Abstract
Despite its central position in oncogenic intracellular signaling networks, the role of mTORC1 in epithelial development has not been studied extensively in vivo. Here, we have used the epidermis as a model system to elucidate the cellular effects and signaling feedback sequelae of mTORC1 loss of function in epithelial tissue. In mice with conditional epidermal loss of the mTORC1 components Rheb or Rptor, mTORC1 loss of function unexpectedly resulted in a profound skin barrier defect with epidermal abrasions, blistering, and early postnatal lethality, due to a thinned epidermis with decreased desmosomal protein expression and incomplete biochemical differentiation. In mice with mTORC1 loss of function, we found that Rho kinase (ROCK) signaling was constitutively activated, resulting in increased cytoskeletal tension and impaired cell-cell adhesion. Inhibition or silencing of ROCK1 was sufficient to rescue keratinocyte adhesion and biochemical differentiation in these mice. mTORC1 loss of function also resulted in marked feedback upregulation of upstream TGF-β signaling, triggering ROCK activity and its downstream effects on desmosomal gene expression. These findings elucidate a role for mTORC1 in the regulation of epithelial barrier formation, cytoskeletal tension, and cell adhesion, underscoring the complexity of signaling feedback following mTORC1 inhibition.
Keywords: Development
Keywords: Skin
Introduction
The mammalian target of rapamycin (mTOR) signaling pathway plays a critical role in integrating upstream nutrient inputs to regulate cell proliferation, growth, and metabolism (1). mTOR kinase exists in 2 complexes, of which mTOR complex 1 (mTORC1) is better studied and is classically inhibited by the macrolide rapamycin (1, 2). mTORC1 activation requires the small Ras-related GTPase Rheb (3), as well as an adaptor protein, Raptor (4). Following activation, mTORC1 triggers cell growth, stimulates cap-dependent protein translation, and regulates glucose uptake, de novo lipid synthesis, and mitochondrial activity and biogenesis (2). Despite its well-characterized role in cellular processes that facilitate tumor progression, mTORC1 inhibitors have been ineffective for the treatment of most solid cancers (5). Though this finding has largely been attributed to relief of feedback inhibition that mTORC1 activity exerts on upstream growth factor receptor signaling (6–11), the in vivo sequelae of mTORC1 inactivation have not been well studied in epithelial tissues that give rise to the majority of cancers in humans. Thus, fully characterizing the role of mTORC1 in epithelial biology is critical before we can devise strategies to circumvent tumor resistance to mTORC1 inhibition.
Among epithelial tissues, the skin is arguably among the best-characterized model systems, and is of additional interest with regard to mTOR function, since mTORC1 inhibitor treatment in transplant or cancer patients is associated with impaired wound healing and a variety of dermatologic toxicity ranging from mild to life-threatening (12–14). The epidermis is a self-renewing, stratified epithelium that serves as a physical and permeability barrier. Cell-cell adhesion — via adherens junctions, desmosomes, and tight junctions — as well as terminal differentiation of keratinocytes into a cornified layer, is essential for epidermal barrier formation (15). Desmosomes are calcium-dependent adhesive structures that anchor to tension-bearing keratin intermediate filaments and provide mechanical strength. Desmosomal isotypes are expressed in a differentiation-dependent pattern in the skin (16), and loss of desmosomal components in humans and mice by genetic or autoimmune mechanisms leads to a spectrum of epidermal barrier defects, from mild defects in cell-cell adhesion and suprabasal acantholysis, to biochemical differentiation defects with severe blistering and lethality (17–21).
In addition to intermediate filaments, the actin cytoskeleton is central to cell-cell adhesion. During adherens junction formation, cadherin-catenin clusters form at nascent cell-cell contacts, coupled with maturation of radial actin filaments into cortical actin bundles (22). Adherens junction assembly requires a fine balance in the activities of 2 members of the Rho family of small GTPases, Rac and Rho, and of the downstream effectors of Rho, mDia and Rho kinase (ROCK) (23–26). In turn, actomyosin contractility and ROCK signaling likely regulate desmosomal junction formation at multiple levels (27). While appropriate RhoA localization and actomyosin contractility are required for membrane translocation of desmosomal precursors, sustained Rho/ROCK activity can hamper this process in vitro (28). In addition, cytoskeletal contractility may exert complex effects on downstream gene expression through mechanotransduction, affecting the activity of multiple transcription factors (29). Despite this wealth of in vitro data, relatively little is known about the in vivo requirement of actomyosin contractility for desmosomal adhesion and epidermal barrier formation.
mTOR signaling perturbation in the epidermis has previously been associated with epithelial carcinogenesis (30), epidermal stem cell senescence (31, 32), and delayed wound healing (33). While this article was in preparation, an additional study demonstrated that mTOR kinase and mTORC1 loss of function is associated with impaired epidermal differentiation and barrier formation; however, the molecular mechanism of this finding was not elucidated, and no effects on cell-cell adhesion were described (34). Here, we used 2 separate genetic models to examine the consequences of conditional epidermal mTORC1 loss of function. Unexpectedly, we find that mTORC1 signaling in the epidermis is required for desmosomal cell-cell adhesion and keratinocyte biochemical differentiation, as exemplified by epidermal blistering, defective skin barrier function, and neonatal lethality of the conditional knockout mice. Mechanistically, these findings are a direct consequence of heightened cytoskeletal tension due to increased ROCK activity, resulting in impaired desmosomal gene expression and adherens junction maturation. Probing further upstream, mTORC1 loss leads to marked feedback upregulation of TGF-β signaling and downstream effects on cytoskeletal contractility, cell adhesion, and biochemical differentiation. These studies reveal a novel in vivo role of mTORC1 function in epithelial biology that has relevance for human disease.
Results
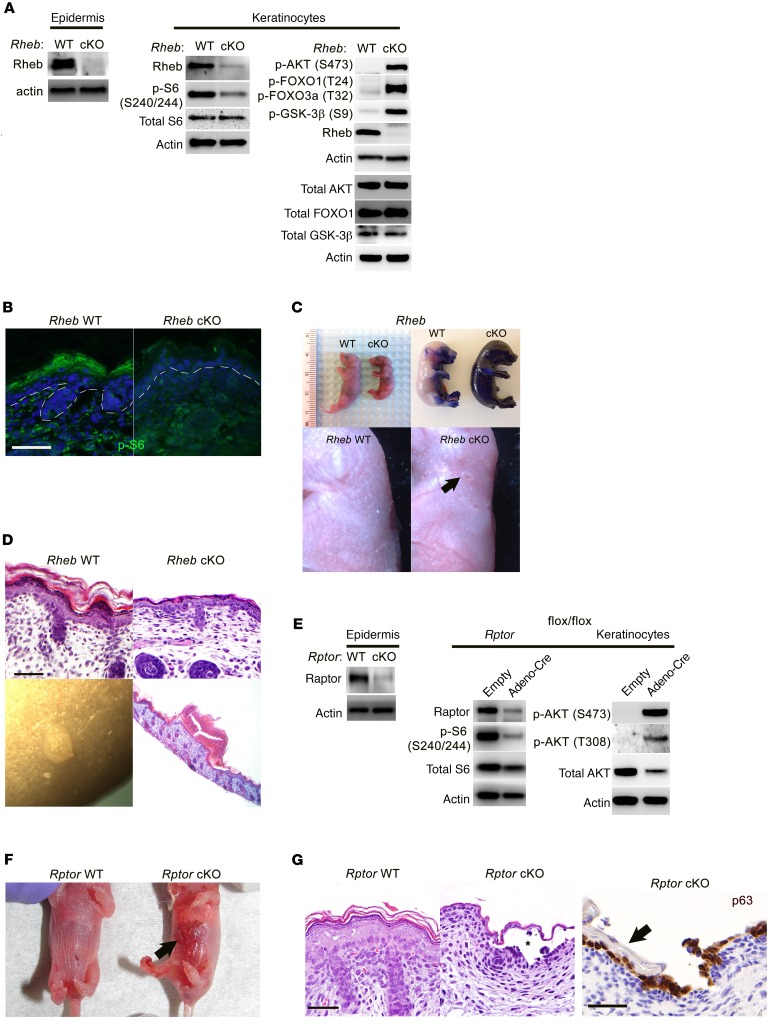
We crossed mice transgenic for Krt14-Cre (35) (which diffusely express Cre recombinase under the control of the keratin 14 promoter in the maturing epidermis by E14.5) to Rhebflox/flox (36) or Rptorflox/flox (37) mice to generate mice with mTORC1 loss of function in the epidermis, hereafter referred to as Rheb- or Rptor-cKO (conditional knockout) mice. As expected, Rheb- and Rptor-cKO epidermis and cultured keratinocytes showed evidence of mTORC1 loss of function and a well-described reciprocal increase in mTORC2 activity (38) by immunoblotting and/or immunofluorescence (Figure 1, A, B, and E). Pups from both cKO models were born at expected Mendelian ratios, but demonstrated uniform perinatal lethality with no pups surviving beyond P1. The lethality and smaller size of the Rheb-cKO pups compared with WT at P0 but not at E18.5 (Figure 1C) were suggestive of postnatal dehydration due to a defective epidermal barrier. This could be demonstrated by abnormally diffuse epidermal toluidine blue uptake at E18.5 in Rheb-cKO pups (Figure 1C). Moreover, both cKO models were characterized by taut, shiny skin with a prominent vascular pattern on gross examination (Figure 1, C and F), and a markedly thin epidermis with absence of full stratification on histology (Figure 1, D and G). Rheb-cKO pups had small subcorneal skin abrasions (Figure 1C) and blisters filled with neutrophilic debris (Figure 1D), findings seen in mouse models with mild desmosomal cell adhesion defects (17), while Rptor-cKO pups showed frank suprabasal epidermal blisters at birth (Figure 1, F and G).
Figure 1. Epidermal-specific mTORC1 loss-of-function models have skin barrier defects and evidence of impaired cell-cell adhesion.
(A) Immunoblotting of WT and Rheb-cKO P0 epidermis and keratinocyte cultures for markers of mTORC1 (p-S6) and mTORC2 (p-AKT, p-FOXO1, p-GSK-3β) activity. Total AKT, total FOXO1, and total GSK-3β were immunoblotted separately using the same biological replicate. (B) Immunofluorescence of WT and Rheb-cKO E18.5 epidermis for p-S6. (C) Rheb-cKO pups are smaller at P0, with thin, shiny skin (top left), and fail to exclude toluidine blue dye at E18.5 (top right), consistent with an epidermal barrier defect. Grossly visible abrasions (arrow, bottom right) and a prominent vascular pattern are seen in P0 cKO epidermis. (D) Representative histologic back skin sections from Rheb-cKO P0 pups reveal markedly thinned epidermis with loss of the upper keratinizing cell layers (top right) and grossly apparent subcorneal pustules (bottom left), filled with neutrophil debris on histology (bottom right). (E) Immunoblotting of WT and Rptor-cKO P0 epidermis and WT and inducible Rptor-cKO keratinocyte cultures for markers of mTORC1 (p-S6) and mTORC2 (p-AKT) activity. (F) Rptor-cKO pups have visible epidermal blisters at P0 (right, arrow). (G) Rptor-cKO pups have a thinned epidermis by histology (right half of left panel, H&E staining) with visible blistering (denoted by an asterisk). The epidermal blisters in Rptor-cKO P0 pups are suprabasal as demonstrated by p63 immunostaining that labels basal cells left behind after superficial epidermal layers (right panel, arrow) are shed. Scale bars: 30 μm throughout.
