Abstract
Uromodulin-associated kidney disease (UAKD) is caused by mutations in the uromodulin (UMOD) gene that result in a misfolded form of UMOD protein, which is normally secreted by nephrons. In UAKD patients, mutant UMOD is poorly secreted and accumulates in the ER of distal kidney epithelium, but its role in disease progression is largely unknown. Here, we modeled UMOD accumulation in mice by expressing the murine equivalent of the human UMOD p.Cys148Trp point mutation (UmodC147W/+ mice). Like affected humans, these UmodC147W/+ mice developed spontaneous and progressive kidney disease with organ failure over 24 weeks. Analysis of diseased kidneys and purified UMOD-producing cells revealed early activation of the PKR-like ER kinase/activating transcription factor 4 (PERK/ATF4) ER stress pathway, innate immune mediators, and increased apoptotic signaling, including caspase-3 activation. Unexpectedly, we also detected autophagy deficiency. Human cells expressing UMOD p.Cys147Trp recapitulated the findings in UmodC147W/+ mice, and autophagy activation with mTOR inhibitors stimulated the intracellular removal of aggregated mutant UMOD. Human cells producing mutant UMOD were susceptible to TNF-α– and TRAIL-mediated apoptosis due to increased expression of the ER stress mediator tribbles-3. Blocking TNF-α in vivo with the soluble recombinant fusion protein TNFR:Fc slowed disease progression in UmodC147W/+ mice by reducing active caspase-3, thereby preventing tubule cell death and loss of epithelial function. These findings reveal a targetable mechanism for disease processes involved in UAKD.
Keywords: Cell Biology, Nephrology
Keywords: Apoptosis pathways, Autophagy, Fibrosis
Introduction
Uromodulin-associated kidney disease (UAKD) is an inherited form of progressive kidney disease, with a prevalence of 8 in 1 million individuals in the general population, but representing as many as 25% of patients with inherited kidney disease, after exclusion of polycystic kidney disease and Alport syndrome (1). Patients with UAKD can begin to show symptoms as early as the teenage years and are characterized throughout disease progression by low uromodulin (UMOD or Tamm-Horsfall protein) levels in urine, polyuria and polydipsia, tubulointerstitial fibrosis, and the presence of hyperuricemia, gout, and hypertension (2). In addition to mutations identified through familial linkage studies of UAKD (3–6), recent GWAS have shown noncoding and coding polymorphisms at the UMOD gene (UMOD) locus to be significantly associated with chronic kidney disease (7–12). While inconclusive, such polymorphisms in the UMOD promoter may point to abnormalities of UMOD expression as being highly associated with the development and progression of kidney disease (13).
UMOD was first isolated from the urine of healthy individuals and characterized as a secreted, high-molecular-weight mucoprotein in 1952 by Igor Tamm and Frank Horsfall (14). More recent studies have revealed UMOD expression to be exclusive to the renal epithelial cells lining the thick ascending limb of the loop of Henle and the early distal tubule (15). UMOD protein is glycosylated at 7 of its 8 potential N-glycosylation sites (16) and is glycosylphosphatidylinositol (GPI) anchored to the luminal surface of tubules, where it is cleaved into the urine at a rate of 50 to 100 milligrams per day (17), making it the most highly secreted protein in the urine of healthy individuals (18). In urine, it is found as a high-molecular-weight polymer (relative molecular weight [Mr] 1 × 106 to 10 × 106) and can be dissociated into monomers of roughly 95 kDa (17). While the function of UMOD is not completely elucidated, knockout animal studies suggest that it plays a role in renoprotection against uropathogenic bacteria and in tightly controlling water transport in the kidney to concentrate urine (19, 20). UMOD has been shown to be critical in preventing crystallization of calcium from tubular filtrate, suggesting a role in the prevention of kidney stone formation (21).
While there are more than 100 inherited mutations in UMOD described (22–27), most result in cysteine residue replacement, which disrupts disulfide bridging at critical nodes of protein folding. The majority of reported mutations reside within the portion of the gene that codes for the N-terminus. Prior studies have revealed impaired trafficking of many of these mutant forms of the protein (28), suggesting that impaired trafficking and loss of secretion into the urine tie the mutations together mechanistically. Recently, investigators created a short transgenic allele expressing a mutant form of mouse UMOD under the Umod short promoter (18). The mouse with this transgene developed kidney disease, and the mutated UMOD protein was shown to localize in the ER. The studies showed a close link between mutant UMOD expression and the regulation of salt and water balance. These previous studies did not closely explore the underlying mechanisms linking ER stress and the pathogenesis of nephron failure.
To explore UAKD further, with the goal of dissecting the signaling pathways triggered by misfolded UMOD protein, we wished to avoid the possible shortcomings of a transgenic model, such as unfaithful expression or unwanted disruption of key coding and noncoding regions, and capitalize on recent technological advances using CRISPR/Cas9 to specifically introduce precise point mutations into the murine Umod locus. We therefore introduced the mouse equivalent of the human UMOD mutation p.Cys148Trp, which lies 1 residue earlier, denoted as p.Cys147Trp, into the endogenous Umod locus of the mouse genome to characterize the disease processes mediated by this mutation.
Results
UmodC147W/+ mice develop chronic kidney disease at 24 weeks and accumulate UMOD in the ER.
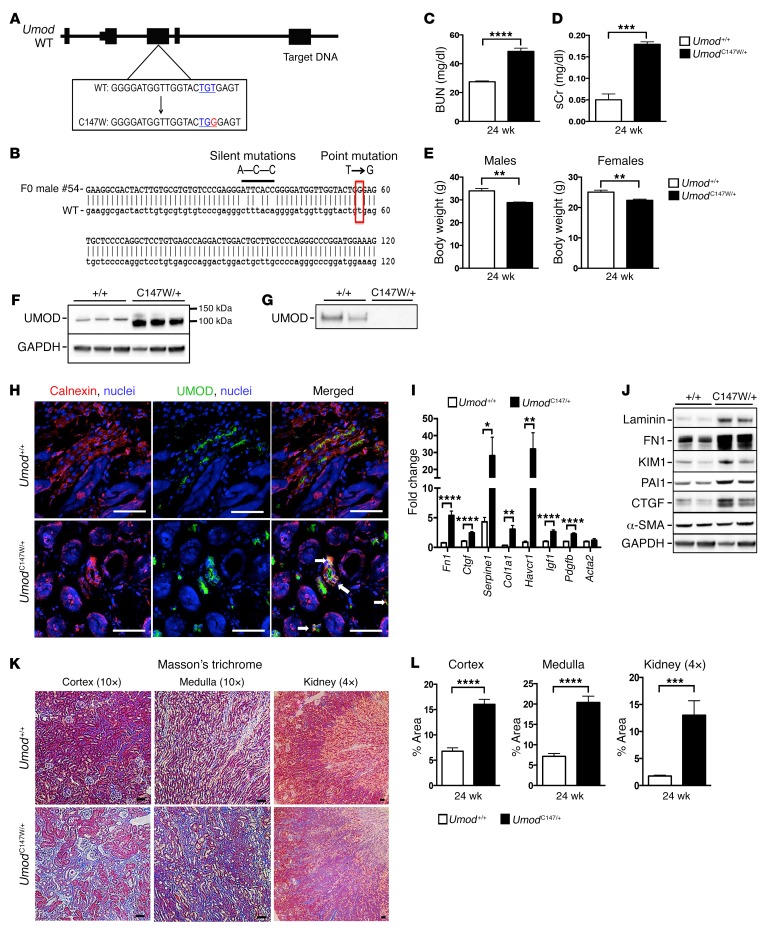
To create a point mutation at the 147th amino acid of the mouse Umod gene, a single CRISPR RNA guide was generated using in vitro transcription (29), purified, and then injected with Cas9 mRNA, along with a 153-bp single-stranded donor DNA fragment containing the mutation, into C57BL/6J mouse embryos at the single-cell stage (30). These embryos were then implanted into foster mothers and brought to term. Sixty mosaic founder males had tissue sampled for screening. PCR was run across the allele of interest, and the amplicon was cloned into a housing vector for sequencing (Figure 1, A and B). Sequencing results identified 5% of founders with the correctly targeted mutations. Positive founders were bred with WT female mice to produce F1 animals. The heterozygous mice of the F1 generation were found to have germline transmission of 50%. A cohort of heterozygous mice and littermate controls were aged and evaluated at 24 weeks. Mice heterozygous for Umod p.Cys147Trp (UmodC147W/+ mice) had elevated blood urea nitrogen (BUN) and serum creatinine (sCr), markers of kidney failure (Figure 1, C and D). Urinary protein levels were also increased but did not reach significance at this 24-week time point (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI93817DS1). The body weights of both males and females were reduced compared with littermate controls (Figure 1E). Consistent with previous reports, levels of UMOD in whole-kidney tissue in mutant animals were elevated and were found to accumulate at a smaller, nonglycosylated pre–Golgi apparatus stage (18). On the other hand, we found that urinary UMOD was markedly reduced in heterozygous mice, consistent with a dominant negative function of this mutant protein (Figure 1, F and G). Loss of UMOD from urine was associated with decreased concentrating capacity, which was determined by measuring urine-specific gravity (Supplemental Figure 1B). We evaluated whole-kidney sections for cellular UMOD expression (Figure 1H). Whereas healthy tubules showed signal for UMOD expression at the luminal surface, mutant mice showed intense signal in intracellular compartments that colocalized with calnexin, an ER membrane marker. Whole-kidney transcripts and protein detection indicated tubular damage in the proximal tubule, which was evidenced by elevated levels of hepatitis A virus cellular receptor 1 (HAVCR1, also known as KIM1). We also observed increases in fibrotic disease markers including laminin, fibronectin (FN1), collagen type I, serpine1 (PAI1), as well as the signaling growth factors CTGF, IGF1, and PDGF-B (Figure 1, I and J). Consistent with these observations, Masson’s trichrome staining revealed significant and widespread interstitial fibrosis in both the cortex and medulla of whole-kidney sections (Figure 1, K and L). Picrosirius red staining revealed marked increases in collagens I and III (Supplemental Figure 1, C and D).
Figure 1. UmodC147W/+ mice exhibit kidney failure at 24 weeks.
(A) Map of murine UMOD locus with annotated WT and mutant p.Cys147Trp sequences (relevant codons are indicated in blue and the point mutation in red). (B) Sequencing results of a representative F0 founder male with a correctly targeted point mutation (in red) and silent mutations (thick black line). (C) BUN results from blood of 24-week-old mice. (D) sCr results from blood of 24-week-old mice. (E) Body weight of 24-week-old male and female mice. (F) Western blot to detect UMOD protein from whole-kidney tissue; the glycosylated band runs larger than the mutant, nonglycosylated band. (G) Western blot to detect UMOD proteins precipitated from urine; levels were undetectable in the mutant urine samples. (H) Immunofluorescence images of kidney sections (7-μm-thick) labeled for the ER marker calnexin (red) and UMOD (green). Arrows indicate an overlap of calnexin and UMOD. Scale bars: 50 μm (original magnification, ×40). (I) Quantitative PCR results for fibrosis-related genes expressed in whole kidneys from 24-week-old mice. (J) Western blot analysis of proteins from whole-kidney tissue to detect markers of fibrosis at the 24-week time point. Note: The blot shown in F was stripped and reprobed for fibrosis markers. (K) Histological images of whole-kidney sections (4-μm-thick) stained with Masson’s trichrome preparation (connective tissue/collagens in blue). Scale bars: 50 μm (original magnification, ×10 [cortex and medulla] and ×4 [kidney]). (L) Quantification of Masson’s trichrome–stained images of collagens. Data represent the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 2-tailed Student’s t test. n = 6–8 per group. +/+, Umod+/+; C147W/+, UmodC147W/+.
