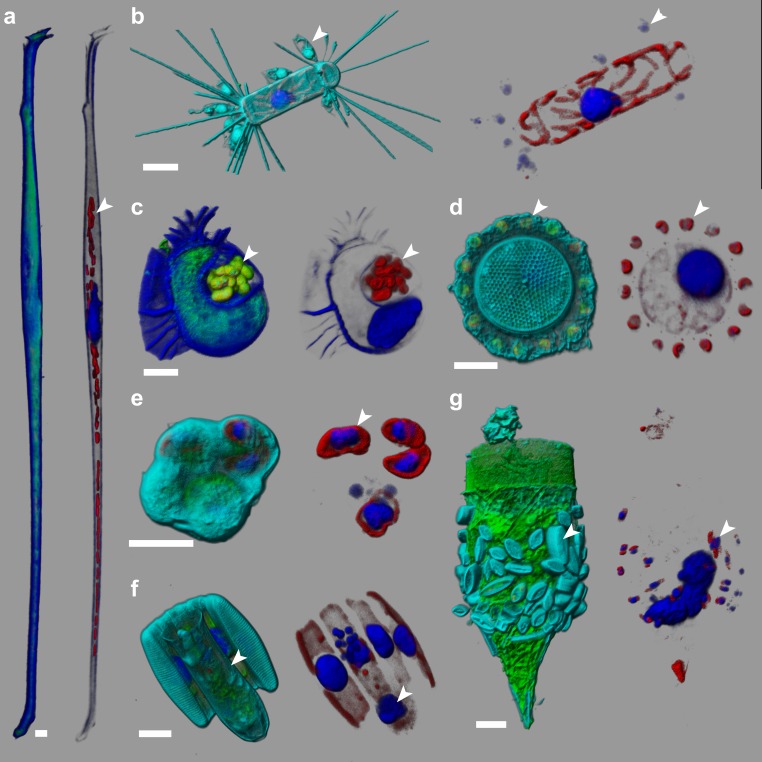
Figure 2. e-HCFM-staining strategy is effective in revealing symbiotic interactions in marine protists.
These seven cells, fixed on board Tara and kept at 4°C for several years, were imaged manually using the e-HCFM workflow (Figure 1). Each cell is illustrated by two panels: the left side overlays all available fluorescent channels whereas the right side displays only the chlorophyll and the Hoechst fluorescence. Four fluorescent channels were recorded: (i) Green: cellular membranes (DiOC6(3)) indicate the core cell bodies; it also stains loricas of tintinnid ciliates (g); (ii) Blue: DNA (Hoechst) identifies nuclei; it also stains the cell-wall of thecate dinoflagellate (a, c); (iii) Red: chlorophyll autofluorescence resolves chloroplasts; (iv) Cyan: PLL-A546 is a generic counterstain for visualizing eukaryotic cells’ surface (not used in a, (c). 3D reconstructions were conducted with the software Imaris (Bitplane). Scale bar is 10 µm. (a) Association between the heterotrophic dinoflagellate Amphisolenia and unidentified cyanobacteria hosted inside the cell wall (arrow head). (b) The diatom Corethron sp. (Figure 2—video 1) harbors several epiphytic nanoflagellates living in small lorica and attached onto the diatom frustule (arrow head). These have been observed in association with different diatom species (see Figure 2—figure supplement 1). (c) The dinoflagellate Citharistes sp. has developed a chamber (phaeosome) for housing cyanobacteria (arrow head). (d) The diatom Thalassiosira sp. is surrounded by a belt chain of 14 coccolithophores (Reticulofenestra sessilis, arrow head). (e) A juvenile pelagic foraminifer hosts endosymbiotic microalgae (arrowhead), likely Pelagodinium dinoflagellates. (f) Colonies of Fragillariopsis sp. diatoms are regularly observed in close interaction with tintinnid Salpingella sp. ciliate (arrowhead). The tintinnid lorica is inserted inside the groove of the barrel formed by the diatom chain. (g) The lorica of the ciliate Tintinnopsis sp. aggregates several epiphyte pennate diatoms, which were still alive prior to fixation as chloroplast and nuclei are visible (arrow head).