Abstract
2,3,7,8-Tetrachlorodibenzo-p-dioxin and related compounds are extraordinarily potent environmental toxic pollutants. Most of the 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicities are mediated by aryl hydrocarbon receptor (AhR), a ligand-dependent transcription factor belonging to the basic helix-loop-helix (bHLH) Per-ARNT-Sim (PAS) family. Upon ligand binding, AhR forms a heterodimer with AhR nuclear translocator (ARNT) and induces the expression of genes involved in various biological responses. One of the genes induced by AhR encodes AhR repressor (AhRR), which also forms a heterodimer with ARNT and represses the activation of AhR-dependent transcription. The control of AhR activation is critical for managing AhR-mediated diseases, but the mechanisms by which AhRR represses AhR activation remain poorly understood, because of the lack of structural information. Here, we determined the structure of the AhRR–ARNT heterodimer by X-ray crystallography, which revealed an asymmetric intertwined domain organization presenting structural features that are both conserved and distinct among bHLH-PAS family members. The structures of AhRR–ARNT and AhR–ARNT were similar in the bHLH-PAS-A region, whereas the PAS-B of ARNT in the AhRR–ARNT complex exhibited a different domain arrangement in this family reported so far. The structure clearly disclosed that AhRR competitively represses AhR binding to ARNT and target DNA and further suggested the existence of an AhRR–ARNT-specific repression mechanism. This study provides a structural basis for understanding the mechanism by which AhRR represses AhR-mediated gene transcription.
Keywords: crystallography, dioxin, structural biology, transcription factor, transcription repressor
Introduction
Aryl hydrocarbon receptor (AhR),3 a ligand-dependent transcription factor, responds to diverse exogenous and endogenous ligands and induces the expression of genes encoding molecules involved in detoxication and metabolism, such as cytochrome P450 1A1, as well as in cell differentiation and proliferation (1–6). A potent exogenous AhR ligand is 2,3,7,8-tetrachlorodibenzo-p-dioxin, a representative environmental contaminant of the halogenated aryl hydrocarbon family. Exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin or other structurally related halogenated aryl hydrocarbons elicits diverse biological and toxicological responses, including teratogenicity, carcinogenicity, neurotoxicity, immune repression, and endocrine disruption (7–10). Halogenated aryl hydrocarbons are chemically and metabolically stable, and thus continuous and inappropriate exposure to these compounds results in chronic toxicities. Because most of these toxicities are mediated by AhR (11–13), understanding the mechanism of AhR activation is crucial for controlling excessive activation of AhR.
In the absence of ligands, AhR resides in the cytoplasm by forming a complex with heat shock protein 90 (14), X-associated protein 2 (15–18), and p23 (19, 20). Upon ligand binding, AhR translocates to the nucleus and forms a heterodimer with AhR nuclear translocator (ARNT), and this AhR–ARNT complex interacts with a specific DNA sequence—the xenobiotic-responsive element (XRE)—to activate transcription (1, 2, 4–7). However, the AhR–ARNT complex concurrently induces the expression of AhR repressor (AhRR), a negative-feedback regulator of AhR signaling that also interacts with ARNT and forms an AhRR–ARNT complex and down-regulates AhR signaling (21). Thus, elucidation of the mechanism of AhRR-mediated transcriptional repression is critical for limiting excessive activation of AhR.
AhR, ARNT, and AhRR belong to the basic helix-loop-helix (bHLH) Per-ARNT-Sim (PAS) family of transcriptional regulators (22–24). Typically, bHLH-PAS family members form a heterodimer with other members of the same family through their N-terminal bHLH-PAS domains (Fig. 1A). Whereas the bHLH domain is responsible for DNA binding, the tandem PAS domains (PAS-A and PAS-B) are involved in protein-protein interaction and ligand binding (22–24). In AhR, ligand binding occurs at the PAS-B domain, but this ligand-binding domain is lacking in AhRR (21, 23, 24). The N-terminal bHLH-PAS region is well-conserved among bHLH-PAS family members, but the C-terminal region, which mediates transcriptional activation or repression through interaction with coactivator or corepressor molecules, is comparatively less conserved (22, 24).
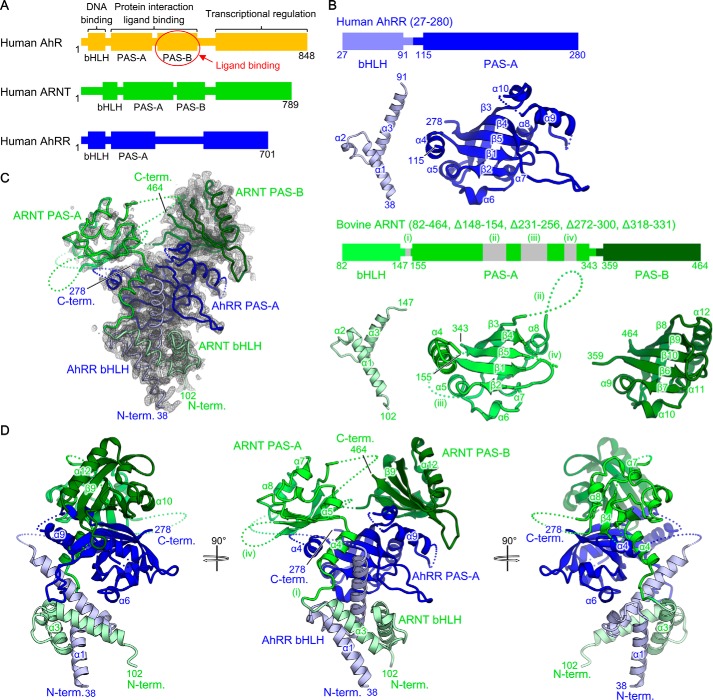
Figure 1.
