role of SIGNR1 in assuaging ΔactA L. m–induced inflammation seen in P. UF1–gavaged Signr1+/+ mice (Figure 8G), which was not evident in P. UF1–gavaged Signr1–/– mice infected with ΔactA L. m (Figure 8G and Supplemental Figure 8). Furthermore, controlled protection by P. UF1 in Signr1+/+ mice was not observed in P. UF1–gavaged Signr1–/– mice infected with ΔactA L. m or L. m3pep, as reduction of ΔactA L. m burden was not detected in any group of ΔactA L. m–infected Signr1–/– mice (Figure 8H). Comparing metabolic pathway analyses of Signr1–/– with Signr1+/+ mice also revealed notable metabolic changes, despite the shared metabolites (e.g., fatty acid metabolism) seen in both mouse strains (Supplemental Figure 9A), while no significant gut microbiota modifications were obvious in Signr1–/– mice compared with Signr1+/+ mice (Supplemental Figure 9B). Collectively, these data support the notion that SIGNR1 regulates protective immunity by potentially activating regulatory signals, including SOCS1 against pathogens (e.g., L. m) (57), whereby its deficiency promotes pathogen persistence. Accordingly, the differential suppressors of Socs1 activation were induced in the colonic tissue of Signr1+/+ mice, but not in Signr1–/– mice infected with ΔactA L. m (Figure 8I). Furthermore, critical to SIGNR1 signaling is the activation of the serine/threonine kinase Raf-1; inhibition of Raf-1 downstream signaling (e.g., MEK) blocks SIGNR1 responses (58). To specifically delineate the signaling induced by the P. UF1 S-layer–SIGNR1 interaction controlling SOCS1 in DCs, MEK was optimally blocked by the PD0325901 inhibitor (59) within 3 hours. This led to significant reduction of Socs1 expression in P. UF1–activated DCs; in contrast, this trend was not observed in Signr1–/– DCs (Figure 8J). These data may highlight the interplay between SIGNR1 and SOCS1 signaling to limit deleterious inflammation (57).
Figure 8. Regulation of intestinal immunity requires SIGNR1.
(A) Depiction of SIGNR1-hFc expression. The cDNA encoding SIGNR1-extracellular domain (exons 4–10) was fused to the Fc of human IgG1. (B) Western blot analysis of SIGNR1-hFc using anti-SIGNR1 antibody. (C) Binding of P. UF1 to SIGNR1 (blue), SIGNR3 (red), control fusion (yellow), or secondary (2nd) antibody (green). (D) Blocking SIGNR1-hFc binding to P. UF1 by zymozan. (E) Relative expression of Signr1 and Signr3 genes in colonic tissue from mice gavaged with P. UF1 or PBS (n = 5 tissues/group). (F) Representative data analyses of SIGNR1+ DCs derived from mice gavaged with P. UF1 or PBS (n = 4 mice/group). (G) Signr1+/+ and Signr1–/– mice were gavaged 4 times with PBS (red), P. UF1 (green), or ΔdlaT P. UF1 (blue) and orally infected with ΔactA L. m. One group of mice was infected with actA L. m3pep (gray), and colonic responses were analyzed 7 days later. Representative data of flow plots, percentages, and total cell counts of Th17 cells, Th1 cells, and IL-10+TGF-β+ Tregs (n = 4–5 mice/group). (H) Determination of ΔactA L. m burden in fecal samples of indicated groups (n = 4–5 fecal samples/group) in Signr1+/+ and Signr1–/– mice. (I) Socs1 expression by qRT-PCR in colonic tissue of ΔactA L. m–infected Signr1+/+ and Signr1–/– mice gavaged with P. UF1 or ΔdlaT P. UF1 (n = 5–7 samples/group). (J) Socs1 expression in BMDCs isolated from Signr1+/+ and Signr1–/– mice (n = 3 samples/group). BMDCs were treated with MEK inhibitor PD0325901 (PD) or PBS. Cells were incubated with P. UF1 (1:2 CFU) or PBS for 3 hours. Data are representative of 2 (D, E and G-J) or 3 independent experiments (B, C and F). Error bars indicate mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, 2-tailed unpaired t test (E and F) or ANOVA plus Tukey post test (G–J).
To evaluate any potential SIGNR1 binding to DlaT, DlaT was overproduced by P. UF1 and purified to demonstrate its interaction with SIGNR1-hFC (Supplemental Figure 9, C and D). No significant binding of DlaT to the SIGNR1-hFc complex was observed (Supplemental Figure 9E). Thus, further studies are required to elucidate the possible implication of P. UF1 S-layer mannosylation for its optimal binding to SIGNR1 to activate signals (e.g., SOCS1) in DCs controlling T cells in inflammatory conditions.
Mitigation of the intestinal inflammation by P. UF1.
