Abstract
The majority of pancreatic ductal adenocarcinomas (PDA) rely on the mRNA stability factor HuR (ELAV-L1) to drive cancer growth and progression. Here we show that CRISPR-Cas9-mediated silencing of the HuR locus increases the relative sensitivity of PDA cells to PARP inhibitors (PARPi). PDA cells treated with PARPi stimulated translocation of HuR from the nucleus to the cytoplasm, specifically promoting stabilization of a new target, polyADP-ribose glycohydrolase (PARG) mRNA, by binding a unique sequence embedded in its 3′ untranslated region (UTR). HuR-dependent upregulation of PARG expression facilitated DNA repair via hydrolysis of polyADP-ribose on related repair proteins. Accordingly, strategies to inhibit HuR directly promoted DNA damage accumulation, inefficient PAR removal, and persistent PARP-1 residency on chromatin (PARP-1 trapping). Immunoprecipitation assays demonstrated that the PARP1 protein binds and post-translationally modifies HuR in PARPi-treated PDA cells. In a mouse xenograft model of human PDA, PARPi monotherapy combined with targeted silencing of HuR significantly reduced tumor growth compared to PARPi therapy alone. Our results highlight the HuR-PARG axis as an opportunity to enhance PARPi-based therapies.
Keywords: Pancreatic ductal adenocarcinoma, PARP inhibitor, PARG, HuR, post-transcriptional gene regulation, chemotherapeutic resistance, DNA damage
Introduction
Pancreatic ductal adenocarcinoma (PDA) is the third leading cause of cancer deaths in the United States (1, 2). Poly(ADP-ribose) polymerase (PARP) inhibitors (PARPi) are the best example of a personalized approach to treating PDA with mutations in the BRCA2/Fanconi anemia (FA) pathway (3–5). The primary target, PARP-1, senses and initiates DNA damage repair (DDR) through auto-modification, by covalently adding poly (ADP-ribose) (PAR) onto itself, and trans-modifying other acceptor proteins (6). PARylated PARP-1 modulates chromatin dynamics, recruits key DNA damage repair factors, and contributes to multiple pathways of DNA strand break repair (7). Poly (ADP-ribose) glycohydrolase (PARG) is a critical DDR-related enzyme that works in concert with PARP-1 to coordinate the efficient repair of DNA lesions. Through exo- and endo-glycolytic activity, PARG removes PAR moieties from PARP-1 and other repair factors, and is critical for restarting replication forks and resolving DDR (8–10). Germline or somatic defects in such DDR and related genes (e.g., BRCA1/2, PALB2, and FA genes) render PDA cells dependent on PARP-1 for Homologous Repair (HR)-driven repair, thereby making PARPi and platinum-based therapies promising strategies to treat a distinct subset of PDA tumors (4, 7, 11).
Despite the promise of PARPi therapies, most responsive tumors develop drug resistance (12, 13). Previous studies highlight adaptive resistance mechanisms such as genomic alterations and copy number variations (e.g., BRCA2 reversion mutations) (14, 15). However, genetic events selected for over time are unlikely to solely contribute to the acute plasticity required by cancer cells to rapidly adapt to anti-cancer agents (16). Beyond mutations, post-transcriptional gene regulation via RNA binding proteins (RBPs) is an adaptable reprogramming mechanism that may drive PARPi resistance. Our group has previously shown that the RBP, HuR [Hu antigen R; embryonic lethal abnormal vision-like 1 (ELAVL1)], promotes a drug- resistant phenotype, through its stress-induced cytoplasmic translocation and stabilization of pro-survival mRNA targets (17–20). Herein, we report for the first time that the anti-tumor response to several clinically-relevant PARPi in PDA is regulated by the HuR-dependent stabilization of PARG.
Materials and Methods
Cell culture
PDA cell lines (MIA PaCa-2, PANC-1, Capan-1, Hs 766T, PL11) were purchased from ATCC (Manassas, VA, 2012). All cell lines were routinely tested for mycoplasma using LookOut® Mycoplasma PCR Detection Kit (MP0035 SIGMA), and only early passage (<10) mycoplasma- negative cell lines were used for in vitro and in vivo experiments. As further validation, genomic DNA extracted, PCR amplified and sent for Sanger sequencing. All cell lines were validated as per the expected KRAS and p53 mutation status (21). Cells were cultured in standard DMEM media supplemented with 10% fetal bovine serum (FBS), 1% L-glutamine and 1% penicillin-streptomycin (Invitrogen) at 37°C and 5%CO2. MIA PaCa-2 and Hs 766T with CRISPR/Cas9 knockout of HuR and MIA PaCa-2 cells with doxycycline inducible silencing of HuR were generated and characterized as previously described (18, 22).
Transfection
Transient siRNA silencing and overexpression of HuR was performed as previously described (20). A Myc-DDK-tagged overexpression plasmid (Origene) and commercially available siRNA (Dharmacon) was used for modulating PARG expression. In all experiments, a fraction of cells were analyzed by RT-qPCR to assess knockdown efficiency, and all functional assays were performed 48 hours after transfection.
RT-qPCR and mRNA expression analysis
Cells transfected with indicated siRNAs for 48 hours were directly harvested (mRNA steady-state level) or treated with 5μg/mL Actinomycin D and harvested at indicated time points. Total RNA extraction, reverse transcription and quantitative PCR (RT-qPCR) performed as previously described (18). Relative quantification was performed using the 2−ΔΔCt method. For detecting PARG isoforms, primers were designed to amplify exclusive regions based on splice sites (available upon request) and a qPCR protocol was modified accordingly to accommodate variations in amplicon size and annealing temperatures.
Immunoblot analysis
Cytoplasmic and nuclear extracts were isolated using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo-Scientific) as per manufacturer’s instructions. Total protein extracts were isolated and immunoblotting was performed as previously described (18). Primary antibodies used are HuR (3A2, 1:10,000; Santa Cruz Biotechnology), glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 1:10,000; Cell Signaling Technology), poly ADP-ribose polymerase (PARP-1; 1:1000; Santa Cruz Biotechnology), PAR (1:1000; Trevigen), PARG (1:1000; Millipore, Abcam), Caspase-3 (1:1000; Cell Signaling Technologies), γH2AX (1:1,000; Millipore), Lamin A/C (1:1,000; Cell Signaling Technology). The membranes were scanned and quantified using Odyssey Infrared Imaging System (LI-COR Biosciences).
Ribonucleoprotein Immunoprecipitation assay (RNP-IP)
PARPi treated cells were fractionated and immunoprecipitated and HuR-bound mRNAs were detected as previously described (17, 20, 23).
Cell growth and survival assays
Cells were seeded at 1000 cells per well in 96-well plates, and treated after 24 hours with increasing concentrations of indicated drugs. Short- and long- term cell survival was assessed by staining with Quant-iT Pico Green (Invitrogen) and soft agar colony formation assays respectively, and as previously described (19). IC50 values were determined through non-linear regression analysis.
Chromatin Tethering
Cells cultured and treated in 150mm dishes were washed three times with ice-cold PBS, collected in 1mL PBS by scraping, and pelleted by spinning at 400g for 5 min. Sequential fractionation was performed with ice-cold 0.1% Triton X-100 in CSK buffer as previously described (24) and the final pellet containing (chromatin-bound proteins) and total cell pellets were lysed in RIPA buffer. Histone H3 is used as a positive control and GAPDH a negative control for the chromatin-bound fraction.
Immunoprecipitation
Cell lysates were extracted using a NP-40 lysis buffer (50mM Tris-HCl, 150nM NaCl, 1% NP-40, protease inhibitors). Sepharose beads coated with primary antibodies (anti-rabbit IgG, Santa Cruz Biotechnology, anti-rabbit HuR, MBL; anti-rabbit N-terminal PARP1, Active Motif) were incubated overnight, added to the precleared lysates, and rotated end-over-end at 4°C for 4–6 hours. Beads were washed 3–5 times with lysis buffer and boiled with Laemmli buffer at 95°C for 10 minutes. Equal amounts of input and immunoprecipitated proteins were analyzed by SDS-PAGE gel electrophoresis and visualized by Licor.
Immunofluorescence
MIA PaCa-2 cells were cultured at 5,000 cells per 8mm coverslip. After treatment, cells were fixed, permeabilized, stained and mounted as previously described (Primary- γH2AX; Millipore; 1:500, HuR; 1:200; Santa Cruz Biotechnology; Secondary- Alexa Fluor 488 F anti-mouse; DAPI ProLong Gold, Life Technologies). Coverslips were imaged with a Zeiss LSM-510 Confocal Laser Microscope and Image J was used for foci counting, as previously reported (17, 20).
PAR ELISA
Total protein lysates were analyzed for PARylation using HT Colorimetric PARP/Apoptosis Assay (Trevigen) as per manufacturer’s instructions (25, 26).
Luciferase reporter assays
Full length PARG 3′UTR and a deletion series of putative HuR binding sites on PARG 3′UTR was sub-cloned into the XhoI and NotI sites of the psiCheck2 vector (Promega). Luciferase activity was performed using Dual-Luciferase Reporter Assay System (Promega).
