Abstract
Violacein is a bisindole antibiotic that is effective against Gram-positive bacteria while the bacterial predator, Bdellovibrio bacteriovorus HD100, predates on Gram-negative strains. In this study, we evaluated the use of both together against multidrug resistant pathogens. The two antibacterial agents did not antagonize the activity of the other. For example, treatment of Staphylococcus aureus with violacein reduced its viability by more than 2,000-fold with or without B. bacteriovorus addition. Likewise, predation of Acinetobacter baumannii reduced the viability of this pathogen by more than 13,000-fold, regardless if violacein was present or not. When used individually against mixed bacterial cultures containing both Gram-positive and Gram-negative strains, violacein and B. bacteriovorus HD100 were effective against only their respective strains. The combined application of both violacein and B. bacteriovorus HD100, however, reduced the total pathogen numbers by as much as 84,500-fold. Their combined effectiveness was also demonstrated using a 4-species culture containing S. aureus, A. baumannii, Bacillus cereus and Klebsiella pneumoniae. When used alone, violacein and bacterial predation reduced the total population by only 19% and 68%, respectively. In conjunction with each other, the pathogen viability was reduced by 2,965-fold (99.98%), illustrating the prospective use of these two antimicrobials together against mixed species populations.
Introduction
Within nature, bacteria are found primarily within polymicrobial communities. This is also true of infections, which may include both pathogens and commensal microbes1,2. One benefit that bacteria within these communities gain is an increased resistance to antibiotics3,4. Two reasons for this include the presence of resistant strains5,6, which can degrade the antibiotic, or because the other strains act as a “sink” for the antibiotic, diluting its impact on the susceptible populations7. To overcome this, researchers have often resorted to using a combination of antibiotics8–10.
Antibiotic resistance on a single species level can be either intrinsic or acquired. Intrinsic resistance is a characteristic that is coded for in the genome of the strain, independent of antibiotic pressure and not a consequence of horizontal gene transfer. An example of intrinsic resistance includes the outer membrane in Gram-negative bacteria, which modulates the cell permeability and limits the entry of hydrophobic antibiotics11. Lacking an outer membrane, Gram-positive strains are generally less protected and antibiotics may enter these bacteria more easily12. The result is that Gram-negative bacteria are less susceptible to many antibiotics that are effective against Gram-positive pathogens.
A case in point is the bisindole antibiotic violacein, which is produced by a number of bacterial species13, including strains of Chromobacterium 14,15, Janthinobacterium 16, Collimonas 17 and Duganella 18. As an antibiotic, violacein is primarily active against Gram-positive strains15,18–20, particularly Staphylococcus aureus 20–22. It was also active against multidrug-resistant S. aureus 23, including one strain that is resistant to seven different antibiotics, i.e., rifampin, ciprofloxacin, clindamycin, oxacillin, erythromycin, gentamycin and tobramycin18. Violacein, however, is generally ineffective against Gram-negative strains.
In addition to conventional chemical-based antibiotics, several groups are also considering the use of predatory bacterial strains as living antibacterial agents24–26. These strains are commonly referred to as Bdellovibrio-and-like-organisms (BALOs). The best characterized BALO is probably Bdellovibrio bacteriovorus HD100, an obligate predator of bacteria that is known to attack more than 100 different human pathogens27–30, including multidrug resistant strains of Acinetobacter baumannii and Klebsiella pneumoniae 28. When it attacks its prey, B. bacteriovorus enters the periplasmic space and consumes its prey from within, where this predator grows, septates and eventually lyses the outer membrane of the prey to facilitate its release. In contrast with violacein, the activity of BALOs is limited to only Gram-negative bacterial strains29–32.
Given the inherent limitations associated with both violacein and B. bacteriovorus, we proposed they might be used in conjunction with one another to expand their individual spectrums of killing. To test this, B. bacteriovorus and violacein were partnered together and their activities evaluated using individual bacterial and polymicrobial populations, with an emphasis on those that included both Gram-negative and Gram-positive pathogens.
Materials and Methods
Bacterial strains and culturing techniques
The bacterial strains used in this study are listed in Table 1. Each of these strains was grown in Lysogeny Broth (LB) broth (Difco BD, USA) overnight at 30 °C and 250 rpm. To examine the impact of the treatment by predatory bacteria or violacein, the bacterial culture were prepared by centrifuging with 2,200 × g for 10 min (5430 R, Eppendorf, USA) and resuspended in HEPES buffer (pH 7.2) to an optical density (OD) of 1.0 at 600 nm wavelength. B. bacteriovorus HD100 was cultured as described previously33,34.
Table 1.
Bacterial strains used in this study.
| Bacterial Strains | Characteristics | Ref |
|---|---|---|
| S. aureus ATCC 25923 | ||
| B. cereus ATCC 14579 | ||
| S. epidermidis ATCC 14990 | ||
| A. baumannii | Clinical isolate | PNUHa |
| E. coli MG1655 | Common lab strain; multidrug resistant | |
| K. pneumoniae | Clinical isolate; multidrug resistant | PNUHa |
| S. aureus CCARM 3090 | Clinical isolate; multidrug resistant | (18)b |
aClinical strains isolated at the Pusan National University Hospital, Yangsan Campus. bObtained from the Culture Collection of Antimicrobial Resistant Microbes (http://knrrb.knrrc.or.kr/index.jsp?rrb=ccarm).
Violacein extraction from Pseudoduganella violaceinigra sp. NI28 cultures
Violacein was extracted from cultures of Pseudoduganella violaceinigra sp. NI28 as described previously18. After dissolving the violacein in ethanol, the solution was filtered using a 0.22um syringe filter (Millipore, USA) and the ethanol was evaporated (EYELA N1110, Tokyo Rikakikai Co., Japan). The violacein was crystallized using an acetone wash designed to remove any contaminating salts35 and dissolved in DMSO (99.7% HPLC Grade, Sigma Aldrich, USA) to a final concentration of 2 g/L.
Antimicrobial treatment
Overnight cultures of the bacterial strains were pelleted (2,200 × g, 10 min) and resuspended in HEPES buffer to an OD of 1.0 (600 nm). Each individual culture was then challenged with predatory bacteria or violacein. For the predatory tests, overnight cultures of B. bacteriovorus HD100 were filtered and added to the mixed bacterial suspension to achieve a predator-to-prey ratio (PPR) of 0.02~0.05. Similarly, for the violacein tests, violacein was added to the mixed bacterial suspension to a final concentration of 20 mg/ml. The cell viability (colony forming units (CFU)) was measured after each treatment. For the dual treatment experiments using predatory bacteria and violacein together, the same conditions were applied.