Crystal structure of AhRR–ARNT complex. A, schematic representation of the domain structures of human AhR, ARNT, and AhRR. B, schematic representation of the crystallization construct and the structure of each domain in the AhRR–ARNT complex. ARNT regions shown in gray ((i)–(iv)) represent the deleted regions. Individual domains are aligned, and the structural elements are labeled. C, electron densities of AhRR–ARNT complex. The 2Fo − Fc difference electron-density map is contoured at the 1.0 σ level with gray mesh. D, crystal structure of AhRR–ARNT complex. AhRR and ARNT are shown in blue and green, respectively, with each domain drawn with slightly different colors as in B. The N (N-term.) and C termini (C-term.) and the structural elements are labeled.
The mechanism of transcriptional repression by AhRR has been described in a few previous reports, but the mechanism remains debated (21, 25–27). Because AhR and AhRR are highly similar, AhRR, like AhR, heterodimerizes with ARNT and interacts with XRE DNA with high affinity. Therefore, AhRR competes with AhR for binding to ARNT and XRE and thus represses AhR signaling (21). Furthermore, SUMOylation of the C-terminal region of AhRR has been reported to be critical for recruiting various corepressor molecules, including ankyrin-repeat protein 2, histone deacetylase (HDAC) 4, and HDAC5, to the promoter region and thereby down-regulating transcription (25, 27). Conversely, Evans et al. (26) have presented contrasting results and have argued that the competitive mechanism for ARNT and XRE binding and the corepressor-mediated mechanism cannot fully explain how AhRR represses transcription; the investigators have proposed the transrepression hypothesis (28), in which AhRR is considered to compete with AhR for binding to unknown coregulatory proteins and promoter-bound transcription factors.
Here, to gain insights into the transcriptional repression mechanism of AhRR, we determined the crystal structure of the AhRR–ARNT complex, which exhibited a spatially distinct asymmetric domain arrangement among the bHLH-PAS family members. Our results provide key structural insights into the mechanisms by which AhRR represses transcription.
Results
Overall structure of AhRR–ARNT heterodimer
To produce a stable AhRR–ARNT complex for crystallization, we coexpressed human AhRR (bHLH-PAS-A) and bovine ARNT with certain flexible loops (residues 148–154, 231–256, 272–300, and 318–331) deleted (bHLH-PAS-A-PAS-B, Δloop) (Fig. 1B). We chose the deleted regions based on the previously reported HIF-1α–ARNT and HIF-2α–ARNT structures (29). We employed reductive ethylation of lysine residues to obtain well-diffracting crystals of the AhRR–ARNT complex and determined its crystal structure at 2.4 Å resolution (Fig. 1, C and D, and Table 1).
Table 1.
Data collection and refinement statistics
| AhRR/ARNT | |
|---|---|
| Data collection | |
| X-ray source | PF-AR NE3A |
| Space group | P41 |
| Cell dimensions | |
| a, b, c (Å) | 78.4, 78.4, 129.8 |
| Resolution (Å) | 50.0–2.4 (2.44–2.40)a |
| Rsym or Rmerge | 5.7 (59.9) |
| I/σI | 28.3 (2.0) |
| Completeness (%) | 100.0 (99.9) |
| Redundancy | 6.8 (6.1) |
| Refinement | |
| Resolution (Å) | 50.0–2.4 |
| No. of reflections | 29,016 |
| Rwork/Rfree | 23.4/26.8 |
| No. of atoms | |
| AhRR | 1,615 |
| ARNT | 2,127 |
| Other | 40 |
| B-factors | |
| AhRR | 58.8 |
| ARNT | 96.3 |
| Other | 57.6 |
| RMSDs | |
| Bond length (Å) | 0.005 |
| Bond angles (°) | 0.97 |
a The highest resolution shell is shown in parentheses.
