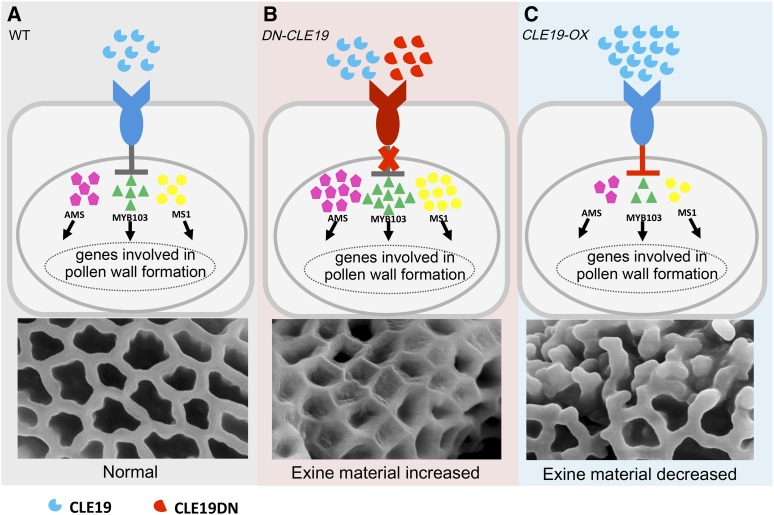
The proper amount of CLE19 is required for the normal formation of pollen exine through regulating the expression of AMS and its downstream networks.
Abstract
The CLAVATA3/ESR-RELATED (CLE) peptide signals are required for cell-cell communication in several plant growth and developmental processes. However, little is known regarding the possible functions of the CLEs in the anther. Here, we show that a T-DNA insertional mutant, and dominant-negative (DN) and overexpression (OX) transgenic plants of the CLE19 gene, exhibited significantly reduced anther size and pollen grain number and abnormal pollen wall formation in Arabidopsis (Arabidopsis thaliana). Interestingly, the DN-CLE19 pollen grains showed a more extensively covered surface, but CLE19-OX pollen exine exhibited clearly missing connections in the network and lacked separation between areas that normally form the lacunae. With a combination of cell biological, genetic, and transcriptomic analyses on cle19, DN-CLE19, and CLE19-OX plants, we demonstrated that CLE19-OX plants produced highly vacuolated and swollen aborted microspores (ams)-like tapetal cells, lacked lipidic tapetosomes and elaioplasts, and had abnormal pollen primexine without obvious accumulation of sporopollenin precursors. Moreover, CLE19 is important for the normal expression of more than 1,000 genes, including the transcription factor gene AMS, 280 AMS-downstream genes, and other genes involved in pollen coat and pollen exine formation, lipid metabolism, pollen germination, and hormone metabolism. In addition, the DN-CLE19(+/+) ams(−/−) plants exhibited the ams anther phenotype and ams(+/−) partially suppressed the DN-CLE19 transgene-induced pollen exine defects. These findings demonstrate that the proper amount of CLE19 signal is essential for the normal expression of AMS and its downstream gene networks in the regulation of anther development and pollen exine formation.
Pollen grains are generated in the male reproductive organ anther of flowering plants and are essential for plant fertility. In the model plant Arabidopsis (Arabidopsis thaliana), pollen grains are ellipsoidal, and the pollen surface is covered by a reticulate exine. In contrast, rice (Oryza sativa) pollen grains are globular and have a smooth surface without the reticulate structure. A well-organized pollen wall structure is essential for the physical and chemical stability of the mature pollen by providing protection for pollen grains from environmental stresses, such as desiccation and microbial attacks (Scott et al., 2004). The pollen wall is composed mainly of three layers: the intine layer, the exine layer, and the pollen coat (Zinkl et al., 1999; Edlund et al., 2004; Blackmore et al., 2007). The intine is the innermost layer of the pollen wall immediately adjacent to the plasma membrane of the pollen vegetative cell and is composed mainly of pectin, cellulose, hemicellulose, hydrolytic enzymes, and hydrophobic proteins (Scott et al., 2004). Both the exine layer and the pollen coat layer are basically of a lipidic nature. The exine is largely formed from acyl lipid and phenylpropanoid precursors, which together form the mixed stable biopolymer known as sporopollenin (Guilford et al., 1988; Bedinger et al., 1994; Thom et al., 1998). The pollen exine has many important functions in the prevention of dehydration of male gametophytes, the facilitation of pollen dispersal, and pollen-stigma recognition and adhesion (Zinkl et al., 1999). The pollen coat fills the cavities of the pollen exine during the late stages of pollen development, and previous studies have demonstrated that developmental defects of pollen wall structure or lack of pollen coat lipids in Arabidopsis lead to a failure or delay of pollen hydration and subsequent male sterility (Preuss et al., 1993; Mayfield and Preuss, 2000; Wilson and Zhang, 2009; Li and Zhang, 2010).
Anther development is precisely regulated temporally by transcriptional networks, receptor-like protein kinase-mediated signal pathways, and other pathways (Ma, 2005; Ge et al., 2010; Chang et al., 2011). The basic helix-loop-helix (bHLH) transcriptional factor DYSFUNCTIONAL TAPETUM1 (DYT1) is among the earliest acting male-specific regulators and is required for the normal development of tapetum by regulating the normal expression of more than 1,000 anther genes, and dyt1 mutants form abnormal prematurely vacuolated tapetal cells and lack mature pollen (Zhang et al., 2006; Feng et al., 2012; Gu et al., 2014; Zhu et al., 2015). In addition, bHLH010, bHLH089, and bHLH091 together are required for normal tapetum development and transcriptome and pollen fertility, forming both feed-forward and feedback loops with DYT1 (Zhu et al., 2015; Cui et al., 2016). Another bHLH transcription factor, ABORTED MICROSPORES (AMS), is a key regulator of anther transcriptome, sporopollenin biosynthesis and secretion, and pollen wall formation (Sorensen et al., 2003; Xu et al., 2010; Feng et al., 2012; Ma et al., 2012; Xu et al., 2014). DEFECTIVE IN TAPETAL DEVELOPMENT AND FUNCTION (TDF1)/MYB35 encodes a putative R2R3 MYB transcription factor, and loss of TDF1 function due to mutation or transgene causes tapetal hypertrophy extending into the locule and results in sporophytic male sterility (Zhu et al., 2008; Feng et al., 2012). MALE STERILITY1 (MS1) is a PHD-finger motif-containing nuclear protein and is essential for tapetum development at the postmeiotic phase and regulates genes important for exine formation (Wilson et al., 2001; Ito et al., 2007). In addition, another R2R3 MYB gene, MS188/MYB103/MYB80, is required for sporopollenin biosynthesis and sexine formation (Phan et al., 2011; Xiong et al., 2016). Further molecular genetic and bioinformatics studies revealed that these transcription factors likely form a genetic pathway as follows: DYT1, MYB35/TDF1, AMS, MS188/MYB103/MYB80, MS1, and MYB99 from upstream to downstream during anther/pollen development (Ito et al., 2007; Feng et al., 2012).
Extracellular ligands and their receptor-like protein kinase (RLK)-mediated signaling pathways play important roles in the regulation of anther development. In particular, the RLKs EMS1/EXS and SERK1/2 and the secreted peptide TPD1 are required for tapetum formation and function (Canales et al., 2002; Zhao et al., 2002; Yang et al., 2003; Albrecht et al., 2005; Colcombet et al., 2005). Furthermore, the TPD1 protein from microsporocyte precursors/microsporocyte interacts with the Leu-rich repeat domain of the RLK EMS1/EXS expressed in the tapetal precursor/tapetal cells to promote the phosphorylation of EMS1, thereby regulating anther tapetum development and function (Canales et al., 2002; Zhao et al., 2002; Yang et al., 2003, 2005; Jia et al., 2008; Huang et al., 2016; Li et al., 2017). In addition, brassinosteroids and the brassinosteroid cell surface receptor BRASSINOSTEROID INSENSITIVE1 are required for pollen exine formation and pollen release through the regulation of key genes in tapetum and pollen development, such as AMS, MYB103, MS1, and MS2 (Ye et al., 2010). In addition, several other RLKs are required for normal anther development, but their ligands remain unknown. For instance, ERECTA, ERECTA-LIKE1 (ERL1), and ERL2 are important for anther lobe formation and normal cell patterning (Torii et al., 1996; Shpak et al., 2003; Hord et al., 2008). BARELY ANY MERISTEM1 (BAM1) and BAM2 regulate the formation of anther somatic cell layers, and the bam1 bam2 double mutant anther forms many pollen mother-like cells but lacks the three subepidermal somatic cell layers (DeYoung et al., 2006; Hord et al., 2006). RECEPTOR-LIKE PROTEIN KINASE2 is essential for the formation of both the tapetum and the middle layer (Mizuno et al., 2007). On the other hand, besides TPD1, no other peptide signals have been implicated in anther or pollen development.
CLAVATA3 (CLV3)/EMBRYO SURROUNDING REGION (ESR)-RELATED (CLE) genes encode a family of putative peptide ligands, with at least 32 CLE members in Arabidopsis (Cock and McCormick, 2001). Mature CLE peptide signals are 12 to 14 amino acids long, and many of them play important roles in various plant developmental processes. For instance, CLV3 is necessary to restrict stem cell numbers in the shoot apical meristem (Fletcher et al., 1999) by promoting the formation of the CLV1/CLV2 protein complex to inhibit the expression of WUSCHEL in the organizing center of the shoot apical meristem (Laux et al., 1996; Fiers et al., 2004). CLE8 regulates the size of embryos and seeds and is expressed in the endosperm and the early embryo to promote WOX8 expression (Fiume and Fletcher, 2012). CLE40 regulates the activity of the root apical meristem and is expressed in differentiating cells in roots and likely acts through the receptor-like kinase CRINKLY4 to restrict the expression and position of WOX5 (Stahl and Simon, 2009). The CLE19 peptide triggers root meristem consumption in a CLV2-dependent manner (Casamitjana-Martínez et al., 2003; Fiers et al., 2005; Al-Refu et al., 2009). However, whether the CLE genes are required for normal anther development still remains unknown.
