GAF1-binding sites are necessary for GA feedback regulation of AtGA20ox2 in transgenic Arabidopsis, indicating that the DELLA-GAF1 complex is a main component of the GA feedback regulation of AtGA20ox2.
Abstract
Gibberellins (GAs) are phytohormones that regulate many aspects of plant growth and development, including germination, elongation, flowering, and floral development. Negative feedback regulation contributes to homeostasis of the GA level. DELLAs are negative regulators of GA signaling and are rapidly degraded in the presence of GAs. DELLAs regulate many target genes, including AtGA20ox2 in Arabidopsis (Arabidopsis thaliana), encoding the GA-biosynthetic enzyme GA 20-oxidase. As DELLAs do not have an apparent DNA-binding motif, transcription factors that act in association with DELLA are necessary for regulating the target genes. Previous studies have identified GAI-ASSOCIATED FACTOR1 (GAF1) as such a DELLA interactor, with which DELLAs act as coactivators, and AtGA20ox2 was identified as a target gene of the DELLA-GAF1 complex. In this study, electrophoretic mobility shift and chromatin immunoprecipitation assays showed that four GAF1-binding sites exist in the AtGA20ox2 promoter. Using transgenic plants, we further evaluated the contribution of the DELLA-GAF1 complex to GA feedback regulation. Mutations in four GAF1-binding sites abolished the negative feedback of AtGA20ox2 in transgenic plants. Our results showed that GAF1-binding sites are necessary for GA feedback regulation of AtGA20ox2, suggesting that the DELLA-GAF1 complex is a main component of the GA feedback regulation of AtGA20ox2.
GAs are tetracyclic diterpenoid phytohormones that regulate many aspects of plant growth and development, including seed germination, leaf expansion, stem elongation, root growth, flower induction, and anther development (Richards et al., 2001). GA biosynthetic enzymes have been characterized using genetic, molecular, and biochemical approaches (Yamaguchi, 2008). In Arabidopsis (Arabidopsis thaliana), six biosynthetic enzymes have been identified. Endogenous levels of GAs are fine-tuned by feedback control of GA biosynthetic enzymes at several steps (Yamaguchi, 2008; Fukazawa et al., 2011; Hedden and Thomas, 2012). The final steps in GA biosynthesis are catalyzed by GA 20-oxidase and GA 3-oxidase, the expression of which is controlled by endogenous GA levels. AtGA20ox1, AtGA20ox2, AtGA20ox3, and AtGA3ox1 are highly up-regulated in GA-deficient mutants, whereas they are down-regulated after the application of GAs (Chiang et al., 1995; Phillips et al., 1995; Rieu et al., 2008). GA-deactivation genes also respond to endogenous GA levels. For example, GA2ox1 and GA2ox2 are up-regulated by GA treatment (Thomas et al., 1999). GA feedback regulation has been shown to depend on GA signaling components, including the GA receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1), DELLA proteins, an F-box adaptor subunit of SCF E3 ubiquitin ligase, and SPINDLY (Sasaki et al., 2003; Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006; Sun, 2011; Hauvermale et al., 2012). Arabidopsis contains five DELLAs, GIBBERELLIN INSENSITIVE (GAI), REPRESSOR OF ga1-3 (RGA), RGA-LIKE1 (RGL1), RGL2, and RGL3, which act as negative regulators of GA signaling (Sun and Gubler, 2004). Binding of GA to GID1 enhances the interaction between GID1 and DELLAs, resulting in the rapid degradation of DELLAs via the ubiquitin-proteasome pathway (Sun, 2011). Under GA-deficient conditions, DELLA proteins accumulate in the nucleus and regulate the expression of target genes. Zentella et al. (2007) have identified 14 early GA-responsive genes, including AtGA20ox2 and AtGA3ox1, as targets of DELLAs, which activate the expression of these genes.
Because DELLAs do not have an apparent DNA-binding motif, it was hypothesized that DNA-binding proteins are involved in the direct regulation of these genes along with DELLAs. Indeed, several transcription factors have been identified as DELLA-interacting proteins. For example, DELLAs regulate hypocotyl elongation by interacting with PHYTOCHROME INTERACTING FACTOR (PIF) and BRASSINAZOLE RESISTANT1 (BZR1), which promote hypocotyl elongation (de Lucas et al., 2008; Feng et al., 2008; Bai et al., 2012). DELLAs inhibit the activities of these transcriptional activators by titrating their DNA-binding activity. GAs trigger the degradation of DELLAs, which release PIFs to activate target genes, including β-EXPANSIN and PACLOBUTRAZOL RESISTANCE, thus promoting hypocotyl elongation. The mechanism of GA-dependent transactivation has been explained by the degradation of DELLAs that titrated transcriptional activators.
However, multiple GA-regulated genes, including AtGA20ox2 and AtGA3ox1, are repressed by GAs. The mechanism of GA feedback regulation is difficult to explain based on the titration of PIF and BZR1 by DELLAs. Moreover, a chromatin immunoprecipitation (ChIP) assay indicated that DELLAs associate with the promoters of several genes (Zentella et al., 2007), while in the titration model, the DELLA complex could not bind to the promoter of target genes. Thus, DELLAs may have another function in the regulation of these genes. DELLAs activate gene expression through transcription factors, including type B ARABIDOPSIS RESPONSE REGULATORS1 (Marín-de la Rosa et al., 2015), SQUAMOSA PROMOTER BINDING PROTEIN-LIKE9 (Yamaguchi et al., 2014), ABSCISIC ACID INSENSITIVE3 (ABI3), and ABI5 (Lim et al., 2013). These findings indicated novel functions for DELLAs. Recently, we identified a DELLA-binding transcription factor, designated GAI-ASSOCIATED FACTOR1 (GAF1), and revealed a new role of DELLAs as transcriptional coactivators (Fukazawa et al., 2014). GAF1 is a transcription factor with zinc finger motifs that shows similarity to maize (Zea mays) INDETERMINATE1 (ID1; Colasanti et al., 1998). The Arabidopsis genome contains 16 ID1-related proteins (IDD, for ID1 domain protein). The maize ID1 and Arabidopsis IDD family proteins bind to the consensus sequence TTTTGTCG (Kozaki et al., 2004). GAF1 also interacts with the corepressor TOPLESS-RELATED (TPR). DELLAs and TPR act as coactivators and a corepressor of GAF1, respectively. By removing DELLA, GAs convert the GAF1 complex from transcriptional activator to repressor. This model can explain the GA feedback regulation. In fact, we have identified GA feedback-regulated genes, including GA20ox2, GA3ox1, and GID1b, as GAF1 target genes. Gel-shift analysis and ChIP assay showed that GAF1 binds directly to the GA20ox2 and GID1b promoters (Fukazawa et al., 2014). These data indicated that the DELLA-GAF1 complex is involved in GA feedback regulation of AtGA20ox2. Various other transcription factors involved in GA feedback regulation, such as YABBY1, AT-HOOK PROTEIN OF GA FEEDBACK1 (AGF1), REPRESSION OF SHOOT GROWTH (RSG), and YABBY4, have been reported (Dai et al., 2007; Matsushita et al., 2007; Fukazawa et al., 2010; Yang et al., 2016).
