Abstract
The mitogen-activated protein kinase (MAPK) cascade is a universal signal transduction module that plays a vital role in regulating growth and development, as well as environmental stress responses in plants. Wheat is one of the most important crops worldwide. Although the MAPK kinase kinase (MAP3K) family in wheat has been investigated, the MAPK and MAPK kinase (MAP2K) gene families remain unknown at present. Here, 54 MAPK and 18 MAPKK genes were identified in wheat using recent genomic information. Phylogenetic analysis of Triticum aestivum L. MAPKs and MAPKKs (TaMAPKs and TaMAPKKs) together with homologous genes from other species classified them into four groups, and the clustering was consistent with the genomic exon/intron structures. Conserved motif analysis found that MAPK proteins contained a typical TXY phosphorylation site and MAPKK proteins contained an S/T-X5-S/T motif. RNA-seq data mapping analysis showed that MAPK and MAPKK genes in group IV had tissue-specific expression profiles, whereas each group I member showed relatively high expression in all organs. Expression patterns of TaMAPK and TaMAPKK genes under stress conditions were also investigated and stress-responsive candidates were identified. Co-expression network analysis identified 11 TaMAPK genes and 6 TaMAPKK genes involved in the interaction network pathway. Overall, this study provided useful information for evolutionary and functional surveys of MAPK and MAPKK gene families in wheat and beyond.
Keywords: abiotic stress, co-expression network, expression profiles, Triticum aestivum
1. Introduction
Wheat is one of the most important cereal crops worldwide, and provides about 30% of the staple food source for humankind [1,2]. Various abiotic stresses, including salt, drought and cold, are major limiting factors to wheat production. Improving the tolerance of wheat in response to diverse stresses holds promise for addressing the ever-increasing human population as well as the threat of climate change.
The self-protection mechanisms of plants activate numerous genes during enduring abiotic stress [3]. Mitogen-activated protein kinase (MAPK) cascade genes code for well-conserved proteins that function as key signal transduction components, consisting of three MAPK members activated by sequential phosphorylation, namely (MAPK kinase kinase) MAP3K- (MAPK kinase) MAP2K-MAPK [4]. Extensive studies have revealed that the MAPKs have indispensable regulatory roles in response to abiotic stresses in plants. MAPK3 and MAPK6 play a critical role in promoting cell division in integuments specifically during ovule development as well as being involved in abiotic stress response [5]. MAPK4, MAPK5, MAPK6 and MAPK7 were specifically induced by abiotic stress treatments such as salt, drought and low temperature in maize [6]. GhMAPK16 might be relevant to abiotic stress signal transduction pathways and its over-expression in Arabidopsis led to significant tolerance to drought stress [7]. The MAPK cascade gene family has been well studied in rice, maize and Brachypodium distachyon [8,9,10]. In wheat, the genome organization, evolutionary features and expression profiles of the MAP3K gene family has also been systematically studied [4]. However, information on wheat MAPK and MAPKK gene families is not well-understood, especially those families involved in abiotic stress response.
In this study, 54 MAPK and 18 MAPKK genes were identified in wheat genome based on a bioinformatics search. The phylogenetic tree, conserved motifs, gene expression pattern and regulatory network of these MAPKs and MAPKKs were further systematically analyzed. Phylogenetic tree and conserved motifs analysis of wheat MAPK and MAPKK families classified them into four groups. Additionally, the expression patterns of 54 MAPK and 18 MAPKK genes under abiotic stress or in various tissues were comprehensively investigated by RNA-seq analyses. Finally, the interaction network of putative wheat MAPK and MAPKK genes was also investigated. Overall, this study provided basic information about the genomic organization of MAPK and MAPKK genes in wheat, which will facilitate further functional studies.
2. Materials and Methods
2.1. Identification of MAPK and MAPKK Genes in Wheat
Potential members of wheat MAPK and MAPKK gene families were identified according to the method described by Wang et al [4]. All available protein sequences for wheat genotype Chinese Spring (CS42) were downloaded from the Ensemble database [11]. The available MAPK and MAPKK genes in Arabidopsis thaliana, Oryza sativa and B. distachyon were used to construct an hidden Markov model (HMM) profile using the hmm-build tool embedded in HMMER 3.0, and then the HMM profile was used to search wheat proteins using the hmmsearch tool embedded in HMMER 3.0 [12]. Conserved domains of wheat MAPK and MAPKK members were confirmed by PFAM [13] and InterProScan database [14]. Finally, all sequences obtained were verified by a BLASTN (Nucleotide BLAST) similarity search against wheat expressed sequence tags (ESTs) deposited in the National Center for Biotechnology Information NCBI database. The theoretical pI (isoelectric point) and Mw (molecular weight) of candidate genes were calculated using compute pI/Mw tool online (http://web.expasy.org/compute_pi/). Subcellular localization of these genes was predicted using the CELLO v2.5 web server [15].
2.2. Multiple Alignments and Phylogenetic Analysis
Multiple sequence alignments were performed using ClustalW [16]. An unrooted phylogenetic tree was generated by MEGA 6.0 software using the neighbor-joining method and the bootstrap test method with 1000 replications [17]. The MEME program [18] was used to predict the conserved motifs of TaMAPKs and TaMAPKKs.