The final structural model contained one complex of AhRR–ARNT in the crystal asymmetric unit. The two bHLH and three PAS domains in the heterodimer were all defined in the electron-density map, although the electron density obtained for the PAS-A domain of ARNT was relatively poor (Fig. 1C). Overall, the AhRR–ARNT heterodimer exhibited an asymmetric intertwined domain organization featuring the shape of an inverted triangle, in which the PAS-A domain of AhRR was positioned at the center, the bHLH domains at the bottom, and the two PAS domains of ARNT at the two apexes at the top (Fig. 1D). The N-terminal regions and some of the flexible loops of both AhRR and ARNT were disordered. Each domain showed the canonical folds found in the bHLH-PAS family: the bHLH domains of both proteins exhibited a helix-loop-helix structure with two α-helices (α1 and α3) connected by a linker region containing a short α-helix (α2), and the three PAS domains (PAS-A of AhRR and PAS-A and PAS-B of ARNT) were composed of a central five-stranded β-sheet flanked by surrounding α-helices (Figs. 1B and 2). Each corresponding domain between AhRR and ARNT was similar, with root-mean-square deviation (RMSD) values of 0.7 and 2.0 Å for bHLH and PAS-A domains, respectively (Fig. 1B). The PAS-B domain of ARNT also resembled the PAS-A domains of AhRR and ARNT (RMSD = 1.0 and 2.9 Å, respectively) (Fig. 1B).
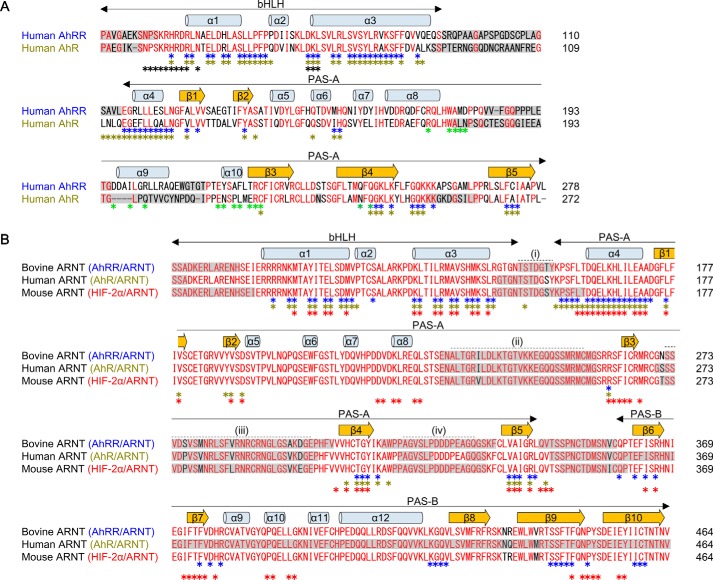
Figure 2.
Sequence alignment of AhRR, AhR, and ARNT. A, sequence alignment of human AhRR and AhR. Secondary structure elements of AhRR are indicated above the alignment. Conserved residues are highlighted in red. AhRR residues interacting with ARNT in the AhRR–ARNT structure (this study) are indicated by blue asterisks (with bHLH-PAS-A) and green asterisks (with PAS-B). AhR residues interacting with ARNT and XRE in the AhR–ARNT–XRE structure (PDB code 5NJ8) (31) are indicated by yellow and black asterisks, respectively. Residues missing in the structural models are surrounded by gray boxes. B, sequence alignment of bovine, human, and mouse ARNT. Secondary structure elements of bovine ARNT are indicated above the alignment. Conserved residues are highlighted in red. ARNT residues interacting with AhRR (this study), AhR (in the AhR–ARNT–XRE structure, PDB code 5NJ8) (31), and HIF-2α (in the HIF-2α–ARNT structure, PDB code 4ZP4) (29) are indicated by blue, yellow, and red asterisks, respectively. The bovine ARNT residues deleted for crystallization in this study are indicated by dashed lines (i)–(iv). Residues missing in the structural models are surrounded by gray boxes.