Transcriptional analysis of UmodC147W/+ kidneys reveals TNF signaling and ER stress–induced apoptotic cell death, but not autophagic signaling pathways.
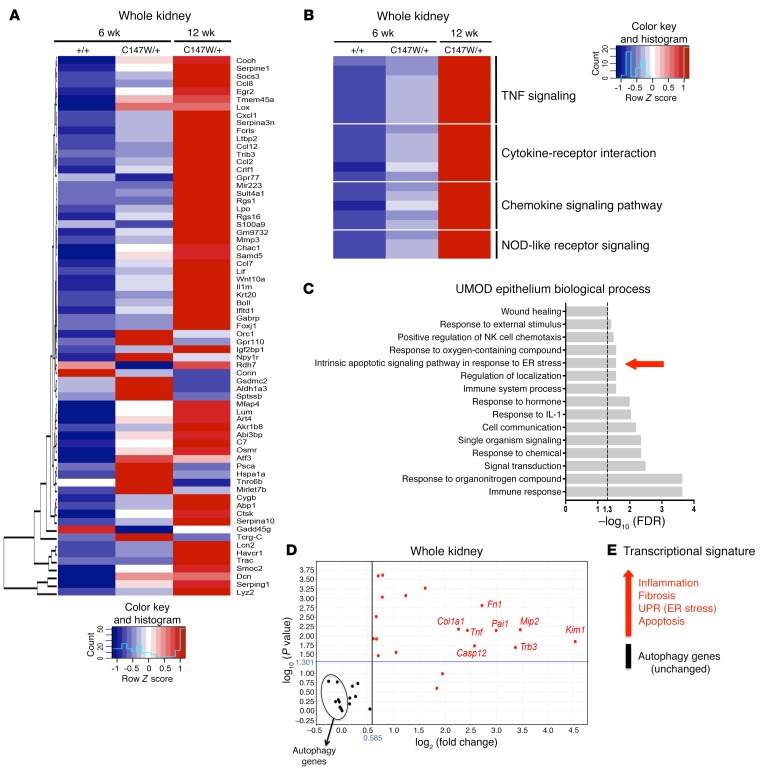
To achieve a greater understanding of the mechanisms linking misfolded protein accumulation to the pathogenesis of disease in UmodC147W/+ mice, we performed RNA-sequencing (RNA-seq) of mRNA isolated from whole-kidney tissue and from UMOD-producing epithelial cells purified from mutant mice and littermate controls at multiple time points, including 6, 12, 24, and 30 weeks (Supplemental Figure 2, A–C) (Gene Expression Omnibus [GEO] database accession no. GSE102566). Because the isolated UMOD-producing epithelial cells lost expression of Umod by day 10 in culture (Supplemental Figure 2D), only RNA from freshly isolated day-0 fractions was sent for sequencing. At early time points in whole-kidney tissue, including 6 and 12 weeks, we observed a marked activation of genes associated with epithelial injury (Lcn2, Havcr1), matrix dysregulation (Serpine1, Mmp3, Smoc2), complement (Gpr77), the JAK/STAT pathway (Lif, Crlf1, Socs3), and the Myd88-dependent pathway (Il1rn, Ccl2), as well as activation of downstream effectors of the ER unfolded protein response (UPR) (Trib3, Casp4, Casp12, Asns, Atf3) (Figure 2A). Gene ontology (GO) analysis of the most dysregulated genes in whole tissue confirmed the activation of innate immune pathways, positive regulation of inflammation, and response to stress (Supplemental Figure 3A). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis similarly revealed innate immune activation, as well as strong engagement of TNF signaling (Figure 2B). Additional upstream analysis of the dysregulated genes specifically in UMOD-producing epithelial cells at the 12-week time point confirmed the regulation of innate immune pathways and TNF signaling and also demonstrated activation of an intrinsic ER stress–mediated cell death pathway (Figure 2C and Supplemental Figure 3, B and C). Given the activation of these pathways early on in disease, we performed a mini-array in a separate cohort of mice at 24 weeks, at the peak of disease, to assess the extent to which these pathways were maintained or exacerbated. Analysis at 24 weeks confirmed the upregulation of matrix-related genes, the epithelial injury marker Havcr1, innate immunity genes, as well as genes for components of the ER stress/UPR pathway. Interestingly, genes related to macroautophagy, the process by which large proteins, aggregates, and organelles are processed for degradation, were not transcriptionally upregulated (Figure 2, D and E, and Supplemental Figure 3D). This was particularly striking, given that we observed aberrant accumulation of mutant UMOD protein in the renal tubular cells of mutant animals (Figure 1).
Figure 2. Transcriptional analysis of UmodC147W/+ kidneys and UMOD-expressing epithelium.
(A) Heatmap of the 40 most DEGs with an adjusted P value of less than 0.05 and a log2 fold change of greater than 1. RNA was isolated from whole-kidney tissue from 12-week-old mice. Scale bar is in fragments per kilobase of transcript per million mapped reads (FPKM). (B) Heatmap of selected enriched terms (adjusted P < 0.05) from KEGG pathway analysis of the 40 most upregulated DEGs. RNA was isolated from whole-kidney tissue from 12-week-old mice. (C) GO enrichment analysis for biological process of the 40 most upregulated DEGs in RNA obtained from isolated UMOD-producing cells from 12-week-old mouse kidneys (adjusted P < 0.05). (D) Volcano plot of genes for inflammation, fibrosis, apoptosis, the UPR, and autophagy (performed with a fold-change cutoff of 1.5 and a P-value cutoff of 0.05). RNA was isolated from 24-week-old whole mouse kidney, and the fold change was assessed using quantitative PCR. (E) Transcriptional signature summary diagram for 24-week-old whole-kidney RNA isolate. n = 8–11 per group.
UmodC147W/+ kidneys undergo ER stress and apoptosis and activate innate immunity.
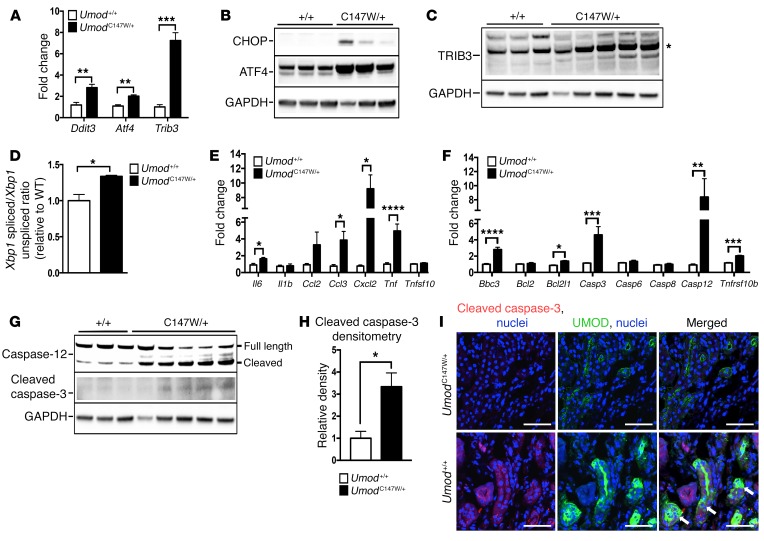
Since the mRNA profiling of mutant kidneys and epithelium revealed UPR and ER stress, innate immunity, and apoptosis as prominent upregulated pathways in the transcriptional signature, we evaluated these candidate disease-contributing processes further. Elevated expression levels of central ER stress pathway regulators Atf4, Ddit3 (encoding CHOP), and Trib3, downstream of the PERK-dependent sensing pathway of the UPR, were confirmed at week 24 (Figure 3A). Similarly, protein levels of CHOP, ATF4, and tribbles-3 (TRIB3) were elevated in whole kidneys from mutant animals (Figure 3, B and C). Measurement of the activation of the IRE1α/XBP1-sensing transcriptional response of the UPR was performed using quantitative PCR to directly compare the levels of active, spliced XBP1 transcript with those of the inactive, unspliced form. This method revealed an increase in the ratio of the active to inactive form of the Xbp1 transcript in mutant kidneys, consistent with induction of the UPR and ER stress mechanisms (Figure 3D). Quantitative PCR of whole-kidney transcripts at week 24 revealed a significant increase in activation of a panel of innate immune and apoptotic genes, including Ccl3 (also known as Mip1a), Cxcl2 (also known as Mip2), Tnf, Bbc3, Casp3, and Casp12, respectively (Figure 3, E and F). We observed elevated levels of the active, cleaved form of caspase-3 protein (Figure 3, G and H). Furthermore, the cleaved version of caspase-12 was also increased in mutant animals (Figure 3G). Caspase-12, which is located in the ER may link ER stress to apoptosis and innate immune signaling (31–38). Immunofluorescence staining revealed increased active caspase-3 distributed within UMOD-positive tubules (arrows) (Figure 3I). Interestingly, active caspase-3 was also observed in non-UMOD–positive tubules, suggesting that distal tubules transmit apoptotic signaling to neighboring tubules of the kidney (Figure 3I).
Figure 3. UmodC147W/+ kidneys activate ER stress, innate immune response, and apoptotic pathways.
(A) Quantitative PCR of whole-kidney tissue at 24 weeks for key ER stress response genes. (B) Western blot of whole-kidney tissue to detect key ER stress mediators. (C) Western blot of whole-kidney tissue to detect TRIB3. Note: The band with the correct size for the predicted molecular weight is marked with a single asterisk. (D) Quantitative PCR of cDNA from whole-kidney tissue for the ratio of spliced to unspliced Xbp1. (E) Quantitative PCR of cDNA from whole-kidney tissue for relevant innate immune mediators. (F) Quantitative PCR of cDNA from whole-kidney tissue for apoptotic mediators. (G) Western blot of whole-kidney tissue to detect cleaved caspase-3 and total caspase-12 (cleaved band runs below the full-length band). (H) Densitometric analysis for cleaved caspase-3 normalized to GAPDH. (I) Immunofluorescence images of kidney sections (7-μm-thick) labeled for active cleaved caspase-3 (red) and UMOD (green). Arrows indicate tubules positive for active caspase-3 and UMOD. Scale bars: 50 μm (original magnification, ×40). Data represent the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 2-tailed Student’s t test. n = 6–8 per group.
UmodC147W/+ kidneys are deficient in autophagy.
