Abstract
Sertoli and granulosa cells are two major types of somatic cells in male and female gonads, respectively. Previous studies have shown that Sertoli and granulosa cells are derived from common progenitor cells and that differentiation of these two cell types is regulated by sex differentiation genes. The signaling pathway including the adhesion and transcription factor Ctnnb1 (cadherin-associated protein, β1, also known as β-catenin) regulates differentiation of granulosa cells in the absence of the transcription factor Sry, and overactivation of β-catenin in the presence of Sry leads to granulosa prior to sex determination. Surprisingly, our previous study found that β-catenin overactivation in Sertoli cells after sex determination can also cause disruption of the testicular cord and aberrant testis development. However, the underlying molecular mechanism was unclear. In this study, we found that constitutive activation of Ctnnb1 in Sertoli cells led to ectopic expression of the granulosa cell-specific marker FOXL2 in testes. Co-staining experiments revealed that FOXL2-positive cells were derived from Sertoli cells, and Sertoli cells were transformed into granulosa-like cells after Ctnnb1 overactivation. Further studies demonstrated that CTNNB1 induced Foxl2 expression by directly binding to transcription factor Tcf/Lef-binding sites in the FOXL2 promoter region. We also found that direct overexpression of Foxl2 decreased the expression of Sertoli cell-specific genes in primary Sertoli cells. Taken together, these results demonstrate that repression of β-catenin (CTNNB1) signaling is required for lineage maintenance of Sertoli cells. Our study provides a new mechanism for Sertoli cell lineage maintenance during gonad development.
Keywords: beta-catenin (B-catenin), cell differentiation, somatic cell genetics, testis, transformation, Ctnnb1, Granulosa-like cells, Sertoli cells, foxl2
Introduction
In mammals, the testes in males or the ovaries in females come from the bipotential gonads, which form as a thickening of the epithelial layer on the ventromedial surface of the mesonephros (1, 2). Development of a testis or ovary is dependent on the differentiation of genital ridge somatic cells, which is regulated by sex-determining genes. The high mobility group (HMG) domain transcription factor Sry is essential for directing Sertoli cell differentiation in XY gonads (3–5). In XX gonads, which lack Sry expression, the somatic cell differentiates into granulosa cell, which is regulated by the RSPO1/WNT4-β-catenin (CTNNB1) signaling pathway (6–8). Inactivation of R-Spondin1 and Wnt4 before sex determination results in partial female-to-male sex reversal in mice (8–11). By contrast, overactivation of Ctnnb1 before sex determination using Sf1-Cre caused male-to-female sex reversal with an increased expression of FOXL2 and reduced expression of SOX9 in the male gonad (12).
Recent studies found that the differentiated Sertoli cells and granulosa cells have the potential to mutually transform after sex commitment. FOXL2 is a forkhead transcription factor specifically expressed in ovarian granulosa cells (13, 14), and deletion of Foxl2 results in aberrant ovarian follicle development and the dysgenesis of ovaries (13). Interestingly, it has been demonstrated that FOXL2 is also required for granulosa cell lineage maintenance. Inactivation of Foxl2 in the granulosa cells of adult ovaries results in an up-regulation of the testis-specific gene Sox9 and the transformation of granulosa cells into Sertoli-like cells along with the formation of a testicular cord-like structure (15). The Dmrt1 gene encodes a nuclear transcription factor, which is abundantly expressed in Sertoli cells. Deletion of Dmrt1 causes the reprogramming of Sertoli cells to granulosa-like cells postnatally, which in turn leads to dysgenesis of the testes (16).
Our previous study (17) found that constitutive activation of Ctnnb1 by deletion of exon 3 in Sertoli cells after sex determination using AMH-Cre transgenic mice caused testicular cord disruption and the loss of expression of Sertoli cell-specific genes. However, the underlying molecular mechanism remains unclear. Interestingly, in the present study, we found that the granulosa cell-specific marker FOXL2 was ectopically expressed in the remnant testicular cords of Ctnnb1 overactivated mice. Lineage tracing experiments revealed that Sertoli cells were transformed into granulosa-like cells after Ctnnb1 overactivation. Further studies demonstrated that CTNNB1 induced Foxl2 expression in the Sertoli cell line by directly interacting with T cell factor/lymphoid enhancer factor (Tcf/Lef)4-binding sites in the Foxl2 promoter region. These results indicate that repression of WNT/β-catenin signaling is essential for Sertoli cell lineage maintenance, and activation of Ctnnb1 causes an up-regulation of FOXL2, which in turn leads to the transformation of Sertoli cells into granulosa-like cells.
Results
Ectopic expression of FOXL2 protein in the testes of Ctnnb1+/flox(ex3) AMH-Cre mice
Our previous studies found that overactivation of Ctnnb1 by deletion of exon 3 in Sertoli cells using AMH-Cre transgenic mice caused testicular cord disruption and loss of Sertoli cell-specific genes' expression (17). To further explore the reason for abnormal testis development in Ctnnb1+/flox(ex3)AMH-Cre mice, the expression of Sertoli cell-specific and granulosa cell-specific genes was examined by immunostaining and real-time PCR assays. In control testes, CTNNB1 protein was localized at the plasma membrane of Sertoli cells and germ cells from E13.5 to P1 (supplemental Fig. S1, A–C). In the Ctnnb1+/flox(ex3) AMH-Cre testes, the accumulation of CTNNB1 protein in the nucleus of Sertoli cells was first observed at E14.5 (supplemental Fig. S1E, white arrowheads), and the number of CTNNB1 accumulated Sertoli cells was significantly increased at P1 (supplemental Fig. S1F, white arrowheads). These results indicated that Ctnnb1 was overactivated in Sertoli cells, which was consistent with the findings obtained in our previous study (17). As shown in supplemental Fig. S2, A–C, WT1-positive Sertoli cells were localized at the periphery region of testicular cords (red asterisks) in control testes from E13.5 to E17.5. Although the abnormality of testicular cords was noted in Ctnnb1+/flox(ex3) AMH-Cre testes from E13.5, the WT1-positive cells were still observed at the periphery region of testicular cords at E15.5 and E17.5 (supplemental Fig. S2, D–F). AMH protein was detected in Sertoli cells of both control (supplemental Fig. S2G, black arrows) and Ctnnb1+/flox(ex3) AMH-Cre testes (supplemental Fig. 2J, black arrowheads) at E13.5, and it was continuously expressed in control Sertoli cells at E15.5 and E17.5 (supplemental Fig. S2, H and I, black arrows). By contrast, AMH protein was completely absent in the testicular cords ofCtnnb1+/flox(ex3) AMH-Cre testes at E15.5 (supplemental Fig. S2K) and E17.5 (supplemental Fig. S2L). DMRT1 protein was detected in germ cells and Sertoli cells of both control (supplemental Fig. S2M, black arrows) and Ctnnb1+/flox(ex3) AMH-Cre testes (supplemental Fig. S2P, black arrowheads) at E13.5, whereas the expression was reduced significantly in Sertoli cells of Ctnnb1+/flox(ex3) AMH-Cre testes at E15.5 and E17.5 (supplemental Fig. S2, Q and R, black arrowheads) but still remained in germ cells.
