Significance
Cells release vesicles containing selectively packaged cargo, including RNA, into the extracellular environment. Prior studies have identified RNA inside extracellular vesicles (EVs), but due to limitations of conventional sequencing methods, highly structured and posttranscriptionally modified RNA species were not effectively captured. Using an alternative sequencing approach (thermostable group II intron reverse transcriptase sequencing, TGIRT-seq), we found that EVs contain abundant small noncoding RNA species, including full-length transfer RNAs and Y RNAs. Using a knockout cell line, we obtained evidence that the RNA-binding protein YBX1 plays a role in sorting small noncoding RNAs into a subpopulation of EVs termed exosomes. These experiments expand our understanding of EV–RNA composition and provide insights into how RNA is sorted into EVs for cellular export.
Keywords: extracellular vesicle, posttranscriptional modification, RNA-binding protein, RNA-seq, thermostable group II intron reverse transcriptase
Abstract
RNA is secreted from cells enclosed within extracellular vesicles (EVs). Defining the RNA composition of EVs is challenging due to their coisolation with contaminants, lack of knowledge of the mechanisms of RNA sorting into EVs, and limitations of conventional RNA-sequencing methods. Here we present our observations using thermostable group II intron reverse transcriptase sequencing (TGIRT-seq) to characterize the RNA extracted from HEK293T cell EVs isolated by flotation gradient ultracentrifugation and from exosomes containing the tetraspanin CD63 further purified from the gradient fractions by immunoisolation. We found that EV-associated transcripts are dominated by full-length, mature transfer RNAs (tRNAs) and other small noncoding RNAs (ncRNAs) encapsulated within vesicles. A substantial proportion of the reads mapping to protein-coding genes, long ncRNAs, and antisense RNAs were due to DNA contamination on the surface of vesicles. Nevertheless, sequences mapping to spliced mRNAs were identified within HEK293T cell EVs and exosomes, among the most abundant being transcripts containing a 5′ terminal oligopyrimidine (5′ TOP) motif. Our results indicate that the RNA-binding protein YBX1, which is required for the sorting of selected miRNAs into exosomes, plays a role in the sorting of highly abundant small ncRNA species, including tRNAs, Y RNAs, and Vault RNAs. Finally, we obtained evidence for an EV-specific tRNA modification, perhaps indicating a role for posttranscriptional modification in the sorting of some RNA species into EVs. Our results suggest that EVs and exosomes could play a role in the purging and intercellular transfer of excess free RNAs, including full-length tRNAs and other small ncRNAs.
Metazoan cells grown in culture release extracellular vesicles (EVs) into the surrounding medium and free vesicles can be found in all bodily fluids (1). EVs can be categorized into multiple classes based on their size, shape, and presumed membrane origin. Exosomes are defined as ∼30–100 nm vesicles that originate from the multivesicular body (MVB) and contain late endosomal markers (2, 3), but there is evidence that biochemically indistinguishable vesicles can bud directly from the plasma membrane (2, 4). Microvesicles or shedding vesicles are generally larger (>200 nm), are more variable in shape and density, and likely originate from the plasma membrane (1, 3, 5). EVs contain various molecular cargos, including proteins, lipids, and RNAs, but how these cargos are sorted into EVs remains unclear.
Since the initial description of RNA in EVs (5–7) and the identification of vesicle-associated circulating transcripts in plasma (8), there has been widespread interest in the possible roles of extracellular RNAs. Seminal EV RNA studies showed the presence of ribonuclease (RNase)-protected mRNA and microRNA (miRNA) species in blood that might be exploited as biomarkers (8) and suggested a role for EVs in the horizontal transfer of genetic information between cells (5–7, 9–11). Several studies implicated miRNAs, in particular, as a mode of intercellular communication based on reporter assays for miRNA function in recipient cells (9–12). Recent studies using sensitive Cre recombinase-based genetic systems in mice have indicated that local and long-range RNA transfer via EVs can occur in vivo (13, 14). High-throughput sequencing of extracellular RNA has identified many other EV-associated RNA biotypes, including various small noncoding RNAs [sncRNA; e.g., small nucleolar RNA (snoRNA), transfer RNA (tRNA), Y RNA, Vault RNA) (15–17), long noncoding RNA (lncRNA) (16, 18), and protein-coding transcripts (16, 19).
The recent application of thermostable group II intron reverse transcriptases (TGIRTs) to sequence low-input RNA from human plasma samples revealed that many circulating sncRNA transcripts are full-length and thus apparently resistant to serum RNases (20). In addition to vesicles, extracellular RNA can be found as circulating ribonucleoprotein particles (RNPs), which cosediment with vesicles during standard EV-isolation protocols culminating in high-speed (>100,000 × g) ultracentifugation (21). The coisolation of RNPs and multiple EV subpopulations has made it difficult to identify the means by which RNA biotypes are exported and how their transfer between cells might occur.
Understanding the mechanisms by which transcripts are sorted into EVs has proven challenging. Argonaute proteins bind mature miRNAs in the cytoplasm, and it has been shown that Argonaute 2 (Ago2)-associated miRNAs can be sorted into exosomes in a process regulated by KRAS signaling (22, 23). We previously used a cell-free reconstitution approach to identify a requirement for the RNA-binding protein (RBP) Y-box protein 1 (YBX1) in the sorting of a non–Ago2-bound miRNA (miR-223) into exosomes (24), suggesting an RBP chaperone-mediated sorting pathway. Other RBPs have been shown to sort miRNAs in other cell types. A sumoylated form of hnRNPA2B1 was shown to bind miRNAs containing a tetranucleotide motif (GGAG) that are exported from T cells (25). SYNCRIP (hnRNPQ) recognizes a GCUG sequence at the 3′ end of miRNAs from hepatocytes for sorting into EVs (26). Also in hepatic cells, HuR (ELAVL1) has been shown to control the export of miR-122 during the starvation stress response (27). Despite several examples of RBP-mediated sorting, it is not clear if RBPs play a broad role in defining the RNA composition of EVs or package only a select few transcripts. Nor is it clear to what extent RBPs play an active role in RNA sorting or are simply present in RNPs that are passively captured by membrane invagination processes that generate EVs. Indeed, to date there has been no description of the role that RBPs play in broadly determining the RNA content of EVs.
Here we present our observations from sequencing RNAs extracted from sucrose density gradient-purified EVs and further purified, CD63-immunoisolated exosomes from HEK293T cells using TGIRT sequencing (TGIRT-seq). We found that sncRNAs, including tRNAs, dominated EV-RNA composition. TGIRT-seq revealed that most of the tRNAs were full-length, mature transcripts, rather than fragments as previously reported, and contain a novel posttranscriptional modification that is not prevalent in cytoplasmic tRNAs. Comparison of the RNA content of exosomes secreted by wild-type and YBX1-null cells indicated that YBX1 plays a role in the export of abundant sncRNA species, suggesting that RBPs play a primary role in defining the total RNA composition of exosomes.
Results
Most EV-Associated Transcripts Are Encapsulated Within EVs.