One difficulty in studying the endogenous functions of these small CLE genes is the shortage of genetic materials, namely the mutant plants. In addition, previous studies revealed that T-DNA insertion mutants of CLE1, CLE7, CLE10, CLE16, CLE18, or CLE19 in Arabidopsis exhibited no visible abnormal phenotype (Fiers et al., 2004; Jun et al., 2010), but overexpression of the CLE genes CLE2, CLE3, CLE4, CLE5, CLE6, CLE7, CLE10, CLE11, and CLE13 resulted in pleiotropic phenotypes similar to those of CLV3 or CLE40 overexpression plants, and in vitro application or overexpression of one of the CLE genes CLE19, CLE21, CLE25, CLE42, and CLE44 caused similar dwarf and short-root phenotypes (Fiers et al., 2004, 2005; Strabala et al., 2006), suggesting a high level of functional redundancy or overlap among CLE members. Fortunately, an antagonistic peptide technology (Song et al., 2013) was developed recently as an effective tool to investigate the endogenous functions of these functionally redundant secreted peptides. Using CLV3 as a test case, Song et al. (2013) examined the antagonistic peptide technology by transformations of wild-type Arabidopsis with constructs carrying the full-length CLV3 with every residue in the peptide-coding region replaced one at a time by Ala to probe the effectiveness of each mutation. They found that the Gly-to-Ala substitution in the core CLE motif caused a dominant-negative (DN) clv3-2-like phenotype. Further substitutions of the Gly residue individually with the other 18 possible proteinaceous amino acids determined that the Gly-to-Thr substitution resulted in the strongest antagonistic effect in the wild type (Song et al., 2012, 2013). With this strategy, the peptide signal CLE19 was revealed to be important for cotyledon establishment in embryos and endosperm development (Xu et al., 2015).
Here, we tested CLE genes for expression in the anther and show that CLE9, CLE16, CLE17, CLE19, CLE41, CLE42, and CLE45 were preferentially expressed in tapetal and reproductive cells in the anther. Using the antagonistic peptide technology, we generated DN mutants for these seven genes, and our analyses of these mutant transgenic plants indicated that they are important for normal pollen development, especially for the development of pollen exine. Using CLE19 as a representative, we further characterized its function in the regulation of pollen exine formation using a combination of morphological molecular and transcriptomic analyses of cle19, DN-CLE19, and CLE19-OX mutants. We finally revealed that CLE19 acts as a negative regulatory element in tapetum development, sporopollenin biosynthesis, and exine formation, likely through the regulation of the transcription factor AMS and downstream genes. Thus, we propose a novel signaling pathway starting from an extracellular peptide signal and eventually affecting the intracellular transcriptional network that is crucial for the regulation of pollen wall development.
RESULTS
Seven CLE Genes Are Expressed Preferentially in Arabidopsis Tapetal and Reproductive Cells
The expression of CLE genes in Arabidopsis was first examined by searching public microarray databases (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi and http://www.arabidopsis.org/portals/expression/microarray/ATGenExpress.jsp) and using quantitative real-time PCR and RNA in situ hybridization analyses to detect the expression of these CLE genes in the developing anther. Among the 14 CLE genes that were included in the microarray expression databases, CLE9, CLE10, CLE21, CLE26, CLE40, CLE44, and CLV3 exhibited relatively high expression levels in the anther, whereas CLE6 and CLE12 were relatively highly expressed in the mature pollen (Supplemental Fig. S1A). Quantitative real-time PCR analyses using Columbia-0 (Col-0) inflorescence and stage 4 to 12 anthers revealed that CLE9, CLE16, CLE17, CLE25, CLE27, CLE41, CLE42, and CLE45 were relatively highly expressed in both inflorescence and stage 4 to 12 anthers (Supplemental Fig. S1B), suggesting that they might be involved in male reproductive development.
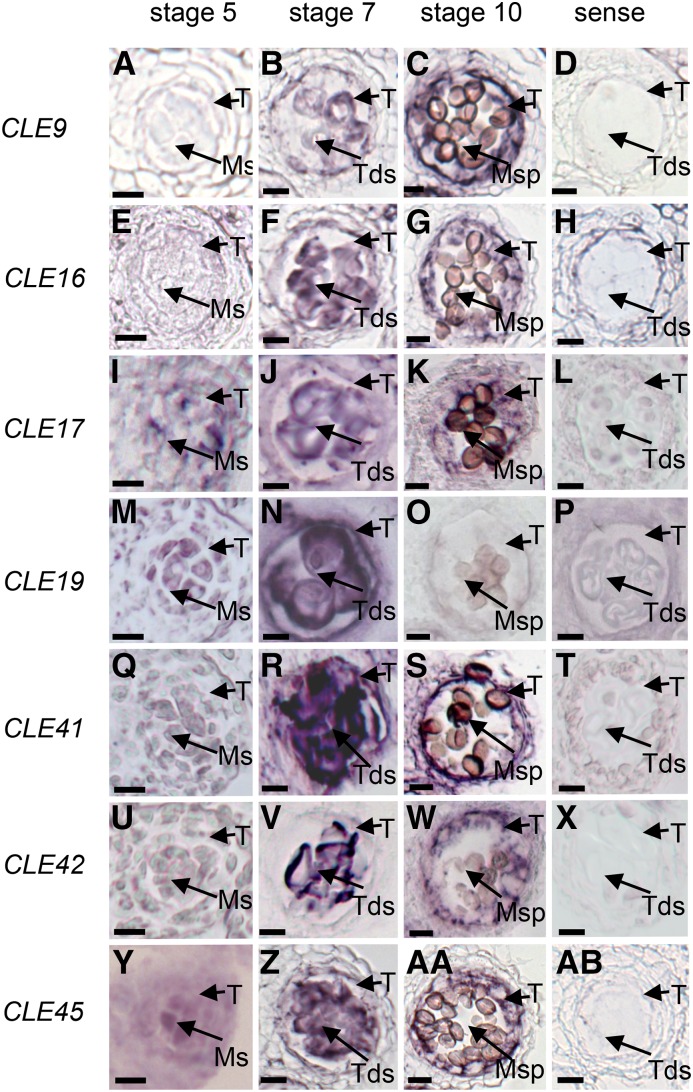
Further RNA in situ hybridization analyses were performed to investigate the expression patterns of these anther-expressed CLE genes. Strong CLE9, CLE16, CLE17, CLE19, CLE41, CLE42, and CLE45 signals were detected in the anther at various developmental stages (Fig. 1). For instance, CLE9 and CLE16 were both expressed in the tapetum and microspores at anther stages 7 to 10 but not in either cell layer at stage 5 (Fig. 1, A–H). CLE19 was expressed preferentially in both tapetal and male reproductive cells at anther stages 5 to 7, and CLE17, CLE41, CLE42, and CLE45 were expressed preferentially in both tapetal and male reproductive cells at anther stages 5 to 10 (Fig. 1, I–AB), suggesting their involvement in regulating tapetum function and pollen development. In addition, their highly similar spatial-temporal expression pattern also suggested that these seven CLE genes possibly participate in similar anther developmental processes.
Figure 1.
Spatial and temporal expression analyses of CLE genes. Expression patterns are shown for CLE9 (A–D), CLE16 (E–H), CLE17 (I–L), CLE19 (M–P), CLE41 (Q–T), CLE42 (U–X), and CLE45 (Y–AB) in wild-type anthers by RNA in situ hybridization. Sections are from anthers of stage 5 (A, E, I, M, Q, U, and Y), stage 7 (B, F, J, N, R, V, and Z), stage 10 (C, G, K, O, S, W, and AA), and controls (D, H, L, P, T, X, and AB) using sense probe on wild-type stage 7 anthers. Ms, Microsporocytes; Msp, microspores; T, tapetum; Tds, tetrads. Bars = 10 μm.
DN Mutant Transgenic Plants of CLE9, CLE16, CLE17, CLE19, CLE41, CLE42, and CLE45 Exhibited Abnormal Anther Development
To further investigate the functions of the CLE genes in anther development, the T-DNA insertional mutants of CLE9, CLE17, CLE19, or CLE42 and the RNA interference (RNAi) mutant of CLE16 were obtained (Supplemental Fig. S2), and the transcript levels of these genes in mutant plants were analyzed using real-time quantitative PCR. The cle9-1, cle16-1, cle17-1, cle19-1 (in the qrt1-2 background), cle19-2, and cle42-1 mutants exhibited drastically reduced expression of the relevant genes (Supplemental Fig. S2) and were further examined phenotypically. Both cle19-1 qrt1-2 and cle19-2 produced abnormally short siliques and had slightly decreased anther size and some aborted pollen grains (Fig. 2, A–E, J–N, S–W, and AB–AF). Considering that cle19-1 was in the qrt1-2 background, we observed the phenotypes of silique, flower, anther, and pollen in the qrt1-2 plants and did not find any defects except for the pollen phenotype (Fig. 2, C, L, U, and AD). The above results suggested that CLE19 is possibly involved in anther development. In contrast, the cle9-1, cle16-1, cle17-1, and cle42-1 single mutants all exhibited normal plant growth and flower and anther development (Supplemental Fig. S3; Supplemental Table S1).
