Abstract
The interaction of a small library of cyclic RGD (Arg-Gly-Asp) peptidomimetics with αVβ6 integrin has been investigated by means of competitive solid phase binding assays to the isolated receptor and docking calculations in the crystal structure of the αVβ6 binding site. To this aim, a rigid receptor-flexible ligand docking protocol has been set up and then applied to predict the binding mode of the cyclic RGD peptidomimetics to αVβ6 integrin. Although the RGD interaction with αVβ6 recapitulates the RGD binding mode observed in αVβ3, differences between the integrin binding pockets can strongly affect the ligand binding ability. In general, the peptidomimetics exhibited IC50 values for integrin αVβ6 (i.e., the concentration of compound required for 50% inhibition of biotinylated fibronectin binding to isolated αVβ6 integrin) in the nanomolar range (77–345 nM), about 10–100 times higher than those for the related αVβ3 receptor, with a single notable ligand displaying a low nanomolar IC50 value (2.3 nM). Insights from the properties of the binding pocket combined with the analysis of the docking poses provided a rationale for ligand recognition and selectivity.
Keywords: RGD peptidomimetics, integrins, molecular docking, binding assays
1. Introduction
Integrins are αβ-heterodimeric transmembrane proteins that are involved in cell adhesion and signaling [1,2]. Because of their central role in a variety of physiological cell functions, as well as in the pathobiology of many diseases, integrins continue to attract interest for the development of therapeutic agents [3,4,5,6,7]. For instance, in cancer, pharmacological research is focused on a group of integrins that play key roles in tumor angiogenesis, progression, and metastasis, and share the property to recognize ligands containing the RGD (Arg-Gly-Asp) sequence, with a specificity determined by the features of the binding pocket and the context of the ligand RGD motif (i.e., flanking residues, conformation) [8,9,10].
Driven by the functional roles and the upregulated expression on various tumor cells of several subtypes from the RGD-binding subfamily, such as α5β1 along with αVβ3 and other αV integrins, extensive chemical research has been carried out to develop RGD-based peptidic and peptidomimetic integrin ligands as inhibitors of integrin functions and as targeting devices for the selective delivery of drugs or imaging probes to tumors [9,10,11,12,13,14,15,16].
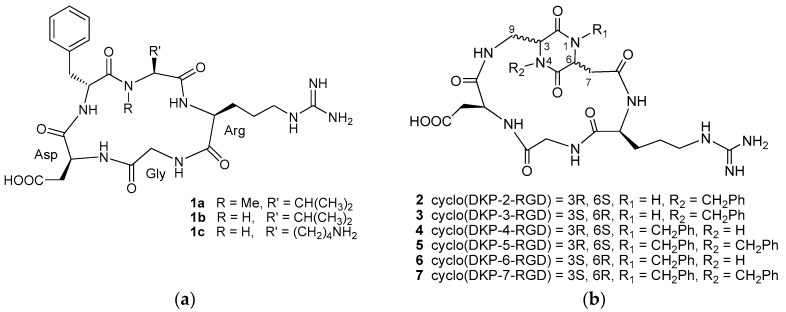
Among them, the best known RGD peptide is the cyclic pentapeptide cyclo[RGDf(N-Me)V] 1a (Cilengitide, Figure 1a) [17], the first integrin antagonist to be tested in clinical trials [18], that is currently undergoing phase II studies for the treatment of several cancer types after its failure in a phase III trial for the treatment of patients with newly diagnosed glioblastoma [19,20]. Other well-known RGD peptides are cyclo[RGDfV] 1b, the parent peptide of Cilengitide, and cyclo[RGDfK] 1c, that has been extensively used as targeting motif for targeted cancer diagnosis and therapy studies (Figure 1a) [10,13,14,15,16,21].
Figure 1.
(a) Cyclic RGD (Arg-Gly-Asp) pentapeptides 1a–c; (b) Cyclic RGD peptidomimetics 2–7 containing DKP scaffolds.
With the aim of developing new small molecule integrin antagonists with improved properties, we have recently synthesized a small library of cyclic RGD peptidomimetic integrin ligands, containing bifunctional diketopiperazine (DKP) scaffolds that differ in the configuration at the two DKP stereocenters and in the substitution at the DKP nitrogen atoms, and that can be viewed as conformationally constrained dipeptide mimics formed by two β-amino acids (Figure 1b) [22,23,24].
In particular, the cyclic RGD peptidomimetics 2–7 derived from trans-DKP scaffolds (DKP2-DKP7) were shown to bind αVβ3, αVβ5, and α5β1 integrins with a preferential affinity towards αVβ3, inhibiting the binding of biotinylated vitronectin to the purified αVβ3 integrin at low- or sub-nanomolar IC50 values [24,25]. The interaction of the cyclic DKP-RGD peptidomimetics with αVβ3 and α5β1 integrins has been investigated by means of integrated spectroscopic and computational studies, gaining insights into the molecular basis of their activity [24,25,26]. In particular, the preferred ligand conformations, displaying an extended arrangement of the RGD motif with a distance of about 9 Å between the Cβ atoms of Asp and Arg, are highly preorganized for the interaction with integrins αVβ3 and α5β1, as demonstrated by the docking studies in the crystal structures and NMR experiments with αVβ3-rich bladder cancer cells and α5β1-rich breast cancer cells [24,25,26].
Recently, ligand 3 (Figure 1b) was reported to display inhibitory effects on the FAK/Akt integrin-activated transduction signaling pathway and on integrin-mediated cell infiltration processes in human glioblastoma cells, thus qualifying as a true integrin antagonist [27]. It was also shown to significantly inhibit the cell adhesion of different cancer cells, and angiogenesis induced by pro-angiogenic growth factors in human endothelial cells [28]. Moreover, after suitable functionalization, it was exploited as a targeting agent for the preparation of conjugates designed to release cytotoxic drugs selectively within cancer cells expressing αVβ3 integrin [29,30,31].
In this context, information on the ability of small RGD molecules to interact with closely related integrin subtypes involved in cancer, is crucial to fully understand the biological activity profiles and to develop suitable integrin ligands for the modulation of integrin functions or the targeted delivery of chemotherapy [9,10,13,14,15,16,21].
Prompted by its pathological relevance in cancer and by the availability of X-ray structural information [32], we became interested in the integrin αVβ6, one of five αV integrins and the unique β6 integrin from the RGD-binding subfamily. αVβ6 is expressed on epithelial cells, especially during development, after injury or inflammation, or on many carcinomas. The ligand binding site of αVβ6 is in the N-terminal head region formed by the interaction of the αV β-propeller domain with the β6 βI domain. αVβ6 preferentially binds to the latency-associated peptide (LAP) of the transforming growth factor-β (TGF-β), but can also recognize the matrix proteins fibronectin and tenascin. In this regard, αVβ6 interacts with the LAP/TGF-β complex by binding with the RGD motif present in the LAP peptide, thus breaking the latency complex and releasing the active form of TGF-β which, in turn, binds to and activates its receptors on the cell surface [32,33]. In particular, similarly to integrin αVβ8, αVβ6 is specialized to activate TGF-β1 and TGF-β3 from large latent complexes. Therefore, high αVβ6 expression in carcinomas may contribute to progression through its effects on TGF-β activity.