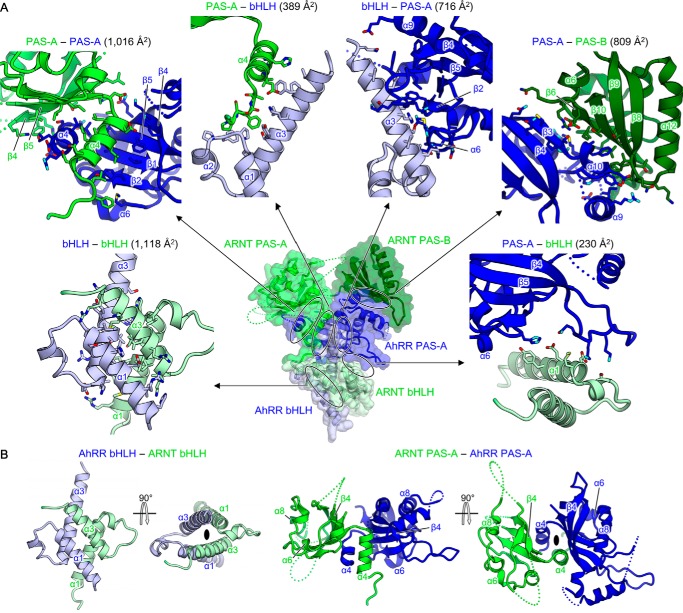
The highly intertwined architecture of the AhRR–ARNT complex was maintained by both intermolecular interactions between AhRR and ARNT and intramolecular interdomain interactions in AhRR (Fig. 3). The intermolecular interfaces could be subdivided into five regions: bHLH (AhRR)–bHLH (ARNT), bHLH (AhRR)–PAS-A (ARNT), PAS-A (AhRR)–bHLH (ARNT), PAS-A (AhRR)–PAS-A (ARNT), and PAS-A (AhRR)–PAS-B (ARNT), which presented contact areas of 1,118, 389, 230, 1,016, and 809 Å2, respectively, and yielded a total contact area of 3,562 Å2 (Fig. 3A). Among these, the interactions between the homotypic domains, bHLH–bHLH and PAS-A–PAS-A, which were arranged in a pseudo-2-fold symmetry, made major contributions to the binding (Fig. 3B). The PAS-A (AhRR)–PAS-B (ARNT) interface, which is specific for the AhRR–ARNT complex (as described in detail in the next subsection), also contributed substantially to the binding (Fig. 3A). Furthermore, the intramolecular interdomain interactions in bHLH (AhRR)–PAS-A (AhRR), presenting a contact area of 716 Å2, enabled a compact folding of AhRR that acted as a scaffold for ARNT binding (Fig. 3A). Because ARNT wrapped around AhRR in an extended conformation, interdomain interactions were not observed in ARNT.
Figure 3.
Domain interfaces in AhRR–ARNT complex. A, domain interfaces in AhRR–ARNT. Each domain interface is enlarged, with residues at the interface shown using stick representations. The contact area of each interface is also shown. B, interactions between homotypic domains, bHLH-bHLH (left) and PAS-A-PAS-A (right), arranged in a pseudo-2-fold symmetry.
Comparison between AhRR–ARNT and ARNT heterodimers formed with other bHLH-PAS family members
Among bHLH-PAS family members, AhRR is unique in that it lacks the PAS-B domain (21, 23–27). Therefore, we compared the structure of AhRR–ARNT with previously reported structures of bHLH-PAS family members. First, we compared the structure of AhRR–ARNT with that of AhR–ARNT encompassing only the bHLH and PAS-A domains, wherein human AhRR and human AhR share 55% sequence identity (30, 31) (Figs. 2A and 4A). Because the residues of AhRR involved in the interaction with ARNT were mostly conserved in AhR (Fig. 2A), the two complexes exhibited similar domain arrangements involving the conserved interfaces, with an RMSD of 1.6 Å (31) (Fig. 4A). The contact area of AhRR and ARNT (2,753 Å2) in this region was comparable to that of AhR and ARNT (2,562 Å2). Moreover, the AhR residues involved in the interaction with XRE DNA were perfectly conserved in AhRR (31) (Fig. 2A). Therefore, the AhRR–ARNT complex would bind to XRE DNA in a similar manner as the AhR–ARNT complex. The structural resemblance of this region provides the basis for the AhRR competition with AhR for interaction with ARNT and XRE DNA.
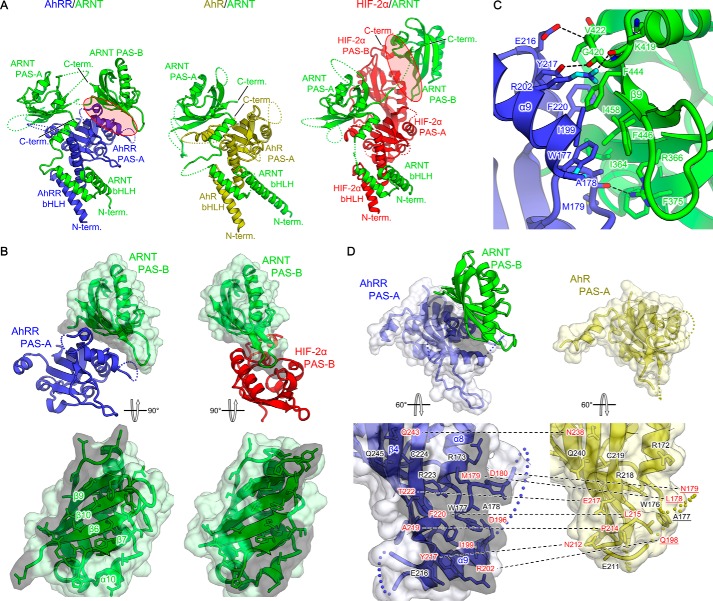
Figure 4.
