Abstract
Corn, a 28-nucleotide RNA, induces yellow fluorescence of its cognate ligand (3,5-difluoro-4-hydroxybenzylidene-imidazolinone-2-oxime, DFHO) by >1000-fold. It was selected in vitro to overcome limitations of other fluorogenic RNAs, particularly rapid photobleaching. We now report the Corn-DFHO co-crystal structure, discovering that the functional species is a quasisymmetric homodimer. Unusually, the dimer interface, where six unpaired adenosines break overall 2-fold symmetry, lacks any intermolecular base pairs. The homodimer encapsulates one DFHO at its inter-protomer interface, sandwiching it with a G-quadruplex from each protomer. Corn and the green-fluorescent Spinach RNA are structurally unrelated. Their convergent use of G-quadruplexes underscores the usefulness of this motif for RNA-induced small-molecule fluorescence. The asymmetric dimer interface of Corn could form the basis for the development of mutants that only fluoresce as heterodimers. Such variants would be analogous to Split GFP, and may be useful in analyzing RNA co-expression or association, or in designing self-assembling RNA nanostructures.
Graphical abstract
INTRODUCTION
Corn1 is an RNA aptamer2 that was recently selected in vitro to bind to 3,5-difluoro-4-hydroxybenzylidene-imidazolinone-2-oxime (DFHO, Fig. 1a), a soluble analog of the intrinsic fluorophore3–5 of red fluorescent protein (RFP). Previously, several RNA aptamers that bind small molecules and markedly enhance their fluorescence have been reported. These include the malachite green (MG) aptamer6, “Spinach” (ref. 7) that binds DFHBI (a soluble analog of the fluorophore of green fluorescent protein, Fig. 1a), and “RNA Mango” (ref. 8, 9) that binds certain thiazole orange derivatives. Like those RNAs, Corn enhances the fluorescence of its bound chromophore more than 1000 times upon binding. In principle, by fusing them to RNAs of interest, these various aptamers can be used to visualize RNA expression or localization in live cells which have been soaked with their cognate chromophores1,6–8. These genetically-encodable RNA tags have the potential to accelerate the study of cellular RNAs much as applications of green fluorescent protein (GFP) revolutionized10 the study of proteins.
Figure 1.
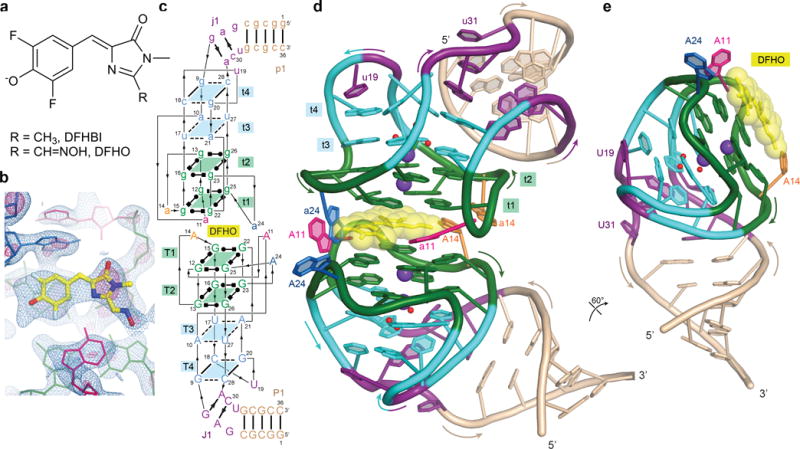
Structure of Corn-DFHO complex. (a) Chemical structures of DFHO and DFHBI (ref. 1,7). (b) Unbiased SAD experimental electron density map overlaid on refined Corn-DFHO structure. Blue and pink meshes correspond to 1.5σ and 4σ contours, respectively. (c) Sequence and secondary structure of Corn-DFHO. Thin lines denote connectivity and Leontis-Westhof symbols44 denote noncanonical base pairs. (d) Cartoon representation of the Corn-DFHO complex, color-coded as in (c). Purple and red spheres represent K+ ions and water molecules, respectively. (e) Cartoon representation of the A protomer of Corn RNA and DFHO, rotated 60° from (d).
Corn-DFHO overcomes three important limitations for in vivo RNA studies of previously reported aptamer-chromophore complexes1. First, unlike the MG chromophore11, the Corn chromophore (DFHO) does not become appreciably cytotoxic when illuminated. Second, unlike malachite green and thiazole orange, DFHO produces only minimal background fluorescence when applied to live cells. Third, and most importantly, Corn-DFHO exhibits markedly increased photostability compared to other aptamer-chromophore complexes12,13, both in vitro and in vivo. Notably, increased photostability is not an intrinsic property of the chromophore1, but a result of the specific interaction of DFHO with the Corn aptamer RNA. Thus, variants of the Spinach RNA aptamer selected to bind to DFHO instead of DFHBI (“Orange Broccoli” and “Red Broccoli”) did not endow the chromophore with the increased photostability exhibited by Corn-DFHO (ref. 1). An additional property of Corn that may facilitate its application to in vivo RNA studies is its small size. Deletion analyses and mutagenesis indicate that 28 nucleotides (nt) suffice for this RNA to bind to DFHO and induce fluorescence1. In contrast, Spinach is nearly 100-nt long7, and even “Baby Spinach”, the result of structure-guided minimization14, is still comprised of 50 nt. A compact aptamer-chromophore complex is less likely to disturb properties of cellular RNAs to which it is appended, and may also have a reduced tendency15 to misfold.
To understand how a small RNA aptamer can induce fluorescence of a chromophore derived from RFP, and as a starting point to analyze and engineer its photophysical properties, we have now determined the crystal structure of the Corn-DFHO complex at 2.35 Å resolution. Unexpectedly, Corn forms a quasisymmetric16 homodimer that binds to one molecule of DFHO, both in the crystalline state and in solution. The dimer interface, which is flanked by a G-quadruplex17,18 from each protomer, encapsulates DFHO and is unusual in lacking any inter-protomer base-pairing. Through structure-guided mutagenesis, we discovered pairs of Corn point mutants that induce DFHO fluorescence only as heterodimers. Such RNAs could form the basis of molecular tools suitable for detecting co-expression and co-localization of RNAs in vivo, analogous to applications of Split GFP and its variants19,20.
RESULTS
Overall structure of the Corn-DFHO complex
A 36-nt RNA construct, comprised of the 28-nt minimal Corn sequence1 (Supplementary Results, Supplementary Fig. 1a) extended proximally with a 4 base-pair stem, co-crystallized with DFHO (Fig. 1a). The unit cell dimensions suggested21 the presence of two RNA molecules per crystallographic asymmetric unit. The structure of the Corn-DFHO complex was solved by the single-wavelength anomalous dispersion (SAD) method and refined at 2.35 Å resolution (Supplementary Table 1 and Online Methods). The unbiased experimental electron density map (Fig. 1b, Supplementary Fig. 1b) was of high quality, allowing the entire RNA to be traced readily. Two molecules of Corn RNA bind a single DFHO molecule as a 2:1 quasisymmetric homodimer (Fig. 1c,d, Supplementary Fig. 1,c,d,e). (Hereafter, the protomers with mean B-factors of 49.0 Å2 and 63.1 Å2 in the Crystal I structure are denoted A and B, respectively, and residues and structural elements of the A and B protomers are identified with upper and lower-case letters e.g., G25 and g25, respectively.) DFHO lies at the interface of the two Corn protomers. After refinement, there was no residual electron density indicative of additional DFHO binding sites.