FOXL2 was expressed in ovarian granulosa cells from E13.5 to P1 (Fig. 1, A–D, black arrows). By contrast, no FOXL2 protein was detected in control testes from E13.5 to P1 (Fig. 1, I–L). Unexpectedly, a small number of FOXL2-positive cells were detected in the remnant testicular cords of Ctnnb1+/flox(ex3) AMH-Cre testes at E14.5 (Fig. 1F, black arrowheads), and the number of FOXL2-positive cells was gradually increased at E17.5 and P1 (Fig. 1, G and H, black arrowheads). The results of the FOXL2 and SOX9 double staining experiment showed that all of the Sertoli cells in the testicular cords of the Ctnnb1+/flox(ex3) AMH-Cre testes at E13.5 were SOX9-positive (supplemental Fig. S3B, white arrowheads), and no FOXL2-positive cells were observed. However, only a few SOX9-positive cells were observed at E15.5 and most of the somatic cells at the periphery region of the testicular cord were FOXL2-positive in Ctnnb1+/flox(ex3) AMH-Cre testes (supplemental Fig. S3E).
Figure 1.
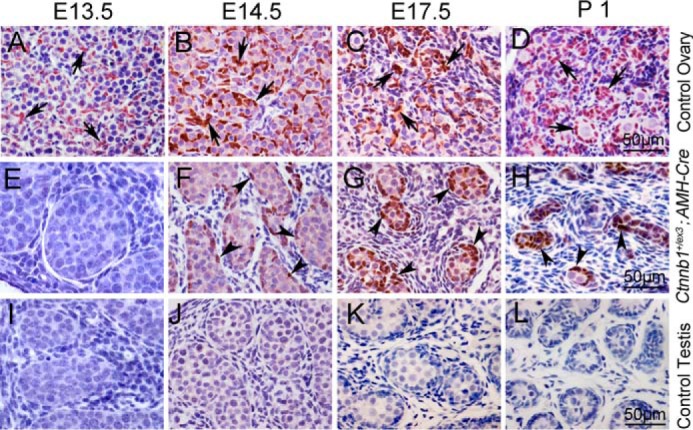
FOXL2 was expressed in the testes of Ctnnb1+/flox(ex3)AMH-Cre mice. IHC was conducted using anti-FOXL2 antibody. FOXL2 protein (brown) was specifically expressed in granulosa cells (black arrows) of control ovaries at E13.5 (A), E14.5 (B), E17.5 (C), and P1 (D). No FOXL2 protein was detected in control testes from E13.5 to P1 (I–L). FOXL2 was not expressed in the testicular cords of Ctnnb1+/flox(ex3) AMH-Cre mice at E13.5 (E), whereas FOXL2-positive cells were found in the remnant seminiferous tubules at E14.5 (F, black arrowheads), and the number of FOXL2-positive cells (black arrowheads) was significantly increased at E17.5 (G) and P1 (H).
FOXL2-positive cells in the testes of Ctnnb1+/flox(ex3)AMH-Cre mice were derived from Sertoli cells
Because FOXL2 is a granulosa cell-specific marker, we speculated that overactivation of Ctnnb1 probably caused Sertoli to granulosa-like cells transformation. To test this hypothesis, a double staining experiment was performed. WT1 was specifically expressed in the Sertoli cells of control testes as shown in Fig. 2, A and C. WT1 was also expressed in the somatic cells of Ctnnb1+/flox(ex3) AMH-Cre testes (Fig. 2D, white arrowheads), and most of these cells were FOXL2-positive (Fig. 2, E and F, white arrowheads), indicating that these FOXL2-positive cells in Ctnnb1+/flox(ex3) AMH-Cre testes were most likely derived from WT1-positive Sertoli cells. To further confirm this result, the GFP-expressed (ROSAmT/mG) transgenic mice were mated with Ctnnb1+/flox(ex3) AMH-Cre mice to generate Ctnnb1+/flox(ex3) ROSAmT/mG AMH-Cre mice (18). In control mice (ROSAmT/mG AMH-Cre), GFP signals were detected in Sertoli cells within the seminiferous tubules, and no GFP signal was detected in the testicular interstitium (Fig. 2, G and I, white arrows), indicating that AMH-Cre is specifically expressed in Sertoli cells. In the Ctnnb1+/flox(ex3) ROSAmT/mG AMH-Cre mice, a FOXL2 signal was co-localized with GFP as shown in Fig. 2L, indicating that these cells were derived from Sertoli cells.
Figure 2.
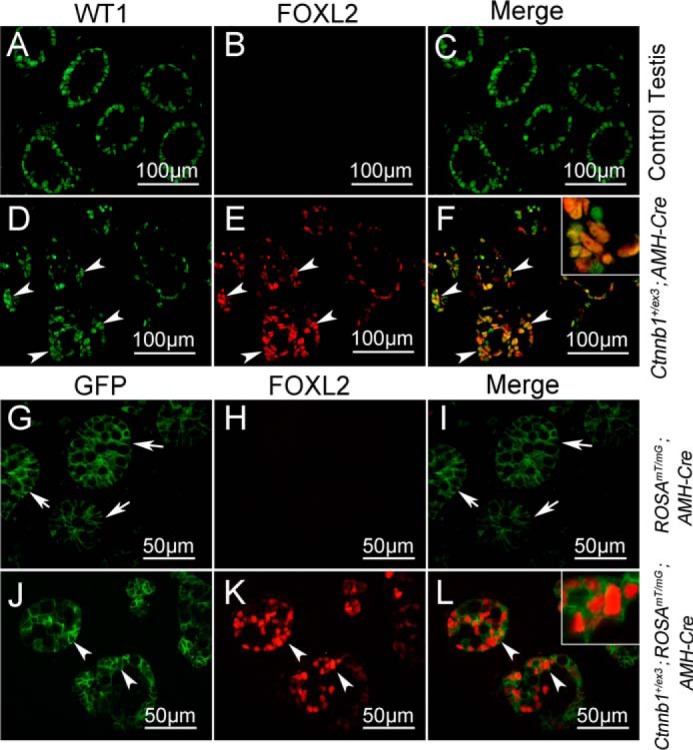
FOXL2 was expressed in Ctnnb1-overactivated Sertoli cells. WT1 and FOXL2 double staining experiment was performed on E17.5 control and Ctnnb1+/flox(ex3) AMH-Cre testes. WT1 protein (green) was expressed in Sertoli cells of control (A and C) and Ctnnb1+/flox(ex3) AMH-Cre (D and F) testes. FOXL2 protein was detected in remnant seminiferous tubules of Ctnnb1+/flox(ex3) AMH-Cre mice (E, white arrowheads), but not in control testes (B). FOXL2 and WT1 were co-localized in Ctnnb1+/flox(ex3) AMH-Cre testes (F, inset). GFP signal was detected in the Sertoli cells of both control (ROSAmT/mG AMH-Cre, G and I, white arrows) and ROSAmT/mG Ctnnb1+/flox(ex3) AMH-Cre mice (J and L, white arrows). FOXL2 protein (14) was only expressed in ROSAmT/mG Ctnnb1+/flox(ex3) AMH-Cre mice (K, white arrowheads), but not in ROSAmT/mG AMH-Cre testes (H), and co-localized with GFP protein (L, inset).
