Abstract
Magnaporthe oryzae is a devastating plant pathogen, which has a detrimental impact on rice production worldwide. Despite its agronomical importance, some newly-emerging pathotypes often overcome race-specific disease resistance rapidly. It is thus desirable to develop a novel strategy for the long-lasting resistance of rice plants to ever-changing fungal pathogens. Brome mosaic virus (BMV)-induced RNA interference (RNAi) has emerged as a useful tool to study host-resistance genes for rice blast protection. Planta-generated silencing of targeted genes inside biotrophic pathogens can be achieved by expression of M. oryzae-derived gene fragments in the BMV-mediated gene silencing system, a technique termed host-induced gene silencing (HIGS). In this study, the effectiveness of BMV-mediated HIGS in M. oryzae was examined by targeting three predicted pathogenicity genes, MoABC1, MoMAC1 and MoPMK1. Systemic generation of fungal gene-specific small interfering RNA (siRNA) molecules induced by inoculation of BMV viral vectors inhibited disease development and reduced the transcription of targeted fungal genes after subsequent M. oryzae inoculation. Combined introduction of fungal gene sequences in sense and antisense orientation mediated by the BMV silencing vectors significantly enhanced the efficiency of this host-generated trans-specific RNAi, implying that these fungal genes played crucial roles in pathogenicity. Collectively, our results indicated that BMV-HIGS system was a great strategy for protecting host plants against the invasion of pathogenic fungi.
Keywords: Magnaporthe oryzae, gene silencing, rice blast, fungal pathogens, brome mosaic virus
1. Introduction
Rice blast disease caused by the ascomycete Magnaporthe oryzae is a serious rice disease in more than 90 countries across the globe [1], which are devastating threats to rice production worldwide [2]. It has been estimated that each year harvest losses caused by rice blast can reach 10% to 50% of the global rice yield [3]. Over years, great progress has been gained to withstand rice blast caused by M. oryzae through chemical treatments. However, fungicide resistance is rapidly developed by the fungus [4]. For decades, constant efforts have been made to find rice cultivars resistant to rice blast because of the strong ability of M. oryzae to evolve new pathotypes. One of the most economical and sustainable routes is genetic resistance that can confront plant diseases effectively, although most genetic resistance is not everlasting in cereals. Some fungal genes play crucial roles in pathogenicity, and knockout of these genes significantly causes less virulence of pathogenic fungus to host plants [5]. A leading model has recently been established for rice blast systems to explore plant-fungus interactions due to genetic accessibility of genome sequences for both the fungus and rice [6]. Thus, there is an urgent need to develop an efficient strategy to confer more long-lasting resistance of rice to ever-changing fungal pathogens.
RNA interference (RNAi) is a conserved biological process across almost all eukaryotic organisms, in which double-stranded RNA (dsRNA) interfering molecules causes degradation of homologous mRNA molecules, and further inhibits the transcription of targeted genes [7]. It has been indicated that RNAi is an important tool for functional genomics researches in most eukaryotes such as worms, protists, animals, and plants [8,9,10,11,12,13]. Increasing evidence has indicated that the RNA silencing technology can be used in crop protection strategies [14,15]. In several insects and nematodes, RNAi reduces the expression of targeted genes by injecting dsRNA [7] and feeding with bacteria that produce the dsRNA [16,17]. For vascular plants, RNAi molecules can quickly transfer from cell to cell, and even systemically throughout the whole plant [8]. This discovery breaks through our traditional thinking that RNAs can just play a part within the cells where they are produced. Similar to other eukaryotes, most fungi are also sensitive to RNAi [18,19,20]. Intriguingly, recent studies have indicated that trafficking of RNAi molecules occurs between fungi and its host plants. The expression of endogenous genes in the infecting fungi significantly decreased in host plants carrying dsRNAs from Blumeria graminis or Fusarium verticillioides genes [21,22], indicating that the host-induced RNAi has a great promise for fighting fungal infection by expression of dsRNA of fungal genes in plants. This is thus desirable to exploit the method for control of rice blast. However, it is time consuming and laborious to generate stable RNAi plants because of long-term selection of transformants and their molecular identification. Besides, the extensive progress of M. oryzae genome data gives rise to the identification of candidate genes associated with pathogenesis. It has gradually been recognized that the development of effective and rapid biotechnologies is crucial to examine the functions of candidate genes responsible for disease development.
Virus-induced gene silencing (VIGS) is a suitable method to deal with these problems mentioned above. Recently, VIGS has become an important tool that triggers RNAi silencing by the use of viral vectors to generate dsRNA of targeted genes [23,24]. Because of its rapid procedure, VIGS has recently been applied to assay gene functions in plants [25]. VIGS biotechnology was primarily successful in dicots until an efficient system was developed for silencing gene expression transiently in wheat and barley mediated by the barley stripe mosaic virus (BSMV) [26]. In addition to BSMV, the brome mosaic virus (BMV) has been identified as suitable for VIGS-induced gene silencing in rice [27]. BMV is the type member of the genus Bromovirus with a positive-strand tripartite RNA genome constituting three RNA segments known as RNA1, RNA2, and RNA3 [28]. A new VIGS system has been established by modifying BMV vector based on the BMV to silence phytoene desaturase (PDS) genes in maize, barley, wheat and rice. Until now, the BMV-VIGS system is utilized to rapidly screen candidate genes in plants that determine the compatible interactions with fungal pathogens [29]. However, a VIGS system that allows efficient and systemic screening of the predicted M. oryzae genes vital for disease development has not been developed for rice-M. oryzae interaction. Nowara et al. [22] have reported that host plants expressing the dsRNAs from B. graminis genes exhibit a marked reduction in the transcription of targeted genes in infecting fungi. Concomitantly, the disease development is apparently suppressed by such host-induced gene silencing (HIGS). This approach is also emulated by Panwar et al. [30] to silence pathogenicity genes in the wheat leaf rust fungus Puccinia triticina. These studies suggest that the small interfering RNA (siRNA) molecules in planta can be transferred into fungal cells during fungal infection. Hence, the HIGS technique could be a great strategy that controls rice blast disease.