The overall fold of each Corn protomer loosely resembles a candy cane, in which the loop of a stem-loop (with U19 at its apex), folds back on itself (Fig. 1e). This folding-back brings four segments of the RNA chain into close proximity, allowing formation of four base quadruples (tetrads). In this quadruplex22, the two tiers (T1 and T2) nearest to the homodimer interface are canonical G-quartets in which the guanine bases make alternating Watson-Crick and Hoogsteen interactions17,18, and the remaining two (tiers T3 and T4) are of mixed base composition. DFHO is surrounded by adenosine residues from both protomers, and is sandwiched between the T1 G-quadruplexes of the two protomers, giving rise to an unprecedented nine-tiered arrangement (four base quadruples from each RNA and DFHO).
Dimeric Corn binds one DFHO in solution
We sought to determine if Corn associates with DFHO with a 1:2 chromophore:RNA stoichiometry in solution using several experimental approaches. Analytical ultracentrifugation (AUC), in an excess of DFHO and over a range of Corn RNA concentrations, showed a dominant (> 90%) species with a sedimentation coefficient consistent with a molecular mass of ~27 kDa (Fig. 2a, Supplementary Fig. 2a, Supplementary Table 2). The calculated monomer molecular mass of our Corn RNA construct is 12 kDa. Gel electrophoretic and HPLC analyses have shown1 that while a small fraction (~10%) of Corn transcripts exist as monomers (corresponding to the ~2.7 S peak in AUC, Fig. 2a), the monomers do not fluoresce in the presence of DFHO. Consistent with this, in size-exclusion chromatography with fluorescence detection (F-SEC) analyses, Corn-DHFO mixtures exhibited a single fluorescent species. This species had an elution volume similar to that of the 76 nt tRNALys3 (Fig. 2b). We performed fluorescence measurements at different DFHO:RNA molar ratios to construct Job plots23. This demonstrated an approximately 1:2 chromophore:RNA stoichiometry (1:1.8 ± 0.2 over three sets of experiments; Fig. 2c, Supplementary Fig. 2b).
Figure 2.
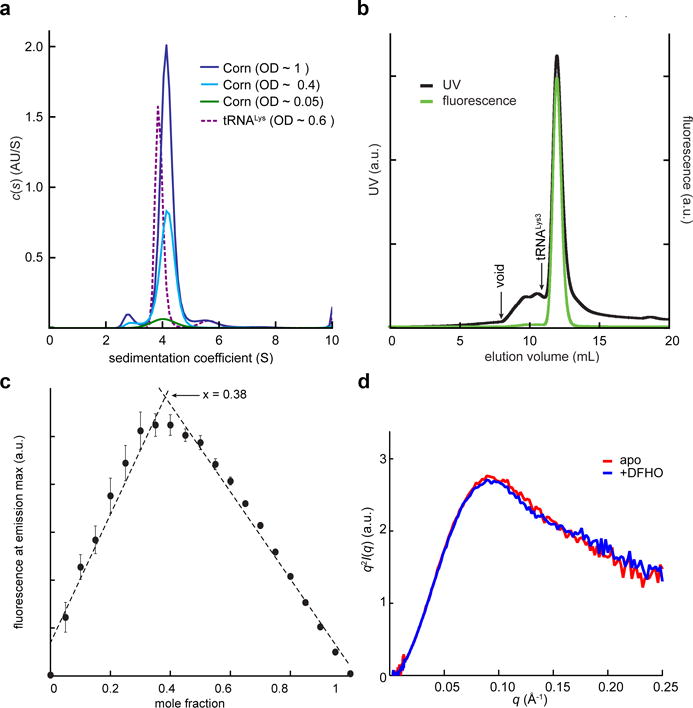
Biophysical analysis of Corn-DFHO dimer. (a) Non-normalized c(s) distributions for Corn RNA at three concentrations and tRNALys3 at a single concentration. (b) Fluorescence-size exclusion chromatogram (F-SEC) for the Corn-DFHO complex. Absorbance was monitored at 260 nm and fluorescence at the emission maximum (543 nm). Arrow denotes void volume (8 mL) as determined by an independent run with blue dextran (MW ~ 6 MDa) under identical conditions. Elution volume of the 76 nt tRNALys3 (10.9 mL) in an independent experiment under identical conditions is also indicated. (c) Job plot23 for DFHO binding to Corn RNA. The fluorescence at the emission maximum (543 nm) was measured as a function of molar fraction [DFHO]/([DFHO]+[Corn RNA]). Mean and standard errors of three independent experiments (Supplementary Fig. 2b). The maximum (0.38), indicates 2:1 stoichiometry of RNA to DFHO. (d) Kratky analysis of experimental free- and DFHO-bound Corn RNA SAXS data.
The preponderance of the dimeric form of Corn over the monomer is consistent with the results of solution small-angle X-ray scattering (SAXS) measurements, which were in good agreement with scattering profiles back-calculated from our homodimeric co-crystal structure (Supplementary Fig. 2c and Supplementary Table 3). Comparison of Kratky plots of SAXS data for free and DFHO-bound Corn (Fig. 2d, Supplementary Fig. 2d) suggest that the RNA is largely prefolded and Corn is a dimer even in the absence of DFHO. To set an upper bound on the dissociation constant (Kd) of the Corn dimer in the absence of DFHO, we analyzed serial dilutions of Corn RNA using native polyacrylamide gel electrophoresis. Even at an RNA concentration of 1.6 nM, we were unable to detect an increase in the amount of the monomeric RNA species. (Supplementary Fig. 3). Thus, the Kd for Corn dimerization is likely less than 1 nM. Since the affinity of Corn for DFHO is considerably weaker1 (Kd ~ 70 nM), this suggests that RNA homodimerization is independent of the chromophore. Altogether, our biophysical analyses indicate that Corn forms a stable homodimer that binds a single molecule of DFHO.
