Despite recent treatment improvements with the approval of cell signaling pathway inhibitors, additional treatment options are needed for patients with relapsed/refractory high-risk chronic lymphocytic leukemia (CLL). An emerging immunotherapeutic strategy for CLL is based on the adoptive transfer of chimeric antigen receptor (CAR)-engineered T cells. CARs enable highly specific targeting of antigens on the surface of tumor cells in a human leukocyte antigen (HLA)-independent manner.1 Recently, CAR-engineered T cells targeting CD19 resulted in sustained responses in some patients with relapsed/refractory CLL.2 Another immunotherapeutic approach is based on the administration of T-cell receptor (TCR)-engineered T cells. Such transgenic TCRs recognize intracellular proteins, which are processed and presented on the surface of tumor cells by HLA molecules.1 However, T-cell epitopes derived from CLL-associated antigens that are suitable targets for TCR-engineered T cells are limited.
One promising candidate for the design of T-cell-based immunotherapeutic strategies is receptor tyrosine kinase-like orphan receptor 1 (ROR1). It has been demonstrated that ROR1 messenger Ribonucleic Acid (mRNA) is highly expressed in CLL cells (Online Supplementary Figure S1) and a uniform expression of ROR1 protein is detected on their cell surface.3,4 In contrast, ROR1 expression was not found in other blood cells or on bone marrow-derived cells, except on a small subset of precursor B cells.3,4 ROR1 mRNA was also not detectable at significant levels in normal adult non-hematopoietic tissues (Online Supplementary Figure S2A).4 When investigating ROR1 protein expression of normal tissues, contradictory results were obtained: according to two studies and data obtained from a public proteomics repository (Human Integrated Protein Expression Database), ROR1 was not or only marginally expressed in normal tissues (Online Supplementary Figure S2B).5,6 However, Balakrishnan et al. reported that ROR1 is expressed in the parathyroid, pancreatic islets, and regions of the gastrointestinal tract.7 Further studies revealed that ROR1 plays an important role in leukemogenesis. Thus, Wnt5a induced the formation of ROR1/ROR2 heterooligomers, which augmented the proliferation and migration of CLL cells.8 In addition, a higher expression of ROR1 on CLL cells was associated with a significantly shorter overall survival of CLL patients, indicating that ROR1 may enhance disease progression.9 Due to the expression profile and important role in CLL pathogenesis, antibodies and CAR-engineered T cells targeting ROR1 have been generated that mediate marked tumor-directed effects.4,10
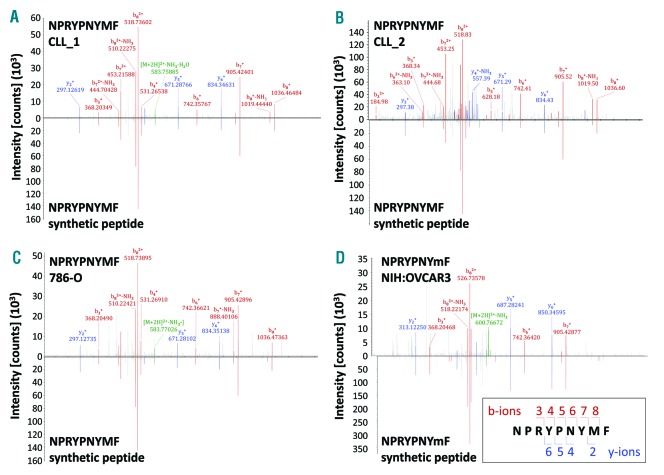
Herein, we searched for a ROR1-derived peptide that may serve as a target structure for T-cell-based immunotherapy of CLL patients. This study was approved by the institutional review board of the University Hospital of Dresden (EK-187052011) and the University of Tübingen (373/2011BO2). Patients and healthy donors gave their written informed consent. Recently, we performed a mass spectrometry-based comparative analysis of non-mutated HLA class I and II ligands naturally presented on peripheral blood mononuclear cells (PBMCs) of CLL patients and healthy donors and identified various CLL-associated antigens.11 T-cell responses against CLL antigen-derived HLA ligands were exclusively detected in the blood of CLL patients. We used this HLA ligandome analysis of CLL cells for the identification of a novel ROR1-derived peptide. Mass spectrometry-based analyses of isolated HLA ligands from PBMCs derived from two CLL patients who share HLA-A*03:01 and HLA-B*07:02 (CLL_1, CLL_2; containing > 80% CLL cells) identified the naturally presented ROR1-derived nonamer NPRYPNYMF (Figure 1A,B and Online Supplementary Table S1). The identification was validated using the synthetic peptide. Altogether, the ROR1 peptide was presented on two out of seven (28.6%) of the analyzed HLA-matched CLL cell samples as well as on the renal cancer cell line 786-0 and the ovarian cancer cell line NIH:OVCAR3 (Figure 1C,D and Online Supplementary Table S1).