In this work, we tested the feasibility of the BMV-HIGS system to silence some putative M. oryzae genes vital for disease development. The rice BMV-VIGS system previously established by Ding et al. [27] was further optimized for adapting to experimental conditions, which allowed the infection with M. oryzae. An ATP-driven efflux pump encoded by MoABC1 has been shown to be essential for pathogenesis for protecting itself against rice defense mechanisms [31]. MoMAC1 encodes a membrane-bound adenylate cyclase (MAC) catalyzing the production of cAMP from ATP, which plays a central role in appressorium formation of M. oryzae [32]. MoPMK1, encoding mitogen-activated protein kinase (MAPK), is involved in the regulation of infectious hyphal growth in M. oryzae [33]. The three genes, MoABC1, MoMAC1 and MoPMK1, were thus selected as candidate genes that were involved in pathogenicity. The inoculation with BMV silencing vectors remarkably repressed fungus invasion and disease development in rice plants. The HIGS efficiency significantly increased when synchronously introducing targeted gene fragments and silencing three fungal pathogenic genes together. Our results provided important evidence that the HIGS technique can be used to control rice blast disease effectively.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
To conduct VIGS experiments, Oryza sativa cv. CO-39 was grown on growth chambers at 25 °C day and 22 °C night, 16 h/8 h light/dark cycles (200 μmol·m−2·s–1). Tobacco (Nicotiana benthamiana) seeds were sown in a soil mixture. At 3 weeks after planting, individual seedlings were transplanted into pots, and were then cultured at 25 °C with a light/dark (16 h/8 h) cycle (200 μmol·m−2·s–1). CO-39 and tobacco seeds were provided by Zhengyi Wang (College of Agriculture and Biotechnology, Zhejiang University).
2.2. Construction of Gene Silencing Vectors
The genome of BMV constitutes three RNA segments, including RNA1, RNA2 and RNA3. Furthermore, the constructs (pF1-11, pF2-2 and pB3m) were used to conduct BMV-mediated gene silencing as described previously by Ding et al. [27] and were provided by R. S. Nelson (Samuel Roberts Noble Foundation, Inc.). A 286-bp fragment of OsPDS was amplified using a pair of primers containing NcoI and AvrII restriction sites, respectively. The products of PCR amplification were digested with NcoI and AvrII, and were then inserted in sense or antisense orientation into the RNA3 component to generate pB3m-OsPDSs and pB3m-OsPDSas, respectively. For M. oryzae pathogenic genes, the primers were designed for MoABC1, MoMAC1 and MoPMK1, respectively. All primers contained NcoI and AvrII restriction sites for integration into pB3m vector (Supplementary Materials Table S1). Gene fragments for inducing VIGS were amplified from the complementary DNA (cDNA) of M. oryzae Guy11 by PCR, and were subsequently inserted into pB3m in sense and antisense orientation, respectively. For in silico analysis of siRNA production, the SIRNA SCAN (http://bioinfo2.noble.org/RNAiScan.htm) was used. All recombinant RNA3 vectors were further confirmed by sequencing.
2.3. In Vitro Transcription of BMV RNAs and BMV Inoculation
To obtain BMV RNA1, RNA2 and RNA3, three plasmids coding for BMV genome were linearized using SpeI (pF1-11) or PshAI (pF2-2, pB3m or recombinant pB3m plasmids) restriction enzymes (New England Biolabs, Beijing, China). The linearized plasmids were used to perform in vitro transcription by the mMessage mMachine T3 in vitro transcription kit (Ambion, Austin, TX, USA). Before BMV inoculation, the synthesized RNAs were verified by agarose gel electrophoresis. To achieve more successful infections, the viral transcripts were initially inoculated into N. benthamiana seedlings that were easily infected and allowed high titers of many viruses to accumulate in leaves. A mixture of 1 µL of each viral RNA component with 60 µL of buffer solutions (1% (w/v) bentonite, 1% (w/v) Celite, 22 mM Na4P2O7·10H2O, 60 mM K2HPO4, and 77 mM glycine) (Sangon Biotech, Shanghai, China) was inoculated into tobacco seedlings. The inoculated plants were then moved to greenhouse at 24/20 °C (day/night). After seven days of inoculation, leaf tissue from the infected N. benthamiana plants was separated and ground in 0.1 M phosphate buffer solution (PBS) (pH 6.5; 1:10, w/v) (Sangon Biotech) at 40 °C. Quantitative real time PCR (qRT-PCR) was conducted to quantify the BMV titer as reported by van der Linde et al. [29]. All crude extracts of N. benthamiana leaves were adjusted to the same virus titer of 104 relative expression units compared with non-inoculated plants. For rice infection, leaves of 2-week-old rice seedlings were dusted with carborundum, and were then inoculated with the crude tobacco extract prepared before. The inoculated rice seedlings were placed in a greenhouse at 25/20 °C (day/night).
2.4. Fungal Inoculation and Phenotypic Analysis
M. oryzae strain Guy11 was used in our experiments and provided by Zhengyi Wang (College of Agriculture and Biotechnology, Zhejiang University). Growth conditions for M. oryzae were carried out, according to the method described by Talbot et al. [3]. For infection with M. oryzae, seedling spray inoculation and cut-leaf assays were used as reported by Wang et al. [34]. Seedling spray inoculation was conducted as follows: conidia were harvested from the plate cultures of fungus grown on complete medium (CM) medium for 12 days. 19-day-old rice seedlings (5 days after BMV inoculation) were inoculated with conidial suspensions of M. oryzae prepared in 0.025% Tween 20 at a concentration of 5 × 104–105 conidia/mL. The inoculated seedlings were then placed in a growth chamber at 14-h light periods and 25 °C with 90% relative humidity until disease symptoms occurred. Disease lesions were monitored after 10 days of fungal incubation, and 5-cm-long leaf tips were randomly selected to calculate lesion densities.
2.5. cDNA Synthesis and qRT-PCR
After centrifugation at 12,000× g for 5 min, 2 µL of leaf extracts was used for virus titer detection. The BMV titers were assayed using qRT-PCR analysis. Tobacco actin was selected as internal reference. qRT-PCR analysis was performed in a ABI 7500 fast real-time PCR system as reported recently by Zhu et al. [35]. To examine whether the recombinant RNA3 vector carrying OsPDS sequence triggered OsPDS silencing, the non-inoculated younger leaves of each inoculated rice plant were separated, and total RNA was isolated from them using Trizol reagent (Takara, Dalian, China). Further, to test the silencing efficiency of OsPDS in different rice leaves, total RNA was isolated from Leaf 4, 5 and 6 of each inoculated rice plant. After extraction, using the PrimeScriptTM RT Kit (Takara), about 1 µg of total RNA was reversely transcribed into cDNA as templates for qRT-PCR. OsPDS mRNA levels in various plants were normalized by the elongation factor 1α (EF-1α). In addition to assay lesion amount and the expression of targeted fungal genes, 5 cm sections of fungal infected leaves were harvested after 10 d of fungal infection (15 d after BMV inoculation). The rice EF-1α and β-tubulin gene (BT) from M. oryzae were selected as respective references to normalize targeted gene expression.
