Significance
Marine microbial communities exert a large influence on ocean ecosystem processes, and viruses in these communities play key roles in controlling microbial abundances, nutrient cycling, and productivity. We show here that dominant viruses in the open ocean persist for long time periods and that many appear tightly locked in coordinated diel oscillations with their bacterial hosts. The persistent structure of viral assemblages, as well as synchronized daily oscillations of viruses and hosts, are in part the result of the regular diurnal coupling of viral and host replication cycles. Collectively, our results suggest that viruses, as key components of marine ecosystems, are intrinsically synchronized with the daily rhythms of microbial community processes in the ocean’s photic zone.
Keywords: marine virus, diel cycles, oligotrophic gyre, phage gene expression, marine bacteriophage
Abstract
Viruses are fundamental components of marine microbial communities that significantly influence oceanic productivity, biogeochemistry, and ecosystem processes. Despite their importance, the temporal activities and dynamics of viral assemblages in natural settings remain largely unexplored. Here we report the transcriptional activities and variability of dominant dsDNA viruses in the open ocean’s euphotic zone over daily and seasonal timescales. While dsDNA viruses exhibited some fluctuation in abundance in both cellular and viral size fractions, the viral assemblage was remarkably stable, with the most abundant viral types persisting over many days. More extended time series indicated that long-term persistence (>1 y) was the rule for most dsDNA viruses observed, suggesting that both core viral genomes as well as viral community structure were conserved over interannual periods. Viral gene transcription in host cell assemblages revealed diel cycling among many different viral types. Most notably, an afternoon peak in cyanophage transcriptional activity coincided with a peak in Prochlorococcus DNA replication, indicating coordinated diurnal coupling of virus and host reproduction. In aggregate, our analyses suggested a tightly synchronized diel coupling of viral and cellular replication cycles in both photoautotrophic and heterotrophic bacterial hosts. A surprising consequence of these findings is that diel cycles in the ocean’s photic zone appear to be universal organizing principles that shape ecosystem dynamics, ecological interactions, and biogeochemical cycling of both cellular and acellular community components.
Viruses are numerically dominant acellular biological entities that play critical roles in global biogeochemical cycles, shape microbial community structure, and potentially influence the physiological status of their hosts (1–3). Although new developments in metagenomics and single-cell genomics have advanced understanding of bacterial and archaeal diversity in nature (4, 5), the temporal dynamics, diversity, and variability of native viral assemblages are not as well documented. This is in part due to the lack of universal phylogenetic markers for viruses as well as the relatively recent development of culture-independent methods to investigate viral genomic diversity in the environment (2, 3, 6, 7).
Studies of marine viruses in particular have had many recent advances (1, 8), including work on viral temporal dynamics. Monthly or interannual time-series studies employing culture-independent methods have shown that seasonality exerts a strong influence on viral diversity in coastal and temperate waters, although some viral groups appear to be able to persist for periods of months or years (9–12). Studies of microbial communities in controlled environments have confirmed that viruses can survive for long time periods, despite fluctuations in abundance (13). Over shorter timescales, field observations and culture-based experiments have provided some evidence of viral diel cycling and suggested that viral diurnal rhythms, if present, might also be important factors structuring mcirobial diversity in natural settings (14–18).
To more deeply explore the diversity and temporal dynamics of viral assemblages in the open ocean, we leveraged large-scale metagenome sequencing and quantitative metatranscriptomics of sympatric marine microbial and viral assemblages. We surveyed these assemblages over both daily and interannual time periods in waters of the North Pacific Subtropical Gyre (NPSG), a habitat representative of oligotrophic oceans that cover ∼40% of the Earth’s surface (19, 20). For diel analyses, we sampled a microbial assemblage surveyed within a coherent water mass over 8 d, using both metagenomics and quantitative transcriptomics to monitor the temporal abundance and transcriptional activities of the most abundant dsDNA viruses (SI Appendix, Fig. S1 and Dataset S1). We also quantified the longer-term abundances (over ∼1.5 y) of the dsDNA viruses observed in this summertime diel survey, leveraging a previous time-series metagenomic effort in the same waters (21). Our sampling and analytical strategy provide a unique opportunity to assess the temporal trends of viral abundances and dynamics in oligotrophic open ocean surface waters over several different time scales.
