Abstract
Similarities on the cellular and neurochemical composition of the amygdaloid subnuclei suggests their clustering into subunits that exhibit unique functional organization. The topological principle of community structure has been used to identify functional subnetworks in neuroimaging data that reflect the brain effective organization. Here we used modularity to investigate the organization of the amygdala using resting state functional magnetic resonance imaging (rsfMRI) data. Our goal was to determine whether such topological organization would reliably reflect the known neurobiology of individual amygdaloid nuclei, allowing for human imaging studies to accurately reflect the underlying neurobiology. Modularity analysis identified amygdaloid elements consistent with the main anatomical subdivisions of the amygdala that embody distinct functional and structural properties. Additionally, functional connectivity pathways of these subunits and their correlation with task-induced amygdala activation revealed distinct functional profiles consistent with the neurobiology of the amygdala nuclei. These modularity findings corroborate the structure–function relationship between amygdala anatomical substructures, supporting the use of network analysis techniques to generate biologically meaningful partitions of brain structures.
Introduction
The amygdala is involved in a series of emotional processes and cognitive functions including learning, memory, attention and perception1. Although small in total size, its complex composition of structurally and functionally heterogeneous subnuclei supports these multiple functions. Histological studies suggest that the amygdala may be composed of at least twenty subnuclei2 that share cytoarchitecture, myeloarchitecture and chemoarchitecture features. Based on these similarities, schemas have been proposed to segregate these nuclei into subunits based on their functional and anatomical characteristic3. Although the boundaries and even the names of these subunits remain unsettled, one of the most widely accepted classification schemas classifies them as the superficial (SF) (corticoid) amygdaloid nucleus, the centromedial (CM) group and the laterobasal (LB) complex4–6. A stereotaxic probabilistic map of the human amygdala, based on the superposition of cytoarchitectonic mapping of cell-body stained histological sections, has been developed by Amunts and colleagues5 and classifies the SF group to include the anterior amygdaloid area, the amygdalopyriform transition area, the amygdaloid-hippocampal area and cortical nuclei (intermediate, dorsal, ventral and posterior), the CM group combines the central and medial nuclei, while the LB group incorporates the lateral, basolateral, basomedial and paralaminar nuclei. This mapping is quite consistent with the cellular and neurochemical composition of the amygdaloid nuclei, since the CM, which is continuous with the bed nucleus of stria terminals, is comprised of cells that are morphologically similar to those in the striatum, while the LB and SF resemble cortical area7–10. Additionally, the LB is the main amygdala division that receives efferent projections from basal forebrain cholinergic neurons projecting mostly to the basolateral nucleus4,11.
The neurobiological composition of these main subdivisions indicates a potentially exclusive functional/anatomical neural interaction that may be captured either in synchronized fluctuation of the fMRI signal, as in rsfMRI, or through anatomical connections, such as, diffusion tensor imaging (DTI). For this reason, different functional and structural MRI based approaches have been proposed to parse the amygdala architecture. As a result, while some studies found the amygdala subdivided in two main regions, which were identified as superior12/superficial-cluster13 and inferior12/deep-cluster13, representing the LB and smaller nuclei (inferior/deep-cluster) and the CM and cortical nuclei (superior/superficial-cluster)14, others found the amygdala subdivided into four main subregions, dividing LB into lateral and basal regions and the CM into central and medial, leaving the cortical nuclei undifferentiated15. Therefore, these findings are not fully in agreement with the amygdala subdivisions shown in the stereotaxic probabilistic map5.
Two recent studies16,17, using imaging approaches, found three subdivisions of the amygdala forecasted by the probabilistic atlas5. The first study proposed a method that started from a hypothetical topographic model, based on the functional-anatomic organization of brain networks subserving social cognition, by defining three social networks: perception, which would be associated with the ventrolateral sector of the amygdala; affiliation, related to the medial section of the amygdala; and aversion, linked with the dorsal sector of the amygdala16. As a result, three amygdala regions were pre-defined in those locations (spherical ROIs) and using an iterative seed-target-seed methodology, the border of these subdivisions were established. The second study proposed an imaging-parcellation method based on meta-analysis, where three amygdala subdivisions were identified by computing whole-brain co-activation patterns for each amygdala seed voxel and grouping these seeds based on similarities between their co-activation patterns17. This method required a systematic analysis of a broad range of associated experiments (6,500 fMRI and PET studies) to increase the robustness of parcellation results and to avoid dependence on any particular user-specified parameter. Therefore, while the first approach16 predefine the initial number of subdivisions, the second approach was a meta-analysis requiring a significant amount of data from different imaging sources.
In this work, we propose to use resting state BOLD data and the topological principle of community structure to identify the main subdivisions of the amygdala. Our approach is based on global topological characteristics that has been shown to produce stable brain structural and functional networks, suggesting that the observed hierarchical modular organization coincide with the underlying anatomical communities at different scales18. Furthermore, by using hierarchical modularity analysis to explore the community structure of brain networks19,20, previous studies have shown that functionally and anatomically related brain regions are more densely interconnected, with relatively few connections between these clusters, indicating that these networks are intimately related and share common topological features, such as modules and hubs21. Therefore, based on these concepts, we employed the principle of modularity to identify subnetworks within the amygdala. We hypothesized that the modular organization of the amygdala will reflect its internal functional segregation and integration consistent with the neurobiological properties of its nuclei. For this purpose, we applied the community structure algorithm to a high spatial and temporal resolution rsfMRI data from the Human Connectome Project (HCP)22,23. We further assessed the functional consequence of each identified subnetwork by determining their functional connectivity and then by correlating these circuits with amygdala activation during an emotional processing task. Our findings validate the association between the modular structure of the amygdala and the biological characteristics of amygdala subdivisions.
Results
Modularity
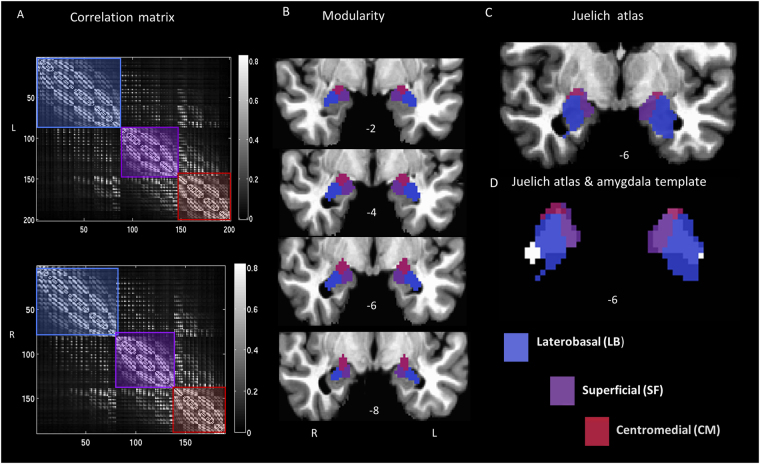
Modular analysis of the amygdala revealed three distinct modules (Fig. 1A and B) that are consistent with the probabilistic anatomical subregion maps5, displayed in the Juelich histological atlas (50% probabilistic mask, FSL - Fig. 1C). However, the borders of the atlas, as implemented in FSL, do not completely coincide with macro-anatomical landmarks of the amygdala5 (Fig. 1D; Juelich atlas volume, left: 2968 mm3, right: 2904 mm3 6, making direct comparison of the modularity results and the anatomical subdivisions difficult, since the amygdala template is smaller (left: 1608 mm3, right: 1512 mm3), but better represents the average amygdala size in healthy adults24. Despite size discrepancies, the synergy between the relative size of the subdivisions found in the Juelich atlas (LB > SF > CM) and the modules obtained from our modularity approach (lateral > medial > dorsal), together with the closeness between the subdivision location, suggest labeling the dorsal module as CM, the medial as SF and the lateral as LB.
Figure 1.
Modularity results for the amygdala subdivisions. (A) Correlation matrix for the amygdala template with modules overlaid in semi-transparent colors, greyscale values indicate r-values, (B) modularity results: LBL (696 mm3), LBR (640 mm3), SFL (472 mm3), SFR (464 mm3), CML (440 mm3), CMR (408 mm3), are displayed on coronal slices located at y-axis values: -2, -4, -6, -8 (L/R suffix = left/right). The maximum modularity factors obtained for this parcellation were QL = 0.26, QR = 0.25; (C) Juelich atlas overlaid on a coronal anatomical image (D) Juelich atlas superimposed on the amygdala template (shown in white, under the Juelich atlas); radiological convention.