Structural comparison of AhRR–ARNT with ARNT heterodimers formed with other bHLH-PAS family members. A, heterodimer structures of AhRR–ARNT (left) (this study), AhR–ARNT (middle) (PDB code 5NJ8) (31), and HIF-2α–ARNT (right) (PDB code 4ZP4) (29). AhRR, AhR, HIF-2α, and ARNT are shown in blue, yellow, red, and green, respectively. B, intermolecular contacts mediated by ARNT PAS-B domain. Top panel, structures of PAS-A-PAS-B (AhRR–ARNT) (left) and PAS-B-PAS-B (HIF-2α–ARNT) (right) interfaces indicated by red circles in A are aligned with ARNT PAS-B. Bottom panel, surfaces of ARNT PAS-B involved in the interaction are shown in gray. C, detailed view of the interactions between AhRR PAS-A and ARNT PAS-B. Residues involved in the interactions are shown using stick representations. Hydrogen bonds are indicated by dashed lines. D, structures of PAS-A domains of AhRR (left) and AhR (right). The AhRR PAS-A residues interacting with ARNT PAS-B in the AhRR–ARNT complex are shown using stick representations and are highlighted in gray (left), and the corresponding residues in AhR are also shown using stick representations (right), with the non-conserved residues labeled in red. The residues unmodeled in the AhR PAS-A structure are underlined.
Next, we compared the AhRR–ARNT structure with the structures of heterodimers containing ARNT and other members of the bHLH-PAS family, including HIF-1α–ARNT, HIF-2α–ARNT, NPAS1–ARNT, and NPAS3–ARNT, which encompassed bHLH, PAS-A, and PAS-B domains (29, 32) (Fig. 4A). Although the domain arrangements from the bHLH to the PAS-A region were conserved among AhRR–ARNT and these structures, with an RMSD of 2.1 Å (for HIF-2α–ARNT), the position and orientation of the ARNT PAS-B domain in AhRR–ARNT differed markedly from those in the other heterodimers (Fig. 4A). The PAS-B (ARNT) in the AhRR–ARNT complex interacted with PAS-A (AhRR), whereas the PAS-B (ARNT) in the other bHLH-PAS family heterodimers interacted with only the PAS-B domain of the partner molecule such as HIF-2α, and the PAS-B (ARNT) surfaces used here for interaction with PAS-A (AhRR) or PAS-B (HIF-2α) were similar but not identical (Fig. 4, A and B). This newly identified protein–protein interface between PAS-A (AhRR) and PAS-B (ARNT) contributed to the formation of the unique quaternary structure of AhRR–ARNT in the bHLH-PAS family.
The unique interface between PAS-A (AhRR) and PAS-B (ARNT) was mainly formed by hydrophobic residues (Fig. 4C). The side chains of Trp-177, Ala-178, Met-179, Ile-199, Tyr-217, and Phe-220 of AhRR and those of Ile-364, Phe-375, Val-422, Phe-444, Phe-446, and Ile-458 of ARNT contributed to the hydrophobic interactions. Furthermore, hydrogen bonds formed between the following residues also contributed to the binding: Ala-178 (AhRR) and Arg-366 (ARNT), Arg-202 (AhRR) and Lys-419 (ARNT), Glu-216 (AhRR) and Val-422 (ARNT), and Tyr-217 (AhRR) and Gly-420 (ARNT) (Fig. 4C). Although the structures of the PAS-A domains of AhRR and AhR were similar, with an RMSD of 0.6 Å (Fig. 4D) (31), most of the residues in the PAS-A (AhRR)–PAS-B (ARNT) interface were not conserved between AhRR and AhR (Figs. 2A and 4D). Notably, the α9 helix of PAS-A (AhRR) made a large contribution to the binding, whereas the corresponding region in AhR was disordered. These observations provide a structural basis for the specific interactions between PAS-A (AhRR) and PAS-B (ARNT), a characteristic feature of the AhRR–ARNT heterodimer.
Discussion
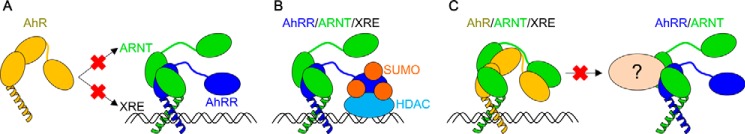
The most widely accepted model of AhRR-mediated repression of AhR signaling is based on the competitive mechanism, where AhRR competes against AhR for heterodimerization with ARNT and thus for binding to XRE DNA (Fig. 5A) (21). Supporting this model, the structure of AhRR–ARNT resembled that of AhR–ARNT in the bHLH-PAS-A region (Fig. 4A), and the residues involved in the interaction with ARNT were mostly conserved between AhRR and AhR (Fig. 2A). Our results further indicated that AhRR–ARNT would efficiently compete with AhR–ARNT for DNA binding: although the structure of the AhRR–ARNT–XRE DNA complex is not available, we found that the AhR residues that interact with XRE DNA were perfectly conserved in AhRR (Fig. 2A), and the bHLH domain was positioned similarly as in the AhR–ARNT complex (Fig. 4A). These findings validate the ability of the competition model to account for AhRR-mediated transcriptional repression to certain extent.
Figure 5.