In the framework of a study investigating the determinants of αVβ6 high specificity for the RGD motif present in the prodomain of TGF-β1 and TGF-β3, the crystal structures of the αVβ6 headpiece with or without a pro-TGF-β3 undecapeptide have been recently solved [32]. The ligand-bound structure revealed that αVβ6 recognizes not only RGD but also the adjacent C-terminal LGRLK motif that folds into an amphipathic α-helix fitting into a hydrophobic pocket in the β6 subunit.
Herein, we report on the interaction of our cyclic DKP-RGD peptidomimetics with αVβ6 integrin by means of competitive solid phase binding assays to the isolated receptor and docking calculations in the crystal structure of the αVβ6 binding site. Starting from the structure of ligand-bound αVβ6, a rigid receptor-flexible ligand docking protocol has been set up and then applied to predict the binding mode of the cyclic RGD peptidomimetics to αVβ6 integrin. The analysis of the properties of the receptor pocket, combined with the examination of the docking poses allowed to rationalize the experimental binding affinities for the αVβ6 integrin, which turned out to be about 10–100 times lower than those for the related αVβ3 receptor. On the basis of docking calculations, the best cyclic RGD peptidomimetic was also identified, displaying a low nanomolar IC50 value.
2. Results
2.1. Integrin Receptor Competitive Binding Assays
The cyclic peptidic (1a–c) and peptidomimetic (2–7) RGD ligands were examined in vitro for their ability to compete with biotinylated fibronectin for binding to the isolated αVβ6 integrin (Table 1). The assay was performed according to previously reported procedures [24,25] with slight modifications. In particular, a concentration of 1 μg/mL of integrin receptor was required for obtaining an efficient coating of the plates and a good reproducibility of the data [21,34]. Various concentrations (10−11–10−4 M) of the RGD ligands in the presence of biotinylated fibronectin (1 μg/mL) were then added to the plates and finally bound fibronectin was revealed by using a streptavidin-biotinylated peroxidase complex (see Section 4 for a detailed description).
Table 1.
Inhibition of biotinylated fibronectin binding to αVβ6 integrin compared with inhibition of biotinylated vitronectin binding to αVβ3.
| Compound | αVβ6 IC50 [nM] 1 | αVβ3 IC50 [nM] 2 | IC50 (αVβ6)/IC50 (αVβ3) |
|---|---|---|---|
|
1a c[RGDf(N-Me)V] |
82.8 ± 4.9 | 0.71 ± 0.06 | 117 |
|
1b c[RGDfV] |
104.7 ± 18.9 | 3.2 ± 1.3 | 33 |
|
1c c[RGDfK] |
52.0 ± 23.8 | 1.4 ± 0.2 | 37 |
| 2 | 345.0 ± 1.0 | 3.2 ± 2.7 | 108 |
| 3 | 95.6 ± 24.6 | 4.5 ± 1.1 | 21 |
| 4 | 95.3 ± 4.9 | 7.6 ± 4.3 | 13 |
| 5 | 173.5 ± 52.5 | 12.6 ± 5.0 | 14 |
| 6 | 76.6 ± 4.2 | 2.1 ± 0.6 | 37 |
| 7 | 2.3 ± 0.8 | 0.2 ± 0.09 | 12 |
|
8 c[DKP-3-RAD] |
4095 ± 1425 | 1500 ± 540 | 3 |
1 IC50 values were calculated as the concentration of compound required for 50% inhibition of biotinylated fibronectin binding as estimated by GraphPad Prism software; all values are the arithmetic mean ± SD of triplicate determinations. 2 Calculated as the concentration of compound required for 50% inhibition of biotinylated vitronectin binding [24].
To validate the binding assay protocol, the well-known cyclopeptidic integrin ligands 1a–c were first assayed. Both compounds 1b (c[RGDfV]) and 1c (c[RGDfK]) showed binding affinities for integrin αVβ6 (expressed as the ligand concentration required for 50% inhibition of endogenous ligand binding) comparable with data recently reported in literature [21] (Table 1). A nanomolar IC50 value similar to that of the other cyclopeptides was observed also for 1a (c[RGDf(N-Me)V], IC50 = 82.8 ± 4.9 nM), in contrast with the micromolar value recently reported [21]. It must be noted that the two IC50 values are obtained by competitive solid phase binding assays by using two different procedures and two different αVβ6 natural ligands: immobilized integrin and soluble fibronectin in the present assay, immobilized latency associated peptide (LAP), and soluble integrin in the other [21]. Noteworthy, the present result is compatible with the outcomes of docking calculations (vide infra).
All of the cyclic DKP-RGD ligands 2–7 showed binding affinities for integrin αVβ6 lower than those for αVβ3, displaying a selectivity ratio (IC50 αVβ6/IC50 αVβ3) ranging from about 10 (4, 5, 7) to nearly 100 (2) (Table 1). The trend is confirmed by the reference cyclopeptides, exhibiting IC50 values for αVβ6 about 35 (1b, 1c)–100 (1a) times higher than those for αVβ3. Among the RGD peptidomimetics, compound 7 proved to be the best αVβ6 ligand, inhibiting the binding of biotinylated fibronectin to αVβ6 at a low nanomolar IC50 value. Interestingly, this ligand was also the most potent αVβ3 ligand of the series, displaying a subnanomolar IC50 value.
As a negative control in the determination of binding activities, a cyclic peptidomimetic containing the Arg-Ala-Asp (RAD) sequence was prepared and tested (compound 8, c[DKP-3-RAD], see Supplementary Materials, Scheme S1 and Figures S1–S6), displaying micromolar IC50 values with both αVβ3 and αVβ6 integrins (Table 1).
2.2. Docking Model of αVβ6 Integrin and X-ray Structure Analysis
The computational model for the interaction of RGD ligands with the αVβ6 integrin was developed by means of docking calculations using Glide V5.7 [35] (see Section 4 for a detailed description), starting from the X-ray structure of the extracellular segment of integrin αVβ6 in complex with the RGD-containing undecapeptide of the TGF-β3 prodomain (PDB code: 4UM9) [32].