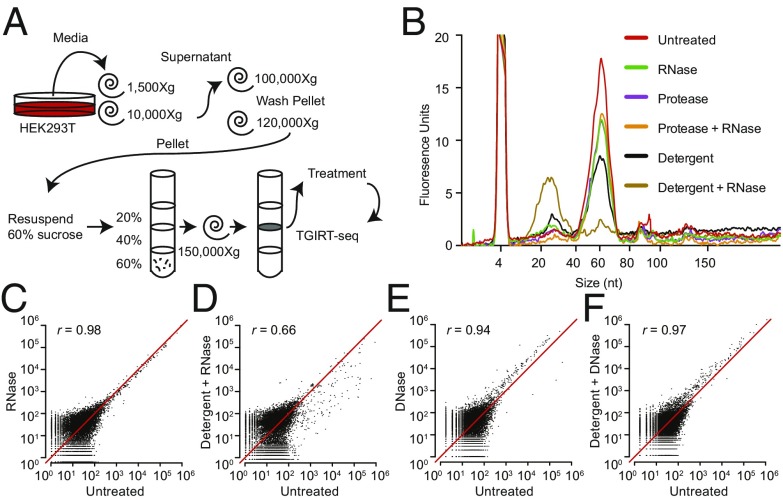
EVs encapsulate RNAs, rendering them resistant to degradation by exogenous RNases (7–9). However, prior studies have focused primarily on the RNase protection of individual transcripts rather than the global RNA content. To broadly define the RNA composition of EVs, we employed TGIRT-seq in combination with RNase protection assays in the presence or absence of a detergent to distinguish transcripts inside or outside of membrane vesicles. EVs were isolated from HEK293T cells by differential centrifugation followed by flotation gradient ultracentrifugation into a discontinuous sucrose gradient to purify vesicles away from RNPs (Fig. 1A). After flotation, EVs were harvested from the 20%/40% sucrose interface and either left untreated or treated with protease and/or RNase in the absence or presence of nonionic detergent (1% Triton X-100; TX-100). Bioanalyzer traces of the RNA after each condition showed a predominant peak of 60–70 nt, which was resistant to RNase under all treatment conditions except in the presence of detergent (Fig. 1B). These results corroborate previous studies showing that the predominant RNAs in EVs are ∼60–70 nt (9, 16). However, previous RNA-seq experiments failed to identify the high abundance transcripts corresponding to this size distribution, indicating that conventional RNA-seq methodology does not efficiently capture these species.
Fig. 1.
EV-associated transcripts before and after treatment of EVs with nucleases and protease in the absence or presence of detergent. (A) Schematic of the workflow for EV isolation, enzyme and detergent treatment, and TGIRT-seq. (B) Bioanalyzer traces (small RNA chip) showing the size distribution of RNAs associated with the EVs before and after the indicated treatments. (C–F) Scatter plots comparing DESeq2-normalized read counts between untreated EVs (no detergent and no nuclease) and those treated with RNase or DNase in the absence or presence of detergent. Correlations were computed as Pearson’s correlation coefficients (r).
Since the retroviral reverse transcriptases (RTs) used routinely to generate RNA-seq libraries underrepresent species with significant secondary structure and are impeded by certain posttranscriptional modifications, we utilized a TGIRT enzyme [TGIRT-III (InGex), a derivative of GsI-IIC RT] to generate cellular and EV libraries for Illumina sequencing (20, 28). The resulting TGIRT-seq datasets for the RNase-protection experiments were denoted EVR1-6 (SI Appendix, Table S1). Scatter plots based on these datasets showed that treatment of the EV fraction with protease and/or RNase in the absence of detergent had little effect on the relative abundance of different transcripts (Fig. 1C and SI Appendix, Fig. S1). In contrast, upon the addition of detergent, the relative abundance of most transcripts was substantially decreased by RNase treatment (Fig. 1D). Pairwise comparisons showed little change under most treatment conditions [Pearson’s correlation coefficient (r) ≥ 0.94], except when detergent was present (untreated vs. detergent only; r = 0.86) and especially when detergent was added together with RNase (r = 0.49–0.70) (SI Appendix, Fig. S1). The decreased correlation between untreated samples and those treated with detergent alone suggested the presence of endogenous RNases in the EV fractions.
To assess DNA contamination in the EV–RNA isolation, the same treatment conditions we used for RNase protection were replicated for DNase protection using a different batch of EVs (datasets EVD1–6; SI Appendix, Table S1). Because the TGIRT enzyme can copy DNA fragments efficiently (20, 29), it was possible that some of the sequencing reads were derived from contaminating DNA. As expected, there was no significant effect of detergent or protease treatment on the relative abundance of the large majority of transcripts (r ≥ 0.97), but there was a minor effect in some datasets when DNase was added in the presence or absence of detergent (r = 0.94–0.97, Fig. 1 E and F; and r = 0.84–0.97 in the larger set of pairwise comparisons, SI Appendix, Fig. S2). Experiments below suggested that the latter reflects cellular DNA that adheres to the surface of the vesicles, even after flotation gradient centrifugation. Together, these results demonstrate that the majority of transcripts recovered from a purification employing a flotation gradient ultracentrifugation step are encapsulated within EVs.
The Most Abundant Transcripts in EVs Are Full-Length tRNAs and Other SncRNAs.
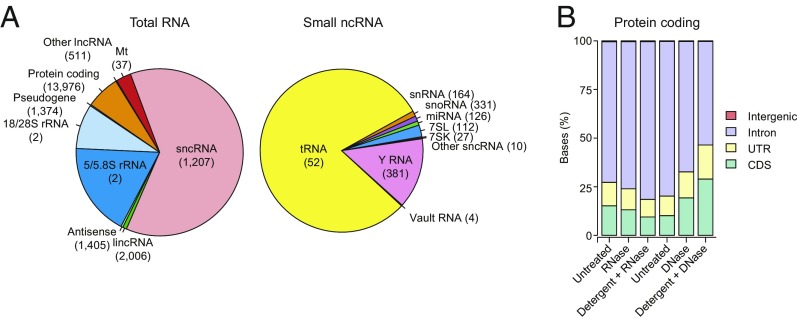
Analysis of the TGIRT-seq datasets revealed that the most abundant transcripts based on read count in the untreated EVs from the RNase-protection experiments were sncRNAs (62%), followed by ribosomal RNA (27%) and transcripts of protein-coding genes (6.6%), with the remaining categories accounting for just 4.4% (Fig. 2A). Among sncRNAs, by far the most abundant species were tRNAs (50% and 80% of the total and sncRNA reads, respectively), followed by Y RNAs (8.7% and 14% of the total and sncRNA reads, respectively; Fig. 2A). Notably, scatter plots showed that the relative abundance of different tRNA species in EVs matched that in the cells from which they were secreted (Spearman’s correlation coefficient ρ = 0.98; SI Appendix, Fig. S3A). Pie charts that were essentially identical to those for the untreated sample were obtained for EVs treated with RNase or with protease and RNase in the absence of detergent, suggesting that there was no substantial RNA or RNP presence on the outside of EVs (SI Appendix, Fig. S4 A and B). TGIRT-seq revealed only low levels of miRNAs in EVs compared with other transcripts (<0.5% of total reads). Due to secondary structure and posttranscriptional modifications, tRNAs and other sncRNAs may have been underrepresented in previous EV small RNA-seq libraries generated by retroviral RTs, thus causing miRNAs to be overrepresented, and previous studies often used extraction methods best suited to small RNAs and/or size selections that enriched for miRNAs. Additionally, the version of TGIRT-seq used here relied on a bead-based library clean-up to remove adapter dimers (30). This method enabled minimal RNA inputs but may have resulted in underrepresentation of miRNAs, which give library products that are close in size to adapter dimers.
Fig. 2.
RNA biotypes associated with EVs and the effect of nuclease and detergent treatment on reads mapping to different regions of protein-coding genes. (A) Pie charts of percentage of mapped total RNA and sncRNA reads (Left and Right, respectively) corresponding to the indicated features for untreated EVs. The number of genes represented for each RNA biotype is shown in parentheses. tRNA gene counts are for tRNA genes grouped by anticodon (n = 52, including two families of suppressive tRNAs that potentially recognize stop codons, SelCysTCA, and iMetCAT; see SI Appendix, SI Methods). SncRNA gene counts include pseudogenes. Pie charts of RNA biotypes within the same EVs after RNase or Protease + RNase treatment are shown in SI Appendix, Fig. S4 A and B. (B) Stacked bar graphs of the percentage of protein-coding gene reads mapping to coding sequences (CDS), untranslated regions (UTR), introns, and intergenic regions before and after different treatments. Different batches of EVs were used for the RNase and DNase protection experiments (left three bars and right three bars, respectively).