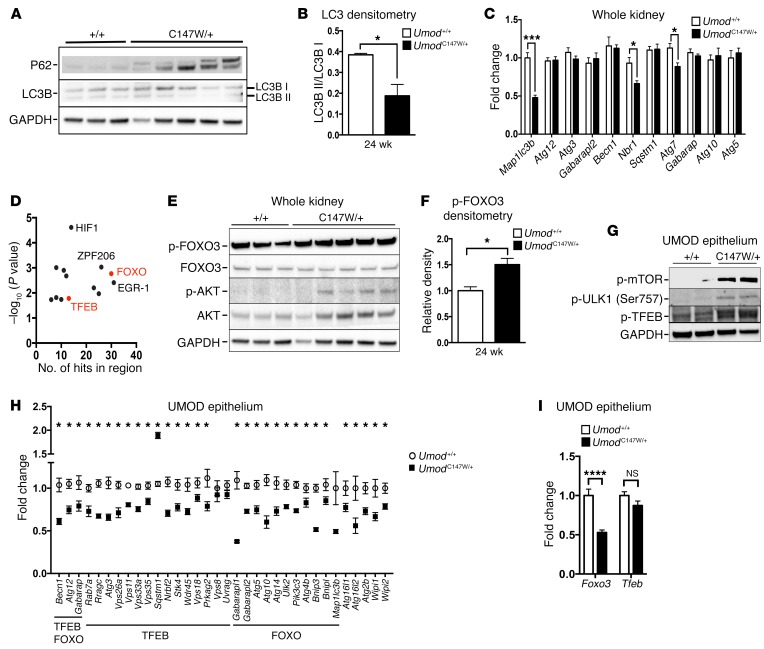
Despite aberrant intracellular accumulation of misfolded, mutant UMOD protein, the RNA-seq analysis demonstrated a lack of upregulation of autophagy genes. Autophagy is an important homeostatic intracellular mechanism for the removal of damaged organelles, cytoplasmic components, and misfolded and aggregated proteins (39–41). To further assess the unexpected lack of upregulation, we evaluated the protein levels of 2 key mediators of autophagy, P62/SQSTM1 and LC3/MAP1LC3B. P62 is a critical ubiquitin-binding scaffold protein that acts as a selective autophagy receptor for ubiquitinated substrates marked for degradation. P62 is recognized by the autophagosome and is itself degraded along with the substrate via autophagy (42, 43). Clearance of autophagic cargo is a hallmark of active autophagy (42). LC3B is a critical component contributing to the integrity of the membrane of the autophagosome. Inactive LC3B I is cleaved into the active LC3B II, and the ratio of these 2 forms reflects on the level of active autophagy (44). In mutant kidneys, we observed both an abundance of P62 and a significantly reduced ratio of active to inactive LC3B (Figure 4, A and B). Not only is this consistent with a lack of engagement of autophagy, but it suggests an active suppression of autophagy mechanisms. Quantitative analysis of a panel of essential autophagy-associated genes also demonstrated a widespread lack of upregulation relative to littermate controls. We observed a significant reduction in gene expression for Map1lc3b, Nbr1, and Atg7 (Figure 4C). An rVista promoter analysis (45) for overrepresented transcription factor binding sites for this panel of selected autophagy genes revealed both FOXO and TFEB binding sites (Figure 4D). The activity levels of TFEB and FOXO3, specifically, have been linked to positive regulation of autophagy (46–51). Protein level analysis of whole-kidney tissues revealed an increased level of active, phosphorylated AKT (p-AKT), which negatively regulates FOXO3 via phosphorylation. Densitometric analysis revealed a 1.5-fold enrichment of inactive p-FOXO3 in mutant kidneys (Figure 4, E and F), which was consistent with suppressed transcriptional regulation of autophagy. To study the impact of mutant UMOD on autophagy specifically in UMOD-producing epithelial cells, we purified primary UMOD-positive murine epithelial cells from mutant and littermate control kidneys at 24 weeks. To further explore whether autophagy is actively suppressed at this time point, we evaluated mTOR, an important negative regulator of the initiation of autophagic phagophore formation via phosphorylation and inactivation of Unc-51–like autophagy-activating kinase 1 (ULK1) (52). Active mTOR also negatively regulates TFEB via phosphorylation, maintaining the transcription factor in the cytosol via conjugation to 14-3-3 proteins (53). In purified UMOD-producing epithelial cells, we detected increased levels of active p-mTOR, as well as inactive p-ULK1 and inactive p-TFEB in the presence of mutant UMOD (Figure 4G). To determine the extent of autophagy suppression at the transcriptional level in the affected UMOD-producing epithelium, we assessed the expression of a broad panel of autophagy-associated genes, with a particular focus on those regulated by TFEB and FOXO (54, 55). We observed a statistically significant, widespread suppression in nearly 90% of all autophagy-associated genes tested (93% of FOXO targets, 83% of TFEB targets), with the single exception of Sqstm1, which was upregulated in mutant cells (Figure 4H). Among the significantly downregulated genes were those encoding autophagic machinery components linked to: autophagic vacuole formation (Atg5, Atg12, Atg16l1, Gabarap Gabarapl1, Gabarapl2, Becn1); targeting proteins to the autophagosome (Atg4b); linking autophagosomes to lysosomes (Vps33a, Vps11); protein transport (Atg10, Atg16l2); and protein ubiquitination (Atg3) (56, 57). Genes associated with transmitting an autophagic response to intracellular signals (Pik3c3, Ulk2), the ATG2-WIPI network (Atg2b, Wipi1, Wipi2, Wdr45), and the cargo-selective complex (CSC) (Vps35, Vps26a) were similarly suppressed (58, 59). Interestingly, transcript expression levels in WT versus mutant UMOD cells of the regulators FOXO3 and TFEB themselves were different. While expression of Foxo3 was significantly downregulated, Tfeb expression was not significantly changed (Figure 4I), suggesting that, in our model, FOXO3 was negatively regulated at both the transcriptional and posttranslational levels, while TFEB was largely regulated at the posttranslational level only in the affected cells.
Figure 4. UmodC147W/+ kidneys are deficient in autophagy.
(A) Western blot of whole-kidney tissue at 24 weeks to detect the macroautophagy mediators p62 and LC3B (LC3B I inactive, LC3B II active). (B) Ratio of active to inactive LC3B by densitometric analysis. (C) Quantitative PCR of whole-kidney tissue at 24 weeks for autophagy-related genes. (D) Promoter analysis for overrepresented transcriptional factor binding sites of relevant autophagy-related genes (P-value cutoff: 0.02). (E) Western blot to detect the transcriptional regulator of autophagy genes FOXO3 and the upstream regulator AKT. Note: The blot from A was stripped and reprobed. (F) Densitometric analysis of p-FOXO3 (inactive). (G and H) Quantitative analysis of isolated UMOD-producing primary murine epithelial cells at 24 weeks for autophagy protein regulators and gene transcripts. (G) Western blot analysis of protein regulators of autophagy. (H) Quantitative PCR for a broad panel of autophagy-related genes, supplemented with additional data points from RNA-seq analysis of isolated UMOD-producing cells at 24 weeks. Gene expression was normalized to Gapdh. (I) Transcript analysis of Foxo3 and Tfeb from RNA-seq of isolated UMOD-producing cells at 24 weeks. Gene expression was normalized to Gapdh. Data represent the mean ± SEM. *P < 0.05, ***P < 0.001, and ****P < 0.0001, by 2-tailed Student’s t test or 2-way ANOVA with post-hoc testing. n = 6–9 per group.
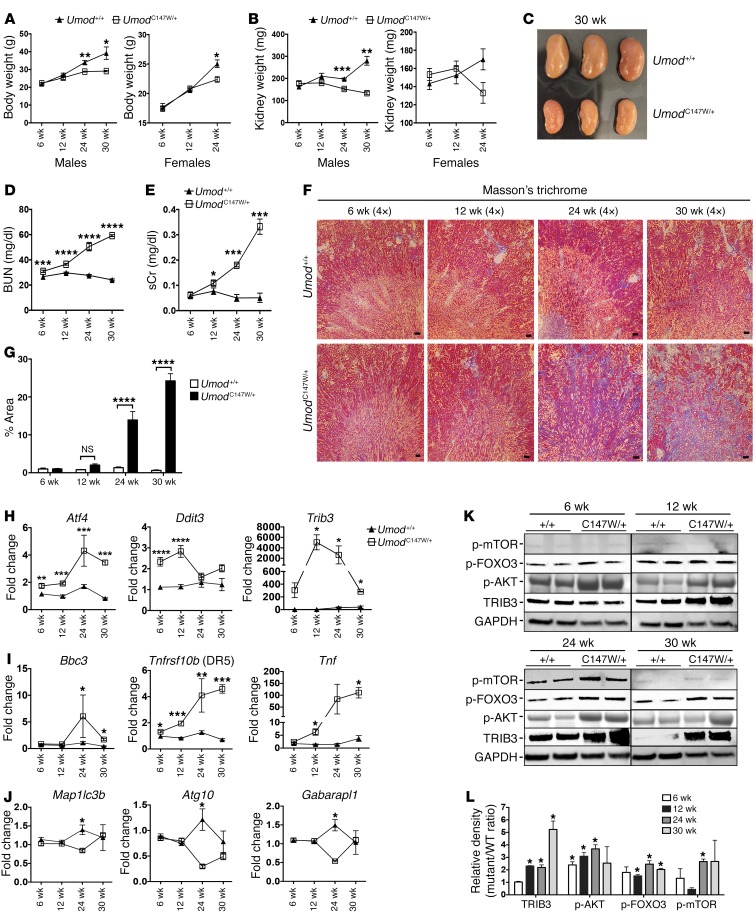
Time course reveals disease progression in UmodC147W/+ mice.
In order to achieve a better understanding of the trajectory of the phenotype in the mutant mice and how that relates to the cell-signaling and homeostatic pathways implicated, we characterized disease from 6 weeks until 30 weeks. Our analysis revealed that the expected increases in body weight with aging did not occur in mutant mice, with a significant divergence in weight for WT mice at 24 weeks. Kidney weights also were significantly lower in mutant mice by 24 weeks. By 30 weeks, we observed a nearly 50% reduction in kidney mass (Figure 5, A–C). Both BUN and sCr levels were significantly elevated in mutant mice by 12 weeks and progressively increased between 12 and 30 weeks (Figure 5, D and E). We observed large differences in Masson’s trichrome staining for deposition of fibrotic matrix by 24 weeks, coincident with highly upregulated matrix protein transcripts (Figure 5, F and G, and Supplemental Figure 4A). To understand the extent of activation of ER stress, cell death, and suppression of autophagic pathways, we purified primary UMOD-producing epithelial cells from mutant and littermate control kidneys. We confirmed significantly elevated expression of Atf4 and Ddit3 (Chop) as early as 6 weeks, while the peak of Trib3 expression occurred at 12 weeks (Figure 5H). It is reported that ER stress–induced expression of Trib3 is regulated by CHOP and ATF4 and that TRIB3 plays a central role in mediating ER stress–induced cell death by sensitizing cells to TNF-α– and TRAIL-mediated apoptosis (60, 61). In line with ER stress activation giving way to a proapoptotic program, UMOD-producing cells revealed elevated Bbc3 (P53-upregulated modulator of apoptosis [PUMA]) at 24 weeks (Figure 5I). PUMA is a central proapoptotic factor elicited by ER stress (62). Both Tnf and Tnfrsf10b (death receptor 5 [DR5]), the receptor for TRAIL, were significantly upregulated in UMOD-producing cells by 12 weeks and continued to be expressed at high levels through 30 weeks (Figure 5I). This pattern was distinct from that of other innate immune cytokines, which had a bimodal distribution (Supplemental Figure 4B). Notably, analysis of autophagy gene expression in cells isolated over this time course revealed no significant differences in expression at 6, 12, and 30 weeks compared with expression in WT kidney, but, as reported above, showed a statistically significant downregulation at 24 weeks, which is when the florid fibrotic disease first appears (Figure 5J and Supplemental Figure 4C). The observed lack of difference in expression, as opposed to downregulation, at 30 weeks could be explained by selective pressure from the disease environment from 24 to 30 weeks favoring cells with more active expression of autophagy genes, affording them enhanced survival to later time points. Furthermore, in isolated UMOD-producing cells, we also observed differential expression of proteins that are key regulators of autophagy, including increased active p-mTOR and inactive p-FOXO3 and its negative regulator p-AKT in mutant cells (Figure 5, K and L). Consistent with the transcript expression levels, we found that TRIB3 protein levels were increased starting at 12 weeks and maintained through 30 weeks (Figure 5, K and L). Overall, these time-course findings suggest that ER stress upregulation precedes cell death pathway signaling and that active suppression of autophagy is a relatively late response to disease.
