Abstract
Melanoma can be stratified into unique subtypes based on distinct pathologies. The acral/mucosal melanoma subtype is characterized by aberrant and constitutive activation of the proto-oncogene receptor tyrosine kinase C-KIT, which drives tumorigenesis. Treatment of these melanoma patients with C-KIT inhibitors has proven challenging, prompting us to investigate the downstream effectors of the C-KIT receptor. We determined that C-KIT stimulates MAP kinase–interacting serine/threonine kinases 1 and 2 (MNK1/2), which phosphorylate eukaryotic translation initiation factor 4E (eIF4E) and render it oncogenic. Depletion of MNK1/2 in melanoma cells with oncogenic C-KIT inhibited cell migration and mRNA translation of the transcriptional repressor SNAI1 and the cell cycle gene CCNE1. This suggested that blocking MNK1/2 activity may inhibit tumor progression, at least in part, by blocking translation initiation of mRNAs encoding cell migration proteins. Moreover, we developed an MNK1/2 inhibitor (SEL201), and found that SEL201-treated KIT-mutant melanoma cells had lower oncogenicity and reduced metastatic ability. Clinically, tumors from melanoma patients harboring KIT mutations displayed a marked increase in MNK1 and phospho-eIF4E. Thus, our studies indicate that blocking MNK1/2 exerts potent antimelanoma effects and support blocking MNK1/2 as a potential strategy to treat patients positive for KIT mutations.
Keywords: Therapeutics
Keywords: Melanoma
Introduction
Early-stage melanoma can be cured by surgery, but patients often develop advanced disease that requires effective systemic treatment. Melanoma arising on acral and mucosal anatomical sites accounts for about 5%–40% of melanoma cases, varying largely between racial groups (1–3). They very rarely have actionable BRAF mutations and have been reported to respond less well to standard-of-care therapy than cutaneous melanoma (4–6). Thus, effective therapeutic options are needed.
Curtin and colleagues first found that melanomas arising in acral, mucosal, and chronic sun-damaged regions harbor a high frequency of activating mutations in the KIT gene, which encodes the C-KIT tyrosine kinase receptor (KIT proto-oncogene receptor tyrosine kinase) (4). More recently, the The Cancer Genome Atlas (TCGA) Network classified cutaneous melanoma into BRAF, NRAS (NRAS proto-oncogene), NF1 (neurofibromin 1), and triple–wild-type groups. About 22% of triple–wild-type melanomas contain KIT aberrations (7). Constitutive activation of C-KIT, via KIT mutation or amplification, leads to the coactivation of downstream RAS/MAPK and PI3K/AKT/mTOR pathways and subsequent promotion of tumorigenesis (8). Current therapeutic strategies for treating KIT-aberrant melanomas include tyrosine kinase inhibitors (TKIs), such as imatinib, dasatinib, and nilotinib (5). However, patients with KIT-aberrant melanoma either are insensitive or rapidly acquire resistance to these TKIs, which thus provide limited clinical benefit (6, 9).
One potential therapeutic target that has not been explored in KIT-driven melanomas is the MNK (MAP kinase–interacting serine/threonine kinase)/eIF4E (eukaryotic translation initiation factor 4E) axis (5). eIF4E is a cap-binding subunit of the eIF4F translation initiation complex, which also includes the helicase eIF4A (eukaryotic translation initiation factor 4A) and the scaffolding protein eIF4G (eukaryotic translation initiation factor 4-γ) (10). eIF4E is overexpressed in a wide variety of human malignancies, including melanoma (11, 12), where it is thought to bolster tumorigenesis and tumor progression by selectively facilitating translation of proliferation and survival promoting mRNAs (e.g., VEGFA, CCNE1, SNAI1, and MCL1). In those metastatic melanomas where eIF4E is found to be highly phosphorylated, this is strongly associated with a reduced life expectancy (12). eIF4E is exclusively phosphorylated by MNK1/2 (MAP kinase–interacting serine/threonine kinases 1 and 2) (13, 14). Although MNK activity and the phosphorylation of eIF4E are dispensable for normal development (15, 16), eIF4E phosphorylation is required for tumorigenesis, lymphomagenesis, and tumor metastasis, thus making MNK1/2 kinases attractive therapeutic targets (15–19). As phospho-eIF4E is the convergence point of the RAS/MAPK and PI3K/AKT/mTOR cascades (20), these data provide support for the prospect of targeting MNK1/2 therapeutically in melanomas harboring KIT mutations and amplifications. We therefore sought to determine the effect of pharmacologically and genetically abrogating MNK1/2 activity in KIT-mutant melanomas. Our in vitro and in vivo data presented herein demonstrate that KIT-mutant melanoma cells are reliant on the MNK/eIF4E axis for survival and metastasis. We propose that MNK1/2 inhibitors could be effective in a molecularly defined cohort of melanoma patients with KIT aberrations, which currently represent a pressing therapeutic challenge.
Results
Elevated MNK1 and eIF4E phosphorylation in melanoma cell lines harboring C-KIT aberrations.
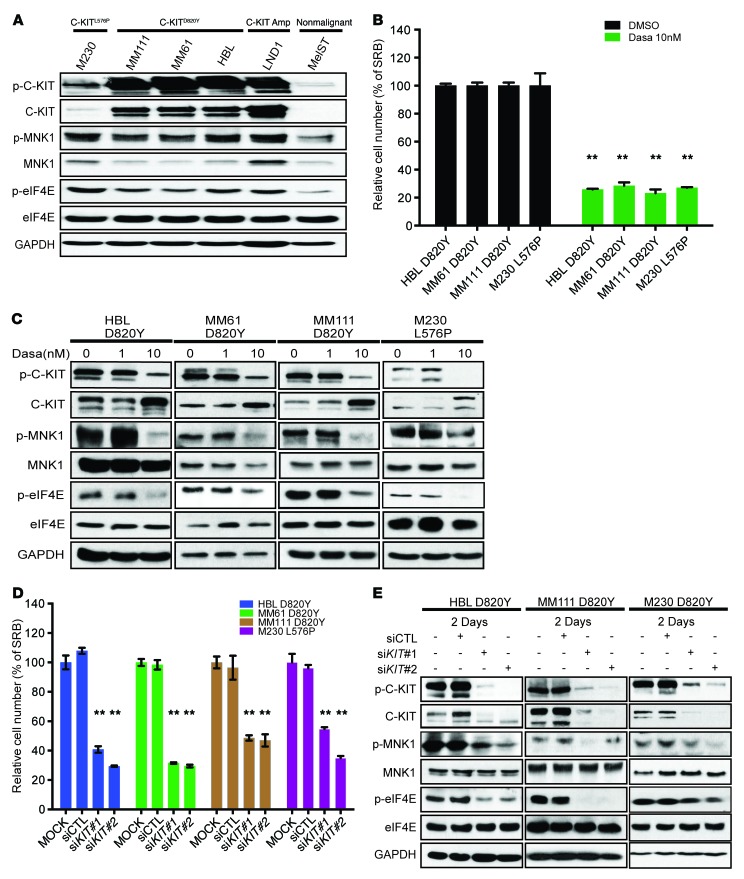
Although expression and activation of MNK1 and MNK2 have been previously demonstrated in human cancer (21, 22), their expression and phosphorylation status in melanoma cell lines has not been previously reported. C-KIT inhibitors are not terribly effective for acral/mucosal melanoma subtypes. We thus decided to profile the expression and phosphorylation of MNK1, and that of its downstream oncogenic substrate eIF4E, in a panel of melanoma cell lines harboring different oncogenic mutations in KIT (Figure 1A). As shown, the expression of phospho-MNK1 and that of phospho-eIF4E are increased in melanomas with aberrant C-KIT, either with KIT point mutation or amplification, compared with the nonmalignant melanocyte line MelST. These cell line results suggest that activation of the MNK/eIF4E axis lies downstream of oncogenic C-KIT signaling.
Figure 1. C-KIT inhibitor dasatinib suppresses cell proliferation and the activation of the MNK/eIF4E axis in KIT-mutant melanomas.
(A) Western blot analysis of phospho-MNK1 (p-MNK1), MNK1, phospho-eIF4E (p-eIF4E), and eIF4E in a panel of melanoma cell lines. (B) Cell proliferation was assessed by SRB staining, 72 hours after vehicle (DMSO) or 10 nM dasatinib treatment in HBL, MM61, MM111, and M230 melanoma cell lines. (C) Western blot analysis of phospho–C-KIT (p-C-KIT), C-KIT, p-eIF4E, eIF4E, p-MNK1, and MNK1 in HBL, MM111, MM61, and M230 melanoma cell lines, following a 24-hour dasatinib treatment. (D) Cell proliferation was assessed by SRB staining, 96 hours after transfection with KIT siRNAs. (E) Western blot analysis of p-C-KIT, C-KIT, p-eIF4E, eIF4E, p-MNK1, and MNK1 in HBL, MM111, and M230 cell lines transfected with KIT siRNAs, at the indicated time points. (B and D) Data represent the mean ± SD, n = 3. **P < 0.01 by 2-way ANOVA. (A, C, and E) GAPDH used as loading control.
Pharmacological or genetic inhibition of C-KIT suppresses the phosphorylation of MNK1 and eIF4E.
