Abstract
Tetratricopeptide repeat (TPR) domains are ubiquitous structural motifs that mediate protein–protein interactions. For example, the TPR domains in the peroxisomal import receptor PEX5 enable binding to a range of type 1 peroxisomal targeting signal motifs. A homolog of PEX5, tetratricopeptide repeat–containing Rab8b-interacting protein (TRIP8b), binds to and functions as an auxiliary subunit of hyperpolarization-activated cyclic nucleotide (HCN)–gated channels. Given the similarity between TRIP8b and PEX5, this difference in function raises the question of what mechanism accounts for their binding specificity. In this report, we found that the cyclic nucleotide–binding domain and the C terminus of the HCN channel are critical for conferring specificity to TRIP8b binding. We show that TRIP8b binds the HCN cyclic nucleotide–binding domain through a 37-residue domain and the HCN C terminus through the TPR domains. Using a combination of fluorescence polarization– and co-immunoprecipitation–based assays, we establish that binding at either site increases affinity at the other. Thus, allosteric coupling of the TRIP8b TPR domains both promotes binding to HCN channels and limits binding to type 1 peroxisomal targeting signal substrates. These results raise the possibility that other TPR domains may be similarly influenced by allosteric mechanisms as a general feature of protein–protein interactions.
Keywords: cooperativity, cAMP, depression, fluorescence anisotropy, ion channel, peroxisome
Introduction
Peroxisomes are membrane-bound organelles involved in a wide variety of processes, including the degradation of fatty acids via α- and/or β-oxidation, synthesis of ether lipids, and the metabolism of H2O2 (1). Although the presence of a membrane sequesters hazardous reactions, this arrangement necessitates the involvement of additional machinery to facilitate the import of newly synthesized proteins (2). To accomplish this function, Peroxin 5 (PEX5) contains a series of tetratricopeptide repeat (TPR)3 domains that bind to a wide array of C-terminal tripeptides, referred to as peroxisomal targeting signal (PTS1) motifs, with a consensus sequence (SACKN)(KRHQN)(SLI) (3). More recently, this consensus sequence has been extended to a stretch of about 12 residues containing both the C-terminal tripeptide making contact with the TPR domains and an unstructured linker region of five residues interacting with the surface of PEX5 (3, 4). This promiscuity in binding is essential for the role of PEX5 in trafficking PTS1 motif-containing proteins to peroxisomes, and PTS1 motifs are known to vary in affinity for PEX5 over several orders of magnitude (5). Despite the progress that has been made in understanding the structure–function relationship of the TPR domains of PEX5, predicting the affinity of the TPR domains for a given ligand remains a difficult task, and the relationship of TPR affinity to the efficiency of peroxisomal import is not well-understood. In some cases, peroxisomal matrix proteins with low-affinity PEX5–PTS1 interactions are efficiently imported into the peroxisome, suggesting that other mechanisms involving PEX5 or interaction with accessory proteins play a critical role in the peroxisomal import function (5). Along these lines, Harano et al. (6) found that Hsp70 binds to PEX5 and enhances its peroxisomal import function, although others attributed the enhancement to its effect on the folding of the PTS1 carrier (7). Interactions of PEX5 with other peroxins can affect PTS1 binding, as shown for PEX13 influencing the import of catalase (ending in a weak PTS1:KANL) (8). For some PTS1 proteins, secondary interactions with PEX5 have been described (9). More recently, redox reactions involving a cysteine group in the N terminus of the protein were shown to modulate the affinity of PEX5 for its targets (10, 11). Thus, fully understanding the functional roles of TPR-containing proteins like PEX5 may require more than defining the affinity of TPR–ligand interactions.
In contrast to the promiscuity of PEX5 for PTS1-containing ligands, tetratricopeptide repeat-containing Rab8b interacting protein (TRIP8b) (12–14) is a structurally related protein that appears to have a unique function in regulating hyperpolarization-activated cyclic nucleotide–gated (HCN) channels (15). Originally described as Pex5l (Pex5-like protein), the TPR domains of TRIP8b contain substantial homology to those of PEX5. Comparisons of the crystal structure of PEX5 in complex with a PTS1 ligand (16) and TRIP8b bound to the C-terminal peptide of an HCN channel pore-forming subunit, HCN2, revealed near-identical binding characteristics (14). It was thought previously that subtle differences in the TRIP8b- and PEX5-binding pockets permitted TRIP8b to specifically bind HCN channel subunits because HCN1, 2, and 4 terminate in SNL, a sequence not thought to bind to PEX5. However, SNL has indeed been identified as a PTS1 motif in at least one protein (17), and, as demonstrated below (Fig. 1), the TPR domains of TRIP8b bind a range of peptide ligands, including canonical PTS1 motifs. Given the substantial structural similarity between PEX5-PTS1 motif binding and the interaction between the TPR domains of TRIP8b and the C terminus of HCN channels, it is unknown what is responsible for the divergence in specificity observed in vivo.
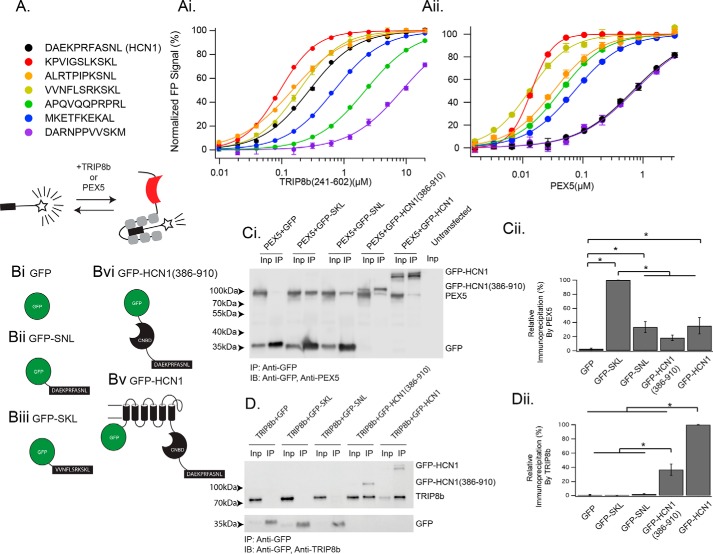
Figure 1.
The TRIP8b TPR domains bind a variety of peptide ligands. A, legend showing the sequence of the peptides used as well as a schematic for the experimental paradigm. Ai, TRIP8b241–602 was titrated into 50 nm of the indicated TAMRA-labeled peptide, with binding detected as a change in fluorescence polarization. See supplemental Table 1 for Kd values. Aii, the same experiment as in Ai was performed with PEX5 substituted for TRIP8b241–602. Error bars represent S.D. for a single run of the experiment, which was performed on three separate occasions and in triplicate on each occasion. Bi–Biv, schematics showing the GFP-tagged constructs used in C and D. Ci and Cii, co-IP experiments were performed with HEK293 cells transfected with the indicated constructs (see “Experimental procedures” for details of the volume loaded in each lane). In each case, GFP was immunoprecipitated, and then PEX5 and GFP were immunoblotted (IB). Note that GFP-HCN1(386–910) is very close in size to PEX5 and appears as the band above the PEX5 band. Also of note is the anomalous migration of the peptide-labeled GFP constructs. Based on our sequencing results, we are confident that this discrepancy is the result of a change in the migration of the protein in the SDS-PAGE gel secondary to the addition of a peptide tag to the C terminus (52). As described in the text, only GFP-SKL is efficiently bound by PEX5. Quantification is provided in Cii and normalized by the interaction between PEX5 and GFP-SKL (one-way ANOVA, F(4,19) = 32.91; GFP-SKL, 100% ± 0%; GFP-SNL, 33.3% ± 7.8%; GFP-HCN1(386–910), 18.4% ± 3.8%; GFP, 2.5% ± 1.1%; GFP-HCN1, 35.4% ± 11.49%; n = 4 distinct experiments). Inp, input. Di, an identical experiment as in C was performed, with the exception that TRIP8b was substituted for PEX5. Note that GFP-SKL fails to bind TRIP8b, whereas both GFP-HCN1(386–910) and GFP-HCN1 efficiently bind TRIP8b. Dii, quantification of the results provided in Di. TRIP8b elutions were scaled to the input fraction and normalized by the interaction between TRIP8b and GFP-HCN1 (see “Experimental procedures”) (F(4,19 = 146.69; GFP-HCN1, 100% ± 0.0%; GFP-HCN1(386–910), 36.6% ± 7.9%; GFP-SNL, 2.1% ± 0.0%; GFP-SKL, 0.2% ± 0.3%; GFP, 0.8% ± 0.6%; n = 4 distinct experiments). See supplemental Fig. 2 for an alternative display setting of the blot in D that highlights the band in the GFP-SNL IP lane, which is nonzero but statistically insignificant. *, p < 0.05 by Tukey's HSD post hoc test.
