Abstract
Wall teichoic acids (WTAs) are the most abundant glycopolymers found on the cell wall of many Gram-positive bacteria, whose diverse surface structures play key roles in multiple biological processes. Despite recent technological advances in glycan analysis, structural elucidation of WTAs remains challenging due to their complex nature. Here, we employed a combination of ultra-performance liquid chromatography-coupled electrospray ionization tandem-MS/MS and NMR to determine the structural complexity of WTAs from Listeria species. We unveiled more than 10 different types of WTA polymers that vary in their linkage and repeating units. Disparity in GlcNAc to ribitol connectivity, as well as variable O-acetylation and glycosylation of GlcNAc contribute to the structural diversity of WTAs. Notably, SPR analysis indicated that constitution of WTA determines the recognition by bacteriophage endolysins. Collectively, these findings provide detailed insight into Listeria cell wall-associated carbohydrates, and will guide further studies on the structure-function relationship of WTAs.
Keywords: carbohydrate structure, carbohydrate-binding protein, high-performance liquid chromatography (HPLC), nuclear magnetic resonance (NMR), surface plasmon resonance (SPR), teichoic acid, Listeria, bacteriophage derived endolysin
Introduction
The genus Listeria consists of Gram-positive bacteria that are currently classified into 17 species, based on their genomic and phenotypic characteristics (1). These species harbor group-specific epitopes, defined by the somatic (O) and flagellar (H) antigens that form the key determinants of serological typing. To date, at least 12 distinct serovars (i.e. 1/2a, 1/2b, 1/2c, 3a, 3b, 3c, 4a, 4b, 4c, 4d, 4e, and 7) have been identified in Listeria monocytogenes, one in Listeria ivanovii (5), and two in Listeria innocua and Listeria welshimeri (6a and 6b) (2). Although L. ivanovii (Liv) is recognized as an animal pathogen, L. monocytogenes (Lmo) is an opportunistic, foodborne human pathogen, which can cause fatal infection in the elderly, newborns, pregnant women, and immunocompromised patients. Lmo serovar patterns are correlated with virulence, as serovars (SVs)4 1/2a, 1/2b, and 4b are responsible for most documented cases of human listeriosis, whereas other SVs typically do not cause the disease (3).
Phenotypic characterization techniques such as serotyping and phagetyping have previously been used for subtyping of Listeria strains (4). However, results are not always reliable due to nonspecific cross-reactivity of the antisera used for typing, and a lack of detailed understanding of the Listeria surface chemistry, especially cell wall-associated carbohydrates (5). The occurrence of various serovars suggests a high degree of cell-wall structural diversity within the Listeria genus. It has been previously established that the major O-antigen determinants in Listeria, whose binding patterns are what determine serovar along with the H-antigens, are cell wall-anchored anionic glycopolymers, termed wall teichoic acids (WTAs) (6). These complex carbohydrates account for up to 60% of the dry weight of the cell wall, and are covalently conjugated to and extended beyond the peptidoglycan (PG) layer (7). Because WTAs are major antigenic determinants, each serovar of Listeria should possess a unique WTA structure. In addition, WTAs are known to mediate multiple other biological roles, such as physiology, immunological recognition, antibiotic resistance, virulence, and interaction with host cells and bacteriophages (6, 8–12).
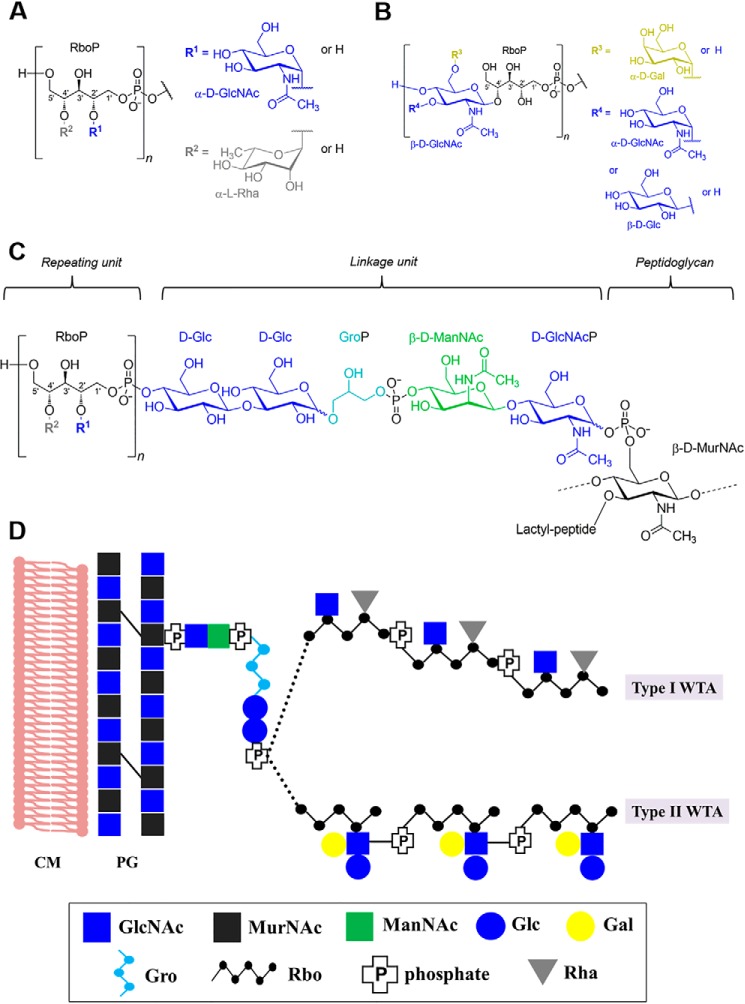
Recent technological advances in glycan analysis have resulted in a somewhat better understanding of the structure and function of bacterial WTAs (13–17). Listeria WTAs are comprised of a polymeric ribitol (Rbo) chain that consists of 20 to 30 repeating units, and a conserved linkage unit that connects the chain to N-acetylmuramic acid (MurNAc) of the PG via a phosphodiester bond. The repeating units are highly variable due to modifications by a variety of glycosidic substitutions (18). Two main structural types exist: type I (Fig. 1A) WTAs adopt [5)-Rbo-1-P-(O→]n chains where the Rbo units are directly interconnected by phosphodiester bonds between C1 and C5, and can be decorated with terminal rhamnose (Rha) or GlcNAc. Type II WTAs bear [4)-GlcNAc-(β1→2/4)-Rbo-1-P-(O→]n (Fig. 1B: only C4 connection to Rbo shown) as the repeating unit, which incorporate the GlcNAc moiety within the polymer chain. The integrated GlcNAc itself may be further decorated with glucose (Glc), galactose (Gal), or with a second GlcNAc residue (19). In contrast, little is known about the linkage unit. To date, only one strain (belonging to type I) has been studied, and it features [4)-Glc-(β1→3)-Glc-(β1→1)-Gro-3-P-(O→4)-ManNAc-(β1→4)-GlcNAc-1-P-(O→6)-MurNAc] as the linkage unit (Fig. 1C) (20). The potentially different linkage unit structures from other Listeria serovars remain unsolved. In addition, several structures and modifications from WTA repeating units of the less frequently occurring serovars within the genus Listeria have not been established. As the correlation between WTAs and serovar is clear and their important biological functions are beginning to emerge (5), a comprehensive view of the diverse WTA structures will further strengthen our knowledge of the Listeria cell wall, beyond what has been defined by serotyping alone.
Figure 1.
A chemical view of two types of Listeria cell wall teichoic acids. A, structure of the type I repeating unit. B, structure of the type II repeating unit. C, structure of the repeating unit, linkage unit, and peptidoglycan in the cell wall of L. monocytogenes serovar 1/2a strain EGD (modified from Kaya et al. (20). R1, GlcNAc; R2, rhamnose. D, a schematic representation of two types of WTAs: serovar 1/2a and serovar 4b were used for illustration of type I and II, respectively. Abbreviations: GlcNAc, N-acetylglucosamine; Rha, rhamnose; Gro, glycerol; Glc, glucose; Gal, galactose; ManNAc, N-acetylmannosamine; MurNAc, N-acetylmuramine.