Results
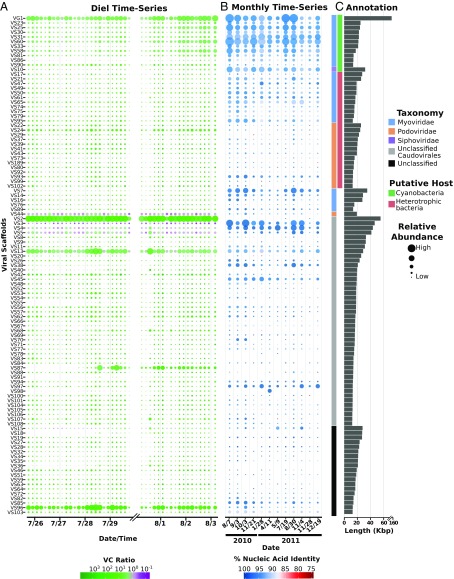
The viral assemblage we sampled over time was very stable, as most viral genomic scaffolds were consistently present throughout the 8-d sampling period (Dataset S2), including 104 of the 107 (97.2%) largest viral scaffolds (Fig. 1A and SI Appendix, Fig. S2). Bacteriophage sharing syntenic gene organization and high protein similarity with known cyanophage were among the most abundant viral sequences (SI Appendix, Fig. S3). Four of the most abundant scaffolds (VS1, 6, 12, and 153) were binned together into a near-complete genome assembly (termed VG1 here) based on their shared synteny and 76% amino acid identity (AAI) to Prochlorococcus phage P-RSM1, as well as their consistent coabundance and tetranucleotide frequency profiles (SI Appendix, Figs. S3 and S4; details in SI Appendix). Also continuously present were viruses similar to SAR11 and SAR116 bacterioplankton phage (here grouped together as heterotrophic bacterial viruses) (Fig. 1C and SI Appendix, Fig. S3 and Dataset S3). Of those viral populations with a host prediction, most shared highest sequence similarity to Myoviridae or Podoviridae, while only one shared significant sequence identity to a Siphoviridae (VS10). Most viral scaffolds could not be assigned a putative host or taxonomic family however, and these included some of the most abundant viral scaffolds recovered (e.g., VS2-5). We compared the viral scaffolds identified here against a custom database that included currently available viral genomes and viral sequences from recent metagenomic studies (22–24) (SI Appendix, Fig. S5; see SI Appendix). Using the established cutoff of 90% AAI for viral taxonomic groupings (23), only 78 scaffolds (16%) could be classified (only eight of which were >15 Kbp; Dataset S4). The majority of the viruses we identified in the diel time series therefore represent previously unidentified groups, underscoring the diverse nature of viruses in natural environments.
Fig. 1.
Abundance and annotation of viral scaffolds recovered over different timescales. (A) Bubble plot of scaffold abundances over the diel time-course reported in this study. Bubble size is proportional to relative abundance of the scaffolds in both the cellular and viral fraction, and the color provides the log10-scaled VC ratio. (B) Bubble plot of scaffold relative abundances in a 1.5-y monthly time-series conducted at Sta. ALOHA, as determined from fragment recruitment analysis. Bubble size is proportional to relative abundance of the scaffolds, and the color provides the average percent identity of the reads mapped. (C) Putative host specificity and taxonomic classifications of the scaffolds are shown in color bars, and scaffold lengths are provided in the bar plot. All scaffolds >15 Kbp in length are shown.
We sequenced metagenomes derived from different size fractions of the same water sample and were therefore able to calculate a ratio of relative abundances between the “cellular” and “viral” size fractions (the >0.2 μm versus the 0.03 μm > 0.2 μm size fractions, respectively) throughout the time series (referred to here as the VC ratio). It should be noted that the “viral fraction” metagenomes potentially contain other contributing sources, for example dissolved or particulate DNA that bound to the filter, or ultrasmall cells that passed through the 0.2-μm prefilters. Likewise, viral DNA present in the “cellular fraction” metagenomes, besides being the result of active intracellular viral infections, could also include other sources such as virion adherence to larger particles or cells, or the retention of large virions on 0.2-μm filters. Nevertheless, ribosomal RNA gene fragment recovery confirmed that the cellular fractions were on average ∼fivefold enriched in cellular genes, relative to the viral fractions that were depleted in cellular gene representation (SI Appendix, Fig. S6 and Dataset S1). Viral scaffold relative abundances were also generally greater in the viral fraction metagenomes compared with the cellular fraction (SI Appendix, Fig. S7). Finally, the active transcription of viral genes in the metatrascriptomes (discussed below) provided further evidence of active intracellular viral infection in resident host populations.
The contrasting VC ratios obtained for different viral scaffolds suggested potential differences in the life history strategies of different viral groups. The majority of the viral scaffolds we observed showed markedly higher relative abundance in the viral versus cellular size fraction metagenomes (Fig. 1A and SI Appendix, Fig. S7 and Dataset S3), likely due to the expected enrichment of viral particles in this fraction. The widespread occurrence of lytic viral life history strategies was also supported by the presence of structural proteins for viral particle assembly and packaging in 366 (>75%) of the viral scaffolds. In contrast, only five potential lysogenic markers (five predicted integrases but no excisionases) were identified among all viral scaffolds (Dataset S5). Intriguingly, some of the largest scaffolds had a higher relative abundance in the cellular size fraction metagenomes over many timepoints (VS3-5, VS44, and VS15; Fig. 1A). No markers indicative of lysogeny were detected in these groups, and all except VS15 contained key structural proteins necessary for the lytic cycle (SI Appendix, Fig. S8). Their persistent abundance in the cellular size fraction suggested that some of these viruses may predominantly reside intracellularly, perhaps as a consequence of pseudolysogeny, which has been postulated previously for viruses in nutrient-depleted environments (25).
Because multiple processes can influence viral abundance in different size fractions, inference of viral life history on the basis of VC ratios alone has important caveats. For example, morphological differences among virions could lead to higher abundance in a specific size fraction irrespective of ecological strategies (26). Despite this caution, the VC ratio provides a useful metric, especially in conjunction with other analyses, in particular information pertaining to viral gene transcription within the same samples (see below). We postulate here that high VC ratios indicate the prevalence of lytic infections, which is consistent with the observed viral scaffold annotations. Conversely, low VC values may suggest the potential for alternative life histories that involve long-term intracellular presence, such as lysogeny or related nonlytic reproductive strategies.