In the crystal structure, the headpiece of αVβ6 adopts a closed conformation [32,36] similar to that adopted by αVβ3 in the X-ray complex with Cilengitide [37]. In both of the crystal complexes the RGD sequence shows an extended conformation characterized by a Cβ(Arg)-Cβ(Asp) distance of 8.9 (αVβ3)–9.4 (αVβ6) Å, and a separation between the charged Arg and Asp side chains of 13.7 (αVβ3)–14.2 Å (αVβ6) (measured between the carbon atoms of the guanidinium and carboxylate groups). As observed in other X-ray structures of integrins in complex with RGD ligands [37,38,39,40], the RGD sequence binds at the interface of the α and β subunits with the carboxylic and guanidine groups acting as an electrostatic clamp, respectively, on a bivalent cation of the β subunit (MIDAS, metal ion-dependent adhesion site) and on specific acid residues of the α subunit. The oxygen atom of RGD aspartate side chain not engaged by MIDAS Mg2+ ion forms hydrogen bonds with backbone NH groups of β6-Asn218 and β6-Ala126. The arginine of RGD makes a bidentate side-on interaction through the guanidinium group to the side chain of Asp218 in the αV subunit, as in binding to αVβ3, but does not interact with αV-Asp150 side chain as for Cilengitide in αVβ3. Other stabilizing hydrogen bond interactions occur between the backbone Gly carbonyl and Asp-NH ligand moieties and β6-Thr221 side chain and β6-Ile219 carbonyl group, respectively. It is worth noting that the salt bridge between αV-Asp219 and β3-Lys253 at the RGD binding site interface cannot be formed in αVβ6 due to β6-Asp256 mutation, making the αV-Asp219 residue more accessible to the interaction with the ligands.
Large differences in the ligand-binding region between αVβ6 and αVβ3 are also represented by point mutations in the β2-β3 loop and in two neighboring interacting loops [32]. The three residues forming π-cation interactions in β3, Tyr166, Arg214, and Arg216, are replaced in β6 by Lys170, Ala217, and Ile219. Furthermore, β3-Tyr122 residue, which is engaged into a π-interaction with the Phe residue of Cilengitide, is mutated into β6-Ala126. In addition to RGD interactions, the immediately following LGRLK sequence of the TGF-β3 peptide forms an amphipatic α-helix that extensively interfaces with the β6 subunit exploiting a hydrophobic pocket close to the RGD binding site. Most of the contacts are formed with Ala126, Ser127, Cys180, Ile183, Tyr185, Ala217 side chains, and Pro179 backbone atoms.
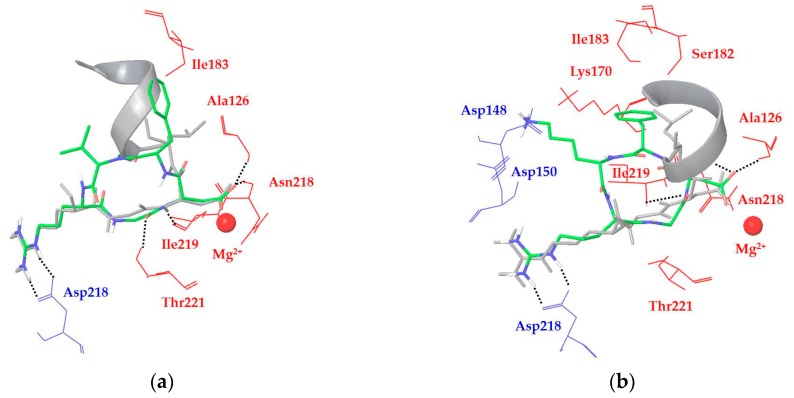
In all of the docking calculations, the X-ray binding mode of the RGD motif with the αVβ6 integrin was taken as a reference model for the analysis of the docking results in terms of ligand–protein interactions. For instance, ligands 1a and 1c are able to reproduce the experimentally determined binding mode of the RGD sequence (Figure 2), even if some hydrogen bond interactions are not optimal in all the calculated docking poses. More importantly, 1a and 1c cannot significantly improve the interaction with the αVβ6 binding site as gained by their D-Phe residue in the αVβ3 pocket (due to β3-Tyr122 mutation into β6-Ala126) or by the TGF-β3 α-helix in the β6-specific hydrophobic pocket. These considerations might explain the reduced binding affinities of these cyclic peptides for integrin αVβ6 when compared to αVβ3. In particular, the docking poses of ligand 1a (Cilengitide) show the D-Phe aromatic group in contact with β6-Ile183 and β6-Ala126, fitting only partially the α-helix region and the corresponding hydrophobic pocket (Figure 2a). Conversely, the docking poses of ligand 1c display electrostatic interactions between the ligand Lys side chain and the integrin αV-Asp150 and αV-Asp148 residues, forcing the ligand D-Phe aromatic moiety in proximity of β6-Lys170, β6-Ser182, and β6-Asn218 residues, far away from the α-helix binding site (Figure 2b).
Figure 2.
Docking best poses of (a) ligand 1a (green) and (b) ligand 1c (green) overlaid to the X-ray structure of the TGF-β3 undecapeptide (grey, α-helix represented as a ribbon) into integrin αVβ6 (from 4UM9.pdb). Only selected integrin residues involved in interactions with the ligand are shown and labeled in blue for αV and red for β6. Non-polar hydrogens are hidden for clarity, while intermolecular hydrogen bonds are shown as black dashed lines.
2.3. Docking of Cyclic DKP-RGD Peptidomimetics into αVβ6 Integrin
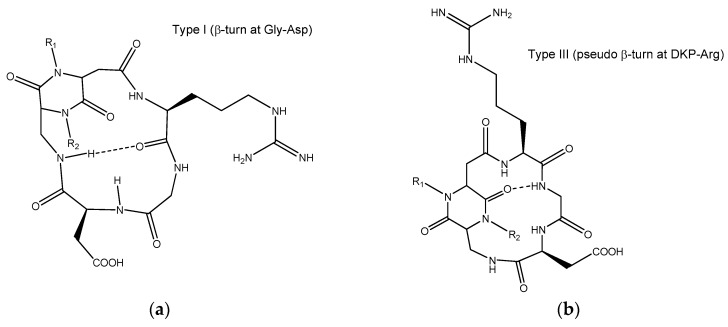
Docking studies were performed starting from the macrocycle conformations of the cyclic DKP-RGD peptidomimetics that have been previously reported [24] and are shown in Appendix A (Figure A1) and in the Supplementary Materials (Figures S7–S9). When compared to the X-ray RGD bound conformation, both type I (characterized by a distorted β-turn at Gly-Asp) and type III (characterized by a pseudo-β-turn at DKP-Arg) geometries preferentially adopted by these ligands, display a similar extended arrangement of the RGD sequence satisfying the pharmocophoric requirements for the binding to integrin αVβ6. Accordingly, docking results show that the top-ranked binding modes of all of the ligands maintain the key interactions observed for the RGD motif into the X-ray complex. The Asp and Arg side chains fit into the corresponding charged regions of the receptor, coordinating the MIDAS ion and forming the bidentate side-on salt bridge with αV-Asp218, respectively. The hydrogen bonds between the oxygen atom of Asp side chain and backbone NH groups of β6-Asn218 and β6-Ala126 are also present, as well as further stabilizing H-bonds involving ligand Gly and Asp backbone moieties and β6-Ile219 and β6-Thr221 residues, even if they are not optimal in all the calculated docking poses.