Other than the structure disruption of the testicular cords, testes tumors were also observed in Ctnnb1+/flox(ex3) AMH-Cre mice at later developmental stages. To identify the feature of tumor cells, testes sections from 8-month-old control and Ctnnb1+/flox(ex3) AMH-Cre mice were stained with anti-WT1, anti-DMRT1, anti-SOX9, and anti-FOXL2 antibodies. As shown in supplemental Fig. S4, WT1, DMRT1, and SOX9 were specifically expressed in Sertoli cells, and no FOXL2 signal was detected in control testes (supplemental Fig. S4, A–D). However, tumor cells in Ctnnb1+/flox(ex3) AMH-Cre mice were WT1- and FOXL2-positive (supplemental Fig. S4, E and H), but DMRT1- and SOX9-negative (supplemental Fig. S4, F and G). Quantitative real-time PCR results also showed that the expression of granulosa cell-specific genes (e.g. Foxl2, Bmp2, Wnt4, Rspo1, Esr1, and Esr2) was significantly increased, whereas the expression of Sertoli cell-specific genes (e.g. Sox9 and Dmrt1) was significantly decreased in the tumor cells of Ctnnb1+/flox(ex3) AMH-Cre mice compared with control Sertoli cells (supplemental Fig. S4I). Taken together, these results indicated that the tumors observed in Ctnnb1+/flox(ex3) AMH-Cre mice were granulosa cell-like tumors.
Sertoli cells were transformed into granulosa-like cells in Ctnnb1+/flox(ex3) AMH-Cre mice
The above experiments showed that Sertoli cells in Ctnnb1-overactivated testes lost Sertoli cell-specific gene expression and began to express the granulosa cell gene FOXL2. To examine the differential expression of other Sertoli and granulosa cell-specific genes, quantitative real-time PCR was performed with transcripts from control ovaries, control testes, and Ctnnb1+/flox(ex3) AMH-Cre testes at E16.5. As shown in Fig. 3, the expression levels of Sertoli cell-specific genes (e.g. Sox9, Cyp26b1, Amh, Gdnf, Dhh, Ptgds, Erbb4, and Shbg) were dramatically reduced in Ctnnb1 over-activated testes (Fig. 3A). By contrast, the mRNA levels of granulosa cell-specific genes (e.g. Wnt4, Rspo1, Foxl2, and Bmp2) were increased in Ctnnb1+/flox(ex3) AMH-Cre testes compared with control testes in varying degrees (Fig. 3B). The expression of the somatic cell gene, Gata4, was not significantly changed in Ctnnb1-overactivated testes.
Figure 3.
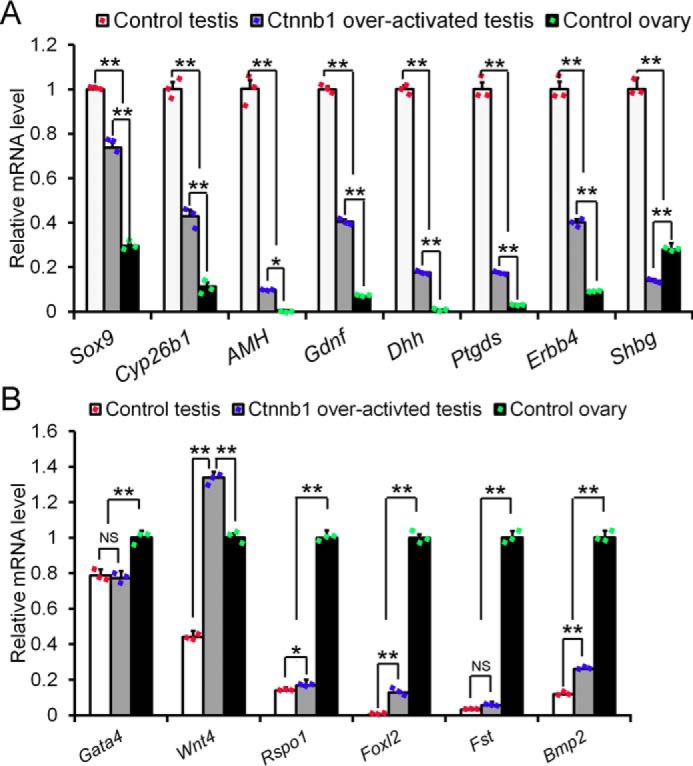
The differential expression of Sertoli and granulosa cell-specific genes in Ctnnb1-overactivated testes was performed with quantitative real-time PCR at E16.5. A, the expression of granulosa cell-specific genes was significantly increased in Ctnnb1-overactivated testes. B, the expression of Sertoli cell-specific genes was significantly decreased in Ctnnb1-overactivated testes. Gonads with the same genotype at each time point were pooled for RNA preparation. The experiments were performed with three independent pools. Data are presented as the mean ± S.E. (t test). *, p < 0.05; **, p < 0.01; NS, not significant.
To view a global gene expression profile in Sertoli cells after Ctnnb1 overactivation, transcriptome sequencing analysis was performed. In control ovary, FOXL2 was abundantly expressed in granulose cells at E12.5 (14). By contrast, FOXL2 was first detected in Ctnnb1+/flox(ex3) ROSAmT/mG AMH-Cre mice at E14.5. To parallel the developmental stages, control granulosa cells from E14.5, and Sertoli cells from E16.5 control and Ctnnb1+/flox(ex3) ROSAmT/mG AMH-Cre mice were isolated and total mRNA was extracted. The results of the RNA-Seq assay showed that 538 genes were up-regulated and 703 genes were down-regulated with at least 2-fold changes in both control granulosa cells and Ctnnb1-overactivated Sertoli cells compared with control Sertoli cells (Fig. 4A). Based on the heat map assay result, the expression of genes in the Ctnnb1-overactivated Sertoli cells was comparable with that in control granulosa cells, but differed from that in control Sertoli cells. Moreover, the expression of a subset of granulosa cell-specific and Sertoli cell-specific genes selected from the 1241 differentially expressed genes also confirmed the result that Sertoli cells in Ctnnb1-overactivated testes were transformed into granulosa-like cells (Fig. 4B).
Figure 4.
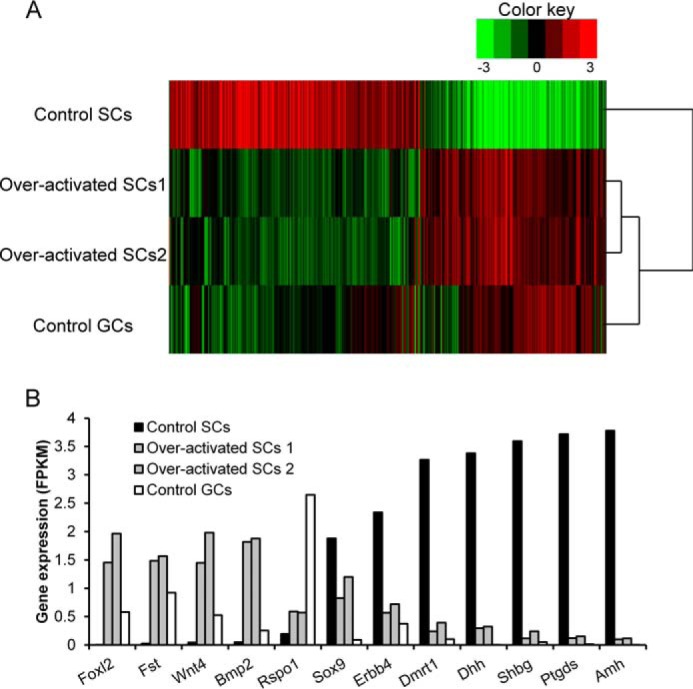
Genome wide expression profiling of control granulosa cells at E14.5, control Sertoli cells, and Ctnnb1-overactivated Sertoli cells at E16.5 was analyzed by transcriptome sequencing analysis. A, heat map of the 1241 genes that were significantly altered (538 up-regulated and 703 down-regulated genes with at least 2-fold changes) from both control granulosa cells (Control GCs) and Ctnnb1-overactivated Sertoli cells (overactivated SCs) compared with control Sertoli cells (Control SCs). The red and green colors represent the relative expression level. B, a subset of granulosa cell-specific and Sertoli cell-specific genes selected from 1241 differentially expressed genes.
