Abstract
Aberrant activation of matrix metalloproteinases (MMPs) is a common feature of pathological cascades observed in diverse disorders, such as cancer, fibrosis, immune dysregulation, and neurodegenerative diseases. MMP-9, in particular, is highly dynamically regulated in several pathological processes. Development of MMP inhibitors has therefore been an attractive strategy for therapeutic intervention. However, a long history of failed clinical trials has demonstrated that broad-spectrum MMP inhibitors have limited clinical utility, which has spurred the development of inhibitors selective for individual MMPs. Attaining selectivity has been technically challenging because of sequence and structural conservation across the various MMPs. Here, through a biochemical and structural screening paradigm, we have identified JNJ0966, a highly selective compound that inhibited activation of MMP-9 zymogen and subsequent generation of catalytically active enzyme. JNJ0966 had no effect on MMP-1, MMP-2, MMP-3, MMP-9, or MMP-14 catalytic activity and did not inhibit activation of the highly related MMP-2 zymogen. The molecular basis for this activity was characterized as an interaction of JNJ0966 with a structural pocket in proximity to the MMP-9 zymogen cleavage site near Arg-106, which is distinct from the catalytic domain. JNJ0966 was efficacious in reducing disease severity in a mouse experimental autoimmune encephalomyelitis model, demonstrating the viability of this therapeutic approach. This discovery reveals an unprecedented pharmacological approach to MMP inhibition, providing an opportunity to improve selectivity of future clinical drug candidates. Targeting zymogen activation in this manner may also allow for pharmaceutical exploration of other enzymes previously viewed as intractable drug targets.
Keywords: allosteric regulation, drug action, drug discovery, enzyme inhibitor, enzyme processing, matrix metalloproteinase (MMP), pharmacology
Introduction
MMPs2 are a family of structurally related zinc-binding proteolytic enzymes that digest extracellular matrix proteins and participate in tissue remodeling and signaling events (1). Currently, ∼23 MMPs have been identified, comprising secreted and membrane-bound forms, and different family members share some common structural and functional domains and have varying degrees of substrate specificity. Abnormal expression and activation of MMPs has been implicated in the pathogenesis and pathological progression of several different human diseases that are centered in many different tissues in the periphery and central nervous system (2, 3). Initial clinical exploration of synthetic MMP inhibitors was focused on oncology indications, as preventing the breakdown of tissue matrices and barriers was viewed as a potential mechanism to limit tumor metastasis.
Despite intensive efforts over many years to develop synthetic MMP inhibitors, only a single MMP inhibitor, Periostat, a tetracycline derivative used in periodontal disease, has progressed into regular clinical use (4). Of the ∼50 other clinical trials conducted with active site MMP inhibitors, all have failed due to the onset of significant dose-limiting musculoskeletal toxicity or lack of efficacy (5). These compounds were based on hydroxamic acid or related chelator chemistry that acts to perturb the critical coordinating zinc in the catalytic domain, which results in loss of enzymatic activity. The high degree of sequence conservation and structural similarity in the catalytic domain across MMPs contributes to the broad-spectrum, nonspecific nature of these compounds.
Among the aforementioned clinical studies that did not meet their primary efficacy end points, there have been results that demonstrate some biological proof of concept for a therapeutic effect of MMP inhibition. In an oncology trial using the MMP active site inhibitor marimastat, patients who developed treatment-related musculoskeletal adverse events had significantly better survival rates as compared with those who did not manifest these adverse events (6). These data confirm the biological hypothesis that attaining a therapeutic exposure of an MMP inhibitor can be efficacious in treating human disease but also highlight the observation that significant dose- and exposure-limiting side effects of broad-spectrum inhibitors limit their clinical utility. Although the molecular basis for MMP inhibitor-induced musculoskeletal toxicity is probably complex and not fully understood, it has been hypothesized that the poor selectivity of active site inhibitors contributes to their side effect profile. Therefore, if selective and tolerable MMP inhibitors could be developed, these therapeutic agents could provide powerful tools for the treatment of many debilitating and often terminal diseases.
Several alternative mechanisms for targeting MMPs have been utilized in which the catalytic zinc is not targeted (7, 8). High-resolution structures have identified a specific subsite within the MMP catalytic domain termed the S1′ pocket, and this domain has been exploited in the discovery of several specific inhibitors for MMP-13 (9–11). A similar approach targeting the S1′ pocket was utilized to create compounds that had some selectivity for MMP-2 over the closely related MMP-9 (12, 13). An alternative approach has targeted the hemopexin domain of membrane type-1 matrix metalloproteinase (MT1-MMP) (14), which is well outside the catalytic domain. In growing efforts to identify inhibitor sites outside of the catalytic domain (15, 16), small peptides have been identified that can interfere with the MMP-2 interaction with collagen substrate (17). Although these new approaches hold promise due to enhanced selectivity, the clinical utility of these compounds and others employing alternative inhibitory strategies has not yet been determined.
The activity of MMP-9 (also known as gelatinase B, 92-kDa gelatinase) is typically associated with the breakdown of type IV and V collagens during tissue remodeling. MMP-9 is also significantly up-regulated in many human diseases, and this aberrant activity is thought to contribute to pathological processes (2). Increases in MMP-9 activity appear to be most deleterious at neurovascular boundaries where the integrity of the blood-brain barrier is compromised in response to disease or injury, such as ischemia (18–20), traumatic brain injury (21–23), hemorrhage (24–26), multiple sclerosis (27–29), and spinal cord injury (30–32). A recent paper (33) has also characterized a role for MMP-9 activity in conferring vulnerability on selected populations of motor neurons, which has implications for pathological neurodegeneration in amyotrophic lateral sclerosis. MMP-9–selective inhibitors could prove useful in ameliorating the pathology associated with these diseases.
As described herein, we have discovered a novel pharmacological approach for highly selective inhibition of MMP-9. This approach has identified a small molecule, known as JNJ0966, which prevents conversion of the MMP-9 zymogen, or proenzyme, into the catalytically active enzyme. We have characterized the molecular mechanism of MMP-9 inhibition through a series of biochemical and structural studies, revealing that the compound interacts directly with the MMP-9 zymogen. The specificity of JNJ0966 for inhibiting MMP-9 activation was also explored, as was potential clinical utility in mouse experimental autoimmune encephalomyelitis (EAE), an in vivo model for human neuroinflammatory disorders such as multiple sclerosis.
Results
Identification of proMMP-9 activation inhibitors
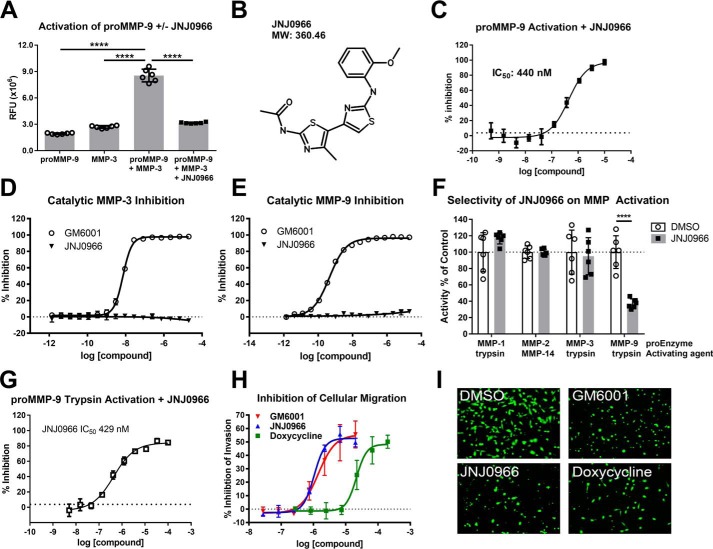
Inhibitors of MMP-9 activation were identified by high-throughput screening using the ThermoFluor® platform to identify compounds that bound to MMP-9 and modified the protein's thermal stability profile (34). Screening against catalytically inactive human MMP-9 (Fig. 1A, amino acids 20–445, termed proMMP-9) purified from bacteria identified several compounds that bound the recombinant protein with varying affinities, one of which, designated JNJ0966 (Fig. 1B), exhibited a ThermoFluor®-determined affinity (KD) of 5.0 μm and was selected for additional profiling. To evaluate the effects of JNJ0966 on proMMP-9 activation, a putative endogenous activation mechanism was utilized, reacting recombinant human proMMP-9 with catalytically active matrix metalloproteinase-3 (catMMP-3). The resultant MMP-9 enzymatic activity was measured by monitoring cleavage of a fluorescent MMP-9–specific substrate, DQ-gelatin, which was not cleaved to a great extent by catMMP-3 or proMMP-9 alone (Fig. 1A). Incubating proMMP-9 with catMMP-3 significantly increased the cleavage of DQ-gelatin relative to proMMP-9 or catMMP-3 alone, indicating that active MMP-9 was generated. However, adding 10 μm JNJ0966 to the reaction before the addition of catMMP-3 significantly reduced subsequent DQ-gelatin cleavage. To evaluate the concentration-response profile of JNJ0966, compound was titrated with a constant concentration of proMMP-9 and catMMP-3, demonstrating an IC50 of 440 nm (95% confidence interval (CI) 341–567 nm) under these assay conditions (Fig. 1C). To exclude the possibility that the effects of JNJ0966 could be explained through a direct inhibition of MMP-3 and/or MMP-9 enzymatic activity, the effects of JNJ0966 on catMMP-3 and active MMP-9 were evaluated. Active MMP-9 (catMMP-9) was generated by first reacting proMMP-9 with catMMP-3. Whereas enzymatic activity could be blocked in both assays by GM6001, a potent, non-selective active site MMP inhibitor (catMMP-3 IC50 = 7.2 nm, 95% CI 6.8–7.7 nm; catMMP-9 IC50 = 0.45 nm, 95% CI 0.42–0.49 nm), JNJ0966 did not exhibit any inhibition of either active enzyme (Fig. 1, D and E). Taken together, these data indicate that JNJ0966 inhibited the conversion of proMMP-9 to active MMP-9 but was not a direct inhibitor of MMP-9 or MMP-3 enzymatic activity.
