Abstract
Rice grain with excessive cadmium (Cd) is a major source of dietary Cd intake and a serious threat to health for people who consume rice as a staple food. The development of elite rice cultivars with consistently low Cd content is challenging for conventional breeding approaches, and new strategies urgently need to be developed. Here, we report the development of new indica rice lines with low Cd accumulation and no transgenes by knocking out the metal transporter gene OsNramp5 using CRISPR/Cas9 system. Hydroponic culture showed that Cd concentrations in shoots and roots of osnramp5 mutants were dramatically decreased, resulting in rescue of impaired growth in high Cd condition. Cd-contaminated paddy field trials demonstrated that Cd concentration in osnramp5 grains was consistently less than 0.05 mg/kg, in contrast to high Cd concentrations from 0.33 mg/kg to 2.90 mg/kg in grains of Huazhan (the wild-type indica rice). In particular, the plant yield was not significantly affected in osnramp5 mutants. Furthermore, we developed promising hybrid rice lines with extremely low Cd content in grains. Our work supplies a practical approach to developing Cd pollution-safe indica rice cultivars that minimizes Cd contamination risk in grains.
Introduction
Cadmium (Cd) is a highly toxic heavy metal for most living organisms1. The biological half-life of Cd in the human body is estimated to be nearly 30 years2. Persistent intake of Cd can lead to chronic kidney toxicity, ‘itai-itai disease’, cancer and other health problems3. Rice is an important staple food for more than half of the world’s population4, and about 90% of the world’s rice is produced in Asia5. A recent survey showed that the mean concentration of Cd in rice grain samples from Asia was higher than in samples from Europe, the Middle East, and North America6. In South and Southeast Asia, indica rice is the main type produced and consumed2. Cd contamination of rice grains in these areas has been reported from China5, India7,8, Thailand9, Indonesia6,10, Bangladesh8, and Sri Lanka8. In general, Cd content in shoots and grains was higher in standard indica rice cultivars than in japonica rice cultivars11–14. Therefore, controlling Cd accumulation in indica rice grains is important for food safety and the health of people who consume rice as a staple of their diet.
Rice grains acquire Cd from contaminated paddy fields. Treatments such as soil removal and replacement, chemical washing, or phytoremediation can repair contaminated soil, but all of these methods are expensive and/or time-consuming15,16. Some measures such as lime and charcoal application, or field flooding for 3 weeks before and after rice heading, can limit the mobility and bioavailability of Cd. However, these methods may also reduce grain yield and are not long-term solutions, because Cd can be remobilized with changes in soil conditions17,18. Selection and breeding of low-Cd accumulation rice cultivars is an economical strategy, and would allow rice production in Cd-polluted soils5,11,14.
Understanding the physiological processes and molecular mechanisms of Cd uptake and transport within rice is essential for low-Cd rice breeding. Cd is uptaken by roots from the soil, transported into shoots through xylem flow, and accumulated in leaves and stems. During the reproductive growth phase, most Cd transported by xylem is transferred to phloem in the nodes and is then preferentially transported into upper nodes and grains rather than into leaf blades. These results are based on visualization of Cd in real time by a positron-emitting tracer imaging system (PETIS)19. Furthermore, Cd stored in leaf blades is remobilized through phloem and transported into grains after redirection in nodes during the ripening stage2,15. Many Cd transporters have been identified and characterized, including: OsIRT1, OsIRT2, and two members of the natural resistance associated macrophage proteins (NRAMP) family, OsNramp5 and OsNramp1, which mediate the root uptake of Cd20–23; OsHMA3, which sequesters Cd into vacuoles of root cells24,25; OsHMA2, which facilitates xylem loading of Cd26,27; and OsLCT1, which plays a role in transferring Cd from xylem to phloem in nodes and transporting Cd by phloem28. Modifying these transporter genes may reduce Cd content in rice. Among them, mutating OsNramp5 causes the largest reduction in Cd content. In a previous study, osnramp5 mutants with a background of the japonica cultivar Koshihikari were isolated from an ion-beam irradiated population. When grown in Cd-contaminated paddy fields, these mutants accumulated only a slight amount of Cd in grains, ranging from 0.01–0.03 mg/kg, less than 3% of the amount in grains of wild type plants. Notably, osnramp5 mutants did not show any significant defects of important agronomical traits such as growth, yield, or taste, although the mutants showed a remarkable reduction of manganese (Mn) content21. The T-DNA insertion mutant of OsNramp5 (with a background of japonica cultivar Zhonghua 11) also showed huge reductions in Cd and Mn accumulation, but the grain yield of this mutant was only 11% of the wild-type plants20. Indica rice tends to accumulate more Cd than japonica cultivars, but the effects of OsNramp5 mutation on accumulation of Cd and other relevant metals, and on major agronomic traits have not yet been clarified in indica rice.
Hybrid rice has a 10–20% yield advantage over conventional rice and has been developed in more than 40 countries worldwide29,30. Indica hybrids dominate hybrid rice production, especially in southern China. However, a considerable proportion of rice grains contaminated with excess Cd have been found in southern China5,31. Thus, genetic improvements in hybrid rice directed at low-Cd accumulation in grains have become a top priority for hybrid rice breeders. So far, there is no Cd pollution-safe rice germplasm in which grains consistently accumulate lower Cd than the national food safety standard of 0.2 mg/kg when grown in high Cd-contaminated paddy fields in China. In 2016, researchers grew 617 indica hybrid varieties from the middle and lower Yangtze River Valley in China and 68 inbred rice cultivars from around the world in paddy fields with soil Cd concentration between 1.80 and 2.80 mg/kg. They found that grain Cd levels ranged from 0.67 to 7.83 mg/kg, much higher than the national standard32. The deficiency of Cd pollution-safe germplasm has limited the breeding of low-Cd rice cultivars and hybrid rice combinations. Thus, new technologies to minimize the Cd content in rice grains are urgently required to reduce the risk of Cd contamination.
In recent years, genome editing technologies have been successfully used to genetically modify plants33–38. Among them, CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated Cas9) has revolutionized genome editing and is widely used because of its simplicity, efficiency, and versatility. However, there are only a few reports to date in which the CRISPR/Cas9 system has been successfully used for rice genetic improvement30,39–44.
In this report, we obtained osnramp5 mutants with backgrounds of Huazhan (HZ) and Longke638S (638S), both of which are extensively used indica parental lines in the current two-line hybrid rice breeding. The transgene-free homozygous mutants were efficiently isolated in the T1 generation. Notably, the mutants had extremely low-Cd content in grains and did not show stunted growth or reduced yield when grown in highly Cd-contaminated paddy fields. Furthermore, the Cd concentration in grains of Long-liang-you-hua-zhan (LLYHZ or 638 S/HZ), a widely cultivated super-hybrid rice in China, was rapidly and considerably reduced by crossing the HZ mutant and 638S mutant.
