Abstract
Shwachman-Diamond syndrome (SDS) (OMIM #260400) is a rare inherited bone marrow failure syndrome (IBMFS) that is primarily characterized by neutropenia and exocrine pancreatic insufficiency. Seventy-five to ninety percent of patients have compound heterozygous loss-of-function mutations in the Shwachman-Bodian-Diamond syndrome (sbds) gene. Using trio whole-exome sequencing (WES) in an sbds-negative SDS family and candidate gene sequencing in additional SBDS-negative SDS cases or molecularly undiagnosed IBMFS cases, we identified 3 independent patients, each of whom carried a de novo missense variant in srp54 (encoding signal recognition particle 54 kDa). These 3 patients shared congenital neutropenia linked with various other SDS phenotypes. 3D protein modeling revealed that the 3 variants affect highly conserved amino acids within the GTPase domain of the protein that are critical for GTP and receptor binding. Indeed, we observed that the GTPase activity of the mutated proteins was impaired. The level of SRP54 mRNA in the bone marrow was 3.6-fold lower in patients with SRP54-mutations than in healthy controls. Profound reductions in neutrophil counts and chemotaxis as well as a diminished exocrine pancreas size in a SRP54-knockdown zebrafish model faithfully recapitulated the human phenotype. In conclusion, autosomal dominant mutations in SRP54, a key member of the cotranslation protein-targeting pathway, lead to syndromic neutropenia with a Shwachman-Diamond–like phenotype.
Keywords: Genetics, Hematology
Keywords: Bone marrow transplantation, Neutrophils
Introduction
Shwachman-Diamond syndrome (SDS) (OMIM #260400), an inherited bone marrow failure syndrome (IBMFS), is a rare multisystem disorder that was first recognized more than 50 years ago (1–4). Its cardinal features include exocrine pancreatic insufficiency and hematological abnormalities in which neutropenia is paramount and can be life threatening. Skeletal abnormalities, neurocognitive deficits, and a variety of other diverse symptoms are also associated with the disease (5). Like many other IBMFSs, patients with SDS have an increased risk of myelodysplasia and malignant transformation, especially acute myeloid leukemia (6). Approximately 75% to 90% of SDS patients have compound heterozygous loss-of-function mutations in the Shwachman-Bodian-Diamond syndrome (sbds) gene (7–9). Since the identification of sbds in 2003, only 2 other causal SDS genes, DNAJC21 (10, 11) and EFL1 (12), have been reported, in contrast to other IBMFSs, for which the situation is the opposite (e.g., >20 loci in Fanconi anemia, >10 loci in Diamond-Blackfan anemia, and several loci in severe congenital neutropenia) (13).
The SBDS protein plays a central role in ribosome biogenesis, as it is involved in the maturation of the 60S subunit (as are DNAJC21 and EFL1) prior to the assembly of the actively translating 80S ribosome (14, 15). During the biogenesis and initiation of protein translation, the ribosome associates with many partner molecules, and of these, the signal recognition particle (SRP) is a major one. SRP is a ribonucleoprotein (RNP) complex that mediates the targeting of nascent signal sequence–carrying polypeptides (including both membrane and secretory proteins) to the translocon at the surface of the ER (16). The mammalian SRP is composed of a single RNA molecule (the ~300-nucleotide-long 7SL RNA) and the following 6 polypeptides: SRP9, SRP14, SRP19, SRP54, SRP68, and SRP72 (17). SRP54 plays a central role in this RNP complex. With its M domain, SRP54 binds to the SRP19-bound 7SL RNA and associates with the signal peptide of the nascent polypeptide chain. Then, via its N and G domains, SRP54 binds to the SRP receptor (SR) on the ER membrane (18, 19). Ultimately, SRP54 forms a link between the SRP and the ER membrane and allows insertion of the signal peptide–carrying polypeptide into the translocation channel. In addition to the SRP RNA, SRP54 is the only other universally conserved SRP component. Here, we demonstrate that mutations in srp54 can lead to syndromic neutropenia with SDS-like features.
Results
Clinical characteristics of patients.
The clinical features of the 3 patients are summarized in Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/JCI92876DS1). Patient AII.1 was a white male and the second child of healthy nonconsanguineous parents (Figure 1A). The patient was born after a full-term, uneventful pregnancy and presented with neonatal transitory respiratory distress, hypotonia, and central apneas. Morphological examination of the newborn revealed small size (–1.5 SD), low weight (–1.5 SD), an occipito-frontal circumference (OFC) of +1.5 SD, low-set, asymmetric ears, thinning hair, frontal angioma, mandibular microretrognathism, a high-arched palate, small teeth, and a pectus carinatum — a series of symptoms that are all uncommon in typical SDS patients. Complete blood counts in the first week of life showed severe neutropenia (0.28 × 109/l and then 0 × 109/l). Hemoglobin (15.8 g/dl) and platelet (265 × 109/l) counts were normal. He received granulocyte-CSF (G-CSF) injections, with no effect on neutrophil counts. A bone marrow smear was hypocellular and showed hypoplasia in the neutrophil lineage with retarded granulocyte maturation. Moreover, numerous cytoplasmic vacuoles were observed in myeloblasts, promyelocytes, and apoptotic cells of the granulocyte lineage (Supplemental Figure 1A). Of note, none of the erythroblasts contained vacuoles, and Perls’ staining revealed no ring sideroblasts, thus excluding Pearson syndrome. Furthermore, iron was detected in macrophages using the Prussian blue reaction. To definitively exclude a mitochondrial DNA disorder (i.e., Pearson syndrome), whole mitochondrial DNA analysis was performed using next-generation sequencing, which revealed no pathogenic mutations or rearrangements (Supplemental Figure 2). The patient’s lymphocyte immunophenotype results and serum Ig levels were normal. There were no alloimmunizations against neutrophils or granulocyte-specific antigens. Cardiac ultrasound showed large interauricular and interventricular septal defects. The cerebral MRI was normal. At 6 months of age, the patient had feeding difficulties and diarrhea. His weight, height, and OFC were 6.5 kg (–1.5 SD), 63 cm (–1.5 SD), and 45 cm (+1.5 SD), respectively. Stool analysis revealed very low elastase levels (<15 μg/g of stool; reference value: >200 μg/g of stool). Fatty infiltration of the pancreas was documented by CT (Supplemental Figure 3A). Blood levels of the fat-soluble vitamins A, D, E, and K were very low. At 11 months of age, the patient’s ventricular and atrial septal defects required surgery. Severe neutropenia led to infections including acute ethmoiditis and perianal abscesses. At 4 years of age, the patient underwent genoidentical allogeneic hematopoietic cell transplantation (HCT) without major complications. His hematologic reconstitution was uneventful. The patient showed psychomotor developmental delay and autistic behavior. At 6 years of age, he continues to receive lipase substitution (see Supplemental Table 1). Direct sequencing of the SBDS gene was performed using genomic DNA isolated from pretransplantation peripheral blood cells and failed to identify any mutations, as did panel sequencing of all known IBMFS genes. Comparative genome hybridization (CGH) also excluded pathogenic structural variants and copy number variations (CNVs) throughout the genome, including inside and in the vicinity of SBDS.
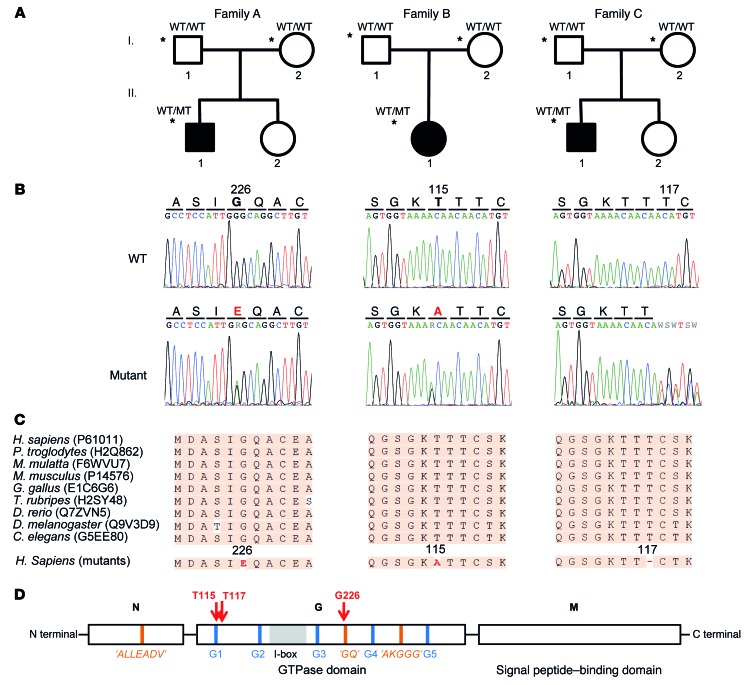
Figure 1. Heterozygous SRP54 mutations identified in 3 families with syndromic neutropenia and SDS-like features.
(A) Pedigrees of the 3 families. Black symbols indicate affected individuals; asterisks indicate subjects who underwent exome sequencing. The mutational status of the SRP54 gene is indicated as WT and mutated (MT). (B) Sample sequence traces showing WT and mutant sequences for each family. (C) Protein sequence conservation among species. (D) Domain structure of the SRP54 GTPase with its N, G, and M domains. The positions of SRP GTPase-specific motifs, the I-box, and the conserved nucleotide-binding elements (G1–G5) are indicated in orange, gray, and blue, respectively. The mutated residues are shown in red.
Patient BII.1 was a 1-year-old white girl who was the only child of healthy nonconsanguineous parents (Figure 1A). At birth, her weight, height, and OFC were 3.26 kg (0 SD), 45.5 cm (–2 SD), and 32.5 cm (–1 SD), respectively. She had no dysmorphic features. Repeated blood counts revealed severe neutropenia (e.g., 0.02 × 109/l), while her other blood parameters were normal, including fetal hemoglobin, cupremia, lymphocyte immunophenotyping, and Ig levels. No alloimmunization against granulocytes was found. Cerebral MRI as well as abdominal and cardiac ultrasound imaging results were normal. Her fecal elastase level was very low (<15 μg/g of stool; reference value: >200 μg/g of stool). Abdominal CT showed a fatty infiltration of the pancreas (Supplemental Figure 3B). At 6 months of age, the patient presented with facial cellulitis and ethmoiditis. She was given G-CSF without success. At 1 year of age, she underwent unrelated cord blood transplantation. Pretransplantation clinical examination revealed small stature (69 cm; –2 SD), low weight (8.5 kg; –0.5 SD), and an OFC of 48 cm (+1 SD) (Supplemental Table 1). No overt developmental delay or psychomotor retardation was observed before she passed away at 16 months of age (see below). A blood cell count revealed a total absence of neutrophils. Her other blood count parameters were normal. Anomalies similar to those in patient AII.1 were observed in the bone marrow smear of this patient, who also had no ring sideroblasts and no mitochondrial DNA anomalies (Supplemental Figure 1B and Supplemental Figure 2). Her ferritin level was normal (37 μg/l). Other cell lineages were normal. HCT and hematopoietic reconstitution were uneventful. Two months after transplantation, the patient developed multiorgan failure, which rapidly led to death. Postmortem pathology showed severe veno-occlusive disease. Direct sequencing of the sbds gene (and panel sequencing of all other IBMFS genes) using genomic DNA isolated from pretransplantation peripheral blood cells failed to identify any mutations. No pathogenic structural variants were detected via CGH array, as was the case with the previous patient.