FC maps
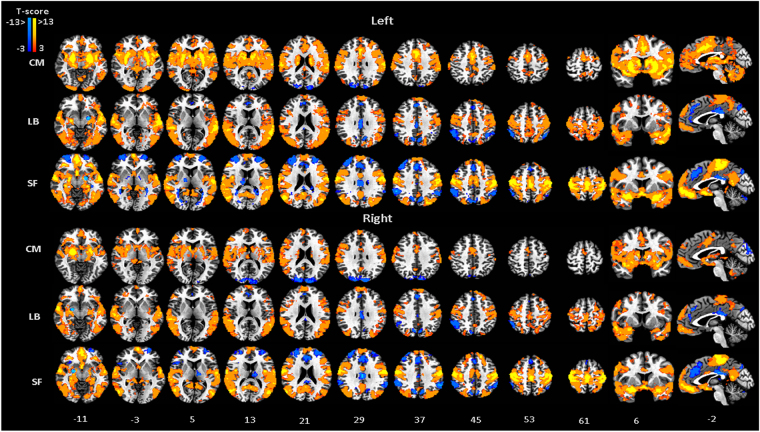
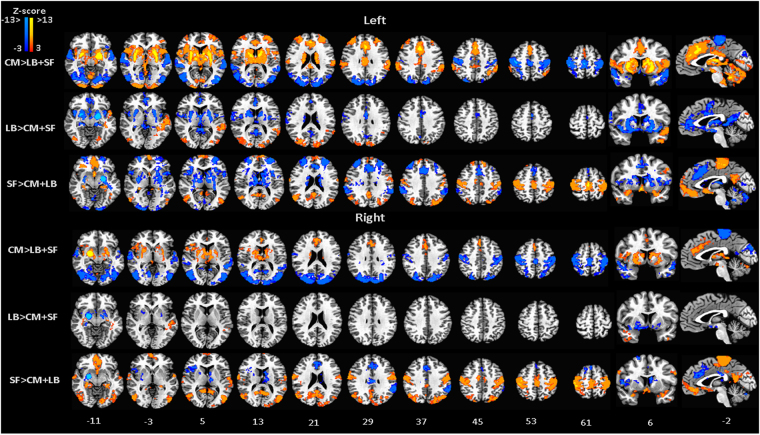
Patterns of significant correlation for each amygdala subdivision reveal unique connectivity circuits. MRI signal at the CM was mainly correlated with the signal in the middle and anterior cingulate cortices, frontal cortex, striatum, insula, cerebellum and precuneus, while a negative correlation was observed at the occipital gyrus (Fig. 2, Table 1). However, an overall reduction in the connectivity strength was observed for the right when compared with the left CM, which was not seen for other amygdala subdivisions (Figs 2 and 2S). In spite of this reduction in strength, the connectivity pattern is preserved between the two sides of this amygdala subdivision. Direct comparison with correlation results from the other seed regions highlight this unique path of connections when contrasting the functional connectivity results of the CM with either the combined results from LB and SF (Fig. 3, Table 2) or the connectivity results from each of the others subdivisions (Fig. 1S, Table 1S).
Figure 2.
FC maps for the amygdala subdivisions, CM, LB and SF, obtained from modularity results. MNI standard space; radiological convention; significance: p < 0.05 FWE corrected. Left/Right indicate seed locations.
Table 1.
Cluster locations for the functional connectivity maps calculated for each amygdala subdivision. T-test results from partial correlation for each subset are corrected for multiple comparisons, p corrected < 0.01; cluster’s peak locations are in Montreal Neurological Institute (MNI) coordinates, locations for the cerebellum subdivisions are from MNI-SUIT space (AFNI-SUIT atlas92), cluster size in number of voxels, XL(R) = X left(right), X = CM, LB, SF.
| Subdivision | x | y | z | Brain region | cluster size | T-score |
|---|---|---|---|---|---|---|
| CML | −24 | −6 | −14 | left Amygdala | 71314 | >13 |
| 22 | −80 | 28 | right Superior occipital gyrus | 477 | −5.9 | |
| −14 | −92 | 24 | left Superior occipital gyrus | 397 | −5.9 | |
| CMR | 24 | −2 | −14 | right Amygdala | 24511 | >15 |
| −8 | 90 | 36 | left cuneus | 2277 | −6.6 | |
| −16 | −68 | −46 | left cerebellum (VIIIa) | 2244 | 6.8 | |
| 2 | 58 | 28 | left superior frontal gyrus | 1488 | 7.7 | |
| 0 | −4 | 48 | cingulate Gyrus | 1394 | 7 | |
| 16 | −72 | −52 | right cerebellum (VIIb) | 533 | 7.6 | |
| 4 | −62 | 32 | right precuneus | 355 | 6.5 | |
| 50 | −46 | −24 | right inferior temporal gyrus | 293 | 5.7 | |
| LBL | −28 | −8 | −20 | left amygdala | 58058 | >13 |
| 0 | 58 | 36 | superior frontal gyrus | 1383 | 8.8 | |
| 50 | −50 | 56 | right inferior parietal lobule | 856 | −7.6 | |
| 4 | 34 | 38 | right medial frontal gyrus | 658 | −7 | |
| 2 | 52 | −16 | right Mid. Orbital gyrus | 629 | 8.1 | |
| −8 | −70 | 34 | left precuneus | 525 | −7.6 | |
| −38 | −56 | 46 | left inferior parietal lobule | 473 | −7.6 | |
| 2 | −22 | 30 | right cingulate gyrus | 454 | −8.7 | |
| 40 | 10 | 24 | right inferior frontal gyrus | 398 | 6.1 | |
| 58 | 28 | 2 | right inferior frontal gyrus | 307 | 6 | |
| LBR | 30 | −6 | −22 | right amygdala | 42326 | >13 |
| 0 | 58 | −10 | medial frontal gyrus | 2564 | 9.2 | |
| 32 | −82 | −36 | right cerebellum (Crus I) | 931 | 8.7 | |
| 50 | −46 | 42 | right inferior parietal lobule | 870 | −6.6 | |
| −26 | −82 | −36 | left cerebellum (Crus I) | 665 | 9.2 | |
| 2 | 32 | 40 | right medial frontal gyrus | 591 | −7.6 | |
| −6 | −72 | 38 | left precuneus | 387 | −6.6 | |
| 0 | −26 | 28 | cingulate gyrus | 310 | −6.6 | |
| SFL | −16 | −6 | −18 | left amygdala | 68153 | >13 |
| 38 | −52 | −32 | right cerebellum (Crus I) | 3118 | −11.5 | |
| 30 | 56 | 18 | right superior frontal gyrus | 2793 | −8.8 | |
| −32 | 58 | 16 | left middle frontal gyrus | 1434 | −9.1 | |
| 2 | 28 | 40 | right medial frontal gyrus | 1286 | −9.1 | |
| 54 | −44 | 42 | right inferior parietal lobule | 1087 | −10.2 | |
| −40 | −30 | −10 | left inferior temporal gyrus | 793 | −7 | |
| −8 | −68 | 38 | left precuneus | 733 | −8.4 | |
| −50 | −52 | 40 | left inferior parietal lobule | 641 | −7.3 | |
| 36 | −46 | 0 | right middle temporal gyrus | 623 | −7.4 | |
| 4 | −24 | 28 | right cingulate gyrus | 451 | −8.5 | |
| 14 | −12 | 6 | right thalamus | 308 | −7.5 | |
| SFR | 20 | −6 | −20 | right amygdala | 65553 | >13 |
| −40 | −60 | −30 | left cerebellum (Crus I) | 1579 | −9.7 | |
| 2 | 38 | 22 | right anterior cingulate | 1541 | −8.4 | |
| −30 | 54 | 24 | left middle frontal gyrus | 1335 | −8 | |
| 30 | 56 | 28 | right superior frontal gyrus | 1166 | −7.7 | |
| 26 | −70 | −26 | right cerebellum (VI) | 1050 | −8.8 | |
| 56 | −44 | 36 | right supramarginal gyrus | 916 | −8.7 | |
| −54 | −48 | 36 | left inferior parietal lobule | 895 | −9.4 | |
| 2 | −22 | 28 | right cingulate gyrus | 639 | −10.5 | |
| −8 | −72 | 40 | left precuneus | 303 | −8.5 | |
| 46 | 18 | 0 | right inferior frontal gyrus | 302 | −6.6 | |
| 12 | −70 | 42 | right precuneus | 297 | −10 | |
| −14 | −12 | 10 | left thalamus | 290 | −7.2 |
Figure 3.
FC-differential maps contrasting the connectivity map of each amygdala subdivision, CM, LB and SF, against the average connectivity pattern of the other two subdivisions, LB + SF, CM + SF, CM + LB. MNI standard space; radiological convention; significance: p < 0.05 FWE corrected.