Within a given serovar, WTA polymers occur as heterogeneous mixtures with various carbohydrate decorations and linkages. The method of choice for WTA structural elucidation is nuclear magnetic resonance (NMR) spectroscopy (13, 18). However, sensitivity is still a hallmark problem for NMR analyses. Recent advances in mass spectrometry (MS)-based approaches also allow for structural elucidation of complex carbohydrates, whereas maintaining a high level of sensitivity, accuracy, acquisition speed, and tolerance for heterogeneous analytes (21). We previously reported the application of electrospray ionization MS for rapid analysis of carbohydrate compositions of Listeria WTAs (22). However, this method is unable to discriminate between diastereoisomeric monosaccharides (e.g. galactose versus glucose substitution), which commonly exist in type II WTAs. Toward this end, we have developed a novel hydrophilic interaction ultra-performance liquid chromatography-coupled electrospray ionization tandem mass spectrometry (UPLC-MS/MS) method, for detailed analysis of WTA structures. Given that certain serovars are responsible for most cases of human listeriosis, the structures we elucidate here are likely to represent secondary pathogenicity factors, rendering them not only interesting subjects for biochemical studies, but also exposing them as possible targets for vaccines, diagnostics, and virulence-related antimicrobial therapy (23). In this light, we also investigate the binding specificity of cell wall-binding domains (CBDs) from Listeria bacteriophage endolysins. As these molecules can recognize and bind to the Listeria cell wall in a serovar-dependent manner (24, 25), and have been proposed as novel tools for diagnostics and biocontrol (26, 27), we evaluated the specificity and binding kinetics of certain CBDs for Listeria WTAs.
Results
Validating UPLC-MS/MS for structural determination of type I Listeria WTAs
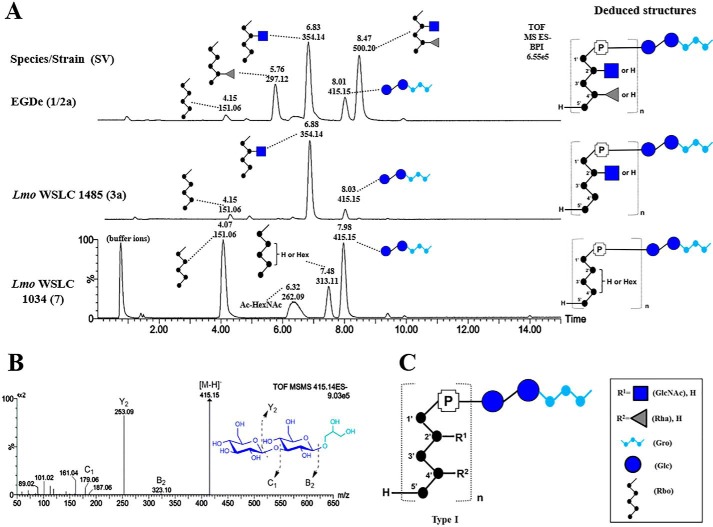
First, purified (supplemental Fig. S1, A and B) and HF-depolymerized (supplemental Fig. S1C) type I WTAs (including SV 1/2a, 3, and 7) were analyzed to validate our methodology, as their structures are already known from previous NMR studies (18). We identified repeating units with their individual carbohydrate constituents based on retention time and mass detection of each peak (Fig. 2A). In SV 1/2a, m/z 151 (4.15 min), m/z 297 (5.76 min), and m/z 500 (8.47 min) correspond to Rbo, Rha-Rbo, and [GlcNAc][Rha]-Rbo, respectively, and m/z 354 (6.83 min) is indicative of a GlcNAc(1→2)-Rbo. In addition, a significant peak of m/z 415 (8.01 min) indicates a species that consists of glucose-glucose-Gro, in agreement with a previous report (20). Fragmentation (MS/MS) of m/z 415 identifies the Glc(1→3)Glc connectivity due to the lack of cross-ring fragments (Fig. 2B). Interestingly, we found this linkage also exists in the infrequent serovars 3a and 7, representing a unique marker for type I WTAs (Fig. 2C). The other portion of the linkage unit, ManNAc-GlcNAc, could not be detected in the chromatogram, likely due to the acid treatment during WTA extractions from the cell wall. Nevertheless, we do not anticipate any change as this disaccharide unit is known to be highly conserved within the Listeria genus (5). In SV 3a, a dominant peak m/z 354 (6.88 min) reveals GlcNAc-Rbo as the major component of the repeating unit (Fig. 2A). This composition was confirmed by NMR spectroscopic analysis of the WTA polymer prior to depolymerization. Its 1H spectrum is shown in supplemental Fig. S2A. Unexpectedly in SV 7, a small percentage of Rbo (about 8%) was found to be substituted with a hexose residue, corresponding to the peak m/z 313. The peak m/z 262 suggests a species of hexosamine with two acetyl groups. However, these two species could not be detected by NMR analysis of corresponding WTA polymers (supplemental Fig. S2B).
Figure 2.
Structure and UPLC-MS/MS analysis of type I Listeria WTAs. A, liquid chromatographic separation and MS-based identification of carbohydrate residues within type I Listeria WTAs from Lmo 1/2a (top); Lmo 3a; Lmo 7 (bottom). Peaks are labeled with their respective retention time (Rt; [min]) and base peak ion [M-H]− (m/z). The deduced structures of the respective WTA type are shown on the right in CFG representation (Consortium for Functional Glycomics; legend box on bottom right). B, structural analysis of m/z 415 by ESI-MS/MS. C, summary of type I WTAs. Abbreviations: GlcNAc, N-acetylglucosamine; Gro, glycerol; P, phosphate; Lmo, L. monocytogenes.
Structural analysis of type II WTAs from a Listeria SV 4b strain by UPLC-MS/MS
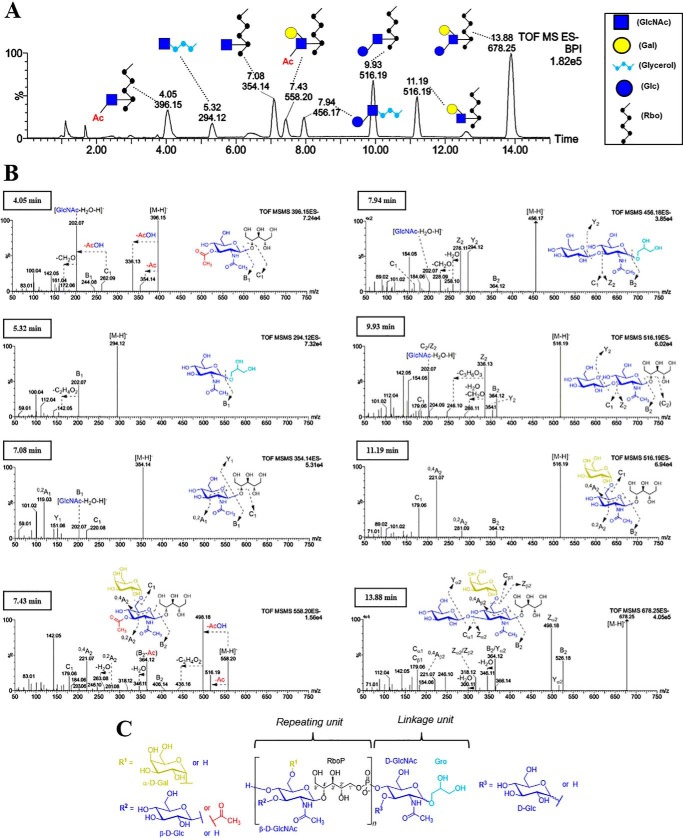
Next, we analyzed the WTAs from the laboratory serovar 4b strain WSLC 1042, the serovar representing the majority of listeriosis outbreaks. The chromatogram suggests that the WTA of 4b is comprised of 8 carbohydrate species with variable retention times that differ by mass (supplemental Fig. S3A) or structure (Fig. 3A). Because the NMR structure of 4b WTA has been previously determined (18), we were able to deduce the 8 individual species' compositions by mass comparison, as well as their monosaccharide connectivity by MS/MS (Fig. 3B). The first two species identified were m/z 354 (7.08 min) and m/z 678 (13.88 min), as their masses correspond to GlcNAc-Rbo and [Gal][Glc]-GlcNAc-Rbo, respectively. Two m/z 516 species were distinguished by their fragmentation patterns. The one eluting at 9.93 min suggests a hexose(1→3)GlcNAc linkage (diagnostic m/z 202 fragment) (28) in contrast to a (1→6)-linkage for the peak at 11.19 min (diagnostic m/z 281 and 221 fragments) (29). In agreement with previous reports, the above two species are denoted as Glc(1→3)GlcNAc-Rbo and Gal(1→6)GlcNAc-Rbo for the peaks at 9.93 and 11.19 min, respectively. In addition to the glycosylation, two O-acetylated species, m/z 396 (4.05 min) and m/z 558 (7.43 min) were detected. Further MS/MS fragmentation of m/z 396 revealed that the aminosugar bears two acetyl (Ac) groups, one on the nitrogen (N-acetyl) and one on an oxygen (O-acetyl). We observed a fragment with m/z 262 corresponding to Ac-GlcNAc, indicating the Ac group is located on the GlcNAc, and not on the Rbo. This is supported by the fragment with m/z 151 (Rbo), and lack of a signal with m/z 193 (Ac-Rbo). Hence, we infer m/z 396 to be Ac-GlcNAc-Rbo. MS/MS of m/z 558 shows a strong presence of the deacetylated fragment m/z 516 and its corresponding cross-ring fragments in line with the Gal(1→6)-GlcNAc-Rbo linkage, and is thus denoted as [Gal(1→6)][3-OAc]-GlcNAc-Rbo. We propose that the O-acetyl group is at C3 as it is the only open position on GlcNAc (C4 is occupied by the phosphodiester bond linking together the WTA polymer). Assuming a conserved O-acetyl position, the mentioned m/z 396 fragment is therefore proposed to be O-acetylated at C3 as well. By comparing the area under the peaks denoting the species with and without O-acetylation (supplemental Fig. S3B), the relative degree of secondary O-acetylation in 4b WTAs was determined to be about 26% (see “Experimental procedures” for Equation 1). In summary, the updated WTA structure of the linkage and repeating units of 4b is shown in Fig. 3C.