Figure 1.
Identification of a novel ROR1-derived peptide naturally presented by tumor cells. Comparison of the mass spectrum of the experimental spectrum (displayed as peaks above the x axis) of the ROR1-derived peptide NPRYPNYMF presented by (A) CLL_1, (B) CLL_2, (C) 786-O, and (D) NIH:OVCAR3 with the synthetic peptide NPRYPNYMF (displayed as peaks below the x axis). The assigned peaks of the b-ions (red) and y-ions (blue) as well as the precursor ions (green) are displayed in color. The masses of b- and y-ion fragments are correlated with the obtained sequence using fragment numbers (right bottom). Analyzing the NIH:OVCAR3 cell line we were able to identify the ROR1 peptide only when applying the parallel reaction monitoring method. Here the peptide was identified with an oxidized methionine, which is indicated by a small ‘m’ (NPRYPNYmF). The spectrum was therefore compared with a spectrum of the synthetic peptide containing also the oxidized methionine. In all other cases data from the data-dependent acquisition method (a top speed collision-induced dissociation fragmentation method) is shown. CLL: chronic lymphocytic leukemia.
The amino acid sequence of the peptide (amino acids 783–791 of the protein) could not be found in any other protein according to the human proteome databases UniProtKB and NCBI non-redundant protein database using Basic Local Alignment Search Tools (BLASTs). As demonstrated in Online Supplementary Table S2, the highest binding score of NPRYPNYMF is predicted for HLA-B*07:02 by all four HLA-binding prediction online-tools (SYFPEITHI, Rankpep, NetMHC 4.0, Immune Epitope Database and Analysis Resource [IEDB]). HLA-B*07:02 was the most frequent HLA-B-allele (approximately 14%) in a cohort of 8862 German stem cell donors.12 The scores for HLA-A*03:01 are not sufficient to expect HLA binding. The proline at position 2 (P2) of the nonamer is a required HLA-B*07:02 anchor position. This peptide has a C-terminus predicted by all three available cleavage models (language models for proteasomal cleavage) of the Rankpep online tool. Furthermore, we investigated whether the ROR1 peptide is also naturally presented by normal cells. When exploring our database encompassing 275 HLA class I ligandomes of various normal tissues, including 45 HLA-B*07+ and 48 HLA-A*03+ samples (PBMCs, bone marrow, spleen, lymph node, kidney, colon, thyroid, lung, liver, brain, testis, bladder, tongue), the identified nonamer could not be found (data not shown). In addition, we evaluated the HLA-ligandome of immunomagnetically isolated B cells from five HLA-B*07:02+ healthy donors. Four of them were also HLA-A*03:01+. Again, the ROR1 peptide could not be detected (data not shown).