Results
Site-specific mutagenesis of OsNramp5 induced by CRISPR/Cas9 system
We chose to target exon IX of OsNramp5 for CRISPR/Cas9-based mutagenesis, as mutations in this exon have been shown to result in low Cd concentration of grains in the japonica Koshihikari background21. We designed two sequence-specific single guide RNA (sgRNA) target sites, OsNramp5-PS1 and OsNramp5-PS2, which are 119 bp apart in the OsNramp5 sequence (Fig. 1a). Then they were ligated into two sgRNA expression cassettes of a Cas9 binary vector45, driven by OsU6 and OsU3 promoters, respectively. The vector was introduced into HZ and 638S by Agrobacterium-mediated transformation. Using site-specific PCR and Sanger sequencing, a total of 14 HZ mutants were recovered from 17 T0 transgenic HZ plants (82.4%) and 8 638S mutants were recovered from 10 T0 transgenic 638S plants (80.0%). Mutation efficiency of individual target sites was varied and depended on different target sites and backgrounds, ranging from 70.0% to 82.4% (Table 1). The majority of mutations were short insertions and deletions (indels), except for one deletion of a 225 bp DNA fragment spanning the two target sites. Every 4th base upstream of the protospacer adjacent motif (PAM) was mutated, and aligned perfectly with the expected Cas9 cleavage site, which is 3 base pairs upstream of the PAM sequence (Fig. 1b).
Figure 1.
CRISPR/Cas9-induced mutations in the OsNramp5 gene. (a) Schematic diagram of OsNramp5 gene structure and two CRISPR/Cas9 target sites. UTRs, exons, and introns are indicated by blank rectangles, black rectangles, and black lines, respectively. The 20-nt target sequences are shown at the bottom of the gene structure, and the following PAMs (NGG) are highlighted in blue. (b) The mutant OsNramp5 genotypes of representative T0 plants are identified by DNA sequencing and alignment. Deletions and insertions are indicated by dashes and red letters, respectively. The numbers on the right side show the sizes of the indels, with “−” and “+” indicating deletion and insertion of the nucleotides involved, respectively. The letters after the numbers represent different bases of the same length.
Table 1.
The mutation rates of T0 transgenic plants.
| Target site | Host cultivar | No. of plants examined | No. of plants with mutations | Mutation rate (%) | Putative homozygous mutations | Putative bi-allelic mutations | Putative heterozygous mutations | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Number | % | Number | % | Number | % | |||||
| OsNramp5-PS1 | Huazhan | 17 | 14 | 82.4 | 3 | 17.6 | 9 | 53.0 | 1 | 5.9 |
| Longke 638S | 10 | 8 | 80.0 | 1 | 10.0 | 6 | 60.0 | 1 | 10.0 | |
| OsNramp5-PS2 | Huazhan | 17 | 12 | 70.6 | 3 | 17.6 | 5 | 29.4 | 1 | 5.9 |
| Longke 638S | 10 | 7 | 70.0 | 1 | 10.0 | 5 | 50.0 | 0 | 0.0 | |
The genotypes of the T0 mutants were analysed in detail by sequencing the PCR products of DNA extracted from at least 5 leaves from different tillers during grain maturation. The genotypes of the mutants could be classified into 4 types: 1) homozygotes with both alleles containing the same mutations, including lines HZ-6–4 and 638S-5-1; 2) heterozygotes with only one allele mutated, such as line 638S-1-1 at target site OsNramp5-PS1; 3) bi-allelic mutants with both copies of the target sequence mutated differently, including lines HZ-7-3 and 638S-4-2 at target site OsNramp5-PS1; and 4) chimeras in which at least 3 distinct target sequences were detected, including lines HZ-7-3 and 638S-4-2 at target site OsNramp5-PS2 (Fig. 1b). Overall, bi-allelic mutations occurred with the highest frequency in the T0 generation (Table 1).
Off-target mutations in the CRISPR/Cas9 system have been reported in previous studies46,47. To determine whether off-target events were produced in our experiments, we examined 4 putative off-target sites in the rice genome that showed the highest sequence similarity with our target sites. Among all 27 T0 transgenic plants and 30 T1 transgenic plants, no off-target events were found (Supplementary Table 1). These results demonstrate that the sgRNA we designed are of high specificity, and thus we have obtained a high gene editing efficiency with undetectable off-target mutations.
Obtaining homozygous knockout lines free of transgenes in the T1 generation
To further understand the inheritance of mutation, 4 T0 plants and their progeny were investigated. For each T0 plant, 20–38 progeny were sampled randomly and genotyped individually at the two target sites (Supplementary Table 2). As expected, all of the T0 putative homozygotes and their offspring had identical genotypes (HZ-6-4 and 638S-5-1), suggesting that the mutations in these T0 plants were faithfully inherited in the next generation. For the T1 generation of putative bi-allelic mutants (HZ-7-3 and 638S-4-2 at OsNramp5-PS1), the genotype segregation ratio was consistent with Mendelian law, indicating that the targeted mutations in T0 plants were inherited normally. For the T1 generation of chimeras (HZ-7-3 and 638S-4-2 at OsNramp5-PS2), some parental mutations were lost, probably because these mutations took place in parental somatic cells that did not participate in gamete production, and some new mutations were generated, probably due to continuous modification of the wild-type (WT) alleles in Cas9 positive T1 lines.
We also tracked the segregation of transgene (T-DNA) in the T1 population of the 4 T0 plants (Supplementary Table 2), based on a PCR assay of Cas9 and HPT, which are the elements of T-DNA (Supplementary Fig. 1). The absence of transgenes was determined by negative PCR results of both Cas9 and HPT. Transgene-free plants were observed in the progeny of all detected T0 plants, with the proportion ranging from 13.6% to 35.0% (Supplementary Table 2). These results indicated that transgene-free homozygous mutants could be easily obtained in the T1 generation, as the inheritance of T-DNA and the targeted gene was relatively independent. We isolated 5 transgene-free homozygous knockout lines with coding frame shifts and premature translational stops, including HZ-6-4-6, HZ-7-3-2, HZ-7-3-12, 638S-5-1-4, and 638S-4-2-37, in the T1 generation to produce the T2 population (Fig. 2).
Figure 2.