Recent clinical trials report limited therapeutic potential of C-KIT inhibitors in melanoma (6, 23–26). To test whether MNK and eIF4E are activated by oncogenic C-KIT, we monitored the phosphorylation of MNK1 and eIF4E in response to 2 C-KIT inhibitors, dasatinib and imatinib. As shown in Figure 1B, dasatinib significantly inhibited cell proliferation of both D820Y (HBL, MM61, MM111) and L576P (M230) KIT-mutant melanoma cell lines. Furthermore, Western blot analysis revealed that dasatinib treatment repressed the expression of phospho-MNK1 and phospho-eIF4E in all tested KIT-mutant melanoma cell lines (Figure 1C). Imatinib, on the other hand, only repressed proliferation of the L576P mutant cell line (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI91258DS1), consistent with previous reports showing that imatinib cannot bind to a C-KIT harboring the D820Y mutation, because of the active conformation of the receptor being stabilized (27, 28). In keeping with the cell proliferation data, imatinib only inhibited the phosphorylation of MNK1 and eIF4E in C-KIT L576P mutant M230 cells (Supplemental Figure 1B). To confirm that dasatinib-induced growth inhibition in KIT-mutant melanoma cells is in part mediated through suppression of MNK activity, we generated HBL cells stably expressing either a constitutively activated MNK1 construct (CA-MNK1) or control vector (pBABE). Overexpression of CA-MNK1 in HBL cells had little effect on cell proliferation in the absence of dasatinib, compared with pBABE-transduced HBL cells (Supplemental Figure 1C). However, CA-MNK1-expressing HBL cells did result in a strong increase of the IC50 of dasatinib, as compared to the control pBABE-expressing HBL cell counterparts (Supplemental Figure 1D). Collectively, these data support our model that the antiproliferative effect of dasatinib acts through repressing MNK1 activity.
In addition to inhibiting C-KIT, dasatinib can also inhibit SRC and PDGFR (29). Thus, we next silenced the expression of C-KIT using siRNA and examined the effect of C-KIT depletion on cell proliferation. As shown in Figure 1, D and E, C-KIT knockdown results in an inhibition of proliferation of KIT-mutant melanoma cell lines and the phosphorylation of MNK1 and eIF4E. These data suggest that MNK1 is activated and eIF4E phosphorylated downstream of C-KIT activating mutations.
MNK1/2 knockdown suppresses metastatic properties of KIT-mutant melanoma.
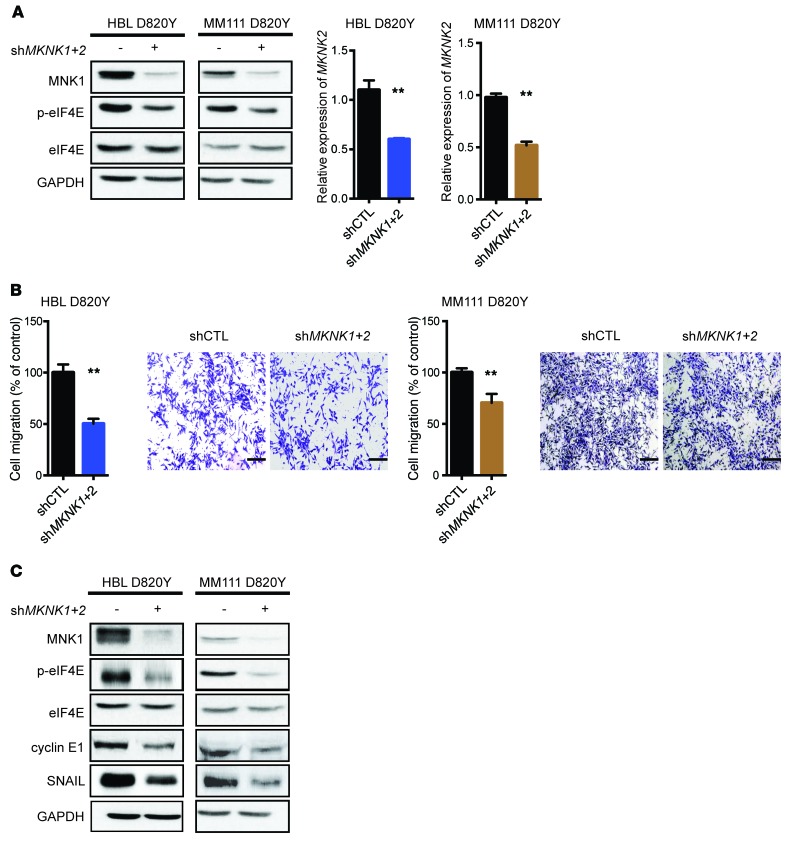
We next investigated the role of MNK1/2 in oncogenic signaling and characterized the effects of blocking these kinases in KIT-mutant melanomas. We stably silenced MKNK1 and MKNK2 in two C-KIT D820Y mutant melanoma cell lines, HBL and MM111, using shRNAs. As shown in Figure 2A (left panel), MNK1 and its substrate phospho-eIF4E were both suppressed in the shMKNK1+2 stable cell lines, compared with the shRNA control counterparts. Because of the low specificity of the currently available MNK2 antibodies, we used quantitative reverse transcriptase PCR (RT-qPCR) to demonstrate the MNK2 depletion in shMKNK1+2 cells (Figure 2A, right panel).
Figure 2. MNK1/2 knockdown in HBL cells suppresses cell migration and the expression of cyclin E1 and SNAIL.
(A) Western blot analysis of MNK1, p-eIF4E, and eIF4E in HBL or MM111 cells expressing shCTL and shMKNK1+2 (left). RT-qPCR was performed to examine the expression level of MKNK2 mRNA in HBL and MM111 cells expressing shCTL and shMKNK1+2 (right). (B) Cell migration was assessed by Transwell assay in shCTL versus shMKNK1+2 HBL and MM111 cells after 48 hours. Representative images are shown. Scale bars: 200 μm; original magnification, ×10. (A and B) Data represent the mean ± SD, n = 3. **P < 0.01 by 2-tailed Student’s t test. (C) Western blot analysis of MNK1, p-eIF4E, eIF4E, cyclin E1, and SNAIL in HBL and MM111 shCTL and shMKNK1+2 cell lines. (A and C) GAPDH is used as loading control.
As the MNK/eIF4E axis is a known facilitator of cell migration (15, 17), we next examined whether inhibition of MNK1/2 could be used as a strategy to block these properties in KIT-mutant melanomas. We performed the Boyden chamber assay, wherein HBL and MM111 shCTL or shMKNK1+2 cells were seeded on top of Transwells to assess cell migration. As shown in Figure 2B, genetic silencing of MNK1 and MNK2 reduced cell migration in KIT-mutant HBL and MM111 melanoma cells. Within the time frame of this in vitro assay (48 hours), there were no discernible differences in cell proliferation between the shCTL and shMKNK1+2 cell lines (Supplemental Figure 1E). These data indicate that suppressing MNK1 and MNK2 can suppress cell migration, independently of its cytostatic effects. Next, we examined the expression of proteins with documented roles in cell migration (17, 30). As shown in Figure 2C, the protein expression of SNAIL and that of cyclin E1 were both reduced in MNK1- and MNK2-depleted KIT-mutant melanoma cell lines. Collectively, our data show that MNK1/2 inhibition reduces cell migration of KIT-mutant melanoma.
mRNA translation of CCNE1 and SNAI1 is regulated downstream of oncogenic C-KIT/MNK/eIF4E.
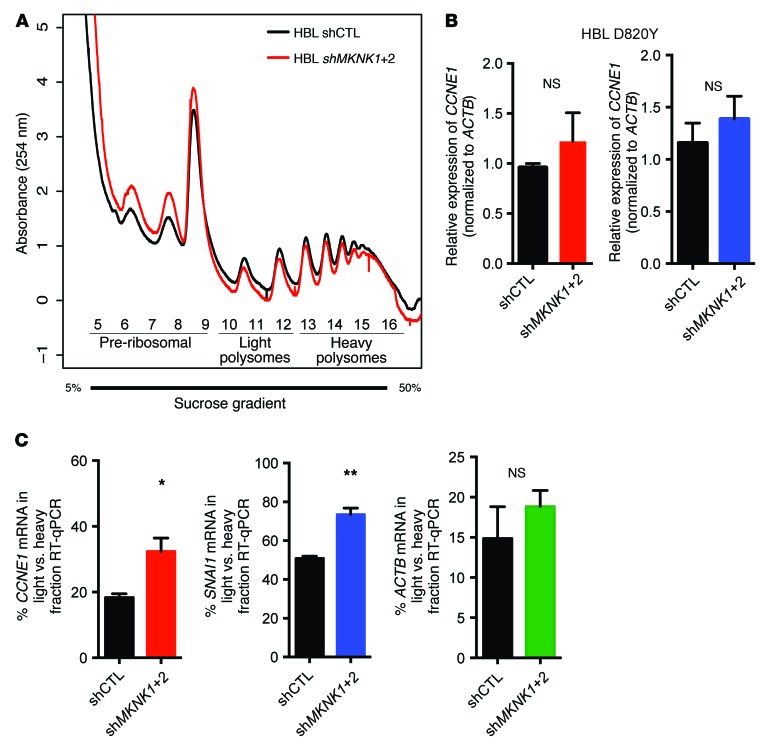
The phosphorylation of eIF4E by MNK1/2 facilitates translation of mRNAs that encode proteins that promote tumor progression to metastasis (15, 17, 31). We hypothesized that one biological outcome in cells with activating C-KIT mutations is aberrant mRNA translation. To uncover the molecular mechanisms underlying the oncogenic activities of the MNK/eIF4E axis downstream of mutant C-KIT, we sought to identify the translational targets of phospho-eIF4E by performing polysome profiling in shMKNK1+2 dual-depleted versus control shRNA HBL cells (Figure 3A). Polysome profiling curves of both MNK1/2-knockdown and control groups overlap, consistent with previous findings that MNK1/2 regulates the translation of a subset of prosurvival and prometastatic mRNA candidates (15, 31). Although the protein levels of cyclin E1 and SNAIL were reduced in MNK1/2-knockdown cell lines compared with controls (Figure 2C), we observed no similar decrease in CCNE1 and SNAI1 mRNA (Figure 3B). Thus, next we tested our hypothesis of increased aberrant protein synthesis in KIT-mutant cells, by examining CCNE1 and SNAI1 mRNA translation in HBL MNK-depleted cells. RT-qPCR analysis of RNA isolated from heavy and light polysome-bound fractions in shCTL and shMKNK1+2 HBL cells indicated that blocking MNK1/2 activity leads to a redistribution of CCNE1 and SNAI1 mRNAs, from heavy (efficiently translated) to light (poorly translated) polysomes (Figure 3C and Supplemental Figure 1F). This is consistent with the tenet that the MNK/eIF4E axis bolsters translation initiation of CCNE1 and SNAI1 mRNA. In conclusion, our data suggest that blocking MNK1/2 activity may inhibit tumor progression, at least in part, by blocking translation initiation of mRNAs encoding proteins that facilitate cell migration.