In this study, we aimed to investigate how important the contribution of the DELLA-GAF1 complex to GA feedback regulation of AtGA20ox2 is compared with that of other factors. We identified multiple GAF1-binding sites in the promoter of AtGA20ox2 and evaluated the effect of mutation of each of these GAF1-binding sites on GA feedback regulation.
RESULTS
The AtGA20ox2 Promoter Is Activated by the DELLA-GAF1 Complex and Is Responsible for GA Feedback Regulation
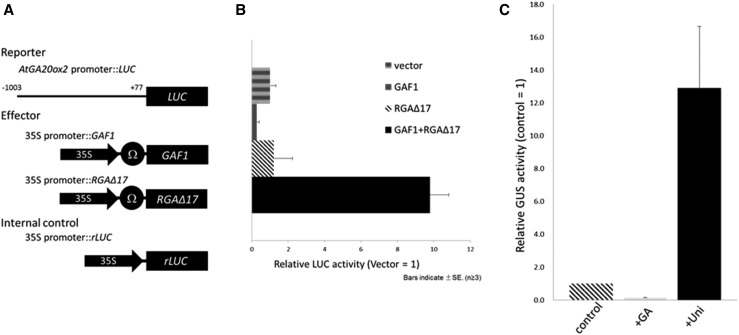
The Arabidopsis genome carries five GA20ox genes, three of which (AtGA20ox1, AtGA20ox2, and AtGA20ox3) function partially redundantly at most developmental stages, including floral transition, while AtGA20ox4 and AtGA20ox5 have very minor roles (Rieu et al., 2008; Plackett et al., 2012). AtGA20ox1, AtGA20ox2, and AtGA20ox3 are under negative feedback regulation by GA, especially, the expression of AtGA20ox2 is most responsive to endogenous GA levels, while AtGA20ox4 and AtGA20ox5 expression is not affected by GA levels. Previously, we found that DELLA-GAF1 is involved in the feedback regulation of AtGA20ox2 (Fukazawa et al., 2014). To determine whether DELLA-GAF1 directly regulates the transcription of AtGA20ox2, we carried out a transactivation assay using Arabidopsis leaves. Effector plasmids expressing GAF1 and/or DELLA under the control of the cauliflower mosaic virus (CaMV) 35S promoter were cobombarded with a reporter plasmid containing a LUC reporter gene driven by the AtGA20ox2 promoter (–1,003 to +77, where +1 indicates the transcription start site; Fig. 1A). Base-position numbering of the AtGA20ox2 promoter was changed from ATG as +1 in our previous study (Fukazawa et al., 2014) to the transcription start site as +1 in this study. DELLA-GAF1 activated the AtGA20ox2 promoter-LUC fusion (Fig. 1B), indicating that the AtGA20ox2 promoter is indeed regulated by the DELLA-GAF1 complex. In this transactivation assay, we used rga-Δ17 as the constitutively active DELLA protein. rga-Δ17 is a mutant protein with a 17-amino acid deletion within the DELLA domain of RGA, rendering it resistant to degradation by GAs (Dill et al., 2001). To investigate whether this promoter region of AtGA20ox2 is sufficient for GA feedback regulation, we generated transgenic plants carrying the promoter fused with a GUS reporter gene and examined the effect of applying GA3 or uniconazole, an inhibitor of GA biosynthesis. Uniconazole-treated transgenic plants exhibited higher GUS activity than did untreated transgenic plants (Fig. 1C). In contrast, GA3 repressed GUS activity. These results indicated that the AtGA20ox2 promoter is regulated by the DELLA-GAF1 complex in a transient assay and GA levels in plants.
Figure 1.
A 1,003-bp region of the AtGA20ox2 promoter is regulated by the DELLA-GAF1 complex and responds to GA. A, Schematic representation of the reporter and effector. A 1,003-bp fragment of the AtGA20ox2 promoter was fused to the LUC gene. The effector plasmid expressed the full-length GAF1 or RGAΔ17 under the control of the CaMV 35S promoter with a viral translation enhancer (Ω). B, Transactivation assay of GAF1 and RGAΔ17. The effector, reporter, and internal control constructs were cobombarded into Arabidopsis leaves. The transfected leaves were incubated for 20 h, and then, LUC and rLUC activities were measured. The results are shown as LUC/rLUC activity. Error bars indicate sd (n = 3). C, Fluorometric GUS assay for GA negative feedback in transgenic Arabidopsis plants carrying the 1,003-bp AtGA20ox2 promoter::GUS construct. Gray and black bars represent the GUS activities of plants treated for 1 week with GA3 and uniconazole (Uni), respectively. The striped bar represents the GUS activity of untreated control plants, which was arbitrarily set to 1. The mean activities of five independent transgenic lines for each construct are shown. Error bars indicate sd.
Identification of the cis-Acting Elements of the AtGA20ox2 Promoter for the DELLA-GAF1 Complex
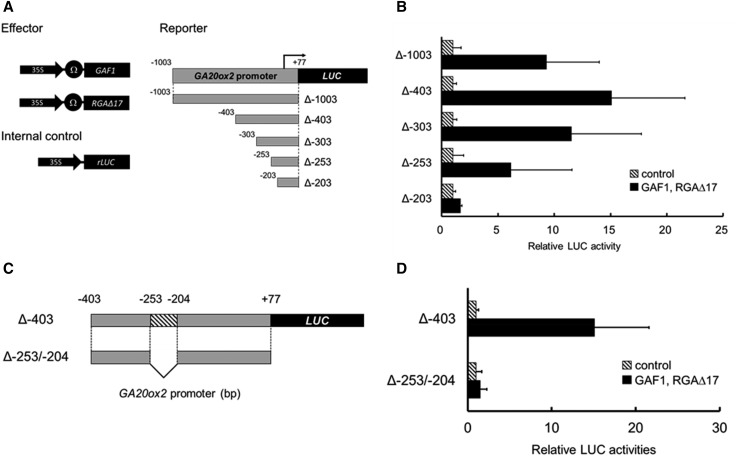
To define the cis-acting elements in the AtGA20ox2 promoter for the DELLA-GAF1 complex, we generated a series of 5′ deletions of the AtGA20ox2 promoter fused with a LUC reporter gene (Δ–403, Δ–303, Δ–253, or Δ–203) and we examined whether DELLA-GAF1 activated these promoters in a transactivation assay using Arabidopsis leaves. The effector plasmids expressing GAF1 and DELLA under the control of the CaMV 35S promoter were cobombarded with a reporter plasmid (Fig. 2A). The DELLA-GAF1 complex activated Δ–1003, Δ–403, Δ–303, and Δ–253 but not Δ–203 (Fig. 2B), and an internal deletion (Δ–253/–204) in the promoter Δ–403 disrupted the activation by DELLA-GAF1 (Fig. 2, C and D). These data suggested that at least a cis-acting element for the DELLA-GAF1 complex is located between –253 and –204. Previously, we identified two GAF1-binding sites, –968 to –949 (cisA) and +6 to +26 (cisD), in the AtGA20ox2 promoter by ChIP assay (Fukazawa et al., 2014). Although Δ–203 and internal deletion (Δ–253/–204) promoters include cisD, DELLA-GAF1 could not activate these promoters. Thus, cisD seemed not to play an important role in the transcriptional activation of AtGA20ox2 by DELLA-GAF1. Therefore, among the four GAF1-binding sites found in the AtGA20ox2 promoter, the cisD region was excluded from further analysis. Taken together, these results suggest that one or more functional cis-elements for the DELLA-GAF1 complex exist in the AtGA20ox2 promoter between –253 and –204.
