Abstract
Endometrial cancer (EC) is one of the most commonly diagnosed gynecologic malignancies in the world, with the morbidity rate of over 7%. The mechanism of the pathogenesis has not been specifically elucidated to date, which is imperative for EC treatment. The aim of our study was to investigate the target relationship between miR-143 and mitogen-activated protein kinase 1 (MAPK1) and explore the effect of miR-143 on the endometrial cancers (EC) cells through targeting MAPK1. We collected EC tissues and adjacent tissues, and transfected miR-143 mimics and MAPK1 siRNA into EC cells with lipofectamine. Reverse transcription-polymerase chain reaction (RT-PCR) and western blot were used to examine the expression of miR-143 and MAPK1 mRNA and the protein expression of MAPK1. Cell counting kit-8, wound healing assay, flow cytometry and transwell assay were applied to examining the alteration of the proliferation, migration, cell cycle and invasion ability of EC cells. We predicted the targeting gene of miR-143 through bioinformatics analysis. MiR-143 was found under-expressed in EC tissues and cells. Overexpression of miR-143 or knockdown of MAPK1 in human EC cell line HEC-1B inhibited the EC cell proliferation, migration and invasion and induced apoptosis. MAPK1 was verified to be a target gene of miR-143. MiR-143 overexpression could effectively inhibit mRNA and protein expression of MAPK1 in HEC-1B cells. Collectively, miR-143 might inhibit the proliferation, migration and invasion of EC cells, and promote the apoptosis of EC cells by suppressing MAPK1. These findings provided a view for new and potential therapeutic method for the clinical treatment of EC.
Keywords: endometrial cancer, miR-143, MAPK1, proliferation, migration
INTRODUCTION
Endometrial cancer (EC) is one of the most commonly diagnosed gynecologic malignancies in the world [1]. EC was considered to have the highest incidence rate (approx. 7%) among the female genital malignant tumors and the incidence has been still on the rise [2]. EC was generally divided into types I (estrogen-dependent) and type II (estrogen-independent) [3]. The vast majority of endometrial carcinomas (80%) belong to type I, only second to atypical endometrial hyperplasia [4]. he EC type I is associated with obesity, infertility, insulin resistance and estrogen therapy, while the EC Type II includes endometrial serous carcinoma, clear cell carcinoma, squamous cell carcinoma, mucinous adenocarcinoma and other types of cancers. High malignancy, poor prognosis, early-stage metastasis and worse clinical outcome often embody in the EC Type II [5]. Previous research has already revealed that several aberrant gene expressions and transduction pathways forebode the initiation of EC [6]. As EC is a multi-factorial disease, the deep mechanism of the pathogenesis has not been specifically elucidated. Therefore, it is of paramount importance to understand the mechanism of EC for the disease prevention and clinical diagnosis and treatment.
MicroRNAs (miRNAs) are a group of small, single-stranded, endogenous and non-coding RNAs [7, 8], which can help regulate different aspects in cancers pathological process, including invasion, migration, proliferation, differentiation, apoptosis and senescence [9–11]. MiRNAs regulate the expression of the target gene by targeting 3’UTR of messenger RNA, ultimately leading to either mRNAs repression or transcription degradation at the posttranscriptional and transcriptional level [12]. In the last few years, substantiated evidence has showed that miRNAs were involved in many cellular processes through the regulation of specific target genes [13]. On the one hand, miRNAs could serve as oncogenes and promoted the process of tumorigenesis [14, 15]. For example, miR-9 could promote tumor metastasis by down-regulating the expression of E-cadherin in esophageal squamous cell carcinoma [16]. On the other, miRNAs also function as tumor suppressors and inhibit the progression of cancers. For example, miR-27a could directly target KRAS to inhibit cells proliferation in esophageal squamous cell carcinoma [17]. MiR-99a could suppress the metastasis of human lung tumor cells by targeting the AKT1 signaling pathway [18]. However, the function and the molecular mechanism of miR-143 in the regulation of EC malignant behavior remain to be further investigated.
Mitogen-activated protein kinase 1 (MAPK1) belongs to the MAP kinase family. MAP kinase, also known as extracellular signal-regulated kinases (ERKs), could act as a binding point for numerous biochemical signals [19, 20]. Besides, ERKs are also involved in a wide range of cellular processes, such as differentiation, proliferation, transcription development and regulation [21]. Moreover, the activation of ERKs requires upstream kinases phosphorylation. Once activated, this kinase could translocate to the nucleus of the activated tumor cells, that is, phosphorylates nuclear targets [22, 23]. Previous study has demonstrated that this protein could function as a transcriptional repressor independently because of its kinase activity [24]. Some studies also revealed that MAPK1 played an important role in tumors progression and different tumors cellular processes [25–27]. In addition, several researchers found that MAPK1 could serve as potential target gene for miRNAs and miR-378 inhibited prostate cancer cell growth through targeting MAPK1 [28]. MiR-143 has been reported to participate in carcinogenesis by influencing MAPK pathway such as pancreatic ductal adenocarcinoma, bladder cancer and acute myeloid leukemia [29–31]. In addition, Ning et al. showed that MAPK1 can be directly targeted by miR-143 during human keratinocyte differentiation [32]. MAPK1 has also been reported to be a common target of miR-143 in renal cell carcinoma [33]. However, few researches were conducted to analyze the relationship between MAPK1 and miR-143 in EC. In our study, we demonstrated that miR-143 could affect EC cells proliferation, migration and invasion through targeting MAPK1.