Figure 2.
Phenotypic analyses of wild-type, cle19 T-DNA insertion mutant, DN-CLE19 transgenic, and CLE19-OX transgenic plants. A to I, Phenotypic analysis of plant growth of wild-type, cle19 T-DNA insertion mutant, DN-CLE19 transgenic, and CLE19-OX transgenic plants. J to R, Phenotypic analysis of flowers of wild-type, CLE19 T-DNA insertion mutant, DN-CLE19 transgenic, and CLE19-OX transgenic plants. S to AA, Phenotypic analysis of anthers of wild-type, CLE19 T-DNA insertion mutant, DN-CLE19 transgenic, and CLE19-OX transgenic plants. AB to AJ, Phenotypic analysis of pollen grains of wild-type, CLE19 T-DNA insertion mutant, DN-CLE19 transgenic, and CLE19-OX transgenic plants. Red arrowheads indicated aborted pollen grains. Bars = 2 cm for plants, 500 μm for flowers, 200 μm for anthers, and 25 μm for pollen grains.
Based on the high amino acid similarity between the functional domains of these Arabidopsis CLE members (Ni and Clark, 2006) and the no/weak defects in anther development observed in the available cle single mutants, we proposed a possible functional redundancy among these anther-expressed CLE genes in the regulation of anther development. Therefore, we generated double and triple mutants of these CLE genes by crossing cle16-1, cle17-1, cle19-1, and cle42-1. The qrt1-2 mutation in cle19-1 plants segregated away during this process. Compared with the cle19-1 mutant, our phenotypic and statistical analyses showed that the cle16 cle19, cle17 cle19, and cle19 cle42 double mutants had fewer pollen grains in the anther (Supplemental Fig. S4, A–AA; Supplemental Table S1). Moreover, the cle17 cle19 cle42 triple mutant showed more barren siliques, less pollen amount, and a larger fraction of defective pollen in the anther (Supplemental Fig. S4, A–AA; Supplemental Table S1). In addition, we generated DN mutant plants for CLE9, CLE16, CLE17, CLE19, CLE41, CLE42, and CLE45 by transforming wild-type plants separately with fusions of the CLE promoter and the coding region for the CLEG6T point mutation (DN-CLE). The DN-CLE transgenic plants exhibited obviously reduced silique length (Fig. 2, F and G; Supplemental Fig. S5, B-G). Alexander staining and scanning electron microscopy experiments revealed abnormal anther development with aborted pollen in the anthers of these DN-CLE plants (Fig. 2, X, Y, AG, and AH; Supplemental Fig. S5, H–U). These results suggested that the normal functions of these genes are important for anther development and that the anther-expressed CLE9, CLE16, CLE17, CLE19, CLE41, and CLE42 might function in a redundant manner. Although the DN transgenic plants of all seven CLE genes exhibited abnormal anther development, only the cle19 knockout mutants showed weak anther developmental defects; therefore, we focused the subsequent detailed functional analyses on CLE19 to gain insights into its role in anther/pollen development.
CLE19 Is Required for Normal Pollen Development, Pollen Germination, and Pollen Tube Elongation
In addition to the above-mentioned cle19 and DN-CLE19 mutants, we also wanted to investigate whether an increase of CLE19 expression affects male reproductive development, using the 35S promoter-driven CLE19 overexpression transgenic lines (CLE19-OX; Fiers et al., 2004). Two CLE19-OX transgenic lines with different CLE19 expression levels, CLE19-OX-S and CLE19-OX-M (Fig. 3D), were selected for the analysis of anther developmental phenotypes. Both lines exhibited obviously reduced fertility, including aborted siliques, small anthers, and abnormal pollen (Fig. 2, H, I, Q, R, Z, AA, AI, and AJ).
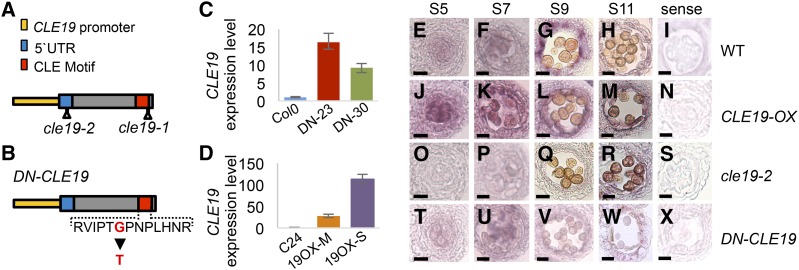
Figure 3.
Spatial and temporal expression of CLE19 in wild-type, CLE19-OX, cle19-2, and DN-CLE19 anthers. A, Diagram showing the T-DNA insertion sites of cle19-1 and cle19-2. UTR, Untranslated region. B, Diagram showing the amino acid mutation site in DN-CLE19. C and D, Quantitative real-time PCR analysis to detect CLE19 expression in stage 4 to 12 anthers of the wild type, DN-CLE19 (C), and CLE19-OX (D) using EF1α as an internal control. DN-23 and DN-30 indicate two individual CLE19::DN-CLE19 transgenic lines. 19OX-M and 19OX-S indicate two independent 35S::CLE19 transgenic lines. Data from three biological replicates were collected. E to X, RNA in situ hybridization results showing the expression of CLE19 in wild-type (WT), CLE19-OX-S, cle19-2, and DN-CLE19-23 mutant anthers. Bars = 10 μm.
As detected using real-time PCR, the expression level of CLE19 was found to be elevated up to over 100-fold in the CLE19-OX-S line (Fig. 3D) and to a lesser extent in DN-CLE19 anthers (Fig. 3C). The anther expression pattern of CLE19 in the transgenic plants was examined further using RNA in situ hybridization analysis (Fig. 3, E–X). Consistently, the expression of CLE19 was obviously increased in CLE19-OX-S and decreased in cle19-2 in both tapetal and male reproductive cells (Fig. 3, J–S). However, the expression in DN-CLE19 anthers showed no obvious difference from that in the wild type (Fig. 3, T–X), suggesting that the 35S promoter is not highly active in the tapetum and that in situ hybridization might not be quantitative enough to detect relatively small differences in expression level.
We further performed statistical analysis for various male fertility processes of the cle19-2, DN-CLE19-23, and CLE19-OX-S plants, including the pollen number in each anther, pollen germination and pollen tube elongation, silique length and silique number in each main stem, and seed number in each silique. Compared with the wild type, DN-CLE19-23 produced obviously fewer pollen grains (48.4% of that in each wild-type anther), a reduced pollen germination rate (15.7% of the rate for the wild type), defective pollen tube elongation, a decreased number of siliques from one reproductive shoot, reduced silique length, and a lower seed number in each silique (Table I). The cle19-1 and cle19-2 plants exhibited similar but weaker phenotypes compared with those of DN-CLE19, suggesting a functional redundancy among the anther-expressed CLE family members, as other members also might be inhibited by the mutated CLE peptide. The CLE19-OX-S anther produced only 9% of the amount of pollen in one wild-type anther (Table I). All these results demonstrated the important function of CLE19 in male reproductive processes and that the correct level of CLE19 expression is important for normal function.
Table I. Phenotypic analysis of cle19-2, DN-CLE19-23, and CLE19-OX-S.
Data are means ± sd. For silique number, three replicates using 30 branches were analyzed. For silique length, three replicates and 30 siliques in each replicate were analyzed. For seed number, three replicates and 30 siliques in each replicate were analyzed. For pollen number, three replicates and more than 30 anthers in each replicate were analyzed. For pollen germination rate and pollen tube length analyses, three replicates and more than 100 pollen grains for each replicate were observed.
| Plant | Silique No. | Silique Length | Seed No. per Silique | Pollen No. per Anther | Pollen Germination Rate | Pollen Tube Length |
|---|---|---|---|---|---|---|
| mm | % | μm | ||||
| Col-0 | 68.9 ± 3.9 | 14.3 ± 0.7 | 54.8 ± 3.6 | 640.3 ± 22.0 | 99.97 ± 5.25 | 208.0 ± 11.8 |
| cle19-2 | 55.9 ± 3.9 | 13.0 ± 1.9 | 41.58 ± 10.1 | 413.6 ± 40.1 | 41.67 ± 5.16 | 137.0 ± 25.0 |
| DN-CLE19-23 | 30.6 ± 3.0 | 10.7 ± 2.5 | 30.6 ± 11.5 | 310.2 ± 99.6 | 15.67 ± 4.72 | 82.8 ± 6.5 |
| C24 | 67.2 ± 3.7 | 14.6 ± 1.0 | 48.8 ± 4.5 | 617.3 ± 34.9 | 99.96 ± 7.20 | 205.1 ± 9.9 |
| CLE19-OX-S | 12.9 ± 3.1 | 9.6 ± 2.4 | 22.0 ± 9.4 | 54.5 ± 26.7 | 98.53 ± 15.18 | 35.5 ± 8.7 |
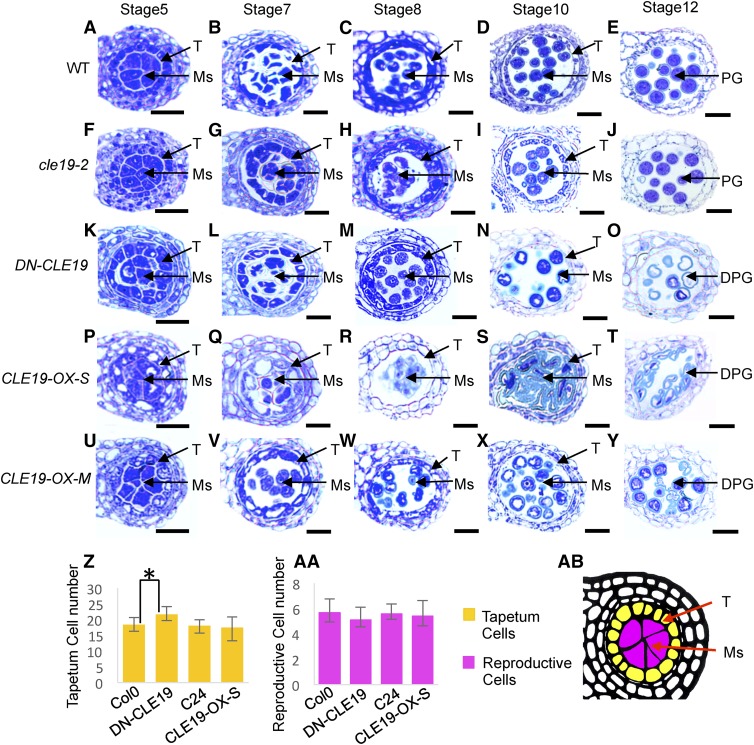
To further investigate the function of CLE19 in anther development, particularly to identify the earliest stages when morphological differences can be observed, anther transverse semithin sections of cle19-2, DN-CLE19-23, and CLE19-OX mutants were examined. At anther stage 5, the wild-type anther formed well-organized five-cell layers in each anther lobe, the epidermis, endothecium, middle layer, tapetum, and microsporocytes, from outer to inner. Afterward, the microsporocytes underwent meiosis and formed the microspores that developed further into the pollen in the wild type at stages 6 to 13.