Table 2.
Locations of the significant clusters obtained from the direct comparison of the functional connectivity pattern of each amygdala subdivision with the average connectivity pattern of the other two subdivisions. Results are corrected for multiple comparisons, p corrected < 0.01; cluster’s peak locations are in MNI coordinates, cluster size in number of voxels, XL(R) = X left (right), X = CM, LB, SF.
| Subdivision | x | y | z | Brain region | cluster size | Z-score |
|---|---|---|---|---|---|---|
| CML > LBL + SFL | −18 | −8 | −10 | left amygdala | 33154 | >13 |
| 10 | −28 | 74 | right paracentral lobule | 20900 | −10.0 | |
| 0 | 26 | 32 | left middle cingulate cortex | 4865 | 10.6 | |
| −36 | 54 | 20 | left middle frontal gyrus | 1691 | 8.0 | |
| −68 | −22 | 24 | left postcentral gyrus | 1656 | 8.7 | |
| 30 | 40 | 28 | right middle frontal gyrus | 1398 | 6.5 | |
| 68 | −32 | 36 | right supramarginal gyrus | 1195 | 6.8 | |
| 28 | −6 | −20 | right hippocampus | 817 | −11.2 | |
| CMR > LBR + SFR | 48 | −74 | 2 | right middle temporal gyrus | 23328 | −8.2 |
| 30 | −4 | −24 | right amygdala | 6105 | <−13 | |
| −52 | −48 | 8 | left middle temporal gyrus | 2288 | −7.3 | |
| 60 | −4 | −10 | right superior temporal gyrus | 2129 | −7.2 | |
| −36 | −46 | −34 | left cerebellum (VI) | 2122 | 7.4 | |
| 2 | 42 | 20 | left anterior cingulate cortex | 1498 | 7.1 | |
| LBL > CML + SFL | −34 | −4 | −26 | left amygdala | 12381 | >13 |
| 0 | 4 | 30 | left anterior cingulate cortex | 1870 | −7.0 | |
| 30 | −84 | 20 | right middle occipital gyrus | 1487 | 6.0 | |
| −18 | −92 | 16 | left middle occipital gyrus | 1079 | 5.4 | |
| LBR > CMR + SFR | 34 | −2 | −24 | right amygdala | 2731 | >13 |
| −30 | −8 | −20 | left hippocampus | 1007 | 12.3 | |
| SFL > CML + LBL | −14 | −4 | −18 | left amygdala | 13575 | >13 |
| −30 | −70 | −24 | left cerebellum (VI) | 9287 | −10.1 | |
| 0 | −20 | 62 | left paracentral lobule | 8517 | 10.1 | |
| −30 | 54 | 16 | left middle frontal gyrus | 7197 | −9.2 | |
| 0 | 40 | −22 | left rectal gyrus | 2460 | 9.9 | |
| 56 | −66 | 22 | right middle temporal gyrus | 1549 | 7.1 | |
| −24 | −80 | 34 | left superior occipital gyrus | 1470 | 6.9 | |
| 16 | −104 | −4 | right calcarine gyrus | 1390 | −6.1 | |
| 0 | −54 | 18 | left precuneus | 1186 | 7.1 | |
| 58 | −44 | 36 | right supramarginal gyrus | 922 | −7.6 | |
| −62 | −42 | 30 | left supramarginal gyrus | 832 | −6.7 | |
| SFR > CMR + LBR | 12 | −20 | 76 | right paracentral lobule | 13240 | 10.1 |
| 14 | −2 | −16 | right amygdala | 11217 | >13 | |
| −10 | −54 | 6 | left calcarine gyrus | 3912 | 8.3 | |
| −20 | −82 | −30 | left cerebellum (Crus I) | 1793 | −8.8 | |
| 26 | −70 | −26 | right cerebellum (VI) | 1517 | −8.1 | |
| −10 | 30 | 26 | left anterior cingulate cortex | 1428 | −6.7 | |
| −12 | −16 | 10 | left thalamus | 885 | −7.1 | |
| −56 | −48 | 36 | left inferior parietal lobule | 828 | −7.3 | |
| 40 | 24 | 10 | right inferior frontal gyrus | 803 | −6.4 |
The LB nuclei was positively correlated with the superior, medial and inferior frontal gyri, precentral gyrus, paracentral lobule, middle temporal gyrus and cerebellum, and negatively correlated with the parietal lobule, precuneus and cingulate gyrus (Fig. 2, Table 1). Further, when compared with the circuit strength from the other two amygdala subdivision, the LB seems to be uniquely associated with inferior and middle temporal gyrus and middle occipital gyrus (Fig. 3, Table 2). Pairwise comparisons also show unique correlations of LB with the inferior temporal gyrus and the middle frontal gyrus when contrasted with CM and with cerebellum, and the middle frontal gyrus and the inferior parietal lobule when compared with SF (Fig. 1S, Table 1S).
Activity in the SF correlated with signal fluctuations in the orbitofrontal cortex, posterior insula, olfactory cortex, precentral gyrus, paracentral lobule and posterior cingulate, and was anti-correlated in the anterior cingulate cortex, superior and middle frontal gyrus, cerebellum, parietal lobule, inferior temporal gyrus and precuneus (Fig. 2, Table 1). Unique connectivity path of the SF subdivision is observed at the paracentral lobule, posterior cingulate cortex (PCC) and orbitofrontal cortex (OFC) when contrasting with the combined correlation results for the other two subdivisions (Fig. 3, Table 2).
Pairwise comparisons show that the connectivity strength of the SF subdivision is higher when compared with CM based circuits in the occipital lobe, PCC, precentral and postcentral gyrus, paracentral lobule, superior frontal gyrus and OFC and higher than the connectivity path of the LB at medial and inferior frontal gyrus, precuneus, PCC, precentral and postcentral gyrus, middle temporal gyrus, precentral gyrus and OFC (Fig. 1S, Table 1S).
Connectivity across sessions
A two-way ANOVA (session x seed location), comparing the connectivity pattern obtained for each amygdala subunit and for each session (subseries), revealed a main effect of the seed location in the striatum, insula, OFC, ACC, superior medial gyrus, occipital gyrus, medial, middle and inferior frontal gyrus, cingulate gyrus, inferior parietal lobule, cerebellum, superior and middle temporal gyrus (Fig. 3SA, Table 3). There was no main effect of session and no interaction between seed location and session. ICC test-retest results of brain connectivity data also show fair to moderate reliability for the functional connectivity across sessions (ICC > 0.4). Intersession consistency was highest from the FC maps obtained for the SF, followed by those from CM and LB (Fig. 3SB).
Table 3.
Significant clusters for the main effect of seed location obtained from the two-way ANOVA. Results are corrected for multiple comparisons, p corrected < 0.01; cluster peak locations are in MNI coordinates, locations for the cerebellum subdivisions are from MNI-SUIT space (AFNI-SUIT atlas92), cluster size in number of voxels.
| x | y | z | Brain region | cluster size | F-score |
|---|---|---|---|---|---|
| −30 | −8 | −24 | right amygdala | 10817 | >20 |
| 2 | 30 | 32 | left superior medial gyrus | 6615 | 19 |
| −50 | −72 | 0 | left middle occipital gyrus | 2498 | 13 |
| 16 | −92 | 22 | right cuneus | 2333 | 12 |
| 2 | 38 | −18 | right medial frontal gyrus | 1449 | 19 |
| 28 | −84 | −32 | right cerebellum (Crus I) | 1438 | 15 |
| −40 | −54 | −30 | Left cerebellum (Crus I) | 1362 | 20 |
| −28 | 52 | 16 | left middle frontal gyrus | 1233 | 13 |
| 30 | 54 | 18 | right middle frontal gyrus | 902 | 14 |
| −58 | −34 | 40 | left inferior parietal lobule | 827 | 9.7 |
| −52 | 0 | −20 | left middle temporal gyrus | 780 | 12 |
| 62 | −36 | 36 | Right inferior parietal lobule | 450 | 10 |
| 62 | −4 | −10 | right superior temporal gyrus | 371 | 17 |
| 40 | 32 | −14 | Right inferior frontal gyrus | 278 | 19 |
| −2 | −28 | 26 | Left cingulate gyrus | 270 | 7.9 |
Altogether, these results indicate a stable pattern of brain connectivity for each modularity based amygdala substructure across sessions (Fig. S2) that generally reproduced those obtained with the entire time series (Fig. 2), highlighting the consistency of the connectivity over time.