RESULTS
MiR-143 expression was downregulated in EC tissues and cells
To study the expression of miR-143 in EC, we measured the expression of miR-143 in 35 EC tissues and the adjacent tissues using RT-PCR. The results indicated that miR-143 expression was significantly downregulated in the EC tissues in comparison with adjacent tissues (P < 0.05, Figure 1A and 1B). Besides, the miR-143 expression in EC cell lines (HEC-1B, AN3CA, KLE, ECC-1and Ishikawa) was significantly lower than normal cells (hESC, P < 0.05, Figure 1C). Given that the expression of miR-143 in HEC-1B cells was relatively higher in comparison with the other four EC cell lines, we selected HEC-1B cell line for our further study.
Figure 1. The expression levels of miR-143 were down-regulated in EC tissues and cell lines.
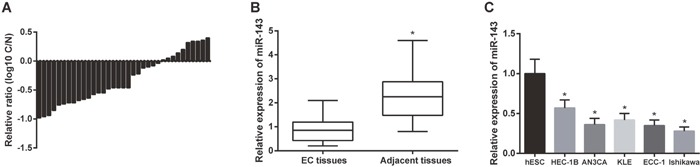
(A) The expression levels of miR-143 in tissue samples were down-regulated compared with normal tissues. C: EC tissues, N: normal tissues. (B) The expression levels of miR-143 in EC tissues were much lower than that in normal tissues. * P<0.05 compared with EC tissues. (C) The expression levels of miR-143 in EC cell lines (HEC-1B, AN3CA, KLE, ECC-1, and Ishikawa) was observably lower than that in normal cell line (hESC). * P<0.05 compared with hESC group.
Overexpression of miR-143 reduced cell viability and cell cycle, inhibited migration and invasion ability of EC cells and promoted cell apoptosis
The expression of miR-143 was detected by RT-PCR. After HEC-1B cells were transfected with miR-143 mimics, miR-143 expressions were upregulated, and miR-143 expression in miR-143 mimics group was higher than that in blank control group and negative control (NC) group (P < 0.05, Figure 2A). Compared with control group and NC group, cell viabilities in miR-143 group were significantly suppressed (P < 0.05, Figure 2B). Cell cycle and apoptosis of EC cells was assessed using FCM. The results have shown that in miR-143 group, cell cycle arrested at G1 phase and a constant accumulation of apoptotic cell was detected (both P < 0.05, Figure 2C and 2D).
Figure 2. Overexpression of miR-143 inhibited cell proliferation and induced apoptosis of the cells.
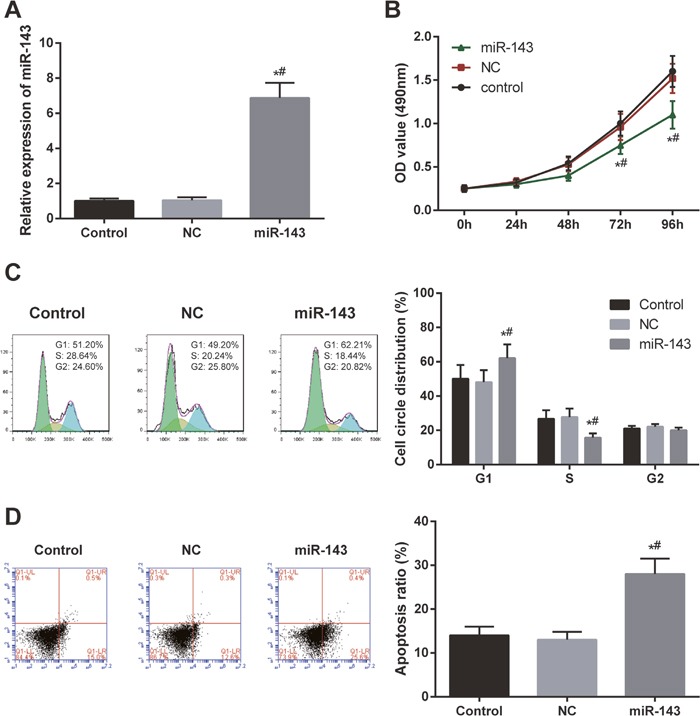
(A) The expression of miR-143 in HEC-1B cells dramatically increased after transfection with miR-143 mimics. (B) Overexpression of miR-143 suppressed HEC-1B cell viability. (C) Overexpression of miR-143 induced EC cell cycle arrest. X axis indicates DNA content and Y axis indicates cell number. Color green: G1 phase; yellow: M phase; blue: G2/M phase. (D) Overexpression of miR-43 promoted EC cell apoptosis observed by FCM. X axis indicates AnnexinV fluorescence intensity and Y axis indicates PI fluorescence intensity. * P<0.05 relative to control group, # P<0.05 relative to NC group.
The migration and invasion abilities of the EC cells were examined in each group by wound healing assay and transwell assay. As shown in the Figure 3, miR-143 mimics group displayed weaker migration and invasion abilities of EC cells in comparison with control group and NC group (both P<0.05).
Figure 3. Overexpression of miR-143 inhibited cell invasion and migration.

(A) The open wound area of EC cells in miR-143 group was much larger than that in control group and NC group. (B) The number of migration cellS in miR-143 group was relatively fewer compared with control group and NC group. (C) The number of invasion cells in miR-143 group was more than that in control group and NC group detected. * P<0.05 compared with control group, # P<0.05 compared with NC group.
Protein and mRNA expression of MAPK1 were observably suppressed by overexpression of miR-143
To validate that MAPK1 was a direct target of miR-143, two types of MAPK1 vectors including wild type (WT) luc-MAPK1 and mutation type luc-MAPK1-mut were constructed (Figure 4A). Overexpression of miR-143 could inhibit the luciferase activity of luc-MAPK1, but had no effect on luc-MAPK1-mut (Figure 4B). This also indicated that miR-143 could directly target 3’UTR of MAPK1 and inhibited the transcriptional activity of MAPK1. The results of RT-PCR and western blot both revealed that protein and mRNA expressions of MAPK1 were suppressed by overexpression of miR-143 (Figure 4C and 4D).