Figure 3.
Compositional and structural analysis of WTA from Lmo serovar 4b WSLC1042. A, liquid chromatographic separation (UPLC-MS) and identification of carbohydrate residues within type II WTAs from Lmo 4b (peaks labeled with retention time [min] and m/z). B, negative mode ESI-MS/MS of all detected carbohydrate-based species for structural elucidation. C, deduced structure of the repeating and the linkage unit in Lmo 4b WTAs. Various substitutions are depicted as different R groups.
Interestingly, a pair of glycerol (Gro)-related species, m/z 294 (5.32 min) and m/z 456 (7.94 min) were detected and identified as the linkage unit (Fig. 3A). MS/MS (Fig. 3B) shows that m/z 294 reflects a GlcNAc-Gro composition, whereas the fragmentation pattern of m/z 456 indicates a Glc(1→3)GlcNAc-Gro connectivity, representing a different type of linkage unit compared with type I WTAs.
Confirmation of connectivity and O-acetylation by NMR spectroscopy
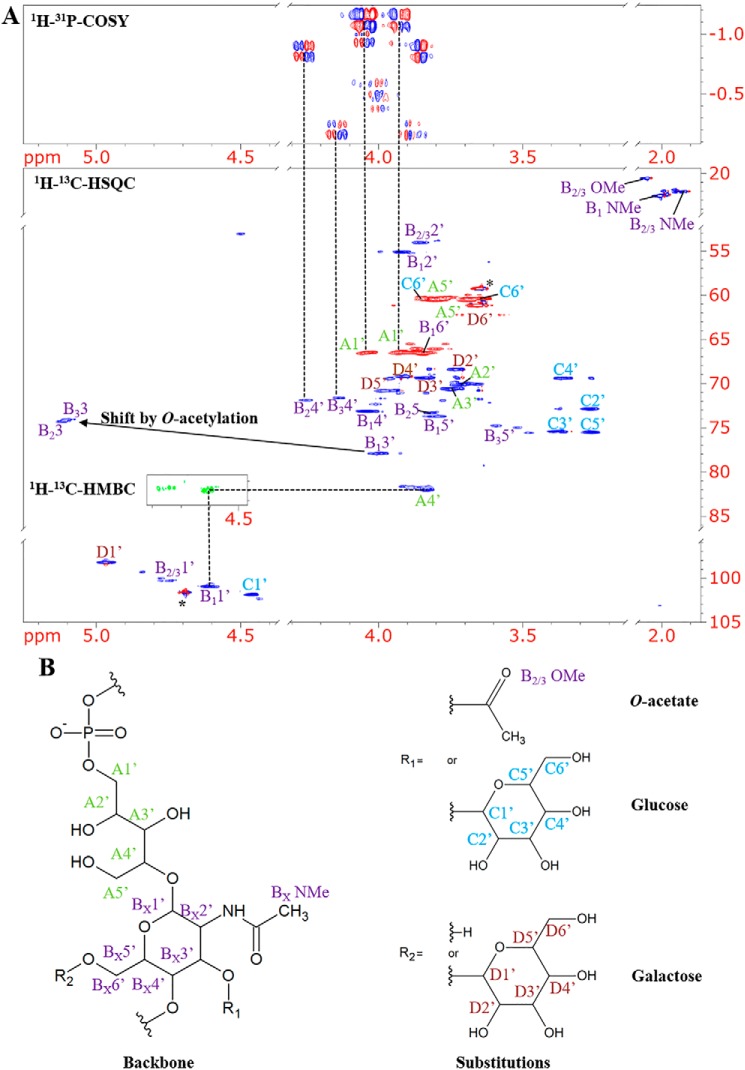
In a parallel approach, a series of NMR experiments including DQF-COSY, 13C-HSQC, 13C-HMBC, and 1H-31P-COSY (Fig. 4) were employed to determine the composition and connectivity of intact WTA polymers of SV 4b, and to rule out any possible confounding by-products resulting from HF digestion. The HP-COSY spectrum of the polymer is consistent with the existence of a phosphodiester linkage from C1′ of Rbo to C4′ of GlcNAc as determined previously for the monomeric units. Cross-peaks in the HSQC spectrum could be assigned to three differently decorated repeating units of the WTA sample (Table 1). H3′ in the variants with the O-acetylated GlcNAc shows a distinct downfield shift to 5.10 ppm. Integration of the respective peaks confirmed that the degree of O-acetylation is similar to what UPLC-MS demonstrated earlier. NMR experiments corroborate that WTA O-acetylation and glucosylation occur at the same C3-OH of GlcNAc and are mutually exclusive. The configuration of the anomeric protons was confirmed by analysis of 3JH1′-H2′ coupling constants (Table 2). For the β-configuration, a value around 8 Hz is expected, whereas for the α-configuration it is about 3 Hz. In agreement with previous reports (17, 18), the configuration of the anomeric center is β for GlcNAc and Glc and α for Gal. Due to the low abundance within the overall polymer it was not possible to identify signals of any Gro-related subunits.
Figure 4.
NMR spectra and structures of the repeating units of WTA from Lmo serovar 4b WSLC1042. A, details of the 1H-13C-HSQC and 1H-31P-COSY spectra. For the major monomeric species the spectra suggest a phosphodiester linkage between Rbo C1′ and GlcNAc C4′ (cf. dotted vertical lines). A small HMBC insert exemplifies the assignment of glycosidic bonds. The HP-COSY spectrum also shows two correlations to lower intensity proton signals with chemical shifts of 4.15 and 4.25 ppm. The corresponding cross-peaks in the HSQC spectrum were assigned to the 4′ positions of 3′ O-acetylated GlcNAc in two minor variants of the monomeric subunit. The GlcNAc 3′ protons in these O-acetylated variants show a significant downfield shift to about 5.1 ppm. The biggest difference in chemical shift between the two minor sets of GlcNAc signals is observed for the 5′ position. This is in agreement with the expected lack of galactose in one of the variants. The asterisk indicates an artifact at the position of the 1H resonance of the residual water signal. B, structures and atom designations of WTA monomeric units (cf. Table 2). A, Rbo (green); B, GlcNAc (purple); C, Glc (blue); D, Gal (red).
Table 1.
1H, 13C, and 31P chemical shifts in Lmo WSLC 1042 WTA in D2O (in ppm)
Assignment of the two minor sets of GlcNAc signals to B2 and B3, respectively, is based on the position of the 5′ cross-peak in the HSQC spectrum, which is expected to be significantly shifted in the absence of Gal. The abbreviations used are: GlcNAc, N-acetylglucosamine; OAc, O-acetyl group; Glc, glucose.
| Unit | 1 | 2 | 3 | 4 | 5 | 6 | NAc | OAc | 31P | |
|---|---|---|---|---|---|---|---|---|---|---|
| Rbo (A) | 1H | 4.03/3.90 | 3.70 | 3.74 | 3.82 | 3.78/3.69 | NAa | NA | NA | |
| 13C | 66.5 | 70.2 | 70.6 | 82.1 | 60.6 | NA | NA | NA | ||
| GlcNAc-3-Glc-4-P-6-Gal (B1) | 1H | 4.74 | 3.92 | 3.99 | 4.04 | 3.97 | 3.84 | 2.00 | −1.1 | |
| 13C | 100.3 | 55.1 | 77.9 | 73.1 | 73.7 | 66.6 | 22.5 | |||
| GlcNAc-3-OAc-4-P-6-Gal (B2) | 1H | 4.73 | 3.85 | 5.10 | 4.25 | 3.82 | NDb | 1.93. | 2.04 | −0.9 |
| 13C | 100.3 | 54.1 | 74.3 | 71.9 | 73.3 | ND | 22.1 | 20.5 | ||
| GlcNAc-3-OAc-4-P (B3) | 1H | 4.73 | 3.85 | 5.09 | 4.12 | 3.59 | ND | 1.93 | 2.04 | −0.2 |
| 13C | 100.3 | 54.1 | 74.1 | 71.6 | 74.8 | ND | 22.1 | 20.5 | ||
| Glc (C) | 1H | 4.46 | 3.26 | 3.36 | 3.36 | 3.26 | 3.82/3.63 | NA | NA | |
| 13C | 101.9 | 72.9 | 75.4 | 69.4 | 75.5 | 60.4 | NA | NA | ||
| Gal (D) | 1H | 4.97 | 3.72 | 3.83 | 3.90 | 3.94 | 3.65 | NA | NA | |
| 13C | 98.3 | 68.4 | 69.4 | 69.2 | 70.8 | 71.2 | NA | NA |
a NA, not applicable.
b ND, not detected.