Epidermal biochemical differentiation and stratification was abnormal in both cKO models. This confirmed the findings of a recent study describing the phenotype of mice with conditional ablation of epidermal mTOR kinase or Rptor, published while this article was in preparation (34). Basal cell markers, including K5, K14, and p63, highlighted a normal to slightly hypertrophic basal layer in both cKO models by immunofluorescence (Supplemental Figure 1, A and C; supplemental material available online with this article; https://doi.org/10.1172/JCI92893DS1) and immunoblotting (Supplemental Figure 1D). In contrast, the spinous (marked by K1, K10) and granular layers (marked by loricrin, filaggrin, involucrin, transglutaminase 1) were dramatically thinned by immunofluorescence (Supplemental Figure 1, A and C) and immunoblotting (Supplemental Figure 1, B and D). Accordingly, for cultured P0 keratinocytes, switching from low (0.05 mM) to high (2.0 mM) calcium (calcium switch assay) failed to induce robust suprabasal biochemical differentiation in the Rheb-cKO model (Supplemental Figure 1E), and similar effects were observed when WT keratinocytes were pretreated with the mTORC1 inhibitor rapamycin before the calcium switch (Supplemental Figure 1F). Taken together, these data demonstrate that mTORC1 is required for keratinocyte biochemical differentiation and the generation of a full-thickness squamous epithelium.
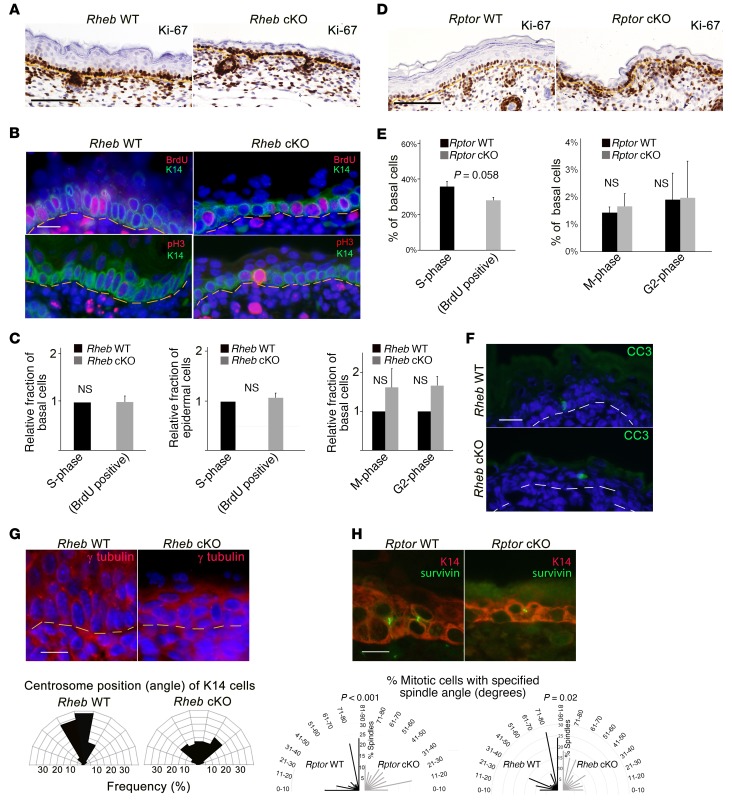
mTORC1 controls proliferation in some contexts (39); thus we tested whether epidermal thinning in Rheb-cKO mice might be due to decreased net proliferation. To avoid measuring a reactive increase in proliferation following birth due to the epidermal barrier defect, we examined expression of Ki-67 in the basal proliferative epidermal compartment at E18.5 and found that Ki-67 labeling was visually equivalent between WT and cKO epidermis for both cKO models (Figure 2, A and D). To exclude the possibility of cell cycle arrest in cKO cells labeling with the Ki-67 proliferation antigen, we also performed BrdU labeling and immunofluorescence for the mitotic marker phospho–histone H3 (pH3) in E18.5 epidermis or cultured keratinocytes and found the proportion of S- and M-phase basal cells to be similar between WT and Rheb-cKO mice (Figure 2, B and C). Similar to recent observations by the Eming group (34), in the Rptor-cKO epidermis, there was an indication of a lower BrdU incorporation rate in the basal layer; however, this just missed statistical significance, and there was no indication of decreased pH3 expression (Figure 2E). Cleaved caspase-3 expression (a marker of apoptosis) was negligible in both WT and Rheb-cKO E18.5 epidermis, labeling only rare cells (Figure 2F). Taken together, these data suggest that there may be a subtle proliferation defect in the Rptor-cKO mice contributing to the thin-skin phenotype that is not seen in the Rheb-cKO mice, and may be potentially attributable to previously described mTORC1-independent effects of Rptor on M-phase progression (40).
Figure 2. mTORC1 loss of function does not significantly alter epidermal proliferation or apoptosis, but is associated with abnormal mitotic polarization.
(A) Ki-67 immunohistochemistry in E18.5 Rheb WT and cKO epidermis. Scale bar: 75 μm. (B) BrdU labeling detected by immunofluorescence (top panels) in E18.5 WT or Rheb-cKO epidermis. Scale bar: 60 μm. Immunofluorescence for phospho–histone H3 (pH3), an M-phase marker (bottom panels), in E18.5 WT and Rheb-cKO epidermis. Scale bar: 60 μm. (C) Mean proportion of basal or total epidermal BrdU-labeled (r = 3, n > 20, ×200 fields each) or pH3-positive cells (r = 3, n > 33, ×200 fields each) in WT versus Rheb-cKO E18.5 epidermis (at least 15 ×200 fields quantified for each; error bars represent SEM; P values indicated are by Student’s t test). (D) Ki-67 immunohistochemistry in E18.5 Rptor WT or cKO epidermis. Scale bar: 75 μm. (E) Mean proportion of basal cells labeled with BrdU (left panel) or pH3 (right panel) in WT versus Rptor cKO E18.5 epidermis (r = 8; at least 20 ×200 fields were quantified for each; error bars represent SEM; P values indicated are by Student’s t test). (F) Representative immunofluorescence for cleaved caspase-3 (CC3) in WT and Rheb-cKO E18.5 epidermis. Scale bar: 60 μm. (G) Immunofluorescence for γ-tubulin in WT and Rheb-cKO E18.5 epidermis (top). Scale bar: 60 μm. Angles between centrosomal axis and plane of basement membrane in Rheb WT versus cKO cells (bottom) (r = 3; minimum of 15 ×200 fields quantified for each). (H) Representative immunofluorescence for the spindle midbody marker survivin (green) and K14 (red) in WT versus Rptor-cKO E17.5 epidermis (top panel). Radial histogram of the angle of basal cell division in WT (r = 8, n = 128) versus Rptor-cKO (r = 9, n = 61) (bottom panel, left) or WT (r = 4, n = 77) versus Rheb-cKO (r = 4, n = 66) (bottom panel, right) (P values indicated by χ2 test).
Since the Rptor-cKO epidermis is more dramatically thinned than the Rheb-cKO epidermis, we cannot exclude a contribution of decreased keratinocyte proliferation to the Rptor-cKO phenotype. However, the apparent lack of a proliferation defect in the Rheb-cKO model suggested the possibility of additional mechanisms contributing to the defective stratification seen in both models with mTORC1 loss of function. By K14 immunostaining, we noted that the basal-layer keratinocytes of the Rheb-cKO epidermis appeared poorly polarized and rounded, in contrast to the columnar polarized basal cells in WT epidermis (Figure 2B). Since cell polarity defects can diminish the number of asymmetric mitoses and impair epidermal stratification (41), we examined a marker of cell polarity in cKO keratinocytes by measuring the centrosome position (42, 43). In contrast to WT cells, where the centrosome polarizes to the apical side of the columnar basal cell nucleus, the centrosomes were haphazardly located in Rheb-cKO cells, consistent with an underlying polarity defect (Figure 2G). Potential interactions between centrosome and mitotic spindle positioning may underlie stratification defects (44). To examine whether the ratio of symmetric and asymmetric mitoses was abnormal in cKO cells, we measured the angle of the mitotic spindle relative to the epidermal-dermal junction in E18.5 Rheb- and Rptor-cKO epidermis compared with controls. While WT mitoses were predominantly oriented perpendicular, with a minority parallel, to the dermal-epidermal junction, cKO mitoses appeared randomly distributed (Figure 2H). Thus, we conclude that impaired polarization and cell stratification likely contribute to the thinned epidermis with mTORC1 loss of function.
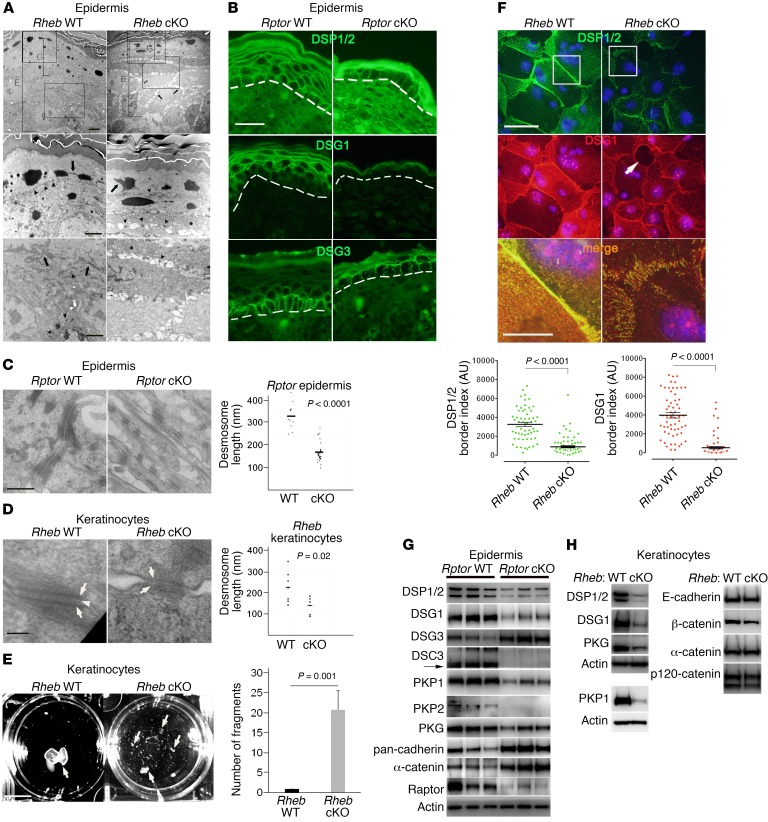
Keratinocyte biochemical differentiation and cell-cell adhesion are reciprocally regulated and calcium-dependent processes (45). The subcorneal pustules in Rheb-cKO mice and suprabasal blisters in Rptor-cKO mice were reminiscent of mice with loss of desmosomal cadherins (17, 18, 46) and suggested an underlying defect in cell-cell adhesion after mTORC1 loss of function. Indeed, on transmission electron microscopy (TEM), there were diffuse intercellular gaps in P0 Rheb-cKO epidermis, with a similar phenotype in keratinocyte culture (Figure 3, A and F). Though overall numbers of desmosomes appeared unchanged with epidermal mTORC1 loss of function (Figure 3A), there was prominent keratin tonofilament retraction, with reduced density of keratin filaments inserting on the desmosomal plaques (Figure 3, A, C, and D). In vivo, membranous levels of desmosomal components including desmoplakin (DSP1/2), desmoglein 1 (DSG1), and DSG3 appeared generally decreased by immunofluorescence (Figure 3B and Supplemental Figure 2A), and desmosomes were significantly shorter both in vivo and in vitro in cKO models by TEM (Figure 3, C and D). In addition, the midline of the desmosomal plaques was poorly formed in cultured keratinocytes from both cKO models (Figure 3D, arrowhead). Taken together, these findings are consistent with failure of in vivo desmosomal expression and maturation in keratinocytes with mTORC1 loss of function.
Figure 3. mTORC1 loss of function in the epidermis results in cell-cell adhesion defects.