Figure 4. MiR-143 directly targeted and regulated MAPK1 in EC cells.
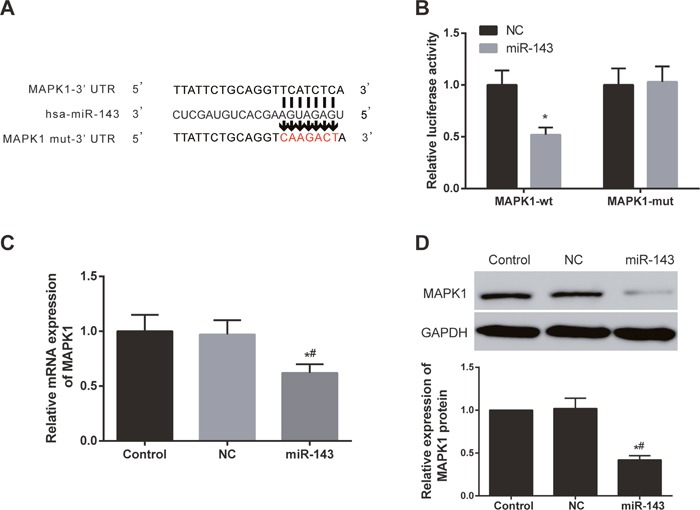
(A) MiR-143 could directly target 3’UTR of MAPK1. (B) The luciferase activity of luc-MAPK1-wt was suppressed by miR-143 overexpression, but there was significant difference in luc-MAPK1-mut. * P<0.05 compared with NC group. (C) Overexpression of miR-143 suppressed MAPK1 expression in EC cells. (D) Overexpression of miR-143 inhibited the expression of MAPK1. * P<0.05 compared with control group, # P<0.05 compared with NC group.
Down-regulation of MAPK1 expression could inhibit the proliferation ability of EC cells and induce apoptosis as well as change the cell cycle distribution
The expression of MAPK1 in HEC-1B cells was knocked down by siRNA. The MAPK1 expression was evaluated using RT-PCR and western blot. The mRNA and protein expressions were down-regulated in HEC-1B cells after transfected with MAPK1-siRNA (P<0.05, Figure 5A and 5B). According to CCK-8 assay results, the proliferation rate of HEC-1B cells in siMAPK1 group was much lower than that in the other two groups at 48 h, 72 h and 96 h (P<0.05, Figure 5C). The cell cycle distribution of HEC-1B cells were changed after inhibiting MAPK1, the number of cells in G1 phase increased and cells in S phase reduced (Figure 5D). In addition, the inhibition of MAPK1 could induce the apoptosis of HEC-1B cells (P<0.05, Figure 5E).
Figure 5. Down-regulation of MAPK1 expression inhibited EC cell proliferation and induced cell cycle arrest and cell apoptosis.
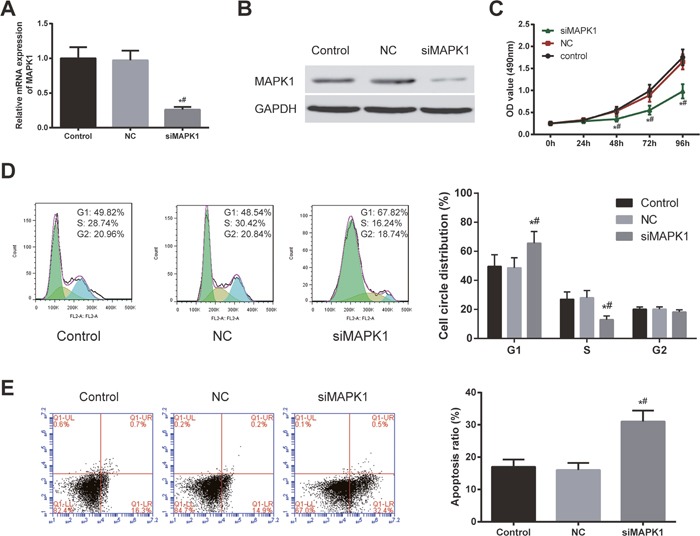
(A) The expression of MAPK1 in HEC-1B cells after transfection with si-MAPK1 significantly decreased. (B) The expression of MAPK1 was down-regulated after transfection with si-MAPK1. (C) The EC cell viability in siMAPK1 group was much lower than that in control group and NC group confirmed. (D) The number of EC cells arrested in G1 stage increased after transfected with si-MAPK1 for 48 h. X axis indicates DNA content and Y axis indicates cell number. Color green: G1 phase; yellow: M phase; blue: G2/M phase. (E) Theapoptosis of EC cells significantly enhanced after transfected with si-MAPK1 for 48 h. X axis indicates AnnexinV fluorescence intensity and Y axis indicates PI fluorescence intensity. * P<0.05 compared with control group, # P<0.05 compared with NC group.
The inhibition of MAPK1 expression in EC cells could suppress the migration and invasion abilities of the cells
In wound healing assay, the wound healing ability of HEC-1B cells was found to be significantly inhibited 24 h after transfection with MAPK1 (both P<0.05, Figure 6A). Furthermore, transwell assay also displayed that inhibition of MAPK1 expression in HEC-1B cells could suppress the migration and invasion abilities of the cells (both P<0.05, Figure 6B and 6C).
Figure 6. The inhibition of MAPK1 expression in EC cells suppressed the migration and invasion abilities of the cells.