Apoptosis assays
Apoptosis was detected by flow cytometry using a fluorogenic substrate for activated caspase-3/7 in live cells (CellEvent® Caspase-3/7 Green Detection Reagent, Life Technologies)
Xenograft Study
Two independent sets of 6-week-old, female, athymic nude mice received 3 x 106 Mia.shHuR cells per flank, prepared in 100μl solution comprised of 80% DPBS and 20% Matrigel, through subcutaneous injections. Tumors were allowed to grow to an average of 50mm3 (Set I: Day7; Set II: Day 23). Mice were randomized into four groups, two of which were started DOX chow (200mg/kg) (Bio-Serv, cat. #S3888) to induce HuR silencing. When tumors reached an average volume of 100mm3 (Set I: Day15, Set II: Day 23) olaparib was administered through intraperitoneal injection (Set I: 100mg/kg/day, Set II: 50mg/kg/day, 5 days a week). Mouse weights and tumors were measured three times per week using an electronic caliper, and tumor volumes were calculated using the formula Volume = (Length x Width2)/2. No mice lost more than 5% of their initial body weight. Mice were sacrificed and tumors harvested, when one of them surpassed 1,500 mm3 (Set I: Day 36; Set II: Day 56). Mouse protocols were approved by the Thomas Jefferson University Institutional Animal Care and Use Committee.
Statistical Analysis
Data and statistical analysis was performed using ISM SPSS (Version 20.0.0, IBM, Armonk, NY). Tumors that did not reach a calculated volume of 20mm3 by day 25 were excluded from the analysis (Set I: one tumor, in the combined olaparib-siHuR treatment group. Set II: two tumors, in the olaparib only group). Individual tumor volume fold changes were used to normalized tumor volume to a set starting volume of 50mm3 (Set I: at the 16. Set II: at day 25). Log2 (Fold Change) function was used to calculate relative tumor duplications and to extract mean tumor duplication time: Δtime/ (tumor duplications). Tumor volumes were analyzed for normality of distribution using Kolmogorov-Smirnov test. Normally distributed continuous parameters were compared using student’s t-test and non-normally distributed parameters compared using a Mann-Whitney U test. Continuous parameters were presented as mean (± S.E). A p-value of less than 0.05 was defined as significant.
RESULTS
Genetic deletion of HuR enhances PARPi sensitivity
To assess PARPi efficacy, the IC50 values for a panel of PDA cell lines were determined. Consistent with previous reports, the DNA repair deficient (DDR-D) cell lines, Capan-1 (loss of BRCA2) and Hs 766T (defective in FANCG) are significantly more sensitive to the PARP inhibitors olaparib (Fig. 1A), veliparib (Supp. Fig. S1A) and rucaparib, than the DNA repair proficient (DDR-P) PDA cell lines, MIA PaCa-2 and PANC-1 (Supp. Table 1) (27–30).
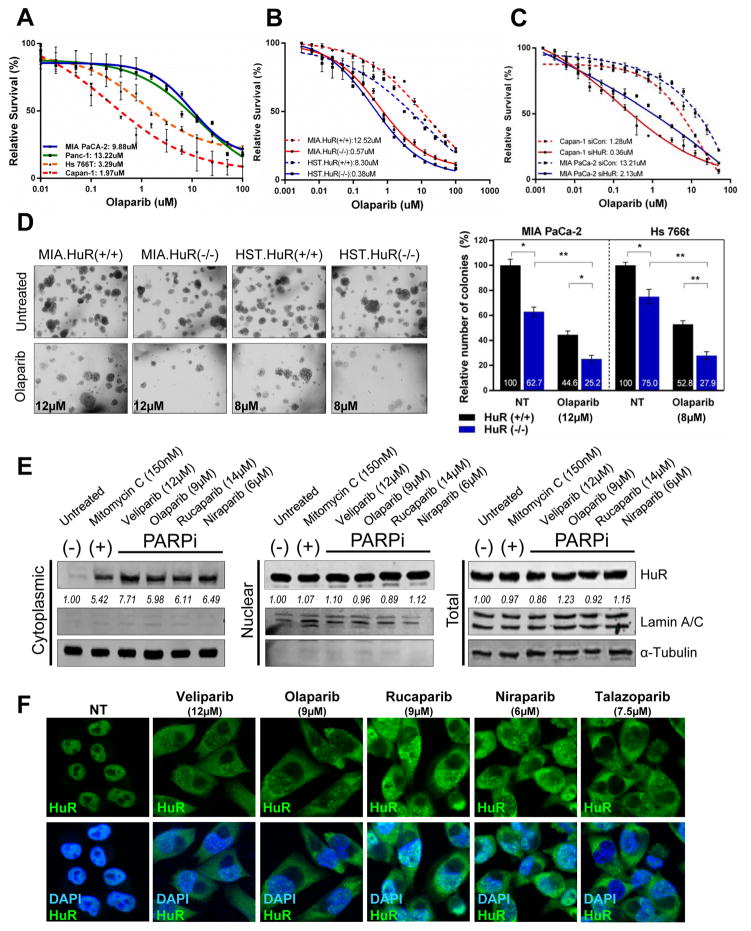
Figure 1. HuR expression regulates sensitivity to PARPi in PDA cells.
Cell survival of PDA cell lines (A), HuR-knockout CRIPSR cell lines, MIA PaCa-2 and Hs 766T [HuR(+/+) vs HuR(−/−)] (B) and HuR-silenced MiaPaCa-2 and Capan-1 cells (C) treated with increasing doses of olaparib for 7 days. (D) Representative images of MIA.HuR(+/+) vs MIA.HuR(−/−) and HST.HuR(+/+) vs HST.HuR(−/−) cells seeded and cultured in soft agar in the presence of respective IC50 doses of olaparib for 4 weeks. (E) HuR expression in MIA PaCa-2 cells treated with indicated IC50 doses of PARPi for 12hr, and fractionated as indicated. Lamin A/C and α-Tubulin used as controls to determine the integrity of nuclear and cytosolic lysates respectively. Mitomycin C used as positive control for cytoplasmic translocation of HuR. (F) Immunofluorescent images of HuR (green) in MIA PaCa-2 cells treated with PARPi for 12hr. Nuclei were stained with DAPI. Magnification 40X.
To evaluate the role of HuR in PARPi response in vitro and in vivo, we used three strategies: 1) siRNA targeting HuR (17, 20); 2) two characterized Clustered Regularly-Interspaced Short Palindromic Repeats (CRISPR)-generated PDA cell lines (DDR-P MIA PaCa-2 and DDR-D Hs 766T) with HuR genetically knocked out (Supp. Fig. S1B) (22); and 3) a doxycycline inducible siHuR cell line MIA.sh290 (18). Dose response curves from cell survival assays in response to several clinically relevant PARPi indicate that CRISPR-knockout of HuR (Table 1) in both MIA PaCa-2 and Hs 766T [HuR (+/+) vs HuR (−/−)] caused a dramatic 20-fold decrease in sensitivity to the PARPi olaparib (Fig. 1B) and veliparib (Supp. Fig. S1C, Table 1). In contrast, we observed a smaller fold change in non-PARPi agents, oxaliplatin (≤7-fold) and gemcitabine (≤3-fold) (Supp. Fig. S1C) (17). We validated these results with siRNA knockdown of HuR in another DDR-D cell line, Capan-1 (Fig. 1C, Supp. Fig. S1D, Table 1). Soft agar growth assays indicated that CRISPR knockout of HuR in MIA PaCa-2 and Hs 766T as well as siRNA silencing of HuR in MIA PaCa-2 and Capan-1 suppresses colony formation and anchorage independent growth under PARPi treatment (Fig. 1D, Supp. Fig. S1E). Accordingly, HuR overexpression promotes resistance to veliparib (2.3- fold change) (Supp. Fig. S1F). Together, these data indicate that HuR expression dramatically modulates the response to PARPi, independent of DDR mutational status.
Table 1. Models to evaluate the role of HuR in PARPi response.
Table indicates the IC50 values of 1) (CRISPR)-generated PDA cell lines DDR- P MIA PaCa-2 and DDR- D Hs 766T with HuR genetically knocked out [HuR (+/+), HuR (−/−)]; 2) siRNA oligos against the HuR coding region as previously described (17, 20); 3) a small molecule inhibitor, MS-444 (40).