2.6. Staining and Confocal Microscopy
Leaf samples were harvested after 10 days of fungal infection (15 days post inoculation with BMV). The samples were treated with 1 M KOH (Sangon Biotech), heated to 70 °C for 20 min, and were then stained in aniline blue solution (0.05%, w/v) in 0.067M K2HPO4 (pH 9.0) (Sangon Biotech) for 5 min. Excessive dye was washed with sterile water. Finally, the staining samples were observed and photographed by epifluorescence microscopy. Fluorescence of aniline blue was monitored using a confocal microscope (Leica, Wetzlar, Germany) with an excitation filter of 405 nm and an emission filter of 500 nm.
2.7. Short Interfering RNA Detection
Small RNA molecules were examined according to the method reported by Panwar et al. [30]. Total RNA was isolated from rice plants at 15 days post inoculation with BMV. Total RNA (about 15 µg) for each sample was separated on a 15% polyacrylamide-7 M urea gel and transferred to neutral Hybond NX membrane (Amersham Pharmacia Biotech, Piscataway, NJ, USA). Probes were labelled with the phosphorus-32 deoxycytidine triphosphate ([α32P]-dCTP) using the DNA labelling kit (Amersham) RNA blot hybridization was performed using PerfectHyb plus Hybridization buffer (Sigma, St. Louis, MO, USA) at 42 °C overnight [36]. DynaMarker prestain (Takara) for small RNA was used as a molecular size marker. The bands were detected using a fluorescent image analyzer.
2.8. Statistical Analyses
Each experiment was conducted with three biological repeats. Each bar represents the mean ± standard deviation (SD), and different letters represent significant differences among different experiment groups using at a Tukey’s test at p < 0.05.
3. Results
3.1. Systemic Gene Silencing of PDS in Rice Mediated by BMV
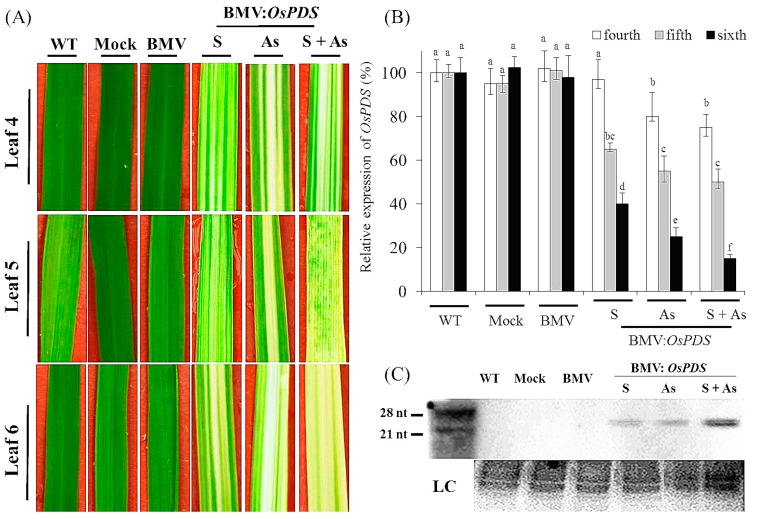
To establish BMV-based HIGS system, Ding’s rice VIGS protocol [27] was first adapted in the lab by using M. oryzae susceptible rice inbred line CO-39 in the experiment, and by using the OsPDS gene as a phenotypic marker for evaluation and optimization of the system. 2-week-old rice CO-39 seedlings at the two-leaf stage were inoculated with BMV carrying derivatives of recombinant RNA3 vector expressing target OsPDS gene segments. The plants treated with buffer (Mock), or treated with non-modified BMV (BMV) as well as plants not treated with BMV (wild type (WT)) were designed as controls. After 15 days of exposure to BMV infection, the plants with inoculation of BMV-OsPDS displayed strong photo-bleaching phenotype in newly emerged upper non-inoculated leaves (Figure 1A). More importantly, BMV infection did not appear to affect the growth of rice cultivar CO-39 (Supplementary Materials Figure S1).
Figure 1.
Silencing of phytoene desaturase (PDS) in rice CO-39 using the Brome mosaic virus (BMV)-induced gene silencing system. (A) Leaf 2 of 2-week-old rice CO-39 plants were inoculated with BMV carrying derivatives of recombinant RNA3 vector expressing gene segments of OsPDSs, either in antisense or as a mixture of sense plus antisense orientation. WT, Mock and BMV (without inserting) plants were considered as the controls. Photographs were taken 15 days post inoculation. OsPDS gene silencing caused visible photo-bleaching symptoms of upper the fourth, fifth and sixth leaf (Leaf 4, 5 and 6). Compared to the plants inoculated with BMV-OsPDS in sense form, more visible photo-bleaching can be seen in the plants inoculated with BMV-OsPDS in sense, antisense and as a mixture of sense plus antisense; (B) Quantitative real time-polymerase chain reaction (qRT-PCR) analyses of OsPDS silencing in rice leaves inoculated with BMV variants; (C) RNA gel blot analyses of OsPDS siRNA accumulation. Small RNAs were detected in rice plants inoculated with BMV variants harboring OsPDS insert in antisense or a mixture of sense plus antisense form. No signal was detected in WT, Mock and BMV inoculated controls. WT, wild type; S, sense; As, antisense; S + As, sense plus antisense; LC, stained rRNAs as loading controls. Each experiment was performed with three biological repeats. Values represented mean ± standard deviation (SD) in three independent experiments, and 15 individual leaves from different plants were collected for each experiment. Different letter indicated a significant difference at p < 0.05.
To identify the best time point of M. oryzae infection, the efficiency of BMV-induced VIGS in different rice leaves was assessed. As shown in Figure 1A, the efficiency of VIGS seemed to depend on the tested leaves. The sixth leaf (Leaf 6) displayed the strongest photo-bleaching phenotype, and the symptoms in Leaf 6 were more apparent than in the fourth and fifth leaf (Leaf 4 and 5). To precisely quantify the silencing efficiency in different leaves, we examined the transcription levels of OsPDS in BMV-infected plants using qRT-PCR. Consistent with the observed phenotype, the silencing of OsPDS was most efficient in Leaf 6 compared with the controls; whereas this was less effective in Leaf 4 and 5 (Figure 1B). Based on the data, we defined the following infection procedure: the second leaf (Leaf 2) of 2-week-d-old rice seedlings was inoculated with BMV, and the plants were infected with M. oryzae for seedling spray inoculation after five days of BMV inoculation.