2.3. Analysis of the Expression Profiles of MAPK and MAPKK Genes by RNA-seq Datasets
To study the expression of MAPK and MAPKK genes in different organs and response to stresses, a total of 28 RNA-seq datasets of wheat variety Chinese Spring obtained from WHEAT URGI (https://urgi.versailles.inra.fr/files/RNASeqWheat/) and NCBI Sequence Read Archive (SRA) database were used to investigate differential expression of TaMAPK and TaMAPKK genes (Table S1). TopHat and Cufflinks software were used to analyze gene expression based on these RNA-seq data with the default parameters [19]. The FPKM (fragments per kilobase of transcript per million fragments mapped) value was calculated for each gene, and the log10-transformed (FPKM + 1) values of MAPK and MAPKK genes were used to generate heat maps.
2.4. Co-expression Network Analysis
Correlation networks are increasingly being used in bioinformatics applications. The WGCNAR_1.49 package was used to construct the co-expression network of wheat MAPK and MAPKK genes by performing weighted correlation network analysis of RNA-seq data [20].
3. Results and Discussion
3.1. Genome-wide Identification of the MAPK and MAPKK Gene Families in Wheat
MAPK and MAPKK gene families are the important protein kinases involved in many physiological processes [21]. In order to identify members of the two families, we performed a HMM search and a total of 54 non-redundant MPAK and 18 MAPKK genes were identified in the wheat genome (Table 1). The predicted genes in the two families were named TaMAPK1 to TaMAPK54 and TaMAPKK1 to TaMAPKK18, respectively. The number of MAPK genes in wheat (54) was much higher than in rice (16), maize (19) and B. distachyon (16) [8,9,10]. The number of TaMAPKK (18) was also slightly higher than in B. distachyon (12) and cucumber (14) [22]. The locations of both groups of MAPK cascade genes were not random across the wheat chromosomes. Sixteen of the TaMAPK genes were located on homoeologous group 7 chromosomes, 14 were located in group 1 chromosomes, and 9, 7, 6 and 2 were in groups 6, 3, 4 and 5, respectively. There was no MAPK gene on a group 2 chromosome. The locations of TaMAPKK genes differed from TaMAPKs. Homologous group 4 contained the most TaMAPKK genes, at 10, followed by group 5 with 5 TaMAPKKs, then groups 3 and 6, harboring 2 and 1, respectively. No TaMAPKK gene was present in groups 1, 2 and 7.
Table 1.
Characteristics of putative wheat mitogen-activated protein kinase (MAPK) and MAPK kinase (MAPKK) genes.
| Gene Family | Gene | pI | Mw (kDa) |
Amino Acid Length | EST Count | Ensemble Wheat Gene ID | Subcellular Location |
|---|---|---|---|---|---|---|---|
| MAPK | TaMAPK1 | 9.71 | 51.6 | 460 | 25 | Traes_6BL_BF59BFB93 | Periplasmic |
| TaMAPK2 | 5.89 | 50.3 | 440 | 10 | Traes_4AL_6F3D0ACCA | Cytoplasmic | |
| TaMAPK3 | 8.83 | 48.4 | 421 | 2 | Traes_7AL_F5620757F | Cytoplasmic | |
| TaMAPK4 | 8.85 | 62.8 | 553 | 24 | Traes_1DS_66BD773BF | Cytoplasmic | |
| TaMAPK5 | 8.51 | 59.1 | 521 | 1 | Traes_1DL_52D511CDD | Cytoplasmic | |