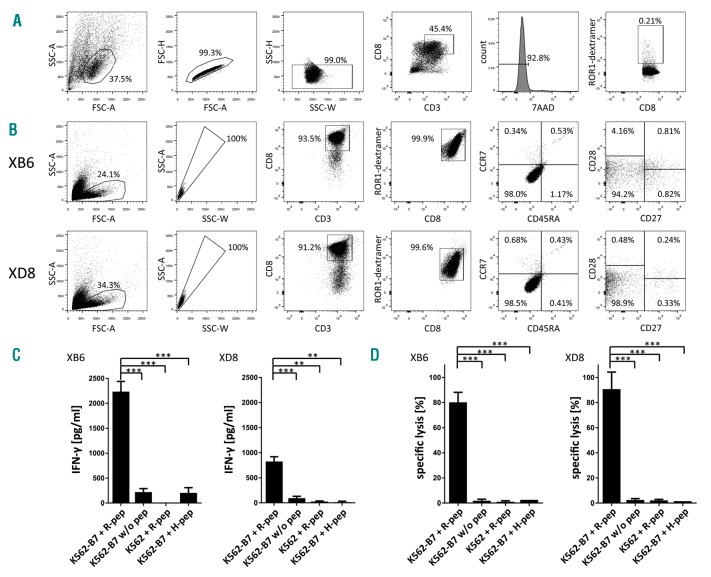
To generate CD8+ T-cell clones, CD8+ T cells were immunomagnetically isolated from peripheral blood of an HLA-B*07:02+ healthy donor and were co-cultured with autologous monocyte-derived dendritic cells (moDCs) pulsed with the ROR1 peptide NPRYPNYMF. After one restimulation, the presence of specific CD8+ T cells was determined by using dextramers. Flow cytometry revealed a frequency of 0.21% ROR1 peptide-specific CD8+ T cells in the polyclonal CD8+ T-cell population (Figure 2A). To establish T-cell clones, CD8+ T cells on single cell level were expanded as previously described.13 The restimulated CD8+ T lymphocytes were fluorescence-activated cell sorter (FACS)-sorted and seeded at single cell level (143 CD8+ T cells in total). After two weeks of co-culture with irradiated allogeneic PBMCs in the presence of interleukin (IL)-2, IL-7, IL-15, and phytohemagglutinin M, CD8+ T-cell clones were tested for ROR1-peptide specificity by dextramer staining and FACS analysis. Four CD8+ T-cell clones, recognizing the HLA-B*07:02-restricted ROR1 peptide, were identified. The fast growing CD8+ T-cell clones XB6 and XD8 with high avidity (based on dextramer staining) were selected for phenotypic and functional analysis. The characterization of the differentiation state of the ROR1 peptide-specific T-cell clones XB6 and XD8 was based on the expression pattern of CD45RA, CCR7, CD27, and CD28, as previously reported.14 Both T-cell clones XB6 and XD8 were predominantly CD45RA−, CCR7−, CD27−, and CD28− (Figure 2B), indicating that they display the differentiated effector-memory 3 phenotype.
Figure 2.
Characterization of differentiation state and function of ROR1 peptide-specific T-cell clones XB6 and XD8. (A) Gating strategy used for the determination of the frequency of ROR1 peptide-specific CD8+ T cells in the polyclonal CD8+ T-cell population after stimulation with peptide-loaded moDCs is demonstrated. Doublets and dead cells (7-AAD positive) were excluded from the analysis. (B) Purity and differentiation state of the ROR1 peptide-specific CD8+ T-cell clones XB6 and XD8 are depicted. (C) ROR1 peptide-specific T-cell clones were co-incubated with ROR1 peptide-loaded, HLA-B*07:02-transduced K562 cells, unloaded HLA-B*07:02-transduced K562 cells, ROR1 peptide-pulsed, HLA-B*07:02− K562 cells, or HLA-B*07:02-transduced K562 cells loaded with the irrelevant peptide GPGHKARVL. After 24 h, supernatants were collected and IFN-γ concentration was determined by enzyme-linked immunosorbent assay (ELISA). The results are presented as mean ± s.d. of triplicate determinations. Statistical significance was calculated by the Student’s t-test. Asterisks indicate a statistically significant difference (*** P<0.001; ** P<0.01). (D) The T-cell clones XB6 and XD8 were co-cultured with 5 × 103 51Cr-labeled, ROR1 peptide-loaded, and HLA-B*07:02-transduced K562 cells per well at an E:T ratio of 10:1 for 4 h. Unloaded HLA-B*07:02-transduced K562 cells, ROR1 peptide-pulsed, HLA-B*07:02− K562 cells, or HLA-B*07:02-transduced K562 cells loaded with the irrelevant peptide GPGHKARVL served as controls. The results of the CD8+ T-cell clones are presented as mean ± s.d. of triplicate determinations. Statistical significance was calculated by the Student’s t-test. Asterisks indicate a statistically significant difference (*** P≤ 0.001).