Likewise, for the mixed species tests, the cultures were prepared by adding equal volumes of each bacterial resuspension prior to addition of violacein and/or B. bacteriovorus HD100. Differences in the colony morphologies were used to differentiate between the two strains when we determined the viabilities of these cultures. For the four strain experiments, individual strains could not be differentiated between based upon the colony morphology and, so, the total viability was determined by plating serial dilutions of the cultures on LB agar plates and enumerating the colonies after 24 h at 37 °C.
Microscopic imaging
Microscopic images were obtained using a Zeiss LSM 780 NLO microscope. To visualize the bacterial strains, dyes were used. Prior to treatment with either violacein or B. bacteriovorus HD100, each of the bacterial cultures were mixed with 6 µM Syto-9 (Invitrogen, USA). This is a live stain and all viable bacterial cells were fluorescently green afterwards. After washing the cells with HEPES to remove any extra dye, they were exposed to either the predatory cells (PPR of 0.1) or 20 mg/ml of violacein in HEPES. After one hour, propidium iodide (Invitrogen, USA) was added to the bacterial cultures to a final concentration of 30 µM. This dye is a dead stain and labels any non-viable bacterial cells red. After 30 minutes at room temperature, the cells were pelleted (16,000 × g, 5 min), washed and resuspended in HEPES before being imaged.
Antibiotic resistance determination
To determine the antibiotic resistant nature of the bacterial strains, we used the same protocol as described previously18. Strain resistance was determined using the latest breakpoint tables available at the European Committee on Antimicrobial Susceptibility Testing (EUCAST) website (http://www.eucast.org/clinical_breakpoints/).
Spot viability assay
To compare the activity of conventional antibiotics and the dual treatment protocol used in this study, we tested representative bacteriostatic and bactericidal antibiotics, i.e., chloramphenicol and gentamycin, respectively. For the assays, a concentrated dosage that was 10-fold higher than the typical dose used in labs was employed. As such, the bacterial community was exposed to either 350 mg/l chloramphenicol, 500 mg/l gentamicin or both. After treatment, the cells viabilities were evaluated using a spot viability plate. For this, the exposed cultures were serially diluted into LB media and 10 µl was spotted on LB agar within a square petridish (SPL, Korea) and incubated for 24 hour at 37 °C. The extent of growth was used as an indication of viability.
Statistical analysis
In this research, all assays were accomplished at least three replicates and the standard deviations among the samples are indicated with error bars on the graphs. Statistical analysis was done using Student’s t-test to compare two sets of results and the statistical significance was marked on the graphs using the marks *, **, *** for p-values of less than 0.05, 0.01, and 0.001 respectively. For comparing three or more data sets, analysis of variance (ANOVA) tests were performed followed by the Tukey post-hoc test. Statistically, significantly different groups using a p-value < 0.05 are shown on the graphs using letters (a, b, c and d).
Results
Activity Spectrum of Violacein and Bacterial Predation
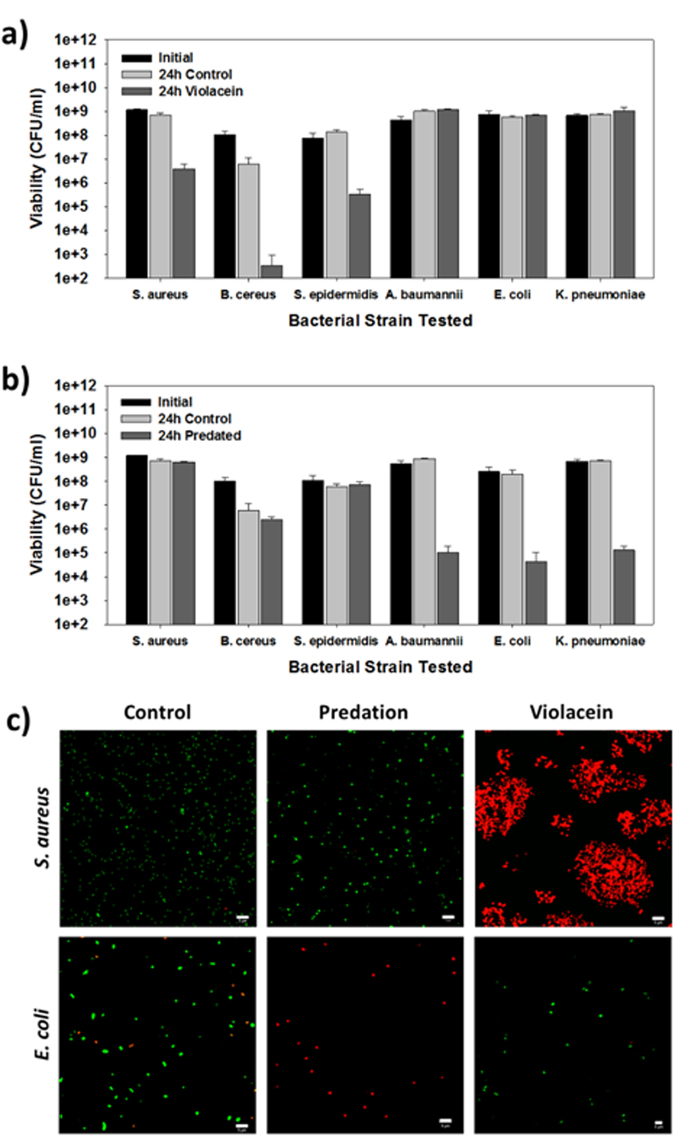
Violacein is quite active against S. aureus, killing more than 99% of the culture when added at a concentration of 20 mg/L or greater18,21,23. Using this concentration, we tested the activity of violacein against six different strains (Tables 1 and 2), including three Gram-positive and three Gram-negative species. As shown in Fig. 1a, only the Gram-positive cultures were sensitive to violacein, with each showing significant viability losses after 24 hours. In contrast, none of the Gram-negative cultures were negatively impacted by violacein (Fig. 1a). The A. baumannii and K. pneumoniae strains used throughout this study were multidrug resistant clinical isolates (Table 2).
Table 2.