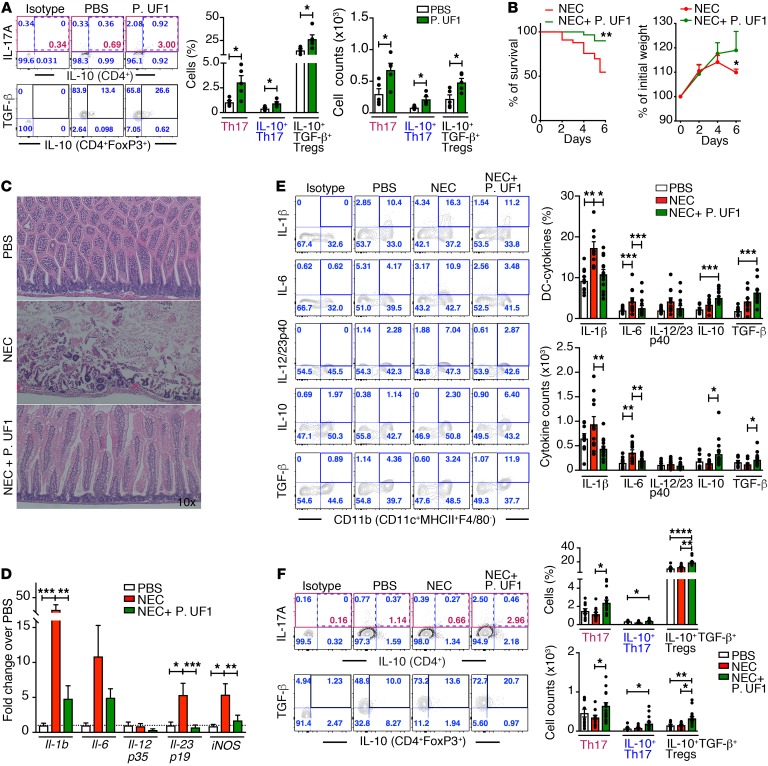
The functional microbiota of the mother shapes the immature immunity of the offspring during gestation (60). In contrast, alteration of the gut microbiota not only affects the immunity of the neonates, but it may also contribute to proinflammatory diseases, including NEC (61, 62). To determine whether P. UF1 regulates immunity in newborn mice, pregnant C57BL/6 dams were gavaged with P. UF1 or PBS. Once these dams had delivered their newborn mice, all newborn mice were euthanized 5 days later to analyze immune responses. Compared with the newborn mice of PBS-gavaged dams, newborn mice of dams gavaged with P. UF1 exhibited regulated TGF-β+IL-10+ DCs (Supplemental Figure 10A) and significantly induced IL-10+ Th17 cells and IL-10+ Tregs (Figure 9A). Additionally, to demonstrate any generation of group 3 innate lymphoid cells (ILC3) in newborn mice delivered by dams gavaged with P. UF1 or PBS, these newborns were further gavaged with P. UF1 or PBS 4 times. Twelve days later, the activation of small intestinal ILC3 of these groups of newborn mice was analyzed. Here, the frequency of IL-17A+IL-22+ ILC3 in P. UF1–gavaged newborn mice was significantly increased when compared with control newborn mice (Supplemental Figure 10B and Supplemental Figure 13). To elucidate whether P. UF1 can mitigate pathologic conditions induced by NEC-like injury (63, 64), newborn mice (5 days of age) delivered by P. UF1–gavaged dams were compared with newborn mice of dams given PBS. The weight and survival rates were significantly increased in the newborn mice of P. UF1–gavaged dams that were also gavaged twice with P. UF1 during NEC-like injury (Figure 9B). Newborn mice subjected to NEC-like injury also demonstrated significant villous sloughing and necrosis within the small intestine when compared with newborn mice gavaged with P. UF1 (Figure 9C). P. UF1 also significantly reduced the expression of iNOS, Il-1b, Il-6, and Il-23 transcripts implicated in NEC and tissue inflammation (14, 65) (Figure 9D). This correlated significantly with the regulation of IL-10+TGF-β+ DCs and elevated frequencies of IL-10+ Tregs, potentially controlling Th17 cells during the height of proinflammatory NEC-like injury, a trend that was less evident in the control newborn mice subjected to NEC-like injury (Figure 9, E and F). Moreover, intestinal IL-17A+IL-22+ ILC3 activation (66–68) was significantly enhanced in newborn mice gavaged with P. UF1 compared with the control group, which could potentially account for the reduction of pathology at the mucosal surface (Supplemental Figure 10C). Further studies are required for understanding the complementary maintenance of regulatory IL17A+IL-22+ ILC3 by P. UF1 in the pathologic conditions of NEC-like injury, in which the interplay between DCs and ILC3 implicated in the maintenance of epithelial barrier function (69–71) and CD4+ T cells (e.g., IL-10+ Th17 cells, Tregs) sustains functional intestinal homeostasis to potentially redirect deleterious signals (e.g., IL-1β) (72) involved in neonatal immature immunity during inflammation (e.g., NEC). Together, these data indeed demonstrate the potency of P. UF1 in controlling the proinflammatory cells (e.g., Th17 cells) (14, 65) toward regulatory cells potentially resisting disease progression.
Figure 9. Mitigation of NEC-like injury by P. UF1 in newborn mice.
(A) C57BL/6 pregnant dams were gavaged with P. UF1 or PBS twice/week during gestation. Five days after birth, newborn mice were sacrificed. Representative data of flow plots, percentages, and total cell counts of Th17 cells and IL-10+TGF-β+ Tregs in newborn mice. Each dot represents 3 pooled small intestinal tissues from each group of newborn mice. (B and C) Five days after birth, newborn mice of dams gavaged with P. UF1 or PBS were separated and subjected to NEC-like injury. Newborn mice gavaged with PBS or P. UF1 (107 CFU/mouse) on days 1, 3, and 5. Newborn mice were sacrificed 6 days later. Survival curve and weight incidence of the newborn mice subjected to NEC-like injury (B), and representative H&E sections of the small intestine of control newborn mice (PBS) or mice subjected to NEC-like injury with or without P. UF1. (C) Original magnification, ×10. (D) qRT-PCR demonstrating expression of proinflammatory genes in the small intestine of P. UF1–gavaged or untreated newborn mice subjected to NEC-like injury. Control newborn mice (PBS) with no NEC-like injury. (E and F) Representative data of flow plots, percentages, and total cell counts of IL-1β+, IL-6+, IL-12/23p40+, IL-10+, and TGF-β+ DCs (E), IL-10+ Th17 cells, and IL-10+ TGF-β+ Tregs (F) in the indicated groups. Data are pooled from 3 experiments in steady state (n = 15 newborn mice/group, A) and for NEC-like injury (PBS, n = 10 newborn mice; NEC, n = 12 newborn mice; NEC + P. UF1, n = 17 newborn mice, E) or 4 experiments (NEC, n = 33 newborn mice; NEC + P. UF1, n = 30 newborn mice, B). Error bars represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, 2-tailed unpaired t test (A and B) or Kruskal-Wallis test (D–F).