The expression of Foxl2 was regulated by CTNNB1 via Tcf/Lef-binding sites
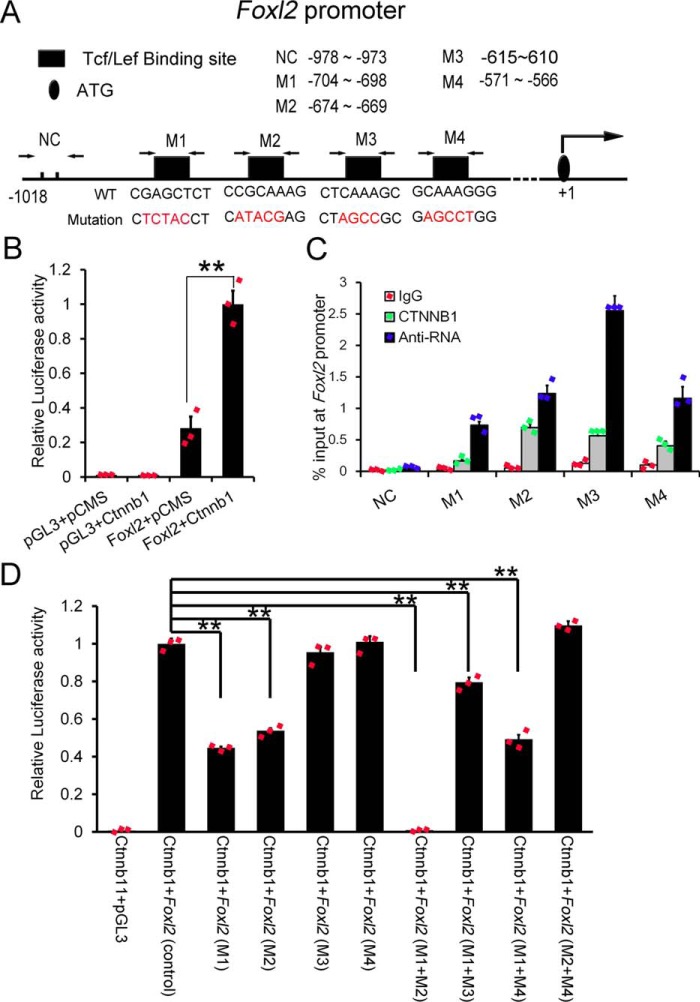
FOXL2 played essential roles in granulosa cell development and lineage maintenance, and loss of Foxl2 caused granulosa to Sertoli-like cell transformation in postnatal ovaries (15). In the present study, we found that overactivation of Ctnnb1 caused an up-regulation of FOXL2 and a transformation of Sertoli cells to granulosa-like cells. To examine whether the expression of Foxl2 is directly regulated by CTNNB1, a luciferase assay was performed using a luciferase reporter plasmid containing the nucleotides from −1018 to +7 in the Foxl2 promoter region (Fig. 5A), which has been reported to be sufficient to direct the expression of Foxl2 (19).
Figure 5.
The Foxl2 promoter was activated by CTNNB1 via an interaction with Tcf/Lef-binding sites in TM4 cells. A, the schematic image showed the predicted Tcf/Lef-binding sites (M1, M2, M3, and M4) within the Foxl2 promoter region and the primers used for the ChIP-qPCR assay. B, the luciferase activity of the Foxl2 promoter reporter in TM4 cells was significantly induced by overexpression of Ctnnb1. C, ChIP-qPCR analysis of four binding sites in Foxl2 promoter in TM4 cell lines by transient transfection of Ctnnb1 vector. D, the M1- and M2-binding sites were essential for the CTNNB1-induced Foxl2 promoter activity. Three experiments was conducted and data are as the mean ± S.D. (one-way analysis of variance). *, p < 0.05; **, p < 0.01.
After transfection with a Ctnnb1-expressing vector in TM4 cells, the luciferase activity was increased ∼3-fold compared with the control vector (Fig. 5B), indicating that the Foxl2 promoter could be activated by CTNNB1. Once the stabilized β-catenin is accumulated, it will be transferred into the nucleus and interact with DNA-bound Tcf/Lef to induce the expression of downstream factors. To test whether the expression of Foxl2 is directly regulated by CTNNB1, the potential Tcf/Lef-binding sites (-(A/T)(A/T)CAAAG- and CGAGCTCT) were searched within the promoter region of Foxl2. As shown in Fig. 5A, four potential Tcf/Lef-binding sites (M1, M2, M3, and M4) were identified in the Foxl2 promoter region (Fig. 5A) based on previous studies (20–22). ChIP-qPCR results showed that all four potential binding sites could be pulled down by CTNNB1 (Fig. 5C). To further examine the function of these four potential binding sites within the Foxl2 promoter, reporter vectors carrying the mutated binding sites were generated. We found that activity of the Foxl2 promoter was significantly reduced when M1 or M2 was mutated, whereas mutation of M3 or M4 did not affect CTNNB1-induced Foxl2 promoter activity. We also found that the activity of the Foxl2 promoter was completely abolished when M1 and M2 were simultaneously mutated (Fig. 5D). Collectively, these results indicated that CTNNB1 induced Foxl2 expression by directly binding to the promoter region via Tcf/Lef-binding sites.
The expression of Sertoli cell-specific genes was repressed by FOXL2
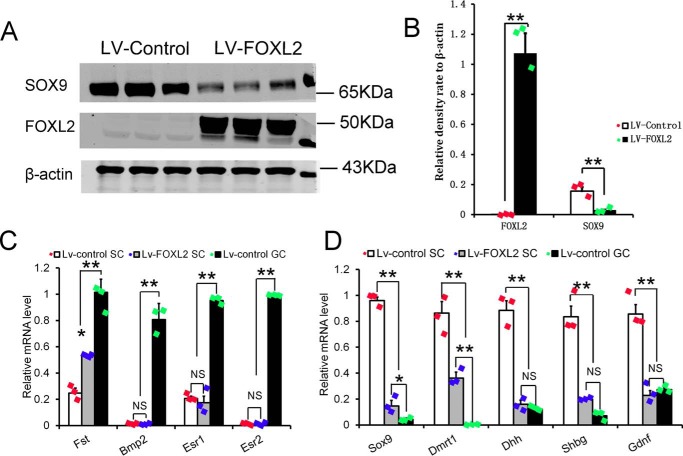
In this study, we found that the expression of Sertoli cell-specific genes was dramatically reduced after overactivation of Ctnnb1, and that CTNNB1 induced Foxl2 expression directly. To test whether the expression of Sertoli cell-specific genes could be repressed by FOXL2, Sertoli cells isolated from 14-day-old mice testes were cultured in vitro and infected with Foxl2-expressing lentivirus. The expression of Sertoli and granulosa cell-specific genes was analyzed by Western blotting and real-time PCR analyses. As shown in Fig. 6, the protein level of SOX9 was dramatically reduced with Foxl2 overexpression (Fig. 6A), and quantitative results showed that the SOX9 protein was reduced ∼50% in Foxl2 overexpressing Sertoli cells (Fig. 6B). Quantitative real-time PCR results showed that the mRNA level of Fst was significantly increased after Foxl2-expressing lentivirus infection, whereas the expression of other granulosa cell-specific genes (e.g. Bmp2, Esr1, and Esr2) was not changed (Fig. 6C). The expression of other Sertoli cell-specific genes (e.g. Dmrt1, Dhh, Shbg, and Gdnf) was also significantly decreased after overexpression of Foxl2 (Fig. 6D). These results indicated that the expression of Sertoli cell-specific genes was inhibited by Foxl2, and Foxl2 prevented the development of Sertoli cells by antagonizing the expression of Sertoli cell-specific genes at the postnatal period.