| DDR status |
Cell lines | Olaparib | Veliparib | Rucaparib | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IC50 μM |
Fold- change |
P-value (two- tailed) |
IC50 μM |
Fold- change |
P-value (two- tailed) |
IC50 μM |
Fold- change |
P-value (two- tailed) |
|||
| CRIPSR knockout | Deficient | HST.HuR(+/+) | 8.30 | 21.8 | >0.0001 | 2.20 | 2.6 | 0.0009 | 4.56 | 12.6 | >0.0001 |
| HST.HuR(−/−) | 0.38 | 0.85 | 0.36 | ||||||||
| Proficient | MIA.HuR(+/+) | 12.52 | 21.9 | >0.0001 | 12.04 | 15.5 | >0.0001 | 10.22 | 20.8 | >0.0001 | |
| MIA.HuR(−/−) | 0.57 | 0.76 | 0.49 | ||||||||
| siRNA silencing | Deficient | Capan-1 siCon |
1.28 | 3.5 | >0.0001 | 4.25 | 10.1 | >0.0001 | 10.40 | 5.3 | >0.0001 |
| Capan-1 siHuR |
0.36 | 0.42 | 1.95 | ||||||||
| Deficient | Hs 766T siCon |
8.40 | 3.8 | >0.0001 | 6.25 | 2.0 | >0.0001 | 9.25 | 2.76 | >0.0001 | |
| Hs 766T siHuR |
2.24 | 3.12 | 3.35 | ||||||||
| Proficient | MIA PaCa-2 siCon |
13.21 | 6.20 | >0.0001 | 13.32 | 5.3 | >0.0001 | 15.60 | 6.24 | >0.0001 | |
| MIA PaCa-2 siHuR |
2.13 | 2.50 | 2.50 | ||||||||
| Deficient | Capan-1 (Vehicle) | 1.87 | 3.0 | 0.0009 | 1.20 | 2.1 | 0.0009 | 1.65 | 3.8 | >0.0001 | |
| Capan-1 (MS-444) | 0.62 | 0.56 | 0.43 | ||||||||
| Small Molecule inhibition (MS-444) | Deficient | Hs 766T (Vehicle) | 4.32 | 2.3 | >0.001 | 3.0 | 4.1 | >0.0001 | 4.18 | 3.2 | >0.001 |
| Hs 766T (MS-444) | 1.87 | 0.73 | 1.31 | ||||||||
| Proficient | MIA PaCa-2 (Vehicle) | 11.67 | 4.4 | >0.0001 | 12.56 | 4.0 | >0.0001 | 11.88 | 3.2 | 0.0009 | |
| MIA PaCa-2 (MS-444) | 2.68 | 3.20 | 3.68 | ||||||||
PARPi induces cytoplasmic translocation of HuR
We previously demonstrated that veliparib causes HuR translocation in a time-dependent manner (17), peaking at 24 hours. (Supp. Fig. S1G). Building upon these data, we treated MIA PaCa-2 cells with IC50 doses of a panel of PARP inhibitors (veliparib, olaparib, rucaparib, niraparib, talazoparib) for 24 hours. Immunoblotting of fractionated lysates (Fig. 1E) and immunofluorescence (Fig. 1F) indicated that cytoplasmic translocation of HuR significantly increased with PARPi stress while total and nuclear expression remained unchanged.
PARP1 binds and PARylates HuR under stress
Ke and colleagues recently demonstrated that under LPS stimulation, PARP1 directly binds HuR, thus resulting in its PARylation and modulating its nucleocytoplasmic shuttling as well as mRNA target binding(31). Though these findings were established in murine macrophages and human kidney cells, they could potentially have profound implications in carcinogenesis and tumor response, particularly in HuR-mediated stress response pathway. Therefore, we treated MIA PaCa2 cells with PARPi olaparib and a non-PARPi DNA damaging agent oxaliplatin. We demonstrated that HuR and PARP1 bind directly through protein-protein interactions, which is further enhanced upon stress; this results in subsequent PARylation of HuR (Supp Fig. S2A). Future studies will define the role of this protein-protein interaction in PDA cells.
HuR binds PARG mRNA under PARPi stress
As an RNA binding protein, HuR promotes PDA cell survival under stress by regulating expression of pro-survival mRNAs (17, 19, 20, 32, 33). We performed a focused screen of DNA repair enzymes critical for regulating PAR turnover to identify potential mRNA targets (34). A 90% knockdown in HuR expression in MIA PaCa-2 cells was validated with a 40% down-regulation of an established HuR target, dCK (Supp. Fig. S2B)(19). The key members of the PARP family, PARPs 1 and 2, are unchanged demonstrating HuR’s selectivity in regulating DDR related transcripts (Supp. Fig. S2A). However, with HuR knockdown, we detected a significant 65% decrease in PARG expression, the main enzyme responsible for PAR degradation through its endo- and exo- glycolytic activity. Other PAR catabolizing enzymes such as terminal (ADP) ribose glylcohydrolase (TARG), ADP-ribosyl-acceptor hydrolase 3 (ARH3), Macro Domain 1 (MacroD1), Ectonucleotide Pyrophosphatase/Phosphodiesterase 1 (ENPP1) and nudix hydrolase 16 (NUDT16) remain unchanged with HuR knockdown. Such HuR-dependent expression changes in PARG were further validated in both MIA PaCa-2 and Hs 766T HuR-CRIPSR cell lines (Supp. Fig S2C).
To determine if these mRNA expression changes are directly due to HuR binding, we performed ribonucleoprotein immunoprecipitation (RNP-IP) assays (23) on cytoplasmic lysates of MIA PaCa-2 cells treated with respective IC50 doses of PARPi, veliparib (12μM/L) and olaparib (9μM/L) for 12 hours (Fig. 2A). HuR binds to PARG mRNA (11.26 and 9.04 fold change, p ≤0.001) in response to PARP inhibition (Fig. 2B), and does not significantly bind to any other established PAR polymerases or hydrolases (Supp. Fig. S2D). These findings were validated through RNP-IP analysis of the HuR knockout MIA PaCa2 cell line, with the isogenic control (Supp. Fig. S2E).
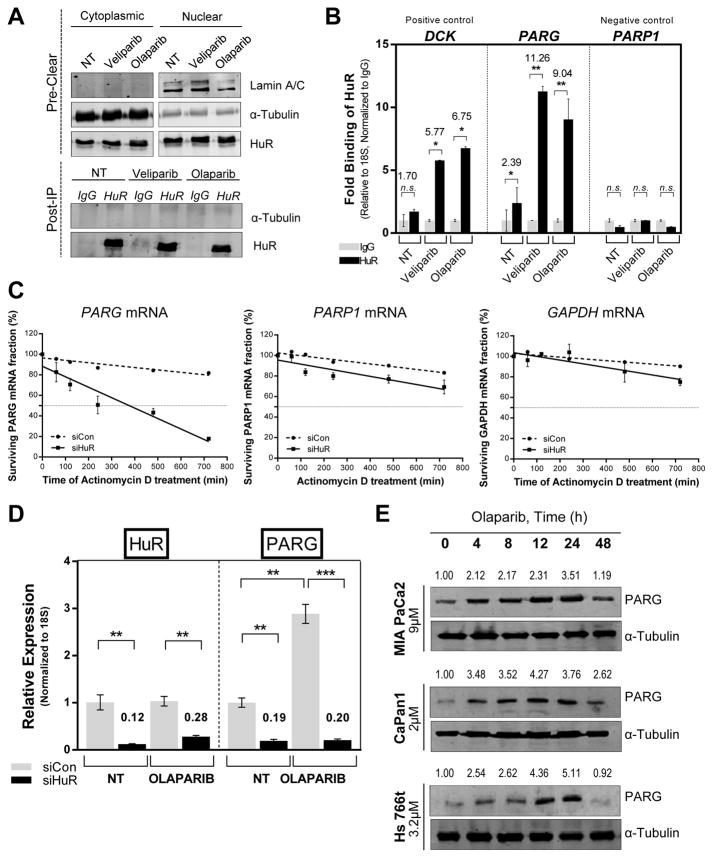
Figure 2. HuR regulates PARG mRNA expression.
(A) mRNP-IP assay performed with cytoplasmic fraction of MIA PaCa-2 cells treated with IC50 doses of veliparib (12μM) and olaparib (9μM) for 12hr, α-Tubulin used as a loading control for the input and a negative control for the IP samples, Lamin A/C used as a control to detect nuclear contamination in the input. (B) The relative binding of PARG mRNA to HuR, normalized to respective IgG controls, as determined by RT-qPCR using 18S rRNA as a loading control, dCK as positive control and PARP-1 as negative control. (C) HuR- silenced MIA PaCa-2 cells were treated with actinomycin D (5 μg/ml) for the indicated times. PARG, GAPDH and PARP-1 mRNA stability was assayed by RT- qPCR using 18S rRNA as a loading control. (D) RT- qPCR indicating HuR and PARG mRNA expression in HuR-silenced MIA PaCa-2 cells incubated in the presence of olaparib for 24hr. (E) PARG expression in DDR- P MIA PaCa-2 and DDR- D Capan-1 and Hs 766T cells treated with veliparib for indicated time points.
PARG is known to undergo alternative splicing resulting in several isoforms (hPARG111, hPARG102 and hPARG 99), which localize to different cellular compartments and maintain PAR homeostasis within the cell. We designed isoform- specific primers of PARG and interrogated HuR- dependent expression changes. HuR knockout MIA PaCa2 cells indicate a significant downregulation of all PARG isoforms (Supp. Fig. S3A), as well as increased mRNA binding in mRNP-IP assays (Supp. Fig. S3B). However, through protein expression assays, we detected and focused on the functional significance of HuR’s regulation of isoform hPARG111, which is primarily nuclear and responsible for the majority of PAR degradation (35). Despite varying levels of hPARG111 mRNA expression, the relevant PDA cell lines, MIA PaCa-2 and PANC-1 (DDR-P) and Capan-1, Hs 766T and PL11 (DDR-D) have similar PARG protein expression (Supp. Fig. 3C).
HuR knockdown decreases PARG mRNA half-life and expression under PARPi stress
HuR silenced MIA PaCa-2 cells (Supp. Fig S2D) treated with a transcriptional inhibitor actinomycin D over a time-course (17, 20, 36) revealed that HuR knockdown resulted in a significant 4–fold decrease in PARG mRNA half-life whereas GAPDH and PARP-1 mRNA stability were not affected (Fig. 2C, Supp. Fig. S2F)(20). Additional RT-qPCR assays confirmed that HuR knockdown decreases PARG expression, both in the presence and absence of PARPi treatment (Fig. 2D). The striking induction of PARG mRNA under olaparib treatment correlates with an increase in PARG protein expression in a time- (Fig. 2E) and dose-dependent manner (Supp. Fig. S3E). Treatment with sub-IC50 doses of non-PARPi DNA damaging agents [gemcitabine (1μM) and oxaliplatin (1μM)] for 24h resulted in cytoplasmic translocation of HuR and corresponding induction of PARG protein expression in MIA PaCa-2 cells (Supp. Fig. S3F). However, for purposes of this study, we sought to explore and focus on the role of PARG expression in regulating PARPi response.