Patient CII.1 was a Hispanic boy and the second child of healthy nonconsanguineous parents (Figure 1A), who was born after a full-term, uneventful pregnancy. His birth weight was 3.24 kg (25th percentile), and no dysmorphic features or bony abnormalities were visible. He did not have any feeding or gastrointestinal problems. At 2 years of age, he was diagnosed with language delay and autism spectrum disorder. At the same time, he was noted as having anemia (hemoglobin 10.5 g/dl) with normal iron stores and neutropenia (0.76 × 109/l). At 4 years of age, the patient was referred to the hematology clinic for evaluation of mild anemia (hemoglobin 9.4–10.5 g/dl), intermittent neutropenia (0.13 × 109/l to 1.88 × 109/l), fevers, and recurrent episodes of mucositis, otitis media, and cellulitis (Supplemental Table 1). Serial complete blood counts showed a cyclical pattern of neutropenia (0 × 109/l to 2.92 × 109/l) lasting 13–15 days. He tested negative for anti-neutrophil antibodies as well as for mutations in an IBMFS gene panel that included SBDS (the analysis was performed using genomic DNA extracted from peripheral blood) (Supplemental Table 1). His initial bone marrow evaluation at 4 years of age showed 80% cellularity with a maturation arrest at the myelocyte stage, while other cell lineages were normal (as reported in the patient’s medical records; images not available). At 8 years of age, he was started on G-CSF at 5 mcg/kg every other day, with improvement in anemia, neutrophil counts, mucositis, and infections. His early growth parameters showed height and weight in the 13th percentile, and these improved, reaching the 30th percentile with G-CSF therapy. He continues to do well on G-CSF therapy and is now 18 years of age.
In summary, all 3 patients presented with neutropenia associated with a host of various other symptoms, including exocrine pancreatic deficiency (2 of 3 patients) and/or autistic behavior (documented in 2 of 3 patients; not assessable in the third, patient BII.1, due to early death), along with a host of other morphological abnormalities and/or bone marrow morphology that are not typically observed in classical SDS (Supplemental Table 1). All 3 patients were negative for any mutations in all known IBMFS genes, including SBDS.
Identification of a candidate gene.
Prior to exome sequencing, sbds single point mutations were excluded in all 3 cases using Sanger sequencing of all coding exons. SBDS CNVs/structural variants were also excluded using CGH microarray analysis, as were mutations in other IBMFS genes using panel sequencing. In the index family (family A) (Figure 1A), exome sequencing was performed in patient AII.1 and his healthy parents. Variants of the index case were ranked on the basis of coding region sequence effect, frequency in public databases, and absence in healthy parents (other modes of transmission were excluded beforehand). A single de novo c.677G>A (p.G226E) missense mutation in the srp54 (signal recognition particle 54 kDa) gene remained after these filtering steps (Table 1 and Figure 1B). A total of 84 additional sbds-negative potential SDS patients were screened for mutations in srp54 using Sanger sequencing and/or WES. The inclusion criteria included SBDS-negative SDS patients and other patients with molecularly undiagnosed severe congenital neutropenia as well as other IBMFSs. This exploratory cohort was composed of the following data sets: (a) Thirty-four patients from the North American Shwachman-Diamond Syndrome Registry and local US Bone Marrow Failure Registries, including thirty-three with neutropenia and/or bone marrow failure and one with no cytopenias. Steatorrhea or abnormal exocrine pancreatic function was noted in 18 of these patients, and failure to thrive was noted in 16 of them. (b) Twenty SBDS-negative patients were included from the Hacettepe Bone Marrow Failure Center (Ankara, Turkey). Of these, 10 had neutropenia and/or bone marrow failure, while 8 had no cytopenias. Steatorrhea or abnormal exocrine pancreatic function was noted in 10 patients, and failure to thrive was noted in 8. (c) Thirty cases were included from a mix of French and European case-by-case referrals. Of the latter cohort, the only available information was the presence of severe congenital neutropenia and/or a clinical SDS phenotype. During the course of this screening, we identified a second family (family B) that harbored a second de novo c.343A>G (p.T115A) missense mutation in SRP54 (Figure 1B and Table 1). To exclude additional candidate genes, we also performed WES of the proband and her parents. Finally, a third family was independently identified at Baylor College of Medicine using trio WES (family C). In this family, we identified a c.349_351del (p.T117del) de novo missense mutation in the SRP54 gene. The variant filtering strategy and the identified variants are summarized in Supplemental Figure 4 and Supplemental Table 2, respectively. All 3 mutations affected highly conserved amino acids located in the GTPase domain of the protein (Figure 1, C and D). The mutations have deleterious predictive values, as indicated by various bioinformatics analyses (Table 1). In addition, none of the 3 variants was found in the 1000 Genomes database (http://www.internationalgenome.org/1000-genomes-browsers/), the Exome Aggregation Consortium (ExAC) Browser (http://exac.broadinstitute.org/), our internal exome database, or various other databases that altogether contain more than 100,000 exomes. All 3 mutations and their de novo nature were validated using Sanger sequencing in all probands and their parents. Finally, the mutations were also validated using Sanger sequencing on genomic DNA extracted from nonhematopoietic cells, i.e., buccal swabs (patient AII.1) and kidney (patient BII.1), thereby confirming the germline nature of these mutations.
Table 1. Details of srp54 sequence alterations in the 3 patients.
3D modeling of srp54 mutations.
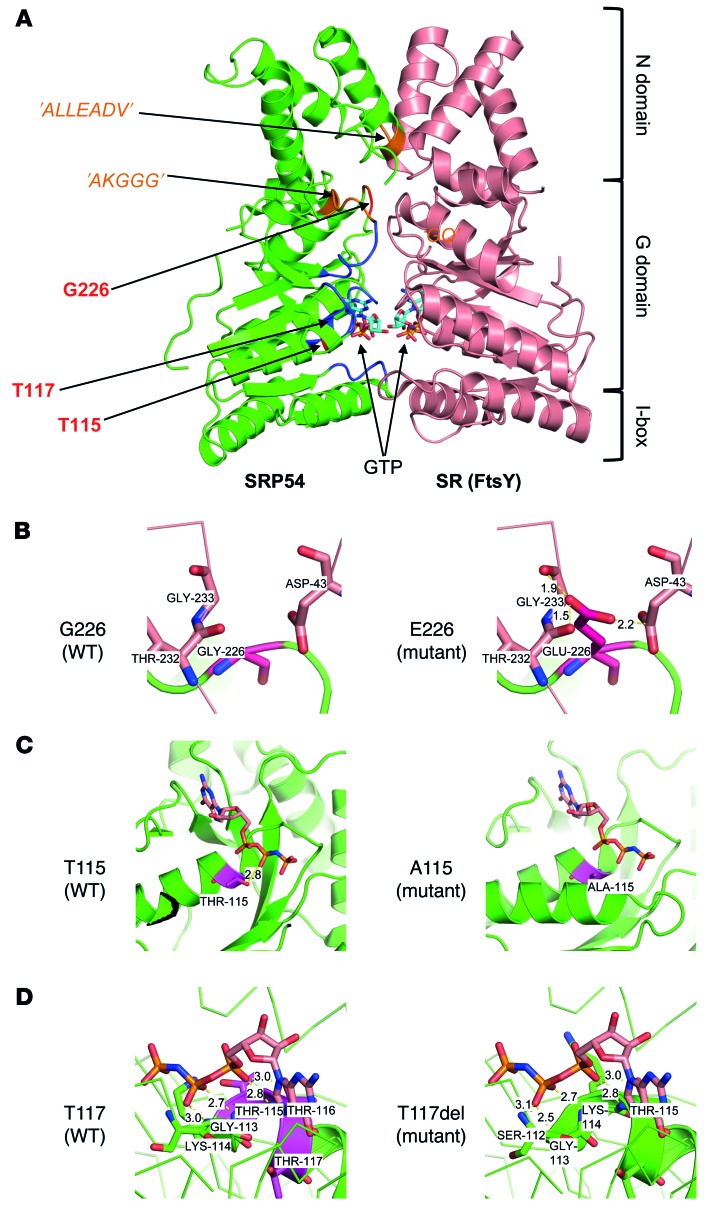
We analyzed the spatial location and predicted functional impact of the 3 identified mutations (p.T115A, p.T117del and p.G226E) using 3D modeling. Figure 2A shows the locations of the 3 affected residues on the structure of SRP54 associated with the SR. The mutated residues are all located around the GTP-binding domain in highly conserved regions (G domain) (20–22). The amino acid change p.G226E, which we observed in family A, is predicted to affect receptor binding, because it would lead to extremely short contacts between E226 in SRP54 and G233, T232, and D43 in the receptor, without major rearrangements of the local protein fold (Figure 2B). This was confirmed by the modification of the electrostatic surface induced by this mutation (Supplemental Figure 5). The mutation p.T115A in family B most likely directly affects GTP binding, because T115 forms an important hydrogen bond with GTP that is lost when the residue is mutated to alanine (Figure 2C). The mutation p.T117del in family C might also impact GTP binding. Residues 115–117 correspond to a threonine triplet. When 1 of these threonines is deleted, the helix structure at Gly113 starts earlier than expected, while the 112–115 region forms extensive H-bonds with the GTP. However, because the region before G113 is a loop region, it is difficult to determine the impact of this mutation on GTP binding (Figure 2D). The p.T117del mutation is therefore likely to have a milder effect than the 2 other mutations (see also below).
Figure 2. Structural impact of SRP54 mutations.
(A) Molecular model of the N and G domains of SRP54 (PDB 2j37 chain W) and its SR (FtsY, PDB 1rj9 chain B). SRP54 and SR are in green and pink, respectively. The positions of SRP GTPase-specific motifs and the conserved nucleotide-binding elements (G1–G5) are indicated in orange and blue, respectively. The mutated residues are shown in red. Each partner binds a GTP molecule. (B) Close-up of the WT residue G226 and side-chain modeling of mutant E226. The very short distances between oxygens of E226 of SRP54 (dark red) and D43, T232, and G233 of the receptor (pink) induce repulsive electrostatic forces that perturb the stability of the complex. Assuming a given protein backbone fold, the observed distances of 1.9 to 2.5 Å are indeed lower than those of standard hydrogen bonds between a hydroxyl group and an oxygen atom (2.8 Å). (C) Close-up of the WT residue T115 and side-chain modeling of mutant A115. According to the model, the hydrogen bond between the hydroxyl side chain of T115 and GTP is lost when mutated to A115. This modification most likely impairs GTP binding. (D) Close-up of the WT and structural model of deletion in T117. As residues 115–117 are all threonine the deletion of threonine introduces an earlier start of the helix structure at G113. According to the structural model, residues 113–116 form extensive hydrogen bonds with GTP. The deletion of T117 may affect GTP binding through rearrangements of residues 113–116, but this is difficult to predict.