Antibiotic resistance of the clinical isolates used in this study. The highlighted values are indicative of resistance.
| AMP | CIP | GEN | TET | CM | RIF | ERY | CC | |
|---|---|---|---|---|---|---|---|---|
| A. baumannii | 0(R) | 0(R) | 0(R) | 18–22(S) | 12–15(I) | 15–17(S) | 15–17(S) | ND |
| K. pneumoniae | 9–10(R) | 29–30(S) | 20–21(S) | 24–25(S) | 23–25(S) | 6–8(R) | 6–8(R) | ND |
| S. aureus CCARM 3090 | 9–10(R) | 0(R) | 23–25(S) | 9–10(R) | 22–25(S) | 30–35(S) | 0(R) | 0(R) |
AMP: ampicillin, CIP: Ciprofloxacin, GEN: Gentamycin, TET: Tetracycline, CM: Chloramphenicol, RIF: Rifampicin, ERY: Erythromycin, CC: Clindamycin. Classifications: (R) – Resistant; (I) – Intermediate; (S) – Susceptible; ND – Not determined.
Figure 1.
Spectrum of activity for violacein and B. bacteriovorus HD100. (a and b) The activity of each antimicrobial was evaluated using three Gram-positive and three Gram-negative bacterial strains. The pathogens were treated with either (a) 20 mg/l violacein or (b) 1 × 107 PFU/ml of B. bacteriovorus HD100. The viabilities of the pathogens were determined initially and after 24 hrs (n = 4). (c) Impact of violacein and B. bacteriovorus HD100 on S. aureus and E. coli. The bacteria were treated with either 20 mg/l violacein or approximately 1 × 107 PFU/ml of B. bacteriovorus HD100 for 1 hr. Using the Live/Dead BacLight kit, the viable and non-viable bacteria were fluorescently labeled and imaged using confocal microscopy. Scale bar – 10 µm.
The results with B. bacteriovorus HD100 stood in stark contrast as only the Gram-negative strains experienced a loss in their viabilities (Fig. 1b). These results clearly illustrate the different activity spectrums for both antimicrobials and highlight the limitations of each. This dichotomy is further demonstrated in Fig. 1c where S. aureus or E. coli were imaged after being exposed to either. In agreement with Fig. 1a and b, violacein only killed S. aureus while B. bacteriovorus HD100 only killed E. coli.
Violacein and B. bacteriovorus Do Not Adversely Impact Each Other’s Activity
The above data shows violacein was not active against the three Gram-negative strains. Given B. bacteriovorus HD100 is also Gram-negative, we speculated that violacein would also not be harmful towards this predator or its activity. Figure 2a shows that this was the case when A. baumannii was used as the prey strain. In both cases, the A. baumannii populations were similarly reduced by more than 4-log due to the activity of B. bacteriovorus HD100. Similarly, the activity of violacein was not thwarted by the presence of B. bacteriovorus HD100, as illustrated in Fig. 2b. This figure shows reduction of the S. aureus populations was comparable whether or not the predator was added.
Figure 2.
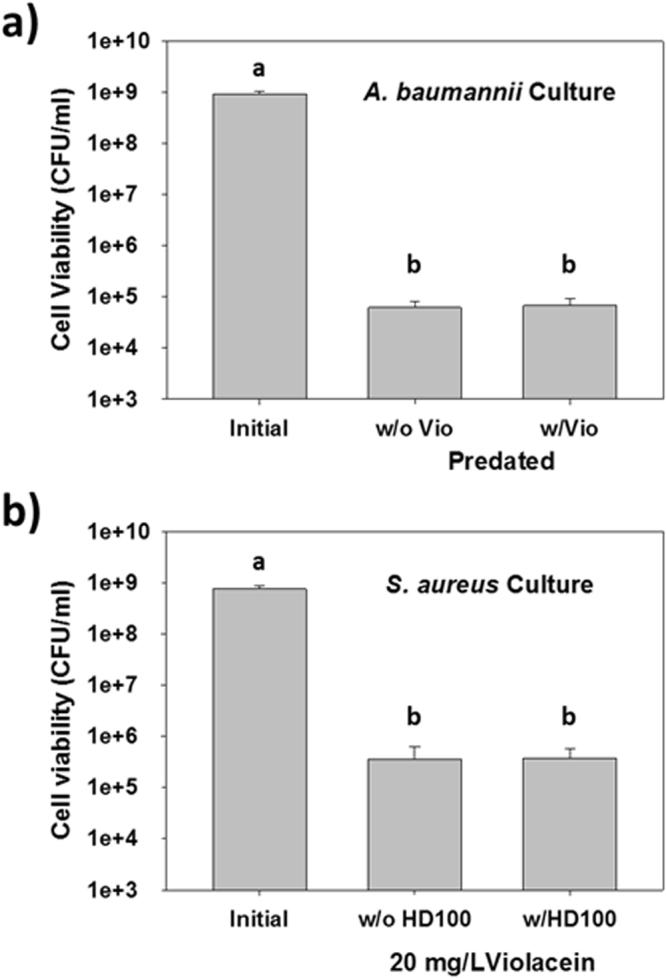
Violacein and B. bacteriovorus HD100 do not hinder the activity of the other. (a) Predation of A. baumannii with or without addition of 20 mg/l violacein. A. baumannii was predated on by B. bacteriovorus HD100 as effectively when violacein was present as when it was omitted. The viabilities were measured after 24 h, as in Fig. 1. a and b = p < 0.05 (n = 3). (b) The activity of violacein against S. aureus with or without addition of B. bacteriovorus. The presence of the predatory bacterium did not negatively impact the activity of violacein against this pathogen. The viabilities were measured after 24 h, as in Fig. 1. a and b = p < 0.05 (n = 3).
The Activities of Violacein and B. bacteriovorus are Compatible with Each Other
We next tested the possibility of using both treatments together against cultures containing both a Gram-positive and a Gram-negative strain. For these tests, we selected S. aureus (average initial population: 4.2 × 108 CFU/ml) and either A. baumannii or K. pneumoniae (average initial population: 7.9 × 108 CFU/ml).
As shown in Fig. 3a, both antimicrobials were active against their respective pathogen: treatment with violacein alone killed only S. aureus while the use of B. bacteriovorus HD100 by itself only reduced the A. baumannii population. As a result, the total pathogen number did not decrease very much, i.e., only 51% with violacein and 69% with B. bacteriovorus HD100. When violacein and B. bacteriovorus HD100 were used together, however, both pathogens were significantly killed and the total pathogen numbers were reduced by 99.8% (620-fold). Similar results were obtained when S. aureus and K. pneumoniae were cultured together (Fig. 3b) as individual treatments with violacein and B. bacteriovorus HD100 led to a 41% and 81% loss in total pathogens, respectively, while a dual treatment caused a 99.999% (84,500-fold) reduction in the pathogen viability.
Figure 3.