To determine if the viruses we characterized were common in the NPSG over longer time periods, we quantified these viral scaffolds in a previously reported 25-m metagenomic time series survey (2010–2011) from the same region (21). A total of 394 (82%) of the viral scaffolds were identified across this seasonal time series (with average amino acid identity >90%), indicating that genetically similar viruses were persistent over yearly periods in this open ocean habitat (Fig. 1B and SI Appendix, Fig. S9 and Dataset S4). Taken together with the persistent viral assemblage we observed over daily periods, this consistency over interannual timescales suggests a stable assemblage structure of viruses in the NPSG, likely mirroring the corresponding stability of the environment and associated host populations (27).
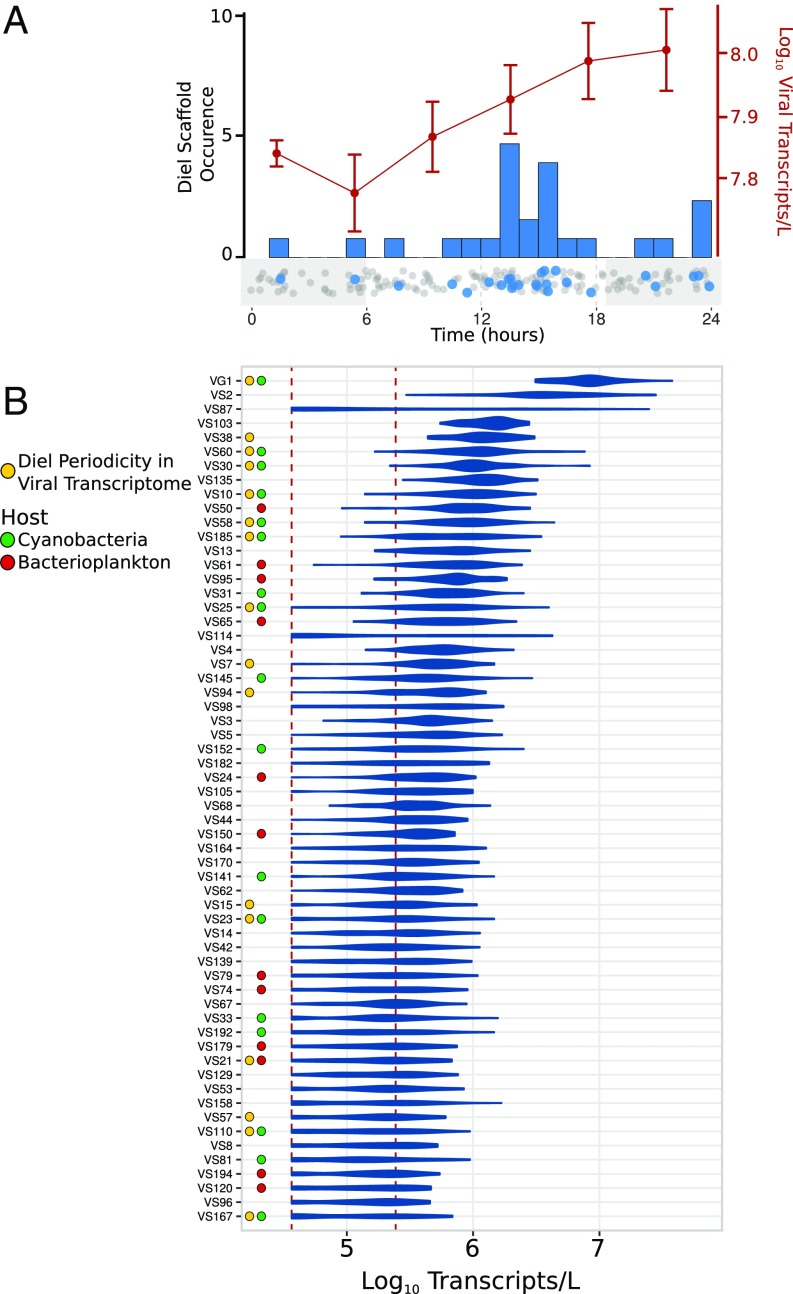
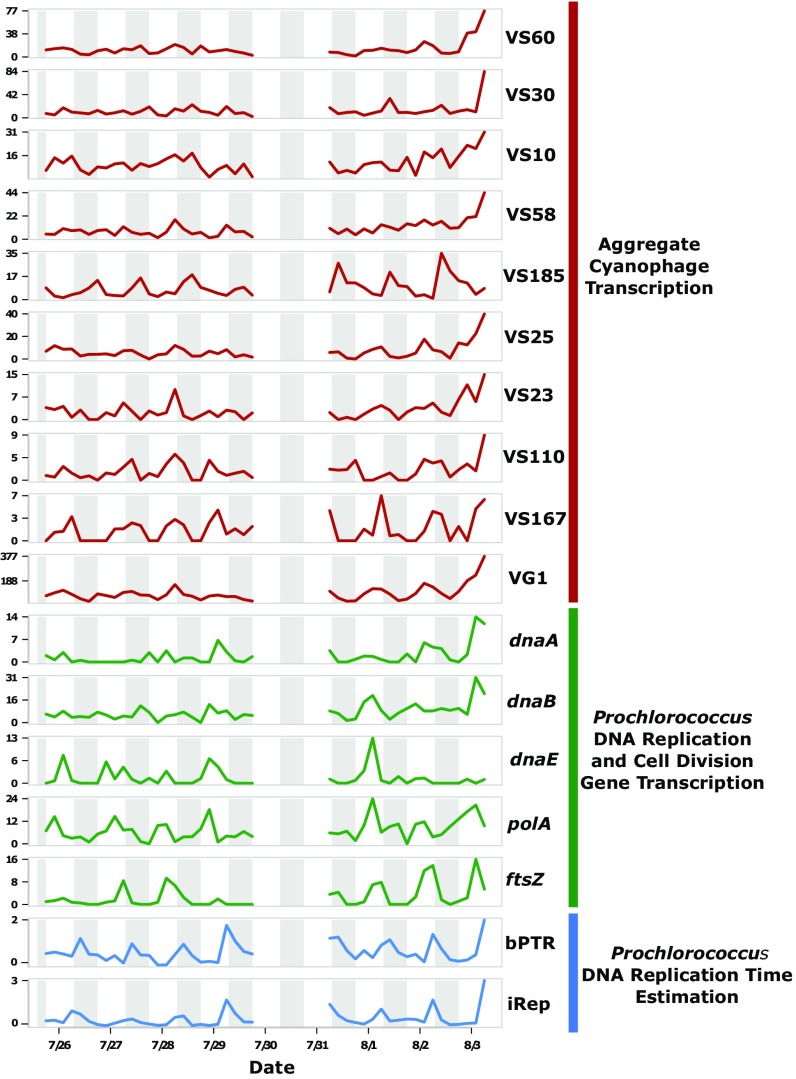
To further characterize the temporal dynamics of viral activities in the open ocean, we measured daily viral gene transcription patterns in the cellular size fractions every 4 h over 8 d. These analyses revealed that genes encoding virus structural proteins were among the most abundant among viral transcripts, consistent with the suggestion of primarily lytic life cycles from the VC ratios (SI Appendix, Fig. S10). Several putative AMGs were also among the most highly expressed viral genes, including photosystem genes and other proteins thought to be involved in the manipulation of cyanobacterial energy metabolism (28) (SI Appendix, Fig. S10). Diel periodicity was detected in 26 viral scaffolds (RAIN periodicity test, corrected P < 0.1; see Methods for details), which likely represents an underestimate due to limitations in detecting low abundance viral transcripts within the total cellular transcript pool (Fig. 2B and Dataset S3). Of the diel viral scaffolds exhibiting diel periodicity, 17 (65%) appeared to be cyanophage (Fig. 2B and SI Appendix, Fig. S11). Most of these cyanophage likely infect surrounding Prochlorococcus hosts. This is supported by their genetic similarity to other Prochlorococcus cyanophage, the high abundance of this cyanobacterium in these samples, and the synchronization of peak cyanophage reproduction and Prochlorococcus DNA replication (estimated from the metagenomic time series; Fig. 3 and SI Appendix, Fig. S12). Additionally, one putative Pelagibacter phage (VS21) and several scaffolds from unclassified groups (VS7, VS13, VS94, and VS105) also exhibited diel transcriptional activity, suggesting that diel viral production occurs across a broad range of different bacteriophage.