(A) TEM of Rheb-cKO P0 epidermis (first row, E) demonstrates diminished granular layer (G) and intercellular gaps (first row, arrows). Desmosomes (second row, arrowheads) are evident (second row, arrow). Tonofilaments are collapsed (second and third rows, arrows) and not easily visible (third row, arrowheads) in cKO cells. Scale bars: 4 μm (top row), 2 μm (second and third rows). (B) Immunofluorescence for desmosomes (dotted line = dermal-epidermal junction). Representative images (r = 3). Scale bar: 30 μm. (C) Desmosomes in Rptor-cKO P0 epidermis have diminished keratin attachment by TEM (left panels; scale bar: 500 nm). Desmosomal length is quantified at right (bar represents mean length in nanometers; n = 12 and 29; P value by Student’s t test). (D) Desmosomes in Rheb-cKO keratinocytes have less dense cytoplasmic plaques (arrows) and poorly formed midline (arrowhead) (left panels; scale bar: 100 nm). Desmosome length is quantified at right (bar represents mean length in nanometers; n = 7 and 6; P value by Student’s t test). (E) Dispase dissociation for epithelial fragments (arrows) in Rheb WT and cKO keratinocyte monolayers (left panel). Scale bar: 1 cm. Quantification of dispase assay (right panel) (n = 4; error bars represent SEM; P value by Student’s t test). (F) Immunofluorescence for desmosomes in Rheb WT and cKO keratinocytes (top images), with intracellular gaps (arrow). Scale bar: 50 μm (top and middle images), 10 μm (bottom images). The third panel of merged staining is inset from the boxed areas in the top 2 panels. Quantification of desmosome protein border fluorescence intensity (bottom panels; r = 3, n = 58). Error bars represent SEM; P < 0.0001 by Mann-Whitney test. (G) Immunoblotting for junctional proteins in Rptor WT versus cKO P0 epidermis. (H) Immunoblotting for junctional proteins in Rheb WT versus cKO keratinocytes. PKP1 was immunoblotted separately using the same biological replicate.
In vitro, cellular adhesive strength of Rheb-cKO compared with WT keratinocytes was reduced, as measured by dispase dissociation assay (Figure 3E). Compared with WT keratinocytes, there was a marked decrease in in vitro levels of desmosomal components on the cell membrane in cKO cells by immunofluorescence (Figure 3F), surface biotinylation (see below), and keratinocyte membrane lysates (see below). Both in vivo (Figure 3G and Supplemental Figure 2, B and C) and in vitro (Figure 3H and see below), the decreased membrane levels of desmosomes reflected an underlying decrease in total levels of most desmosome proteins in both cKO models, including desmoplakin (DSP1/2), desmoglein 1 (DSG1), desmocollin 3 (DSC3), plakophilin 1 (PKP1), plakophilin 2 (PKP2), and plakophilin 3 (PKP3). Similarly, treatment of WT keratinocytes with rapamycin or mTOR kinase inhibitors (AZD8055 and torin1) resulted in decreased levels of DSP1/2 and DSG1 (see below). A subset of desmosome proteins undergo differential expression during keratinocyte maturation (47); for example, DSG1 is expressed in the superficial epidermis, while DSG3 is predominantly expressed in basal cells (Figure 3B). Since there was defective keratinocyte differentiation in the cKO models, the decrease in some desmosomal components (such as DSG1) could be exacerbated by this maturation defect. Similarly, total levels of DSG3, predominantly expressed in basal epidermis, were not decreased in Rptor-cKO epidermis (Figure 3G and Supplemental Figure 2B). Lower expression of DSG3 may have been potentially masked by a defect in differentiation in cKO cells or perhaps represented an in vivo compensatory effect not seen in vitro, since DSG3 levels were decreased in cKO keratinocytes in culture (see below). Such a response has previously been described in Netherton syndrome (48). Overall, however, the uniform decrease in most protein levels, including DSP1/2 and PKP3, which are expressed by keratinocytes throughout differentiation (Supplemental Figure 2B and see below), suggested that the failure of desmosome protein expression in keratinocytes with mTORC1 loss of function was at least partly independent of cellular differentiation.
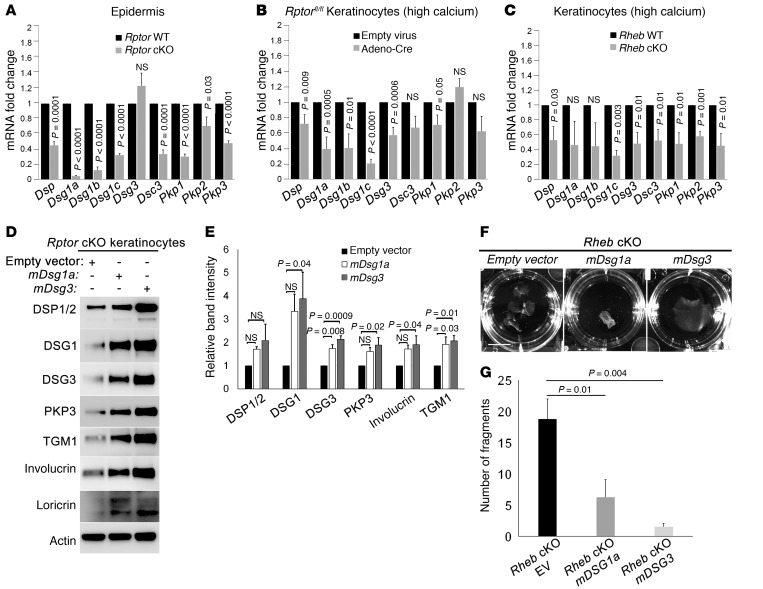
Desmosomal protein expression is regulated at multiple levels, including at the level of gene expression as well as by protein turnover (49). We examined mRNA expression for desmosomal genes in both cKO models. In vivo (Figure 4A) and in vitro in both high (Figure 4, B and C) and low (Supplemental Figure 4J) calcium concentrations, desmosomal gene expression was significantly and fairly uniformly decreased in cKO cells. One exception to this was Dsg3, which was expressed at normal levels in vivo (Figure 4A), though significantly decreased in vitro (Figure 4, B and C, and Supplemental Figure 4, J and K) consistent with DSG3 protein expression described above. If altered gene expression is contributing significantly to regulation of desmosomal adhesion in keratinocytes with mTORC1 loss of function, then transfection with exogenous desmosomal components should be sufficient to rescue the adhesion phenotype in cKO cells. Transient transfection of Rptor-cKO cells with either Dsg1a or Dsg3 significantly increased protein levels of other desmosomal components (Figure 4, D and E), potentially because of protein stabilization in nascent adhesion complexes. Expression of either construct was sufficient to rescue in vitro adhesion in Rheb-cKO cells by dispase dissociation assay (Figure 4, F and G). Because keratinocyte differentiation and cell-cell adhesion are reciprocally regulated and calcium-dependent processes (21, 45), we also tested whether rescue of the cell adhesion defect was sufficient to restore normal biochemical differentiation in vitro. Consistent with our hypothesis, expression of either Dsg1 or Dsg3 resulted in increased expression of involucrin, loricrin, and TGM1, markers of granular and spinous differentiation in keratinocytes (Figure 4, D and E). Taken together, these results indicate that mTORC1 regulates desmosomal adhesion at least in part via gene transcription and that failure of biochemical differentiation with mTORC1 loss of function is largely due to loss of normal desmosomal adhesion.
Figure 4. mTORC1 loss of function is associated with reduced desmosomal mRNA expression and is compensated by transfection of exogenous desmosomal cadherins.
(A–C) Quantitative real-time reverse transcriptase PCR of desmosomal gene transcripts in P0 Rptor WT versus cKO epidermis (A); in WT versus inducible Rptor-cKO keratinocytes grown in high calcium (B); and in Rheb WT versus cKO keratinocytes grown in high calcium (C) (r = 3 for each; error bars represent SEM; P values indicated are by Student’s t test). (D) Immunoblotting of inducible Rptor-cKO keratinocytes transiently transfected with empty vector, DSG1 mouse cDNA ORF, or DSG3 mouse cDNA ORF for desmosomal proteins (DSP, DSG1, DSG3, and PKP3) and markers of biochemical differentiation (TGM1, involucrin, and loricrin). (E) Densitometry quantification of representative immunoblot experiments shown in D (r = 3; error bars represent SEM; P values by 1-way ANOVA). (F) Dispase dissociation assay for Rheb-cKO keratinocytes after transient transfection with empty vector, DSG1 mouse cDNA ORF, or DSG3 mouse cDNA ORF. Scale bar: 1 cm. (G) Quantification of representative dispase assay experiments shown in F (r = 4; error bars represent SEM; P values by 1-way ANOVA). EV, empty vector.
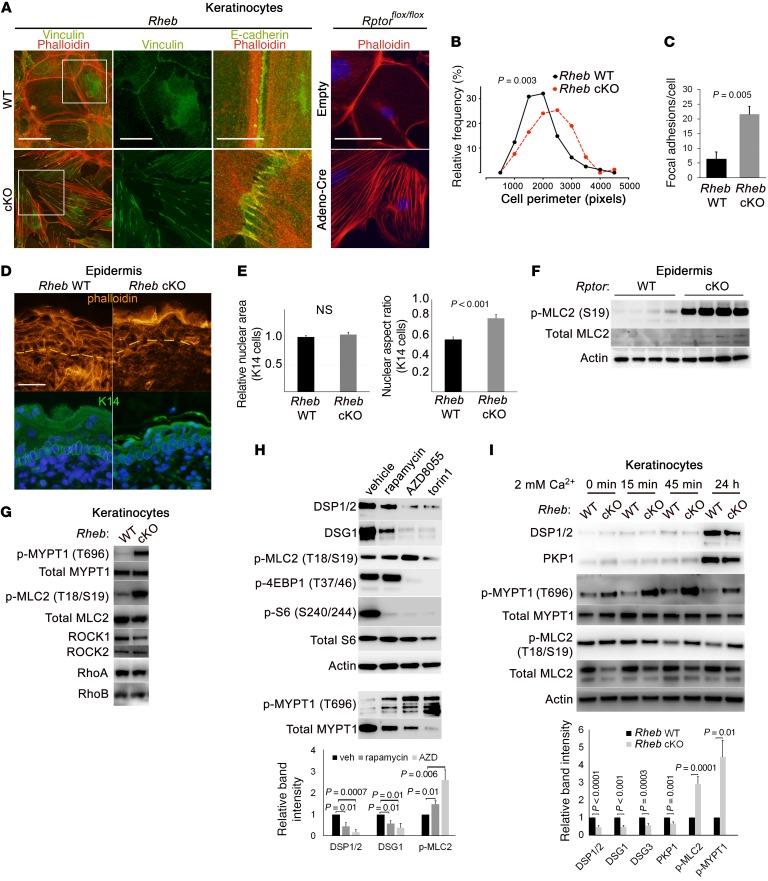
Next, we examined whether the cell-cell adhesion defect with mTORC1 loss of function was specific to desmosomal junctions. We queried expression and localization of other cell junction proteins in both cKO models. In contrast to desmosomes, adherens and tight junction components, such as E-cadherin, β-catenin, α-catenin, afadin, and ZO-1, showed normal or even higher total and membrane levels by immunoblotting and immunofluorescence in the cKO models in vivo (Figure 3G and Supplemental Figure 2E) and in vitro (Figure 3H, Supplemental Figure 2D, and see below) when compared with WT mice. However, immunofluorescence revealed that adherens junctions failed to mature into continuous adhesion belts and remained stalled in punctate adhesion zippers with radially oriented filamentous actin (F-actin) filaments (Figure 5A and Supplemental Figure 2F). Both maturation of adherens junctions (50, 51) and desmoplakin translocation into nascent desmosomes (27, 28) require appropriately regulated actomyosin contractility. We noted that keratinocytes with mTORC1 loss of function showed evidence of abnormally high cytoskeletal tension, with prominent F-actin stress fibers with absence of cortical actin belts (Figure 5A); increased cell size, likely due to cell spreading (Figure 5B); and increased focal adhesion formation (Figure 5C and Supplemental Figure 3, A and B). This abnormal cytoskeletal organization was reflected in vivo by lack of cortical F-actin belts in the cKO epidermis (Figure 5D), as well as by abnormalities in basal cell nuclear shape seen in the Rheb-cKO model (Figure 5, D and E), since nuclear shape is regulated in part by cytoplasmic actin contractility (52). Though nuclear area was unchanged in Rheb-cKO basal cells, the nuclear aspect ratio (ratio of nuclear width to nuclear height) was significantly increased in comparison with WT cells (Figure 5E). Collectively, these data suggest that mTORC1 loss of function results in abnormally increased actin contractility in keratinocytes.