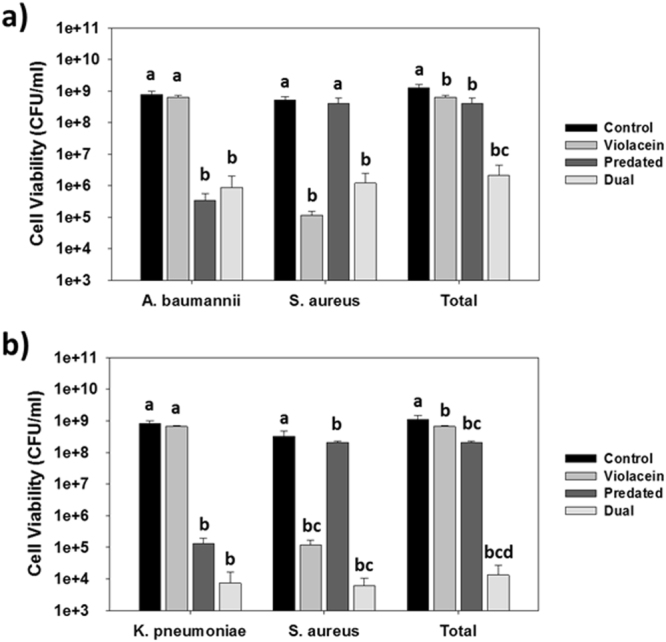
Treatment of two-strain cultures containing S. aureus with violacein and B. bacteriovorus HD100. (a) Impact of 20 mg/l violacein alone, 1 × 107 PFU/ml B. bacteriovorus HD100 alone or both together against a mixed culture containing S. aureus and A. baumannii. The viabilities were measured after 24 hr. Each of the antimicrobials was effective against their respective pathogen when used alone, but together led to a significant loss in the total pathogen numbers. a, b and c = p < 0.05 (n = 3). (b) Impact of 20 mg/l violacein alone, 1 × 107 PFU/ml B. bacteriovorus HD100 alone or both together against a mixed culture containing S. aureus and K. pneumoniae. The viabilities were measured after 24 hr. Each of the antimicrobials was effective against their respective pathogen when used alone, but together led to a significant loss in the total pathogen numbers. a, b, c and d = p < 0.05 (n = 3).
Dual treatment tests were also performed using either A. baumannii or K. pneumoniae alongside a different Gram-positive bacteria, i.e., S. epidermidis (Fig. 4). In both cultures, the viability of each pathogen was reduced by more than 99.3%.
Figure 4.
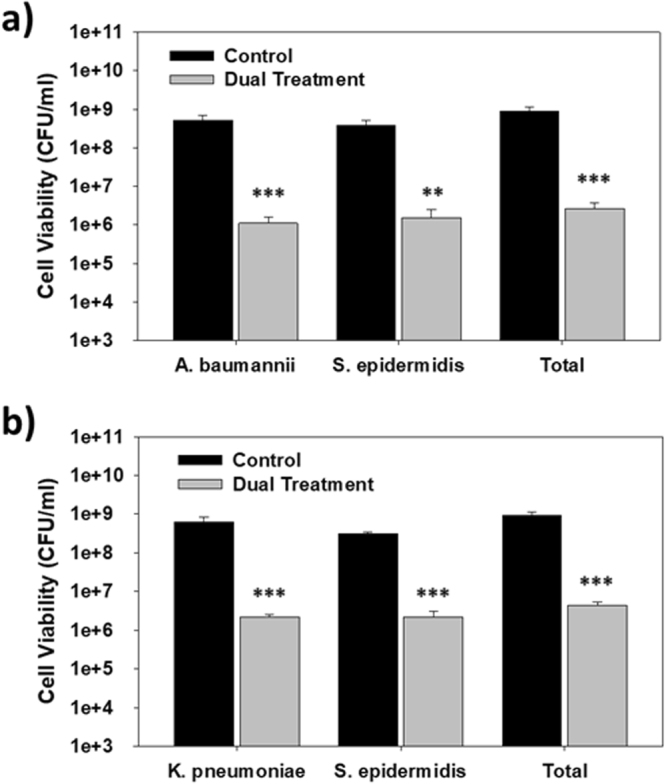
Treatment of two-strain cultures containing S. epidermidis with violacein and B. bacteriovorus HD100. (a) Dual treatment of a mixed culture containing S. epidermidis and A. baumannii using 20 mg/l violacein and 1 × 107 PFU/ml B. bacteriovorus HD100. The viabilities were measured after 24 hr (**p < 0.001; ***p < 0.001) (n = 3). (b) Dual treatment of a mixed culture containing S. epidermidis and K. pneumoniae using 20 mg/l violacein and 1 × 107 PFU/ml B. bacteriovorus HD100. The viabilities were measured after 24 hr (***p < 0.001) (n = 3).
Application of the Dual Treatment to a Complex Microbial Community
Subsequently, we tested if a dual treatment is also effective against a complex microbial consortium harboring four different human pathogens, i.e., S. aureus, A. baumannii, B. cereus and K. pneumoniae (Fig. 5). The population of each pathogenic strain is listed in Table 3, with a total initial population of 5.9 × 108 CFU/ml. After treatment with either B. bacteriovorus HD100 or violacein alone, the Gram-negative and Gram-positive strain viabilities decreased, respectively, but we could not reliably measure the impact due to the overwhelming presence of the surviving strains. The use of violacein alone resulted in a 19% reduction of the total number of pathogens while B. bacteriovorus HD100 led to a 68% loss (3.1-fold). As shown in Fig. 5, when both antimicrobials were used, the number pathogens was reduced by 99.96% (2,970-fold).
Figure 5.
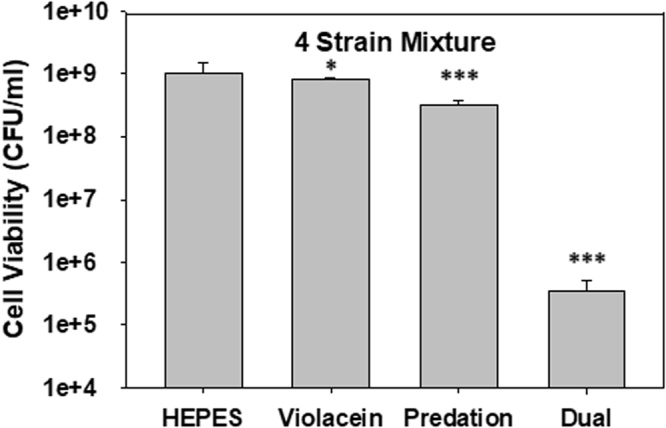
Dual treatment of polymicrobial culture containing four different pathogens. Impact of 20 mg/l violacein alone, 1 × 107 PFU/ml B. bacteriovorus HD100 alone or both antimicrobials together against a culture containing four pathogens (S. aureus, B. cereus, A. baumannii and K. pneumoniae, Table 3). The viabilities of the culture as a whole were measured after 24 hr. Each of the antimicrobials led to mild but significant losses when used alone, but together led to a 99.96% loss in the total pathogen numbers (*p < 0.05; ***p < 0.001) (n = 3).