Discussion
Collectively, nutritional HBM and the bacterial species within (73, 74) not only govern both the development and the health of newborn infants (75, 76), but also affect optimal bacterial composition and related metabolites of the neonatal gut (77), thus influencing intestinal immunity and disease risk later in life (12). Yet mechanistic insights highlighting the critical role of HBM feeding are limited and require further investigation for understanding the underlying inputs promoting protective and regulated immune signaling that may be fine-tuned by gut microbiota and their induced metabolites. With this notion in mind, our studies demonstrate that HBM directs the fate of the bacterial composition of preterm infants and promotes the propagation of beneficial bacteria, including P.UF1, within preterm infants’ gut microbiota; this critical trend was lacking in preterm infants fed formula. Importantly, this deficiency in the levels of beneficial bacteria undoubtedly contributes to uncontrolled inflammation that may potentially result in serious proinflammatory diseases such as NEC, wherein functional gut homeostasis is significantly impaired (14). This was evidenced by the fact that transfaunation of GF mice with FF fecal microbiota augmented proinflammation; however, addition of P. UF1 to this transfaunation mitigated the inflammatory responses and increased protective Th17 cells and Tregs. Such immune regulation was also shown in GF mice monoassociated with P. UF1, where, in addition to regulated innate and T cell responses, particularly those of Th17 cells and Tregs, critical regulatory metabolites (e.g., tryptophan, phenylalanine) (78) were also induced.
It was recently demonstrated that the differentiation of intestinal Th17 cells is highly dependent on their constant interaction with the gut microbiota, including SFB, a delicate association that determines the nature of immune responses in steady state and with disease (79). Likewise, we demonstrate that bacterial-dependent Th17 cell formation and its regulation is tightly intertwined with the bacterial DlaT gene product and is significantly fortified by functional Tregs and potentially key metabolites (e.g., tryptophan, kynurenine) (80). Importantly, the formation of protective DlaT-specific Th17 cells was robustly sustained during L. m infection, a trend that was not observed using SFB-specific peptides expressed by L. m that skewed these cells toward Th1 responses (35). Here, the nature of the bacterial species (34) may influence T cell plasticity to either differentiate into gut regulatory Th17 cells (e.g., P. UF1) or proinflammatory Th1 cell responses (e.g., SFB). Further, P. UF1 not only regulated protective IFN-γ+ Th1 cells and IL-10+ Th17 cells (81) and sustained IL-10+ Tregs to successfully eliminate ΔactA L. m (82), but it also resisted microbial dysbiosis potentially induced by L. m infection. P. UF1 also elicited critical metabolites (e.g., citrulline) involved in immune protection. In this regard, DlaT not only influences Th17 cells, but also directs the bacterial molecular machinery, as its deletion profoundly changes the bacterial transcriptome orchestrating metabolomic cycles, resulting in further inflammation deleterious to protection against pathogen infection. Yet, does IL-17 play a role against intracellular pathogens such as L. m? Classically protective responses against L. m were considered to require IFN-γ+ Th1 cells (48). However, IFN-γ deficiency does not exclude the activation of CD4+ T cells against this pathogen (83). Moreover, protective Th17 cells were induced against several intracellular pathogens, including Salmonella typhimurium (84) and L. m (85), indicating that Th17 cells are not redundant against intracellular pathogens. Thus, synergistically induced Th1 and Th17 cell responses will swiftly resolve the infection and limit tissue damage (86). Here, we demonstrate this side of protective immunity directed by P. UF1 against L. m infection, a model in which both Th1 and Th17 cells were highly regulated. Studies are required to further elucidate underlying mechanisms implicated in protective Th17 cells that may bridge protective responses to fortify Th1 cell activation against intracellular pathogens.
CLRs, particularly SIGNR1, expressed by phagocytes (e.g., DCs) are critical pattern-recognition receptors interacting with microbial carbohydrate recognition domains (CRDs) (87) to efficiently internalize and phagocytize microbes in concert with other molecules, including complement receptors (88), and to potentiate the oxidative responses to pathogens (e.g., Candida albicans) via dectin-1/SyK-dependent signaling (89). One can speculate that the signaling pathways of SIGNR1 involve regulated immune complexes (e.g., SOCS1) whose activation may be dependent on a mannosylated S-layer binding to SIGNR1 to control proinflammation in DCs that influence regulatory Th17 cells (90, 91). Here, we show that SIGNR1 signaling participates in DC regulation by activating Socs1 to control proinflammatory signaling (e.g., IL-1β) (92), promoting IL-10+ Th17 cells by suppressing STAT1 signaling, thus limiting IFN-γ signaling in these cells (93). Disruption of SIGNR1 signaling (e.g., SOCS1) results in augmented pathogenic conditions, leading to further disequilibrium and lack of a functional homeostasis required for resolving L. m infection. Accordingly, future investigations are warranted to assess the impact of signals directed by SIGNR1 interaction with P. UF1 that mediate regulation during inflammatory conditions and whether mannose-containing S-layer constituents of P. UF1, including lipomannan synthesized by glycolipid pathways involving mannosyltransferases PimA and MptA (94), initiate regulatory signals (SOCS1) in intestinal DCs that skew pathogen-induced inflammation toward regulation.
Although the underlying pathological mechanisms involved in NEC are currently elusive and what is commonly termed NEC in human infants likely represents several different mechanistic pathways, induced pathogenic inflammation implicated in disease manifestation is of significant relevance (6, 95). To investigate whether P. UF1 ameliorates pathogenic inflammation induced in newborn mice, an experimental NEC-like injury was employed whereby severe epithelial damage can be observed (63, 64). In this murine model, P. UF1 regulated deleterious signals (e.g., iNOS, IL-1β) involved in the pathogenesis of NEC-like injury by controlling DCs and sustaining IL-10+ Tregs to potentially regulate IL-10+ Th17 cells (81, 96) and IL-17A+IL-22+ ILC3, all of which render a condition to oppose the manifestation of NEC-like injury in newborn mice. Such regulatory conditions were also established by feeding P. UF1 to pregnant dams compared with feeding it to control newborn mice after birth. In this permutation, P. UF1 coordinates critical signals in DCs to produce protective IL-10 (97) and TGF-β; a trend that can potentially promote IL-10+ Tregs and their mobilization against ensuing pathogenic inflammation, as seen in newborn mice subjected to NEC-like injury. These encouraging data may pave the way to further elucidate critical transcriptic and metabolic factors embedded within the P. UF1 genomic machinery and their relevance in the control of pathogenic inflammation occurring with NEC in preterm infants and potential immunotherapeutic translation in the clinic setting in the near future.
