Abstract
Anthrax is a life-threatening disease caused by infection with Bacillus anthracis, which expresses lethal factor and the receptor-binding protective antigen. These two proteins combine to form anthrax lethal toxin (LT), whose proximal targets are mitogen-activated kinase kinases (MKKs). However, the downstream mediators of LT toxicity remain elusive. Here we report that LT exposure rapidly reduces the levels of c-Jun, a key regulator of cell proliferation and survival. Blockade of proteasome-dependent protein degradation with the 26S proteasome inhibitor MG132 largely restored c-Jun protein levels, suggesting that LT promotes degradation of c-Jun protein. Using the MKK1/2 inhibitor U0126, we further show that MKK1/2–Erk1/2 pathway inactivation similarly reduces c-Jun protein, which was also restored by MG132 pre-exposure. Interestingly, c-Jun protein rebounded to normal levels 4 h following U0126 exposure but not after LT exposure. The restoration of c-Jun in U0126-exposed cells was associated with increased c-Jun mRNA levels and was blocked by inactivation of the JNK1/2 signaling pathway. These results indicate that LT reduces c-Jun both by promoting c-Jun protein degradation via inactivation of MKK1/2–Erk1/2 signaling and by blocking c-Jun gene transcription via inactivation of MKK4–JNK1/2 signaling. In line with the known functions of c-Jun, LT also inhibited cell proliferation. Ectopic expression of LT-resistant MKK2 and MKK4 variants partially restored Erk1/2 and JNK1/2 signaling in LT-exposed cells, enabling the cells to maintain relatively normal c-Jun protein levels and cell proliferation. Taken together, these findings indicate that LT reduces c-Jun protein levels via two distinct mechanisms, thereby inhibiting critical cell functions, including cellular proliferation.
Keywords: anthrax toxin, c-Jun transcription factor, cell proliferation, MAPK, protein degradation, transcription factor
Introduction
Anthrax is a life-threatening disease caused by infection with Bacillus anthracis (1, 2). B. anthracis expresses lethal factor (LF)2 and the receptor-binding protective antigen (PA) on its pXO1 virulence plasmid (3–6). The combination of LF and PA forms lethal toxin (LT). LF is a zinc-dependent metalloprotease with specific activity against certain mitogen-activated kinase kinases (MKKs) (7) and NACHT leucine-rich repeat protein 1 (NALP1) (8, 9). The MKKs lie in the middle of three-tiered signaling cascades that comprise a mitogen-activated protein kinase kinase kinase (MKKK), an MKK, and an MAPK (10, 11). Extracellular stimuli such as growth factors or cytokines initiate activation of MKKKs that phosphorylate MKKs, which, in turn, phosphorylate MAPKs. LF cleavage of MKKs at their docking sites (D sites) disrupts the activation of MAPKs, including Erk1/2, which are activated by MKK1 and MKK2; p38 MAPKs, which are activated by MKK3 and MKK6; and JNKs, which are activated by MKK4 and MKK7 (7, 12–14).
Studies from our laboratory and others have revealed that LT treatment induces cell cycle arrest and inhibition of cell proliferation (15–20). However, it has remained elusive whether the antiproliferative effect of LT was only due to a blockade in the activation/phosphorylation of transcriptional factors of the MKK signaling cascades or whether other mechanisms were involved. Here we provide data demonstrating that LT treatment causes a rapid reduction in the levels of c-Jun protein, a major member of the transcription factor activator protein 1 (AP-1) family that is a key regulator of cell proliferation, cell survival, and tumorigenesis. LT induces the reduction of c-Jun protein by promoting its degradation via inactivation of the MKK1/2–Erk1/2 signaling pathway and by blocking its gene transcription via inactivation of the MKK4–JNK1/2 signaling pathway. Our findings support a pathogenic role for LT in reducing c-Jun protein levels via two distinct mechanisms inhibiting critical cellular functions.
Results
Anthrax lethal toxin treatment causes a rapid reduction in c-Jun protein
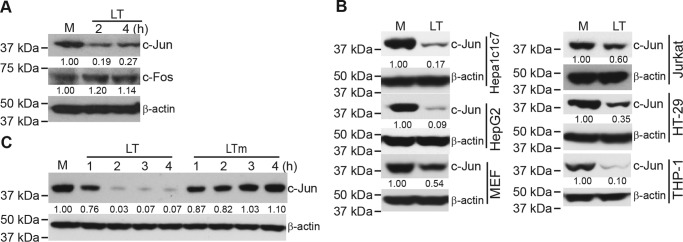
A previous study from our laboratory showed that anthrax LT inhibits the proliferation of T cells, which correlates with reduced AP-1 activity in LT-treated T cells (15). AP-1 is a transcription factor family that is composed of dimeric basic region–leucine zipper proteins belonging to the Jun, Fos, Maf, and ATF subfamilies (21). To understand how LT inhibits AP-1 activity, we first measured, by Western blotting, the levels of c-Jun and c-Fos proteins, two major members of the AP-1 transcription factor family. As shown in Fig. 1A, LT treatment caused a rapid reduction in the levels of c-Jun but not c-Fos protein in Hepa1c1c7 cells. The inhibitory effect of LT on c-Jun is not cell- or species-specific because the levels of c-Jun protein were also reduced following LT treatment in mouse embryonic fibroblasts (MEFs), human hepatocellular carcinoma cells (HepG2), human immortalized T cells (Jurkat), human colorectal adenocarcinoma cells (HT-29), and human monocytic leukemia cells (THP-1) (Fig. 1B). Reduction of c-Jun by LT requires its proteolytic activity, as an LT mutant lacking proteolytic function failed to induce the reduction of c-Jun protein (Fig. 1C).
Figure 1.
Anthrax LT treatment causes a rapid reduction in the levels of c-Jun protein. Cells were extracted with SDS sample buffer after treatment as indicated below. The expression levels of the indicated proteins in the cells lysates were assessed by Western blotting. A, Hepa1c1c7 cells were either left untreated (M) or treated with LT for 2 and 4 h. B, cells were left untreated or treated with LT for 4 h. C, Hepa1c1c7 cells were left untreated or treated with LT or an LT mutant that lacks proteolytic activity (LTm) for 1, 2, 3, and 4 h. β-Actin levels were used as loading controls. Data shown are representative of at least two independent experiments. The intensities of protein bands were quantified using the Image Studio software of the Odyssey system and normalized by the amounts of β-actin; relative amounts are shown below each lane.