Figure 2.
Identification of the DELLA-GAF1-regulated region of the AtGA20ox2 promoter using a transactivation assay of GAF1 and RGAΔ17. A and C, Reporter, effector, and internal control constructs used in the assay are shown in A. The constructs were cobombarded into Arabidopsis leaves. B and D, The transfected leaves were incubated for 20 h, and then, LUC and rLUC activities were measured. The results are shown as LUC/rLUC activity. Error bars indicate sd (n = 3).
Identification of GAF1-Binding Sites in the AtGA20ox2 Promoter
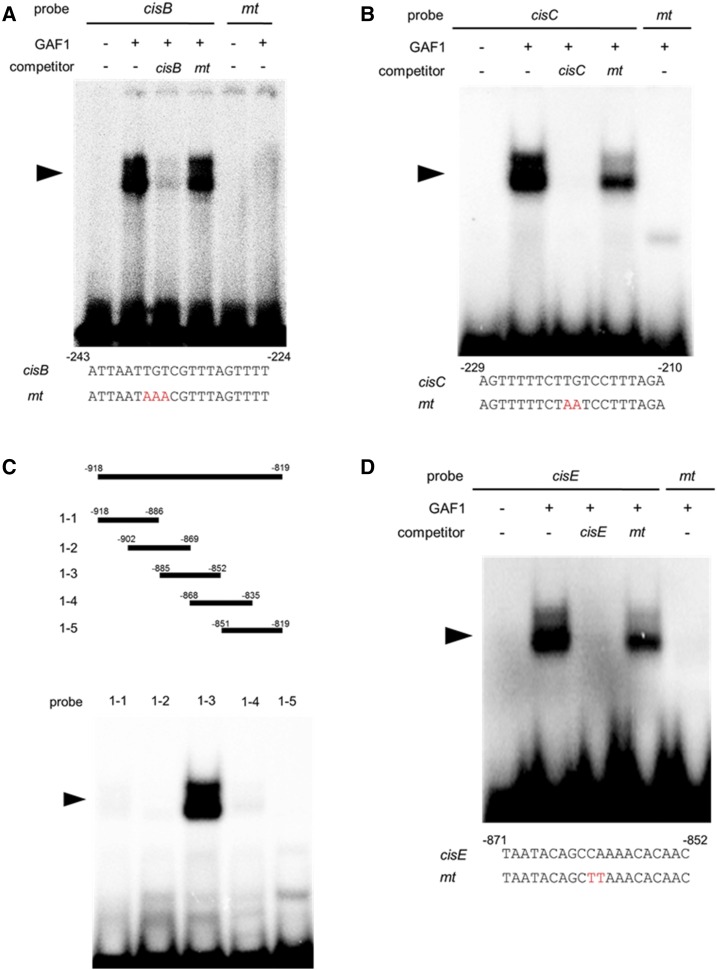
GAF1 belongs to the IDD transcription factor family and binds to ID1-cis, a consensus motif TTTTGTCG (Fukazawa et al., 2014). Two sequences similar to ID1-cis, AATTGTCG and TCTTGTCC, were found in the region –253 to –204 of the AtGA20ox2 promoter (+1, transcription start site). To investigate whether GAF1 binds to these sequences, a gel-shift assay was performed. Recombinant GAF1 protein specifically bound to AATTGTCG and TCTTGTCC sequences, named cisB and cisC, respectively (Fig. 3, A and B). The newly identified GAF1-binding sequences had substitutions at 2 bp of the 8-bp ID1-cis sequence, suggesting that a yet unidentified GAF1-binding sequence might exist in the AtGA20ox2 promoter. Therefore, we searched for novel GAF1-binding sites in the region –1,003 to –403 using a comprehensive gel-shift assay. In this analysis, we examined which 100-bp DNA fragments between –1,003 and –403 inhibit the binding of GAF1 to ID1-cis. Complex formation of the ID1-cis probe with GAF1 was inhibited by the DNA fragment from –918 to –819 (Supplemental Fig. S1), indicating the presence of an unidentified GAF1-binding site in this region of the AtGA20ox2 promoter. Further gel-shift assays revealed a novel GAF1-binding site, located from –885 to –852 (Fig. 3C). And we identified a new GAF1-binding site, named cisE (Fig. 3D).
Figure 3.
Identification of GAF1-binding regions in the AtGA20ox2 promoter in vitro. A and B, Gel retardation assays using recombinant GAF1 protein. A, Oligonucleotides containing cisB (–243 to –224, wild type; lanes 1–4) or mtcisB (mt; lanes 5 and 6) were used as probes. B, Oligonucleotides containing cisC (–229 to –210, wild type; lanes 1–4) or mtcisC (mt; lane 5) were used as probes. Red letters indicate mutated bases. Wild type and mt indicate competition with a 200-fold excess of unlabeled wild-type and mutated probe, respectively. The specific GAF1-DNA complexes are indicated by arrowheads. +, Addition to the reaction mixtures; –, omission from the reaction mixtures. C, Gel retardation assay using recombinant GAF1 protein. Thirty-base-pair regions of the AtGA20ox2 promoter were used as probes. D, Oligonucleotides containing cisE (–871 to –852, wild type; lanes 1–4) or mtcisE (mt; lane 5) were used as probes. Red letters indicate mutated bases. Wild type and mt indicate competition with a 200-fold excess of unlabeled wild-type and mutated probe, respectively. The specific GAF1-DNA complexes are indicated by arrowheads. +, Addition to the reaction mixtures; –, omission from the reaction mixtures.