Figure 1.
JNJ0966 inhibited activation of proMMP-9 but exhibited no effect on catalytic activity or maturation of other MMPs. A, proMMP-9 activation assay utilizing the fluorescent substrate DQ-gelatin. proMMP-9, catMMP-3, or a combination of the two in the presence or absence of 10 μm JNJ0966 was incubated together then with DQ-gelatin. proMMP-9 activated by catMMP-3 exhibited significant increases in activity as compared with either enzyme alone, and this was significantly inhibited by JNJ0966 (n = 6). RFU, relative fluorescence units; ****, p < 0.0001, one-way ANOVA with Bonferroni multiple-comparison post-test. B, molecular structure of JNJ0966 and molecular weight (MW). C, proMMP-9 activation assay characterizing JNJ0966 concentration response with catMMP-3 activation (IC50 = 440 nm). D and E, activity assay in which concentration responses of JNJ0966 and GM6001 were incubated with catMMP-3 (D) or catMMP-9 (E). GM6001 inhibited catMMP-3 (D, IC50 = 7.2 nm) and catMMP-9 (E, IC50 = 0.5 nm), but JNJ0966 had no effect at any concentration (n = 6). F, activation assay on proMMP-1, proMMP-2, proMMP-3, and proMMP-9 activated by trypsin or catMMP-14, as indicated, in the presence (black bars) or absence (white bars) of 10 μm JNJ0966. JNJ0966 only significantly inhibited the activation of proMMP-9 by trypsin. Data were normalized to 100% for each enzyme maximal activity (n = 6; ****, p < 0.001, two-tailed t test). G, proMMP-9 activation assay characterizing JNJ0966 concentration response with trypsin activation (IC50 = 429 nm). H, HT1080 cellular invasion assay demonstrating the concentration-response curves of doxycycline (green squares; IC50 = 21 μm), GM6001 (blue triangles; IC50 = 1.4 μm), and JNJ0966 (red circle; IC50 = 1.0 μm) inhibiting cellular transmigration across a MatrigelTM layer (n = 4). I, representative images from the bottom side of transwells from the indicated treatments illustrate calcein AM–labeled cells that have migrated through Matrigel to the bottom filter insert layer. In all graphs (A, C, D, E, F, and G), data are presented as means ± S.D. (error bars). All curves are fit by nonlinear regression.
The utility of this novel mechanism for MMP inhibition resides in the potential for specificity, as the physiologic pathways for activation of different MMPs appear to be distinct (35). To test the selectivity of JNJ0966 for proMMP-9 versus other MMP family members, proenzyme versions of MMP-1 (proMMP-1), MMP-3 (proMMP-3), and proMMP-9 zymogens were reacted with trypsin as an alternative activating enzyme, and the proenzyme of MMP-2 (proMMP-2) was reacted with a catalytic fragment of MMP-14 (36, 37). In this assay, the activations of proMMP-1, proMMP-2, and proMMP-3 were not significantly different in the presence or absence of 10 μm JNJ0966, whereas proMMP-9 activation by trypsin was significantly attenuated (Fig. 1F). Additional concentration-response studies demonstrated that JNJ0966 inhibited trypsin-dependent proMMP-9 activation to an extent similar to that observed for inhibition of proMMP-9 activation by catMMP-3 (IC50 = 429 nm, 95% CI 405–602 nm; Fig. 1G). These data also indicate that JNJ0966 at 10 μm did not inhibit the enzymatic activity of MMP-1, MMP-2, or MMP-14.
To explore the consequences of inhibiting proMMP-9 activation in a functional cellular context, we utilized an invasion assay with HT1080 cells. Cellular invasion through a MatrigelTM extracellular matrix layer was measured in control cells or in cells treated with increasing concentrations of JNJ0966, GM6001, or doxycycline, the latter of which is a low-potency, non-selective MMP inhibitor. Doxycycline inhibited the invasion of HT1080 cells across the MatrigelTM layer (IC50 = 21.4 μm, 95% CI 13.7–33.5 μm); however, JNJ0966 (IC50 = 1.0 μm, 95% CI 0.8–1.4 μm) and GM6001 (IC50 = 1.4 μm, 95% CI 0.7–2.8 μm) were 21- and 15-fold more potent, respectively (Fig. 1, H and I). These results were consistent with previous reports demonstrating reduced invasion through simulated extracellular matrices in cells with reduced MMP-9 catalytic activity (38–40) and suggested that JNJ0966 inhibited native cellular activation of proMMP-9 in a manner that resulted in reduced capability of treated cells to digest and permeate through the MatrigelTM layer.
JNJ0966 inhibits proMMP-9 maturation
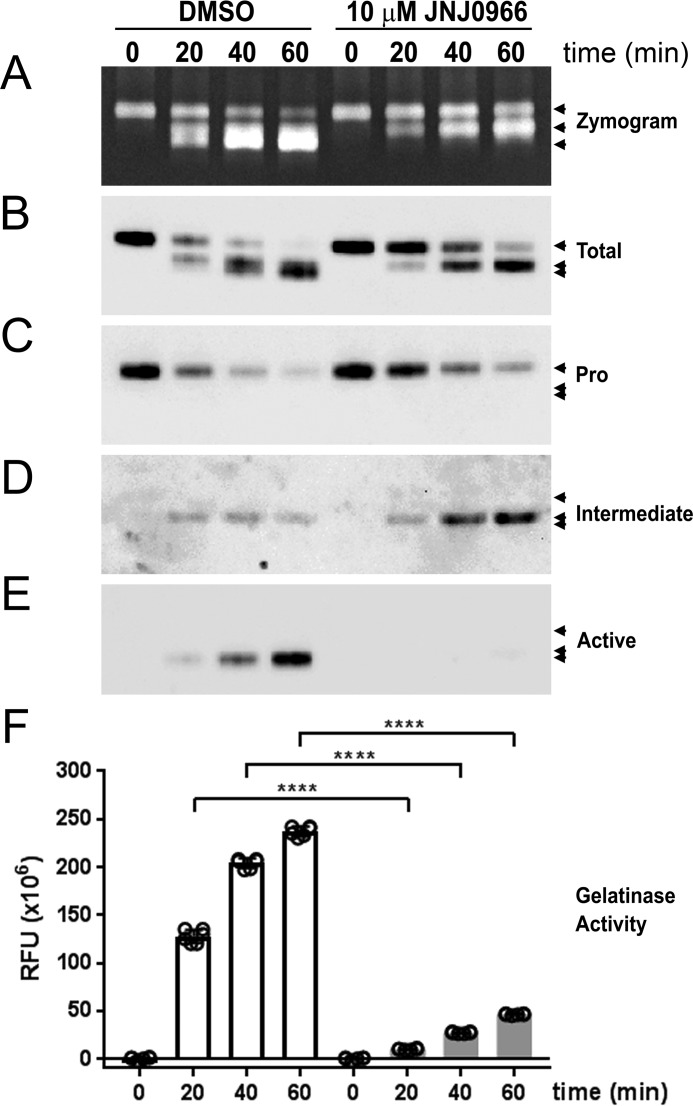
Activation of proMMP-9 is a sequential two-step process in which the full-length proenzyme (92 kDa) is cleaved at Glu-59 to an intermediate form (86 kDa), followed by a second cleavage event at Arg-106 to generate the mature, lower-molecular mass (82 kDa) catalytically active enzyme (41, 42). To confirm and extend the observed biochemical and functional effects of JNJ0966, the activation of proMMP-9 was characterized utilizing a parallel approach, with simultaneous analysis of replicate samples by gelatin zymography, immunoblotting, and DQ-gelatin activity assays measured over time. The immunoblotting studies utilized an antibody that bound to all forms of the protein (pan-MMP-9, or total MMP-9) and antibodies that specifically recognized only proMMP-9, the intermediate form, or fully processed MMP-9. The latter two antibodies were generated against the N-terminal neo-epitopes generated after each of the proMMP-9 cleavage events at Glu-59 and Arg-106 and were specific to those forms, respectively. The anti-proMMP-9 antibody was generated against an epitope residing at the N terminus only in the intact full-length protein. In the absence of JNJ0966, incubating proMMP-9 with catMMP-3 resulted in a time-dependent conversion of proMMP-9 to the intermediate and fully processed forms (Fig. 2, A–E). The addition of JNJ0966 to in the reaction resulted in a significant reduction in fully processed MMP-9 (Fig. 2, A, B, and E) and an apparent accumulation of the intermediate species (Fig. 2, A, B, and D). Analyzing replicate samples in a DQ-gelatin activity assay demonstrated increasing amounts of MMP-9 activity in control conditions that correlated with the accumulation of fully processed MMP-9, whereas in samples that contained JNJ0966, a significant reduction in this lower molecular weight species and enzymatic activity was observed (Fig. 2, E and F).