Table 2.
3JH1′-H2′ coupling constants of WTA polymers from selected strains for determination of anomeric configuration
| Strain (WSLC) | 1042 (4b) | 1020 (4a) | 2012 (6b) | 3009 (5) | 1485 (3a) | |
|---|---|---|---|---|---|---|
| GlcNAc | JHH [Hz] | 8.0 | 8.3 | 8.0 | 7.9 | 3.3 |
| Glc | JHH [Hz] | 7.6 | NAa | 7.4 | 7.2 | NA |
| Gal | JHH [Hz] | 3.7 | NA | NA | NA | NA |
a NA, not applicable.
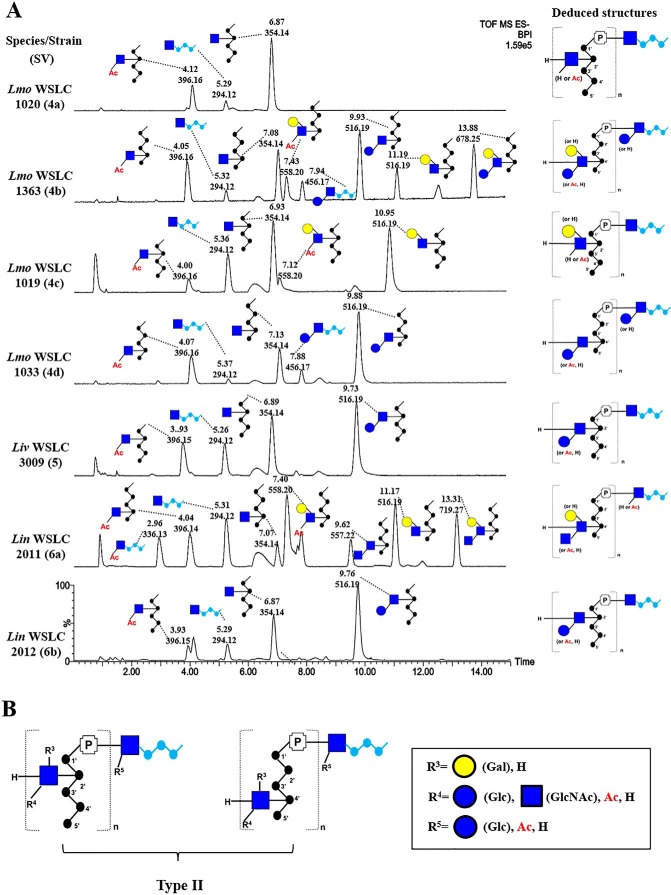
Structural diversity and distinct modifications of type II Listeria WTAs
Various type I and II WTA polymers were analyzed by native PAGE (separates by charge, size, and shape), followed by Alcian blue staining (supplemental Fig. S4). Our results demonstrate that there are clear differences in mass of the WTA polymers among different SVs, likely due to differences in decoration, but also overall polymer chain length (or number of repeating units n), for which there is some variation among different strains (18). Using the chromatogram of 4b WTAs as a reference, we next sought to investigate the correlation between the WTA structure and serovar (Fig. 5A). The chromatogram of 4a reveals a peak with m/z 354 (6.87 min) that elutes differently from 4b (7.08 min), suggesting two diastereoisomers. Despite the presence of two stereogenic centers, ribitol itself is an achiral meso compound due to the internal plane of symmetry through C3. However, substitution (at C2 or C4) breaks this symmetry, resulting in a diastereomorphic relationship between the m/z 354 species in 4a and 4b. In UPLC-MS analysis, the different arrangement of the two disaccharides results in a minor shift (∼12 s) in retention time. In agreement with previous work, NMR analysis (supplemental Fig. S5A) confirmed that the difference lies in the connectivity between GlcNAc and Rbo: GlcNAc(β1→2)-Rbo in 4a and GlcNAc(β1→4)-Rbo in 4b, whereas the linkage between GlcNAc and the phosphate stays the same for both serovars. The other two dominant peaks m/z 396 (4.12 min) and m/z 294 (5.29 min) were assigned as Ac-GlcNAc-Rbo and GlcNAc-Gro, respectively. Hence, the comprehensive WTA structure of the 4a repeating unit is denoted as: [4)-[3-O-Ac]-d-GlcNAc-(β1→2)-d-Rbo-1-P-(O→]n; the linkage unit as GlcNAc-(1→3)-Gro-P.
Figure 5.
A structural summary of type II WTAs isolated from various Listeria serovars. A, comparison of type II UPLC-MS base peak chromatograms of Lmo 4a (top); Lmo 4b; Lmo 4c; Lmo 4d; Liv 5; Lin 6a; Lin 6b (bottom; peaks labeled with retention time [min] and m/z). B, bottom-up summary of diverse type II WTA structures. Abbreviations: P, phosphate; Lmo, L. monocytogenes; Liv, L. ivanovii; Lin, L. innocua.
In Fig. 5A, analysis of the WTA from another SV 4b strain named 1363 revealed a similar chromatographic pattern, with a slightly different degree of O-acetylation (Table 3). SV 4c shows an m/z 354 peak at 6.93 min, suggesting the presence of GlcNAc(β1→2)-Rbo. Significant differences in retention times are visible for m/z 558 (7.12 versus 7.48 min in 4b) and m/z 516 (10.95 versus 11.19 min in 4b). The structure of 4c WTA is thus deduced as [4)-[Gal-(α1→6)][3-O-Ac]-GlcNAc-(β1→2)-Rbo-1-P-(O→]n, with a linkage unit as GlcNAc(1→3)-Gro. Both SV 4d and 4e adopt the same WTA structure as 4b, except the lack of a Gal substitution on GlcNAc.
Table 3.
Summary of composition and constitution of WTAs from different Listeria species and serovars
The abbreviations used are: GlcNAc, N-acetylglucosamine; Rha, rhamnose; Rbo, ribitol; OAc, O-acetyl group; Gro, glycerol; Glc, glucose; Gal, galactose.
| Species | Strain (WSLC) | Source | SV | WTA decorationa | GlcNAc substitution at Rbo | Degree of O-acetylation of GlcNAc | Linkage unit |
|---|---|---|---|---|---|---|---|
| % | |||||||
| L. monocytogenes | EGDe | ATCC BAA-679 | 1/2a | Pendant GlcNAc/Rha, Rbo | C2 | 0 | Glc-Glc-glycerol |
| L. monocytogenes | 10403S | D. Portnoy | 1/2a | Pendant GlcNAc/Rha, Rbo | C2 | 0 | Glc-Glc-glycerol |
| L. monocytogenes | 1485 | Soft cheese | 3a | Pendant GlcNAc, Rbo | C2 | 0 | Glc-Glc-glycerol |
| L. monocytogenes | 1034 | SLCC2482 | 7 | Pendant hexose, Rbo | None | 0 | Glc-Glc-glycerol |
| L. monocytogenes | 1020 | ATCC19114 | 4a | Incorporated GlcNAc, Rbo | C2 | 25 | GlcNAc-glycerol |
| L. monocytogenes | 1042 | ATCC23074 | 4b | Incorporated GlcNAc, Gal, Glc, Rbo | C4 | 26 | Glc-GlcNAc-glycerol |
| L. monocytogenes | 1363 | This study | 4b | Incorporated GlcNAc, Gal, Glc, Rbo | C4 | 28 | Glc-GlcNAc-glycerol |
| L. monocytogenes | 1019 | ATCC19116 | 4c | Incorporated GlcNAc, Gal, Rbo, | C2 | 21 | GlcNAc-glycerol |
| L. monocytogenes | 1033 | ATCC19117 | 4d | Incorporated GlcNAc, Glc, Rbo | C4 | 26 | Glc-GlcNAc-glycerol |
| L. ivanovii | 3009 | SLCC 4769 | 5 | Incorporated GlcNAc, Glc, Rbo | C2 | 15 | GlcNAc-glycerol |
| L. innocua | 2011 | ATCC33090 | 6a | Incorporated GlcNAc, GlcNAc, Gal, Rbo | C4 | 48 | OAc-GlcNAc-glycerol |
| L. innocua | 2012 | ATCC33091 | 6b | Incorporated GlcNAc, Glc, Rbo | C2 | 14 | GlcNAc-glycerol |
a a, the incorporated GlcNAc refers to a GlcNAc residue that is integrated into polyribitol-phosphate while forming the repeating unit and b, pendant GlcNAc refers to a GlcNAc residue without any substitution.