Compared with the wild type (Fig. 4, A–E), the cle19-2 mutant anther showed no obvious cytological defects during anther stages 5 to 10, except for fewer pollen grains in the anther at stage 12 (Fig. 4, F–J). However, the DN-CLE19-23 anthers tended to produce more than normal tapetum cells surrounding the microsporocytes at stage 5, and aborted pollen was observed at stage 10 and onward (Fig. 4, K–O). In comparison, change of the tapetal cell number was not obvious in either CLE19-OX-S or CLE19-OX-M anthers; nevertheless, most of the tapetal cells were enlarged and excessively vacuolated at stages 5 to 7, and the CLE19-OX-S tapetum soon degenerated at stage 8 (Fig. 4, P–R). Therefore, the development of most pollen grains was defective (Fig. 4, S and T). The CLE19-OX-M tapetum exhibited a weaker vacuolated morphology compared with CLE19-OX-S but also disappeared at stage 10. A small portion of defective pollen was observed at stage 10 and onward (Fig. 4, U–Y). These results further strengthened the idea that a proper amount of CLE19 is required for normal anther development.
Figure 4.
Cell biological analyses of wild-type, cle19-2, DN-CLE19, and CLE19-OX anthers. A to E, Wild-type (WT) anther morphology using semithin transverse sections. F to J, Morphology of cle19-2 anthers using semithin transverse sections. K to O, Morphology of DN-CLE19 anthers using semithin transverse sections. P to T, Morphology of CLE19-OX-S anthers using semithin transverse sections. U to Y, Morphology of CLE19-OX-M anthers using semithin transverse sections. Z to AA, Statistical analyses of the tapetal or reproductive cell numbers in DN-CLE19, CLE19-OX, and wild-type anthers, respectively. *P < 0.05 by Student's t-test. AB, How the numbers of tapetal or reproductive cells in each lobe in transverse sections were calculated. DPG, Degenerated pollen grains; Ms, meiocytes; PG, pollen grains; T, tapetum. Bars = 20 μm.
Pollen Exine Formation Was Affected in CLE19 Mutants
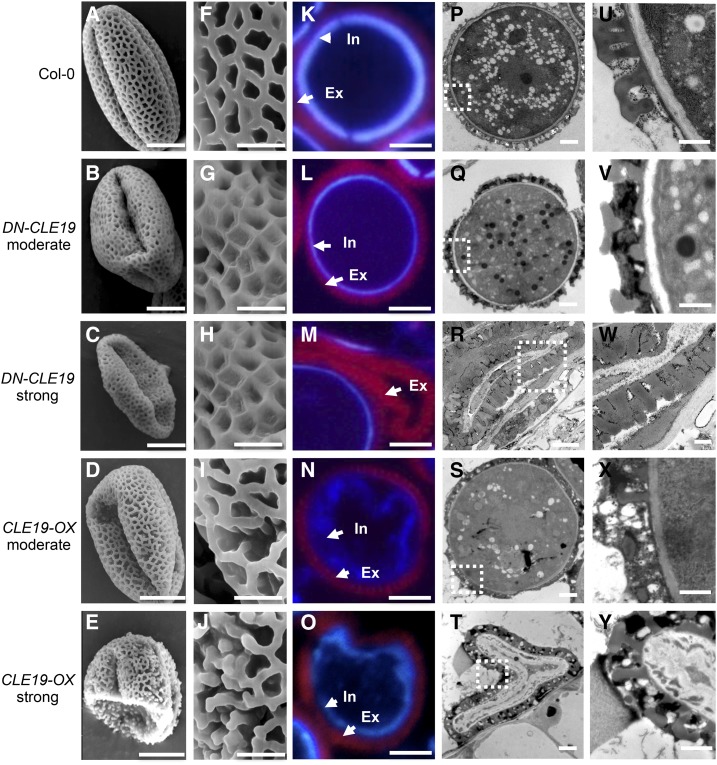
We then further investigated whether CLE19 is involved in pollen exine formation. The morphological pollen exine structure of wild-type, DN-CLE19-23, and CLE19-OX-S plants was first observed using scanning electron microscopy. Pollen grains with either moderate or severe defects were analyzed in both DN-CLE19-23 and CLE19-OX-S plants. The results showed that the external surface of wild-type pollen exine had a network-like structure formed with well-organized lacunae and three narrow apertures (Fig. 5, A and F). In comparison, both the moderately defective and the severely defective pollen of DN-CLE19-23 exhibited more extensively covered surfaces with smaller lacunae filled with additional materials (Fig. 5, B, C, G, and H). In contrast, the CLE19-OX pollen exine exhibited clearly missing connections in the network and lack of separation between the spaces that would form the lacunae (Fig. 5, D, E, I, and J), suggesting that the proper amount of CLE19 is required for the formation of normal pollen exine, possibly promoting the proper organization of the network and lacunae.
Figure 5.
Microscopic analyses of wild-type, DN-CLE19, and CLE19-OX pollen exine. A to J, Pollen grains from wild-type, DN-CLE19, and CLE19-OX plants visualized by scanning electron microscopy. K to O, Cytochemical staining of semithin sections of wild-type, DN-CLE19, and CLE19-OX pollen. P to Y, Wild-type, DN-CLE19, and CLE19-OX pollen visualized by TEM. A, F, K, P, and U, Wild-type pollen. B, G, L, Q, and V, DN-CLE19-M pollen. C, H, M, R, and W, DN-CLE19-S pollen. D, I, N, S, and X, CLE19-OX-M pollen. E, J, O, T, and Y, CLE19-OX-S pollen. Ex, Exine; In, intine. Bars = 10 μm for A to E, 2 μm for F to J, 5 μm for K to O, 2 μm for P to T, and 0.5 μm for U to Y.
Given that the pollen wall is composed of the exine and the intine layers, we further examined the characteristics of these two pollen wall layers through a diethyloxadicarbocyanine iodide- and Tinopal-based staining method. As shown in Figure 5K, the wild-type pollen exine was stained as red and the intine was stained as purple. In comparison, stronger red exine signals but minimal purple intine signals were observed in the DN-CLE19-23 pollen grains (Fig. 5, L and M), especially in the DN-CLE19 severe pollen grain, where little or no intine signal was observed (Fig. 5M), suggesting that CLE19 is required for the normal formation of both pollen exine and intine. Consistently, the CLE19-OX pollen exhibited discrete red exine signals and irregular inward-folded purple intine fluorescence (Fig. 5, N and O). These data suggested that CLE19 possibly negatively and positively regulates pollen exine and intine formation, respectively.
To further analyze the function of CLE19 in pollen exine formation, transmission electron microscopy (TEM) analyses were performed. Wild-type pollen grains exhibited a uniformly structured cell wall with clearly visible exine (T-shaped bacula and tectum; Fig. 5, P and U). However, although the exine T-shaped structure of the DN-CLE19-23 pollen appeared normal, the lacunae were filled with extra sporopollenin-like materials (Fig. 5, Q, R, V, and W). In contrast, the CLE19-OX exine-like layer lacked the complete bacula/tectum structure (Fig. 5, S, T, X, and Y), demonstrating that CLE19 is a negative regulator of pollen exine biogenesis, especially for the construction of the T-shaped structure.
DN-CLE19 Transgenic Plants Are Deficient in Pollen Coat Formation
As the DN-CLE19 mutant pollen grains exhibited germination defects (Table I) and the pollen coat was filled with extra unknown substances, we assessed whether these substances result from additional synthesis or defective degradation/exploitation. Thus, the pollen coat structures of wild-type and DN-CLE19 plants at anther stages 10 to 13 were examined. Both wild-type and DN-CLE19 pollen in stage 10 to 11 anthers were covered with pollen coat materials, which filled the reticulate networks that surrounded the wild-type pollen; such materials were reduced and almost invisible in stage 12 to 13 wild-type pollen. Therefore, the well-organized reticulate exine networks were clearly visible on the surface of the wild-type pollen. However, these pollen coat materials were still present on DN-CLE19 mutant pollen at anther stages 12 to 13 (Supplemental Fig. S6C), suggesting that the defect was in the degradation or removal of these materials.