Since DKP-RGD compounds are highly preorganized for binding to αVβ6, the modulation of their experimental affinity can be explained by considering the different orientation of the aromatic rings of the DKP scaffold within the binding site. Indeed, docking results showed that, depending on DKP scaffold (i.e., on endocyclic nitrogen N-1/N-4 substitution and carbon C-3/C-6 stereochemistry) and macrocycle conformation, the aromatic moieties differently fit into the integrin pocket.
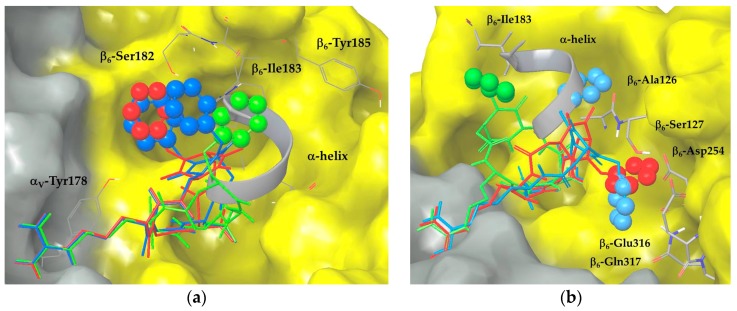
Docking calculations starting from type III geometry of ligand 2 place the N-4 benzyl group of the scaffold between α and β subunits interacting with the aromatic side chain of αV-Tyr178 and with β6-Ser182. This benzyl position could prevent optimal RGD interactions and perturb the whole ligand binding, thus explaining its high-nanomolar experimental binding affinity (IC50 = 345.0 ± 1.0 nM). A different binding mode of the aromatic moiety is observed in the docking poses of ligand 4 that displays an improved binding ability (IC50 = 95.3 ± 4.9 nM). The N-1 benzyl substitution and the type I macrocycle geometry generate docking poses with similar docking scores and RGD interactions with respect to ligand 2, but with a different orientation of the benzyl group that interacts with β6-Ile183, a ‘hot spot’ residue of the TGF-β3 α-helix pocket. By placing the aromatic ring in the α-helix region, ligand 4 seems to be more effective in displacing the natural ligand in the competitive binding assay. In line with these considerations, in the docking poses of the di-benzylated compound 5 (IC50 = 173.5 ± 52.5 nM), the aromatic moieties partially fit both of the benzyl binding regions identified by ligands 2 and 4. When compared to the mono-benzylated compounds, the addition of a second aromatic group improves the experimental binding affinity only with respect to ligand 2, while it has a perturbing effect with respect to ligand 4. In fact, the docking poses of ligand 5, adopting the type III geometry, show one benzyl group placed between αV-Tyr178 and β6-Ser182 (as in ligand 2), and the second one exposed to the solvent or in contact with β6-Ile183 (as in ligand 4). The absence of a stable interaction of the benzyl moieties with key residues of the hydrophobic β6 pocket is probably responsible for the intermediate affinity of 5 for the receptor. The superimposition of the best poses of the three ligands (featuring the same 3R, 6S scaffold configuration) is shown in Figure 3a.
Figure 3.
Docking best poses of (a) ligands 2 (red), 4 (green) and 5 (blue) and (b) ligands 3 (red), 6 (green) and 7 (blue) into integrin αVβ6 (αV surface in grey, β6 surface in yellow). The X-ray structure of the TGF-β3 α-helix portion is shown as a grey ribbon. Ligand aromatic rings are represented as space-filling spheres.
In the docking poses of ligand 3, featuring the type III macrocycle geometry and the N-4 benzyl substitution, the aromatic moiety is shifted towards β6-Glu316, β6-Gln317, and β6-Asp254 residues close to the ADMIDAS (adjacent to MIDAS) ion, forming a large number of favorable contacts with the protein. When compared to ligand 4, the benzyl group fits a different protein region that stabilizes the binding of the RGD motif, as suggested by the experimental receptor affinity (IC50 = 95.6 ± 24.6 nM). For ligand 6, the switch to type I macrocycle conformation and to N-1 benzyl substitution produces ligand poses with the aromatic ring in contact with β6-Ile183 of the α-helix pocket. Such a binding mode corresponds to the docking pose observed for ligand 4 and nicely agrees with the experimental IC50 value of 76.6 ± 4.2 nM, suggesting that the position of the aromatic ring close to α-helix hydrophobic region has a stabilizing effect on ligand binding. The docking poses of ligand 7, the most active compound of the library exhibiting a low nanomolar IC50 value, show some similarities with both ligands 3 and 6. The presence of two aromatic rings improves the experimental integrin affinity as compared to the mono-benzylated analogs, because both benzyl groups productively contribute to ligand stabilization. In particular, ligand 7, adopting the type I geometry, docks one benzyl group in the protein region explored by ligand 3, forming contacts with β6-Glu316 and β6-Gln317 residues, and the other one in the α-helix hydrophobic pocket, nicely overlapping the α-helix region and forming contacts with β6-Ile183, β6-Ala126, and β6-Ser127. Interestingly, ligand 7 displays the best docking score among all of the investigated compounds (see the Supplementary Materials, Table S1) and the highest number of docking poses maintaining all the crystallographic RGD interactions. The superimposition of the best poses of the three ligands featuring the same 3S, 6R scaffold configuration is shown in Figure 3b.
Finally, docking calculations on compound 8 (c[DKP-3-RAD], used as a negative control), starting from the type III geometry, fail in reproducing the X-ray key interactions of Asp and Arg residues. In the best pose of the stereoisomer containing the (S)-Ala amino acid the carboxylate group coordinates the ADMIDAS Ca2+ ion, while the guanidinium group form a π-cation interaction with αV-Tyr178 side chain. In the best pose of the stereoisomer containing the (R)-Ala residue, the coordination of the Asp side chain to MIDAS Mg2+ is kept (without hydrogen bonds to β6 residues), while the Arg side chain interacts with αV-Asp148 and αV-Asp150. Accordingly, the docking scores of the RAD peptidomimetics are about 1–3 kcal/mol worse than those calculated for the RGD ligands (see the Supplementary Materials, Table S1).
3. Discussion
In view of the roles played by integrin αVβ6 in cancer growth and progression [5,6,7,8,9,41], small-molecule integrin antagonists may be a valuable tool to modulate these processes. Investigating the interaction of cyclic RGD peptidomimetics with this integrin subtype represents the first essential step towards their exploitation as inhibitors of integrin functions and as receptor ligands for targeted therapy and imaging of tumors. The cyclic DKP-RGD peptidomimetics 2–7 have been designed to target the αVβ3 integrin, qualifying as excellent ligands (from low- to sub-nanomolar IC50 values in the inhibition of biotinylated vitronectin binding to αVβ3) of this RGD-recognizing heterodimer [24]. Prompted by the similarity of the ligand RGD binding observed in the crystal structures of Cilengitide to αVβ3 and TGF-β3 peptide to αVβ6 [32,37], we decided to investigate the interaction of the cyclic peptidomimetics with the αVβ6 integrin by means of competitive cell-free binding assays and docking studies. Indeed, the favored geometries of these ligands are characterized by an extended arrangement of the RGD sequence comparable to the X-ray αVβ6-bound RGD conformation and suitable to establish useful interactions with αVβ6 integrin.