In vivo and in vitro functional impact of the mutations.
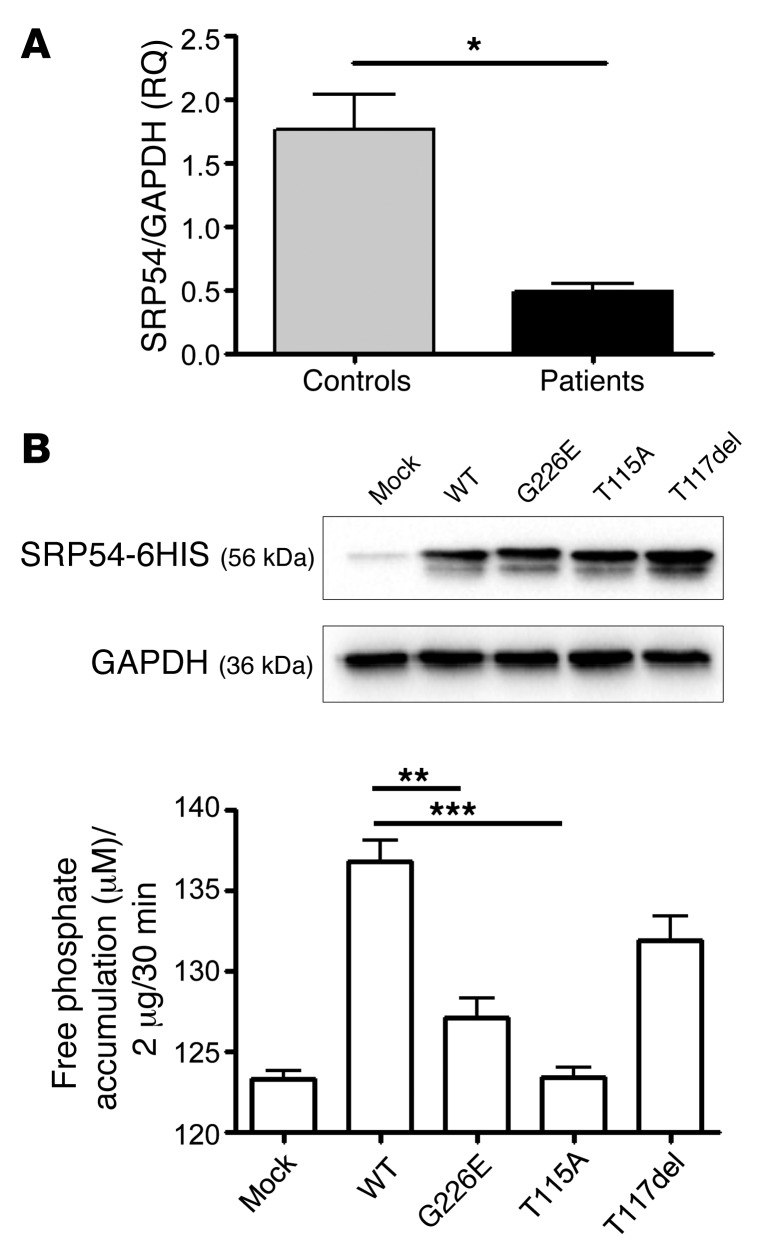
The level of SRP54 mRNA in (pretransplantation) bone marrow mononuclear cells from patients AII.1 and BII.1 was 3.6-fold lower than those in healthy bone marrow donors (Figure 3A). We assessed the in vitro GTPase activity of recombinant WT and mutant SRP54 using a malachite green assay. The p.G226E mutation reduced the activity by a factor of 3.5. In the p.T115A mutant, phosphate release was nearly completely abolished, whereas the p.T117del (third patient) mutant showed a clear (although not attaining statistical significance; P = 0.0579), yet moderate, 1.6-fold decrease in enzymatic activity (Figure 3B). This result is consistent with the above-mentioned structural modeling, which also predicted a milder functional impact for this mutation. Finally, when SRP54 was silenced in 2 cell lines (HeLa and HL60), it had no impact on the expression of 2 other proteins known to be mutated in SDS (i.e., SBDS and DNAJC21) (Supplemental Figure 6) or on the mTOR pathway, which was previously linked to SDS pathophysiology (23, 24).
Figure 3. In vivo and in vitro functional impact of SRP54 mutations.
(A) Quantitative RT-PCR analysis of SRP54 expression in bone marrow cells from patients AII.1 and BII.1 compared with bone marrow cells from healthy controls (see Methods for the source of healthy bone marrow cells). Measurements were performed in triplicate. RQ, relative quantitation. (B) GTPase activity of recombinant WT and mutated SRP54 proteins. Representative Western blot analysis of whole cellular extract of HEK293 cells transiently transfected with a mammalian expression vector containing WT human SRP54 cDNA or the mutated forms (G226E, T115A, and T117del). Western blot analyses were performed with an anti-SRP54 antibody (top lane). Loading controls were performed with an antibody against GAPDH (bottom lane). Graph demonstrates GTPase activity of recombinant WT and mutated SRP54 proteins. Phosphate release was measured with the Sigma-Aldrich ATPase/GTPase Activity Assay Kit (MAK113) using malachite green reagent. Each experiment was repeated 3 times, and error bars indicate the average values with SD. ***P < 0.001, **P < 0.01, and *P < 0.05, by 2-tailed t test.
Zebrafish model.
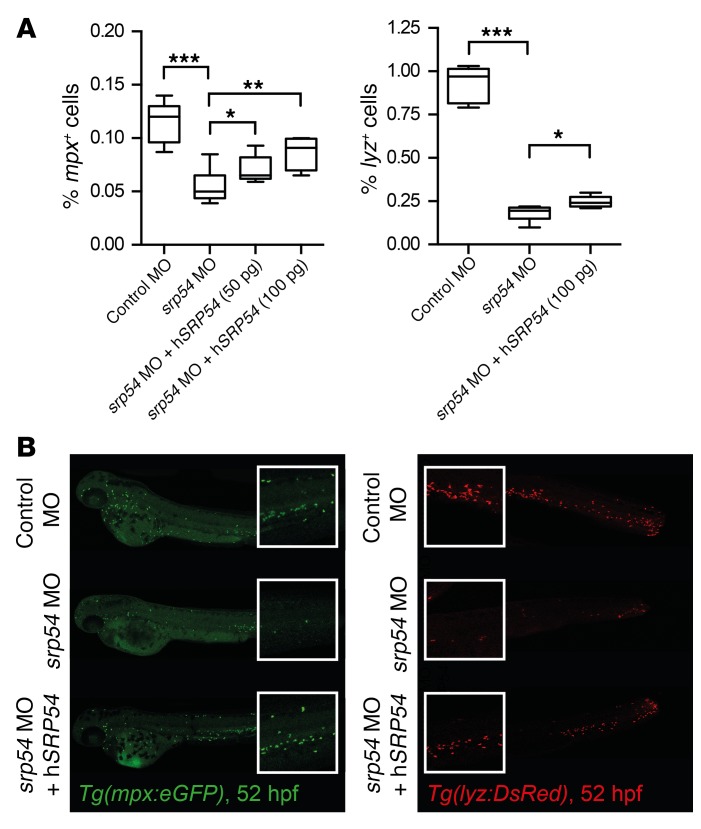
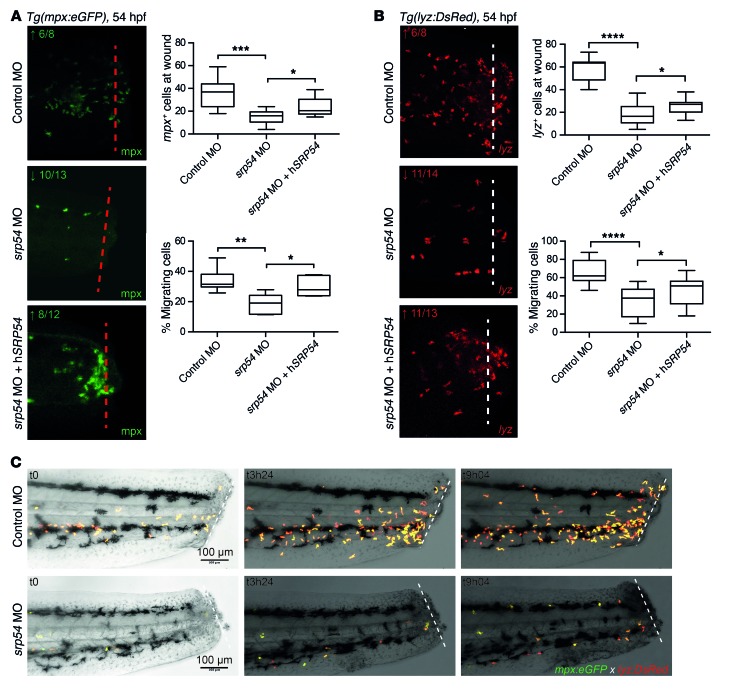
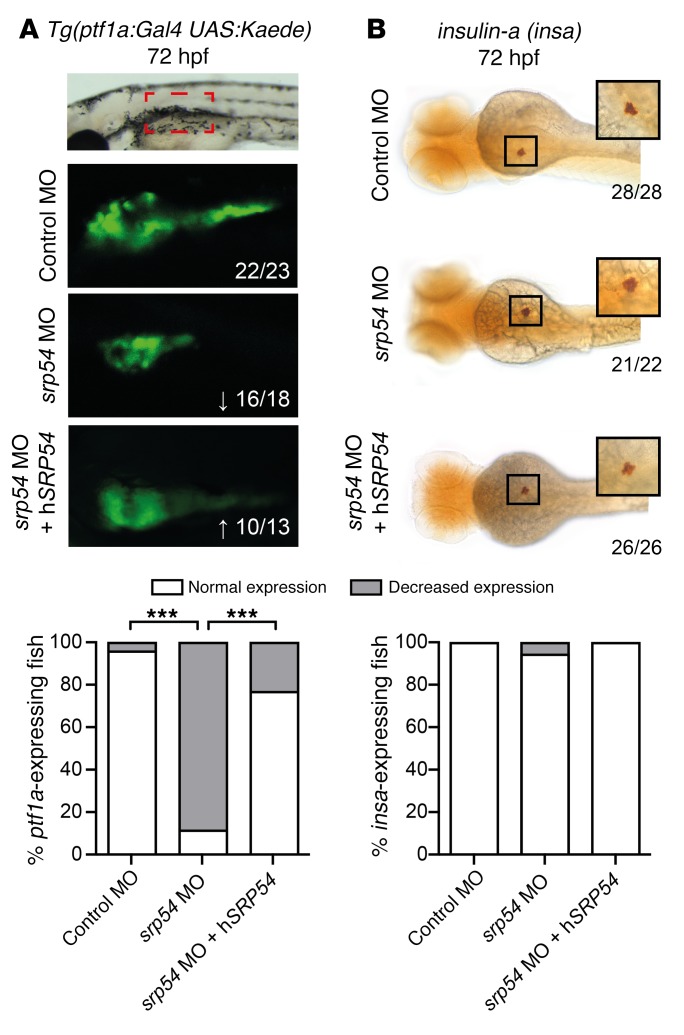
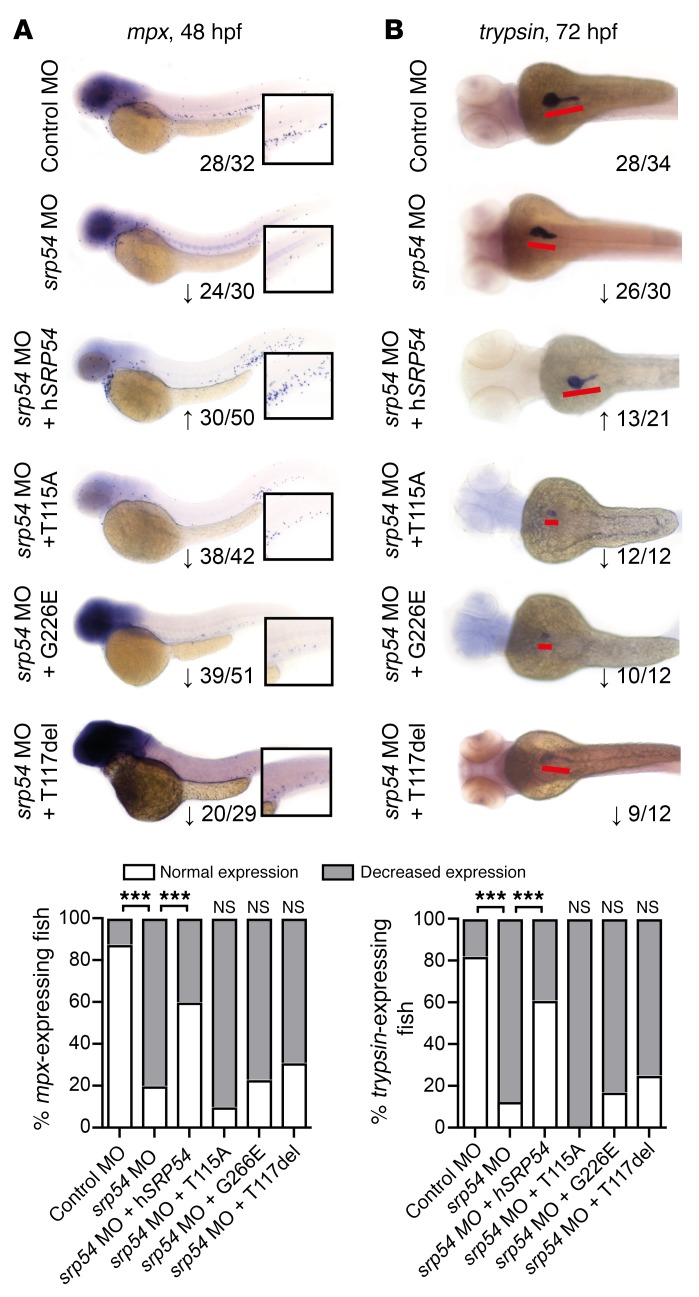
The zebrafish is an established model for studying hematopoiesis and development (25, 26). At 26 hours post fertilization (hpf), expression data indicated ubiquitous staining for the zebrafish homolog srp54 (bottom image, Supplemental Figure 7A). Later in development, srp54 expression was more pronounced in the anterior part of the zebrafish (top image, Supplemental Figure 7A). A similar expression pattern has been shown for the zebrafish homolog of the sbds gene at these developmental time points (27). To investigate the role of srp54 in zebrafish embryonic development, we performed in vivo loss-of-function experiments by treating zebrafish embryos with 2 different antisense morpholino oligonucleotides (MOs), to inhibit srp54 pre-mRNA splicing. Both MOs were designed to induce intron retention. We found that transcripts in morphants were indeed misspliced (Supplemental Figure 7B). To analyze the neutrophil numbers in developing zebrafish embryos, control and srp54 MO–injected transgenic Tg(mpx:eGFP) and Tg(lyz:DsRed) fish were first analyzed using flow cytometry, which revealed respectively fewer mpx+ and lyz+ neutrophils in dissociated srp54 morphants than in the control-injected fish, which could be partially rescued by coinjection of human srp54 mRNA (Figure 4) (28). This suggests a conserved functional identity between the 2 orthologs, which is consistent with their high evolutionary conservation (96% sequence identity between zebrafish and human proteins) (Supplemental Figure 8). Following tail fin injury, we further observed fewer numbers of neutrophils migrating to the site of injury in srp54 morphants than in the embryos injected with the control MO (Figure 5, A and B), which again could be rescued by coinjection of human SRP54 mRNA. Time-lapse imaging in double-transgenic embryos further documented both the quantitative reduction in neutrophils — visualized as migrating lyz:DsRed/mpx:eGFP cells — and the chemotaxis defect in response to injury (Figure 5C and Supplemental Videos 1 and 2). Pancreas development was also impaired upon srp54 knockdown, as shown by altered ptf1a expression at 72 hpf, indicating exocrine pancreatic defects (Figure 6A), while the endocrine pancreas appeared unaltered as monitored by whole-mount ISH (WISH) of insulin-a (insa) (Figure 6B). Both the neutrophil and the pancreas phenotypes were confirmed by mpx and trypsin WISH, which showed reduced numbers of neutrophils (Figure 7A and Supplemental Figure 9A) and incompletely surrounded islets (Figure 7B) in srp54 morphants versus control MO–injected embryos. We obtained similar results for both phenotypes with a second MO (Supplemental