HuR binds to two discrete AU-rich elements in PARG 3′UTR
HuR binds to its target mRNAs through distinct AU-rich elements (AREs) in their 3′-untranslated regions (37). Reporter assays indicated an increase in luciferase activity in cells co-expressing full-length PARG 3′UTR (Luc+3′UTR) and an HuR overexpression plasmid (Fig. 3A), likely due to an increase in HuR’s regulation of PARG via its 3′UTR. Accordingly, this regulatory induction in the presence of veliparib treatment was lost when HuR was silenced (Fig. 3B). Computational sequence predictions identified 3 putative AREs within PARG 3′UTR. To further identify the minimal regulatory HuR-binding sequence, a deletion series of constructs derived from PARG 3′UTR (Supp. Fig. S3G) was co-transfected with HuR overexpression plasmid in MIA PaCa-2 and Hs 766T cells (Supp. Fig. S3H, S3I). Deletion of either or both sites 1 (41bp) and 3 (43bp) caused significant reduction in luciferase activity suggesting that both contribute to HuR’s regulation of PARG 3′UTR.
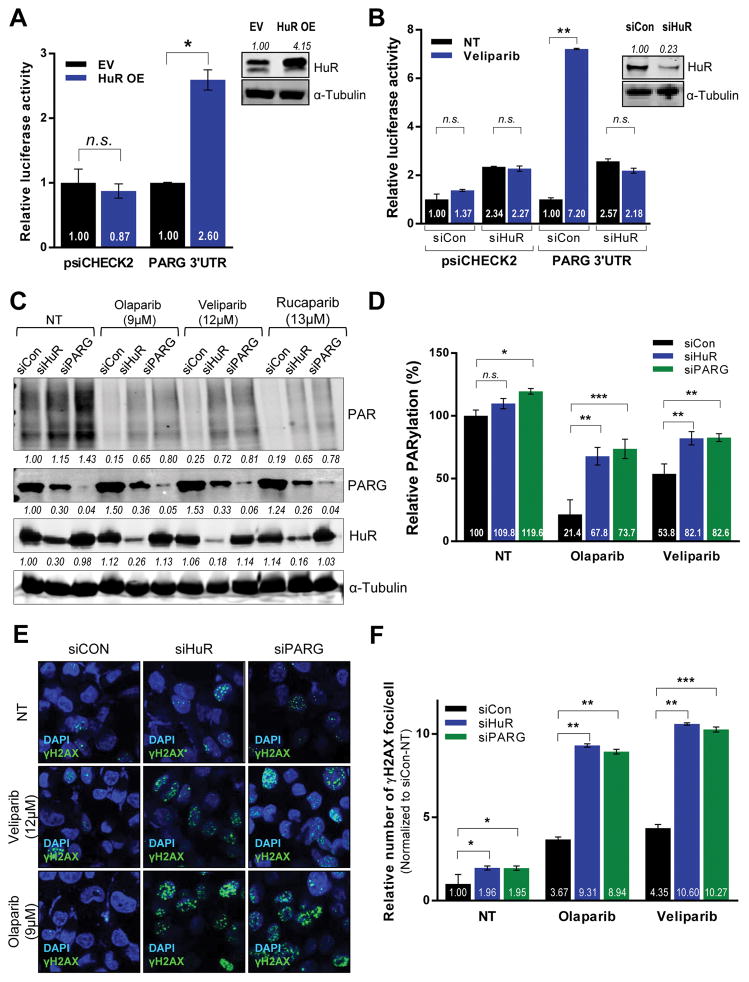
Figure 3. HuR regulates PARG protein expression and function.
Luciferase activity in MIA PaCa-2 cells co-expressing a luciferase reporter construct with PARG 3′UTR and (A) HuR overexpression or (B) HuR silencing (C) HuR, PARG and PAR protein expression in total lysates from HuR- and PARG-silenced MIA PaCa-2 cells treated with IC50 doses of indicated PARPi for 24hours, using α-Tubulin as a loading control. (D) ELISA indicating relative PARylation in MIA PaCa-2 cells transfected and treated as above. The indicated fold changes are means of three independent experiments, normalized to control transfected sample under no treatment (NT). (E) DSBs assessed by immunofluorescence staining for γH2AX (green) in MIA PaCa-2 cells transfected and treated as described above. (F) DNA damage foci were quantified and plotted ± SD.
HuR regulates PARG protein expression
Irrespective of their DDR status, PDA cell lines treated with respective IC50 doses of olaparib showed a significant increase in basal PARG expression (as previously shown, Fig. 2E). MIA PaCa-2 cells were transfected with HuR and PARG siRNAs for 48 hours, followed by treatment with IC50 doses of 3 clinically relevant PARP inhibitors for 24 hours (Fig. 3C). As expected, PARP inhibition induced a mild increase in PARG protein expression in control cells. However, HuR silencing significantly decreased PARG protein expression under no treatment, as well as the corresponding PARPi- treated conditions in both MIA PaCa-2 cells (Fig. 3C) and Hs 766T (Fig. S4A), demonstrating that PARG expression is mediated by HuR even in the absence of stress and independent of DDR status.
HuR regulates PARylation through PARG
Downregulation of PARG, either through HuR silencing or via a PARG-specific siRNA, directly affects the extent of PAR-degradation, therefore causing persistence of total PAR polymers i.e. PARylation, as assessed by immunoblotting (Fig. 3C, top panel) and ELISA (Fig. 3D). Similar results were obtained in DDR-D Hs 766T cells (Supp. Fig. S4A, S4B). Extensive protein expression studies also showed that PARG is significantly downregulated with HuR knockout in both DNA repair proficient MIA PaCa2 and deficient Hs 766T cells, while expression of other PAR catabolizing enzymes such as TARG1, ARH3, ENPP1, and MarcoD1 is not affected(Supp. Fig. S4C). Concurrently, HuR overexpression resulted in PARG upregulation causing a decrease in overall PAR levels in MIA PaCa-2 cells (Supp. Fig. S3D). These data show that HuR regulates PARG expression as well as its downstream function of PAR degradation.
HuR-mediated upregulation of PARG affects DNA damage response and apoptosis
To assess the effects of HuR-mediated PARG regulation on DDR, we performed relative quantification of γH2AX foci, a marker of DSBs in DNA. In control cells, the basal level of DNA damage is increased markedly upon PARP inhibition (Fig. 3E, 3F) and as previously shown (4). However, both HuR and PARG silencing further increased veliparib- and olaparib-induced DNA damage foci in DDR-P MIA PaCa-2 cell line.
The enhanced DNA damage due to HuR and PARG silencing correlated with a dramatic increase in apoptosis upon PARPi treatment, as indicated by staining the apoptotic population with a highly sensitive probe for activated caspase-3/-7 in MIA PaCa-2 and Hs766T cells (Fig. 4A, Supp. S5A). Our results indicate that HuR and/or PARG silencing enhanced PARPi- induced DNA damage and apoptosis regardless of DDR proficiency.
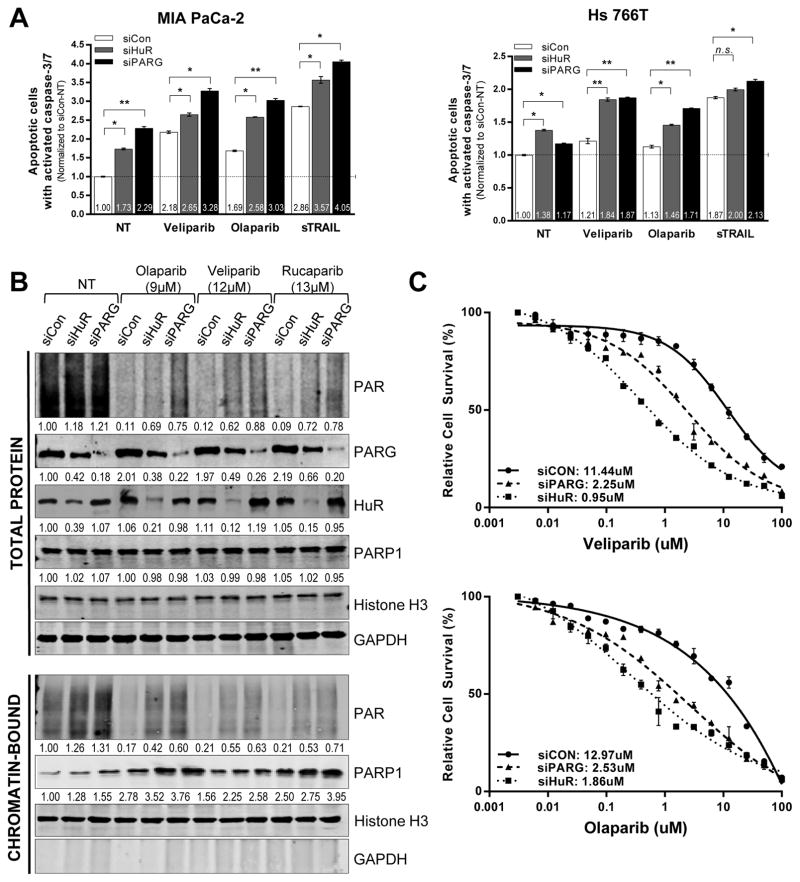
Figure 4. HuR and PARG inhibition enhances PARPi-induced apoptosis and PARP-1 trapping on chromatin and increases PARPi efficacy.