The complete WTA structures of SV 5 and 6b were determined for the first time. Their chromatograms (Fig. 5A) and NMR spectra (supplemental Fig. S5, B and C) are almost identical and are indicative of a composition similar to 4d, but with a C2-connectivity like in 4a, suggesting that other determinants differentiate SV 5 and 6b, 4d and 4e. SV 6a was found to possess three new peaks with m/z 336 (9.62 min), m/z 557 (9.62 min), and m/z 719 (13.31 min). MS/MS analysis (supplemental Fig. S6) revealed that m/z 336 corresponds to Ac-GlcNAc-Gro, suggesting a novel modification of the linkage unit. Fragmentation of m/z 557 demonstrated a GlcNAc-(1→3)-GlcNAc linkage, and fragmentation of m/z 719 shows a pattern similar to the MS/MS of m/z 516 with Gal, suggesting that m/z 719 bears (1→6)-linked Gal. Notably, all Listeria type II strains (SVs 4, 5, and 6) appear to feature O-acetylation as a unique modification in their WTAs. A summary of the diverse type II WTA structures is presented in Fig. 5B and Table 3.
The structure of Listeria WTAs determines the recognition specificity of bacteriophage endolysins
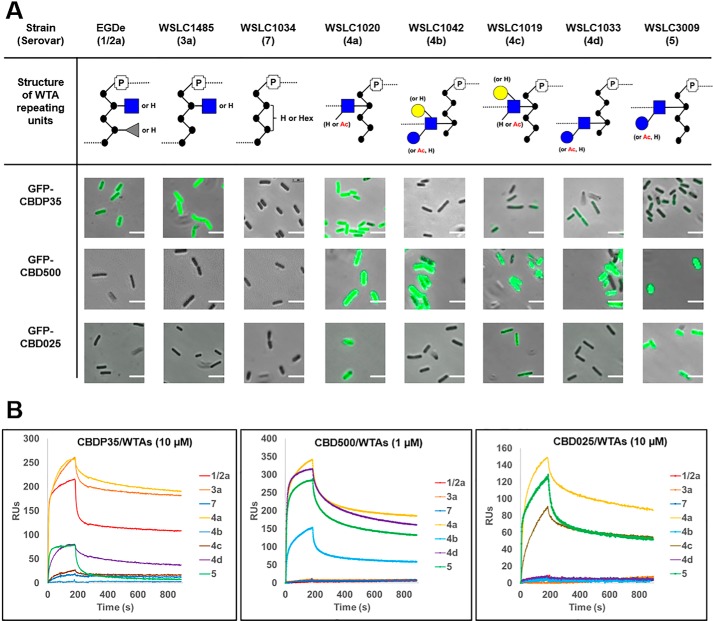
The structure of Listeria WTAs are known to be recognized as binding ligands by bacteriophage tail fiber proteins and the encoded endolysins (11) (30). Endolysins are peptidoglycan hydrolases that function to degrade the bacterial cell wall from within, to release progeny virions at the end of the phage lytic cycle. As bacteriolytic enzymes, they can also lyse susceptible Gram-positive bacteria from the outside. These enzymes are typically composed of an N-terminal catalytic domain responsible for proteolytic cleavage of the PG, and a C-terminal CBD that confers target specificity by recognizing and binding to diverse carbohydrate epitopes on the bacterial cell wall, such as WTAs (31). To elucidate the structural specificity of CBDs for WTA, the binding interactions of three fluorescently-labeled Listeria-specific endolysin CBDs (CBDP35, CBD500, and CBD025) were investigated due to the correlation of their binding patterns with serovar (Fig. 6A). Quantitative surface plasmon resonance analysis demonstrated that CBDs indeed possess discriminative binding specificity for purified WTAs (Fig. 6B). CBDP35 strongly binds to WTA units substituted with an undecorated GlcNAc residue (SVs 1/2a, 3a, and 4a); and only weakly to the other subtypes of SV 4 and SV 5. In contrast, CBD500 recognizes WTAs equipped with O-acetylated GlcNAc (SVs 4 and 5) even when they are glycosylated by different monosaccharides such as glucose and/or galactose. CBD025 exclusively recognizes GlcNAc bound via the C2 position of the ribitol backbone (SV 4a, 4c, 5, and 6b), but not C4-bound GlcNAc, such as in SVs 4b, 4d, and 6a (data not shown). Furthermore, we demonstrated that CBDs interact with the WTA partner in a dose-dependent manner (supplemental Fig. S7), and found that the binding affinity generally lies in the submicromolar range (Table 4). In summary, we show that CBDs recognize the Listeria cell surface based on the specific constitution of their WTA ligands. Together, this demonstrates both the extreme sensitivity and specificity with which endolysins recognize and bind to their ligands, and promotes their use for rapidly probing select WTA structures.
Figure 6.
WTA structure determines the binding specificity of bacteriophage endolysins. A, comparing the binding patterns of GFP-tagged CBDs to Listeria strains with diverse WTA structures. The images are made by merging both phase-contrast and fluorescence channel in the same field. Scale bar is 2 μm. CFG representation of monosaccharides: blue ●, Glc; blue ■, GlcNAc; grey ▴, Rha; yellow ●, Gal. B, binding specificity of immobilized CBDP35 (left), CBD500 (middle) and CBD025 (right) to respective Listeria WTAs by BIAcore analysis. The serovars used are shown in the legend. RU, relative units.
Table 4.
Kinetic and affinity data for the binding of WTAs to immobilized CBDs
| A+B ⇆ AB |
A+B ⇆ AB* |
Equilibirum association constant KA | Equilibirum dissociation constant KD | x2 | |||
|---|---|---|---|---|---|---|---|
| Association rate constant ka1 (1/Ms) | Dissociation rate constant kd1 (1/s) | Forward rate constant ka2 (1/s) | Backward rate constant kd2 (1/s) | ||||
| m−1 | m | ||||||
| CBDP35/ WTAWSLC1485 | 4.77 × 103 | 0.0201 | 9.60 × 10−3 | 3.36 × 10−4 | 7.02 × 106 | 2.27 × 10−7 | 19.5 |
| CBD500/WTAWSLC1042 | 5.31 × 104 | 0.0141 | 3.59 × 10−3 | 8.75 × 10−3 | 1.92 × 107 | 5.21 × 10−8 | 2.64 |
| CBD025/WTAWSLC1020 | 2.15 × 103 | 0.0368 | 8.95 × 10−3 | 4.87 × 10−3 | 1.13 × 106 | 8.85 × 10−7 | 20.4 |
Discussion
The cell wall of most Gram-positive bacteria contains two major complex glycopolymers: peptidoglycan and teichoic acids (including both wall teichoic acids and lipoteichoic acids). A recent report has demonstrated the usefulness of ultra-performance liquid chromatography to uncover the significant complexity and variability in the composition and structure of peptidoglycan (32). To better understand the intricacies of the Gram-positive cell wall, we focus on the WTAs of Listeria, which are structurally heterogeneous, polydisperse, and highly negatively charged mixtures of glycan polymers. Similar to peptidoglycan, WTAs play important roles in maintaining the integrity of the bacterial cell, and contribute to various processes during growth and division. It has been recently suggested that the absence of WTA or loss of its compositional identity or integrity leads to attenuated virulence of Lmo (16, 17), demonstrating that virulence is also dependent upon maintenance of a specific WTA structure. This hypothesis is in congruence with the understanding that WTAs are the major determinants of the antigenic pattern known as serovar, a phenotypic identifier for strain diversity and an indicator of virulence. Given that subtle constitutional and configurational differences are known to produce molecules with very different biological activities, and that WTA repeating units can be glycosylated by different isomeric monosaccharides, full structural elucidation is warranted.
MS/MS has previously been used to discriminate between constitutional variants based on patterns of diagnostic fragment ions (33). Similarly, identification of isomeric monosaccharides and their mode of attachment has been traditionally determined by GC-MS, following methylation, hydrolysis, and derivatization of purified target molecules (34). However, this does not permit clarification of the sequence order, nor the linkage configuration of the monosaccharides within the oligosaccharide. To elucidate the WTA diversity within the genus Listeria, we describe a novel bioanalytical approach that allows for analysis of both compositional and structural features of WTA polymers. We show that UPLC-MS/MS can be employed with an accuracy on par with NMR, requiring lower sample amount and less time, whereas maintaining excellent reproducibility. Additionally, we have compiled a database (supplemental Table S1) with the masses of [M-H]− for all identified WTA fragments. This database can be further developed as a diagnostic toolbox for probing the surface chemistry of unknown Listeria strains, providing valuable information on serovar and associated pathogenicity or virulence of a given strain or isolate. This could be especially relevant, given that the most pathogenic serovars 1/2 and 4b harbor unique and characteristic WTA structures and glycosidic substitutions.