The Expression of AMS and Downstream Transcription Factor Genes Was Affected in cle19 Mutants
As the transcription factor genes DYT1, MYB35/TDF, AMS, MS188/MYB103/MYB80, and MS1 are required for normal anther development in a DYT1-MYB35/TDF1-AMS-MS188/MYB103/MYB80-MS1 regulatory pathway (Ge et al., 2010; Chang et al., 2011; Zhu et al., 2011) and AMS was reported to act as a master regulator coordinating pollen wall development (Xu et al., 2014), we further examined the expression levels of genes encoding these five transcription factors in DN-CLE19 and CLE19-OX anthers using quantitative real-time PCR analyses. The expression levels of AMS, MS188/MYB103/MYB80, and MS1 showed over 2-fold increases in DN-CLE19 but over 2-fold decreases in CLE19-OX anthers (Fig. 6, A and B), suggesting that these genes are negatively regulated by CLE19. However, the expression of either DYT1 or MYB35 exhibited no obvious difference between the DN-CLE19 and wild-type anthers, but both showed 40% reductions in CLE19-OX anthers (Fig. 6, A and B), suggesting that the expression of these two genes possibly was not or was only slightly affected by CLE19. These results suggested that CLE19 probably functions as a negative regulator of the AMS-dependent transcriptional networks for normal pollen wall formation.
Figure 6.
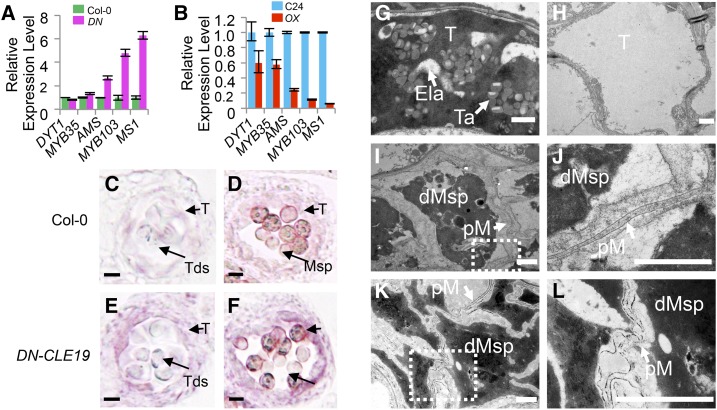
Quantitative real-time PCR and RNA in situ hybridization analyses to test the expression of AMS and downstream transcription factor genes in DN-CLE19 and CLE19-OX transgenic anthers, and TEM observation of CLE19-OX and ams tapetal cells and microspores. A and B, Relative expression levels of the transcription factor genes DYT1, MYB35, AMS, MYB103, and MS1 in DN-CLE19 and CLE19-OX mutants compared with related wild-type plants, with ACTIN expression as an internal control. C and D, Expression of AMS in wild-type anthers. E and F, Expression of AMS in DN-CLE19 anthers. C and E show anther stage 7, and D and F show anther stage 10. G to L, Tapetal cells of wild-type and CLE19-OX-S anthers visualized by TEM. G, The wild type. H, CLE19-OX-S. I to L, Microspore cells of CLE19-OX-S and ams anthers visualized by TEM. I and J, CLE19-OX-S. K and L, ams. dMsp, Degenerated microspores; Ela, elaioplasts; pM, primexine; T, tapetum; Ta, tapetosomes. Bars = 1 μm.
The expression of AMS in DN-CLE19 and CLE19-OX anthers was tested further using RNA in situ hybridization analysis. The results showed that the AMS signal was hardly seen at anther stage 5 (Fig. 6, C and D) and was detected in both the tapetum and microsporocytes at stages 7 to 10, albeit weakly (Supplemental Fig. S7, B and C); the signal then disappeared at anther stage 12 (Supplemental Fig. S7, B and C). In comparison, AMS expression was obviously increased in the DN-CLE19 transgenic anthers at anther stages 7 to 10 (Fig. 6, E and F), suggesting that inhibition of the CLE19 function caused an increase in AMS expression. However, no AMS signal was detected in CLE19-OX transgenic anthers at all examined stages, similar to the results in the ams mutant (Supplemental Fig. S7, G–P), revealing that the increased CLE19 function suppresses AMS expression. These data demonstrated that CLE19 acts as a negative upstream regulator of AMS.
CLE19-OX Anthers Had ams-Like Swollen Tapetal Cells
It is known that tapetal cells secrete sporopollenin precursors onto the primexine of the developing pollen for the formation of the pollen exine and that AMS plays an important role in this process, with ams showing severely swollen tapetal cells with large vacuoles and few lipidic tapetosomes and elaioplasts (Xu et al., 2014). We postulated that, if CLE19 acts as a negative upstream regulator of AMS, tapetal cells of the CLE19-OX anther would exhibit similar abnormal phenotypes to that of ams. Therefore, we performed TEM analysis of CLE19-OX tapetal cells. As expected, in contrast to the condensed and degenerated cytoplasm of wild-type tapetal cells, with evident disintegration of cell membrane and accumulation of lipidic tapetosomes and elaioplasts (Fig. 6G), CLE19-OX tapetal cells were highly vacuolated and swollen, with large vacuoles and lack of lipidic tapetosomes and elaioplasts (Fig. 6H). In addition, sporopollenin precursors on the primexine were clearly observed on the outer surface of wild-type pollen grains; however, no obvious accumulation of sporopollenin precursors was seen on the primexine of the CLE19-OX (Fig. 6, I and J) and ams (Fig. 6, K and L) pollen grains.
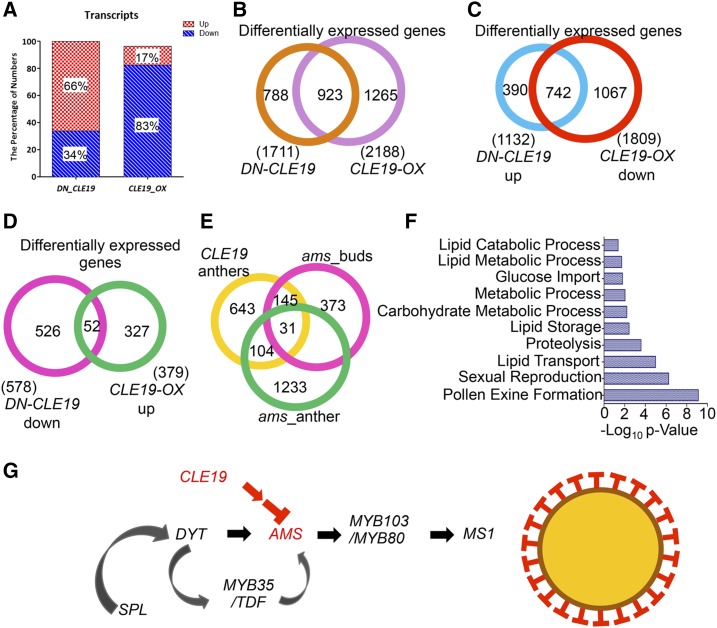
Altered Expression of 1,711 and 2,188 Genes in DN-CLE19 and CLE19-OX Anthers, Respectively
To further investigate the CLE19 effects on anther transcriptomes, those of stage 4 to 10 anthers were assessed (Supplemental Fig. S8, A and B). Compared with the wild type, 578 (34% of the total of 1,711 differentially expressed genes) were down-regulated and 1,133 (66%) were up-regulated in the DN-CLE19 anthers (Fig. 7, A and B; Supplemental Data Set S1). Moreover, the expression of 1,809 genes was down-regulated and that of 379 genes was up-regulated (83% and 17% of the 2,188 total altered genes, respectively) in the CLE19-OX anthers (Fig. 7, A and B; Supplemental Data Set S2). The fact that most of the differentially expressed genes were up-regulated in the loss-of-function DN-CLE19 mutant and down-regulated in the gain-of-function CLE19-OX mutant suggests that CLE19 negatively regulates anther gene expression. Interestingly, the expression of 742 genes was both up-regulated in DN-CLE19 and down-regulated in CLE19-OX (Fig. 7C; Supplemental Data Set S3), while only 52 genes were both down-regulated in DN-CLE19 and up-regulated in CLE19-OX transcriptomes (Fig. 7D; Supplemental Data Set S4), further supporting CLE19 functioning as a negative regulator of many anther genes.
Figure 7.
Transcriptomic comparison analyses between CLE19 and AMS. A, Percentages of up- and down-regulated genes in the DN-CLE19 and CLE19-OX transcriptomes, respectively. B, Comparison of differentially expressed genes between DN-CLE19 and CLE19-OX transcriptomes. C, Comparison of differentially expressed genes between DN-CLE19 up-regulated and CLE19-OX down-regulated gene groups. D, Comparison of differentially expressed genes between DN-CLE19 down-regulated and CLE19-OX up-regulated gene groups. E, Venn diagram showing the overlap between genes affected in CLE19 anthers, ams floral buds, and ams anthers. F, Enriched biological process categories for transcriptome analysis of genes affected in both CLE19 and ams mutants. G, CLE19 genetic pathways controlling anther development.