Figure 2.
JNJ0966 reduced proMMP-9 maturation through intermediate states to active fully processed enzyme. A–F, proMMP-9 samples were reacted with catMMP-3 (0, 20, 40, and 60 min) in the presence or absence of 10 μm JNJ0966. Sample aliquots from the same time points were loaded on replicate gels in each panel (A–E). Three arrowheads in each panel denote the migration of proMMP-9 at 92 kDa, intermediate MMP-9 at 86 kDa, and active MMP-9 at 82 kDa. A, gelatin zymograph demonstrating maturation of proMMP-9 through activation and the effects of reducing active MMP-9 in the presence of JNJ0966. B–E, replicate immunoblots probed with antibodies for total MMP-9 (B), proMMP-9 (C), intermediate MMP-9 (D), and fully processed MMP-9 (E). Combined, the immunoblots demonstrate the accumulation of fully processed MMP-9 in DMSO controls and the reduced conversion of MMP-9 to the mature species in the presence of JNJ0966. F, activity assay to monitor development of gelatinase activity in the same sample aliquots analyzed in A–E (n = 3 for each assay time point; data are represented as means ± S.D. (error bars); ****, p < 0.0001, two-tailed t test).
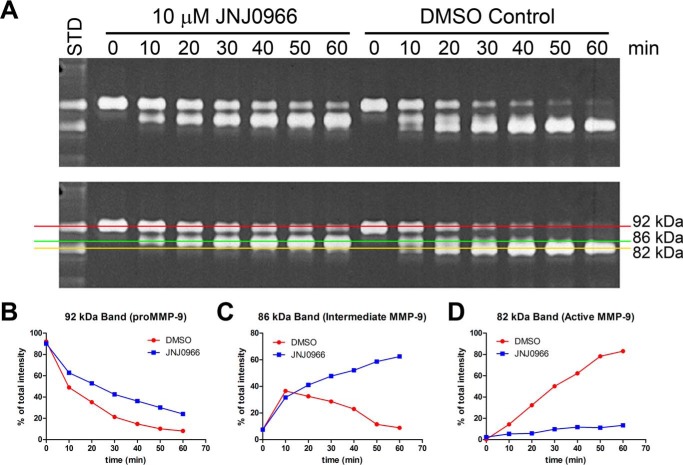
To fully explore the kinetics of MMP-9 maturation in the presence and absence of 10 μm JNJ0966, a more detailed time course was conducted, and the relative abundance of different MMP-9 species was quantified by densitometry of a gelatin zymogram (Fig. 3, A–D). Processing of the 92-kDa full-length proMMP-9 form was reduced by JNJ0966, as more of this species remained at each time point relative to the controls; however, after an initial delay in processing, the conversion from 92-kDa proMMP-9 appeared to proceed at a similar rate in both control and JNJ0966-containing conditions (Fig. 3, A and B). In control conditions, abundance of the 86-kDa intermediate form peaked at the 10-min time point and then declined thereafter; however, in the presence of JNJ0966, the intermediate form accumulated over time (Fig. 3, A and C). The 82-kDa fully processed form of MMP-9 accumulated over time in control conditions, whereas in the presence of JNJ0966, the rate of accumulation was substantially reduced (Fig. 3, A and D). These results indicated that JNJ0966 had a minor effect on reducing the initial processing step in proMMP-9 activation (from 92 to 86 kDa); however, a more robust effect was observed on inhibiting the processing of the intermediate form to the catalytically active species (from 86 to 82 kDa). Interestingly, in ThermoFluor studies, the affinity (KD) of JNJ0966 for proMMP-9 improved from 5 to 0.33 μm when a shorter construct containing amino acids 67–445 was utilized as compared with the longer construct containing amino acids 20–445. No binding of JNJ0966 to catalytically active MMP-9 could be detected. Together, these data indicate that JNJ0966 binds preferentially to the intermediate MMP-9 species and with a reduced affinity for the unprocessed full-length zymogen.
Figure 3.
Analysis of proMMP-9 maturation kinetics by gelatin zymography in the presence and absence of 10 μm JNJ0966. A, gelatin zymogram of proMMP-9 samples that have been reacted with catMMP-3 in the presence and absence of 10 μm JNJ0966 for 0, 10, 20, 30, 40, 50, and 60 min. Zymograms are duplicate pictures; the bottom panel is overlaid with graphical lines to illustrate the three different MMP-9 molecular species (92, 86, and 82 kDa). B–D, graphs of relative abundance of the individual MMP-9 forms as a percentage of total enzyme abundance in each condition as measured by densitometric quantitation of the zymogram. In all graphs, relative abundance of proMMP-9 species is depicted from DMSO control (red lines) and 10 μm JNJ0966 (blue lines) conditions. B, graph of the unprocessed 92-kDa proMMP-9 with (blue lines) and without (red line) JNJ0966. C, graph of the 86-kDa intermediately processed proMMP-9. D, graph of the 82-kDa fully processed MMP-9.
Mechanistic and structural basis of JNJ0966 activity
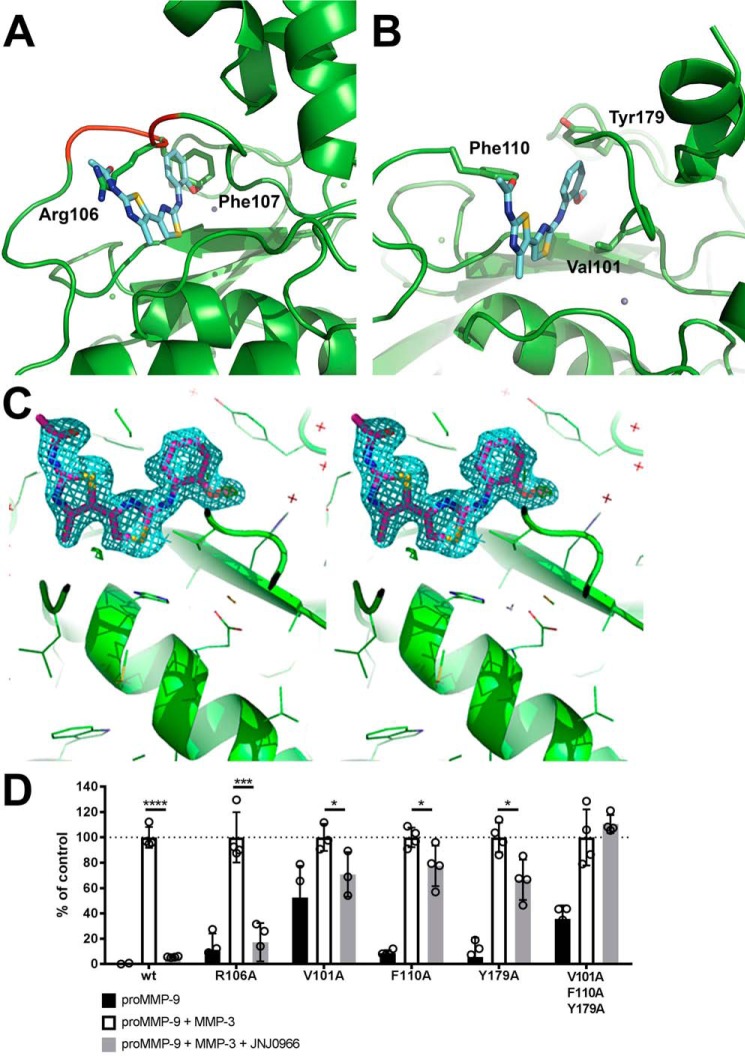
In an effort to characterize the mechanistic basis for JNJ0966 inhibitory activity, crystallization experiments were initiated on proMMP-9 with and without the addition of JNJ0966. A construct was designed starting after the first observable electron density seen in previous MMP-9 structures and extending to the end of the metalloproteinase domain (amino acids 29–444) (43). This construct lacked the fibronectin II domains (amino acids 216–390; proMMP-9desFnII), demonstrated similar JNJ0966-binding affinities in ThermoFluor® assays (KD = 3.3 μm), and exhibited similar structural characteristics of the catalytic and activation domains as compared with constructs that contained the fibronectin II domains (43, 44). Examination of the proMMP-9desFnII crystal structure complexed with JNJ0966 revealed that the JNJ0966 phenoxy moiety bound in a region of space that was occupied by Phe-107 in the unbound proMMP-9desFnII, and the JNJ0966 acetamide group was located in the same location as the Arg-106 guanadino group in the unbound proMMP-9desFnII (Fig. 4, A–C). Both of these differences suggested that proMMP-9desFnII residues near the activation cleavage site were reoriented to accommodate JNJ0966 binding. Residues 107–109 around the activation site (residues 103–108) were disordered in the presence of JNJ0966, with poor observed electron density for this region. No hydrogen bond was observed for the JNJ0966 oxygen, suggesting that this group may impart steric orientation or alter the aromatic ring electronics, both of which may be important for compound binding. The proMMP-9desFnII residues that were proximal to the JNJ0966 phenoxy group include Val-101, Pro-102, Tyr-179, His-190 (coordinated to the structural zinc), and Phe-192. The JNJ0966 inner thialzole ring was located near proMMP-9desFnII residues Phe-110 and His-405 (coordinated to the catalytic zinc). A 2.8-Å contact was also observed between the aniline nitrogen and the backbone carbonyl of Ala-191. In the absence of inhibitor, several solvent molecules occupied the space where the two thiazole rings in the JNJ0966 structure were situated. The terminal methyl thiazole ring was located near residues Arg-106, Leu-114, and Asp-410. Interestingly, the interactions of Cys-99 remained consistent between the JNJ0966-bound and -unbound proMMP-9desFnII structures. This residue in the pro-domain coordinates with zinc bound in the catalytic domain and is involved in zymogen activation for virtually all MMPs. No differences were observed in the zinc coordination of either the catalytic or structural zinc ions, indicating JNJ0966-induced structural perturbation appeared to be limited to the region between proMMP-9desFnII residues 103 and 108. Crystallographic and refinement statistics are provided in Table 1.
