Abstract
Escherichia coli and other Enterobacteriaceae are among the most common pathogens of the human urinary tract. Among the genetic gains of function associated with urinary E. coli isolates is the Yersinia high pathogenicity island (HPI), which directs the biosynthesis of yersiniabactin (Ybt), a virulence-associated metallophore. Using a metabolomics approach, we found that E. coli and other Enterobacteriaceae expressing the Yersinia HPI also secrete escherichelin, a second metallophore whose chemical structure matches a known synthetic inhibitor of the virulence-associated pyochelin siderophore system in Pseudomonas aeruginosa. We detected escherichelin during clinical E. coli urinary tract infection (UTI) and experimental human colonization with a commensal, potentially probiotic E. coli bacteriuria strain. Escherichelin production by colonizing enterobacteria may help human hosts resist opportunistic infections by Pseudomonas and other pyochelin-expressing bacteria. This siderophore-based mechanism of microbial antagonism may be one of many elements contributing to the protective effects of the human microbiome. Future UTI-preventive probiotic strains may benefit by retaining the escherichelin biosynthetic capacity of the Yersinia HPI while eliminating the Ybt biosynthetic capacity.
Keywords: Infectious disease
Keywords: Bacterial infections, UTI/pyelonephritis
Introduction
Urinary tract infections (UTIs) are among the most common infectious diseases treated by physicians and are increasingly associated with antibiotic-resistant bacteria (1, 2). Although bacteriuria, the presence of bacteria in the urine, is a key feature of UTI, it also occurs frequently in the absence of symptoms. Bacteriuria seeded from the intestinal microbiome may self-resolve, progress to UTI, or persist asymptomatically, in which case antibiotic treatment is not recommended (3). Indeed, recent studies support the existence of urinary tract microbiomes in otherwise healthy individuals (4). Multiple lines of evidence suggest that the microbiome can prevent infections. Paradoxically, antibiotic treatment has been observed to increase the risk of subsequent UTIs (5) and to increase the risk of UTIs caused by atypical uropathogens such as Pseudomonas (6). Members of the family Enterobacteriaceae, particularly E. coli, inhabit the intestinal microbiome and are most commonly associated with asymptomatic bacteriuria and UTI. The early bacteriuric state from which UTIs can develop is poorly understood and challenging to study, but it is known that UTI-associated E. coli isolates frequently possess nonconserved genetic systems related to virulence (7, 8).
Prominent among virulence-associated adaptations in uropathogenic E. coli (UPEC) isolates are siderophores, specialized metabolites able to bind extracellular Fe(III) ions for nutritional use in low-iron conditions (7, 9). High-affinity Fe(III) binding by siderophores helps bacteria resist host-mediated iron deprivation, a prototypical example of nutritional immunity (10, 11). Siderophores are widespread among medically important bacteria, which often express chemically distinctive siderophore types. A recent quantitative metabolomics study of E. coli isolates from patients with UTI revealed that the resident intestinal strain in each patient that successfully infected the urinary tract preferentially secreted the siderophores yersiniabactin (Ybt) and salmochelin (12). These results suggest that the early stages of ascending UTI are competitive and favor isolates with specific siderophore profiles. In this and other studies, Ybt, a siderophore encoded by the Yersinia high pathogenicity island (HPI), was associated with urinary tract virulence (7, 13–17).
The Yersinia HPI is a complex, 4-operon mobile genetic element encoding proteins involved in Ybt biosynthesis, import, and transcriptional regulation. It is highly prevalent among clinical extraintestinal pathogenic E. coli isolates and is also found with varying frequencies in other Enterobacteriaceae, including the eponymous Yersinia pestis (18). Yersinia HPI genes are highly upregulated during UTI, and Ybt is detectable in patients’ urine specimens (12, 14, 17). Ybt, the recognized product of Yersinia HPI–positive bacteria to date, has been distinguished from other E. coli siderophores by its ability to form stable Cu(II) complexes. This broad-spectrum metal-binding property enables Ybt to scavenge Fe(III) for nutritional use, detoxify copper, and catalyze superoxide dismutation (14, 15, 19). The multiple functions of Ybt illustrate the importance of diverse metal ion interactions during microbial pathogenesis.
A hybrid nonribosomal peptide synthetase/polyketide synthase (NRPS/PKS) pathway encoded on the Yersinia HPI directs the biosynthesis of Ybt. There is remarkable natural diversity in microbial NRPS/PKS systems, which are notable in infectious diseases as the source of antibiotics such as mupirocin, tetracyclines, and macrolides, but also the source of many virulence-associated siderophores such as pyochelin in Pseudomonas aeruginosa and mycobactin in Mycobacterium tuberculosis (20–22). NRPS and PKS pathways are characterized by their thiotemplating, assembly line mechanism, whereby all substrates and intermediates are covalently attached to phosphopantetheinylated carrier protein (CP) domains as they proceed through the catalytic domains of large, multimodular biosynthetic enzymes. The Ybt biosynthetic substrates are chorismate, 3 molecules of cysteine, malonate from malonyl-CoA, and methyl groups derived from S-adenosyl-methionine (SAM). During Ybt biosynthesis, YbtS converts chorismate to salicylate (23), which is activated and transferred onto the first NRPS, HMWP2 (irp2), by YbtE. After condensing salicylate and 2 molecules of cysteine, the last intermediate synthesized by HMWP2 is transferred onto the hybrid NRPS/PKS HMWP1 (irp1), where malonate, another molecule of cysteine, and methyl groups are incorporated. At this point, YbtU catalyzes an NADPH-dependent reduction of an HMWP1-bound intermediate. Once the molecule is complete, Ybt is released from the final CP domain of HMWP1 by a terminal thioesterase domain (20, 24). Although Ybt biosynthesis has been elegantly characterized in vitro, an untargeted survey of this pathway’s products in bacterial culture has not been reported.
In this study, comparative metabolomic profiling revealed escherichelin, a newly appreciated cellular product of the Ybt biosynthetic pathway in E. coli and other urinary Enterobacteriaceae isolates. Structural characterization identified escherichelin as 2-(2-hydroxyphenyl)-4,5-dihydro-[2,4]bisthiazolyl-4-carboxylic acid, (abbreviated as HPTT-COOH from hydroxyphenyl-thiazolyl-thiazolinyl-carboxylic acid), an Fe(III) chelator that was previously synthesized and found to inhibit pyochelin-mediated iron uptake in P. aeruginosa (25). Escherichelin purified from E. coli lacks the siderophoric activity of Ybt, but inhibits P. aeruginosa growth in a pyochelin-dependent culture condition. Consistent with this activity, we observed that P. aeruginosa minimizes escherichelin release compared with E. coli, despite generating the same biosynthetic precursors to escherichelin during pyochelin biosynthesis. This bioactivity and its production during human E. coli bacteriuria raises the possibility that escherichelin may help human hosts avoid developing symptomatic UTIs from pyochelin-producing organisms. Escherichelin was detectable in a human subject who was experimentally colonized with a derivative of the bacterial interference strain E. coli 83972 that has been studied in several human trials as a potential agent to prevent UTI (26, 27). These findings reveal how a second metallophore product of the Yersinia HPI biosynthetic system may participate in interspecies competition during polymicrobial bacteriuria.
Results
Metabolomic profiling identifies multiple Yersinia HPI–associated metabolites.
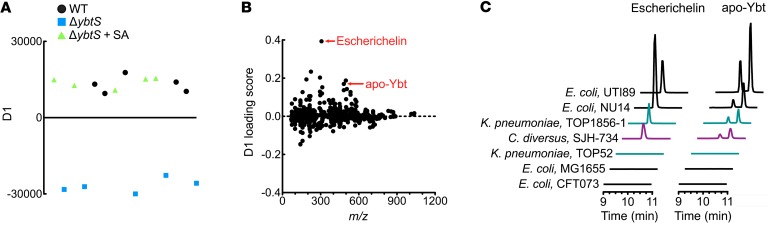
To identify secreted metabolites associated with the Yersinia HPI, we used liquid chromatography–mass spectrometry (LC-MS) to compare culture supernatants from UTI89, a Ybt-producing model UPEC strain (28), and its isogenic mutant UTI89ΔybtS that cannot perform the first committed step of Ybt biosynthesis, salicylate synthesis (12, 23). As a control, the biosynthetic block in UTI89ΔybtS was experimentally overcome by chemical complementation with salicylate-supplemented medium. The metabolomic profiles from Ybt producers (WT and UTI89ΔybtS plus salicylate) and the Ybt nonproducer (UTI89ΔybtS) were compared using supervised principal component analysis–discriminant analysis (PCA-DA) (Figure 1A). Uniform separation of these 2 groups along the y axis of the PCA-DA plot confirmed that extracellular metabolites can differentiate Ybt-producing from nonproducing strains (Figure 1A).
Figure 1. Metabolomic analysis reveals escherichelin as an additional product of Ybt-producing E. coli.
(A) Two-group PCA-DA of LC-MS data demonstrate consistent variance in the secreted metabolomes of the Ybt producers, WT UTI89 and UTI89ΔybtS chemically complemented with salicylic acid (SA), and the nonproducer UTI89ΔybtS. Each point indicates the average of 3 technical replicates. Five biological replicates were analyzed from each experimental group. (B) The PCA-DA loadings plot, where each point is a molecular feature identified in the LC-MS analysis, reveals a molecular feature with m/z 307 (escherichelin) that best distinguishes Ybt producers from nonproducers. (C) LC-MS/MS chromatograms of escherichelin (left) and apo-Ybt (right) from culture supernatants of Enterobacteriaceae isolates grown in iron-restricted medium. All of the strains that produced Ybt also produced escherichelin.