Figure 10). Interestingly, while injection with human srp54 mRNA was able to rescue the number of neutrophils and their migration capacity (Figure 4, Figure 5, Figure 7A, and Supplemental Figure 9, A and B) as well as pancreas development in srp54 morphants (Figure 6 and Figure 7B), coinjection of human mutant srp54 RNA, expressing either the p.T115A, the p.G226E, or the p.T117del mutation, did not rescue the neutrophil or the pancreas phenotypes caused by knocking down zebrafish srp54, and in all cases showed phenotypes statistically indistinguishable from those observed in srp54 morphants (Figure 7 and Supplemental Figure 9, A and B). Of note, we found that expression levels of the hematopoietic stem cell genes runx1 and c-myb and the erythrocyte marker globin were not altered by srp54 knockdown (Supplemental Figure 11, A and B), consistent with the observation that quantitative and qualitative defects in neutrophils were the most prominent hematologic defects. Finally, srp54 knockdown did not affect embryo body length in assays performed up until 5 days after fertilization (Supplemental Figure 11C).
Figure 4. srp54 gene knockdown reduces neutrophils in zebrafish embryos.
(A) Flow cytometric quantification of dissociated Tg(mpx:eGFP) (left) and Tg(lyz:DsRed) embryos (right) at 52 hpf, respectively. Percentages of positive fluorescence in dissociated control–injected, srp54 MO–injected, and MO plus hSRP54 mRNA–coinjected transgenic embryos are shown. ***P < 0.001, **P < 0.01, *P < 0.05, by 1-way ANOVA for multiple comparisons. Error bars indicate the mean ± SD. (B) Representative images of control-, srp54 MO–, and srp54 MO plus 100 pg hSRP54 mRNA–injected Tg(mpx:eGFP) and Tg(lyz:DsRed) embryos. Lateral views are shown, with anterior to the left, dorsal up. Whole embryo size: 3.1 mm; original magnification for insets, x2 (images were taken with a x5 objective on a Leica DM 2000 LED microscope). A minimum of 4 biological replicate experiments with at least 5 embryos per group in each biological replicate experiment were performed. hSRP54, human SRP54.
Figure 5. srp54 gene knockdown reduces neutrophil chemotaxis in zebrafish embryos.
(A and B) Tail fin wounding experiments and detection of neutrophils using anti-mpx immunostaining (A) and Tg(lyz:DsRed) embryos (B), respectively. Shown are representative images and corresponding quantitation of neutrophils, indicating reduced numbers at the wound in srp54 morphants compared with control-injected or srp54 MO plus hSRP54 mRNA–coinjected embryos. Dotted lines indicate the localization of the wound. Graphs summarize the counts of all visible neutrophils in the region of the wound and, respectively, the percentages of migrating neutrophils to the wound region (neutrophils at the wound relative to total numbers in the tail region, from the yolk sac extension to the tip of the wounded tail). Arrows indicate downregulation (↓) or rescued phenotype (↑) for each gene. Numbers indicate the sum of embryos with the respective phenotype/total number of embryos analyzed. Data from 3 biological replicates with 2 or more animals per group for each individual biological replicate experiment are shown. ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05, by 1-way ANOVA with multiple comparisons. Error bars indicate the mean ± SD. (C) Still images of confocal time-lapse live imaging of tail fin amputation in Tg(mpx:eGFP lyz:DsRed) transgenic embryos. Shown are 3 representative time points for control-injected and spr54 MO–injected transgenic embryos, in which the migration of double-positive neutrophils to the wound area was visible over time. For each time point, merged images are shown. Dotted lines indicate the localization of the wound. Scale bars: 100 μm (A–C).
Figure 6. Effects of srp54 gene knockdown on pancreas development in zebrafish embryos.
(A) Confocal images of control-injected, MO-injected, and MO plus 100 pg hSRP54 mRNA–injected transgenic Tg(ptf1a:Gal4 UAS:Kaede) embryos at 72 hpf. Lateral views are shown, with anterior to the left, dorsal up. Arrows indicate downregulation (↓) or rescued expression (↑) for each gene. The red boxed area corresponds to 300 µm (confocal images were taken with a ×10 objective). Original magnification, ×3, for the images below, which show enlarged views of the red boxed area. (B) WISH of insa at 72 hpf in control-injected, MO-injected and MO plus 100 pg hSRP54 mRNA–injected embryos. Dorsal views are shown, with anterior to the left. Shown are representative images from 3 biological replicate experiments with 4 or more embryos per group for each individual biological replicate experiment. Embryo size depicted: 1.75 mm (whole embryo size is 3.5 mm); original magnification of insets, ×2 (images were taken with a ×5 objective on a Leica DM 2000 LED microscope). The graphs below each panel in A and B display the percentages of embryos with normal versus decreased expression in all embryos analyzed across the biological replicates. Numbers indicate the amount of embryos with the respective phenotype/total number of embryos analyzed. ***P < 0.001, by Fisher’s exact test.