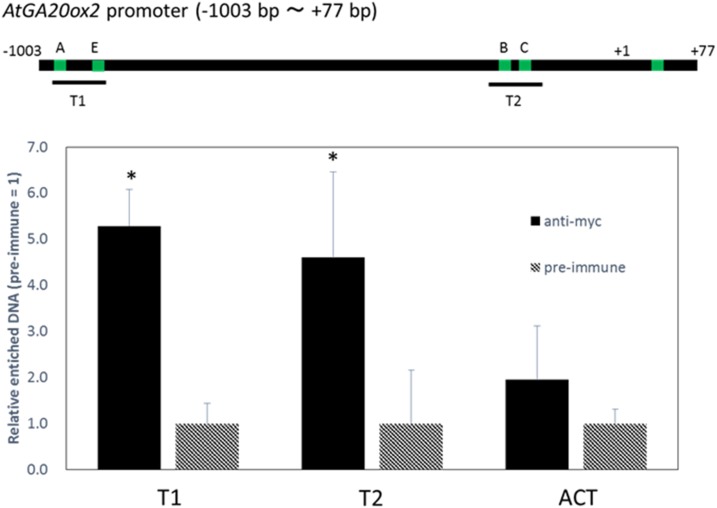
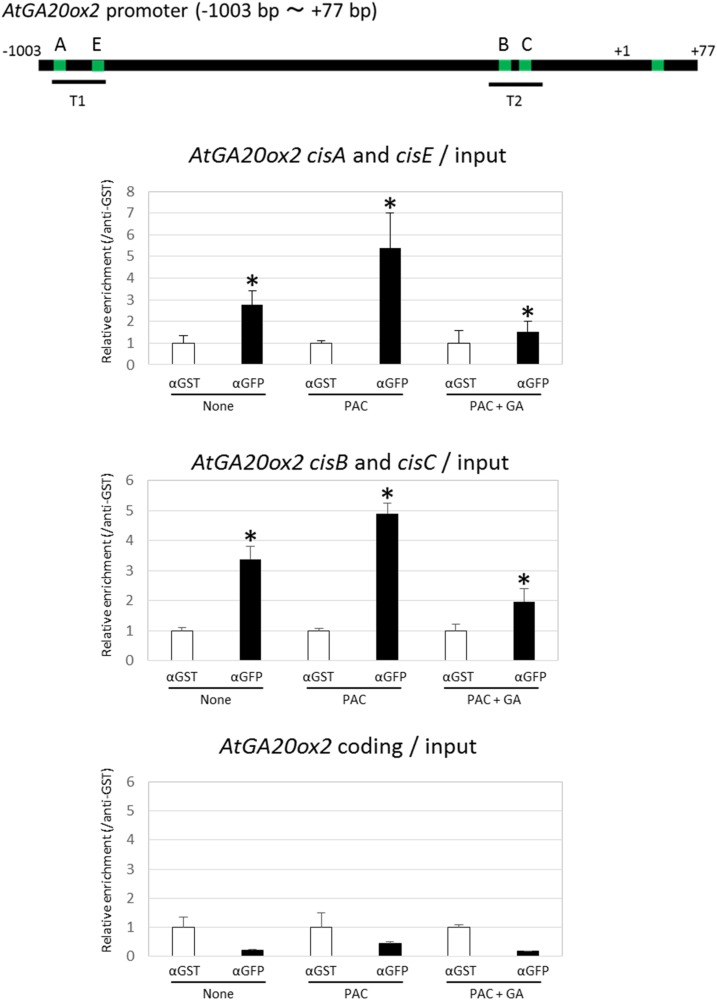
To investigate the binding of GAF1 to the AtGA20ox2 promoter in vivo, we carried out ChIP analysis using transgenic plants expressing myc-tagged GAF1 under the control of the CaMV 35S promoter. The results showed that GAF1 binds to two regions, T1 (cisA and cisE) and T2 (cisB and cisC; Fig. 4). The ChIP assay did not allow distinguishing between cisA and cisE and between cisB and cisC because the binding sites are too close; however, these results indicated that GAF1 binds directly to at least two regions in the AtGA20ox2 promoter in vivo.
Figure 4.
GAF1 binds to multiple regions of the AtGA20ox2 promoter in vivo. ChIP assays were performed with anti-GST or anti-GFP in RGApro:RGA-GFP transgenic plants. Green bars indicate GAF1-binding sites of the AtGA20ox2 promoter. The coprecipitated level of each DNA fragment was quantified by real-time PCR and normalized to the input DNA. The relative coprecipitated levels of each DNA fragment using anti-GST were set to 1, and relative enrichments of each DNA fragment using anti-GFP compared with a DNA fragment using anti-GST are shown in the graph. ACT, ACTIN. Error bars indicate the sd of three biological replicates (n = 3). *, P < 0.05 versus preimmune DNA fragment enrichment by Student’s t test.
Mutations in GAF1-Binding Sites of the AtGA20ox2 Promoter Disrupt Transcriptional Activation by DELLA-GAF1
To determine whether the GAF1-binding sites identified in vitro are functional in vivo in the activation of the AtGA20ox2 promoter by the DELLA-GAF1 complex, we constructed a series of mutant promoter-LUC fusions, in which mutations were introduced to eliminate the GAF1-binding sequences (Fig. 5, A and B). Internal deletion analysis showed that the region –253 to –204 of the AtGA20ox2 promoter, including cisB and cisC, is important for promoter activation by DELLA-GAF1 (Fig. 2C). We identified cisE as a new GAF1-binding site in the AtGA20ox2 promoter. We investigated whether the four GAF1-binding sites (i.e. cisA, cisB, cisC, and cisE) in the AtGA20ox2 promoter are involved in transcriptional activation by DELLA-GAF1. Promoters with a single cis-element mutation (mtE) or two combinations of triple cis-element mutations (mtABC or mtBCE) of the four GAF1-binding sites were still activated by DELLA-GAF1, although the transactivation activity was lower than that of the wild-type promoter (Fig. 5C). Mutations in all four GAF1-binding sites (mtABCE) completely abolished transcriptional activation by DELLA-GAF1 (Fig. 5D). This loss-of-function experiment showed that at least one GAF1-binding site is necessary for DELLA-GAF1-mediated activation of AtGA20ox2.
Figure 5.
Mutation of four GAF1-binding sites completely abolishes DELLA-GAF1 activation of the GA20ox2 promoter. A, Comparison between ID1-cis and GAF1-binding sites in the AtGA20ox2 promoter. Green letters show conserved sequence among these GAF1-binding sites. Arrows indicate the direction of each GAF1-binding site. B, Transactivation assays of GAF1 and RGA. Reporter constructs of each AtGA20ox2 promoter mutant fused with LUC used in the assay are shown. Red bars indicate mutations in GAF1-binding sites of the AtGA20ox2 promoter. C and D, The constructs were cobombarded into Arabidopsis leaves. The transfected leaves were incubated for 20 h, after which the LUC and rLUC activities were measured. The results are shown as LUC/rLUC activity. WT, Wild type. Error bars indicate sd (n = 6). *, P < 0.05 versus control by Student’s t test.