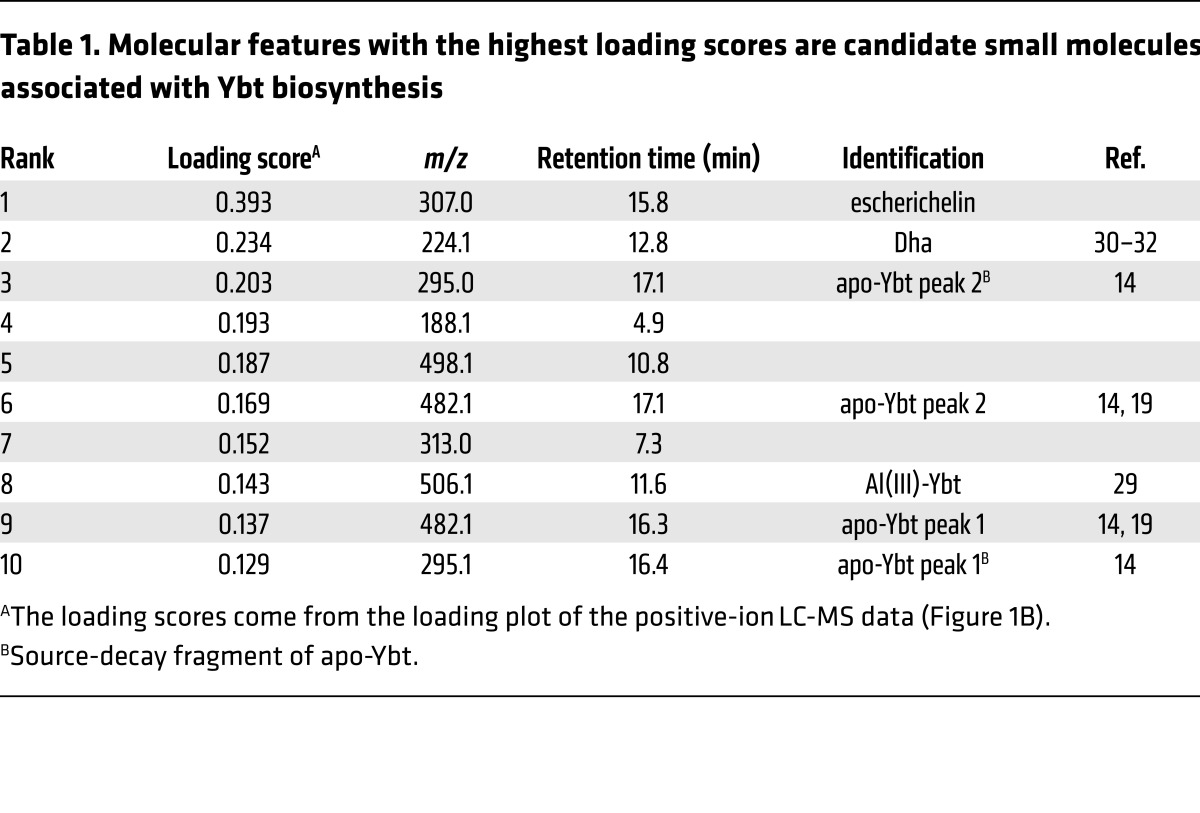
To identify the specific extracellular metabolites that distinguish Ybt producers from nonproducers, we performed a loadings plot analysis (Figure 1B). In this plot, features with the highest D1 loading scores are most strongly associated with Ybt producers and are, therefore, candidates for follow-up analysis. Ybt in the metal-free form (apo) and bound to Al(III) were among the molecular features with the highest loading scores, as expected (Table 1) (29). We also observed dihydroaeruginoic acid (Dha), a known metabolite of pyochelin-producing strains of P. aeruginosa (Table 1 and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI92464DS1) (30, 31). Dha is the free form of an early phosphopanthetheine-bound intermediate in both the pyochelin and Ybt biosynthetic pathways (20, 32). Unexpectedly, the molecular feature with the highest loading score was an ion with m/z 307, which is lower than apo-Ybt (m/z 482) (Figure 1B and Table 1). Like Ybt, the molecular feature at m/z 307, herein called escherichelin, was present in the WT and salicylate-complemented UTI89ΔybtS supernatants but entirely absent in ΔybtS supernatants. These results are consistent with the production of a previously unappreciated secreted metabolite by E. coli with an intact Yersinia HPI.
Table 1. Molecular features with the highest loading scores are candidate small molecules associated with Ybt biosynthesis.
Clinical Enterobacteriaceae isolates coproduce escherichelin and Ybt.
To determine whether escherichelin production is specific to UTI89 or is present in other bacteria encoding the Yersinia HPI, we measured its production in a series of clinical urinary isolates and model strains of the Enterobacteriaceae family (Figure 1C). The UPEC model strains UTI89 and NU14 produced both Ybt and escherichelin, whereas CFT073, a widely used UPEC model strain with known mutations in the Yersinia HPI, and the K12 strain MG1655 produced neither (33). Klebsiella pneumoniae and Citrobacter diversus isolates that produced Ybt also produced escherichelin (Figure 1C). These observations suggest that escherichelin production is pervasive among clinically significant Ybt-producing Enterobacteriaceae family members.
Escherichelin production is associated with early Ybt biosynthetic enzymes.
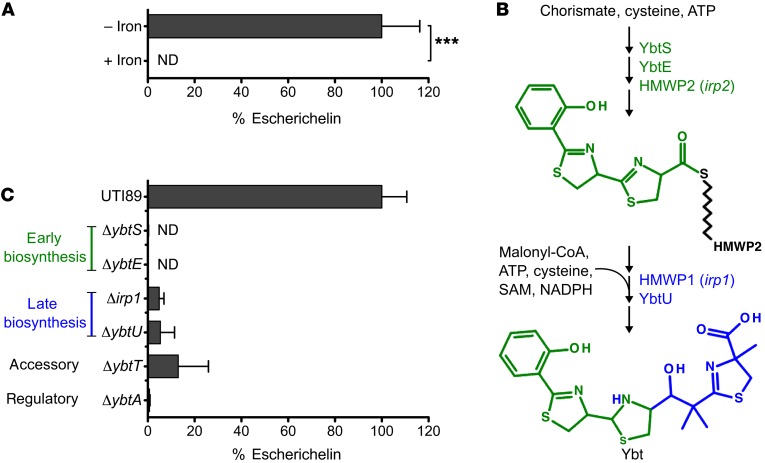
The Yersinia HPI and other siderophore systems are regulated by Fur, an iron-responsive transcription factor, such that Ybt biosynthesis is downregulated in iron-rich conditions. To determine whether escherichelin is similarly regulated, we measured its secretion in a high-iron medium (complete M63 supplemented with 16.2 mg/l ferric chloride) (34). The high-iron medium abolished escherichelin production, suggesting that it is regulated by iron in a manner similar to that of Ybt (Figure 2A).
Figure 2. Escherichelin production is iron repressible and dependent on early Ybt biosynthetic enzymes.
Escherichelin levels in culture supernatants were quantified by LC-MS/MS relative to a 13C-IS. (A) UTI89 grown in complete M63 medium with or without 16.2 mg/l FeCl3 (34). Data represent the mean of 3 replicates, with SD plotted. ***P < 0.001, by unpaired t test. (B) Schematic of the Ybt biosynthetic pathway. Black zig-zag line designates a phosphopantetheine group covalently attached to the biosynthetic enzyme. Green designates chemical moieties built by early biosynthetic enzymes (YbtS, YbtE, and HMWP2), and blue designates chemical moieties built by late biosynthetic enzymes (HMWP1 and YbtU). (C) Isogenic mutants of UTI89 in select genes of the Yersinia HPI grown in iron-restricted medium. Data represent the mean of 8 replicates, with SD plotted.
To better understand the relationship between escherichelin secretion and Ybt biosynthetic genes (Figure 2B), we screened a panel of Yersinia HPI gene deletion mutants for altered escherichelin production (Figure 2C). Mutants lacking the biosynthetic genes ybtS, ybtE, irp1, or ybtU do not produce Ybt (Supplemental Figure 2) (23, 35–38). A mutant lacking the Yersinia HPI transcriptional regulator ybtA also produces no Ybt (38, 39). Deletion of ybtT, a putative editing enzyme, exhibits an intermediate phenotype with reduced Ybt levels (Supplemental Figure 2) (37, 38, 40). These isogenic mutants are not attenuated for growth in iron-restricted medium (38). Among Ybt-null strains, extracellular escherichelin was undetectable in the culture supernatants of UTI89ΔybtS, ΔybtE, and ΔybtA (the 2 early biosynthetic genes and transcriptional regulator, respectively). In contrast, escherichelin was detectable in supernatants from UTI89Δirp1 and ΔybtU, the late Ybt biosynthetic pathway deletion mutants (Figure 2C). Intracellular escherichelin levels followed a trend nearly identical to that of the extracellular levels between strains (Supplemental Figure 3). Overall, these patterns favor biosynthesis, rather than cellular export, as the major determinant of escherichelin secretion.
The requirement of early Ybt biosynthetic genes for escherichelin production is consistent with it being a Yersinia HPI product. The observation that late biosynthetic mutants (UTI89Δirp1 and ΔybtU) produce escherichelin, but not Ybt, further suggests that escherichelin is a de novo metabolite and not a Ybt breakdown product. The diminished, but not absent, escherichelin levels in late biosynthesis mutants (UTI89Δirp1 and ΔybtU, Figure 2C) mirror the diminished irp2 expression observed with analogous Y. pestis mutants (36, 37). Although the precise reason for this altered gene expression in the mutants remains unclear, it has been proposed that feedback regulation from a late biosynthetic gene product is reduced, leading to the downregulation. The feedback regulation is believed to be mediated by YbtA, a member of the AraC transcription factor family whose members typically respond to allosteric activation by metabolites.
Escherichelin is HPTT-COOH.
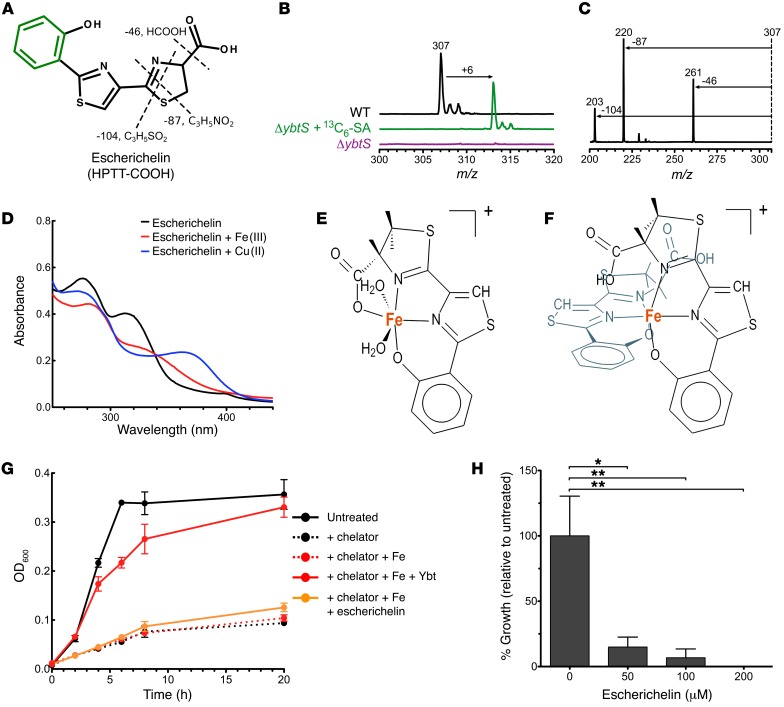
Multiple structural analyses were used to determine the chemical structure of escherichelin. These analyses were guided in part by previous in vitro investigations of Ybt biosynthesis that noted a product with a positive-ion m/z of 307 that was identified as 2-(2-hydroxyphenyl)-4,5-dihydro-[2,4]bisthiazolyl-4-carboxylic acid (abbreviated as HPTT-COOH from hydroxylphenyl-thiazolyl-thiazolinyl-carboxylic acid) (24, 41–44). HPTT-COOH is a truncated version of Ybt that arises when a biosynthetic intermediate (Figure 2B, top structure) derived from salicylate and 2 cysteines is released from the first NRPS protein, HMWP2 (irp2), and spontaneously oxidized. High-resolution MS of escherichelin yielded an exact mass of 307.0216 for the [M+H]+ ion, supporting an empiric formula of C13H10N2O3S2 (calculated mass: 307.0206 for C13H11N2O3S2+). To determine whether escherichelin is synthesized from salicylate, we chemically complemented UTI89ΔybtS with 13C6-salicylate and analyzed the supernatant by LC-MS. Upon 13C6-salicylate complementation, escherichelin at m/z 307 disappeared, and a new molecular feature at m/z 313 appeared at the same retention time, consistent with incorporation of the six 13C atoms (Figure 3, A and B). The empiric formula and heavy isotope labeling are consistent with a biosynthetic product derived from salicylate and 2 cysteines.