Figure 3. Inhibition of MNK1/2 impairs CCNE1 and SNAI1 mRNA translation in KIT-mutant melanoma cells.
(A) Absorbance profiles (254 nm) of sucrose gradients loaded with HBL shCTL (black) and shMKNK1+2 (red) cytosolic extracts. (B) RT-qPCR was performed to monitor SNAI1 and CCNE1 mRNA in HBL shCTL and shMKNK1+2 cell lines. Data represent the mean ± SD, n = 3. P > 0.05 by 2-tailed Student’s t test. (C) RT-qPCR was performed to determine the distribution of ACTB, SNAI1, and CCNE1 mRNAs in light versus heavy polysome fractions obtained from the sucrose gradient described in A. Data represent the mean ± SD, n = 3. *P < 0.05, **P < 0.01 by 2-tailed Student’s t test.
MNK1 activity is elevated in KIT-mutant human melanoma patients.
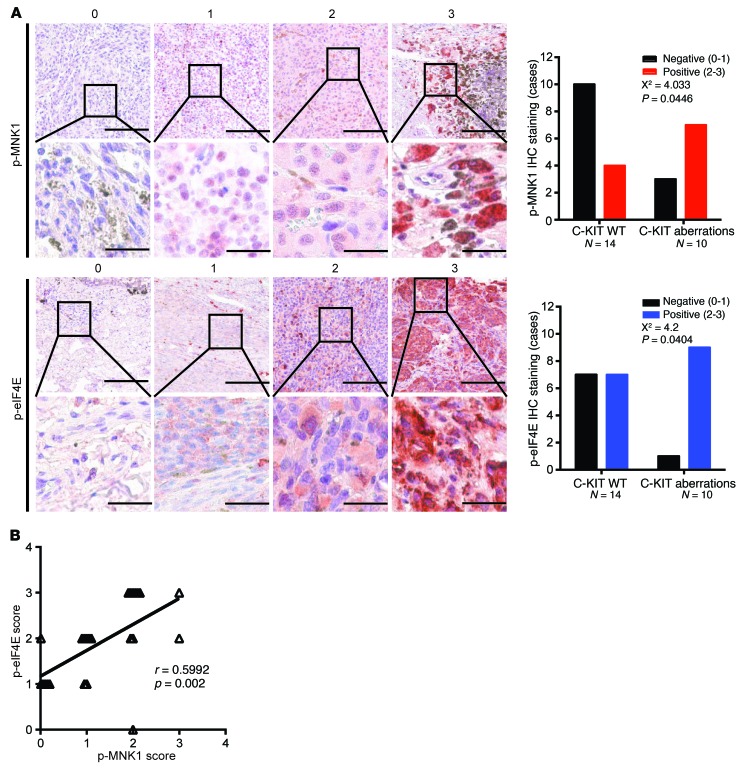
We previously demonstrated that MNK1 phosphorylation and eIF4E phosphorylation are elevated in C-KIT–aberrant melanoma cell lines (Figure 1A). To test whether these observations are clinically relevant, we used immunohistochemistry (IHC) to evaluate MNK1, phospho-MNK1, and phospho-eIF4E expression in archival surgical patient specimens of melanomas wherein KIT was either wild-type or mutated (Supplemental Table 1). We found that KIT-mutant melanomas expressed significantly higher levels of phospho-MNK1 (trend test via χ2, P = 0.0446) compared with C-KIT wild-type melanomas (Figure 4A). KIT-mutant melanomas also expressed higher levels of MNK1 compared with C-KIT wild-type melanomas (Supplemental Figure 2). We then determined the phosphorylation state of eIF4E, one of the best-described substrates of MNK1/2. In keeping with higher MNK1/2 activity in melanomas driven by oncogenic C-KIT, phospho-eIF4E was also higher in KIT-mutant melanomas versus wild-type C-KIT melanomas (trend test via χ2, P = 0.0404) (Figure 4A). As expected, phospho-MNK1 and phospho-eIF4E levels positively correlated within a given patient sample (trend test via Pearson correlation, P = 0.002) (Figure 4B). These data show that MNK1 and eIF4E phosphorylation is elevated in human melanomas harboring C-KIT aberrations. Together, our data indicate that MNK1 and MNK2 have a critical function in KIT-mutant melanoma and suggest that these kinases are a therapeutic target in this hard-to-treat disease.
Figure 4. KIT-mutant melanoma patients express high phospho-MNK1 and phospho-eIF4E.
(A) Representative images of p-MNK1 and p-eIF4E IHC staining. Bar graphs of p-MNK1 and p-eIF4E IHC scores in melanoma patients are shown in the right panel. χ2 test, P values shown in the figure. For p-MNK1 and p-eIF4E top panels, scale bars: 200 μm; original magnification, ×4. For p-MNK1 and p-eIF4E bottom panels, scale bars: 40 μm; original magnification: ×20. (B) The correlation between p-eIF4E and p-MNK1 IHC scores in melanoma patients. Pearson correlation test, r and P values are shown.
Development of SEL201, a novel MNK1/2 inhibitor.
Multiple C-KIT inhibitors have been tested in patients whose melanomas harbor C-KIT aberrations, but their effects are dismal for reasons that are addressed in the Discussion. Our data suggest targeting downstream of oncogenic C-KIT, prompting us to block MNK1/2 kinase activity.
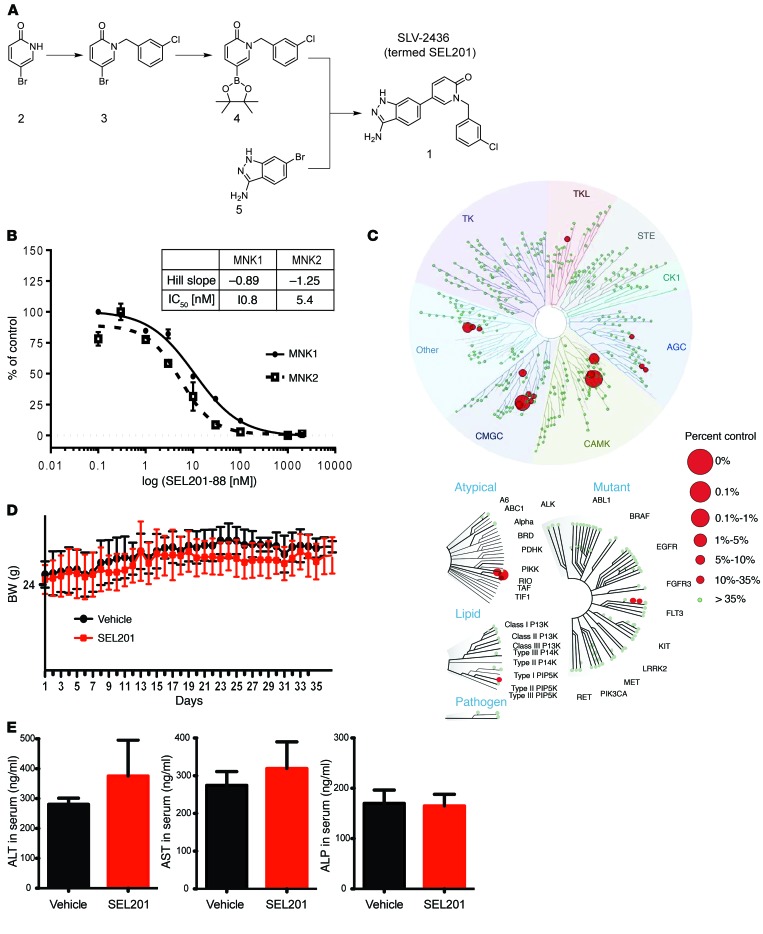
In efforts to improve the biophysical properties of existing MNK1/2 inhibitors, we report herein the development of SLV-2436 (SEL201-88), a novel ATP-competitive inhibitor of MNK1 and MNK2. Using a structure-driven approach (detailed in Methods and Supplemental Figure 3), we produced a novel class of amino-1H-indazol-1,2-dihydropyridin-2-one derivative as a potent inhibitor of MNK1/2 (Figure 5A). In brief, 5-(3-amino-1H-indazol-6-yl)-1-[(3-chlorophenyl)methyl]-1,2-dihydropyridin-2-one (also known as SEL201-88) (panel 1, Figure 5A) was derived by a 3-step synthesis starting from commercially available 5-bromopyridin-2-ol (panel 2) as shown in Figure 5A. Nucleophilic substitution of pyridinone (panel 2) with 3-chlorobenzyl bromide resulted in an intermediate compound (panel 3), which we used for the Miyaura borylation reaction enabling the synthesis of boronate ester (panel 4). Boronate (panel 4) and 6-bromo-1H-indazol-3-amine (panel 5) served as coupling partners in the Suzuki reaction, resulting in the final compound SEL201-88 (termed SEL201) (panel 1). We next performed in vitro kinase assays using recombinant MNK1 and MNK2 protein and increasing concentrations of SEL201, to show that SEL201 is highly potent, with an IC50 of 10.8 nM and 5.4 nM for MNK1 and MNK2, respectively (Figure 5B). To confirm the kinome selectivity of SEL201, we performed the broad KINOMEscan (DiscoverX) competitive binding assay at 1 μM, which included 450 distinct kinases (32). Figure 5C shows the observed binding profile for SEL201, which was significantly concentrated in the CAMK family of kinases that comprises MNK1 and MNK2.