TRIP8b influences the gating and subcellular distribution of HCN channels, which, in the brain, are important regulators of neuronal excitability and function (13). HCN channels are noninactivating and open in response to hyperpolarization to mediate a nonspecific cationic conductance (18). In addition to their regulation by voltage, HCN channel function is also influenced by the binding of cyclic nucleotides to an intracellular cyclic nucleotide–binding domain (CNBD) (18). Four different genes encode HCN pore-forming subunits (HCN1–4), and HCN1 and HCN2 are the predominant subunits of HCN channels in the mammalian brain (19). These two subunits form both hetero- and homomeric channels in CA1 pyramidal neurons (20, 21), where they are expressed at higher levels in the distal dendrites (22). This pattern of expression is TRIP8b-dependent and facilitates the role of the channel in regulating synaptic integration (23) and calcium signaling (24). Recently, the interaction of HCN channels with TRIP8b has been identified as a therapeutic target for the treatment of depression, and mice lacking TRIP8b show an increase in nondepressed behavior (13, 25). Therefore, there is considerable interest in understanding the structure–function relationship of this interaction with the goal of developing new antidepressants.
Given the overwhelming similarity of the TPR domains of TRIP8b and PEX5, we reasoned that differences in the binding specificities of these two proteins could involve allosteric interactions between domains of the two molecules that are not conserved. In addition to the TRIP8b–HCN interaction involving the TRIP8b TPR domains described above, a second intermolecular TRIP8b–HCN interaction occurs between an 80-residue “core” domain of TRIP8b (26, 27) and the CNBD of HCN channels. In a recent series of structural studies, this core domain of TRIP8b was observed to bind to the CNBD at residues that are distinct from those involved in binding cAMP (28, 29). The relationship between the binding of TRIP8b and cAMP to the CNBD has been described as a mutually antagonistic allosteric interaction, with the binding of either ligand reducing the affinity of the CNBD for the other (28). Despite binding at different locations on the CNBD, certain mutations of the CNBD (such as R538E in HCN1 (30) and R591E in HCN2 (27, 30)) affect the binding of both TRIP8b and cAMP.
In this report, we establish that allosterism between the two TRIP8b–HCN interaction sites plays a crucial role in both limiting the interaction of TRIP8b with PTS1 substrates and in promoting the binding of TRIP8b to HCN channels. Furthermore, our study provides an additional context for understanding how allosterism influences the functional specificity of TPR domain–containing proteins such PEX5 and TRIP8b.
Results
The TPR domains of TRIP8b bind to a wide range of peptides
Despite their substantial homology, comparisons of the crystal structure of TRIP8b and PEX5 suggested that key differences in the residues that coordinate the −2 position of peptide ligands (Lys in the canonical PTS1 motif but Asn in HCN1, 2, and 4) were responsible for producing the specificity of these protein-protein interactions (14). However, an earlier study using qualitative methods suggested that the binding specificities of the TPR domains of TRIP8b and PEX5 may in fact be overlapping (31). Given this apparent discrepancy, we first sought to quantitatively determine whether TRIP8b would bind to PTS1 motifs. Toward that end, we employed a fluorescence polarization-based paradigm and synthesized a series of 11-residue peptides labeled with tetramethyl-6-carboxyrhodamine (TAMRA) and chosen to ensure that a diversity of PTS1 sequences was represented (5) (Fig. 1 and supplemental Table 1). Fluorescence polarization refers to the excitation of a fluorophore with polarized light and measurement of the polarization of the emitted fluorescence (32). In the absence of a large binding partner (such as a protein), a small fluorophore will emit unpolarized light (32). In contrast, when bound by a binding partner that restricts the motion of the fluorophore, the emitted light becomes polarized. We next purified a large TRIP8b fragment corresponding to residues 241–602 of the 1a-4 isoform of TRIP8b, which contains all TPR domains and is sufficient for binding the C-terminal tail of HCN (33). To determine whether the TPR domains of TRIP8b were capable of binding the different peptides, we titrated increasing concentrations of TRIP8b241–602 into a fixed concentration (50 nm) of each TAMRA-labeled peptide(Fig. 1Ai). Interestingly, we noted that TRIP8b241–602 was capable of binding to all PTS1 motifs, some with higher affinity than HCN1. These results indicate that PTS1 peptides compete with HCN1 for binding the TPR domains of TRIP8b.
We next purified the long isoform of PEX5 (see “Experimental procedures”) and titrated increasing concentrations of PEX5 into a fixed concentration (50 nm) of each TAMRA-labeled PTS1-containing or HCN1 peptide (Fig. 1Aii). Unlike TRIP8b, which bound both HCN1 and PTS1 motifs with similar affinity, PEX5 exhibited a significantly lower affinity for HCN1 compared with PTS1-containing peptides. We next generated a mutant HCN1 peptide ending in SKL, the canonical PTS1 motif, rather than SNL (supplemental Fig. 1). Interestingly, the affinity of PEX5 for the mutant HCN1 peptide ending in SKL was ∼70-fold higher than wild-type HCN1 ending in SNL, suggesting that the asparagine residue of HCN subunit tripeptides may be conserved to minimize interaction with PEX5.
We were surprised to note that the PTS1 peptides were being efficiently bound by TRIP8b, so we next set out to determine whether the full-length protein would behave similarly. Toward that end, we generated a series of GFP-tagged constructs for co-IP experiments (Fig. 1B). We generated an untagged construct (GFP), one terminating with a high-affinity PTS1 motif (the C terminus of Δ3, Δ2-enoyl-CoA isomerase, VVNFLSRKSKL, referred to as GFP-SKL), one terminating with the C terminus of HCN1 (DAEKPRFASNL, referred to as GFP-SNL), one terminating in the cytoplasmic domain of HCN1 (GFP-HCN1(386–910)), and a full-length N-terminally tagged HCN1 construct (GFP-HCN1). Co-IP experiments demonstrated that PEX5 was efficiently immunoprecipitated only with the GFP-SKL construct, although weak binding also occurred with the HCN1 fragments (Fig. 1C). In contrast, co-IP with TRIP8b showed an efficient interaction with both GFP-HCN1(386–910) and GFP-HCN1 but not GFP-SKL (Fig. 1D). We also noted that TRIP8b only minimally interacted with GFP-SNL (supplemental Fig. 2), consistent with the importance of the bipartite interaction for TRIP8b–HCN binding. To ensure that the failure of TRIP8b to bind the GFP-SKL construct was not the result of interference from the N-terminal portion of TRIP8b, we also evaluated a truncated TRIP8b construct (TRIP8b259–602) and obtained similar results (supplemental Fig. 3). These experiments indicate that, despite binding PTS1 motifs in vitro, full-length TRIP8b does not bind PTS1 motif–containing proteins.
To determine the physiological relevance of the interaction we observed between the HCN1 cytoplasmic domain and PEX5, we next performed overexpression experiments in COS cells. Although our GFP-SKL construct was efficiently targeted to peroxisomes, HCN1-GFP was expressed diffusely in a pattern that was similar to GFP (supplemental Fig. 4). This experiment suggests that, although PEX5 may bind the cytoplasmic domain in vitro, this interaction is not sufficient to target the membrane-bound HCN1 channel to peroxisomes.
A 37-residue domain of TRIP8b is sufficient for binding the CNBD of HCN
Because TRIP8b binds numerous distinct peptides in vitro but preferentially binds HCN subunits in vivo, we reasoned that an allosteric interaction between the two HCN-binding sites may confer specificity. To investigate this possibility, we first sought to identify the domain of TRIP8b that is involved in binding to the CNBD of HCN pore-forming subunits. We took advantage of the observation that cAMP and TRIP8b stabilize distinct conformations of the CNBD (15, 27–29) to study the binding of TRIP8b using a series of fluorescence polarization–based assays. We then purified a series of protein fragments to study the binding of TRIP8b to the CNBD of HCN in vitro. In particular, we employed a larger TRIP8b fragment incorporating the entire 80-residue core domain (TRIP8b219–602) as well as the CNBD of HCN2 (CNBD443–645), the crystal structure of which has been solved previously (34). Titrating increasing concentrations of TRIP8b219–602 into a fixed concentration of CNBD443–645 (0.1 μm) and FITC-tagged cAMP (8-f-cAMP, 10 nm) decreased the polarization as 8-f-cAMP was displaced from CNBD443–645 (Kd = 2.78 ± 0.09 μm, mean ± S.E.; Fig. 2, A–C). We then repeated this experiment using a smaller TRIP8b construct that was missing a portion of the 80-residue domain shown previously to interact with the CNBD (TRIP8b241–602), and again obtained a binding curve (Kd = 6.72 ± 0.56 μm). To ensure that these observations corresponded to binding of TRIP8b to the CNBD of HCN (and were not the result of an artifact), we repeated these experiments using a smaller TRIP8b construct in which most of the 80-residue minimal domain was missing (TRIP8b259–602). Using this smaller TRIP8b construct, we did not observe any displacement of 8-f-cAMP.