Rapid LT-induced reduction of c-Jun does not occur at the level of protein synthesis
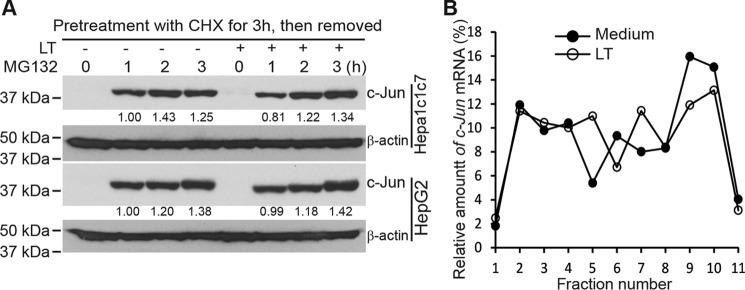
We have reported previously that LT inhibits the accumulation of hypoxia-inducible factor (HIF) 1α, the master regulator of cellular responses to hypoxia, by blocking its translation (22). To investigate whether LT inhibits c-Jun by a similar mechanism, we first investigated the effect of LT treatment on de novo c-Jun protein synthesis. We treated HepG2 and Hepa1c1c7 cells with cycloheximide (CHX) for 3 h to block c-Jun translation (with or without wild-type LT for the last 1 h), washed away the CHX, and then cultured the cells in fresh medium to resume protein synthesis. As shown in Fig. 2A, MG132-induced accumulation of de novo synthesized c-Jun in HepG2 and Hepa1c1c7 cells was not affected by LT treatment. To confirm that LT treatment has no effect on c-Jun translation, we next examined the relative distribution of c-Jun mRNA in individual fractions collected from polysome sucrose gradients. In these sucrose gradient experiments (from top to bottom), fractions 1–4 include mRNAs that are not associated with components of the translational machinery or co-sediment with individual ribosomal subunits (monosomes), so they are not considered to be undergoing translation. Fractions 5–7 include mRNAs that bind to single ribosomes or formed polysomes of low molecular weight, and they are considered to be translated at low to moderate levels. Fractions 8–10 include the mRNAs that are associated with polysomes of high molecular weight, and they are thus considered to be actively translated. As shown in Fig. 2B, unlike its effects on HIF-1α, LT treatment did not appreciably change the distribution of c-Jun mRNA, further supporting the conclusion that LT does not inhibit c-Jun by blocking its translation.
Figure 2.
Anthrax LT treatment does not inhibit c-Jun protein synthesis. A, Western blot analysis of c-Jun protein levels in HepG2 and Hepa1c1c7 cells, which were treated with CHX for 3 h in the presence or absence of LT for the last 1 h. Cells were subsequently washed with PBS twice and then incubated with MG132 for 0–3 h. The intensities of protein bands were quantified using the Image Studio software of the Odyssey system and normalized by the amounts of β-actin; relative amounts are shown below each lane. B, polysomal distribution of c-Jun mRNA in HepG2 cells cultured in the presence or absence of LT for 3 h. The treated cells were extracted with a polysome extraction buffer, and the supernatants were fractioned through a 10% to 50% linear sucrose gradient. The level of c-Jun mRNA in each gradient fraction was determined by quantitative PCR and plotted as a percentage of the total c-Jun mRNA levels in each sample. The peak in fraction 4 corresponds to monosomes, whereas the peak in fractions 8–10 corresponds to polysomes with active translation.
LT treatment promotes c-Jun protein degradation
Regulation of c-Jun has been reported to occur at two other major levels: gene transcription and protein stability (21, 23). To investigate whether LT reduces c-Jun through promoting its proteasome-dependent degradation, we pretreated cell cultures with MG132 (a 26S proteasome inhibitor) 30 min prior to LT treatment to block protein degradation. Western blotting showed that inhibition of protein degradation largely corrected the LT-induced c-Jun reduction in both HepG2 and Hepa1c1c7 cells (Fig. 3A), suggesting that LT treatment promotes c-Jun protein degradation. Moreover, a higher protein turnover rate was observed in LT-treated compared with untreated Hepa1c1c7 cells (Fig. 3B), further substantiating that LT treatment enhances c-Jun protein degradation. Together, these data indicate that promotion of protein degradation is a major mechanism by which LT reduces the levels of c-Jun protein.
Figure 3.
Anthrax LT treatment promotes c-Jun protein degradation. A, Western blot analysis of c-Jun protein levels in Hepa1c1c7 and HepG2 cells pretreated with DMSO or MG132 for 30 min and then incubated with (LT) or without (M) LT for 4 h. The intensities of protein bands were quantified using the Image Studio software of the Odyssey system and normalized by the amounts of β-actin; relative amounts are shown below each lane. B, turnover of c-Jun protein in Hepa1c1c7 cells cultured in the presence or absence of LT. Cells were cultured with or without LT for 1 h and then treated with CHX for 0, 1, 2, 3, and 4 h as indicated. c-Jun protein levels were determined by Western blotting (top panel). The intensities of the protein bands were quantified using the Image Studio software of the Odyssey system (bottom panel). The relative amount of c-Jun protein at each time point was normalized by the amount of β-actin. Data shown are representative of at least two independent experiments.
LT treatment reduces c-Jun protein by inhibiting the MKK1/2–Erk1/2 and MKK4/7–JNK signaling pathways
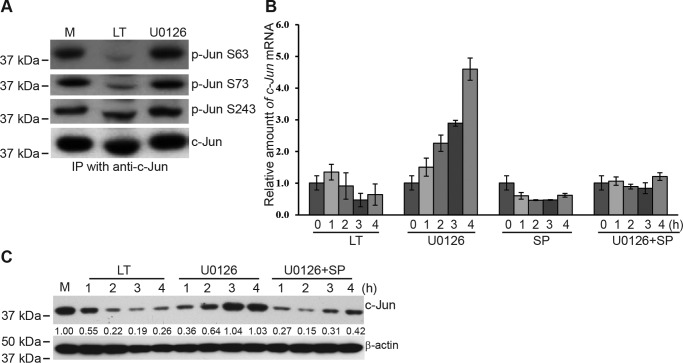
LF is a zinc-dependent metalloprotease with specific activity against MKKs. Consistent with our previous studies (22, 24), LT treatment led to cleavage and degradation of MKK1, MKK2, MKK3, MKK4, and MKK6, but not MKK7, in Hepa1c1c7 cells (Fig. 4A). The cleavage of MKKs led to inactivation of their downstream signaling molecules, including Erk1/2, p38, and JNK1/2 (Fig. 4B). Of note, JNK is phosphorylated both at Tyr-185 by MKK4 and at Thr-183 by MKK7, which is required for full activation (25, 26). The antibody used in this study detects JNK1/2 with dual phosphorylation of Thr-183 and Tyr-185. Even though MKK7 remains largely intact in LT-treated cells, our studies demonstrate that cleavage of MKK4 is sufficient to disrupt dual phosphorylation and optimal activation of JNK1/2. To dissect the signaling pathways underlying the LT-dependent reduction in c-Jun protein levels, we treated cells with specific inhibitors that inhibit MKK1/2–Erk1/2 (U0126), JNK1/2 (SP600125), or p38 (SB203580). As shown in Fig. 4C, inactivation of MKK1/2–Erk1/2 led to a rapid but temporary reduction of c-Jun that returned to baseline levels at 3 h. Inhibition of JNK1/2 also caused a marked reduction of c-Jun, but this effect persisted at least 4 h. In contrast, inactivation of p38 did not affect the levels of c-Jun (Fig. 4C). These data suggest that LT may reduce c-Jun by inactivating the MKK1/2–Erk1/2 and MKK4/7–JNK pathways.