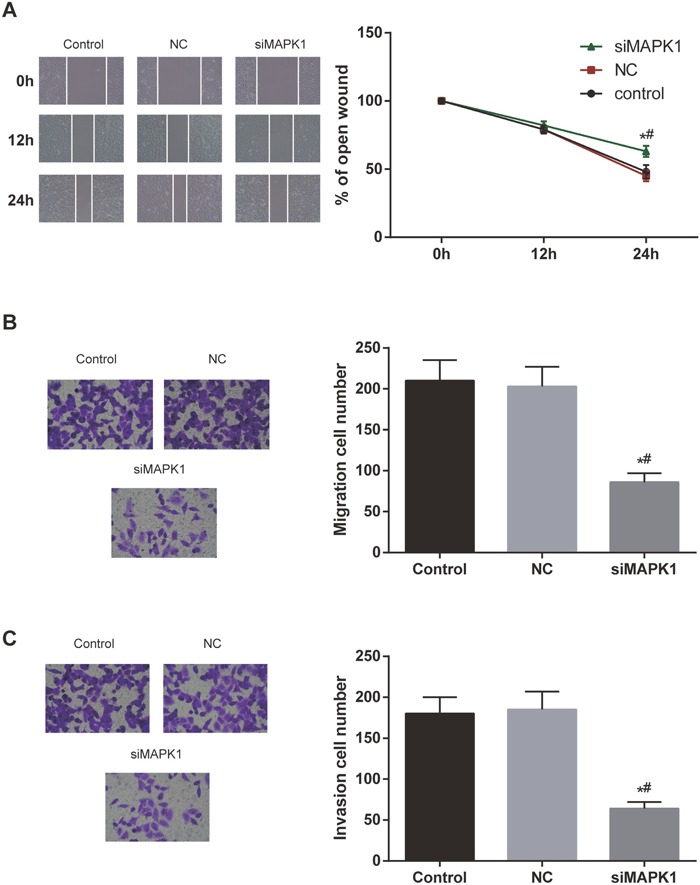
(A) The open wound area in si-MAPK1 group was larger than that in control group and NC group. (B, C) The migration and invasion ability of EC cells significantly decreased after transfection with si-MAPK1. * P<0.05 compared with control group, # P<0.05 compared with NC group.
DISCUSSION
In the current study, miR-143 was found under-expressed in EC tissues and cells. Overexpression of miR-143 or knockdown of MAPK1 in human EC cell line HEC-1B inhibited the EC cell proliferation, migration and invasion and induced apoptosis. MAPK1 was verified to be a target gene of miR-143. MiR-143 overexpression could effectively inhibit mRNA and protein expression of MAPK1 in HEC-1B cells.
EC is one of the most widespread gynecologic tumors among women around the world [34]. Many types of aberrant molecular expressions, including oncogenes and tumor suppressors, are related to progression and tumorigenesis of EC [35]. For a molecular mechanism, multiple regulators of gene expression have been founded and miRNA plays a vital role in the regulation of gene expressions. Recent findings have found that miR-143 family members exerted influence on multiple features of tumor cells through different pathways and targets. For instance, Liu et al. found that miR-143 could mediate the apoptotic process in osteosarcoma cells by manipulating the expression of Bcl-2. Mao et al. revealed that miR-143, as a tumor suppressor, could inhibit the expression of family with sequence similarity 83 (FAM83F) in esophageal squamous cell carcinoma (ESCC). Besides, Liu et al. found that miR-143 could inhibit cell invasion and proliferation through down-regulating TLR2 expression in hepatoma cells [36–38]. These studies indicate that miR-143 could possibly suppress EC progression or development by silencing some gene. In our study, the down-regulation of miR-143 in EC tissues and cells indicated that miR-143 could function as a tumor suppressor in EC. We have confirmed that miR-143 could efficiently down-regulate MAPK1 expression in EC cells. Thus, we speculate that miR-143 could probably hinder EC carcinogenesis by inhibiting MAPK1.
MAPK1 can be targeted by various miRNAs in various cancers. For instance, Kouhkan et al. found that cell cycle arrest of glioblastoma multiforme (GBM) cancer cells was induced by miR-192-1's targeting MAPK1. Chen et al. revealed that miR-378 suppressed prostate cancer cell growth through down-regulation of MAPK1 in vitro and in vivo. Yan et al. demonstrated that deregulated miR-335 that targets MAPK1 was implicated in poor outcome of pediatric acute lymphoblastic leukemia [28, 39, 40]. MAPK1 is the core protein of the MAPK/ERK pathway. As we know, MAPK1 could manipulate cell biological function by modulating downstream substrates of the MAPK/ERK signaling pathway, such as Cyclin D1 and c-MYC [41]. Up to now, nearly one-third of human cancers revealed that MAPK/ERK pathway was abnormally activated and played an essential role in their pathogenesis [42]. Since the MAPK/ERK signaling pathway plays a vital role in the development of tumor, numerous researches have focused on the potential therapeutic implication. For instance, Sorafenib, a tyrosine kinase inhibitor (TKI), could inhibit papillary thyroid carcinoma growth by down-regulating MAPK/ERK signaling pathway [43]. Moreover, miRNAs have been confirmed to regulate target genes of MAPK/ERK pathway, to some extent, demonstrating their important roles in manipulation of the signaling cascade. Liu et al. revealed that miR-524-5p could inhibit cell migration and tumor proliferation and suppressed their activities in melanoma by targeting MAPK1 and BRAF through MAPK/ERK signaling pathway [44]. Another research showed that miR-133a inhibited the MAPK/ERK cascade by targeting the upstream protein LASP1, which could suppress tumor metastasis and growth in colorectal cancer [45]. All in all, we could conclude that miR-143 could suppress EC cell migration, invasion and proliferation by targeting down MAPK1, which therefore suppresses MAPK/ERK signaling.