The Expression of 280 AMS-Downstream Genes Was Affected in Both DN-CLE19 and CLE19-OX Mutants
Considering the similar developmental defects of tapetum of CLE19-OX and the ams mutant, we further compared the 923 genes that were commonly regulated in DN-CLE19 and CLE19-OX in our transcription data sets with the microarray-based ams transcriptomic data of the stage 4 to 7 anther and buds of four anther developmental stages (Xu et al., 2010; Feng et al., 2012; Ma et al., 2012). The results revealed that the expression of 135 out of the 923 CLE19 downstream genes also was altered in the ams stage 4 to 7 anthers and that of 201 also was altered in various stages of ams buds (Fig. 6E; Supplemental Data Set S5). The combined set included a total of 280 genes commonly affected by both CLE19 and AMS; the putative functions of these genes are enriched in various biological processes, including pollen exine formation, sexual reproduction, lipid transport, lipid storage, and lipid metabolism (Fig. 7F), suggesting that CLE19 likely functions through AMS-MYB103/MYB80-MS1 pathways, but probably not through the more upstream regulators DYT1 and/or MYB35, to regulate the transcriptional networks important for pollen exine formation (Fig. 7G).
CLE19 Controls the Synthesis of Lipidic and Phenolic Components and Flavonoids Required for Pollen Wall Formation
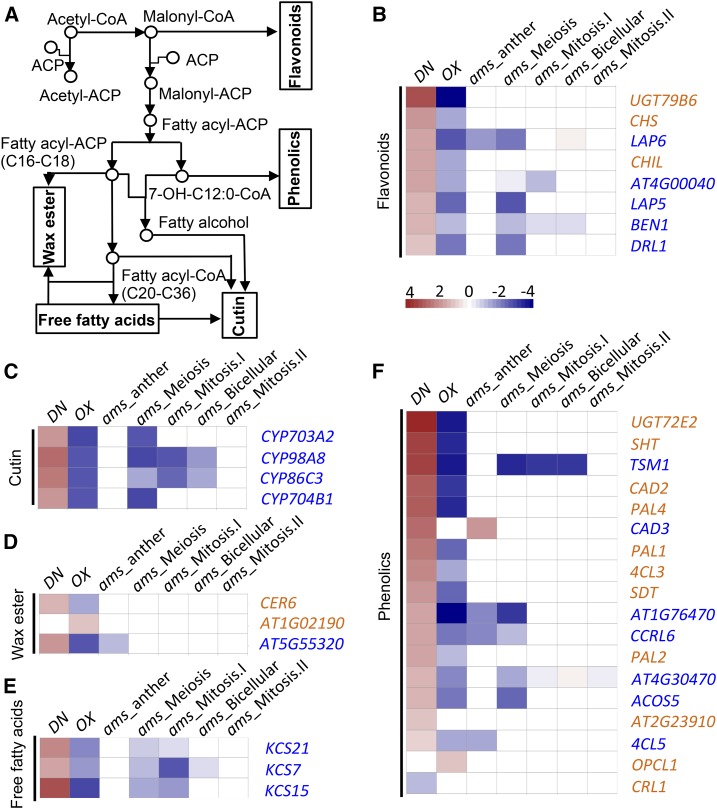
Sporopollenins are the major components of pollen exine. Increasing evidence indicates that sporopollenin consists mainly of complex aliphatic monomers, including very-long chain fatty acids and their polyhydroxylated derivatives, as well as phenolic compounds (Kim and Douglas, 2013). Lipid metabolism and transport genes are highly expressed in the pollen compared with vegetative tissues, suggesting important roles of lipid metabolism in pollen development (Honys and Twell, 2004). Among the differentially expressed genes, many are involved in the pollen coat and exine formation, as determined by previous studies (Wilson and Zhang, 2009; Ariizumi and Toriyama, 2011), contributing to the synthesis of pollen coat precursors and sporopollenin. It is generally understood that sterol esters and saturated acyl groups are the major components of the pollen coat, similar to those of lipid or oil droplets but different from lipids of biological membranes.
As developmental defects were observed in both tapetal cells and pollen wall in CLE19 mutants, we analyzed the expression of genes implicated in lipid acyl metabolism, including the metabolism of flavonoids, phenolics, free fatty acids, wax ester, and cutin (Fig. 8A), and found that 36 lipid acyl metabolism-related genes showed more that 2-fold differences in expression between DN-CLE19 and/or CLE19-OX mutant anthers. Among these, 20 genes also were altered in expression in ams anthers and/or floral buds (Ma et al., 2012; Xu et al., 2014). Importantly, 19 out of the 20 coregulated genes showed changes in expression in the opposite direction: up-regulated in the CLE19-DN mutant but down-regulated in CLE19-OX and ams mutants (Fig. 8, B–F). These results suggest that CLE19 mediates the tapetum and pollen wall developmental signaling pathways by negatively regulating the expression of these genes, with at least some dependence on AMS function.
Figure 8.
Transcriptomic analyses of genes involved in the metabolism of flavonoids, phenolics, wax, cutin, and free fatty acids in the wild type and the CLE19 mutant. A, Metabolic processes of the main compounds of the pollen wall. B to F, Fold changes in expression levels of genes involved in flavonoid metabolic, cutin metabolic, wax ester metabolic, free fatty acid metabolic, and phenolic metabolic processes.
The ams Mutation Partially Suppressed DN-CLE19-Induced Abnormal Anther Development and Pollen Exine Formation
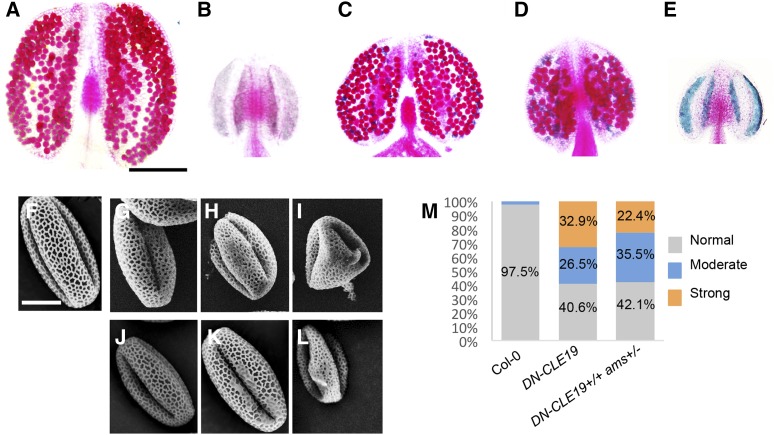
To test the hypothesis that CLE19 functions with at least some dependence on AMS function, we generated DN-CLE19(+/+) ams(+/−) and DN-CLE19(+/+) ams(−/−) plants by crossing the DN-CLE19-23 homozygous plant [DN-CLE19-23(+/+)] with the ams mutant and observed the pollen exine structure of each genotype. Compared with the wild-type anthers containing pollen with a well-organized pollen exine structure (Fig. 9, A and F), ams produced smaller anthers without any pollen (Fig. 9B) and DN-CLE19-23 produced slightly smaller anthers and fewer pollen grains, with some pollen grains being aborted (Fig. 9, C and G–I). Interestingly, the DN-CLE19(+/+) ams(+/−) anthers were even smaller than the DN-CLE19 anthers, with few pollen grains (Fig. 9D); the DN-CLE19(+/+) ams(−/−) anthers were similar to the ams anthers (Fig. 9E).
Figure 9.
Phenotypic analysis of anthers from wild-type, ams mutant, DN-CLE19 transgenic, DN-CLE19(+/+) ams(+/−), and DN-CLE19(+/+) ams(−/−) plants. A to E, Alexander-stained anthers from wild-type (A), ams mutant (B), DN-CLE19 transgenic (C), DN-CLE19(+/+) ams(+/−) (D), and DN-CLE19(+/+) ams(−/−) (E) plants. F to L, Pollen grains from wild-type (F), DN-CLE19 transgenic (G–I), and DN-CLE19(+/+) ams(+/−) (J–L) plants. M, Statistical analysis data to show the percentage of pollen grains with abnormal exine in wild-type, DN-CLE19 transgenic, and DN-CLE19(+/+) ams(+/−) plants. Bars = 200 μm for anthers and 10 μm for pollen grains.
To further investigate the genetic interaction between CLE19 and AMS, we then compared the pollen exine structure between pollen grains from the DN-CLE19 and DN-CLE19(+/+) ams(+/−) plants. As both the DN-CLE19 and DN-CLE19(+/+) ams(+/−) plants produced pollen grains with pollen exine defects with different degrees of severity, we classified the defects into three levels: slight (Fig. 9, G and J), moderate (Fig. 9, H and K), and severe (Fig. 9, I and L) defects. Statistical analysis showed that, after the ams(+/−) genotype was introduced into the DN-CLE19 plants, the percentage of severely defective pollen was obviously reduced (from 32.9% to 22.4%; Fig. 9M) and the percentages of pollen with moderate defects and slight defects were increased (from 26.5% to 35.5% and from 40.6% to 42.1%, respectively; Fig. 9M), demonstrating that CLE19 works in a way dependent on the AMS function.