Fig. 2.
Diel timing and abundance of viral transcription. (A) The red line shows average transcripts per liter of total viral transcripts analyzed in this study, with error bars denoting SE. The histogram shows the peak time of aggregate viral transcripts for individual scaffolds, and the dotplot below it provides the peak times of all scaffolds, with significantly diel scaffolds in blue and all other scaffolds in gray. (B) Viral scaffolds abundant in the transcriptomes are shown on the y axis. The shape of the plot denotes the distribution of abundances recovered in all 44 time-points. The x axis shows transcripts per liter. The dotted red lines denote the upper and lower bounds of thresholds of detection. Colored dots next to the scaffolds denote putative host designations and scaffolds for which significantly diel transcription was determined.
Fig. 3.
Diel synchrony of cyanophage and Prochlorococcus activities. Temporal profiles for diel cyanophage aggregate transcription (red), Prochlorococcus DNA replication and cell division marker gene transcription (green), and two metrics for estimating the timing of Prochlorococcus DNA replication (iRep and bPTR; blue). Units for the transcriptional profiles are ×105 transcripts per L. Night-time periods are shaded gray.
Overall viral transcript abundance increased in the late afternoon and evening time-points (Fig. 2A; Mann–Whitney u test, P < 0.005). Interestingly, total viral transcript abundance increased throughout the nighttime hours, while the peak transcription of the majority of diel viral scaffolds occurred between 1200 and 1800 hours. Analysis of the aggregate transcription of diel viral scaffolds confirmed a peak at 1800 hours, consistent with the peak time of transcription of the majority of diel scaffolds (SI Appendix, Fig. S13). Conversely, aggregate transcription of nondiel viral scaffolds increased throughout the evening, with a peak at 2200 hours and trough at 0600 hours. This suggested that the increase in total viral transcription throughout the night was primarily due to the aggregate activities of viruses with no clear diel periodicity, perhaps also reflecting the presence of viral diel cycles below our detection limits. In summary, while total viral transcriptional activity appeared to increase during the night, those scaffolds exhibiting significant diel periodicity (mostly cyanophage) peaked in the afternoon to early evening.