Figure 4.
Anthrax LT treatment reduces c-Jun by disrupting MAPK signaling pathways. A and B, Hepa1c1c7 cells were left untreated (M) or treated with LT for 1, 2, and 4 h. Cell lysates were subjected to Western blot analysis for assessing protein levels of various MKKs (A) and total and phosphorylated Erk1/2, p38, and JNK1/2 (B). C, Hepa1c1c7 cells were left untreated or treated with the MAPK inhibitors U0126, SB203580, and SP600125 for 1, 2, 3, and 4 h. The levels of c-Jun protein in treated cells were determined by Western blotting. β-Actin was used as a loading control. Data shown are representative of three independent experiments. The intensities of protein bands were quantified using the Image Studio software of the Odyssey system and normalized by the amounts of β-actin; relative amounts are shown below each lane.
LT inactivation of MKK1/2–Erk1/2 promotes c-Jun protein degradation by a Fra-1–independent pathway
The rapid LT-induced reduction of c-Jun is coincident with Erk1/2 deactivation (Figs. 1C and 3B). Pretreatment of cells with MG132 prevented the U0126-induced c-Jun reduction (Fig. 5A), suggesting that LT may promote c-Jun degradation by inhibiting the MKK1/2–Erk1/2 pathway. To further investigate how Erk1/2 inactivation leads to c-Jun degradation, we measured the protein levels of Fra-1, which is a target of Erk1/2 and has been shown to form a heterodimer with c-Jun and stabilize c-Jun protein (27, 28). As shown in Fig. 5B, LT treatment also caused a time-dependent reduction in Fra-1 protein levels. Erk1/2 stabilizes Fra-1 by phosphorylating its two serine residues at the C terminus (Ser-252 and Ser-265) (29). We next generated a constitutively active Fra-1 mutant with substitution of Ser-252 and Ser-265 with Asp (Fra-12D), which mimics phosphorylated serine. The Fra-1 mutant (HA-Fra-12D) was resistant to LT treatment, maintaining a similar expression level in LT-treated cells (Fig. 5C). However, LT-induced c-Jun degradation was not prevented by the Fra-1 mutant, suggesting that LT-induced c-Jun degradation does not depend on Fra-1 destabilization. To further confirm that Fra-1 destabilization is not involved in LT-induced c-Jun degradation, we knocked down the expression of Fra-1 using a specific siRNA. As shown in Fig. 5D, knockdown of Fra-1 had negligible effects on the levels of c-Jun at steady state or upon LT- and U0126-induced c-Jun degradation. Taken together, these results suggest that c-Jun degradation by LT inactivation of the MKK1/2–Erk1/2 pathway is not due to destabilization of Fra-1.
Figure 5.
Anthrax LT-induced c-Jun degradation is not due to destabilization of Fra-1. Cells were extracted with SDS sample buffer after treatment as indicated below. The expression levels of the indicated proteins in the cells lysates were assessed by Western blotting. A, Hepa1c1c7 cells were left untreated (M) or treated with U0126 for 1 h. B, Hepa1c1c7 cells were left untreated or treated with LT for 1, 2, and 4 h. C, Hepa1c1c7 cells expressing GFP (Control), HA-Fra-1/GFP, or HA-Fra-12D/GFP were left untreated or treated with LT for 4 h. D, Hepa1c1c7 cells were transfected with control siRNA or Fra-1–specific siRNA. Three days after transfection, cells were left untreated or treated with LT for 2 and 4 h or U0126 for 1 h. β-Actin was used as a loading control. Data shown are representative of at least two independent experiments. The intensities of protein bands were quantified using the Image Studio software of the Odyssey system and normalized by the amounts of β-actin; relative amounts are shown below each lane.
LT treatment blocks c-Jun gene transcription by inhibiting the MKK4–JNK1/2 pathway
Inactivation of MKK1/2–Erk1/2 by U0126 induced a rapid reduction of c-Jun, which returned to normal levels by 3 h (Fig. 4C). However, this restoration did not occur in LT-treated cells. We reasoned that this difference was due to the intact JNK1/2 activity in U0126-treated cells, which phosphorylates and activates c-Jun. c-Jun, in turn, has been shown to promote its own transcription through positive feedback regulation (30). To test this hypothesis, we examined the phosphorylation of c-Jun in LT- and U0126-treated cells at the Ser-63 and Ser-73 sites, which are mediated by JNKs (31), as well as at Ser-243, which is induced by GSK3 (32). As predicted, phosphorylation of c-Jun at Ser-63 and Ser-73 was drastically reduced in LT-treated but not in U0126-treated cells (Fig. 6A). In contrast, phosphorylation of c-Jun at Ser-243 was not affected by LT treatment (Fig. 6A). In line with the levels of JNK1/2-mediated c-Jun phosphorylation, c-Jun gene transcription was inhibited in LT-treated cells but dramatically increased in U0126-treated cells. The U0126-induced increase of c-Jun gene transcription was dependent on the activity of JNKs because it was blocked by SP600125, a specific inhibitor of JNKs (Fig. 6B). Moreover, combined treatment with U0126 and SP600125 largely prevented the restoration of c-Jun protein observed in U0126-treated cells (Fig. 6C), mimicking the effect of LT treatment.
Figure 6.