Figure 7. Mutated versions of SRP54 identified in patients cannot rescue neutropenia or exocrine pancreas development in srp54-deficient zebrafish.
WISH of mpx at 48 hpf (A) and trypsin at 72 hpf (B) in control-injected, MO-injected, MO plus 100 pg hSRP54 mRNA–injected, MO plus 100 pg T115A mRNA–injected, MO plus 100 pg G226E mRNA–injected, and MO plus 100 pg T117del mRNA–injected zebrafish embryos. Shown are representative images and summarized data from experiments performed in 3 or more biological replicates with at least 4 fish per group for each independent replicate experiment. Numbers indicate the sum of embryos with the respective phenotype per total number of embryos analyzed in all replicate experiments for the respective condition. Arrows indicate downregulation (↓) or rescued expression (↑) for each gene. (A) Whole embryo size is 3.1 mm; original magnification of insets: ×2. (B) Embryo size depicted is 1.75 mm (whole embryo size at this age is 3.5 mm); the red lines highlight the pancreas in each embryo. Images were taken with a ×5 objective on a Leica DM 2000 LED microscope. The graphs below display the percentages of embryos with normal versus decreased expression in all embryos analyzed across the biological replicates. Note that upon coinjection of mutated mRNA, the phenotype was statistically indistinguishable from srp54 MO–injected embryos, indicating that, as opposed to WT SRP54, the mutated versions of the protein cannot rescue the phenotype. ***P < 0.001, by Fisher’s exact test.
Discussion
IBMFSs are a heterogeneous group of disorders that share complex overlapping phenotypes, including dysmorphogenesis, a partial or total lack of mono- or multilineage blood cell production, and an increased risk of malignant transformation. Although traditionally classified according to characteristic clinical symptoms, a new disease nosology based on molecular etiology and pathway biology is emerging. Surprisingly, however, most, if not all, underlying mutations associated with IBMFS affect genes that are not specifically expressed in hematopoietic cells but that are instead engaged in fundamental and ubiquitous cellular processes, such as ribosome biogenesis, DNA repair, or telomere maintenance. Here, we add a key cellular entity to the genes known as mutated in IBMFS. We show that de novo mutations in SRP54 cause syndromic congenital neutropenia with a Shwachman-Diamond–like phenotype, a phenotype that was completely reproduced by knocking down the zebrafish homolog of this gene. In the cases presented here, several clinical and biological characteristics differ from those typically observed in classical SDS (summarized in Supplemental Table 1). Among them is the cell morphology of myeloid precursors that contain cytoplasmic vacuoles. This unusual feature indicates that the mechanism leading to neutropenia may be different in patients with SRP54 mutations compared with that in typical SDS patients.
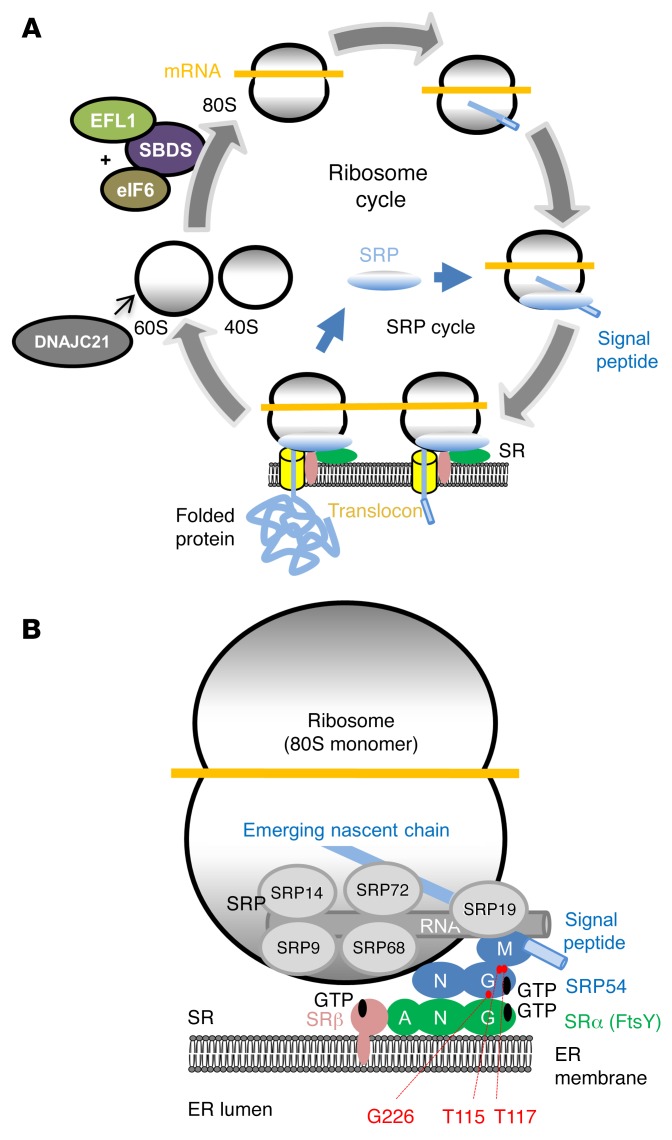
Several other IBMFSs are also caused by an abnormal translational machinery. These include Diamond-Blackfan anemia and the canonical SDS (29). While all these disorders (and others) are associated with mutations that affect 40S or 60S ribosomal subunits and/or their assembly (with the exception of 2 sporadic cases of aplastic anemia that were mutated in another SRP subunit [SRP72] [ref. 30]), the SRP complex has thus far remained unaffected by any disease-causing mutations. SRP54 is therefore the second SRP component to be associated with a monogenic disease. Although several potentially pathogenic CNVs encompassing SRP54 have been reported in studies referring to unrelated phenotypes (Supplemental Table 3), to our knowledge, no pathogenic single nucleotide variant or indel has previously been reported in this gene. Several heterozygous variants predicted by bioinformatics tools to be damaging have been reported in public databases (Supplemental Table 4) (as is the case for many genes), but in contrast to the 3 de novo mutations reported here, none of them affects the conserved domains of the protein. Of note, with the exception of the SRP RNA, SRP54 is the only one of the six SRP polypeptides that is universally conserved throughout evolution (31, 32), with 96% sequence identity between human and zebrafish molecules. This high level of conservation is explained by the central role played by SRP54 in the cotranslational process that affects over one-third of the human proteome (see below). Indeed, SRP54 defines the business end of the signal recognition particle, because it links 3 key elements of the SRP complex, i.e., the 7SL RNA and the signal sequence of the nascent ribosome-bound polypeptide chain, to the SR on the ER membrane in a GTP-regulated manner (Figure 8) (33). It is therefore highly probable that modification of SRP54 function, as suggested by 3D modeling and enzymatic activity assessments and confirmed in zebrafish rescue experiments, impairs protein translocation in at least a quantitative manner.
Figure 8. Schematic positioning of SRP54 in the eukaryotic translational machinery.
(A) Ribosomal and SRP cycles. Proteins involved in SDS are depicted: SBDS and EFL1 are involved in the assembly of the 60S and 40S subunits, and DNAJC21 is involved in maturation of the 60S subunit. (B) Close-up of the ribosomal complex bound to the SR on the ER membrane. SRP54 (blue) is part of the SRP composed of 6 proteins (SRP9, SRP14, SRP19, SRP68, SRP72) and of the approximately 300-nucleotide-long 7SL RNA molecule. The signal peptide of the emerging nascent peptide chain is recognized by the M domain of SRP54 and the 7SL RNA. The N and G domains of SRP54 are interacting with the homologous N and G domains of SRα (FtsY). The mutated residues are shown in red.
We used the zebrafish model to explore the relevance of srp54 gene expression in vivo. Interestingly, srp54-knockdown fish faithfully phenocopied the hallmarks of SDS that are observed in patients, including profound neutropenia and defective exocrine pancreas development. Neutrophils of SDS patients have been reported to show impaired chemotaxis and migration deficiency (34, 35), which lead to repeated infections, as was also observed in all 3 patients reported here. In accordance with the results of these studies, we observed a decrease in the capacity of neutrophils to migrate toward the injury site in the zebrafish model (Figure 5).
Given our results, the underlying disease mechanism could be, at this point, either haploinsufficiency or a dominant negative effect. Haploinsufficiency is supported by: (a) decreased expression of SRP54 in the bone marrow of patients AII.1 and BII.2 as compared with that seen in healthy controls (Figure 3A); (b) the phenotype of the SRP54-knockdown zebrafish (Figures 4–6); and (c) the intolerance of the SRP54 gene to loss-of-function mutations, according to public databases (pLi score = 1.0 in the ExAC database). A dominant negative mechanism is supported by: (a) the presence of CNVs in the general population and in patients with unrelated diseases (Supplemental Table 3); (b) the presence of 1 loss-of-function variant in ExAC; and (c) the lower GTPase activity of the mutated proteins compared with that of the WT protein and the differences among mutated proteins observed in the zebrafish rescue experiments. How do SRP54 mutations lead to syndromic neutropenia with SDS-like features? SBDS is specifically required for the translation of C/EBP α and ß mRNAs, both of which are key regulators of granulocytic differentiation (36). A similar mechanism may explain the selective effect of SRP54 mutations on neutrophils, because SRP54 expression levels influence TRAIL (37), which is itself involved in neutrophil apoptosis (38). However, these data do not explain the phenotype observed in the pancreas or, indeed, the potential involvement of many other organs (e.g., the skeleton and the brain). An alternative, more global explanation that is in line with the near-pleiotropic phenotype observed in SDS might be the following: nearly 40% of the almost 20,000 human protein–coding genes are predicted to be secreted or membrane bound (39). These proteins have a de facto need for SRP54 for their synthesis. Although homozygous mutations in SRP54 are probably lethal, as suggested by an increase in early lethality in zebrafish exposed to higher MO doses (data not shown), pathogenic heterozygous SRP54 mutations probably lead to both quantitative and qualitative disruption of secreted and membrane-bound protein synthesis. Although this would affect many organs, it may particularly disturb cells and tissues that are highly dependent on such proteins, such as the pancreas, which contains the highest number of secreted proteins in the body, as well as hematopoietic cells, which contain a high number of transcripts for secreted proteins (39). Indeed, mature cells, such as neutrophils, constantly reshuffle surface proteins to adapt to the cell’s life cycle activities such as chemotaxis, extravasation and microbe destruction. Consistently, it was recently shown that the quantity of available SRP54 within a cell is critically important for cellular homeostasis, as its knockdown in mammalian cells led to both quantitative and qualitative changes in the cellular protein content (40). Finally, we show here that SRP54 mutations are not restricted to patients with SDS and could be more generally the origin of syndromic neutropenia. The fact that our single patient (patient CII.1) with no apparent exocrine pancreatic deficiency and milder neutropenia (both in quantitative terms and with respect to the age of first clinical manifestations) harbored an equally less severe mutation — on the basis of structural and functional analyses (including a lower impact on GTPase activity and an observed milder phenotype in zebrafish rescue experiments as compared with the other 2 mutations) — hints that SRP54 mutations could lead to diverse clinical phenotypes that may be based on the status of each patient’s specific mutation.