Table 3.
Impact of predation or violacein on the survival of the four pathogens within a mixed population.
| Initial | HEPES (24 hr) | Violacein (24 hr) | Predation (24 hr) | |
|---|---|---|---|---|
| B. cereus | 8.9 ± 1.4 × 106 | 1.8 ± 0.7 × 107 | N.D.a | 3.6 ± 1.3 × 107 |
| S. aureus ATCC 25923 | 2.7 ± 0.4 × 108 | 3.1 ± 0.2 × 108 | N.D. | 2.9 ± 0.7 × 108 |
| K. pneumoniae | 9.7 ± 0.7 × 107 | 3.5 ± 0.2 × 108 | 4.2 ± 0.1 × 108 | N.D. |
| A. baumannii | 2.1 ± 0.5 × 108 | 3.5 ± 0.8 × 108 | 4.2 ± 0.3 × 108 | N.D. |
| Total (Average) | 5.9 × 108 | 1.03 × 109 | 8.4 × 108 (81%) | 3.2 × 108 (32%) |
aN.D. – Not detected on the agar plates with a 107 dilution.
Dual Treatment Impacts on Multidrug Resistant Bacterial Populations
As a final test using both predatory bacteria and violacein together, a test culture containing multidrug resistant S. aureus (Table 2) was prepared. The other strain was the same A. baumannii as above, which is also multidrug resistant. Both of these strains are resistant to numerous antibiotics, as listed in Table 2, and the initial population of each pathogen within the cultures was 5.2 × 108 and 3.2 × 108 CFU/ml, respectively. For comparison, we also treated the mixed pathogen culture with a concentrated blend of gentamicin with chloramphenicol (Fig. 6). The image show the use of B. bacteriovorus HD100 and violacein was much more effective at killing the pathogens than the use of gentamicin and chloramphenicol, even though the concentrations of the two antibiotics were much higher than what is typically used.
Figure 6.
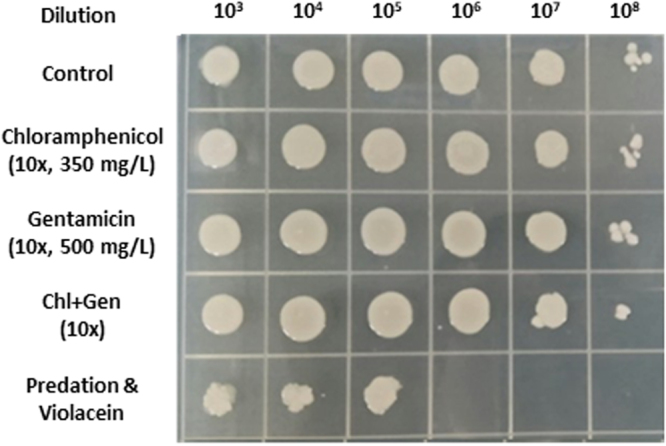
Dual treatment of a culture containing multidrug resistant strains of A. baumannii and S. aureus. A mixed culture containing A. baumannii alongside S. aureus CCARM 3090 was treated with either chloramphenicol, gentamicin, these two antibiotics together (Chl + Gen) or the dual treatment (Predation & Violacein). The concentrations of the two antibiotics were 10x higher than commonly used in lab experiments. After 24 hr, the cultures were serially diluted and 10 µl spotted onto an agar plate. The image here shows the resulting growth, illustrating the much higher degree of killing obtained with the dual treatment protocol.
Discussion
Due to the universal spread of antibiotic resistance, it is projected that the number of human mortalities each year resulting from antimicrobial resistant pathogens will surpass that caused by cancer by 205036. Two alternative antimicrobials currently being studied by various groups are violacein and predatory bacteria18,23,25,28,37,38. In addition to their antibiotic activities, both also show promising toxicology results, a key characteristic for broad spectrum antibiotics8,39; in tests with mice, violacein was non-toxic at concentrations as high as 1 mg/kg40 while B. bacteriovorus was not toxic towards several different animal hosts41–43 or in human cell cultures44–46.
Treatment of S. aureus with violacein not only led to a significant loss of viability but also caused a substantial degree of aggregation. Similar results were seen in other studies where S. aureus was exposed to either violacein22 or galangin47. In the former study, violacein was shown to disrupt the membrane integrity of S. aureus, which led to a rupturing of the cell membrane and leakage of the cellular components. It was hypothesized by Cushnie et al. (2007) that galangin similarly damages the membrane and exposes the hydrophobic regions located within the phospholipid bilayer, which then interact and cause aggregation47. The same process likely occurs with violacein, which would account for the cell aggregates seen here. The activity of violacein also helps to explain the inherent resistance of Gram-negative bacterial strains since the outer membrane would act like a sponge and absorb violacein, protecting the inner membrane. With B. bacteriovorus, however, only E. coli was killed while the S. aureus cells remained viable (Fig. 1c). This is a clear demonstration that S. aureus is not a prey for B. bacteriovorus and reaffirms the long held knowledge that BALOs only predate and consume Gram-negative strains29–32, despite claims in one study that S. aureus is also attacked48. Although the mode of activity for violacein has recently been resolved, much still remains unanswered about how predatory bacteria recognize, enter and kill their prey and the processes involved in each of these steps.
The results above illustrate inherent limitations for both violacein and B. bacteriovorus HD100; violacein is largely effective against Gram-positive bacteria while B. bacteriovorus HD100 attacks only Gram-negative strains. Given the different activity spectrums for these two antimicrobials, we proposed they might be complementary and used alongside each other against polymicrobial populations. However, it was not known if predatory bacteria and violacein negatively impact the activity of the other. Here we show that this was not the case – violacein was just as effective against S. aureus regardless of whether B. bacteriovorus was present while predation of A. baumannii proceeded just as successfully in the presence or absence of violacein. This suggested that they can be used together and hinted at their potential use to eradicate polymicrobial populations.