To further investigate whether transgene orientation affected gene silencing efficiency, the BMV carrying the sense or antisense orientation of OsPDS fragments (BMV-OsPDSs or-OsPDSas) was inoculated into rice plants, respectively. The OsPDS silencing phenotype caused by BMV-OsPDSs did not noticeably differ from that induced by BMV-OsPDSas (Figure 1A). In accordance with this, the results of qRT-PCR analyses indicated no significant difference of the OsPDS expression between plants inoculated with BMV-OsPDSs and -OsPDSas (Figure 1B). Furthermore, we examined the effects of the insertion of both sense and antisense OsPDS fragments on the silencing efficiency of OsPDS. For this purpose, plants were inoculated with BMV-OsPDSs and BMV-OsPDSas simultaneously. As shown in Figure 1A, the OsPDS silencing phenotype was even further evident in plants carrying both the sense and antisense sequence compared to the previous two treatments. qRT-PCR analyses further confirmed it: the Leaf 6 of plants inoculated with the BMV-OsPDSs plus BMV-OsPDSas constructs exhibited a reproducible and efficient silencing of average 90% compared with the controls, which was associated with the occurrence of photo-bleaching. It has previously been indicated that the generation of small RNAs is responsible for RNAi-induced gene silencing [7]. Herein, northern blotting was used to detect siRNA molecules specific to OsPDS in Leaf 6 of non-inoculated plants. The results showed the band of relevant siRNA production targeted, which was triggered by the inoculation of BMV (Figure 1C).
3.2. BMV-HIGS Allows Functional Analysis of Fungal Pathogenic Genes during the Rice-M. oryzae Interaction
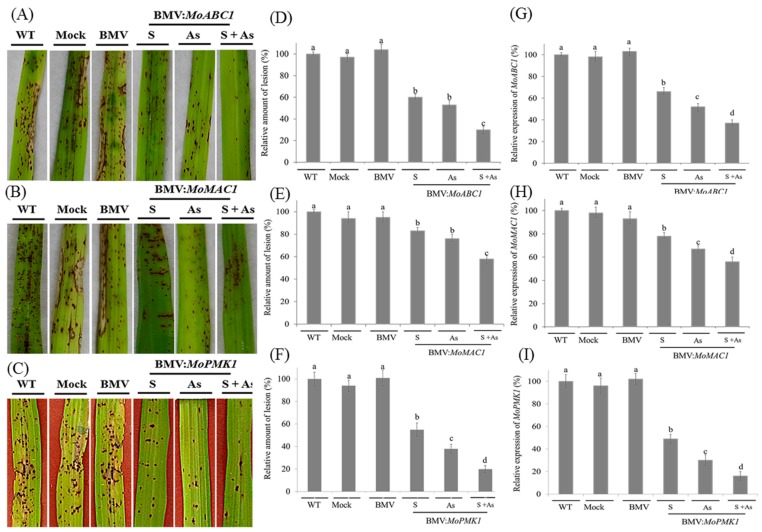
Based on the established system for BMV-induced gene silencing, several putative M. oryzae genes were chosen as targeted genes. Pathogen genes responsible for fungal invasions were of interest because the inhibited expression of these pathogenicity-determining genes would potentially lead to reduction of disease development. Targeting such genes would test whether the BMV-HIGS system was suitable for analyzing the functions of fungal genes. In this study, three pathogenic genes were chosen: MoABC1, MoMAC1 and MoPMK1, respectively. Sense or antisense fragments of MoABC1, MoMAC1 and MoPMK1 coding regions were inserted into the BMV RNA3 component to generate silencing constructs. Leaf 2 of 2-week-old CO-39 plants was inoculated with BMV vectors carrying targeted gene fragments in sense or antisense orientation. By five days post inoculation, when virus symptoms were seen in upper non-inoculated leaves, seedling spray was used to perform the infection of M. oryzae to rice leaves, and rice blast disease symptoms were observed in the days following the inoculation (Figure 2A–C). Compared with the controls, the number of lesions was observed to distinctly decrease in rice plants inoculated with BMV vectors carrying the targeted gene fragments (Figure 2A–C).
Figure 2.
Effects of BMV-derived host-induced gene silencing (HIGS) of targeted M. oryzae genes on disease development in rice. Leaf 2 of 2-week-old rice CO-39 leaves were infected with BMV carrying derivatives of recombinant RNA3 construct carrying target MoABC1, MoMAC1, or MoPMK1 gene fragments, either in sense (S), antisense (As) or as a mixture of sense plus antisense (S + As) form. WT, Mock and BMV-treated plants were selected as the control groups. After 5 days of BMV or buffer inoculation, M. oryzae was used to infect rice plants. (A–C) Rice blast disease phenotype in the control and silenced rice plants. Plants were treated with BMV vectors harboring the MoABC1, MoMAC1, or MoPMK1 gene fragments. After 10 days of fungal infection, the 6th leaves had photographs taken. (D–F) Quantification of lesion density on both the BMV-infected plants carrying the targeted gene fragments and controls. After 10 days of M. oryzae infection, lesions in leaves from 15 independent plants were counted. (G–I) qRT-PCR analysis of the MoABC1, MoMAC1, and MoPMK1 transcripts in rice leaves inoculated with BMV variants. Silencing was induced by BMV carrying derivatives of recombinant RNA3 vector expressing the targeted gene fragments, either in sense, antisense or as a mixture of sense plus antisense form. S, sense; As, antisense; S + As, sense plus antisense. Each experiment was performed with three biological repeats. Values represented mean ± SD in three independent experiments, and 15 individual leaves from different plants were collected for each experiment. Different letter indicated a significant difference at p < 0.05.
To examine if the efficiency of M. oryzae gene silencing was enhanced by introducing both sense and antisense forms of the corresponding gene fragments, rice leaves were infected with the BMV carrying derivatives of recombinant RNA3 vector expressing the pathogenic gene segments in a mixture of sense plus antisense form. In contrast to the previous two treatments, combined introduction of the sense plus antisense sequence even further suppressed rice blast symptom development. In the case of MoABC1, reduction of fungal disease phenotype generated by the BMV-MoABC1as construct did not markedly differ from that caused by the BMV-MoABC1s (Figure 2A), whereas the combined use of sense plus antisense forms significantly reduced the number of lesions in rice leaves compared to the previous two treatments. Compared with MoABC1, the reduction of fungal disease phenotype was seen less clearly in the case of MoMAC1 (Figure 2B). The most significant reduction of fungal disease phenotype appears in the plants inoculated by the BMV carrying derivatives of recombinant RNA3 vector expressing the MoPMK1 gene segments, especially in a mixture of sense plus antisense form (Figure 2C).