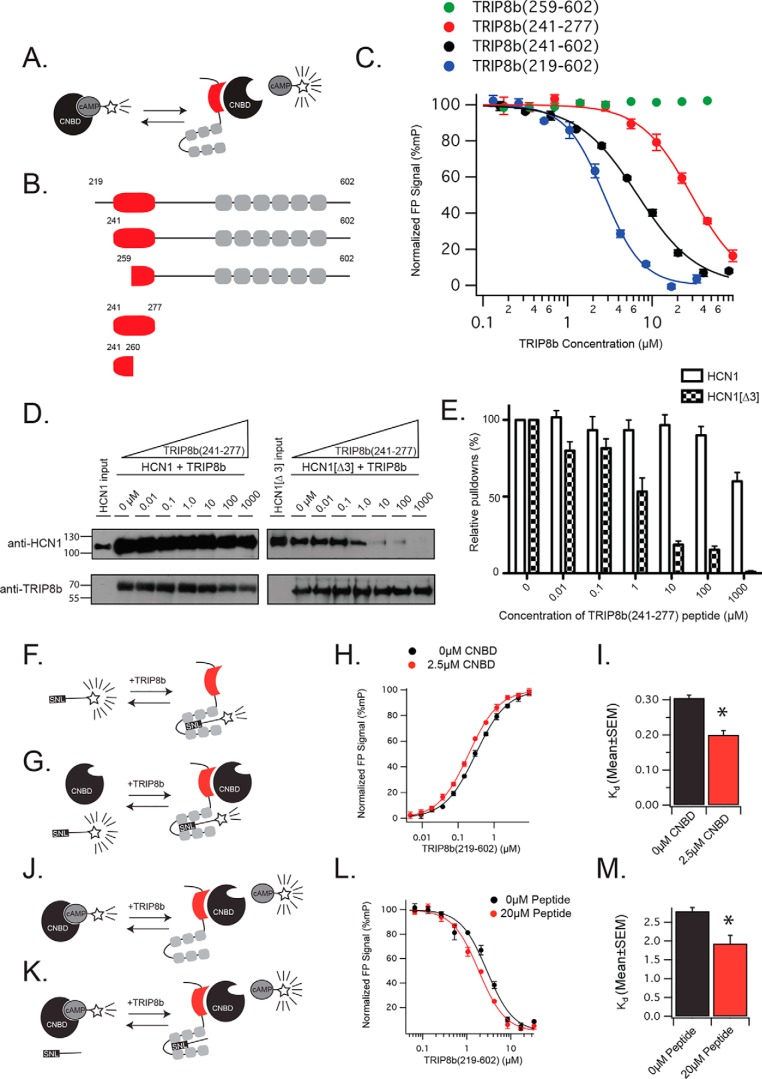
Figure 2.
Identification of a 37-residue fragment of TRIP8b that is sufficient to disrupt the CNBD binding site. A, schematic of the paradigm for examining the binding of TRIP8b to the CNBD of HCN channels. TRIP8b is titrated into a fixed concentration of CNBD443–645 and 8-f-cAMP. B, schematic of the different TRIP8b constructs used. Gray boxes denote TPR domains, whereas the red shape represents the 37-residue fragment that we identify as minimal for displacing cAMP. C, titration of different TRIP8b constructs into a fixed concentration of CNBD443–645 and 8-f-cAMP produces a binding curve. Note that the TRIP8b construct containing the entire 80-residue fragment known to bind the CNBD has the highest affinity (TRIP8b219–602, blue), whereas a sufficiently small TRIP8b fragment (TRIP8b259–602, green) fails to produce detectable binding. D, the 37-residue fragment is sufficient to disrupt the CNBD binding between TRIP8b and HCN1. GST-TRIP8b is able to bind wild-type HCN1 in the presence of increasing concentrations of the 37-residue fragment because the binding between the TPR domains of TRIP8b and the C-terminal tail remains intact. HCN1ΔSNL lacks the substrate for binding the TPR domains and requires the CNBD interaction to bind TRIP8b. In the presence of increasing concentrations of the 37-residue peptide, HCN1ΔSNL fails to bind TRIP8b. E, quantification of the results in D. The binding curve generated by this experiment indicates an IC50 = 1.7 ± 0.8 μm; data are plotted as mean ± S.E. F, schematic showing the binding of TRIP8b219–602 to the TAMRA-labeled HCN1 peptide. G, schematic showing the binding of TRIP8b219–602 in the presence of CNBD443–645. H, representative fluorescence polarization–based assay showing an increase in the affinity of TRIP8b219–602 for HCN1 in the presence of CNBD443–645. I, quantification of three distinct replications of the results from C (0 μm CNBD443–645, 0.29 ± 0.00 μm; 2.5 μm CNBD443–645, 0.19 ± 0.01 μm; mean ± S.E.; n = 3 distinct experiments, t = 6.7; *, p < 0.01). J, schematic for examining the binding of TRIP8b to CNBD443–645. As in Figs. 3–5, increasing concentrations of TRIP8b bind to CNBD443–645 and displace 8-f-cAMP. K, schematic for the experiment performed in the presence of unlabeled HCN1 peptide. L, representative experiment showing the displacement of 8-f-cAMP from CNBD443–645 by the presence of increasing concentrations of TRIP8b. Note that the presence of the unlabeled HCN1 peptide increases the affinity of TRIP8b for CNBD443–645. M, quantification of three distinct replications of the experiment in G (0 μm peptide, 2.78 ± 0.10 μm; 20 μm peptide, 1.92 ± 0.22 μm; mean ± S.E.; n = 4 distinct experiments; t = 3.5; *, p < 0.05). Error bars represent S.D. *, p < 0.05 by two-tailed t test.
Based on the observation that TRIP8b241–602 displaced 8-f-cAMP but TRIP8b259–602 did not, we reasoned that the N-terminal residues spanning 241–259 may be involved in binding. To test this hypothesis, we synthesized an unlabeled 20-residue peptide spanning residues 241–260. As a control, we also generated a scrambled 20-residue peptide with identical composition but a randomized primary structure. We then titrated each 20-residue peptide into a fixed concentration of CNBD443–645 and 8-f-cAMP but did not observe binding in either case (data not shown). These results indicate that, although the 20-residue segment spanning 241–260 is necessary for binding, it is not sufficient.
We next identified a 37-residue fragment spanning TRIP8b241–277. This fragment begins at the same residue in the TRIP8b construct that produced binding (TRIP8b241–602) but ends prior to the TPR domains of TRIP8b (14). We titrated increasing concentrations of this unlabeled 37-residue peptide into a fixed concentration of CNBD443–645 and 8-f-cAMP and this time observed displacement of 8-f-cAMP (Fig. 2C, Kd = 29.2 ± 4.8 μm).
To confirm that the 37-residue fragment is sufficient for binding the CNBD of HCN, we subsequently transfected HEK cells with either HCN1 or HCN1ΔSNL. HCN1ΔSNL is a mutant HCN1 construct lacking the final three C-terminal residues that we established previously disrupts binding to the TPR domains of TRIP8b (26, 30, 35) without interfering with the CNBD interaction. We then used purified, full-length, GST-tagged TRIP8b to bind either HCN1 or HCN1ΔSNL from cell lysate in the presence of increasing concentrations of the 37-residue peptide (Fig. 2, D and E). Regardless of the concentration of the 37-residue peptide, HCN1 was efficiently immunoprecipitated by TRIP8b, consistent with the presence of two interaction sites. However, in the absence of the C-terminal tail interaction (HCN1ΔSNL), increasing concentrations of the 37-residue peptide efficiently disrupted binding (Fig. 2, D and E). These results demonstrate that the 37-residue sequence is necessary and sufficient for binding the CNBD of HCN.
The two TRIP8b-HCN–binding sites exhibit positive allosterism
Having established an assay that allows us to also examine the CNBD-binding site in vitro, we next sought to determine whether the two binding sites influenced one another. First, we titrated increasing concentrations of TRIP8b219–602 into TAMRA-labeled HCN1 tail peptide in the presence or absence of 2.5 μm CNBD443–645 (Fig. 2, F–I). Here we noted that, in the presence of CNBD443–645, the affinity of TRIP8b219–602 for the HCN1 tail increased (Fig. 2, F–I). We then performed the complementary experiment by titrating increasing concentrations of TRIP8b219–602 into a fixed concentration of CNBD and unlabeled HCN1 peptide while monitoring the displacement of 8-f-cAMP (Fig. 2, J–M). Here we noted that the binding of TRIP8b to the unlabeled HCN1 peptide increased the affinity of TRIP8b for CNBD443–645 (Fig. 2, J–M). These results establish that the two TRIP8b-HCN–binding sites exhibit positive allosterism, with binding at either site increasing the affinity at the other. Moreover, because we employed a large TRIP8b fragment that included both binding sites, whereas the HCN components were broken into pieces, we conclude that this allosterism is the result of a conformational change within TRIP8b.