Amygdala activation and rsfMRI
As expected, the entire amygdala was significantly activated when subjects performed the face/shape matching task (Fig. 4A1). However, raising the threshold (T > 13) indicated that the most significant activated voxels were located within the CM and the SF subdivisions (Fig. 4A2). Average BOLD signal in each amygdala subdivision shows the LB as the least activated during the fear/anger faces task, while the superficial subdivision was the most active during the task (Fig. 4B); there were no significant differences in activation between subdivisions in the left and right hemisphere. Notably, only the CM showed significant correlation between the average BOLD signal in this amygdala subdivision and its mean connectivity strength with the entire brain (Figs 4C and 5S ), while no significant correlation between BOLD and corresponding connectivity maps was observed for the others two amygdala subdivisions.
Figure 4.
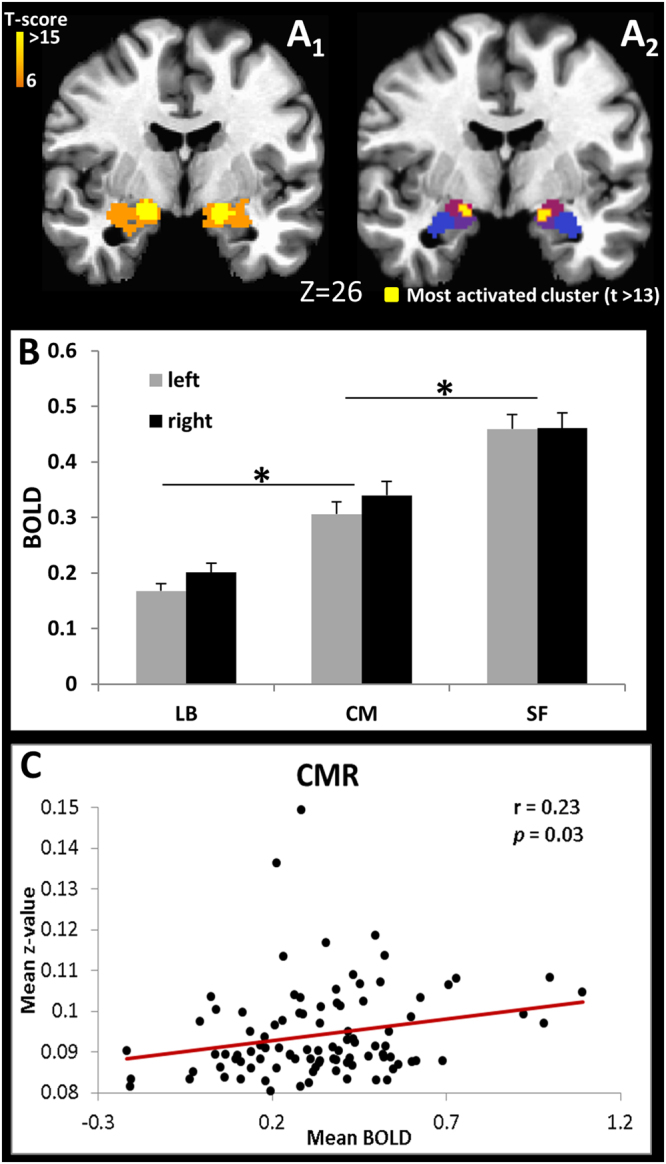
(A) Amygdala activation for the threshold of T ≥ 6 (A 1), location of the most significant activated voxels (T > 13, peak value T = 17) (A 2) overlaid over the amygdala subdivisions, CM, LB and SF (color pattern from Fig. 1), radiological convention; (B) Average BOLD signal for each amygdala subdivision (*p < 0.0001); (C) ROI analysis correlating the mean BOLD signal at the CMR with the mean positive and significant (p < 0.05) Fisher transformed correlation values of the CMR connectivity pathway.
Discussion
By using the community structure algorithm to evaluate the topological organization of the amygdala, we identified three subdivisions, which is congruent with previous findings16,17, but with a method that did not require an extended meta-analysis. Moreover, modularity has shown to be very reliable, even with small datasets; the results from the HCP dataset (98 subjects) reproduced our previous pilot findings from a group of just 18 subjects, acquired in our center with different EPI sequences and different set of parameters (see Supplemental Material Fig. 4S). Additionally, our method is free of initial hypotheses related to the number of modules, which is automatically established by the algorithm based on the principle of modularity. Finally, our method was able to identify the CM and SF based only on information about node connections, despite the poor separation of these modules as illustrated in Fig. 1A, which may explain why previous data driven work12,13 were not able to separate the CM from SF.
Our work further supports that topological properties may have an impact on the study of structural-function relations in brain networks. Whilst it helps to understand the fundamental architecture of connections within and between brain regions, it also provides a way to elucidate how this architecture supports neurophysiological dynamics. Modularity approaches can be either top-down, such as the Newman’s method25, where a network is repeatedly subdivides into smaller portions until there is nothing to be gained in modularity value (Q), or bottom-up, as in Louvain’s method26,27, which is a hierarchical clustering approach, where nodes are progressively merged to others nodes becoming larger and larger sets of nodes, a process that is repeated iteratively until no increase of modularity is possible28. By using the Louvain’s method, we were able to identify amygdala modules that resemble the most accepted classification schemes for its anatomical subdivision5, suggesting a degree of functional segregation of these subunits that is consistent with the biological characteristics of the amygdala substructures.
Different methods have been proposed to find small communities in large networks, such as the Surprise method29 and Optimal Compression method30, each having particular strengths and weaknesses. For example the Surprise method29 is best suited for the study of binarized networks. However, it is vulnerable to noise errors that affect small modules, and in the case of the amygdala, located in a brain area with low SNR, it may not necessarily improve the quality of the partitions. The Optimal Compression method30, on the other hand, is not recommended when “community structure are considered as statistical deviation from the null model in which the degree sequence is held constant but links are otherwise equiprobable among all nodes, wherein the modularity optimization methods by definition provides the optimal partitioning”30. In contrast, by construction, the Louvain method26 is able to unfold a complete hierarchical community structure for the network, with each level of the hierarchy being given by the intermediate partitions found at each pass. In our work, we considered only the top level of this hierarchy, namely the final partition found by the algorithm, since no further meaningful sub-module was found in the amygdala (additional sub-modules mostly consisted of single voxels, as tested on dataset from Fig. 4S) when lower levels of modular hierarchy were evaluated.
Regional connectivity
Temporally correlated patterns of low-frequency fluctuations during rest revealed regions of distinct functional networks associated with each of the three amygdala subdivisions, which were maintained across sessions, demonstrating the robustness and stability of our findings. Notably, their combination reproduces the previously reported16,31 connectivity pattern for the entire amygdala, as areas from the ventro-temporal-limbic network (described by Glerean and colleagues31) are identified in the connectivity pathway of the amygdala subdivisions.
Signal variation at the LB subdivision was positively correlated with activity in the superior temporal gyrus, middle frontal and precentral gyri, supporting the involvement of similar circuits in associative learning and emotion regulation32,33. In particular, the connection between the LB and the temporal lobes was previously observed using rsFC6,12,16 and probabilistic DTI34, and is supported by axonal tracing studies in non-human primates, where a connection between the superior temporal gyrus and the ventrolateral nucleus of the amygdala was identified35.
Significant correlation was found between the SF region and posterior insula, OFC, subgenual ACC, inferior parietal lobule, olfactory cortex, middle temporal gyrus and PCC. This correlation pattern suggests an involvement of SF in olfaction-related affective processes, consistent with a meta-analysis of human imaging data17 and rat anatomy36,37. Similarities of the connectivity pattern of the LB and SF mainly with the temporal lobe38, suggests that these two amygdaloid nuclei may share a common functional organization. Comparative architectonic studies have also suggested a separation of the SF from the CM group, assigning it to the LB group39, which is justified by the fact that these two nuclei also contain similar cortical –like neuronal composition7,9.
Despite the biological resemblance between the SF and LB, we also observed unique connectivity patterns between SF and areas of the default mode network (DMN), although anatomical projections from the amygdala to the PCC has not been verified40, suggesting a possible indirect connection between these regions through the sACC, as previously reported in monkeys41. Nevertheless, a functional connection between the amygdala and the PCC has also been reported42, suggesting an involvement of SF in self-awareness related emotional processes, autobiographical memory, past self-relevant stimuli and future prospection43. The SF was also anti-correlated with the more dorsal part of the precuneus, which is consistent with the pattern of positive correlations, previously observed, between the DMN and the ventral, but not the dorsal precuneus44. In addition, a decrease in regional cerebral blood flow in the PCC, medial frontal cortex and ventral but not dorsal precuneus during a working memory task45, suggests that dorsal precuneus (BA 7) may not be part of the DMN46. Furthermore, the connection between the SF and the precuneus may corroborate an amygdala-precuneus pathway that has been anatomically observed in tract tracing studies in monkeys47,48. Lastly, our findings showing the SF amygdala as the most activated in response to an emotional task that elicited either fear or anger, corroborates its involvement in directing attention towards affective stimuli49.