Homozygous osnramp5 mutants induced by CRISPR/Cas9. (a) DNA sequence alignments for the 5 homozygous osnramp5 mutants identified in the T1 generation, together with a wild-type (WT) control. The numbers on the right side are the sizes of the indels, with “−” and “+” showing deletion and insertion of nucleotides involved, respectively. (b) Deduced OsNramp5 amino acid sequence alignments for the 5 homozygous mutants and WT. Each of the mutant alleles codes for truncated and disrupted OsNramp5 proteins.
Effect of knocking out OsNramp5 on metal accumulation and major agronomic traits in indica rice
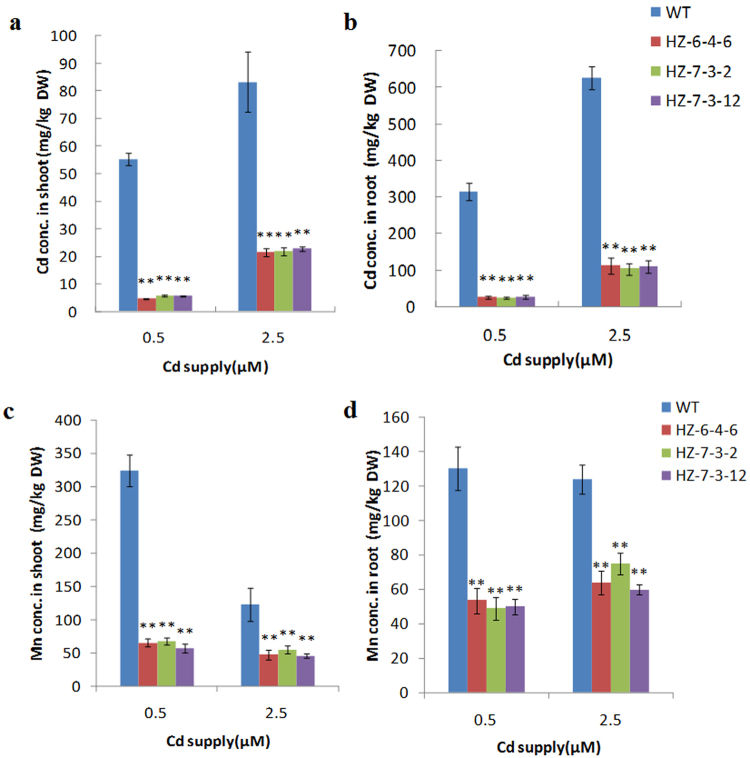
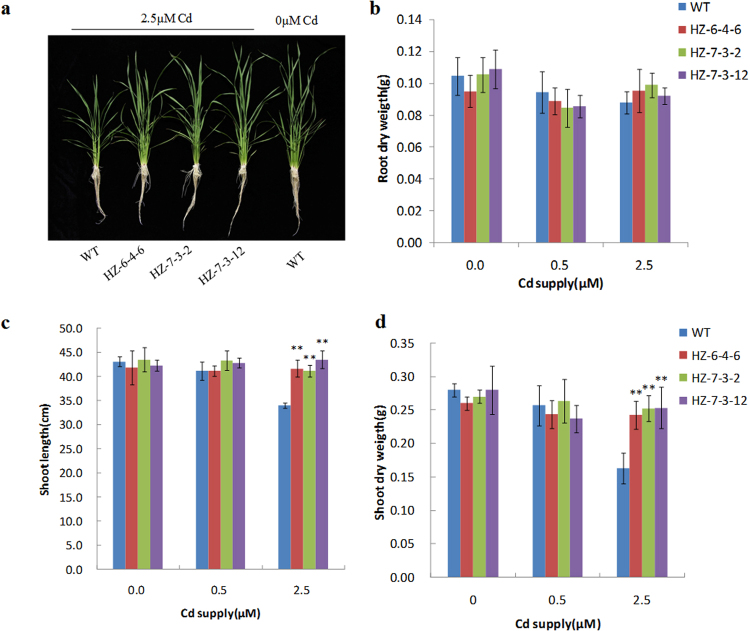
To measure metal accumulation and agronomic traits in the OsNramp5 knockout mutants, the progeny of HZ-6-4-6, HZ-7-3-2, HZ-7-3-12, and WT plants were cultivated in nutrient solution supplemented with Cd or in soil contaminated by Cd. Metal analysis of seedlings grown in hydroponic culture with 0.5 µM or 2.5 µM Cd showed that both the Cd and Mn concentration in shoots and roots was much lower in osnramp5 mutant lines than in WT plants (3.7-13 fold difference of Cd and 1.7–5.6 fold difference of Mn; Fig. 3). Under the toxic condition of 2.5 µM Cd, growth of WT plants was severely inhibited compared with that under no Cd stress, whereas growth of mutant lines was close to that of WT plants under no Cd stress (Fig. 4), indicating that the decrease of Cd in mutants could greatly rescue the reduced growth in the presence of normal-level Mn. In contrast, the concentration of other relevant metals in roots, including iron (Fe), zinc (Zn) and copper (Cu), was not different between the mutants and WT plants (Supplementary Fig. 2d,e,f). In shoots of mutants, the Zn concentration remained essentially unchanged, while the Fe concentration was lower and the Cu concentration was higher compared with that in shoots of WT plants (Supplementary Fig. 2a,b,c). However, the change in concentrations of Fe and Cu was much less than that for Cd and Mn.
Figure 3.
Concentrations of Cd and Mn in shoots and roots at different Cd levels. Three osnramp5 mutant lines (HZ-6-4-6, HZ-7-3-2, HZ-7-3-12) and WT plants (HZ) were grown in normal nutrient solution for 2 weeks and then transferred to the nutrient solution containing 0.5 or 2.5 µM Cd for another 2 weeks. The shoots and roots were harvested and subjected to mineral analysis by inductively coupled plasma optical emission spectrometry (ICP-OES). (a and b) Cd concentrations in the shoots (a) and roots (b). (c and d) Mn concentrations in the shoots (c) and roots (d). DW, dry weight. Data are means ± SD of three biological replicates, and three plants were mixed in one replication for metal determination. Two asterisks indicate statistically significant difference in comparison to WT at P < 0.01 by Student’s t test.
Figure 4.
Phenotypic analysis of osnramp5 mutants at different Cd levels. Three mutant lines (HZ-6-4-6, HZ-7-3-2, HZ-7-3-12) and WT plants (HZ) were cultivated in normal nutrient solution for 2 weeks and then transferred to the nutrient solution containing 0.5 or 2.5 µM Cd for another 2 weeks. (a) Plant morphologies of the mutant lines and WT in response to 2.5 µM Cd exposure. (b) Root dry weight. (c) Shoot length. (d) Root dry weight. Data are means ± SD of three biological replicates, and three plants were mixed in one replication. Two asterisks indicate statistically significant difference in comparison to WT at P < 0.01 by Student’s t test.