| TaMAPK6 | 9.48 | 57.5 | 519 | 12 | Traes_1BL_7FA80EF00 | Periplasmic | |
| TaMAPK7 | 8.34 | 51.9 | 450 | 1 | Traes_7DL_FE0ECD387 | Cytoplasmic | |
| TaMAPK8 | 8.94 | 68.2 | 598 | 33 | Traes_3DL_5D82311EA | Cytoplasmic | |
| TaMAPK9 | 6.41 | 36.3 | 315 | 35 | Traes_4BS_9285C0809 | Cytoplasmic | |
| TaMAPK10 | 5.7 | 31.2 | 272 | 22 | Traes_6BS_17C1E5829 | Cytoplasmic | |
| TaMAPK11 | 8.74 | 62.3 | 549 | 26 | Traes_1AS_C9CFD7AC8 | Cytoplasmic | |
| TaMAPK12 | 8.78 | 26.7 | 239 | 19 | Traes_7AL_4AF13CC8B | Cytoplasmic | |
| TaMAPK13 | 9.71 | 51.6 | 460 | 28 | Traes_6DL_F48A5E31E | Periplasmic | |
| TaMAPK14 | 5.75 | 42.9 | 377 | 36 | Traes_1AL_89DDB4243 | Cytoplasmic | |
| TaMAPK15 | 5.46 | 42.8 | 369 | 61 | Traes_4BL_2CEFDE904 | Cytoplasmic | |
| TaMAPK16 | 9.32 | 67.2 | 586 | 22 | TRAES3BF058500020CFD_g | Periplasmic Cytoplasmic | |
| TaMAPK17 | 8.2 | 31.4 | 275 | 0 | Traes_6BS_2D0054D1F | InnerMembrane Cytoplasmic | |
| TaMAPK18 | 9.51 | 44.3 | 382 | 9 | Traes_7DS_C268073F4 | Periplasmic Cytoplasmic | |
| TaMAPK19 | 6.13 | 25.5 | 218 | 5 | Traes_7DS_1D8A8BFA2 | Cytoplasmic | |
| TaMAPK20 | 7.05 | 65.1 | 578 | 57 | Traes_7DL_FB75EA9C3 | Cytoplasmic | |
| TaMAPK21 | 8.82 | 30.8 | 269 | 22 | Traes_7DL_99CBA4440 | Cytoplasmic | |
| TaMAPK22 | 7.19 | 42.7 | 373 | 25 | Traes_7DL_73DF29BF0 | Cytoplasmic | |
| TaMAPK23 | 7.24 | 47.9 | 417 | 1 | Traes_3DL_281B6BCBF | Cytoplasmic | |
| TaMAPK24 | 9.18 | 33.4 | 289 | 9 | Traes_3DL_38B762939 | Cytoplasmic | |
| TaMAPK25 | 5.46 | 42.8 | 369 | 61 | Traes_4DL_15045954F | Cytoplasmic | |
| TaMAPK26 | 5.55 | 50.1 | 440 | 10 | Traes_5DL_8DC610F26 | Cytoplasmic | |
| TaMAPK27 | 8.92 | 62.4 | 549 | 25 | Traes_1BS_26C55B2B1 | Cytoplasmic | |
| TaMAPK28 | 5.75 | 42.9 | 376 | 36 | Traes_1BL_C5CD09285 | Cytoplasmic | |
| TaMAPK29 | 8.99 | 52.5 | 457 | 2 | Traes_1BL_9360D8CEC | Cytoplasmic | |
| TaMAPK30 | 6.5 | 55.4 | 484 | 63 | Traes_4AS_5015DF7A2 | Cytoplasmic | |
| TaMAPK31 | 9.5 | 54.9 | 485 | 66 | Traes_4AL_E432524A0 | Periplasmic | |
| TaMAPK32 | 5.19 | 32.8 | 280 | 28 | Traes_1DL_319058D6B | Cytoplasmic | |
| TaMAPK33 | 9.25 | 36.3 | 321 | 1 | Traes_1DL_8C6B737E9 | Periplasmic | |
| TaMAPK34 | 9.01 | 61.0 | 539 | 27 | Traes_1DL_73AAC8631 | Cytoplasmic | |
| TaMAPK35 | 6.36 | 57.2 | 500 | 0 | Traes_6AS_50BE5D59F | Cytoplasmic | |
| TaMAPK36 | 7.2 | 43.5 | 380 | 22 | Traes_6AS_8225741A6 | Cytoplasmic | |
| TaMAPK37 | 9.71 | 51.5 | 460 | 28 | Traes_6AL_BE97161B9 | Periplasmic | |
| TaMAPK38 | 8.27 | 24.1 | 207 | 17 | Traes_3AL_F88B0A8E6 | Cytoplasmic | |
| TaMAPK39 | 8.99 | 35.5 | 307 | 21 | Traes_3AL_669FE0293 | Cytoplasmic | |
| TaMAPK40 | 6.2 | 21.4 | 184 | 1 | Traes_3AL_532CA9EE7 | Cytoplasmic | |
| TaMAPK41 | 9.14 | 61.7 | 543 | 3 | Traes_1AL_30C4B017F | Cytoplasmic | |
| TaMAPK42 | 8.52 | 56.4 | 496 | 2 | Traes_1AL_14DCD6020 | Cytoplasmic | |
| TaMAPK43 | 9.5 | 51.6 | 467 | 7 | Traes_1AL_A7173BBAE | Periplasmic | |
| TaMAPK44 | 6.48 | 57.5 | 500 | 1 | Traes_6DS_6424C38F2 | Cytoplasmic | |
| TaMAPK45 | 7.63 | 43.8 | 382 | 21 | Traes_6DS_A61864DB2 | Cytoplasmic | |
| TaMAPK46 | 9.46 | 55.1 | 484 | 60 | Traes_7AS_E1135D559 | Periplasmic Cytoplasmic | |