Figure 5. Loss of desmosomal cell adhesion with epidermal mTORC1 loss of function is accompanied by increased cytoskeletal tension and ROCK activity.
(A) Immunofluorescence in Rheb and Rptor WT and cKO keratinocytes for desmosomal, focal adhesion (vinculin), and adherens junction markers (E-cadherin, phalloidin staining for actin filaments). Second panel of vinculin staining is inset from the boxed areas in the first panel. Scale bar: 50 μm (first panel), 25 μm (second panel), 10 μm (third panel), 50 μm (fourth panel). (B) Distribution of cell perimeter length (r = 6, n = 79) in Rheb WT versus cKO keratinocytes (P value by Mann-Whitney test). (C) Quantification of mean number of focal adhesions per cell (r = 3, n = 50) in Rheb WT versus cKO keratinocytes (P value by Student’s t test). (D) Immunofluorescence in Rheb WT and cKO P0 epidermis for actin (phalloidin) and basal cells (K14). Scale bar: 30 μm. (E) Mean nuclear area and mean nuclear aspect ratio (the ratio of nuclear width to nuclear height) for basal cells in P0 Rheb WT versus cKO epidermis (n > 100 cells each). (F) Immunoblotting of Rptor WT and cKO P0 epidermal lysates for ROCK activation. (G) Immunoblotting of Rheb WT and cKO keratinocyte lysates for ROCK activation. (H) Immunoblotting of WT cultured keratinocytes with or without mTORC1 (rapamycin, 200 nM) or mTORC1/2 inhibitors (AZD8055, 500 nM, and torin1, 1 μM) for desmosomes and ROCK activation (top). p-4EBP1, p-S6, and total S6 were immunoblotted in parallel, contemporaneously with DSP, DSG1, and p-MLC2. p-MYPT1 and total MYPT1 were immunoblotted separately using the same biological replicate. Densitometry quantification of immunoblots (bottom) (r = 3; P values are by Student’s t test). (I) Immunoblotting of Rheb WT and cKO keratinocytes for desmosomes and ROCK activation (top). Densitometry quantification of immunoblots, with additional desmosomal markers DSG3 and PKP1 (bottom) (r > 4; P values indicated are by Student’s t test). Error bars represent SEM.
Actomyosin contractility is principally orchestrated by small GTPases, including Rho GTPase and its downstream effector ROCK (53). Consistent with the prominent actin stress fibers in cKO keratinocytes, both cKO models also showed evidence of increased keratinocyte ROCK activity in vivo and in vitro, with higher levels of phosphorylated myosin phosphatase (p-MYPT1) and myosin light chain (p-MLC2) in both epidermis (Figure 5F and Supplemental Figure 3C) and keratinocytes (Figure 5G and see below). Increased Rho activity was also evidenced by increased levels of membrane-localized RhoA,B,C and decreased ROCK inhibitor RhoE in cKO keratinocytes (Supplemental Figure 3, D and E). Increased ROCK activity was also seen with pharmacologic mTORC1 inhibition, and the extent of MYPT1 and MLC2 phosphorylation was correlated with the differing efficacy of mTORC1 inhibition documented for these inhibitors (Figure 5H and ref. 5). Notably, both mTORC1 loss-of-function models showed expected increases in mTORC2 signaling (Figure 1, A and E), and mTORC2 has a known role in the regulation of Rho GTPases (54). However, the fact that the mTOR kinase inhibitors AZD8055 and torin1, which inhibit both mTORC1 and mTORC2, failed to rescue increased ROCK activity argues against the possibility that increased mTORC2 signaling was mediating the effects of mTORC1 loss of function on cytoskeletal tension in this system. Cumulatively, these data suggest that increased ROCK activity occurs downstream of mTORC1 loss of function in epidermal keratinocytes and independent of mTORC2 activation.
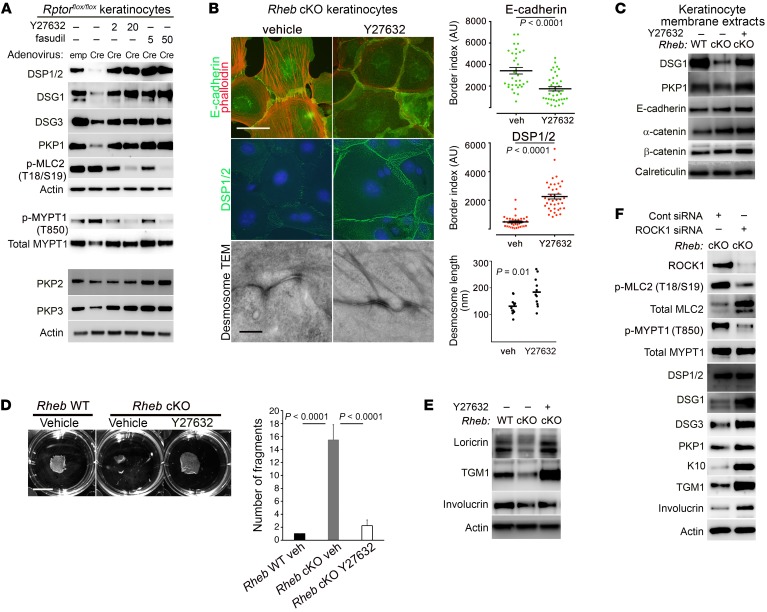
Loss of desmosomal components such as plakophilins can contribute to actomyosin contractility via RhoA activation (28). However, the increase in ROCK activity could be seen even in cKO keratinocytes cultured in low-calcium media, where desmosomal adhesions are largely unformed (Figure 5I), suggesting that ROCK activation occurs independent of desmosome loss in the context of mTORC1 loss of function. ROCK itself has a well-described role in the regulation of gene transcription networks, and is a key effector of cellular mechanotransduction in this capacity (29). Thus, we hypothesized that decreased desmosomal gene expression in cKO cells could itself be due to increased cytoskeletal tension mediated by ROCK activity. Treatment of cKO cells with ROCK inhibitors (Y27632 or fasudil) restored MYPT1 and MLC2 phosphorylation to normal levels in cKO cells (Figure 6A and Supplemental Figure 3F), eliminated actin stress fiber formation, and restored cortical actin belts in adhesive cells (Figure 6B). Moreover, ROCK inhibitors completely rescued total (Figure 6A and Supplemental Figure 3F) and membrane levels (Figure 6, B and C, and Supplemental Figure 3G) of desmosome proteins in both mTORC1 loss-of-function models. This rescue of desmosomal protein levels by ROCK inhibitors likely occurred at the gene expression level, because treatment with fasudil was sufficient to rescue desmosomal gene expression in Rheb-cKO keratinocytes, increasing levels significantly above those in WT keratinocytes (Supplemental Figure 4, J and K). In addition, Y27632 restored keratin filament insertion to desmosomal plaques (Figure 6B) and desmosomal size (Figure 6B) by TEM and enabled maturation of adherens zippers into continuous adherens junctions (Figure 6B). Finally, functional rescue of cell adhesion upon ROCK inhibition was evidenced by restoration of cKO keratinocyte monolayer integrity in dispase dissociation assays (Figure 6D).
Figure 6. ROCK inhibition or depletion rescues adhesion and biochemical differentiation defects in keratinocytes with mTORC1 loss of function.
(A) Immunoblotting of WT and inducible Rptor-cKO cell lysates with or without ROCK inhibition using Y27632 (2 or 20 μM) or fasudil (5 or 50 μM) for desmosomal components and ROCK activity markers. p-MYPT1/total MYPT1 and PKP2/PKP3 were immunoblotted separately from the upper panel using the same biological replicates. (B) Immunofluorescence for adherens junction components (top panels, left) and DSP1/2 (middle panels, left) in Rheb-cKO keratinocytes following Y27632 (10 μM) treatment. TEM (bottom panels, left) of Rheb-cKO desmosomes with or without Y27632 treatment. Scale bar: 50 μm (top 2 panels, left) and 500 nm (bottom panels, left). Quantification of DSP1/2 immunofluorescence (middle panel, right) and E-cadherin (top panel, right) (r = 3, n > 33 cell borders each) in Rheb-cKO keratinocytes with or without Y27632 (P value by Mann-Whitney test, DSP, or Student’s t test, E-cadherin). Quantification of desmosomal length on TEM for Rheb-cKO keratinocytes with or without Y27632 (bottom panel, right) (bar represents mean length in nanometers, n = 11 and 12, P values by Student’s t test). (C) Immunoblotting of membrane extracts from Rheb WT and cKO keratinocytes with or without Y27632 (20 μM) for desmosomal and adherens junction components. (D) Dispase dissociation (left panel) for Rheb WT or cKO keratinocytes with or without Y27632 (20 μM). Scale bar: 1 cm. Quantification of dispase assay experiments (right panel) (r = 4; P values by 1-way ANOVA). (E) Immunoblotting of Rheb WT or cKO keratinocytes with or without Y27632 (20 μM) for biochemical differentiation markers. (F) Immunoblotting of Rheb-cKO cells with or without ROCK1 siRNA (50 nm) for ROCK1 and markers of ROCK activity, desmosomal and biochemical differentiation markers. ROCK1, total MLC2, and total MYPT1 were immunoblotted in parallel, contemporaneously with the other markers. Error bars represent SEM throughout.
To confirm the specificity of the effects of pharmacologic ROCK inhibition, we also performed siRNA-mediated depletion of ROCK1 in Rheb-cKO keratinocyte cultures. ROCK1 depletion was sufficient to phenocopy the effects of pharmacologic ROCK inhibition, with decreased evidence of ROCK activity by p-MLC2 and p-MYPT1 immunoblotting and increased levels of total desmosomal proteins in Rheb-cKO cells (Figure 6F). We next tested whether rescue of cell adhesion by ROCK inhibition or depletion was sufficient to restore expression of calcium-induced differentiation markers (loricrin, transglutaminase 1, and involucrin) in cKO cells in culture. Indeed, ROCK inhibition by Y27632 or depletion by ROCK1 siRNA rescued Rheb-cKO keratinocyte biochemical differentiation in vitro (Figure 6, E and F, and Supplemental Figure 3H). Interestingly, fasudil treatment (Supplemental Figure 3F) or ROCK1 depletion (Supplemental Figure 3I) in WT keratinocytes also raised desmosomal protein levels and increased expression of markers of biochemical differentiation, suggesting that basal ROCK activity plays a role in regulating desmosomal levels and biochemical differentiation as well. Indeed, this effect of ROCK on biochemical differentiation has been reported previously (55, 56). Taken together, these results indicate that mTORC1 loss of function results in increased cytoskeletal contractility mediated by ROCK, which impairs desmosomal gene expression, cell-cell adhesion, and biochemical differentiation in keratinocytes.
Finally, we sought to elucidate which signaling pathways function upstream of ROCK activation in keratinocytes with mTORC1 loss of function by performing microarray-based differential expression analysis of E18.5 epidermis from Rheb WT and Rheb-cKO mice. Out of 39,601 gene-annotated transcripts we found 1,198 genes significantly (greater than 2 SD log2 fold change) upregulated and 815 downregulated in Rheb-cKO compared with Rheb WT epidermis, including downregulation of expected genes involved in keratinocyte maturation (KRT10, filaggrin, involucrin, corneodesmosin) as well as upregulation of those involved in wound healing/migration (KRT6A, KRT16, S100A8, S100A9, MMP3) and barrier loss response (many of the small proline-rich proteins) (Supplemental Table 1). Interestingly, only a subset of desmosomal genes were significantly altered in this analysis, for reasons that remain unclear. We used these genes to identify significantly enriched canonical pathways in the cKO epidermis using Ingenuity Pathway Analysis (Qiagen; https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/). Among the differentially expressed genes, the 2 top upstream regulator pathways predicted to be activated in Rheb-cKO epidermis included MYC (P = 7.80 × 10–32) and TGF-β signaling (P = 9.86 × 10–30) (Supplemental Figure 4B). Interestingly, we independently observed before expression analysis that c-MYC protein levels were dramatically increased in the Rheb-cKO epidermis by immunohistochemistry, potentially related to failure of keratinocyte maturation with mTORC1 loss of function (Supplemental Figure 4A). However, the prediction that TGF-β signaling was activated with Rheb cKO was intriguing given that TGF-β is a well-known upstream activator of ROCK signaling (57), and mice and humans with aberrant TGF-β activation in the skin have a thin, translucent epidermis similar to that seen with the Rheb-cKO mice (58, 59). In addition, some previous studies have suggested that mTORC1 may inhibit the related BMP signaling pathway in murine skin (60).