(A) Relative number of apoptotic cells quantified and normalized to control- (NT) MIA PaCa-2 and Hs 766T cells. A 3hr treatment with soluble TNF-related apoptosis-inducing ligand (sTRAIL) is used as a positive control. (B) HuR- and PARG- silenced DDR- P MIA PaCa-2 cells treated with IC50 doses of indicated PARPi for 6h were harvested and fractionated to isolate soluble and chromatin- tethered proteins. HuR, PARG, PARP-1 and PAR expression analyzed, with GAPDH (total protein extract) and Histone H3 (nuclear chromatin- tethered fraction) as the loading controls. A representative image of one of three independent experiments is shown. (C) Cell survival in HuR- and PARG- silenced MIA PaCa-2 cells were treated with increasing doses of olaparib and veliparib for 5 days.
HuR and PARG inhibition enhances PARP trapping on chromatin by PARP inhibitors
In addition to preventing PAR production, a crucial step in DDR, PARP inhibitors can also behave as ‘poisons’ that induce cytotoxic accumulation of inactivated PARP-1-DNA complexes tethered to chromatin (38, 39), thus preventing PARP-1 release from unrepaired DNA strand breaks. We hypothesized that HuR stabilization of PARG in stressed cells could reduce PARPi-induced ‘trapping’ of PARP-1 on chromatin, which potentially allows successful resolution of DNA repair and replication fork progression. We silenced HuR and PARG in MIA PaCa-2 cells followed by treatment with IC50 doses of olaparib, veliparib and rucaparib for 6 hours, and isolated soluble and chromatin-associated proteins. As above, HuR silencing downregulated PARG expression, and silenced HuR and PARG expression resulted in persistent PARylation in the presence of PARP inhibition (Fig. 4B, total protein).
Consistent with previous reports, all three PARPis resulted in increased PARP-1-DNA complexes (trapped PARP-1) (Fig. 4B, chromatin-bound), with olaparib and rucaparib exhibiting a higher PARP trapping potency. Furthermore, HuR and PARG silencing significantly enhanced the extent of trapped PARP-1 on chromatin, both under no treatment (NT) and PARPi treated conditions. Similar results in DDR- D Hs 766T (Supp. S4B) indicated that once again, irrespective of the presence of DNA repair mutations, HuR and PARG silencing enhanced PARPi cytotoxicity; in both cases, this was associated with increasing PARP-1 trapping on chromatin.
Prioritizing the importance of HuR and PARG expression on PARPi efficacy in PDA cells
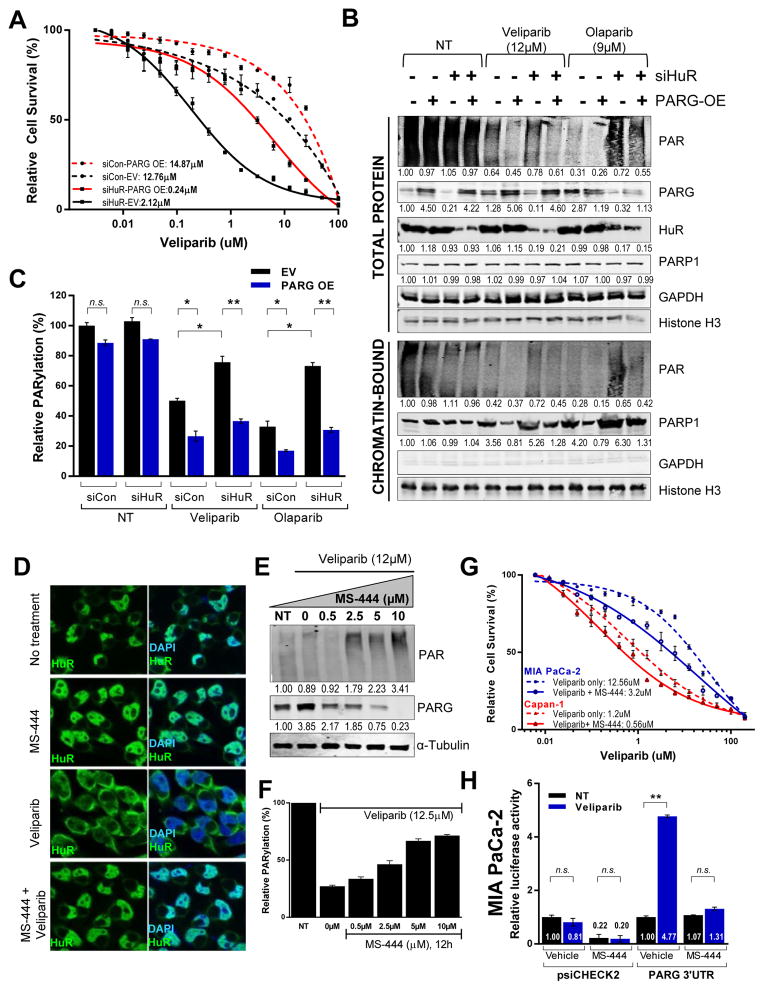
The role of PARG expression in regulating response to PARP inhibition is further highlighted by over a 5-fold decrease in IC50 values of olaparib and veliparib with PARG silencing in MIA PaCa-2 cells respectively (Fig. 4C). Further, PARG overexpression alone in MIA PaCa-2 cells caused increased resistance to veliparib (Fig. 5A) and olaparib (Supp. Fig. S6A). Although HuR knockdown enhances sensitivity to PARPi, a rescue of PARG expression in HuR- silenced cells partially restores PARPi resistance. Rescuing PARG expression in the presence or absence of HuR, indicated efficient removal of PARylation, particularly in the presence of PARP inhibition shown via immunoblotting and ELISA (Fig. 5B, 5C). As shown before (Fig. 3C, 4C, Supp. Fig. S4A, S4B), HuR inhibition in the presence of PARPi treatment resulted in persistence of PARylation and increased chromatin-trapped PARP-1. Importantly, PARG rescue facilitated PARP-1 release from chromatin, potentially recycling PARP-1 for enhanced repair and thus contributing to a resistant phenotype (Fig. 5B).
Figure 5. PARG overexpression rescues HuR’s regulation of PARPi response.
(A) Cell survival of MIA PaCa-2 cells co-transfected with HuR siRNA and PARG overexpression plasmid and treated with olaparib for 7days. (B) PARG rescue and HuR, PARP-1 and PAR expression validated, with GAPDH (total protein extract) and Histone H3 (nuclear chromatin- tethered fraction) as the loading controls. (C) ELISA to quantitate relative PARylation with PARG rescue in HuR silenced MIA PaCa-2 cells. (D) Immunofluorescence of HuR (green) in MIA PaCa-2 cells treated with veliparib for 12hr, with or without a 6hr pre-treatment of small-molecule HuR inhibitor, MS-444. Nuclei stained with DAPI (blue). Magnification 40X. (E) Relative PARylation and immunoblotting of total protein lysates of MIA PaCa-2 cells treated with increasing dosage of MS-444, in the presence of veliparib for 12h. (F) Cell survival of MIA PaCa-2 and Capan-1 cells treated with indicated doses of veliparib, with or without 5μM/L MS-444. (G) Luciferase activity in MIA PaCa-2 cells transfected with luciferase reporter constructs with PARG 3′UTR and incubated in the presence of MS-444 for 24 hr.
Small molecule HuR inhibitor MS-444 affects PARG expression and re-sensitizes PDA cells to PARPi
HuR function was perturbed using a small molecule inhibitor MS-444 that prevents HuR dimerization, a step critical for its stress- induced translocation to the cytoplasm (40, 41). Immunoblotting (Supp. Fig. S6B) and immunofluorescence (Fig. 5D) show that veliparib-induced translocation is blocked effectively by MS-444 at concentrations as low as 2.5 μM/L. HuR inhibition via MS-444 correlates with a strong decrease in overall PARG expression (Fig. 5E) and an associated accumulation of total PARylation (Fig. 5F). Concurrently, co-treatment with a sub-lethal dose of MS-444 (5μM)(20) that prevents HuR translocation, but does not affect cell survival, enhanced sensitivity to veliparib (Fig. 5G) and olaparib (Supp. Fig. S6C, Table 1) in both MIA PaCa-2 and Capan-1 cells. Concurrently, MS-444 also abrogates the PARPi induced stabilization of PARG mRNA in both DDR-D and DDR-P PDA cells (Fig. 5H, Supp. Fig. S6D). Taken together, these data indicate that small molecule inhibition of HuR inhibits PARG upregulation and function (i.e. PARylation), and could be potentially used to increase efficacy of PARPi.