Discussion
Our collective results indicate that over monthly and interannual time scales, many of the abundant virus types in the NPSG are consistently present and actively infecting cells on a daily basis. Recent analysis of marine viruses cultured over several years revealed genomic similarity that was attributed to long-term persistence of specific viral ecotypes (29), and work on laboratory systems and controlled environments has shed light on the incidence and potential mechanisms which allow for long-term phage–host coexistence (13, 30). Moreover, a recent large-scale metagenomic comparison recovered certain abundant viral types in distant locales, consistent with the continuous presence of certain viral types across both space and time (24). Time-series analyses have also revealed the persistence of certain viral groups in temperate or coastal systems despite the presence of robust seasonal transitions that play a large role in structuring overall viral diversity (9–11, 18, 31, 32). The relatively stable environmental conditions of the NPSG, together with the overall stability of host populations there (27), help explain the consistent long-term presence of many different viral types we observed, compared with environments with more pronounced seasonality. These results could be viewed as broadly consistent with the “Bank” model (33), wherein viral assemblages in total are composed of a smaller subset of active and abundant viruses along with a larger subset of rarer viruses. An important nuance here is that the pool of abundant viruses may be substantially larger and persist for longer time periods in environments having less pronounced seasonal transitions in host abundances, such as the NPSG.
As with any metagenomic study, the viral sequences reported here likely represent an underestimate of actual dsDNA viral diversity, due to difficulties in assembling low abundance viral genomes as well as in identifying novel sequences as viral in origin. Our study, together with other large-scale metagenomic analyses (22–24, 34–37), further expands knowledge of viral genomic diversity and dynamics in the environment. Further advances in cataloguing viral diversity and improvement of methods to identify viral sequences are still required, however, to better interpret viral genotypic diversity and dynamics in natural settings.
Another outcome of our study is the documentation of diel cycling in the transcriptional activities of different pelagic viruses that likely infect both phototrophic and heterotrophic bacterial hosts. Of the 170 viral scaffolds showing transcriptional activity throughout the sampling period, 26 (15.3%) exhibited diel periodicity in their aggregate transcriptional profiles. Because mRNA from dsDNA viruses should only be detected during the viral replication cycle, we interpret viral assemblage transcriptional patterns as indicators of the timing of intracellular viral reproductive activities. Since structural genes were among the most highly expressed diel viral transcripts (SI Appendix, Fig. S10), many viruses were likely undergoing diurnal lytic cycles. Previous laboratory studies have suggested that cyanophage genes exhibit a defined “expression program,” which takes place across variable time intervals depending on the virus (38). While we could detect diel periodicity in many viral transcripts, our regular 4-h sampling periods could not well resolve finer-scale temporal patterns (e.g., early genes versus late genes) over the course of our measurements. Nevertheless, many of the viruses reported here likely also underwent defined gene expression programs, although not necessarily immediately upon infection. Specifically, our results suggest that for most of the diel viruses we observed, early transcriptional activities peaked between 1200 and 1400 hours. For future viral assemblage studies, higher temporal resolution sampling strategies targeting in the midday to late evening may better resolve different stages in the viral replication cycle in situ. Gene-targeted approaches, for example using qPCR, will also be useful to differentiate the different stages of the viral replicative cycle among different viruses in complex populations.
Diel cycling in marine virus reproduction has been suspected for some time (39), but direct evidence from field populations has remained elusive. The discovery of photosynthetic genes in cyanophage, as well as increased cyanophage reproduction in laboratory cultures during host photosynthesis, has provided some evidence that viral diel cycles might play a role in viral fitness (40–42). Culture-based infection studies and direct counts have also provided tantalizing clues of potential diel cycles in marine viruses (14, 15, 17).
We demonstrate here that diel cycling does occur in natural viral populations in different viral groups. We suggest that diel viral activities result from the direct coupling of virion replication with host diel replication cycles, and that they are selectively advantageous for several reasons. First, diel cycling of microbial activities is prevalent in the NPSG (43) with the greatest photosynthetic energy being available midday, in theory providing maximal energetic resources for virion production during the same time. Second, the similar timing of cyanophage activity with Prochlorococcus diel DNA replication (Fig. 3 and SI Appendix, Fig. S12) indicates that viral cooption of host replication cycles also plays a role in the timing of viral diurnal cycles. Finally, the late afternoon to evening burst of viral reproductive activity we observed may enhance viral progeny survival, via avoidance of UV damage and photodegradation that is expected to occur during peak daylight hours (44). Depending on the duration of the lytic cycle, this strategy could facilitate postburst infection of new hosts over the nighttime period, before the initiation of a new viral replication cycles the following afternoon. An alternative hypothesis here is that light-dependent adsorption, shown to occur in some cyanophage (45), could lead to increased daytime infection and, therefore, peak transcription in the afternoon.