Three-dimensional structure of Corn RNA
Of the four tetrads in the Corn structure, T1 and T2 are canonical17, near-planar G-quartets (Fig. 1 and 3). An octacoordinate metal ion (MA; K+ in our structures) lies on the 4-fold symmetry axis, equidistant from the T1 and T2 planes (Fig. 3b). The mixed tetrad T3 is comprised of two non-canonical, water-mediated A•U base pairs (Fig. 3d). A21 and U27 make a single direct hydrogen bond and one water-mediated interaction, while A10 and U17 make two water-mediated interactions only. A heptacoordinate metal ion (MB; K+ in our structures) lies between the planes of T2 and T3 (Fig. 3c,d). MB coordinates the four O6 atoms of the guanines that form T2, the O2 atom of U17 from T3, and two water molecules in the plane of T3. T4 is comprised of two Watson-Crick G•C base pairs (Fig. 3e), each of which exhibits a pronounced negative24 roll. Despite their angular offset of ~70°, these two G•C pairs hydrogen bond with each other, giving rise to a severely buckled tetrad. T4 is held together by the exocyclic amine of each guanine, which hydrogen bonds with the O4′ of the cytosine from the adjacent G•C pair. A water molecule lies between the T3 and T4 planes. It hydrogen bonds with the O2 of U27, as well as two of the water molecules in the T3 plane. On their face distal from T3, the two G•C pairs of T4 are further buttressed by the stacking with U19 from the apex of the loop and A29 from the junction region J1 (Fig. 3a,e,f). Overall, the quadruplex core of Corn is characterized by the increasing non-planarity of its tetrads concomitant with distance from the dimer interface, and its mixed, complex connectivity (Fig. 1c, 3f). While the two G-quartets T1 and T2 are parallel, the connectivity between T2, T3 and T4 is antiparallel.
Figure 3.
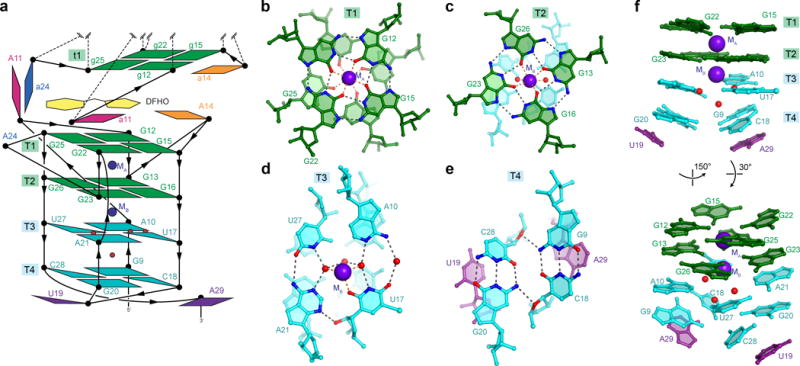
Structure of Corn protomer. (a) Schematic of the connectivity and stereochemistry of a protomer of the A protomer of Corn RNA, the DFHO binding site, and the t1 G-quartet of the B protomer. (b) The T1 G-quartet and cation MA. Black and orange dashed lines represent hydrogen bonds and inner-sphere coordination, respectively. (c) The T2 G-quartet and cation MB. (d) The mixed tetrad T3 and cation MB. (e) The mixed tetrad T4. (f) Side and tilted views of the T1-T4 nucleobases. Lower tiers deviate from planarity.
A junction (J1) transitions between the mixed connectivity T1-T4 quadruplex to the A-form, antiparallel duplex of paired region P1 (Fig. 1e). J1 is comprised of three nucleotide planes. Most distal from the quadruplex are G6 and U31, the latter of which is extrahelical. These two nucleotides do not hydrogen bond with each other. Next are the A7•C30 and G8•A29 planes. Neither of these are conventional base pairs. Instead, the minor groove edges of the nucleotides in each plane are closely apposed. The resulting narrowing of the minor groove is stabilized by ribose zipper-type interactions (Supplementary Fig. 4). Thus, the 2′-OH of C30 makes bifurcated hydrogen bonds with the 2′-OH and N3 of A7, and similarly the 2′-OH of G8 makes bifurcated hydrogen bonds with the 2′-OH and N3 of A29. The unusually narrow minor groove continues into the quadruplex, where two additional ribose zipper-type interactions are present. However, instead of taking place within a tier, as in J1, the ribose zipper hydrogen bonds in the quadruplex are between successive quartet planes. The 2′-OH of U27 (part of T3) makes bifurcated hydrogen bonds with the 2′-OH and N3 of G9 (part of T4). Similarly, the 2′-OH of A10 (part of T3) makes bifurcated hydrogen bonds with the 2′-OH and N3 of G26 (part of T2). The unique geometry of J1 enables continuous base stacking between P1, J1 and the quadruplex on the 5′ side of Corn.
Quasisymmetry in the DFHO binding site
The overall 3D structures of the two homodimeric Corn protomers complexed 2:1 with DFHO are essentially the same. The eight nucleotides of the T1 and T2 G-quartets in the two protomers superimpose almost exactly with a root mean square difference (rmsd) of 0.21 Å for eight C1′ atom pairs (the mean precision of our crystal structure is ~0.2 Å, Online Methods). Excluding the three adenosines at the dimer interface (A11, A14, A24), the rmsd for the entire RNA is 0.79 Å (33 C1′ atom pairs). However, if the three adenosines from each protomer are included (36 C1′ atom pairs), the rmsd increases to 1.2 Å. This is because these six nucleotides adopt distinctly different conformations in the two protomers of identical sequence. Thus, the RNA in the Corn-DFHO complex exhibits quasisymmetry16,25,26.
In the co-crystal structure, the two heterocycles of DFHO and the benzylidene (methine) carbon connecting them are essentially coplanar. DFHO is sandwiched between the T1 and t1 quartets of the two protomers, and hemmed in by five unpaired adenosines (Fig. 4a). In the A protomer, DFHO stacks directly over two nucleobases of T1; the phenolic and imidazolone rings of the chromophore stack on G25 and G12, respectively (Fig. 4b). In the B protomer, DFHO stacks less extensively; while the phenolic ring is primarily under the nucleobase of g25, the imidazolone ring points to the center of the t1 quartet (Fig. 4b). The 4-fold symmetry axes of the two quadruplexes are displaced 5 Å, and make an angle of 4° relative to each other. Nonetheless, because of slight buckling within the T1 and t1 quartets, the nucleobases of G12 and G25 in both protomers form surfaces parallel to the plane of DFHO. This is in contrast to the other two residues of T1 and t1 (G15 and G22), which are offset from this plane by 20°. The planar stack between DFHO, G12 and G25 extends in both directions, including G13 and G26 in T2 and t2, and A10 in T3 and t3 (Supplementary Fig. 4).
Figure 4.