GAF1-Binding Sites Are Necessary for GA Feedback Regulation of AtGA20ox2
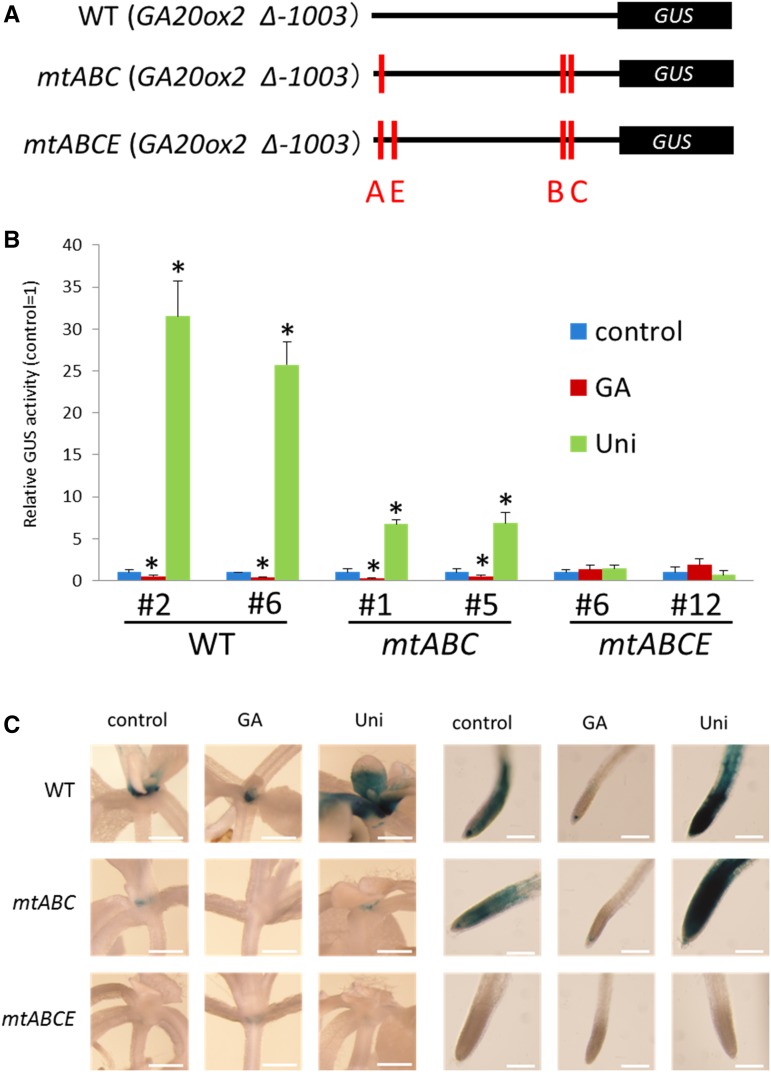
To determine whether the four GAF1-binding sites are involved in the GA negative feedback regulation of AtGA20ox2, we generated transgenic plants carrying mutant promoter-GUS constructs, in which mutations were introduced to eliminate the GAF1-binding sites (Fig. 6A), and GUS activity of each transgenic plant was confirmed by fluorometric GUS enzyme analysis. As expected, these mutations affected the GA negative feedback regulation of AtGA20ox2. Two combinations of triple cis-element mutations (mtABC or mtBCE) among four functional GAF1-binding sites in the AtGA20ox2 promoter decreased its response to GA (Fig. 6B). Mutation of all four GAF1-binding sites (mtABCE) completely inhibited GA feedback regulation of the promoter (Fig. 6B). Thus, at least one GAF1-binding site in the AtGA20ox2 promoter is necessary for GA negative feedback, suggesting that the DELLA-GAF1 complex is a main component in GA feedback regulation of AtGA20ox2.
Figure 6.
Mutation of four GAF1-binding sites completely abolishes the GA responsibility of the AtGA20ox2 promoter. A, Reporter constructs of each AtGA20ox2 promoter mutant fused with GUS used in the assay. Red bars indicate mutations in GAF1-binding sites of the AtGA20ox2 promoter. B, Fluorometric GUS assay for GA negative feedback in transgenic Arabidopsis plants carrying 1,003 bp of AtGA20ox2 promoter::GUS, mtABCAtGA20ox2 promoter::GUS (including three mutations), or mtABCEAtGA20ox2 promoter::GUS (including four mutations). The red and green bars represent the GUS activities of plants treated for 1 week with GA3 and uniconazole (Uni), respectively. The blue bars represent the GUS activity of untreated control plants, which was arbitrarily set to 1 (control = 1). The mean activities of eight independent transgenic lines for each construct are shown. Error bars indicate sd. *, P < 0.05 versus control by Student’s t test. C, Comparison of GUS staining patterns in transgenic plants carrying the wild-type (WT), mtAtGA20ox2 promoter::GUS (mtABC), and mtAtGA20ox2 promoter::GUS (mtABCE) constructs with those of the mutant versions. Meristematic regions of shoot and root tip in seedlings were observed at 12 d. Bars = 0.05 mm (shoot) or 0.2 mm (root tip).
GAF1-Binding Sites Are Involved in Tissue-Specific Expression in Root Tip and Shoot Apices
To evaluate the contribution of the GAF1-binding sites of the AtGA20ox2 promoter to the tissue-specific expression of AtGA20ox2, we compared GUS expression patterns in transgenic plants carrying AtGA20ox2 promoter-GUS and mutant AtGA20ox2 promoter-GUS constructs. Histochemical analysis indicated that AtGA20ox2 is expressed mainly in the root tip and shoot apices (Fig. 6C), consistent with the expression pattern of GAF1 promoter:GUS (Fukazawa et al., 2014). These observations indicated that GAF1 could regulate the expression of AtGA20ox2 in root tip and shoot apices. The expression of mutant AtGA20ox2 promoter-GUS (mtABC) and mutant AtGA20ox2 promoter-GUS (mtABCE) in shoot apices and in root tip decreased and abolished the response of AtGA20ox2 to GAs, respectively. The responses to GAs and uniconazole in root apices of mutant AtGA20ox2 promoter-GUS (mtABC) transgenic plants were higher than those in shoot apices (Fig. 6C). Together, these results indicate that binding of GAF1 to the AtGA20ox2 promoter is necessary for GA feedback regulation of AtGA20ox2 and its tissue-specific expression in the root tip and shoot apices.
GA-Induced DELLA Degradation Decreases the Binding of DELLA to GAF1-Binding Sites of the AtGA20ox2 Promoter
In GA feedback regulation, endogenous GA levels affect the amount of DELLA protein, which is reflected in the expression level of AtGA20ox2 (Phillips et al., 1995; Middleton et al., 2012). We hypothesized that a decrease in GAs would promote DELLA accumulation, after which DELLA would bind to the GAF1-binding sites of the AtGA20ox2 promoter in vivo, whereas exogenous GA treatment would promote the degradation of DELLAs, resulting in decreased availability of DELLA for binding to GAF1 sites on the AtGA20ox2 promoter. To confirm this, we performed a ChIP assay using transgenic plants carrying RGApro:RGA-GFP. A ChIP assay using anti-GFP antibody showed that that RGA-GFP binds to two regions in the AtGA20ox2 promoter, T1 (cisA and cisE) and T2 (cisB and cisC). In contrast, RGA-GFP did not bind to the coding region of AtGA20ox2. Moreover, the binding of RGA-GFP to the AtGA20ox2 promoter was increased by treatment with paclobutrazol, an inhibitor of GA biosynthesis, and decreased by GAs (Fig. 7).
Figure 7.
DELLAs bind to the GAF1-binding region of the AtGA20ox2 promoter in a GA-dependent manner. The DELLA complex binds to the AtGA20ox2 promoter in vivo. ChIP assays were performed with anti-GST or anti-GFP using transgenic plants carrying with RGApro:RGA-GFP that were treated with paclobutrazol (PAC) or PAC + GA3 or left untreated for 1 week. The coprecipitated level of each DNA fragment T1, including AtGA20ox2, cisA, and cisE, T2, including AtGA20ox2, cisB, and cisC, or AtGA20ox2 coding, was quantified by real-time PCR and normalized to the input DNA. The relative coprecipitated levels of each DNA fragment using anti-GST were set to 1, and relative enrichments of each DNA fragment using anti-GFP versus a DNA fragment using anti-GST are shown in the graphs. Error bars indicate the sd of three biological replicates (n = 3). *, P < 0.05 versus control by Student’s t test.