Figure 6.
The expression of SOX9 and other Sertoli cell-specific genes was significantly repressed by FOXL2. Sertoli cells from 14-day-old mice were infected with Foxl2 overexpression lentivirus and after 48 h infection cells were collected for gene expression assay. A, the protein levels of SOX9 and FOXL2 were analyzed by Western blotting. B, density analytical results showed that the expression of SOX9 protein was dramatically reduced in Sertoli cells after Foxl2 overexpression. C, the differential expression of granulosa cell-specific genes in Foxl2 overexpressed Sertoli cells. D, the expression of Sertoli cell specific genes was significantly decreased after overexpression of Foxl2 in Sertoli cells. Data are presented as the mean ± S.D. (one-way analysis of variance) measured in triplicate. *, p < 0.05; **, p < 0.01; NS, not significant.
Discussion
Previous studies have demonstrated that Foxl2 and Dmrt1 play important roles in granulosa cell and Sertoli cell lineage maintenance. Inactivation of Foxl2 in granulosa cells of adult ovaries results in granulosa to Sertoli-like cell transdifferentiation (15). By contrast, deletion of Dmrt1 results in Sertoli to granulosa-like cell transformation (16). In the present study, we found that overactivation of Ctnnb1 in Sertoli cells also resulted in Sertoli to granulosa-like cell transdifferentiation, implying that repression of WNT/β-catenin signaling is essential for Sertoli cell lineage maintenance.
WNT/β-catenin signaling is known to play important roles in multiple developmental processes and tumorigenesis. It has been reported that inactivation of Ctnnb1 in somatic cells of undifferentiated genital ridge using Sf1-Cre transgenic mice results in the formation of a testis-specific coelomic vessel, the appearance of adrenal-like steroidogenic cells, and the loss of germ cells in female gonads. However, Sertoli cell differentiation and testis development are not affected (23). A similar phenotype is also observed in Rspo1 and Wnt4 knock-out mouse models (8–11), indicating that RSPO1-WNT/β-catenin signaling is indispensable for somatic cell differentiation in XX gonads development. By contrast, overactivation of Ctnnb1 in somatic cells of the undifferentiated genital ridge mediated by Sf1-Cre results in aberrant somatic cell differentiation and partial male-to-female sex reversal in XY gonads, whereas the development of XX gonads is not affected (12). All these results suggest that WNT/β-catenin signaling plays critical roles in somatic cell differentiation in female gonads during sex determination.
Interestingly, our previous study found that CTNNB1 was also expressed in Sertoli cells after sex determination, mainly on the cell membrane. Conditional knock-out of Ctnnb1 in Sertoli cells by AMH-Cre did not affect the development of the testis, whereas constitutive stabilization of Ctnnb1 in Sertoli cells resulted in testicular cord disruption and germ cell loss during embryonic stage and tumor development at later developmental stage (17). Other studies have also reported that overactivation of WNT/β-catenin signaling in Sertoli cells using Amhr2-Cre transgenic mice caused germ cell apoptosis and male infertility. However, in this mouse model, the defect of testis development was not detected until ∼5 weeks of age, and no testis tumor was observed (24, 25). The discrepancy between our and other mouse models is most likely due to the relatively weak efficiency of AMHR2-Cre or a wider spectrum of AMHR2-Cre activity. Testis tumors were observed in mouse models by activation of WNT/β-catenin signaling and simultaneous inactivation of phosphatidylinositol 3-kinase (PI3K)/AKT or activation of RAS pathway using Amhr2-Cre. The granulosa cell-specific marker FOXL2 was detected in the tumor cells of these mouse models (26, 27), suggesting that the interaction of WNT/β-catenin signaling with the PI3K/AKT and RAS pathway is probably involved in Sertoli cell development.
In this study, we demonstrated that constitutive stabilization of Ctnnb1 resulted in Sertoli to granulosa-like cell transformation, which in turn caused the loss of Sertoli cell-specific gene expression and the disruption of testicular cords. Our study suggested that repression of β-catenin signaling was required for Sertoli cell lineage maintenance. Previous studies found that deletion of Ctnnb1 caused abnormal granulosa cell differentiation during sex determination (12). We speculated that WNT/β-catenin signaling is probably also required for granulosa cell lineage maintenance after sex determination, which requires further investigation.
FOXL2 is a forkhead transcription factor that is specifically expressed in the ovarian granulosa cells from approximately E12.5 in mice (13, 14). Previous studies have demonstrated that Foxl2 plays important roles in ovary development and deletion of this gene results in aberrant ovarian follicle development and the dysgenesis of ovaries (9, 13). However, as a key factor for granulosa cell development, the regulation of Foxl2 expression during gonad development has not been studied previously. In this study, we demonstrated that CTNNB1 induced Foxl2 expression by directly binding to Tcf/Lef-binding sites in the promoter region. The up-regulation of other granulosa cell-specific genes in Ctnnb1-overactivated Sertoli cells was most likely a consequence of an up-regulation of Foxl2. However, we could not exclude the possibility that some of these genes were also directly induced by CTNNB1. These results also suggest that WNT/β-catenin signaling is involved in female sex determination most likely by inducing Foxl2 expression.
The expression of Sertoli cell-specific genes was dramatically reduced after overactivation of Ctnnb1. Whether the expression of Sertoli cell-specific genes were directly repressed by CTNNB1 or indirectly inhibited by FOXL2 still needs to be addressed. Our results showed that the expression of Sertoli cell genes in cultured primary Sertoli cells was inhibited by Foxl2 overexpression. Previous in vitro studies have demonstrated that FOXL2 interacted with Sox9-regulatory sequences that are required for its testis-specific expression (15), thereby inhibiting Sox9 expression. Collectively, based on these results and those obtained in our in vitro study, we speculated that the expression of Sox9 and other Sertoli cell-specific genes is most likely indirectly inhibited by β-catenin signaling via Foxl2.
In summary, our results demonstrated that repression of WNT/β-catenin signaling was essential for Sertoli cell lineage maintenance, and overactivation of Ctnnb1 resulted in a Sertoli cell to granulosa-like cell transformation. We also found that Ctnnb1 induced Foxl2 expression by binding to the Foxl2 promoter region directly via Tcf/Lef-binding sites (-(A/T)(A/T)CAAAG- and CGAGCTCT). These results also suggest that WNT/β-catenin signaling is involved in female sex determination most likely by inducing Foxl2 expression. It may be required for maintenance of the granulosa cells, after their specification. Collectively, our study provides a novel mechanism for Sertoli cell lineage maintenance, which will shed light on better understanding the regulation of somatic cells differentiation during testis development.