In the current study, we first demonstrated the molecular mechanism of miR-143 in influencing EC cells through targeting MAPK1. MiR-143 was found to be capable of suppressing proliferation, migration and invasion and cell cycle process of HEC-1B cells, and inducing apoptosis via downregulation of MAPK1, which not only proved our previous conjuncture was right, but provided some valuable evidence for the intensive study on the subject. However, it was not sufficient to completely reach the conclusion through in vitro experiments only, which was also a limitation of our study. Therefore, our later study would further substantiate the conclusion by conducting in vivo experiments.
In conclusion, our study showed that overexpression of miR-143 could suppress proliferation, migration and invasion and cell cycle process of HEC-1B cells, and induce apoptosis by targeting MAPK1. These results revealed that miR-143 functions as a tumor suppressor and might play an important role in the inhibition of EC cell proliferation and metastasis. These findings not only laid foundation for the in-depth study on the molecular mechanism of mRNA, but may provide a new and potential therapeutic method for the clinical treatment of EC.
MATERIALS AND METHODS
Samples
We collected tissue samples from 35 EC patients in the First Affiliated Hospital of Zhengzhou University. Before surgery, radiotherapy or chemotherapy was not applied to any one of the patients. The adjacent non-neoplastic tissue was over 5 cm distance from the tumor's margin. All tissue specimens were frozen immediately using liquid nitrogen, after which tissues were conserved under 80 degree centigrade. The collection of EC tissue samples in this study was approved by the First Affiliated Hospital of Zhengzhou University Ethics Committee. Meanwhile, human endometrial stromal cell line (hESC) and endometrial carcinoma cell lines (HEC-1B AN3CA, KLE, ECC-1, and Ishikawa) cells in the study were bought from the Cell Center of Chinese Academy of Medical Sciences. The cells were cultured in dulbecco's modified eagle medium (DMEM; Sigma, St. Louis, MO, USA) culture medium with 10% fetal bovine serum (FBS; Invitrogen, Gaithersburg, MD, USA), 95% humidity and 5% CO2 at 37 degrees centigrade.
Cell transfection
The vectors containing negative control, miR-143 mimics and MAPK1 siRNA were bought from Dharmacon Corporation in USA. Human EC cells were incubated in culture plate with 50%-70% density of DMEM without fetal calf serum (FCS; Invitrogen, Gaithersburg, MD, USA) and cultured one day before transfection. Vectors were transferred into cells through lipofectamine Dharma FECT I in accordance with the manufacturer's protocols.
Cell counting kit-8 (CCK8)
Cells were collected and dissolved with 0.25% pancreatic enzymes after transfection for 16 h. Then the cells were resuspended in complete medium and plated at a density of 0.8×105 cells/well on 96-well plates, and repeated five times in every group. CCK8 (Beyotime, Jiangsu, China) was added into the plates along the wall of the wells, shaken for blending and cultured at 37 degree centigrade for 2 h. Wavelength absorbance of optical density (OD) value was set at 490 nm for each well on automatic enzyme-linked immune detector. Cell proliferation activity was determined with CCK8 method after transfection for 0 h, 24 h, 48 h, 72 h and 96 h.
Flow cytometry (FCM) assay
After transfected for 16 h, HEC-1B cells were collected. The cells were washed twice with phosphate buffered saline (PBS) and then were resuspended with PBS. Later, 100 μg/ml RNase A was dropped into cells, which were incubated at 37 degree centigrade for 30 min. Finally, a total of 50 μg/ml phosphate (PI) was added and cell cycle was analyzed through FCM (BD Biosciences, New York, USA).
After being transfected for 48 h, the cells were digested by trypsin, and then the cells were fixed and washed twice with PBS. The EC apoptosis was evaluated using a PE Annexin V apoptosis detection kit I. 5×105 EC cells were incubated with binding buffer and respectively added annexin V-PE/7-ADD. After being cultured 15 min at room temperature, 200 μL binding buffer was added and cell apoptosis was evaluated by means of FCM (BD Biosciences, New York, USA).
Wound healing assay
Wound healing assay was performed to examine and evaluate the migration ability of the cells. Cells were seeded in twelve-well plates at the density of 2×105 cells per well. At 80-90% confluence, the monolayer of cells was scratched with a sterile 200 ml micropipette tip. The scratched cells were washed with serum-free medium three times. After being removed from the cellular debris, the cells were incubated for 0 h, 12 h, 24 h, and closed area of the wound was observed under an optical microscope (Nikon, Japan).
Transwell assay
Matrigel was melted at 4 °C overnight, and diluted with precooling serum-free medium to 1 mg/mL. Then 40 μL Matrigel was added to the upper one of chambers and incubated at 37 degree centigrade for 4-5 h. In the upper chamber, we added a total of 2×105 EC cells in 200 μL serum-free medium. In the lower chamber, we added 600 μL of DMEM medium containing 10% FBS. After incubated at 37 degree centigrade and 5% CO2 for 24 h, cotton swabs were used to slightly wipe off the cells which failed to pass through and still remained on the membrane surface. The cells which invaded or migrated to the lower chamber were fixed with methanol for 10 min and stained with 0.1% crystal violet for 5 min. Finally, the number of invasion cells was observed and counted on the lower surface under a microscope.
Dual luciferase reporter assay
MAPK1 3’UTR with miR-143 binding site was amplified to psiCHECK-2 luciferase vector (Promega, Madison, WI, USA) as luc-MAPK1 vector. T Luc-MAPK1-mut vector with MAPK1 3’UTR mutation without miR-143 binding site was constructed by XL Site-directed Mutagenesis Kit (Qiagen, Hilden, Germany). Cells were co-transfected with Luc-MAPK1-wt/Luc-MAPK1-mut and miR-143/negative. After transfected for 48 h, cell gene activity was tested by dual luciferase assay kit (Promega, Wisconsin, USA). Turner Designs Spreadsheet Interface Version 2.0.1 was used for calculation and analysis.