| TaMAPK47 | 8.74 | 26.9 | 232 | 0 | Traes_7AS_71B4C13A6 | Cytoplasmic | |
| TaMAPK48 | 5.31 | 31.9 | 275 | 8 | Traes_7AS_74B19D5B6 | Cytoplasmic | |
| TaMAPK49 | 6.55 | 31.6 | 276 | 15 | Traes_7AL_81A545A54 | Cytoplasmic | |
| TaMAPK50 | 6.04 | 50.4 | 443 | 10 | Traes_5BL_A39EC2FCD | Cytoplasmic | |
| TaMAPK51 | 5.31 | 31.9 | 275 | 8 | Traes_7BS_F5BB0A116 | Cytoplasmic | |
| TaMAPK52 | 8.5 | 51.5 | 449 | 2 | Traes_7BL_44A22C7FC | Cytoplasmic | |
| TaMAPK53 | 6.55 | 48.3 | 426 | 52 | Traes_7BL_992E1F443 | Cytoplasmic | |
| TaMAPK54 | 7.2 | 43.2 | 377 | 29 | Traes_7BL_95B1705A2 | Cytoplasmic | |
| MAPKK | TaMAPKK1 | 5.91 | 20.7 | 188 | 4 | Traes_6DL_8D2D914D7 | Periplasmic Cytoplasmic |
| TaMAPKK2 | 7.88 | 53.5 | 480 | 0 | Traes_5BL_BB574E77D | Cytoplasmic | |
| TaMAPKK3 | 5.37 | 41.9 | 377 | 3 | Traes_5DL_0886CB561 | Cytoplasmic | |
| TaMAPKK4 | 5.89 | 33.6 | 299 | 1 | Traes_5AL_5EFCEDAFB | Cytoplasmic | |
| TaMAPKK5 | 7.7 | 36.2 | 337 | 0 | Traes_4BS_EE048CC01 | Cytoplasmic | |
| TaMAPKK6 | 8.95 | 36.2 | 343 | 0 | Traes_4BS_59F3F68D1 | Periplasmic Cytoplasmic | |
| TaMAPKK7 | 8.28 | 31.8 | 303 | 0 | Traes_4BS_84D913A2E | Outer Membrane Periplasmic Cytoplasmic | |
| TaMAPKK8 | 8.62 | 35.9 | 334 | 0 | Traes_4BS_BDB56BC90 | Cytoplasmic | |
| TaMAPKK9 | 7.72 | 36.3 | 338 | 0 | TRAES3BF024700110CFD_g | Cytoplasmic | |
| TaMAPKK10 | 7.72 | 36.3 | 338 | 0 | TRAES3BF024900030CFD_g | Cytoplasmic | |
| TaMAPKK11 | 6.9 | 36.2 | 336 | 0 | Traes_4DS_7F2F2671B | Cytoplasmic | |
| TaMAPKK12 | 8.82 | 35.4 | 336 | 0 | Traes_4DS_7A016C2E4 | Periplasmic Cytoplasmic | |
| TaMAPKK13 | 6.69 | 71.6 | 647 | 1 | Traes_5DL_F89F21E65 | Cytoplasmic | |
| TaMAPKK14 | 7.15 | 36.1 | 337 | 0 | Traes_4AL_75E7BE9EE | Cytoplasmic | |
| TaMAPKK15 | 8.21 | 36.3 | 337 | 0 | Traes_4AL_429683D72 | Cytoplasmic | |
| TaMAPKK16 | 8.36 | 30.3 | 283 | 0 | Traes_4AL_B7C432896 | Cytoplasmic | |
| TaMAPKK17 | 8.88 | 35.6 | 337 | 0 | Traes_4AL_84DFF6A541 | Cytoplasmic | |
| TaMAPKK18 | 5.64 | 53.8 | 476 | 3 | Traes_5BL_EC9896AB3 | Cytoplasmic |
EST, expressed sequence tag; pI, isoelectric point; Mw, molecular weight
To support the actual existence of these putative genes, we performed a BLASTN search against the wheat EST and UniGene database using the MAPK and MAPKK genes as queries (Table 1). Fifty-one MAPK genes and 5 MAPKK genes had EST support, representing 94.4% and 27.8% of the predicted genes, respectively. We speculated that the TaMAPKs and TaMAPKKs with no EST support might have very low expression that could not be detected experimentally, or was not expressed under any conditions we used. The lengths of putative TaMAPK proteins ranged from 184 to 598 amino acids, with putative molecular weights (Mw) ranging from 21.4 to 68.2 kDa and theoretical pI ranging from 5.19 to 9.17, respectively. The lengths of TaMAPKK proteins ranged from 188 to 647 amino acids, Mw from 20.7 to 71.6 kDa and pI from 5.37 to 8.95, respectively. Subcellular localization analysis found that most of TaMAPKs and TaMAKKs were localized in cytoplasmic (Table 1).