In conclusion, we report that heterozygous mutations in SRP54, a key component of the cotranslational protein machinery, lead to syndromic neutropenia with SDS-like features. This is directly relevant not only to patients with molecularly undiagnosed IBMFS, including SBDS-negative SDS patients, but also to the molecular nosology of IBMFS. These findings also open a window into the pathophysiology of the SRP.
Methods
Subjects and exome sequencing
The subjects reported in this study were members of 2 unrelated families of European descent (families A and B; Figure 1A) and 1 North American family of Hispanic (Mexican) origin (family C; Figure 1A). In all 3 families, the parents were nonconsanguineous and healthy, as were other siblings. Exome sequencing was performed for the parents and probands of all 3 families. Genomic DNA was extracted and purified from peripheral blood using standard protocols. For WES of family A, sequencing libraries were prepared using the Ion Ampliseq Exome Kit (Thermo Fisher Scientific) according to the manufacturer’s recommendations and sequenced on an Ion Proton Sequencer (Thermo Fisher Scientific). Sequencing libraries for families B and C were prepared using the TruSeq Exome Kit (Illumina) and NimbleGen SeqCap EZ probes (VCRome; Roche Sequencing), respectively, and WES was performed on Illumina NextSeq 500 and HiSeq 2500 platforms, respectively. A minimum of 6.5 Gb of sequence data per individual were obtained and mapped to the hg19 reference genome using the Ion Torrent Server (Thermo Fisher Scientific) for family A and the Isaac Genome Alignment Software 2.1.0 (Illumina) for family B. The mean coverage was at least 60-fold, and more than 95 % of target sequences were covered a minimum of 10 times. CNVs were identified using Conifer software (41). Other variants were called using the Torrent Variant Caller 5.0 with low-stringency calling settings (family A) or the Isaac Variant Caller 2.1.0 (family B). Only variants covered by more than 5 variant reads were considered. Annotation was performed using the KGGSeq software package (42). Bioinformatics analysis of family C was performed as previously described (43). We focused only on protein-altering variants (i.e., missense, nonsense, and splice-site variants and coding indels) with fewer than 0.01 alternative allele frequencies in dbSNP (build 135), 1000 Genomes Project, ExAC, an in-house database, and databases at Institut Imagine (Necker Hospital, Paris, France), Baylor College of Medicine, and Radboud University Medical Center (Nijmegen, Netherlands). To identify potential causal mutations, we focused on de novo mutations in the index cases of each family. Other modes of transmission — recessive and X-linked — were considered and excluded for all 3 families. The complete variant filtering strategy is presented in Supplemental Figure 4. Sequence variants reported here were deposited in the ClinVar (www.ncbi.nlm.nih.gov/clinvar/) public database under accession numbers SCV000583969, SCV000583970, and SCV000583971.
Targeted sequencing of SRP54
The candidate srp54 variant identified by WES was validated using conventional capillary Sanger sequencing in the same families including the proband and his or her parents. Additional patients were screened using Sanger sequencing of all 15 exons of srp54. Primers used for PCR amplification and sequencing are presented in Supplemental Table 5. Briefly, srp54 exons were amplified using the Expand Long Template PCR System (Roche Diagnostics) according to the manufacturer’s recommendations. After purification with the Exostar Kit (GE Healthcare Life Sciences), PCR products were bidirectionally sequenced using the same amplification primers — except for exon 14 (Supplemental Table 5) — using the Big Dye Terminator Kit v3.1 (Thermo Fisher Scientific). Sequencing reactions were run on an ABI PRISM 3730xl sequencer (Thermo Fisher Scientific). Sanger sequencing for patient CII.1 was performed at Baylor Genetics clinical laboratories, and the primer sequences are listed in Supplemental Table 5.
3D modeling of SRP54 mutations
The structure of the SRP54 protein was modeled using PDB 2j37 chain W as a template (44). The GTP-binding site was modeled according to the superimposition of the SRP54 model and PDB 2j7p. The position of the SR (FtsY) was determined according to the structure of PDB 1rj9 (45). The mutant side-chain structures were modeled using the mutagenesis wizard of the PyMOL program (46). Electrostatic surfaces were calculated using PDB2PQR (47) and APBS (48). All figures were generated using PyMOL (46).
Quantification of SRP54 transcripts
Total RNA was extracted from mononuclear bone marrow cells (cells from 5 healthy bone marrow donors were obtained from Caltag Medsystems Ltd.) using TRIzol Reagent (Invitrogen, Thermo Fisher Scientific) and reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturer’s instructions. Real-time quantitative PCR was performed in a total volume of 20 μl using the SsoAdvanced SYBR Green Supermix Kit (Bio-Rad) and the following gene-specific primers: SRP54, 5′-TGATAATGGTGGCATTGCTC-3′ and 5′-GGCTTTAGCGGTGTCTTTTG-3′; GAPDH, 5′-GGTGAAGGTCGGAGTCAACGGA-3′ and 5′-GAGGGATCTCGCTCCTGGAAGA-3′. After an initial denaturing at 96°C for 10 minutes, the temperatures and incubation times used for cycling were 95°C for 10 seconds and 60°C for 30 seconds using an ABI Prism 7000 real-time PCR machine (Thermo Fisher Scientific). The results were obtained using SDS Series Software (Thermo Fisher Scientific) and evaluated using GraphPad Prism 6 (GraphPad Software). Melting-curve analysis was performed to assess the specificity of the PCR products.
Cloning and site-directed mutagenesis of SRP54 cDNA
Human srp54 cDNA was first cloned into the pCS2– expression vector (RZPD). For this purpose, the human SRP54 gene (GenBank U51920.1) was amplified by PCR on cDNA synthetized from RNA of peripheral blood mononuclear cells obtained from a healthy donor, using the following primer pairs: 5′-GCGCGCTCGAGATGGTTCTAGCAGACCTTGGAAGAAAAATAAC-3′ and 5′-GCGCGTCTAGATTACATATTATTGAATCCCATCATGCCTT-3′. The PCR fragment was then cloned with the restriction enzymes XhoI and XbaI into pCS2- for zebrafish experiments (see next paragraph). This construct was then used to generate the mutants G226E (c.677G>A), T115A (c.343A>G), and T117del (c.349_351del) by site-directed mutagenesis according to the manufacturer’s instructions (QuikChange II XL Site-Directed Mutagenesis Kit; Agilent Technologies). The primers used to generate the G226E mutant were: 5′-TGGATGCCTCCATTGAGCAGGCTTGTGAAGC-3′ and 5′-GCTTCACAAGCCTGCTCAATGGAGGCATCCA-3′. The primers used to generate the T115A mutant were: 5′-GATTGCAAGGGAGTGGTAAAGCAACAACATGTTCAAAGCT-3′ and 5′-AGCTTTGAACATGTTGTTGCTTTACCACTCCCTTGCAATC-3′. The primers used to generate the T117del mutant were: 5′-gttggattgcaagggagtggtaaaacaacatgttcaaag-3′ and 5′-ctttgaacatgttgttttaccactcccttgcaatccaac-3′. The SRP54 gene and the 3 mutated forms (mutations G226E, T115A, and T117del) were amplified by PCR from pCS2– plasmids with the forward 5′-GCGCGACCGGTATGGTTCTAGCAGACCTTGGAAGAAAAATAAC-3′ and reverse 5′-GCGCGTTAATTAATTAATGGTGATGGTGATGATGCATATTATTGAATCCCATCATGCCTT-3′ primer pairs and then cloned using restriction enzymes AgeI and PacI into the pQCXIP plasmid (Clontech) with the addition of a 6-histidine tag at the C-terminus.
GTPase activity assay
pQCXIP plasmids containing the WT or the 3 mutated SRP54 cDNAs were transiently transfected into the HEK293 cell line (Sigma-Aldrich). Forty-eight hours after transfection, proteins were extracted with NP-40 lysis buffer (150 mM sodium chloride, 1% NP-40, 50 mM Tris, pH 8.0, 1% NP-40) complemented with a protease inhibitor cocktail (cOmplete, Roche). Proteins were quantified using a Bradford protein assay according to the manufacturer’s instructions (Bio-Rad). Protein (20 μg) was analyzed by Western blotting using an anti-SRP54 antibody (sc393855; Santa Cruz Biotechnology) at 1:1,000 dilution. Uniformed loading was monitored using an antibody against GAPDH (MAB 374; Merck-Millipore). GTPase activity was assessed using 2 μg cell lysate and a 30-minute incubation with the ATPase/GTPase Activity Assay Kit (Sigma-Aldrich; malachite green assay, MAK113) according to the manufacturer’s instructions. Results were obtained by dosing triplicates of 3 independent transfections and analyzed using GraphPad Prism 6.
Zebrafish experiments
Zebrafish husbandry and genetic strains.
Zebrafish were bred and maintained at 28°C as described by Nüsslein-Volhard and Dahm (49). Staging was performed in hpf as summarized by Warga and Kimmel (50) and according to Federation of European Laboratory Animal Science Associations (FELASA) and Swiss federal law guidelines. The following lines were used in this study: WT Tübingen strains, Tg(mpx:eGFP) (51), Tg(lyz:DsRed) (52), and Tg(ptf1a:Gal4;UAS:Kaede) (53).
MO design and validation.
Human and zebrafish SRP54 share a high (96%) degree of sequence homology (Supplemental Figure 8). Two SRP54 splice MOs to prevent pre-mRNA splicing and a standard control MO were synthesized by Gene Tools (Gene Tools LLC): MO1, GTTTAACTTTCGTTACTTACTTGAC; MO2, ACTACCCAATAATGGCTCACCTTAG, and the control MO, CCTCTTACCTCAGTTACAATTTATA. Embryos were injected at the single-cell stage. Phenol red (0.05%) (Sigma-Aldrich) was added as an injection tracer. Embryos were raised to appropriate stages and fixed in 4% paraformaldehyde (PFA) in 1× PBS for gene expression analysis. For validation, control-injected and MO-injected embryos were collected, and mRNA was isolated using the RNeasy Mini Kit (QIAGEN). cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche) according to the manufacturer’s protocol. Real-time PCR (RT-PCR) was performed using the following primers to verify splice modification on agarose gels: MO1, TAGCTGATCTGGGGAGGAAA and GTTCCTTGAAAACGGCATGT; MO2, CATCATGTTTGTGGGTCTGC and ACCACTTGTGTCCACGATGA. For rescue experiments, human WT and mutated srp54 cDNAs cloned into the pCS2– expression vector (RZPD) were used (refer to the previous paragraph on cloning and site-directed mutagenesis of SRP54 cDNA). Capped RNA from each of the 4 constructs (WT, G226E, T115A, and T117del) was synthesized using the mMessage mMachine SP6 Kit (Ambion, Life Technologies, Thermo Fisher Scientific). RNA (50–100 pg) was injected into the yolk of single-cell-stage embryos together with the srp54 MO to rescue the MO phenotype.