Anthrax LT treatment inhibits c-Jun gene transcription via inactivation of JNKs. A, untreated (M) and LT- and U0126-treated Hep1c1c7 cells were lysed with radioimmune precipitation assay buffer. c-Jun protein in the cell lysates was immunoprecipitated with c-Jun antibody and protein G–conjugated magnetic beads. The levels of phosphorylated and total c-Jun were assessed by Western blotting. Data shown are representative of at least two independent experiments. B, quantitative PCR analysis of c-Jun mRNA expression in Hepa1c1c7 cells treated with LT, U0126, SP600125 (SP), or U0126 + SP600125 for 0, 1, 2, 3, and 4 h. The relative amounts of c-Jun mRNA were normalized by the levels of β-actin mRNA. Shown is one representative experiment, with error bars depicting the standard deviation derived from the results from triplicates for each experimental condition. C, Hepa1c1c7 cells were left untreated or treated with LT, U0126, or U0126+SP600125 for 1, 2, 3, and 4 h. The levels of c-Jun and β-actin were measured by Western blotting. Data shown are representative of at least two independent experiments. The intensities of protein bands were quantified using the Image Studio software of the Odyssey system and normalized by the amounts of β-actin; relative amounts are shown below each lane.
Restoration of c-Jun protein levels and cell proliferation by expression of LT-resistant MKK2 and MKK7-4 mutants
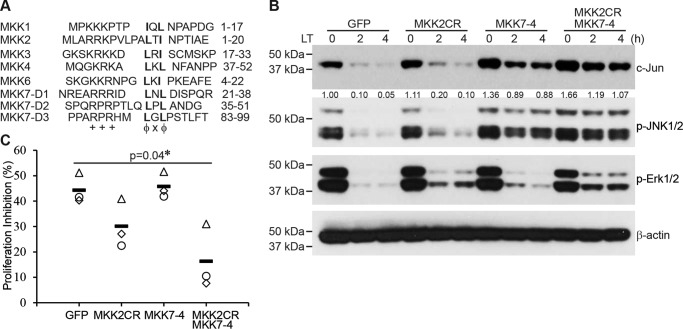
Having shown that inactivation of the MKK1/2–Erk1/2 and MKK4–JNK1/2 signaling pathways was responsible for c-Jun reduction in LT-treated cells, we next generated LT-resistant MKKs to restore those signaling pathways, as described under “Experimental procedures.” LF cleavage of MKKs occurs within a specific consensus sequence (++++XφX↓φ) (7); basic and hydrophobic residues are indicated by + and φ respectively, X indicates any amino acid, and the cleavage site is indicated by ↓, most of which overlaps with the high-affinity MAPK D site, +++X1–5φXφ) (7, 33) (Fig. 7A). The cleavage site of MKK1 (Ile-9) is essential for its docking site, and although an Ile-9 to Asp mutation made MKK1 resistant to LT treatment, it also totally disrupted its function to phosphorylate Erk1/2 (data not shown). The LF cleavage site consensus sequence of MKK2 partially overlaps with its docking sequence. A mutation of the LF cleavage site of MKK2 (Ala-11) to Asp made it resistant to LF cleavage, and the mutant partially maintained its function to activate Erk1/2 (Fig. 7B). MKK7 is resistant to LF cleavage (Fig. 4A). Replacement of the D site of MKK4 with the D site of MKK7 (MKK7-4) markedly restored the function of MKK4 in LT-treated cells (Fig. 7A). In line with the restoration of Erk1/2 and JNK1/2 function, cells ectopically dually expressing the LF-resistant MKK2 (MKK2CR) and MKK7-4 showed higher levels of c-Jun protein after LT treatment compared with controls (Fig. 7B). Consistent with the role of c-Jun in regulating cell proliferation, MKK2CR and MKK7-4 rendered cells resistant to LT-induced inhibition of cell growth (Fig. 7C). Collectively, these data support the conclusion that LT inhibits c-Jun by targeting both the MKK1/2–Erk1/2 and MKK4/7–JNK1/2 pathways, leading to arrest of cell proliferation.
Figure 7.
Combination of LT-resistant MKK2 and MKK4 reverses LT-induced c-Jun reduction and cell growth arrest. A, sequence alignment of the docking sites of MKKs, including the three reported docking sites of MKK7. B, Hepa1c1c7 cells expressing GFP (control), LT-resistant MKK2 (MKK2CR), MKK4 with a replacement of the MKK7 D site (MKK7-4), or MKK2CR + MKK7-4 were treated with LT for 0, 2, and 4 h. The levels of c-Jun, p-JNK1/2, p-Erk1/2, and β-actin were determined by Western blotting. The intensities of protein bands were quantified using the Image Studio software of the Odyssey system and normalized by the amounts of β-actin; relative amounts are shown below each lane. C, proliferation of Hepa1c1c7 cells expressing GFP (control), LT-resistant MKK2, MKK4 with a replacement of the MKK7 D site, or MKK2CR + MKK7-4, which were treated with or without LT for 48 h. Shown are the cell proliferation inhibition percentages from three independent experiments. Open symbols represent three independent experiments; solid bars indicate the average, and the asterisk indicates a statistically significant difference.
Discussion
Although it is well-known that anthrax LT cleaves MKKs (7), the effect of LT on downstream transcription factors remained poorly understood. In this study, we show that LT treatment causes a rapid reduction in the levels of c-Jun. The reduction is caused by enhancing c-Jun protein degradation via inactivation of the MKK1/2–Erk1/2 signaling pathway and by blocking its gene transcription via inactivation of the MKK4–JNK1/2 signaling pathway. c-Jun is a major member of the AP-1 transcription factor family, which comprises dimeric basic region–leucine zipper proteins belonging to the Jun (c-Jun, JunB, and JunD), Fos (c-Fos, FosB, Fra-1, and Fra-2), Maf (c-Maf, MafB, MafA, MafG/F/K, and Nrl), and ATF (ATF2, LRF1/ATF3, B-ATF, JDP1, and JDP2) subfamilies (21, 23). LT treatment leads to a rapid reduction of c-Jun but not c-Fos, with which it forms the AP-1 heterodimer. LT treatment also markedly reduces the levels of JunB and JunD (data not shown). As this study focused on the regulation of c-Jun, the effects of LT on other AP-1 members remain to be investigated.