DISCUSSION
A Proper Amount of CLE19 Is Important for Pollen Wall Formation
Cell-cell communication is often mediated by the extracellular ligands and their receptors and is essential for the development of multicellular organisms, including anther development in angiosperms (Zhao et al., 2002; Yang et al., 2003, 2016; Jia et al., 2008; Huang et al., 2016). During pollen development, the adjacent tapetal layer provides metabolites and sporopollenin precursors for pollen coat and exine formation as well as signals essential for pollen development (Mariani et al., 1990; Goldberg et al., 1993; Sanders et al., 1999). The pollen mother cells surrounded by the tapetum undergo meiosis and form a tetrad enveloped by a thick protective callose (1,3-β-glucan) wall that is digested subsequently by glucanases secreted from the tapetum, resulting in the release of microspores. Then, the microspores develop into mature pollen grains with the deposition of lipidic precursors/components secreted by the tapetal cells onto the pollen surface to form the pollen exine and pollen coats (Mascarenhas, 1975; Stieglitz, 1977; Pacini and Juniper, 1979). The signaling communication between tapetal cells and the reproductive cells (including pollen mother cells and developing pollen grains) is important for normal anther development and pollen formation. For instance, Arabidopsis reproductive cell-preferential TPD1 and the tapetum-expressed EMS1 and SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1/2 (SERK1/2) complex interact to regulate normal tapetum differentiation and pollen development, and plants with mutations in TPD1, EMS1, or SERK1/2 produce no microspores (Zhao et al., 2002; Yang et al., 2003; Albrecht et al., 2005; Huang et al., 2016; Li et al., 2017). However, the functions of extracellular ligand and RLK-mediated signaling pathways in the regulation of pollen development are rarely reported.
In this study, using combined strategies of cell biology, genetics, and transcriptomics, we found that CLE19 is required for the normal development of pollen, especially for the normal biogenesis of pollen exine. All CLE19 mutant lines exhibited abnormal pollen development with a portion of pollen grains aborted; however, pollen exine structures exhibited differences among mutants with different CLE19 expression levels. Unlike the wild-type pollen grains that are covered by a well-organized network-like lacunae structure, the cle19-2 and DN-CLE19 pollen grains had a more extensively covered surface with broader muri and smaller lacunae units, and most of the lacunae units were filled with extra sporopollenin-like substances (Fig. 5). In contrast, the CLE19-OX pollen grains exhibited a shortage of the characteristic reticulate structures composed of bacula and tectum (Fig. 5), suggesting that the lacunae of pollen exine were incomplete. Further transcriptomic analyses revealed that the expression of numerous genes involved in pollen wall assembly, lipid metabolism, transport, and localization were up-regulated in DN-CLE19 but down-regulated in CLE19-OX anthers (Figs. 7 and 8), demonstrating that CLE19 functions as a negative regulator of these genes. These data suggested that a proper amount of the CLE19 signal is essential for the normal organization of the pollen wall, and either more or less than the normal level of CLE19 would destroy the balance of both the expression of pollen developmental genes and the construction of normal pollen exine.
CLE19 Functions as a Negative Regulator of AMS-Regulated Pollen Wall Developmental Pathways
Previous studies have demonstrated that the transcription factors DYT1, MYB35/TDF1, AMS, MYB103/MYB80, and MS1 are required for anther development and that they form a genetically defined pathway/regulatory cascade: DYT1-MYB35/TDF1-AMS-MYB103/MYB80-MS1 (Zhu et al., 2011). AMS is a key regulator of sporopollenin biosynthesis, secretion, and pollen wall formation and can form a complex with MS188/MYB103/MYB80 that likely activates the expression of CYP703A2 and MS1 encoding a transcription factor (Xu et al., 2010, 2014; Zhu et al., 2011; Feng et al., 2012; Gu et al., 2014; Xiong et al., 2016; Ferguson et al., 2017). DYT1 and MYB35/TDF1 are required for the development of tapetum and reproductive cells at early stages, as supported by the findings that DYT1 and MYB35/TDF1 mutants exhibit abnormal tapetal cells at anther stage 5 and onward and that their anthers lack any pollen (Zhang et al., 2006; Zhu et al., 2008; Gu et al., 2014). These two genes are important for normal anther and pollen development, at least in part by activating the normal expression of AMS. In our study, we showed that CLE19 negatively regulates the expression of AMS and the downstream genes MYB103/MYB80 and MS1 to prevent their deleterious overexpression, but it does not obviously affect the normal expression of DYT1 and MYB35/TDF1 (Fig. 6), demonstrating that CLE19 probably functions to modulate the proper level of the activity of the AMS-regulated network and the appropriate amount of sporopollenin biosynthesis and pollen wall formation. The cytological results that both CLE19-OX transgenic anthers and the ams mutant anthers showed similar abnormal tapetum cells and pollen primexine developmental defects, and the genetic result that ams(+/−) partially suppressed the DN-CLE19 transgene-induced pollen exine defects (Fig. 9), together support the ideas that AMS works downstream of the CLE19 signaling pathway and that the full effect of CLE19 depends on the normal level of AMS.
Therefore, we propose that the CLE19 peptide, probably through unknown plasma membrane-localized receptor(s), negatively regulates the AMS-MYB103-MS1 transcriptional cascades to maintain their functional homeostasis and ensure the normal development of pollen (Fig. 10A). However, in the DN-CLE19 mutants, the reduced function of CLE19 releases the transcriptional inhibition and causes the deleterious overexpression of AMS and downstream networks; such overexpression then subsequently affects the normal organization of pollen exine (Fig. 10B). In contrast, in the CLE19-OX anthers, CLE19 synthesized at abnormally high levels excessively inhibits the expression and function of AMS and downstream networks, thereby inducing abnormal pollen development (Fig. 10C).
Figure 10.
Proposed working model for CLE19 in the regulation of anther development. A, In wild-type anthers, CLE19 peptides interact with their unknown receptor to negatively regulate the AMS-MYB103-MS1 transcriptional cascade to prevent their deleterious overexpression, which triggers the normal expression of downstream exine formation-related genes, and subsequently regulate the formation of normally well-organized pollen exine. B, In DN-CLE19 anthers, CLE19 peptides without function competitively interact with the unknown CLE19 receptor, which blocks or obviously reduces signal transduction from the functional CLE19 to its downstream molecules, leading to the deleterious overexpression of the AMS-MYB103-MS1 transcriptional cascade and downstream exine formation-related genes, and subsequently affect normal pollen exine formation. C, In CLE19-OX anthers, overexpression of CLE19 peptide strengthens the suppression of transcription factor gene expression, resulting in abnormal pollen exine formation.
Functional Redundancy between Anther-Expressed CLE Genes
As an extracellular peptide hormone, CLE19 was expressed at relatively low levels in the inflorescence and stage 4 to 12 anthers in our quantitative real-time PCR analysis using EF1α as the internal control (Supplemental Fig. S1B). Nevertheless, both CLE19-OX and DN-CLE19 transgenic plants showed obviously male fertility defects, even though the cle19 single mutants showed weak anther developmental defects (Fig. 2). We would like to offer the following thoughts. First, RNA in situ hybridization analysis results showed that CLE19 was expressed only in the tapetal and male reproductive cells at anther stages 5 to 7 (Fig. 1, M–P), suggesting strong cell specificity and a short duration of CLE19 expression in the anther. We believe that both the high cell specificity and the short duration of CLE19 expression contribute to its signal being diluted by nonexpressing cells in the inflorescence or stage 4 to 12 anthers. Second, as supported by the results here, CLE19 acts as a negative regulator of AMS and additional downstream genes, which are required for normal tapetum function and pollen development. If CLE19 is allowed to be expressed at very high levels, normal pollen development would be inhibited, as seen in the CLE19-OX plants.
It is challenging to obtain mutants of CLE genes because they are all small genes; in addition, the anther-expressed CLE genes might have overlapping (redundant) functions. In this study, knockout mutants of five anther-expressed CLE genes, CLE9, CLE16, CLE17, CLE19, and CLE42, were characterized briefly, and only CLE19 knockout mutants exhibited very weak anther developmental defects, whereas mutants of the other four genes showed no obvious developmental or fertility defects. Nevertheless, our statistical analysis results showed that, compared with the wild-type plants, their double and triple mutants showed several detectable defects: increased barren silique number, reduced total pollen number but increased aborted pollen number in each anther, and increased percentage of pollen with abnormal pollen exine structure (Supplemental Fig. S4; Supplemental Table S1). In addition, the DN transgenic mutants of these seven anther-expressed CLE members, DN-CLE9, DN-CLE16, DN-CLE17, DN-CLE19, DN-CLE41, DN-CLE42, and DN-CLE45, all showed severe anther developmental and pollen exine formation defects (Supplemental Fig. S5). All these results suggested that these anther-expressed CLE members possibly function in a redundant way in the regulation of male fertility, especially in the regulation of pollen exine formation. We believe that further studies with various multiple mutants may uncover the details of redundant and distinctive functions on different developmental aspects between these CLE members.
In addition, further studies also are needed to uncover additional signaling components in the complex network that regulates pollen wall formation. For example, what is the RLK that perceives the CLE peptide signal and transduces the signal into the cytoplasm? What are the downstream effectors, such as kinase and other factors, that respond to the CLE-RLK signaling module, and how do they activate the AMS transcription factor? What is the proper amount of CLE peptide? Further investigations using a combination of omics, biochemistry, cell biology, and genetic strategies may answer these questions and further advance the study of male fertility.
CONCLUSION
In summary, our observations demonstrated that CLE19 is essential for normal anther development and pollen wall formation, likely and in large part by negatively regulating the expression of AMS and the downstream genes MYB103/MYB80 and MS1 to prevent their deleterious overexpression, and to modulate the proper level of the activity of the AMS-regulated network and the appropriate anther development and pollen wall formation.