Figure 3. Identification of escherichelin as HPTT-COOH, a metallophore inhibitor of Pseudomonas iron uptake.
(A) Multiple chemical characterizations identify escherichelin as HPTT-COOH, a compound shown to inhibit pyochelin-mediated iron uptake in Pseudomonas. The positions of 13C salicylate incorporation are indicated in green, while MS/MS neutral losses are indicated as dashed lines. (B) Mass spectra at the retention time for escherichelin from WT UTI89, UTI89ΔybtS, and 13C6-SA–supplemented UTI89ΔybtS culture supernatants revealed that 6 carbons in escherichelin are derived from salicylate. (C) MS/MS product ion spectrum of escherichelin. The 46-Da neutral loss indicates the presence of a carboxylic acid group, and the 87- and 104-Da losses correspond to fragmentation within the thiazoline ring. (D) UV-visible absorption spectra of purified escherichelin incubated with equimolar FeCl3 or CuSO4. (E and F) DFT simulations of escherichelin bound to Fe(III) in a 1:1 (E) and 2:1 (F) complex. The simulations predict stable Fe(III)-escherichelin complexes with hexacoordinate octahedral geometry. (G) UTI89 growth curve in the presence or absence of a ferric ion chelator (0.5 mM EDDHA) and/or ferric ion supplementation (1 μM FeCl3). Growth was monitored by OD at 600 nm (OD600). Data represent the mean of 3, with SD plotted. (H) PW5011 (pvdA-E02::ISlacZ/hah), a P. aeruginosa strain MPAO1 transposon mutant (55, 56) expressing pyochelin as its sole siderophore, was grown in succinate medium for 20 hours in the presence of increasing escherichelin concentrations relative to vehicle control. Bacterial growth was quantified by enumerating CFU/ml and is expressed as a percentage of the vehicle control. Data represent the mean of 3, with the SEM plotted. *P < 0.01 and **P < 0.001, by 1-way ANOVA with Dunnett’s multiple comparisons test.
Information about the connectivity of escherichelin’s atoms was obtained using tandem MS (MS/MS) and 1H-NMR. MS/MS of the escherichelin [M+H]+ ion yielded unique fragments arising from neutral losses of 46, 78, and 104 Da (Figure 3C). The 46-Da neutral loss is consistent with an equivalent loss of HCOOH (formic acid or sequential loss of H2O and CO) and supports the presence of a carboxylic acid. The atypical neutral losses of 78 and 104 Da are consistent with equivalent cleavages through the thiazoline ring to lose neutral fragments containing the carboxylic acid and either nitrogen [M-78]+ or sulfur [M-104]+ (Figure 3A). To further investigate the structure, we performed one-dimensional 1H and two-dimensional 1H correlation spectroscopy (COSY) NMR experiments. The chemical shifts and assignments were as follows: 1H NMR (500 MHz, CDCl3, δ), 8.15 (s, 1H,-CH=); 6.97, 7.1, 7.4, and 7.63 (s, 1H, aromatic), 5.05 (m, ~1H, CH-COOH), and 3.2 (dd, 2H, -CH2). The 5.05 multiplet was partially obscured by a solvent impurity, preventing an accurate signal assignment. The chemical shifts at 6.97, 7.1, 7.4, and 7.63 ppm that each integrate to 1 proton and correspond to aromatic protons further support the presence of a salicylate-derived aromatic ring. A singlet at 8.15 ppm is consistent with the vinylic proton of the thiazole, and coupled resonances at 3.2 and 5.05 ppm are consistent with the presence of the methylene protons and the C-H proton of the thiazoline ring, respectively (Supplemental Figure 4). Together, the molecular formula from high-resolution MS, the MS/MS fingerprint, and the 1H-NMR chemical shifts support HPTT-COOH as the escherichelin structure. Escherichelin corresponds to an oxidized form of a Ybt biosynthetic intermediate and is structurally similar to the P. aeruginosa siderophore pyochelin. Escherichelin and pyochelin differ in the oxidation state of their first heterocyclic ring, and pyochelin is also further modified by the reduction of the second thiazoline to a thiazole and by N-methylation (45).
Escherichelin binds Fe(III).
Given that escherichelin is produced by a siderophore biosynthetic pathway, regulated by Fur, and retains some of the functional groups required for metal binding by Ybt and pyochelin, we assessed the ability of purified escherichelin to form an Fe(III) complex in aqueous solution with UV-visible spectroscopy. As reported in previous studies, Fe(III) addition caused notable shifts in the UV-visible absorption spectra of Ybt and pyochelin, consistent with the formation of Fe(III) complexes (19, 46, 47). The escherichelin UV-visible absorption spectrum was similarly shifted following exposure to equimolar Fe(III) (Figure 3D). Cu(II) addition caused a qualitatively distinct shift from Fe(III) (Figure 3D). These results demonstrate that escherichelin, like Ybt and pyochelin, is a bacterial metallophore that forms Fe(III) and Cu(II) complexes.
To determine whether escherichelin binds metal ions in a manner similar to Ybt and pyochelin, we conducted quantum-based calculations using density functional theory (DFT). These calculations predict stable 1:1 and 1:2 complexes of Fe(III)-escherichelin in a hexacoordinate octahedral geometry (Figure 3, E and F). In the 1:1 complex, Fe(III) is bound by the salicylate oxygen, the 2 heterocyclic nitrogens, and the carboxylic acid group in the equatorial positions and by 2 water molecules in the axial positions. In the 1:2 complex, the carboxylic acids of the 2 escherichelin molecules and water do not participate as ligands. Instead, the nitrogen atoms and phenolic oxygen atoms of 2 the escherichelins occupy all of the liganding positions. Pyochelin binds Fe(III) in similar 1:1 and 1:2 complexes with the same ligands (48, 49). As with Ybt, escherichelin is also predicted to form a stable Cu(II) complex (Supplemental Figure 5). Overall, the calculated metal ion coordination in the Fe(III)- and Cu(II)-escherichelin complexes is similar to that observed for metal-Ybt complexes and Fe(III)-Pch (15, 19, 48, 50). These results support the idea that escherichelin is an Fe(III)-binding molecule.
Escherichelin does not support iron-dependent E. coli growth.
To determine whether escherichelin, like Ybt, can promote iron-dependent growth in E. coli possessing Yersinia HPI-encoded transport genes, we measured UTI89 growth in a previously described siderophore-dependent growth assay (Figure 3G) (16, 19). Addition of the iron chelator EDDHA to the test medium causes substantial growth restriction. As previously observed, addition of Ybt that has been preincubated with equimolar Fe(III) restores growth to a density comparable to that of the unchelated medium. In contrast, addition of escherichelin that has been preincubated with equimolar Fe(III) resulted in no significant growth enhancement (Figure 3G). Although both escherichelin and Ybt bind Fe(III), these results show a nonequivalent ability to support iron-limited growth. This discrepancy may arise from differences in Fe(III) affinity or from differential recognition of the complexes by bacterial transport proteins.
P. aeruginosa growth inhibition by escherichelin.
The deficient siderophoric activity of escherichelin raises the possibility that this molecule performs an alternative function. In prior work by Mislin et al., escherichelin (previously designated HPTT-COOH) was synthesized as a pyochelin analog (51) and found to act as a competitive inhibitor of pyochelin-mediated iron import through the TonB-dependent outer membrane transporter FptA in P. aeruginosa (25, 51, 52). Escherichelin did not mediate iron uptake and inhibited Fe(III)-pyochelin transport through FptA with a Ki of 27 nM. In silico docking simulations could rationalize these results through occupancy of the Fe(III)-pyochelin binding site of FptA by a 1:2 Fe(III)-escherichelin complex [Fe(III):escherichelin] (25, 48). Given that escherichelin can bind Fe(III) in solution but does not possess canonical siderophoric activity, we hypothesized that it may be produced by UPEC to inhibit pyochelin uptake by competing P. aeruginosa strains in the urinary tract.
To determine whether escherichelin can inhibit ferric pyochelin uptake, we examined the effect of escherichelin on growth of the P. aeruginosa strain PAO1. Although pyochelin enhances P. aeruginosa growth during infections (53, 54), it is functionally redundant with pyoverdin, a second siderophore, during in vitro growth in standard culture media. Escherichelin addition to a defined, iron-restricted minimal medium caused a dose-dependent reduction in growth of a pyoverdin-deficient P. aeruginosa mutant strain, PW5011 (Figure 3H) (55–57). These results are consistent with the escherichelin-mediated inhibition of iron uptake in P. aeruginosa noted by Mislin et al. (25).
P. aeruginosa minimizes escherichelin production.
Production of a P. aeruginosa iron uptake inhibitor from the enzyme-bound precursor hydroxyphenyl-thiazolinyl-thiazolinyl-S-ppant (Figure 2B, top structure) presents a potential paradox, because this enzyme-bound species is also a biosynthetic intermediate of pyochelin (20). Escherichelin release from this intermediate in P. aeruginosa would be expected to autoantagonize pyochelin-mediated iron uptake. To determine whether P. aeruginosa minimizes escherichelin release relative to E. coli, we compared escherichelin production between UTI89 and PAO1 in low-iron medium. Despite comparable growth of both strains and substantial pyochelin production by PAO1, extracellular and intracellular levels of escherichelin were markedly lower in PAO1 than in UTI89 (Figure 4 and Supplemental Figures 6 and 7). This may reflect selective pressure on the P. aeruginosa pyochelin biosynthetic pathway to avoid inhibiting pyochelin-mediated iron uptake, as well as an opposing selective pressure on E. coli to suppress competing Pseudomonas strains in polymicrobial settings.