Our findings serve to provide insights into the mechanisms of action of LT, a toxin critical for the virulence factor of B. anthracis. Anthrax LT acts to reduce the levels of c-Jun protein, a key component of all AP-1 complexes (23, 34). The transcription of c-Jun is activated by many stimuli, including growth factors, cytokines, and UV irradiation (23). c-Jun gene induction is generally mediated by the c-Jun 12-O-tetradecanoylphorbol-13-acetate (TPA) response element, which is recognized by c-Jun–ATF2 heterodimers (35, 36). The transcriptional activity of c-Jun activity, in turn, is regulated by JNKs, which specifically phosphorylate it at two positive regulatory serine residues located within its amino-terminal activation domain (Ser-63 and Ser-73) (31, 37). ATF2 activity is regulated by JNKs and p38, which phosphorylate the same positive regulatory sites of ATF2 (35, 38). Our results from loss-of-function and gain-of-function studies clearly demonstrate that JNK1/2 activity is essential for c-Jun restoration in U0126-treated cells. The rebound in c-Jun protein levels is associated with increased levels of c-Jun mRNA, suggesting that intact JNKs in U0126-treated cells activate c-Jun by phosphorylating its Ser-63 and Ser-73 and promoting its gene transcription. Phosphorylation of Ser-63 and Ser-73 by JNKs has also been reported to increase c-Jun protein stability (39). Although we cannot fully exclude a role for JNK1/2 in increasing c-Jun protein stability, the correlation of increased c-Jun protein with increased mRNA levels supports increased c-Jun gene transcription as the major mechanism underlying the recovery of c-Jun protein levels in U0126-treated cells.
In addition, our results reveal new complexities in the regulation of c-Jun stability. LT treatment promotes c-Jun protein degradation by inactivation of the MKK1/2–Erk1/2 pathway but not by the most predictable pathway. Fra-1, a member of the Fos family has been shown to form a heterodimer with c-Jun that stabilizes c-Jun (27). Furthermore, Erk1/2 activation leads to phosphorylation of the Fra-1 C-terminal domain, which positively regulates Fra-1 protein stability (27–29). Consistent with previous studies, inactivation of Erk1/2 by LT treatment caused a reduction in the levels of Fra-1 that was restored by mutations of the two C-terminal serine residues (Ser-252 and Ser-265) to Asp. Although the levels of Fra-1 were restored by the Ser-to-Asp substitutions, the levels of c-Jun protein were not restored, indicating that LT-induced c-Jun degradation is not due to Fra-1 destabilization. The Fra-1 knockdown experiment further excluded the possibility that LT promotes c-Jun degradation by targeting Fra-1. c-Jun protein is degraded by a ubiquitination-dependent proteasome pathway. Previous studies showed that FBW7, Itch, COP1, MEKK1, and SAG/ROC2 can act as the ubiquitin ligase (E3) to induce c-Jun ubiquitination. Our preliminary experiments using siRNA knockdown suggest that COP1 may be the E3 mediating c-Jun degradation in LT-treated cells (data not shown), which is consistent with recent findings that Erk1/2 signaling suppresses COP1 activity (40).
The effect of anthrax LT on c-Jun has important clinical implications as well, especially with respect to oncological indications. The c-Jun transcription factor was originally discovered as an oncoprotein encoded by a cellular insert in the genome of avian sarcoma virus, an oncogenic retrovirus isolated from spontaneous chicken tumor 17 (41, 42). The c-Jun protein is a key member of the AP-1 transcription factor family that plays a critical role in regulating cell proliferation, cell survival, and tumorigenesis (34). In line with the functions of c-Jun, LT treatment inhibits cell proliferation. The merit of LT as a potential therapeutic reagent for human cancers is highlighted by the frequent mutations in the RAS–RAF–MKK1/2–ERK pathway that are associated with human cancers (43). To this end, investigators have sought to avoid systemic toxicity and to increase the tumor-targeting specificity of anthrax LT by changing the furin-sensitive cleavage site of PA to a site that can be cleaved by matrix metalloproteases (44) or urokinase plasminogen activator (45), two enzymes produced abundantly by tumor cells (46). These LT variants exhibit robust anti-tumor activity regardless of BRAF mutations in mouse models in vivo (44, 46–48). Engineered LT acts by directly inhibiting the growth of tumors harboring BRAF mutations that causes hyperactivation of MKK1/2–Erk1/2 (49–51) or by suppressing the proliferation of tumor endothelial cells and disrupting tumor angiogenesis (44, 47, 52, 53). Our findings in this and previous studies that LT reduces the levels of c-Jun and HIF-1 support LT-based therapies for cancer treatment, as they would act through multiple distinct mechanisms. Moreover, by blocking JNK-mediated c-Jun restoration in MKK1/2-inactivated cells, LT-based therapies might have more sustained anti-tumor effects compared with chemical inhibitors only targeting MKK1/2 or Erk1/2, which are either approved or being studied in clinical trials (54, 55).
Experimental procedures
Cells and reagents
Murine liver hepatoma Hepa1c1c7 cells, MEFs, human hepatocellular carcinoma HepG2 cells, the human immortalized T cell line Jurkat, human colorectal adenocarcinoma HT-29 cells, and human monocytic leukemia THP-1 cells were used in this study. Hepa1c1c7 cells were cultured with α minimal essential medium; HepG2 cells were cultured with Eagle's minimal essential medium; MEFs and HT-29 cells were cultured with Dulbecco's modified Eagle's medium; and Jurkat and THP-1 cells were cultured with RPMI 1640 medium. All media were supplemented with 10% FBS, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 2 mm l-glutamine. Cultures were conducted at 37 °C in an incubator with 5% CO2. Lyophilized recombinant PA and LF were purchased from List Biological Laboratories, Inc. (Campbell, CA) and reconstituted in sterile water to make stock solutions with final concentrations of 1 mg/ml. MG132 (an inhibitor of the 26S proteasome), U0126 (an inhibitor of MKK1/2), SB203580 (an inhibitor of p38), SP600125 (an inhibitor of JNKs), and CHX were purchased from EMD Millipore (Billerica, MA). Antibodies recognizing MKK1, MKK2, MKK3, MKK4, MKK6, and MKK7 were purchased from Santa Cruz Biotechnology (Dallas, TX). Antibodies recognizing p-Erk1/2, Erk1/2, p-p38, p38, p-JNK1/2, JNK1/2, c-Jun, p-c-Jun, Fra-1, p-Fra-1, c-Fos, and HA were purchased from Cell Signaling Technology. Anti-β-actin was purchased from Sigma. TaqMan gene expression assay primers and probes for c-Jun and β-actin were purchased from Invitrogen.