Figure 4.
Structural characterization of JNJ0966 and proMMP-9 complex. A, superposition of JNJ0966 structure on the proMMP-9 coordinates. A stick diagram of JNJ0966 (carbon backbone is represented in cyan, oxygen in red, nitrogen in blue, and sulfur in yellow) is superimposed on the ribbon diagram of uncomplexed proMMP-9 (green), demonstrating how JNJ09066 occupies space typically occupied by the side chains of Arg-106 and Phe-107 (green sticks). Residues 105–109 are shown in red on the proMMP-9 backbone. B, structure of JNJ0966 in a complex with proMMP9. In the ribbon diagram of proMMP9, residues near the interface with JNJ0966 are labeled in black (Val-101, Phe-110, and Tyr-179). The activation loop (residues 103–108) was disordered in the JNJ0966-MMP-9 structure. C, electron density of the proMMP9 JNJ0966 complex. The figure is in wall-eyed stereo. The 2|Fo| − |Fc| map (cyan) at the 1σ contour level shows the unambiguous binding mode of JNJ0966. D, activation assay comparing wild-type and amino acid substitution mutant versions of proMMP-9 activated by catMMP-3 in the presence or absence of 10 μm JNJ0966, as measured by DQ-gelatin cleavage. Basal activity (black bars), basal activity plus catMMP-3 activation (open bars), and basal activity plus catMMP-3 with 10 μm JNJ0966 (hatched bars) are shown for each MMP-9 mutant. Data are normalized to 100% for each MMP-9 mutant plus catMMP-3 levels. The R106A mutation in MMP-9 had little impact on the activity of JNJ0966, whereas V101A, F110A, and Y179A reduced the magnitude of the inhibitory effect. The triple mutant of the latter three residues abolished the effects of JNJ0966 in inhibiting activation. Data are represented as means ± S.D. (error bars), n = 4. *, p < 0.05; ***, p < 0.001; ****, p < 0.0001, two-tailed t test.
Table 1.
Crystallographic and refinement statistics for unbound proMMP-9 and proMMP-9 complexed with JNJ0966
| Parameter | Unbound proMMP-9 | proMMP-9 + JNJ0966 |
|---|---|---|
| PDB code | 5UE3 | 5UE4 |
| Space group | C2 | C2 |
| Unit cell dimensions | ||
| a, b, c (Å) | 90.28, 73.24, 77.51 | 89.82, 72.95, 77.54 |
| α, β, γ (degrees) | 90.00, 106.26, 90.00 | 90.00, 106.91, 90.00 |
| Molecules per asymmetric unit | 2 | 2 |
| Mosaicity | 0.37 | 1.24 |
| Resolution range | 49.19–1.60 (1.66–1.60)a | 43.00–1.80 (1.86–1.80) |
| Total no. of reflections (F > 0) | 200,188 | 144,023 |
| No. of unique reflections | 62,722 | 44,322 |
| Average redundancy | 3.19 (3.19) | 3.25 (3.37) |
| Completeness (%) | 98.1 (97.2) | 99.7 (99.9) |
| Rmerge | 0.048 (0.181) | 0.052 (0.335) |
| Reduced χ2 | 0.99 (1.09) | 0.95 (0.79) |
| Output 〈I/σI〉 | 13.4 (5.2) | 10.5 (2.4) |
| Wilson B-factor | 14.28 | 29.7 |
| Reflections used in refinement | 62,702 (6158) | 44,303 (4430) |
| Reflections used for Rfree | 6345 (606) | 2000 (201) |
| Rwork | 0.1545 (0.1689) | 0.1991 (0.2902) |
| Rfree | 0.1838 (0.2139) | 0.2243 (0.3064) |
| Protein atoms | 3591 | 3553 |
| Ligand atoms | 0 | 48 |
| Protein residues | 458 | 456 |
| Root mean square deviation | ||
| Bonds (Å) | 0.011 | 0.007 |
| Angles (degrees) | 1.24 | 1.075 |
| Ramachandran statistics (%) | ||
| Favored | 97 | 97 |
| Allowed | 2.7 | 2.5 |
| Outliers | 0.23 | 0.23 |
| Rotamer outliers (%) | 0 | 1.7 |
| Clashscore | 4.3 | 6.18 |
| B-factor | ||
| Average | 21.06 | 32.37 |
| Average protein | 18.87 | 37.08 |
| Average ligand | 34.35 | |
| Average water | 33.49 | 37.42 |
a Data for the highest-resolution shell are shown in parentheses.
High-resolution structural analysis predicted several amino acids within proMMP-9 that were important for interaction with JNJ0966. To test this hypothesis and further confirm the molecular nature of the interaction site, several amino acid point substitution mutants were generated near the Arg-106 activation site and within the putative JNJ0966 binding pocket identified through structural studies. Purified MMP-9 proteins containing the amino acid substitutions were tested in DQ-gelatin activation assays to assess basal activity of the zymogen, activation by catMMP-3, and potential inhibition of activation by JNJ0966 (Fig. 4D). An alanine mutation at the proMMP-9 Arg-106 cleavage site did not prevent activation by catMMP-3, and activation was significantly inhibited by JNJ0966 to a degree comparable with wild-type proMMP-9 (17.2 ± 7.6 and 5.6 ± 0.2% of activated control, respectively). A proMMP-9 construct containing an alanine mutation at Val-101 exhibited a higher basal activity level (52.5 ± 12.5% of activated control) but was still able to be activated further by catMMP-3, and this activation was significantly inhibited to near basal levels (for this construct) by JNJ0966 (70.8 ± 10.2% of activated control). In contrast, alanine mutations at Phe-110 and Tyr-179 reduced the effectiveness of JNJ0966. Both proteins exhibited low basal zymogen activity levels and could be activated by catMMP-3, yet JNJ0966 reduced activity to only 77.5 ± 8.0 and 66.6 ± 7.9% of activated control, respectively (Fig. 4C). Triple alanine mutations (Val-101, Phe-110, and Tyr-179) in the identified binding pocket completely prevented JNJ0966 from blocking proMMP-9 activation (110.6 ± 3.5% of activated control; Fig. 4D).
JNJ0966 reduces motor deficits in EAE
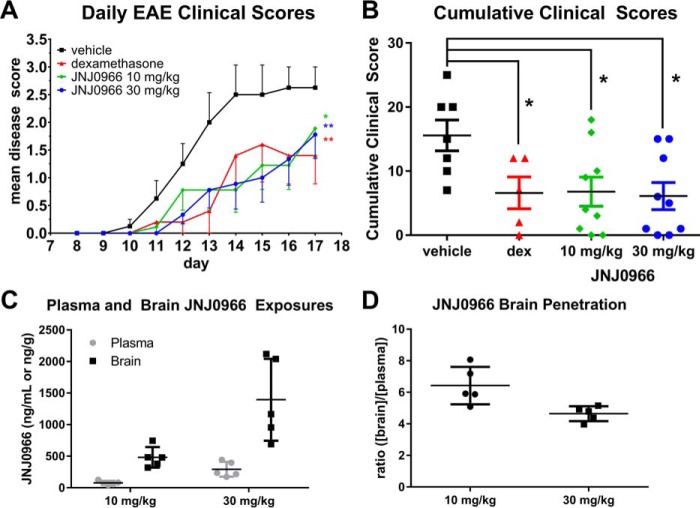
To investigate the potential use of proMMP-9 activation inhibitors as therapeutic agents, JNJ0966 was evaluated in the mouse EAE model, a widely used animal model of human neuroinflammatory disease, including multiple sclerosis. EAE was induced on day 0 via injection of myelin oligodendrocyte glycoprotein synthetic peptide. On day 8 following immunization, mice received a twice daily doses of vehicle, dexamethasone (1 mg/kg; a positive control in this model), 10 mg/kg JNJ0966, or 30 mg/kg JNJ0966 by oral gavage. Animals were scored based on a clinical observation scale of motor performance with increasing disability indicated by a higher disease score. Animals treated with vehicle exhibited initial symptoms on day 10, with mean disability scores increasing daily until study termination on day 17 (Fig. 5A). Dexamethasone-treated animals exhibited significantly reduced clinical disability as compared with the vehicle group, as did animals treated with either dose of JNJ0966 (Fig. 5A). The JNJ0966-treated groups did not differ significantly from dexamethasone-positive controls. The therapeutic effects of JNJ0966 and dexamethasone were confirmed in evaluating the cumulative clinical scores from the study, in which clinical scores from all three treated groups were significantly different from vehicle (Fig. 5B).
Figure 5.