Accordingly, docking calculations of the cyclic peptidomimetics in the αVβ6 crystal structure predicted RGD binding modes reproducing the key interactions found in the X-ray complex of TGF-β3 peptide to αVβ6. In particular, the electrostatic clamp of ligand Arg and Asp side chains with the corresponding charged regions in the αV and β6 receptor subunits is properly formed, in combination with a stabilizing network of hydrogen bonds whose specific features depend on the structural properties of each particular ligand (e.g., scaffold substitution and stereochemistry).
However, similar to what is observed for cyclic RGD peptides, the peptidomimetics exhibited binding affinities for the αVβ6 integrin (measured as the concentration of compound required for 50% inhibition of biotinylated fibronectin binding to isolated αVβ6 integrin) about 10–100 times lower than those for the related αVβ3 receptor (Table 1). Although the RGD binding mode found in αVβ6 integrin recapitulates the RGD interaction with αVβ3, differences between integrin binding pockets can affect the ligand recognition and binding ability. For instance, β3-Tyr122 residue, which is engaged into a π-interaction with the D-Phe residue of Cilengitide [37] or with a suitable aromatic moiety of the cyclic DKP-RGD peptidomimetics 2–7 [24], is mutated into β6-Ala126, hampering an important contribution to the complex stabilization.
More importantly, further sequence differences between the β subunits create a β6-specific hydrophobic pocket that was shown to play a key role in the elucidation of specificity determinants of integrin β subunits [32]. In particular, three βI-domain loops contribute to the main difference in the ligand-binding region between αVβ6 and αVβ3 [32]. The amphipathic α-helix of the TGF-β3 undecapeptide makes extensive contacts with these three loops, fully exploiting the hydrophobic pocket composed only of residues from the β6 subunit, and acting to stabilize the RGD interaction. Interestingly, in contrast to the αVβ6 complex, complexes of αVβ3, αIIbβ3, and α5β1 exhibit little interaction beyond that with the RGD motif [37,38,39,40].
As shown by docking results, the cyclic DKP-RGD peptidomimetics 2–7 can only partially take advantage of the structural peculiarity of αVβ6 integrin, fitting the hydrophobic pocket to an incomplete extent thanks to the structural features of specific ligands. In particular, the analysis of the docking poses suggests that the interactions of the DKP benzyl moieties with key residues of the hydrophobic β6 pocket as well as their overlap with the α-helix region of the TGF-β3 peptide, correlate well with the ability of the cyclic peptidomimetics to displace the natural ligand in competitive binding assays. Indeed, ligand 7, displaying the best fit to the TGF-β3 peptide and the most favorable interactions with αVβ6 integrin for both the RGD and not-RGD portions in the calculated binding modes, appears the best αVβ6 ligand of the library, inhibiting the binding of biotinylated fibronectin to αVβ6 at a low nanomolar IC50 value (2.3 ± 0.8 nM).
Recently, a cyclic RGD peptide endowed with sub-nanomolar binding affinity toward the αVβ6 integrin and a remarkable selectivity against other integrins has been reported as a result of a strategy based on the grafting of the epitope from the αVβ6 binding helix onto a cyclic β-sheet structure [42]. Although, in contrast to linear peptides [43], the DLXXL-motif was not essential for the αVβ6 activity of the cyclic peptides, three RGD-flanking hydrophobic residues were shown to significantly contribute to the interaction, fitting the wide hydrophobic pocket and projecting their side chains in the same direction as the key residues of the TGF-β3 helical motif [42].
In conclusion, experimental and computational tools for the evaluation of αVβ6 integrin ligands have been set up and then applied to investigate the interaction of cyclic RGD peptidomimetics with αVβ6 integrin. In particular, a docking protocol was defined and then exploited to predict the binding mode of the cyclopeptides to the αVβ6 integrin, generating poses that fairly reflect the results of the competitive binding assays to the isolated receptor. Insights from the features of the binding pocket combined with the analysis of the docking poses provided a rationale for ligand recognition and enabled to outline the molecular bases of ligand binding affinities. This understanding might in turn be exploited to develop αVβ6-selective ligands and improved targeting agents for biomedical applications.
4. Materials and Methods
4.1. Integrin Ligands
The cyclic peptidic and peptidomimetic RGD ligands used in this work were purchased or synthesized according to published procedures [17,24]. The synthesis and the characterization of the new cyclic peptidomimetic c[DKP-3-RAD] 8 are reported in the Supplementary Materials (Scheme S1, Figures S1–S6).
4.2. Solid-Phase Receptor Binding Assay
Recombinant human integrin αVβ6 receptor (R&D Systems, Minneapolis, MN, USA) was diluted to 1.0 μg/mL in coating buffer containing 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM MnCl2, 2 mM CaCl2, and 1 mM MgCl2. An aliquot of diluted receptor (100 μL/well) was added to 96-well microtiter plates (Nunc MaxiSorp, Thermo Fisher Scientific, Roskilde, Denmark) and incubated overnight at 4 °C. The plates were incubated with blocking solution (coating buffer plus 1% bovine serum albumin) for additional 2 h at room temperature to block nonspecific binding. After washing twice with blocking solution, plates were incubated shaking in the dark for 3 h at room temperature, with various concentrations (10−4–10−11 M) of test compounds in the presence of 1 μg/mL biotinylated fibronectin (Molecular Innovations, Novi, MI, USA). Biotinylation was performed using an EZ-Link Sulfo-NHS-Biotinylation kit (Pierce, Rockford, IL, USA). After washing three times, the plates were incubated shaking for 1 h, at room temperature, with streptavidin-biotinylated peroxidase complex (Amersham Biosciences, Uppsala, Sweden). After washing 3 times with blocking solution, plates were incubated with 100 μL/well of Substrate Reagent Solution (R&D Systems) for 30 min shaking in the dark, before stopping the reaction with the addition of 50 μL/well 2N H2SO4. Absorbance at 415 nm was read in a SynergyTM HT Multi-Detection Microplate Reader (BioTek Instruments, Inc., Winooski, VT, USA). Each data point represents the average of triplicate wells; data analysis was carried out by nonlinear regression analysis with GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA, USA). Each experiment was repeated in triplicate.