Models of AhRR transcriptional repression mechanism. A, competitive repression model. AhRR competes with AhR for heterodimerization with ARNT and binding to XRE DNA. B, corepressor-mediated repression model. AhRR–ARNT heterodimer binds to XRE DNA and recruits corepressor molecules such as HDAC4 and HDAC5, which leads to transcriptional repression. C, transrepression model. AhRR–ARNT heterodimer competes with AhR–ARNT heterodimer for binding to unknown interaction partners.
In contrast to the conservation of structural features between AhR–ARNT and AhRR–ARNT in the bHLH-PAS-A region, the PAS-B (ARNT) domain in AhRR–ARNT was located at a distinct position as in other complexes (Fig. 4A). Although the structure of the AhR–ARNT heterodimer across the entire bHLH-PAS-A-PAS-B region remains to be solved, the structures of AhRR–ARNT and AhR–ARNT are likely to differ because of the lack of PAS-B in AhRR, which might be relevant to the transcriptional repression activity of AhRR–ARNT. The SUMOylation of both AhRR (Lys-542, Lys-583, and Lys-660 in the C-terminal region) (25) and ARNT (Lys-245 in the PAS-A domain) (33) is critical for the recruitment of corepressor molecules and the resultant transcriptional repression activity of AhRR–ARNT (Fig. 5B). AhRR and ARNT enhance the SUMOylation of each other through heterodimerization, whereas AhR does not enhance the SUMOylation of ARNT (25). In the transrepression hypothesis proposed by Evans et al. (26), which assumes the existence of unknown interaction partners common for AhR and AhRR, AhRR functions in a manner independent of its competition for ARNT and XRE or functions without its C-terminal region (Fig. 5C). Considering these findings together with the results of the structural analysis, it is tempting to speculate that the unique quaternary architecture identified in this study plays roles in recruiting SUMO E3 ligase for the SUMOylation of AhRR–ARNT or in interacting with the unknown molecules proposed in the transrepression model. However, further studies are required to test and validate this hypothesis.
In summary, we determined the crystal structure of the AhRR–ARNT complex, which revealed structural features of the heterodimer that are conserved and distinct among bHLH-PAS family members. Our findings advance the current understanding of the mechanism by which AhR activation is repressed by AhRR and thus should contribute to the development of a therapeutic strategy for limiting excessive activation of AhR.
Experimental procedures
Preparation of recombinant proteins
Gene sequences encoding hAhRR (bHLH-PAS-A) (residues 27–280) and bARNT (bHLH-PAS-A-PAS-B Δloop) (residues 82–464, with residues 148–154, 231–256, 272–300, and 318–331 deleted), attached with an N-terminal hexahistidine-FLAG tag followed by PreScission Protease recognition sequences, were inserted into pFastBac Dual vector (Thermo Fisher Scientific Inc.). Spodoptera frugiperda Sf9 cells (Thermo Fisher Scientific Inc.) were coinfected with recombinant baculoviruses expressing hAhRR (bHLH-PAS-A) and bARNT (bHLH-PAS-A-PAS-B Δloop) and incubated for 60 h at 300 K. The cells were collected by centrifugation and lysed through sonication in a buffer containing 25 mm Tris-HCl, pH 8.0, 500 mm NaCl, 10% (v/v) glycerol, and 25 mm imidazole HCl, pH 8.0. The recombinant proteins were purified from the cleared lysate by using nickel-nitrilotriacetic acid resin (Qiagen) and a HisTrap column (GE Healthcare), after which the tag was cleaved using PreScission Protease. The proteins were further purified using a Superdex 200 gel-filtration column (GE Healthcare) equilibrated with 10 mm Tris-HCl, pH 8.0, 500 mm NaCl, and 10% (v/v) glycerol.
Crystallization, data collection, and structure determination
The purified AhRR–ARNT complex was lysine-ethylated by using a Reductive Alkylation Kit (Hampton Research) according to the manufacturer's instructions and then purified using a Superdex 200 gel-filtration column. The protein solution used for crystallization contained ∼10 mg/ml lysine-ethylated AhRR–ARNT in a buffer consisting of 10 mm Tris-HCl, pH 8.0, 350 mm NaCl, and 7% (v/v) glycerol. Crystals were grown at 293 K using the sitting-drop vapor-diffusion method; the crystallization droplets were prepared by mixing the protein solution (0.4 μl), reservoir solution (0.4 μl; 12.3% (w/v) PEG 20000 and 100 mm Hepes-NaOH, pH 7.3), and additive solution (0.1 μl; 0.5 m dimethylethylammonium propane sulfonate).
X-ray diffraction data were collected (λ = 1.0000 Å) on a Beamline PF-AR NE3A (Ibaraki, Japan) under cryogenic conditions at 100 K. Prior to flash-cooling, crystals were equilibrated in a cryoprotectant solution consisting of 12.5% (w/v) PEG 20000, 100 mm Hepes-NaOH, pH 7.3, 350 mm NaCl, and 25% (v/v) glycerol. X-ray diffraction data were processed using HKL-2000 (34).