Allosterism promotes specificity of TRIP8b binding
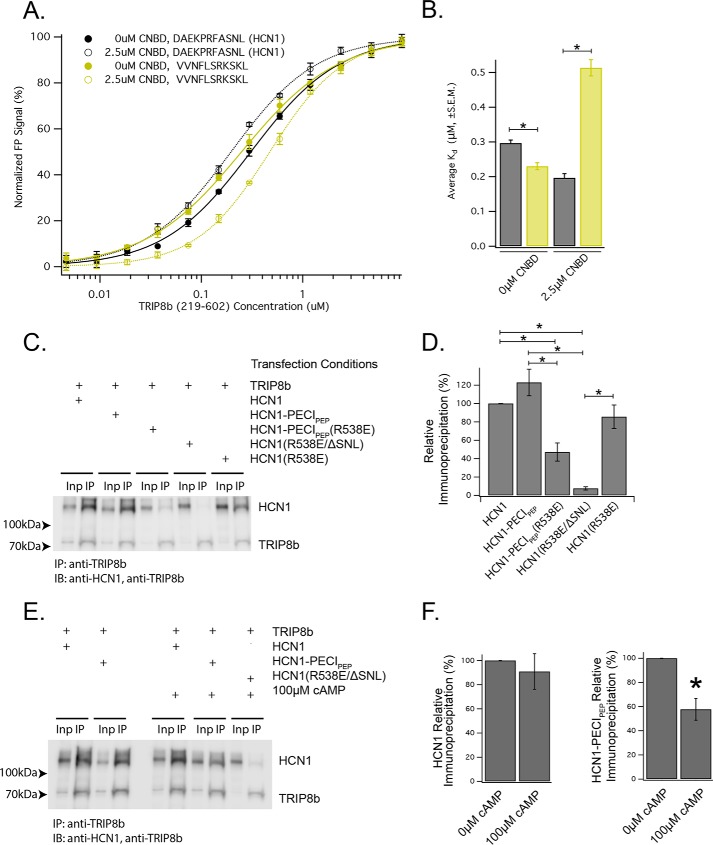
Having observed that each TRIP8b-HCN–binding site is capable of allosterically modulating the other, we reasoned that allosteric modulation of the TPR domains by the CNBD-binding domain of TRIP8b could inhibit the binding of PTS1 motifs by the TPR domains of TRIP8b. We selected the C-terminal 11 residues of a high-affinity PTS1 motif (VVNFLSRKSKL) as a canonical PTS1 sequence (referred to as PECIpep, for the gene name). We titrated TRIP8b219–602 into a fixed concentration of TAMRA-labeled PECIpep in the presence or absence of 2.5 μm CNBD443–645 (Fig. 3A). Interestingly, although the presence of CNBD443–645 increased the affinity of TRIP8b219–602 for the C-terminal tail of HCN1, it decreased the affinity of TRIP8b219–602for PECIpep. These results suggest that the TRIP8b-CNBD interaction impairs the binding of TRIP8b to PTS1 peptides (Fig. 3B).
Figure 3.
The interaction of TRIP8b with the CNBD of HCN limits binding of non-HCN peptides. A, representative experiment showing the results of titrating TRIP8b219–602 into a fixed concentration of the indicated TAMRA-labeled peptides in the presence or absence of CNBD443–645. B, quantification of three distinct experiments shown in A. HCN1 (0 μm CNBD443–645, 0.29 ± 0.00 μm; 2.5 μm CNBD443–645, 0.19 ± 0.01 μm; mean ± S.E.) and VVNFLSRKSKL (0 μm CNBD443–645, 0.23 ± 0.01 μm; 2.5 μm CNBD443–645, 0.51 ± 0.02 μm; mean ± S.E.). Two-way ANOVA revealed an effect of peptide (F(1,8) = 72.11; *, p < 0.05), an effect of the presence of CNBD443–645 (F(1,8) = 38.78; *, p < 0.05), and an interaction between peptide and the presence of CNBD443–645 (F(1,8) = 169.55; *, p < 0.05). C, co-immunoprecipitation experiments reveal a weaker interaction between TRIP8b and a chimeric HCN1 construct. HEK293T cells were transiently transfected with the constructs described at the top. After an input (Inp) fraction was removed, TRIP8b was immunoprecipitated from cell lysates. HCN1-PECIPEP is the chimeric HCN1 construct with the C-terminal 11 residues of HCN1 replaced by VVNFLSRKSKL. Note from A that the VVNFLSRKSKL peptide binds the TPR domains of TRIP8b with a higher affinity than the wild-type C terminus of HCN1. IB, immunoblot. D, quantification of the results from three distinct experiments shown in C. *, p < 0.05 denotes the results of Tukey's post hoc tests. E, representative co-immunoprecipitation experiments performed in the presence or absence of 100 μm cAMP in the lysis and wash buffers. HEK293T cells were transfected with the indicated constructs, and an anti-TRIP8b antibody was used as specified below. Note that HCN1 is efficiently bound in the presence or absence of cAMP (compare the two sets of bands presented), whereas HCN1-PECIPEP is less efficiently bound in the presence of 100 μm cAMP. F, quantification of six distinct immunoprecipitation experiments revealed no difference in the immunoprecipitation of HCN1 by TRIP8b (0 μm cAMP, 100% ± 0%; 100 μm cAMP, 90.9% ± 14.7%; t test; *, p > 0.6). In contrast, immunoprecipitation of HCN1-PECIPEP was significantly reduced in the presence of cAMP (0 μm cAMP, 100% ± 0%; 100 μm cAMP, 57.7% ± 8.9%; t test; *, p < 0.05).
Although our prior results indicate that there is a unique conformation of the TPR domains of TRIP8b that is induced by binding the C terminus of HCN, the magnitude of the change in affinity cast the physiological significance of these observations into doubt. To determine whether the effects might be more pronounced using full-length proteins, we next generated a chimeric HCN1 construct that terminates in the 11-residue peptide (VVNFLSRKSKL), which we refer to as HCN1-PECIPEP. We have shown previously that TRIP8b can immunoprecipitate HCN1 in the absence of the C-terminal tail because of binding at the CNBD (30). To ensure that we examined only the effect of the TPR domains binding to the C terminus of HCN, we also generated a double mutant HCN1 construct in which the CNBD binding by TRIP8b was ablated (HCN1(R538E) (30)) and the chimeric tail is in place (HCN1-PECIPEP(R538E)). The arginine located at position 538 is located in the CNBD and plays an important role in binding cAMP (30). Moreover, although the precise structural determinants of the interaction are unknown, mutating this residue to Glu disrupts CNBD binding by TRIP8b (30). As a negative control for the co-immunoprecipitation, we also used an HCN1 construct in which the CNBD binding site was ablated and the C-terminal tripeptide was lost (HCN1(R538E/∆SNL)). We have shown previously that this double mutant construct fails to bind TRIP8b.
Consistent with our prior results (30), we noted that TRIP8b efficiently binds HCN1 (Fig. 4C, lanes 1 and 2) as well as HCN1-PECIPEP (Fig. 4C, lanes 3 and 4). However, HCN1-PECIPEP(R538E) was much less efficiently bound than HCN1(R538E), suggesting that the CNBD-binding site is responsible for binding HCN1-PECIPEP (Fig. 3D). We were concerned that the R538E mutation may have altered binding in some non-physiologic way, so we next considered a more relevant physiologic manipulation. In particular, we took advantage of the fact that cAMP binding to the CNBD of HCN channels is known to reduce the affinity of the CNBD for TRIP8b (30). We next repeated our immunoprecipitation experiments in the presence or absence of 100 μm cAMP in the lysis and wash buffers. Consistent with prior reports (30), this did not affect immunoprecipitation of HCN1 by TRIP8b. However, HCN1-PECIPEP was less efficiently bound in the presence of 100 μm cAMP (Fig. 3, E and F). These results establish that, in the context of full-length HCN1, the TPR domains of TRIP8b bind to the chimeric C terminus with significantly lower affinity than to the wild-type HCN1 tail.
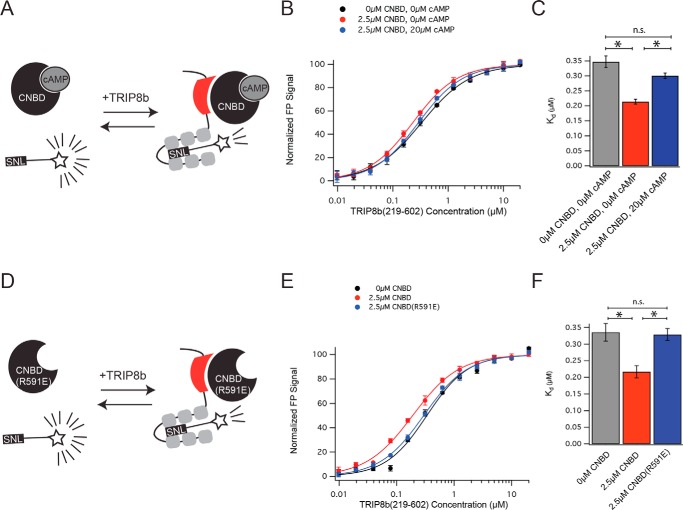
Figure 4.