Experimental procedures
Mice
All animal work was performed according to the regulations of the Institutional Animal Care and Use Committee of the Institute of Zoology, Chinese Academy of Sciences. All mice were maintained on a C57BL/6;129/SvEv mixed background. Ctnnb1+/flox(ex3) ROSAmT/mG AMH-Cre mice were obtained by crossing Ctnnb1flox(ex3)/flox(ex3) (28); ROSAmT/mG (18) female mice with AMH-Cre transgenic males (29). DNA isolated from adult tail tips and fetal tissues was used for genotyping. Genotyping was performed by PCR as previously described (28, 30). There is no randomization used to determine how animals were allocated to experimental groups and processed.
Tissue collection and histological analysis
Testes were dissected from mutant and control mice at different developmental stages immediately after euthanasia and were fixed in 4% paraformaldehyde for up to 24 h, stored in 70% ethanol, and embedded in paraffin. Five-micrometer thick sections were cut and mounted onto glass slides. After deparaffinization, the sections were processed for immunohistochemistry (IHC) and immunofluorescent analysis. There is no blinding done to the group allocation during the experiment.
Immunohistochemistry analysis
Immunohistochemistry analysis of tissues from at least three mice for each genotype was performed using a Vectastain ABC (avidin-biotin-peroxidase) kit (Vector Laboratories, Burlingame, CA) as recommended. The antibodies were diluted as follows: WT1 (1:800, EPITOMICS, 2797-1), AMH (1:200, Santa, sc-6886), DMRT1 (1:50, Santa Cruz, sc-98341), SOX9 (1:400, Millipore, AB5535), and FOXL2 (1:100, Abcam, ab5096). The IHC procedure was performed as previously described (30). Stained sections were examined using Nikon Microscopy, and images were captured with a Nikon DS-Ri1 CCD camera.
Immunofluorescence analysis
After rehydration and antigen retrieval, 5-μm sections were incubated with 5% donkey serum in 0.3% Triton X-100 for 1 h. Next, the sections were immunolabeled with primary antibodies for 1.5 h. Antibodies were diluted as follows: CTNNB1 (1:400, Abcam, ab6302), FOXL2 (1:100, Abcam, ab5096), SOX9 (1:200, Millipore, AB5535), WT1 (1:200, EPITOMICS, 2797-1), and GFP (1:100, Santa Cruz Biochemistry, sc-9996). After three washes in PBS, fluorescence-labeled secondary antibodies (1:200, Jackson ImmunoResearch) were used for detection. DAPI was used to label the nuclei. Finally, the sections were analyzed using confocal laser scanning microscopy (Carl Zeiss Inc., Thornwood, NY).
Somatic cell isolation
Control Sertoli cells and Ctnnb1-overactivated Sertoli cells from ROSAmT/mG AMH-Cre and Ctnnb1+/flox(ex3) ROSAmT/mG AMH-Cre mice at E16.5 were sorted using flow cytometry. At least three pregnant mice were used for each group. Briefly, testes were dissected and washed 3 times with PBS. Testes with GFP fluorescence were pooled together and digested with 0.25% trypsin-EDTA. After neutralization with FBS, a single cell solution was made by pipette. The cell suspension was washed with 3 times with PBS and transferred into a 1.5-ml conical tube through a 70-μm cell strainer. The supernatant was removed after centrifugation at 300 × g for 5 min at room temperature. The cells were resuspended with FACS buffer and purified by BD FACSAria. The cells were stored at −80 °C for RNA extraction.
Control granulosa cells at E14.5 and 28-day-old mice were isolated using the adherent culture method as previously described (31). After mechanical dissection, ovaries from E14.5 or follicles from 28-day-old mice were digested with 1 mg/ml of collagenase I (Sigma, C0130), 0.25% trypsin-EDTA, and 0.5 mg/ml of DNase I (Sigma, D5025) for 10 min at 37 °C. After two washes, the cells were seeded in DMEM/F-12 culture medium supplemented with 10% FBS, 1% penicillin/streptomycin and incubated at 37 °C and 5% CO2 for adhesion. For E14.5, the cell suspension was cultured for 4–6 h. For 28-day-old, the cell suspension was cultured overnight. After most of the somatic cells attached, the supernatant was removed and the adherent cells were digested with 0.25% trypsin-EDTA. The cells were collected and stored at −80 °C for RNA extraction.
Sertoli cells from 14-day-old and adult mice were isolated as previously described (32). Briefly, after removal of the testes' tunica albuginea, seminiferous tubules were enzymatically digested in DMEM/F-12 with 1 mg/ml of collagenase IV (Sigma, C5138) and 1 mg/ml of hyaluronidase type III (Sigma, H3506) and incubated in a shaking water bath at 100 oscillations per minute for 15 min at 37 °C. Next, the samples were washed twice with DMEM/F-12 and further digested with 2 mg/ml of collagenase I (Sigma, C0130), 0.5 mg/ml of DNase I (Sigma, D5025), and 1 mg/ml of hyaluronidase type III for 20–30 min at 37 °C. Tubules were precipitated, washed twice with DMEM/F-12, and incubated with 2 mg/ml of collagenase I, 0.5 mg/ml of DNase I, 2 mg/ml of hyaluronidase, and 1 mg/ml of trypsin (Sigma, T6763) for 40–60 min at 37 °C. The dispersed cells, containing primarily Sertoli cells and type A spermatogonia, were then washed twice with DMEM/F-12 and placed into culture dishes in DMEM/F-12 with 10% FBS, 1% penicillin/streptomycin and incubated at 37 °C and 5% CO2. After culturing overnight, cells were exposed to 20 mm Tris (pH 7.4) to remove spermatogonia by hypotonic treatment. Cells were cultured in an incubator at 37 °C and 5% CO2. The testicular tumor cells from 4-month-old Ctnnb1+/flox(ex3) AMH-Cre mice were dissected using forceps under the microscopy.
Nucleic acid isolation and quantitative real-time PCR
Total RNA was extracted using a Qiagen RNeasy kit according to the manufacturer's instructions. Two micrograms of total RNA was used to synthesize first-strand cDNA. To quantify gene expression, a real-time SYBR Green assay was performed. Gapdh were used as an endogenous control for gene expression analysis. The relative mRNA level of candidate genes was calculated using the formula 2−ΔΔCT as described in the SYBR Green user manual. The primers used for real-time PCR were listed in supplemental Table S1.
RNA sequencing analysis
Total RNA was extracted from isolated cells, including control granulosa cells, control Sertoli cells, and Ctnnb1-overactivated Sertoli cells from E16.5 embryos using the RNeasy Mini Kit (Qiagen, 74104) according to the manufacturer's instructions. The main reagents and instruments used for RNA library construction were from the New England Biolabs Next Ultra RNA Library Prep Kit (New England Biolabs, E7530L). RNA-Seq was performed as previously described (33, 34). The sequenced raw data were filtered to remove adaptor reads, low quality reads, and reads with a single copy number. Clean reads were classified according to their copy number and saturation of the library was analyzed. All aligned RNA-Seq reads were mapped to Mus musculus the USCS Genome Browser mm9 references using Tophat software, and the Fragments Per Kilobase of exon model per Million mapped reads (FPKM) of each gene was calculated using Cufflinks. Differentially expressed genes between control and Ctnnb1-overactivated Sertoli cells were identified as previously described using FPKM > 0.001 and a threshold absolute 2-fold change for the normalized gene expression level.
Plasmid construction
A Foxl2 promoter fragment encompassing nucleotides −1018 to +7 (19) was cloned into the pGL3basic luciferase reporter vector. Four potential Tcf/Lef-binding sites were identified in the promoter region of the Foxl2 gene based on previous studies (22, 35), including nucleotides −704 to −698, −674 to −669, −615 to −610, and −571 to −566. The Foxl2 promoter luciferase reporter vectors carrying the mutant Tcf/Lef-binding sites were generated by site-directed mutagenesis using the Q5® High Fidelity DNA Polymerase (New England Biolabs, M0491S).