Figure 4. Escherichelin release is suppressed in P. aeruginosa relative to E. coli.
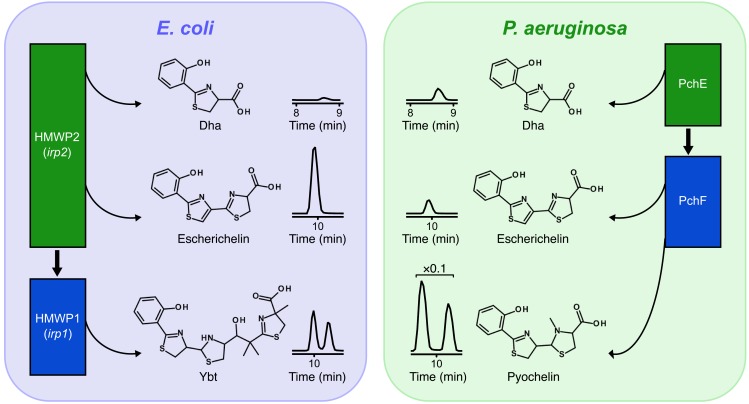
LC-MS/MS chromatograms of extracellular metabolites released from Ybt and pyochelin biosynthetic enzymes in the E. coli strain UTI89 (left) and the P. aeruginosa strain PA01 (right) cultured in iron-deficient media are displayed in the center. Chromatograms are identically scaled, except for P. aeruginosa pyochelin, which is at ×0.1 magnification. The accompanying illustrations depict the sequential release of Dha, escherichelin, pyochelin, and apo-Ybt from multidomain NRPS/PKS enzymes associated with early (green rectangles) and late (blue rectangles) biosynthesis of Ybt (left) and pyochelin (right). Thick black arrows designate the biosynthetic steps at which metabolite intermediates transfer from early to late proteins.
Escherichelin is produced during human E. coli cystitis.
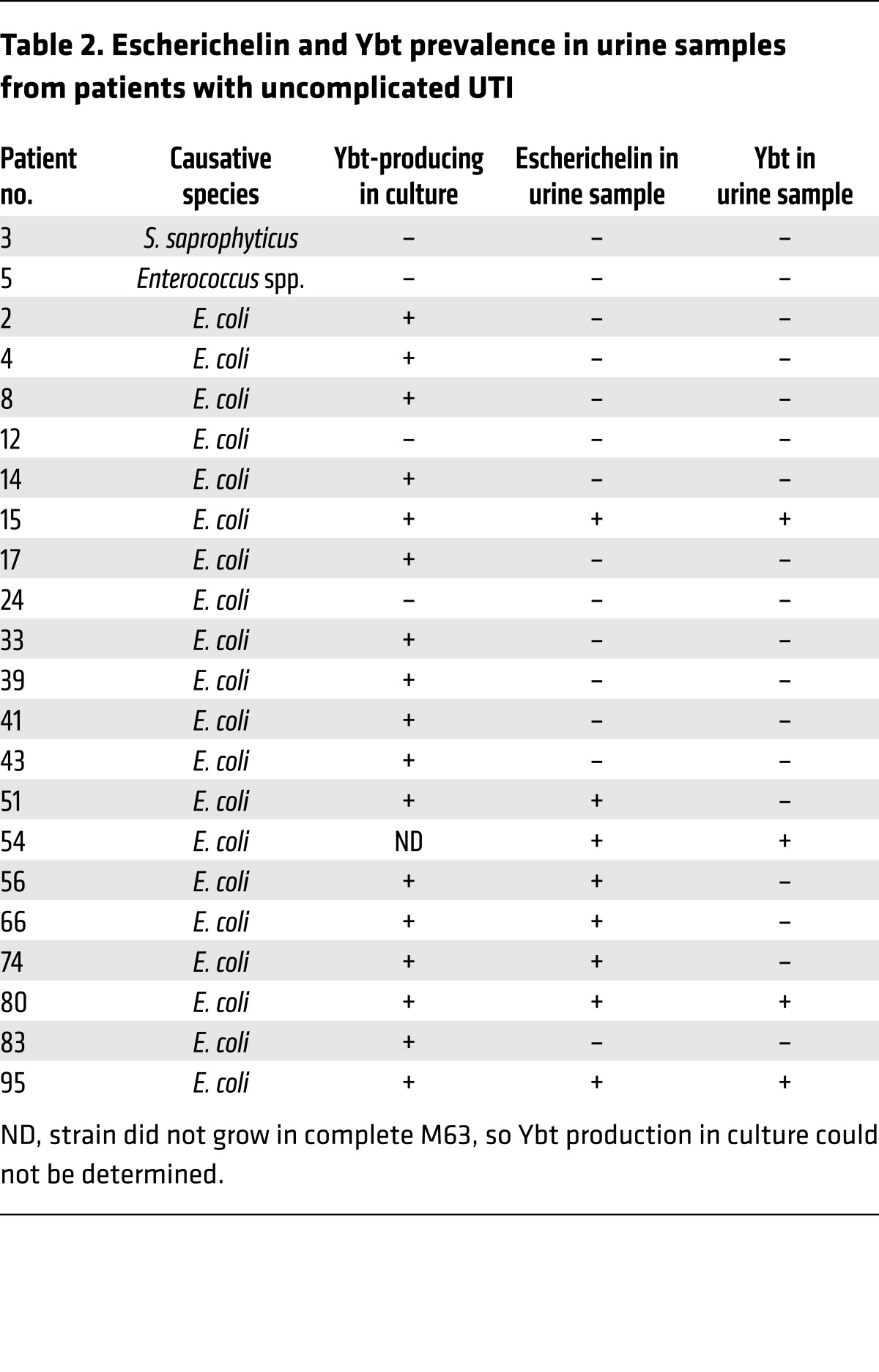
Enterobacterial bacteriuria may be a frequent occurrence in the human urinary tract and is frequently accompanied by other flora (58–60). When native enterobacteria in the microbiome are suppressed or eliminated by antibiotic treatment, P. aeruginosa UTIs become more frequent (6). Therefore, we hypothesize that escherichelin release during enterobacterial bacteriuria prevents P. aeruginosa UTI. To determine whether Enterobacteriaceae produce escherichelin in the human urinary tract, we used LC-MS/MS to detect escherichelin in urine from 18 patients with uncomplicated cystitis caused by Ybt-producing E. coli. Escherichelin was detectable in 8 of the 18 samples (Figure 5A and Table 2). In four escherichelin-positive specimens, Ybt was undetectable. As expected, escherichelin was undetectable in patients with cystitis caused by Gram-positive species (Staphylococcus saprophyticus or Enterococcus sp.) or Ybt-null E. coli. These results demonstrate that E. coli can produce escherichelin in the human bladder.
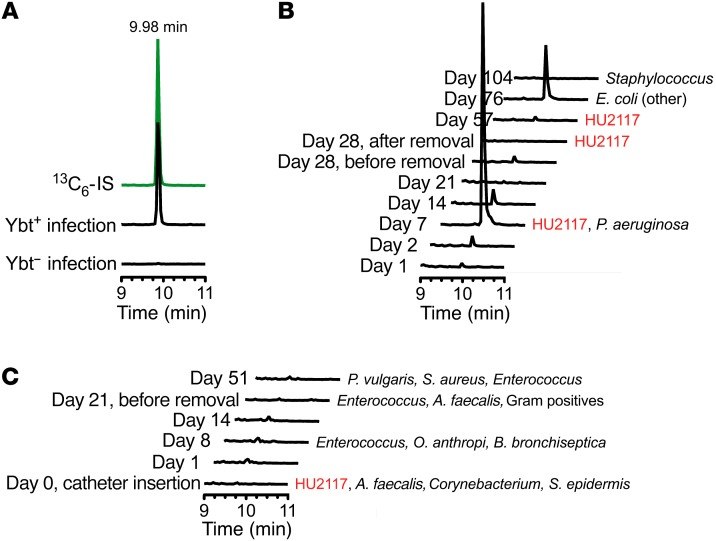
Figure 5. Escherichelin is produced during clinical E. coli UTI and experimental human colonization with the asymptomatic bacteriuria strain E. coli HU2117.
(A) Representative LC-MS/MS chromatogram for escherichelin in urine from a patient with an uncomplicated UTI caused by a Ybt-producing UPEC strain (results from 18 urine samples are shown in Table 1). Escherichelin coeluted with the 13C-IS (green). Escherichelin was undetectable in urine from a patient with a UTI caused by a Ybt-nonproducing UPEC strain. (B and C) LC-MS/MS chromatograms for escherichelin in urine from subjects who were successfully (B) or unsuccessfully (C) colonized with E. coli HU2117. The days after insertion of the HU2117-coated study catheter are indicated on the left. Where microbiological culture data were available, the results are listed to the right of the chromatograms. The colonized subject (B, subject 9) was colonized with HU2117 for 57 days and then developed a febrile UTI caused by another strain of E. coli on day 76, after colonization had ended. On day 7, P. aeruginosa was also present in this subject’s urine. Escherichelin was detectable throughout the colonization period and during the symptomatic E. coli UTI. The noncolonized subject (C) did not show a presence of HU2117 after day 0, when the study catheter was inserted, and escherichelin was not detected in the longitudinal samples. P. vulgaris, Proteus vulgaris; A. faecalis, Alcaligenes faecalis; O. anthropi, Ochrobactrum anthropi; B. bronchiseptica, Bordetella bronchiseptica; S. epidermis, Staphylococcus epidermidis.
Table 2. Escherichelin and Ybt prevalence in urine samples from patients with uncomplicated UTI.
Escherichelin is produced during experimental bladder colonization with an E. coli 83972 strain.
The concept that one bacterial strain or species can prevent infection by another — termed bacterial interference — has been of longstanding interest in infectious diseases (61–63). In this context, an avirulent strain or species that outcompetes more pathogenic strains may be useful as a probiotic to prevent infections. E. coli 83972, which asymptomatically colonized a young Swedish girl for over 2 years (64), has been used in multiple UTI prevention studies (26, 27, 63, 65–70). Among these studies are anecdotal reports of protection against symptomatic P. aeruginosa UTIs, including 1 patient with asymptomatic cocolonization of 83972 and P. aeruginosa that progressed to UTI following loss of 83972 (26, 66). E. coli 83972 possesses the Yersinia HPI (71, 72) and was observed to produce escherichelin in iron-restricted medium (data not shown).