Mn concentration was mostly decreased among essential micronutrients in osnramp5 mutants, and the great reduction in Mn concentration may affect plant growth. To gain insight into the effect of low Mn supply to growth of osnramp5 mutants, both mutant and WT plants were grown in normal hydroponic culture for 12 d, and then transferred to nutrient solutions with different low concentrations of Mn for 18 d. Under the condition of no Mn supply, the mutant lines showed severely stunted growth and yellowing leaves (typical symptoms of Mn deficiency), in contrast to the phenotypes of WT plants. However, the mutants grew similarly to WT plants in the presence of 2 µM or higher Mn (Supplementary Fig. 3). These results imply that mutants are tolerant of low Mn conditions to a certain extent.
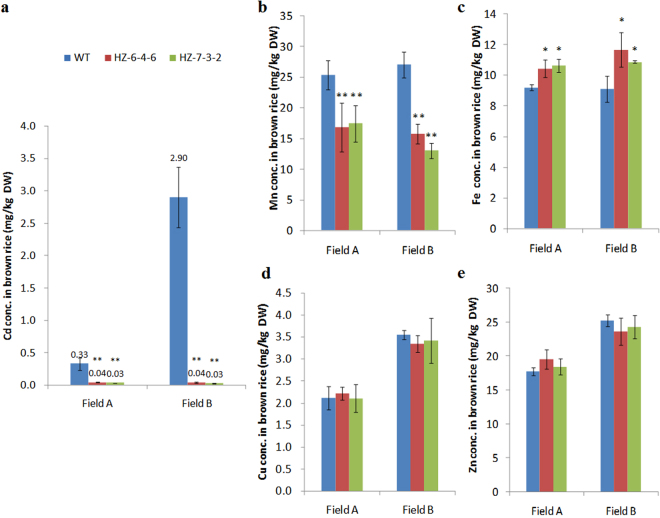
When mutant plants were grown until maturity in two restricted experimental paddy fields contaminated by different Cd levels (1.69 mg/kg and 2.37 mg/kg), the Cd concentration in grains (brown rice) was <0.05 mg/kg, compared with an average of 0.33 mg/kg and 2.90 mg/kg in the grains of WT plants (Fig. 5a). Similar results were found in the pot with Cd-polluted soil, where the Cd concentration in grains of mutants was also <0.05 mg/kg, and that of WT was 1.06 mg/kg (Supplementary Fig. 4). These results suggest that knockout of OsNramp5 could produce low Cd-accumulating rice in high Cd-polluted fields. The analysis of other relevant metals in grains showed that mutants accumulate 34–53% less Mn than in WT plants (Fig. 5b). Unexpectedly, the concentration of Fe was higher in the mutants (Fig. 5c), but no significant differences in Cu or Zn concentration were observed between mutants and WT plants (Fig. 5d and e). In straw, the concentrations of Cd and Mn were much lower in the mutants than in WT plants, and the concentrations of Fe, Zn, and Cu were similar between the mutants and WT plants (Supplementary Table 3 and Supplementary Table 4).
Figure 5.
Metal concentrations in the brown rice of osnramp5 mutants. Two mutant lines (HZ-6-4-6, HZ-7-3-2) and WT plants (HZ) were cultivated together in two restricted experimental paddy fields containing 1.69 mg/kg (Field A) and 2.37 mg/kg (Field B) Cd in the soil, respectively. (a) Cd, (b) Mn, (c) Fe, (d) Cu and (e) Zn concentrations in brown rice were determined by ICP-OES. DW, dry weight. Data are means ± SD of three biological replicates, and grains of three plants were mixed in one replication. One or two asterisks indicate statistically significant difference in comparison to WT at P < 0.05 or P < 0.01 by Student’s t test.
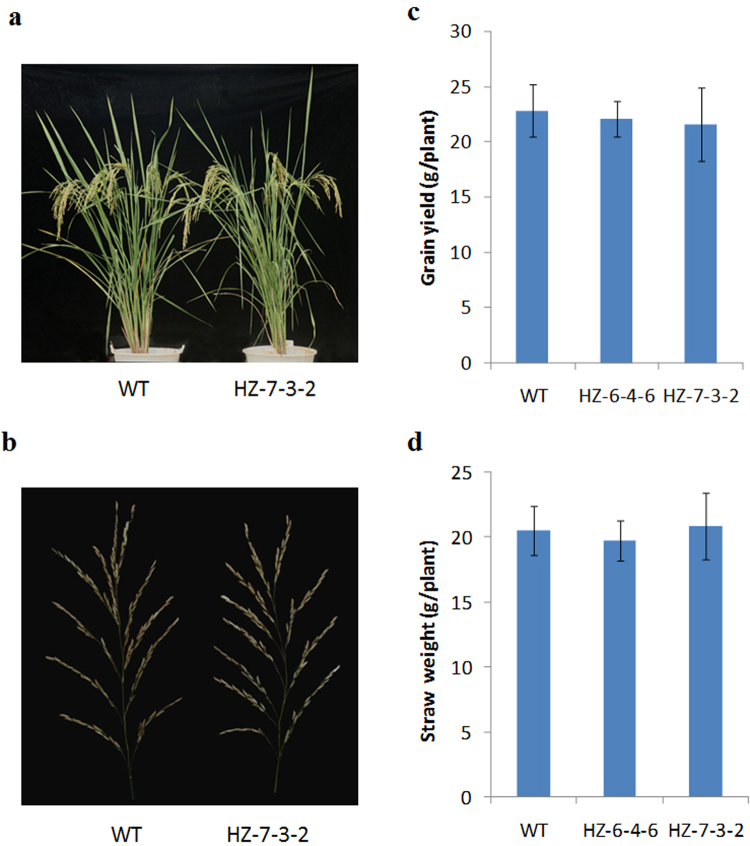
Phenotypic analysis showed that there were no visible differences in plant growth or panicle morphologies between the mutant lines and the WT plants (Fig. 6a and b; Supplementary Fig. 5). Moreover, mutant lines exhibited no significant differences in grain yield, straw weight (Fig. 6c and d), grain quality (Supplementary Table 5), or other main agronomic traits (Supplementary Table 6), compared with WT plants. These results demonstrate that knocking out OsNramp5 could have negligible effects on growth, biomass, and main agronomic traits, under our cultivation conditions.
Figure 6.