In summary, the current studies elucidate mechanisms involved in the attenuation of inflammatory responses affecting cellular homeostasis via the interaction of P. UF1 with SIGNR1. This results in the regulation of proinflammatory signals (e.g., IL-1β) promoting functional T cell homeostasis (regulatory Th17 cells) to efficiently clear pathogens and to limit disease progression. Errors in these molecular mechanisms mobilize pathologic signals, resulting in destruction of intestinal homeostasis, leading to devastating proinflammatory diseases, including NEC. Thus, the current investigation may address some of the knowledge gap related to HBM feeding and its impact on the signaling complexes that directly reduce intestinal inflammation as well as distal tissue damage associated with intestinal disorders (e.g., NEC). The impact of P. UF1–induced metabolites on gut homeostasis remains to be determined in order to draw a clear conclusion on the critical metabolites involved in regulated immunity against pathogens and disease. Independent of such considerations, our data distinctly support the notion that HBM, the bacteria within (e.g., P. UF1), and their gene products (e.g., DlaT) critically regulate the proinflammatory environment potentially implicated in NEC affecting preterm infants. Accordingly, candidates for modulators of the local response with translational importance would be beneficial bacteria such as P. UF1. Future strategies to restore the balance of regulatory lymphocytes may provide novel approaches to dampen inflammation and to prevent inflammatory disease progression such as that seen with NEC.
Methods
Human fecal sample collection.
Preterm infants with gestational ages less than or equal to 32 completed weeks and birth weights less than or equal to 1,800 g were eligible for our study. Infants with major congenital anomalies or malformations were excluded. Infants from 3 University of Florida–affiliated hospitals who met the inclusion criteria were enrolled in the study shortly after birth. Stool samples were collected weekly from the study infants starting with meconium and continued until discharge. The samples were immediately stored at –80°C. The analyzed selected samples were derived from control infants who did not develop NEC or culture positive for sepsis (Supplemental Table 1). Two groups, each with 20 subjects, were selected and either received an HBMF diet or an FF diet. For each group, subject samples from 2 different time points were analyzed: the first group of samples was collected at 13 ± 2 to 3 days after birth with 11 ± 3 days of feeding, and the second group of samples was collected at 21 ± 3 days after birth with 19 ± 4 days of feeding.
Animals.
C57BL/6, B6.PL-Thy1a/CyJ, B6.129(Cg)-Foxp3tm3(DTR/GFP)Ayr/J (FoxP3DTR), B6.129S2-H2dlAb1-Ea/J (H2-Ab1–/–), Rag1–/–, and BALB/c mice were obtained from Jackson Laboratory. Signr1–/– mice and B6.129P2(Cg)-Rorctm2Litt/J (Rorc(γt)GFP/GFP) mice were provided by Huang Shau-Ku (Johns Hopkins University School of Medicine, Baltimore, Maryland, USA) and Liang Zhou (University of Florida). C57BL/6 GF mice were provided by the Animal Care Facility of the University of Florida and maintained there until the experiments were performed. Mice (5 days old or 6 to 8 weeks old) were maintained under specific pathogen–free, Helicobacter-free conditions.
First draft genome analyses of P. UF1.
Fecal samples from HBMF preterm infants (100 mg) were dissolved in sterile lectin-binding buffer (98), and biotinylated Concanavalin A (ConA, 100 μg) (Vector Laboratories) was added to the suspension. The fecal contents were incubated at 4°C for 3 hours and subsequently washed with lectin-binding buffer 3 times. The streptavidin-ferrous beads (Thermo Fisher Scientific) were then added to the washed fecal contents, allowing the ConA-bacteria complex to bind to the beads. The ferrous beads were washed 6 times with lectin-binding buffer in the presence of a magnet. The enriched bacteria, via bead complex, were spread over several MRS-lactate agar plates (MRS agar with 1% sodium lactate; w/v) to grow the separated bacteria for 3 days. Bacterial DNA of the colonies was isolated, and bacterial identity was determined by the amplification of 16S rRNA gene and sequencing analyses. One of the isolates was designated as P. UF1. Genomic DNA of the P. UF1 was isolated, purified, and subjected to PACBIO RS II for whole genome sequencing (99). The sequenced contigs by PACBIO RS II were assembled into 1 unique contig using SMRT Portal (Pacific Biosciences), which was then compared with known sequences of P. freudenreichii subsp. shermanii CIRM-BIA1 and of P. freudenreichii subsp. freudenreichii DSM20271T. A distinguishable prophage identified by PHAST (Phage Search Tool) was also shown in the comparison to demonstrate the key bacterial strain differences and the missing gaps visualized in a circular genome map. The circular map was generated using Blast Ring Image Generator (BRIG) (100). The genome of the P. UF1 was annotated and combined using Rapid Annotation Server (RAST), KEGG Orthology And Links Annotation (KOALA) (101), and Rapid Prokaryotic Genome Annotation (PROKKA) (102). The subsystem categories were summarized and visualized by using RAST. The genome sequence of P. UF1 was submitted to the NCBI’s Genome database (CP018002). The P. UF1 isogenic strains were constructed as described in Supplemental Experimental Procedures, using primers presented in Supplemental Table 2.
Ultra–high resolution metabolomic analyses.