To determine whether 83972 secretes escherichelin in the human urinary tract, we analyzed urine specimens following experimental colonization of catheter-dependent patients with an 83972-derived E. coli strain lacking the virulence-associated papG adhesin (E. coli HU2117) (27). Bladder colonization was established by inserting urinary catheters that had been precoated with E. coli HU2117. Longitudinal urine specimens were available from 1 subject (104 days for subject 9) who became colonized with HU2117 and another subject who was unsuccessfully colonized (51 days for subject 10). Both subjects showed evidence of polymicrobial urinary tract colonization during the collection periods. In subject 9, escherichelin was detected throughout the 57 days of documented HU2117 colonization, reaching a maximum on day 7 after catheterization (Figure 5B). P. aeruginosa was detected by culture in the urine on day 7 but was unassociated with symptoms and became undetectable in subsequent specimens (27). On day 76, HU2117 colonization had ended, and the subject developed a febrile UTI caused by another E. coli strain, which was accompanied by detectable escherichelin. Subject 10 was not colonized with HU2117 or any other E. coli strains after day 0, and escherichelin was undetectable in all specimens (Figure 5C). These results are consistent with escherichelin production by HU2117 in conditions in which P. aeruginosa is encountered, which may contribute to the ability of this strain to suppress symptomatic P. aeruginosa UTI.
Discussion
In this study, we used isogenic mutants in the Yersinia HPI and untargeted LC-MS–based metabolomics to identify a previously unappreciated biosynthetic product of bacteria carrying the Yersinia HPI. Escherichelin had previously been observed during the in vitro reconstitution of Ybt biosynthesis (24), but we now identify it as a physiologically relevant product of the Ybt biosynthetic pathway. The name escherichelin was chosen to recognize its identification as a natural product from pathogenic E. coli (escheri-) and its metal chelating properties (-chelin). Escherichelin may have eluded discovery in the 20 years since the identification of Ybt because its reverse-phase chromatographic and UV-visible spectroscopic characteristics are nearly identical to those of Ybt, making them difficult to resolve. Although escherichelin lacks the siderophoric activity of Ybt, the fortuitous synthesis of escherichelin prior to the present discovery (51) and the demonstration of its ability to inhibit pyochelin-mediated iron uptake by P. aeruginosa (25) suggests an alternative functional rationale for escherichelin’s secretion by Enterobacteriaceae: to inhibit iron uptake in a competing species. This niche exclusion function is consistent with clinical observations regarding Pseudomonas UTI, asymptomatic bacteriuria, and protective influences of a healthy human microbiome.
Other microbes have demonstrated an ability to interfere with pyochelin function. Pseudomonas fluorescens can synthesize an enantiomeric form of pyochelin (enantio-pyochelin), which preserves the coordinating properties of pyochelin while resisting FptA-mediated uptake by pyochelin-producing competitor strains. The fungal commensal and opportunistic pathogen C. albicans suppresses pyochelin biosynthesis, interfering with P. aeruginosa virulence, but not colonization, in a murine model (73). A detailed study by Mislin et al. (25) revealed that escherichelin acts as a drug-like inhibitor of pyochelin-mediated iron uptake. Specifically, escherichelin competitively inhibited radioactive 55Fe iron uptake through the pyochelin outer membrane transporter FptA, with a Ki of 27 nM. A molecular docking simulation supports an inhibitory mechanism based on occupancy of the pyochelin-binding site with a 1:2 Fe(III)-escherichelin complex. Here, we experimentally confirmed that escherichelin forms an Fe(III) complex and found by quantum calculations that this complex favors the molecular geometry used in the docking model of Mislin et al. (25). We also experimentally confirmed that escherichelin purified from E. coli inhibits P. aeruginosa pyochelin–dependent growth. Overall, the results are consistent with a noncanonical siderophore function for escherichelin as an effector of interspecies bacterial antagonism. To our knowledge, this is the first report of a microbial natural product that is a direct inhibitor of a competing species’ siderophore transport system.
Multiple studies have implicated the pyochelin siderophore system in virulence (74–76), including genetic abrogation of pyochelin-mediated iron uptake that diminished P. aeruginosa virulence in murine infection models (53, 54). In this context, escherichelin-producing Enterobacteriaceae at colonized devices or mucosal surfaces may prevent P. aeruginosa from progressing to symptomatic infection by chemically inhibiting pyochelin-mediated iron uptake. This scenario may occur at epithelial surfaces during transient or persistent colonization of the urinary tract. The apparent ability of asymptomatic bacteriuria E. coli strain 83972 (or its derivative HU2117) to produce escherichelin during experimental human colonization is consistent with a model in which E. coli in the urinary tract secretes an antivirulence compound against P. aeruginosa (26, 27). Expression of the virulence-associated Yersinia HPI in colonizing Enterobacteriaceae such as E. coli strain 83972 may thus confer both risk (from Ybt production) and benefit (escherichelin production) to patients (77). Engineering an E. coli strain that colonizes the bladder and retains escherichelin, but not Ybt, production may be a desirable bacterial interference strategy in the future. Lack of a recognized asymptomatic bacteriuria mouse model and mouse-specific bacterial tissue tropisms in the urinary tract suggest that these future engineered strains may be best evaluated in human studies.
Whereas the Ybt and pyochelin biosynthetic machineries both generate the immediate enzyme-bound precursor to escherichelin (Figure 2B, top structure), the pyochelin machinery in P. aeruginosa releases less escherichelin than does the Ybt machinery in E. coli. This is consistent with the evolutionary hypothesis that the pyochelin biosynthetic pathway evolved to avoid autoinhibition of Fe(III)-pyochelin uptake. The architectures of these pathways provide a potential explanation for this difference in escherichelin production. Specifically, the escherichelin precursor in the Ybt pathway forms at the terminus of the first NRPS protein (HMWP2), where it must be transferred in trans to the second NRPS/PKS protein (HMWP1). In the pyochelin pathway, this same intermediate is instead passed in cis to a subsequent domain on the same protein (PchF). In the Ybt biosynthetic pathway, this handoff between proteins appears to be particularly susceptible to spontaneous hydrolysis, thereby allowing it to become a branch point in which the intermediate can proceed to form either escherichelin or Ybt. In contrast, this immediate precursor to escherichelin may be less vulnerable to hydrolysis off the pyochelin biosynthetic machinery, because it does not occur at a handoff between proteins. During pyochelin biosynthesis, Dha is passed in trans from PchE to PchF, and it is Dha, rather than escherichelin, that accumulates in P. aeruginosa. Intriguingly, Dha has been shown to possess antimicrobial activity against fungal and bacterial species (31). Applied generally, this suggests that the NRPS/PKS pathways may evolve to segregate modules onto separate proteins at steps in the pathway where a “leak” yields a product that benefits the organism. Whether the branch point in Ybt biosynthesis that yields escherichelin and the similar branch points in other NRPS/PKS pathways reflect uncontrolled “leaks” or regulated processes is unclear. Further biochemical and genetic characterization of these pathways may address this possibility.
Advances in MS and the application of powerful computational and statistical analyses to the data have led to the fruitful application of metabolomics to secondary metabolism (78). MS-based metabolomics is enabling the rapid discovery of secondary metabolites from newly sequenced bacterial strains as well as novel products from well-studied pathways (79). This approach moves beyond the functional screens that were historically used to discover siderophores and other natural products. With this unbiased approach, one is open to discovering any molecule that is chemically compatible with the instrumentation, regardless of the molecule’s function. Continued application of LC-MS–based metabolomics to other siderophore biosynthetic pathways will further our understanding of iron acquisition by pathogens.
Methods
Bacterial strains and culture conditions.
The well-characterized, cystitis-derived model UPEC strain UTI89 was used for these studies (28). The creation of the isogenic mutants in ybtS, ybtE, irp1, ybtU, ybtT, and ybtA was previously described (12, 38). UTI89 Δirp1/entB::kan was generated in the same manner, using the lambda red recombinase method (80, 81). For growth under iron-restricted conditions, strains were first grown in Luria-Bertani (LB) broth (BD Biosciences) overnight at 37 °C under shaking conditions. They were then back-diluted 1:1,000 into M63 minimal medium supplemented with 10 mg/ml niacin and 0.2% (v/v) glycerol (Sigma-Aldrich; complete M63). For salicylate complementation experiments, 50 μM salicylate or 13C6-salicylate (Sigma-Aldrich) was also added. To achieve iron-replete conditions, 16.2 mg/l FeCl3 (Sigma-Aldrich) was added to the complete M63. For comparison of E. coli and P. aeruginosa metabolites, the model P. aeruginosa strain PAO1 was used (82). In these experiments, UTI89 and PAO1 were grown in complete M63 supplemented with 1% (w/v) casamino acids (BD Biosciences). For E. coli siderophore–dependent growth assays, the iron-limited and nutrient-rich yeast extract–casamino acids (YESCA) medium was used (1% casamino acids and 0.12% yeast extract; BD Biosciences). All cultures were grown at 37°C under shaking conditions for approximately 18 hours. Other model E. coli strains (MG1655 [ref. 83], CFT073 [ref. 84], NU14 [ref. 85], and 83972 [ref. 64]) as well as clinical isolates of UPEC, K. pneumoniae, and C. diversus were grown in the same manner.
Metabolomic analysis.
For metabolomics experiments, 5 biological replicate cultures were cultivated from 5 distinct single colonies under iron-restricted conditions, as described above. Cultures were centrifuged at 7,142 g for 10 minutes to pellet the cells. The spent supernatants were collected and filtered through 0.22-μM filters (EMD Millipore). A quality control sample was generated by combining an equal volume of each sample. The samples were analyzed in technical triplicates and in a randomized order on a Shimadzu Prominence UFLC-coupled AB Sciex 4000 QTrap mass spectrometer with a Turbo V electrospray ionization (ESI) source. The LC separation was performed on an Ascentis Express phenyl-hexyl column (100 × 2.1 mm, 2.7 μM; Sigma-Aldrich) with a 32-minute linear gradient from 2% A (HPLC-grade water plus 0.1% [v/v] formic acid; Sigma-Aldrich) to 98% B (90% acetonitrile plus 0.1% [v/v] formic acid; EMD Millipore) at 0.35 ml/min. The MS was operated in positive ion–enhanced MS mode, scanning from 50 to 1,200 m/z. The quality control sample was injected first to precondition the column and every 10 samples thereafter to assess instrument stability.
MarkerView, version 1.2.0 (Sciex) was used for the generation of an aligned peak list and statistical analysis. The triplicate runs were averaged, and the data were Pareto scaled for supervised PCA-DA with 2 groups: (a) UTI89 and ΔybtS plus salicylate; and (b) ΔybtS. Candidate molecular features were identified on the PCA-DA loading plots and then verified by visual inspection of the LC-MS data.
Metabolite detection and quantification in bacterial culture.