The significant correlation between resting activity of the CM nuclei and signal fluctuation from areas attributed to the salience network (SN), e.g., anterior insula, ACC and middle cingulate cortex (MCC)50, is consistent with the central nucleus involvement in facilitating attention to salient stimuli51. In addition, the CM was the only amygdala subdivision in which the rsFC pattern predicted activation in response to fear and angry faces, thus indicating its association with adverse feelings. Even though the SF had the highest average activation for this task, its BOLD signal did not correlate with the mean functional connectivity pattern of this subdivision. Moreover, the most activated voxels occurred not only in the SF, but also in the CM subdivision. Therefore, our findings suggest an involvement of CM and its connected areas (SN and striatum) with negative emotion and is consistent with previous findings. For example, the strongest amygdala functional connectivity within the SN is seen in those with the greatest amygdala activation to and aroused by negative pictures52, which is sustained by the role of the amygdala, SN and ventral striatum on the identification of the emotional significance of environmental stimuli and the production of affective states53. Multiple fMRI studies have also reported activation in the ACC/MCC in anger and fear emotional tasks54, while monkey studies show that this part of the cingulate cortex receives input from the amygdala41,55 and has been implicated in fear56. The anterior insula has also been linked with such emotional experiences as fear and anger57, supported by a projection from the central nucleus of the amygdala35. The striatum has been associated with anger58, such that functional connections between the CM and the striatum are enhanced during stress59. The CM participates not only in the expression of conditioned fear10 but also in the learning and consolidation of fear conditioning60. Experiments in rats show that lesions of the central, medial and cortical amygdala nuclei markedly increase the number of contacts a rat will make with a sedated cat, demonstrating a decrease of fear in these lesioned animals61. The involvement of CM in anger has also been demonstrated in cats, with stimulation of the central nucleus suppressing defensive rage, while stimulation of the medial nucleus enhances aggressive behavior62. Overall, our findings reinforce the previously observed participation of the CM and connected areas (SN and striatum) in fear and anger.
Validation and reproducibility
The functional connectivity circuits for the modularity defined amygdala subdivisions reproduce the major findings obtained by Bickart et al.16 and by Roy et al.6, which used the Juelich atlas as a template. Both found that the CM nucleus correlated with the striatum and insula, that the LB complex correlated with the temporal lobe, and activity of the SF mainly correlated with activity of the OFC. Additionally, Roy and colleagues6 also reported laterality differences of the CM connections, as observed here, that is consistent with those previously reported63. For example, right amygdala lesions in rats have been shown to generate greater deficits in contextual fear than left sided lesions64. Previous electrophysiological studies also showed hemispheric lateralization of pain processing by CM neurons. Specifically, neurons in the left latero-capsular division of the central nucleus of the amygdala (CeLC) did not develop increased responsiveness in a rodent model of arthritis pain, while the right CeLC played a major role in the processing of prolonged nociceptive inputs and develops sensitization65. This suggests a tonic inhibitory mechanism from the prefrontal cortical areas over the left CeLC, which exert a top-down inhibitory influence on the amygdala66. In our work, the lower connectivity strength observed for the CM, besides being consistent with previous work, may also indicate a diminished inhibitory effect from the prefrontal cortical areas over the right CM, since this was the amygdala subregion most connected to these cortical areas. This may explain the unique correlation of its connectivity pattern and the activity on this area during the faces task, once more being consistent with a predominant role of the right amygdala in negative emotions. Finally, while the anatomical and functional basis for this lateralization remains unclear, taken together, these findings highlight the role of the CM on amygdala lateralization.
The functional connectivity between the LB and SF with the SMA differ from the results obtained by Roy and colleagues6, although they are consistent with another resting state study that identified connectivity between the LB and SF with motor areas38. Structural connections between the amygdala and motor areas has also been previously observed in a DTI study34. Studies in cats67 and monkeys68 have shown that the projections to the motor system arise from the magnocellular division of the basal nucleus of the amygdaloid complex. In addition, disruption of the amygdala-motor pathway has been suggested to be responsible for the inability of those with Autism to react to social stimuli69, which is consistent with data from both monkeys70 and humans71 reporting that the LB and SF subregions are especially sensitive to social stimuli. Therefore, together with previous findings, our work indicate that a direct amygdala-motor pathway might provide a mechanism by which the amygdala can influence more complex motor behaviors34.
Finally our results benefited from using high spatial and temporal resolution data from the HCP22. The high spatial resolution improves the assessment of small brain regions, such as the amygdala, by limiting partial volume effects, reducing dephasing artifacts and improving in-plane MRI signal uniformity72, while the high scan rate has the further advantage of minimizing the aliasing of physiological artifacts over the low frequencies of interest73. Even though smaller voxels always carry the disadvantage of lower SNR, it was compensated by the large sample size and multiple long rsfMRI runs.
Conclusions
In conclusion, we have applied a community structure approach to characterize the modular organization of the amygdala using resting state fMRI data from the HCP. The anatomical and functional specificity of the results provide compelling evidence of the structure–function association between the subnuclei identified herein and histological anatomical substructures, demonstrating that graph theory based network analysis techniques can generate biologically meaningful partitions of brain structures. Finally, our findings suggest that characterization of regional modular organization may be useful to evaluate disease- or age-induced abnormalities.
Methods
Subjects
Analyses were performed on 100 adult healthy volunteer (46 males and 54 females; age range 22 to 36 years; mean age 29.4 ± 3.6 years), randomly selected through the 100-unrelated option from the WU-Minn Consortium HCP (WU-Minn HCP 500 Subjects + MEG2 Data Release), obtained through the Open Access agreement. Data is publicly available at the HCP online database (http://www.humanconnectome.org/documentation/S500/). All experiments were performed in accordance with the relevant guidelines and regulations of the Human Connectome Project74. Detailed description on the standard operating procedures of the protocol is available at the WU-Minn HCP 500 Subjects + MEG2 Data Release: Reference Manual Appendix IV. Subject recruitment procedures and informed consent forms, including consent to share de-identified data, were approved by the Washington University (WU) institutional review board75. The use of the HCP data was approved by the Office of Human Subjects Research Protections at the NIH. All data presented in this paper is not identifiable, only group results are presented overlaid on a MNI template.
Data acquisition
MRI
Images were acquired on a 3 Tesla Skyra Siemens system using a 32-channel head coil, a customized SC72 gradient insert (100 mT/m) and a customized body transmit coil. Resting state fMRI data were acquired in four runs on two different days (two runs per day); task-evoked fMRI was acquired in two runs on the same day after completion of the rsfMRI. Both used a multi-band gradient-echo EPI sequence22,76 (TE/TR 33.1/720 ms, resolution 2 mm isotropic, 72 oblique-axial slices, 1200 images/rsfMRI run, 176 images/task-evoked fMRI run, MB acceleration factor = 8, BW = 2290 Hz/Pix). Within each day, acquisitions alternated between phase encoding in the right-to-left (RL) direction in one run and the left-to-right (LR) direction in the other run. Anatomical images were acquired with a high-resolution (0.7 mm isotropic) T1-weighted magnetization-prepared rapid gradient echo (3D-MPRAGE) sequence. Anatomical and functional imaging sequences covered the whole brain. Subjects were given a simple instruction to rest and keep their eyes open with relaxed fixation on a projected bright cross-hair on a dark background (presented in a darkened room)77.
fMRI task – Emotion Processing
Participants performed a block design modified face-shape matching task33 wherein they were asked to match which of two faces/shapes presented on the bottom of the screen with the face/shape at the top of the screen; faces exhibited either an angry or fearful expression. Each of the two runs includes 6 blocks (3 face blocks and 3 shape blocks), each block was composed of 6 trials of the same task type (face or shape), with trials presented for 2000 ms and an inter-trial interval of 1000 ms. Shape blocks were presented interleaved with face blocks and each block was preceded by a 3000 ms task cue (“shape” or “face”); 8 seconds of fixation was presented at the end of each run77.