JNJ0966 reduced motor symptoms in mouse EAE and penetrates into the brain. A, mice that had induced EAE were treated twice daily by oral gavage with vehicle (black squares), JNJ0966 at 10 mg/kg (green diamonds), or 30 mg/kg (blue circles) or dexamethasone (1 mg/kg; red triangles). Animals receiving either dose of JNJ0966 or dexamethasone evidenced delayed onset of motor disability and also had significantly reduced disease courses relative to the vehicle controls. Data were analyzed via repeated-measures ANOVA with Bonferroni multiple-comparison post-test to examine differences between the group (n = 7 for vehicle group, n = 5 for dexamethasone group, n = 9 for JNJ0966 10 mg/kg group, and n = 9 for JNJ0966 30 mg/kg group (*, p < 0.05; **, p < 0.01). Points, mean clinical scores; bars, S.E. B, cumulative clinical scores for individual animals plotted for vehicle, dexamethasone, JNJ0966 10 mg/kg, and JNJ0966 30 mg/kg. All treatment groups were significantly lower than vehicle (one-way ANOVA with Bonferroni multiple-comparison post-test; *, p < 0.05). C, plasma (gray circles) and brain (black squares) concentrations of JNJ0966 after 10- or 30-mg/kg dosing in five animals on the last day of dosing (day 17) in the EAE study. D, ratio of brain to plasma concentrations of JNJ0966 after 10-mg/kg (circles) or 30-mg/kg (squares) dosing for animal exposures shown in C. For C and D, individual values are plotted, with bars for means and S.D.
To investigate JNJ0966 penetration into the central nervous system, terminal plasma and brain samples were analyzed, and the amount of JNJ0966 in each compartment was determined. The exposures of JNJ0966 were dose-dependent, with plasma and brain concentrations for the 10-mg/kg dose of 77.5 ± 31.1 ng/ml (215 nm) and 481.6 ± 162.5 ng/g (∼1336 nm), respectively, whereas the 30-mg/kg dose achieved 293.6 ± 118.4 ng/ml (815 nm) in plasma and 1394.0 ± 649.1 ng/g (∼3867 nm) in brain (Fig. 5C). JNJ0966 was preferentially partitioned in brain, with brain/plasma ratios of 6.2 for the 10-mg/kg dose and 4.7 for the 30-mg/kg dose (Fig. 5D). Concentrations of JNJ0966 in the brain at either dose were consistent with exposure levels that were necessary to inhibit proMMP-9 activation based on in vitro IC50 values (440 nm; Fig. 1C) and ThermoFluor-determined affinity (KD = 0.33 μm for the 67–445 construct, similar to the intermediate form).
Discussion
Several decades of intense investigation have not yielded therapeutically viable MMP inhibitors. This has been historically attributed to the generally poor specificity of active site–directed MMP inhibitors, leading to dose-limiting toxicities and adverse side effects. Here we describe the discovery and characterization of a highly selective MMP-9 inhibitor that functions through a novel molecular mechanism targeting zymogen activation. This new approach may allow for the development of selective inhibitors while exploring a structural space on MMP enzymes outside of the prototypical substrate binding and catalytic domains. Such inhibitors with enhanced selectivity may allow for identification of clinical candidates unencumbered by the toxicities associated with previous-generation MMP inhibitors. The studies performed here indicate that JNJ0966 was selective (Fig. 1F), able to penetrate the central nervous system (Fig. 5C), and efficacious in reducing EAE disease severity (Fig. 5, A and B). Future work aimed at more fully characterizing JNJ0966 and related compounds may therefore produce therapeutics useful for targeting the plethora of diseases impacted by pathological MMP-9 activity.
The endogenous physiological activation mechanism for MMP-9 remains unclear. We and others have shown MMP-9 can be readily activated by MMP-3 (Fig. 1A) (41, 42) and trypsin (Fig. 1, F and G) (42, 45) or by other serine proteases (46). Other studies have suggested a role for plasmin in MMP-3–independent activation of MMP-9 (47), although this remains controversial, as other reports indicate that plasmin is only a poor direct activator (48) and probably exerts an effect on proMMP-9 activation through activation of latent MMP-3 (49). Urokinase plasminogen activator has also recently been proposed to play a role in direct MMP-9 activation (50). Clearly, it remains an open question as to which protease or proteases activate MMP-9 in vivo, and it is possible that this may vary in different tissues and disease states. It is interesting that divergent enzymes, such as MMP-3 and trypsin, produce similar cleavage patterns in MMP-9 and generate the same neo-epitope starting at amino acid Phe-107 (41, 51), suggesting that Arg-106 in the MMP-9 activation site is generally susceptible to protease cleavage. These results, combined with our structural and functional data, indicate that molecules like JNJ0966 may inhibit proMMP-9 activation regardless of which enzyme is responsible for activation through a direct interaction with the proMMP-9 zymogen activation process. Additional studies examining JNJ0966 inhibition of proMMP-9 activation with other potential activating proteases or activation through other biochemical mechanisms (52) need to be conducted to substantiate this possibility.
Given the nature of the interaction between JNJ0966 and proMMP-9 and the structural reorientation observed near the Arg-106/Phe-107 activation site (Fig. 4, A–C), the reduction in initial cleavage of full-length proMMP-9 at Glu-59/Met-60 to generate the intermediate form is somewhat surprising (Figs. 2C and 3B). It is possible that JNJ09066 binding causes an allosteric perturbation that reduces cleavage at the Glu-59/Met-60 site; however, we did not observe any evidence for this in our structural studies. For the second cleavage site (Arg-106/Phe-107), the mechanism of inhibition was better resolved. The region that we have termed the “activation loop” (amino acids 103–108) was clearly displaced by the binding of JNJ0966 in the structure as compared with unbound proMMP-9desFnII (Fig. 4, A–C). The reorientation of this region caused by compound binding or the physical effect of compound binding near this region appears to render the activation loop a less preferred substrate for cleavage. It is also possible that JNJ0966 functions by limiting the mobility of residues near the cleavage site such that they can no longer engage in the activating enzyme catalytic pocket. Whereas the exact molecular mechanisms of how JNJ0966 may inhibit cleavage of the proMMP-9 activation loop remain somewhat unclear, whatever the nature of this effect, JNJ0966 appears to dramatically reduce the efficiency at which this domain is cleaved.
Oral administration of JNJ0966 significantly diminished the clinical disease score observed in the mouse EAE model, demonstrating the potential clinical utility of inhibiting proMMP-9 activation as a therapeutic approach. This observation is consistent with previous studies that have demonstrated efficacy of MMP active site inhibitors in the EAE model (53, 54). Although a beneficial effect of some unknown off-target pharmacology cannot be excluded, it can be reasonably concluded that the therapeutic effects in animals treated with JNJ0966 result from reduced overall MMP-9 activity. Despite the compelling nature of the EAE efficacy observations in this neuroinflammatory model, future studies in models for other indications, such as oncology, fibrosis, and different neurodegenerative diseases, would be required to demonstrate effectiveness and proof of concept in these diverse indications.
These data in toto represent a significant advance in the field of MMP inhibition that may facilitate the design of future clinical candidates with improved selectivity and safety profiles as compared with previous generations of MMP inhibitors. If therapeutic levels of specific MMP inhibitors can be safely achieved in patients, these compounds could address potential pathogenic features of several human diseases.
Experimental procedures
Materials and animal welfare
All reagents were purchased from Sigma unless otherwise indicated. All compounds were dissolved in DMSO for in vitro studies and diluted further in DMSO for all concentration-response studies before dilution in assay-appropriate aqueous buffers. Final DMSO concentrations in all in vitro assays were <0.1%, matched in all tested samples, and did not interfere with assay performance. Incubations were performed at room temperature unless otherwise specified. Animal protocols were approved by the Janssen Research and Development institutional animal care and use committee and conform to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
JNJ0966 compound information
The compound known as JNJ0966 (Fig. 1B) was identified from a commercially available compound library sourced from ChemBridge Corp. (San Diego, CA). The chemical name for JNJ096 is N-{2-[(2-methoxyphenyl)amino]-4′-methyl-4,5′-bi-1,3-thiazol-2′-yl}acetamide (ChemBridge catalogue no. 5935943). The Chemical Abstracts Service registry number is 315705-75-0.
MMP protein reagent generation
Human proMMP-9 cDNA was transfected into COS-1 cells (ATCC, Manassas, VA) seeded in a cell factory. Cells were grown for 48 h, and then conditioned media (CM) was collected and cleared by centrifugation (5000 × g for 20 min at 4 °C). CM was concentrated on a stirred cell (10,000 molecular weight cut-off) and dialyzed overnight in assay buffer (AB; 50 mm Hepes, pH 7.5, 10 mm CaCl2, 0.05% Brij-35). proMMP-9 was purified from CM utilizing a gelatin-Sepharose 4B column (GE Healthcare). The column was washed with AB, and proMMP-9 was eluted in AB supplemented with 10% DMSO. Column eluate was dialyzed overnight at 4 °C against AB and then aliquoted and frozen at −80 °C. Recombinant catalytic human MMP-3 (catMMP-3) was purchased from Life Diagnostic, Inc. (West Chester, PA). Human proMMP-1, proMMP-2, proMMP-3, and active MMP-14 were purchased from EMD Millipore (Billerica, MA). Mutant versions of proMMP-9 were generated using standard primer-based PCR mutagenesis strategies, and all constructs were confirmed by sequencing. Mutant versions of proMMP-9 were produced and purified from transfected COS-1 cells as described above.