Figure 5. Time course reveals a pattern of disease progression in UmodC147W/+ mice.
(A) Body weight over time in male and female mice. (B) Left kidney weight over time in male and female mice. (C) Image of 30-week-old kidneys showing a marked reduction the size of the kidneys with mutation. (D) BUN measurements over a 30-week period. (E) sCr over a 30-week period. (F) Masson’s trichrome–stained images from 6 to 30 weeks. Scale bars: 50 μm (original magnification, ×4). (G) Quantification of collagen area (blue stain) from Masson’s trichrome–stained images. (H–J) Quantitative PCR of directly isolated UMOD-producing primary murine tubular epithelial cells over time. (H) Time course showing levels of ER stress genes. (I) Time course showing levels of apoptosis/innate immune genes. (J) Time course showing levels of autophagy-related genes. (K) Representative Western blots to detect proteins in isolated UMOD-producing primary murine epithelial cells from 6 to 30 weeks showing TRIB3 and key autophagy regulators. (L) Densitometric analysis of TRIB3, p-AKT, p-FOXO3, and p-mTOR. Data represent the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 2-way ANOVA with post-hoc testing. n = 8 per group for 6 to 24 weeks; n = 3 per group for 30 weeks.
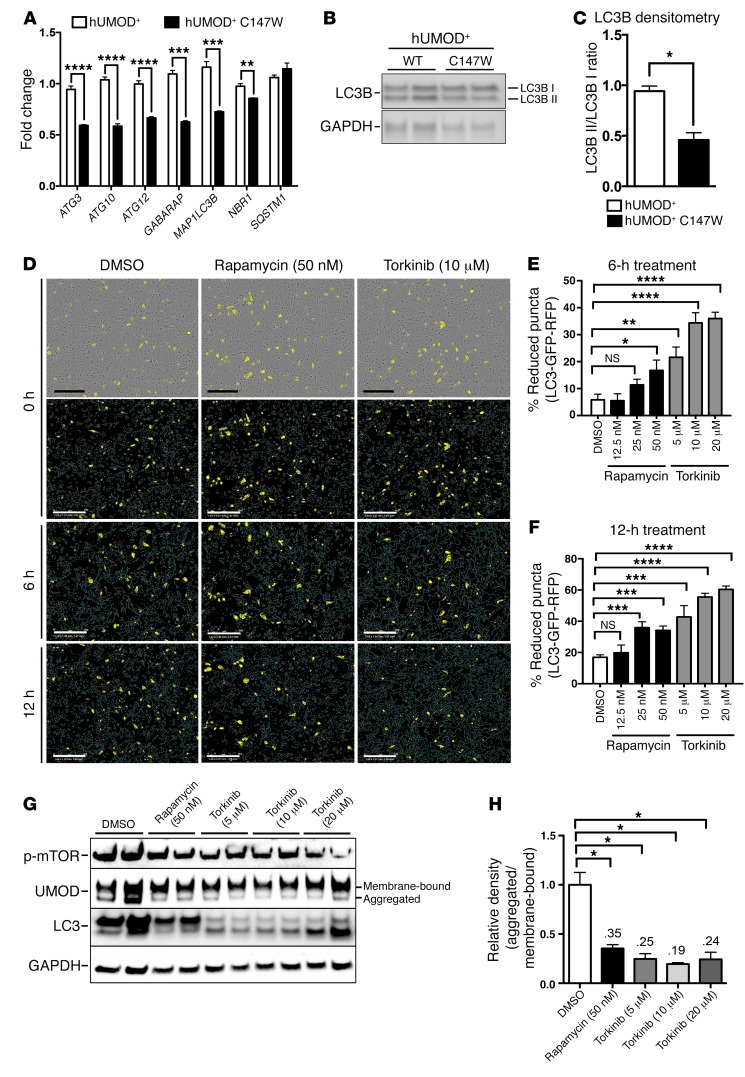
Autophagy enhancement removes mutant UMOD in primary human renal epithelial cells.
We uncovered an overall lack of upregulation of autophagy across all time points tested and a paradoxical suppression of autophagy during the established disease state at 24 weeks in primary purified murine UMOD-producing cells. We sought, therefore, to explore whether autophagy enhancement could benefit cell health by promoting the removal of aggregated protein to the lysosomal machinery. To screen for the efficacy of relevant compounds in removing aggregated mutant protein in an in vitro system, we established primary, immortalized human UMOD-producing cell lines expressing either WT or mutant UMOD. Human UMOD cells that were directly isolated were highly enriched for UMOD expression as well as expression of 2 markers of distal renal epithelium, SLC12A1 (NKCC2) and KCNJ1 (ROMK) (13, 20, 63, 64) (Supplemental Figure 2, E–G). Expression of the proximal tubule marker HAVCR1 (KIM1) was largely absent from UMOD-producing cell isolates compared with purified proximal tubule control fractions (Supplemental Figure 2H). Like the mouse UMOD cells, human UMOD cells lost robust expression of UMOD by day 10 in culture (Supplemental Figure 2E). We therefore used lentiviral vectors to establish stable transgenic expression of the WT and mutant forms of UMOD in 2 independent cell lines. The induced expression levels of WT and mutant UMOD were highly similar in the respective cell lines (Supplemental Figure 2I). Characterization of these independent cell lines indicated that expression of mutant UMOD directly reduced expression of important autophagy genes including ATG3, ATG12, and MAP1LC3B (Figure 6A). Consistent with our mouse model, we also observed a significant reduction in the ratio of active to inactive LC3B in the mutant human cell line (Figure 6, B and C). Therefore, production of mutant UMOD protein specifically suppresses autophagy. To study this further, we transiently expressed a transgenic LC3B fusion protein tagged with both red fluorescent protein (RFP) and GFP fluorophores, whereby maturation from the autophagosome to autolysosome via fusion with an acidic lysosome results in loss of the acid-sensitive GFP fluorescence from tagged LC3B, and thus loss of colocalized GFP-RFP puncta. Treatment of the UMOD-mutant cell line with the autophagy-activating compounds rapamycin or torkinib enhanced autophagy flux, detected by loss of LC3B-GFP-RFP (yellow) puncta as early as 6 hours after treatment with the compound, with the effect continuing to 12 hours (Figure 6, D–F). Consistent with this activation of autophagy flux, we observed a statistically significant clearance of the intracellular, aggregated form of mutant UMOD following treatment with either rapamycin or torkinib (Figure 6, G and H).
Figure 6. Autophagy enhancement degrades accumulated mutant UMOD in primary human kidney epithelial cells.
(A) Quantitative PCR of human UMOD-producing cell lines (hUMOD+) for key autophagy-related genes. (B) Western blot analysis to detect LC3B (LC3B I inactive, LC3B II active). (C) Ratio of active to inactive LC3B by densitometric analysis. (D) Images of LC3B-GFP-RFP puncta in hUMOD+ C147W-mutant cell line with either DMSO (0.1%), rapamycin (50 nM), or torkinib (10 μM) at 0, 6, and 12 hours. Scale bars: 300 μm (original magnification, ×10). Note the bright-field representation at the top with yellow puncta. Blue lines in the images indicate the cell outline. (E and F) Quantification of the percentage of reduction in the number of LC3B-GFP-RFP puncta from 0 to 6 hours and 0 to 12 hours. (G) Western blot analysis of p-mTOR, UMOD, and LC3B (LC3B I inactive, LC3B II active) in the hUMOD+ C147W-mutant cell line. Note the clearance of the lower, aggregate-sized band of UMOD with treatment of the autophagy enhancers rapamycin and torkinib. (H) Densitometric analysis of the ratio of aggregated to membrane-bound forms of UMOD. Representative data from 1 of 3 (A–F) or 1 of 5 (G and H) experiments are shown. Data represent the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, by 2-tailed Student’s t test or 2-way ANOVA with post-hoc testing. n = 3–6 per group.
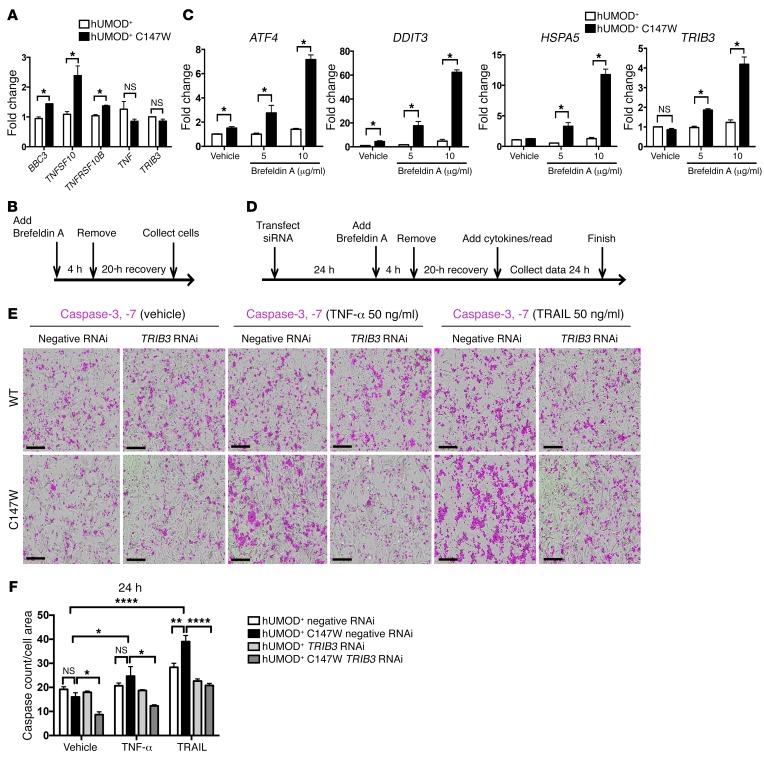
Silencing of TRIB3 in primary human renal epithelial cells rescues ER stress–induced sensitivity to TNF-α– and TRAIL-mediated apoptosis.
To study the role of potential signaling pathways downstream of ER stress activation on cell fate, we used the human UMOD-producing cell lines to establish an in vitro human cell–based assay and tested the effect of induced, transient ER stress on cell response. In steady-state conditions, mutant UMOD–producing cell lines expressed higher levels of transcripts for the proapoptotic factors BBC3, TNFSF10 (encoding TRAIL), and TNFRSF10B (encoding death receptor 5, or DR5), whereas the expression levels of TNF and TRIB3 were no different (Figure 7A). We stimulated mild, transient ER stress by short exposure to low-dose brefeldin A (an inhibitor of trafficking from the ER to the Golgi apparatus) and observed a dose-dependent increase in expression of ATF4, DDIT3 (CHOP), HSPA5 (BIP), and TRIB3 in the UMOD-mutant line that was not seen in the control cell lines expressing WT UMOD (Figure 7, B and C). Interestingly ATF4 and DDIT3 were significantly upregulated at baseline; however, transcripts for the chaperone proteins HSPA5 and TRIB3 were not. This would suggest that some elements of ER stress are moderately activated in steady-state conditions in mutant cells, but the level of stress is not substantial enough to elicit expression of the downstream genes. A threshold level of ER stress must be met before the cell death–promoting gene TRIB3 is upregulated. In addition, when the cell lines were subjected to transient ER stress, we observed that the mutant UMOD-producing cell lines were more susceptible to caspase-3 and caspase-7 activation when subsequently treated with exogenous TNF-α or TRAIL for 24 hours (Figure 7, D–F). To determine the role of TRIB3 in mediating the ER stress–induced susceptibility to cell death, we silenced TRIB3 with siRNA (Supplemental Figure 5). Once the method of silencing was established, the cell lines were pretreated with siRNA and then exposed to ER stress followed by exogenous cytokines. The level of caspase-3 and caspase-7 activity upon TNF-α and TRAIL treatment in mutant cell lines was markedly suppressed when TRIB3 was silenced (Figure 7, D–F).