Real-time PCR (RT-PCR)
Total RNA from the EC cells or tissues was extracted using Trizol Reagent (Invitrogen, Carlsbad, CA) following the instructions of manufacturer. Total RNA was reversely transcribed into cDNA using TIANScript RT Kit (Tiangen biotech, Beijing, China). Real-time PCR was carried out on a THUNDERBIRD SYBR® qPCR Mix (Toyobo, Japan). The RT-PCR was performed with CFX96 Touch RT-PCR Detection System (Bio-Rad). U6 and GAPDH were employed as reference genes to normalize the expression of miRNA and mRNA, respectively. The primers used for RT-PCR were listed in Table 1.
Table 1. The sequences of primers used in RT-PCR.
| cDNA | Primer sequences | |
|---|---|---|
| miR-143 | F | 5’-AGTGCGTGTCGTGGAGTC-3’ |
| R | 5’-GCCTGAGATGAAGCACTGT-3’ | |
| U6 | F | 5’-CTCGCTTCGGCAGCACA-3’ |
| R | 5’-AACGCTTCACGAATTTGCGT-3’ | |
| MAPK1 | F | 5’-AGGCTGTTCCCAAATGCT-3’ |
| R | 5’-CGTCACTCGGGTCGTAAT-3’ | |
| GAPDH | F | 5’-GAAATCCCATCACCACTTCCAGG-3’ |
| R | 5’-GAGCCCCAGCCTTCTCCATG-3’ |
F: forward primer; R: reverse primer.
Western blot
Total proteins were extracted from the cells by RIPA buffer (Sigma-Aldrich, St. Louis, MO). BCA kit (Beyotime Biotechnology, Haimen, China) was employed to examine the protein concentrations. Then the proteins were separated from sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE; Bio-Rad, Hercules, CA, USA) and transferred into polyvinylidene fluoride PVDF (Invitrogen, Gaithersburg, MD, USA) membranes. The membranes were washed off with TBST for 2 min and blocked in 5% non-fat milk for 2 h at room temperature. After blocked, the membranes were incubated with mouse anti-MPK1 and mouse anti-GAPDH primary antibodies (Abcam, Southampton, UK) followed by incubation with HRP-conjugated secondary antibody (Abcam, Southampton, UK) for 1 h at room temperate. The relative band density was determined using an enhanced chemiluminescence (ECL) detection system (Amersham Corp., Amersham, U.K.). Lab Work4.5 was used to examine the OD of the protein bands.
Statistical analysis
SPSS 21.0 software (IBM Corporation, New York, NY, USA) was used for performing statistical analyses. All data are presented as the mean ± SD. Differences were analyzed by t-test or one-way analysis of variance. P<0.05 was considered statistically significant.
Acknowledgments
This study was supported by the First Affiliated Hospital of Zhengzhou University Foundation Funded Project and Henan Province University Major Research Project (18A320072).
Abbreviations
- CCK8
Cell counting kit-8
- DMEM
dulbecco's modified eagle medium
- EC
endometrial cancer
- FCS
fetal calf serum
- FCM
flow cytometry
- GBM
glioblastoma multiforme
- miRNAs
microRNAs
- NC
negative control
- PBS
phosphate buffered saline
- PI
phosphate
- WT
wild type
Author contributions
Study design and conception: Lei Chang; data collection: Lei Chang, Huirong Shi, Dongya Zhang; data analysis: Lei Chang, Yangyang Bian; manuscript drafting: Lei Chang; manuscript critical revision: Ruixia Guo; approving article publishing: all authors.
CONFLICTS OF INTEREST
There are no conflicts of interest to report.
FUNDING
The First Affiliated Hospital of Zhengzhou University Foundation Funded Project
Henan Province University Major Research Project (18A320072)
ETHICAL STATEMENT AND INFORMED CONSENT
The collection of EC tissue samples in this study was approved by the First Affiliated Hospital of Zhengzhou University Ethics Committee. Informed consent was obtained from all individual participants included in the study.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.de Haydu C, Black JD, Schwab CL, English DP, Santin AD. An update on the current pharmacotherapy for endometrial cancer. Expert Opin Pharmacother. 2016;17:489–499. doi: 10.1517/14656566.2016.1127351. [DOI] [PubMed] [Google Scholar]
- 3.Chen HX, Xu XX, Tan BZ, Zhang Z, Zhou XD. MicroRNA-29b inhibits angiogenesis by targeting VEGFA through the MAPK/ERK and PI3K/Akt signaling pathways in endometrial carcinoma. Cell Physiol Biochem. 2017;41:933–946. doi: 10.1159/000460510. [DOI] [PubMed] [Google Scholar]
- 4.Hrzenjak A, Dieber-Rotheneder M, Moinfar F, Petru E, Zatloukal K. Molecular mechanisms of endometrial stromal sarcoma and undifferentiated endometrial sarcoma as premises for new therapeutic strategies. Cancer Lett. 2014;354:21–27. doi: 10.1016/j.canlet.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 5.Yang Q, Su Q, Wang G, Bi F, Sa R. Effect of AT1R knockdown on ishikawa cell proliferation induced by estrogen. Arch Gynecol Obstet. 2012;286:481–487. doi: 10.1007/s00404-012-2305-7. [DOI] [PubMed] [Google Scholar]
- 6.Ruan H, Liang X, Zhao W, Ma L, Zhao Y. The effects of microRNA-183 promots cell proliferation and invasion by targeting MMP-9 in endometrial cancer. Biomed Pharmacother. 2017;89:812–818. doi: 10.1016/j.biopha.2017.02.091. [DOI] [PubMed] [Google Scholar]