MMP activation and activity assays
Activation and activity assays were conducted in Proxiplate-384 Plus F plates (PerkinElmer Life Sciences). For activation assays, compounds were mixed with proMMP-9 (20 nm) for 30 min, and then catMMP-3 (10 nm) was added and incubated for 30 min at 37 °C. DQ-gelatin (Molecular Probes/Thermo Fisher Scientific) was then added to a final concentration of 10 μg/ml. After 10 min, EDTA (10 mm) was added to stop the reaction, and DQ-gelatin fluorescence was measured. Background fluorescence (wells containing only catMMP-3 and DQ-gelatin) was subtracted from all wells. Mean percentage inhibition (% Inh) was calculated (% Inh = (PosCtrl-sample)/PosCtrl), where PosCtrl represents values from wells containing proMMP-9, catMMP-3, and DQ-gelatin. For activity assays, catMMP-3 (10 nm) or activated MMP-9 (30 nm) was mixed with compounds for 30 min. A fluorescence-quench synthetic peptide substrate (200 nm acetyl-Cys(Eu)-Pro-Leu-Gly-Leu-Lys(QSY7)-Ala-Arg-amide) or DQ-gelatin was added and incubated for 15 min and then measured. Substrate background was subtracted from all wells, and mean percentage inhibition was calculated as above. IC50 values were determined by nonlinear regression analysis in Prism 5 (sigmoidal dose response, variable slope; GraphPad Software, San Diego, CA). Activation assays that included the use of trypsin were conducted similarly, with the exception of adding bovine pancreatic trypsin inhibitor to the assay before DQ-gelatin incubation to limit trypsin digestion of the substrate.
Cellular migration assay and gelatin zymography
Cellular invasion assays were performed in FluoroBlokTM 96-well plates (BD Biosciences) precoated with MatrigelTM, as described previously (38–40). HT1080 cells (ATCC) were added (13,000 cells/well) in 10 μl of serum-free medium to the top chamber with test compound. The bottom feeder tray was filled with serum-free medium supplemented with 6% fetal bovine serum and test compounds and incubated at 5% CO2, 37 °C for 24 h. Calcein AM (4 μg/ml; Molecular Probes/ThermoFisher Scientific) was added to the feeder layer to label cells, and fluorescence intensity from cells that had migrated to the bottom layer was quantified. Mean percentage inhibition of invasion and IC50 values were calculated as above. Images of migrated cells were acquired on a Zeiss (Thornwood, NY) AX10 inverted microscope with a Plan-APOCHROMAT ×20 objective (0.75 numeric aperture) with a Hamamatsu (Bridgewater, NJ) ORCA-R2 digital camera.
Gelatin zymography was done essentially as described previously (55). Briefly, MMP-9 recombinant protein from activation assays was size-separated via electrophoresis through polyacrylamide gels impregnated with gelatin (Invitrogen/Thermo Fisher Scientific). Gels were then stained with Coomassie Blue and destained overnight to reveal cleared, non-stained regions that resulted from in-gel gelatin hydrolysis by MMP-9.
Antibody generation
Monoclonal antibodies against total MMP-9 were developed in collaboration with Dr. James S. Trimmer (University of California, Davis, CA) according to standard protocols (56). The L51/82 hybridoma was derived from mice immunized with human MMP-9 residues 107–445. L51/82 is available from NeuroMab (Davis, CA). The MMP-9 pro-domain and both cleavage site–specific antibodies were generated at New England Peptide (Gardner, MA). Anti-proMMP-9 was raised and purified against amino acids 39–51 (LTDRQLAEEYLYRC). Anti-intermediate antibodies were raised and purified against residues 60–65 (MRGESKC) and cross-purified against a read-through peptide consisting of residues 53–65 (CGYTRVAEMRGESK). Anti-activated MMP-9 antibodies were raised and purified against MMP-9 residues 107–113 (FQTFEGDC) and cross-purified against a read-through peptide of residues 99–113 (CGVPDLGRFQTFEGD). Read-through peptides were used to deplete any antibodies that were not specific for the very amino terminus of the immunogen. Immunoblots were run and developed through standard protocols (56).
Crystallography
Utilizing an overlapping PCR strategy, human proMMP9 constructs were designed to encode for amino acids 29–444 that lacked the fibronectin II (FnII) domain, amino acids 216–390 (proMMP-9desFnII), and subcloned into pET-11a (EMD Millipore). The FnII domain is distal to the pro-domain cleavage site, and similar constructs have been successfully crystallized (PDB code 1GKC) (44). The construct was transformed into BL21(DE3) RIL cells (Agilent Technologies, Santa Clara, CA), grown overnight at 37 °C, and then subcultured until an OD of 0.6 was achieved. Expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h. Purification was performed according to published methods (43). Briefly, inclusion bodies were solubilized in extraction buffer (6 m urea, 20 mm Tris, 1 mm DTT, 5 mm EDTA), captured by anion exchange chromatography, and eluted in a linear NaCl gradient. Pooled protein was refolded in 20 mm Tris, pH 7.5, 150 mm NaCl, 10 mm CaCl2, 10 mm ZnCl2, 10 mm l-arginine, 10 mm reduced and oxidized glutathione and then concentrated and dialyzed against 20 mm Tris, pH 7.5, 150 mm NaCl, 10 mm 1,10-phenanthroline and further purified on size exclusion chromatography. Crystals of proMMP-9desFnII were grown by adding 1 μl of protein to 1 μl of crystallization buffer (25% PEG 8000, 1% glycerol, 0.2 m ammonium sulfate, and 100 mm sodium cacodylate, pH 5.5). X-ray data of proMMP-9desFnII crystals were collected at the ESRF synchrotron beamline ID23 1 (Grenoble, France). Crystals diffracted to 1.7 Å, and data were processed with d*trek (57). The structure was solved by molecular replacement using the program EPMR (58) with the published MMP-9 structure as the search molecule (PDB code 1L6J) (43). Two molecules of proMMP-9desFnII were found in the asymmetric unit. The data were refined with the programs CNX (Accelrys) and Phenix. The first residue visible in electron density at the N terminus was Asp-41. Co-crystallization trials with activation inhibitors did not produce crystals; however, soaking proMMP-9desFnII crystal for 24 h with 0.8 mm JNJ0966 was successful. Data on JNJ0966-soaked crystals were collected at IMCA-CAT at the Advanced Photon Source (Argonne, IL). Data were processed with the d*trek program (58) and refined with the program Phenix (59). Relevant collection statistics for all data sets are found in Table 1. Atomic coordinates for proMMP-9desFnII alone (5UE3) and proMMP-9desFnII complexed with JNJ0966 (5UE4) have been deposited in the PDB.
EAE in mice
EAE studies were conducted in female C57Bl/6 mice age 6–8 weeks. Complete Freund's adjuvant (CFA) was prepared by mixing Mycobacterium tuberculosis H37 Ra (8 mg/ml, Difco/BD Biosciences) in incomplete Freund's adjuvant (Difco/BD Biosciences). Synthetic myelin oligodendrocyte glycoprotein (MOG) peptide (4 mg/ml) was emulsified in CFA in a 1:1 ratio (v/v). EAE was induced on day 0 by subcutaneous injection of 100 μl of MOG/CFA in the tail and IP injection of 200 ng of pertussis toxin. On day 2, animals received an additional IP injection of pertussis toxin (200 ng). Animals were assessed daily and scored according to clinical severity: 0, no abnormality; 1, limp tail; 2, limp tail and hind limb weakness; 3, partial hind limb paralysis; 4, complete hind limb paralysis. Raters for motor scores were blinded to treatment and treatment group. Before the onset of symptoms on day 8, mice were randomly assigned to groups. Vehicle-treated animals received 20% hydroxypropyl-β-cyclodextrin via oral gavage. JNJ0966 and dexamethasone were dissolved in 20% hydroxypropyl-β-cyclodextrin, such that animals received 10 or 30 mg of drug per kg of body weight (mg/kg) of JNJ0966 and 1 mg/kg of dexamethasone; both drugs were administered by oral gavage. All groups were dosed twice daily every day until day 17 of the study, at which point animals were sacrificed, and plasma and brain tissue were collected to analyze JNJ0966 levels using mass spectroscopy.
Author contributions
R. H. S. and K. J. R. jointly conceived of the idea for identifying activation inhibitors. R. H. S., K. J. R., M. T., L. C. K., and R. A. wrote and edited the manuscript. R. H. S., S. L. B., M. S., C. H., F. L., E. D., D. M., J. S., I. D., K. I. C., K. D., B. T., C. M., S. B., R. W., and C. S.-H. conceived and developed biochemical reagents, conducted experiments, and performed subsequent analyses. K. L., Y.-M. Z., and P. J. conceived and developed all small molecule chemistry. M. T. and T. M. H. conceived and conducted biophysical screening. R. A., F. L., C. M., J. S., M. T., and L. C. K. conceived, conducted, and analyzed crystallography studies.
Acknowledgments
We sincerely thank H. Moore Arnold for thoughtful assistance in reviewing and editing the manuscript. Use of the IMCA-CAT beamline 17-ID (or 17-BM) at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with the University of Chicago. Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357.
This research was fully supported by Janssen Research and Development, LLC. During the time this work was performed, all authors were full-time employees of Janssen Research and Development.
This article was selected as one of our Editors' Picks.