Figure 7. Silencing of TRIB3 in primary human renal epithelial cells rescues them from ER stress–induced sensitivity to TNF-α– and TRAIL–mediated apoptosis.
(A) Quantitative PCR of human UMOD-producing cell lines for the expression of genes related to ER stress–mediated apoptosis. (B and C) In vitro protocol for inducing mutant UMOD protein–mediated apoptosis. (B) Timeline diagram indicating the experimental setup for transient treatment of UMOD-producing cell lines with the ER stress inducer brefeldin A (5 and 10 μg/ml). (C) Quantitative PCR of human UMOD-producing cell lines treated as indicated in panel B for key genes related to ER stress. Note the dose responsiveness to increasing concentrations of brefeldin A. (D–F) In vitro platform for assessing the role of TRIB3 in ER stress–mediated apoptosis in primary human UMOD-producing cell lines. (D) Schema of the experimental protocol for TRIB3 silencing, transient induction of ER stress with brefeldin A, recovery, stimulation with cytokines to induce apoptosis, and quantification of caspase-3 and caspase-7 activity. (E) Images of caspase-3, -7 activity (in purple) of human UMOD-producing cell lines after 24 hours of treatment with vehicle, TNF-α (50 ng/ml), or TRAIL (50 ng/ml). Scale bars: 300 μm. (F) Quantification of caspase-3, -7 activity normalized to the cell area. Note the marked reduction in response to TNF-α and TRAIL treatment in the mutant cell line with TRIB3 silencing. Representative data from 1 of 3 experiments are shown (A, C, E, and F). Data represent the mean ± SEM. *P < 0.05, **P < 0.01, and ****P < 0.0001, by 2-tailed Student’s t test or 2-way ANOVA with post-hoc testing. n = 3–6 per group.
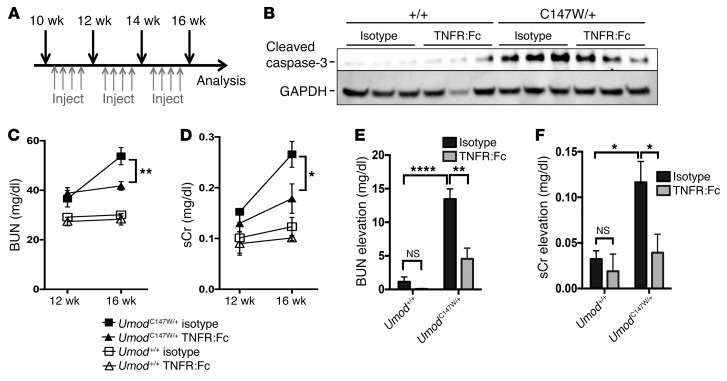
Suppression of TNF-α signaling slows the progression of kidney disease.
UMOD-mutant kidneys have increased expression of critical proapoptotic mediators, including TRIB3, DR5, TNF-α, and BCL2-binding component-3, and have elevated active caspase-3 by 24 weeks. UMOD-mutant human cell lines are more susceptible to TNF-α– and TRAIL-mediated apoptosis following induced ER stress. We therefore reasoned that inhibition of TNF-α signaling might be an attractive approach to reduce the activation of the apoptotic signaling cascade in susceptible distal epithelium and thereby preserve organ function. To test this hypothesis, we stimulated mutant UMOD human cell lines with TNF-α and subsequently treated them with the soluble recombinant fusion protein TNFR:Fc or isotype IgG control antibody. TNFR:Fc reduced caspase-3, -7 activity to below baseline levels, indicating that the inhibitory activity could suppress responses to exogenous TNF-α, but also probably suppresses the effect of endogenously produced TNF-α (Supplemental Figure 6A). Next, we treated a cohort of UmodC147W/+ mice and their littermate controls twice weekly from 10 to 16 weeks of age with isotype control antibody or soluble TNFR:Fc to antagonize TNF-α (Figure 8A). This time frame is consistent with the first signs of an ER stress–regulated apoptotic response, such as TRIB3 upregulation. While whole kidneys from mutant mice treated with IgG control showed substantial enrichment of the active, cleaved form of caspase-3 at 16 weeks, kidneys from TNFR:Fc-treated mice showed a marked reduction of active caspase-3 (Figure 8B). Consistent with a reduction of apoptotic cell death mechanisms, we observed a statistically significant improvement in renal function in TNFR:Fc-treated mutant mice compared with that in isotype-treated animals, demonstrated by reduced levels of BUN and sCr (Figure 8, C–F). Furthermore, even at 16 weeks, prior to the florid fibrosis observed at 24 weeks, trends in reduced expression of the profibrotic genes Fn1, Col1a1, Serpine1 (Pai1), and Ctgf and the inflammatory and apoptotic mediators Ccl3, Cxcl2, Il6, Tnf and Bbc3, Casp12, and Tnfrsf10b (Dr5), respectively, were observed in TNFR:Fc-treated mutant mice (Supplemental Figure 6B). This approach also reduced protein levels of the profibrotic signaling factor CTGF (Supplemental Figure 6, C and D) and the extracellular matrix proteins fibronectin and laminin (Supplemental Figure 6, E–G).
Figure 8. TNF-α blockade prevents a functional decline in UmodC147W/+ mice.
(A) Schema showing the therapeutic protocol for delivery of TNFR:Fc (10 mg/kg/dose) and evaluation of disease. (B) Western blot analysis to detect cleaved caspase-3 (active form) in whole-kidney tissue from 16-week-old mice. Note the reduction of active caspase-3 with TNFR:Fc treatment. (C) BUN measurements from 12 to 16 weeks. (D) sCr levels from 12 to 16 weeks. (E) Quantification of longitudinal elevation in BUN per individual animal. (F) Quantification of longitudinal elevation in sCr per individual animal. Data represent the mean ± SEM. *P < 0.05, **P < 0.01, and ****P < 0.0001, by 2-way ANOVA with post-hoc testing. n = 4–6 per group.
Discussion
The UPR, which participates in the broader ER stress response, is an adaptive mechanism that can protect cells from the consequences of acute protein misfolding, engage chaperones such as BiP and p58IPK, and reduce protein synthesis and energy expenditure. However, in conditions of chronic ER stress, in which the UPR does not effectively restore proteostasis of the cell, its persistent activation potentiates innate immune responses, increases inflammatory signaling, and can ultimately lead to cell death (62, 65–69). This series of events has been demonstrated in the setting of misfolded proteins in the CNS in diseases such as amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease (70, 71). Similarly, the studies presented here highlight the idea that misfolded UMOD activates UPR mechanisms and triggers ER stress–dependent cell death in renal tubule epithelial cells.
Recent studies have suggested that mutations in the UMOD gene, which are found in UAKD, account for up to 25% of inherited chronic kidney diseases following exclusion of polycystic kidney disease and Alport nephropathy (1). Although UAKD is considered quite rare in the broader community, the lack of a phenotype to distinguish it from other forms of chronic kidney disease, the presentation delayed often until adulthood, and the lack of a diagnostic test suggest that the prevalence may be higher than originally thought. Moreover, polymorphisms in the UMOD locus have been identified with high frequency in hypertensive and diabetic kidney diseases, but the function of those polymorphisms is currently unclear (11, 72). Although there are multiple mutations in the coding regions of the UMOD gene, the majority of those studied disrupt critical cysteine residues, preventing the proper folding of the protein and ultimately leading to impaired trafficking and ER retention (28). It is likely, therefore, that many of these mutations resulting in ER retention also cause ER stress–mediated tubular apoptosis. Importantly, in the clinic, apoptosis of affected distal epithelium from a renal biopsy of a patient with UAKD has also been reported (73). Here, we show for the first time to our knowledge that using a widely prescribed inhibitor of TNF-α to abrogate pro-death signaling is a potentially effective therapeutic approach for such patients with this disease.
Although exacerbated ER stress has been recognized to cause cell death, the molecular mechanisms connecting the UPR to ER stress pathway activation and thence cell death have recently been proposed (36, 37, 62, 74). Notably, TRIB3, a pseudokinase with docking or scaffold functions that can regulate complex signaling pathways, has been shown to negatively feed back on the UPR. It has been reported in neurons and pancreatic β cells to play an important role in eliciting programmed cell death in conditions of chronic ER stress by sensitizing cells to TNF-α– and TRAIL-induced apoptosis (61, 75).
In our in vivo model of UAKD, using an unbiased approach, we identified Trib3 as a highly upregulated factor early in the course of disease in both whole tissue and specifically in mutant UMOD-producing cells. We separately identified the ER stress pathway mediators ATF4 and CHOP engaged early in disease pathogenesis as well. Moreover, we identified increasing positive regulation of TNF-α signaling as disease progressed and subsequent activation of ER-tethered caspase-12 and executioner caspase-3 at the late stage of disease. We confirmed an important role for TRIB3 in human distal tubule cells expressing mutant UMOD p.Cys147Trp in the transduction of ER stress signaling to sensitize the cell to apoptosis. First, there was marked expression of TRIB3 upon induction of ER stress with brefeldin A treatment in the mutant cell line, which led to increased sensitivity to exogenous TNF-α– and TRAIL-induced caspase-3, -7 activation and apoptosis compared with that observed in WT cells. Silencing of TRIB3 using siRNA, after pretreatment with brefeldin A, substantially reduced ER stress–mediated caspase-3, -7 activation upon treatment with TNF or TRAIL. Importantly, the steady-state condition in vitro yielded only mild elevations of ER stress genes including ATF4 and CHOP and was insufficient to induce TRIB3. Expression of mutant UMOD alone did not result in greater susceptibility to cell death. Only upon brefeldin A pretreatment as an additional ER stress insult did we find differential effects on apoptosis in mutant cells compared with WT cells. This finding suggests that mild UPR responses are compensatory and that second hits from the environment are required to overwhelm the compensatory ER stress mechanisms and drive distal epithelium toward an exacerbated, chronic ER stress response, ultimately pushing the distal epithelium toward cell death. It should be noted that a recent report described a different point mutation knocked into the mouse Umod gene (UmodC125R). In that model, the phenotype was much milder than that seen in this report, and no increase in apoptosis, detected by nick-end labeling, was demonstrated at 6 months (76).