The apo conformation of the CNBD promotes TRIP8b allosterism. A, schematic demonstrating the experimental paradigm. TRIP8b219–602 was titrated into a fixed concentration of HCN1-TAMRA in the presence or absence of CNBD443–645 and cAMP. B, representative results from a single experiment. Error bars represent S.D. C, quantification across three experiments, shown as mean ± S.E. One-way ANOVA was significant (F(2,6) = 25.69; p < 0.01, n = 3 distinct experiments). *, p < 0.05 by Tukey's test. n.s., not significant. D, schematic for the experiments shown in E and F. TRIP8b219–602 was titrated into a fixed concentration of HCN1-TAMRA in the presence of either CNBD443–645 or CNBD(R591E)443–645. E, representative experiment as in B. F, quantification from three distinct experiments as in C. One-way ANOVA was significant (F(2,6) = 9.93, p < 0.05, n = 3 distinct experiments). *, p < 0.05 on Tukey's post-hoc test.
The apo conformation of the CNBD is required for positive allosterism
We next asked whether the conformation of the CNBD could influence the binding of the TPR domains of TRIP8b to the C terminus of HCN. Given that cAMP binding to the CNBD limits the interaction of TRIP8b with the CNBD (30), we reasoned that the cAMP-bound CNBD may not have the same allosteric effect as the apo conformation. To test this hypothesis, we again titrated TRIP8b219–602 into a fixed concentration of the TAMRA-labeled HCN1 tail in the presence or absence of CNBD443–645 and 20 μm cAMP (Fig. 4, A–C). As above, the presence of the apo conformation of CNBD443–645 increased the affinity of TRIP8b219–602 for the TAMRA-labeled HCN1 tail. However, this effect was lost in the presence of cAMP, indicating that the cAMP-bound conformation of CNBD443–645does not increase the affinity of TRIP8b219–602 for the TAMRA-labeled HCN1 tail (0 μm CNBD, 0 μm cAMP: 0.34 ± 0.01 μm; 2.5 μm CNBD, 0 μm cAMP: 0.21 ± 0.00 μm; 2.5 μm CNBD, 20 μm cAMP: 0.30 ± 0.00 μm; F(2,6) = 25.69, p < 0.001, mean ± S.E. across three distinct experiments). To confirm this result, we next considered a cAMP-insensitive version of CNBD443–645 with a key arginine residue mutated to a glutamic acid (named CNBD(R591E)443–645 for the numbering based on the full-length protein) (20). Previous work has established that this cAMP-insensitive mutant also reduces the binding of TRIP8b to the CNBD (27, 30), hence we reasoned that it would not increase the affinity of TRIP8b219–602 for the TAMRA-labeled HCN1 tail. Consistent with this reasoning, we did not observe an increase in the affinity of TRIP8b219–602 for the TAMRA-labeled HCN1 tail in the presence of 2.5 μm CNBD(R591E)223–30 (0 μm CNBD443–645: 0.33 ± 0.02 μm; 2.5 μm CNBD443–645: 0.21 ± 0.01 μm; 2.5 μm CNBD(R591E)443–645: 0.32 ± 0.01 μm; F(2,6) = 9.9, p < 0.05). Identical results were also obtained for a TAMRA-labeled HCN2 tail (supplemental Fig. 5). Therefore, we conclude that only the apo conformation of the CNBD increases the affinity of TRIP8b for the cytoplasmic tail of HCN.
Discussion
In this report, we have established that allosterism between the two TRIP8b-HCN–binding sites is important for limiting the interaction of TRIP8b with non-HCN proteins (schematized in Fig. 5). Our results suggest that, in the case of TRIP8b, it is not possible to extrapolate from the affinity of the TPR domains for a given peptide to predicting binding partners of TRIP8b. Although we did not explicitly consider the case of PEX5, a related hypothesis has been considered whereby allosteric modulation of the PEX5 TPR domains would facilitate binding PTS1 motif–containing proteins and later allow for the release of these proteins into peroxisomes (6, 7, 36). For example, PEX14-binding of PEX5 leads to the release of monomeric catalase (37), suggesting a role for negative allosterism. Moreover, some PTS1 motif–containing proteins also bind PEX5 through a second interaction site (9), raising the possibility of positive allosterism, as we have seen here for TRIP8b-HCN.
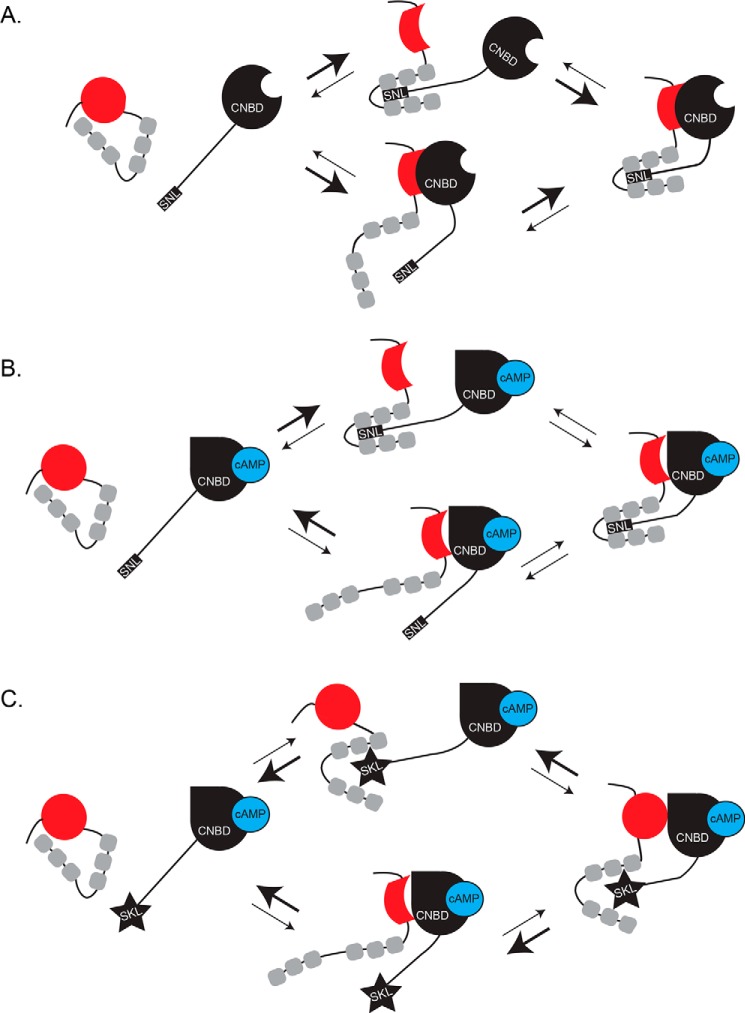
Figure 5.
Schematic illustrating the proposed model. A, TRIP8b is unbound from the cytoplasmic domain of HCN (with CNBD (black) and C-terminal SNL tail shown). Binding between the two proteins initially occurs at either the cytoplasmic C-terminal tail (top) or at the CNBD (bottom). Note that binding at either of the two binding sites causes a conformational change in the other portion of TRIP8b that favors binding to HCN (represented as a change in the red CNBD-binding domain and a shift in the orientation of the TPR domains). On the right, TRIP8b is stably bound to HCN. B, left, shows TRIP8b and the cytoplasmic C terminus of HCN bound to cAMP. Binding occurs first at the TPR domains (above) or at the CNBD (below). Note that TRIP8b binding to the CNBD in the presence of cAMP is less stable (bottom) and fails to elicit a conformational change in the TPR domains (compare with A). In the presence of cAMP, binding at either binding site does not promote binding of the other binding site (represented by the weight of the arrows). C, TRIP8b is unbound from chimeric HCN1 protein in the presence of cAMP. Binding is unfavorable at either the CNBD binding site (below) or the cytoplasmic tail (above), consistent with our experimental results in Fig. 3.
Previous efforts to understand the differences in substrate specificities of PEX5 and TRIP8b have been based on the crystal structures of each of the two proteins (14, 16). These studies presumed that the observed conformation of the TPR domains was required for binding the ligands with which their respective proteins were crystallized (14). More recent evidence demonstrates that the TPR domains of PEX5 are capable of adopting distinct conformations to accommodate different binding partners (38, 39). In an intriguing study, Fodor et al. (38) crystallized a dimer of alanine glyoxylate aminotransferase in complex with PEX5 in a 1:2:1 arrangement. Surprisingly, the PEX5 residues involved in binding the PTS1 motif of each alanine glyoxylate aminotransferase monomer were not identical despite binding the same substrate. These results suggest that TPR domains are capable of a substantial amount of flexibility in binding, with distinct conformations of PEX5 achieved even when binding the same ligand. We reason that, based on their significant structural homology, TRIP8b functions similarly to PEX5 in that different residues may be involved in binding different substrates. This difference in residue involvement almost certainly translates into differences in the conformation of the TPR domains. Moreover, in the presence of the CNBD interaction with TRIP8b, the interaction of the TPR domains with non-HCN peptides is reduced (Fig. 3). These results indicate that allosterism between the TPR domains and the core domain of TRIP8b is crucial for mediating specificity in cargo binding.