DISCUSSION
The DELLA-GAF1 Complex Is Involved in GA Feedback Regulation of AtGA20ox2
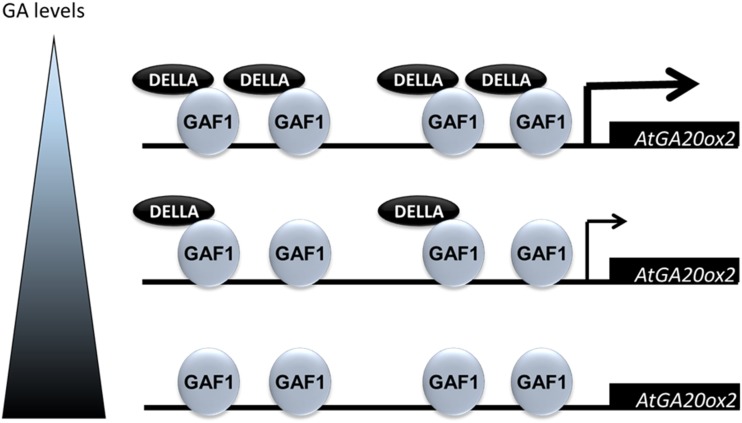
Previously, we generated transgenic plants expressing a mutant form of GAF1 that cannot bind to DELLAs (ΔPAM) under the control of the CaMV 35S promoter in the ga1-3 background, a GA-deficient mutant. The up-regulation of AtGA20ox2 in ga1-3 was repressed by overexpression of ΔPAM, indicating that the DELLA-GAF1 complex is involved in GA feedback regulation (Fukazawa et al., 2014). In this study, we identified four GAF1-binding sites in the AtGA20ox2 promoter, and these GAF1-binding sites were found to be necessary for GA feedback regulation of AtGA20ox2. In addition, we showed that the stabilization and localization of GAF1-GFP protein does not change with or without GA and that ΔPAM could bind to DNA (Fukazawa et al., 2014). These results indicated that DELLA is not required for binding of GAF1 to DNA, but DELLA is required for the activation of target genes, such as AtGA20ox2. Based on these data and ChIP analysis, we provide a model in which a decrease in GAs promotes the accumulation of DELLAs, upon which DELLA binds to GAF1-binding sites of the AtGA20ox2 promoter in vivo, whereas exogenous GA treatment promotes the degradation of DELLAs, reducing the amount of DELLA available to bind to the AtGA20ox2 promoter. Because DELLA proteins do not have an apparent DNA-binding motif, they associate with GAF1 to bind to the AtGA20ox2 promoter and might control the feedback regulation of AtGA20ox2 (Fig. 8).
Figure 8.
Proposed model of GA feedback regulation by the DELLA-GAF1 complex in a GA-dependent manner. In GA feedback regulation, endogenous GA levels affect the amount of DELLA protein, which is reflected in the expression level of AtGA20ox2. A decrease in GAs promotes DELLA accumulation, resulting in increased DELLA binding to GAF1-binding sites on the AtGA20ox2 promoter and the activation of AtGA20ox2 expression, whereas exogenous GA treatment suppresses the accumulation of DELLA, thus decreasing the amount of DELLA on the AtGA20ox2 promoter and AtGA20ox2 expression. DELLAs associate with GAF1 on the AtGA20ox2 promoter and might control the feedback regulation of AtGA20ox2.
GAF1-Binding Sequences in the Promoter of GA Feedback-Regulated Genes
GAF1 belongs to the IDD family of transcription factors, which bind to the consensus sequences TTTTGTCG and TTTGTCGTATT (Kozaki et al., 2004). Four out of five GAF1-binding sites identified in the AtGA20ox2 promoter, cisA, cisB, cisC, and cisD, include the TTGTC sequence. GAF1 also binds to cisE, but this site apparently does not have a TTGTC sequence; however, it does have a similar sequence, TTGGC. The maize ID1 protein also binds to the TTTTGGCGTTAT sequence, which does not include a complete consensus sequence (Kozaki et al., 2004). Comparison of these binding sequences suggests that TTTTG(G/T)CNNTA might be a more appropriate consensus sequence for GAF1 binding. AtGA3ox1 and GID1b also are activated by the DELLA-GAF1 complex (Fukazawa et al., 2014). TTGTC or TTGGC was found in the AtGA3ox1 and GID1b promoter, suggesting that the DELLA-GAF1 complex also is involved in the GA feedback regulation of these genes.
GAF1-Binding Sequences in the AtGA20ox2 Promoter Are Necessary for Tissue-Specific Expression of AtGA20ox2 in the Shoot Apices and Root Tip
The expression of AtGA20ox1 and AtGA20ox2 in seedlings is strongly down-regulated by GA (Phillips et al., 1995). In contrast, the expression of these genes is up-regulated in the ga1-3 GA-deficient mutant. The DELLA proteins GAI and RGA are major repressors of GA-promoted vegetative growth (Dill and Sun, 2001; King et al., 2001). mRNAs of RGA and GAI are detected in all tissues throughout plant development (Silverstone et al., 1998; Lee et al., 2002; Tyler et al., 2004). Rieu et al. (2008) investigated the effect of the gai-t6 and rga-24 loss-of-function mutations on the expression of GA20ox in the ga1-3 background. The expression of AtGA20ox1 and AtGA20ox2 was decreased significantly in gai-t6 rga-24 ga1-3, suggesting that both proteins are involved in GA feedback regulation of AtGA20ox1 and AtGA20ox2; however, the expression patterns of GAI and RGA are different from those of AtGA20ox1 and AtGA20ox2 in seedlings. GAI and RGA are expressed ubiquitously, whereas the expression of AtGA20ox1 and AtGA20ox2 is restricted to the meristematic tissue (Plackett et al., 2012). Because the expression pattern of GAF1 seemed similar to that of AtGA20ox1 and AtGA20ox2 (Fukazawa et al., 2014), GAF1 may be a determinant of the tissue-specific expression of AtGA20ox1 and AtGA20ox2 in GA-deficient conditions. AtGA20ox1 and AtGA20ox2 are the most highly expressed GA20ox genes during vegetative and early reproductive development, and both genes act redundantly to promote hypocotyl and internode elongation as well as flowering time. Because the mutants ga20ox1 ga20ox2 and gaf1 gaf2 exhibit short-hypocotyl and late-flowering phenotypes (Rieu et al., 2008; Plackett et al., 2012; Fukazawa et al., 2014), DELLA-GAF1 might be involved in the expression of AtGA20ox2 in the hypocotyl and shoot apex.