When evaluating the functional properties of the T-cell clones XB6 and XD8, we found that both T-cell clones secrete interferon (IFN)-γ after stimulation with ROR1 peptide-loaded, HLA-B*07:02-transduced K562 cells15 (Figure 2C). Only marginal IFN-γ release by the T-cell clones was observed after contact with ROR1 peptide-pulsed, HLA-B*07:02− K562 cells, HLA-B*07:02-trans-duced K562 cells loaded with the irrelevant human immunodeficiency virus gag polyprotein derived peptide GPGHKARVL or unloaded HLA-B*07:02-transduced K562 cells. In addition, a profound cytotoxicity of both T-cell clones against ROR1 peptide-loaded, HLA-B*07:02-transduced K562 cells was observed (Figure 2D). In contrast, ROR1 peptide-pulsed, HLA-B*07:02− K562 cells, HLA-B*07:02-transduced K562 cells loaded with GPGHKARVL, or unloaded HLA-B*07:02-transduced K562 cells were only marginally killed.
Knowledge of the ROR1-specific TCR sequences might be valuable for generating TCR-engineered CD8+ T cells, or to track ROR1-specific T cells after administration. Here, sequences of the CDR3 regions of the α and β TCR chain were determined at a single cell level. RT-PCR amplification and sequencing of TCRα and TCRβ chains from single T cells as well as subsequent cloning of PCR fragments was performed as previously described.16 Analysis of TCRα and TCRβ sequences and junction peptide amino acid sequence extraction was conducted with reference to the international ImMunoGeneTics information system (IMGT) database. Junction peptides were analyzed using KNIME 2.5.2. Nucleotide sequences were in silico translated into amino acid sequences, which are presented in Online Supplementary Table S3. TRAV, TRAJ, TRBV, TRBD and TRBJ gene usage was different in the T-cell clones XB6 and XD8, and CDR3 amino acid sequences differed in length and did not obviously share motifs.
Recently, it has been reported that the ROR1 protein is expressed on the surface of CLL cells, but not on normal mature B cells.3,4 In agreement with these studies, we identified the HLA-B*07:02-restricted ROR1 peptide NPRYPNYMF as a component of the HLA ligandome of CLL samples (Figure 1 A,B and Online Supplementary Table S1). In contrast, the ROR1 peptide could not be found in the HLA ligandome of normal cells (data not shown). Following these findings, the cytotoxic activity of the ROR1 peptide-specific T-cell clones XB6 and XD8 against primary CLL cells was determined by a flow cytometry-based killing assay.
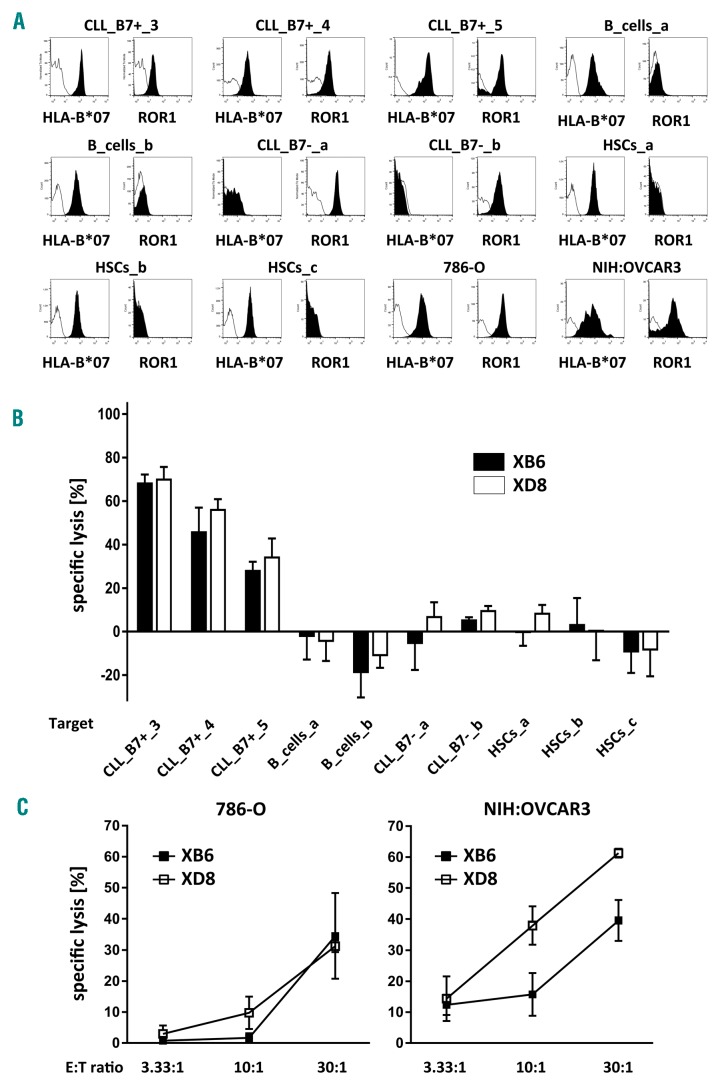