Plasmids and cell transfection
Plasmids containing the cDNA expressing human MKK2, MKK2A11D, MKK4, and MKK7 were generously provided by Dr. Nicholas S. Duesbery (Van Andel Research Institute, Grand Rapids, MI) (56). These plasmids were used as templates for PCR to amply the fragments encoding MKK2, MKK2A11D, MKK4 without the docking region, and the docking region of MKK7. Human Fra-1 cDNA was generated by RT-PCR. Fragments of MKK2, MKK2A11D, MKK4 and MKK7, and Fra-1 cDNA were cloned into the thymidine-adenine (TA) cloning vector pDRIVE, confirmed by DNA sequencing, and then subcloned into the pMIG-w retroviral vector with a HA tag at the N terminus. The Fra-1 S252DS265D mutant was generated using the QuikChange site-directed mutagenesis kit from Agilent (Santa Clara, CA). Retroviral plasmids were transfected into 293T cells together with packaging plasmids using the standard calcium phosphate method to produce pseudovirus. Supernatants were harvested 48 h following the transfection, filtered through 0.45-μm filters (Millipore), and then used to transduce cells. To knock down the expression of Fra-1, Fra-1–specific siRNA was transfected into cells using Lipofectamine RNAiMAX reagent (Invitrogen) following the protocol suggested by the manufacturer.
Western blotting
Cells were lysed with NuPAGE LDS sample buffer (Invitrogen). Cell extracts were then separated on 4–12% NuPAGE BisTris gels (Invitrogen) and transferred to nitrocellulose membranes (Bio-Rad). The membranes were probed with antibodies of interest using standard Western blotting techniques as described previously (22).
Quantitative PCR
Total RNA was extracted from cells with TRIzol (Invitrogen) following the protocol suggested by the manufacturer. RNA was then reverse-transcribed to cDNA using the Omniscript RT kit manufactured by Qiagen (Valencia, CA). The levels of c-Jun mRNA were measured by quantitative PCR performed using standard techniques with a TaqMan probe (Mm00495062_s1) purchased from Applied Biosystems (Foster City, CA) and normalized to the levels of β-actin (Mm02619580_g1).
Polysome analysis
Polysome analysis was performed as described previously (22, 57). In brief, HepG2 cells were pretreated with or without LT for 3 h. The treated cells were then incubated with 0.1 mg/ml CHX for 5 min, detached by scraping into 1 ml of polysome extraction buffer (0.3 m NaCl, 15 mm MgCl2, 15 mm Tris-HCl (pH 7.6), 1% Triton X-100, 1 mg/ml heparin, and 0.1 mg/ml CHX), and lysed on ice for 10 min. Nuclei were pelleted (10,000 × g for 10 min), and the resulting supernatant was fractionated through a 10–50% linear sucrose gradient to fractionate cytoplasmic components according to their molecular weights. The eluted fractions were prepared using a fraction collector (Brandel, Gaithersburg, MD), and their quality was monitored at 254 nm by using a UV-6 detector (ISCO, Lincoln, NE). RNA in each fraction was isolated using TRIzol LS (Invitrogen) and reverse-transcribed to cDNA. The levels of c-Jun and β-actin mRNA in each of the fractions were measured by quantitative PCR, and their abundance was calculated as a percent of the total mRNA in the gradient.
Cell proliferation analysis
Eighty thousand Hepa1c1c7 cells were added to individual wells of 12-well plates. Six replicates were used for each group. The cells were treated with or without LT for 48 h. The cell number in each well was counted after the treatment using a TC20 automated cell counter (Bio-Rad). LT-induced cell proliferation inhibition was calculated using the following formula: proliferation inhibition (%) = (cell number of untreated group − cell number of LT-treated group)/cell number of untreated group × 100%.
Author contributions
W. O. conceived and designed the study, performed the majority of the experiments, analyzed the data, and prepared the manuscript. P. G. and H. F. performed some of the experiments. D. F. supervised the study, provided experimental advice, and prepared the manuscript. All authors approved the final version of the manuscript.
Acknowledgments
We thank Dr. Nicholas Duesbery (Laboratory of Cancer and Developmental Cell Biology, Van Andel Research Institute) for providing critical plasmid reagents and Drs. Brian Roelofs and Tao Wang for thoughtful review of the manuscript.
This work was partially supported by an internal Food and Drug Administration medical countermeasures grant. The authors declare that they have no conflicts of interest with the contents of this article. This manuscript reflects the views of the authors and should not be construed to represent FDA's views or policies.
- LF
- lethal factor
- PA
- protective antigen
- LT
- lethal toxin
- MKK
- mitogen-activated kinase kinase
- MKKK
- mitogen-activated kinase kinase kinase
- D site
- docking site
- MEF
- mouse embryonic fibroblast
- HIF
- hypoxia-inducible factor
- CHX
- cycloheximide
- ATF
- activating transcription factor.