- 7.Yanokura M, Banno K, Iida M, Irie H, Umene K, Masuda K, Kobayashi Y, Tominaga E, Aoki D. MicroRNAS in endometrial cancer: recent advances and potential clinical applications. EXCLI J. 2015;14:190–198. doi: 10.17179/excli2014-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banno K, Yanokura M, Kisu I, Yamagami W, Susumu N, Aoki D. MicroRNAs in endometrial cancer. Int J Clin Oncol. 2013;18:186–192. doi: 10.1007/s10147-013-0526-9. [DOI] [PubMed] [Google Scholar]
- 9.Qiao J, Lee S, Paul P, Theiss L, Tiao J, Qiao L, Kong A, Chung DH. miR-335 and miR-363 regulation of neuroblastoma tumorigenesis and metastasis. Surgery. 2013;154:226–233. doi: 10.1016/j.surg.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 11.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 12.Bae J, Won M, Kim DY, Kim JH, Kim YM, Kim YT, Nam JH, Suh DS. Identification of differentially expressed microRNAs in endometrial cancer cells after progesterone treatment. Int J Gynecol Cancer. 2012;22:561–565. doi: 10.1097/IGC.0b013e31824927db. [DOI] [PubMed] [Google Scholar]
- 13.Wilczynski M, Danielska J, Dzieniecka M, Malinowski A. [The role of miRNA in endometrial cancer in the context of miRNA 205]. [Article in Polish] Ginekol Pol. 2015;86:856–861. [PubMed] [Google Scholar]
- 14.Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: current status and prospective. Int J Cancer. 2007;120:953–960. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- 15.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 16.Song Y, Li J, Zhu Y, Dai Y, Zeng T, Liu L, Li J, Wang H, Qin Y, Zeng M, Guan XY, Li Y. MicroRNA-9 promotes tumor metastasis via repressing E-cadherin in esophageal squamous cell carcinoma. Oncotarget. 2014;5:11669–11680. doi: 10.18632/oncotarget.2581. https://doi.org/10.18632/oncotarget.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y, Duan Y, Zhou H. MicroRNA-27a directly targets KRAS to inhibit cell proliferation in esophageal squamous cell carcinoma. Oncol Lett. 2015;9:471–477. doi: 10.3892/ol.2014.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu SH, Zhang CL, Dong FS, Zhang YM. miR-99a suppresses the metastasis of human non-small cell lung cancer cells by targeting AKT1 signaling pathway. J Cell Biochem. 2015;116:268–276. doi: 10.1002/jcb.24965. [DOI] [PubMed] [Google Scholar]
- 19.Wu LK, Liu YC, Ma G, Shi LL, He XM. High levels of glucose promote the activation of hepatic stellate cells via the p38-mitogen-activated protein kinase signal pathway. Genet Mol Res. 2016:15. doi: 10.4238/gmr.15038419. [DOI] [PubMed] [Google Scholar]
- 20.Jung YC, Han S, Hua L, Ahn YH, Cho H, Lee CJ, Lee H, Cho YY, Ryu JH, Jeon R, Kim WY. Kazinol-E is a specific inhibitor of ERK that suppresses the enrichment of a breast cancer stem-like cell population. Biochem Biophys Res Commun. 2016;470:294–299. doi: 10.1016/j.bbrc.2016.01.066. [DOI] [PubMed] [Google Scholar]
- 21.Bai J, Zheng Y, Wang G, Liu P. Protective effect of D-limonene against oxidative stress-induced cell damage in human lens epithelial cells via the p38 pathway. Oxid Med Cell Longev. 2016;2016:5962832. doi: 10.1155/2016/5962832. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Wang G, Jiao H, Zheng JN, Sun X. HSP27 regulates TGF-beta mediated lung fibroblast differentiation through the Smad3 and ERK pathways. Int J Mol Med. 2017;39:183–190. doi: 10.3892/ijmm.2016.2813. [DOI] [PubMed] [Google Scholar]
- 23.Zhao J, Liu J, Pang X, Zhang X, Wang S, Wu D. Epigallocatechin-3-gallate inhibits angiotensin II-induced C-reactive protein generation through interfering with the AT1-ROS-ERK1/2 signaling pathway in hepatocytes. Naunyn Schmiedebergs Arch Pharmacol. 2016;389:1225–1234. doi: 10.1007/s00210-016-1279-6. [DOI] [PubMed] [Google Scholar]
- 24.Hong SK, Park JR, Kwon OS, Kim KT, Bae GY, Cha HJ. Induction of integrin beta3 by sustained ERK activity promotes the invasiveness of TGFbeta-induced mesenchymal tumor cells. Cancer Lett. 2016;376:339–346. doi: 10.1016/j.canlet.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Xie Y, Cui D, Ma Y, Sui L, Zhu C, Kong H, Kong Y. Osteopontin promotes invasion, migration and epithelial-mesenchymal transition of human endometrial carcinoma cell HEC-1A through AKT and ERK1/2 signaling. Cell Physiol Biochem. 2015;37:1503–1512. doi: 10.1159/000438518. [DOI] [PubMed] [Google Scholar]
- 26.Ali I, Damdimopoulou P, Stenius U, Halldin K. Cadmium at nanomolar concentrations activates Raf-MEK-ERK1/2 MAPKs signaling via EGFR in human cancer cell lines. Chem Biol Interact. 2015;231:44–52. doi: 10.1016/j.cbi.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Guo Q, Wang C, Yan L, Fu Y, Fan M, Zhao X, Li M. Dual-specificity phosphatase 6 (Dusp6), a negative regulator of FGF2/ERK1/2 signaling, enhances 17beta-estradiol-induced cell growth in endometrial adenocarcinoma cell. Mol Cell Endocrinol. 2013;376:60–69. doi: 10.1016/j.mce.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Chen QG, Zhou W, Han T, Du SQ, Li ZH, Zhang Z, Shan GY, Kong CZ. MiR-378 suppresses prostate cancer cell growth through downregulation of MAPK1 in vitro and in vivo. Tumour bBol. 2016;37:2095–2103. doi: 10.1007/s13277-015-3996-8. [DOI] [PubMed] [Google Scholar]