The observation that mutant epithelium in our model expressed TNF superfamily members and was more susceptible to TNF-α– and TRAIL-mediated cell death led us to test whether TNF-α blockade could be a rational and safe approach to protect the nephron from cell death and thereby preserve organ function. In a proof-of-concept experiment, systemic delivery of TNFR:Fc to mutant mice over a 6-week period reduced active caspase-3 in whole kidney and substantially reduced loss of organ function. Therefore, an anti–TNF-α approach may be effective in preventing nephron loss in patients with UAKD, particularly if activation of TNF-α signaling in the diseased kidney can be established, such as from a urinary biomarker. It is noteworthy that in hypertensive and diabetic kidney diseases compared with normally functioning kidneys, KEGG pathway analysis of differentially expressed genes (DEGs) reveals highly significant enrichment of TNF signaling, suggesting that further study of susceptibility of epithelium to TNF-α–dependent cell death in other forms of chronic kidney disease merits investigation (Supplemental Figure 7).
Many chronic diseases are characterized by misfolded proteins, which are caused either by mutations/polymorphisms in coding sequences, or by aberrant protein production or clearance. These include neurodegenerative diseases like Alzheimer’s, Parkinson’s, Huntington’s, ALS, and prion encephalopathies; lung diseases such as cystic fibrosis and α1–anti-trypsin deficiency; and hematological diseases including sickle cell anemia. An important contribution of autophagy to the manifestation of several of these diseases has been identified (40, 77). More specifically, the body of research around aggrephagy (autophagy of aggresomes/aggregated proteins) describes a conserved endogenous mechanism for the removal of harmful protein aggregates too large to be processed effectively by the proteasome (39, 78, 79) and highlights the idea that insufficient autophagy due to genetic predisposition may underlie the manifestation of disease. In this new, genetically defined model of UAKD, autophagy was not engaged at the transcript level and was actively suppressed at the protein level, despite toxic accumulation of mutant UMOD protein. Mechanistically, we identified negative regulation of 2 critical transcription factors necessary for autophagy gene transcription (47, 50): FOXO3 and TFEB in mutant tissues and cells. Consistent with this finding, we observed increased levels of their known negative regulators, AKT and mTOR, respectively. The level of inactive, p-ULK1, also negatively regulated by mTOR, was increased in whole-kidney tissue. The accumulation of P62 and the reduction of active LC3B II, hallmarks of suppressed autophagy, were clearly demonstrated in the diseased kidney. Thus, autophagy suppression in our model can be directly attributed to enhanced AKT and mTOR signaling as a likely result of increased signaling from growth factors such as PDGF, connective tissue growth factor (CTGF), and IGF-1 (80–85), which are abundant in settings of inflammation and fibrosis and elevated levels of which we observed in whole kidney tissue at 24 weeks (Figure 1I). In particular, chronic exposure to IGF-1 has been demonstrated to result in the reduction of both autophagy and cell viability (82). More specifically, our model suggests that as the mutant animals age, accumulation of the misfolded UMOD protein and environmental ER stress “second hits” progressively incur a pathological ER stress–mediated apoptosis through TNF superfamily members and TRIB3, ultimately resulting in exacerbated inflammation and fibrosis. The growth factors that accompany this fibrotic response, including CTGF, PDGF, and IGF-1, promote AKT and mTOR signaling, thus negatively regulating the checkpoints for autophagy induction at both the transcriptional level, through TFEB and FOXO3 inactivation, and the protein level, by inhibition of ULK1 (Supplemental Figure 8). Importantly, in human UMOD-producing cells expressing mutant UMOD p.Cys147Trp, we successfully abrogated this suppressive autophagy phenotype by treatment with the mTOR inhibitors rapamycin or torkinib to increase autophagy flux. Both drugs enhanced clearance of the aggregated mutant form of UMOD p.Cys147Trp in the in vitro assay and therefore may be an attractive approach for treating UAKD. Pharmacological approaches to augment autophagy and regulate mTOR are in development for many disease indications, including neuropathies, kidney disease, cardiovascular disease, immune thrombocytopenia, and colorectal cancer, among others (86–91). Of note, patients with autosomal dominant polycystic kidney disease (ADPKD) in the SIRENA study who received a 6-month treatment of sirolimus (rapamycin) added to conventional therapy experienced halted growth of kidney cysts, with no observed serious adverse events (91). Nevertheless, long-term treatment with mTOR inhibitors may not be sufficiently well tolerated in patients with chronic diseases such as UAKD. Efforts are currently underway to identify mTOR-independent mechanisms to enhance autophagy (92, 93).
In addition to strategies that inhibit downstream cell death mediators like TNF-α or activate endogenous autophagy programs to alleviate the cellular burden of misfolded protein accumulation, an emergent therapeutic approach to treating chronic states of ER stress and UPR activation, particularly in the field of neurodegeneration (94, 95), has yielded several small-molecule modulators of components of the UPR itself. In particular, compounds such as GSK2606414 and ISRIB act at different levels of the PERK/eIF2α axis to inhibit the UPR and have proven effective in murine models of neurodegeneration (96–98). Nevertheless, the current studies using these modulators highlight important concerns that question the translatability of these compounds into effective treatments for human disease: the lack of long-term tolerability due to toxicity to highly secretory organs like the liver and pancreas, in particular for the PERK inhibitor GSK2606414, and the lack of solubility of ISRIB (96, 97, 99). Concern for systemic, long-term administration of these inhibitors also stems from the understanding that transient UPR activation is indeed beneficial to cells and has evolved as an adaptive mechanism to protect cells against instances of acute protein misfolding, only becoming pathological in chronically activated states that trigger downstream events, such as TRIB3 upregulation and TNF signaling, that can lead to cell death (60, 100–104). Therefore, we believe that, although modulation of the UPR is a sound strategy, mechanistically speaking, to address the underlying pathology of UAKD in the mutant UMOD-expressing epithelium, the necessity for systemic, long-term administration of the available UPR inhibitors would incur unwanted toxicity. Future studies will be required to identify and test viable UPR modulators whose long-term, systemic administration can be better tolerated in the case of chronic diseases such as UAKD.
Overall, we conclude that the UmodC147W/+ mouse model described here recapitulates many features of UAKD. We believe our model provides critical insights into the relevant signaling pathways related to UAKD pathogenesis and informs on the temporal progression of the disease in order of the underlying pathological events (Supplemental Figure 8). By creating an RNA-seq database of a time course of whole-kidney as well as UMOD-producing epithelial cell–specific RNAs, we have enabled an unbiased approach to identifying an important role for chronically activated ER stress responses that promote the production of innate immune signaling molecules, leading to a hyperinflammatory state and ultimately tubular cell death. By perturbing human kidney epithelial cells expressing mutant UMOD protein, we have validated the role of these relevant cellular responses in a human system. In addition, we have identified maladaptive suppression of autophagy in settings of fibrosis, which may also serve to amplify the toxic consequences of misfolded protein accumulation. Finally, we provide initial validation in vivo that suppression of TNF signaling with drugs that are widely used in clinical practice may benefit patients with UAKD.
Methods
Development of the UmodC147W/+ mouse line.
The CRISPR-Cas9 system was used to insert the p.Cys147Trp mutation into the UMOD gene at residue 147. To reduce the likelihood of off-target cutting by Cas9 and to maximize on-target cleavage efficiency, established guidelines for single-guide RNA (sgRNA) design were followed (29, 30, 105, 106). To further maximize guide specificity, we used the online tool ZiFiT to select for a truncated, 17-bp guide with the fewest off-target sites with fewer than 3 mismatches (107). Truncated guides (tru-gRNAs) are more intolerant of mismatches at off-target sites, allowing for greater specificity of guided Cas9 cleavage (108). Off-target mutations are reportedly very rare in Cas9-generated mice (109). Backcrossing founder animals with WT inbred strains will effectively segregate out any unlinked variants that arise from off-target Cas9 cleavage (109). Ultimately, a single sgRNA molecule was designed with a 17-bp target sequence (5′-GTCCCGAGGGCTTTACA-3′). The sgRNA was synthesized by first annealing and cloning oligonucleotide pairs harboring 5-bp overhangs into the vector provided by the GeneArt CRISPR Nuclease Vector Kit (Thermo Fisher Scientific) and then performing in vitro runoff transcription (IVT) off the U6 promoter using a HiScribe T7 High Yield RNA Synthesis Kit (New England BioLabs) according to the manufacturer’s instructions. sgRNA was purified using an RNeasy Mini Kit (QIAGEN). A single 153-nt single-stranded oligonucleotide (ssODN) donor template harboring the required mutation as well as 3 silent mutations, included to prevent repeated nuclease activity of Cas9 at the recombined locus, was ordered from Integrated DNA Technologies (IDT) (5′-CTGGCCACCTGTGTCAACACAGAA GGCGACTACTTGTGcgtgtgtcccgagggAttCacCggggatggttggtacTGGGAGTGCTCCCCAGGCTCCTGTGAGCCAGGACTGGACTGCTTG CCCCAGGGCCCGGATGGAAAGCTGGTGTGTCAA-3′). Together, the sgRNA (50 ng/μl), donor ssODN (100 ng/μl), and Cas9 mRNA (100 ng/μl) were microinjected into single-cell-stage embryo pronuclei derived from C57BL/6J mice at the Genome Modification Facility of Harvard University (Cambridge, Massachusetts, USA). Sixty founder males were screened for recombination and sequenced across the relevant locus. Three positive F0 founders were confirmed by sequence alignment and bred with C57BL/6J females purchased from The Jackson Laboratory. A single independent mouse line was generated with 2 rounds of backcrossing the positive F1 generation with C57BL/6J mice, and a TaqMan SNP genotyping protocol was designed to identify heterozygous, homozygous, and WT offspring: forward, 5′-GGGCTCAGTAACTGTCATGC-3′; reverse, 5′-AGTCCAGTCCTGGCTCACA-3′; reporter 1 WT, 5′-TTGGTACTGTGAGTGCTC-3′; reporter 2 mutant, 5′-TGGTACTGGGAGTGCTC-3′.
Further details on the methods can be found in the Supplemental Experimental Procedures.
Statistics.
All data are presented as the mean ± SEM. Significance of difference between groups was evaluated by 2-way ANOVA with Bonferroni’s post-hoc analysis or Student’s t test where appropriate (GraphPad Prism 7). A P value of less than 0.05 was considered statistically significant.
Study approval.
All animal studies were performed under protocols approved by the IACUCs of Biogen (protocol 0489–2013) and the University of Washington (protocol 4244). Human kidney tissue was obtained from kidneys discarded during nephrectomy procedures performed at Tufts New England Medical Center (Boston, Massachusetts, USA) under a research agreement. All patients consented to the use of their discarded tissue for research purposes, and all samples were deidentified. A pathologist scored kidney tissue for the presence of disease, and basic clinical and laboratory data were provided.
Author contributions
Conceptualization: BGJ and JSD; methodology: BGJ, LTD, GM, DR, LF, ZL, and JSD; validation: BGJ, DR, and JSD; formal analysis: BGJ and LTD; investigation: BGJ, LTD, GM, DR, AM, LF, AMR, ZL, and JSD; resources: JSD and LF; data curation: BGJ, LTD, and JSD; writing of the manuscript, review, and editing: BGJ and JSD; visualization: BGJ and JSD; supervision: JSD, BGJ, DR, and LF; project administration: JSD; and funding acquisition: JSD.
Supplementary Material
Acknowledgments
The studies were funded by Biogen and NIH grants DK093493 and DK094768. The authors thank Timothy Zheng (Biogen) for assistance with dose determination for soluble TNFR:Fc; Wenning Qin (Biogen) and Lynn Valley (Charles River Laboratories) for managing animal husbandry; Suzanne Szak (Biogen) for submission of RNA-seq data to the NCBI’s GEO database; and Chris Ehrenfels (Biogen) for management of the microscopy core.