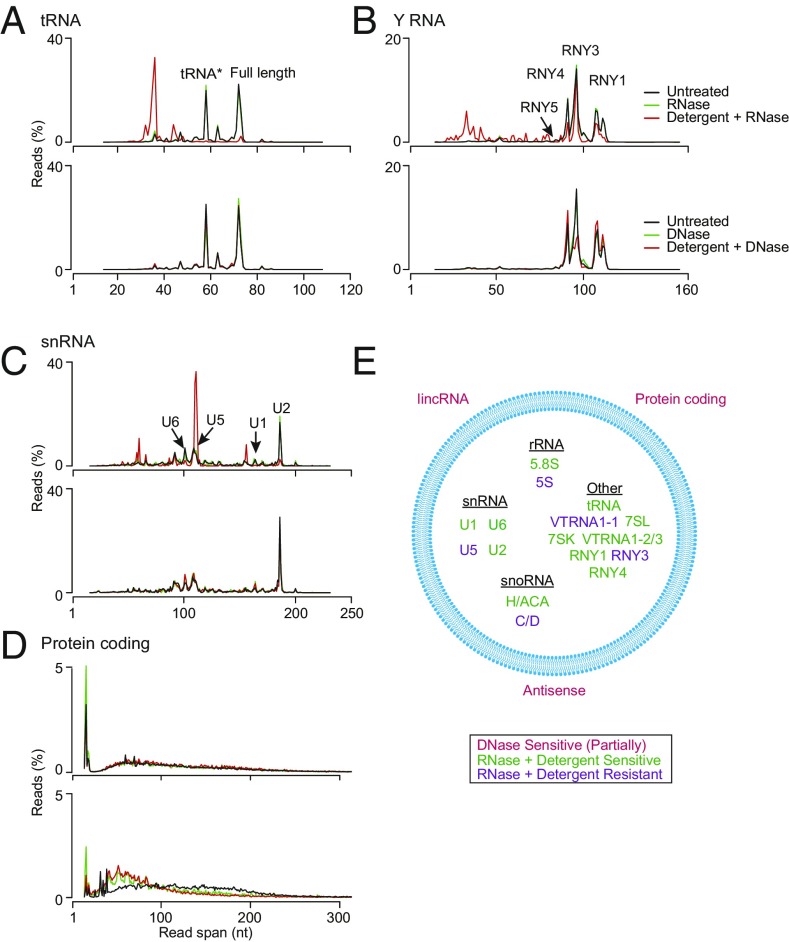
Significantly, read-span analysis before and after different treatments and Integrative Genomics Viewer (IGV) coverage plots and alignments showed that most of the tRNAs and other sncRNAs present within EVs were full-length, mature transcripts (Fig. 3A and SI Appendix, Figs. S5 and S6). In the case of tRNAs, the most abundant RNA species in EVs, read-span analysis showed two prominent peaks, one corresponding to full-length tRNAs and the other to 5′-truncated reads beginning in the d-loop (denoted tRNA*), which results below suggested corresponds to a tRNA modification that impedes reverse transcription by GsI-IIC RT. IGV alignments showed that the full-length tRNA reads began precisely at the processed 5′ end of the mature tRNA (or with an extra noncoded G residue in the case of histidyl tRNA), had posttranscriptional modifications expected for mature tRNAs, and in most cases ended with the posttranscriptionally added 3′ CCA (SI Appendix, Fig. S6). Both the full-length tRNAs and tRNA* species were fully sensitive to RNase in the presence but not absence of detergents and insensitive to DNase, confirming that most tRNAs were enclosed within vesicles (Fig. 3A).
Fig. 3.
Characterization of EV–RNA biotypes by read-span analysis. (A–D) Read spans from paired-end sequences of tRNA (A), Y RNA (B), small nuclear RNA (snRNA) (C), and reads mapping to protein-coding genes (D) before and after the indicated treatments (keys at upper right). The plots show the percentage of total reads of different read spans for the mapped paired-end reads for each biotype. Peaks for different subspecies of Y RNA and snRNA are indicated on the plots. Results for other RNA biotypes (7SL RNA, snoRNA, 7SK RNA, Vault RNA, 5/5.8S rRNA, and 18/28S rRNA) are shown in SI Appendix, Fig. S5. (E) Summary schematic showing the identity and location of each biotype inside or outside of EVs (blue circle) inferred from nuclease and detergent sensitivity of the TGIRT-seq reads. The peak corresponding to tRNA reads with a strong stop at position 16 in the D-loop is denoted tRNA*.
In addition to full-length tRNAs, read-span analysis and IGV alignments showed the presence of full-length Y RNAs (RNY1, 3, 4, and 5), 7SL and 7SK RNA, both H/ACA- and C/D-box snoRNAs, all species of snRNA, and three species of Vault RNA (VTRNA1-1, 1-2, and 1-3), whereas only short read spans were found for 18S and 28S rRNA, as expected for Illumina sequencing (Fig. 3 and SI Appendix, Figs. S5 and S6). The presence of nuclear snRNAs and 7SK RNA and nucleolar snoRNAs within cytoplasmically derived EVs parallels previous observations for RNAs associated with the cytoplasmic interferon-induced innate immunity protein IFIT5 as well as for host cell RNAs packaged in retroviral virions and may reflect the packaging of aberrantly localized or nonfunctional nuclear RNAs that are not associated with their normal partner proteins (31–33). Most species of sncRNAs were degraded by RNase in the presence of detergent, but several (RNY3, VTRNA1-1, C/D-box snoRNAs, 5S rRNA, and U5 snRNA) were resistant to RNase, raising the possibility that they were present in vesicles as RNPs. All of these sncRNA reads were resistant to DNase in the presence or absence of detergent as expected (Fig. 3 and SI Appendix, Fig. S5).
tRNA and Y RNA fragments, some of which result from cleavage by RNases that leave 3′ phosphates, have been suggested to play important roles in regulating cellular processes and in intercellular signaling (34–36). Because 3′ phosphates impede the TGIRT template-switching reaction used for RNA-seq adapter addition (28), it was possible that they were underrepresented in the TGIRT-seq libraries. To check specifically for such RNA fragments, we compared EV RNAs before and after treatment using the phosphatase activity of T4 polynucleotide kinase to remove 3′ phosphates (datasets GEV and GEV-3′P, respectively; SI Appendix, Table S2). This treatment resulted in a substantial increase in the reads mapping to 18S and 28S rRNA fragments, as well as long RNA fragments that overlap miRNA loci (SI Appendix, Fig. S4 C and D). However, it had no effect on the read-span profile for cellular tRNAs (SI Appendix, Fig. S5G) and only a minor effect on the read-span profiles for Y RNAs (SI Appendix, Fig. S5H). These findings confirmed that HEK293T EVs contained predominantly full-length tRNAs and Y RNAs and that tRNA and Y RNA fragments were not abundant in these EVs.
Our nuclease sensitivity experiments identified reads mapping to protein-coding genes (6.6–10% in the untreated controls for the RNase- and DNase-protection experiments). However, most of these reads mapped to introns rather than coding sequences, and the proportion of reads mapping to introns decreased after DNase treatment in the presence or absence of detergent, suggesting that a significant proportion of these reads came from contaminating DNA (Fig. 2B). By read-span analysis, the reads mapping to protein-coding genes appeared as a broad distribution from ∼50–200 nt, which was degraded to shorter fragments after DNase but not RNase treatment (Fig. 3D). Further, this DNase sensitivity was observed irrespective of the presence or absence of detergent in the reaction, indicating DNA contamination on the surface of vesicles. This surface DNA contamination also accounted for a substantial proportion of the reads mapping to long intergenic noncoding RNAs (lincRNAs), antisense RNAs, and other lncRNAs (SI Appendix, Fig. S5 I–K).