Figure 5. Chemical synthesis, in vitro kinome selectivity, and biosafety of SEL201.
(A) Chemical synthesis procedure of SEL201. (B) The in vitro ADP-Glo assay was performed by incubation of SEL201 with recombinant MNK1 and MNK2, peptide substrates, and ATP. After the kinase reaction, luminescence intensity generated by the remaining ADP was measured. (C) Kinome selectivity of SEL201 was assessed using KINOMEscan (DiscoverX) panel, which consists of 450 kinases. Circles represent targets that interacted with SEL201 at the concentration of 1 μM. (D) Body weight kinetics of tumor-bearing mice (6 animals per group) was assessed throughout the study (endpoint on day 37). Vehicle or SEL201 was administered p.o. to animals at the dosage of 50 mg/kg twice daily (100 mg/kg/d; mean ± SD). (E) Assessment of blood biochemistry in mice (3 animals per group) given SEL201 at the dosage of 50 mg/kg twice daily (100 mg/kg/d) was performed at the study endpoint (day 37). AST, aspartate aminotransferase; ALT, alanine transaminase; ALP, alkaline phosphatase.
To investigate the pharmacodynamic properties of SEL201, 5 consecutive oral doses of 10, 25, and 50 mg/kg were administered to mice every 12 hours (twice-daily schedule). At the 10 mg/kg twice-daily dosage, 4 hours after the fifth administration, we determined a low plasma concentration of 125 ng/ml SEL201. However, dosing at 25 and 50 mg/kg twice daily, equivalent to 50 and 100 mg/kg/d of SEL201, yielded substantially increased dose-dependent plasma exposure, reaching an average level of 1,299 ng/ml and 2,075 ng/ml, respectively. At the 24-hour time point, SEL201 was still detectable in the plasma, with dose-dependent concentrations of 9, 73, and 124 ng/ml in the 10, 25, and 50 mg/kg twice-daily treatment groups (Supplemental Figure 4A). Oral (p.o.) administration of SEL201 at the dosage of 50 mg/kg twice daily, that is, 100 mg/kg/d, for 37 days was well tolerated in mice. There were no abnormalities in body weight kinetics, which was comparable in control and SEL201-treated animals throughout the study (Figure 5D). Furthermore, 37 days of consecutive administration of SEL201, at the dosage of 100 mg/kg/d, did not result in any overt clinical signs of toxicity. A blood chemistry panel was performed, including hematological and biochemical parameters, at the study endpoint and confirmed that multiple dosings with SEL201 at 100 mg/kg/d is safe (Supplemental Table 2, B and C). Importantly, unlike the hepatotoxicity accompanying dasatinib treatment (33–36), SEL201 administration is not associated with liver toxicity as measured by serum levels of aspartate aminotransferase, alanine transaminase, and alkaline phosphatase, which were not significantly different from those in the vehicle-treated group (Figure 5E). Finally, the pharmacokinetic profile of SEL201 in mice after p.o. and i.v. administration (Supplemental Figure 4B and Supplemental Table 2A) revealed a relatively short half-life of the compound in plasma (about 1.44 hours). Nevertheless, SEL201 showed very good oral bioavailability, with a maximum plasma concentration of 1,078 ng/ml reached 0.25 hours after p.o. administration. In summary, SEL201 is well tolerated in mice.
MNK1/2 inhibitor SEL201 decreases clonogenicity, cell migration, and melanoma metastasis.
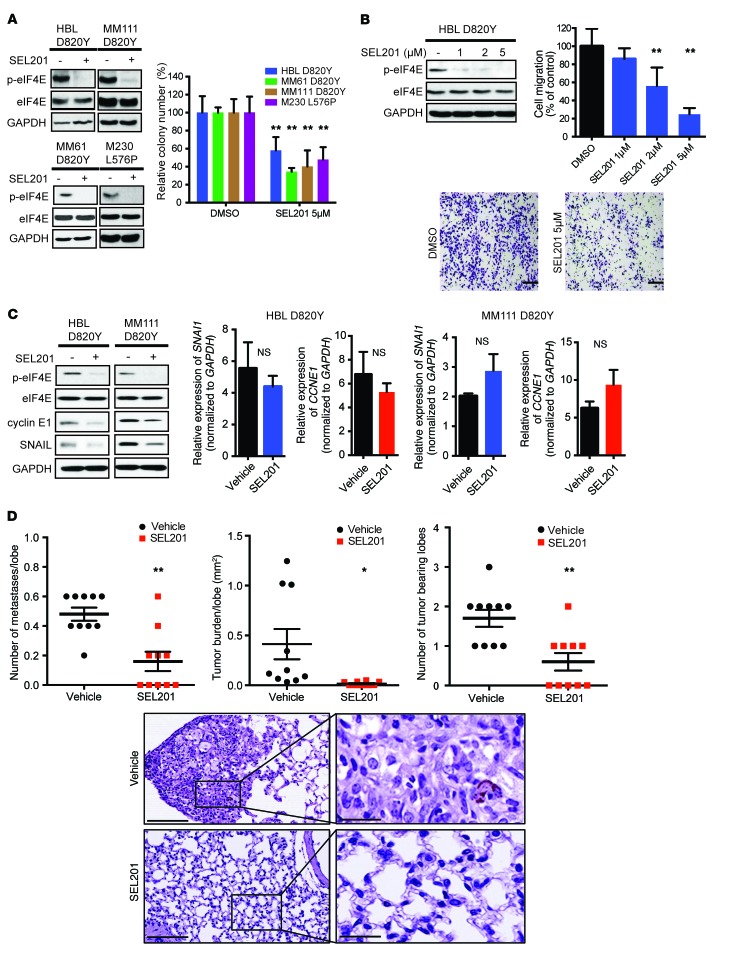
We next explored the effects of blocking MNK1/2 kinases pharmacologically in KIT-mutant melanomas. SEL201 potently suppressed MNK1/2 activity in our panel of KIT-mutant melanoma cell lines, as determined by repressed phosphorylation of its substrate eIF4E (Figure 6A). In addition, SEL201 significantly decreased the colony-forming ability of both C-KIT D820Y and L576P mutant melanoma cell lines (Figure 6A). Moreover, SEL201 did not inhibit the proliferation of normal, nonmalignant MelST melanocytes at the doses tested in the KIT-mutant cell lines (Supplemental Figure 5A). We next investigated the effects of SEL201 on cell migration, in an attempt to validate our findings in KIT-mutant melanoma cells that are devoid of both MNK1 and MNK2. As shown, pharmacologically inhibiting MNK1/2 activity using the inhibitor SEL201 reduced the cell migration ability of HBL melanoma cells (Figure 6B). Notably, SEL201 did not decrease cell proliferation within the time frame of the migration assay (48 hours), which indicated that SEL201 suppressed migration, independently of its effect on cell proliferation (Supplemental Figure 5B).
Figure 6. MNK1/2 inhibitor SEL201 suppresses clonogenicity, cell migration, and in vivo tumor metastasis.
(A) Graph of 14-day clonogenic assay: HBL, MM61, MM111, and M230 cell lines were treated with or without 5 μM SEL201 (right). Data represent the mean ± SD, n = 3. **P < 0.01 by 2-way ANOVA. Western blot analysis of p-eIF4E and eIF4E; HBL, MM61, MM111, and M230 cell lines were treated with 5 μM SEL201 for 24 hours (left). (B) Western blot analysis of p-eIF4E and eIF4E in HBL cells following SEL201 treatment (48 hours). Cell migration was assessed by Transwell assay. Representative images are shown. Scale bars: 200 μm; original magnification, ×10. Data represent the mean ± SD, n = 3. **P < 0.01 by 1-way ANOVA. (C) HBL and MM111 cell lines were treated with 5 μM SEL201 for 24 hours (left). Western blot analysis of p-eIF4E, eIF4E, cyclin E1, and SNAIL. HBL and MM111 cell lines were treated with 5 μM SEL201 for 24 hours (right). RT-qPCR analysis for SNAI1 and CCNE1 mRNA expression. Data represent the mean ± SD, n = 3. P > 0.05 by 2-tailed Student’s t test. (D) 8 × 106 MM111 cells were injected in the tail vein of NOD/SCID mice. Tumor burden was defined as the percentage of lung area occupied by tumor cells. Representative images are shown. For the left panel, scale bars: 300 μm. For the right panel, scale bars: 50 μm. Data represent the mean ± SD, n = 10. *P < 0.05, **P < 0.01 by 2-way ANOVA. (A–C) GAPDH is used as a loading control.
Next, we examined whether SEL201 suppressed the expression of MNK1/2 translational targets cyclin E1 and SNAIL. We demonstrated that cyclin E1 and SNAIL protein expression was decreased in SEL201-treated HBL and MM111 cells (Figure 6C, left panel), with no similar reduction in their mRNA levels (Figure 6C, right panel). These data suggest that the novel MNK1/2 inhibitor SEL201 may elicit its antioncogenic effects by blocking mRNA translation in KIT-mutant melanoma cells.