The crystal structure of the AhRR–ARNT complex was solved through molecular replacement performed with Phaser (35) by using the coordinates of HIF-2α–ARNT (Protein Data Bank (PDB) code 4ZP4) (29) and AhR (PDB code 4M4X) (36) as search models. The model was subject to iterative cycles of manual model building by using the program COOT (37) and restrained refinement by using REFMAC (38) (Table 1). The quality of the refined model was evaluated using MolProbity (39), and the structural figures were prepared using CueMol. The coordinate and structure-factor data of the AhRR–ARNT complex have been deposited to Protein Data Bank under PDB code 5Y7Y.
Author contributions
S. S. and U. O. designed the experiments. S. S. prepared recombinant proteins and performed crystallization and structure determination with assistance from U. O. S. S., U. O., and T. S. wrote the manuscript. U. O. and T. S. supervised the project.
Acknowledgments
We thank the beamline staff members at the Photon Factory and SPring-8 for assistance with data collection.
This work was supported by a Grant-in-Aid from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (to U. O. and T. S.), the Takeda Science Foundation (to U. O. and T. S.), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (to U. O.), the Daiichi Sankyo Foundation of Life Science (to U. O.), and the Naito Foundation (to U. O.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 5Y7Y) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- AhR
- aryl hydrocarbon receptor
- bHLH
- basic helix-loop-helix
- PAS
- Per-ARNT-Sim
- ARNT
- AhR nuclear translocator
- AhRR
- AhR repressor
- XRE
- xenobiotic-responsive element
- HDAC
- histone deacetylase
- RMSD
- root-mean-square deviation
- PDB
- Protein Data Bank.
References
- 1. Mulero-Navarro S., and Fernandez-Salguero P. M. (2016) New trends in aryl hydrocarbon receptor biology. Front. Cell Dev. Biol. 4, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abel J., and Haarmann-Stemmann T. (2010) An introduction to the molecular basics of aryl hydrocarbon receptor biology. Biol. Chem. 391, 1235–1248 [DOI] [PubMed] [Google Scholar]
- 3. Denison M. S., and Nagy S. R. (2003) Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 43, 309–334 [DOI] [PubMed] [Google Scholar]
- 4. Denison M. S., Pandini A., Nagy S. R., Baldwin E. P., and Bonati L. (2002) Ligand binding and activation of the Ah receptor. Chem.-Biol. Interact. 141, 3–24 [DOI] [PubMed] [Google Scholar]
- 5. Hahn M. E. (1998) The aryl hydrocarbon receptor: a comparative perspective. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 121, 23–53 [DOI] [PubMed] [Google Scholar]
- 6. Schmidt J. V., and Bradfield C. A. (1996) Ah receptor signaling pathways. Annu. Rev. Cell Dev. Biol. 12, 55–89 [DOI] [PubMed] [Google Scholar]
- 7. Mandal P. K. (2005) Dioxin: a review of its environmental effects and its aryl hydrocarbon receptor biology. J. Comp. Physiol. B 175, 221–230 [DOI] [PubMed] [Google Scholar]
- 8. Pohjanvirta R., and Tuomisto J. (1994) Short-term toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in laboratory animals: effects, mechanisms, and animal models. Pharmacol. Rev. 46, 483–549 [PubMed] [Google Scholar]
- 9. Whitlock J. P. (1990) Genetic and molecular aspects of 2,3,7,8-tetrachlorodibenzo-p-dioxin action. Annu. Rev. Pharmacol. Toxicol. 30, 251–277 [DOI] [PubMed] [Google Scholar]
- 10. Poland A., and Knutson J. C. (1982) 2,3,7,8-Tetrachlorodibenzo-P-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 22, 517–554 [DOI] [PubMed] [Google Scholar]
- 11. Shimizu Y., Nakatsuru Y., Ichinose M., Takahashi Y., Kume H., Mimura J., Fujii-Kuriyama Y., and Ishikawa T. (2000) Benzo[a]pyrene carcinogenicity is lost in mice lacking the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. U.S.A. 97, 779–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mimura J., Yamashita K., Nakamura K., Morita M., Takagi T. N., Nakao K., Ema M., Sogawa K., Yasuda M., Katsuki M., and Fujii-Kuriyama Y. (1997) Loss of teratogenic response to 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) in mice lacking the Ah (dioxin) receptor. Genes Cells 2, 645–654 [DOI] [PubMed] [Google Scholar]
- 13. Fernandez-Salguero P. M., Hilbert D. M., Rudikoff S., Ward J. M., and Gonzalez F. J. (1996) Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol. Appl. Pharmacol. 140, 173–179 [DOI] [PubMed] [Google Scholar]
- 14. Perdew G. H. (1988) Association of the Ah receptor with the 90-kDa heat-shock protein. J. Biol. Chem. 263, 13802–13805 [PubMed] [Google Scholar]