Intriguingly, one scaffold (VS21) with homology to a SAR11 virus (Pelagibacter phage HTVC008M) also exhibited diel aggregate transcriptional activity, suggesting that viral diel activity is not limited to cyanophage. Heterotrophic bacteria exhibit pronounced diel cycles in the NPSG (43), and energy acquisition for these bacterioplankton may also be greatest in the day due to cross-feeding from photoautotrophs (43, 46). Diel cycling in heterotrophic bacterial viruses might therefore be just as beneficial as it is for cyanophage. Several other viral scaffolds we analyzed (eight in total) also displayed diel transcriptional activities, but their hosts at present remain unidentified. Notably, three of these viruses were distinct from all of the other viruses characterized in that their peak transcriptional activity occurred between midnight and noon. The life history strategies of these “night active” viruses are unlike those that infect Prochlorococcus, and they presumably infect different hosts.
Our observation of diel viral activity indicates that viruses in the ocean’s photic zone have evolved to coopt the diel cycles of their hosts, implying that viral-driven mortality and metabolic manipulation of viral hosts might also exhibit diurnal oscillations. Perpetual (diel) lytic activities and the viral host evolutionary “arms races” are evident in these data, and may help explain recent reports of consistent diel fluctuations in Prochlorococcus cell abundance, as well as the extensive genomic microdiversity in cooccurring Prochlorococcus populations (47, 48). These data suggest that some key emergent properties of euphotic zone microbial communities are manifestations of diel cycling not only of microbial cells, but also of viruses entrained by the diurnal oscillations of their hosts.
Materials and Methods
We collected water samples in two periods of diel sampling from July 27 to July 31 and August 1 to August 3 of 2015. Throughout these periods, we sampled water at a depth of 15 m every 4 h (SI Appendix, Fig. S1A). Detailed information regarding the sampling regime on the cruise has been described in a previous study (49), and general cruise information and associated biogeochemical and oceanographic measurements can be found online (hahana.soest.hawaii.edu/hoelegacy/hoelegacy.html). Samples were collected from a total of 44 time-points, and metagenomes were sequenced from the >0.2 μm and the 0.2 > 0.03 μm size fractions (the cellular and viral size fractions, respectively). Quantitative transcriptomes were also generated from the >0.2 μm size fraction samples using spike-in molecular standards, consistent with methods previously described (50).
Library preparation for the cellular fraction metagenomes was performed using the Illumina TruSeq Nano LT library preparation kit, while for the viral fraction, metagenome library preparation was performed using the Illumina Neoprep library automation instrument and a Neoprep compatible TruSeq Nano LT kit (Illumina). Metatranscriptomic samples were prepared through the addition of 5–50 ng of total RNA to the ScriptSeq cDNA V2 library preparation kit (Epicentre). For the transcriptomes, quantitative standards were also spiked-in after extraction but before library preparation, consistent with previous methods (50). All sequencing was performed on an Illumina Nextseq500 system, yielding an average of 37.6 million sequence reads for the cellular metagenomes, 43.5 million reads for the viral metagenomes, and 25.0 million reads for the metatranscriptomes (read length ∼ 150 bp). The cellular fraction metagenomes, viral fraction metagenomes, and metatranscriptomes were each multiplexed on two runs each (six runs in total). Detailed sequencing statistics are provided in Dataset S1.
To accommodate the large size of the metagenomic datasets, a step-wise assembly and reassembly workflow was implemented to recover viral sequences (SI Appendix, Fig. S1B). This workflow implemented the tool VirSorter (51) to extract viral sequences from metagenomic assemblies of both size fractions. Ultimately, 483 viral scaffolds were recovered and given numerical identifiers with the prefix “VS.” Scaffold relative abundances were determined using a read-mapping workflow that implemented BWA v0.7.5a-r405 (52) and BEDTools v2.17.0 (53). For transcriptomic analyses, an end-joining, quality-trimming, and read-mapping workflow similar to that previously described was implemented (49). Diel periodicity analyses of the scaffolds in the metagenomic and transcriptome time-series were conducted using the R package RAIN (54). Details for all methods can be found in SI Appendix.
Raw data from metagenomic and transcriptomic experiments are available at the NCBI Sequence Read Archive (SRA) under BioProject accession PRJNA358725. Viral scaffolds have been deposited at DDBJ/ENA/GenBank under the accession NTLX00000000. The version described in this paper is version NTLX01000000.
Supplementary Material
Acknowledgments
We thank the captain and crew of the R/V Kilo Moana and chief scientist Sam Wilson for their effort and assistance on the Hawaii Ocean Experiment Legacy II cruise, Paul Den Uyl for help with metagenomic library preparation, and Joshua Weitz and his laboratory group for comments on an earlier version of this manuscript. This work was supported by Simons Foundation Grant 329108 (to E.F.D.) and Gordon and Betty Moore Foundation Grant 3777 (to E.F.D.). In addition, we acknowledge support and assistance from David M. Karl and Matt J. Church through the Hawaii Ocean Time-series program (supported by NSF Grant OCE1260164) and Center for Microbial Oceanography: Research and Education Grant EF0424599. This is a contribution of the Simons Collaboration in Ocean Processes and Ecology.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. PRJNA358725).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714821114/-/DCSupplemental.