Finally, to enhance the translational aspect of this work, potentially facilitating clinical trials, we tested the ability of SEL201 to block KIT-mutant melanoma metastasis. We established a xenograft model wherein KIT-mutant MM111 cells were injected via tail vein (day 0), and then randomized on day 1 to vehicle control or SEL201 treatment groups. Mice were treated with either vehicle or SEL201 (75 mg/kg/d, 5 consecutive days per week) for 35 days. At the endpoint, animals were euthanized, and lungs were harvested, H&E-stained, step-sectioned, and quantified for the presence of metastases (scoring pathologist was blinded to treatment group). As shown in Figure 6D, SEL201 significantly decreased both the number and the size of metastases in the lung. Furthermore, we tested the expression and phosphorylation of eIF4E as a surrogate for SEL201-mediated inhibition of MNK1/2 activity in the lungs of treated mice. As shown in Supplemental Figure 6A, SEL201 suppressed the expression of phospho-eIF4E in the lung metastases, compared with the lungs of vehicle-treated animals. Tumors present in the lungs of vehicle and SEL201 treatment groups expressed comparable levels of eIF4E (Supplemental Figure 6B). Collectively, our data show that the MNK1/2 inhibitor SEL201 reduces clonogenicity, migration, and lung metastasis of KIT-mutant melanoma cells.
Discussion
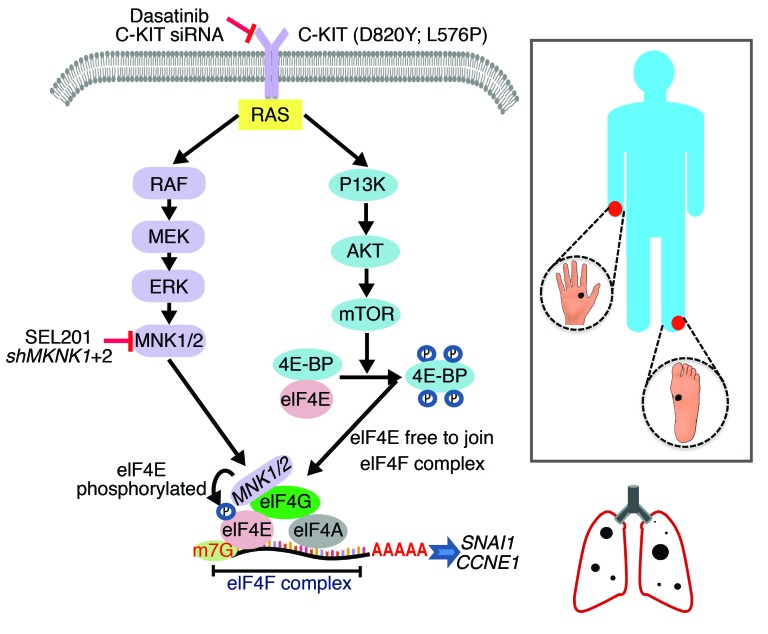
The data described herein add melanoma to the growing list of malignancies with aberrant MNK1/2 activity (37–40). This work has led to several insights into the biology of melanomas wherein KIT is mutated. First, phospho-MNK1 and phospho-eIF4E expression is significantly elevated in KIT-mutant/-amplified melanomas compared with specimens wherein KIT is wild-type or not amplified. Genetic and pharmacological inhibition of MNK1/2 suppresses cell survival, cell migration, and metastasis. At the molecular level, blocking MNK1/2 suppresses the expression of SNAIL and cyclin E1 in C-KIT melanoma, which are overexpressed in metastatic melanoma and other cancers (Figure 7 and refs. 41, 42).
Figure 7. Schematic diagram of the MNK/eIF4E axis in KIT-mutant acral or mucosal melanoma.
Clinical studies examining the efficacy of receptor tyrosine kinase inhibitors in patients with acral and mucosal melanoma, in which KIT alterations are frequent, have been underwhelming, and only 20%–30% of acral and mucosal melanoma patients respond to anti–PD-1 immunotherapy (5, 24, 43, 44). For example, only melanoma patients with specific KIT mutations (L576P) respond to the C-KIT inhibitor imatinib, while patients who possessed KIT amplifications or NRAS mutations received no benefit from this treatment (24). Furthermore, secondary mutations in the kinase domain of C-KIT, such as A829P and T670I, confer acquired resistance to the C-KIT inhibitor imatinib (9). In addition, the median progression-free survival after anti–PD-1 immunotherapy is reportedly short, approximately 4 months, in patients with acral and mucosal melanoma (44). Our data support inhibiting MNK1/2 as a novel therapeutic strategy for these incurable KIT-driven melanomas. Encouragingly, though severe liver toxicity is an adverse event associated with numerous C-KIT inhibitors, including dasatinib and imatinib (33–36), we observed no clinical signs of liver dysfunction with p.o. administration of SEL201 in our in vivo xenograft studies. Importantly, although we focused on blocking MNK1/2 activity in KIT-mutant melanoma in the present study, other subtypes of melanoma are expected to also be amenable to MNK1/2 inhibition. Indeed, 2 BRAFV600E/PTEN-null melanoma cell lines analyzed in our cohort also overexpressed phospho-eIF4E (data not shown), and are the subject of ongoing studies in our laboratory. Recently, melanomas with loss-of-function mutations in the tumor suppressor neurofibromin 1 (NF1) have been found, accounting for 15% in patients with cutaneous melanoma (7). A recent report shows that MNKs are therapeutic targets for NF1-deficient malignant peripheral nerve sheath tumors (39), suggesting that MNK1/2 may also be druggable in NF1-mutant melanomas. Moreover, our work highlights the need to test MNK1/2 inhibitors in diseases wherein KIT is commonly mutated, such as gastrointestinal stromal tumors (GISTs). In GISTs, secondary mutations or amplification of KIT contributes to resistance to C-KIT inhibitors, and in these cases MNK1/2 therapeutic intervention would be predicted to overcome resistance (9, 24).
eIF4E, a critical regulator of protein synthesis, is one of the best-studied substrates of MNK1/2 (13, 14, 45). The translation initiation complex eIF4F comprises the oncoprotein eIF4E, the helicase eIF4A, and the scaffolding protein eIF4G. Previous studies, including our own work, have shown that disrupting the eIF4F complex significantly potentiated the effectiveness of BRAF inhibitor, and overcame acquired drug resistance in BRAF-mutant melanoma (46, 47). Our profiling of MNK1/2 activity in patient samples suggests that the phosphorylation state of eIF4E has the potential to be used as a biomarker to screen for tumors that are driven by MNK1/2 kinases. We predict that activating mutations in KIT alter mRNA translation, and our data support this prediction. Mechanistically, we reveal that cyclin E1 and SNAIL protein synthesis is regulated by MNK1/2 kinases in KIT-mutant melanomas. In future work, it will be important to uncover the molecular mechanisms underlying the oncogenic activities of the MNK/eIF4E axis, downstream of mutant KIT. We are currently identifying genome-wide translational targets of phospho-eIF4E in KIT-mutant melanomas, with the goal of translating basic research findings into potential therapeutic targets for correcting the defects caused by mutant C-KIT.
Our findings show increased MNK1/2 kinase activity in KIT-mutant/-amplified melanomas, and we find that blocking MNK1/2 exerts potent antimelanoma effects in vitro and in vivo. Our study, and the work of others (38, 39, 48, 49), strongly support the preclinical development of MNK1/2 inhibitors for the treatment of patients with melanoma, and potentially other malignancies harboring KIT aberrations.
Methods
Reagents.
Dasatinib was obtained from Bristol-Myers Squibb. SLV-2436 (SEL201) was derived (see below) and provided by Selvita. Drugs were dissolved in DMSO, and 10-mM aliquots were stored at –80°C. The following commercially available antibodies were purchased from Cell Signaling Technology: phospho-MNK1 (Thr197/202, catalog 2111), phospho-eIF4E (S209, catalog 9741), phospho–C-KIT (Tyr703, catalog 3073), C-KIT (catalog 3308), MNK1 (catalog 2195), and GAPDH (catalog 2118). eIF4E antibody (catalog 610269) was purchased from BD Biosciences and cyclin E1 (catalog sc-481) from Santa Cruz Biotechnology. Transfection reagents Lipofectamine 2000 and Lipofectamine RNAiMax were purchased from Invitrogen.
Cell culture.
All cell lines used in this study were a gift from G.E. Ghanem (Institute Jules Bordet, Brussels, Belgium), except M230. M230 was a gift from Antoni Ribas (UCLA, Los Angeles, California, USA). MelST was cultured in DMEM containing 10% FBS and 1% penicillin/streptomycin; MM61, MM111, HBL, and LND1 cells were cultured in Ham’s F10 containing 10% FBS and 1% penicillin/streptomycin. M230 cells were cultured in RPMI-164 containing 10% FBS and 1% penicillin/streptomycin. Stable cell lines (HBL pBABE, CA-MNK1, shCTL, shMKNK1+2; and MM111 shCTL, shMKNK1+2) were cultured in Ham’s F10 containing 10% FBS and 1% penicillin/streptomycin supplemented with 1 μg/ml of puromycin for selection. All cell lines were maintained at 37°C in a humidified incubator with 5% CO2.
Proliferation assay.
Cell proliferation was tested by sulforhodamine B (SRB) assay. Cells were plated in 96-well plates 24 hours before treatment, and then treated with indicated drugs for 24, 48, and 96 hours. After each treatment, 96-well plates were harvested and fixed with 10% trichloroacetic acid for 1 hour. Plates were then washed and sterilized with water 3 times and allowed to air-dry overnight. Once the fixation was completed, each well was stained with 100 μl of 0.4% (wt/vol) SRB dissolved in 1% acetic acid for at least 30 minutes. After staining, plates were washed with 1% acetic acid 3 times and air-dried overnight. Bound SRB was solubilized by addition of 100 μl/well of 10 mM unbuffered Tris base, pH 10.5, for 10 minutes. Absorbance at 564 nm was read using a FLUOstar OPTIMA plate reader.