Data analysis
Preprocessing - 1st level fMRI analysis
The datasets underwent an initial preprocessing by the HCP consortium. Anatomical images were distortion corrected, co-registered and averaged across runs, AC-PC registered, brain extracted, B1 bias field corrected and normalized to MNI152 space. FMRI runs (rsfMRI and task-evoked) underwent gradient distortion correction, motion correction, registration to the T1w image, spatial normalization to the MNI standard space, 4D global mean-based intensity normalization, B1 bias field correction; rsfMRI were also subject to independent component analysis (ICA)-based artifact removal. Finally, both task and resting data were whole brain masked23,77. Local data processing was performed in AFNI78 and MATLAB (The MathWorks Inc, Natick, Massachusetts). The pre-processed task-evoked fMRI time series were smoothed (FWHM = 4 mm) to improve signal-to-noise ratio (SNR)79, censored for time points that exceed a Euclidean distance of 0.3 mm and then modeled in a fixed effect analysis, two runs per subject, with canonical hemodynamic responses time-locked to the shape-face epochs, contrasting face with shape condition; head motion parameters (3 translations and 3 rotations) were also entered into the model as regressors of no interest. Time-series datasets were scaled so that beta weights could be interpreted as percent signal-change. Each pre-processed rsfMRI run had the first four EPI volumes removed to ensure signal equilibrium, following by band-pass filtering (0.01–0.10 Hz) to minimize instrument induced drifts80 and physiological noise81. Multi-linear regression with the 6 time-varying realignment parameters was performed to minimize motion related fluctuations in the MRI signals. The first three principal components (using PCA) of white matter (WM) and cerebrospinal fluid (CSF) signals were also regress out to preclude non-neuronal induced signal fluctuations82,83. The WM and CSF masks were generated by segmenting the preprocessed high-resolution anatomical images using SPM884 and down-gridding the obtained masks to the same resolution as the pre-processed functional data. Individual subject rsfMRI time series were smoothed (FWHM = 4 mm) to improve SNR79, and censored for time points that exceed a framewise displacement (FD) threshold >0.5 mm and the root mean square variance across voxels (DVARS) > 0.5%85. Time series with excessive number of time points censored (more than 30% for rsfMRI, 20% for task-evoked fMRI) were discarded; two subjects were excluded from resting BOLD and another two from the task BOLD data.
Modularity
Modularity analysis of the amygdala was performed using a weighted-connectivity conserving null model in fully connected, undirected networks with positive and negative weights27 from the Brain Connectivity Toolbox (https://sites.google.com/site/bctnet/) on the remaining 98 subjects. More specifically, Pearson correlation coefficient was computed for each voxel (node) inside of the anatomically pre-defined left and right amygdala templates in MNI space (from Jerne Volumes of Interest database86,87), which was resampled to 2 mm isotropic to reproduce the image resolution of the processed functional data. Individuals’ correlation matrices were then averaged across subjects and runs to obtain the final connectivity matrix, M. Following, after removing the auto-correlation values from M, a Louvain algorithm26, was used to find the community affiliation vector (C) corresponding to the undirected and weighted correlation matrix (M)27, accounting for the contribution of positive and negative edges. The algorithm finds the optimal number of clusters by aggregating the nodes in the network into groups of modules. More specifically, Louvain method starts from a set of nodes that, through subsequent passes of the algorithm, are clustered to others nodes becoming larger and larger sets of nodes (it is optimized by allowing only local changes of communities); next the found communities are aggregated in order to build a new network of communities, finally it is repeated iteratively until it reaches a number of modules that have a maximal possible number of within group connections, and a minimal possible number of between-group connections88, i.e., until the modularity factor, Q, is maximized26,27. Therefore, no initial information regarding the number of modules is needed. However, the algorithm is non-deterministic, and may produce different solutions each run, so to address this limitation, the calculation was repeated 100 times (the calculation of Q is independent in each repetition) and the maximum modularity factor, QM, was registered (by considering precision of 4 decimal places only one Q value was found), then it was repeated another 100 times and another modularity factor was selected, Qm; these two values were compared and if they are different the latest is kept to compare with the results from the next iteration. This process was repeated until no change to the maximum Q was observed (QM = Qm). Note that for every step (every QM and Qm) the number of modules remained constant varying only their size. Next the final community affiliation, vector (C) was entered into the fine-tuning27 modularity algorithm, in order to refine the modules borders. Here, again the process was repeated iteratively, as before, until no change in the maximum modularity factor, Q, was observed; this allowed for the number of optimal partitions (modules) to be determined automatically. The modularity factor, Q, ranges between 0 and 1, and represents the goodness in which a network is optimally partitioned into functional modules27.
Functional Connectivity (FC) maps
Seed-voxel partial correlation was used to calculate the FC maps. The final subdivisions of left and right amygdala were used as seeds and correlated with the entire brain for each subject, regressing out the signal contribution from the neighbor seeds. The cross-correlation coefficient maps were then converted to z-score maps using the Fisher’s r-to-z transformation. Final FC maps for each subject were generated by averaging the results across runs. In order to evaluate temporal variations in the connectivity pattern generated for each amygdala seed, the original time-series were divided into four equal subseries (sessions) and FC maps were calculated for each session and averaged across runs for each subject.
One sample t-tests were performed independently on the FC maps generated for each amygdala subset. Differential assessments of the FC maps were conducted to evaluate differences in functional connectivity across amygdala subdivisions by contrasting the connectivity path of one seed against the combination of the others and by pairwise comparison (see results in the supplemental materials) using a linear mixed-effects modeling approach (3dLME)89. The FC pathways for the different sessions (subseries) and each amygdala subdivision were calculated with t-test, and the difference between sessions across seed location (amygdala subdivisions) was evaluated with two-way analysis of variance (ANOVA) using 3dLME89. Results were corrected for multiple comparisons using 3dClustSim, based on Monte Carlo simulations90, including voxels of the entire brain and considering a conservative imaging smoothing (FWHM) of 10 mm; statistical significance was set for p corrected < 0.05 (see Supplemental material).
The reliability of the connectivity maps obtained for the four sessions (subseries) was also evaluated using a two-way mixed single measures intra-class correlation (ICC(3,1))91; specifically the between subject and residuals mean square values were computed for each voxel using the function 3dICC_REML89 on the FC maps generated for each amygdala subdivision, considering all sessions (see Supplemental material).
fMRI group analysis
One sample t-tests were performed on the beta weights generated for each subject providing a group activation map for the angry/fear faces contrasted against shapes. In addition, region-of-interest (ROI) analyses were carried out on the beta weights using the modularity results as a mask to extract the average BOLD signal in each amygdala subdivision for each subject. Results from the ROI analyses were compared with the average of the positive and significant (p < 0.05) Fisher transformed correlations obtained from the FC maps generated from seed-voxel partial correlations for each amygdala subdivision. Both analyses were carried out for the 96 subjects that survived head motion criteria for both resting and task-evoked datasets.
Availability of materials and data
All data analyzed in this manuscript were obtained from the WU-Minn Consortium HCP (WU-Minn HCP 500 Subjects + MEG2 Data Release), which is publicly available at the HCP online database (http://www.humanconnectome.org/). Detailed description of the analysis is included in the methods section and in the supplemental material.
Electronic supplementary material
Acknowledgements
This work was supported by the National Institute on Drug Abuse Intramural Research Program. Data were provided by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University.
Author Contributions
Elisabeth C. Caparelli carried all the data analysis and wrote the manuscript. Thomas J. Ross and Yihong Yang provided some guidelines for the data analysis and reviewed the manuscript. Hong Gu also provided some guidelines for the data analysis and helped with script writing. Xia Liang provided some guide on using the Brain Connectivity Toolbox. Elliot A. Stein provided important comments on the scientific significance of the work and reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14613-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Baxter MG, Murray EA. The amygdala and reward. Nat Rev Neurosci. 2002;3:563–573. doi: 10.1038/nrn875. [DOI] [PubMed] [Google Scholar]
- 2.Freese, J. L. & Amaral, D. G. In The human amygdala (eds P J Whalen & s E A Phelp) 3–42 (2009).
- 3.Swanson LW, Petrovich GD. What is the amygdala? Trends Neurosci. 1998;21:323–331. doi: 10.1016/S0166-2236(98)01265-X. [DOI] [PubMed] [Google Scholar]
- 4.Heimer, L. et al. The human basal forebrain Part II. The primate nervous system Part III. (1999).
- 5.Amunts K, et al. Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: intersubject variability and probability maps. Anatomy and embryology. 2005;210:343–352. doi: 10.1007/s00429-005-0025-5. [DOI] [PubMed] [Google Scholar]
- 6.Roy AK, et al. Functional connectivity of the human amygdala using resting state fMRI. Neuroimage. 2009;45:614–626. doi: 10.1016/j.neuroimage.2008.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sah P, Faber ES, Lopez De Armentia M, Power J. The amygdaloid complex: anatomy and physiology. Physiol Rev. 2003;83:803–834. doi: 10.1152/physrev.00002.2003. [DOI] [PubMed] [Google Scholar]
- 8.Puelles L. Thoughts on the development, structure and evolution of the mammalian and avian telencephalic pallium. Philos Trans R Soc Lond B Biol Sci. 2001;356:1583–1598. doi: 10.1098/rstb.2001.0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heimer, L., Van Hoesen, G. W., Trinmble, M. & Zahm, D. S. Anatomy of Neuropsychiatry. 1–171 (2008).