Mouse fecal samples and P. UF1 bacterial cultures were collected and analyzed at the Department of Medicine Clinical Biomarkers Laboratory at Emory University (27, 103), as described in Supplemental Experimental Procedures.
Experimental NEC-like injury.
C57BL/6 female (dam) mice were gavaged during pregnancy with P. UF1 (109 CFU/dam) or PBS twice weekly. On day 5, newborn mice were subjected to experimental NEC-like injury. Briefly, the cecal contents of 3 healthy adult mice (6 weeks old) were cultured overnight at 37°C in regular bacterial culture media (1:1) (LB Broth Base, GIBCO, Invitrogen). The CFU of stock mixture bacteria was then determined. Multiple aliquots were frozen at –80°C. The day prior to inducing NEC-like injury, an aliquot was cultured for 16 hours at 37°C to gavage newborn mice with 107 CFU-cultured bacteria. Newborn mice were gavaged twice daily with 107 CFU of a standardized adult commensal bacterial mixture (104) for 4 days. Newborn mice were gavaged 6 times daily with formula (Similac Advance infant formula, Abbott Nutrition; Esbilac canine milk replacer, PetAg; at the ratio 2:1). Newborn mice were exposed to episodes of hypoxia (300 seconds in 95% N2, 5% O2) twice daily for 4 days. Newborn mice were observed closely for clinical signs, including severe abdominal distension, apnea, cyanosis, lethargy, and weight loss. Newborn mice from P. UF1–gavaged dams were also gavaged with P. UF1 (107 CFU/mouse) during experimental procedures on days 1, 3, and 5. Newborn mice were sacrificed 6 days later. PBS-gavaged newborn mice that remained with their mothers were used as controls.
Histopathology.
The severity of intestinal injury was determined by histopathology. The entire small intestine was fixed, sectioned, and stained with H&E. Stained sections were blindly evaluated by a boarded veterinary pathologist.
Statistics.
Statistical analyses were performed using GraphPad Prism (version 6.0 for Mac OS X). Mean and SEM values and statistical significance between 2 variables were determined by 2-tailed unpaired t tests. Normality was tested using the Shapiro-Wilk normality test. Where the groups follow a Gaussian distribution, parametric analyses were performed (2-tailed unpaired t test for 2 variables or 1-way ANOVA followed by Tukey’s post test for 3 or more variables). Where the groups did not follow a Gaussian distribution, nonparametric analyses were performed (Mann-Whitney U test for 2 variables or Kruskal-Wallis test followed by Dunn’s post test for 3 or more variables). Differences were considered to be significant at P < 0.05. For microbiota analyses, the Kruskal-Wallis test was used in LEfSe to perform statistical testing for microbiota compositions. Differences were considered to be significant at P values of less than 0.05, and only features with an LDA score of more than 2 were shown. For tests in differential gene expression analyses, the negative binomial (NB) model was applied as the statistical model to compute P values, followed by the Benjamini-Hochberg procedure to calculate q values (105). For metabolomic analyses, the differential expression of fecal metabolites among different groups was determined with t tests or ANOVA tests using Python, and pathway and network analyses were performed using Mummichog, a set of algorithms specifically designed for high-throughput metabolomics (106).
Study approval.
Fecal samples were collected after informed written consent from a parent and approval by the IRB (no. 386-2008) at the University of Florida. All animal studies were approved by the IACUC of the University of Florida under the following protocol numbers: 201406559, 201408484, and 201609388. Mice were maintained in accordance with the Animal Welfare Act and the Public Health Policy on Humane Care.
Author contributions
MM directed and designed the cellular and molecular experiments, which were executed by NC, YG, BS, MG, MZ, JLO, and FA. NC, MZ, YG, and BS performed the mouse work. YG, MG, and FA performed the molecular studies, gene deletion and complementation, transcriptomic studies, and generation of ΔactA L. m and ΔactA L. m3pep. SL and DJ performed, analyzed, and directed metabolomic studies. SL, KL, and MG conducted all bioinformatic work for metabolic data analyses. WGF analyzed the genome of P. UF1. MG and YG performed experiments involving microbiota, transcriptomic analyses, and P. UF1 genome-wide analyses. JN collected and provided the fecal samples. NC, YG, MG, BS, JLO, JN, FA, SL, DPJ, and MM analyzed and interpreted the data. MM wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH R01 DK109560 (to MM), the NIH/NCRR Clinical and Translational Science Award (to MM), and Gatorade Trust Funds Florida (to MM). We thank Melissa N. Valletti and Max R. Van Belkum for excellent technical assistance.
Version 1. 09/25/2017
Electronic publication
Version 2. 11/01/2017
Print issue publication
Footnotes
Conflict of interest: M. Mohamadzadeh and B. Sahay are the inventors of P. UF1 (US provisional application no. US2016/032096).
Reference information: J Clin Invest. 2017;127(11):3970–3986.https://doi.org/10.1172/JCI95376.
Contributor Information
Natacha Colliou, Email: collioun@ufl.edu.
Yong Ge, Email: gey@ufl.edu.
Bikash Sahay, Email: sahayb@ufl.edu.
Minghao Gong, Email: gmhhope@ufl.edu.
Mojgan Zadeh, Email: zadehm@ufl.edu.
Ken Liu, Email: ken.liu@emory.edu.
Dean P. Jones, Email: dpjones@emory.edu.
Shuzhao Li, Email: sli49@emory.edu.
Mansour Mohamadzadeh, Email: m.zadeh@ufl.edu.