Plasmids, virus production, and stable cell selection.
Human shMKNK1 and shMKNK2 plasmids were obtained from Sidong Huang. Individual shRNA vectors used were collected from the Mission TRC library (Sigma-Aldrich): shMKNK1 (TRCN0000314803); shMKNK2 (TRCN0000199855). shRNAs were cotransfected with PLP1, VSVG, and PLP2 packaging constructs into 293FT cells using Lipofectamine 2000 (Invitrogen) to generate viral particles. Viral supernatants were harvested 48 hours after transfection and spun at 173 g for 5 minutes. Control or shMKNK1+2 viral supernatants were added to HBL cells overnight to establish shMKNK1, shMKNK2, and dual shMKNK1+2 stable knockdown cell lines. After 2 rounds of infection, cells were treated with puromycin (1 μg/ml) for 48 hours for subclone selection.
Polysome profiling.
Polysome profiling was performed as previously described (50). For sucrose gradient fractionation and polysome isolation, HBL shCTL and HBL shMKNK1+2 cells were used. Cells were treated with cycloheximide (100 μg/ml) 5 minutes before harvesting, then washed in cold PBS containing 100 μg/ml cycloheximide, followed by centrifugation for 5 minutes at 390 g. Cell pellets were lysed in hypotonic buffer: 5 mM Tris-HCl (pH 7.5), 2.5 mM MgCl2, 1.5 mM KCl, and 1X protease inhibitor cocktail (EDTA-free), containing 1 mM DTT and RNase inhibitor (100 U). Samples were kept on ice for 12 minutes, then centrifuged at 13,523 g for 7 minutes. The supernatants were loaded onto 10%–50% sucrose gradient followed by centrifugation at 260,110 g for 2 hours at 4°C. Fractions were collected (24 fractions, 12 drops each) using a Foxy JR ISCO collector, and data (absorbance, 254 nm) were collected.
Western blot analysis.
Cells were treated with dasatinib, imatinib, or SEL201 at the indicated times, and pellets were harvested to obtain protein extracts. Briefly, cell pellets were lysed in RIPA buffer (50 mM Tris-HCl, pH 8.0, with 150 mM sodium chloride, 1.0% Igepal CA-630 [NP-40], 0.5% sodium deoxycholate, and 0.1% SDS). After sonication, cell lysates were centrifuged at 15,871 g for 15 minutes. The supernatants were collected, and protein concentrations were quantified. Equal amounts of protein were loaded and separated on a 10% SDS-PAGE. After transferring to a nitrocellulose membrane (Bio-Rad), 5% milk/TBS was used to block for 1 hour, and then probed for target antibodies overnight at 4°C. After incubation with HRP-conjugated secondary antibodies for 1 hour at room temperature, the signals of targeted protein were developed with chemiluminescence substrate ECL Western blotting detection reagent (Amersham, GE Healthcare).
RNA interference.
Cells, other than M230, were seeded in 10-cm dishes at 80% confluence. C-KIT or control siRNAs were added into dishes 20 minutes after incubation with transfection reagent Lipofectamine RNAiMAX following the manufacturer’s instructions. M230 cells were electroporated (Amaxa) to introduce siRNAs. Briefly, 1 million M230 cells were added into electroporation cuvettes along with C-KIT or control siRNAs and electroporated using Amaxa program W-001. The electroporated cells were seeded into 10-cm dishes and allowed to attach overnight. After 16 hours, cells were washed with 1X PBS and fresh medium was added. Two days after transfection, cell pellets were harvested for Western blotting. The sequences of the previously validated C-KIT siRNA pair were as follows: number 1, 5′-AGCAGGAAAUAAAGUAUAGGUUUAG-3′ and 5′-CUAAACCUAUACUUUAUUUCCUGCUAC-3′; number 2, 5′-CGAUUCUAAGUUCUACAAGAUGATC-3′ and 5′-GAUCAUCUUGUAGAACUUAGAAUCGAC-3′. AllStars Negative Control siRNA (Qiagen) was used as nonsilencing control.
RNA isolation.
To isolate mRNAs from each polysome fraction, Trizol (Invitrogen) was added in each fraction tube, followed by 5 minutes of incubation at room temperature. Then 200 μl of chloroform (VWR International) was added to each tube and mixed well for 15 seconds. Following centrifugation at 12,000 g for 15 minutes at 4°C, the clear phase was carefully obtained and placed into an RNase-free tube. Five hundred microliters of isopropanol (Sigma-Aldrich) was added to the clear phase, and the mixture was centrifuged for 30 minutes (12,000 g, 4°C). The isopropanol was then removed, and the remaining pellets were washed with 1 ml of 75% ethanol (in diethylpyrocarbonate water) followed by centrifugation for 5 minutes (12,000 g, 4°C). The remaining liquid was then carefully aspirated, and the pellets were allowed to air-dry. Twenty microliters of diethylpyrocarbonate water was added to dissolve RNA pellets, which were then quantitated (NanoDrop 1000, Thermo Fisher Scientific).
Clonogenic assay.
One thousand cells per well were seeded in 6-well plates, and the cells were allowed to adhere overnight. After overnight incubation, the cells were treated with either DMSO (control) or SEL201 at the indicated concentrations. After 14 days, media were removed from the wells, and the cells were stained with 0.5% (wt/vol) crystal violet in 70% ethanol. After 1 hour of incubation at room temperature, staining dye was washed, and the colony numbers were counted by GelCount (Oxford Optronix). The experiment was done in triplicate.
Animal studies.
All animals were handled in strict accordance with good animal practice, and maintained according to the standards of pathogen-free conditions. The pharmacokinetic profile of SEL201 was assessed in 6-week-old female CD-1 mice (outbred colony, Jagiellonian University Medical College, Kraków, Poland) (3 animals per time point). SEL201 was freshly dissolved in DMSO and then diluted in Captisol (Ligand) for administration with a volume of 10 μl per 1 g of body weight via the oral (p.o.; 5 mg/kg) or i.v. (2 mg/kg) route. Animals were sacrificed at 8 time points (5, 15, and 30 minutes and 1, 2, 4, 6, and 24 hours) and blood samples harvested. Plasma samples were collected and stored at –80°C for further analysis. To evaluate the pharmacodynamic properties of SEL201, 10- to 16-week-old male C57BL/6 mice (stock 000664, The Jackson Laboratory) were divided into a control group and 3 dosing groups. Animals were given either vehicle (DMSO + N,N-Dimethylacetamide + Captisol) or SEL201 at 10-, 25-, and 50-mg/kg doses (freshly dissolved). Drugs were administered p.o. in a volume of 10 μl per 1 g of body weight. Each animal received a total of 5 doses with twice-daily schedule (i.e., every 12 hours). Body weight was assessed once daily. Six animals per experimental group supported sample collection at 2 time points (i.e., 4 hours and 24 hours) after the last, fifth administration, with 3 animals per time point. Plasma samples were collected and stored at –80°C for further analysis. For safety assessment of SEL201, 7- to 8-week-old tumor-bearing female Hsd:Athymic Nude-Foxn1nu mice (strain code 069, Envigo) were used. Before use, SEL201 was freshly dissolved, and doses of 50 mg/kg were administered twice daily p.o. in a volume of 10 μl per 1 g of body weight. Body weight was assessed every day. At the end of the experiment on day 37, mice were anesthetized and blood samples for total cell counts and biochemistry were obtained.
For metastasis studies, 8-week-old female NOD/SCID mice (strain code 394, Charles River Laboratories) were injected in the tail vein with 8 × 106 MM111 cells that had been resuspended in PBS. Animals were orally gavaged with vehicle or 75 mg/kg/d of SEL201 for 35 days. At the endpoint, mice were euthanized, and lungs were harvested and fixed in 10% formalin for H&E staining, step sectioning, and IHC assessment for tumor metastasis.
Immunohistochemistry and scoring.
Human melanoma tissue samples were acquired from 24 patients (including acral, mucosal, or chronic sun-damaged melanoma) hospitalized between January 2015 and January 2016 at Peking University Cancer Hospital. During this period, a total of 888 patients were hospitalized, of whom 112 were identified as having KIT mutations in their melanomas. Paraffin-embedded samples were stained with H&E and tested for melanoma markers (S-100B, HMB-45, or MART-1) to confirm the diagnosis of melanoma by the pathology department in Peking University Cancer Hospital, or by the pathology department of hospitals where patients were initially diagnosed. Clinical data, including age, sex, thickness (Breslow), and ulceration, were collected in 16 of 24 patients.
IHC analysis for phospho-eIF4E (Ser209, Abcam catalog ab76256), phospho-MNK1 (Thr197/202, Cell Signaling Technology catalog 2111), and MNK1 (Sigma-Aldrich, catalog SAB4503423) was conducted on formalin-fixed, paraffin-embedded tumor sections at 1:50 dilution, followed by a standard avidin-biotin detection protocol. Hematoxylin-counterstained slides were mounted with coverslips, and staining intensity was determined by a clinically certified pathologist at Peking University Cancer Hospital & Institute and 2 other pathologists at the Segal Cancer Centre, Jewish General Hospital. Staining intensity was scored as 0, 1, 2, and 3 (0 as negative staining, 3 as the strongest staining intensity). All pathologists were blinded to all clinical data and antibodies used for IHC. Representative images of the intensity scores are shown in Figure 1. Five images of melanoma lesions were taken from each stained slide, and the scoring was given according to the average intensity of the red staining in these images. Less than 10% of positive (red) staining for the targeted protein in a sample was considered negative staining, and was scored as 0; score 1 was given to those samples that showed weak red staining; score 2 was given to the samples that revealed mild red staining; and score 3 was given to those samples that had the darkest red staining.