To examine the effects of M. oryzae gene silencing on disease phenotypes, we looked at the number of lesions on Leaf 6 of the rice plants after 10 days of fungal infection. Compared with the controls, the fungal density was remarkably decreased in rice leaves silenced for all the tested genes (Figure 2D–F). Up to statistical data, the strongest disease suppression occurred in the inoculated leaves mixed with recombinant BMV RNA3 components carrying targeted genes in sense and antisense orientation, especially in the case of MoPMK1. Reduction of lesion number in the rice plants inoculated with BMV-MoPMK1s + as construct reached above 85% compared with the controls. After 10 days of M. oryzae infection, the efficiency of targeted fungal genes was also detected using qRT-PCR analysis. As shown in Figure 2G–I, endogenous transcript abundance of all three corresponding M. oryzae genes was significantly reduced. Silencing of MoPMK1 was the strongest when BMV-MoPMK1s and BMV-MoPMK1as were introduced simultaneously compared with introduction of the sense or antisense form alone. In the case of MoABC1, MoMAC1 and MoPMK1, the efficiency of fungal gene silencing reached about 65%, 44% and 89%, respectively.
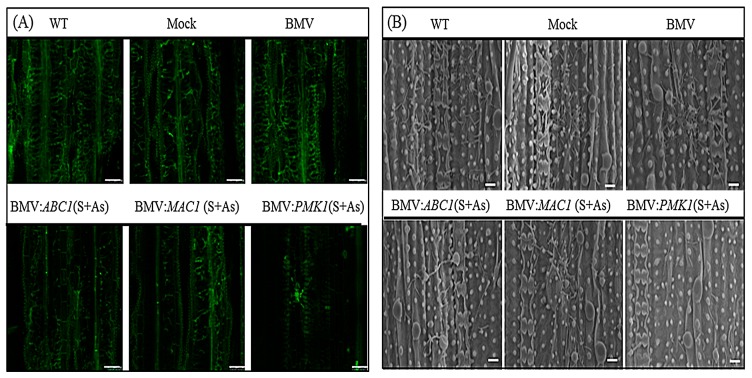
Reduction of leaf lesions might result from a significant inhibition of fungal growth. Thus, we further investigated the effects of BMV-induced gene silencing on the accumulation of fungi inside leaf tissue. Compared with the controls, the inoculation with BMV vectors targeting M. oryzae genes significantly inhibited the fungal growth and development. Considerable reduction of the lesion amount was seen in the BMV-infected leaves carrying both the sense and antisense forms of targeted sequences simultaneously. The data revealed that the growth of M. oryzae inside host plants was markedly impaired, which was likely attributed to the silencing of endogenous MoABC1, MoMAC1 and MoPMK1 genes. Moreover, we examined the effects of gene silencing on the fungal development in leaves; chosen sites from Leaf 6 of BMV-silenced and control plants, 10 days after fungal infection, were assayed by scanning electron microscope (SEM) and confocal microscopy. The BMV silencing vectors were used to treat rice leaves, in which M. oryzae infection was remarkably suppressed, indicating severe arrest of mycelial growth. In contrast, widespread fungal growth was observed in the controls. As shown in Figure 3A,B, the most significant reduction of fungal growth in the leaves appeared in the case of MoPMK1 gene.
Figure 3.
Observation of fungal development in rice CO-39 leaves. (A) Fungal development in rice CO-39 leaves was observed by confocal microscope. Plates represent a projection of scans taken from inside Leaf 6 of rice plants 10 days after M. oryzae infection but previously inoculated with BMV carrying derivatives of recombinant RNA3 vector expressing target MoABC1, MoMAC1 and MoPMK1 gene segments in as a mixture of sense plus antisense form. WT, Mock and BMV (without inserting) plants were used as controls. Silencing of the targeted M. oryzae genes induced by host-generated RNAi restricts fungal development whereas control plants show extensive mycelia growth. Scale bars are 10 µm. (B) Under a scanning electron microscope (SEM), fungal development in rice leaves inoculated with BMV variants were observed. Scale bars are 50 µm.
3.3. Effective Control of Rice Blast Disease by Co-Silencing of M. oryzae Pathogenic Genes
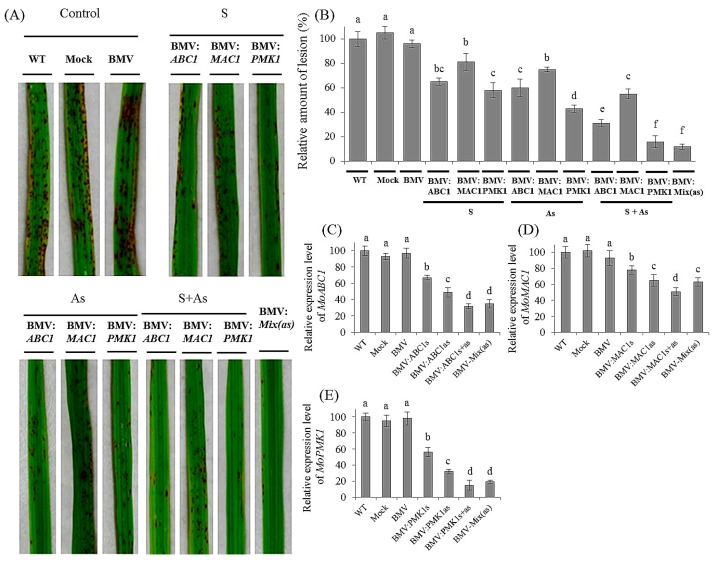
Based on the individual gene silencing study with the BMV-HIGS system, we were curious about whether the effects of the growth inhibition of M. oryzae were more obvious when silencing three fungal pathogenic genes together. Rice plants were also inoculated with BMV-MoABC1as, BMV-MoMAC1as and BMV-MoPMK1as simultaneously. Meanwhile, we repeated all the experimental groups in the case of MoABC1, MoMAC1 and MoPMK1 genes. As shown in Figure 4A, the development of rice blast symptoms was even further reduced in the plants that had simultaneously introduced antisense forms of three targeted gene fragments compared to the previous individual experiments (Figure 4B). Silencing efficiency of targeted fungal genes was also determined by qRT-PCR analyses, 10 days after M. oryzae infection. In the silenced seedlings inoculated with BMV-MoABC1as, BMV-MoMAC1as and BMV-MoPMK1as simultaneously, a marked reduction in endogenous gene expression was observed for all three corresponding M. oryzae genes (Figure 4C–F).