In summary, by combining nuclease treatments with read-span analysis, we found that EVs contained predominantly full-length sncRNAs, particularly tRNAs, and were able to categorize EV-encapsulated RNAs into two distinct classes (Fig. 3E): those sensitive to RNase in the presence of detergent (full-length tRNAs, 5.8S rRNA, RNY1 and -4, VTRNA1-2 and -1-3, 7SL and 7SK RNA, H/ACA-box snoRNA, and U1, U2, and U6 snRNAs), and those resistant to RNase in the presence of detergent, consistent with protection by bound proteins in RNPs (RNY3, 5S rRNA, C/D-box snoRNA, U5 snRNA, and VTRNA1-1). Our experiments also indicated previously unrecognized cellular DNA contamination that adhered to the surface of EVs even after flotation gradient centrifugation and contributed a substantial proportion of the reads mapping to protein-coding genes, lncRNAs, and antisense RNAs.
HEK293T Cell Exosomes Contain Spliced mRNAs.
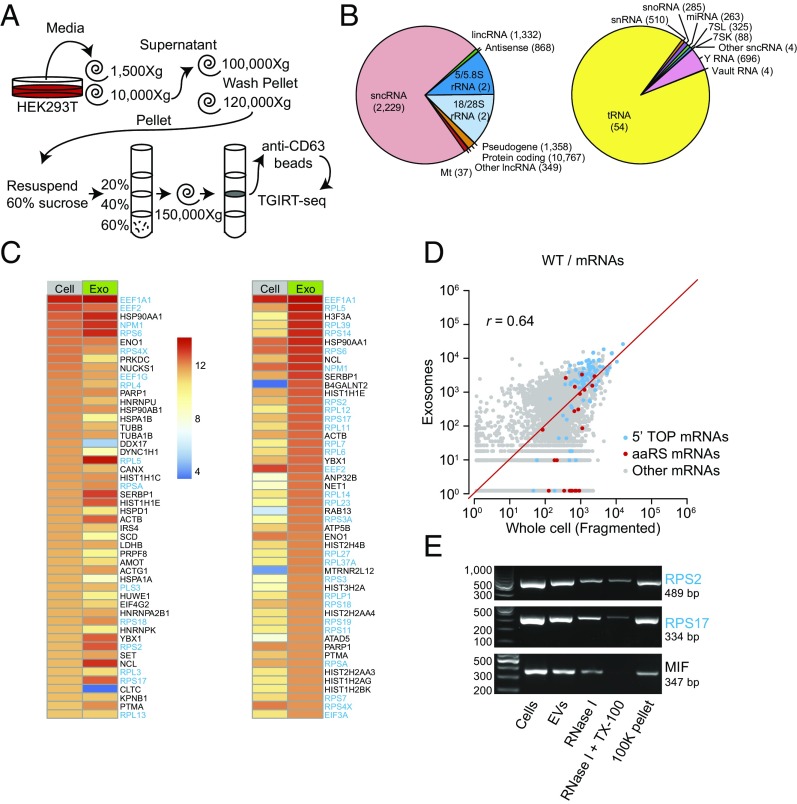
Because cells release multiple populations of EVs, we focused further analysis on a subpopulation of EVs that contain the tetraspanin CD63, likely originating from an endosome-derived MVB, the classical definition of an exosome (24, 37). To isolate CD63+ vesicles (exosomes), an additional anti-CD63 antibody affinity purification step was included after the flotation gradient centrifugation (Fig. 4A). For the experiments below, we isolated whole-cell and CD63+ exosomal RNAs from the same cultures and treated the RNAs with DNase before TGIRT-seq library preparation. Whole-cell RNAs were depleted of rRNA and either fragmented before RNA-seq for quantitative comparisons of RNA levels or analyzed directly without fragmentation to preserve intact sncRNAs (30) (datasets WC, WCF, and WEX; SI Appendix, Table S3).
Fig. 4.
Characterization of RNA biotypes and spliced mRNAs present in CD63+ exosomes. (A) Exosome (CD63+ EV) purification and TGIRT-seq schematic. (B) Pie charts showing the percentage of different RNA biotypes in DNase-treated exosomal RNAs from wild-type HEK293T cells. The pie charts show the percentage of mapped total RNA and sncRNA reads (Left and Right, respectively) corresponding to the indicated features. The number of genes represented for each biotype is shown in parentheses. tRNA gene counts are for tRNA genes grouped by anticodon (see Fig. 2 legend and SI Appendix, SI Methods), and sncRNA gene counts include pseudogenes. (C and D) Heat maps (C) and scatter plots (D) showing the relative abundance of different mRNAs in DNase-treated whole-cell (fragmented) and exosomal RNAs based on normalized read counts. For this analysis, reads that mapped to protein-coding genes in the initial mapping were remapped to the human transcriptome reference sequence (Ensemble GRCh38 Release 76). The heat maps show the 50 most abundant mRNAs ranked by normalized read count in whole cells (left two columns) and exosomes (right two columns), whereas the scatter plots compare all mRNAs. 5′ TOP-containing mRNAs, defined as 73 of 80 ribosomal protein mRNAs plus EEF1A1, EEF1B2, EEF1D, EEF1G, EEF2, EIF3A, EIF3E, EIF3F, EIF3H, EIF4B, HNRNPA1, NAP1L1, NPM1, PABPC1, RACK1, TPT1, and VIM mRNAs (57), are denoted with blue dots; aaRS mRNAs are denoted with red dots. r = Pearson’s correlation coefficient. (E) RT-PCR products obtained from RNA isolated from EVs treated with RNase I in the absence or presence of detergent (TX-100). The expected amplicon sizes for RPS2, RPS17, and MIF mRNAs are given. The left lane shows molecular weight markers.
We confirmed that the RNA content of CD63+ exosomes was broadly similar to that of the total EV population, with TGIRT-seq showing a similar profile of RNA biotypes, including a high proportion of sncRNAs (73% of mapped reads), with the most abundant being tRNAs (66% and 91% of total RNA and sncRNA reads, respectively) and Y RNAs (4.0% and 5.4% of total RNA and sncRNA reads, respectively) and only 1.8% of the reads mapping to protein-coding genes (Fig. 4B). As for EVs, the relative abundance of different tRNA species in exosomes matched that in the cells from which they were secreted (ρ = 0.97; SI Appendix, Fig. S3B).
Our nuclease sensitivity experiments on total EV populations indicated that many of the reads mapping to protein-coding genes reflected DNA contamination on the surface of the vesicles but left open the possibility that EVs contained smaller amounts of mRNAs or fragments thereof (Fig. 3D). To address this possibility for CD63+ exosomes, we analyzed protein-coding reads that mapped to a human transcriptome reference sequence (Ensembl GRCh38) and compared them to similarly mapped reads for whole-cell RNAs from the same cultures (Fig. 4 C–E). In the case of exosomes, the reads mapping to the human transcriptome corresponded to only 0.6% of the total mapped reads (Fig. 4B) but contained only 0.5–1.2% intron sequences (SI Appendix, Fig. S7A), indicating that most if not all corresponded to mRNAs or fragments thereof.
Heat maps of the top 50 protein-coding genes showed both similarities and differences between the most abundant mRNA species in cells and exosomes (Fig. 4C). Notably, many of the most abundant mRNAs by normalized read count in the whole-cell and exosomal RNA preparations encode ribosomal proteins or other proteins involved in translation and belong to a family containing a 5′ terminal oligopyrimidine (5′ TOP) sequence, which allows coordinated translational repression of these transcripts (Fig. 4C, gene names highlighted in blue) (38–41).
Scatter plots comparing all putative mRNA species showed some correlation between the relative abundance of different mRNA species in whole cells and exosomes (r = 0.64) (Fig. 4D). The majority of the 5′ TOP mRNAs appeared to be overrepresented in exosomes, but some were underrepresented regardless of whether or not the whole-cell RNAs were fragmented before sequencing (Fig. 4D and SI Appendix, Fig. S7 B and C). By contrast, reads mapping to mRNAs for aminoacyl-tRNA synthetases (aaRSs), which encode proteins that function in protein synthesis but lack a 5′ TOP sequence, did not appear to be similarly overrepresented (Fig. 4D and SI Appendix, Fig. S7 B and C). Since most 5′ TOP mRNAs are relatively short (mean transcript length ∼ 1 kb), we looked to see if there was a general relationship between transcript length and abundance in exosomes compared with cells. We found that exosomes were enriched in transcripts <1.5 kb, possibly contributing to the enrichment for 5′ TOP mRNA sequences (SI Appendix, Fig. S8).