3.2. Multiple Alignments, Phylogenetic and Conserved Motif Analysis of TaMAPKs and TaMAPKKs
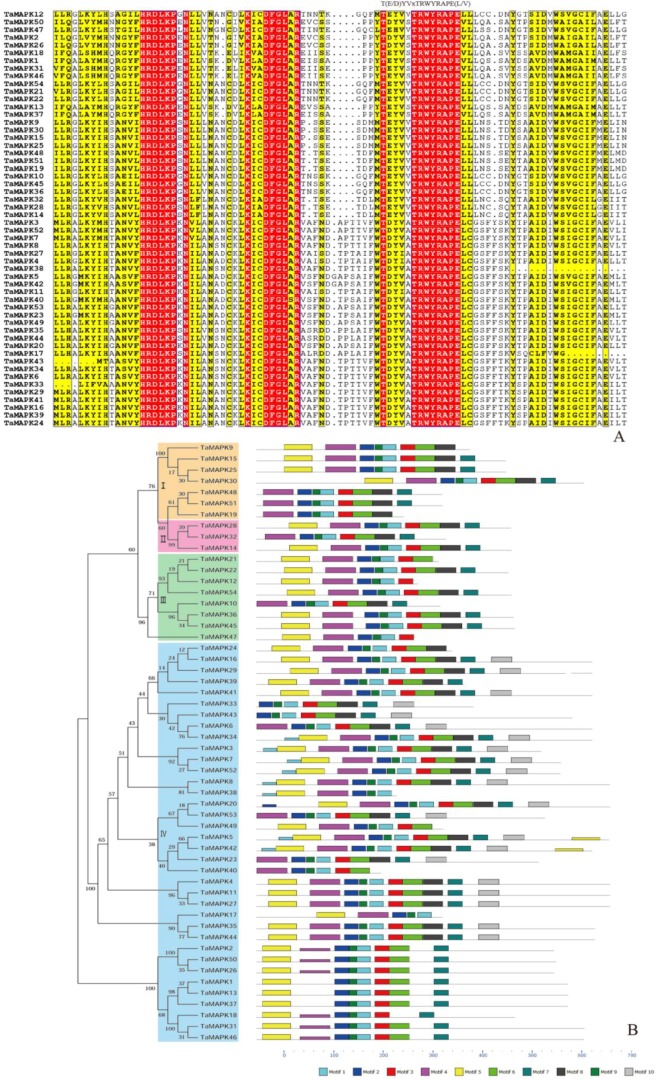
To further evaluate the phylogenetic relationships of the wheat MAPK and MAPKK genes, the full-length protein sequences of the 54 TaMAPKs and 18 TaMAPKKs were aligned using ClustalW software [16] and phylogenetic trees were constructed using the neighbor joining (NJ) method in MEGA 6.0 [17]. As shown in Figure 1A, the sequences of the TaMAPK genes in the motif regions were highly conserved. Through phylogenetic analyses relative to homologs in Arabidopsis and cucumber, we classified the TaMAPKs into four groups (Figure 1B), each group including 7 (Group I), 3 (II), 8 (III), 36 (IV) members. The results indicated evolutionary differences in wheat MAPK genes, consistent with those in B. distachyon [8]. Each MAPK cluster classified by phylogenetic analysis shared similar conserved motif compositions. A total of ten motifs were identified in TaMAPK proteins. Motif TXY is an important phosphorylation site for MAPK activation. Groups I-III have a TEY motif in their activation loops, except TaMAPK47 where THE replaced the TEY. The THE motif was not found in rice, maize, M. domestica or B. distachyon [9,10,23,24] suggesting that it might be wheat-specific.
Figure 1.
Phylogenetic relationship and conserved motifs composition of the wheat MAPK family. (A) Multiple alignments to identify the conserved signature motif T(E/D)YVxTRWYRAPE(L/V) in the protein sequences of wheat MAPK family; (B) Phylogenetic analysis (left) and conserved domain (right) compositions of wheat MAPK family. The unrooted phylogenetic tree was constructed using MEGA 6.0 tool with the neighbor-joining method and 1,000 bootstrap replications. Conserved motifs were predicated by MEME tool and each motif was represented with different colored boxes.
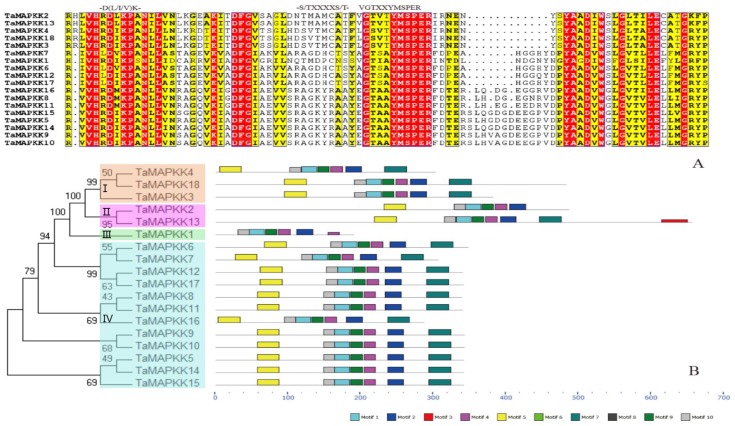
As shown in Figure 2A, members of the TaMAPKK family also clustered into four groups. Groups I, II and III contained a S/T-X5-S/T motif, which functions as a phosphorylation site, but was not present in Group IV. This motif is not only found in rice, Arabidopsis and B. distachyon, but also in some animals and fungi [24]. Most interestingly, the MAPKKs that include the S/T-X5-S/T motif were highly expressed in many tissues or responded to multiple stresses.
Figure 2.
Phylogenetic relationship and conserved motifs compositions of the wheat MAPKK family. (A) Multiple alignments and conserved signature motifs found in the protein sequences of wheat MAPKK family; (B) Phylogenetic analysis (left) and conserved domain compositions (right) in wheat MAPKK family. The unrooted phylogenetic tree was constructed by MEGA 6.0 using the neighbor-joining method and the bootstrap test method with 1000 replications. Conserved motifs were predicated by MEME tool and each motif was represented with different colored boxes.