General procedure for the synthesis of SEL-201 (SLV-2436).
All chemicals were purchased at the highest commercial quality and used without further purification, unless otherwise stated. Reactions were monitored by TLC carried out on Sigma-Aldrich silica gel plates (60 F254) using UV light as visualizing agent. Sigma-Aldrich silica gel, 63–200 μm mesh size, was used for column chromatography. Melting points were obtained using a BUCHI Melting Point M-560 instrument and are uncorrected. 1H (400 MHz), 13C (101 MHz) nuclear magnetic resonance spectra were recorded at ambient temperature on Varian Mercury VX. Chemical shifts (δ) are reported in parts per million and coupling constants (J) are given in hertz. The following abbreviations were used in Supplemental Figure 3 to explain the multiplicities: s, singlet; d, doublet; dd, doublet of doublets; m, multiplet. Liquid chromatography–mass spectrometry analyses were performed on Bruker Amazon SL. Analytical separation was carried out on Waters Symmetry column C18 3.9 × 150 mm 5 μm using a flow rate of 1.2 ml/min with detection at 254 nm. The mobile phase was water (solvent A) and acetonitrile (solvent B) both containing 0.1% formic acid. High-resolution mass spectrometry analysis was performed on a Q Exactive Focus Hybrid Quadrupole-Orbitrap Mass Spectrometer. Yields refer to chromatographically and spectroscopically pure compounds unless otherwise stated. The detailed protocol for chemical synthesis and characterization of SEL201-88 can be found in Supplemental Figure 3.
In vitro kinase assay and selectivity kinome panel.
In vitro kinase assay was conducted using ADP-Glo kinase assay (Promega) according to the manufacturer’s protocol. MNK1 or MNK2 recombinant proteins were purchased from Carna. In brief, MNK1 or MNK2 recombinant proteins first were preincubated with serial dilutions of SEL201 or 2% DMSO (buffer used in MNK1 kinase assay: 60 mM HEPES, pH 7.5, 3 mM MgCl2, 3 mM MnCl2, 50 μg/ml PEG20.000; buffer used in MNK2 kinase assay: 5 mM MOPS, pH 7.5, 3 mM MgCl2, 0.4 mM EDTA, 0.25 mM DTT). ATP was added at a concentration of 21 μM or 200 μM in MNK1 or MNK2 kinase assay, respectively, to allow the kinase reactions to proceed for 2 hours at room temperature. Secondly, the remaining ADP was converted to ATP by addition of ADP-Glo kinase detection reagent. ATP then reacts with luciferase and luciferin contained in the kinase detection reagent, to generate luminescence. The intensity of luminescence generated from the MNK1 or MNK2 kinase assays was measured using a plate reader (Biotek). Data were normalized to DMSO and were plotted against inhibitor concentrations presented on logarithmic scale and fitted to dose-response curves using the 4-parameter model in GraphPad Prism software. In vitro kinome selectivity of SEL201-88 was investigated using the KINOMEscan (DiscoverX) assay to quantitatively measure interactions between test compounds and 450 kinase assays and disease-relevant mutant variants, and presented using TREEspot Compound Profile Visualization Tool (DiscoverX).
Statistics.
All in vitro experiments were performed in triplicate, and quantitative data are shown as the average of all biological replicates. In vivo experiments were repeated twice, with the number of animals indicated in the figures. All statistical analyses were performed using GraphPad Prism (GraphPad Software). The details of each statistical test used are as follows: 2-way ANOVA (Figure 1, B and D; Figure 6, A and D; Supplemental Figure 1, A, C, E, and F); 2-tailed Student’s t test (Figure 2, A and B; Figure 3, B and C; Figure 6C); χ2 test (Figure 4A and Supplemental Figure 2); 1-way ANOVA (Figure 6B and Supplemental Figure 5B). P values less than 0.05 and 0.01 were considered significant. Error bars on all graphs are represented as the SD of the mean. NS indicates P greater than 0.05 by 2-tailed Student’s t test.
Study approval.
All animal care and experiments were carried out according to rules and regulations established by the Canadian Council of Animal Care, and protocols were approved by the McGill University Animal Care Committee. Consent was obtained for human tissue acquisition according to guidelines set by Peking University Cancer Hospital & Institute and their affiliated local medical authorities. The IHC study on human melanoma samples was approved by the medical ethics committee of Peking University Cancer Hospital & Institute and was conducted according to the Declaration of Helsinki principles.
Author contributions
YZ, SVDR, and WHM designed the research. YZ performed all the experiments except those included in Figure 5. AD, EŻ and M Mikulski synthesized and characterized SEL201-88, and performed in vitro kinase assay and kinome selectivity profiling, and AG performed pharmacokinetic analysis of SEL201-88. TR and K Brzózka designed experiments, developed SEL201-88, and edited relevant manuscript sections. YZ, SVDR, and WHM wrote and edited the paper. WY and JS helped with manuscript editing and in vivo experiments. FH helped with manuscript editing. JG, YK, and M Ma provided all melanoma patient samples and technical support for immunohistochemistry. BH helped establish stable cell lines. QG helped with in vivo experiments. SDdS and JT helped with pathological analysis of C-KIT patient samples. SDdS helped with pathological analysis of all animal lung sections in this study. GEG and FJ provided KIT-mutant melanoma cell lines and edited the manuscript. SH provided reagents and technical support and helped with manuscript editing. CG, K Bijian and MAAJ provided general technical support and manuscript editing. YC and IT provided technical consulting for polysome pro-filing, and IT helped with manuscript editing.
Supplementary Material
Acknowledgments
This article is dedicated to the memory of Mademoiselle Caroline Chagnon. This research is funded by the Canadian Institutes for Health Research (grant MOP-142281, WHM), and (grant MOP-130540, SH), and the Canadian Cancer Society (grant 703811, WHM and SH). This research was supported by the Rossy Cancer Network. SH is supported by a CRC Chair in Functional Genomics. This study also received travel grants from Québec/Wallonie-Bruxelles International for scientific exchange between Jewish General Hospital, McGill University (SVDR), and Institut Jules Bordet, Université Libre de Bruxelles (GEG). Development of MNK1/2 inhibitors by Selvita S.A. has been cofinanced by the National Centre for Research and Development, INNOTECH Program (INNOTECH-K1/HI1/16/157483/NCBR/12). The authors thank Audrey Emond for technical advice, and Michael Witcher and Leon C. van Kempen for helpful discussion.
Version 1. 10/16/2017
Electronic publication
Version 2. 11/01/2017
Print issue publication
Funding Statement
This study also received travel grants from Québec/Wallonie-Bruxelles International for scientific exchange between Jewish General Hospital, McGill University and Institut Jules Bordet, Université Libre de Bruxelles.
Footnotes
Conflict of interest: A. Dreas, E. Żyłkiewicz, T. Rzymski, K. Brzózka, and A. Golas are employees of Selvita S.A.
Reference information: J Clin Invest. 2017;127(11):4179–4192.https://doi.org/10.1172/JCI91258.
Contributor Information
Yao Zhan, Email: yao.zhan@mail.mcgill.ca.
Jun Guo, Email: guoj307@126.com.
William Yang, Email: zhuolin.yang@mail.mcgill.ca.
Christophe Goncalves, Email: goncalves.christophe@gmail.com.
Tomasz Rzymski, Email: tomasz.rzymski@selvita.com.
Agnieszka Dreas, Email: agnieszka.dreas@selvita.com.
Eliza Żyłkiewicz, Email: eliza.majewska@selvita.com.
Maciej Mikulski, Email: maciej.mikuiski@selvita.com.
Krzysztof Brzózka, Email: krzysztof.brzozka@selvita.com.
Aniela Golas, Email: aniela.golas@selvita.com.
Yan Kong, Email: k-yan08@163.com.
Meng Ma, Email: mameng1223@sina.com.
Fan Huang, Email: fan.huang@mail.mcgill.ca.
Bonnie Huor, Email: bonnie.huor@mail.mcgill.ca.
Qianyu Guo, Email: qianyu.guo@rocketmail.com.
Sabrina Daniela da Silva, Email: sabrina.wurzba@gmail.com.
Jose Torres, Email: yosefmeir18@gmail.com.
Yutian Cai, Email: cyt456@gmail.com.
Ivan Topisirovic, Email: ivan.topisirovic@mcgill.ca.
Jie Su, Email: jacksu2003@yahoo.ca.
Krikor Bijian, Email: krikor.bijian@mail.mcgill.ca.
Sidong Huang, Email: sidong.huang@mcgill.ca.
Fabrice Journe, Email: fabrice.journe@bordet.be.
Sonia V. del Rincón, Email: sonia.delrincon@mcgill.ca.