Inducible shHuR silencing in vivo enhances olaparib-mediated suppression of PDA xenograft growth
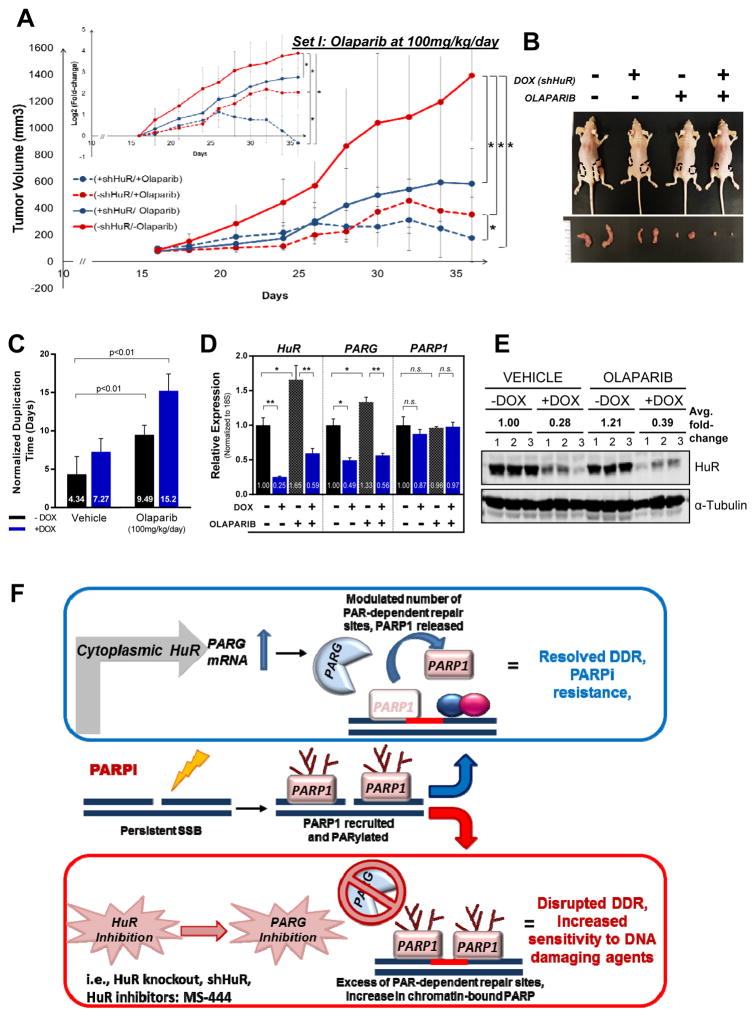
Based on our in vitro findings and previously published studies emphasizing the role of HuR in tumor development and growth (18), we sought to investigate the role of HuR in PDA xenografts under PARP inhibition. We used previously characterized MIA PaCa-2 cells (DDR-P) in which HuR silencing can be induced upon doxycycline (DOX) treatment (MIA.shHuR, previously reported as MIA.sh290) (18). In vitro characterization indicated a decrease in sensitivity to olaparib (18-fold, p< 0.001) with DOX treatment (Supp. Fig. S7A, S7B). Athymic nude female mice were injected subcutaneously in their hind flanks with MIA.shHuR and respective groups were treated with DOX chow and olaparib (100mg/kg/day, 5days a week). In the vehicle treated arms, the effect of DOX chow was significantly evident as early as day 21 (P<0.05) and continued this trend, ending with a 3.6-fold decrease in median normalized tumor volume as compared to mice on a normal diet (no DOX) at the end of the study (1212±472mm3 vs. 336±104mm3, p<0.05) (Fig. 6A, 6B, Supp. Fig. S7C). Olaparib treatment resulted in a significantly noticeable retardation in growth for all time points (P<0.05) with a final 5.6-fold decrease in tumor volume when compared to vehicle only (1212±472mm3 vs. 216±41mm3, P<0.01). Moreover, this effect further progressed to a 9.3 - fold change in tumor volume when HuR is silenced (1212±472mm3 vs. 131±76mm3 mm3, p<0.001). Tumor volumes indicate a significant reduction with HuR silencing, in both vehicle, as previously described (18), and olaparib treatment arms (starting at days 34 and 24, respectively, Supp. Fig. S7C). Additionally, olaparib treatment caused a 3- fold increase in the duplication time of tumors, further aggravated to a ≥5-fold (p<0.001) increase with HuR silencing (Fig 6C). In an independent experiment at a lower dosage of olaparib (50mg/kg/day) treatment, similar trends of growth delay were observed in xenografted tumors (Supp. Fig. S7C, S7D). While low dose olaparib (50mg/kg/day) treatment did not significantly affect tumor growth rate (400±44 mm3 vs. 394±23 mm3, P=NS), the addition of HuR inhibition resulted in a significant growth delay (2.3 fold increase in duplication time, P<0.01) and relative decrease in tumor volume (400±44 mm3 vs. 236±24 mm3,P<0.01).
Figure 6. HuR silencing in vivo enhances olaparib- mediated suppression of PDA xenograft growth.
Mia.shHuR xenografts in athymic, nude mice were randomized into DOX and olaparib treatment groups. (A) Tumor volumes are plotted, with each point representing the mean ± 2SE of each group, *P<0.05. Inset shows differences in number of duplications. (B) Representative image of mice and tumor per group. (C) Tumor duplication time (days) per group (D) HuR, PARG and PARP-1 mRNA expression in extracted tumors, relative to vehicle- treated –DOX group. Each bar represents the mean ± SEM (n = 3 per group). (E) HuR protein expression when tumors were harvested (day 36, n=3). (F) Working model: In response to PARPi stress, cytoplasmic HuR binds to and stabilizes PARG mRNA, thereby increasing PARG expression and modulating PARP1-chromatin dynamics. HuR and PARG inhibition breaks such acute resistance by enhancing chromatin- trapped PARP-1 and accumulation of damaged DNA and apoptosis.
Expression analysis of tumors harvested on day 36 (Set I) and day 56 (Set II) validated a significant decrease in HuR and PARG expression upon DOX induction at the mRNA and protein levels (Fig. 6D, 6E, Supp Fig. S7E) in both vehicle and olaparib treatment groups. The overall findings indicate that HuR inhibition enhances olaparib-mediated reduction of PDA tumor growth in vivo. These findings support the notion that HuR inhibition can sensitize PDA cells to PARPi therapy in vivo; even in a DDR-proficient PDA cell line.
Discussion
To date, the best personalized strategy for PDA is the synthetic lethal approach to treat patients’ tumors with DNA repair gene mutations. Recent Next Generation Sequencing and copy number variation studies estimate that a portion of PDAs may have a DDR molecular signature which may render these tumors sensitive to PARPi and platinum-based therapies (42). In fact, ongoing clinical trials demonstrate that selected BRCA-mutated PDAs have progression-free survival times of 12 months or more, with response rates of over 50%(43). Collectively, these data are intriguing, but also point to the sobering reality that: 1) even in the best setting where patients are identified with BRCA2 or related gene mutations, many patients respond to therapy but ultimately succumb to disease (43); and 2) the majority of PDA patients (DDR-proficient) will most likely not benefit from PARPi therapy.
Our study directly addresses the above two unexplored points. It should be noted that even the model drug for personalized oncology, the tyrosine kinase inhibitor, imatinib (Gleevec), which targets the BCR-ABL translocation in cancer required further development of next generation compounds because the cancer cells frequently develop resistance to therapy (44). Even though mutations in BCR-ABL have been found that confer resistance to imatinib, many other proposed and unknown molecular mechanisms can also account for the relapse of disease (45). Similarly, the general mechanism by which PARPi resistance occurs is still unknown, though some published instances highlight reversion mutations in the BRCA2 gene as the proposed mechanism (14, 15, 43). Moreover, even in patients with drug resistance mutations, it is unknown how the cancer cell survives while selecting for a reversion mutation (e.g., BRCA2) (46, 47).
Based on our data, we propose that PDA cells hijack an innate rapid stress response pathway governed by the RBP, HuR (Fig. 6). This is the first study to show that DNA damage triggers activation of PARG, which is directly related to the ability of HuR to rapidly stabilize specific mRNA (i.e., PARG) (17, 18, 20, 32). The link between PARG activation and DNA repair is emerging (48–50). Our data indicate that in response to (or during) DNA damage, an HuR-dependent increase in PARG expression and activity (i.e. reduced PAR levels) (Fig. 2, 3) may serve as a buffer on the total number of PAR-dependent signal factories that form in the cell. We hypothesize that a repair system with greater PARG activity and correspondingly diminished PAR production could modulate the number of PARP-1- dependent PAR binding sites on chromatin and improve PDA cell survival in the face of damage. Inversely, with diminished PARG (i.e. HuR silencing), PARP-1 could potentially form an excess of repair complexes that are difficult to resolve, leading to an increase in chromatin-bound PARP-1 (Figure 4B, Supp. Fig. S5B). This would negate an efficient DNA repair response. We believe that inhibition of the HuR/PARG axis enhances PARP-trapping on chromatin, and can be translated to improve PARPi efficacy in all PDAs, regardless of DNA repair status. In fact, we inhibited PARG expression using MS-444, a previously characterized tool for HuR inhibition (32, 41, 51). We consequently observed increased PARPi efficacy both in vitro and in vivo, independent of the cell line used (Fig 3C, 5A). Ongoing DNA repair mechanistic studies will depict the importance of the HuR/PARG axis on: 1) recruitment of downstream repair factors (RAD51, XRCC1) to sites of damage; and 2) the overall efficiency of specific repair pathways (e.g., homologous recombination; DNA inter-strand cross-link repair) by introducing exogenous nicks and DSBs (52).