To assess whether 5′ TOP mRNA sequences were inside of vesicles, we compared the read coverage across two abundant exosomal mRNAs, EEF1A and RPS2, after DNase or RNase treatment of EVs in the presence or absence of detergent. We found that reads corresponding to spliced transcripts map across EEF1A and RPS2 exons, unless treated with RNase and detergent, indicating that the mRNA sequences were enclosed in exosomes (SI Appendix, Fig. S9). These results did not distinguish whether these species were full-length mRNA or a series of fragments but minimally demonstrated the presence of multiply spliced mRNA segments inside EVs with an enrichment for 5′ TOP-containing transcripts.
To further confirm that spliced mRNAs were present in EVs, we performed reverse transcription PCR using SuperScript IV and primers that spanned at least two splice junctions for three putative mRNAs (RPS2, RPL17, MIF). Spliced mRNAs were detected in exosomes and showed partial sensitivity to RNase alone that was increased by adding detergent (Fig. 4E). In summary, we provide evidence for the presence of mRNAs in HEK293T cell exosomes with transcripts containing a 5′ TOP sequence motif being particularly abundant.
YBX1-Dependent Exosomal RNAs.
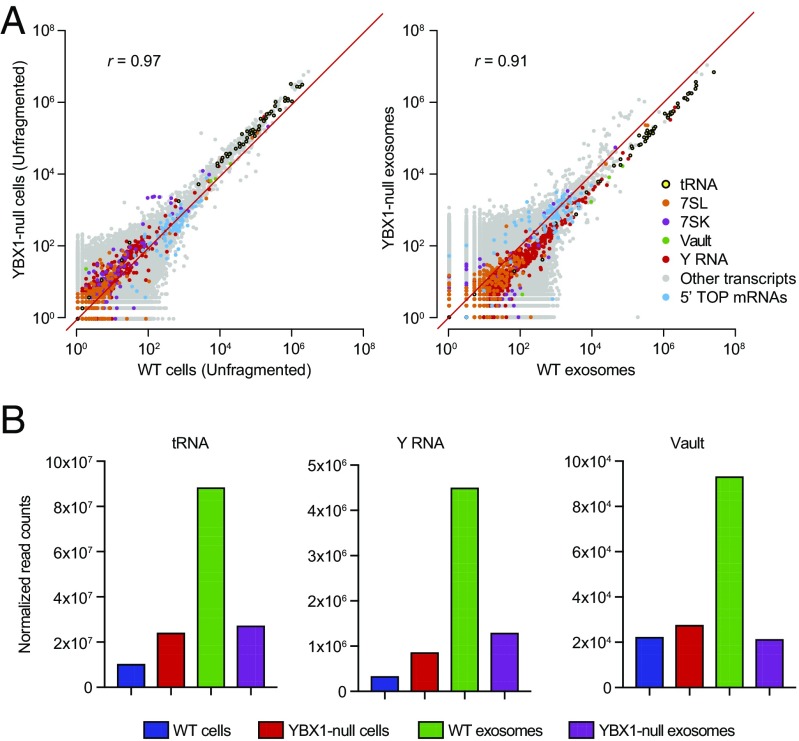
We previously identified an RBP, YBX1, that is present in purified exosomes and directs the selective sorting of some miRNAs (24). We therefore sought to more comprehensively define the YBX1-dependent exosomal RNA composition by performing TGIRT-seq on DNase-treated RNAs from purified wild-type and YBX1-null cell exosomes (datasets WC, WCF, and WEX for wild-type cells and YC, YCF, and YEX for YBX1-null cells; SI Appendix, Table S3). Scatter plots comparing DESeq2-normalized mapped read counts for WT and YBX1-null cellular RNAs did not show dramatic changes in the relative abundance of major exosomal RNA biotypes, except for an increase in moderately expressed cellular tRNAs (Fig. 5A, Left). In contrast, exosomes from YBX1-null cells showed substantially decreased abundance of the large majority of cellular tRNAs, Y RNAs, and Vault RNAs (Fig. 5A, Right), whereas other RNA biotypes were not significantly affected (shown by the composite scatter plot in Fig. 5A, Right and by separate scatter plots for different RNA biotypes in SI Appendix, Fig. S10A). An exception was mitochondrial tRNAs, a minor component of the total reads (Fig. 4B), which appeared to be overrepresented in both YBX1-null cells and exosomes (SI Appendix, Fig. S10B). Bar graphs of normalized read counts showed that tRNAs were 69% decreased, Y RNAs were 71% decreased, and Vault RNAs were 77% decreased in YBX1-null exosomes (Fig. 5B). Conversely, YBX1-null cells showed an increase in the proportion of reads that mapped to cellular tRNAs (2.3-fold), Y RNAs (2.5-fold), and Vault RNAs (1.2-fold) (Fig. 5B), suggesting that blocking cellular export via the exosome pathway resulted in accumulation of these transcripts in cells.
Fig. 5.
Sequencing of cellular and CD63+ exosomal RNA from wild-type and YBX1-null cells. (A) Scatter plots comparing DESeq2-normalized mapped read counts for the indicated biotypes for DNase-treated WT and YBX1-null cellular RNA (unfragmented; Left) and exosomal RNA (Right). (B) Bar graph showing total normalized read counts mapping to tRNA, Y RNA, and Vault RNA in each library. Normalized read counts represent DESeq2-normalized read counts summed for all transcripts annotated for each indicated biotype, including those mapping to pseudogenes, in the GENCODE gene set. Bar graphs showing normalized read counts for different subspecies of Y and Vault RNA are shown in SI Appendix, Fig. S11.
Further analysis showed that the relative abundance of reads mapping to nearly all cellular tRNA genes was decreased by 50% or more in YBX1-null exosomal RNAs, with most decreased by 70–90% (mean proportion KO/WT = 0.31; Fig. 6A). Individual tRNA isoacceptor levels in cells were increased ∼twofold (mean proportion KO/WT = 2.34) (Fig. 6A), further indicating that blocking YBX1-dependent secretion of tRNAs resulted in tRNA accumulation in cells. Thus, the substantial decrease in the proportion of exosomal reads mapping to cellular tRNAs was not confined to a small group of highly expressed tRNA genes. Similarly, different subspecies of Y RNA (RNY1, -3, -4, and -5) and Vault RNA (VTRNA1-1, -1-2, and -1-3) all showed decreases in YBX1-null exosomes (SI Appendix, Fig. S11). Similar decreases in the relative abundance of tRNAs, Y RNAs, and Vault RNAs in YBX1-null exosomes were also observed in two earlier versions of this experiment using non–DNase-treated exosomal RNAs. Together, these results suggest a broad role for YBX1 in packaging cellular tRNA as well as some other sncRNA species.
Fig. 6.
Effect of the YBX1-null mutation on the sorting of different tRNA species into exosomes. (A) Histogram showing the number of tRNA species grouped by anticodon (n = 49) with the indicated ratio of normalized reads in DNase-treated YBX1-null versus wild-type exosomal RNA preparations. (B–D) Read-span (B), stop site (C), and start site (D) distributions for tRNA mapped reads as a percentage of total tRNA mapped reads. The peak representing the tRNA* species with a strong stop at position 16 in the D-loop is indicated (B and D).