- 15. Petrulis J. R., Hord N. G., and Perdew G. H. (2000) Subcellular localization of the aryl hydrocarbon receptor is modulated by the immunophilin homolog hepatitis B virus X-associated protein 2. J. Biol. Chem. 275, 37448–37453 [DOI] [PubMed] [Google Scholar]
- 16. Bell D. R., and Poland A. (2000) Binding of aryl hydrocarbon receptor (AhR) to AhR-interacting protein: the role of hsp90. J. Biol. Chem. 275, 36407–36414 [DOI] [PubMed] [Google Scholar]
- 17. Meyer B. K., Pray-Grant M. G., Vanden Heuvel J. P., and Perdew G. H. (1998) Hepatitis B virus X-associated protein 2 is a subunit of the unliganded aryl hydrocarbon receptor core complex and exhibits transcriptional enhancer activity. Mol. Cell Biol. 18, 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carver L. A., LaPres J. J., Jain S., Dunham E. E., and Bradfield C. A. (1998) Characterization of the Ah receptor-associated protein, ARA9. J. Biol. Chem. 273, 33580–33587 [DOI] [PubMed] [Google Scholar]
- 19. Cox M. B., and Miller C. A. 3rd (2002) The p23 co-chaperone facilitates dioxin receptor signaling in a yeast model system. Toxicol. Lett. 129, 13–21 [DOI] [PubMed] [Google Scholar]
- 20. Kazlauskas A., Poellinger L., and Pongratz I. (1999) Evidence that the co-chaperone p23 regulates ligand responsiveness of the dioxin (aryl hydrocarbon) receptor. J. Biol. Chem. 274, 13519–13524 [DOI] [PubMed] [Google Scholar]
- 21. Mimura J., Ema M., Sogawa K., and Fujii-Kuriyama Y. (1999) Identification of a novel mechanism of regulation of Ah (dioxin) receptor function. Genes Dev. 13, 20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu D., and Rastinejad F. (2017) Structural characterization of mammalian bHLH-PAS transcription factors. Curr. Opin. Struct. Biol. 43, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kewley R. J., Whitelaw M. L., and Chapman-Smith A. (2004) The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 36, 189–204 [DOI] [PubMed] [Google Scholar]
- 24. Gu Y. Z., Hogenesch J. B., and Bradfield C. A. (2000) The PAS superfamily: sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol. 40, 519–561 [DOI] [PubMed] [Google Scholar]
- 25. Oshima M., Mimura J., Sekine H., Okawa H., and Fujii-Kuriyama Y. (2009) SUMO modification regulates the transcriptional repressor function of aryl hydrocarbon receptor repressor. J. Biol. Chem. 284, 11017–11026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Evans B. R., Karchner S. I., Allan L. L., Pollenz R. S., Tanguay R. L., Jenny M. J., Sherr D. H., and Hahn M. E. (2008) Repression of aryl hydrocarbon receptor (AHR) signaling by AHR repressor: role of DNA binding and competition for AHR nuclear translocator. Mol. Pharmacol. 73, 387–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oshima M., Mimura J., Yamamoto M., and Fujii-Kuriyama Y. (2007) Molecular mechanism of transcriptional repression of AhR repressor involving ANKRA2, HDAC4, and HDAC5. Biochem. Biophys. Res. Commun. 364, 276–282 [DOI] [PubMed] [Google Scholar]
- 28. Pascual G., and Glass C. K. (2006) Nuclear receptors versus inflammation: mechanisms of transrepression. Trends Endocrinol. Metab. 17, 321–327 [DOI] [PubMed] [Google Scholar]
- 29. Wu D., Potluri N., Lu J., Kim Y., and Rastinejad F. (2015) Structural integration in hypoxia-inducible factors. Nature 524, 303–308 [DOI] [PubMed] [Google Scholar]
- 30. Seok S. H., Lee W., Jiang L., Molugu K., Zheng A., Li Y., Park S., Bradfield C. A., and Xing Y. (2017) Structural hierarchy controlling dimerization and target DNA recognition in the AHR transcriptional complex. Proc. Natl. Acad. Sci. U.S.A. 114, 5431–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schulte K. W., Green E., Wilz A., Platten M., and Daumke O. (2017) Structural basis for aryl hydrocarbon receptor-mediated gene activation. Structure 25, 1025–1033 [DOI] [PubMed] [Google Scholar]
- 32. Wu D., Su X., Potluri N., Kim Y., and Rastinejad F. (2016) NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors. Elife 5, e18790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tojo M., Matsuzaki K., Minami T., Honda Y., Yasuda H., Chiba T., Saya H., Fujii-Kuriyama Y., and Nakao M. (2002) The aryl hydrocarbon receptor nuclear transporter is modulated by the SUMO-1 conjugation system. J. Biol. Chem. 277, 46576–46585 [DOI] [PubMed] [Google Scholar]
- 34. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 35. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu D., Potluri N., Kim Y., and Rastinejad F. (2013) Structure and dimerization properties of the aryl hydrocarbon receptor PAS-A domain. Mol. Cell Biol. 33, 4346–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 38. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 39. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]