Polymicrobial infections are a serious problem since complex bacterial communities can benefit from the properties of each of the members. An example of this is the protection afforded to a susceptible population by a resistant member of the community, such as through enzymatic inactivation of the antibiotic5,6. Another possibility is the “inoculum effect”, whereby the antibiotic activity is diluted out by the presence of other bacteria, which act as “sinks” for the antibiotic and limit its effectiveness against susceptible microbe populations7. We were concerned that this may be a problem for violacein. Violacein is a hydrophobic compound and attacks cellular membranes, implying the presence of naturally-resistant Gram-negative bacterial cells may act as a “sink” for this antibiotic and dilute its impact on S. aureus or other Gram-positive pathogens. The results, however, suggest that this is not a problem as violacein at the concentration used was as effective at killing S. aureus regardless if A. baumannii or K. pneumoniae were present.
Likewise, although they are not attacked by B. bacteriovorus, Gram-positive strains may act as decoys and slow the predation of susceptible Gram-negative bacterial strains49. As B. bacteriovorus does not display a clear chemotaxis towards prey cells50, it is generally thought that interactions between B. bacteriovorus HD100 and its prey are random events that occur as the predator swims within the media. By extrapolation, this means B. bacteriovorus will also come in contact and interact with non-prey bacterial cells. Such interactions have been observed with both Neisseria gonorrhoeae 51 and Bacillus subtilis 49, although neither is predated upon. Stemming from this reasoning, Wilkinson (2001) developed the first mathematical model to describe the impact decoys may have on B. bacteriovorus predation rates and efficacies using a double-Monod framework based upon a continuous flow system52. Hobley et al. (2006) followed this with experimental data using B. subtilis as a decoy in predation cultures with E. coli S17-1 as the prey, and developed their own model based upon the Lotka-Volterra equations49. They found predation rates were slower during the first seven hours when B. subtilis was present as a decoy at a ratio of approximately 1:2 (decoy:prey). In contrast, the study by Van Essche et al. (2010) found the presence of a decoy, i.e., Gram-positive Actinomyces naeslundii ATCC 12104, at a 14-fold excess had no impact on predation of Aggregatibacter actinomycetemcomitans ATCC 4371853. Similar results were reported by Loozen et al. (2014) in their study with a six-member bacterial community54. The six species present within the community were all oral bacteria and included four Gram-negative and two Gram-positive species. Along with the Gram-positive bacterial strains, two of the Gram-negative strains, i.e., Porphyromonas gingivalis and Prevotella intermedia, were not predated upon by B. bacteriovorus HD100. However, A. actinomycetemcomitans and Fusobacterium nucleatum are both prey and the presence of the four other strains did not hinder their predation.
Clearly some non-prey bacteria can act as decoys and slow down predation initially49, while others have no impact53,54. Here, we found the presence of S. aureus did not negatively impact predation. Similar results were seen with S. epidermidis. One clear difference between S. aureus and S. epidermidis, though, was the viabilities after a dual treatment with K. pneumoniae. With S. epidermidis, the viabilities were comparable with those seen after single treatments in Fig. 1. With S. aureus, however, the viabilities of both pathogens were reduced further by the dual treatment (Fig. 3).
The activities of violacein and B. bacteriovorus HD100 were also evaluated using a four-member pathogen culture, with positive results. As with the two-strain cultures, the presence of the four different strains did not significantly deter the activities of either antimicrobial when used alone (Table 3) nor when used together (Fig. 5). In the latter case, the number of viable pathogens was reduced by more than 3-log and stands as a clear demonstration the potential these two antimicrobials have when used together.
Not only are violacein and B. bacteriovorus HD100 effective against their respective classes of bacteria, they are also both active against multidrug resistant pathogens. For violacein, the minimum inhibitory concentrations for a wild-type S. aureus and four other strains, including a clinical isolate that is resistant to seven different antibiotics, were identical18. Along the same lines, the antibiotic resistant nature of the prey had no obvious impact on the ability of B. bacteriovorus to attack28, while predation reduces the presence of the antibiotic resistance marker by degrading the prey DNA55. Consequently, violacein and B. bacteriovorus are both active against multidrug resistant pathogens. This was illustrated here in experiments with a mixed culture containing multidrug resistant strains of S. aureus and A. baumannii. For comparison, we also exposed this culture in parallel to high doses of chloramphenicol, gentamicin or both of these antibiotics. With chloramphenicol and/or gentamicin, the culture viabilities were not significantly impacted, even though the concentration of each antibiotic was 10-fold higher than commonly employed in the lab. In contrast, a dual treatment with violacein and B. bacteriovorus HD100 led to a 2- to 3-log loss, a result that is indistinguishable with that in Fig. 3 with the wild-type S. aureus.
In conclusion, this study evaluated the activities of violacein and B. bacteriovorus HD100 when used together against polymicrobial cultures. Violacein, being active primarily against Gram-positive strains such as S. aureus, and B. bacteriovorus HD100, which attacks only Gram-negative bacteria, were found to be compatible and not diminish the activity of the other. This was demonstrated in cultures containing two and four different bacterial species, including both Gram-positive and Gram-negative strains. When used alone, the overall viabilities decreased marginally (less than 1-log reduction) but, when violacein and B. bacteriovorus HD100 were used together the number of viable pathogens was reduced by 3-log or greater, even when multidrug resistant pathogens were present.
Acknowledgements
We would like to thank the UNIST Central Research Facilities (ucrf.unist.ac.kr), specifically Jin-Hoe Hur at the UNIST Olympus Bioimaging Center, for use of the microscopes and imaging software. Also, we would like to thank Edouard Jurkevitch at the Hebrew University of Jerusalem for providing B. bacteriovorus HD100. Funding for this research was sponsored by the National Research Foundation of Korea within the General Research Program (Grant No. 2016R1D1A1A09919912) and under the Space Core Technology Development Project (Grant No. 2017M1A3A3A02016642). We appreciate the support.