Morphology and yield analysis of osnramp5 mutants. Two mutant lines (HZ-6-4-6, HZ-7-3-2) and WT plants (HZ) were cultivated in the Cd contaminated experimental field (Field A). (a) Plant morphology. (b) Panicle morphology. (c) Grain yield. (d) Straw dry weight. There is no significant difference between WT and mutant lines in grain yield and straw dry weight according to Student’s t test.
Development of promising hybrid rice lines with extremely low Cd content in grains
We developed two independent mutant hybrid rice lines (P12-1, P12-2) by crossing the 638S mutants (638S-5-1-4, 638S-4-2-37) and HZ mutants (HZ-6-4-6, HZ-7-3-12), respectively. The hybrid rice line P12-C generated by crossing WT 638S and HZ mutant (HZ-6-4-6) served as a control. These mutants and WT LLYHZ plants were grown in the experimental paddy field with 2.37 mg/kg Cd in soil. Metal analysis showed that Cd concentration in grains of mutant lines (P12-1, P12-2) was decreased by more than 98% (<0.05 mg/kg) and the Mn concentration was reduced by 41% compared with WT plants. However, there was no significant difference in the concentrations of Fe, Zn, or Cu in grains between the mutant lines (P12-1, P12-2) and WT plants (Fig. 7a). In contrast, concentrations of all five metals in the grains of P12-C were similar to that of WT plants (Fig. 7a), suggesting that metal content was changed only when both of the parental lines were mutated. In addition, there was no obvious difference in plant morphology, grain yield, or straw weight between the mutant lines and WT plants (Fig. 7b–d). These results demonstrate that the knockout of OsNramp5 by the CRISPR/Cas9 system could be applied to two-line indica hybrid breeding, developing Cd pollution-safe hybrids suitable for planting in high Cd paddy fields.
Figure 7.
Morphology and yield analysis of mutant hybrid rice lines. Two osnramp5 mutant hybrid rice lines (P12-1, P12-2), control line (P12 C) and WT plants (LLYHZ) were cultivated in the highly Cd-contaminatedexperimental field (Field B). (a) Metal concentration in brown rice. (b) Plant morphology. (c) Grain yield. (d) Straw dry weigh. DW, dry weight. Two asterisks indicate statistically significant difference in comparison to WT at P < 0.01 by Student’s t test.
Discussion
There is a worldwide concern about Cd contamination in rice grains, and thus lowering grain Cd content is critical, but still challenging. In the present study, OsNramp5 was knocked out using the CRISPR/Cas9 system, in the popular male sterile line 638S and restorer line HZ of indica hybrid rice. In addition, the mutant hybrid LLYHZ was developed by crossing the mutant parental lines. The osnramp5 mutants and their hybrids had nearly undetectable levels of Cd in the grains, and yield and main agronomic traits were not affected when they were grown in Cd-contaminated paddy fields. These phenotypes were similar to that of the osnramp5 mutant with the Koshihikari background21. In contrast, the growth and yield of the T-DNA insertion mutant of OsNramp5 with Zhonghua 11 background were heavily influenced20. The inconsistent effect of OsNramp5 mutation on plant yield probably arises from different Mn concentration in mutants caused by differences in mutation sites, genetic backgrounds, or supplies of Mn.
Mn is an essential micronutrient for plant growth and development, and thus Mn deficiency could result in lower yield in osnramp5. Mn concentrations in the shoots/straws were much higher in HZ and LLYHZ knockout mutants than in the Zhonghua11 knockout mutant, in both normal hydroponic culture and soil culture (Fig. 3c, Supplementary Table 3 and Supplementary Table 4)20. In addition, Mn concentrations in straws of HZ and LLYHZ knockout mutants grown in the paddy fields were decreased by 80.6–82.6% compared with WT plants, and were either higher than or close to 150 mg/kg (Supplementary Table 3 and Supplementary Table 4). This level of Mn concentration was high enough to cause toxic symptoms in barley48. In contrast, the Mn concentration in straw of the Zhonghua 11 knockout mutant grown in soil culture was approximately 95% less than that in WT plants20. On the other hand, the HZ knockout mutants probably require less Mn for normal growth than the Zhonghua 11 knockout mutant. Here, we performed hydroponic culture with a method similar to that used by Yang et al.49, except with a different Mn concentration gradient. Mn concentration in the shoots of the HZ knockout mutant (2 µM Mn supplied) was close to that in the Zhonghua 11 knockout mutant (1.6 µM Mn supplied) (Supplementary Table 7)49. However, the HZ knockout mutant had a similar phenotype to WT plants, whereas the Zhonghua 11 knockout mutant showed reduced growth (Supplementary Fig. 3)49. It has been described that the critical deficiency concentrations of Mn in plants are similar, varying between 10 and 20 mg/kg dry weight in fully expanded leaves50. Mn concentrations in the shoots of both the HZ knockout mutant and Zhonghua 11 knockout mutant were within this range. It is likely that the higher Mn concentration in straws of HZ knockout mutants and their lower sensitivity to low-Mn concentration, caused the different influence on their yield compared with that of Zhonghua11 knockout mutant.
In contrast to other crops, rice is usually cultivated under flooded conditions, where is anaerobic and Mn concentration is generally high. Rice has developed strategies to accumulate high Mn without symptoms of toxicity48. It is possible that the growth of osnramp5 mutants would not be inhibited, when mutants were cultivated in Cd-polluted soils with a low Mn concentration, by applying Mn fertilizer or prolonging flooded time properly, due to greatly increased bioavailability of Mn under flooded conditions51. The effects of knocking out OsNramp5 on Mn concentration in shoots, on plant growth, and on main agronomic traits under gradient low-Mn field conditions need to be studied further in various rice cultivars with multiple genetic backgrounds.
The markedly lower accumulation of Mn and Cd in the osnramp5 mutant in the present study supports the crucial role of OsNramp5 for Mn and Cd uptake20,52. In addition, increasing Cd in the external solution decreased Mn accumulation in shoots of WT plants, while this effect was not obvious in osnramp5 mutants (Fig. 3c). This result implies that Mn and Cd compete for uptake mediated by the OsNramp5 transporter, and an abundance of one metal could disturb the uptake of the other. Rice is a staple food crop with high Mn accumulation, and the majority of Mn is taken up by OsNramp520. It seems likely that external Cd competitively enters the root cells during the process of OsNramp5 mediating uptake of Mn from the soil, as OsNramp5 has limited substrate specificity. Genotypic variation of Cd accumulation between two rice subspecies, japonica and indica, has been reported11–14, but little is known about the molecular mechanism. As OsNramp5 contributes greatly to Cd accumulation in rice, further analysis is needed to determine whether there is a difference in the expression level of OsNramp5 between indica and japonica rice, and whether there is a positive correlation between the expression level of OsNramp5 and the Cd content in rice.