Lentivirus packaging
Lentivirus containing Foxl2 cDNA was generated using the Lentivector Expression Systems. Briefly, 293T-packaging cells were cultured and transfected with VsVg, Δ8.9, and Foxl2 overexpression vectors by VigoFect transfection reagent (Vigorous Biotechnology, T001). After overnight incubation at 37 °C and 5% CO2, 10 ml of fresh DMEM was supplemented with 10% FBS and 1% penicillin/streptomycin. The virus was harvested at 48 and 72 h after transfection with 50-ml capped conical centrifuge tubes. The samples were centrifuged at 3,000 × g for 15 min at 4 °C to remove cell debris. Viral particles were concentrated by centrifugation at 12,000 × g at 4 °C for 2 h and re-dissolved with fresh DMEM. The concentrated virus particles were stored at −80 °C for subsequent use. Sertoli cells from 14-old-day mice were infected with Foxl2 overexpression lentivirus and collected at 48 h after infection and prepared for the gene expression assay.
Luciferase assay
The robust stable TM4 cell line tested for mycoplasma contamination came from the Group of Bioinformatics, the State Key Laboratory of Stem cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences. Luciferase activity was measured using a dual luciferase reporter assay system according to the manufacturer's instructions (Promega, E1910). Briefly, TM4 cells were transfected with Foxl2 control or mutant promoter luciferase reporter plasmids (200 ng) and Ctnnb1 overexpression plasmids (250 ng) using Lipofectamine 3000 transfection reagent (Invitrogen, L3000001) in a 24-well culture plate. Cells were harvested and lysed after a 48-h transfection. After the supernatant of the cell lysate was transferred into a 96-well plate (20 μl per assay), the luciferase assays were performed on a GloMax® 96 Microplate Luminometer (Promega, E6511) using standard reagents.
Western blot analyses
Cells were lysed with radioimmune precipitation assay lysis buffer (RIPA) containing a complete Mini protease-inhibitor mixture (Roche Applied Science, 04693116001). The homogenates were centrifuged at 13,000 × g for 15 min at 4 °C, and the protein concentrations were determined using the Bio-Rad protein assay (Bio-Rad Laboratories). The protein lysates (approximately 25 μg) were electrophoresed under reducing conditions in SDS-PAGE gels and transferred onto nitrocellulose membranes. After incubating in primary antibody, immunoblotting was performed using a fluorescent dye-labeled secondary antibody (Invitrogen), and the blots were scanned using an Odyssey infrared imager.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using the EZ-ChIPTM Chromatin Immunoprecipitation Kit (Millipore, 17-371) according to the manufacturer's instructions. Briefly, TM4 cells were cultured in vitro and transfected with the Ctnnb1 overexpression vector for 48 h using Lipofectamine 3000. After incubation at 37 °C with 5% CO2 for 48 h, cells were collected and cross-linked with 1% formaldehyde. Glycine was added to quench unreacted formaldehyde. Sonication was performed to shear the chromatin to an average DNA fragment size of 200∼1000 bp. Immunoprecipitations were performed using a positive control antibody (anti-RNA Polymerase II), negative control antibody (CST, normal rabbit IgG), and CTNNB1 antibody (1:250, Abcam, ab 6302) followed by Protein G-conjugated agarose beads as the secondary reagent. Protein-DNA cross-links were reversed during incubation at 65 °C in 0.2 m NaCl and DNA was purified to remove the chromatin proteins and analyzed by quantitative PCR. Primers for four potential Tcf/Lef-binding site regions were designed and are listed in supplemental Table S1. The % input of DNA from each immunoprecipitation was calculated using the following formula: % Input = 2 (−ΔCt [normalized ChIP]), ΔCt [normalized ChIP] = (Ct [ChIP] − (Ct [Input] − Log2 (input dilution factor))), where input dilution factor = (fraction of the input chromatin saved) − 1. The average normalized ChIP Ct values for replicate samples were also obtained.
Statistical analysis
Experiments were repeated at least three times. Three to five control or Ctnnb1-mutant testes at each time point were used for immunostaining. The quantitative results were presented as the mean ± S.D. The data were evaluated for significant differences using Student's t test with a paired two-tailed distribution, and one-way analysis of variance. p values < 0.05 were considered to be significant. For every figure, all the data meet normal distribution.
Author contributions
Y. Q. L., L. J. Z., and F. G. designed the research; Y. Q. L., L. J. Z., Y. Q. H., and M. C.(2) performed the experiments, Y. Q. L., L. J. Z., and Y. Q. H. analyzed the data; M. C.(1), F. H., Y. Q., X. H. C., and F. C. T. contributed new reagents or analytic tools; and Y. Q. L., L. J. Z., and F. G. wrote the paper.
Supplementary Material
Acknowledgment
We thank Hua Qin for assistance in fluorescence-activated cell sorting.
This work was supported by National Key R&D program of China Grant 2016YFA0500901, National Natural Science Funds for Distinguished Young Scholar Grant 81525011, and National Natural Science Foundation of China Grants 31471348, 31601193, and 31671496. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Table S1 and Figs. S1–S4.
- Tcf/Lef
- T cell factor/lymphoid enhancer factor
- IHC
- immunohistochemistry
- qPCR
- quantitative PCR.