References
- 1. Mock M., and Fouet A. (2001) Anthrax. Annu. Rev. Microbiol. 55, 647–671 [DOI] [PubMed] [Google Scholar]
- 2. Sweeney D. A., Hicks C. W., Cui X., Li Y., and Eichacker P. Q. (2011) Anthrax infection. Am. J. Respir. Crit. Care Med. 184, 1333–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bhatnagar R., and Batra S. (2001) Anthrax toxin. Crit. Rev. Microbiol. 27, 167–200 [DOI] [PubMed] [Google Scholar]
- 4. Moayeri M., Leppla S. H., Vrentas C., Pomerantsev A. P., and Liu S. (2015) Anthrax pathogenesis. Annu. Rev. Microbiol. 69, 185–208 [DOI] [PubMed] [Google Scholar]
- 5. Liu S., Moayeri M., and Leppla S. H. (2014) Anthrax lethal and edema toxins in anthrax pathogenesis. Trends Microbiol. 22, 317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu L., and Frucht D. M. (2007) Bacillus anthracis: a multi-faceted role for anthrax lethal toxin in thwarting host immune defenses. Int. J. Biochem. Cell Biol. 39, 20–24 [DOI] [PubMed] [Google Scholar]
- 7. Bardwell A. J., Abdollahi M., and Bardwell L. (2004) Anthrax lethal factor-cleavage products of MAPK (mitogen-activated protein kinase) kinases exhibit reduced binding to their cognate MAPKs. Biochem. J. 378, 569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Levinsohn J. L., Newman Z. L., Hellmich K. A., Fattah R., Getz M. A., Liu S., Sastalla I., Leppla S. H., and Moayeri M. (2012) Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PLoS Pathog. 8, e1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frew B. C., Joag V. R., and Mogridge J. (2012) Proteolytic processing of Nlrp1b is required for inflammasome activity. PLoS Pathog. 8, e1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang L., and Karin M. (2001) Mammalian MAP kinase signalling cascades. Nature 410, 37–40 [DOI] [PubMed] [Google Scholar]
- 11. Yang S. H., Sharrocks A. D., and Whitmarsh A. J. (2013) MAP kinase signalling cascades and transcriptional regulation. Gene 513, 1–13 [DOI] [PubMed] [Google Scholar]
- 12. Chopra A. P., Boone S. A., Liang X., and Duesbery N. S. (2003) Anthrax lethal factor proteolysis and inactivation of MAPK kinase. J. Biol. Chem. 278, 9402–9406 [DOI] [PubMed] [Google Scholar]
- 13. Tonello F., and Montecucco C. (2009) The anthrax lethal factor and its MAPK kinase-specific metalloprotease activity. Mol. Aspects Med. 30, 431–438 [DOI] [PubMed] [Google Scholar]
- 14. Duesbery N. S., Webb C. P., Leppla S. H., Gordon V. M., Klimpel K. R., Copeland T. D., Ahn N. G., Oskarsson M. K., Fukasawa K., Paull K. D., and Vande Woude G. F. (1998) Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science 280, 734–737 [DOI] [PubMed] [Google Scholar]
- 15. Fang H., Cordoba-Rodriguez R., Lankford C. S., and Frucht D. M. (2005) Anthrax lethal toxin blocks MAPK kinase-dependent IL-2 production in CD4+ T cells. J. Immunol. 174, 4966–4971 [DOI] [PubMed] [Google Scholar]
- 16. Sun C., Fang H., Xie T., Auth R. D., Patel N., Murray P. R., Snoy P. J., and Frucht D. M. (2012) Anthrax lethal toxin disrupts intestinal barrier function and causes systemic infections with enteric bacteria. PLoS ONE 7, e33583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Depeille P., Young J. J., Boguslawski E. A., Berghuis B. D., Kort E. J., Resau J. H., Frankel A. E., and Duesbery N. S. (2007) Anthrax lethal toxin inhibits growth of and vascular endothelial growth factor release from endothelial cells expressing the human herpes virus 8 viral G protein coupled receptor. Clin. Cancer Res. 13, 5926–5934 [DOI] [PubMed] [Google Scholar]
- 18. Fang H., Xu L., Chen T. Y., Cyr J. M., and Frucht D. M. (2006) Anthrax lethal toxin has direct and potent inhibitory effects on B cell proliferation and immunoglobulin production. J. Immunol. 176, 6155–6161 [DOI] [PubMed] [Google Scholar]
- 19. Comer J. E., Chopra A. K., Peterson J. W., and König R. (2005) Direct inhibition of T-lymphocyte activation by anthrax toxins in vivo. Infect. Immun. 73, 8275–8281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moayeri M., and Leppla S. H. (2009) Cellular and systemic effects of anthrax lethal toxin and edema toxin. Mol. Aspects Med. 30, 439–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karin M., Liu Z., and Zandi E. (1997) AP-1 function and regulation. Curr. Opin. Cell Biol. 9, 240–246 [DOI] [PubMed] [Google Scholar]
- 22. Ouyang W., Torigoe C., Fang H., Xie T., and Frucht D. M. (2014) Anthrax lethal toxin inhibits translation of hypoxia-inducible factor 1α and causes decreased tolerance to hypoxic stress. J. Biol. Chem. 289, 4180–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kappelmann M., Bosserhoff A., and Kuphal S. (2014) AP-1/c-Jun transcription factors: regulation and function in malignant melanoma. Eur. J. Cell Biol. 93, 76–81 [DOI] [PubMed] [Google Scholar]
- 24. Xu L., Fang H., and Frucht D. M. (2008) Anthrax lethal toxin increases superoxide production in murine neutrophils via differential effects on MAPK signaling pathways. J. Immunol. 180, 4139–4147 [DOI] [PubMed] [Google Scholar]
- 25. Tournier C., Dong C., Turner T. K., Jones S. N., Flavell R. A., and Davis R. J. (2001) MKK7 is an essential component of the JNK signal transduction pathway activated by proinflammatory cytokines. Genes Dev. 15, 1419–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang X., Destrument A., and Tournier C. (2007) Physiological roles of MKK4 and MKK7: insights from animal models. Biochim. Biophys. Acta 1773, 1349–1357 [DOI] [PubMed] [Google Scholar]
- 27. Talotta F., Mega T., Bossis G., Casalino L., Basbous J., Jariel-Encontre I., Piechaczyk M., and Verde P. (2010) Heterodimerization with Fra-1 cooperates with the ERK pathway to stabilize c-Jun in response to the RAS oncoprotein. Oncogene 29, 4732–4740 [DOI] [PubMed] [Google Scholar]
- 28. Basbous J., Chalbos D., Hipskind R., Jariel-Encontre I., and Piechaczyk M. (2007) Ubiquitin-independent proteasomal degradation of Fra-1 is antagonized by Erk1/2 pathway-mediated phosphorylation of a unique C-terminal destabilizer. Mol. Cell. Biol. 27, 3936–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pakay J. L., Diesch J., Gilan O., Yip Y. Y., Sayan E., Kolch W., Mariadason J. M., Hannan R. D., Tulchinsky E., and Dhillon A. S. (2012) A 19S proteasomal subunit cooperates with an ERK MAPK-regulated degron to regulate accumulation of Fra-1 in tumour cells. Oncogene 31, 1817–1824 [DOI] [PubMed] [Google Scholar]
- 30. Angel P., Hattori K., Smeal T., and Karin M. (1988) The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell 55, 875–885 [DOI] [PubMed] [Google Scholar]