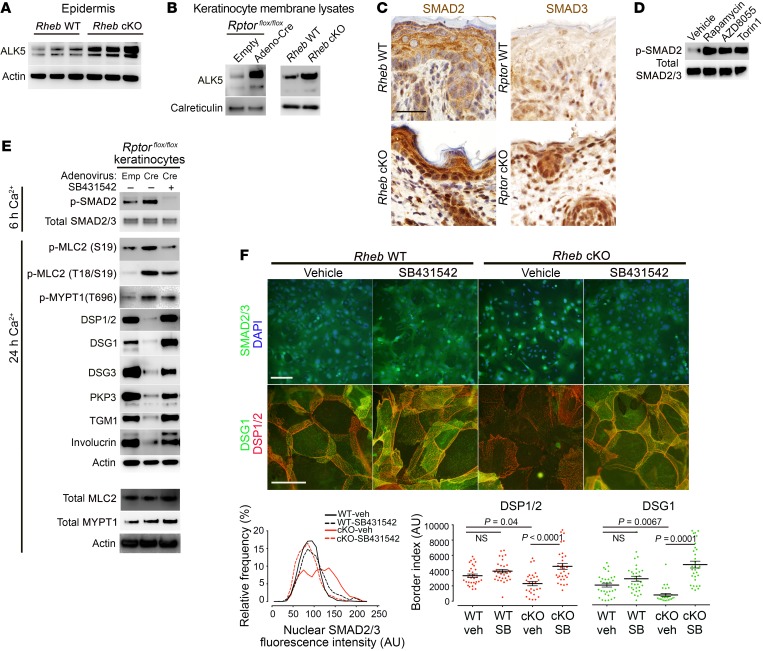
To validate the microarray predictions, we first examined expression of ALK5 (TGF-β receptor I) in Rheb- and Rptor-cKO P0 epidermis by immunoblotting and immunohistochemistry and found ALK5 levels to be increased relative to WT epidermis (Figure 7A and Supplemental Figure 4, C and D). Using cell membrane–enriched protein lysates from Rheb- and Rptor-cKO cells for immunoblotting, we found that membrane levels of ALK5 were also increased in cKO keratinocytes (Figure 7B). Downstream TGF-β signaling was activated with mTORC1 loss of function based on increased nuclear SMAD2 and/or SMAD3 levels in the cKO epidermis (Figure 7C) and keratinocyte cultures (see below in Figure 7F and Supplemental Figure 4G), and increased phosphorylated SMAD2 (p-SMAD2) levels in keratinocyte cultures (Figure 7E and Supplemental Figure 4F). Pharmacologic inhibition of mTORC1 with rapamycin, AZD8055, or torin1 also increased p-SMAD2 levels in WT keratinocytes (Figure 7D). Importantly, TGF-β treatment was sufficient to induce ROCK signaling (measured by p-MYPT1 and p-MLC2 levels) (Supplemental Figure 4E) and suppress desmosomal protein expression (DSP1/2, DSG1, DSG3, and PKP3) in WT keratinocytes (Supplemental Figure 4H).
Figure 7. TGF-β receptor (ALK) expression and downstream signaling are upregulated in vivo and in vitro with mTORC1 loss of function and mediate increased ROCK activity.
(A) Immunoblotting of P0 Rheb WT and cKO epidermis for ALK5. (B) Immunoblotting of membrane lysates from Rheb WT and cKO keratinocytes and keratinocytes with or without inducible Rptor-cKO for ALK5. Calreticulin levels are used to normalize for membrane protein. ALK5 and calreticulin were immunoblotted in parallel, contemporaneously. (C) Immunohistochemistry for ALK activity markers in WT, Rheb-cKO, and inducible Rptor-KO P0 epidermis. Scale bar: 30 μm. (D) Immunoblotting of WT keratinocytes with or without mTORC1 (rapamycin, 200 nM) or mTORC1/2 inhibitors (AZD8055, 500 nM, or torin1, 1 μM) for markers of ALK activity. (E) Immunoblotting of lysates from WT or inducible Rptor-KO keratinocytes with or without ALK inhibitor treatment (SB431542, 10 μM) for markers of ALK activity, ROCK activity, and desmosome and differentiation markers. Total MLC2 and total MYPT1 were immunoblotted separately using the same biological replicate. (F) Immunofluorescence for SMAD2/3 localization (top row; scale bar: 100 μm) and desmosome proteins (middle row; scale bar: 50 μm) in Rheb WT and cKO keratinocytes with or without SB431542 (10 μM). Quantification of nuclear SMAD2/3 fluorescence (bottom row, left) from experiments above (r = 3, n > 1,874 cells, P < 0.0001 by Kruskal-Wallis test). Quantification of desmosome protein border fluorescence (bottom row, right) (r = 3, n > 28 cells, P values indicated are by 1-way ANOVA, DSP, or Kruskal-Wallis test, DSG1).
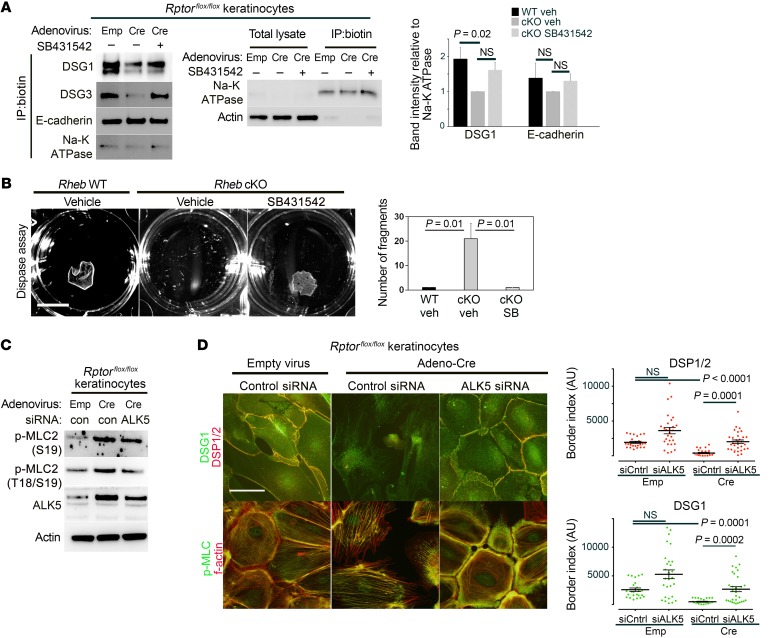
Finally, we tested whether ALK5/TGF-β signaling was required to activate ROCK signaling downstream of mTORC1 loss of function. We used SB431542, a pharmacologic ALK5 inhibitor, the efficacy of which was confirmed by a decrease in p-SMAD2 levels (Figure 7E) and a decrease in nuclear localization of SMAD2/3 (Figure 7F) in cKO keratinocytes. Strikingly, ALK5 inhibition with SB431542 was sufficient to suppress increased ROCK activity in cells with Rheb or Rptor loss, as measured by decreased p-MLC2 levels by immunoblotting (Figure 7E) and decreased F-actin stress fibers (data not shown). In addition, SB431542 rescued total desmosomal protein levels in cKO cells by immunoblotting (Figure 7E) and membranous desmosomal protein levels in cKO cells by immunofluorescence (Figure 7F) and surface biotinylation assays (Figure 8A). Inhibition of ALK5 was also sufficient to rescue expression of biochemical differentiation markers (Figure 7E) in keratinocytes with mTORC1 loss of function. Dispase assays on cKO keratinocyte monolayers demonstrated functional rescue of cell adhesion strength upon agitation with ALK5 inhibition (Figure 8B). Finally, to verify the specificity of the effects of ALK5 pharmacologic inhibition, we treated cells with ALK5 siRNA. Surprisingly, even relatively modest ALK5 knockdown by siRNA reduced p-MLC2 levels in keratinocytes with mTORC1 loss of function (Figure 8C and Supplemental Figure 5, A and B), as well as actin stress fiber formation (Figure 8D and Supplemental Figure 5D). ALK5 knockdown in cKO cells was sufficient to rescue cell-cell adhesion as evidenced by total (Supplemental Figure 5, A and B) and membranous (Figure 8D and Supplemental Figure 5D) desmosomal protein levels (DSP1/2, DSG1, DSG3, PKP3), and some measures of biochemical differentiation (loricrin, TGM1) were also rescued (Supplemental Figure 5A). Interestingly, similar effects were seen in WT cells with ALK5 inhibition (Supplemental Figure 4I) and ALK5 knockdown (Supplemental Figure 5C), suggesting a role for basal ALK5 signaling in the regulation of adhesion and differentiation. Cumulatively, these data support a model in which mTORC1 silencing leads to increased TGF-β receptor levels and signaling, which in turn mediates ROCK hyperactivity in keratinocytes. Increased cytoskeletal tension downstream of ROCK results in decreased desmosomal gene expression and decreased cell-cell adhesion, which contributes to impaired biochemical differentiation in the epidermis (Supplemental Figure 6).
Figure 8. TGF-β receptor (ALK) mediates increased ROCK activity and decreased desmosomal cell-cell adhesion.
(A) Immunoblotting following surface biotinylation and immunoprecipitation (IP) in WT or inducible Rptor-KO keratinocyte lysates with or without SB431542 treatment (10 μM) for membrane desmosome and adherens junction levels (left panel). Na-K ATPase was used to normalize for membrane protein. Enrichment of cell surface proteins in biotin immunoprecipitates, using Na-K ATPase as a control for membrane protein (middle panel). Densitometry quantification of immunoblot analysis of surface biotinylation experiments demonstrates significantly decreased membrane expression of DSG1 but not E-cadherin in Rptorflox/flox keratinocyte cultures treated with adenoviral Cre, compared with those treated with empty adenovirus. Treatment with SB431542 is sufficient to rescue membrane DSG1 expression (n = 3, error bars represent SEM, P values are by 1-way ANOVA). (B) Dispase assay (left) in Rheb WT or cKO keratinocytes with or without SB431542 (10 μM) (scale bar: 1 cm). Quantification of dispase assay experiments (right) (n = 3, error bars represent SEM, P values by 1-way ANOVA). (C) Immunoblotting of WT or inducible Rptor-KO keratinocytes with or without ALK5 siRNA for markers of ROCK activity. (D) Immunofluorescence of WT or inducible Rptor-KO keratinocytes with or without ALK5 siRNA for desmosomal protein levels and ROCK activity (left panels) (scale bar: 50 μm). Quantification of DSP and DSG1 border fluorescence (right panels) from experiments on the left (r = 3, n > 24 cells, P values by Kruskal-Wallis test).
Discussion
By virtue of 2 independent loss-of-function models, we have characterized the role played by mTORC1 signaling in epidermal morphogenesis. Abrogation of mTORC1 signaling, via deletion of Rheb or Rptor, resulted in a profound loss of epidermal stratification and barrier function, culminating in neonatal lethality. mTORC1 function was necessary for several aspects of barrier function: cell adhesion secondary to desmosomal maturation, biochemical differentiation, and cell stratification via mitotic spindle polarization. Mechanistically, Rheb or Rptor loss elicited marked feedback upregulation of ALK5/TGF-β receptor I expression and SMAD2/3 activity resulting in enhanced ROCK signaling and actomyosin contractility, which led to downregulation of desmosomal gene expression and subsequent failure of biochemical differentiation. Genetic rescue of desmosomal expression or genetic or pharmacologic inhibition of ALK5 or ROCK activity completely rescued desmosomal protein expression, keratinocyte biochemical differentiation, and cell adhesion in vitro. These findings highlight a unique and previously unreported role for mTORC1 in regulating desmosomal function and cell adhesion.