References
- 1. Byskov A. G. (1986) Differentiation of mammalian embryonic gonad. Physiol. Rev. 66, 71–117 [DOI] [PubMed] [Google Scholar]
- 2. Capel B., Albrecht K. H., Washburn L. L., and Eicher E. M. (1999) Migration of mesonephric cells into the mammalian gonad depends on Sry. Mech. Dev. 84, 127–131 [DOI] [PubMed] [Google Scholar]
- 3. Gubbay J., Collignon J., Koopman P., Capel B., Economou A., Münsterberg A., Vivian N., Goodfellow P., and Lovell-Badge R. (1990) A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 346, 245–250 [DOI] [PubMed] [Google Scholar]
- 4. Koopman P., Gubbay J., Vivian N., Goodfellow P., and Lovell-Badge R. (1991) Male development of chromosomally female mice transgenic for Sry. Nature 351, 117–121 [DOI] [PubMed] [Google Scholar]
- 5. Koopman P., Bullejos M., and Bowles J. (2001) Regulation of male sexual development by Sry and Sox9. J. Exp. Zool. 290, 463–474 [DOI] [PubMed] [Google Scholar]
- 6. Bernard P., and Harley V. R. (2007) Wnt4 action in gonadal development and sex determination. Int. J. Biochem. Cell Biol. 39, 31–43 [DOI] [PubMed] [Google Scholar]
- 7. Chassot A. A., Gillot I., and Chaboissier M. C. (2014) R-spondin1, WNT4, and the CTNNB1 signaling pathway: strict control over ovarian differentiation. Reproduction 148, R97–110 [DOI] [PubMed] [Google Scholar]
- 8. Tomizuka K., Horikoshi K., Kitada R., Sugawara Y., Iba Y., Kojima A., Yoshitome A., Yamawaki K., Amagai M., Inoue A., Oshima T., and Kakitani M. (2008) R-spondin1 plays an essential role in ovarian development through positively regulating Wnt-4 signaling. Hum. Mol. Genet. 17, 1278–1291 [DOI] [PubMed] [Google Scholar]
- 9. Vainio S., Heikkilä M., Kispert A., Chin N., and McMahon A. P. (1999) Female development in mammals is regulated by Wnt-4 signalling. Nature 397, 405–409 [DOI] [PubMed] [Google Scholar]
- 10. Heikkilä M., Prunskaite R., Naillat F., Itäranta P., Vuoristo J., Leppäluoto J., Peltoketo H., and Vainio S. (2005) The partial female to male sex reversal in Wnt-4-deficient females involves induced expression of testosterone biosynthetic genes and testosterone production, and depends on androgen action. Endocrinology 146, 4016–4023 [DOI] [PubMed] [Google Scholar]
- 11. Chassot A. A., Ranc F., Gregoire E. P., Roepers-Gajadien H. L., Taketo M. M., Camerino G., de Rooij D. G., Schedl A., and Chaboissier M. C. (2008) Activation of β-catenin signaling by Rspo1 controls differentiation of the mammalian ovary. Hum. Mol. Genet. 17, 1264–1277 [DOI] [PubMed] [Google Scholar]
- 12. Maatouk D. M., DiNapoli L., Alvers A., Parker K. L., Taketo M. M., and Capel B. (2008) Stabilization of β-catenin in XY gonads causes male-to-female sex-reversal. Hum. Mol. Genet. 17, 2949–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ottolenghi C., Omari S., Garcia-Ortiz J. E., Uda M., Crisponi L., Forabosco A., Pilia G., and Schlessinger D. (2005) Foxl2 is required for commitment to ovary differentiation. Hum. Mol. Genet. 14, 2053–2062 [DOI] [PubMed] [Google Scholar]
- 14. Schmidt D., Ovitt C. E., Anlag K., Fehsenfeld S., Gredsted L., Treier A. C., and Treier M. (2004) The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development 131, 933–942 [DOI] [PubMed] [Google Scholar]
- 15. Uhlenhaut N. H., Jakob S., Anlag K., Eisenberger T., Sekido R., Kress J., Treier A. C., Klugmann C., Klasen C., Holter N. I., Riethmacher D., Schütz G., Cooney A. J., Lovell-Badge R., and Treier M. (2009) Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 139, 1130–1142 [DOI] [PubMed] [Google Scholar]
- 16. Matson C. K., Murphy M. W., Sarver A. L., Griswold M. D., Bardwell V. J., and Zarkower D. (2011) DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 476, 101–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chang H., Gao F., Guillou F., Taketo M. M., Huff V., and Behringer R. R. (2008) Wt1 negatively regulates β-catenin signaling during testis development. Development 135, 1875–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muzumdar M. D., Tasic B., Miyamichi K., Li L., and Luo L. (2007) A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 [DOI] [PubMed] [Google Scholar]
- 19. Tran S., Wang Y., Lamba P., Zhou X., Boehm U., and Bernard D. J. (2013) The CpG island in the murine foxl2 proximal promoter is differentially methylated in primary and immortalized cells. PloS One 8, e76642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jho E. H., Zhang T., Domon C., Joo C. K., Freund J. N., and Costantini F. (2002) Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 22, 1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bar A., Cohen A., Eisner U., Risenfeld G., and Hurwitz S. (1978) Differential response of calcium transport systems in laying hens to exogenous and endogenous changes in vitamin D status. J. Nutr. 108, 1322–1328 [DOI] [PubMed] [Google Scholar]
- 22. Blauwkamp T. A., Chang M. V., and Cadigan K. M. (2008) Novel TCF-binding sites specify transcriptional repression by Wnt signalling. EMBO J. 27, 1436–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu C. F., Bingham N., Parker K., and Yao H. H. (2009) Sex-specific roles of β-catenin in mouse gonadal development. Hum. Mol. Genet. 18, 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tanwar P. S., Kaneko-Tarui T., Zhang L., Rani P., Taketo M. M., and Teixeira J. (2010) Constitutive WNT/β-catenin signaling in murine Sertoli cells disrupts their differentiation and ability to support spermatogenesis. Biol. Reprod. 82, 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boyer A., Hermo L., Paquet M., Robaire B., and Boerboom D. (2008) Seminiferous tubule degeneration and infertility in mice with sustained activation of WNT/CTNNB1 signaling in sertoli cells. Biol. Reprod. 79, 475–485 [DOI] [PubMed] [Google Scholar]
- 26. Clark P. E., Polosukhina D., Love H., Correa H., Coffin C., Perlman E. J., de Caestecker M., Moses H. L., and Zent R. (2011) β-Catenin and K-RAS synergize to form primitive renal epithelial tumors with features of epithelial Wilms' tumors. Am. J. Pathol. 179, 3045–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Richards J. S., Fan H. Y., Liu Z., Tsoi M., Laguë M. N., Boyer A., and Boerboom D. (2012) Either Kras activation or Pten loss similarly enhance the dominant-stable CTNNB1-induced genetic program to promote granulosa cell tumor development in the ovary and testis. Oncogene 31, 1504–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harada N., Tamai Y., Ishikawa T., Sauer B., Takaku K., Oshima M., and Taketo M. M. (1999) Intestinal polyposis in mice with a dominant stable mutation of the β-catenin gene. EMBO J. 18, 5931–5942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lécureuil C., Fontaine I., Crepieux P., and Guillou F. (2002) Sertoli and granulosa cell-specific Cre recombinase activity in transgenic mice. Genesis 33, 114–118 [DOI] [PubMed] [Google Scholar]
- 30. Gao F., Maiti S., Alam N., Zhang Z., Deng J. M., Behringer R. R., Lécureuil C., Guillou F., and Huff V. (2006) The Wilms tumor gene, Wt1, is required for Sox9 expression and maintenance of tubular architecture in the developing testis. Proc. Natl. Acad. Sci. U. S. A. 103, 11987–11992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pan B., Chao H., Chen B., Zhang L., Li L., Sun X., and Shen W. (2011) DNA methylation of germ-cell-specific basic helix-loop-helix (HLH) transcription factors, Sohlh2 and Figlalpha during gametogenesis. Mol. Hum. Reprod. 17, 550–561 [DOI] [PubMed] [Google Scholar]
- 32. Li X. X., Chen S. R., Shen B., Yang J. L., Ji S. Y., Wen Q., Zheng Q. S., Li L., Zhang J., Hu Z. Y., Huang X. X., and Liu Y. X. (2013) The heat-induced reversible change in the blood-testis barrier (BTB) is regulated by the androgen receptor (AR) via the partitioning defective protein (Par) polarity complex in the mouse. Biol. Reprod. 89, 12. [DOI] [PubMed] [Google Scholar]
- 33. Yu C., Ji S. Y., Dang Y. J., Sha Q. Q., Yuan Y. F., Zhou J. J., Yan L. Y., Qiao J., Tang F., and Fan H. Y. (2016) Oocyte-expressed yes-associated protein is a key activator of the early zygotic genome in mouse. Cell Res. 26, 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dang Y., Yan L., Hu B., Fan X., Ren Y., Li R., Lian Y., Yan J., Li Q., Zhang Y., Li M., Ren X., Huang J., Wu Y., Liu P., Wen L., Zhang C., Huang Y., Tang F., and Qiao J. (2016) Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 17, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Easwaran V., Pishvaian M., Salimuddin, and Byers S. (1999) Cross-regulation of beta-catenin-LEF/TCF and retinoid signaling pathways. Curr. Biol. 9, 1415–1418 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.