The atomic coordinates and structure factors (codes 5UE3 and 5U4E) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- MMP
- matrix metalloproteinase
- MT1-MMP
- membrane type-1 matrix metalloproteinase
- CM
- conditioned medium
- FnII
- fibronectin II
- EAE
- experimental autoimmune encephalomyelitis
- CFA
- Complete Freund's adjuvant
- MOG
- myelin oligodendrocyte glycoprotein
- CI
- confidence interval
- PDB
- Protein Data Bank
- AB
- assay buffer
- ANOVA
- analysis of variance.
References
- 1. Page-McCaw A., Ewald A. J., and Werb Z. (2007) Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 8, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fingleton B. (2007) Matrix metalloproteinases as valid clinical targets. Curr. Pharm. Des. 13, 333–346 [DOI] [PubMed] [Google Scholar]
- 3. Baranger K., Rivera S., Liechti F. D., Grandgirard D., Bigas J., Seco J., Tarrago T., Leib S. L., and Khrestchatisky M. (2014) Endogenous and synthetic MMP inhibitors in CNS physiopathology. Prog. Brain Res. 214, 313–351 [DOI] [PubMed] [Google Scholar]
- 4. Golub L. M., Sorsa T., Lee H. M., Ciancio S., Sorbi D., Ramamurthy N. S., Gruber B., Salo T., and Konttinen Y. T. (1995) Doxycycline inhibits neutrophil (PMN)-type matrix metalloproteinases in human adult periodontitis gingiva. J. Clin. Periodontol. 22, 100–109 [DOI] [PubMed] [Google Scholar]
- 5. Skiles J. W., Gonnella N. C., and Jeng A. Y. (2004) The design, structure, and clinical update of small molecular weight matrix metalloproteinase inhibitors. Curr. Med. Chem. 11, 2911–2977 [DOI] [PubMed] [Google Scholar]
- 6. King J., Zhao J., Clingan P., and Morris D. (2003) Randomised double blind placebo control study of adjuvant treatment with the metalloproteinase inhibitor, Marimastat in patients with inoperable colorectal hepatic metastases: significant survival advantage in patients with musculoskeletal side-effects. Anticancer Res. 23, 639–645 [PubMed] [Google Scholar]
- 7. Pirard B. (2007) Insight into the structural determinants for selective inhibition of matrix metalloproteinases. Drug Discov. Today 12, 640–646 [DOI] [PubMed] [Google Scholar]
- 8. Fields G. B. (2015) New strategies for targeting matrix metalloproteinases. Matrix. Biol. 44, 239–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Engel C. K., Pirard B., Schimanski S., Kirsch R., Habermann J., Klingler O., Schlotte V., Weithmann K. U., and Wendt K. U. (2005) Structural basis for the highly selective inhibition of MMP-13. Chem. Biol. 12, 181–189 [DOI] [PubMed] [Google Scholar]
- 10. Gege C., Bao B., Bluhm H., Boer J., Gallagher B. M., Korniski B., Powers T. S., Steeneck C., Taveras A. G., and Baragi V. M. (2012) Discovery and evaluation of a non-Zn chelating, selective matrix metalloproteinase 13 (MMP-13) inhibitor for potential intra-articular treatment of osteoarthritis. J. Med. Chem. 55, 709–716 [DOI] [PubMed] [Google Scholar]
- 11. Schnute M. E., O'Brien P. M., Nahra J., Morris M., Howard Roark W., Hanau C. E., Ruminski P. G., Scholten J. A., Fletcher T. R., Hamper B. C., Carroll J. N., Patt W. C., Shieh H. S., Collins B., Pavlovsky A. G., et al. (2010) Discovery of (pyridin-4-yl)-2H-tetrazole as a novel scaffold to identify highly selective matrix metalloproteinase-13 inhibitors for the treatment of osteoarthritis. Bioorg. Med. Chem. Lett. 20, 576–580 [DOI] [PubMed] [Google Scholar]
- 12. Fabre B., Filipiak K., Díaz N., Zapico J. M., Suárez D., Ramos A., and de Pascual-Teresa B. (2014) An integrated computational and experimental approach to gaining selectivity for MMP-2 within the gelatinase subfamily. Chembiochem 15, 399–412 [DOI] [PubMed] [Google Scholar]
- 13. Zapico J. M., Puckowska A., Filipiak K., Coderch C., de Pascual-Teresa B., and Ramos A. (2015) Design and synthesis of potent hydroxamate inhibitors with increased selectivity within the gelatinase family. Org. Biomol. Chem. 13, 142–156 [DOI] [PubMed] [Google Scholar]
- 14. Remacle A. G., Golubkov V. S., Shiryaev S. A., Dahl R., Stebbins J. L., Chernov A. V., Cheltsov A. V., Pellecchia M., and Strongin A. Y. (2012) Novel MT1-MMP small-molecule inhibitors based on insights into hemopexin domain function in tumor growth. Cancer Res. 72, 2339–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sela-Passwell N., Rosenblum G., Shoham T., and Sagi I. (2010) Structural and functional bases for allosteric control of MMP activities: can it pave the path for selective inhibition? Biochim. Biophys. Acta 1803, 29–38 [DOI] [PubMed] [Google Scholar]
- 16. Udi Y., Fragai M., Grossman M., Mitternacht S., Arad-Yellin R., Calderone V., Melikian M., Toccafondi M., Berezovsky I. N., Luchinat C., and Sagi I. (2013) Unraveling hidden regulatory sites in structurally homologous metalloproteases. J. Mol. Biol. 425, 2330–2346 [DOI] [PubMed] [Google Scholar]
- 17. Xu X., Chen Z., Wang Y., Bonewald L., and Steffensen B. (2007) Inhibition of MMP-2 gelatinolysis by targeting exodomain-substrate interactions. Biochem. J. 406, 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aoki T., Sumii T., Mori T., Wang X., and Lo E. H. (2002) Blood-brain barrier disruption and matrix metalloproteinase-9 expression during reperfusion injury: mechanical versus embolic focal ischemia in spontaneously hypertensive rats. Stroke 33, 2711–2717 [DOI] [PubMed] [Google Scholar]
- 19. Fujimura M., Gasche Y., Morita-Fujimura Y., Massengale J., Kawase M., and Chan P. H. (1999) Early appearance of activated matrix metalloproteinase-9 and blood-brain barrier disruption in mice after focal cerebral ischemia and reperfusion. Brain Res. 842, 92–100 [DOI] [PubMed] [Google Scholar]
- 20. Yang Y., Estrada E. Y., Thompson J. F., Liu W., and Rosenberg G. A. (2007) Matrix metalloproteinase-mediated disruption of tight junction proteins in cerebral vessels is reversed by synthetic matrix metalloproteinase inhibitor in focal ischemia in rat. J. Cereb. Blood Flow Metab. 27, 697–709 [DOI] [PubMed] [Google Scholar]
- 21. Shibayama M., Kuchiwaki H., Inao S., Ichimi K., Yoshida J., and Hamaguchi M. (1997) Induction of matrix metalloproteinases following brain injury in rats. Acta Neurochir. Suppl. 70, 220–221 [DOI] [PubMed] [Google Scholar]
- 22. Suehiro E., Fujisawa H., Akimura T., Ishihara H., Kajiwara K., Kato S., Fujii M., Yamashita S., Maekawa T., and Suzuki M. (2004) Increased matrix metalloproteinase-9 in blood in association with activation of interleukin-6 after traumatic brain injury: influence of hypothermic therapy. J. Neurotrauma 21, 1706–1711 [DOI] [PubMed] [Google Scholar]
- 23. Wang X., Jung J., Asahi M., Chwang W., Russo L., Moskowitz M. A., Dixon C. E., Fini M. E., and Lo E. H. (2000) Effects of matrix metalloproteinase-9 gene knock-out on morphological and motor outcomes after traumatic brain injury. J. Neurosci. 20, 7037–7042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abilleira S., Montaner J., Molina C. A., Monasterio J., Castillo J., and Alvarez-Sabín J. (2003) Matrix metalloproteinase-9 concentration after spontaneous intracerebral hemorrhage. J. Neurosurg. 99, 65–70 [DOI] [PubMed] [Google Scholar]
- 25. Power C., Henry S., Del Bigio M. R., Larsen P. H., Corbett D., Imai Y., Yong V. W., and Peeling J. (2003) Intracerebral hemorrhage induces macrophage activation and matrix metalloproteinases. Ann. Neurol. 53, 731–742 [DOI] [PubMed] [Google Scholar]
- 26. Tejima E., Zhao B. Q., Tsuji K., Rosell A., van Leyen K., Gonzalez R. G., Montaner J., Wang X., and Lo E. H. (2007) Astrocytic induction of matrix metalloproteinase-9 and edema in brain hemorrhage. J. Cereb. Blood Flow Metab. 27, 460–468 [DOI] [PubMed] [Google Scholar]
- 27. Cossins J. A., Clements J. M., Ford J., Miller K. M., Pigott R., Vos W., Van der Valk P., and De Groot C. J. (1997) Enhanced expression of MMP-7 and MMP-9 in demyelinating multiple sclerosis lesions. Acta Neuropathol. 94, 590–598 [DOI] [PubMed] [Google Scholar]
- 28. Leppert D., Ford J., Stabler G., Grygar C., Lienert C., Huber S., Miller K. M., Hauser S. L., and Kappos L. (1998) Matrix metalloproteinase-9 (gelatinase B) is selectively elevated in CSF during relapses and stable phases of multiple sclerosis. Brain 121, 2327–2334 [DOI] [PubMed] [Google Scholar]