Epidermal maturation consists of multiple interconnected events, including cellular stratification and biochemical differentiation, both of which depend on aspects of cellular adhesion. We found that both biochemical differentiation and stratification were abnormal upon mTORC1 silencing. The failure of biochemical differentiation appeared to be at least in part downstream of the cell adhesion defect following mTORC1 loss of function, since exogenous desmosomal protein expression was sufficient to rescue the expression of some maturation markers in cKO keratinocytes in vitro. However, biochemical differentiation also requires appropriate cellular stratification, which was perturbed in mice with mTORC1 loss of function. Abnormal stratification was evidenced by the thin skin phenotype and was likely secondary to abnormal mitotic spindle polarization, resulting in an imbalance in symmetric and asymmetric mitosis. This phenotype is commonly seen in murine models with perturbed keratinocyte polarity and can be recapitulated by disruption of adherens junction cell-cell adhesion or cell-matrix adhesion, but, interestingly, not by desmoplakin inactivation (41). It remains uncertain whether the mitotic polarity and cell stratification defects in mice with mTORC1 loss of function are mechanistically related to the observed defects in desmosomal adhesion, cytoskeletal hypercontractility, and TGF-β signaling. In principle, the polarity and stratification defects could result from a parallel pathway downstream of mTORC1 silencing, contributing to the biochemical differentiation defects, and this will be a subject of future study.
Cell-cell contact in response to calcium first initiates adherens junction assembly, which in turn provides cues for desmosomal assembly. While adherens junction marker expression was largely normal, we observed a dramatic decrease in total expression levels and membrane localization of all desmosomal isotypes in both mTORC1 loss-of-function models, in vitro and in vivo. This defect correlated with the severity of the phenotype since the degree of blistering and neonatal survival correlated roughly with the extent of desmosomal protein expression in vivo in the 2 models. For reasons potentially attributable to differences in genetic background or non–mTORC1-dependent effects, Rheb-cKO pups exhibited a lesser degree of desmosomal protein loss in vivo, correlating with superficial epidermal pustules, rather than frank suprabasal blistering as seen in the Rptor-cKO pups. However, despite these phenotypic differences, both models showed a profound functional defect in keratinocyte cell adhesion, with smaller desmosomes and poor adhesion in vitro.
It is possible that differences in genetic background of the mouse strains examined also explain why the recent study examining Rptor and mTOR kinase loss of function by the Eming group did not note skin blistering, though profound defects in epidermal differentiation were described (34). Although both cell adhesion and differentiation are calcium-dependent processes, cell adhesion is known to precede biochemical differentiation in vitro (61). Desmosomal proteins have also emerged as important regulators of signaling pathways involved in differentiation (62). Accordingly, we found that rescue of desmosomal protein expression was sufficient to rescue biochemical keratinocyte differentiation in Rptor-cKO keratinocytes in vitro, strongly suggesting that the biochemical differentiation defect is downstream of the cell adhesion abnormality in these mice. Given these data, it is likely that desmosomal levels were decreased in the Eming Rptor loss-of-function mice; however, perhaps because of strain background or environmental differences, there may have been only subclinical evidence of desmosomal dysfunction (i.e., no visible blistering or very subtle acantholysis). The trauma of birth as a potential contributing factor in blistering/peeling of the skin (19) coupled with the short lifespan of Rptor-cKO pups may be an additional reason why subclinical desmosomal dysfunction may remain undetected.
Rho kinases are principal effectors of the small GTPase Rho, and play important roles in regulating actin cytoskeletal organization, cell adhesion, and gene expression via mechanotransduction (29, 63). Previous studies have demonstrated that Rho activity has a complex relationship with adherens junction formation; Rho signaling through mDia promotes peripheral localization of E-cadherin/α-catenin and maintains stable adherens junctions, while signaling via ROCK disrupts adherens junctions in epithelial cells (24, 25, 64). Lower levels of active Rho may preferentially activate mDia, while higher levels of active Rho favor signaling through ROCK (65). Thus, higher levels of active, membrane-bound Rho might favor ROCK hyperactivity in Rheb- and Rptor-cKO keratinocytes. In contrast, the role of the actin cytoskeleton and its regulators in desmosomal protein expression, assembly, and dissolution has only recently begun to be elucidated. It is known that DSP incorporation into junctions takes place in 3 distinct phases, with the last phase being cytochalasin-sensitive and actin-dependent (27). Desmosomal components can in turn reciprocally regulate actin dynamics: loss of PKP2 (28) and loss of plakoglobin (66) are associated with a global increase in RhoA activity, actomyosin contractility, loss of cortical actin bundling, and an increase in stress fibers and focal adhesions (67).
Our work adds an additional mechanism by which cytoskeletal regulators may alter desmosomal adhesion: via ROCK-mediated suppression of desmosomal gene expression. Though the downstream transcriptional regulators of this effect remain to be discovered, ROCK activity plays a key role in mechanotransduction, regulating transcription factors such as those involved in Hippo signaling and SRF. It is therefore conceivable that aberrant activation of ROCK may lead to suppression of desmosomal gene expression with or without mislocalization of key desmosomal components, which in turn can exacerbate actomyosin contractility, thereby initiating a cycle of events culminating in profound adhesive dysfunction.
Although the 2 isoforms of ROCK, ROCK1 and ROCK2, share highly conserved kinase domains, they exhibit vastly different biological functionalities. ROCK1 is critical for formation of stress fibers, focal adhesions, and phosphorylation of MLC2, a major substrate involved in actomyosin contractility, while ROCK2 is dispensable for and may even dampen these responses (55, 68–70). In proliferating WT and Rheb-cKO keratinocytes grown in low calcium, ROCK1 expression was greater than ROCK2 (data not shown); moreover, ROCK1 knockdown completely inhibited phosphorylation of MLC2 and MYPT1. RhoE is a known negative regulator of ROCK signaling that specifically inhibits ROCK1-induced stress fiber formation (71), and its membrane localization was significantly decreased in Rptor-cKO keratinocytes. We therefore conclude that under conditions of mTORC1 inhibition, ROCK1 is likely the principal driver of actomyosin contractility in murine keratinocytes. Accordingly, treatment of Rheb- and Rptor-cKO cells with ROCK inhibitors or ROCK1 siRNA rescued the expression of desmosomal proteins and increased terminal differentiation markers. In agreement with our findings, it has previously been reported that ROCK1 knockdown promotes terminal differentiation while ROCK2 knockdown inhibits terminal differentiation in human and murine keratinocytes (55, 56).
We examined differentially expressed genes and identified signaling pathways/upstream regulators that were activated following mTORC1 loss of function. One of the top regulators was TGF-β1 signaling, which was significantly activated in the context of Rheb loss. In mouse epidermis, TGF-β signaling regulates wound healing and hair follicle development (72, 73), and can either stimulate or inhibit epidermal proliferation, keratinocyte differentiation, and tumor formation, depending on the biological context (74–78). Interestingly, induction of active TGF-β1 during development also results in thinning of the epidermis with a shiny, taut erythemic skin and neonatal lethality (58, 79). A similar phenotype has also been noted in Loeys-Dietz syndrome in humans, characterized by a net increase in TGF-β signaling (59). Notably, mTORC1 inhibition by rapamycin or silencing of Rptor can induce phosphorylation of SMAD3 or SMAD1/5/8 in normal or tumorigenic prostate epithelial cells (80, 81) and hair follicle stem cells (60). However, a conclusive mechanism linking mTORC1 loss of function to TGF-β/BMP signaling has been elusive. In validating our microarray results, we found that both total and membrane-localized ALK5 expression was enhanced in epidermis and keratinocytes of Rheb- and Rptor-cKO mice, with a corresponding increase in SMAD2/3 phosphorylation and nuclear localization. The mechanisms underlying ALK5 upregulation remain unclear and are the subject of further study.
TGF-β signaling regulates Rho/ROCK signaling, actomyosin contractility, and stress fiber formation via multiple mechanisms (82, 83). We tested the hypothesis that TGF-β to ROCK signaling requires ALK5 activity in the context of mTORC1 loss of function, a finding that has been documented in other contexts (84). ALK5 inhibition or knockdown inhibited stress fiber formation and improved cortical organization of actin and phosphorylated MLC2. Importantly, ALK5 inhibition rescued total and membrane-localized desmosomal expression, keratinocyte differentiation marker expression, and cellular adhesive strength. Significantly, PKP2 loss can also stimulate TGF-β signaling, and this reciprocal regulation may reinforce the effects on both noted in our system (85). TGF-β signaling plays a key role in epithelial-mesenchymal transition, in which SLUG-induced desmosomal dissociation is a necessary and initial step (86). Our findings highlight a potential mechanistic basis for this effect.
Finally, since TGF-β is a known and potent stimulant of PI3K/mTOR signaling, elucidating the effects of mTORC1 inactivation on TGF-β signaling further broadens our understanding of the role of mTORC1 in growth factor receptor feedback suppression. Pharmacologic or genetic mTORC1 silencing in MEFs directly or indirectly stabilizes docking (IRS) (6, 7) and adaptor proteins (GRB10) (10, 11) required for insulin receptor/insulin growth factor receptor signaling and leads to upregulation of platelet-derived growth factor receptor (PDGFR) expression (87). Here, we describe a novel in vivo role for mTORC1 silencing in the regulation of epithelial TGF-β signaling, which could also promote tumor progression. Indeed, via increased TGF-β signaling, mTORC1 inactivation in the normal epidermis is sufficient to substantially inhibit cell differentiation, compromise desmosomal cell-cell adhesion, and heighten cytoskeletal contractility, cellular processes known to potentiate cell migration or even metastatic spread in epithelial tumors. Though these studies were performed in the mouse skin, their potential relevance to humans is further evidenced by the fact that mTORC1 inhibitor therapy for renal transplantation in humans is associated with delayed wound healing (12) and skin eruptions, sometimes requiring discontinuation of the drug (13). Precisely how mTORC1 silencing modulates TGF-β receptor levels, and whether this promotes tumor progression in carcinogenesis models, are important areas of future study and may be critical to developing strategies to circumvent tumor cell resistance to mTORC1 inhibitors.
Methods
Mice.
Mice expressing Cre recombinase under control of the human keratin 14 (K14) promoter (Krt14-Cre) [stock no. 004782, STOCK Tg(KRT14-Cre)1Amc/J] (35) and mice carrying loxP sites flanking exon 6 of Rptor (stock no. 013188, B6.Cg-Rptortm1.1Dmsa/J) (37) were from The Jackson Laboratory. Mice with loxP-flanked Rheb1 alleles were generated in the laboratory of P.F. Worley (36). Genomic DNA was isolated from tail snips and genotyping performed with primers as described in Supplemental Methods.
Barrier function assay.
To evaluate the integrity of the epidermal barrier, a toluidine blue penetration assay was performed on E18.5 embryos as described in Supplemental Methods.
BrdU labeling.
To measure in vivo proliferation, pregnant female mice were injected i.p. with a single dose of BrdU (100 mg/kg body weight) 3 hours prior to sacrificing. BrdU incorporation was detected by immunohistochemistry of paraffin-embedded sections using an anti-BrdU monoclonal antibody (mouse, 1:2,000; Fitzgerald Industries). The number of BrdU-positive cells was plotted as a relative fraction of K14-positive cells in each section.
Primary mouse keratinocyte cultures.
Primary mouse keratinocytes were isolated from newborn (P0) skin of pups using standard procedures as described in detail in Supplemental Methods. To knock out Rptor, Rptorflox/flox keratinocytes were infected with Cre recombinase–expressing or empty adenoviral vectors (Vector Biolabs) before plating. To induce cell-cell contacts, desmosomal maturation, and differentiation, the calcium concentration was elevated to 2 mM.
Reagents and antibodies.