References
- 1.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371(9606):75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romero R, Espinoza J, Gonçalves LF, Kusanovic JP, Friel L, Hassan S. The role of inflammation and infection in preterm birth. Semin Reprod Med. 2007;25(1):21–39. doi: 10.1055/s-2006-956773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groer MW, Luciano AA, Dishaw LJ, Ashmeade TL, Miller E, Gilbert JA. Development of the preterm infant gut microbiome: a research priority. Microbiome. 2014;2:38. doi: 10.1186/2049-2618-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim JC, Golden JM, Ford HR. Pathogenesis of neonatal necrotizing enterocolitis. Pediatr Surg Int. 2015;31(6):509–518. doi: 10.1007/s00383-015-3697-9. [DOI] [PubMed] [Google Scholar]
- 5.Besner GE. A pain in the NEC: research challenges and opportunities. J Pediatr Surg. 2015;50(1):23–29. doi: 10.1016/j.jpedsurg.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 6.Neu J, Walker WA. Necrotizing enterocolitis. N Engl J Med. 2011;364(3):255–264. doi: 10.1056/NEJMra1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donovan SM, Wang M, Li M, Friedberg I, Schwartz SL, Chapkin RS. Host-microbe interactions in the neonatal intestine: role of human milk oligosaccharides. Adv Nutr. 2012;3(3):450S–455S. doi: 10.3945/an.112.001859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murgas Torrazza R, Neu J. The developing intestinal microbiome and its relationship to health and disease in the neonate. J Perinatol. 2011;31 Suppl 1:S29–S34. doi: 10.1038/jp.2010.172. [DOI] [PubMed] [Google Scholar]
- 9.Morelli L. Postnatal development of intestinal microflora as influenced by infant nutrition. J Nutr. 2008;138(9):1791S–1795S. doi: 10.1093/jn/138.9.1791S. [DOI] [PubMed] [Google Scholar]
- 10.Bode L. Human milk oligosaccharides: prebiotics and beyond. Nutr Rev. 2009;67 Suppl 2:S183–S191. doi: 10.1111/j.1753-4887.2009.00239.x. [DOI] [PubMed] [Google Scholar]
- 11.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urbaniak C, et al. Effect of chemotherapy on the microbiota and metabolome of human milk, a case report. Microbiome. 2014;2:24. doi: 10.1186/2049-2618-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140(6):845–858. doi: 10.1016/j.cell.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 14.Egan CE, et al. Toll-like receptor 4-mediated lymphocyte influx induces neonatal necrotizing enterocolitis. J Clin Invest. 2016;126(2):495–508. doi: 10.1172/JCI83356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8(1):15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535(7610):85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimm V, Radulovic K, Riedel CU. Colonization of C57BL/6 mice by a potential probiotic bifidobacterium bifidum strain under germ-free and specific pathogen-free conditions and during experimental colitis. PLoS ONE. 2015;10(10):e0139935. doi: 10.1371/journal.pone.0139935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim YG, et al. Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science. 2017;356(6335):315–319. doi: 10.1126/science.aag2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011;23(4):473–480. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14(7):685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamada N, Seo SU, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13(5):321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 23.Coccia M, et al. IL-1β mediates chronic intestinal inflammation by promoting the accumulation of IL-17A secreting innate lymphoid cells and CD4(+) Th17 cells. J Exp Med. 2012;209(9):1595–1609. doi: 10.1084/jem.20111453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Negrete A, Majdalani N, Phue JN, Shiloach J. Reducing acetate excretion from E. coli K-12 by over-expressing the small RNA SgrS. N Biotechnol. 2013;30(2):269–273. doi: 10.1016/j.nbt.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnes MJ, Powrie F. Regulatory T cells reinforce intestinal homeostasis. Immunity. 2009;31(3):401–411. doi: 10.1016/j.immuni.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Veldhoen M, Hirota K, Christensen J, O’Garra A, Stockinger B. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med. 2009;206(1):43–49. doi: 10.1084/jem.20081438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S, et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol. 2013;9(7):e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Libert C, Dejager L. How steroids steer T cells. Cell Rep. 2014;7(4):938–939. doi: 10.1016/j.celrep.2014.04.041. [DOI] [PubMed] [Google Scholar]
- 29.Ulvik A, Midttun Ø, Pedersen ER, Nygård O, Ueland PM. Association of plasma B-6 vitamers with systemic markers of inflammation before and after pyridoxine treatment in patients with stable angina pectoris. Am J Clin Nutr. 2012;95(5):1072–1078. doi: 10.3945/ajcn.111.029751. [DOI] [PubMed] [Google Scholar]
- 30.Murakami K, Korbsrisate S, Asahara N, Hashimoto Y, Murooka Y. Cloning and characterization of the glutamate 1-semialdehyde aminomutase gene from Xanthomonas campestris pv. phaseoli. Appl Microbiol Biotechnol. 1993;38(4):502–506. doi: 10.1007/BF00242945. [DOI] [PubMed] [Google Scholar]
- 31.Lightfoot YL, et al. SIGNR3-dependent immune regulation by Lactobacillus acidophilus surface layer protein A in colitis. EMBO J. 2015;34(7):881–895. doi: 10.15252/embj.201490296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478(7368):250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sefik E, et al. Mucosal immunology. individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science. 2015;349(6251):993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geva-Zatorsky N, et al. Mining the human gut microbiota for immunomodulatory organisms. Cell. 2017;168(5):928–943.e11. doi: 10.1016/j.cell.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510(7503):152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24(2):179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Zhou ZH, McCarthy DB, O’Connor CM, Reed LJ, Stoops JK. The remarkable structural and functional organization of the eukaryotic pyruvate dehydrogenase complexes. Proc Natl Acad Sci U S A. 2001;98(26):14802–14807. doi: 10.1073/pnas.011597698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goto Y, et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity. 2014;40(4):594–607. doi: 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mucida D, Park Y, Cheroutre H. From the diet to the nucleus: vitamin A and TGF-beta join efforts at the mucosal interface of the intestine. Semin Immunol. 2009;21(1):14–21. doi: 10.1016/j.smim.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.ten Broeke T, Wubbolts R, Stoorvogel W. MHC class II antigen presentation by dendritic cells regulated through endosomal sorting. Cold Spring Harb Perspect Biol. 2013;5(12):a016873. doi: 10.1101/cshperspect.a016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohamadzadeh M, et al. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4623–4630. doi: 10.1073/pnas.1005066107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68(5):869–877. doi: 10.1016/0092-8674(92)90030-G. [DOI] [PubMed] [Google Scholar]