Another PEX5-related protein that was identified in yeast (40), and recently renamed PEX9 (41, 42) may also shed light onto the binding characteristics of TRIP8b. Initial work showed that it did not bind the classical PTS1 motifs, nor was it able to complement PEX5. However, PEX9 was recently shown to be an import receptor for a limited subset of peroxisomal proteins; namely, a glutathione transferase, malate synthase 1, and malate synthase 2, and to be strongly induced in the presence of oleate (41, 42). Although the transferase ends in SRL and both malate synthases contain the SKL motif (and are also recognized by PEX5), PEX9 does not bind to other SKL or PTS1 proteins. Hence, there must be other contact sites underlying the PEX9 binding stringency, and it is likely that a similar situation occurs in TRIP8b.
In this report, we have noted that the TPR domains of PEX5 are capable of binding the C terminus of HCN (Fig. 1) with an affinity that is nearly that of genuine PTS1 motifs. Despite this, HCN1 is not transported to peroxisomes by PEX5, likely because it is membrane-bound (supplemental Fig. 4).
We observed that the CNBD of HCN is capable of promoting binding of the TPR domains of TRIP8b to the HCN C terminus but that this appears to only hold true for the apo state of the CNBD (Fig. 4). The R591E mutation in the CNBD is known to reduce (but not entirely eliminate (27, 30)) the interaction of the CNBD with TRIP8b and disrupted the positive allosteric effect (Fig. 4, D–F), as did the addition of cAMP (Fig. 4, A–C). Our evidence that the cAMP-bound conformation of the CNBD fails to influence the conformation of TPR domains of TRIP8b suggests an interesting hypothesis in light of recent work showing that both TRIP8b-HCN–binding sites are required for dendritic trafficking of the ion channel in CA1 pyramidal neurons (25, 43). In particular, cAMP signaling in vivo may disrupt the interaction between the CNBD of HCN and TRIP8b with consequences for subcellular trafficking. Future work may examine whether cAMP plays a role in regulating HCN channel function not only by directly agonizing channel activity but also by regulating its affinity for TRIP8b.
The interaction between TRIP8b and HCN channels has been identified as a therapeutic target for the treatment of major depressive disorder (25, 33, 43, 44). The TPR domains of TRIP8b were initially considered an attractive target because of the accessible ligand-binding pocket (14), but the observations provided here indicate that disrupting the CNBD binding site will also inhibit TRIP8b function.
Experimental procedures
Cloning
All oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). All restriction enzymes were purchased from New England Biotechnologies (Ipswich, MA). Throughout the manuscript, TRIP8b residues are referred to using the 1a-4 isoform as a reference. TRIP8b219–602 was generated using PCR amplification of the TRIP8b insert from a plasmid template using 5′ atagcgccatggctcggctgacc 3′ and 5′ cgccgcctcgagcccgggtcaaggatccaaattgaaag 3′ followed by Nco1/Xho1 digestion and ligated into a modified pGS21a bacterial expression vector (described previously (33)). The CNBD fragment used in our study (CNBD443–645) has been referred to previously as HCN2J (34) and spans residues 443–645 of the mouse HCN2 isoform with a maltose-binding protein tag (a generous gift from Dr. Eric Accili, University of British Columbia). In all experiments, the long isoform of human PEX5 was used (NM_000319.4). For protein expression, a plasmid template was used to produce a PCR product using primers 5′ gtgcccatggcaatgcgggagctg 3′ and 5′ gtgcgtcgac tcactggggcaggccaaa 3′, followed by NcoI/SalI digestion and ligation into pGS21-a. All PTS1 motif peptides correspond to human proteins. Mouse TRIP8b and HCN constructs were used, although the C-terminal 11 residues of HCN1 and HCN2 are identical in the mouse and human proteins. HCN1-PECIpep and HCN1-PECIpep(R538E) were both generated by digesting their respective templates with DraIII/EcoR1 and performing oligonucleotide insertion.
Co-immunoprecipitation from cells
HEK293T cells were transiently transfected (Mirus Bio LLC, Madison, WI) with the indicated plasmids according to the instructions of the manufacturer. Cells were 80–90% confluent at the time of transfection. Plasmids were used in a 1:3 ratio of GFP-tagged construct to TRIP8b, PEX5, or TRIP8b(259–602). 24 h after transfection, cells were washed with PBS and then lysed in 500 μl of TEEN-TX buffer (50 mm Tris, 1 mm EDTA, 1 mm EGTA, 150 mm NaCl, and 0.1% Triton X-100) supplemented with protease inhibitor mixture (Thermo Fisher, Waltham, MA). The lysate was then sonicated briefly (three 1-s pulses) and centrifuged at 21,000 × g for 15 min. For the experiments described in Fig. 1, 50 μl of the supernatant was then added to 50 μl of sample buffer (Bio-Rad, supplemented with 100 mm dithiothreitol), boiled for 5 min, and frozen at −80 °C for later use as an input fraction. The immunoprecipitation was carried out with 400 μl of supernatant and 5 μl of custom guinea pig anti-GFP antibody. The lysate/antibody mixture was nutated at 4 °C for 2 h before adding 50 μl of prewashed protein A/G bead slurry. The bead/lysate mixture was nutated for 1 h at 4 °C before being washed in TEEN-TX buffer (five 5-min washes with nutation at 4 °C) and eluted by boiling in 50 μl of sample buffer. 15 μl of input and 15 μl of eluate were then used for Western blotting to evaluate the results. Rabbit anti-Pex5 (Abcam, ab192322), mouse anti-TRIP8b (Neuromab, N212/17), custom rabbit anti-GFP, and custom rabbit anti-TRIP8b antibodies were used for protein detection. For the experiments described in Fig. 4, 30 μl of lysate was set aside as an input fraction, and the remainder was incubated with custom guinea pig anti-TRIP8b antibody (see Refs. 45–47 for validation of the antibody) and 70 μl of protein A/G bead slurry (Thermo Fisher Scientific). After overnight incubation at 4 °C, the beads were vigorously washed five times for 5 min in 1 ml of lysis buffer at 4 °C. The protein was then eluted using 100 μl of 2× sample buffer (1610737, Bio-Rad). 2 μl of the input fraction and 10 μl of the elution fraction were then loaded onto a 15-well 10% polyacrylamide gel (Bio-Rad) for immunoblotting.
Protein expression
All protein constructs were purified using nickel-nitrilotriacetic acid chromatography (Qiagen, Hilden, Germany) as described elsewhere (25, 43). The purity of the protein constructs was verified by Coomassie staining, and concentration was determined by Bradford assay (33). No additional purification steps were performed to remove any endogenous cAMP that may have been bound to the CNBD constructs used in our experiments (48).
Peptides
All peptides were purchased from GenScript (Piscataway, NJ). Peptides were dissolved in PBS and stored at −80 °C. The 37 residue peptide referred to in the text is as follows: EFERAKAAVESDTEFWDKMQAEWEEMARRNWISENQE.
Fluorescence polarization
Fluorescence polarization assays were performed as described previously (25, 43). Briefly, to examine the binding of the TPR domains of TRIP8b to the C terminus of HCN1 and HCN2, a series of 2-fold dilutions of TRIP8b219–602 was titrated into 50 nm labeled peptide in the presence or absence of either 2.5 μm CNBD443–645 or 2.5 μm CNBD(R591E)443–645, as specified in the text. All experiments were performed using sterile filtered PBS supplemented with 1 mm DTT, and measurements were taken in triplicate in black 384-well microtiter plates (Corning, Corning, NY). In our previous report (33), we used a different buffer for a similar experiment and attribute differences in the affinity of TRIP8b for the HCN C-terminal peptides to this difference. The polarization measurements were obtained using a Tecan microplate reader (Tecan, Zurich, Switzerland) at the Structural Biology facility at Northwestern University. All curve fitting and data analyses were performed using Igor Pro (WaveMetrics). Curves were initially fit by the Hill equation (49) based on a protocol described previously (32). However, given that many of the Kd values were found to be near the concentration of fluorophore-labeled ligand (50 nm), we re-evaluated our data using a quadratic equation that corrects for errors associated with ligand depletion (50) with [LR]/[L] = (([R]+[L]+Kd)-(([R]+[L]+Kd)[caret]2)-4×[R]×[L])[caret].5))/2×[L], where [L] is the concentration of the fluorophore-labeled peptide, [R] is the concentration of the titrated protein, and [LR] represents the protein–peptide complex.
In a subset of experiments, we performed displacement-based assays with TRIP8b binding displacing 8-f-cAMP from the CNBD of HCN (Fig. 2, C and J–M). Although we recognize that this interaction is complex, it was beyond the scope of this manuscript to develop a more sophisticated model for examining this interaction. As we were only concerned with comparing the relative affinities under different conditions, we fit the data for these experiments with the Hill equation (49).