Escherichelin, Dha, pyochelin, and Ybt were detected in culture supernatants and cellular extracts from UTI89, PAO1, and clinical Enterobacteriaceae strains by a multiple reaction monitoring (MRM) method on the Shimadzu Prominence UFLC-coupled AB Sciex QTrap 4000 with MS/MS transitions for escherichelin (307.0/219.9), Dha (224.0/178.0), pyochelin (325.0/190.0), apo-Ybt (482.2/295.1), and Fe-Ybt (535.6/349.5) and a collision energy of 37 V. The above column, mobile phases, and flow rate were used with the following LC gradient: solvent B was held constant at 5% for 2 minutes, increased to 56% over 8 minutes, and then increased to 98% over 2 minutes, all with a flow rate of 0.4 ml/min.
Stable isotope dilution was used to quantify escherichelin levels. A 13C6-escherichelin internal standard (IS) was generated by culturing UTI89 ΔybtS in complete M63 minimal medium supplemented with 50 μM 13C6-salicylate. For quantification of extracellular escherichelin, strains were grown under iron-restricted conditions, as described above, and 1 ml of culture supernatant was spiked with 25 μl 13C6-escherichelin IS and 6 μl FeCl3. The samples were incubated at room temperature for 10 minutes, centrifuged at 15,996 g for 3 minutes, and extracted on C18 solid-phase extraction 96-well plates (United Chemical Technologies) with 80% methanol (Sigma-Aldrich). Samples were then analyzed by the escherichelin-specific MRM, and escherichelin levels were assessed by taking the peak area ratio of the native isotope escherichelin to the IS 13C6-escherichelin. Viable CFU were measured to ensure that all strains grew to an equal final density.
Intracellular escherichelin was extracted from 50-ml cultures grown under iron-restricted conditions, as described above. The cultures were centrifuged at 7,412 g for 10 minutes to pellet the cells. The supernatant was discarded, and the cells were washed 3 times with 25 ml PBS (Sigma-Aldrich). The pellets were then resuspended in 1.2 ml ice-cold 80% methanol, briefly vortexed, and incubated on dry ice for 30 minutes. After thawing, the samples were centrifuged at 15,996 g for 10 minutes to pellet the cell debris. The supernatant was retained, and 800 μl acetonitrile was added. The mixture was incubated on ice for 15 minutes and then centrifuged at 15,996 g for 2 minutes to remove any remaining debris. 13C6-escherichelin (10 μl) IS was added to 250 μl of each sample, and escherichelin levels were quantified by LC-MRM, as described above.
MS for escherichelin identification.
A Shimadzu UFLC-coupled Agilent 6550 Q-TOF mass spectrometer with a Dual AJS ESI ion source was used to determine the accurate mass, and therefore molecular formula, of escherichelin in UTI89 supernatant that had been prepared as described above. LC separation was performed on an Ascentis Express phenyl-hexyl column (100 × 2.1 mm, 2.7 μM) with the same mobile phases as described above. The gradient was as follows: solvent B was held constant at 2% for 1 minute, then increased to 50% over 6 minutes, then increased to 98% over 2 minutes, and then held constant at 98% for 1 minute, all with a flow rate of 0.6 ml/min. The MS was operated in positive-ion mode, scanning a mass range from 75 to 1,100 Da. The calculated mass for HPTT-COOH is [M+H]+ (C13H11N2O3S2+) = 307.0206, and 307.0216 was found.
MS/MS was performed on the Shimadzu Prominence UFLC-coupled AB Sciex QTrap 4000. Supernatant from UTI89 grown under iron-restricted conditions was prepared as described above for detection and quantification. Data were collected in the positive-ion, enhanced product ion (EPI) mode, with the parent ion m/z set at 307 and the collision energy set at 37 V.
Escherichelin purification.
Escherichelin was purified from UTI89Δirp1/entB::kan grown under iron-restricted conditions, as described above, with the addition of 20 μM 2,2′-dipyridiyl (Sigma-Aldrich) and 0.1% casamino acids (w/v) to the complete M63. Culture supernatant was applied to a methanol-conditioned C18 silica column (Sigma-Aldrich). The column was washed with water and then eluted with 80% methanol. The eluate was dried down with a lyophilizer (Labconco), resuspended in 20% methanol, and further purified on a Bio-Rad BioLogic DuoFlow 10 system equipped with a QuadTec UV-Vis detector and a Kromasil Eternity-5-phenylhexyl column (AkzoNobel). The column was run at 0.7 ml/min with water plus 0.1% formic acid (solvent A) and acetonitrile plus 0.1% formic acid (solvent B) and the following gradient: solvent B held steady at 0% for 2.5 ml, then increased to 30% over 2 ml, then increased to 70% over 20 ml, and finally increased to 100% over 2 ml. Fractions containing escherichelin were identified via UV-Vis detection at 318 nm, pooled together, and dried down by lyophilization. Purification was confirmed by LC-MS.
NMR.
Purified escherichelin was resuspended in CDCl3. One-dimensional 1H and two-dimensional 1H COSY spectra were collected at 20°C on a Varian Unity Inova-500 MHz instrument.
UV-Vis spectroscopy.
Purified escherichelin was reconstituted to 1 mM in 50% ethanol (Sigma-Aldrich). For experiments to monitor metal binding, escherichelin was diluted to 100 μM in water and combined with an equimolar amount of FeCl3 or CuSO4 (Sigma-Aldrich). The samples were incubated at room temperature for 2 hours, and then a wavelength scan was collected from 200 to 800 nm on a UV-Vis spectrophotometer (Beckman Coulter; DU800). For each escherichelin-plus-metal scan, the instrument was blanked with a metal-only control. For escherichelin alone, the instrument was blanked with 5% ethanol.
E. coli siderophore–dependent growth assay.
The assay was adapted from a previously published Fe(III)-Ybt–dependent growth assay, and apo-Ybt was purified as previously described (16, 19). UTI89 was grown overnight in LB broth at 37°C under shaking conditions and then washed 3 times by centrifugation and resuspension in a volume of PBS equal to the culture volume. The washed bacteria were inoculated 1:100 into YESCA or YESCA supplemented with 0.5 mM ethylenediamine-N,N′-bis(2-hydroxyphenylacetic acid) (EDDHA; Complete Green Company). Prior to their addition, apo-Ybt and escherichelin were preincubated for 2 hours with an equimolar amount of FeCl3 to allow for complex formation. The Fe(III) complexes were added at a final concentration of 1 μM, and the cultures were grown at 37°C under shaking conditions. Bacterial growth was monitored over time by measuring the OD at 600 nm.
Escherichelin activity assay on P. aeruginosa.
PW5011 (pvdA-E02::ISlacZ/hah) (55, 56) was grown in LB broth overnight at 37°C under shaking conditions and then back-diluted into succinate medium (25) at 108 CFU/ml. The growth experiments were performed in a final volume of 100 μl in 96-well plates. Escherichelin, solubilized in ethanol, was added to the cultures at increasing concentrations. The final ethanol (vehicle) concentration in all cultures was 5% (v/v). The cultures were grown at 37°C under shaking conditions for 20 hours, at which point they were serially diluted in PBS and plated on LB agar for CFU quantification.
Theoretical calculations.
The theoretical calculations to identify thermodynamically stable escherichelin-metal complexes were performed with the DFT method as previously described (15, 19). This DFT approach was previously used to model a structure of Fe(III)-Ybt that matched the published crystal structure (50). The simulated Fe(III)-escherichelin complexes agree with a previously described model of Fe(III)-escherichelin bound to FptA (25).
Escherichelin detection in urine.
The clinical uncomplicated cystitis isolates were grown under iron-restricted conditions, and their supernatants were analyzed for Ybt by LC-MRM. Seventeen of the nineteen selected isolates were Ybt producers. The 2 Ybt nonproducers were used as controls. Two additional strains (Staphylococcus saprophyticus and Enterococcus sp.) were selected from the cohort to serve as non–E. coli, Ybt-negative controls. After determining whether the causative strain was a Ybt producer, the corresponding urine samples were thawed on ice and then centrifuged at 15,996 g for 3 minutes to remove any debris. To each 500 μl of urine, 12.5 μl 13C6-escherichelin IS and 3 μl 0.5 M FeCl3 were added. The samples were incubated at room temperature for 15 minutes and then centrifuged at 15,996 g for 5 minutes. The supernatant was then extracted on 96-well C18 solid-phase extraction plates with 80% methanol. Escherichelin was detected by LC-MRM using the method described above. Escherichelin was detected in the bacterial interference trial specimens using the same extraction and LC-MRM methods.
Statistics.
GraphPad Prism 4 (GraphPad Software) was used to perform statistical analysis and generate graphs for this study. All P values were determined by unpaired, 2-tailed t test or 1-way ANOVA with Dunnett’s multiple comparisons test. A P value of 0.05 or less was considered statistically significant.
Study approval.
The urine and bacterial specimens from patients with uncomplicated cystitis were collected with the approval of the Human Subjects Review Committee of the University of Washington. All participants provided written informed consent for the sample collection and following analysis, prior to inclusion in the study. Study design, inclusion and exclusion criteria, and sample preparation were previously described (14, 86). β-Hemolytic E. coli isolates from urine cultures with 105 or more CFU/ml were selected from the cohort for analysis.
The HU2117 experimental human colonization study protocol was approved by the FDA (investigational new drug [IND] 14007), the IRB of Baylor College of Medicine, and the Houston VA Research and Development committee and registered as a clinical trial (ClinicalTrials.gov identifier: NCT00554996. All participants provided written informed consent prior to inclusion in the study. The study design, inclusion and exclusion criteria, and specimen preparation were previously reported (27).