Author Contributions
H.I., S.Y.C. and R.J.M. conceived the idea; H.I., S.Y.C. and S.S. performed the experiments; H.I., S.Y.C. and R.J.M. analyzed the data and wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Hansol Im and Seong Yeol Cho contributed equally to this work
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dalton T, et al. An In Vivo Polymicrobial Biofilm Wound Infection Model to Study Interspecies Interactions. Plos One. 2011;6:e27317. doi: 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME. Polymicrobial Interactions: Impact on Pathogenesis and Human Disease. Clin Microbiol Rev. 2012;25:193. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weimer KE, et al. Divergent mechanisms for passive pneumococcal resistance to beta-lactam antibiotics in the presence of Haemophilus influenzae. J Infect Dis. 2011;203:549–555. doi: 10.1093/infdis/jiq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vega NM, Gore J. Collective antibiotic resistance: mechanisms and implications. Curr Opin Microbiol. 2014;21:28–34. doi: 10.1016/j.mib.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brook I. The role of beta-lactamase-producing-bacteria in mixed infections. BMC Infect Dis. 2009;9:202. doi: 10.1186/1471-2334-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yaguchi T, et al. Aqueous two-phase system-derived biofilms for bacterial interaction studies. Biomacromolecules. 2012;13:2655–2661. doi: 10.1021/bm300500y. [DOI] [PubMed] [Google Scholar]
- 7.Tan C, et al. The inoculum effect and band-pass bacterial response to periodic antibiotic treatment. Mol Syst Biol. 2012;8:617. doi: 10.1038/msb.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorbach SL. Piperacillin/tazobactam in the treatment of polymicrobial infections. Intensive Care Med. 1994;20(Suppl 3):S27–34. doi: 10.1007/BF01745248. [DOI] [PubMed] [Google Scholar]
- 9.Rogers GB, et al. Revealing the dynamics of polymicrobial infections: implications for antibiotic therapy. Trends Microbiol. 2010;18:357–364. doi: 10.1016/j.tim.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akers, K. S. et al. Biofilms and persistent wound infections in United States military trauma patients: a case-control analysis. Bmc Infect Dis14, 10.1186/1471-2334-14-190 (2014). [DOI] [PMC free article] [PubMed]
- 11.Denyer SP, Maillard JY. Cellular impermeability and uptake of biocides and antibiotics in Gram-negative bacteria. J Appl Microbiol. 2002;92(Suppl):35S–45S. doi: 10.1046/j.1365-2672.92.5s1.19.x. [DOI] [PubMed] [Google Scholar]
- 12.Lambert PA. Cellular impermeability and uptake of biocides and antibiotics in Gram-positive bacteria and mycobacteria. J Appl Microbiol. 2002;92(Suppl):46S–54S. doi: 10.1046/j.1365-2672.92.5s1.7.x. [DOI] [PubMed] [Google Scholar]
- 13.Choi, S. Y., Yoon, K. H., Lee, J. I. & Mitchell, R. J. Violacein: Properties and Production of a Versatile Bacterial Pigment. Biomed Res Int, 10.1155/2015/465056 (2015). [DOI] [PMC free article] [PubMed]
- 14.Moss MO, Ryall C. Distribution of chromobacteria in a lowland river. Microb Ecol. 1981;7:139–149. doi: 10.1007/BF02032496. [DOI] [PubMed] [Google Scholar]
- 15.Lichstein HC, Vandesand VF. Violacein, an Antibiotic Pigment Produced by Chromobacterium violaceum. J Infect Dis. 1945;76:47–51. doi: 10.1093/infdis/76.1.47. [DOI] [Google Scholar]
- 16.Jude, B. A., Tanner, J., Koko, T. & McLaughlin, E. C. Analysis, characterization, and synthesis of violacein from Janthinobacterium isolate extracts. Abstr Pap Am Chem S244 (2012).
- 17.Hakvag S, et al. Violacein-producing Collimonas sp. from the sea surface microlayer of costal waters in Trondelag, Norway. Mar Drugs. 2009;7:576–588. doi: 10.3390/md7040576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi, S. Y., Kim, S., Lyuck, S., Kim, S. B. & Mitchell, R. J. High-level production of violacein by the newly isolated Duganella violaceinigra str. NI28 and its impact on Staphylococcus aureus. Sci Rep-Uk5, 10.1038/srep15598 (2015). [DOI] [PMC free article] [PubMed]
- 19.Nakamura Y, Sawada T, Morita Y, Tamiya E. Isolation of a psychrotrophic bacterium from the organic residue of a water tank keeping rainbow trout and antibacterial effect of violet pigment produced from the strain. Biochem Eng J. 2002;12:79–86. doi: 10.1016/S1369-703X(02)00079-7. [DOI] [Google Scholar]
- 20.Nakamura Y, Asada C, Sawada T. Production of antibacterial violet pigment by psychrotropic bacterium RT102 strain. Biotechnol Bioproc E. 2003;8:37–40. doi: 10.1007/BF02932896. [DOI] [Google Scholar]
- 21.Subramaniam S, Ravi V, Sivasubramanian A. Synergistic antimicrobial profiling of violacein with commercial antibiotics against pathogenic micro-organisms. Pharm Biol. 2014;52:86–90. doi: 10.3109/13880209.2013.815634. [DOI] [PubMed] [Google Scholar]
- 22.Aruldass CA, Masalamany SR, Venil CK, Ahmad WA. Antibacterial mode of action of violacein from Chromobacterium violaceum UTM5 against Staphylococcus aureus and methicillin-resistant Staphylococcus aureus (MRSA) Environ Sci Pollut Res Int. 2017 doi: 10.1007/s11356-017-8855-2. [DOI] [PubMed] [Google Scholar]
- 23.Cazoto LL, Martins D, Ribeiro MG, Duran N, Nakazato G. Antibacterial activity of violacein against Staphylococcus aureus isolated from bovine mastitis. J Antibiot (Tokyo) 2011;64:395–397. doi: 10.1038/ja.2011.13. [DOI] [PubMed] [Google Scholar]
- 24.Dwidar M, Monnappa AK, Mitchell RJ. The dual probiotic and antibiotic nature of Bdellovibrio bacteriovorus. BMB Rep. 2012;45:71–78. doi: 10.5483/BMBRep.2012.45.2.71. [DOI] [PubMed] [Google Scholar]
- 25.Shatzkes K, Connell ND, Kadouri DE. Predatory bacteria: a new therapeutic approach for a post-antibiotic era. Future Microbiol. 2017;12:469–472. doi: 10.2217/fmb-2017-0021. [DOI] [PubMed] [Google Scholar]
- 26.Tyson J, Sockett RE. Predatory Bacteria: Moving from Curiosity Towards Curative. Trends Microbiol. 2017;25:90–91. doi: 10.1016/j.tim.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 27.Dashiff A, Kadouri DE. Predation of oral pathogens by Bdellovibrio bacteriovorus 109J. Mol Oral Microbiol. 2011;26:19–34. doi: 10.1111/j.2041-1014.2010.00592.x. [DOI] [PubMed] [Google Scholar]