Read-span analysis showed little difference in the size distribution of any of these sncRNAs between wild-type and YBX1-null exosomes (SI Appendix, Fig. S12). In both wild-type and YBX1-null exosomes, a high proportion (35%) of tRNA reads spanned the full-length mature tRNA sequence (∼75 nt; Fig. 6B). Most of the tRNA reads began at the 5′ end of the mature tRNA (position 1), and nearly all tRNA reads terminated at the 3′ end of the mature tRNA (positions 72–75; Fig. 6C). The remaining peaks present in Fig. 6B represented 5′ truncated tRNA reads, the most prominent being the tRNA* species with a reverse transcription stop at position 16 in the D-loop (Fig. 6D).
In contrast to sncRNAs, exosomes from YBX1-null cells did not show a decrease in 5′ TOP mRNAs compared with their levels in cells (Fig. 5A, blue dots, compare Left and Right). Therefore, although we cannot conclusively eliminate the possibility of a secondary mutation, our results strongly suggest that YBX1 plays a specific role in defining the RNA content of exosomes by influencing the packaging of tRNAs and other sncRNAs.
An Exosome-Specific tRNA Modification.
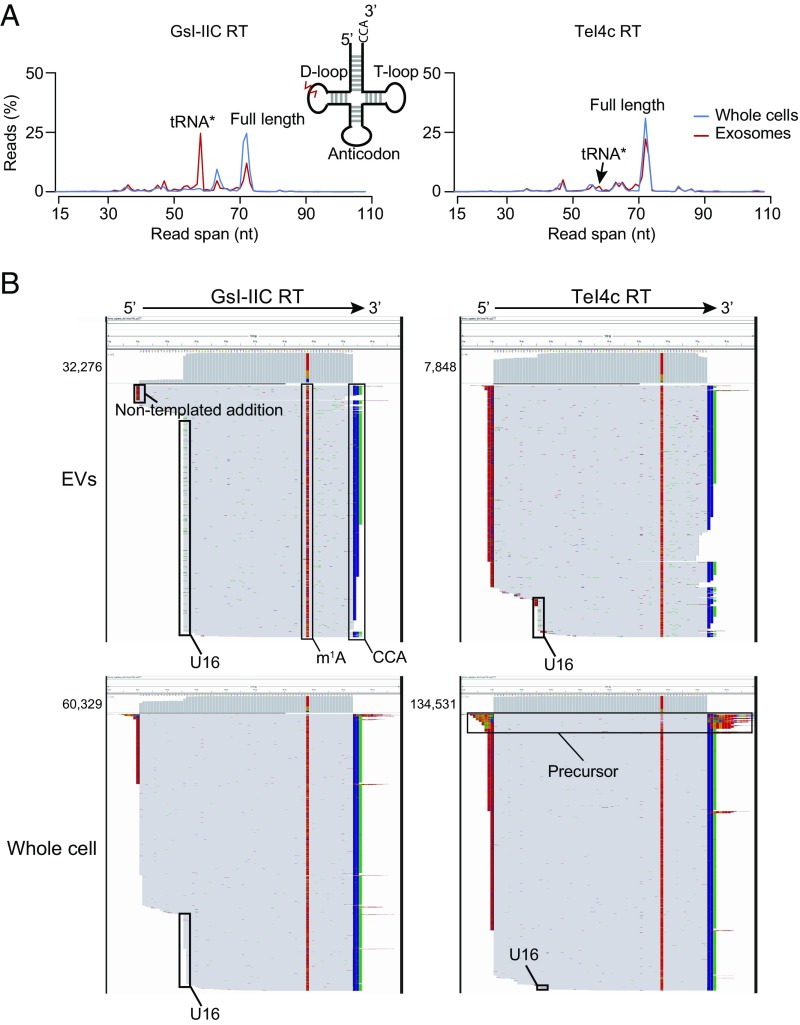
The results above showed that in addition to full-length tRNAs, EVs and exosomes contained a 15-nt shorter species (tRNA*), which could reflect a D-loop truncation or a posttranscriptional modification that caused a reverse transcription stop. This D-loop stop was not universal for all exosomal tRNAs, with ArgCCT, LysCTT, LysTTT, MetCAT, PheGAA, and ValTAC as the primary contributors to this peak (SI Appendix, Fig. S13). TGIRT enzymes have been shown previously to read through transcripts with stable secondary structure and posttranscriptional modification (20, 28) and do not stop at the known D-loop modification dihydrouridine, which is prevalent in all tRNAs. Nevertheless, it remained possible that exosomal tRNAs contain a previously unknown modification that results in a more stringent reverse transcription stop.
Since TGIRT enzymes show differential ability to read through modified transcripts, we purified EVs from conditioned medium (according to the protocol in Fig. 1A) and split them into two reverse transcription reactions using either GsI-IIC RT (the enzyme used for the previous analyses in this study) or an alternative TGIRT enzyme, TeI4c RT (20, 28) (datasets GC and GEV for GsI-IIC RT and TC and TEV for TeI4c RT; SI Appendix, Table S2). The GsI-IIC enzyme gave a read profile with a tRNA* peak in EVs that was greatly diminished in cellular tRNAs from the same culture, demonstrating that the D-loop stop is EV-specific (Fig. 7A, Left). In contrast, TeI4c RT showed only a very small peak at the position corresponding to the tRNA* peak in the same EV RNA preparation and no peak in the same whole-cell RNA preparation (Fig. 7A, Right). IGV alignments confirmed that the tRNA* species had a strong reverse transcription stop at the dihydrouridine site for the majority of reads obtained for EV tRNAs with GsI-IIC RT but not with TeI4c RT and was not abundant in reads obtained for cellular tRNAs with either enzyme (shown for LysCTT and ValTAC in Fig. 7B and SI Appendix, Fig. S14, respectively). Because the tRNA* species was highly enriched in EV compared with cellular RNAs, it likely represented an as yet unidentified additional or alternative modification at the dihydrouridine site.
Fig. 7.
Evidence for an exosome-specific tRNA modification. (A) Read-span distributions for all reads mapping to tRNAs in TGIRT-seq libraries prepared from unfragmented whole-cell RNA and exosomal RNA with either GsI-IIC RT (TGIRT-III; Left) or TeI4c RT (Right). The peaks corresponding to full-length mature tRNA and the tRNA* species with a strong stop at position 16 in the D-loop are indicated. The Inset shows a tRNA schematic indicating the approximate position of the putative exosome-specific tRNA modification (jagged red line). (B) IGV screen shots showing read coverage across the tRNA coding sequence for a tRNA lysine isoacceptor (CTT) from TGIRT-seq of EV and unfragmented whole-cell RNAs with GsI-IIC RT or TeI4c RT. Coverage plots are above read alignments to the tRNA coding sequence with reads sorted by top strand position. The numbers of reads are indicated to the left of the coverage plot and were down sampled to 1,000 reads in IGV for visualization. Nucleotides in reads matching nucleotides in the annotated reference are colored gray. Nucleotides in reads that do not match the annotated reference are color coded by nucleotide (A, green; C, blue; G, brown; and T, red). 1-methyladenine (m1A) and the 3′ CCA posttranscriptional modifications, extra nontemplated nucleotides added to the 3′ end of the cDNA by the RT, and the D-loop modification (stop at position U16) are indicated. The reads identified as tRNA precursors have short 5′- and 3′-end extensions that map to genomic loci encoding this tRNA (SI Appendix, Fig. S14B).
Discussion
In this report, we describe the use of RNA-seq, employing a highly processive TGIRT, to characterize the RNA content of HEK293T cell EVs and exosomes and to investigate the role of YBX1 in packaging these RNAs. RNase and DNase protection experiments in the presence and absence of detergent showed that the most abundant RNA species in both EVs and exosomes are tRNAs and other sncRNAs and that a high proportion of these sncRNAs are present as full-length, mature transcripts. tRNAs and most other sncRNAs are fully sensitive to RNase in the presence of detergent, but some (5S rRNA, U5 snRNA, and some snoRNA, Y RNA, and Vault RNA species) are RNase-resistant, suggesting that they may be protected in RNPs associated with EVs. Results of previous studies with multiple cell types have shown a dominant peak corresponding to full-length tRNA as evaluated by bioanalyzer size distribution (9, 11, 15, 16, 42), but studies that present sequencing results report that the majority of tRNAs are present as fragments (15, 16, 19, 36, 42, 43), presumably reflecting the fact that conventional RTs do not give full-length reads of tRNAs. In contrast, a recent study sequencing cell-free RNA from human plasma by TGIRT-seq revealed the presence of mostly full-length tRNAs and other structured sncRNAs (20). Our findings using TGIRT-seq clearly demonstrate that EVs likewise contain a high proportion of full-length tRNAs and other sncRNA species (Figs. 3 and 7) and indicate that conventional RNA-seq methods are not sufficient to characterize the RNA content of EVs.