The DELLA-GAF1 Complex Is a Major Component of GA Feedback Regulation
The levels of bioactive GAs in plants are maintained by feedback regulation. In Arabidopsis, AtGA20ox1, AtGA20ox2, AtGA20ox3, and AtGA3ox1 are down-regulated by exogenous GAs. Feedback regulation of these genes did not occur in the penta della mutant (Livne et al., 2015), indicating that DELLA proteins are necessary for GA feedback regulation. Various other transcription factors involved in GA feedback regulation have been reported (Yamaguchi, 2008; Fukazawa et al., 2011). For example, RSG is involved in the GA feedback regulation of NtGA20ox1 (Fukazawa et al., 2000, 2010). The 14-3-3 proteins and NtCDPK1 (for Ca2+-dependent kinase1) control the localization of RSG in a GA-dependent manner (Igarashi et al., 2001; Ishida et al., 2004, 2008, Ito et al., 2010, 2014). AGF1 was identified as a regulator of feedback regulation of AtGA3ox1 in Arabidopsis (Matsushita et al., 2007). These transcription factors are involved in regulating the expression of GA20ox and GA3ox. They might act synergistically with the DELLA-GAF1 complex in GA feedback regulation. However, the relationship between these transcription factors and the DELLA-GAF1 complex is not clear at present.
Recently, it was reported that brassinosteroids (BRs) also regulate GA biosynthesis. The expression of AtGA20ox1 and AtGA3ox1 genes is lower in cpd, a BR-deficient mutant, and bri1-1, a BR-insensitive mutant, than in the wild type (Unterholzner et al., 2015). In addition, treatment with BR strongly increased AtGA20ox1 expression in cpd. BES/BZR1, a transcription factor regulating BR signaling, binds directly to AtGA20ox1 and AtGA3ox1 promoters. Although the GA levels are decreased in cpd and bri1-1 (Unterholzner et al., 2015), GA feedback regulation does not occur in these mutants, suggesting that BR signaling can affect GA feedback regulation. However, GA application reduced AtGA20ox1 expression in bri1-1, suggesting that the feedback repression of GA production does not rely on BR signaling. Moreover, AtGA20ox2 is not a target of BZR1 (Unterholzner et al., 2015). In GA-deficient conditions, accumulated DELLA interacts with BZR1 and inhibits its DNA-binding ability (Bai et al., 2012), thus inhibiting BZR1-dependent activation of its target genes, including GA biosynthetic genes. These data indicate that the regulation of GA biosynthetic genes by BZR1 is not involved in GA feedback regulation.
In this study, we showed that the DELLA-GAF1 complex is a main component of the GA feedback regulation of AtGA20ox2. Further investigation of how the DELLA-GAF1 complex regulates other target genes will help to reveal the molecular mechanisms of the fine-regulation of GA negative feedback and provide insights into the interaction between the control of endogenous levels of GAs and GA signaling.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All transgenic lines in this study were derived from Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0) wild type. To generate transgenic plants overexpressing MYC-tagged GAF1, a 4×MYC tag was amplified and cloned into the NotI site of pBIJ4-GAF1. To generate mutant plants, AtGA20ox2 promoter and mutated AtGA20ox2 promoters were cloned into the SalI-BglII site of the binary vector pBI101. The primers are listed in Supplemental Table S1. Agrobacterium tumefaciens-mediated Arabidopsis transformation was carried out by the floral dip method (Clough and Bent, 1998). The primer sets for cloning are listed in Supplemental Table S1. Plants were grown in a controlled growth chamber at 22°C under white light illumination (16 h of light/8 h of dark).
Application of GA3 to AtGA20ox2 or mtAtGA20ox2 Promoter:GUS Transgenic Plants
Seven-day-old seedlings of transgenic plants grown on one-half-strength Murashige and Skoog (1/2 MS) agar medium were transferred to 1/2 MS agar plates containing uniconazole or GA3 and grown for an additional 1 week. To investigate GA sensitivity, plants were treated or not with 1 mg L−1 uniconazole, 1 μm paclobutrazol, or 10 mg L−1 GA3.
Transactivation Assay
GA20ox2 promoters with a deletion from positions –1,003 to +77 (+1, transcription start site) were cloned into the SalI-BglII site of the p-less GUS vector, which is a pUC18-based plasmid that contains the GUS gene cassette of pBI101 (Takahashi et al., 1995). All primers used for transient assay analysis are shown in Supplemental Table S1. GAF1 and RGAΔ17, an internal deletion mutant of RGA, were cloned into the NotI-XhoI site of the pJ4 vector, which carries the CaMV 35S promoter with a viral translation enhancer, the Ω sequence (Fukazawa et al., 2000), to be used as effectors. Transactivation assays were performed as described previously (Ohta et al., 2001). Reporter plasmid (1.2 μg) and 1.6 μg of each effector plasmid were used for each bombardment. For the normalization of reporter gene activity, 0.4 μg of reference plasmid was cobombarded. Bombarded leaves were incubated in darkness for 20 h, after which luciferase activity was quantified. The data are presented are averages of three independent biological replicates.
Gel Retardation Assay
Gel retardation assays were performed following the procedure described previously (Fukazawa et al., 2000, 2010). GAF1 was cloned into the NotI-XhoI site of the pET30b vector (Novagen). Recombinant protein 6xHis-GAF1 was expressed and affinity purified from Escherichia coli BL21 (DE3) pLysE using Ni+ resin (Novagen). The nucleotide sequences of the double-stranded oligonucleotides used for the gel mobility shift assays are described in Supplemental Table S1. The oligonucleotides were annealed and then labeled using [α-32P]dCTP and the Klenow fragment of DNA polymerase I. Binding mixtures contained 50 fmol of labeled probe, 1 μg of purified recombinant GAF1 or 1 μg of control extract of E. coli, and 2 μg of poly(dI/dC). DNA competitor was used at 100-fold excess molar concentration. The binding buffer consisted of 20 mm Tris-HCl, pH 7.5, 3 mm MgCl2, 50 mm KCl, 1 mm EDTA, 10% (v/v) glycerol, and 2 μm ZnCl2. Reactions were incubated at 4°C for 30 min and loaded onto 4% (w/v) polyacrylamide gels containing 6.7 mm Tris-HCl, pH 7.5, 1 mm EDTA, and 3.3 mm sodium acetate.
Fluorometric GUS Enzyme Analysis
Transgenic plants were homogenized in the extraction buffer described by Jefferson et al. (1987). After removal of the cell debris by centrifugation at 15,000g for 10 min at 15°C, GUS activities were determined using 1 mm 4-methylumbelliferyl glucuronide as a substrate at 37°C. Before the feedback assay experiment, we confirmed GUS activity in each transgenic plant.
Histochemical Staining
Kanamycin-resistant transgenic plants were histochemically stained to detect GUS activity by immersing seedlings in a staining solution (100 mm sodium phosphate buffer, pH 7, with 50 mm NaCl, 1 mm potassium ferricyanide, 0.1% [v/v] Triton X-100, and 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide) overnight at 37°C. After staining, the samples were immersed in a fixing solution (5% [v/v] formaldehyde, 5% [v/v] acetic acid, and 20% [v/v] ethanol), followed by dechlorophylation in 70% (v/v) ethanol.