- 29. Lindberg R. L., De Groot C. J., Montagne L., Freitag P., van der Valk P., Kappos L., and Leppert D. (2001) The expression profile of matrix metalloproteinases (MMPs) and their inhibitors (TIMPs) in lesions and normal appearing white matter of multiple sclerosis. Brain 124, 1743–1753 [DOI] [PubMed] [Google Scholar]
- 30. de Castro R. C. Jr, Burns C. L., McAdoo D. J., and Romanic A. M. (2000) Metalloproteinase increases in the injured rat spinal cord. Neuroreport 11, 3551–3554 [DOI] [PubMed] [Google Scholar]
- 31. Duchossoy Y., Horvat J. C., and Stettler O. (2001) MMP-related gelatinase activity is strongly induced in scar tissue of injured adult spinal cord and forms pathways for ingrowing neurites. Mol. Cell. Neurosci. 17, 945–956 [DOI] [PubMed] [Google Scholar]
- 32. Goussev S., Hsu J. Y., Lin Y., Tjoa T., Maida N., Werb Z., and Noble-Haeusslein L. J. (2003) Differential temporal expression of matrix metalloproteinases after spinal cord injury: relationship to revascularization and wound healing. J. Neurosurg. 99, 188–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaplan A., Spiller K. J., Towne C., Kanning K. C., Choe G. T., Geber A., Akay T., Aebischer P., and Henderson C. E. (2014) Neuronal matrix metalloproteinase-9 is a determinant of selective neurodegeneration. Neuron 81, 333–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cummings M. D., Farnum M. A., and Nelen M. I. (2006) Universal screening methods and applications of ThermoFluor. J. Biomol. Screen. 11, 854–863 [DOI] [PubMed] [Google Scholar]
- 35. Ra H. J., and Parks W. C. (2007) Control of matrix metalloproteinase catalytic activity. Matrix Biol. 26, 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hernandez-Barrantes S., Toth M., Bernardo M. M., Yurkova M., Gervasi D. C., Raz Y., Sang Q. A., and Fridman R. (2000) Binding of active (57 kDa) membrane type 1-matrix metalloproteinase (MT1-MMP) to tissue inhibitor of metalloproteinase (TIMP)-2 regulates MT1-MMP processing and pro-MMP-2 activation. J. Biol. Chem. 275, 12080–12089 [DOI] [PubMed] [Google Scholar]
- 37. Strongin A. Y., Collier I., Bannikov G., Marmer B. L., Grant G. A., and Goldberg G. I. (1995) Mechanism of cell surface activation of 72-kDa type IV collagenase: isolation of the activated form of the membrane metalloprotease. J. Biol. Chem. 270, 5331–5338 [DOI] [PubMed] [Google Scholar]
- 38. Björklund M., Heikkilä P., and Koivunen E. (2004) Peptide inhibition of catalytic and noncatalytic activities of matrix metalloproteinase-9 blocks tumor cell migration and invasion. J. Biol. Chem. 279, 29589–29597 [DOI] [PubMed] [Google Scholar]
- 39. Maquoi E., Munaut C., Colige A., Lambert C., Frankenne F., Noël A., Grams F., Krell H. W., and Foidart J. M. (2002) Stimulation of matrix metalloproteinase-9 expression in human fibrosarcoma cells by synthetic matrix metalloproteinase inhibitors. Exp. Cell Res. 275, 110–121 [DOI] [PubMed] [Google Scholar]
- 40. Park B. C., Thapa D., Lee Y. S., Kwak M. K., Lee E. S., Choi H. G., Yong C. S., and Kim J. A. (2007) 1-Furan-2-yl-3-pyridin-2-yl-propenone inhibits the invasion and migration of HT1080 human fibrosarcoma cells through the inhibition of proMMP-2 activation and down regulation of MMP-9 and MT1-MMP. Eur. J. Pharmacol. 567, 193–197 [DOI] [PubMed] [Google Scholar]
- 41. Ogata Y., Enghild J. J., and Nagase H. (1992) Matrix metalloproteinase 3 (stromelysin) activates the precursor for the human matrix metalloproteinase 9. J. Biol. Chem. 267, 3581–3584 [PubMed] [Google Scholar]
- 42. Ogata Y., Itoh Y., and Nagase H. (1995) Steps involved in activation of the pro-matrix metalloproteinase 9 (progelatinase B)-tissue inhibitor of metalloproteinases-1 complex by 4-aminophenylmercuric acetate and proteinases. J. Biol. Chem. 270, 18506–18511 [DOI] [PubMed] [Google Scholar]
- 43. Elkins P. A., Ho Y. S., Smith W. W., Janson C. A., D'Alessio K. J., McQueney M. S., Cummings M. D., and Romanic A. M. (2002) Structure of the C-terminally truncated human ProMMP9, a gelatin-binding matrix metalloproteinase. Acta Crystallogr. D Biol. Crystallogr. 58, 1182–1192 [DOI] [PubMed] [Google Scholar]
- 44. Rowsell S., Hawtin P., Minshull C. A., Jepson H., Brockbank S. M., Barratt D. G., Slater A. M., McPheat W. L., Waterson D., Henney A. M., and Pauptit R. A. (2002) Crystal structure of human MMP9 in complex with a reverse hydroxamate inhibitor. J. Mol. Biol. 319, 173–181 [DOI] [PubMed] [Google Scholar]
- 45. Bu C. H., and Pourmotabbed T. (1995) Mechanism of activation of human neutrophil gelatinase B: discriminating between the role of Ca2+ in activation and catalysis. J. Biol. Chem. 270, 18563–18569 [DOI] [PubMed] [Google Scholar]
- 46. Okada Y., Gonoji Y., Naka K., Tomita K., Nakanishi I., Iwata K., Yamashita K., and Hayakawa T. (1992) Matrix metalloproteinase 9 (92-kDa gelatinase/type IV collagenase) from HT 1080 human fibrosarcoma cells: purification and activation of the precursor and enzymic properties. J. Biol. Chem. 267, 21712–21719 [PubMed] [Google Scholar]
- 47. Lijnen H. R., Silence J., Van Hoef B., and Collen D. (1998) Stromelysin-1 (MMP-3)-independent gelatinase expression and activation in mice. Blood 91, 2045–2053 [PubMed] [Google Scholar]
- 48. Ramos-DeSimone N., Hahn-Dantona E., Sipley J., Nagase H., French D. L., and Quigley J. P. (1999) Activation of matrix metalloproteinase-9 (MMP-9) via a converging plasmin/stromelysin-1 cascade enhances tumor cell invasion. J. Biol. Chem. 274, 13066–13076 [DOI] [PubMed] [Google Scholar]
- 49. Nagase H., Enghild J. J., Suzuki K., and Salvesen G. (1990) Stepwise activation mechanisms of the precursor of matrix metalloproteinase 3 (stromelysin) by proteinases and (4-aminophenyl)mercuric acetate. Biochemistry 29, 5783–5789 [DOI] [PubMed] [Google Scholar]
- 50. Zhao Y., Lyons C. E. Jr, Xiao A., Templeton D. J., Sang Q. A., Brew K., and Hussaini I. M. (2008) Urokinase directly activates matrix metalloproteinases-9: a potential role in glioblastoma invasion. Biochem. Biophys. Res. Commun. 369, 1215–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Duncan M. E., Richardson J. P., Murray G. I., Melvin W. T., and Fothergill J. E. (1998) Human matrix metalloproteinase-9: activation by limited trypsin treatment and generation of monoclonal antibodies specific for the activated form. Eur. J. Biochem. 258, 37–43 [DOI] [PubMed] [Google Scholar]
- 52. Gu Z., Kaul M., Yan B., Kridel S. J., Cui J., Strongin A., Smith J. W., Liddington R. C., and Lipton S. A. (2002) S-Nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science 297, 1186–1190 [DOI] [PubMed] [Google Scholar]
- 53. Clements J. M., Cossins J. A., Wells G. M., Corkill D. J., Helfrich K., Wood L. M., Pigott R., Stabler G., Ward G. A., Gearing A. J., and Miller K. M. (1997) Matrix metalloproteinase expression during experimental autoimmune encephalomyelitis and effects of a combined matrix metalloproteinase and tumour necrosis factor-α inhibitor. J. Neuroimmunol. 74, 85–94 [DOI] [PubMed] [Google Scholar]
- 54. Gijbels K., Galardy R. E., and Steinman L. (1994) Reversal of experimental autoimmune encephalomyelitis with a hydroxamate inhibitor of matrix metalloproteases. J. Clin. Invest. 94, 2177–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Toth M., Sohail A., and Fridman R. (2012) Assessment of gelatinases (MMP-2 and MMP-9) by gelatin zymography. Methods Mol. Biol. 878, 121–135 [DOI] [PubMed] [Google Scholar]
- 56. Bekele-Arcuri Z., Matos M. F., Manganas L., Strassle B. W., Monaghan M. M., Rhodes K. J., and Trimmer J. S. (1996) Generation and characterization of subtype-specific monoclonal antibodies to K+ channel α- and β-subunit polypeptides. Neuropharmacology 35, 851–865 [DOI] [PubMed] [Google Scholar]
- 57. Pflugrath J. W. (1999) The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55, 1718–1725 [DOI] [PubMed] [Google Scholar]
- 58. Kissinger C. R., Gehlhaar D. K., and Fogel D. B. (1999) Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D Biol. Crystallogr. 55, 484–491 [DOI] [PubMed] [Google Scholar]
- 59. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]