We further speculate that PARG inhibitors may work better against cancer than PARP inhibitors (49, 53, 54). First, due to HuR’s established overabundance in cancer (19, 55–58), an HuR-dependent increase in PARG in tumor versus normal cells provides a therapeutic window. Second, PARG has a high specific activity for PAR degradation and helps maintain ADP- ribosylation dynamics within the cell (6), and thus could be a selective target. Third, despite their opposing enzymatic activities, PARP-1 and PARG localize to target promoters and regulate several common DDR- and metabolism-related genes (53, 59). Therefore, inhibiting PARG could potentially also target genes regulated by PARP-1, including those involved in cell structure, stress response, maintaining genetic stability and damage repair, metabolism, and GTPase regulation. Fourth, most PARPis do not selectively hit PARP-1 activity and thus may have unwanted off target effects (60). Meanwhile, PARG is the primary enzyme for hydrolyzing PARylation, and thus inhibiting this enzyme in the context of the HuR regulated- DNA repair process (17) could potentially increase specificity and reduce toxicity compared to currently studied pan-PARP inhibitors. With increasing evidence for PARG’s role in the DDR pathway, future studies will aim to study PARG inhibition in PDA with small molecule inhibitors (54) and gene silencing methods. These studies will ultimately reveal whether targeting PARG is a better therapeutic strategy than targeting PARP in cancer cells.
HuR has been independently identified by multiple studies as a PAR-binding protein in response to DNA damage (under H2O2 or methyl methane sulfonate stimulation), indicating PARylation as a means of coordinating HuR-specific RNA metabolic processes (61, 62). The role of PARylation in facilitating nuclear export, especially in CRM1-dependent pathways has been well documented (63, 64) which further indicates that HuR-CRM1 nuclear export could be modulated with PARP activity and expression. In addition to the striking similarities between PAR and nucleic acids, the ability of RNA recognition motifs (RRMs) to function as alternative PAR binding motifs (PBMs) adds an additional layer of complexity wherein PAR could compete with RNA and thus prevent protein functions such as localization, stability, splicing etc. (65). Herein, we provide support for a new mechanistic insight into PARP1’s regulation of HuR (Supp. Fig. S2A)(31). PARP1 activation, upon genotoxic stress, results in PARylated HuR which not only facilitates its cytoplasmic translocation, but also regulates its target binding(31). Cytoplasmic HuR selectively binds to several target mRNAs, which could presumably be affected by the degree of PARylation (as well as other PTMs such as phosphorylation, ubiquitination etc.). PARylation potentially contributes to HuR’s function by affecting: i) its specificity, wherein extent (length, branching, etc) of ADPribose polymers regulates binding affinities and ii) its selectivity, wherein the extent of PARylation allows differential binding to disparate pools of target mRNAs. HuR’s stabilization of PARG mRNA and protein expression, in addition to enhancing DNA repair, also supports a putative feedback loop wherein PARG dePARylates HuR, thus facilitating its release from target mRNAs and shuttling back into the nucleus. Further studies will investigate the specific PARG isoforms that regulate HuR’s function and vice versa, as well as further elucidate the timing and spatiotemporal organization of this complex process.
Finally, directly targeting HuR in PDA cells may remain our best strategy to enhance clinical effectiveness of PARPi, as it regulates a cadre of pro-survival transcripts (17, 19, 20, 32, 33). Therefore, promising attempts to target HuR are ongoing via small molecule inhibitors or a siHuR nanoparticle strategy (41, 66, 67) (Fig. 5) in combination with DNA damaging agents. Complementary studies will define and target the specific upstream mechanisms (e.g. kinases) that facilitate HuR translocation to the cytoplasm in PDA (17). Finally, it will be interesting to determine if the HuR/PARG axis has an essential role in DNA repair in normal cellular and developmental biology, or if this HuR-regulated repair mechanism is unique to cancer cells.
Supplementary Material
Acknowledgments
Financial support: This work was supported by a seed grant from the Hirshberg Foundation for Pancreatic Cancer Research (J.R. Brody and J.M. Pascal), NIH-NCI R21 CA182692 01A1 (J.R. Brody) and 1R01CA212600-01 (J.R. Brody), American Cancer Society MRSG-14-019-01-CDD (J.M. Winter, J.R. Brody), the Mary Halinski Pancreatic Cancer Research Fund (J.R. Brody and A. Nevler), and Fund A Cure and the Michele Barnett Rudnick Fund (J.R. Brody and S.N. Chand).
Footnotes
Disclosures: The authors declare no potential conflicts of interest.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research. 2014;74(11):2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Society AC. American Cancer Society: Cancer Facts and Figures. 2015 http://wwwcancerorg/research/cancerfactsfigures/index.
- 3.Carnevale J, Ashworth A. Assessing the Significance of BRCA1 and BRCA2 Mutations in Pancreatic Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33(28):3080–1. doi: 10.1200/JCO.2015.61.6961. [DOI] [PubMed] [Google Scholar]
- 4.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 5.Forster MD, Dedes KJ, Sandhu S, Frentzas S, Kristeleit R, Ashworth A, et al. Treatment with olaparib in a patient with PTEN-deficient endometrioid endometrial cancer. Nature reviews Clinical oncology. 2011;8(5):302–6. doi: 10.1038/nrclinonc.2011.42. [DOI] [PubMed] [Google Scholar]
- 6.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. The Biochemical journal. 1999;342( Pt 2):249–68. [PMC free article] [PubMed] [Google Scholar]
- 7.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 8.Brochu G, Duchaine C, Thibeault L, Lagueux J, Shah GM, Poirier GG. Mode of action of poly(ADP-ribose) glycohydrolase. Biochimica et biophysica acta. 1994;1219(2):342–50. doi: 10.1016/0167-4781(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 9.Fisher AE, Hochegger H, Takeda S, Caldecott KW. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol Cell Biol. 2007;27(15):5597–605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barkauskaite E, Brassington A, Tan ES, Warwicker J, Dunstan MS, Banos B, et al. Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities. Nature communications. 2013 doi: 10.1038/ncomms3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tutt A, Bertwistle D, Valentine J, Gabriel A, Swift S, Ross G, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. The EMBO journal. 2001;20(17):4704–16. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouwman P, Jonkers J. Molecular pathways: how can BRCA-mutated tumors become resistant to PARP inhibitors? Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(3):540–7. doi: 10.1158/1078-0432.CCR-13-0225. [DOI] [PubMed] [Google Scholar]
- 13.Johnson N, Johnson SF, Yao W, Li YC, Choi YE, Bernhardy AJ, et al. Stabilization of mutant BRCA1 protein confers PARP inhibitor and platinum resistance. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(42):17041–6. doi: 10.1073/pnas.1305170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakai W, Swisher EM, Jacquemont C, Chandramohan KV, Couch FJ, Langdon SP, et al. Functional restoration of BRCA2 protein by secondary BRCA2 mutations in BRCA2-mutated ovarian carcinoma. Cancer research. 2009;69(16):6381–6. doi: 10.1158/0008-5472.CAN-09-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451(7182):1111–5. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 16.Oplustilova L, Wolanin K, Mistrik M, Korinkova G, Simkova D, Bouchal J, et al. Evaluation of candidate biomarkers to predict cancer cell sensitivity or resistance to PARP-1 inhibitor treatment. Cell cycle. 2012;11(20):3837–50. doi: 10.4161/cc.22026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lal S, Burkhart RA, Beeharry N, Bhattacharjee V, Londin ER, Cozzitorto JA, et al. HuR posttranscriptionally regulates WEE1: implications for the DNA damage response in pancreatic cancer cells. Cancer research. 2014;74(4):1128–40. doi: 10.1158/0008-5472.CAN-13-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jimbo M, Blanco FF, Huang YH, Telonis AG, Screnci BA, Cosma GL, et al. Targeting the mRNA-binding protein HuR impairs malignant characteristics of pancreatic ductal adenocarcinoma cells. Oncotarget. 2015;6(29):27312–31. doi: 10.18632/oncotarget.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costantino CL, Witkiewicz AK, Kuwano Y, Cozzitorto JA, Kennedy EP, Dasgupta A, et al. The role of HuR in gemcitabine efficacy in pancreatic cancer: HuR Up-regulates the expression of the gemcitabine metabolizing enzyme deoxycytidine kinase. Cancer research. 2009;69(11):4567–72. doi: 10.1158/0008-5472.CAN-09-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanco FF, Jimbo M, Wulfkuhle J, Gallagher I, Deng J, Enyenihi L, et al. The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells. Oncogene. 2015 doi: 10.1038/onc.2015.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deer EL, Gonzalez-Hernandez J, Coursen JD, Shea JE, Ngatia J, Scaife CL, et al. Phenotype and Genotype of Pancreatic Cancer Cell Lines. Pancreas. 2010;39(4):425–35. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lal S, Cheung EC, Zarei M, Preet R, Chand SN, Mambelli-Lisboa NC, et al. CRISPR Knockout of the HuR Gene Causes a Xenograft Lethal Phenotype. Molecular cancer research : MCR. 2017 doi: 10.1158/1541-7786.MCR-16-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cozzitorto JA, Jimbo M, Chand S, Blanco F, Lal S, Gilbert M, et al. Studying RNA-binding protein interactions with target mRNAs in eukaryotic cells: native ribonucleoprotein immunoprecipitation (RIP) assays. Methods Mol Biol. 2015;1262:239–46. doi: 10.1007/978-1-4939-2253-6_14. [DOI] [PubMed] [Google Scholar]