Previous efforts primarily focused on identification of the WTA repeating unit (18, 22). However, our findings described here also allow for elucidation of the linkage unit structure. For the first time, we demonstrate that two types of linkage units exist in Listeria WTA: Glc-Glc-Gro in type I (SVs 1/2, 3, and 7), and [Glc]xGlcNAc-Gro (x = 0 or 1) in type II (SVs 4, 5, and 6). In theory, these linkage units should be denoted as [Hex]xHexNAc-Gro (x = 0 or 1), because our UPLC-MS/MS could not discriminate between monosaccharide isomers. However, we are confident in our assumption because Glc-Gro has been reported to be the linkage unit of type I WTA (20), and GlcNAc is the sole constituent utilized within all type II WTAs. It would be unlikely (and inefficient) to utilize a different substrate (e.g. GalNAc or ManNAc) for WTA synthesis. Nonetheless, the presence of HexNAc (or GlcNAc as we speculate) represents a novel linkage type that significantly differs from the other type previously known. In addition, structural modification of the linkage unit (through glycosylation and acetylation) can further diversify the overall composition of the WTA molecule.
Our methodology also revealed that O-acetylation of the incorporated GlcNAc in the repeating unit is a unique modification of all type II WTAs (SVs 4, 5, and 6). Interestingly, O-acetylation, along with N-acetylation have been found to be involved in numerous biological functions in Gram-positive bacteria, including the ability to confer intrinsic resistance to lysozyme through O-acetylation of MurNAc in the peptidoglycan backbone (35). Additionally, O-acetylation of GlcNAc in PG of Lmo may confer resistance to antimicrobial compounds and limit innate immune responses (36). Although O-acetylation on MurNAc of PG is a widely known modification in Gram-positive bacteria, O-acetylation of WTAs has only recently been reported (17). Here, we were able to demonstrate that (i) this modification only occurs in type II WTAs, and (ii) that the modification is not ubiquitous, but only affects a fraction (20% to 40%) of the WTA repeating units. The two classes of Listeria WTAs also fundamentally differ in the anomeric configuration of their repeating GlcNAc unit. The UPLC-MS/MS method is unable to discern the configuration of the GlcNAc-Rbo linkage, as both m/z 354 in 3a (shown as α-GlcNAc: Fig. 2A) and m/z 354 in 4a elute at the same retention time (∼6.88 min: Fig. 5A). However, NMR analysis clearly revealed that the configuration of GlcNAc differs between two types, e.g. pendant α-GlcNAc in type I and backbone-integrated β-GlcNAc in type II.
Despite existing knowledge on the genetic background of WTA biosynthesis in Listeria (14, 17, 37, 38), very little is known about the mechanism of the biochemical synthesis pathway. Previous studies on Staphylococcus and Bacillus may be relevant, as these genera also utilize polyribitolphosphate as the WTA repeating unit. However, their modification patterns are different, with Staphylococcus featuring α-GlcNAc, β-GlcNAc, and d-alanine (8), and Bacillus containing α-glucose and d-alanine (39). Based on the aforementioned data, Listeria appears to possess specific enzymes to recognize (α- or β-linked) GlcNAc to produce the corresponding type of WTAs, which might be functionally similar to the glycosyltransferase TarM of Staphylococus (40). Given the extreme structural diversity of WTAs, even within a single bacterial species, understanding the intricacies of the biochemical synthesis will certainly aid in understanding differences in structure, and their biological roles. On this note, we have here determined the WTA structures of SV 4c, 5, 6a, and 6b, whose WTA compositions and constitutions have not been previously described. Interestingly, we find that 4c shares the same repeating unit structure as a previously reported 4-“non-b” strain (17), suggesting that they probably represent the same serovar. We also reveal that SV 7 appears to possess a unique hexose substitution, unlike the GlcNAc residue in all type I and II WTAs. Overall, this new profile of WTA structures expands our knowledge about the diversity of Listeria WTAs, and further strengthens the already established theory that WTA structure is the basis for O-antigen diversity. Further research will focus on elucidating the biochemical pathways involved in synthesis of Listeria WTAs, to better understand the source of this structural diversity.
For the first time, we report the binding specificity and affinity of Listeria endolysins to purified WTA polymers, which have previously been shown to have a nanomolar affinity only to whole cells (25). We found that the interaction between CBD and WTA is purely dependent upon electrostatic forces, as 1 m NaCl alone was able to regenerate the functionalized SPR chip surface after each measurement. Taken together, these data provide key information for further understanding the molecular mechanisms underlying the WTA–endolysin interactions, supporting the potential application of these CBDs for detection and characterization of Listeria.
Experimental procedures
Bacterial strains and growth conditions
Listeria strains were routinely grown in brain heart infusion medium, with constant shaking at 30 °C. All strains used in this study are listed in Table 3.
Extraction and purification of Listeria cell walls and WTAs
Cell walls from Listeria strains were prepared for extraction and purification of WTAs as previously described (24). Purified WTA polymers were depolymerized into monomeric repeating units by hydrolysis of the phosphodiester bonds using 48% hydrofluoric acid for 20 h at 0 °C (7). The degraded products were subject to a Superdex 200 size exclusion column (GE Healthcare, Glattbrugg, Switzerland) in distilled water at 25 °C (flow rate, 0.4 ml/min). Corresponding fractions (smaller than 1 kDa) were identified by a UV detector at 205 nm, collected, and pooled for dialysis (MWCO 100–500 Da, Spectra laboratories, Inc.) in distilled water. The WTA monomers were then lyophilized and subjected to UPLC-MS/MS for compositional and structural analysis.
UPLC-MS/MS
A Waters Acquity UPLC system equipped with an Acquity UPLC BEH Amide column (2.1 × 100 mm, 1.7 μm) was used, coupled to a Synapt G2 MS system composed of an electrospray ionization (ESI) source and a quadrupole time-of-flight (qTOF) analyzer (Waters Corp., Milford, MA). The MS was used in the negative mode exclusively, and calibrated with a sodium formate solution with leucine-enkephalin (m/z 554.2615) as the lock mass, which was acquired every 60 s during UPLC-MS measurements (correction applied with 5 scans averaged).
Chromatographic conditions
ACN and H2O were used as eluents with a flow rate of 0.17 ml/min, both with 0.1% NH3 additive. To avoid strong solvent effects during injection, mixtures of ACN/H2O, 8:2 and 2:8 (v/v), were used as weak and strong needle wash solutions, respectively. The dialyzed WTA monomer materials (see above) were dissolved in 50 μl of ACN/H2O, 1:1 (v/v), and 0.1–1 μl were injected for the UPLC-MS analysis (partial loop mode; column temperature 35 °C). The gradient began with 20% H2O and 80% ACN, and was linearly increased to 40% H2O over 18 min, followed by a 2-min re-equilibration at the initial conditions.
MS conditions
The spectra were acquired in resolution mode. The voltages of capillary, sample cone, and extraction cone were set at 2000, 25, and 4 V, respectively. The desolvation gas flow rate was 850 liters/h at 350 °C. The cone gas flow was 20 liters/h and the source temperature 120 °C. Full scan mass spectra were acquired from m/z 50 to 1200 with a 0.3-s scan time in centroid mode. Besides the full scan, exact masses were selected to perform MS/MS through collision-induced dissociation. Argon was used as the collision gas, the low mass resolution was set to 15 for a very narrow m/z isolation width of the selected ions, and the transfer collision energies were ramps ranging from 5–15 to 10–30 V for molecules with m/z around 300 (lower end) and 700 (higher end). All data were collected and processed using MassLynx software, version 4.1 (Waters Corp., Milford, MA) and MS spectra were background-corrected by subtracting the signals between 0 and 1 min of their respective chromatograms. Retention time shifts due to column aging were corrected in the base peak ion chromatograms on the basis of the major peaks from a WTA 1042 (4b) monomer-sample, which was always included in each sequence batch as a positive control (stored at −20 °C between batches; samples analyzed over a time course of several months).