Figure 4.
Effect of BMV-derived HIGS of targeted MoABC1, MoMAC1 and MoPMK1 on disease development in rice. Leaf 2 of 2-week-old rice CO-39 plants were infected with BMV harboring derivatives of recombinant RNA3 vector expressing MoABC1, MoMAC1 and MoPMK1 gene fragments, in sense (S), antisense (As) or as a mixture of sense plus antisense form (S + As). In addition, some plants were also inoculated with a mixture of BMV carrying derivatives of recombinant RNA3 vector expressing the targeted gene segments in antisense. WT, Mock and BMV (without inserting) plants were selected as the control groups. After 10 days of BMV infection, rice plants were treated with M. oryzae. (A) Rice blast disease phenotype in the control and silenced rice plants. Plants were treated with BMV vectors harboring candidate M. oryzae gene fragments. After 10 days of fungal infection, the 6th leaves had photographs taken. (B) Quantification of lesion density on rice plants treated with BMV vectors carrying the targeted gene fragments and controls. After 10 days of M. oryzae infection, lesions in leaves from 15 independent plants were counted. Furthermore, qRT-PCR analysis of the expression of MoABC1 (C) MoMAC1 (D) and MoPMK1 (E) in rice leaves inoculated with BMV variants, including S, sense; As, antisense; S + As, sense plus antisense, and BMV-Mix(as). Each experiment was performed with three biological repeats. Values represented mean ± SD in three independent experiments, and 15 individual leaves from different plants were collected for each experiment. Different letter indicated a significant difference at p < 0.05.
4. Discussion
Chemical mutagenesis and Agrobacterium T-DNA or transposons insertions are the most known strategies for studying loss-of-function in fungi, which have been widely used and are also a pivotal choice for the model fungi M. oryzae [37,38,39]. By contrast, RNAi is a reverse genetics tool for studying functional genomics that presents many advantages and avoids many disadvantages of traditional approaches to functional analyses in M. oryzae [24,30,35]: (1) it is rapid; (2) it can avoid time-consuming stable fungi transformation; and (3) it can work for the studies of M. oryzae. In addition, it has been proved that siRNA can across the species boundary between plants to fungus and silence the expression of fungal target genes [29,30]. The knowledge about siRNA has inspired us to develop an efficient, in planta system for studying the interaction of M. oryzae and its rice host via use of virus vector in rice to produce designed siRNA.
The prerequisite for such systems is a virus vector for efficient siRNA production in rice. Ding et al. [27] has developed a BMV-based system for the purpose of rice VIGS work, which is easy to operate and can systematically produce designed siRNA against the internal rice target genes. Nevertheless, it is essential to optimize Ding’s BMV system for the experimental conditions that could allow pathogenic invasion of both M. oryzae and BMV. In this study, 2-week-old rice plant (cultivar CO-39) at the two-leaf stage rice was used as the materials; the constant temperature and high humidity was applied to favor both BMV virus and fungal infection. Since secondary pathogen infection and pathogenic development of the fungus may be influenced by host defense [40], it is always a concern that BMV responding to host defense may intervene with the growth of host plant. It has been reported that BMV infection did not affect the susceptibility of maize plants to fungi [29,41]. Consistent with these results, we did not detect any effects on M. oryzae infection of BMV-inoculated plants: the defined virus titer in the experiments caused mild BMV-infected symptoms, but the symptoms of BMV-inoculated rice leaves had not been considered to influence the investigation of targeted fungal genes, depending on the assays of host PDS gene silencing, a practice that was implicated by Scofield and Nelson [42]. Fungal disease symptoms were observed much similar in both BMV-inoculated and control plants. These findings provided an important basis for the screening system of gene functions.
Previous studies have indicated that silencing signals stably produced from host plants can transfer into fungi from plants and inhibit the transcription of their corresponding genes in infecting fungi [22,30]. However, it is unknown if a transient expression system like BMV can produce sufficient silencing signals, and then transport into pathogens. It would dramatically facilitate the study of fungal and host plant interaction if the BMV system can possess a similar capability and be successful applied as the HIGS system. Three M. oryzae genes including MoABC1, MoMAC1 and MoPMK1 responsible for disease development were chosen in the study. The transcription of targeted M. oryzae gene fragments in rice using BMV vectors caused gene silencing of the targeted genes in the infecting fungi and blast disease suppression, indicating that the transfer of silencing signals from rice plants into M. oryzae cells may disturb the fungus–plant interaction.
Interestingly, the efficiency of gene silencing was distinctly different among the three fungal genes. Our results indicated that the silencing efficiency of MoMAC1 was greatly lower than MoABC1 and MoPMK1. The possible explanation was that the extent of gene silencing detected was associated with the expression patterns of the genes. MoPMK1 is involved in the regulation of appressorium formation and infectious hyphal growth in M. oryzae, and MoABC1 plays an essential role in protecting M. oryzae against plant defense mechanisms after penetrating epidermal cells of host plants by infectious hyphae. However, MoMAC1 gene is crucial for vegetative growth, conidiation, and conidial germination [42,43,44,45]. Thus, it was postulated that the silencing signal was transferred from host plant into fungi through infectious hyphae more easily, which led to the higher silencing efficiency of MoPMK1 and MoABC1. A common characteristic of the fungal genes for which silencing was observed obviously was that they were transcribed highly in the infectious hyphae than that in the fungal conidia of the infecting leaves. Therefore, based on the BMV-HIGS system, selection of certain pathogenic genes transcribed highly in the infectious hyphae may be beneficial to control rice blast disease effectively.
One of the universal concerns is whether VIGS inadvertently causes the suppression of non-target genes, so-called ‘off-target’ effects [46]. The sequences that had been selected as silencing inserts were searched against all available rice genomic database and the other pathogenic genes of M. oryzae, and no extensive homology was detected. In fact, no obviously aberrant phenotypes occurred in the rice plants with targeted siRNA molecules. Thus, careful selection of targeted gene sequences for the insertion of the BMV vectors may be crucial for achieving efficient and specific gene silencing. The success of pathogen gene suppression is dependent on the dynamic interplays between BMV spread and fungi infection.