Sources of primary antibodies are described in Supplemental Methods. The following reagents were used: rapamycin, AZD8055, and fasudil (LC Laboratories); Y27632 and torin1 (Cell Signaling Technology); SB431542 (Sigma-Aldrich); calcium (Lonza); Silencer Select Negative Control siRNA, ROCK1 siRNA, ALK5 siRNA, and Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific); and Dispase II protease (Sigma-Aldrich).
Histology and immunostaining.
Mouse skins were fixed in 10% neutral buffered formalin (Sigma-Aldrich), embedded in paraffin, sectioned at 4 μm, and used for immunohistochemistry. Immunostaining or immunofluorescence was performed as previously described (88) and is detailed in Supplemental Methods.
Protein lysate preparation and immunoblotting.
Epidermis was separated from the dermis following incubation of P0 pup skin with 3.8% ammonium thiocyanate for 10 minutes at room temperature and lysates prepared and immunoblotted as described previously (88) and in Supplemental Methods. Membrane lysates were prepared using the Subcellular Protein Fractionation kit for Cultured Cells (78840; Thermo Fisher Scientific). Cell surface biotinylation was performed using the Pierce Cell Surface Protein Isolation Kit (89881; Thermo Fisher Scientific).
siRNA-mediated gene silencing.
siRNAs (Silencer Select) for ROCK1, ALK5, and negative control were purchased from Thermo Fisher Scientific. Primary keratinocytes were transfected with 50 nm siRNA using Lipofectamine RNAiMAX reagent according to the transfection guidelines.
Plasmid transfection.
Rheb- and Rptor-cKO cells were transiently transfected with 2 μg of empty vector cDNA, mouse DSG1a cDNA (MR217075; Origene), or mouse DSG3 cDNA (MMM1013-211691952; Dharmacon) in 6-well plates, using Lipofectamine 3000 (L300008; Thermo Fisher Scientific), according to transfection guidelines.
RNA isolation and quantitative real-time reverse transcriptase PCR.
Total RNA was extracted using TRIzol (15596026; Invitrogen) for epidermis or RNeasy (74104; Qiagen) for keratinocytes according to guidelines. RNA was converted to cDNA using SuperScript III (18080051; Thermo Fisher Scientific) according to guidelines. mRNA levels were quantified using an ABI Prism 7900HT Real-Time PCR system (Applied Biosystems) with primers and probes detailed in Supplemental Methods. Threshold cycle (Ct) was obtained from PCR reaction curves and mRNA levels quantitated using the comparative Ct method with actin serving as reference.
Immunocytochemistry.
Primary keratinocytes were seeded on coverslips coated with fibronectin. Cells were fixed in 100% methanol at –20°C for 30 minutes or 4% paraformaldehyde for 15 minutes at room temperature, according to antibody specifications and immunocytochemistry performed using standard protocols (Supplemental Methods), and visualized using an Olympus BX41 epifluorescence microscope. Image analysis for border intensity, cell perimeter, nuclear immunostaining, and quantification was done in ImageJ (NIH) as detailed in Supplemental Methods.
Transmission electron microscopy.
Following standard fixation and embedding conditions detailed in Supplemental Methods, ultrathin sections were stained with uranyl acetate and lead citrate and examined with a transmission electron microscope (Hitachi 7600, equipped with Dual AMT CCD camera system).
Dispase dissociation assays.
Rheb WT and cKO cells were grown on uncoated 6-well plates. Confluent keratinocyte monolayers were pretreated with inhibitors for 24 hours prior to switch in 1.2 mM calcium for 18 hours. At assay, cells were rinsed with PBS and incubated in 2.5 U/ml dispase solution (Sigma-Aldrich) in PBS for 15 minutes at 37°C. Detached sheets were transferred to 15-ml conical tubes in PBS and stressed by mechanical rotations. Resulting fragments were visualized and images captured.
Microarray analysis.
Microarray analysis was performed as detailed in Supplemental Methods (GEO accession number GSE102026).
Statistics.
For image analysis, normal distribution was assessed using the D’Agostino and Pearson normality test. If normally distributed, statistical significance was determined with Student’s t test when comparing 2 experimental groups, or with 1-way ANOVA with Dunnett’s correction when comparing 3 or more experimental groups. If not normally distributed, statistical significance was determined with the Mann-Whitney test when comparing 2 experimental groups, or with the Kruskal-Wallis test with Dunn’s correction when comparing 3 or more experimental groups. For RNA and protein quantification, cell proliferation experiments, and dispase assays, statistical analysis was performed using unpaired Student’s t test or 1-way ANOVA with Tukey’s test for multiple comparisons. All tests assumed a 2-tailed deviation and were performed in Prism 7 (GraphPad). Where data are represented as a scatter plot or histogram, bars represent mean ± SEM. P values less than 0.05 were considered statistically significant.
Study approval.
All animal studies were carried out following protocol approval from the Johns Hopkins Animal Care and Use Committee.
Author contributions
Conception and design were performed by KA, TLL, BX, and PW. Data acquisition was performed by KA, PP, AD, LE, AJE, CCT, TLL, AS, AT, and HK. Data analysis was performed by KA, TLL, CCT, AD, DG, HK, AS, and AT. Drafting of the manuscript was performed by KA, TLL, AJE, and PW.
Supplementary Material
Acknowledgments
The authors thank Barbara Smith and Tyler Stephens for their assistance with the transmission electron microscopy. Funding for this research was provided in part by the Tuberous Sclerosis Research Program of the Congressionally Directed Medical Research Programs (CDMRP) (W81XWH-12-1-0186) (to TLL), and the National Cancer Institute (R01CA200858) and National Institute on Drug Abuse (DA00266).
Version 1. 09/25/2017
Electronic publication
Version 2. 11/01/2017
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(11):4001–4017.https://doi.org/10.1172/JCI92893.
Contributor Information
Kaushal Asrani, Email: kaushal.asrani@gmail.com.
Akshay Sood, Email: sood.86@gmail.com.
Alba Torres, Email: atorre16@jhmi.edu.
Dan Georgess, Email: georgess.dan@gmail.com.
Pornima Phatak, Email: pphatak@som.umaryland.edu.
Harsimar Kaur, Email: harsimar.bhatia@gmail.com.
Amber Dubin, Email: amberdub1nc@gmail.com.
C. Conover Talbot, Jr., Email: cct@jhmi.edu.
Loubna Elhelu, Email: loubna.elhelu@udc.edu.
Bo Xiao, Email: neuroscience365@gmail.com.
Paul Worley, Email: pworley@jhmi.edu.
References
- 1.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40(2):310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17(15):1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110(2):163–175. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 5.Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci Signal. 2009;2(67):pe24. doi: 10.1126/scisignal.267pe24. [DOI] [PubMed] [Google Scholar]
- 6.Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol. 2004;14(18):1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 7.Harrington LS, et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166(2):213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, et al. PDGFRs are critical for PI3K/Akt activation and negatively regulated by mTOR. J Clin Invest. 2007;117(3):730–738. doi: 10.1172/JCI28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carracedo A, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118(9):3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu PP, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332(6035):1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu Y, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332(6035):1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valente JF, et al. Comparison of sirolimus vs. mycophenolate mofetil on surgical complications and wound healing in adult kidney transplantation. Am J Transplant. 2003;3(9):1128–1134. doi: 10.1034/j.1600-6143.2003.00185.x. [DOI] [PubMed] [Google Scholar]
- 13.Liu LS, McNiff JM, Colegio OR. Palmoplantar peeling secondary to sirolimus therapy. Am J Transplant. 2014;14(1):221–225. doi: 10.1111/ajt.12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balagula Y, et al. Clinical and histopathologic characteristics of rash in cancer patients treated with mammalian target of rapamycin inhibitors. Cancer. 2012;118(20):5078–5083. doi: 10.1002/cncr.27505. [DOI] [PubMed] [Google Scholar]
- 15.Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6(4):328–340. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- 16.Delva E, Tucker DK, Kowalczyk AP. The desmosome. Cold Spring Harb Perspect Biol. 2009;1(2):a002543. doi: 10.1101/cshperspect.a002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chidgey M, et al. Mice lacking desmocollin 1 show epidermal fragility accompanied by barrier defects and abnormal differentiation. J Cell Biol. 2001;155(5):821–832. doi: 10.1083/jcb.200105009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasioukhin V, Bowers E, Bauer C, Degenstein L, Fuchs E. Desmoplakin is essential in epidermal sheet formation. Nat Cell Biol. 2001;3(12):1076–1085. doi: 10.1038/ncb1201-1076. [DOI] [PubMed] [Google Scholar]
- 19.Sumigray KD, Lechler T. Cell adhesion in epidermal development and barrier formation. Curr Top Dev Biol. 2015;112:383–414. doi: 10.1016/bs.ctdb.2014.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koch PJ, et al. Targeted disruption of the pemphigus vulgaris antigen (desmoglein 3) gene in mice causes loss of keratinocyte cell adhesion with a phenotype similar to pemphigus vulgaris. J Cell Biol. 1997;137(5):1091–1102. doi: 10.1083/jcb.137.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Getsios S, et al. Desmoglein 1-dependent suppression of EGFR signaling promotes epidermal differentiation and morphogenesis. J Cell Biol. 2009;185(7):1243–1258. doi: 10.1083/jcb.200809044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris TJ, Tepass U. Adherens junctions: from molecules to morphogenesis. Nat Rev Mol Cell Biol. 2010;11(7):502–514. doi: 10.1038/nrm2927. [DOI] [PubMed] [Google Scholar]
- 23.Takeichi M. Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat Rev Mol Cell Biol. 2014;15(6):397–410. doi: 10.1038/nrm3802. [DOI] [PubMed] [Google Scholar]
- 24.Zandy NL, Playford M, Pendergast AM. Abl tyrosine kinases regulate cell-cell adhesion through Rho GTPases. Pråoc Natl Acad Sci U S A. 2007;104(45):17686–17691. doi: 10.1073/pnas.0703077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahai E, Marshall CJ. ROCK and Dia have opposing effects on adherens junctions downstream of Rho. Nat Cell Biol. 2002;4(6):408–415. doi: 10.1038/ncb796. [DOI] [PubMed] [Google Scholar]
- 26.Fang WB, Ireton RC, Zhuang G, Takahashi T, Reynolds A, Chen J. Overexpression of EPHA2 receptor destabilizes adherens junctions via a RhoA-dependent mechanism. J Cell Sci. 2008;121(pt 3):358–368. doi: 10.1242/jcs.017145. [DOI] [PubMed] [Google Scholar]
- 27.Godsel LM, et al. Desmoplakin assembly dynamics in four dimensions: multiple phases differentially regulated by intermediate filaments and actin. J Cell Biol. 2005;171(6):1045–1059. doi: 10.1083/jcb.200510038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Godsel LM, et al. Plakophilin 2 couples actomyosin remodeling to desmosomal plaque assembly via RhoA. Mol Biol Cell. 2010;21(16):2844–2859. doi: 10.1091/mbc.E10-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajakylä EK, Vartiainen MK. Rho, nuclear actin, and actin-binding proteins in the regulation of transcription and gene expression. Small GTPases. 2014;5:e27539. doi: 10.4161/sgtp.27539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu ZH, et al. Mammalian target of rapamycin activator RHEB is frequently overexpressed in human carcinomas and is critical and sufficient for skin epithelial carcinogenesis. Cancer Res. 2010;70(8):3287–3298. doi: 10.1158/0008-5472.CAN-09-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castilho RM, Squarize CH, Chodosh LA, Williams BO, Gutkind JS. mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell. 2009;5(3):279–289. doi: 10.1016/j.stem.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iglesias-Bartolome R, et al. mTOR inhibition prevents epithelial stem cell senescence and protects from radiation-induced mucositis. Cell Stem Cell. 2012;11(3):401–414. doi: 10.1016/j.stem.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feldmeyer L, Hofbauer GF, Böni T, French LE, Hafner J. Mammalian target of rapamycin (mTOR) inhibitors slow skin carcinogenesis, but impair wound healing. Br J Dermatol. 2012;166(2):422–424. doi: 10.1111/j.1365-2133.2011.10591.x. [DOI] [PubMed] [Google Scholar]