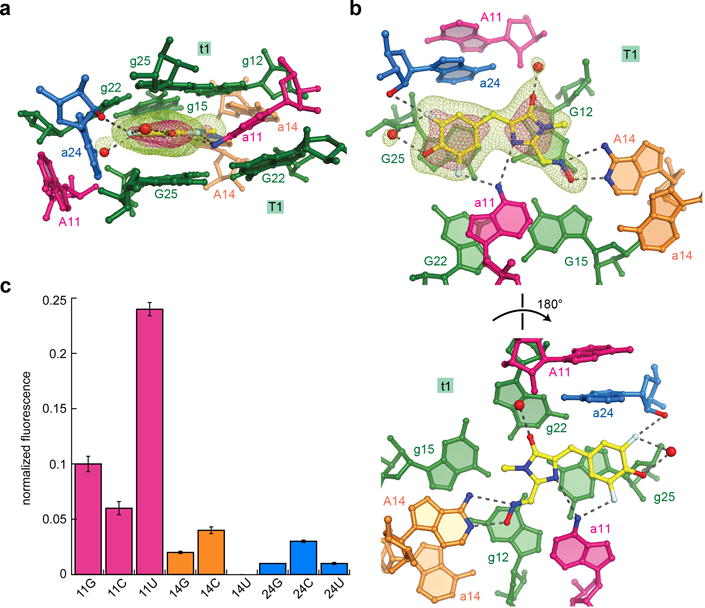
Architecture and functional importance of quasisymmetric DFHO binding site. (a) Side view of the DFHO binding site, color-coded as in Fig. 1. Water molecules are depicted as red spheres. Mesh depicts a portion of the |Fo| − |Fc| electron density map, calculated before addition of DFHO and associated water molecules to the crystallographic model; green and pink mesh are contoured at 3σ and 7σ, respectively. (b) Axial views of the DFHO binding site, depicting DFHO above the T1 G-quartet, looking down on the T1 and t1 G-quartets (top and bottom, respectively) (c) DFHO fluorescence activation by point mutants of the three interfacial adenosines of Corn. Fluorescence of each mutant is normalized to that of the wild-type. Error bars represent s.d. from three independent experiments.
Three adenosines, A14 from the A protomer and a11 and a24 from the B protomer, directly and asymmetrically interact with DFHO (Fig 4a,b). A14 makes two hydrogen bonds with the oxime moiety of DFHO through its Watson-Crick face. The exocyclic amine of a11 donates hydrogen bonds to both the N3 imine of the oxazolone ring and one of the fluorines of the phenolic ring of DFHO. The nucleobases of A11 and a24 stack on each other and are almost perpendicular to the t1-DFHO-T1 planes This arrangement allows the nucleobase of a24 to be in van der Waals contact with both the phenolic ring and the benzylidene carbon of DFHO. In addition, the 2′-OH of a24 donates a hydrogen bond to one of the fluorines of the chromophore. Binding to Corn buries 98% of the total solvent accessible area of DFHO (470 Å2), and results in a fluorescence lifetime of over 4 ns (Supplementary Fig. 5). At the current resolution limit, two ordered water molecules can be identified that are associated with DFHO. They hydrogen bond to the phenolate and carbonyl oxygen atoms of the chromophore, respectively.
Importance of Corn structural features
The importance of the G-quadruplex structure of Corn for its function is consistent with the loss of fluorescence1 in the presence of Li+, which is known to destabilize other G-quadruplexes27–29. In addition to our crystal structure, the ability of Corn RNA to bind to thioflavin T (ThT) and induce its fluorescence is also indicative of the presence of a G-quadruplex1,30. The interaction of Corn with ThT differs from its association with DFHO in that the ligand:RNA stoichiometry is not 1:2 (Supplementary Fig. 6).
During the selection of Corn, it was found that the mutation G8A reduces fluorescence by about a third1. In our structure, the exocyclic amine of G8 hydrogen bonds to G9, presumably helping to stabilize the buckled tetrad T4 (Supplementary Fig. 4a,c). While truncation of Corn to exclude the four distal base pairs of the 5-bp P1 in our crystallization construct does not significantly affect fluorescence (not shown), further truncation by deleting C5 severely reduces fluorescence1, highlighting the importance of the stability of P1 and J1 for Corn function. Site-directed mutagenesis supports the importance of the three interfacial adenosines for Corn function (Fig. 4c). Mutations of A14 and A24 are severely deleterious, while mutations of A11 retain some fluorescence. In particular, the A11U mutant is ~25% as fluorescent as the parental Corn sequence. Our structure shows that in protomer A, nucleotide A11 is extrahelical (Fig. 4a,b), while in protomer B it hydrogen bonds to the two rings of DFHO through its N6 amine. The relatively high activity of the A11U mutant suggests that the hydrogen bonding interactions are not essential for fluorescence activation of DFHO, and steric crowding (possibly by the O4 oxygen atom of an uracil) can substitute for them.
Our discovery that Corn binds DFHO as a quasisymmetric homodimer hinted that interfacial sequence variants may exist that elicit fluorescence only as heterodimers. Such RNAs may serve as probes for RNA co-expression or association. In a preliminary search for such variants, we assayed the fluorescence of all pairs of mutants at the interfacial adenosines, and found three pairings that were markedly fluorescent (Supplementary Fig. 7, Supplementary Table 4). These pairs could form the basis for the future development of RNA analogs of Split GFPs, which are proteins that have been separated into two fragments that are inactive in isolation, but when mixed together, regain fluorescence19. Split GFPs have found numerous applications in fluorescence complementation experiments to examine protein interactions20.
Despite the chemical similarity of DFHO and DFHBI, Spinach-DFHBI and Corn-DFHO are orthogonal. Spinach-DFHBI is nearly 20-fold brighter than Spinach-DFHO, and Corn-DFHO is more than 400-fold brighter than Corn-DFHBI (Fig. 5a, Supplementary Fig. 8). The preference of Spinach for DFHBI likely results from the oxime substituent of DFHO clashing with A69 of that aptamer (Fig. 5b). For Corn, the smaller DFHBI would result in loss of two hydrogen bonds between the ligand and the RNA made by A14 (Fig. 5c). Presumably, if DFHBI is bound, it is no longer constrained enough to fluoresce much above background. Notably, Corn is more selective for its cognate chromophore (DFHO) than Spinach is for DFHBI (Supplementary Fig. 8).
Figure 5.
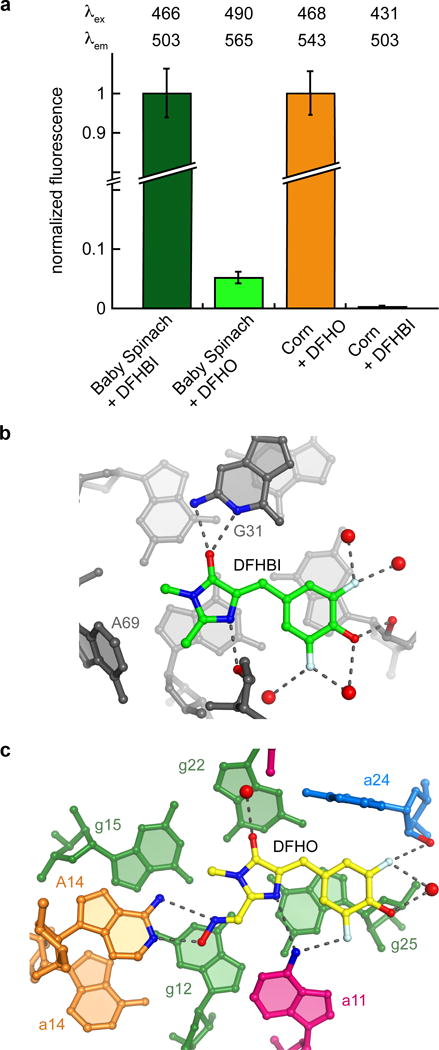
Comparison of chromophore binding and fluorescence activation by Spinach and Corn. (a) Fluorescence of Baby Spinach (ref. 14) or Corn RNA with DFHBI or DFHO. Fluorescence measurements at excitation and emission maxima for each complex, λex and λem, respectively, were corrected by subtracting the fluorescence of the fluorophore at the corresponding λex and λem. Baby Spinach experiments are normalized to Baby-Spinach-DFHBI fluorescence and Corn experiments are normalized to Corn-DFHO experiments. (b) Interactions between DFHBI and Spinach RNA (PDB 4TS0). (c) Interactions between DFHO and Corn RNA.