The rice seedling starts to show significant reductions in internal gene expression after 3 days of BMV inoculation and the silencing effect persists until about 21 days post inoculation [27], indicating the period of BMV mediated siRNA production in rice plants. The window for the inoculation of rice plants with M. oryzae, therefore, was quite wide; 15 days after BMV infection was a proper period to wait before inoculating with M. oryzae to achieve gene silencing. We also found that, based on the BMV-HIGS system, the silencing efficiency of targeted M. oryzae genes was greater with the mixture of fungal genes in sense and antisense orientation than that in sense or antisense orientation alone, indicating that such self-complementary sequences significantly enhanced the capability of dsRNA generation to increase the efficiency of BMV silencing. Thus, these data can be applied to design more effective BMV vectors for the HIGS system.
5. Conclusions
Based on our data, the rice blast control was more successful when silencing more pathogenic genes in M. oryzae than silencing single genes. The improved understanding will be conducive to developing more effective tools against fungal infection. The results showed that some M. oryzae genes were markedly silenced using the HIGS system. We also proposed the BMV-HIGS as a novel strategy for studying gene function in M. oryzae, which owned the potential to control fungal disease. If gene silencing was confined to some particular cells, this may restrict the genes that can be assayed for their functions, and would be an increasing concern in designing constructs for engineering RNAi-triggered defenses. Taken together, the BMV-HIGS is a rapid process to promote the understanding of fungus biology and the pathogenicity of these pathogens during plant-fungus interaction.
Acknowledgments
This work was mainly supported by KWS SAAT SE and partially supported by the National Natural Science Foundation of China (31600210). The BMV vectors were provided by R. Nelson (Samuel Roberts Noble Foundation, Inc.) and the M. oryzae strain Guy11 was provided by Zhengyi Wang (College of Agriculture and Biotechnology, Zhejiang University). We are grateful to them for helpful advice.
Supplementary Materials
The following are available online at www.mdpi.com/2073-4425/8/10/241/s1. Figure S1: Effects of BMV infection on the growth of rice CO-39 plants. 2-week-old rice CO-39 plants were inoculated with BMV vector without inserting gene fragments for 15 days. Growth phenotypes of WT, Mock and BMV (without inserting) plants were monitored (A). In addition, fresh weight (B) and length (C) of shoots and roots from these plants was examined. Each experiment was performed with three biological repeats. Values represented mean ± SD in at least three independent sample collections (n = 10). Different letter indicated a significant difference at p < 0.05. Table S1: Primers used in this study.
Author Contributions
H.W., Z.W., J.Z. conceived and designed the experiment; L.Z. performed the experiment; C.Z. and Z.L. analyzed the data; L.Z. and H.W. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- 1.Couch B.C., Kohn L.M. A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae from M. grisea. Mycologia. 2002;94:683–693. doi: 10.1080/15572536.2003.11833196. [DOI] [PubMed] [Google Scholar]
- 2.Galhano R., Talbot N.J. The biology of blast: Understanding how Magnaporthe oryzae invades rice plants. Fungal Biol. Rev. 2011;25:61–67. doi: 10.1016/j.fbr.2011.01.006. [DOI] [Google Scholar]
- 3.Talbot N.J. On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 2003;57:177–202. doi: 10.1146/annurev.micro.57.030502.090957. [DOI] [PubMed] [Google Scholar]
- 4.Ceasar S.A., Ignacimuthu S. Genetic engineering of crop plants for fungal resistance: Role of antifungal genes. Biotechnol. Lett. 2012;34:995–1002. doi: 10.1007/s10529-012-0871-1. [DOI] [PubMed] [Google Scholar]
- 5.Li Y., Yue X., Que Y., Yan X., Ma Z., Talbot N.J., Wang Z. Characterization of four LIM protein-encoding genes involved in infection-related development and pathogenicity by the rice blast fungus Magnaporthe oryzae. PLoS One. 2014;9:e88246. doi: 10.1371/journal.pone.0088246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean R.A., Talbot N.J., Ebbole D.J., Farman M.L., Mitchell T.K., Orbach M.J., Thon M., Kulkarni R., Xu J.R., Pan H., et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- 7.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 8.Haywood V., Kragler F., Lucas W.J. Plasmodesmata: Pathways for protein and ribonucleoprotein signaling. Plant Cell. 2002;14:S303–S325. doi: 10.1105/tpc.000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klahre U., Crete P., Leuenberger S.A., Iglesias V.A., Meins F., Jr. High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proc. Natl. Acad. Sci. USA. 2002;99:11981–11986. doi: 10.1073/pnas.182204199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding B., Wang Y. Viroids: Uniquely simple and tractable models to elucidate regulation of cell-to-cell trafficking of RNA. DNA Cell. Biol. 2009;28:51–56. doi: 10.1089/dna.2008.0811. [DOI] [PubMed] [Google Scholar]
- 11.Tomilov A.A., Tomilova N.B., Wroblewski T., Michelmore R., Yoder J.I. Transspecific gene silencing between host and parasitic plants. Plant J. 2008;56:389–397. doi: 10.1111/j.1365-313X.2008.03613.x. [DOI] [PubMed] [Google Scholar]
- 12.Huang G., Allen R., Davis E.L., Baum T.J., Hussey R.S. Engineering broad rootknot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. USA. 2006;103:14302–14306. doi: 10.1073/pnas.0604698103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bakhetia M., Charlton W.L., Urwin P.E., McPherson M.J., Atkinson H.J. RNA interference and plant parasitic nematodes. Trends Plant Sci. 2008;10:362–367. doi: 10.1016/j.tplants.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Nunes C.C., Dean R.A. Host-induced gene silencing: A tool for understanding fungal host interaction and for developing novel disease control strategies. Mol. Plant Pathol. 2012;13:519–529. doi: 10.1111/j.1364-3703.2011.00766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vinay P., McCallum B., Jordan M., Loewen M., Fobert P., McCartney C., Bakkeren G. RNA silencing approaches for identifying pathogenicity and virulence elements towards engineering crop resistance to plant pathogenic fungi. CAB Rev. 2016;11:1–13. [Google Scholar]