WISH, flow cytometry, and tail fin wounding.
WISH was performed as described in Konantz et al. (54). Briefly, c-myb riboprobes were generated using pBK-CMV as a plasmid template (a gift of Caroline E. Burns, Massachusetts General Hospital, Boston, Massachusetts, USA), EcoRI for DNA linearization, and T7 RNA polymerase for further antisense probe production. The insa probe was generated by PCR using the following primers: forward, 5′-atggcagtgtggcttcaggc-3′ and reverse, 5′-gaattctcagttacagtagt-3′. After PCR products were purified from agarose gels using the QIAquick Gel Extraction Kit (QIAGEN), the products were subcloned using the Dual Promoter TA Cloning Kit (Invitrogen, Thermo Fisher Scientific), and the plasmid was then linearized using the appropriate enzyme. RT-PCR was used to generate riboprobes for globin, mpx, srp54, and trypsin using the following gene-specific primers: srp54 (forward, gttttgcttgccaaaaccat, and reverse, gggagacattgagggactga); mpx (forward, CTTTAGCAGTGGCAGGAAGG, and reverse, GAAAATCCGAGATGGCGATA); trypsin (forward, CAGCGCAGAACATGTTAGA, and reverse, GTCTCTGAACAGCGGCTACC); and globin (forward, CCAGCAGATTGTGGTTGATG, and reverse, GCCTTCTTTGACAAGGTTG). Each antisense primer was designed with the T7-promoter sequence tagged to its 5′ end. Riboprobes were labeled with digoxygenin- or fluorescein 12–labeled (insa-labeled) UTP (Roche). Stained embryos mounted in 89% glycerol were photographed using a Leica digital MC170 HD camera attached to a Leica DM 2000 LED (Leica Microsystems). For flow cytometry, dechorionated transgenic embryos were dissociated into single cells by incubating them with Liberase enzyme (Roche) at 30°C and at 200 g as previously described (55). After dissociation was complete, an equal amount of 5% BSA was added to inactivate the enzyme. Cells were washed once in PBS and passed through a 35-μM filter. The number of fluorescence-labeled cells was then determined using flow cytometry on a BD Accuri C6 Flow Cytometer (BD Biosciences), and SYTOX Blue (Thermo Fisher Scientific) was used to identify dead cells. For the tail fin wounding experiments, larvae were anesthetized using tricaine. The tail fins were cut at the end of the spinal cord using a sterile scalpel blade, and the embryos were then allowed to recover at 29°C in embryo medium for 6 to 8 hours. Embryos were then either fixed overnight in 4% PFA with PBS or mounted in 0.8% low-melting agarose on glass-bottomed slides for live imaging. Immunostaining was performed to detect neutrophils at the wound site according to standard protocols, using a rabbit polyclonal antibody against mpx (GTX128379; GeneTex International Corp.). Stained embryos were then mounted in 0.8% low-melting agarose. Images and videos were obtained using a Zeiss LSM 720 (Carl Zeiss) or a Leica SP5-II-Matrix (Leica Microsystems) confocal microscope. Mpx-positive cells from both WISH and immunostaining were semiautomatically counted using Fiji software (56). Finally, confocal time-lapse live imaging was performed in Tg(mpx:eGFP lyz:DsRed) embryos for 9 hours starting 30 minutes after the tail fin was amputated at 48 hpf.
Additional details regarding Methods can be found in the supplemental material.
Statistics
Data shown represent the mean ± SEM or the percentage of expression. P values were derived using a 1-way ANOVA for multiple comparisons, followed by a Newman-Keuls test for pairwise comparisons, a Mann-Whitney U test, a 2-tailed t test, or Fisher’s exact test for comparison of 2 groups. A P value of less than 0.05 was considered statistically significant. Statistical analyses were performed using GraphPad Prism 6.
Study approval
All participants (and their parents) gave written informed consent for genetic analyses in this study, which was approved by the IRBs of Strasbourg University Hospitals (protocol DC-2013-1911) and Baylor College of Medicine (protocol H-22769).
Author contributions
RC and SB designed the study, analyzed the data, and wrote the manuscript. MK and CL designed zebrafish experiments, performed the experiments, and analyzed the data. JSM, EA, and LS performed zebrafish experiments. ZM and EW designed, performed, and analyzed the 3D modeling data. VR, AM, NO, OL, and JGG performed protein experiments. AP, LN, AH, AS, MDF, SU, and NAA performed sequencing experiments. AK, NP, and YY performed NGS data analyses. MSL analyzed the exome-sequencing data and discussed the results. MPC and MDF performed and analyzed the pathology results. CSS interpreted radiological, CT-scan, and ultrasound images. PAB and VP performed and analyzed mitochondrial DNA sequencing by NGS. CM, AM, GA, NO, OL, and JGG performed the cloning and expression of WT and mutant proteins. ALMB and NM performed the CGH array experiments. CP, EBL, BI, AS, MF, NAA, and SU identified patients, provided samples, and analyzed and interpreted clinical and experimental data. MR, FR, AG, MCB, ME, MV, JG, LF, CTR, JT, KM, KLB, DHM, and DLS interpreted clinical data and discussed the results. All authors contributed to the writing and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Taco (TW) Kuijpers (Academic Medical Center, Amsterdam, Netherlands), Charlotte Niemeyer and Marcin Wlodarski (University of Freiburg, Germany), and Jamel Chelly (University of Strasbourg) for discussions and critical reading of this manuscript and/or providing us with samples. We thank Dean R. Campagna (Boston Children’s Hospital, Boston, Massachusetts, USA) and Deniz Ceylan (Hacettepe University, Sihhiye, Ankara, Turkey) for their technical assistance. We thank the following groups for sending us patients’ samples and/or for ongoing collaboration on the genetics of severe congenital neutropenia: Myungshin Kim and Sung Sup Park (Seoul National University Hospital, Seoul, South Korea); Sabine Mellor-Heineke, Cornelia Zeidler, Karl Heinrich Welte, Saskia Biskup, and Heinz Gabriel (Center for Genomics and Transcriptomics, Tübingen, Germany); Dirk Lebrecht (University Medical Center, Freiburg, Germany); Albert Catala Temprano and Juan I. Aróstegui (Hospital Sant Joan de Deu, Barcelona, Spain); Julián Sevilla, Eva Gálvez de la Villa, and Luis I. Gonzalez-Granado (Hospital Nino Jesus, Madrid, Spain); Marielle Alders (Academisch Medisch Centrum Universiteit, Amsterdam, Netherlands); Sofia Douzgou and Nicola Marchank (Central Manchester University Hospitals, Manchester, United Kingdom); Edward G. Brooks (South Texas Pediatric Blood and Cancer Center, San Antonio, Texas, USA); Dean John (University of Aberdeen, School of Biological Sciences, Aberdeen, United Kingdom); and Mirella Filocamo and the “Cell Line and DNA Biobank from Patients Affected by Genetic Diseases,” member of the Telethon Network of Genetic Biobanks (project GTB12001), which is funded by Telethon Italy. We are grateful to Christine Bole-Feyssot and Alexander Hoischen for screening the databases of Institut Imagine (Necker Hospital, Paris, France) and Radboud University Medical Center (Nijmegen, Netherlands), respectively, for the presence of SRP54 variants. We thank Chunjing Qu for providing quality control data for the exome data produced at Baylor College of Medicine. Additionally, we thank Judith Konantz (Center for Regenerative Therapies [CRT], Dresden, Germany) for her help with the zebrafish pancreas data. We would also like to thank the patients and their families for their continued engagement in this research. This work was supported by grants from the Agence Nationale de la Recherche (ANR) (ANR-11-LABX-0070_TRANSPLANTEX), INSERM UMR – S1109; and the Institut Universitaire de France (IUF) (all to SB); the University of Strasbourg (IDEX UNISTRA, to CP and SB); the INTERREG V European regional development fund (European Union) program (project 3.2 TRIDIAG, to RC, SB, and CL); and the Swiss National Science Foundation (SNF) (149735, to CL).
Version 1. 10/03/2017
Electronic publication
Version 2. 11/01/2017
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(11):4090–4103. https://doi.org/10.1172/JCI92876.
Contributor Information
Raphael Carapito, Email: carapito@unistra.fr.
Martina Konantz, Email: martina.konantz@unibas.ch.
Catherine Paillard, Email: catherine.paillard@chru-strasbourg.fr.
Angélique Pichot, Email: apichot@unistra.fr.
Yaping Yang, Email: yapingy@bcm.edu.
Ghada Alsaleh, Email: galfarhan@unistra.fr.
Lydie Naegely, Email: naegely@unistra.fr.
Aline Kolmer, Email: alinekolmer777@gmail.com.
Nicodème Paul, Email: npaul@unistra.fr.
Antoine Hanauer, Email: ahanauer@unistra.fr.
Véronique Rolli, Email: vrolli@unistra.fr.
Joëlle S. Müller, Email: joelle.mueller@stud.unibas.ch.
Elisa Alghisi, Email: elisa.alghisi@unibas.ch.
Loïc Sauteur, Email: loic.sauteur@unibas.ch.
Cécile Macquin, Email: cecile.macquin@inserm.fr.
Aurore Morlon, Email: am@biomica.fr.
Consuelo Sebastia Sancho, Email: consuelo.sebastiasancho@chru-strasbourg.fr.
Patrizia Amati-Bonneau, Email: pabonneau@chu-angers.fr.
Vincent Procaccio, Email: Viprocaccio@chu-angers.fr.
Anne-Laure Mosca-Boidron, Email: anne-laure.mosca-boidron@chu-dijon.fr.
Nathalie Marle, Email: nathalie.marle@chu-dijon.fr.
Naël Osmani, Email: osmani@unistra.fr.
Olivier Lefebvre, Email: lefebvre@unistra.fr.
Sule Unal, Email: suleunal2003@hotmail.com.
Mirjana Radosavljevic, Email: mirjana@unistra.fr.
Marie-Pierre Chenard, Email: Marie-Pierre.CHENARD@chru-strasbourg.fr.
Fanny Rialland, Email: Fanny.RIALLAND@chu-nantes.fr.
Audrey Grain, Email: audrey.grain@chu-nantes.fr.
Marie-Christine Béné, Email: mariecbene@gmail.com.
Marion Eveillard, Email: marion.eveillard@chu-nantes.fr.
Marie Vincent, Email: marie.vincent@chu-nantes.fr.
Julien Guy, Email: julien.guy@chu-dijon.Fr.