DISCUSSION
GFP, RFP and their many variants have divergent amino acid sequences and exhibit different fluorescence spectra, yet share a conserved three-dimensional structure, the canonical 11-stranded β-barrel10,31,32. In contrast, when aptamer RNAs that bind to chromophores derived from the intrinsic chromophores of the fluorescent proteins were selected in vitro, they had sequences that suggested multiple independent structural classes7. The crystal structures of Spinach (ref. 14,33) and Corn demonstrate that, indeed, two fluorogenic RNAs that bind to chemically similar chromophores can have unrelated overall structures. Both aptamers organize their otherwise distinctly different chromophore binding sites around G-quadruplexes, indicating that they have converged independently on this structural motif, and supporting the hypothesis that G-quadruplexes are uniquely well suited to construct RNAs that induce fluorescence from GFP-derived chromophores14. The structural diversity of fluorogenic RNAs may facilitate the design of fluorescent tags with a range of functions that exceeds what has been achieved for fluorescent proteins within the structural confines of the 11-stranded β-barrel.
The homodimer interface of Corn is remarkable in two ways. First, despite its stability (Kd < 1 nM), there are no inter-protomer Watson-Crick base pairs. Kissing loops34,35, perhaps the most widespread strategy for homodimerization of RNAs (other than simple duplex formation), result from base-pairing of the complementary loop sequences of two stem-loops, and even association of larger RNAs and RNPs typically involves the formation of some intermolecular Watson-Crick pairs (e.g., between tRNA and the T-box riboswitch36,37, or between U2 and U6 snRNAs in the spliceosome38). Second, despite the global 2-fold symmetry of the Corn heterodimer, the arrangement of the three interfacial adenosines residues is asymmetric (Figs. 1d, 4a,b). DFHO is bound in a flat conformation, and thus exhibits a plane of mirror symmetry. Since Corn RNA is chiral, and therefore cannot exhibit mirror symmetry, its homodimeric binding site to a mirror-symmetric ligand must necessarily break symmetry. Previously characterized homodimeric RNAs (e.g., ref. 34,39,40), exhibit two-fold rotational symmetry (see ref. 41 for an example of a protein-malachite green complex with two-fold rotational symmetry); to our knowledge, this is the first reported example of quasisymmetry in an RNA homodimer. Quasisymmetry is an important organizing principle for proteins in large assemblies such as viruses25,26, and our finding that one RNA sequence can adopt multiple stable conformations in one particle adds a new principle to the design of RNA nanostructures. Future structural characterization of chromophore-free Corn will be needed to establish whether the Corn dimer is intrinsically asymmetric, or if the asymmetry is induced by binding of DFHO, and what RNA-RNA interactions give rise to the stability of the ligand-free homodimer.
The rapid photobleaching exhibited by the Spinach-DFHBI complex7,12,13 limits its applications, and motivated1 in vitro selection of Corn. Crystallographic studies14,33 of Spinach revealed a binding site that makes only three hydrogen-bonding interactions with the imidazolone of the DFHBI fluorophore. Since mobility is linked to photodamage in several fluorophores42,43, it was hypothesized that functionalizing the imidazolone with additional hydrogen bonding groups that could be bound by an RNA would result in a more photostable RNA-fluorophore complex1. Comparison of the Spinach and Corn ligand binding sites (Fig. 5b,c) demonstrates that, consistent with the greatly increased photostability of Corn, this RNA makes additional hydrogen bonds to the imidazolone, exploiting the oxime moiety that distinguishes DFHO from DFHBI. The structures of Corn and Spinach are distinctly different. Corn sandwiches DFHO between G-quadruplexes from the two RNA protomers, while Spinach constrains DFHBI between a G-quadruplex and a base triple14,33. Spinach hydrogen bonds with DFHBI using the Watson-Crick face of a guanine and two ribose 2′-OHs (Fig. 5b). Corn forms hydrogen bonds with DFHBI using unpaired adenine nucleobases from both protomers (Fig. 5c).
The crystal structure of the Corn RNA in complex with DFHO reveals that the aptamer RNA binds the chromophore with 2:1 stoichiometry, showing how an RNA homodimer can induce fluorescence of a small molecule by binding it at an asymmetric dimer interface. That Corn, RNA Mango9 and Spinach14,33 all employ G-quadruplexes at their structural cores speaks to the ubiquity of quadruplexes in RNA aptamers that promote small-molecule fluorescence, while their distinctly different structures suggest the versatility of this nucleic acid structural motif. Structure-guided mutagenesis of Corn suggests that it may be possible to generate obligate heterodimeric variants that may serve as new fluorescence-based tools for detecting and quantifying RNA association. By virtue of their directionality and small size, such heterodimeric RNAs may also find use in the design of novel fluorescent RNA nanostructures.
ONLINE METHODS
RNA and chromophore preparation
The RNA constructs used in this study are listed in Supplementary Table 5. RNAs 3–12 were prepared by in vitro transcription from PCR templates, essentially as described45. Transcript RNAs were purified by electrophoresis on 10% polyacrylamide, 1x TBE, 8 M urea gels (29:1 acrylamide/bisacrylamide) and were electroeluted, washed with 1 M KCl, desalted by centrifugal ultrafiltration (Amicon, Millipore), filtered (Amicon Ultrafree-MC) and stored at 4 °C. RNAs 1 and 2 were chemically synthesized (Dharmacon, GE Life Sciences) deprotected according to manufacturer’s instructions, desalted by ultrafiltration as above, stored at 4 °C and used without further purification. DFHO was synthesized and purified as described1, dissolved in neat DMSO at 20 mM, and stored at −20°C until use.
Crystallization and diffraction data collection
RNA (crystal I, RNA 1; crystal II, RNA 2) was heated to 95 °C for 2 min in Hepes-NaOH, pH 7.0, and KCl and cooled on ice for 2 min. MgCl2 and DFHO were added, and the solution was heated to 65 °C for 5 min and cooled to 25 °C over 15 min. Final concentrations were 200 μM RNA, 50 mM Hepes-NaOH pH 7.0, 0.1 M KCl, 250 μM DFHO, 10 mM MgCl2 and 2% (v/v) DMSO. For crystallization by the hanging-drop vapor-diffusion method, 200 nl of RNA-DFHO complex was mixed with 100 nl of a reservoir solution consisting of 0.1 M Tris-HCl pH 8.05 and PEG 550 MME (crystal I, 30%; crystal II, 32.5%). Crystals grew in 1–4 days at 21°C to maximum dimensions of 150 × 150 × 100 μm3 and fluoresced intensely above background under ultraviolet light (not shown). Crystal I was vitrified by mounting in a nylon loop and plunging directly into liquid nitrogen. Crystal II was transferred to the reservoir solution supplemented with 0.1 mM iridium hexamine and equilibrated for one hour prior to vitrification. Crystals I and II contain one 2:1 Corn:DFHO complex per crystallographic asymmetric unit. SAD data from crystal I were collected at 100 K with 1.5498 Å X-radiation at beamline 5.0.2 of the Advanced Light Source (ALS). Anomalous data from crystal II were collected at 100 K with 1.1050 Å X-radiation at ALS. Data were indexed, integrated, and scaled with HKL2000 (ref. 46). Data collection statistics are summarized in Supplementary Table 1.