References
- 1.Forman SB, et al. Is superficial spreading melanoma still the most common form of malignant melanoma? J Am Acad Dermatol. 2008;58(6):1013–1020. doi: 10.1016/j.jaad.2007.10.650. [DOI] [PubMed] [Google Scholar]
- 2.Chan KK, Chan RC, Ho RS, Chan JY. Clinical patterns of melanoma in Asians: 11-year experience in a tertiary referral center. Ann Plast Surg. 2016;77(suppl 1):S6–S11. doi: 10.1097/SAP.0000000000000731. [DOI] [PubMed] [Google Scholar]
- 3.Kong Y, et al. Large-scale analysis of KIT aberrations in Chinese patients with melanoma. Clin Cancer Res. 2011;17(7):1684–1691. doi: 10.1158/1078-0432.CCR-10-2346. [DOI] [PubMed] [Google Scholar]
- 4.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24(26):4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 5.Carvajal RD, et al. KIT as a therapeutic target in metastatic melanoma. JAMA. 2011;305(22):2327–2334. doi: 10.1001/jama.2011.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo J, et al. Phase II, open-label, single-arm trial of imatinib mesylate in patients with metastatic melanoma harboring c-Kit mutation or amplification. J Clin Oncol. 2011;29(21):2904–2909. doi: 10.1200/JCO.2010.33.9275. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Network Genomic Classification of Cutaneous Melanoma. Cell. 2015;161(7):1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Todd JR, Scurr LL, Becker TM, Kefford RF, Rizos H. The MAPK pathway functions as a redundant survival signal that reinforces the PI3K cascade in c-Kit mutant melanoma. Oncogene. 2014;33(2):236–245. doi: 10.1038/onc.2012.562. [DOI] [PubMed] [Google Scholar]
- 9.Todd JR, Becker TM, Kefford RF, Rizos H. Secondary c-Kit mutations confer acquired resistance to RTK inhibitors in c-Kit mutant melanoma cells. Pigment Cell Melanoma Res. 2013;26(4):518–526. doi: 10.1111/pcmr.12107. [DOI] [PubMed] [Google Scholar]
- 10.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khosravi S, Tam KJ, Ardekani GS, Martinka M, McElwee KJ, Ong CJ. eIF4E is an adverse prognostic marker of melanoma patient survival by increasing melanoma cell invasion. J Invest Dermatol. 2015;135(5):1358–1367. doi: 10.1038/jid.2014.552. [DOI] [PubMed] [Google Scholar]
- 12.Carter JH, et al. Phosphorylation of eIF4E serine 209 is associated with tumour progression and reduced survival in malignant melanoma. Br J Cancer. 2016;114(4):444–453. doi: 10.1038/bjc.2015.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16(8):1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16(8):1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furic L, et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci U S A. 2010;107(32):14134–14139. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueda T, et al. Combined deficiency for MAP kinase-interacting kinase 1 and 2 (Mnk1 and Mnk2) delays tumor development. Proc Natl Acad Sci U S A. 2010;107(32):13984–13990. doi: 10.1073/pnas.1008136107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robichaud N, et al. Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene. 2015;34(16):2032–2042. doi: 10.1038/onc.2014.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Topisirovic I, Ruiz-Gutierrez M, Borden KL. Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Res. 2004;64(23):8639–8642. doi: 10.1158/0008-5472.CAN-04-2677. [DOI] [PubMed] [Google Scholar]
- 19.Wendel HG, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21(24):3232–3237. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou J, Lam F, Proud C, Wang S. Targeting Mnks for cancer therapy. Oncotarget. 2012;3(2):118–131. doi: 10.18632/oncotarget.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grzmil M, et al. MAP kinase-interacting kinase 1 regulates SMAD2-dependent TGF-β signaling pathway in human glioblastoma. Cancer Res. 2011;71(6):2392–2402. doi: 10.1158/0008-5472.CAN-10-3112. [DOI] [PubMed] [Google Scholar]
- 22.Fan S, Ramalingam SS, Kauh J, Xu Z, Khuri FR, Sun SY. Phosphorylated eukaryotic translation initiation factor 4 (eIF4E) is elevated in human cancer tissues. Cancer Biol Ther. 2009;8(15):1463–1469. doi: 10.4161/cbt.8.15.8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyman K, et al. Multicenter Phase II trial of high-dose imatinib mesylate in metastatic melanoma: significant toxicity with no clinical efficacy. Cancer. 2006;106(9):2005–2011. doi: 10.1002/cncr.21834. [DOI] [PubMed] [Google Scholar]
- 24.Hodi FS, et al. Imatinib for melanomas harboring mutationally activated or amplified KIT arising on mucosal, acral, and chronically sun-damaged skin. J Clin Oncol. 2013;31(26):3182–3190. doi: 10.1200/JCO.2012.47.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodman SE, et al. Activity of dasatinib against L576P KIT mutant melanoma: molecular, cellular, and clinical correlates. Mol Cancer Ther. 2009;8(8):2079–2085. doi: 10.1158/1535-7163.MCT-09-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyle M, Long GV. Diagnosis and treatment of KIT-mutant metastatic melanoma. J Clin Oncol. 2013;31(26):3176–3181. doi: 10.1200/JCO.2013.50.4662. [DOI] [PubMed] [Google Scholar]
- 27.Antonescu CR, et al. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin Cancer Res. 2005;11(11):4182–4190. doi: 10.1158/1078-0432.CCR-04-2245. [DOI] [PubMed] [Google Scholar]
- 28.DiNitto JP, Wu JC. Molecular mechanisms of drug resistance in tyrosine kinases cAbl and cKit. Crit Rev Biochem Mol Biol. 2011;46(4):295–309. doi: 10.3109/10409238.2011.578612. [DOI] [PubMed] [Google Scholar]
- 29.Lindauer M, Hochhaus A. Dasatinib. Recent Results Cancer Res. 2014;201:27–65. doi: 10.1007/978-3-642-54490-3_2. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, et al. ZYG11A serves as an oncogene in non-small cell lung cancer and influences CCNE1 expression. Oncotarget. 2016;7(7):8029–8042. doi: 10.18632/oncotarget.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485(7396):55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabian MA, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23(3):329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 33.Harbaum L, Marx A, Goekkurt E, Schafhausen P, Atanackovic D. Treatment with dasatinib for chronic myeloid leukemia following imatinib-induced hepatotoxicity. Int J Hematol. 2014;99(1):91–94. doi: 10.1007/s12185-013-1474-x. [DOI] [PubMed] [Google Scholar]
- 34.Yang X, et al. Autophagy protects against dasatinib-induced hepatotoxicity via p38 signaling. Oncotarget. 2015;6(8):6203–6217. doi: 10.18632/oncotarget.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong JH, et al. Early imatinib-mesylate-induced hepatotoxicity in chronic myelogenous leukaemia. Acta Haematol. 2007;118(4):205–208. doi: 10.1159/000111092. [DOI] [PubMed] [Google Scholar]
- 36.Jabbour E, Deininger M, Hochhaus A. Management of adverse events associated with tyrosine kinase inhibitors in the treatment of chronic myeloid leukemia. Leukemia. 2011;25(2):201–210. doi: 10.1038/leu.2010.215. [DOI] [PubMed] [Google Scholar]
- 37.Li P, et al. Inhibition of Mnk enhances apoptotic activity of cytarabine in acute myeloid leukemia cells. Oncotarget. 2016;7(35):56811–56825. doi: 10.18632/oncotarget.10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell JB, et al. MNK inhibition disrupts mesenchymal glioma stem cells and prolongs survival in a mouse model of glioblastoma. Mol Cancer Res. 2016;14(10):984–993. doi: 10.1158/1541-7786.MCR-16-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lock R, et al. Cotargeting MNK and MEK kinases induces the regression of NF1-mutant cancers. J Clin Invest. 2016;126(6):2181–2190. doi: 10.1172/JCI85183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wheater MJ, Johnson PW, Blaydes JP. The role of MNK proteins and eIF4E phosphorylation in breast cancer cell proliferation and survival. Cancer Biol Ther. 2010;10(7):728–735. doi: 10.4161/cbt.10.7.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flørenes VA, Faye RS, Maelandsmo GM, Nesland JM, Holm R. Levels of cyclin D1 and D3 in malignant melanoma: deregulated cyclin D3 expression is associated with poor clinical outcome in superficial melanoma. Clin Cancer Res. 2000;6(9):3614–3620. [PubMed] [Google Scholar]
- 42.Bales E, et al. The low molecular weight cyclin E isoforms augment angiogenesis and metastasis of human melanoma cells in vivo. Cancer Res. 2005;65(3):692–697. [PubMed] [Google Scholar]
- 43.Kluger HM, et al. A phase 2 trial of dasatinib in advanced melanoma. Cancer. 2011;117(10):2202–2208. doi: 10.1002/cncr.25766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shoushtari AN, et al. The efficacy of anti-PD-1 agents in acral and mucosal melanoma. Cancer. 2016;122(21):3354–3362. doi: 10.1002/cncr.30259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roux PP, Topisirovic I. Regulation of mRNA translation by signaling pathways. Cold Spring Harb Perspect Biol. 2012;4(11):a012252. doi: 10.1101/cshperspect.a012252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng Y, et al. SBI-0640756 attenuates the growth of clinically unresponsive melanomas by disrupting the eIF4F translation initiation complex. Cancer Res. 2015;75(24):5211–5218. doi: 10.1158/0008-5472.CAN-15-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhan Y, et al. The role of eIF4E in response and acquired resistance to vemurafenib in melanoma. J Invest Dermatol. 2015;135(5):1368–1376. doi: 10.1038/jid.2015.11. [DOI] [PubMed] [Google Scholar]
- 48.Kosciuczuk EM, et al. Merestinib blocks Mnk kinase activity in acute myeloid leukemia progenitors and exhibits antileukemic effects in vitro and in vivo. Blood. 2016;128(3):410–414. doi: 10.1182/blood-2016-02-698704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kosciuczuk EM, Saleiro D, Platanias LC. Dual targeting of eIF4E by blocking MNK and mTOR pathways in leukemia. Cytokine. 2017;89:116–121. doi: 10.1016/j.cyto.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gandin V, et al. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J Vis Exp. 2014;(87):51455. doi: 10.3791/51455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.