Author contributions
SIO and JPH conceived and designed the experiments. SIO, DEG, and DAD performed the experiments. AES and BWT collected human specimens. SIO, DEG, DAD, and JPH analyzed the data. SIO and JPH wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Chloe Pinkner, Jan Crowley, and Hung Tran at Washington University for their technical assistance. JPH holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund and acknowledges receiving grants from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (R01DK099534 and R56DK111930) and a Prevention Epicenters Grant from the Centers for Disease Control and Prevention (CDC U54CK000482). SIO is supported by a training grant from the National Institute of Allergy and Infectious Diseases (5T32AI007172-35) and the Graduate Research Fellowship Program (DGE-1143954) from the National Science Foundation (NSF). Computations were supported by a grant from the United States Public Health Service (P41-RR00954); some computations were performed at the Washington University Center for High Performance Computing and the Washington University Computational Chemistry Facility, funded by a grant from the NSF (CHE-0443501). The high-resolution MS performed in this project was supported by a grant from the National Institute of General Medical Sciences (8 P41 GM103422) from the NIH. The P. aeruginosa transposon mutant PW5011 was obtained from the Manoil laboratory at the University of Washington (NIH grant P30 DK089507). Specimens from patients with uncomplicated UTI were obtained with support from NIH grant P50DK064540. Experimental patient colonization studies were supported by a grant from the NIDDK, NIH (R21 DK092293) and by resources of the Houston VA Center for Innovations in Quality, Effectiveness, and Safety (CIN13-413) at the Michael E. DeBakey Veterans Affairs Medical Center. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Version 1. 09/25/2017
Electronic publication
Version 2. 11/01/2017
Print issue publication
Funding Statement
Graduate Research Fellowship Program
National Institute of Allergy and Infectious Diseases T32 training grant
National Institute of Diabetes and Digestive and Kidney Diseases
National Institute of Diabetes and Digestive and Kidney Diseases
Prevention Epicenters Grant
Washington University Center for High Performance Computing and Washington University Computational Chemistry Facility
National Institute of General Medical Sciences, Biomedical Mass Spectrometry Resource at Washington University in St. Louis
National Institute of Diabetes and Digestive and Kidney Diseases
National Institute of Diabetes and Digestive and Kidney Diseases
Michael E. DeBakey VA Medical Center
Footnotes
DAdA’s present address is: Department of Medicine, University of Minnesota, Minneapolis, Minnesota, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(11):4018–4030.https://doi.org/10.1172/JCI92464.
References
- 1.Foxman B. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am. 2014;28(1):1–13. doi: 10.1016/j.idc.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Zowawi HM, et al. The emerging threat of multidrug-resistant Gram-negative bacteria in urology. Nat Rev Urol. 2015;12(10):570–584. doi: 10.1038/nrurol.2015.199. [DOI] [PubMed] [Google Scholar]
- 3.Nicolle LE, et al. Infectious Diseases Society of America guidelines for the diagnosis and treatment of asymptomatic bacteriuria in adults. Clin Infect Dis. 2005;40(5):643–654. doi: 10.1086/427507. [DOI] [PubMed] [Google Scholar]
- 4.Brubaker L, Wolfe AJ. The new world of the urinary microbiota in women. Am J Obstet Gynecol. 2015;213(5):644–649. doi: 10.1016/j.ajog.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith HS, et al. Antecedent antimicrobial use increases the risk of uncomplicated cystitis in young women. Clin Infect Dis. 1997;25(1):63–68. doi: 10.1086/514502. [DOI] [PubMed] [Google Scholar]
- 6.Bitsori M, Maraki S, Koukouraki S, Galanakis E. Pseudomonas aeruginosa urinary tract infection in children: risk factors and outcomes. J Urol. 2012;187(1):260–264. doi: 10.1016/j.juro.2011.09.035. [DOI] [PubMed] [Google Scholar]
- 7.Parker KS, Wilson JD, Marschall J, Mucha PJ, Henderson JP. Network analysis reveals sex- and antibiotic resistance-associated antivirulence targets in clinical uropathogens. ACS Infect Dis. 2015;1(11):523–532. doi: 10.1021/acsinfecdis.5b00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson JR. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev. 1991;4(1):80–128. doi: 10.1128/CMR.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71(3):413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol. 2012;10(8):525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arezes J, et al. Hepcidin-induced hypoferremia is a critical host defense mechanism against the siderophilic bacterium Vibrio vulnificus. Cell Host Microbe. 2015;17(1):47–57. doi: 10.1016/j.chom.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henderson JP, et al. Quantitative metabolomics reveals an epigenetic blueprint for iron acquisition in uropathogenic Escherichia coli. PLoS Pathog. 2009;5(2):e1000305. doi: 10.1371/journal.ppat.1000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marschall J, Zhang L, Foxman B, Warren DK, Henderson JP, CDC Prevention Epicenters Program Both host and pathogen factors predispose to Escherichia coli urinary-source bacteremia in hospitalized patients. Clin Infect Dis. 2012;54(12):1692–1698. doi: 10.1093/cid/cis252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaturvedi KS, Hung CS, Crowley JR, Stapleton AE, Henderson JP. The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat Chem Biol. 2012;8(8):731–736. doi: 10.1038/nchembio.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaturvedi KS, et al. Cupric yersiniabactin is a virulence-associated superoxide dismutase mimic. ACS Chem Biol. 2014;9(2):551–561. doi: 10.1021/cb400658k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koh EI, Hung CS, Henderson JP. The yersiniabactin-associated ATP binding cassette proteins YbtP and YbtQ enhance Escherichia coli fitness during high-titer cystitis. Infect Immun. 2016;84(5):1312–1319. doi: 10.1128/IAI.01211-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brumbaugh AR, et al. Blocking yersiniabactin import attenuates extraintestinal pathogenic Escherichia coli in cystitis and pyelonephritis and represents a novel target to prevent urinary tract infection. Infect Immun. 2015;83(4):1443–1450. doi: 10.1128/IAI.02904-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bach S, de Almeida A, Carniel E. The Yersinia high-pathogenicity island is present in different members of the family Enterobacteriaceae. FEMS Microbiol Lett. 2000;183(2):289–294. doi: 10.1111/j.1574-6968.2000.tb08973.x. [DOI] [PubMed] [Google Scholar]
- 19.Koh EI, Hung CS, Parker KS, Crowley JR, Giblin DE, Henderson JP. Metal selectivity by the virulence-associated yersiniabactin metallophore system. Metallomics. 2015;7(6):1011–1022. doi: 10.1039/C4MT00341A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66(2):223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quadri LE, Keating TA, Patel HM, Walsh CT. Assembly of the Pseudomonas aeruginosa nonribosomal peptide siderophore pyochelin: In vitro reconstitution of aryl-4, 2-bisthiazoline synthetase activity from PchD, PchE, and PchF. Biochemistry. 1999;38(45):14941–14954. doi: 10.1021/bi991787c. [DOI] [PubMed] [Google Scholar]
- 22.Reddy PV, et al. Disruption of mycobactin biosynthesis leads to attenuation of Mycobacterium tuberculosis for growth and virulence. J Infect Dis. 2013;208(8):1255–1265. doi: 10.1093/infdis/jit250. [DOI] [PubMed] [Google Scholar]
- 23.Pelludat C, Brem D, Heesemann J. Irp9, encoded by the high-pathogenicity island of Yersinia enterocolitica, is able to convert chorismate into salicylate, the precursor of the siderophore yersiniabactin. J Bacteriol. 2003;185(18):5648–5653. doi: 10.1128/JB.185.18.5648-5653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller DA, Luo L, Hillson N, Keating TA, Walsh CT. Yersiniabactin synthetase: a four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem Biol. 2002;9(3):333–344. doi: 10.1016/S1074-5521(02)00115-1. [DOI] [PubMed] [Google Scholar]
- 25.Mislin GL, Hoegy F, Cobessi D, Poole K, Rognan D, Schalk IJ. Binding properties of pyochelin and structurally related molecules to FptA of Pseudomonas aeruginosa. J Mol Biol. 2006;357(5):1437–1448. doi: 10.1016/j.jmb.2006.01.080. [DOI] [PubMed] [Google Scholar]
- 26.Prasad A, Cevallos ME, Riosa S, Darouiche RO, Trautner BW. A bacterial interference strategy for prevention of UTI in persons practicing intermittent catheterization. Spinal Cord. 2009;47(7):565–569. doi: 10.1038/sc.2008.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horwitz D, et al. Decreased microbiota diversity associated with urinary tract infection in a trial of bacterial interference. J Infect. 2015;71(3):358–367. doi: 10.1016/j.jinf.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen SL, et al. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci U S A. 2006;103(15):5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambers CE, McIntyre DD, Mouck M, Sokol PA. Physical and structural characterization of yersiniophore, a siderophore produced by clinical isolates of Yersinia enterocolitica. Biometals. 1996;9(2):157–167. doi: 10.1007/BF00144621. [DOI] [PubMed] [Google Scholar]
- 30.Serino L, Reimmann C, Visca P, Beyeler M, Chiesa VD, Haas D. Biosynthesis of pyochelin and dihydroaeruginoic acid requires the iron-regulated pchDCBA operon in Pseudomonas aeruginosa. J Bacteriol. 1997;179(1):248–257. doi: 10.1128/jb.179.1.248-257.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carmi R, Carmeli S, Levy E, Gough FJ. (+)-(S)-dihydroaeruginoic acid, an inhibitor of Septoria tritici and other phytopathogenic fungi and bacteria, produced by Pseudomonas fluorescens. J Nat Prod. 1994;57(9):1200–1205. doi: 10.1021/np50111a002. [DOI] [PubMed] [Google Scholar]
- 32.Gehring AM, Mori I, Perry RD, Walsh CT. The nonribosomal peptide synthetase HMWP2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron-chelating virulence factor of Yersinia pestis. Biochemistry. 1998;37(33):11637–11650. doi: 10.1021/bi9812571. [DOI] [PubMed] [Google Scholar]
- 33.Brzuszkiewicz E, et al. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci U S A. 2006;103(34):12879–12884. doi: 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lv H, Hung CS, Henderson JP. Metabolomic analysis of siderophore cheater mutants reveals metabolic costs of expression in uropathogenic Escherichia coli. J Proteome Res. 2014;13(3):1397–1404. doi: 10.1021/pr4009749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gehring AM, et al. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem Biol. 1998;5(10):573–586. doi: 10.1016/S1074-5521(98)90115-6. [DOI] [PubMed] [Google Scholar]