- 16.Timmons L., Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 17.Huvenne H., Smagghe G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J. Insect Physiol. 2010;56:227–235. doi: 10.1016/j.jinsphys.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Nakayashiki H. RNA silencing in fungi: Mechanisms and applications. FEBS Lett. 2005;579:5950–5957. doi: 10.1016/j.febslet.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 19.Nakayashiki H., Kadotani N., Mayama S. Evolution and diversification of RNA silencing proteins in fungi. J. Mol. Evol. 2006;63:127–135. doi: 10.1007/s00239-005-0257-2. [DOI] [PubMed] [Google Scholar]
- 20.Nakayashiki H., Nguyen Q.B. RNA interference: Roles in fungal biology. Curr. Opin. Microbiol. 2008;11:494–502. doi: 10.1016/j.mib.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Tinoco M.L.P., Dias B., Dall’Astta R.C., Pamphile J.A., Aragão F.J.L. In vivo trans-specific gene silencing in fungal cells by in planta expression of a double-stranded RNA. BMC Biol. 2010;8:27. doi: 10.1186/1741-7007-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nowara D., Gay A., Lacomme C., Shaw J., Ridout C., Douchkov D., Hensel G., Kumlehn J., Schweizer P. HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell. 2010;22:3130–3141. doi: 10.1105/tpc.110.077040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purkayastha A., Dasgupta I. Virus-induced gene silencing: A versatile tool for discovery of gene functions in plants. Plant Physiol. Biochem. 2009;47:967–976. doi: 10.1016/j.plaphy.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Becker A., Lange M. VIGS-genomics goes functional. Trends Plant Sci. 2009;15:1–4. doi: 10.1016/j.tplants.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Senthil-Kumar M., Mysore K.S. New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 2011;16:656–665. doi: 10.1016/j.tplants.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 26.Yin C., Jurgenson J.E., Hulbert S.H. Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol. Plant Microbe Interact. 2011;24:554–561. doi: 10.1094/MPMI-10-10-0229. [DOI] [PubMed] [Google Scholar]
- 27.Ding X.S., Schneider W.L., Chaluvadi S.R., Mian M.A., Nelson R.S. Characterization of a Brome mosaic virus strain and its use as a vector for gene silencing in monocotyledonous hosts. Mol. Plant Microbe Interact. 2006;19:1229–1239. doi: 10.1094/MPMI-19-1229. [DOI] [PubMed] [Google Scholar]
- 28.De Jong W., Ahlquist P. Host-specific alterations in viral RNA accumulation and infection spread in a brome mosaic virus isolate with an expanded host range. J. Virol. 1995;69:1485–1492. doi: 10.1128/jvi.69.3.1485-1492.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Linde K., Kastner C., Kumlehn J., Kahmann R., Doehlemann G. Systemic virus-induced gene silencing allows functional characterization of maize genes during biotrophic interaction with Ustilago maydis. New Phytol. 2011;189:471–483. doi: 10.1111/j.1469-8137.2010.03474.x. [DOI] [PubMed] [Google Scholar]
- 30.Panwar V., McCallum B., Bakkeren G. Host-induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the Barley stripe mosaic virus. Plant Mol. Biol. 2013;81:595–608. doi: 10.1007/s11103-013-0022-7. [DOI] [PubMed] [Google Scholar]
- 31.Urban M., Bhargava T., Hamer J.E. An ATP-driven efflux pump is a novel pathogenicity factor in rice blast disease. EMBO J. 1999;18:512–521. doi: 10.1093/emboj/18.3.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi W., Dean R.A. The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell. 1997;9:1973–1983. doi: 10.1105/tpc.9.11.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu J.R., Hamer J.E. MAP kinase and cAMP signaling regulate infection structure formation, and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 1996;10:696–706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- 34.Wang H., Hao J., Chen X., Hao Z., Wang X., Lou Y., Peng Y., Guo Z. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol. Biol. 2007;65:799–815. doi: 10.1007/s11103-007-9244-x. [DOI] [PubMed] [Google Scholar]
- 35.Zhu L., Guo J., Zhu J., Zhou C. Enhanced expression of EsWAX1 improves drought tolerance with increased accumulation of cuticular wax and ascorbic acid in transgenic Arabidopsis. Plant Physiol. Biochem. 2014;75:24–35. doi: 10.1016/j.plaphy.2013.11.028. [DOI] [PubMed] [Google Scholar]
- 36.Pall G.S., Hamilton A.J. Improved northern blot method for enhanced detection of small RNA. Nat. Protocol. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 37.Rodríguez-Ortiz R., Michielse C., Rep M., Limón M.C., Avalos J. Genetic basis of carotenoid overproduction in Fusarium oxysporum. Fungal Genet. Biol. 2012;49:684–696. doi: 10.1016/j.fgb.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 38.Meng Y., Patel G., Heist M., Betts M.F., Tucker S.L., Galadima N., Donofrio N.M., Brown D., Mitchell T.K., Li L., et al. A systematic analysis of T-DNA insertion events in Magnaporthe oryzae. Fungal Genet. Biol. 2007;44:1050–1064. doi: 10.1016/j.fgb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Rashmi S., Maurya S., Upadhyay R.S. The improvement of competitive saprophytic capabilities of Trichoderma species through the use of chemical mutagens. Braz. J. Microbiol. 2016;47:10–17. doi: 10.1016/j.bjm.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tufan H.A., Stefanato F.L., McGrann G.R., MacCormack R., Boyd L.A. The Barley stripe mosaic virus system used for virus induced gene silencing in cereals differentially affects susceptibility to fungal pathogens in wheat. J. Plant Physiol. 2011;168:990–994. doi: 10.1016/j.jplph.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 41.van der Linde K., Doehlemann G. Utilizing virus-induced gene silencing for the functional characterization of maize genes during infection with the fungal pathogen Ustilago maydis. Methods Mol. Biol. 2013;975:47–60. doi: 10.1007/978-1-62703-278-0_4. [DOI] [PubMed] [Google Scholar]
- 42.Scofield S.R., Nelson R.S. Resources for virus-induced gene silencing in the grasses. Plant Physiol. 2009;149:152–157. doi: 10.1104/pp.108.128702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jansen M., Jarosch B., Schaffrath U. The barley mutant emr1 exhibits restored resistance against Magnaporthe oryzae in the hypersusceptible mlo-genetic background. Planta. 2007;225:1381–1391. doi: 10.1007/s00425-006-0447-1. [DOI] [PubMed] [Google Scholar]
- 44.Jansen M., Slusarenko A.J., Schaffrath U. Competence of roots for race-specific resistance and the induction of acquired resistance against Magnaporthe oryzae. Mol. Plant Pathol. 2006;7:191–195. doi: 10.1111/j.1364-3703.2006.00331.x. [DOI] [PubMed] [Google Scholar]
- 45.Liu X.H., Lin F.C. Investigation of the biological roles of autophagy in appressorium morphogenesis in Magnaporthe oryzae. J. Zhejiang Univ. Sci. B. 2008;9:793–796. doi: 10.1631/jzus.B0860013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.