Structure determination and refinement
Substructure determination, SAD phasing and density modification on Crystal II data was with AutoSol in Phenix (ref. 47). The best maps were from a heavy atom substructure consisting of two Ir atoms, using the full resolution of the data, and a solvent content of 0.4. The electron density maps allowed immediate tracing48 of 70 nucleotides in the asymmetric unit (Supplementary Fig. 1b). Rounds of manual rebuilding were interspersed with maximum-likelihood restrained refinement against SAD data with Refmac5 (ref. 49). ERRASER (ref. 50) improved RNA geometry. Metal ions were identified based on inspection of anomalous difference Fourier syntheses and by B-factor analysis. The Ir co-crystal structure (Crystal II) consists of 72 RNA residues, 1 DFHO molecule, 4 K+ and 2 Mg2+ ions, 2 Ir atoms, 1 DMSO and 13 water molecules. The structure of Crystal I was solved by molecular replacement51 using a partial model build with Crystal II data as a search model (TFZ = 31.0, LLG = 2647). The sequence register was confirmed by the location of two strong peaks (>10 σ) in an anomalous difference map calculated with Crystal I data, which located the site-specific uracil to 5-iodouracil substitution of residue 17 in each protomer. The iodouracil co-crystal structure (Crystal I) consists of 72 RNA residues, 1 DFHO molecule, 4 K+ and 4 Mg2+ ions, and 52 water molecules. Refinement statistics are summarized in Supplementary Table 1. Mean coordinate precision values reported are maximum likelihood estimates47. Structural figures were prepared with PyMOL (http://www.pymol.org/) and the refined Crystal I atomic coordinates.
Analytical ultracentrifugation
RNA (RNA 3) was heated to 95 °C for 2 min in Tris-HCl, pH 7.5, and KCl and cooled on ice for 2 min. MgCl2 and DFHO were added, and the solution was heated to 65°C for 5 min and cooled to 25°C over 15 min. Final concentrations were 3.42 μM RNA and 6.84 μM DFHO (OD ~ 0.9), 1.37 μM RNA and 2.74 μM DFHO (OD ~ 0.45), and 171 nM RNA and 342 nM DFHO (OD ~ 0.05) in 1x AUC buffer (50 mM Tris-HCl pH 7.5, 0.1 M KCl, 10 mM MgCl2). The reference cell sample for each Corn sample was comprised of the matched concentration of DFHO in 1x AUC buffer. Control tRNALys3 was heated to 80 °C for 2 min in Tris-HCl, pH 7.5, and KCl, and heated for 60 °C for 2 min. MgCl2 was added and the solution was incubated on ice for 15 min. The final concentration was 0.98 μM (OD ~ 0.6). The reference cell sample for the tRNA sample was 1x AUC buffer. AUC analyses are summarized in Supplementary Table 2. Absorbance was measured at 260 nm with a run speed of 60,000 RPM at 20 °C.
Fluorescence spectroscopy
For Split Corn measurements, RNA corresponding to one Corn complex (wt, RNA 3; 11G, RNA 4; 11C, RNA 5; 11U, RNA 6; 14G, RNA 7; 14C, RNA 8; 14U, RNA 9; 24G, RNA 10; 24C, RNA 11; 24U, RNA 12) was heated to 95°C for 2 min in Tris-HCl, pH 7.5, and KCl and cooled on ice for 2 min. MgCl2 and DFHO were added, and the solution was heated to 65°C for 5 min and cooled to 25°C over 15 min. Final concentrations for Split Corn experiments were 0.5 μM of each RNA for measurements of two mutants or 1 μM of a single mutant RNA, 5 μM DFHO in 1x F-SEC buffer (0.1 M Tris-HCl, pH 7.5, 0.1 M KCl, 10 mM MgCl2). For fluorescence measurements for the construction of Job plots, the total concentration of the RNA and DFHO was held constant (2.5 μM) while their mole fractions were varied; fluorescence was measured in 1x AUC buffer (50 mM Tris-HCl, pH 7.5, 0.1 M KCl, 10 mM MgCl2). For measurements of Baby Spinach and Corn fluorescence with DFHBI or DFHO, RNA corresponding to one Corn or Baby Spinach complex (Corn, RNA 3; Baby Spinach, RNA 13) was folded as above. Final concentrations were 1 μM Baby Spinach RNA or 0.5 μM Corn dimer (1 μM of monomer construct), 2.5 μM DFHBI or DFHO in 1x AUC buffer (50 mM Tris-HCl, pH 7.5, 0.1 M KCl, 10 mM MgCl2). Fluorescence spectra were collected with a PTI fluorimeter at 20°C. For competition measurements on Corn, excitation was at 450 nm, integration time was 1 s nm−1 and excitation and emission slits were 0.5 mm. The two fluorophores were added together to Corn during folding at 65 °C, and their total maximum concentration was 10 μM. For competition measurements on Baby Spinach14, 1 μM RNA folded by incubating 2 min at 90 °C, 2 min on ice, the two fluorophores and 5 mM MgCl2 were added, and the mixture incubated for 5 min at 65 °C, and cooled to 25 °C over 15 min. Maximum total concentration of the two fluorophores was 10 μM, excitation was 450 nm, and integration time was 2 sec. All results represent average of three independent experiments and the error bars indicate standard deviation. For ThT measurements, 4-(3,6-Dimethyl-1,3-benzothiazol-3-ium-2-yl)-N, N-dimethylaniline chloride (thioflavin T, ThT; Abcam) was dissolved in water at 2 mM concentration and then diluted with 100 mM KCl, 10 mM MgCl2, 100 mM TRIS pH 7.5 to yield a 200 μM stock solution. 0.5μM refolded Corn (100 mM KCl, 10 mM MgCl2, 100 mM TRIS pH 7.5) was incubated with 2.5 μM ThT for 10 minutes in the dark; fluorescence was excited at 450 nm and emission spectra collected with an integration speed of 1 s nm−1.
F-SEC
RNA (RNA 3) was heated to 95 °C for 2 min in Tris-HCl, pH 7.5, and KCl and cooled on ice for 2 min. MgCl2 and DFHO were added, and the solution was heated to 65°C for 5 min and cooled to 25°C over 15 min. Final concentrations were 33.3 μM RNA in 1x FSEC buffer (100 mM Tris-HCl pH 7.5, 0.1 M KCl, 10 mM MgCl2) supplemented with 66.6 μM DFHO. The Corn-DFHO complex was separated in 1x FSEC buffer at 0.5 mL/min on a Superdex 75 size-exclusion column (GE Life Sciences) using a Shimadzu HPLC at 4°C. Absorbance was measured at 260 nm and fluorescence was monitored using excitation of 468 nm and emission of 543 nm.