- 44.Ge Y, Zhao N, Hu X, Shi T, Cai Q, Yuan Z. A novel transcriptional activator, tubX, is required for the stability of Bacillus sphaericus mosquitocidal plasmid pBsph. J Bacteriol. 2014;196(24):4304–4314. doi: 10.1128/JB.01855-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang SC, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203(10):2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 47.Chung H, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149(7):1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O’Garra A, Murphy KM. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260(5107):547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 49.Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992;68(3):521–531. doi: 10.1016/0092-8674(92)90188-I. [DOI] [PubMed] [Google Scholar]
- 50.Zhou L, Littman DR. Transcriptional regulatory networks in Th17 cell differentiation. Curr Opin Immunol. 2009;21(2):146–152. doi: 10.1016/j.coi.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benson A, et al. Microbial infection-induced expansion of effector T cells overcomes the suppressive effects of regulatory T cells via an IL-2 deprivation mechanism. J Immunol. 2012;188(2):800–810. doi: 10.4049/jimmunol.1100769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ravindran R, et al. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science. 2014;343(6168):313–317. doi: 10.1126/science.1246829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol. 2007;8(2):191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 54.Wang Z, et al. Regulatory T cells promote a protective Th17-associated immune response to intestinal bacterial infection with C. rodentium. Mucosal Immunol. 2014;7(6):1290–1301. doi: 10.1038/mi.2014.17. [DOI] [PubMed] [Google Scholar]
- 55.Geijtenbeek TB, van Vliet SJ, Engering A, ‘t Hart BA, van Kooyk Y. Self- and nonself-recognition by C-type lectins on dendritic cells. Annu Rev Immunol. 2004;22:33–54. doi: 10.1146/annurev.immunol.22.012703.104558. [DOI] [PubMed] [Google Scholar]
- 56.Lightfoot YL, et al. SIGNR3-dependent immune regulation by Lactobacillus acidophilus surface layer protein A in colitis. EMBO J. 2015;34(7):881–895. doi: 10.15252/embj.201490296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Srivastava V, et al. Toll-like receptor 2 and DC-SIGNR1 differentially regulate suppressors of cytokine signaling 1 in dendritic cells during Mycobacterium tuberculosis infection. J Biol Chem. 2009;284(38):25532–25541. doi: 10.1074/jbc.M109.006221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.den Dunnen J, Gringhuis SI, Geijtenbeek TB. Dusting the sugar fingerprint: C-type lectin signaling in adaptive immunity. Immunol Lett. 2010;128(1):12–16. doi: 10.1016/j.imlet.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 59.Yang J, et al. Pathological axonal death through a MAPK cascade that triggers a local energy deficit. Cell. 2015;160(1-2):161–176. doi: 10.1016/j.cell.2014.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gomez de Agüero M, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351(6279):1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 61.Torrazza RM, Neu J. The altered gut microbiome and necrotizing enterocolitis. Clin Perinatol. 2013;40(1):93–108. doi: 10.1016/j.clp.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neu J. The microbiome during pregnancy and early postnatal life. Semin Fetal Neonatal Med. 2016;21(6):373–379. doi: 10.1016/j.siny.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 63.Egan CE, et al. Toll-like receptor 4-mediated lymphocyte influx induces neonatal necrotizing enterocolitis. J Clin Invest. 2016;126(2):495–508. doi: 10.1172/JCI83356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian R, et al. Characterization of a necrotizing enterocolitis model in newborn mice. Int J Clin Exp Med. 2010;3(4):293–302. [PMC free article] [PubMed] [Google Scholar]
- 65.Underwood MA, et al. Bifidobacterium longum subsp. infantis in experimental necrotizing enterocolitis: alterations in inflammation, innate immune response, and the microbiota. Pediatr Res. 2014;76(4):326–333. doi: 10.1038/pr.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen L, et al. IL-23 activates innate lymphoid cells to promote neonatal intestinal pathology. Mucosal Immunol. 2015;8(2):390–402. doi: 10.1038/mi.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sonnenberg GF, Artis D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat Med. 2015;21(7):698–708. doi: 10.1038/nm.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hepworth MR, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498(7452):113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rankin LC, et al. Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nat Immunol. 2016;17(2):179–186. doi: 10.1038/ni.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12(5):383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 71.Sugimoto K, et al. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. 2008;118(2):534–544. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coccia M, et al. IL-1β mediates chronic intestinal inflammation by promoting the accumulation of IL-17A secreting innate lymphoid cells and CD4(+) Th17 cells. J Exp Med. 2012;209(9):1595–1609. doi: 10.1084/jem.20111453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Díaz-Ropero MP, et al. Two Lactobacillus strains, isolated from breast milk, differently modulate the immune response. J Appl Microbiol. 2007;102(2):337–343. doi: 10.1111/j.1365-2672.2006.03102.x. [DOI] [PubMed] [Google Scholar]