Laurence Faivre, Email: laurence.faivre@chu-dijon.fr.
Christel Thauvin-Robinet, Email: christel.thauvin@chu-dijon.fr.
Julien Thevenon, Email: julien.thevenon@chu-dijon.fr.
Kasiani Myers, Email: kasiani.myers@cchmc.org.
Akiko Shimamura, Email: Akiko.Shimamura@childrens.harvard.edu.
Elodie Bottollier-Lemallaz, Email: elodie.bottollier-lemallaz@chu-dijon.fr.
Eric Westhof, Email: eric.westhof@ibmc-cnrs.unistra.fr.
Claudia Lengerke, Email: claudia.lengerke@unibas.ch.
Bertrand Isidor, Email: Bertrand.ISIDOR@chu-nantes.fr.
Seiamak Bahram, Email: siamak@unistra.fr.
References
- 1.Bartholomew LG, Baggenstoss AH, Morlock CG, Comfort MW. Primary atrophy and lipomatosis of the pancreas. Gastroenterology. 1959;36(5):563–572. [PubMed] [Google Scholar]
- 2.Nezelof C, Watchi M. [Lipomatous congenital hypoplasia of the exocrine pancreas in children. (2 cases and review of the literature)] Arch Fr Pediatr. 1961;18:1135–1172. [PubMed] [Google Scholar]
- 3.Bodian M, Sheldon W, Lightwood R. Congenital hypoplasia of the exocrine pancreas. Acta Paediatr. 1964;53:282–293. doi: 10.1111/j.1651-2227.1964.tb07237.x. [DOI] [PubMed] [Google Scholar]
- 4.Shwachman H, Diamond LK, OskiI FA, Khaw KT. The syndrome of pancreatic insufficiency and bone marrow dysfunction. J Pediatr. 1964;65:645–663. doi: 10.1016/S0022-3476(64)80150-5. [DOI] [PubMed] [Google Scholar]
- 5.Burroughs L, Woolfrey A, Shimamura A. Shwachman-Diamond syndrome: a review of the clinical presentation, molecular pathogenesis, diagnosis, and treatment. Hematol Oncol Clin North Am. 2009;23(2):233–248. doi: 10.1016/j.hoc.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babushok DV, Bessler M, Olson TS. Genetic predisposition to myelodysplastic syndrome and acute myeloid leukemia in children and young adults. Leuk Lymphoma. 2016;57(3):520–536. doi: 10.3109/10428194.2015.1115041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boocock GR, et al. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33(1):97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- 8.Kuijpers TW, Alders M, Tool AT, Mellink C, Roos D, Hennekam RC. Hematologic abnormalities in Shwachman Diamond syndrome: lack of genotype-phenotype relationship. Blood. 2005;106(1):356–361. doi: 10.1182/blood-2004-11-4371. [DOI] [PubMed] [Google Scholar]
- 9.Woloszynek JR, et al. Mutations of the SBDS gene are present in most patients with Shwachman-Diamond syndrome. Blood. 2004;104(12):3588–3590. doi: 10.1182/blood-2004-04-1516. [DOI] [PubMed] [Google Scholar]
- 10.Tummala H, et al. DNAJC21 Mutations link a cancer-prone bone marrow failure syndrome to corruption in 60S ribosome subunit maturation. Am J Hum Genet. 2016;99(1):115–124. doi: 10.1016/j.ajhg.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhanraj S, et al. Biallelic mutations in DNAJC21 cause Shwachman-Diamond syndrome. Blood. 2017;129(11):1557–1562. doi: 10.1182/blood-2016-08-735431. [DOI] [PubMed] [Google Scholar]
- 12.Stepensky P, et al. Mutations in EFL1, an SBDS partner, are associated with infantile pancytopenia, exocrine pancreatic insufficiency and skeletal anomalies in aShwachman-Diamond like syndrome. J Med Genet. 2017;54(8):558–566. doi: 10.1136/jmedgenet-2016-104366. [DOI] [PubMed] [Google Scholar]
- 13.Khincha PP, Savage SA. Genomic characterization of the inherited bone marrow failure syndromes. Semin Hematol. 2013;50(4):333–347. doi: 10.1053/j.seminhematol.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menne TF, et al. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat Genet. 2007;39(4):486–495. doi: 10.1038/ng1994. [DOI] [PubMed] [Google Scholar]
- 15.Wong CC, Traynor D, Basse N, Kay RR, Warren AJ. Defective ribosome assembly in Shwachman-Diamond syndrome. Blood. 2011;118(16):4305–4312. doi: 10.1182/blood-2011-06-353938. [DOI] [PubMed] [Google Scholar]
- 16.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 17.Akopian D, Shen K, Zhang X, Shan SO. Signal recognition particle: an essential protein-targeting machine. Annu Rev Biochem. 2013;82:693–721. doi: 10.1146/annurev-biochem-072711-164732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gowda K, Chittenden K, Zwieb C. Binding site of the M-domain of human protein SRP54 determined by systematic site-directed mutagenesis of signal recognition particle RNA. Nucleic Acids Res. 1997;25(2):388–394. doi: 10.1093/nar/25.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pool MR, Stumm J, Fulga TA, Sinning I, Dobberstein B. Distinct modes of signal recognition particle interaction with the ribosome. Science. 2002;297(5585):1345–1348. doi: 10.1126/science.1072366. [DOI] [PubMed] [Google Scholar]
- 20.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 21.Bange G, Petzold G, Wild K, Parlitz RO, Sinning I. The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound GTP. Proc Natl Acad Sci U S A. 2007;104(34):13621–13625. doi: 10.1073/pnas.0702570104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wild K, Rosendal KR, Sinning I. A structural step into the SRP cycle. Mol Microbiol. 2004;53(2):357–363. doi: 10.1111/j.1365-2958.2004.04139.x. [DOI] [PubMed] [Google Scholar]
- 23.Ravera S, et al. Evaluation of energy metabolism and calcium homeostasis in cells affected by Shwachman-Diamond syndrome. Sci Rep. 2016;6:25441. doi: 10.1038/srep25441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bezzerri V, et al. New insights into the Shwachman-Diamond Syndrome-related haematological disorder: hyper-activation of mTOR and STAT3 in leukocytes. Sci Rep. 2016;6:33165. doi: 10.1038/srep33165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jagannathan-Bogdan M, Zon LI. Hematopoiesis. Development. 2013;140(12):2463–2467. doi: 10.1242/dev.083147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowe RG, Mandelbaum J, Zon LI, Daley GQ. Engineering hematopoietic stem cells: lessons from development. Cell Stem Cell. 2016;18(6):707–720. doi: 10.1016/j.stem.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Provost E, et al. Ribosomal biogenesis genes play an essential and p53-independent role in zebrafish pancreas development. Development. 2012;139(17):3232–3241. doi: 10.1242/dev.077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. 2011;117(4):e49–e56. doi: 10.1182/blood-2010-10-314120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruggero D, Shimamura A. Marrow failure: a window into ribosome biology. Blood. 2014;124(18):2784–2792. doi: 10.1182/blood-2014-04-526301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirwan M, et al. Exome sequencing identifies autosomal-dominant SRP72 mutations associated with familial aplasia and myelodysplasia. Am J Hum Genet. 2012;90(5):888–892. doi: 10.1016/j.ajhg.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yurist S, Dahan I, Eichler J. SRP19 is a dispensable component of the signal recognition particle in Archaea. J Bacteriol. 2007;189(1):276–279. doi: 10.1128/JB.01410-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose RW, Pohlschröder M. In vivo analysis of an essential archaeal signal recognition particle in its native host. J Bacteriol. 2002;184(12):3260–3267. doi: 10.1128/JB.184.12.3260-3267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zwieb C, Bhuiyan S. Archaea signal recognition particle shows the way. Archaea. 2010;2010:485051. doi: 10.1155/2010/485051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orelio C, Kuijpers TW. Shwachman-Diamond syndrome neutrophils have altered chemoattractant-induced F-actin polymerization and polarization characteristics. Haematologica. 2009;94(3):409–413. doi: 10.3324/haematol.13733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stepanovic V, Wessels D, Goldman FD, Geiger J, Soll DR. The chemotaxis defect of Shwachman-Diamond Syndrome leukocytes. Cell Motil Cytoskeleton. 2004;57(3):158–174. doi: 10.1002/cm.10164. [DOI] [PubMed] [Google Scholar]
- 36.In K, Zaini MA, Müller C, Warren AJ, von Lindern M, Calkhoven CF. Shwachman-Bodian-Diamond syndrome (SBDS) protein deficiency impairs translation re-initiation from C/EBPα and C/EBPβ mRNAs. Nucleic Acids Res. 2016;44(9):4134–4146. doi: 10.1093/nar/gkw005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren YG, Wagner KW, Knee DA, Aza-Blanc P, Nasoff M, Deveraux QL. Differential regulation of the TRAIL death receptors DR4 and DR5 by the signal recognition particle. Mol Biol Cell. 2004;15(11):5064–5074. doi: 10.1091/mbc.E04-03-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamohara H, et al. Regulation of tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) and TRAIL receptor expression in human neutrophils. Immunology. 2004;111(2):186–194. doi: 10.1111/j.0019-2805.2003.01794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki R, Kawahara H. UBQLN4 recognizes mislocalized transmembrane domain proteins and targets these to proteasomal degradation. EMBO Rep. 2016;17(6):842–857. doi: 10.15252/embr.201541402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485(7397):246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li MX, Gui HS, Kwan JS, Bao SY, Sham PC. A comprehensive framework for prioritizing variants in exome sequencing studies of Mendelian diseases. Nucleic Acids Res. 2012;40(7):e53. doi: 10.1093/nar/gkr1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369(16):1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halic M, et al. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006;444(7118):507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- 45.Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427(6971):215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- 46. The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC. https://www.pymol.org/ Accessed August 29, 2017.
- 47.Dolinsky TJ, et al. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35(Web Server issue):W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nüsslein-Volhard C, Dahm R, eds. Zebrafish: A Practical Approach. Oxford, England: Oxford University Press; 2002. [Google Scholar]
- 50.Warga RM, Kimmel CB. Cell movements during epiboly and gastrulation in zebrafish. Development. 1990;108(4):569–580. doi: 10.1242/dev.108.4.569. [DOI] [PubMed] [Google Scholar]
- 51.Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108(13):3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- 52.Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev Biol. 2007;7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parsons MJ, et al. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech Dev. 2009;126(10):898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konantz M, et al. Evi1 regulates Notch activation to induce zebrafish hematopoietic stem cell emergence. EMBO J. 2016;35(21):2315–2331. doi: 10.15252/embj.201593454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Esain V, Cortes M, North TE. Enumerating hematopoietic stem and progenitor cells in zebrafish embryos. Methods Mol Biol. 2016;1451:191–206. doi: 10.1007/978-1-4939-3771-4_13. [DOI] [PubMed] [Google Scholar]
- 56.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 58.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.