Integration
For extracted ion chromatograms of specific analytes, the corresponding exact masses were selected with a 0.05 Da window. Integration was conducted with enabled smoothing (window size (scans): ± 4; number of smooths: 2; Savitzky Golay smoothing method) and automatic peak detection parameters. The relative degree of O-acetylation was determined by the ratio in Equation 1.
| (Eq. 1) |
For example, in the WTA of 1042, the numerator contains the O-acetylated peaks m/z 396 and 558; the denominator contains peaks m/z 396, 558, 354, 516 (two peaks), and 678, which correspond to all identified repeating units in the polymer, both with and without O-acetylation. Potential differences in ionization efficiency of the involved species were ignored for this rough estimation. A dilution series of sample WTA 1042 (4b) confirmed that the relative signal ratios remained constant in the range of relevant signal abundances that were observed for the major species in the various samples (ratios stable up to a signal height of ∼3e5 for the major species, or an area of ∼60,000 (arbitrary units); injection volumes lowered if value exceeded).
NMR spectroscopy
The WTA polymers of WSLC1034, 1485, 1042, 1020, 3009, and 2012 were analyzed by NMR spectroscopy for detailed structural elucidation (41). A series of experiments, including DQF-COSY, 1H-13C-HSQC, 1H-13C-HMBC, and a 1H-31P-COSY (42) were acquired in D2O at 25 °C. NMR spectra were recorded on a Bruker Avance III HD 600 MHz spectrometer equipped with a Prodigy triple-resonance probe and a Bruker Avance III HD 500MHz spectrometer equipped with a BBFO broadband probe.
Protein purification
Expression and purification of His-tagged CBDP35, CBD500, and CBD025 of respective phage endolysins fused to a green fluorescence protein (GFP) were carried out as previously described (25).
Fluorescence binding assay
The abilities of GFP-tagged CBDs to bind to Listeria cell surface were tested using fluorescence binding assay as previously reported (24). Briefly, Listeria cells from log phase cultures (A600 nm = ∼0.5) were harvested by centrifugation (10,000 × g, 1 min), and resuspended in 1/5 volume of PBS (pH 7.4). 100 μl of cells were incubated with 5 μl of 1 mg/ml of GFP-CBDs and incubated for 5 min at room temperature. The cells were spun down, washed three times, and finally resuspended in PBS buffer. The samples were then subjected to confocal laser scanning microscopy.
Surface plasmon resonance analysis
Sensorgrams of binding of purified Listeria WTA polymers to immobilized CBDs were obtained using surface plasmon resonance (Biacore X, GE Healthcare, Glattbrugg, Switzerland) as previously described (43, 44), with slight modifications. First, the carboxymethylated surface of a CMD500L chip (Xantec bioanalytics GmbH, Duesseldorf, Germany) was coated with 0.2 mg/ml of CBDP35 (or CBD500 or CBD025) at a flow rate of 5 μl/min; 10 mm sodium acetate, pH 5.0), using the amine coupling procedure according to the manufacturer's manual. To control nonspecific binding, a second flow cell was treated in the same manner, but without immobilization of CBD proteins. For specificity studies, the WTA samples (analyte) isolated from different Listeria strains were flowed through both cells in running buffer (10 mm BisTris, 100 mm NaCl, 3.4 mm EDTA, 0.005% Tween 20, pH 6) at 10 μl/min at 25 °C. For each strain, association was measured for 180 s and dissociation was measured for 720 s. After each injection, the surface was regenerated with a 45-s injection of regeneration buffer (1 m NaCl, 50 mm Tris, pH 9), at a flow rate of 10 μl/min. This cycle was repeated after each measurement. For kinetics studies, the interaction between CBDs and various WTA concentrations (ranging from 0 to 50000 nm of the respective WTAs) were performed at 10 μl/min at 25 °C. For each concentration, association was measured for 180 s, and dissociation was monitored for 720 s. The surface was then regenerated by regeneration buffer prior to the next measurement. For all curves, the “two-state confirmation change” model gave the best fit and was therefore used for calculation of KA and KD.
Native PAGE analysis
PAGE of WTAs was performed similarly to previously described protocols (17). Briefly, 10 μg of purified WTAs were mixed with sample buffer (40% glycerol, 62.5 mm Tris-HCl, pH 6.8, supplemented with 0.01% bromphenol blue), and then analyzed on 4–20% precast TBE gels (Bio-Rad). Gels were then developed in TBE running buffer using constant voltage (200 V) for 30 min, washed twice with distilled water, stained with 1% Alcian blue stain (0.1% Alcian Blue8GX, 5% acetic acid, 40% EtOH) for 2 h, followed by destaining (10% acetic acid, 40% EtOH) overnight. A DNA ladder (100 bp, Thermo Fisher Scientific) was used for verification of the migration of WTA polymers.
Author contributions
Conception and design, acquisition of data, analysis and interpretation of data, drafting or revising the article was performed by Y. S., S. B., E. S., and M. O. E., acquisition of data, analysis, and interpretation of data was performed by B. G., A. J. R., M. R. E., and conception and design, drafting, or revising the article was performed by L. F., L. N., and M. J. L.
Supplementary Material
Acknowledgments
We appreciate Dr. Mathias Schmelcher for helpful suggestions in the preparation of this manuscript. We thank Prof. Herbert Hächler at the Institute for Food Safety and Hygiene, University of Zurich, for Listeria serotyping service.
This work was supported in part by Swiss National Science Foundation Grant 310030_156947/1. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Table S1 and Figs. S1–S7.
- SV
- serovar
- WTA
- wall teichoic acid
- PG
- peptidoglycan
- Rbo
- ribitol
- UPLC
- ultra-performance liquid chromatography
- Ac
- acetyl
- HSQC
- heteronuclear single quantum coherence
- HMBC
- heteronucleear multiple bond correlation
- ESI
- electrospray ionization
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Orsi R. H., and Wiedmann M. (2016) Characteristics and distribution of Listeria spp., including Listeria species newly described because 2009. Appl. Microbiol. Biotechnol. 100, 5273–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gasanov U., Hughes D., and Hansbro P. M. (2005) Methods for the isolation and identification of Listeria spp., and Listeria monocytogenes: a review. FEMS Microbiol. Rev. 29, 851–875 [DOI] [PubMed] [Google Scholar]
- 3. Vázquez-Boland J. A., Kuhn M., Berche P., Chakraborty T., Domínguez-Bernal G., Goebel W., González-Zorn B., Wehland J., and Kreft J. (2001) Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14, 584–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu D. (2006) Identification, subtyping and virulence determination of Listeria monocytogenes, an important foodborne pathogen. J. Med. Microbiol. 55, 645–659 [DOI] [PubMed] [Google Scholar]
- 5. Brown S., Santa Maria J. P. Jr., and Walker S. (2013) Wall teichoic acids of Gram-positive bacteria. Annu. Rev. Microbiol. 67, 313–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kamisango K., Fujii H., Okumura H., Saiki I., Araki Y., Yamamura Y., and Azuma I. (1983) Structural and immunochemical studies of teichoic acid of Listeria monocytogenes. J. Biochem. 93, 1401–1409 [DOI] [PubMed] [Google Scholar]
- 7. Fiedler F. (1988) Biochemistry of the cell surface of Listeria strains: a locating general view. Infection 16, S92–97 [DOI] [PubMed] [Google Scholar]
- 8. Brown S., Xia G., Luhachack L. G., Campbell J., Meredith T. C., Chen C., Winstel V., Gekeler C., Irazoqui J. E., Peschel A., and Walker S. (2012) Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc. Natl. Acad. Sci. U.S.A. 109, 18909–18914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Winstel V., Liang C., Sanchez-Carballo P., Steglich M., Munar M., Bröker B. M., Penadés J. R., Nübel U., Holst O., Dandekar T., Peschel A., and Xia G. (2013) Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nat. Commun. 4, 2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Winstel V., Kühner P., Salomon F., Larsen J., Skov R., Hoffmann W., Peschel A., and Weidenmaier C. (2015) Wall teichoic acid glycosylation governs Staphylococcus aureus nasal colonization. mBio 6, e00632–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bielmann R., Habann M., Eugster M. R., Lurz R., Calendar R., Klumpp J., and Loessner M. J. (2015) Receptor binding proteins of Listeria monocytogenes bacteriophages A118 and P35 recognize serovar-specific teichoic acids. Virology 477, 110–118 [DOI] [PubMed] [Google Scholar]