4.3. Computational Studies
Protein setup. The crystal structure of the extracellular domain of the integrin αVβ6 in complex with the HGRGDLGRLKK undecapeptide of the TGF-β3 prodomain (PDB code: 4UM9) [32] was used for docking studies. Docking was performed only on the globular head of the integrin, because the headgroup of integrin has been identified in the X-ray structure as the ligand-binding region. The protein was truncated to residue sequences 1–439 for chain α (chain C of crystal asymmetric unit) and 114–355 for chain β (chain D of crystal asymmetric unit). According to the X-ray structure, the bivalent cation at MIDAS has been modeled as Mg2+ ion, whereas all of the other metal cations were modeled as Ca2+ ions. All waters molecules were deleted except for the three water molecules coordinating the MIDAS cation and the single water molecule found around ADMIDAS ion. The structure was then prepared by using the Protein Preparation Wizard of the graphical user interface Maestro and the OPLSAA force field [44]. Hydrogen bonds were optimized according to the exhaustive sampling option and the entire complex was optimized by using a restrained minimization with convergence on heavy atoms to a RMSD (root-mean-square deviation) of 0.30 Å.
Ligand docking calculations. The automated docking calculations were performed by using Glide V5.7 in the standard precision (SP) mode [35]. The grids were generated for the RGD-integrin αVβ6 complex structure prepared as described in the protein setup section. The center of the grid-enclosing box was defined by the center of the bound ligand. For the grid generation step, the size of the inner cubic box for placing the ligand center was set to 12 Å, and a value of 26 Å was used for the outer cubic box. The outer box dimensions fit the entire active site. No further modifications were applied to the default settings. For the docking calculations, the GlideScore function was used to select 20 poses for each ligand after a post-minimization step. The flexible docking option was selected and the SP modality was used with amide bonds set to trans configurations. No Epik state penalty was added to the docking score and all of the ligands were considered in their zwitterionic form (and protonated Lys residue for 1c). To validate the docking protocol, a known αVβ6 ligand was selected, i.e., the cyclic pentapeptide c[RGDfK] 1c, showing an IC50 value to the isolated receptor of 52.0 ± 23.8 nM (see Table 1). In fact, due to the high conformational flexibility, the X-ray ligand (the undecapeptide of the TGF-β3 prodomain) is not suitable for standard docking calculations. For compound c[RGDfK] 1c, Glide succeeded in reproducing the experimentally determined binding mode of the RGD motif, as it corresponds to the best-scored pose (see Figure 2b).
Ligand conformations. The conformations of the ligands used in docking studies are described in the Appendix A. To avoid incomplete sampling of macrocycle conformations during docking analyses, the assessment of the preferred conformations of the cyclic systems has been performed as a separate step before docking [45].
Acknowledgments
We thank the University of Milan for a fellowship (to Simone Zanella) and Ministero dell’Università e della Ricerca (PRIN 2015 project 20157WW5EH) for financial support.
Supplementary Materials
The following are available online at http://www.mdpi.com/2072-6694/9/10/128/s1. Scheme S1: Synthesis of compound 8, Figures S1–S6: HPLC traces and NMR spectra of compound 8, Figures S7–S9: Preferred conformations identified for the cyclic [DKP-RGD] peptidomimetics (2D and 3D representations), Table S1: Glide docking score values of the best poses.
Appendix A
Docking studies were performed starting from the preferred macrocycle conformations of the cyclic DKP-RGD peptidomimetics previously determined [24,46]. Four different geometries (denoted as type I–IV) were identified in the free state conformational analysis of the cyclic RGD ligands containing the DKP scaffolds, by means of computational and spectroscopic NMR studies, as summarized in the Supplementary Materials. Depending on the configuration and substitution of the DKP scaffold, the cyclic DKP-RGD ligands showed different intramolecular H-bonding patterns as characterized by specific β- and γ-turns and diverse arrangements of the RGD sequence. In Figure A1, the type I and III patterns are shown as they correspond to the preferred macrocycle conformations adopted by the compounds under investigation. Docking calculations of ligand 6 were also run from type IV geometry, obtaining results worse than those provided by type I.
Figure A1.
(a) 2D representation of type I geometry; (b) 2D representation of type III geometry.
As the NMR-based solution structures of 1a (Cilengitide) exhibit conformations closely resembling the crystal structure bound to the head group of integrin αVβ3, docking calculations were performed from the X-ray αVβ3-bound geometry [47]. Interestingly, the type I conformation of the cyclic DKP-RGD peptidomimetics is very similar to this geometry.
Metropolis Monte Carlo/Stochastic Dynamics (MC/SD) simulations [48], using the implicit water GB/SA solvation model [49], were performed on ligand 1c within the framework of MacroModel V9.9 [50]. MC/SD simulations were performed at 300 K using the all-atoms AMBER* force field and a time step of 1 fs for 20 ns of simulation time. Side-chain dihedral angles were defined as internal coordinate degrees of freedom in the Monte Carlo part of the algorithm. A total of 5000 conformations were stored for analysis. To assess the convergence of the calculations, three independent MC/SD simulations were run starting from different macrocycle conformations. The analysis of the sampled structures showed that during all of the simulations compound 1c preferentially adopts an extended arrangement of the RGD sequence, characterized by a β-turn centered on Gly-Asp residues, comparable to the X-ray αVβ3–bound geometry of Cilengitide. Macrocycle conformations featuring a β-turn at D-Phe-Lys or a β-turn at Lys-Arg were also sampled during the MC/SD simulations. Three representative macrocycle conformations were then used for docking calculations and the best results were obtained starting from the preferred Gly-Asp β-turn conformation.