- 74.Lara-Villoslada F, Olivares M, Sierra S, Rodríguez JM, Boza J, Xaus J. Beneficial effects of probiotic bacteria isolated from breast milk. Br J Nutr. 2007;98 Suppl 1:S96–100. doi: 10.1017/S0007114507832910. [DOI] [PubMed] [Google Scholar]
- 75.Daniels MC, Adair LS. Breast-feeding influences cognitive development in Filipino children. J Nutr. 2005;135(11):2589–2595. doi: 10.1093/jn/135.11.2589. [DOI] [PubMed] [Google Scholar]
- 76.German JB, Dillard CJ, Ward RE. Bioactive components in milk. Curr Opin Clin Nutr Metab Care. 2002;5(6):653–658. doi: 10.1097/00075197-200211000-00007. [DOI] [PubMed] [Google Scholar]
- 77.Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4653–4658. doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sikalidis AK. Amino acids and immune response: a role for cysteine, glutamine, phenylalanine, tryptophan and arginine in T-cell function and cancer? Pathol Oncol Res. 2015;21(1):9–17. doi: 10.1007/s12253-014-9860-0. [DOI] [PubMed] [Google Scholar]
- 79.Ivanov, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126(6):1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 80.Romani L, Zelante T, De Luca A, Fallarino F, Puccetti P. IL-17 and therapeutic kynurenines in pathogenic inflammation to fungi. J Immunol. 2008;180(8):5157–5162. doi: 10.4049/jimmunol.180.8.5157. [DOI] [PubMed] [Google Scholar]
- 81.McGeachy MJ, et al. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat Immunol. 2007;8(12):1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- 82.Khader SA, Gopal R. IL-17 in protective immunity to intracellular pathogens. Virulence. 2010;1(5):423–427. doi: 10.4161/viru.1.5.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harty JT, Bevan MJ. Specific immunity to Listeria monocytogenes in the absence of IFN gamma. Immunity. 1995;3(1):109–117. doi: 10.1016/1074-7613(95)90163-9. [DOI] [PubMed] [Google Scholar]
- 84.Thiemann S, et al. Enhancement of IFNγ production by distinct commensals ameliorates salmonella-induced disease. Cell Host Microbe. 2017;21(6):682–694.e5. doi: 10.1016/j.chom.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 85.Chung Y, et al. Epstein Barr virus-induced 3 (EBI3) together with IL-12 negatively regulates T helper 17-mediated immunity to Listeria monocytogenes infection. PLoS Pathog. 2013;9(9):e1003628. doi: 10.1371/journal.ppat.1003628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Veldhoen M. Interleukin 17 is a chief orchestrator of immunity. Nat Immunol. 2017;18(6):612–621. doi: 10.1038/ni.3742. [DOI] [PubMed] [Google Scholar]
- 87.Gringhuis SI, den Dunnen J, Litjens M, van der Vlist M, Geijtenbeek TB. Carbohydrate-specific signaling through the DC-SIGN signalosome tailors immunity to Mycobacterium tuberculosis, HIV-1 and Helicobacter pylori. Nat Immunol. 2009;10(10):1081–1088. doi: 10.1038/ni.1778. [DOI] [PubMed] [Google Scholar]
- 88.Kang YS, et al. A dominant complement fixation pathway for pneumococcal polysaccharides initiated by SIGN-R1 interacting with C1q. Cell. 2006;125(1):47–58. doi: 10.1016/j.cell.2006.01.046. [DOI] [PubMed] [Google Scholar]
- 89.Takahara K, Tokieda S, Nagaoka K, Takeda T, Kimura Y, Inaba K. C-type lectin SIGNR1 enhances cellular oxidative burst response against C. albicans in cooperation with Dectin-1. Eur J Immunol. 2011;41(5):1435–1444. doi: 10.1002/eji.200940188. [DOI] [PubMed] [Google Scholar]
- 90.Esplugues E, et al. Control of TH17 cells occurs in the small intestine. Nature. 2011;475(7357):514–518. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ohnmacht C, Marques R, Presley L, Sawa S, Lochner M, Eberl G. Intestinal microbiota, evolution of the immune system and the bad reputation of pro-inflammatory immunity. Cell Microbiol. 2011;13(5):653–659. doi: 10.1111/j.1462-5822.2011.01577.x. [DOI] [PubMed] [Google Scholar]
- 92.Ha YJ, et al. SOCS1 suppresses IL-1β-induced C/EBPβ expression via transcriptional regulation in human chondrocytes. Exp Mol Med. 2016;48:e241. doi: 10.1038/emm.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dalpke AH, Opper S, Zimmermann S, Heeg K. Suppressors of cytokine signaling (SOCS)-1 and SOCS-3 are induced by CpG-DNA and modulate cytokine responses in APCs. J Immunol. 2001;166(12):7082–7089. doi: 10.4049/jimmunol.166.12.7082. [DOI] [PubMed] [Google Scholar]
- 94.Tanne A, et al. A murine DC-SIGN homologue contributes to early host defense against Mycobacterium tuberculosis. J Exp Med. 2009;206(10):2205–2220. doi: 10.1084/jem.20090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Niño DF, Sodhi CP, Hackam DJ. Necrotizing enterocolitis: new insights into pathogenesis and mechanisms. Nat Rev Gastroenterol Hepatol. 2016;13(10):590–600. doi: 10.1038/nrgastro.2016.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Torchinsky MB, Garaude J, Martin AP, Blander JM. Innate immune recognition of infected apoptotic cells directs T(H)17 cell differentiation. Nature. 2009;458(7234):78–82. doi: 10.1038/nature07781. [DOI] [PubMed] [Google Scholar]
- 97.Emami CN, et al. Role of interleukin-10 in the pathogenesis of necrotizing enterocolitis. Am J Surg. 2012;203(4):428–435. doi: 10.1016/j.amjsurg.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Eriksson M, Johannssen T, von Smolinski D, Gruber AD, Seeberger PH, Lepenies B. The C-type lectin receptor SIGNR3 binds to fungi present in commensal microbiota and influences immune regulation in experimental colitis. Front Immunol. 2013;4:196. doi: 10.3389/fimmu.2013.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim KE, et al. Long-read, whole-genome shotgun sequence data for five model organisms. Sci Data. 2014;1:140045. doi: 10.1038/sdata.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428(4):726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 102.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 103.Yu T, Park Y, Li S, Jones DP. Hybrid feature detection and information accumulation using high-resolution LC-MS metabolomics data. J Proteome Res. 2013;12(3):1419–1427. doi: 10.1021/pr301053d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tian R, et al. Characterization of a necrotizing enterocolitis model in newborn mice. Int J Clin Exp Med. 2010;3(4):293–302. [PMC free article] [PubMed] [Google Scholar]
- 105.McClure R, et al. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41(14):e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li S, et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol. 2013;9(7):e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.