- 36.Bearden SW, Fetherston JD, Perry RD. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65(5):1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geoffroy VA, Fetherston JD, Perry RD. Yersinia pestis YbtU and YbtT are involved in synthesis of the siderophore yersiniabactin but have different effects on regulation. Infect Immun. 2000;68(8):4452–4461. doi: 10.1128/IAI.68.8.4452-4461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lv H, Henderson JP. Yersinia high pathogenicity island genes modify the Escherichia coli primary metabolome independently of siderophore production. J Proteome Res. 2011;10(12):5547–5554. doi: 10.1021/pr200756n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fetherston JD, Bearden SW, Perry RD. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol Microbiol. 1996;22(2):315–325. doi: 10.1046/j.1365-2958.1996.00118.x. [DOI] [PubMed] [Google Scholar]
- 40.Miller MC, et al. Reduced synthesis of the Ybt siderophore or production of aberrant Ybt-like molecules activates transcription of yersiniabactin genes in Yersinia pestis. Microbiology (Reading, Engl) 2010;156(Pt 7):2226–2238. doi: 10.1099/mic.0.037945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keating TA, Miller DA, Walsh CT. Expression, purification, and characterization of HMWP2, a 229 kDa, six domain protein subunit of Yersiniabactin synthetase. Biochemistry. 2000;39(16):4729–4739. doi: 10.1021/bi992923g. [DOI] [PubMed] [Google Scholar]
- 42.Miller DA, Walsh CT. Yersiniabactin synthetase: probing the recognition of carrier protein domains by the catalytic heterocyclization domains, Cy1 and Cy2, in the chain-initiating HWMP2 subunit. Biochemistry. 2001;40(17):5313–5321. doi: 10.1021/bi002905v. [DOI] [PubMed] [Google Scholar]
- 43.Suo Z. Thioesterase portability and peptidyl carrier protein swapping in yersiniabactin synthetase from Yersinia pestis. Biochemistry. 2005;44(12):4926–4938. doi: 10.1021/bi047538s. [DOI] [PubMed] [Google Scholar]
- 44.Suo Z, Walsh CT, Miller DA. Tandem heterocyclization activity of the multidomain 230 kDa HMWP2 subunit of Yersinia pestis yersiniabactin synthetase: interaction of the 1-1382 and 1383-2035 fragments. Biochemistry. 1999;38(42):14023–14035. doi: 10.1021/bi991574n. [DOI] [PubMed] [Google Scholar]
- 45.Patel HM, Walsh CT. In vitro reconstitution of the Pseudomonas aeruginosa nonribosomal peptide synthesis of pyochelin: characterization of backbone tailoring thiazoline reductase and N-methyltransferase activities. Biochemistry. 2001;40(30):9023–9031. doi: 10.1021/bi010519n. [DOI] [PubMed] [Google Scholar]
- 46.Cox CD, Graham R. Isolation of an iron-binding compound from Pseudomonas aeruginosa. J Bacteriol. 1979;137(1):357–364. doi: 10.1128/jb.137.1.357-364.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brandel J, Humbert N, Elhabiri M, Schalk IJ, Mislin GL, Albrecht-Gary AM. Pyochelin, a siderophore of Pseudomonas aeruginosa: physicochemical characterization of the iron(III), copper(II) and zinc(II) complexes. Dalton Trans. 2012;41(9):2820–2834. doi: 10.1039/c1dt11804h. [DOI] [PubMed] [Google Scholar]
- 48.Cobessi D, Celia H, Pattus F. Crystal structure at high resolution of ferric-pyochelin and its membrane receptor FptA from Pseudomonas aeruginosa. J Mol Biol. 2005;352(4):893–904. doi: 10.1016/j.jmb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 49.Tseng CF, et al. Bacterial siderophores: the solution stoichiometry and coordination of the Fe(III) complexes of pyochelin and related compounds. J Biol Inorg Chem. 2006;11(4):419–432. doi: 10.1007/s00775-006-0088-7. [DOI] [PubMed] [Google Scholar]
- 50.Miller MC, Parkin S, Fetherston JD, Perry RD, Demoll E. Crystal structure of ferric-yersiniabactin, a virulence factor of Yersinia pestis. J Inorg Biochem. 2006;100(9):1495–1500. doi: 10.1016/j.jinorgbio.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 51.Mislin GL, Burger A, Abdallah MA. Synthesis of new thiazole analogues of pyochelin, a siderophore of Pseudomonas aeruginosa and Burkholderia cepacia. A new conversion of thiazolines into thiazoles. Tetrahedron. 2004;60(52):12139–12145. doi: 10.1016/j.tet.2004.10.030. [DOI] [Google Scholar]
- 52.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minandri F, et al. Role of iron uptake systems in Pseudomonas aeruginosa virulence and airway infection. Infect Immun. 2016;84(8):2324–2335. doi: 10.1128/IAI.00098-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sokol PA. Surface expression of ferripyochelin-binding protein is required for virulence of Pseudomonas aeruginosa. Infect Immun. 1987;55(9):2021–2025. doi: 10.1128/iai.55.9.2021-2025.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacobs MA, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2003;100(24):14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Held K, Ramage E, Jacobs M, Gallagher L, Manoil C. Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. J Bacteriol. 2012;194(23):6387–6389. doi: 10.1128/JB.01479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schalk IJ, Guillon L. Pyoverdine biosynthesis and secretion in Pseudomonas aeruginosa: implications for metal homeostasis. Environ Microbiol. 2013;15(6):1661–1673. doi: 10.1111/1462-2920.12013. [DOI] [PubMed] [Google Scholar]
- 58.Hooton TM, et al. A prospective study of asymptomatic bacteriuria in sexually active young women. N Engl J Med. 2000;343(14):992–997. doi: 10.1056/NEJM200010053431402. [DOI] [PubMed] [Google Scholar]
- 59.Grigoryan L, Abers MS, Kizilbash QF, Petersen NJ, Trautner BW. A comparison of the microbiologic profile of indwelling versus external urinary catheters. Am J Infect Control. 2014;42(6):682–684. doi: 10.1016/j.ajic.2014.02.028. [DOI] [PubMed] [Google Scholar]
- 60.Kang MS, Lee BS, Lee HJ, Hwang SW, Han ZA. Prevalence of and risk factors for multidrug-resistant bacteria in urine cultures of spinal cord injury patients. Ann Rehabil Med. 2015;39(5):686–695. doi: 10.5535/arm.2015.39.5.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reid G, Howard J, Gan BS. Can bacterial interference prevent infection? Trends Microbiol. 2001;9(9):424–428. doi: 10.1016/S0966-842X(01)02132-1. [DOI] [PubMed] [Google Scholar]
- 62.Falagas ME, Rafailidis PI, Makris GC. Bacterial interference for the prevention and treatment of infections. Int J Antimicrob Agents. 2008;31(6):518–522. doi: 10.1016/j.ijantimicag.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 63.Darouiche RO, Hull RA. Bacterial interference for prevention of urinary tract infection. Clin Infect Dis. 2012;55(10):1400–1407. doi: 10.1093/cid/cis639. [DOI] [PubMed] [Google Scholar]
- 64.Andersson P, et al. Persistence of Escherichia coli bacteriuria is not determined by bacterial adherence. Infect Immun. 1991;59(9):2915–2921. doi: 10.1128/iai.59.9.2915-2921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hull R, et al. Urinary tract infection prophylaxis using Escherichia coli 83972 in spinal cord injured patients. J Urol. 2000;163(3):872–877. doi: 10.1016/S0022-5347(05)67823-8. [DOI] [PubMed] [Google Scholar]
- 66.Darouiche RO, Donovan WH, Del Terzo M, Thornby JI, Rudy DC, Hull RA. Pilot trial of bacterial interference for preventing urinary tract infection. Urology. 2001;58(3):339–344. doi: 10.1016/S0090-4295(01)01271-7. [DOI] [PubMed] [Google Scholar]
- 67.Darouiche RO, Thornby JI, Cerra-Stewart C, Donovan WH, Hull RA. Bacterial interference for prevention of urinary tract infection: a prospective, randomized, placebo-controlled, double-blind pilot trial. Clin Infect Dis. 2005;41(10):1531–1534. doi: 10.1086/497272. [DOI] [PubMed] [Google Scholar]
- 68.Trautner BW, Hull RA, Thornby JI, Darouiche RO. Coating urinary catheters with an avirulent strain of Escherichia coli as a means to establish asymptomatic colonization. Infect Control Hosp Epidemiol. 2007;28(1):92–94. doi: 10.1086/510872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sundén F, Håkansson L, Ljunggren E, Wullt B. Escherichia coli 83972 bacteriuria protects against recurrent lower urinary tract infections in patients with incomplete bladder emptying. J Urol. 2010;184(1):179–185. doi: 10.1016/j.juro.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 70.Darouiche RO, et al. Multicenter randomized controlled trial of bacterial interference for prevention of urinary tract infection in patients with neurogenic bladder. Urology. 2011;78(2):341–346. doi: 10.1016/j.urology.2011.03.062. [DOI] [PubMed] [Google Scholar]
- 71.Watts RE, et al. Contribution of siderophore systems to growth and urinary tract colonization of asymptomatic bacteriuria Escherichia coli. Infect Immun. 2012;80(1):333–344. doi: 10.1128/IAI.05594-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zdziarski J, et al. Host imprints on bacterial genomes--rapid, divergent evolution in individual patients. PLoS Pathog. 2010;6(8):e1001078. doi: 10.1371/journal.ppat.1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lopez-Medina E, et al. Candida albicans Inhibits Pseudomonas aeruginosa virulence through suppression of pyochelin and pyoverdine biosynthesis. PLoS Pathog. 2015;11(8):e1005129. doi: 10.1371/journal.ppat.1005129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Konings AF, et al. Pseudomonas aeruginosa uses multiple pathways to acquire iron during chronic infection in cystic fibrosis lungs. Infect Immun. 2013;81(8):2697–2704. doi: 10.1128/IAI.00418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang J, Mushegian A, Lory S, Jin S. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc Natl Acad Sci U S A. 1996;93(19):10434–10439. doi: 10.1073/pnas.93.19.10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Visca P, Chiarini F, Mansi A, Vetriani C, Serino L, Orsi N. Virulence determinants in Pseudomonas aeruginosa strains from urinary tract infections. Epidemiol Infect. 1992;108(2):323–336. doi: 10.1017/S0950268800049797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Köves B, et al. Rare emergence of symptoms during long-term asymptomatic Escherichia coli 83972 carriage without an altered virulence factor repertoire. J Urol. 2014;191(2):519–528. doi: 10.1016/j.juro.2013.07.060. [DOI] [PubMed] [Google Scholar]
- 78.Krug D, Müller R. Secondary metabolomics: the impact of mass spectrometry-based approaches on the discovery and characterization of microbial natural products. Nat Prod Rep. 2014;31(6):768–783. doi: 10.1039/c3np70127a. [DOI] [PubMed] [Google Scholar]
- 79.Vinayavekhin N, Saghatelian A. Regulation of alkyl-dihydrothiazole-carboxylates (ATCs) by iron and the pyochelin gene cluster in Pseudomonas aeruginosa. ACS Chem Biol. 2009;4(8):617–623. doi: 10.1021/cb900075n. [DOI] [PubMed] [Google Scholar]
- 80.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murphy KC, Campellone KG. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol Biol. 2003;4:11. doi: 10.1186/1471-2199-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stover CK, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406(6799):959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 83.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 84.Welch RA, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A. 2002;99(26):17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hultgren SJ, Schwan WR, Schaeffer AJ, Duncan JL. Regulation of production of type 1 pili among urinary tract isolates of Escherichia coli. Infect Immun. 1986;54(3):613–620. doi: 10.1128/iai.54.3.613-620.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shields-Cutler RR, et al. Human urinary composition controls antibacterial activity of siderocalin. J Biol Chem. 2015;290(26):15949–15960. doi: 10.1074/jbc.M115.645812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.