- 28.Kadouri DE, To K, Shanks RMQ, Doi Y. Predatory Bacteria: A Potential Ally against Multidrug-Resistant Gram-Negative Pathogens. Plos One. 2013;8:e63397. doi: 10.1371/journal.pone.0063397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monnappa, A. K., Dwidar, M., Seo, J. K., Hur, J. H. & Mitchell, R. J. Bdellovibrio bacteriovorus Inhibits Staphylococcus aureus Biofilm Formation and Invasion into Human Epithelial Cells. Sci Rep-Uk4, 10.1038/srep03811 (2014). [DOI] [PMC free article] [PubMed]
- 30.Dashiff A, Junka RA, Libera M, Kadouri DE. Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J Appl Microbiol. 2011;110:431–444. doi: 10.1111/j.1365-2672.2010.04900.x. [DOI] [PubMed] [Google Scholar]
- 31.Stolp H, Starr MP. Bdellovibrio bacteriovorus Gen. Et Sp. N., a Predatory, Ectoparasitic, and Bacteriolytic Microorganism. Antonie Van Leeuwenhoek. 1963;29:217–248. doi: 10.1007/BF02046064. [DOI] [PubMed] [Google Scholar]
- 32.Chu WH, Zhu W. Isolation of Bdellovibrio as Biological Therapeutic Agents Used For the Treatment of Aeromonas hydrophila Infection in Fish. Zoonoses Public Hlth. 2010;57:258–264. doi: 10.1111/j.1863-2378.2008.01224.x. [DOI] [PubMed] [Google Scholar]
- 33.Im H, Son S, Mitchell RJ, Ghim CM. Serum albumin and osmolality inhibit Bdellovibrio bacteriovorus predation in human serum. Sci Rep. 2017;7:5896. doi: 10.1038/s41598-017-06272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dwidar, M., Im, H., Seo, J. K. & Mitchell, R. J. Attack-Phase Bdellovibrio bacteriovorus Responses to Extracellular Nutrients Are Analogous to Those Seen During Late Intraperiplasmic Growth. Microb Ecol, 10.1007/s00248-017-1003-1 (2017). [DOI] [PubMed]
- 35.Rodrigues AL, et al. Systems metabolic engineering of Escherichia coli for production of the antitumor drugs violacein and deoxyviolacein. Metab Eng. 2013;20:29–41. doi: 10.1016/j.ymben.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Blondeau JM, Vaughan D. A review of antimicrobial resistance in Canada. Can J Microbiol. 2000;46:867–877. doi: 10.1139/w00-076. [DOI] [PubMed] [Google Scholar]
- 37.Choi, S. Y., Im, H. & Mitchell, R. J. Violacein and bacterial predation: promising alternatives for priority multidrug resistant human pathogens. Future Microbiol, 10.2217/fmb-2017-0090 (2017). [DOI] [PubMed]
- 38.Chanyi RM, Koval SF, Brooke JS. Stenotrophomonas maltophilia biofilm reduction by Bdellovibrio exovorus. Env Microbiol Rep. 2016;8:343–351. doi: 10.1111/1758-2229.12384. [DOI] [PubMed] [Google Scholar]
- 39.Tamma PD, Cosgrove SE, Maragakis LL. Combination therapy for treatment of infections with gram-negative bacteria. Clin Microbiol Rev. 2012;25:450–470. doi: 10.1128/CMR.05041-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bromberg N, et al. Growth inhibition and pro-apoptotic activity of violacein in Ehrlich ascites tumor. Chem Biol Interact. 2010;186:43–52. doi: 10.1016/j.cbi.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 41.Romanowski EG, et al. Predatory bacteria are nontoxic to the rabbit ocular surface. Sci Rep. 2016;6:30987. doi: 10.1038/srep30987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atterbury RJ, et al. Effects of orally administered Bdellovibrio bacteriovorus on the well-being and Salmonella colonization of young chicks. Appl Environ Microbiol. 2011;77:5794–5803. doi: 10.1128/AEM.00426-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willis AR, et al. Injections of Predatory Bacteria Work Alongside Host Immune Cells to Treat Shigella Infection in Zebrafish Larvae. Curr Biol. 2016;26:3343–3351. doi: 10.1016/j.cub.2016.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta S, Tang C, Tran M, Kadouri DE. Effect of Predatory Bacteria on Human Cell Lines. Plos One. 2016;11:e0161242. doi: 10.1371/journal.pone.0161242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monnappa AK, Bari W, Choi SY, Mitchell RJ. Investigating the Responses of Human Epithelial Cells to Predatory Bacteria. Sci Rep. 2016;6:33485. doi: 10.1038/srep33485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dwidar M, Leung BM, Yaguchi T, Takayama S, Mitchell RJ. Patterning bacterial communities on epithelial cells. PLoS One. 2013;8:e67165. doi: 10.1371/journal.pone.0067165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cushnie TPT, Hamilton VES, Chapman DG, Taylor PW, Lamb AJ. Aggregation of Staphylococcus aureus following treatment with the antibacterial flavonol galangin. J Appl Microbiol. 2007;103:1562–1567. doi: 10.1111/j.1365-2672.2007.03393.x. [DOI] [PubMed] [Google Scholar]
- 48.Iebba, V. et al. Bdellovibrio bacteriovorus directly attacks Pseudomonas aeruginosa and Staphylococcus aureus Cystic fibrosis isolates. Front Microbiol5, 10.3389/fmicb.2014.00280 (2014). [DOI] [PMC free article] [PubMed]
- 49.Hobley L, King JR, Sockett RE. Bdellovibrio predation in the presence of decoys: Three-way bacterial interactions revealed by mathematical and experimental analyses. Appl Environ Microb. 2006;72:6757–6765. doi: 10.1128/AEM.00844-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Straley SC, Conti SF. Chemotaxis by Bdellovibrio bacteriovorus toward prey. J Bacteriol. 1977;132:628–640. doi: 10.1128/jb.132.2.628-640.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drutz DJ. Response of Neisseria gonorrhoeae to Bdellovibrio species. Infect Immun. 1976;13:247–251. doi: 10.1128/iai.13.1.247-251.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilkinson MH. Predation in the presence of decoys: an inhibitory factor on pathogen control by bacteriophages or bdellovibrios in dense and diverse ecosystems. J Theor Biol. 2001;208:27–36. doi: 10.1006/jtbi.2000.2197. [DOI] [PubMed] [Google Scholar]
- 53.Van Essche M, et al. Killing of anaerobic pathogens by predatory bacteria. Mol Oral Microbiol. 2011;26:52–61. doi: 10.1111/j.2041-1014.2010.00595.x. [DOI] [PubMed] [Google Scholar]
- 54.Loozen G, et al. Effect of Bdellovibrio bacteriovorus HD100 on multispecies oral communities. Anaerobe. 2015;35:45–53. doi: 10.1016/j.anaerobe.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 55.Monnappa AK, Dwidar M, Mitchell RJ. Application of bacterial predation to mitigate recombinant bacterial populations and their DNA. Soil Biol Biochem. 2013;57:427–435. doi: 10.1016/j.soilbio.2012.09.010. [DOI] [Google Scholar]