References
- 1.Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 2.Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 3.Breitbart M. Marine viruses: Truth or dare. Annu Rev Mar Sci. 2012;4:425–448. doi: 10.1146/annurev-marine-120709-142805. [DOI] [PubMed] [Google Scholar]
- 4.Lasken RS, McLean JS. Recent advances in genomic DNA sequencing of microbial species from single cells. Nat Rev Genet. 2014;15:577–584. doi: 10.1038/nrg3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharon I, Banfield JF. Microbiology. Genomes from metagenomics. Science. 2013;342:1057–1058. doi: 10.1126/science.1247023. [DOI] [PubMed] [Google Scholar]
- 6.Danovaro R, et al. Marine viruses and global climate change. FEMS Microbiol Rev. 2011;35:993–1034. doi: 10.1111/j.1574-6976.2010.00258.x. [DOI] [PubMed] [Google Scholar]
- 7.Cobián Güemes AG, et al. Viruses as winners in the game of life. Annu Rev Virol. 2016;3:197–214. doi: 10.1146/annurev-virology-100114-054952. [DOI] [PubMed] [Google Scholar]
- 8.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 9.Pagarete A, et al. Strong seasonality and interannual recurrence in marine myovirus communities. Appl Environ Microbiol. 2013;79:6253–6259. doi: 10.1128/AEM.01075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow C-ET, Kim DY, Sachdeva R, Caron DA, Fuhrman JA. Top-down controls on bacterial community structure: Microbial network analysis of bacteria, T4-like viruses and protists. ISME J. 2014;8:816–829. doi: 10.1038/ismej.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldsmith DB, Parsons RJ, Beyene D, Salamon P, Breitbart M. Deep sequencing of the viral phoH gene reveals temporal variation, depth-specific composition, and persistent dominance of the same viral phoH genes in the Sargasso Sea. PeerJ. 2015;3:e997. doi: 10.7717/peerj.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waterbury JB, Valois FW. Resistance to co-occurring phages enables marine synechococcus communities to coexist with cyanophages abundant in seawater. Appl Environ Microbiol. 1993;59:3393–3399. doi: 10.1128/aem.59.10.3393-3399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez-Brito B, et al. Viral and microbial community dynamics in four aquatic environments. ISME J. 2010;4:739–751. doi: 10.1038/ismej.2010.1. [DOI] [PubMed] [Google Scholar]
- 14.Clokie MRJ, Millard AD, Mehta JY, Mann NH. Virus isolation studies suggest short-term variations in abundance in natural cyanophage populations of the Indian Ocean. J Mar Biol Assoc UK. 2006;86:499. [Google Scholar]
- 15.Bettarel Y, et al. Strong, weak, and missing links in a microbial community of the N.W. Mediterranean Sea. FEMS Microbiol Ecol. 2002;42:451–462. doi: 10.1111/j.1574-6941.2002.tb01034.x. [DOI] [PubMed] [Google Scholar]
- 16.Tsai AY, et al. Viral lysis and nanoflagellate grazing as factors controlling diel variations of Synechococcus spp. Summer abundance in coastal waters of Taiwan. Aquat Microb Ecol. 2012;66:159–167. [Google Scholar]
- 17.Winget DM, Wommack KE. Diel and daily fluctuations in virioplankton production in coastal ecosystems. Environ Microbiol. 2009;11:2904–2914. doi: 10.1111/j.1462-2920.2009.02038.x. [DOI] [PubMed] [Google Scholar]
- 18.Wommack KE, Ravel J, Hill RT, Chun J, Colwell RR. Population dynamics of chesapeake bay virioplankton: Total-community analysis by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1999;65:231–240. doi: 10.1128/aem.65.1.231-240.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karl DM, Church MJ. Microbial oceanography and the Hawaii Ocean time-series programme. Nat Rev Microbiol. 2014;12:699–713. doi: 10.1038/nrmicro3333. [DOI] [PubMed] [Google Scholar]
- 20.Polovina JJ, Howell EA, Abecassis M. Ocean’s least productive waters are expanding. Geophys Res Lett. 2008;35:L03618. [Google Scholar]
- 21.Mende DR, et al. Environmental drivers of a microbial genomic transition zone in the ocean’s interior. Nat Microbiol. 2017 doi: 10.1038/s41564-017-0008-3. [DOI] [PubMed] [Google Scholar]
- 22.Mizuno CM, Rodriguez-Valera F, Kimes NE, Ghai R. Expanding the marine virosphere using metagenomics. PLoS Genet. 2013;9:e1003987. doi: 10.1371/journal.pgen.1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paez-Espino D, et al. Uncovering Earth’s virome. Nature. 2016;536:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 24.Roux S, et al. Tara Oceans Coordinators Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature. 2016;537:689–693. doi: 10.1038/nature19366. [DOI] [PubMed] [Google Scholar]
- 25.Los M, Wegrzyn G. 2012. Pseudolysogeny. Advances in Virus Research, Bacteriophages, Part A, eds Lobocka M, Szybalski WT (Elsevier, Amsterdam), Vol 82, pp 339–349.