- 34.Ding X, et al. mTORC1 and mTORC2 regulate skin morphogenesis and epidermal barrier formation. Nat Commun. 2016;7:13226. doi: 10.1038/ncomms13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127(22):4775–4785. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- 36.Zou J, et al. Rheb1 is required for mTORC1 and myelination in postnatal brain development. Dev Cell. 2011;20(1):97–108. doi: 10.1016/j.devcel.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468(7327):1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- 38.Manning BD, Logsdon MN, Lipovsky AI, Abbott D, Kwiatkowski DJ, Cantley LC. Feedback inhibition of Akt signaling limits the growth of tumors lacking Tsc2. Genes Dev. 2005;19(15):1773–1778. doi: 10.1101/gad.1314605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Javier AF, Bata-Csorgo Z, Ellis CN, Kang S, Voorhees JJ, Cooper KD. Rapamycin (sirolimus) inhibits proliferating cell nuclear antigen expression and blocks cell cycle in the G1 phase in human keratinocyte stem cells. J Clin Invest. 1997;99(9):2094–2099. doi: 10.1172/JCI119382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramírez-Valle F, Badura ML, Braunstein S, Narasimhan M, Schneider RJ. Mitotic raptor promotes mTORC1 activity, G(2)/M cell cycle progression, and internal ribosome entry site-mediated mRNA translation. Mol Cell Biol. 2010;30(13):3151–3164. doi: 10.1128/MCB.00322-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437(7056):275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luxenburg C, Pasolli HA, Williams SE, Fuchs E. Developmental roles for Srf, cortical cytoskeleton and cell shape in epidermal spindle orientation. Nat Cell Biol. 2011;13(3):203–214. doi: 10.1038/ncb2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hebert AM, DuBoff B, Casaletto JB, Gladden AB, McClatchey AI. Merlin/ERM proteins establish cortical asymmetry and centrosome position. Genes Dev. 2012;26(24):2709–2723. doi: 10.1101/gad.194027.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kulukian A, Fuchs E. Spindle orientation and epidermal morphogenesis. Philos Trans R Soc Lond B Biol Sci. 2013;368(1629):20130016. doi: 10.1098/rstb.2013.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bikle DD, Xie Z, Tu CL. Calcium regulation of keratinocyte differentiation. Expert Rev Endocrinol Metab. 2012;7(4):461–472. doi: 10.1586/eem.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen E, Yu QC, Fuchs E. Mice expressing a mutant desmosomal cadherin exhibit abnormalities in desmosomes, proliferation, and epidermal differentiation. J Cell Biol. 1996;133(6):1367–1382. doi: 10.1083/jcb.133.6.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson JL, Najor NA, Green KJ. Desmosomes: regulators of cellular signaling and adhesion in epidermal health and disease. Cold Spring Harb Perspect Med. 2014;4(11):a015297. doi: 10.1101/cshperspect.a015297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hachem JP, et al. Serine protease activity and residual LEKTI expression determine phenotype in Netherton syndrome. J Invest Dermatol. 2006;126(7):1609–1621. doi: 10.1038/sj.jid.5700288. [DOI] [PubMed] [Google Scholar]
- 49.Ferone G, et al. p63 control of desmosome gene expression and adhesion is compromised in AEC syndrome. Hum Mol Genet. 2013;22(3):531–543. doi: 10.1093/hmg/dds464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100(2):209–219. doi: 10.1016/S0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- 51.Vaezi A, Bauer C, Vasioukhin V, Fuchs E. Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev Cell. 2002;3(3):367–381. doi: 10.1016/S1534-5807(02)00259-9. [DOI] [PubMed] [Google Scholar]
- 52.Versaevel M, Grevesse T, Gabriele S. Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat Commun. 2012;3:671. doi: 10.1038/ncomms1668. [DOI] [PubMed] [Google Scholar]
- 53.Amano M, et al. Formation of actin stress fibers and focal adhesions enhanced by Rho-kinase. Science. 1997;275(5304):1308–1311. doi: 10.1126/science.275.5304.1308. [DOI] [PubMed] [Google Scholar]
- 54.Jacinto E, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6(11):1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 55.Lock FE, Hotchin NA. Distinct roles for ROCK1 and ROCK2 in the regulation of keratinocyte differentiation. PLoS One. 2009;4(12):e8190. doi: 10.1371/journal.pone.0008190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grossi M, et al. Negative control of keratinocyte differentiation by Rho/CRIK signaling coupled with up-regulation of KyoT1/2 (FHL1) expression. Proc Natl Acad Sci U S A. 2005;102(32):11313–11318. doi: 10.1073/pnas.0505011102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moustakas A, Heldin CH. Dynamic control of TGF-beta signaling and its links to the cytoskeleton. FEBS Lett. 2008;582(14):2051–2065. doi: 10.1016/j.febslet.2008.03.027. [DOI] [PubMed] [Google Scholar]
- 58.Sellheyer K, et al. Inhibition of skin development by overexpression of transforming growth factor beta 1 in the epidermis of transgenic mice. Proc Natl Acad Sci U S A. 1993;90(11):5237–5241. doi: 10.1073/pnas.90.11.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Laer L, Dietz H, Loeys B. Loeys-Dietz syndrome. Adv Exp Med Biol. 2014;802:95–105. doi: 10.1007/978-94-007-7893-1_7. [DOI] [PubMed] [Google Scholar]
- 60.Deng Z, et al. mTOR signaling promotes stem cell activation via counterbalancing BMP-mediated suppression during hair regeneration. J Mol Cell Biol. 2015;7(1):62–72. doi: 10.1093/jmcb/mjv005. [DOI] [PubMed] [Google Scholar]
- 61.Charest JL, Jennings JM, King WP, Kowalczyk AP, García AJ. Cadherin-mediated cell-cell contact regulates keratinocyte differentiation. J Invest Dermatol. 2009;129(3):564–572. doi: 10.1038/jid.2008.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Green KJ, Simpson CL. Desmosomes: new perspectives on a classic. J Invest Dermatol. 2007;127(11):2499–2515. doi: 10.1038/sj.jid.5701015. [DOI] [PubMed] [Google Scholar]
- 63.Riento K, Ridley AJ. Rocks: multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol. 2003;4(6):446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- 64.Samarin SN, Ivanov AI, Flatau G, Parkos CA, Nusrat A. Rho/Rho-associated kinase-II signaling mediates disassembly of epithelial apical junctions. Mol Biol Cell. 2007;18(9):3429–3439. doi: 10.1091/mbc.E07-04-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Narumiya S, Tanji M, Ishizaki T. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 2009;28(1–2):65–76. doi: 10.1007/s10555-008-9170-7. [DOI] [PubMed] [Google Scholar]
- 66.Todorović V, et al. Plakoglobin regulates cell motility through Rho- and fibronectin-dependent Src signaling. J Cell Sci. 2010;123(pt 20):3576–3586. doi: 10.1242/jcs.070391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koetsier JL, Amargo EV, Todorović V, Green KJ, Godsel LM. Plakophilin 2 affects cell migration by modulating focal adhesion dynamics and integrin protein expression. J Invest Dermatol. 2014;134(1):112–122. doi: 10.1038/jid.2013.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoneda A, Multhaupt HA, Couchman JR. The Rho kinases I and II regulate different aspects of myosin II activity. J Cell Biol. 2005;170(3):443–453. doi: 10.1083/jcb.200412043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shimizu Y, et al. ROCK-I regulates closure of the eyelids and ventral body wall by inducing assembly of actomyosin bundles. J Cell Biol. 2005;168(6):941–953. doi: 10.1083/jcb.200411179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thumkeo D, et al. Targeted disruption of the mouse rho-associated kinase 2 gene results in intrauterine growth retardation and fetal death. Mol Cell Biol. 2003;23(14):5043–5055. doi: 10.1128/MCB.23.14.5043-5055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riento K, Guasch RM, Garg R, Jin B, Ridley AJ. RhoE binds to ROCK I and inhibits downstream signaling. Mol Cell Biol. 2003;23(12):4219–4229. doi: 10.1128/MCB.23.12.4219-4229.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ashcroft GS, et al. Mice lacking Smad3 show accelerated wound healing and an impaired local inflammatory response. Nat Cell Biol. 1999;1(5):260–266. doi: 10.1038/12971. [DOI] [PubMed] [Google Scholar]
- 73.Foitzik K, et al. Control of murine hair follicle regression (catagen) by TGF-β1 in vivo. FASEB J. 2000;14(5):752–760. doi: 10.1096/fasebj.14.5.752. [DOI] [PubMed] [Google Scholar]
- 74.Blessing M, Schirmacher P, Kaiser S. Overexpression of bone morphogenetic protein-6 (BMP-6) in the epidermis of transgenic mice: inhibition or stimulation of proliferation depending on the pattern of transgene expression and formation of psoriatic lesions. J Cell Biol. 1996;135(1):227–239. doi: 10.1083/jcb.135.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Markell LM, Masiuk KE, Blazanin N, Glick AB. Pharmacologic inhibition of ALK5 causes selective induction of terminal differentiation in mouse keratinocytes expressing oncogenic HRAS. Mol Cancer Res. 2011;9(6):746–756. doi: 10.1158/1541-7786.MCR-11-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cui W, et al. TGFbeta1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell. 1996;86(4):531–542. doi: 10.1016/S0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- 77.Amendt C, Schirmacher P, Weber H, Blessing M. Expression of a dominant negative type II TGF-β receptor in mouse skin results in an increase in carcinoma incidence and an acceleration of carcinoma development. Oncogene. 1998;17(1):25–34. doi: 10.1038/sj.onc.1202161. [DOI] [PubMed] [Google Scholar]
- 78.Huang JJ, Blobe GC. Dichotomous roles of TGF-β in human cancer. Biochem Soc Trans. 2016;44(5):1441–1454. doi: 10.1042/BST20160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu X, Alexander V, Vijayachandra K, Bhogte E, Diamond I, Glick A. Conditional epidermal expression of TGFβ1 blocks neonatal lethality but causes a reversible hyperplasia and alopecia. Proc Natl Acad Sci U S A. 2001;98(16):9139–9144. doi: 10.1073/pnas.161016098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Song K, Wang H, Krebs TL, Danielpour D. Novel roles of Akt and mTOR in suppressing TGF-β/ALK5-mediated Smad3 activation. EMBO J. 2006;25(1):58–69. doi: 10.1038/sj.emboj.7600917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wahdan-Alaswad RS, et al. Insulin-like growth factor I suppresses bone morphogenetic protein signaling in prostate cancer cells by activating mTOR signaling. Cancer Res. 2010;70(22):9106–9117. doi: 10.1158/0008-5472.CAN-10-1119. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 82.Bhowmick NA, et al. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol Biol Cell. 2001;12(1):27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Edlund S, Landström M, Heldin CH, Aspenström P. Transforming growth factor-beta-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Mol Biol Cell. 2002;13(3):902–914. doi: 10.1091/mbc.01-08-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fleming YM, et al. TGF-beta-mediated activation of RhoA signalling is required for efficient (V12)HaRas and (V600E)BRAF transformation. Oncogene. 2009;28(7):983–993. doi: 10.1038/onc.2008.449. [DOI] [PubMed] [Google Scholar]
- 85.Dubash AD, et al. Plakophilin-2 loss promotes TGF-β1/p38 MAPK-dependent fibrotic gene expression in cardiomyocytes. J Cell Biol. 2016;212(4):425–438. doi: 10.1083/jcb.201507018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Savagner P, Yamada KM, Thiery JP. The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J Cell Biol. 1997;137(6):1403–1419. doi: 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang H, et al. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J Clin Invest. 2003;112(8):1223–1233. doi: 10.1172/JCI17222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ghosh S, Varela L, Sood A, Park BH, Lotan TL. mTOR signaling feedback modulates mammary epithelial differentiation and restrains invasion downstream of PTEN loss. Cancer Res. 2013;73(16):5218–5231. doi: 10.1158/0008-5472.CAN-13-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.