ChIP Assay
The ChIP experiment was performed following the procedure described previously (Fukazawa et al., 2010) with some modifications. In brief, 2-week-old 4×myc-GAF1 transgenic or Col-0 plants were cross-linked in 1% (v/v) formaldehyde by vacuum filtration for 10 min and incubated at 4°C for 1 h. Aliquots of each protein sample were immunoprecipitated with anti-GST (Santa Cruz Biotechnology), anti-myc (MBL International), and anti-GAI (generated in house) antibodies for 12 h at 4°C. Chromatin-antibody complexes were precipitated with salmon sperm DNA/protein G agarose beads at 4°C for 2 h. Primers used for ChIP analysis are listed in Supplemental Table S1. The coprecipitated level of each DNA fragment was quantified by real-time PCR using specific primer sets and normalized to input DNA. The coprecipitated levels of preimmune serum or anti-GST antibody (immunoprecipitated DNA/input DNA) were set to 1. The results are shown as relative DNA enrichment. Error bars indicate sd (n = 3).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: GAF1 (At3g50700), RGA (At2g01570), AtGA20ox2 (At5g51810), and ACTIN2 (At3g18780).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of the GAF1-binding region in the AtGA20ox2 promoter.
Supplemental Table S1. Primers used in this work.
Acknowledgments
We thank Dr. Belay T. Ayele (University of Manitoba) for helpful comments on the article.
Footnotes
This study was supported in part by grants from the Japan Society for the Promotion of Science (JSPS) to J.F. (26440148) and Y.T. (15H04392) and by grants from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan to J.F. (24116525) and Y.T. (24118004).
References
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY (2012) Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HH, Hwang I, Goodman HM (1995) Isolation of the Arabidopsis GA4 locus. Plant Cell 7: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colasanti J, Yuan Z, Sundaresan V (1998) The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603 [DOI] [PubMed] [Google Scholar]
- Dai M, Zhao Y, Ma Q, Hu Y, Hedden P, Zhang Q, Zhou DX (2007) The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol 144: 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dill A, Jung HS, Sun TP (2001) The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci USA 98: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Sun T (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Nakata M, Ito T, Matsushita A, Yamaguchi S, Takahashi Y (2011) bZIP transcription factor RSG controls the feedback regulation of NtGA20ox1 via intracellular localization and epigenetic mechanism. Plant Signal Behav 6: 26–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Nakata M, Ito T, Yamaguchi S, Takahashi Y (2010) The transcription factor RSG regulates negative feedback of NtGA20ox1 encoding GA 20-oxidase. Plant J 62: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y (2000) Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12: 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Teramura H, Murakoshi S, Nasuno K, Nishida N, Ito T, Yoshida M, Kamiya Y, Yamaguchi S, Takahashi Y (2014) DELLAs function as coactivators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in Arabidopsis. Plant Cell 26: 2920–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauvermale AL, Ariizumi T, Steber CM (2012) Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol 160: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochem J 444: 11–25 [DOI] [PubMed] [Google Scholar]
- Igarashi D, Ishida S, Fukazawa J, Takahashi Y (2001) 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13: 2483–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Fukazawa J, Yuasa T, Takahashi Y (2004) Involvement of 14-3-3 signaling protein binding in the functional regulation of the transcriptional activator REPRESSION OF SHOOT GROWTH by gibberellins. Plant Cell 16: 2641–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Yuasa T, Nakata M, Takahashi Y (2008) A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor REPRESSION OF SHOOT GROWTH in response to gibberellins. Plant Cell 20: 3273–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Nakata M, Fukazawa J, Ishida S, Takahashi Y (2010) Alteration of substrate specificity: the variable N-terminal domain of tobacco Ca2+-dependent protein kinase is important for substrate recognition. Plant Cell 22: 1592–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Nakata M, Fukazawa J, Ishida S, Takahashi Y (2014) Scaffold function of Ca2+-dependent protein kinase: tobacco Ca2+-DEPENDENT PROTEIN KINASE1 transfers 14-3-3 to the substrate REPRESSION OF SHOOT GROWTH after phosphorylation. Plant Physiol 165: 1737–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KE, Moritz T, Harberd NP (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159: 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki A, Hake S, Colasanti J (2004) The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res 32: 1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Park J, Lee N, Jeong J, Toh S, Watanabe A, Kim J, Kang H, Kim DH, Kawakami N, et al. (2013) ABA-insensitive3, ABA-insensitive5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25: 4863–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livne S, Lor VS, Nir I, Eliaz N, Aharoni A, Olszewski NE, Eshed Y, Weiss D (2015) Uncovering DELLA-independent gibberellin responses by characterizing new tomato procera mutants. Plant Cell 27: 1579–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-de la Rosa N, Pfeiffer A, Hill K, Locascio A, Bhalerao RP, Miskolczi P, Grønlund AL, Wanchoo-Kohli A, Thomas SG, Bennett MJ, et al. (2015) Genome wide binding site analysis reveals transcriptional coactivation of cytokinin-responsive genes by DELLA proteins. PLoS Genet 11: e1005337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita A, Furumoto T, Ishida S, Takahashi Y (2007) AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol 143: 1152–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton AM, Úbeda-Tomás S, Griffiths J, Holman T, Hedden P, Thomas SG, Phillips AL, Holdsworth MJ, Bennett MJ, King JR, et al. (2012) Mathematical modeling elucidates the role of transcriptional feedback in gibberellin signaling. Proc Natl Acad Sci USA 109: 7571–7576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NE, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P (1995) Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol 108: 1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plackett AR, Powers SJ, Fernandez-Garcia N, Urbanova T, Takebayashi Y, Seo M, Jikumaru Y, Benlloch R, Nilsson O, Ruiz-Rivero O, et al. (2012) Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 24: 941–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards DE, King KE, Ait-Ali T, Harberd NP (2001) How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol 52: 67–88 [DOI] [PubMed] [Google Scholar]
- Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, Griffiths J, Powers SJ, Gong F, Linhartova T, Eriksson S, Nilsson O, Thomas SG, et al. (2008) The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J 53: 488–504 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, et al. (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP. (2011) The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol 21: R338–R345 [DOI] [PubMed] [Google Scholar]
- Sun TP, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Sakai T, Ishida S, Nagata T (1995) Identification of auxin-responsive elements of parB and their expression in apices of shoot and root. Proc Natl Acad Sci USA 92: 6359–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA 96: 4698–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP (2004) DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Unterholzner SJ, Rozhon W, Papacek M, Ciomas J, Lange T, Kugler KG, Mayer KF, Sieberer T, Poppenberger B (2015) Brassinosteroids are master regulators of gibberellin biosynthesis in Arabidopsis. Plant Cell 27: 2261–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Winter CM, Wu MF, Kanno Y, Yamaguchi A, Seo M, Wagner D (2014) Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science 344: 638–641 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yang C, Ma Y, Li J (2016) The rice YABBY4 gene regulates plant growth and development through modulating the gibberellin pathway. J Exp Bot 67: 5545–5556 [DOI] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, et al. (2007) Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]