The presence of mRNA inside EVs has been controversial in part due to the cosedimentation of contaminating RNA associated with RNPs present in exosome pellets obtained by high-speed ultracentrifugation or hydrophilic polymer precipitation (e.g., PEG, ExoQuick). A recent report using a multistage EV isolation procedure including density gradient flotation identified lincRNA, antisense RNA, and protein-coding RNAs as being associated with a particular EV subpopulation (42). In contrast, our nuclease sensitivity results indicated that a substantial proportion of the reads mapping to protein-coding genes, lincRNAs, and antisense RNAs are due to DNA contamination on the surface of vesicles (Figs. 2 and 3 and SI Appendix, Fig. S5). These disparate results underscore the importance of both upstream (i.e., EV isolation) and downstream (i.e., RNA isolation) purification procedures when characterizing EV composition.
Nevertheless, in addition to sncRNAs, we find smaller amounts of spliced mRNAs (<1% of mapped reads) in flotation gradient-purified, DNase-treated RNA from HEK293T cell exosomes (Fig. 4). These transcripts are enriched for 5′ TOP mRNA sequences (Fig. 4D and SI Appendix, Figs. S7B and S8). 5′ TOP mRNAs are translationally repressed via recruitment to RNA granules in response to starvation (40), suggesting a link between cellular metabolism and EV–RNA loading. If so, the mRNA content of EVs and exosomes may be cell type-specific and change under different physiological conditions. Our results also confirm a previous report that aaRS mRNA sequences are present in exosomes, of potential interest because of the orthogonal extracellular signaling function of these enzymes (44, 45).
Accumulating evidence supports a role for RBPs in the sorting of specific transcripts into EVs derived from various cell types. These studies have primarily focused on the sorting of select miRNAs into EVs from multiple cells types (see Introduction). To date, there has been no attempt to understand how individual RBPs define the total RNA composition of EVs beyond specific miRNAs. YBX1 has been shown to bind an array of coding and noncoding cellular transcripts, including directly binding tRNA sequences via its cold-shock domain (46–49) and other RNAs nonspecifically via its positively charged, intrinsically disordered C-terminal region (50). We find that HEK293T cells devoid of YBX1 show decreased secretion via CD63+ exosomes of various abundant sncRNAs, including tRNAs, Y RNAs, and Vault RNAs relative to other RNA biotypes (Fig. 5). YBX1 is involved in the formation of stress granules in cells (49, 51), and many other RBPs are associated with RNA granules via intrinsically disordered sequences (52). These observations together with those described above for 5′ TOP mRNAs could link RNA granule formation to the RNA composition of EVs. Translational repression during the generalized stress response may result in an accumulation of free tRNAs that are bound by YBX1 and exported via exosomes.
A recent study of breast cancer cells reported that the cleavage of tRNAs induced by cellular stress results in the production of a class of tRNA fragments containing an oligoguanine motif, which binds YBX1 in a sequence-specific manner (47). It was suggested that these tRNA fragments then displace YBX1 from the 3′ untranslated region of multiple mRNAs, leading to the destabilization of transcripts that promote breast cancer progression. Similarly, competition for YBX1 between different cellular RNAs may be a mechanism to modulate exosomal RNA content (e.g., miRNA vs. tRNA). We note, however, that the previous study used conventional RNA-seq methods, which have difficulty fully reverse-transcribing tRNAs, and that our results indicate that YBX1 in HEK293T cells functions in the cellular export of virtually all cellular tRNAs and not just those containing an oligoguanine motif.
Using two distinct TGIRT enzymes, we find that the major class of truncated tRNA reads (tRNA*) from our sequencing libraries is likely due to a posttranscriptional modification rather than a 5′ tRNA truncation (Fig. 7). The tRNA* species appears highly enriched in EVs, suggesting a possible role of posttranscriptional modification in the sorting of tRNAs into or specialized processing of tRNAs within EVs. The putative modification appears at a nucleotide position normally occupied by dihydrouridine, but the identity of this modification is currently unknown, as is whether it occurs before, after, or instead of uridine reduction in cells or EVs, perhaps as a result of exposure of sequences normally bound by a protein partner. The proportion of tRNA* to full length is similar between WT and YBX1-null cells even though YBX1-null cells have fewer total tRNAs, possibly indicating that the modification occurs in EVs. Recent evidence indicates that Y RNA fragments can be generated in EVs, indicating that vesicles may enable specialized biochemical conversions (53). The low yield of RNA in purified exosomes presents a challenge to the identification of the exosome-specific modification. Nonetheless, that TeI4c RT reads through the site of modification without base misincorporation suggests that the modification does not affect Watson–Crick base pairing and therefore may be a bulky group that is added to the ribose moiety or the non-Watson–Crick face of the nucleotide base.
Finally, how might the relationship between YBX1, RNA granules, and exosome RNA composition contribute to cellular physiology? YBX1 might bind damaged, nonfunctional, or excess free tRNAs and other sncRNAs that are not associated with their normal protein partners as an early step that targets these RNAs for sorting into exosomes. EVs have been suggested as possible vehicles for the purging of circular RNAs, which are byproducts of RNA splicing (54). Such purging of unbound RNAs may be particularly relevant under stress conditions, where the accumulation of excess free RNA polymerase III (PolIII) transcripts may be detrimental to individual cells. As has been shown recently for the 7SL RNA component of the signal recognition particle, the intracellular accumulation or intercellular transfer of an abundant free PolIII transcript via membrane vesicles can activate cellular pathways responsible for the detection and defense against viral RNAs (55). The same may be true for tRNAs, Y RNAs, and Vault RNAs, with the purged RNAs protected inside EVs able to travel to nearby cells and contribute to or regulate recipient cell metabolism, including by triggering of innate immune responses. In this way, YBX1 may play a role in resource sharing and/or signaling between cells within tissue microenvironments. Because YBX1 is overexpressed in many malignancies (56), these processes may be particularly important for the survival and growth of cancer cells in the context of the tumor microenvironment. Direct examination and modulation of the relationship between YBX1 localization, EV-RNA content, RNA granules, and growth/stress conditions along with the ability to distinguish full-length tRNAs from tRNA fragments using TGIRT enzymes should help to unravel how these relationships influence cellular physiology in health and disease.
Methods
Detailed materials and methods describing the cell lines used in this study, growth conditions, EV purification, TGIRT-seq and bioinformatics analysis, and all experimental procedures are available in SI Appendix.
Supplementary Material
Acknowledgments
We acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources that contributed to the research results reported within this paper. Funding for this work was from the Howard Hughes Medical Institute (R.S.) and National Institutes of Health R01 Grant GM37949 (to A.M.L.).
Footnotes
Conflict of interest statement: Thermostable group II intron reverse transcriptase (TGIRT) enzymes and methods for their use are the subject of patents and patent applications that have been licensed by the University of Texas and East Tennessee State University to InGex, LLC. A.M.L., some former and present members of the A.M.L. laboratory, and the University of Texas are minority equity holders in InGex, LLC and receive royalty payments from the sale of TGIRT enzymes and kits and from the sublicensing of intellectual property to other companies.
Data deposition: The TGIRT-seq datasets described in this manuscript have been deposited in the National Center for Biotechnology Information Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. SRP108712).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712108114/-/DCSupplemental.