- 24.Sever-Chroneos Z, Angus SP, Fribourg AF, Wan H, Todorov I, Knudsen KE, et al. Retinoblastoma tumor suppressor protein signals through inhibition of cyclin-dependent kinase 2 activity to disrupt PCNA function in S phase. Mol Cell Biol. 2001;21(12):4032–45. doi: 10.1128/MCB.21.12.4032-4045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kummar S, Chen A, Ji J, Zhang Y, Reid JM, Ames M, et al. Phase I study of PARP inhibitor ABT-888 in combination with topotecan in adults with refractory solid tumors and lymphomas. Cancer research. 2011;71(17):5626–34. doi: 10.1158/0008-5472.CAN-11-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murai J, Huang SN, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Differential trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer research. 2012;72(21):5588–99. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCabe N, Lord CJ, Tutt AN, Martin NM, Smith GC, Ashworth A. BRCA2-deficient CAPAN-1 cells are extremely sensitive to the inhibition of Poly (ADP-Ribose) polymerase: an issue of potency. Cancer biology & therapy. 2005;4(9):934–6. doi: 10.4161/cbt.4.9.2141. [DOI] [PubMed] [Google Scholar]
- 28.Gallmeier E, Kern SE. Targeting Fanconi anemia/BRCA2 pathway defects in cancer: the significance of preclinical pharmacogenomic models. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13(1):4–10. doi: 10.1158/1078-0432.CCR-06-1637. [DOI] [PubMed] [Google Scholar]
- 29.Gallmeier E, Calhoun ES, Rago C, Brody JR, Cunningham SC, Hucl T, et al. Targeted disruption of FANCC and FANCG in human cancer provides a preclinical model for specific therapeutic options. Gastroenterology. 2006;130(7):2145–54. doi: 10.1053/j.gastro.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 30.van der Heijden MS, Brody JR, Dezentje DA, Gallmeier E, Cunningham SC, Swartz MJ, et al. In vivo therapeutic responses contingent on Fanconi anemia/BRCA2 status of the tumor. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11(20):7508–15. doi: 10.1158/1078-0432.CCR-05-1048. [DOI] [PubMed] [Google Scholar]
- 31.Ke Y, Han Y, Guo X, Wen J, Wang K, Jiang X, et al. PARP1 promotes gene expression at the post-transcriptiona level by modulating the RNA-binding protein HuR. Nature communications. 2017;8:14632. doi: 10.1038/ncomms14632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romeo C, Weber MC, Zarei M, DeCicco D, Chand SN, Lobo AD, et al. HuR Contributes to TRAIL Resistance by Restricting Death Receptor 4 Expression in Pancreatic Cancer Cells. Molecular cancer research : MCR. 2016 doi: 10.1158/1541-7786.MCR-15-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pineda DM, Rittenhouse DW, Valley CC, Cozzitorto JA, Burkhart RA, Leiby B, et al. HuR’s post-transcriptional regulation of Death Receptor 5 in pancreatic cancer cells. Cancer biology & therapy. 2012;13(10):946–55. doi: 10.4161/cbt.20952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nature reviews Molecular cell biology. 2012;13(7):411–24. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 35.Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Experimental cell research. 2004;297(2):521–32. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 36.Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, et al. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000;20(3):760–9. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(9):2987–92. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murai J, Huang SY, Renaud A, Zhang Y, Ji J, Takeda S, et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Molecular cancer therapeutics. 2014;13(2):433–43. doi: 10.1158/1535-7163.MCT-13-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer research. 2012;72(21):5588–99. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meisner NC, Hintersteiner M, Mueller K, Bauer R, Seifert JM, Naegeli HU, et al. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nature chemical biology. 2007;3(8):508–15. doi: 10.1038/nchembio.2007.14. [DOI] [PubMed] [Google Scholar]
- 41.Blanco FF, Preet R, Aguado A, Vishwakarma V, Stevens LE, Vyas A, et al. Impact of HuR inhibition by the small molecule MS-444 on colorectal cancer cell tumorigenesis. Oncotarget. 2016 doi: 10.18632/oncotarget.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pishvaian MJ, Biankin AV, Bailey P, Chang DK, Laheru D, Wolfgang CL, et al. BRCA2 secondary mutation-mediated resistance to platinum and PARP inhibitor-based therapy in pancreatic cancer. British journal of cancer. 2017;116(8):1021–6. doi: 10.1038/bjc.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer cell. 2002;2(2):117–25. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 45.Lamontanara AJ, Gencer EB, Kuzyk O, Hantschel O. Mechanisms of resistance to BCR-ABL and other kinase inhibitors. Biochimica et biophysica acta. 2013;1834(7):1449–59. doi: 10.1016/j.bbapap.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 46.Swisher EM, Sakai W, Karlan BY, Wurz K, Urban N, Taniguchi T. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer research. 2008;68(8):2581–6. doi: 10.1158/0008-5472.CAN-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashworth A. Drug resistance caused by reversion mutation. Cancer research. 2008;68(24):10021–3. doi: 10.1158/0008-5472.CAN-08-2287. [DOI] [PubMed] [Google Scholar]
- 48.Shirai H, Poetsch AR, Gunji A, Maeda D, Fujimori H, Fujihara H, et al. PARG dysfunction enhances DNA double strand break formation in S-phase after alkylation DNA damage and augments different cell death pathways. Cell death & disease. 2013;4:e656. doi: 10.1038/cddis.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakadate Y, Kodera Y, Kitamura Y, Tachibana T, Tamura T, Koizumi F. Silencing of poly(ADP-ribose) glycohydrolase sensitizes lung cancer cells to radiation through the abrogation of DNA damage checkpoint. Biochemical and biophysical research communications. 2013;441(4):793–8. doi: 10.1016/j.bbrc.2013.10.134. [DOI] [PubMed] [Google Scholar]
- 50.Ray Chaudhuri A, Ahuja AK, Herrador R, Lopes M. Poly(ADP-ribosyl) glycohydrolase prevents the accumulation of unusual replication structures during unperturbed S phase. Mol Cell Biol. 2015;35(5):856–65. doi: 10.1128/MCB.01077-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blanco FF, Jimbo M, Wulfkuhle J, Gallagher I, Deng J, Enyenihi L, et al. The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells. Oncogene. 2016;35(19):2529–41. doi: 10.1038/onc.2015.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bindra RS, Goglia AG, Jasin M, Powell SN. Development of an assay to measure mutagenic non-homologous end-joining repair activity in mammalian cells. Nucleic acids research. 2013;41(11):e115. doi: 10.1093/nar/gkt255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim IK, Stegeman RA, Brosey CA, Ellenberger T. A quantitative assay reveals ligand specificity of the DNA scaffold repair protein XRCC1 and efficient disassembly of complexes of XRCC1 and the poly(ADP-ribose) polymerase 1 by poly(ADP-ribose) glycohydrolase. The Journal of biological chemistry. 2015;290(6):3775–83. doi: 10.1074/jbc.M114.624718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.James DI, Smith KM, Jordan AM, Fairweather EE, Griffiths LA, Hamilton NS, et al. First-in-Class Chemical Probes against Poly(ADP-ribose) Glycohydrolase (PARG) Inhibit DNA Repair with Differential Pharmacology to Olaparib. ACS chemical biology. 2016;11(11):3179–90. doi: 10.1021/acschembio.6b00609. [DOI] [PubMed] [Google Scholar]
- 55.Richards NG, Rittenhouse DW, Freydin B, Cozzitorto JA, Grenda D, Rui H, et al. HuR status is a powerful marker for prognosis and response to gemcitabine-based chemotherapy for resected pancreatic ductal adenocarcinoma patients. Annals of surgery. 2010;252(3):499–505. doi: 10.1097/SLA.0b013e3181f1fd44. discussion -6. [DOI] [PubMed] [Google Scholar]
- 56.Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, et al. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22(46):7146–54. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 57.Nabors LB, Gillespie GY, Harkins L, King PH. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer research. 2001;61(5):2154–61. [PubMed] [Google Scholar]
- 58.Young LE, Sanduja S, Bemis-Standoli K, Pena EA, Price RL, Dixon DA. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology. 2009;136(5):1669–79. doi: 10.1053/j.gastro.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frizzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y, et al. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J Biol Chem. 2009;284(49):33926–38. doi: 10.1074/jbc.M109.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steffen JD, Brody JR, Armen RS, Pascal JM. Structural Implications for Selective Targeting of PARPs. Frontiers in oncology. 2013;3:301. doi: 10.3389/fonc.2013.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic acids research. 2008;36(22):6959–76. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, Nielsen ML. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Molecular cell. 2013;52(2):272–85. doi: 10.1016/j.molcel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- 63.Zerfaoui M, Errami Y, Naura AS, Suzuki Y, Kim H, Ju J, et al. Poly(ADP-ribose) polymerase-1 is a determining factor in Crm1-mediated nuclear export and retention of p65 NF-kappa B upon TLR4 stimulation. Journal of immunology (Baltimore, Md : 1950) 2010;185(3):1894–902. doi: 10.4049/jimmunol.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M, et al. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nature cell biology. 2007;9(10):1175–83. doi: 10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- 65.Teloni F, Altmeyer M. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic acids research. 2016;44(3):993–1006. doi: 10.1093/nar/gkv1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu X, Lan L, Wilson DM, Marquez RT, Tsao WC, Gao P, et al. Identification and validation of novel small molecule disruptors of HuR-mRNA interaction. ACS chemical biology. 2015;10(6):1476–84. doi: 10.1021/cb500851u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang YH, Peng W, Furuuchi N, Gerhart J, Rhodes K, Mukherjee N, et al. Delivery of Therapeutics Targeting the mRNA-Binding Protein HuR Using 3DNA Nanocarriers Suppresses Ovarian Tumor Growth. Cancer research. 2016;76(6):1549–59. doi: 10.1158/0008-5472.CAN-15-2073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.