- 31. Dérijard B., Hibi M., Wu I. H., Barrett T., Su B., Deng T., Karin M., and Davis R. J. (1994) JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76, 1025–1037 [DOI] [PubMed] [Google Scholar]
- 32. Wei W., Jin J., Schlisio S., Harper J. W., and Kaelin W. G. Jr. (2005) The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8, 25–33 [DOI] [PubMed] [Google Scholar]
- 33. Ho D. T., Bardwell A. J., Grewal S., Iverson C., and Bardwell L. (2006) Interacting JNK-docking sites in MKK7 promote binding and activation of JNK mitogen-activated protein kinases. J. Biol. Chem. 281, 13169–13179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meng Q., and Xia Y. (2011) c-Jun, at the crossroad of the signaling network. Protein Cell 2, 889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Dam H., Wilhelm D., Herr I., Steffen A., Herrlich P., and Angel P. (1995) ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 14, 1798–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van Dam H., Duyndam M., Rottier R., Bosch A., de Vries-Smits L., Herrlich P., Zantema A., Angel P., and van der Eb A. J. (1993) Heterodimer formation of cJun and ATF-2 is responsible for induction of c-jun by the 243 amino acid adenovirus E1A protein. EMBO J. 12, 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Whitmarsh A. J., and Davis R. J. (1996) Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J. Mol. Med. 74, 589–607 [DOI] [PubMed] [Google Scholar]
- 38. Gupta S., Campbell D., Dérijard B., and Davis R. J. (1995) Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science 267, 389–393 [DOI] [PubMed] [Google Scholar]
- 39. Musti A. M., Treier M., and Bohmann D. (1997) Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science 275, 400–402 [DOI] [PubMed] [Google Scholar]
- 40. Zhang Z., Newton K., Kummerfeld S. K., Webster J., Kirkpatrick D. S., Phu L., Eastham-Anderson J., Liu J., Lee W. P., Wu J., Li H., Junttila M. R., and Dixit V. M. (2017) Transcription factor Etv5 is essential for the maintenance of alveolar type II cells. Proc. Natl. Acad. Sci. U.S.A. 114, 3903–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Maki Y., Bos T. J., Davis C., Starbuck M., and Vogt P. K. (1987) Avian sarcoma virus 17 carries the jun oncogene. Proc. Natl. Acad. Sci. U.S.A. 84, 2848–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cavalieri F., Ruscio T., Tinoco R., Benedict S., Davis C., and Vogt P. K. (1985) Isolation of three new avian sarcoma viruses: ASV 9, ASV 17, and ASV 25. Virology 143, 680–683 [DOI] [PubMed] [Google Scholar]
- 43. Samatar A. A., and Poulikakos P. I. (2014) Targeting RAS-ERK signalling in cancer: promises and challenges. Nat. Rev. Drug Discov. 13, 928–942 [DOI] [PubMed] [Google Scholar]
- 44. Liu S., Wang H., Currie B. M., Molinolo A., Leung H. J., Moayeri M., Basile J. R., Alfano R. W., Gutkind J. S., Frankel A. E., Bugge T. H., and Leppla S. H. (2008) Matrix metalloproteinase-activated anthrax lethal toxin demonstrates high potency in targeting tumor vasculature. J. Biol. Chem. 283, 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu S., Bugge T. H., and Leppla S. H. (2001) Targeting of tumor cells by cell surface urokinase plasminogen activator-dependent anthrax toxin. J. Biol. Chem. 276, 17976–17984 [DOI] [PubMed] [Google Scholar]
- 46. Peters D. E., Hoover B., Cloud L. G., Liu S., Molinolo A. A., Leppla S. H., and Bugge T. H. (2014) Comparative toxicity and efficacy of engineered anthrax lethal toxin variants with broad anti-tumor activities. Toxicol. Appl. Pharmacol. 279, 220–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu S., Liu J., Ma Q., Cao L., Fattah R. J., Yu Z., Bugge T. H., Finkel T., and Leppla S. H. (2016) Solid tumor therapy by selectively targeting stromal endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 113, E4079–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Phillips D. D., Fattah R. J., Crown D., Zhang Y., Liu S., Moayeri M., Fischer E. R., Hansen B. T., Ghirlando R., Nestorovich E. M., Wein A. N., Simons L., Leppla S. H., and Leysath C. E. (2013) Engineering anthrax toxin variants that exclusively form octamers and their application to targeting tumors. J. Biol. Chem. 288, 9058–9065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Abi-Habib R. J., Urieto J. O., Liu S., Leppla S. H., Duesbery N. S., and Frankel A. E. (2005) BRAF status and mitogen-activated protein/extracellular signal-regulated kinase kinase 1/2 activity indicate sensitivity of melanoma cells to anthrax lethal toxin. Mol. Cancer Ther. 4, 1303–1310 [DOI] [PubMed] [Google Scholar]
- 50. Huang D., Ding Y., Luo W. M., Bender S., Qian C. N., Kort E., Zhang Z. F., VandenBeldt K., Duesbery N. S., Resau J. H., and Teh B. T. (2008) Inhibition of MAPK kinase signaling pathways suppressed renal cell carcinoma growth and angiogenesis in vivo. Cancer Res. 68, 81–88 [DOI] [PubMed] [Google Scholar]
- 51. Davies H., Bignell G. R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M. J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., et al. (2002) Mutations of the BRAF gene in human cancer. Nature 417, 949–954 [DOI] [PubMed] [Google Scholar]
- 52. Alfano R. W., Leppla S. H., Liu S., Bugge T. H., Meininger C. J., Lairmore T. C., Mulne A. F., Davis S. H., Duesbery N. S., and Frankel A. E. (2009) Matrix metalloproteinase-activated anthrax lethal toxin inhibits endothelial invasion and neovasculature formation during in vitro morphogenesis. Mol. Cancer Res. 7, 452–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Alfano R. W., Leppla S. H., Liu S., Bugge T. H., Duesbery N. S., and Frankel A. E. (2008) Potent inhibition of tumor angiogenesis by the matrix metalloproteinase-activated anthrax lethal toxin: implications for broad anti-tumor efficacy. Cell Cycle 7, 745–749 [DOI] [PubMed] [Google Scholar]
- 54. King J. W., and Nathan P. D. (2014) Role of the MEK inhibitor trametinib in the treatment of metastatic melanoma. Future Oncol. 10, 1559–1570 [DOI] [PubMed] [Google Scholar]
- 55. Zhu Z., Liu W., and Gotlieb V. (2016) The rapidly evolving therapies for advanced melanoma: towards immunotherapy, molecular targeted therapy, and beyond. Crit. Rev. Oncol. Hematol. 99, 91–99 [DOI] [PubMed] [Google Scholar]
- 56. Lee C. S., Dykema K. J., Hawkins D. M., Cherba D. M., Webb C. P., Furge K. A., and Duesbery N. S. (2011) MEK2 is sufficient but not necessary for proliferation and anchorage-independent growth of SK-MEL-28 melanoma cells. PLoS ONE 6, e17165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liu L., Rao J. N., Zou T., Xiao L., Wang P. Y., Turner D. J., Gorospe M., and Wang J. Y. (2009) Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol. Biol. Cell 20, 4885–4898 [DOI] [PMC free article] [PubMed] [Google Scholar]