SAXS
RNA (RNA 3) in 1x SAXS buffer (50 mM Hepes-KOH pH 7.5, 0.1 M KCl, 10 mM MgCl2) was purified by size-exclusion chromatography (Superdex 75, GE Life Sciences). RNA samples were then exhaustively exchanged into buffer (1x SAXS or 1x SAXS supplemented with 200 μM DFHO and 2% (v/v) DMSO) with Amicon centrifugal microconcentrators (Millipore). RNA samples were diluted to a final concentration of 0.45 g/L. SAXS experiments were performed at beamline 12-ID-B of the Advanced Photon Source (APS). Scattering data were reduced to a one-dimensional scattering plot with IGOR PRO (WaveMetrics). Rg values were calculated from a Guinier plot in the q range, such that qmax × Rg ~ 1.3. PRIMUS (ref. 52) was used to generate the pair probability distributions (P(r)). Kratky and P(r) plots are presented normalized to I0. CRYSOL (ref. 53) was used to back-calculate the scattering profile for the refined Crystal I structure. SAXS analyses are summarized in Supplementary Table 3.
Fluorescence lifetime measurements
Fluorescence lifetime measurements were performed at 20 °C on an EasyLife-LS (Photon Technology International) system equipped with an EL450 diode (Horiba), 535/43 and 445/45 filter sets, integration time was 1 s nm−1 and each measurement was averaged 7 times. A 10 mm path-length quartz cuvette was used. 5 μM Corn was refolded by incubation for 2 min at 90 °C, 2 min on ice, followed by 5 min incubation at 65 °C in the presence of 4 μM DFHO and 10 mM MgCl2, and cooled from 65 °C to 25 °C over 15 min. The instrument response function (IRF) was determined using 5% Ludox (Sigma). The data shown are the average of three independent experiments. Data was fitted to one, two and three exponentials using software provided by the manufacturer.
Synthesis of 32P-labeled RNA
To body-label Corn RNA transcripts, Corn RNA (RNA 3) was in vitro transcribed from a PCR essentially as described45, but in a reaction containing 0.1 mM unlabeled ATP, 0.5 mM GTP, 0.5 mM UTP, 0.5 mM CTP, and 7.5 μL [α-32P]ATP (Perkin-Elmer, 3000 Ci/mmol, 10 μCi/μL) in a 50 μL reaction. After 3 h at 37°C, reactions were treated with RQ1 RNase-free DNase (Promega) for 15 min at 37°C. 50 μL of formamide stop buffer (90% formamide, 1x TBE) was added and the RNA was purified on a 10% polyacrylamide, 1x TBE 8 M urea gel (29:1 acrylamide/bisacrylamide). Labeled RNA was visualized by phosphorimaging. Bands were excised from the gel and soaked for 3 h in 2 mL 0.5 M ammonium acetate (pH 6.9), 1 mM EDTA at room temperature. The eluted RNA was washed with 1 M KCl, desalted and concentrated with a centrifugal concentrator (Amicon, Millipore) with a 10 kDa cut-off, filtered (Amicon Ultrafree-MC) and stored at −20°C. The concentration of RNA was determined by Cherenkov counting and using as reference a 32P-containing NTP of known specific activity.
Gel-shift dimerization assay
A constant amount of body-labeled Corn RNA (15.0 fmol, ~ 100 cpm total) was incubated with increasing concentration of unlabeled Corn RNA in 1x gel-shift buffer (90 μg ml−1 BSA, 5% (w/v) PEG 8000, 50 mM Hepes-KOH pH 7.5, 0.1 M KCl) at 37 °C for 15 min. MgCl2 to a final concentration of 10 mM was added last. BSA and PEG have been shown to alleviate non-specific aggregation of dilute nucleic acids54. Samples were mixed immediately with loading buffer, or incubated for 12 h at 20 °C prior to addition of loading buffer. 2 μl of native loading buffer (50% (v/v) glycerol, 82.5 mM Tris, 164.5 mM Hepes, 0.25 mM EDTA) was added to each 10 μl reaction. For the sample subjected to snap cooling (Supplementary Fig. 3c,f), BSA, PEG and MgCl2 were added after the RNA had been heated to 90 °C for 3 min and placed on ice for 3 min. Samples were loaded onto a 15% 19:1 acrylamide/bisacrylamide gel cast and pre-equilibrated with native running buffer (16.5 mM Tris, 33 mM Hepes, 0.05 mM EDTA, 20 mM KCl, 5 mM MgCl2). Gels (34 cm wide, 25 cm long, 0.05 cm thick) were pre-run for one hour at 32 W. Sample volumes of 12 μL were loaded onto the gel and run for 2 h at 32 W and 3.5 h at 25 W. The anodic and cathodic buffers were mixed every hour. The gel was fixed for 15 min (5% glycerol, 5% acetic acid, 10% methanol), dried (70° for 2 h) and bands visualized by phosphorimaging.
Data Availability
Protein Data Bank
Atomic coordinates and structure factor amplitudes for Corn-DFHO co-crystal structures have been deposited with accession codes 1BJO and 1BJP.
Supplementary Material
Acknowledgments
We thank the staff at beamlines 5.0.1 and 5.0.2 of the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory, and beamline 24-ID-E of the Advanced Photon Source (APS), Argonne National Laboratory, for crystallographic data collection; the staff at APS beamline ID-12-B for SAXS; G. Piszczek (Biophysics Core, US National Heart, Lung and Blood Institute, NHLBI, National Institutes of Health (NIH)) for analytical ultracentrifugation; D. Lee and R. Levine (NHLBI) for mass spectrometry; and S. Bachas, M. Chen, C. Fagan, C. Jones, R. Trachman, M. Warner, and J. Zhang for discussions, and an anonymous referee for proposing the experiment in Supplementary Fig. 3. This work was partly conducted at the ALS, on the Berkeley Center for Structural Biology Beamlines, and at the APS on the 24-ID-E (NE-CAT) and 12-ID-C beamlines, which are supported by the NIH. Use of ALS and APS was supported by the US Department of Energy. This work was supported in part by the NIH (R01 NS064516 S.R.J.), the NIH-Oxford-Cambridge Research Scholars Program (K.D.W.), and the intramural program of the NHLBI, NIH.
Footnotes
Accession Codes
Atomic coordinates and structure factor amplitudes for Corn-DFHO co-crystal structures have been deposited with accession codes 5BJO and 5BJP.
Author contributions
A.R.F. and S.R.J. conceived the project; W.S. and G.S.F. synthesized the chromophore and initial aptamers; K.D.W. designed and carried out crystallographic experiments; K.D.W. and L.S. performed biochemical and biophysical experiments, and K.D.W. and A.R.F. prepared the manuscript with input from all authors.