GST pulldown
GST pulldown was performed as described previously (30). Briefly, GST-tagged TRIP8b was purified from BL21 bacteria (Stratagene, San Diego, CA) using glutathione-Sepharose affinity chromatography (Amersham Biosciences, Arlington Heights, IL). HEK293 cells were transfected with the indicated HCN1 plasmid constructs, and after removal of an input fraction, the lysate was incubated with 5 μg of GST-tagged TRIP8b for 2 h at 4 °C. The beads were then washed six times with lysis buffer (see above), and protein was eluted from the beads using SDS sample buffer.
Immunocytochemistry
COS-7 cells (ATCC, Manassas, VA) were maintained as described previously (51). All transient transfections were performed using Mirus TransIT-LT1 transfection reagent using 1.5 μl of transfection reagent, 0.5 μg cDNA, and 50 μl of serum-free media per coverslip in a 24-well plate. 24 h following transfection, the cells were washed once using PBS and then fixed for 10 min at room temperature in 4% paraformaldehyde. The cells were then washed three times in PBS and kept in PBS in a 4 °C refrigerator, covered, until staining. On the day of staining, coverslips were blocked for 1 h at room temperature in blocking buffer (PBS with 5% normal goat serum and 0.03% Triton X-100). The coverslips were then incubated overnight in primary antibodies at 4 °C with gentle shaking. The next day, the coverslips were washed three times for 5 min in PBS-T (PBS with 0.03% Triton X-100) and then incubated for 1 h in secondary antibody at room temperature (with gentle shaking). Coverslips were then washed three times for 5 min in PBS-T, with 1 μm DAPI added to the final wash. The coverslips were then transferred onto microscope slides using PermaFluor (Thermo Fisher Scientific). After a 30-min incubation at room temperature, the coverslips were sealed with clear fingernail polish. 1:1000 goat anti-rabbit conjugated to Alexa Fluor 647 was used to minimize spectral overlap with GFP and was purchased from Thermo Fisher Scientific. Rabbit anti-PMP70 (P0497) was purchased from Sigma-Aldrich (St. Louis, MO) and used at 1:1000 dilution.
Microscopy
Imaging work was performed at the Northwestern University Center for Advanced Microscopy on a Nikon C2 confocal microscope using a ×100 lens.
Author contributions
K. A. L., Y. H., and D. M. C. conceived the study. K. A. L., Y. H., X. C., and R. E. L. performed the experiments. K. A. L., Y. H., R. J. H., J. E. K., P. P. V. V., and D. M. C. wrote the paper. All authors approved the final version of the paper.
Supplementary Material
Acknowledgments
The Structural Biology facility at Northwestern University and the Northwestern University Center for Advanced Microscopy are generously supported by NCI, National Institutes of Health CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center.
This work was supported by National Institutes of Health Grants 2R01NS059934, R01MH106511, and R21MH104471 (to D. M. C.); Brain Research Foundation Grant SG 2012-01 (to D. M. C.); Chicago Biomedical Consortium Grant HTS-004 (to Y. H. and D. M. C.); National Institutes of Health Grant 2T32MH067564 (to K. A. L.), Northwestern University Clinical and Translational Sciences Institute Grants 8UL1TR000150 and UL1TR001422 (to Y. H.), and Brain and Behavior Research Foundation Grant NARSAD 25138 (to Y. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. 1–5 and Table 1.
- TPR
- tetratricopeptide repeat
- HCN
- hyperpolarization-activated cyclic nucleotide
- CNBD
- cyclic nucleotide–binding domain
- TAMRA
- tetramethyl-6-carboxyrhodamine
- IP
- immunoprecipitation
- ANOVA
- analysis of variance
- 8-f-camp
- 8-thioacetamido-fluorescein-cAMP.
References
- 1. Van Veldhoven P. P. (2010) Biochemistry and genetics of inherited disorders of peroxisomal fatty acid metabolism. J. Lipid Res. 51, 2863–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Emmanouilidis L., Gopalswamy M., Passon D. M., Wilmanns M., and Sattler M. (2016) Structural biology of the import pathways of peroxisomal matrix proteins. Biochim. Biophys. Acta 1863, 804–813 [DOI] [PubMed] [Google Scholar]
- 3. Brocard C., and Hartig A. (2006) Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim. Biophys. Acta 1763, 1565–1573 [DOI] [PubMed] [Google Scholar]
- 4. Maynard E. L., Gatto G. J. Jr, and Berg J. M. (2004) Pex5p binding affinities for canonical and noncanonical PTS1 peptides. Proteins 55, 856–861 [DOI] [PubMed] [Google Scholar]
- 5. Ghosh D., and Berg J. M. (2010) A proteome-wide perspective on peroxisome targeting signal 1 (PTS1)-Pex5p affinities. J. Am. Chem. Soc. 132, 3973–3979 [DOI] [PubMed] [Google Scholar]
- 6. Harano T., Nose S., Uezu R., Shimizu N., and Fujiki Y. (2001) Hsp70 regulates the interaction between the peroxisome targeting signal type 1 (PTS1)-receptor Pex5p and PTS1. Biochem. J. 357, 157–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harper C. C., Berg J. M., and Gould S. J. (2003) PEX5 binds the PTS1 independently of Hsp70 and the peroxin PEX12. J. Biol. Chem. 278, 7897–7901 [DOI] [PubMed] [Google Scholar]
- 8. Otera H., Setoguchi K., Hamasaki M., Kumashiro T., Shimizu N., and Fujiki Y. (2002) Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner: conserved Pex5p WXXXF/Y motifs are critical for matrix protein import. Mol. Cell Biol. 22, 1639–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hagen S., Drepper F., Fischer S., Fodor K., Passon D., Platta H. W., Zenn M., Schliebs W., Girzalsky W., Wilmanns M., Warscheid B., and Erdmann R. (2015) Structural insights into cargo recognition by the yeast PTS1 receptor. J. Biol. Chem. 290, 26610–26626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ma C., Hagstrom D., Polley S. G., and Subramani S. (2013) Redox-regulated cargo binding and release by the peroxisomal targeting signal receptor, Pex5. J. Biol. Chem. 288, 27220–27231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Apanasets O., Grou C. P., Van Veldhoven P. P., Brees C., Wang B., Nordgren M., Dodt G., Azevedo J. E., and Fransen M. (2014) PEX5, the shuttling import receptor for peroxisomal matrix proteins, is a redox-sensitive protein. Traffic 15, 94–103 [DOI] [PubMed] [Google Scholar]
- 12. Santoro B., Wainger B. J., and Siegelbaum S. A. (2004) Regulation of HCN channel surface expression by a novel C-terminal protein-protein interaction. J. Neurosci. 24, 10750–10762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lewis A. S., Vaidya S. P., Blaiss C. A., Liu Z., Stoub T. R., Brager D. H., Chen X., Bender R. A., Estep C. M., Popov A. B., Kang C. E., Van Veldhoven P. P., Bayliss D. A., Nicholson D. A., Powell C. M., et al. (2011) Deletion of the hyperpolarization-activated cyclic nucleotide-gated channel auxiliary subunit TRIP8b impairs hippocampal Ih localization and function and promotes antidepressant behavior in mice. J. Neurosci. 31, 7424–7440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bankston J. R., Camp S. S., DiMaio F., Lewis A. S., Chetkovich D. M., and Zagotta W. N. (2012) Structure and stoichiometry of an accessory subunit TRIP8b interaction with hyperpolarization-activated cyclic nucleotide-gated channels. Proc. Natl. Acad. Sci. U.S.A. 109, 7899–7904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zolles G., Wenzel D., Bildl W., Schulte U., Hofmann A., Müller C. S., Thumfart J. O., Vlachos A., Deller T., Pfeifer A., Fleischmann B. K., Roeper J., Fakler B., and Klöcker N. (2009) Association with the auxiliary subunit PEX5R/Trip8b controls responsiveness of HCN channels to cAMP and adrenergic stimulation. Neuron 62, 814–825 [DOI] [PubMed] [Google Scholar]
- 16. Gatto G. J. Jr, Geisbrecht B. V., Gould S. J., and Berg J. M. (2000) Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 7, 1091–1095 [DOI] [PubMed] [Google Scholar]
- 17. Amery L., Brees C., Baes M., Setoyama C., Miura R., Mannaerts G. P., and Van Veldhoven P. P. (1998) C-terminal tripeptide Ser-Asn-Leu (SNL) of human D-aspartate oxidase is a functional peroxisome-targeting signal. Biochem. J. 336, 367–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Biel M., Wahl-Schott C., Michalakis S., and Zong X. (2009) Hyperpolarization-activated cation channels: from genes to function. Physiol. Rev. 89, 847–885 [DOI] [PubMed] [Google Scholar]
- 19. Notomi T., and Shigemoto R. (2004) Immunohistochemical localization of Ih channel subunits, HCN1–4, in the rat brain. J. Comp. Neurol. 471, 241–276 [DOI] [PubMed] [Google Scholar]