- 26.Brum JR, Steward GF. Morphological characterization of viruses in the stratified water column of alkaline, hypersaline Mono Lake. Microb Ecol. 2010;60:636–643. doi: 10.1007/s00248-010-9688-4. [DOI] [PubMed] [Google Scholar]
- 27.Bryant JA, et al. Wind and sunlight shape microbial diversity in surface waters of the North Pacific Subtropical Gyre. ISME J. 2016;10:1308–1322. doi: 10.1038/ismej.2015.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson LR, et al. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci USA. 2011;108:E757–E764. doi: 10.1073/pnas.1102164108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marston MF, Martiny JBH. Genomic diversification of marine cyanophages into stable ecotypes. Environ Microbiol. 2016;18:4240–4253. doi: 10.1111/1462-2920.13556. [DOI] [PubMed] [Google Scholar]
- 30.Schwartz DA, Lindell D. Genetic hurdles limit the arms race between Prochlorococcus and the T7-like podoviruses infecting them. ISME J. 2017;11:1836–1851. doi: 10.1038/ismej.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang K, Wommack KE, Chen F. Abundance and distribution of Synechococcus spp. and cyanophages in the Chesapeake Bay. Appl Environ Microbiol. 2011;77:7459–7468. doi: 10.1128/AEM.00267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandaa R-A, Larsen A. Seasonal variations in virus-host populations in Norwegian coastal waters: Focusing on the cyanophage community infecting marine Synechococcus spp. Appl Environ Microbiol. 2006;72:4610–4618. doi: 10.1128/AEM.00168-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breitbart M, Rohwer F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005;13:278–284. doi: 10.1016/j.tim.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Mizuno CM, Ghai R, Saghaï A, López-García P, Rodriguez-Valera F. Genomes of abundant and widespread viruses from the deep ocean. MBio. 2016;7:e00805-16. doi: 10.1128/mBio.00805-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hurwitz BL, Sullivan MB. The Pacific Ocean virome (POV): A marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS One. 2013;8:e57355. doi: 10.1371/journal.pone.0057355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurwitz BL, Westveld AH, Brum JR, Sullivan MB. Modeling ecological drivers in marine viral communities using comparative metagenomics and network analyses. Proc Natl Acad Sci USA. 2014;111:10714–10719. doi: 10.1073/pnas.1319778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brum JR, et al. Tara Oceans Coordinators Ocean plankton. Patterns and ecological drivers of ocean viral communities. Science. 2015;348:1261498. doi: 10.1126/science.1261498. [DOI] [PubMed] [Google Scholar]
- 38.Doron S, et al. Transcriptome dynamics of a broad host-range cyanophage and its hosts. ISME J. 2016;10:1437–1455. doi: 10.1038/ismej.2015.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clokie MRJ, Mann NH. Marine cyanophages and light. Environ Microbiol. 2006;8:2074–2082. doi: 10.1111/j.1462-2920.2006.01171.x. [DOI] [PubMed] [Google Scholar]
- 40.Mann NH, Cook A, Millard A, Bailey S, Clokie M. Marine ecosystems: Bacterial photosynthesis genes in a virus. Nature. 2003;424:741–741. doi: 10.1038/424741a. [DOI] [PubMed] [Google Scholar]
- 41.Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. 2005;438:86–89. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]
- 42.Benson R, Martin E. Effects of photosynthetic inhibitors and light-dark regimes on the replication of cyanophage SM-2. Arch Microbiol. 1981;129:165–167. [Google Scholar]
- 43.Ottesen EA, et al. Ocean microbes. Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages. Science. 2014;345:207–212. doi: 10.1126/science.1252476. [DOI] [PubMed] [Google Scholar]
- 44.Suttle CA, Chen F. Mechanisms and rates of decay of marine viruses in seawater. Appl Environ Microbiol. 1992;58:3721–3729. doi: 10.1128/aem.58.11.3721-3729.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jia Y, Shan J, Millard A, Clokie MRJ, Mann NH. Light-dependent adsorption of photosynthetic cyanophages to Synechococcus sp. WH7803. FEMS Microbiol Lett. 2010;310:120–126. doi: 10.1111/j.1574-6968.2010.02054.x. [DOI] [PubMed] [Google Scholar]
- 46.Aylward FO, et al. Microbial community transcriptional networks are conserved in three domains at ocean basin scales. Proc Natl Acad Sci USA. 2015;112:5443–5448. doi: 10.1073/pnas.1502883112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kashtan N, et al. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science. 2014;344:416–420. doi: 10.1126/science.1248575. [DOI] [PubMed] [Google Scholar]
- 48.Ribalet F, et al. Light-driven synchrony of Prochlorococcus growth and mortality in the subtropical Pacific gyre. Proc Natl Acad Sci USA. 2015;112:8008–8012. doi: 10.1073/pnas.1424279112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson ST, et al. Coordinated regulation of growth, activity and transcription in natural populations of the unicellular nitrogen-fixing cyanobacterium Crocosphaera. Nat Microbiol. 2017;2:17118. doi: 10.1038/nmicrobiol.2017.118. [DOI] [PubMed] [Google Scholar]
- 50.Gifford SM, Becker JW, Sosa OA, Repeta DJ, DeLong EF. Quantitative transcriptomics reveals the growth- and nutrient-dependent response of a streamlined marine methylotroph to methanol and naturally occurring dissolved organic matter. MBio. 2016;7:e01279-16. doi: 10.1128/mBio.01279-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roux S, Enault F, Hurwitz BL, Sullivan MB. VirSorter: Mining viral signal from microbial genomic data. PeerJ. 2015;3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quinlan AR. BEDTools: The Swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics. 2014;47:11.12.1–11.12.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thaben PF, Westermark PO. Detecting rhythms in time series with RAIN. J Biol Rhythms. 2014;29:391–400. doi: 10.1177/0748730414553029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.