- 29.Huang FT, Peng JF, Cheng WJ, Zhuang YY, Wang LY, Li CQ, Tang J, Chen WY, Li YH, Zhang SN. MiR-143 targeting TAK1 attenuates pancreatic ductal adenocarcinoma progression via MAPK and NF-κB pathway in vitro. Dig Dis Sci. 2017;62:944–957. doi: 10.1007/s10620-017-4472-7. [DOI] [PubMed] [Google Scholar]
- 30.Hartmann JU, Braeuer-Hartmann D, Gerloff D, Katzerke C, Wurm AA, Müller-Tidow C, Schwind S, Tenen DG, Niederwieser D, Behre G. The G-CSF induced MiR-143 targets MAPK-family proteins and is a prognostic factor for RIC-transplanted AML patients. Blood. 2014;124:2200–2200. [Google Scholar]
- 31.Noguchi S, Yasui Y, Iwasaki J, Kumazaki M, Yamada N, Naito S, Akao Y. Replacement treatment with microRNA-143 and -145 induces synergistic inhibition of the growth of human bladder cancer cells by regulating PI3K/Akt and MAPK signaling pathways. Cancer Lett. 2013;328:353–361. doi: 10.1016/j.canlet.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 32.Wu N, Sulpice E, Obeid P, Benzina S, Kermarrec F, Combe S, Gidrol X. The miR-17 family links p63 protein to MAPK signaling to promote the onset of human keratinocyte differentiation. PLoS One. 2012;7:e45761. doi: 10.1371/journal.pone.0045761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshino H, Enokida H, Itesako T, Kojima S, Kinoshita T, Tatarano S, Chiyomaru T, Nakagawa M, Seki N. Tumor-suppressive microRNA-143/145 cluster targets hexokinase-2 in renal cell carcinoma. Cancer Sci. 2013;104:1567–1574. doi: 10.1111/cas.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu YY, Tian J, Hao Q, Yin LR. MicroRNA-495 downregulates FOXC1 expression to suppress cell growth and migration in endometrial cancer. Tumour Biol. 2016;37:239–251. doi: 10.1007/s13277-015-3686-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruchim I, Sarfstein R, Werner H. The IGF hormonal network in endometrial cancer: functions, regulation, and targeting approaches. Front Endocrinol. 2014;5:76. doi: 10.3389/fendo.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H, Wang H, Liu H, Chen Y. Effect of miR-143 on the apoptosis of osteosarcoma cells. Int J Clin Exp Pathol. 2015;8:14241–14246. [PMC free article] [PubMed] [Google Scholar]
- 37.Mao Y, Liu J, Zhang D, Li B. miR-143 inhibits tumor progression by targeting FAM83F in esophageal squamous cell carcinoma. Tumour Biol. 2016;37:9009–9022. doi: 10.1007/s13277-015-4760-9. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Gong J, Xu B. miR-143 down-regulates TLR2 expression in hepatoma cells and inhibits hepatoma cell proliferation and invasion. Int J Clin Exp Pathol. 2015;8:12738–12747. [PMC free article] [PubMed] [Google Scholar]
- 39.Kouhkan F, Mobarra N, Soufi-Zomorrod M, Keramati F, Hosseini Rad SM, Fathi-Roudsari M, Tavakoli R, Hajarizadeh A, Ziaei S, Lahmi R, Hanif H, Soleimani M. MicroRNA-129-1 acts as tumour suppressor and induces cell cycle arrest of GBM cancer cells through targeting IGF2BP3 and MAPK1. J Med Genet. 2016;53:24–33. doi: 10.1136/jmedgenet-2015-103225. [DOI] [PubMed] [Google Scholar]
- 40.Yan J, Jiang N, Huang G, Tay JL, Lin B, Bi C, Koh GS, Li Z, Tan J, Chung TH, Lu Y, Ariffin H, Kham SK, et al. Deregulated MIR335 that targets MAPK1 is implicated in poor outcome of paediatric acute lymphoblastic leukaemia. Br J Haematol. 2013;163:93–103. doi: 10.1111/bjh.12489. [DOI] [PubMed] [Google Scholar]
- 41.Deschenes-Simard X, Kottakis F, Meloche S, Ferbeyre G. ERKs in cancer: friends or foes? Cancer Res. 2014;74:412–419. doi: 10.1158/0008-5472.CAN-13-2381. [DOI] [PubMed] [Google Scholar]
- 42.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 43.Carneiro RM, Carneiro BA, Agulnik M, Kopp PA, Giles FJ. Targeted therapies in advanced differentiated thyroid cancer. Cancer Treat Rev. 2015;41:690–698. doi: 10.1016/j.ctrv.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 44.Liu SM, Lu J, Lee HC, Chung FH, Ma N. miR-524-5p suppresses the growth of oncogenic BRAF melanoma by targeting BRAF and ERK2. Oncotarget. 2014;5:9444–9459. doi: 10.18632/oncotarget.2452. https://doi.org/10.18632/oncotarget.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H, An H, Wang B, Liao Q, Li W, Jin X, Cui S, Zhang Y, Ding Y, Zhao L. miR-133a represses tumour growth and metastasis in colorectal cancer by targeting LIM and SH3 protein 1 and inhibiting the MAPK pathway. Eur J Cancer. 2013;49:3924–3935. doi: 10.1016/j.ejca.2013.07.149. [DOI] [PubMed] [Google Scholar]