3.3. Tissue-Specific Expression Patterns of TaMAPK and TaMAPKK Genes
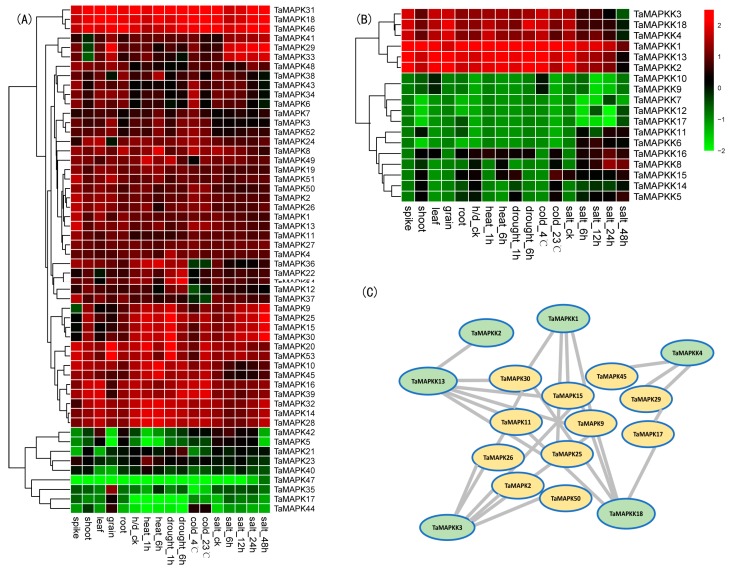
The expression patterns of TaMAPKs in various wheat tissues were investigated using RNA-seq data for five tissues. As shown in Figure 3A, the heat map indicates that all 54 detected genes were involved in various biological processes and were expressed in the majority of wheat tissues, but their expression levels were highly variable. TaMAPK14, 18, 28, 31, 32 and 46 were expressed at high levels in all five tissues. Some TaMAPKs were highly expressed in specific tissues. For example, TaMAPK17 and 35 were highly expressed in developing grain, suggesting that those genes may play indispensable roles in grain development. TaMAPK5 and 42 were relatively highly expressed in leaf and root tissues. TaMAPKs highly expressed in specific organs may have specific roles in growth and development of the corresponding tissues. It is noteworthy that tissue expression specificity of MAPK genes has been reported in B. distachyon, maize and cucumber [9,15,24]. All members of group II were highly expressed in multiple tissues, consistent with CsMAPK13 in cucumber and BdMAPK11 in B. distachyon [15,24].
Figure 3.
Expression patterns and interaction network of MAPK/MAPKK genes in wheat. (A) Relative expression levels of MAPK genes in various tissues and under different abiotic stresses determined using RNA-seq. Red represents increased and green represents decreased expression level; (B) Relative expression level of wheat MAPKK genes in various tissues and under abiotic stresses determined using RNA-seq; (C) The interaction network of MAPK/MAPKK genes in wheat.
Expression profiles of TaMAPKK genes in different tissues showed that most genes were expressed in five different tissues. Interestingly, MAPKKs with the S/T-X5-S/T motif show high expression in specific tissues (Figure 3B). For example, TaMAPKK9 and TaMAPKK10 showed relatively higher expression in leaves than in other tissues, indicating that they positively controlled leaf development. Members of Groups I, II and III with S/T-X5-S/T motifs had higher expression in all organs, consistent with BdMAPKK3 and BdMAPKK6 in B. distachyon and suggesting that this motif was an important phosphorylation site in relation to gene expression level [24]. Conserved domains identification and analysis may facilitate identification of functional units in these kinase genes and accelerate an understanding of their roles in plant growth and development as well as stress response.
3.4. Expression Patterns of TaMAPK and TaMAPKK Genes under Abiotic Stress
There is increasing data showing that the MAPK and MAPKK gene families have key roles in determining response to abiotic stresses [21]. For example, ZmMAPK1, GhMAPK4 and SiMAPK7 were reported to regulate abiotic stress response [25,26,27]. We analyzed the expression patterns of TaMAPK genes to determine whether they were responsive to heat, drought, cold or salt stress. As shown in Figure 3A, 84% and 16% TaMAPKs displayed up- or down-regulation following stress treatments. TaMAPK38 was up-regulated after 6 h of heat stress, whereas TaMAPK5 and TaMAPK42 were suppressed. TaMAPK10 and TaMAPK45 were suppressed whereas TaMAPK29, TaMAPK33 and TaMAPK41 were induced by salt stress. This was consistent with previous research reporting that TaMAPK4 was strongly induced under high salinity stress [28], and that over-expression of OsMAPK33 in rice led to enhanced salt sensitivity [29]. However, multiple numbers of clustered genes, such as TaMAPK32, TaMAPK14 and TaMAPK28, had similar expression profiles under abiotic stress, indicating that these genes might have analogous physiological functions. The expression levels of clustered genes were not significantly altered by stress treatment as also found in B. distachyon [24]. Finally, the TaMAPK47 gene was specifically expressed after 24 h of salt treatment, indicating it might be a salt-responsive candidate. Genes that are expressed under abiotic stress conditions may be the specific genes responsible for regulation of stress response and tolerance. Compared with TaMAPK genes TaMAPKKs show more significant differences under various stress conditions. For example, transcripts of TaMAPKK8 and TaMAPKK15 were down-regulated under heat treatment for 1 h, but were induced after 6 h. This was contrary to reports for SbMAPKK in S. brachiata in which there was up-regulation followed by down-regulation under dehydration treatment [30]. TaMAPKK12 and 16 were induced and TaMAPKK7 and 17 were reduced under all stress treatments, whereas other genes were reduced or induced following just one or two stress treatments. TaMAPKK5 and TaMAPKK14 showed opposite expression patterns following heat and salt treatments, suggesting that functional differentiation had occurred in these genes and that they were involved in regulating different stress signaling pathways. In summary, wheat MAPK/MAPKK genes likely play vital roles in different biological process and various abiotic stresses.