References
- 1.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 2.Bobrie A, Colombo M, Krumeich S, Raposo G, Théry C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J Extracell Vesicles. 2012;1:18397. doi: 10.3402/jev.v1i0.18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gould SJ, Raposo G. As we wait: Coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles. 2013;2:20389. doi: 10.3402/jev.v2i0.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booth AM, et al. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol. 2006;172:923–935. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ratajczak J, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 7.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosaka N, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meckes DG, Jr, et al. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci USA. 2010;107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pegtel DM, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Squadrito ML, et al. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8:1432–1446. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 13.Ridder K, et al. Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol. 2014;12:e1001874. doi: 10.1371/journal.pbio.1001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zomer A, et al. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 2015;161:1046–1057. doi: 10.1016/j.cell.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40:10937–10949. doi: 10.1093/nar/gks832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nolte-’t Hoen EN, et al. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272–9285. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogawa Y, Taketomi Y, Murakami M, Tsujimoto M, Yanoshita R. Small RNA transcriptomes of two types of exosomes in human whole saliva determined by next generation sequencing. Biol Pharm Bull. 2013;36:66–75. doi: 10.1248/bpb.b12-00607. [DOI] [PubMed] [Google Scholar]
- 18.Batagov AO, Kuznetsov VA, Kurochkin IV. Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles. BMC Genomics. 2011;12(Suppl 3):S18. doi: 10.1186/1471-2164-12-S3-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vojtech L, et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42:7290–7304. doi: 10.1093/nar/gku347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin Y, et al. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA. 2016;22:111–128. doi: 10.1261/rna.054809.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cha DJ, et al. KRAS-dependent sorting of miRNA to exosomes. Elife. 2015;4:e07197. doi: 10.7554/eLife.07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenzie AJ, et al. KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep. 2016;15:978–987. doi: 10.1016/j.celrep.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife. 2016;5:e19276. doi: 10.7554/eLife.19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villarroya-Beltri C, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santangelo L, et al. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016;17:799–808. doi: 10.1016/j.celrep.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 27.Mukherjee K, et al. Reversible HuR-microRNA binding controls extracellular export of miR-122 and augments stress response. EMBO Rep. 2016;17:1184–1203. doi: 10.15252/embr.201541930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohr S, et al. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA. 2013;19:958–970. doi: 10.1261/rna.039743.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu DC, Lambowitz AM. Facile single-stranded DNA sequencing of human plasma DNA via thermostable group II intron reverse transcriptase template switching. Sci Rep. 2017;7:8421. doi: 10.1038/s41598-017-09064-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nottingham RM, et al. RNA-seq of human reference RNA samples using a thermostable group II intron reverse transcriptase. RNA. 2016;22:597–613. doi: 10.1261/rna.055558.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katibah GE, et al. Broad and adaptable RNA structure recognition by the human interferon-induced tetratricopeptide repeat protein IFIT5. Proc Natl Acad Sci USA. 2014;111:12025–12030. doi: 10.1073/pnas.1412842111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eckwahl MJ, Sim S, Smith D, Telesnitsky A, Wolin SL. A retrovirus packages nascent host noncoding RNAs from a novel surveillance pathway. Genes Dev. 2015;29:646–657. doi: 10.1101/gad.258731.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eckwahl MJ, et al. Analysis of the human immunodeficiency virus-1 RNA packageome. RNA. 2016;22:1228–1238. doi: 10.1261/rna.057299.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dhahbi JM, et al. 5′-YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiol Genomics. 2013;45:990–998. doi: 10.1152/physiolgenomics.00129.2013. [DOI] [PubMed] [Google Scholar]
- 35.Kumar P, Kuscu C, Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs) Trends Biochem Sci. 2016;41:679–689. doi: 10.1016/j.tibs.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tosar JP, et al. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res. 2015;43:5601–5616. doi: 10.1093/nar/gkv432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 38.Avni D, Biberman Y, Meyuhas O. The 5′ terminal oligopyrimidine tract confers translational control on TOP mRNAs in a cell type- and sequence context-dependent manner. Nucleic Acids Res. 1997;25:995–1001. doi: 10.1093/nar/25.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avni D, Shama S, Loreni F, Meyuhas O. Vertebrate mRNAs with a 5′-terminal pyrimidine tract are candidates for translational repression in quiescent cells: Characterization of the translational cis-regulatory element. Mol Cell Biol. 1994;14:3822–3833. doi: 10.1128/mcb.14.6.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Damgaard CK, Lykke-Andersen J. Translational coregulation of 5′TOP mRNAs by TIA-1 and TIAR. Genes Dev. 2011;25:2057–2068. doi: 10.1101/gad.17355911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iadevaia V, Caldarola S, Tino E, Amaldi F, Loreni F. All translation elongation factors and the e, f, and h subunits of translation initiation factor 3 are encoded by 5′-terminal oligopyrimidine (TOP) mRNAs. RNA. 2008;14:1730–1736. doi: 10.1261/rna.1037108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lässer C, et al. Two distinct extracellular RNA signatures released by a single cell type identified by microarray and next-generation sequencing. RNA Biol. 2017;14:58–72. doi: 10.1080/15476286.2016.1249092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guzman N, et al. Breast cancer-specific miR signature unique to extracellular vesicles includes “microRNA-like” tRNA fragments. Mol Cancer Res. 2015;13:891–901. doi: 10.1158/1541-7786.MCR-14-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang F, et al. Regulated capture by exosomes of mRNAs for cytoplasmic tRNA synthetases. J Biol Chem. 2013;288:29223–29228. doi: 10.1074/jbc.C113.490599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lo WS, et al. Human tRNA synthetase catalytic nulls with diverse functions. Science. 2014;345:328–332. doi: 10.1126/science.1252943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blenkiron C, Hurley DG, Fitzgerald S, Print CG, Lasham A. Links between the oncoprotein YB-1 and small non-coding RNAs in breast cancer. PLoS One. 2013;8:e80171. doi: 10.1371/journal.pone.0080171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodarzi H, et al. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu TT, et al. Noncoding RNAs that associate with YB-1 alter proliferation in prostate cancer cells. RNA. 2015;21:1159–1172. doi: 10.1261/rna.045559.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lyons SM, Achorn C, Kedersha NL, Anderson PJ, Ivanov P. YB-1 regulates tiRNA-induced stress granule formation but not translational repression. Nucleic Acids Res. 2016;44:6949–6960. doi: 10.1093/nar/gkw418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lyabin DN, Eliseeva IA, Ovchinnikov LP. YB-1 protein: Functions and regulation. Wiley Interdiscip Rev RNA. 2014;5:95–110. doi: 10.1002/wrna.1200. [DOI] [PubMed] [Google Scholar]
- 51.Somasekharan SP, et al. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J Cell Biol. 2015;208:913–929. doi: 10.1083/jcb.201411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin Y, Protter DS, Rosen MK, Parker R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chakrabortty SK, Prakash A, Nechooshtan G, Hearn S, Gingeras TR. Extracellular vesicle-mediated transfer of processed and functional RNY5 RNA. RNA. 2015;21:1966–1979. doi: 10.1261/rna.053629.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lasda E, Parker R. Circular RNAs co-precipitate with extracellular vesicles: A possible mechanism for circRNA clearance. PLoS One. 2016;11:e0148407. doi: 10.1371/journal.pone.0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nabet BY, et al. Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell. 2017;170:352–366.e13. doi: 10.1016/j.cell.2017.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lasham A, Print CG, Woolley AG, Dunn SE, Braithwaite AW. YB-1: Oncoprotein, prognostic marker and therapeutic target? Biochem J. 2013;449:11–23. doi: 10.1042/BJ20121323. [DOI] [PubMed] [Google Scholar]
- 57.Meyuhas O, Kahan T. The race to decipher the top secrets of TOP mRNAs. Biochim Biophys Acta. 2015;1849:801–811. doi: 10.1016/j.bbagrm.2014.08.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.