Version 1. 10/09/2017
Electronic publication
Version 2. 11/01/2017
Print issue publication
Footnotes
Conflict of interest: B.G. Johnson, L.T. Dang, G. Marsh, A.M. Roach, A. Monti, D. Reyon, L. Feigenbaum, and J.S. Duffield hold stock in Biogen.
Reference information: J Clin Invest. 2017;127(11):3954–3969.https://doi.org/10.1172/JCI93817.
Contributor Information
Bryce G. Johnson, Email: brycej4@gmail.com.
Graham Marsh, Email: graham.marsh@biogen.com.
Allie M. Roach, Email: allie.roach@biogen.com.
Zebulon G. Levine, Email: zebulonlevine@fas.harvard.edu.
Anthony Monti, Email: amonti05@gmail.com.
Deepak Reyon, Email: deepak.reyon@gmail.com.
Lionel Feigenbaum, Email: lionel.feigenbaum@gmail.com.
Jeremy S. Duffield, Email: jeremysd@u.washington.edu.
References
- 1.Gast C, Marinaki T, Arenas-Hernandez M, Campbell S, Venkat-Raman G. SuO028 genetic testing reveals increased prevalence of uromodulin associated kidney disease. Nephrol Dial Transplant. 2015;30(suppl_3):iii56 [Google Scholar]
- 2.Gardner KD. Evolution of clinical signs in adult-onset cystic disease of the renal medulla. Ann Intern Med. 1971;74(1):47–54. doi: 10.7326/0003-4819-74-1-47. [DOI] [PubMed] [Google Scholar]
- 3.Hart TC, et al. Mutations of the UMOD gene are responsible for medullary cystic kidney disease 2 and familial juvenile hyperuricaemic nephropathy. J Med Genet. 2002;39(12):882–892. doi: 10.1136/jmg.39.12.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rampoldi L, et al. Allelism of MCKD, FJHN and GCKD caused by impairment of uromodulin export dynamics. Hum Mol Genet. 2003;12(24):3369–3384. doi: 10.1093/hmg/ddg353. [DOI] [PubMed] [Google Scholar]
- 5.Dahan K, et al. A cluster of mutations in the UMOD gene causes familial juvenile hyperuricemic nephropathy with abnormal expression of uromodulin. J Am Soc Nephrol. 2003;14(11):2883–2893. doi: 10.1097/01.ASN.0000092147.83480.B5. [DOI] [PubMed] [Google Scholar]
- 6.Hodanová K, et al. Mapping of a new candidate locus for uromodulin-associated kidney disease (UAKD) to chromosome 1q41. Kidney Int. 2005;68(4):1472–1482. doi: 10.1111/j.1523-1755.2005.00560.x. [DOI] [PubMed] [Google Scholar]
- 7.Böger CA, et al. Association of eGFR-related loci identified by GWAS with incident CKD and ESRD. PLoS Genet. 2011;7(9):e1002292. doi: 10.1371/journal.pgen.1002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudbjartsson DF, et al. Association of variants at UMOD with chronic kidney disease and kidney stones-role of age and comorbid diseases. PLoS Genet. 2010;6(7):e1001039. doi: 10.1371/journal.pgen.1001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Köttgen A, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet. 2009;41(6):712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Köttgen A, et al. Association of estimated glomerular filtration rate and urinary uromodulin concentrations with rare variants identified by UMOD gene region sequencing. PLoS ONE. 2012;7(5):e38311. doi: 10.1371/journal.pone.0038311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padmanabhan S, et al. Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet. 2010;6(10):e1001177. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wuttke M, Köttgen A. Insights into kidney diseases from genome-wide association studies. Nat Rev Nephrol. 2016;12(9):549–562. doi: 10.1038/nrneph.2016.107. [DOI] [PubMed] [Google Scholar]
- 13.Trudu M, et al. Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med. 2013;19(12):1655–1660. doi: 10.1038/nm.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamm I, Horsfall FL. A mucoprotein derived from human urine which reacts with influenza, mumps, and Newcastle disease viruses. J Exp Med. 1952;95(1):71–97. doi: 10.1084/jem.95.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachmann S, Koeppen-Hagemann I, Kriz W. Ultrastructural localization of Tamm-Horsfall glycoprotein (THP) in rat kidney as revealed by protein A-gold immunocytochemistry. Histochemistry. 1985;83(6):531–538. doi: 10.1007/BF00492456. [DOI] [PubMed] [Google Scholar]
- 16.van Rooijen JJ, Voskamp AF, Kamerling JP, Vliegenthart JF. Glycosylation sites and site-specific glycosylation in human Tamm-Horsfall glycoprotein. Glycobiology. 1999;9(1):21–30. doi: 10.1093/glycob/9.1.21. [DOI] [PubMed] [Google Scholar]
- 17.Cavallone D, Malagolini N, Serafini-Cessi F. Mechanism of release of urinary Tamm-Horsfall glycoprotein from the kidney GPI-anchored counterpart. Biochem Biophys Res Commun. 2001;280(1):110–114. doi: 10.1006/bbrc.2000.4090. [DOI] [PubMed] [Google Scholar]
- 18.Bernascone I, et al. A transgenic mouse model for uromodulin-associated kidney diseases shows specific tubulo-interstitial damage, urinary concentrating defect and renal failure. Hum Mol Genet. 2010;19(15):2998–3010. doi: 10.1093/hmg/ddq205. [DOI] [PubMed] [Google Scholar]
- 19.Bates JM, et al. Tamm-Horsfall protein knockout mice are more prone to urinary tract infection: rapid communication. Kidney Int. 2004;65(3):791–797. doi: 10.1111/j.1523-1755.2004.00452.x. [DOI] [PubMed] [Google Scholar]
- 20.Renigunta A, Renigunta V, Saritas T, Decher N, Mutig K, Waldegger S. Tamm-Horsfall glycoprotein interacts with renal outer medullary potassium channel ROMK2 and regulates its function. J Biol Chem. 2011;286(3):2224–2235. doi: 10.1074/jbc.M110.149880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mo L, Liaw L, Evan AP, Sommer AJ, Lieske JC, Wu XR. Renal calcinosis and stone formation in mice lacking osteopontin, Tamm-Horsfall protein, or both. Am J Physiol Renal Physiol. 2007;293(6):F1935–F1943. doi: 10.1152/ajprenal.00383.2007. [DOI] [PubMed] [Google Scholar]
- 22.Wei X, et al. Novel uromodulin mutation in familial juvenile hyperuricemic nephropathy. Am J Nephrol. 2012;36(2):114–120. doi: 10.1159/000339752. [DOI] [PubMed] [Google Scholar]
- 23.Vylet’al P, et al. Alterations of uromodulin biology: a common denominator of the genetically heterogeneous FJHN/MCKD syndrome. Kidney Int. 2006;70(6):1155–1169. doi: 10.1038/sj.ki.5001728. [DOI] [PubMed] [Google Scholar]
- 24.Rezende-Lima W, Parreira KS, García-González M, Riveira E, Banet JF, Lens XM. Homozygosity for uromodulin disorders: FJHN and MCKD-type 2. Kidney Int. 2004;66(2):558–563. doi: 10.1111/j.1523-1755.2004.00774.x. [DOI] [PubMed] [Google Scholar]
- 25.Moskowitz JL, et al. Association between genotype and phenotype in uromodulin-associated kidney disease. Clin J Am Soc Nephrol. 2013;8(8):1349–1357. doi: 10.2215/CJN.11151012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lens XM, Banet JF, Outeda P, Barrio-Lucía V. A novel pattern of mutation in uromodulin disorders: autosomal dominant medullary cystic kidney disease type 2, familial juvenile hyperuricemic nephropathy, and autosomal dominant glomerulocystic kidney disease. Am J Kidney Dis. 2005;46(1):52–57. doi: 10.1053/j.ajkd.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 27. Bleyer AJ, Hart PS, Kmoch S. Autosomal Dominant Tubulointerstitial Kidney Disease, UMOD-Related. In: Pagon RA, et al, eds. GeneReviews(R). Seattle (WA): University of Washington, Seattle; 1993–2017. [PubMed] [Google Scholar]
- 28.Bernascone I, et al. Defective intracellular trafficking of uromodulin mutant isoforms. Traffic. 2006;7(11):1567–1579. doi: 10.1111/j.1600-0854.2006.00481.x. [DOI] [PubMed] [Google Scholar]
- 29.Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32(12):1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kouranova E, et al. CRISPRs for optimal targeting: Delivery of CRISPR components as DNA, RNA, and protein into cultured cells and single-cell embryos. Hum Gene Ther. 2016;27(6):464–475. doi: 10.1089/hum.2016.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Fazio P, Ocker M, Montalbano R. New drugs, old fashioned ways: ER stress induced cell death. Curr Pharm Biotechnol. 2012;13(11):2228–2234. doi: 10.2174/138920112802501962. [DOI] [PubMed] [Google Scholar]
- 32.Kalai M, et al. Regulation of the expression and processing of caspase-12. J Cell Biol. 2003;162(3):457–467. doi: 10.1083/jcb.200303157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamkanfi M, Kalai M, Vandenabeele P. Caspase-12: an overview. Cell Death Differ. 2004;11(4):365–368. doi: 10.1038/sj.cdd.4401364. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa T, et al. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403(6765):98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 35.Quiroz-Baez R, Ferrera P, Rosendo-Gutiérrez R, Morán J, Bermúdez-Rattoni F, Arias C. Caspase-12 activation is involved in amyloid-β protein-induced synaptic toxicity. J Alzheimers Dis. 2011;26(3):467–476. doi: 10.3233/JAD-2011-110326. [DOI] [PubMed] [Google Scholar]
- 36.Shiraishi H, Okamoto H, Yoshimura A, Yoshida H. ER stress-induced apoptosis and caspase-12 activation occurs downstream of mitochondrial apoptosis involving Apaf-1. J Cell Sci. 2006;119(Pt 19):3958–3966. doi: 10.1242/jcs.03160. [DOI] [PubMed] [Google Scholar]
- 37.Szegezdi E, Fitzgerald U, Samali A. Caspase-12 and ER-stress-mediated apoptosis: the story so far. Ann N Y Acad Sci. 2003;1010:186–194. doi: 10.1196/annals.1299.032. [DOI] [PubMed] [Google Scholar]
- 38.Vande Walle L, et al. Does caspase-12 suppress inflammasome activation? Nature. 2016;534(7605):E1–E4. doi: 10.1038/nature17649. [DOI] [PubMed] [Google Scholar]
- 39.Lystad AH, Simonsen A. Assays to monitor aggrephagy. Methods. 2015;75:112–119. doi: 10.1016/j.ymeth.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 40.Jiang P, Mizushima N. Autophagy and human diseases. Cell Res. 2014;24(1):69–79. doi: 10.1038/cr.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443(7113):780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 42.Bjørkøy G, Lamark T, Pankiv S, Øvervatn A, Brech A, Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Meth Enzymol. 2009;452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 43.Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015;282(24):4672–4678. doi: 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- 44.Tanida I, Ueno T, Kominami E. LC3 and autophagy. Methods Mol Biol. 2008;445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- 45.Dubchak I, et al. Whole-Genome rVISTA: a tool to determine enrichment of transcription factor binding sites in gene promoters from transcriptomic data. Bioinformatics. 2013;29(16):2059–2061. doi: 10.1093/bioinformatics/btt318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martina JA, Puertollano R. TFEB and TFE3: The art of multi-tasking under stress conditions. Transcription. 2017;8(1):48–54. doi: 10.1080/21541264.2016.1264353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milan G, et al. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat Commun. 2015;6:6670. doi: 10.1038/ncomms7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandri M. FOXOphagy path to inducing stress resistance and cell survival. Nat Cell Biol. 2012;14(8):786–788. doi: 10.1038/ncb2550. [DOI] [PubMed] [Google Scholar]