HLA-B*07:02+ ROR1+ CLL cells and HLA-B*07:02− ROR1+ CLL cells (purity: >80%) were obtained from blood samples of five patients (Figure 3A). HLA-B*07:02+ ROR1− B cells (purity: >90%) were immunomagnetically isolated from PBMCs of two healthy donors (Figure 3A). HLA-B*07:02+ ROR1− CD34+ hematopoietic stem cells (HSCs, purity: >90%) were immunomagnetically isolated from leukapheresis products of healthy stem cell donors (Figure 3A). Whereas HLA-B*07:02− ROR1+ CLL cells, HLA-B*07:02+ ROR1− B cells, and HLA-B*07:02+ ROR1− HSCs were not or only marginally killed by the T-cell clones XB6 and XD8, they efficiently lysed HLA-B*07:02+ ROR1+ CLL cells (Figure 3B). The cytotoxic potential of the T-cell clones XB6 and XD8 against normal non-hematopoietic cells remains to be determined.
Figure 3.
Lysis of primary CLL cells and cancer cell lines 786-O and NIH:OVCAR3 by ROR1 peptide-specific T-cell clones. (A) The expression of HLA-B*07:02 and ROR1 protein by CLL cells, B cells, CD34+ hematopoietic stem cells (HSCs), and the cancer cell lines 786-O and NIH:OVCAR3 was determined by flow cytometry. Percentage of cells staining positive for each molecule (filled) compared to the staining intensity of the isotype control antibody are shown. (B) The cytotoxic potential of the CD8+ T-cell clones XB6 and XD8 against HLA-B*07:02+ ROR1+ CLL cells, HLA-B*07:02− ROR1+ CLL cells, HLA-B*07:02+ ROR1− HSCs, and HLA-B*07:02+ ROR1− B cells was evaluated by flow cytometry. Therefore, ROR1 specific CD8+ T cells were added to eFluor670-labeled target cells (2 × 104 cells/well) at an E:T ratio of 10:1 for 24 h. Isolated CD8+ T cells from the same donor as the generated ROR1 peptide specific T-cell clones served as controls. After flow cytometry-based counting of viable target cells, the percentage of lysis was assessed. Data obtained by co-culturing target cells in the presence of unspecific CD8+ T cells was defined as the 0% lysis reference point. The results for the CD8+ T-cell clones XB6 and XD8 are presented as mean ± s.d. (C) The T-cell clones XB6 and XD8 were co-cultured with 5×103 51Cr-labeled, 786-O or NIH:OVCAR3 cells at different E:T ratios for 4 h. The results for the CD8+ T-cell clones are presented as mean ± s.d. of triplicate determinations. CLL: chronic lymphocytic leukemia.
In addition to CLL cells, ROR1 has been identified in various human cancer tissues.17 Therefore, we explored the cytotoxic potential of the T-cell clones XB6 and XD8 against the HLA-B*07:02 and ROR1 expressing cancer cell lines 786-0 and NIH:OVCAR3 (Figure 3C), which have been shown to naturally present the ROR1 peptide NPRYPNYMF (Figure 1C,D and Online Supplementary Table S1). As depicted in Figure 3C, the T-cell clones efficiently lysed both cancer cell lines.
In conclusion, we identified the first naturally presented ROR1 peptide in the HLA ligandome of CLL samples by mass spectrometry. This ROR1 peptide could not be found in 275 HLA class I ligandomes of various normal tissues. We also demonstrated that ROR1 peptide-specific CD8+ T-cell clones efficiently lysed HLA-B*07:02+ROR1+ CLL cells and the cancer cell lines 786-0 and NIH:OVCAR3. This ROR1 peptide may represent an attractive candidate for T cell-based immunotherapy of tumor patients.