Competing financial interests
S.R.J. is the co-founder of Lucerna Technologies and has equity in this company. Lucerna has licensed commercialization of technology related to Spinach and other RNA-flourophore complexes. The other authors declare no competing financial interests.
References
- 1.Song W, Filonov GS, Jaffrey SR. Imaging RNA polymerase III transcription dynamics using a photostable RNA-fluorophore complex. Nature Chem Biol. doi: 10.1038/nchembio.2477. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA. 2000;97:11990–11995. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wall MA, Socolich M, Ranganathan R. The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nature Struct Biol. 2000;7:1133–1138. doi: 10.1038/81992. [DOI] [PubMed] [Google Scholar]
- 5.Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-Å resolution. Proc Natl Acad Sci USA. 2001;98:462–467. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baugh C, Grate D, Wilson C. 2.8 Å crystal structure of the malachite green aptamer. J Mol Biol. 2000;301:117–128. doi: 10.1006/jmbi.2000.3951. [DOI] [PubMed] [Google Scholar]
- 7.Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolgosheina EV, et al. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem Biol. 2014;9:2412–2420. doi: 10.1021/cb500499x. [DOI] [PubMed] [Google Scholar]
- 9.Trachman RJI, et al. Structural basis for high-affinity fluorophore binding and activation by RNA Mango. Nature Chem Biol. 2017;13:807–813. doi: 10.1038/nchembio.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Day RN, Davidson MW. The Fluorescent Protein Revolution. CRC Press; Boca Raton: p. 2014. [Google Scholar]
- 11.Grate D, Wilson C. Laser-mediated, site-specific inactivation of RNA transcripts. Proc Natl Acad Sci USA. 1999;96:6131–6136. doi: 10.1073/pnas.96.11.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang PC, et al. Photochemical properties of Spinach and its use in selective imaging. Chemical Sci. 2013;4:2865–2873. [Google Scholar]
- 13.Han KY, Leslie BJ, Fei JY, Zhang JC, Ha T. Understanding the photophysics of the Spinach-DFHBI RNA aptamer-fluorogen complex to improve live-cell RNA Imaging. J Am Chem Soc. 2013;135:19033–19038. doi: 10.1021/ja411060p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warner KD, et al. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nature Struct Mol Biol. 2014;21:658–663. doi: 10.1038/nsmb.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strack RL, Disney MD, Jaffrey SR. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nature Methods. 2013;10:1219–1224. doi: 10.1038/nmeth.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 17.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci U S A. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh I, Hamilton AD, Regan L. Antiparallel leucine zipper-directed protein reassembly: application to the green fluorescent protein. J Am Chem Soc. 2000;122:5658–5659. [Google Scholar]
- 20.Shekhawat SS, Ghosh I. Split-protein systems: beyond binary protein-protein interactions. Curr Op Chem Biol. 2011;15:789–797. doi: 10.1016/j.cbpa.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 22.Laughlan G, et al. The high-resolution crystal structure of a parallel-stranded guanine tetraplex. Science. 1994;265:520–524. doi: 10.1126/science.8036494. [DOI] [PubMed] [Google Scholar]
- 23.Job P. Formation and stability of inorganic complexes in solution. Annali di Chimica Applicata. 1928;9:113–203. [Google Scholar]
- 24.Dickerson RE, et al. Definitions and nomenclature of nucleic acid structure parameters. EMBO J. 1989;8:1–4. doi: 10.1002/j.1460-2075.1989.tb03339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 26.Jones CP, Ferré-D’Amaré AR. RNA quaternary structure and global symmetry. Trends Biochem Sci. 2015;40:211–220. doi: 10.1016/j.tibs.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard FB, Miles HT. Poly(inosinic acid) helices: essential chelation of alkali metal ions in the axial channel. Biochemistry. 1982;21:6736–6745. doi: 10.1021/bi00269a019. [DOI] [PubMed] [Google Scholar]
- 28.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 29.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 30.Xu S, et al. Thioflavin T as an efficient fluorescence sensor for selective recognition of RNA G-quadruplexes. Scientific Reports. 2016;6:24793. doi: 10.1038/srep24793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ormö M, et al. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 32.Yang F, Moss LG, Phillips GN. The molecular structure of green fluorescent protein. Nature Biotech. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 33.Huang H, et al. A G-quadruplex–containing RNA activates fluorescence in a GFP-like fluorophore. Nature Chem Biol. 2014;10:686–691. doi: 10.1038/nchembio.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westhof E, Dumas P, Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985;184:119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- 35.Marino JP, Gregorian RS, Csankovski G, Crothers DM. Bent helix formation between RNA hairpins with complementary loops. Science. 1995;268:1448–1454. doi: 10.1126/science.7539549. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Ferré-D’Amaré AR. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature. 2013;500:363–366. doi: 10.1038/nature12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grigg JC, Ke A. Structural determinants for geometry and information decoding by T box leader RNA. Structure. 2013;21:2025–2032. doi: 10.1016/j.str.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burke JE, Sashital DG, Zuo X, Wang YX, Butcher SE. Structure of the yeast U2/U6 snRNA complex. RNA. 2012;18:673–683. doi: 10.1261/rna.031138.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis JH, et al. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J Mol Biol. 2005;351:371–82. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 40.Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nature Struct Biol. 2001;8:1064–1068. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 41.Szent-Gyorgyi C, et al. Malachite green mediates homodimerization of antibody VL domains to form a fluorescent ternary complex with singular symmetric interfaces. J Mol Biol. 2013;425:4595–4613. doi: 10.1016/j.jmb.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong J, et al. Isomerization in fluorescent protein chromophores involves addition/elimination. J Am Chem Soc. 2008;130:14096–14098. doi: 10.1021/ja803416h. [DOI] [PubMed] [Google Scholar]
- 43.Shank NI, Pham HH, Waggoner AS, Armitage BA. Twisted cyanines: a non-planar fluorogenic dye with superior photostability and its use in a protein-based fluoromodule. J Am Chem Soc. 2013;135:242–251. doi: 10.1021/ja308629w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao H, Edwards TE, Ferré-D’Amaré AR. Structural basis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch. Chem Biol. 2008;15:1125–1137. doi: 10.1016/j.chembiol.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otwinowski Z, Minor W. Processing of diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 47.Adams P, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murshudov GN, AAV, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 50.Chou FC, Sripakdeevong P, Dibrov SM, Hermann T, Das R. Correcting pervasive errors in RNA crystallography through enumerative structure prediction. Nature Methods. 2013;10:74–76. doi: 10.1038/nmeth.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCoy A, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. Journal of Applied Crystallography. 2003;36:1277–1282. [Google Scholar]
- 53.Svergun DI, Bargerato C, Koch MHJ. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst. 1995;28:768–773. [Google Scholar]
- 54.Tsai C, Smider V, Hwang BJ, Chu G. Electrophoretic mobility shift assays for protein–DNA complexes involved in DNA repair. Meth Mol Biol. 2012;920:53–78. doi: 10.1007/978-1-61779-998-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Protein Data Bank
Atomic coordinates and structure factor amplitudes for Corn-DFHO co-crystal structures have been deposited with accession codes 1BJO and 1BJP.