3.5. Interactions between TaMAPK and TaMAPKK Family Members
In general, MAPK cascade kinase genes form conserved signaling modules, involving three functionally linked protein kinases: MAPKs, MAPKKs and MAP3Ks [8]. In this study, we constructed the regulatory network of the MAPK, MAPKK cascade based upon different tissues and stress treatments in wheat. The WGCNAR package [20] provides a comprehensive set of functions for performing weighted correlation network analysis by RNA-seq data. As shown in Figure 3C, 6 MAPKK genes and 11 MAPK genes were involved in a single cascade. The 6 MAPKK genes contained S/T-X5-S/T motifs showed high expression levels. Furthermore, 4 of the 11 MAPK genes in Group I, including TaMAPK9, TaMAPK15, TaMAPK25 and TaMAPK30, showed close associations with MAPKK genes. In Arabidopsis, Group I members AtMAPK3 and AtMAPK6 play important roles in anther cell differentiation and normal anther lobe formation, and AtMAPK6 is involved in seed formation and the modulation of lateral and primary root development [5,31], suggesting that TaMAPK9, TaMAPK15, TaMAPK25 and TaMAPK30 might be involved in the regulatory network that controls organ differentiation or development. The constructed interaction network between TaMAPK and TaMAPKK provides information for further studies on MAPK regulatory pathways in wheat and other species.
Acknowledgments
This work was mainly supported by the State Key Project of Research and Development Plan of China (Grant No. 2016YFD0101004 and 2016YFD0100302), and partially supported by the Fundamental Research Funds for the Central Universities of China (Grant No. 2452015003).
Supplementary Materials
The following can be found online at www.mdpi.com/2073-4425/8/10/284/s1, Table S1. Accession number and sample information of RNA-seq datasets used in this study.
Author Contributions
H.Z. and H.Y. performed all experiments, and prepared the paper. X.Z. contributed to data collection. M.W. contributed to data analysis. W.S. provided experimental coordination and reviewed the manuscript. X.N. conceived the study and revised the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Mayer K.F.X., Rogers J., Doležel J., Pozniak C., Eversole K., Feuillet C., Gill B., Friebe B., Lukaszewski A.J., Sourdille P., et al. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- 2.Oono Y., Kobayashi F., Kawahara Y., Yazawa T., Handa H., Itoh T., Matsumoto T. Characterisation of the wheat (Triticum aestivum L.) transcriptome by de novo assembly for the discovery of phosphate starvation-responsive genes: Gene expression in Pi-stressed wheat. BMC Genom. 2013;14:77. doi: 10.1186/1471-2164-14-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rong W., Qi L., Wang A., Ye X., Du L., Liang H., Xin Z., Zhang Z. The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 2014;12:468. doi: 10.1111/pbi.12153. [DOI] [PubMed] [Google Scholar]
- 4.Wang M., Yue H., Feng K., Deng P., Song W., Nie X. Genome-wide identification, phylogeny and expressional profiles of mitogen activated protein kinase kinase kinase (MAPKKK) gene family in bread wheat (Triticum aestivum L.) BMC Genom. 2016;17:668. doi: 10.1186/s12864-016-2993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H., Liu Y., Bruffett K., Lee J., Hause G., Walker J.C., Zhang S. Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell. 2008;20:602–613. doi: 10.1105/tpc.108.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu T., Kong X.P., Zong X.J., Li D.P., Li D.Q. Expression analysis of five maize MAP kinase genes in response to various abiotic stresses and signal molecules. Mol. Biol. Rep. 2011;38:3967–3975. doi: 10.1007/s11033-010-0514-3. [DOI] [PubMed] [Google Scholar]
- 7.Shi J., Zhang L., An H. GhMPK16, a novel stress-responsive group D MAPK gene from cotton, is involved in disease resistance and drought sensitivity. BMC Mol. Biol. 2011;12:1–36. doi: 10.1186/1471-2199-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang M., Feng W., Cao J., Li P., She J., Chu Z. Genome-wide exploration of the molecular evolution and regulatory network of mitogen-activated protein kinase cascades upon multiple stresses in Brachypodium distachyon. BMC Genom. 2015;16:228. doi: 10.1186/s12864-015-1452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y., Zhang D., Wang L., Li D. Genome-wide analysis of mitogen-activated protein kinase gene family in maize. Plant Mol. Biol. Rep. 2013;31:1446–1460. doi: 10.1007/s11105-013-0623-y. [DOI] [Google Scholar]