Author Contributions
Monica Civera, Daniela Arosio and Laura Belvisi conceived and designed the experiments; Monica Civera, Daniela Arosio, Francesca Bonato, Leonardo Manzoni, Luca Pignataro and Simone Zanella performed the experiments; Monica Civera, Daniela Arosio, Francesca Bonato, Cesare Gennari, Umberto Piarulli and Laura Belvisi analyzed and interpret the data; Monica Civera, Daniela Arosio and Laura Belvisi wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References and Notes
- 1.Takada Y., Ye X., Simon S. The integrins. Genome Biol. 2007;8:215.1–215.9. doi: 10.1186/gb-2007-8-5-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hynes R.O. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/S0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 3.Avraamides C.J., Garmy-Susini B., Varner J.A. Integrins in angiogenesis and lymphangiogenesis. Nat. Rev. Cancer. 2008;8:604–617. doi: 10.1038/nrc2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimaoka M., Springer T.A. Therapeutic antagonists and conformational regulation of integrin function. Nat. Rev. Drug Discov. 2003;2:703–716. doi: 10.1038/nrd1174. [DOI] [PubMed] [Google Scholar]
- 5.Ley K., Rivera-Nieves J., Sandborn W.J., Shattil S. Integrin-based therapeutics: Biological basis, clinical use and new drugs. Nat. Rev. Drug Discov. 2016;15:173–183. doi: 10.1038/nrd.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodman S.L., Picard M. Integrins as therapeutic targets. Trends Pharmacol. Sci. 2012;33:405–412. doi: 10.1016/j.tips.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Cox D., Brennan M., Moran N. Integrins as therapeutic targets: Lessons and opportunities. Nat. Rev. Drug Discov. 2010;9:804–820. doi: 10.1038/nrd3266. [DOI] [PubMed] [Google Scholar]
- 8.Desgrosellier J.S., Cheresh D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheldrake H.M., Patterson L.H. Strategies to inhibit tumor associated integrin receptors: Rationale for dual and multi-antagonists. J. Med. Chem. 2014;57:6301–6315. doi: 10.1021/jm5000547. [DOI] [PubMed] [Google Scholar]
- 10.Mas-Moruno C., Fraioli R., Rechenmacher F., Neubauer S., Kapp T.G., Kessler H. αVβ3- or α5β1-integrin-selective peptidomimetics for surface coating. Angew. Chem. Int. Ed. Engl. 2016;55:7048–7068. doi: 10.1002/anie.201509782. [DOI] [PubMed] [Google Scholar]
- 11.Paolillo M., Russo M.A., Serra M., Colombo L., Schinelli S. Small molecule integrin antagonists in cancer therapy. Mini-Rev. Med. Chem. 2009;9:1439–1446. doi: 10.2174/138955709789957404. [DOI] [PubMed] [Google Scholar]
- 12.Auzzas L., Zanardi F., Battistini L., Burreddu P., Carta P., Rassu G., Curti C., Casiraghi G. Targeting αVβ3 integrin: Design and applications of mono- and multifunctional RGD-based peptides and semipeptides. Curr. Med. Chem. 2010;17:1255–1299. doi: 10.2174/092986710790936301. [DOI] [PubMed] [Google Scholar]
- 13.Marelli U.K., Rechenmacher F., Sobahi T.R.A., Mas-Moruno C., Kessler H. Tumor targeting via integrin ligands. Front. Oncol. 2013;3:222. doi: 10.3389/fonc.2013.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arosio D., Casagrande C. Advancement in integrin facilitated drug delivery. Adv. Drug Deliv. Rev. 2016;97:111–143. doi: 10.1016/j.addr.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Arosio D., Manzoni L., Corno C., Perego P. Integrin-targeted Peptide- and peptidomimetic-drug conjugates for the treatment of tumors. Recent Pat. Anticancer Drug Discov. 2017;12:148–168. doi: 10.2174/1574892812666170203151930. [DOI] [PubMed] [Google Scholar]
- 16.Dal Corso A., Pignataro L., Belvisi L., Gennari C. αVβ3 Integrin-targeted peptide/peptidomimetic-drug conjugates: In-depth analysis of the linker technology. Curr. Top. Med. Chem. 2016;16:314–329. doi: 10.2174/1568026615666150701114343. [DOI] [PubMed] [Google Scholar]
- 17.Dechantsreiter M.A., Planker E., Mathä B., Lohof E., Hölzemann G., Jonczyk A., Goodman S.L., Kessler H. N-methylated cyclic RGD peptides as highly active and selective αVβ3 integrin antagonists. J. Med. Chem. 1999;42:3033–3040. doi: 10.1021/jm970832g. [DOI] [PubMed] [Google Scholar]
- 18.Mas-Moruno C., Rechenmacher F., Kessler H. Cilengitide: The first anti-angiogenic small molecule drug candidate. Design, synthesis and clinical evaluation. Anticancer Agents Med. Chem. 2010;10:753–768. doi: 10.2174/187152010794728639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stupp R., Hegi M.E., Gorlia T., Erridge S.C., Perry J., Hong Y.-K., Aldape K.D., Lhermitte B., Pietsch T., Grujicic D., et al. Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071-22072 study): A multicenter, randomized, open-label, phase 3 trial. Lancet Oncol. 2014;15:1100–1108. doi: 10.1016/S1470-2045(14)70379-1. [DOI] [PubMed] [Google Scholar]
- 20.Vansteenkiste J., Barlesi F., Waller C.F., Bennouna J., Gridelli C., Goekkurt E., Verhoeven D., Szczesna A., Feurer M., Milanowski J., et al. Cilengitide combined with cetuximab and platinum-based chemotherapy as first-line treatment in advanced non-small-cell lung cancer (NSCLC) patients: Results of an open-label, randomized, controlled phase II study (CERTO) Ann. Oncol. 2015;26:1734–1740. doi: 10.1093/annonc/mdv219. [DOI] [PubMed] [Google Scholar]
- 21.Kapp T.G., Rechenmacher F., Neubauer S., Maltsev O.V., Cavalcanti-Adam E.A., Zarka R., Reuning U., Notni J., Wester H.-J., Mas-Moruno C., et al. A comprehensive evaluation of the activity and selectivity profile of ligands for RGD-binding integrins. Sci. Rep. 2017;7:39805. doi: 10.1038/srep39805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ressurreição A.S.M., Bordessa A., Civera M., Belvisi L., Gennari C., Piarulli U. Synthesis and conformational studies of peptidomimetics containing a new bifunctional diketopiperazine scaffold acting as a β-hairpin inducer. J. Org. Chem. 2008;73:652–660. doi: 10.1021/jo702072z. [DOI] [PubMed] [Google Scholar]
- 23.Ressurreição A.S.M., Vidu A., Civera M., Belvisi L., Potenza D., Manzoni L., Ongeri S., Gennari C., Piarulli U. Cyclic RGD-peptidomimetics containing bifunctional diketopiperazine scaffolds as new potent integrin ligands. Chem. Eur. J. 2009;15:12184–12188. doi: 10.1002/chem.200902398. [DOI] [PubMed] [Google Scholar]
- 24.Marchini M., Mingozzi M., Colombo R., Guzzetti I., Belvisi L., Vasile F., Potenza D., Piarulli U., Arosio D., Gennari C. Cyclic RGD peptidomimetics containing bifunctional diketopiperazine scaffolds as new potent integrin ligands. Chem. Eur. J. 2012;18:6195–6207. doi: 10.1002/chem.201200457. [DOI] [PubMed] [Google Scholar]