- 20. Chen S., Wang J., and Siegelbaum S. A. (2001) Properties of hyperpolarization-activated pacemaker current defined by coassembly of Hcn1 and Hcn2 subunits and basal modulation by cyclic nucleotide. J. Gen. Physiol. 117, 491–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zha Q., Brewster A. L., Richichi C., Bender R. A., and Baram T. Z. (2008) Activity-dependent heteromerization of the hyperpolarization-activated, cyclic-nucleotide gated (HCN) channels: role of N-linked glycosylation. J. Neurochem. 105, 68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lörincz A., Notomi T., Tamás G., Shigemoto R., and Nusser Z. (2002) Polarized and compartment-dependent distribution of HCN1 in pyramidal cell dendrites. Nat. Neurosci. 5, 1185–1193 [DOI] [PubMed] [Google Scholar]
- 23. Magee J. (1999) Dendritic Ih normalizes temporal summation in hippocampal CA1 neurons. Nat. Neurosci. 2, 848. [DOI] [PubMed] [Google Scholar]
- 24. Tsay D., Dudman J. T., and Siegelbaum S. A. (2007) HCN1 channels constrain synaptically evoked Ca2+ spikes in distal dendrites of CA1 pyramidal neurons. Neuron 56, 1076–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han Y., Heuermann R. J., Lyman K. A., Fisher D., Ismail Q. A., and Chetkovich D. M. (2017) HCN-channel dendritic targeting requires bipartite interaction with TRIP8b and regulates antidepressant-like behavioral effects. Mol. Psychiatry 22, 458–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Santoro B., Hu L., Liu H., Saponaro A., Pian P., Piskorowski R. A., Moroni A., and Siegelbaum S. A. (2011) TRIP8b regulates HCN1 channel trafficking and gating through two distinct C-terminal interaction sites. J. Neurosci. 31, 4074–4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu L., Santoro B., Saponaro A., Liu H., Moroni A., and Siegelbaum S. (2013) Binding of the auxiliary subunit TRIP8b to HCN channels shifts the mode of action of cAMP. J. Gen. Physiol. 142, 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saponaro A., Pauleta S. R., Cantini F., Matzapetakis M., Hammann C., Donadoni C., Hu L., Thiel G., Banci L., Santoro B., and Moroni A. (2014) Structural basis for the mutual antagonism of cAMP and TRIP8b in regulating HCN channel function. Proc. Natl. Acad. Sci. U.S.A. 111, 14577–14582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DeBerg H. A., Bankston J. R., Rosenbaum J. C., Brzovic P. S., Zagotta W. N., and Stoll S. (2015) Structural mechanism for the regulation of HCN ion channels by the accessory protein TRIP8b. Structure 23, 734–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Han Y., Noam Y., Lewis A. S., Gallagher J. J., Wadman W. J., Baram T. Z., and Chetkovich D. M. (2011) Trafficking and gating of hyperpolarization-activated cyclic nucleotide-gated channels are regulated by interaction with tetratricopeptide repeat-containing Rab8b-interacting protein (TRIP8b) and cyclic AMP at distinct sites. J. Biol. Chem. 286, 20823–20834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fransen M., Amery L., Hartig A., Brees C., Rabijns A., Mannaerts G. P., and Van Veldhoven P. P. (2008) Comparison of the PTS1- and Rab8b-binding properties of Pex5p and Pex5Rp/TRIP8b. Biochim. Biophys. Acta 1783, 864–873 [DOI] [PubMed] [Google Scholar]
- 32. Rossi A. M., and Taylor C. W. (2011) Analysis of protein-ligand interactions by fluorescence polarization. Nat. Protoc. 6, 365–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Han Y., Lyman K., Clutter M., Schiltz G. E., Ismail Q. A., Prados D. B., Luan C. H., and Chetkovich D. M. (2015) Identification of small-molecule inhibitors of hyperpolarization-activated cyclic nucleotide–gated channels. J. Biomol. Screen 20, 1124–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zagotta W. N., Olivier N. B., Black K. D., Young E. C., Olson R., and Gouaux E. (2003) Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature 11, 200–205 [DOI] [PubMed] [Google Scholar]
- 35. Piskorowski R., Santoro B., and Siegelbaum S. A. (2011) TRIP8b splice forms act in concert to regulate the localization and expression of HCN1 channels in CA1 pyramidal neurons. Neuron 70, 495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lanyon-Hogg T., Hooper J., Gunn S., Warriner S. L., and Baker A. (2014) PEX14 binding to Arabidopsis PEX5 has differential effects on PTS1 and PTS2 cargo occupancy of the receptor. FEBS Lett. 588, 2223–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Freitas M. O., Francisco T., Rodrigues T. A., Alencastre I. S., Pinto M. P., Grou C. P., Carvalho A. F., Fransen M., Sá-Miranda C., and Azevedo J. E. (2011) PEX5 protein binds monomeric catalase blocking its tetramerization and releases it upon binding the N-terminal domain of PEX14. J. Biol. Chem. 286, 40509–40519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fodor K., Wolf J., Erdmann R., Schliebs W., and Wilmanns M. (2012) Molecular requirements for peroxisomal targeting of alanine-glyoxylate aminotransferase as an essential determinant in primary hyperoxaluria type 1. PLoS Biol. 10, e1001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fodor K., Wolf J., Reglinski K., Passon D. M., Lou Y., Schliebs W., Erdmann R., and Wilmanns M. (2015) Ligand-induced compaction of the PEX5 receptor-binding cavity impacts protein import efficiency into peroxisomes. Traffic 16, 85–98 [DOI] [PubMed] [Google Scholar]
- 40. Amery L., Sano H., Mannaerts G. P., Snider J., Van Looy J., Fransen M., and Van Veldhoven P. P. (2001) Identification of PEX5p-related novel peroxisome-targeting signal 1 (PTS1)-binding proteins in mammals. Biochem. J. 357, 635–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Effelsberg D., Cruz-Zaragoza L. D., Schliebs W., and Erdmann R. (2016) Pex9p is a novel yeast peroxisomal import receptor for PTS1-proteins. J. Cell Sci. 129, 4057–4066 [DOI] [PubMed] [Google Scholar]
- 42. Yifrach E., Chuartzman S. G., Dahan N., Maskit S., Zada L., Weill U., Yofe I., Olender T., Schuldiner M., and Zalckvar E. (2016) Characterization of proteome dynamics in oleate reveals a novel peroxisome targeting receptor. J. Cell Sci. 129, 4067–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Han Y., Lyman K. A., Clutter M., Schiltz G. E., Ismail Q.-A., Cheng X., Luan C.-H., and Chetkovich D. M. (2016) Method for identifying small molecule inhibitors of the protein-protein interaction between HCN1 and TRIP8b. J. Vis. Exp. e54540–e54540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lyman K. A., Han Y., and Chetkovich D. M. (2017) Animal models suggest the TRIP8b-HCN interaction is a therapeutic target for major depressive disorder. Expert. Opin. Ther. Targets 21, 235–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shin M., Simkin D., Suyeoka G. M., and Chetkovich D. M. (2006) Evaluation of HCN2 abnormalities as a cause of juvenile audiogenic seizures in Black Swiss mice. Brain Res. 1083, 14–20 [DOI] [PubMed] [Google Scholar]
- 46. Shin M., and Chetkovich D. M. (2007) Activity-dependent regulation of h channel distribution in hippocampal CA1 pyramidal neurons. J. Biol. Chem. 282, 33168–33180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chung W. K., Shin M., Jaramillo T. C., Leibel R. L., LeDuc C. A., Fischer S. G., Tzilianos E., Gheith A. A., Lewis A. S., and Chetkovich D. M. (2009) Absence epilepsy in apathetic, a spontaneous mutant mouse lacking the H channel subunit, HCN2. Neurobiol. Dis. 33, 499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lolicato M., Nardini M., Gazzarrini S., Möller S., Bertinetti D., Herberg F. W., Bolognesi M., Martin H., Fasolini M., Bertrand J. A., Arrigoni C., Thiel G., and Moroni A. (2011) Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels. J. Biol. Chem. 286, 44811–44820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Goutelle S., Maurin M., Rougier F., Barbaut X., Bourguignon L., Ducher M., and Maire P. (2008) The Hill equation: a review of its capabilities in pharmacological modelling. Fundam. Clin. Pharmacol. 22, 633–648 [DOI] [PubMed] [Google Scholar]
- 50. Pollard T. D. (2010) A guide to simple and informative binding assays. Mol. Biol. Cell 21, 4061–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lewis A. S., Schwartz E., Chan C. S., Noam Y., Shin M., Wadman W. J., Surmeier D. J., Baram T. Z., Macdonald R. L., and Chetkovich D. M. (2009) Alternatively spliced isoforms of TRIP8b differentially control H channel trafficking and function. J. Neurosci. 29, 6250–6265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shi Y., Mowery R. A., Ashley J., Hentz M., Ramirez A. J., Bilgicer B., Slunt-Brown H., Borchelt D. R., and Shaw B. F. (2012) Abnormal SDS-PAGE migration of cytosolic proteins can identify domains and mechanisms that control surfactant binding. Protein Sci. 21, 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.