- 10.LeDoux J. The amygdala. Curr Biol. 2007;17:R868–R874. doi: 10.1016/j.cub.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Unal CT, Pare D, Zaborszky L. Impact of basal forebrain cholinergic inputs on basolateral amygdala neurons. J Neurosci. 2015;35:853–863. doi: 10.1523/JNEUROSCI.2706-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra A, Rogers BP, Chen LM, Gore JC. Functional connectivity-based parcellation of amygdala using self-organized mapping: a data driven approach. Hum. Brain Mapping. 2014;35:1247–1260. doi: 10.1002/hbm.22249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bach DR, Behrens TE, Garrido L, Weiskopf N, Dolan RJ. Deep and superficial amygdala nuclei projections revealed in vivo by probabilistic tractography. J Neurosci. 2011;31:618–623. doi: 10.1523/JNEUROSCI.2744-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pitkänen, A. Connectivity of the rat amygdaloid complex. 31–116 (Oxford University Press, 2000).
- 15.Saygin ZM, Osher DE, Augustinack J, Fischl B, Gabrieli JD. Connectivity-based segmentation of human amygdala nuclei using probabilistic tractography. Neuroimage. 2011;56:1353–1361. doi: 10.1016/j.neuroimage.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bickart KC, Hollenbeck MC, Barrett LF, Dickerson BC. Intrinsic Amygdala-Cortical Functional Connectivity Predicts Social Network Size in Humans. J Neurosci. 2012;32:14729–14741. doi: 10.1523/JNEUROSCI.1599-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bzdok D, Laird AR, Zilles K, Fox PT, Eickhoff SB. An investigation of the structural, connectional, and functional subspecialization in the human amygdala. Hum Brain Mapp. 2013;34:3247–3266. doi: 10.1002/hbm.22138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou C, Zemanova L, Zamora G, Hilgetag CC, Kurths J. Hierarchical organization unveiled by functional connectivity in complex brain networks. Physical review letters. 2006;97:238103. doi: 10.1103/PhysRevLett.97.238103. [DOI] [PubMed] [Google Scholar]
- 19.Ferrarini L, et al. Hierarchical functional modularity in the resting-state human brain. Hum. Brain Mapping. 2009;30:2220–2231. doi: 10.1002/hbm.20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meunier D, Achard S, Morcom A, Bullmore E. Age-related changes in modular organization of human brain functional networks. Neuroimage. 2009;44:715–723. doi: 10.1016/j.neuroimage.2008.09.062. [DOI] [PubMed] [Google Scholar]
- 21.Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nature reviews. Neuroscience. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 22.Uğurbil K, et al. Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. Neuroimage. 2013;80:80–104. doi: 10.1016/j.neuroimage.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glasser MF, et al. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brierley B, Shaw P, David AS. The human amygdala: a systematic review and meta-analysis of volumetric magnetic resonance imaging. Brain Res Brain Res Rev. 2002;39:84–105. doi: 10.1016/S0165-0173(02)00160-1. [DOI] [PubMed] [Google Scholar]
- 25.Newman ME. Modularity and community structure in networks. Proc Natl Acad Sci USA. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blondel, V., Guillaume, J., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J Stat Mech, P10008, 10.1088/1742-5468/2008/10/P10008 (2008).
- 27.Rubinov M, Sporns O. Weight-conserving characterization of complex functional brain networks. Neuroimage. 2011;56:2068–2079. doi: 10.1016/j.neuroimage.2011.03.069. [DOI] [PubMed] [Google Scholar]
- 28.Eickhoff SB, Thirion B, Varoquaux G, Bzdok D. Connectivity-based parcellation: Critique and implications. Hum Brain Mapp. 2015;36:4771–4792. doi: 10.1002/hbm.22933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicolini C, Bifone A. Modular structure of brain functional networks: breaking the resolution limit by Surprise. Scientific Reports. 2016;6:19250. doi: 10.1038/srep19250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosvall M, Bergstrom CT. An information-theoretic framework for resolving community structure in complex networks. Proc Natl Acad Sci USA. 2007;104:7327–7331. doi: 10.1073/pnas.0611034104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glerean E, et al. Reorganization of functionally connected brain subnetworks in high-functioning autism. Hum Brain Mapp. 2016;37:1066–1079. doi: 10.1002/hbm.23084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phelps EA, LeDoux JE. Contributions of the amygdala to emotion processing: from animal models to human behavior. Neuron. 2005;48:175–187. doi: 10.1016/j.neuron.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 33.Hariri AR, Bookheimer SY, Mazziotta JC. Neuroreport. 2000. Modulating emotional responses: effects of a neocortical network on the limbic system; pp. 43–48. [DOI] [PubMed] [Google Scholar]
- 34.Grezes J, Valabregue R, Gholipour B, Chevallier C. A direct amygdala-motor pathway for emotional displays to influence action: A diffusion tensor imaging study. Hum Brain Mapp. 2014;35:5974–5983. doi: 10.1002/hbm.22598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stefanacci L, Amaral DG. Some observations on cortical inputs to the macaque monkey amygdala: an anterograde tracing study. J Comp Neurol. 2002;451:301–323. doi: 10.1002/cne.10339. [DOI] [PubMed] [Google Scholar]
- 36.Kemppainen S, Jolkkonen E, Pitkänen A. Projections from the posterior cortical nucleus of the amygdala to the hippocampal formation and parahippocampal region in rat. Hippocampus. 2002;12:735–755. doi: 10.1002/hipo.10020. [DOI] [PubMed] [Google Scholar]
- 37.Moreno N, Gonzalez A. Evolution of the amygdaloid complex in vertebrates, with special reference to the anamnio-amniotic transition. J Anat. 2007;211:151–163. doi: 10.1111/j.1469-7580.2007.00780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Qin W, Jiang T, Zhang Y, Yu C. Sex-dependent correlations between the personality dimension of harm avoidance and the resting-state functional connectivity of amygdala subregions. PLoS One. 2012;7:e35925. doi: 10.1371/journal.pone.0035925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephan H, Andy OJ. Acta Anat (Basel) 1977. Quantitative comparison of the amygdala in insectivores and primates; pp. 130–153. [DOI] [PubMed] [Google Scholar]
- 40.Vogt, B. A. Cingulate Neurobiology and Disease. (Oxford University Press Inc., 2009).