It has been reported that knockdown of OsNramp5 could lead to more Cd accumulation in shoots of RNAi lines than WT plants, making it a promising candidate for phytoremediation in paddy fields52,53. However, OsNramp5 RNAi knockdown lines generated by Sasaki et al.20, as well as OsNramp5 knockout lines in our study and other previous studies20,21,49, accumulated much less Cd in shoots than WT plants. This contradictory effect is not likely to arise from the direct role of OsNramp5. There is strong evidence that Cd and Fe share transporters in rice, such as OsIRT1, OsIRT2, OsNramp1, and OsNramp5 20,22,23. The expression of some Fe/Cd transporter genes were highly induced in OsNramp5 knockout or knockdown lines, especially under the condition of Fe (II) deficiency49,52,53. The increased expression of these Fe/Cd transporters enhanced Cd translocation from root to shoot22,52. The expression of OsNramp5 in Anjana Dhan RNAi lines was suppressed only by 1/3 to 1/2 of that in WT plants53, so the decreased amount of Cd in shoots by suppression of OsNramp5 would be less than the increased amount of Cd by induction of other Fe/Cd transporters, giving rise to higher Cd concentrations in shoots of RNAi lines. But the degree of decrease in Cd amount in shoots was very serious, when the structure of OsNramp5 protein was broken down in knockout lines21, or when the expression of OsNramp5 was extremely suppressed in knockdown lines20, which cannot be complemented by an increase in Cd translocation through other Fe/Cd transporters.
In conclusion, we evaluated the effects of OsNramp5 mutation on accumulation of Cd and relevant metals, and on main agronomic traits in indica rice using CRISPR/Cas9-mediated mutagenesis. The high Cd accumulation trait of indica rice was modified quickly, effectively, and conveniently, whereas yield and main agronomic traits were not impaired. Transgenes were segregated out through selfing, creating genome-edited mutant plants that were free of transgenes. These plants were not different from plants carrying spontaneous mutations or mutations induced by physical and chemical treatment, in addition to the advantage that the precise mutation reduced the risk of introducing undesired traits41,54. Thus, these genome-edited mutants could circumvent public and governmental concerns about introduced persistent transgenes or other unwanted genomic fragments. Our study developed promising indica rice lines and their hybrids with extremely low Cd content in grains and without compromising yield using the CRISPR/Cas9 mediated OsNramp5 editing system. This approach can be used to reduce grain Cd levels in other widespread indica rice cultivars, facilitating the development of precision breeding targeted at Cd pollution-safe rice.
Methods
Vector construction and rice transformation
The Cas9 plant expression vector (pYLCRISPR/Cas9Pubi-H) and sgRNA expression vector (pYLgRNA) were provided by Prof. Yao-Guang Liu (South China Agricultural University). According to the design principles of the target sequences in the CRISPR/Cas9 system, 19 to 20 bases upstream of the PAM motif were selected as candidate target sequences. A BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) of the target sequences (including PAM) against the rice genome was carried out to confirm their targeting specificity in the genome. CRISPR/Cas9 plant expression vectors were constructed as previously described45, based on Golden Gate cloning. Binary constructs were then introduced into Agrobacterium tumefaciens strain EHA105. Popular parental lines of indica hybrid rice (Oryza sativa L. cv. Huazhan and Longke 638S) were used for plant transformation. Agrobacterium-mediated transformations of embryogenic calli were performed by Wuhan Biorun biological technology Co., Ltd. After 4 weeks of rooting, regenerated rice plants were transferred to plastic buckets in a greenhouse maintained at 30 °C during the day and 26 °C at night.
Mutation detection and assay of transgene-free plant lines
Genomic DNA was extracted from at least 5 leaves of different tillers during the rice maturity period using the cetyltrimethyl ammonium bromide (CTAB) method55. PCR was performed to amplify the genomic region containing the CRISPR/Cas9 target sites using specific primers (Supplementary Table 8). The PCR products were purified and directly sequenced. If the sample produced superimposed sequence chromatograms, it was cloned into the pEASY-Blunt (Trans Gen Biotech, Beijing, China), and at least 10 clones were sequenced to identify the genotype of the mutation. Multiple amino acid sequence alignments were performed using ClustalX256 and displayed using GeneDoc software (http://www.softpedia.com/get/Science-CAD/GeneDoc.shtml#download).
The identification of transgene-free plants was conducted using T1 generation plants. The plants were analysed by PCR using HPT-specific and Cas9-specific primers (Supplementary Table 8) and agarose gel electrophoresis. The CRISPR/Cas9 plasmid targeting OsNramp5 and the T0 transgenic plants were selected as positive controls, and HZ or 638S and H2O were used as negative controls. HPT- and Cas9-negative plants were considered transgene-free plants.
Hydroponic experiments
Seedlings of three independent mutant lines and WT plants were grown hydroponically in rice culture solution described by Yoshida et al.57. The plants were grown in a greenhouse at 26 °C to 30 °C under natural light. The culture solution was renewed every 3 d. In the Cd treatment experiments, plants were cultivated in normal nutrient solution for 14 d and then transferred to the nutrient solution containing 0.5 or 2.5 µM Cd (added as CdCl2) for another 14 d. The roots were washed three times with deionized water and separated from the shoots, and subjected to metal concentration determination as described below. In the low Mn concentration gradient experiments, plants were grown in normal nutrient solution for 12 d, and then transferred to the nutrient solution with 0, 2, 4, or 6 µM Mn (lower than normal culture) for 18 d. Three biological replicates were used for each treatment.
Field and pot experiments
The independent mutant lines and WT plants were cultivated in different Cd-polluted experimental paddy fields in China. The soil Cd concentrations were 1.69 mg/kg in field A, 2.37 mg/kg in field B and 1.53 mg/kg in field C; the soil Mn concentrations were 246 mg/kg in field A, 162 mg/kg in field B and 849 mg/kg in field C; and the soil pH values were 5.1 in field A, 5.5 in field B and 6.1 in field C. 28-day-old seedlings were transplanted into the flooded field plots, with one plant per hill, spaced at 20 cm × 20 cm. The plot for each line included 4 rows, with a total of 40 plants. The fields were irrigated intermittently until grain maturity. We applied inorganic fertilizers containing N, K, and P using standard methods. After grains were harvested, metal concentration and major agronomic traits, including grain yields and straw weight, were analysed. Rice quality was measured using the standard of NY/T 593-2013 published by the Rice Product Quality Supervision and Inspection Centre, Ministry of Agriculture, China (http://www.zbgb.org/27/StandardDetail1476335.htm). The field experiments were arranged in a randomized complete block design with three plot replications. In pot experiments, 28-day-old seedlings were transplanted into plastic pots filled with polluted paddy soil with 1.31 mg/kg Cd and 259 mg/kg Mn concentration, with a pH value of 5.7. Plants were grown in a greenhouse at 26 to 30 °C under natural light, with three biological replicates. Tap water was supplied intermittently throughout the cultivation.