- 50.Warr MR, et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494(7437):323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webb AE, Kundaje A, Brunet A. Characterization of the direct targets of FOXO transcription factors throughout evolution. Aging Cell. 2016;15(4):673–685. doi: 10.1111/acel.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nazio F, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15(4):406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 53.Roczniak-Ferguson A, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5(228):ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Webb AE, Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci. 2014;39(4):159–169. doi: 10.1016/j.tibs.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palmieri M, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011;20(19):3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 56.Wartosch L, Günesdogan U, Graham SC, Luzio JP. Recruitment of VPS33A to HOPS by VPS16 is required for lysosome fusion with endosomes and autophagosomes. Traffic. 2015;16(7):727–742. doi: 10.1111/tra.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Velikkakath AK, Nishimura T, Oita E, Ishihara N, Mizushima N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell. 2012;23(5):896–909. doi: 10.1091/mbc.E11-09-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zavodszky E, Seaman MN, Rubinsztein DC. VPS35 Parkinson mutation impairs autophagy via WASH. Cell Cycle. 2014;13(14):2155–2156. doi: 10.4161/cc.29734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 2005;24(6):1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu M, Xu LG, Zhai Z, Shu HB. SINK is a p65-interacting negative regulator of NF-kappaB-dependent transcription. J Biol Chem. 2003;278(29):27072–27079. doi: 10.1074/jbc.M209814200. [DOI] [PubMed] [Google Scholar]
- 62.Zou CG, et al. The molecular mechanism of endoplasmic reticulum stress-induced apoptosis in PC-12 neuronal cells: the protective effect of insulin-like growth factor I. Endocrinology. 2009;150(1):277–285. doi: 10.1210/en.2008-0794. [DOI] [PubMed] [Google Scholar]
- 63.Kohda Y, et al. Localization of the ROMK potassium channel to the apical membrane of distal nephron in rat kidney. Kidney Int. 1998;54(4):1214–1223. doi: 10.1046/j.1523-1755.1998.00120.x. [DOI] [PubMed] [Google Scholar]
- 64.Kaplan MR, Plotkin MD, Lee WS, Xu ZC, Lytton J, Hebert SC. Apical localization of the Na-K-Cl cotransporter, rBSC1, on rat thick ascending limbs. Kidney Int. 1996;49(1):40–47. doi: 10.1038/ki.1996.6. [DOI] [PubMed] [Google Scholar]
- 65.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7(12):1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 66.Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol. 2008;8(9):663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 67.Liu YP, et al. Endoplasmic reticulum stress regulates the innate immunity critical transcription factor IRF3. J Immunol. 2012;189(9):4630–4639. doi: 10.4049/jimmunol.1102737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muralidharan S, Mandrekar P. Cellular stress response and innate immune signaling: integrating pathways in host defense and inflammation. J Leukoc Biol. 2013;94(6):1167–1184. doi: 10.1189/jlb.0313153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115(10):2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walker AK, Atkin JD. Stress signaling from the endoplasmic reticulum: A central player in the pathogenesis of amyotrophic lateral sclerosis. IUBMB Life. 2011;63(9):754–763. doi: 10.1002/iub.520. [DOI] [PubMed] [Google Scholar]
- 71.Huang HC, Tang D, Lu SY, Jiang ZF. Endoplasmic reticulum stress as a novel neuronal mediator in Alzheimer’s disease. Neurol Res. 2015;37(4):366–374. doi: 10.1179/1743132814Y.0000000448. [DOI] [PubMed] [Google Scholar]
- 72.Prudente S, et al. The rs12917707 polymorphism at the UMOD locus glomerular filtration rate in individuals with type 2 diabetes: evidence of heterogeneity across two different European populations. Nephrol Dial Transplant. 2017;32(10):1718–1722. doi: 10.1093/ndt/gfw262. [DOI] [PubMed] [Google Scholar]
- 73.Nasr SH, Lucia JP, Galgano SJ, Markowitz GS, D’Agati VD. Uromodulin storage disease. Kidney Int. 2008;73(8):971–976. doi: 10.1038/sj.ki.5002679. [DOI] [PubMed] [Google Scholar]
- 74.Schönthal AH. Endoplasmic reticulum stress: its role in disease and novel prospects for therapy. Scientifica (Cairo) 2012;2012:857516. doi: 10.6064/2012/857516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fang N, et al. TRIB3 alters endoplasmic reticulum stress-induced β-cell apoptosis via the NF-κB pathway. Metab Clin Exp. 2014;63(6):822–830. doi: 10.1016/j.metabol.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 76.Piret SE, et al. A mouse model for inherited renal fibrosis associated with endoplasmic reticulum stress. Dis Model Mech. 2017;10(6):773–786. doi: 10.1242/dmm.029488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Menzies FM, Fleming A, Rubinsztein DC. Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci. 2015;16(6):345–357. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- 78.Lamark T, Johansen T. Aggrephagy: selective disposal of protein aggregates by macroautophagy. Int J Cell Biol. 2012;2012:736905. doi: 10.1155/2012/736905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hyttinen JM, Amadio M, Viiri J, Pascale A, Salminen A, Kaarniranta K. Clearance of misfolded and aggregated proteins by aggrephagy and implications for aggregation diseases. Ageing Res Rev. 2014;18:16–28. doi: 10.1016/j.arr.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 80.Walsh PT, Smith LM, O’Connor R. Insulin-like growth factor-1 activates Akt and Jun N-terminal kinases (JNKs) in promoting the survival of T lymphocytes. Immunology. 2002;107(4):461–471. doi: 10.1046/j.1365-2567.2002.01525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma X, Bai Y. IGF-1 activates the P13K/AKT signaling pathway via upregulation of secretory clusterin. Mol Med Rep. 2012;6(6):1433–1437. doi: 10.3892/mmr.2012.1110. [DOI] [PubMed] [Google Scholar]
- 82.Bitto A, et al. Long-term IGF-I exposure decreases autophagy and cell viability. PLoS ONE. 2010;5(9):e12592. doi: 10.1371/journal.pone.0012592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rauch BH, Weber A, Braun M, Zimmermann N, Schrör K. PDGF-induced Akt phosphorylation does not activate NF-kappa B in human vascular smooth muscle cells and fibroblasts. FEBS Lett. 2000;481(1):3–7. doi: 10.1016/S0014-5793(00)01957-8. [DOI] [PubMed] [Google Scholar]
- 84.Wahab NA, Weston BS, Mason RM. Connective tissue growth factor CCN2 interacts with and activates the tyrosine kinase receptor TrkA. J Am Soc Nephrol. 2005;16(2):340–351. doi: 10.1681/ASN.2003100905. [DOI] [PubMed] [Google Scholar]
- 85.Crean JK, Furlong F, Mitchell D, McArdle E, Godson C, Martin F. Connective tissue growth factor/CCN2 stimulates actin disassembly through Akt/protein kinase B-mediated phosphorylation and cytoplasmic translocation of p27(Kip-1) FASEB J. 2006;20(10):1712–1714. doi: 10.1096/fj.05-5010fje. [DOI] [PubMed] [Google Scholar]
- 86.Hoang B, et al. Targeting TORC2 in multiple myeloma with a new mTOR kinase inhibitor. Blood. 2010;116(22):4560–4568. doi: 10.1182/blood-2010-05-285726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Towers CG, Thorburn A. Therapeutic targeting of autophagy. EBioMedicine. 2016;14:15–23. doi: 10.1016/j.ebiom.2016.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shan NN, Dong LL, Zhang XM, Liu X, Li Y. Targeting autophagy as a potential therapeutic approach for immune thrombocytopenia therapy. Crit Rev Oncol Hematol. 2016;100:11–15. doi: 10.1016/j.critrevonc.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 89.Schiattarella GG, Hill JA. Therapeutic targeting of autophagy in cardiovascular disease. J Mol Cell Cardiol. 2016;95:86–93. doi: 10.1016/j.yjmcc.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Riquelme JA, et al. Therapeutic targeting of autophagy in myocardial infarction and heart failure. Expert Rev Cardiovasc Ther. 2016;14(9):1007–1019. doi: 10.1080/14779072.2016.1202760. [DOI] [PubMed] [Google Scholar]
- 91.Perico N, et al. Sirolimus therapy to halt the progression of ADPKD. J Am Soc Nephrol. 2010;21(6):1031–1040. doi: 10.1681/ASN.2009121302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kuo SY, et al. Small-molecule enhancers of autophagy modulate cellular disease phenotypes suggested by human genetics. Proc Natl Acad Sci U S A. 2015;112(31):E4281–E4287. doi: 10.1073/pnas.1512289112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sarkar S, et al. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat Chem Biol. 2007;3(6):331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rivas A, Vidal RL, Hetz C. Targeting the unfolded protein response for disease intervention. Expert Opin Ther Targets. 2015;19(9):1203–1218. doi: 10.1517/14728222.2015.1053869. [DOI] [PubMed] [Google Scholar]
- 95.Ma T, Klann E. PERK: a novel therapeutic target for neurodegenerative diseases? Alzheimers Res Ther. 2014;6(3):30. doi: 10.1186/alzrt260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moreno JA, et al. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci Transl Med. 2013;5(206):206ra138. doi: 10.1126/scitranslmed.3006767. [DOI] [PubMed] [Google Scholar]
- 97.Halliday M, et al. Partial restoration of protein synthesis rates by the small molecule ISRIB prevents neurodegeneration without pancreatic toxicity. Cell Death Dis. 2015;6:e1672. doi: 10.1038/cddis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Radford H, Moreno JA, Verity N, Halliday M, Mallucci GR. PERK inhibition prevents tau-mediated neurodegeneration in a mouse model of frontotemporal dementia. Acta Neuropathol. 2015;130(5):633–642. doi: 10.1007/s00401-015-1487-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Halliday M, et al. Repurposed drugs targeting eIF2alpha-P-mediated translational repression prevent neurodegeneration in mice. Brain. 2017;140(6):1768–1783. doi: 10.1093/brain/awx074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13(3):184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta. 2013;1833(12):3460–3470. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moore KA, Hollien J. The unfolded protein response in secretory cell function. Annu Rev Genet. 2012;46:165–183. doi: 10.1146/annurev-genet-110711-155644. [DOI] [PubMed] [Google Scholar]
- 103.Liu Z, Lv Y, Zhao N, Guan G, Wang J. Protein kinase R-like ER kinase and its role in endoplasmic reticulum stress-decided cell fate. Cell Death Dis. 2015;6:e1822. doi: 10.1038/cddis.2015.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hu P, Han Z, Couvillon AD, Kaufman RJ, Exton JH. Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1alpha-mediated NF-kappaB activation and down-regulation of TRAF2 expression. Mol Cell Biol. 2006;26(8):3071–3084. doi: 10.1128/MCB.26.8.3071-3084.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ren X, et al. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 2014;9(3):1151–1162. doi: 10.1016/j.celrep.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38(Web Server issue):W462–W468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Iyer V, et al. Off-target mutations are rare in Cas9-modified mice. Nat Methods. 2015;12(6):479. doi: 10.1038/nmeth.3408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.