- 41.Vogt BA, Pandya DN. Cingulate cortex of the rhesus monkey: II. Cortical afferents. J Comp Neurol. 1987;262:271–289. doi: 10.1002/cne.902620208. [DOI] [PubMed] [Google Scholar]
- 42.Chase HW, Moses-Kolko EL, Zevallos C, Wisner KL, Phillips ML. Disrupted posterior cingulate-amygdala connectivity in postpartum depressed women as measured with resting BOLD fMRI. Soc Cogn Affect Neurosci. 2014;9:1069–1075. doi: 10.1093/scan/nst083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchetti I, Koster EH, Sonuga-Barke EJ, De Raedt R. The default mode network and recurrent depression: a neurobiological model of cognitive risk factors. Neuropsychol Rev. 2012;22:229–251. doi: 10.1007/s11065-012-9199-9. [DOI] [PubMed] [Google Scholar]
- 44.Cauda, F. et al. Functional connectivity of the posteromedial cortex. PLoS One5, 10.1371/journal.pone.0013107 (2010). [DOI] [PMC free article] [PubMed]
- 45.Pfefferbaum A, et al. Cerebral blood flow in posterior cortical nodes of the default mode network decreases with task engagement but remains higher than in most brain regions. Cerebral cortex (New York, N. Y.: 1991) 2011;21:233–244. doi: 10.1093/cercor/bhq090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang S, Li CS. Functional connectivity mapping of the human precuneus by resting state fMRI. Neuroimage. 2012;59:3548–3562. doi: 10.1016/j.neuroimage.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parvizi J, Van Hoesen GW, Buckwalter J, Damasio A. Neural connections of the posteromedial cortex in the macaque. Proc Natl Acad Sci USA. 2006;103:1563–1568. doi: 10.1073/pnas.0507729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leichnetz GR. Connections of the medial posterior parietal cortex (area 7m) in the monkey. Anat Rec. 2001;263:215–236. doi: 10.1002/ar.1082. [DOI] [PubMed] [Google Scholar]
- 49.Van Schuerbeek P, Baeken C, Luypaert R, De Raedt R, De Mey J. Does the amygdala response correlate with the personality trait ‘harm avoidance’ while evaluating emotional stimuli explicitly? Behav Brain Funct. 2014;10:18. doi: 10.1186/1744-9081-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seeley WW, et al. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kapp BS, Supple WFJ, Whalen PJ. Effects of electrical stimulation of the amygdaloid central nucleus on neocortical arousal in the rabbit. Behav Neurosci. 1994;108:81–93. doi: 10.1037/0735-7044.108.1.81. [DOI] [PubMed] [Google Scholar]
- 52.Touroutoglou A, Bickart KC, Barrett LF, Dickerson BC. Amygdala task-evoked activity and task-free connectivity independently contribute to feelings of arousal. Hum Brain Mapp. 2014;35:5316–5327. doi: 10.1002/hbm.22552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phillips ML, Drevets WC, Rauch SL, Lane R. Neurobiology of emotion perception I: The neural basis of normal emotion perception. Biol Psychiatry. 2003;54:504–514. doi: 10.1016/S0006-3223(03)00168-9. [DOI] [PubMed] [Google Scholar]
- 54.Vogt BA. Pain and emotion interactions in subregions of the cingulate gyrus. Nat Rev Neurosci. 2005;6:533–544. doi: 10.1038/nrn1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aggleton JP. The contribution of the amygdala to normal and abnormal emotional states. Trends Neurosci. 1993;16:328–333. doi: 10.1016/0166-2236(93)90110-8. [DOI] [PubMed] [Google Scholar]
- 56.Whalen PJ, et al. Masked presentations of emotional facial expressions modulate amygdala activity without explicit knowledge. J Neurosci. 1998;18:411–418. doi: 10.1523/JNEUROSCI.18-01-00411.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Craig AD. How do you feel-now? The anterior insula and human awareness. Nat Rev Neurosci. 2009;10:59–70. doi: 10.1038/nrn2555. [DOI] [PubMed] [Google Scholar]
- 58.Muhlberger A, et al. Stop looking angry and smile, please: start and stop of the very same facial expression differentially activate threat- and reward-related brain networks. Soc Cogn Affect Neurosci. 2011;6:321–329. doi: 10.1093/scan/nsq039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vogel S, et al. Blocking the mineralocorticoid receptor in humans prevents the stress-induced enhancement of centromedial amygdala connectivity with the dorsal striatum. Neuropsychopharmacology. 2015;40:947–956. doi: 10.1038/npp.2014.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilensky AE, Schafe GE, Kristensen MP, LeDoux JE. Rethinking the fear circuit: the central nucleus of the amygdala is required for the acquisition, consolidation, and expression of Pavlovian fear conditioning. J Neurosci. 2006;26:12387–12396. doi: 10.1523/JNEUROSCI.4316-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blanchard DC, Blanchard RJ. Innate and conditioned reactions to threat in rats with amygdaloid lesions. J Comp Physiol Psychol. 1972;81:281–290. doi: 10.1037/h0033521. [DOI] [PubMed] [Google Scholar]
- 62.Gregg TR, Siegel A. Brain structures and neurotransmitters regulating aggression in cats: implications for human aggression. Prog Neuropsychopharmacol Biol Psychiatry. 2001;25:91–140. doi: 10.1016/S0278-5846(00)00150-0. [DOI] [PubMed] [Google Scholar]
- 63.Ball T, et al. Response properties of human amygdala subregions: evidence based on functional MRI combined with probabilistic anatomical maps. PLoS One. 2007;2:e307. doi: 10.1371/journal.pone.0000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baker KB, Kim JJ. Amygdalar lateralization in fear conditioning: evidence for greater involvement of the right amygdala. Behav Neurosci. 2004;118:15–23. doi: 10.1037/0735-7044.118.1.15. [DOI] [PubMed] [Google Scholar]
- 65.Ji G, Neugebauer V. Hemispheric Lateralization of Pain Processing by Amygdala Neurons. J Neurophysiol. 2009;102:2253–2264. doi: 10.1152/jn.00166.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quirk GJ, Beer JS. Prefrontal involvement in the regulation of emotion: convergence of rat and human studies. Curr Opin Neurobiol. 2006;16:723–727. doi: 10.1016/j.conb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 67.Llamas A, Avendano C, Reinoso-Suarez F. Amygdaloid projections to prefrontal and motor cortex. Science. 1977;195:794–796. doi: 10.1126/science.836591. [DOI] [PubMed] [Google Scholar]
- 68.Amaral DG, Price JL. Amygdalo-cortical projections in the monkey (Macaca fascicularis) J Comp Neurol. 1984;230:465–496. doi: 10.1002/cne.902300402. [DOI] [PubMed] [Google Scholar]
- 69.Gotts SJ, et al. Fractionation of social brain circuits in autism spectrum disorders. Brain. 2012;135:2711–2725. doi: 10.1093/brain/aws160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoffman KL, Gothard KM, Schmid MC, Logothetis NK. Facial-expression and gaze-selective responses in the monkey amygdala. Curr Biol. 2007;17:766–772. doi: 10.1016/j.cub.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 71.Ball T, et al. Anatomical specificity of functional amygdala imaging of responses to stimuli with positive and negative emotional valence. J Neurosci Methods. 2009;180:57–70. doi: 10.1016/j.jneumeth.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 72.Reichenbach JR, et al. Theory and application of static field inhomogeneity effects in gradient-echo imaging. J Magn Reson Imaging. 1997;7:266–279. doi: 10.1002/jmri.1880070203. [DOI] [PubMed] [Google Scholar]
- 73.Cordes D, Nandy RR, Schafer S, Wager TD. Characterization and reduction of cardiac- and respiratory-induced noise as a function of the sampling rate (TR) in fMRI. Neuroimage. 2014;89:314–330. doi: 10.1016/j.neuroimage.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jacob Y, et al. Dependency Network Analysis (DEPNA) Reveals Context Related Influence of Brain Network Nodes. Sci Rep. 2016;6:27444. doi: 10.1038/srep27444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Glasser MF, et al. The Human Connectome Project’s neuroimaging approach. Nat Neurosci. 2016;19:1175–1187. doi: 10.1038/nn.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moeller S, et al. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn Reson Med. 2010;63:1144–1153. doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.WU-Minn HCP 500 Subjects + MEG2 Data Release: Reference Manualhttp://www.humanconnectome.org/documentation/S500/
- 78.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 79.Smith, S. M. In Functional MRI: an introduction to methods (eds P. P. M. Jezzard, Matthews, & S. Smith, M.) (Oxford University Press, 2003).
- 80.Foerster B, Tomasi D, Caparelli EC. Magnetic field shift due to mechanical vibration in functional magnetic resonance imaging. Magn. Reson. Med. 2005;54:1261–1267. doi: 10.1002/mrm.20695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cordes D, et al. Frequencies Contributing to Functional Connectivity in the Cerebral Cortex in “Resting-state” Data. AJNR. 2001;22:1326–1333. [PMC free article] [PubMed] [Google Scholar]
- 82.Lund TE, Madsen KH, Sidaros K, Luo WL, Nichols TE. Non-white noise in fMRI: does modelling have an impact? Neuroimage. 2006;29:54–66. doi: 10.1016/j.neuroimage.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 83.Chai XJ, Castanon AN, Ongur D, Whitfield-Gabrieli S. Anticorrelations in resting state networks without global signal regression. Neuroimage. 2012;59:1420–1428. doi: 10.1016/j.neuroimage.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 85.Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nielsen, F. A. & Hansen, L. K. Automatic anatomical labelling of Talairach coordinates and generation of volumes of interest via the BrainMap database, http://neuro.imm.dtu.dk/services/jerne/ninf/voi (2002).
- 87.Fox, P. T., Mikiten, S., Davis, G. & Lancaster, J. L. In Functional Neuroimaging: Technical Foundations (eds R. W. Thatcher et al.) Ch. 9, 95–105 (Academic Press, 1994).
- 88.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 89.Chen G, Saad ZS, Britton JC, Pine DS, Cox RW. Linear mixed-effects modeling approach to FMRI group analysis. Neuroimage. 2013;73:176–190. doi: 10.1016/j.neuroimage.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ward, B. D. Simultaneous inference for FMRI data. http://afni.nimh.nih.gov/pub/dist/doc/manual/AlphaSim.pdf (2000).
- 91.Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull. 1979;86:420–428. doi: 10.1037/0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- 92.Diedrichsen J, Balsters JH, Flavell J, Cussans E, Ramnani N. A probabilistic MR atlas of the human cerebellum. Neuroimage. 2009;46:39–46. doi: 10.1016/j.neuroimage.2009.01.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.