Supplementary Material
Acknowledgments
The authors would like to thank Bärbel Löbel, Karin Günther, Petra Lorenz, and Maria Schmiedgen for technical assistance, and Lorenz Jahn for kindly providing the HLA-B*07:02-transduced K562 cell line.
Footnotes
Funding: this work was supported by two grants from the Bundesministerium für Bildung und Forschung and by the ERC Advanced Grant 339842: MUTAEDITING and by the Graduiertenakademie TU Dresden.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Johnson LA, June CH. Driving gene-engineered T cell immunotherapy of cancer. Cell Res. 2017;27:38–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porter DL, Hwang W, Frey NV, et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci Transl Med. 2015;7(303):303ra139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baskar S, Kwong KY, Hofer T, et al. Unique cell surface expression of receptor tyrosine kinase ROR1 in human B-cell chronic lymphocytic leukemia. Clin Cancer Res. 2008;14(2):396–404. [DOI] [PubMed] [Google Scholar]
- 4.Hudecek M, Schmitt TM, Baskar S, et al. The B-cell tumor-associated antigen ROR1 can be targeted with T cells modified to express a ROR1-specific chimeric antigen receptor. Blood. 2010;116(22):4532–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukuda T, Chen L, Endo T, et al. Antisera induced by infusions of autologous Ad-CD154-leukemia B cells identify ROR1 as an oncofetal antigen and receptor for Wnt5a. Proc Natl Acad Sci USA. 2008;105(8):3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dave H, Anver MR, Butcher DO, et al. Restricted cell surface expression of receptor tyrosine kinase ROR1 in pediatric B-lineage acute lymphoblastic leukemia suggests targetability with therapeutic monoclonal antibodies. PLoS One. 2012;7(12):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balakrishnan A, Goodpaster T, Randolph-Habecker J, et al. Analysis of ROR1 protein expression in human cancer and normal tissues. Clin Cancer Res. 2017;23(12):3061–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu J, Chen L, Cui B, et al. Wnt5a induces ROR1/ROR2 heterooligomerization to enhance leukemia chemotaxis and proliferation. J Clin Invest. 2016;126(2):585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui B, Ghia EM, Chen L, et al. High-level ROR1 associates with accelerated disease-progression in chronic lymphocytic leukemia. Blood. 2016;128(25):2931–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daneshmanesh AH, Hojjat-Farsangi M, Khan AS, et al. Monoclonal antibodies against ROR1 induce apoptosis of chronic lymphocytic leukemia (CLL) cells. Leukemia. 2012;26(6):1348–1355. [DOI] [PubMed] [Google Scholar]
- 11.Kowalewski J, Schuster H, Backert L, et al. HLA ligandome analysis identifies the underlying specificities of spontaneous antileukemia immune responses in chronic lymphocytic leukemia (CLL). Proc Natl Acad Sci U S A. 2015;112(2):E166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt AH, Baier D, Solloch UV, et al. Estimation of high-resolution HLA-A, -B, -C, -DRB1 allele and haplotype frequencies based on 8862 German stem cell donors and implications for strategic donor registry planning. Human Immunol. 2009;70:895–902. [DOI] [PubMed] [Google Scholar]
- 13.Tunger A, Wehner R, von Bonin M, et al. Generation of high-avidity, WT1-reactive CD8+ cytotoxic T cell clones with anti-leukemic activity by streptamer technology. Leuk Lymphoma. 2017;58(5):1246–1249. [DOI] [PubMed] [Google Scholar]
- 14.Romero P, Zippelius A, Kurth I, et al. Four functionally distinct populations of human effector-memory CD8+ T lymphocytes. J Immunol. 2007;178(7):4112–4119. [DOI] [PubMed] [Google Scholar]
- 15.Jahn L, Hombrink P, Hassan C, et al. Therapeutic targeting of the BCR associated protein CD79b in a TCR-based approach is hampered by aberrant expression of CD79b. Blood. 2015;125(6):949–958. [DOI] [PubMed] [Google Scholar]
- 16.Eugster A, Lindner A, Heninger A-K, et al. Measuring T cell receptor and T cell gene expression diversity in antigen-responsive human CD4+ T cells. J Immunol Methods. 2013;400–401:13–22. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S, Chen L, Wang-Rodriguez J, et al. The onco-embryonic antigen ROR1 is expressed by a variety of human cancers. Am J Pathol. 2012;181(6):1903–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.