- 12. Wanner S., Schade J., Keinhörster D., Weller N., George S. E., Kull L., Bauer J., Grau T., Winstel V., Stoy H., Kretschmer D., Kolata J., Wolz C., Bröker B. M., and Weidenmaier C. (2017) Wall teichoic acids mediate increased virulence in Staphylococcus aureus. Nat. Microbiol. 2, 16257. [DOI] [PubMed] [Google Scholar]
- 13. Tomita S., Furihata K., Tanaka N., Satoh E., Nukada T., and Okada S. (2012) Determination of strain-specific wall teichoic acid structures in Lactobacillus plantarum reveals diverse α-d-glucosyl substitutions and high structural uniformity of the repeating units. Microbiology 158, 2712–2723 [DOI] [PubMed] [Google Scholar]
- 14. Eugster M. R., Morax L. S., Hüls V. J., Huwiler S. G., Leclercq A., Lecuit M., and Loessner M. J. (2015) Bacteriophage predation promotes serovar diversification in Listeria monocytogenes. Mol. Microbiol. 97, 33–46 [DOI] [PubMed] [Google Scholar]
- 15. Tomita S., Furihata K., Nukada T., Satoh E., Uchimura T., and Okada S. (2009) Structures of two monomeric units of teichoic acid prepared from the cell wall of Lactobacillus plantarum NRIC 1068. Biosci. Biotechnol. Biochem. 73, 530–535 [DOI] [PubMed] [Google Scholar]
- 16. Carvalho F., Atilano M. L., Pombinho R., Covas G., Gallo R. L., Filipe S. R., Sousa S., and Cabanes D. (2015) l-Rhamnosylation of Listeria monocytogenes wall teichoic acids promotes resistance to antimicrobial peptides by delaying interaction with the membrane. PLoS Pathog. 11, e1004919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spears P. A., Havell E. A., Hamrick T. S., Goforth J. B., Levine A. L., Abraham S. T., Heiss C., Azadi P., and Orndorff P. E. (2016) Listeria monocytogenes wall teichoic acid decoration in virulence and cell-to-cell spread. Mol. Microbiol. 101, 714–730 [DOI] [PubMed] [Google Scholar]
- 18. Uchikawa K., Sekikawa I., and Azuma I. (1986) Structural studies on teichoic acids in cell walls of several serotypes of Listeria monocytogenes. J. Biochem. 99, 315–327 [DOI] [PubMed] [Google Scholar]
- 19. Fujii H., Kamisango K., Nagaoka M., Uchikawa K., Sekikawa I., Yamamoto K., and Azuma I. (1985) Structural study on teichoic acids of Listeria monocytogenes types 4a and 4d. J. Biochem. 97, 883–891 [DOI] [PubMed] [Google Scholar]
- 20. Kaya S., Araki Y., and Ito E. (1985) Characterization of a novel linkage unit between ribitol teichoic acid and peptidoglycan in Listeria monocytogenes cell walls. Eur. J. Biochem. 146, 517–522 [DOI] [PubMed] [Google Scholar]
- 21. Kailemia M. J., Ruhaak L. R., Lebrilla C. B., and Amster I. J. (2014) Oligosaccharide analysis by mass spectrometry: a review of recent developments. Anal. Chem. 86, 196–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eugster M. R., and Loessner M. J. (2011) Rapid analysis of Listeria monocytogenes cell wall teichoic acid carbohydrates by ESI-MS/MS. PloS One 6, e21500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weidenmaier C., and Peschel A. (2008) Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 6, 276–287 [DOI] [PubMed] [Google Scholar]
- 24. Eugster M. R., Haug M. C., Huwiler S. G., and Loessner M. J. (2011) The cell wall binding domain of Listeria bacteriophage endolysin PlyP35 recognizes terminal GlcNAc residues in cell wall teichoic acid. Mol. Microbiol. 81, 1419–1432 [DOI] [PubMed] [Google Scholar]
- 25. Schmelcher M., Shabarova T., Eugster M. R., Eichenseher F., Tchang V. S., Banz M., and Loessner M. J. (2010) Rapid multiplex detection and differentiation of Listeria cells by use of fluorescent phage endolysin cell wall binding domains. Appl. Environ. Microbiol. 76, 5745–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schmelcher M., Donovan D. M., and Loessner M. J. (2012) Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 7, 1147–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hagens S., and Loessner M. J. (2014) Phages of Listeria offer novel tools for diagnostics and biocontrol. Front. Microbiol. 5, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pfenninger A., Karas M., Finke B., and Stahl B. (2002) Structural analysis of underivatized neutral human milk oligosaccharides in the negative ion mode by nano-electrospray MS(n) (part 1: methodology). J. Am. Soc. Mass Spectrom. 13, 1331–1340 [DOI] [PubMed] [Google Scholar]
- 29. Wang Y., Black B. A., Curtis J. M., and Gänzle M. G. (2014) Characterization of α-galacto-oligosaccharides formed via heterologous expression of α-galactosidases from Lactobacillus reuteri in Lactococcus lactis. Appl. Microbiol. Biotechnol. 98, 2507–2517 [DOI] [PubMed] [Google Scholar]
- 30. Loessner M. J., Kramer K., Ebel F., and Scherer S. (2002) C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44, 335–349 [DOI] [PubMed] [Google Scholar]
- 31. Loessner M. J. (2005) Bacteriophage endolysins: current state of research and applications. Curr. Opin. Microbiol. 8, 480–487 [DOI] [PubMed] [Google Scholar]
- 32. Espaillat A., Forsmo O., El Biari K., Björk R., Lemaitre B., Trygg J., Cañada F. J., de Pedro M. A., and Cava F. (2016) Chemometric analysis of bacterial peptidoglycan reveals atypical modifications that empower the cell wall against predatory enzymes and fly innate immunity. J Am Chem Soc 138, 9193–9204 [DOI] [PubMed] [Google Scholar]
- 33. Fang T. T., and Bendiak B. (2007) The stereochemical dependence of unimolecular dissociation of monosaccharide-glycolaldehyde anions in the gas phase: a basis for assignment of the stereochemistry and anomeric configuration of monosaccharides in oligosaccharides by mass spectrometry via a key discriminatory product ion of disaccharide fragmentation, m/z 221. J. Am. Chem. Soc. 129, 9721–9736 [DOI] [PubMed] [Google Scholar]
- 34. Ciucanu I. (2006) Per-O-methylation reaction for structural analysis of carbohydrates by mass spectrometry. Anal. Chim. Acta 576, 147–155 [DOI] [PubMed] [Google Scholar]
- 35. Bernard E., Rolain T., David B., André G., Dupres V., Dufrêne Y. F., Hallet B., Chapot-Chartier M. P., and Hols P. (2012) Dual role for the O-acetyltransferase OatA in peptidoglycan modification and control of cell septation in Lactobacillus plantarum. PloS One 7, e47893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aubry C., Goulard C., Nahori M. A., Cayet N., Decalf J., Sachse M., Boneca I. G., Cossart P., and Dussurget O. (2011) OatA, a peptidoglycan O-acetyltransferase involved in Listeria monocytogenes immune escape, is critical for virulence. J. Infect. Dis. 204, 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dubail I., Bigot A., Lazarevic V., Soldo B., Euphrasie D., Dupuis M., and Charbit A. (2006) Identification of an essential gene of Listeria monocytogenes involved in teichoic acid biogenesis. J. Bacteriol. 188, 6580–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cheng Y., Promadej N., Kim J. W., and Kathariou S. (2008) Teichoic acid glycosylation mediated by gtcA is required for phage adsorption and susceptibility of Listeria monocytogenes serotype 4b. Appl. Environ. Microbiol. 74, 1653–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bhavsar A. P., Erdman L. K., Schertzer J. W., and Brown E. D. (2004) Teichoic acid is an essential polymer in Bacillus subtilis that is functionally distinct from teichuronic acid. J. Bacteriol. 186, 7865–7873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sobhanifar S., Worrall L. J., Gruninger R. J., Wasney G. A., Blaukopf M., Baumann L., Lameignere E., Solomonson M., Brown E. D., Withers S. G., and Strynadka N. C. (2015) Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid α-glycosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 112, E576–E585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Duus J., Gotfredsen C. H., and Bock K. (2000) Carbohydrate structural determination by NMR spectroscopy: modern methods and limitations. Chem. Rev. 100, 4589–4614 [DOI] [PubMed] [Google Scholar]
- 42. Sklenár V., Miyashiro H., Zon G., Miles H. T., and Bax A. (1986) Assignment of the 31P and 1H resonances in oligonucleotides by two-dimensional NMR spectroscopy. FEBS Lett. 208, 94–98 [DOI] [PubMed] [Google Scholar]
- 43. Ganguly J., Low L. Y., Kamal N., Saile E., Forsberg L. S., Gutierrez-Sanchez G., Hoffmaster A. R., Liddington R., Quinn C. P., Carlson R. W., and Kannenberg E. L. (2013) The secondary cell wall polysaccharide of Bacillus anthracis provides the specific binding ligand for the C-terminal cell wall-binding domain of two phage endolysins, PlyL and PlyG. Glycobiology 23, 820–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Briers Y., Schmelcher M., Loessner M. J., Hendrix J., Engelborghs Y., Volckaert G., and Lavigne R. (2009) The high-affinity peptidoglycan binding domain of Pseudomonas phage endolysin KZ144. Biochem. Biophys. Res. Commun. 383, 187–191 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.