- 10.Liu Q., Xue Q. Computational identification and phylogenetic analysis of the MAPK gene family in Oryza sativa. Plant Physiol. Biochem. 2007;45:6. doi: 10.1016/j.plaphy.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Bolser D.M., Kerhornou A., Walts B., Kersey P. Triticeae resources in ensembl plants. Plant Cell Physiology. 2015;56:e3. doi: 10.1093/pcp/pcu183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wheeler T.J., Eddy S.R. NHMMER: DNA homology search with profile HMMs. Bioinformatics. 2013;29:2487–2489. doi: 10.1093/bioinformatics/btt403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., et al. The PFAM protein families database: towards a more sustainable future. Nucl. Acids Research. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zdobnov E.M., Apweiler R. InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–850. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 15.Yu C.S., Chen Y.C., Lu C.H., Hwang J.K. Prediction of protein subcellular localization. Proteins. 2006;64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- 16.Larkin M., Blackshields G., Brown N., Chenna R., McGettigan P., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 17.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evo. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey T.L., Bodén M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J.Y., Li W.W., Noble W.S. MEME SUITE: Tools for motif discovery and searching. Nucl. Acid. Research. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2014;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langfelder R.B.P., Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong X., Pan J., Zhang D., Jiang S., Cai G., Wang L., Li D. Identification of mitogen-activated protein kinase kinase gene family and MKK–MAPK interaction network in maize. Biochem. Biophys. Res. Commun. 2013;441:964–969. doi: 10.1016/j.bbrc.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Wang J., Pan C., Wang Y., Ye L., Wu J., Chen L., Zou T., Lu G. Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genom. 2015;16:386. doi: 10.1186/s12864-015-1621-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S., Xu R., Luo X., Jiang Z., Shu H. Genome-wide identification and expression analysis of the MAPK and MAPKK gene family in Malus domestica. Gene. 2013;531:377–387. doi: 10.1016/j.gene.2013.07.107. [DOI] [PubMed] [Google Scholar]
- 24.Chen L., Hu W., Tan S., Wang M., Ma Z., Zhou S., Deng X., Zhang Y., Huang C., Yang G., et al. Genome-wide identification and analysis of MAPK and MAPKK gene families in Brachypodium distachyon. PLoS ONE. 2012;7:e46744. doi: 10.1371/journal.pone.0046744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu L., Yan J., Yang Y., He L., Zhu W. Enhanced tolerance to chilling stress in tomato by overexpression of a mitogen-activated protein kinase, SlMPK7. Plant Mol. Biol. Rep. 2006;34:76–88. doi: 10.1007/s11105-015-0897-3. [DOI] [Google Scholar]
- 26.Wu L., Zu X., Zhang H., Wu L., Xi Z., Chen Y. Overexpression of ZmMAPK1 enhances drought and heat stress in transgenic Arabidopsis thaliana. Plant Mol. Biol. 2015;88:429–443. doi: 10.1007/s11103-015-0333-y. [DOI] [PubMed] [Google Scholar]
- 27.Wang N.N., Zhao L.L., Lu R., Li Y., Li X.B. Cotton mitogen-activated protein kinase4 (GhMPK4) confers the transgenic Arabidopsis hypersensitivity to salt and osmotic stresses. Plant Cell Tissue Organ Cult. 2015;123:619–632. doi: 10.1007/s11240-015-0865-5. [DOI] [Google Scholar]
- 28.Hao L., Wen Y., Zhao Y., Lu W., Xiao K. Wheat mitogen-activated protein kinase gene TaMPK4 improves plant tolerance to multiple stresses through modifying root growth, ROS metabolism, and nutrient acquisitions. Plant Cell Rep. 2015;34:2081–2097. doi: 10.1007/s00299-015-1853-2. [DOI] [PubMed] [Google Scholar]
- 29.Lee S.K., Kim B.G., Kwon T.R., Jeong M.J., Park S.R., Lee J.W., Byun M.O., Kwon H.B., Matthews B.F., Hong C.B., et al. Overexpression of the mitogen-activated protein kinase gene OsMAPK33 enhances sensitivity to salt stress in rice (Oryza sativa L.) J. Biosci. 2011;36:139. doi: 10.1007/s12038-011-9002-8. [DOI] [PubMed] [Google Scholar]
- 30.Agarwal P.K., Gupta K., Jha B. Molecular characterization of the Salicornia brachiata SbMAPKK gene and its expression by abiotic stress. Mol. Biol. Rep. 2010;37:981–986. doi: 10.1007/s11033-009-9774-1. [DOI] [PubMed] [Google Scholar]
- 31.López-Bucio J.S., Dubrovsky J.G., Raya-González J., Ugartechea-Chirino Y., López-Bucio J., de Luna-Valdez L.A., Ramos-Vega M., León P., Guevara-García A.A. Arabidopsis thaliana mitogen-activated protein kinase 6 is involved in seed formation and modulation of primary and lateral root development. J. Exp. Bot. 2014;65:169–183. doi: 10.1093/jxb/ert368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.