- 25.Guzzetti I., Civera M., Vasile F., Arosio D., Tringali C., Piarulli U., Gennari C., Pignataro L., Belvisi L., Potenza D. Insights into the binding of cyclic RGD peptidomimetics to α5β1 integrin by using live-cell NMR and computational studies. ChemistryOpen. 2017;6:128–136. doi: 10.1002/open.201600112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guzzetti I., Civera M., Vasile F., Araldi E.M.V., Belvisi L., Gennari C., Potenza D., Fanelli R., Piarulli U. Determination of the binding epitope of RGD-peptidomimetics to αVβ3 and αIIbβ3 integrin-rich intact cells by NMR and computational studies. Org. Biomol. Chem. 2013;11:3886–3893. doi: 10.1039/c3ob40540k. [DOI] [PubMed] [Google Scholar]
- 27.Panzeri S., Zanella S., Arosio D., Vahdati L., Dal Corso A., Pignataro L., Paolillo M., Schinelli S., Belvisi L., Gennari C., et al. Cyclic isoDGR and RGD peptidomimetics containing bifunctional diketopiperazine scaffolds are integrin antagonists. Chem. Eur. J. 2015;21:6265–6271. doi: 10.1002/chem.201406567. [DOI] [PubMed] [Google Scholar]
- 28.Fanelli R., Schembri L., Piarulli U., Pinoli M., Rasini E., Paolillo M., Galiazzo M.C., Cosentino M., Marino F. Effects of a novel cyclic RGD peptidomimetic on cell proliferation, migration and angiogenic activity in human endothelial cells. Vasc. Cell. 2014;6:11. doi: 10.1186/2045-824X-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colombo R., Mingozzi M., Belvisi L., Arosio D., Piarulli U., Carenini N., Perego P., Zaffaroni N., De Cesare M., Castiglioni V., et al. Synthesis and biological evaluation (in vitro and in vivo) of cyclic arginine-glycine-aspartate (RGD) peptidomimetic-paclitaxel conjugates targeting integrin αVβ3. J. Med. Chem. 2012;55:10460–10474. doi: 10.1021/jm301058f. [DOI] [PubMed] [Google Scholar]
- 30.Dal Corso A., Caruso M., Belvisi L., Arosio D., Piarulli U., Albanese C., Gasparri F., Marsiglio A., Sola F., Troiani S., et al. Synthesis and biological evaluation of RGD peptidomimetic-paclitaxel conjugates bearing lysosomally cleavable linkers. Chem. Eur. J. 2015;21:6921–6929. doi: 10.1002/chem.201500158. [DOI] [PubMed] [Google Scholar]
- 31.Pina A., Dal Corso A., Caruso M., Belvisi L., Arosio D., Zanella S., Gasparri F., Albanese C., Cucchi U., Fraietta I., et al. Targeting integrin αVβ3 with theranostic RGD-camptothecin conjugates bearing a disulfide linker: Biological evaluation reveals a complex scenario. ChemistrySelect. 2017;2:4759–4766. doi: 10.1002/slct.201701052. [DOI] [Google Scholar]
- 32.Dong X., Hudson N.E., Lu C., Springer T.A. Structural determinants of integrin β-subunit specificity for latent TGF-β. Nat. Struct. Mol. Biol. 2014;21:1091–1096. doi: 10.1038/nsmb.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paolillo M., Serra M., Schinelli S. Integrins in glioblastoma: Still an attractive target? Pharmacol. Res. 2016;113:55–61. doi: 10.1016/j.phrs.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Goodman S.L., Hölzemann G., Sulyok G.A., Kessler H. Nanomolar small molecule inhibitors for αVβ6, αVβ5, and αVβ3 integrins. J. Med. Chem. 2002;45:1045–1051. doi: 10.1021/jm0102598. [DOI] [PubMed] [Google Scholar]
- 35.Glide. Schrödinger, LLC; New York, NY, USA: 2011. version 5.7. [Google Scholar]
- 36.For the sake of precision, in [32] the ligand-free αVβ6 structure is defined similar to the state 1 and the ligand-bound αVβ6 similar to the state 2 of the eight intermediate states identified for integrin αIIbβ3 opening in Zhu J., Zhu J., Springer T.A. Complete integrin headpiece opening in eight steps. J. Cell Biol. 2013;201:1053–1068. doi: 10.1083/jcb.201212037.
- 37.Xiong J.P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S.L., Arnaout M.A. Crystal Structure of the Extracellular Segment of Integrin αVβ3 in Complex with an Arg-Gly-Asp Ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 38.Springer T.A., Zhu J., Xiao T. Structural basis for distinctive recognition of fibrinogen γC peptide by the platelet integrin αIIbβ3. J. Cell Biol. 2008;182:791–800. doi: 10.1083/jcb.200801146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia W., Springer T.A. Metal ion and ligand binding of integrin α5β1. Proc. Natl. Acad. Sci. USA. 2014;111:17863–17868. doi: 10.1073/pnas.1420645111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagae M., Re S., Mihara E., Nogi T., Sugita Y., Takagi J. Crystal structure of α5β1 integrin ectodomain: Atomic details of the fibronectin receptor. J. Cell Biol. 2012;197:131–140. doi: 10.1083/jcb.201111077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bandyopadhyay A., Raghavan S. Defining the role of integrin αVβ6 in cancer. Curr. Drug Targets. 2009;10:645–652. doi: 10.2174/138945009788680374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maltsev O.V., Marelli U.K., Kapp T.G., Di Leva F.S., Di Maro S., Nieberler M., Reuning U., Schwaiger M., Novellino E., Marinelli L., et al. Stable peptides instead of stapled peptides: Highly potent αvβ6-selective integrin ligands. Angew. Chem. Int. Ed. Engl. 2016;55:1535–1539. doi: 10.1002/anie.201508709. [DOI] [PubMed] [Google Scholar]
- 43.Data on the role of the post-RGD helix in stabilizing the RGD interaction and increasing affinity and potency of RGDLXX(L/I) peptides are reported in DiCara D., Rapisarda C., Sutcliffe J.L., Violette S.M., Weinreb P.H., Hart I.R., Howard M.J., Marshall J.F. Structure-Function Analysis of Arg-Gly-Asp Helix Motifs in αVβ6 Integrin Ligands. J. Biol. Chem. 2007;282:9657–9665. doi: 10.1074/jbc.M610461200.
- 44.Maestro. Schrödinger, LLC; New York, NY, USA: 2011. version 9.2. [Google Scholar]
- 45.Allen S.E., Dokholyan N.V., Bowers A.A. Dynamic Docking of Conformationally Constrained Macrocycles: Methods and Applications. ACS Chem. Biol. 2016;11:10–24. doi: 10.1021/acschembio.5b00663. [DOI] [PubMed] [Google Scholar]
- 46.Vasile F., Civera M., Belvisi L., Potenza D., Tiana G. Thermodynamically-weighted conformational ensemble of cyclic RGD peptidomimetics from NOE data. J. Phys. Chem. B. 2016;120:7098–7107. doi: 10.1021/acs.jpcb.6b04941. [DOI] [PubMed] [Google Scholar]
- 47.Marelli U.K., Frank A.O., Wahl B., La Pietra V., Novellino E., Marinelli L., Herdtweck E., Groll M., Kessler H. Receptor-bound conformation of cilengitide better represented by its solution-state structure than the solid-state structure. Chem. Eur. J. 2014;20:14201–14206. doi: 10.1002/chem.201403839. [DOI] [PubMed] [Google Scholar]
- 48.Guarnieri F., Still W.C. A rapidly convergent simulation method: Mixed monte carlo/stochastic dynamics. J. Comput. Chem. 1994;15:1302–1310. doi: 10.1002/jcc.540151111. [DOI] [Google Scholar]
- 49.Still W.C., Tempczyk A., Hawley R.C., Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J. Am. Chem. Soc. 1990;112:6127–6129. doi: 10.1021/ja00172a038. [DOI] [Google Scholar]
- 50.MacroModel. Schrödinger, LLC; New York, NY, USA: 2011. version 9.9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.