Metal concentration determination
Shoots, roots, and brown rice samples were harvested and dried at 70 °C for 2 d. The dried samples were ground into powder and digested with an acid mixture of HNO3-HClO4 (6:1,v/v), and then placed on an electric heating plate for digestion until they were almost completely evaporated. After cooling, the residues were dissolved in 1% HNO3 (1:10, v/v) and filtered. The filtered solutions were diluted to 10 mL for brown rice samples, and 25 mL for shoot and root samples. Then, the metal concentration was determined by inductively coupled plasma optical emission spectrometry (SPS3 ICP-720 OES; Agilent Technologies) at the following wavelengths: 226.502 (Cd), 293.305 (Mn), 238.204 (Fe), 206.200 (Zn), and 324.754 (Cu) nm.
Electronic supplementary material
Acknowledgements
We thank Prof. Yao-Guang Liu (South China Agricultural University, Guangzhou) for kindly providing the plant binary vector pYLCRISPR/Cas9Pubi-H and the pYLgRNA plasmids. This work was supported by the Major Scientific Research Project of Hunan Province (Grant No. XCNZ [2014] 180), Hunan Provincial Natural Science Fund for Distinguished Young Scholars (Grant No.11JJ1007), the Natural Science Foundation of Hunan Province (Grant No. 14JJ7084) and the Earmarked Fund for Modern Agro-Industry Technology System of China (Grant No. CARS-01-04B).
Author Contributions
H.L., S.X., and B.Z. designed the study. L.T., B.M., Y.L., Q.L., L.Z., H.H., W.W., X.Z., Y.S., Y.P., Y.H., and Y.P. performed the experiments. L.T. prepared the figures and tables, and wrote the manuscript. C.C., X.F., H.L., S.X., and B.Z. discussed the results, supervised and edited the paper. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14832-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hongqing Li, Email: hqli@scnu.edu.cn.
Shitou Xia, Email: xstone0505@hunau.net.
Bingran Zhao, Email: brzhaorice@163.com.
References
- 1.Clemens S, Aarts MG, Thomine S, Verbruggen N. Plant science: the key to preventing slow cadmium poisoning. Trends Plant Sci. 2013;18:92–99. doi: 10.1016/j.tplants.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Uraguchi S, Fujiwara T. Rice breaks ground for cadmium-free cereals. Current opinion in plant biology. 2013;16:328–334. doi: 10.1016/j.pbi.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Bertin G, Averbeck D. Cadmium: cellular effects, modifications of biomolecules, modulation of DNA repair and genotoxic consequences (a review) Biochimie. 2006;88:1549. doi: 10.1016/j.biochi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Yuan LP. Development of Hybrid Rice to Ensure Food Security. Rice Science. 2014;21:1–2. doi: 10.1016/S1672-6308(13)60167-5. [DOI] [Google Scholar]
- 5.Hu Y, Cheng H, Tao S. The Challenges and Solutions for Cadmium-contaminated Rice in China: A Critical Review. Environment international. 2016;92–93:515–532. doi: 10.1016/j.envint.2016.04.042. [DOI] [PubMed] [Google Scholar]
- 6.Jallad KN. Heavy metal exposure from ingesting rice and its related potential hazardous health risks to humans. Environmental science and pollution research international. 2015;22:15449–15458. doi: 10.1007/s11356-015-4753-7. [DOI] [PubMed] [Google Scholar]
- 7.Shraim, A. M. Rice is a potential dietary source of not only arsenic but also other toxic elements like lead and chromium. Arabian Journal of Chemistry, 1–10 (2014).
- 8.Meharg AA, et al. Variation in rice cadmium related to human exposure. Environmental science & technology. 2013;47:5613–5618. doi: 10.1021/es400521h. [DOI] [PubMed] [Google Scholar]
- 9.Honda R, et al. Cadmium induced renal dysfunction among residents of rice farming area downstream from a zinc-mineralized belt in Thailand. Toxicology Letters. 2010;198:26–32. doi: 10.1016/j.toxlet.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe T, Shimbo S, Moon CS, Zhang ZW, Ikeda M. Cadmium contents in rice samples from various areas in the world. Science of the Total Environment. 1996;184:191–196. doi: 10.1016/0048-9697(96)05100-5. [DOI] [PubMed] [Google Scholar]
- 11.Arao T, Ae N. Genotypic variations in cadmium levels of rice grain. Soil Science and Plant Nutrition. 2003;49:473–479. doi: 10.1080/00380768.2003.10410035. [DOI] [Google Scholar]
- 12.Arao T, Ishikawa S. Genotypic differences in cadmium concentration and distribution of soybean and rice. Jarq Japan Agricultural Research Quarterly. 2006;40:21–30. doi: 10.6090/jarq.40.21. [DOI] [Google Scholar]
- 13.Uraguchi S, et al. Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. Journal of Experimental Botany. 2009;60:2677. doi: 10.1093/jxb/erp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant CA, Clarke JM, Duguid S, Chaney RL. Selection and breeding of plant cultivars to minimize cadmium accumulation. Science of the Total Environment. 2008;390:301–310. doi: 10.1016/j.scitotenv.2007.10.038. [DOI] [PubMed] [Google Scholar]
- 15.Uraguchi S, Fujiwara T. Cadmium transport and tolerance in rice: perspectives for reducing grain cadmium accumulation. Rice. 2012;5:5. doi: 10.1186/1939-8433-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebbs SD, et al. Phytoextraction of cadmium and zinc from a contaminated soil. Journal of Environmental Quality. 1997;26:1424–1430. doi: 10.2134/jeq1997.00472425002600050032x. [DOI] [Google Scholar]
- 17.Arao T, Ishikawa S, Murakami M, Abe K. Heavy metal contamination of agricultural soil and countermeasures in Japan. Paddy and Water Environment. 2010;8:247–257. doi: 10.1007/s10333-010-0205-7. [DOI] [Google Scholar]
- 18.Huang, S. et al. Evaluation of the effects of lime-bassanite-charcoal amendment on the immobilization of cadmium in contaminated soil. Bull Environ Contam Toxicol 1–6 (2016). [DOI] [PubMed]
- 19.Fujimaki S, et al. Tracing cadmium from culture to spikelet: noninvasive imaging and quantitative characterization of absorption, transport, and accumulation of cadmium in an intact rice plant. Plant physiology. 2010;152:1796–1806. doi: 10.1104/pp.109.151035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasaki A, Yamaji N, Yokosho K, Ma JF. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell. 2012;24:2155–2167. doi: 10.1105/tpc.112.096925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa S, et al. Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19166–19171. doi: 10.1073/pnas.1211132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimo H, et al. Low cadmium (LCD), a novel gene related to cadmium tolerance and accumulation in rice. Journal of Experimental Botany. 2011;62:5727–5734. doi: 10.1093/jxb/err300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senoura T, Shimo H, Ishikawa S, Nishizawa NK. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. Journal of Experimental Botany. 2011;62:4843–4850. doi: 10.1093/jxb/err136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueno D, Koyama E, Yamaji N, Ma JF. Physiological, genetic, and molecular characterization of a high-Cd-accumulating rice cultivar, Jarjan. Journal of Experimental Botany. 2011;62:2265–2272. doi: 10.1093/jxb/erq383. [DOI] [PubMed] [Google Scholar]
- 25.Ueno D, et al. Gene limiting cadmium accumulation in rice. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16500. doi: 10.1073/pnas.1005396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satoh-Nagasawa N, et al. Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant & cell physiology. 2012;53:213–224. doi: 10.1093/pcp/pcr166. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi R, et al. The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant, cell & environment. 2012;35:1948–1957. doi: 10.1111/j.1365-3040.2012.02527.x. [DOI] [PubMed] [Google Scholar]
- 28.Uraguchi S, et al. Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20959–20964. doi: 10.1073/pnas.1116531109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su N, et al. Disruption of a Rice Pentatricopeptide Repeat Protein Causes a Seedling-Specific Albino Phenotype and Its Utilization to Enhance Seed Purity in Hybrid Rice Production. Plant physiology. 2012;159:227. doi: 10.1104/pp.112.195081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H, et al. Development of Commercial Thermo-sensitive Genic Male Sterile Rice Accelerates Hybrid Rice Breeding Using the CRISPR/Cas9-mediated TMS5 Editing System. Scientific reports. 2016;6:37395. doi: 10.1038/srep37395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao FJ, Ma Y, Zhu YG, Tang Z, McGrath SP. Soil contamination in China: current status and mitigation strategies. Environmental science & technology. 2015;49:750–759. doi: 10.1021/es5047099. [DOI] [PubMed] [Google Scholar]
- 32.Sun L, et al. Genetic Diversity, Rather than Cultivar Type, Determines Relative Grain Cd Accumulation in Hybrid Rice. Frontiers in plant science. 2016;7:1407. doi: 10.3389/fpls.2016.01407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng Z, et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Research. 2013;23:1229. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hyun Y, et al. Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta. 2015;241:271. doi: 10.1007/s00425-014-2180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li T, Liu B, Spalding MH, Weeks DP, Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nature Biotechnology. 2012;30:390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- 36.Nekrasov V, Staskawicz B, Weigel D. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol. Nature Biotechnology. 2013;31:691–693. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- 37.Svitashev S, et al. Targeted Mutagenesis, Precise Gene Editing, and Site-Specific Gene Insertion in Maize Using Cas9 and Guide RNA. Plant physiology. 2015;169:931. doi: 10.1104/pp.15.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Townsend JA, et al. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shan Q, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nature Biotechnology. 2013;31:686. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 40.Xu R, et al. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. Journal of genetics and genomics. 2016;43:529–532. doi: 10.1016/j.jgg.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Li M, et al. Reassessment of the Four Yield-related Genes Gn1a, DEP1, GS3, and IPA1 in Rice Using a CRISPR/Cas9 System. Frontiers in plant science. 2016;7:377. doi: 10.3389/fpls.2016.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F, et al. Enhanced Rice Blast Resistance by CRISPR/Cas9-Targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PloS one. 2016;11:e0154027. doi: 10.1371/journal.pone.0154027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Y, et al. Engineering Herbicide-Resistant Rice Plants through CRISPR/Cas9-Mediated Homologous Recombination of Acetolactate Synthase. Molecular Plant. 2016;9:628–631. doi: 10.1016/j.molp.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Huang S, Weigel D, Beachy RN, Li J. A proposed regulatory framework for genome-edited crops. Nature genetics. 2016;48:109–111. doi: 10.1038/ng.3484. [DOI] [PubMed] [Google Scholar]
- 45.Ma X, et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Molecular Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 46.Endo M. Multigene Knockout Utilizing Off-Target Mutations of the CRISPR/Cas9 System in Rice. Plant & cell physiology. 2014;56:41–47. doi: 10.1093/pcp/pcu154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puchta H. Applying CRISPR/Cas for genome engineering in plants: the best is yet to come. Current opinion in plant biology. 2016;36:1–8. doi: 10.1016/j.pbi.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 48.Sasaki A, Yamaji N, Xia J, Ma JF. OsYSL6 Is Involved in the Detoxification of Excess Manganese in Rice. Plant physiology. 2011;157:1832–1840. doi: 10.1104/pp.111.186031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang M, et al. OsNRAMP5 contributes to manganese translocation and distribution in rice shoots. Journal of Experimental Botany. 2014;65:4849–4861. doi: 10.1093/jxb/eru259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marschner, P. Marschner’s Mineral Nutrition of Higher Plants, 3rd ed. (2013).
- 51.Xu XY, Mcgrath SP, Meharg AA, Zhao FJ. Growing rice aerobically markedly decreases arsenic accumulation. Environmental science & technology. 2008;42:5574–5579. doi: 10.1021/es800324u. [DOI] [PubMed] [Google Scholar]
- 52.Ishimaru Y, et al. Characterizing the role of rice NRAMP5 in Manganese, Iron and Cadmium Transport. Scientific reports. 2012;2:286. doi: 10.1038/srep00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi R, et al. From laboratory to field: OsNRAMP5-knockdown rice is a promising candidate for Cd phytoremediation in paddy fields. PloS one. 2014;9:e98816. doi: 10.1371/journal.pone.0098816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pyott DE, Sheehan E, Molnar A. Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Molecular plant pathology. 2016;17:1276–1288. doi: 10.1111/mpp.12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rowland LJ, Nguyen B. Use of polyethylene glycol for purification of DNA from leaf tissue of woody plants. Biotechniques. 1993;14:734. [PubMed] [Google Scholar]
- 56.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida, S., Forno, D. A., Cock, J. H. & Gomez, K. A. Laboratory manual for physiological studies of rice, 3rd ed. (1976).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.