MATERIALS AND METHODS
Plant Growth and Genotyping
Arabidopsis (Arabidopsis thaliana) plants were grown on soil in greenhouse conditions (21°C) under long-day conditions (16 h of light/8 h of dark). T-DNA insertion mutant alleles of cle19 (CS879453 and CS321816), cle9 (SALK_018122C), cle17 (SALK_103714C), and cle42 (SALK_057407C) mutants and RNAi plants of CLE16 (CS201209) in the Col-0 background were all obtained from Arabidopsis Biological Resource Center stocks. The T-DNA insertion of CLE19 alleles was verified using the T-DNA border primer SALKLB1.3 in combination with the gene-specific primers listed in Supplemental Table S2.
Molecular Cloning and Generation of Transgenic Plants and Genotyping
Genomic sequences of CLE9, CLE16, CLE17, CLE19, CLE41, CLE42, and CLE45 were cloned into the pDONR221 vector (Life Technologies) with the primers list in Supplemental Table S3. Using a PCR-based site-directed mutagenesis kit (Transgene) and specific primers (Supplemental Table S3), the Gly codon at the sixth position of the CLE motif of these CLE genes was replaced by the Thr codon, producing pDONR221-CLE9G6T, pDONR221-CLE16G6T, pDONR221-CLE17G6T, pDONR221-CLE19G6T, pDONR221-CLE41G6T, pDONR221-CLE42G6T, and pDONR221-CLE45G6T entry clones. These entry clones were then subcloned into the pBGWFS7 binary vector (Karimi et al., 2002) to produce CLE19G6T, CLE19G6T, CLE16G6T, CLE17G6T, CLE41G6T, CLE42G6T, and CLE45G6T. Transformation was performed via an Agrobacterium tumefaciens-mediated floral dip method (Clough and Bent, 1998). Transgenic plants were obtained under the selection of 25 mg of Basta (Sigma-Aldrich).
Quantitative Real-Time PCR Analysis
To analyze the expression levels of the CLEs and other downstream genes, stage 1 to 11 flower buds of Col-0, each related T-DNA or RNAi mutant, and DN-CLE transgenic plants were collected, and total RNA was extracted according to a Trizol-based (Sigma-Aldrich) method. After DNase I digestion and first-strand cDNA synthesis, quantitative real-time PCR was performed with SYBR premix Ex Taq II (Takara) on the ABI StepOnePlus real-time system (Life Technologies) using the primers listed in Supplemental Table S4 and with EF1a (AT5G60390) as the internal control.
RNA in Situ Hybridization
RNA in situ hybridization analysis was performed using the Digoxigenin RNA Labeling Kit (Roche) and the PCR DIG Probe Synthesis Kit (Roche). cDNA fragments of target genes about 400 bp were amplified using the specific primers listed in Supplemental Table S5. PCR products were cloned into the pGEM-T vector and confirmed by sequencing. Completely digested plasmid DNAs were used as the template for transcription with T7 or SP6 RNA polymerase.
Phenotypic Analysis of Flowers and Anthers
Plants were photographed with a digital camera (Canon). Flower images were taken with the SPOT FLEX digital camera (Diagnostic Instruments) using the SteREO Discovery V8 dissecting microscope (Carl Zeiss Microimaging).
Stage 12 anthers were collected and stained overnight at room temperature using the Alexander solution prepared following a published protocol (Alexander, 1969), and additional anthers were pressed to release the stained pollen grains and photographed by an AXIO ScopeA1 microscope (Carl Zeiss Microimaging) with a Axio Cam HRc camera (Carl Zeiss Microimaging).
Light and Electron Microscopy Observation
Inflorescences of wild-type and mutant plants were collected and fixed in FAA (formaldehyde–acetic acid–ethanol) solution and embedded in Technovit 7100 resin (Heraeus Kulzer) as described (Jin et al., 2013). Semithin sections were prepared by cutting inflorescence materials into 1-µm-thick sections using a motorized RM2265 rotary microtome (Leica Microsystems) and then were stained with 0.05% Toluidine Blue O for 15 to 30 min and photographed by bright-field microscopy.
For pollen wall observation, sections were stained with Toluidine Blue for 5 min (10 mg mL−1), Tinopal for 15 min (10 μg μL−1; Sigma-Aldrich), and diethyloxadicarbocyanine iodide for 5 min (5 μL mL−1; Sigma-Aldrich), and sample photographs were taken using an AXIO ScopeA1 microscope with a 390- to 440-nm excitation filter and a 478-nm blocking filter.
For scanning electron microscopy examination, fresh pollen grains released from stage 13 anthers were coated with gold and observed with an SU8010 microscope (Hitachi).
For TEM observation, different stage buds were fixed in glutaric dialdehyde buffer and embedded into the fresh mixed resin. Ultrathin sections (70-100 nm thick) were observed with a Tecnai G2 Spirit TWIN transmission electron microscope (FEI).
RNA-seq and Data Analysis
More than 1,000 stage 4 to 10 anthers from wild-type and mutant plants were collected and immediately frozen in liquid nitrogen. Total RNA from each sample was extracted and purified using the ZR Plant RNA Miniprep Kit (Zymo Research). Two micrograms of total RNA of each sample was used for deep sequencing by an Illumina HiSeq 2000 system. All sequencing data were mapped and analyzed following the previously reported method (Zhu et al., 2015).
Accession Numbers
The original RNA-seq data from this article have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE94607. Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: CLE9 (At1g26600), CLE16 (At2g01505), CLE17 (At1g70895), CLE19 (At3g24225), CLE42 (At2g34925), CLE45 (At1g69588), DYT1 (At4g21330), AMS (At2g16910), MYB35/TDF1 (At3g28470), MS1 (At5g22260), MYB103 (At1g63910), EF1α (At5g60390), and ACT2 (At3g18780). Germplasm used included cle9-1 (SALK_018122C), cle16-1 (CS201209), cle17-1 (SALK_103714C), cle19-1 (CS879453), qrt1-2 (SALK_CS8846), cle19-2 (CS321816), and cle42-1 (SALK_057407C).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression patterns of CLE genes in various Arabidopsis organs.
Supplemental Figure S2. T-DNA locations or RNAi targeting sites and relative expression of detected genes in the cle9-1, cle16-1, cle17-1, qrt1-2, cle19-1 qrt1-2, cle19-2, and cle42-1 mutants.
Supplemental Figure S3. Phenotypic analyses of plant growth, anthers, and pollen of wild-type and cle9-1, cle16-1, cle17-1, qrt1-2, cle19-1 qrt1-2, cle19-2, and cle42-1 plants.
Supplemental Figure S4. Phenotypic analyses of siliques, flowers, anthers, and pollen grains in the wild type and the cle single, double, and triple mutants.
Supplemental Figure S5. Phenotypic analyses of plant growth, anthers, and pollen grains of wild-type and DN-CLE transgenic plants.
Supplemental Figure S6. Comparison of exine patterns among cle19-2, DN-CLE19, DN-CLE41, DN-CLE42, and the wild type.
Supplemental Figure S7. RNA in situ hybridization analysis to detect the spatial and temporal expression of the AMS gene in wild-type, ams, DN-CLE19, and CLE19-OX transgenic anthers.
Supplemental Figure S8. Scatterplots for replicates of transcriptome data.
Supplemental Table S1. Statistical phenotypic analysis results of the cle single, double, and triple mutants.
Supplemental Table S2. Primers used for genotyping.
Supplemental Table S3. Primers used for DN-CLE transgenic constructs in this work.
Supplemental Table S4. Primers used for quantitative real-time PCR analysis.
Supplemental Table S5. Primers used for in situ constructs in this work.
Supplemental Data Set S1. List of 578 genes down-regulated and 1,133 genes up-regulated in DN-CLE19-23 transcriptomic analysis in this study.
Supplemental Data Set S2. List of 1,809 genes down-regulated and 379 genes up-regulated in CLE19-OX-S transcriptomic analysis in this study.
Supplemental Data Set S3. List of 742 genes both up-regulated in DN-CLE19-23 and down-regulated in CLE19-OX-S transcriptomic data.
Supplemental Data Set S4. List of 52 genes both down-regulated in DN-CLE19-23 and up-regulated in CLE19-OX-S transcriptomic data.
Supplemental Data Set S5. List of overlap genes differentially expressed in both DN-CLE19-23 and CLE19-OX-S transcription data and the microarray-based transcriptomic data of the ams stage 4 to 7 anthers and 404 RNA-seq-based transcriptomic data of ams buds.
Acknowledgments
We thank the Salk Institute Genomic Analysis Laboratory and the Ohio State University Arabidopsis Biological Resource Center for providing the sequence-indexed Arabidopsis T-DNA insertion mutants.
Footnotes
This work was supported by grants from the National Natural Science Foundation of China (31130006, 31470282, 31670316, and 31370029) and the State Key Laboratory of Genetic Engineering at Fudan University.
Articles can be viewed without a subscription.
References
- Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S (2005) The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 17: 3337–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander MP. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Al-Refu K, Edward S, Ingham E, Goodfield M (2009) Alterations in the basement membrane zone in cutaneous lupus erythematosus (CLE) as demonstrated by immunohistochemistry. Arch Dermatol Res 301: 47 [Google Scholar]
- Ariizumi T, Toriyama K (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Bedinger PA, Hardeman KJ, Loukides CA (1994) Travelling in style: the cell biology of pollen. Trends Cell Biol 4: 132–138 [DOI] [PubMed] [Google Scholar]
- Blackmore S, Wortley AH, Skvarla JJ, Rowley JR (2007) Pollen wall development in flowering plants. New Phytol 174: 483–498 [DOI] [PubMed] [Google Scholar]
- Canales C, Bhatt AM, Scott R, Dickinson H (2002) EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr Biol 12: 1718–1727 [DOI] [PubMed] [Google Scholar]
- Casamitjana-Martínez E, Hofhuis HF, Xu J, Liu CM, Heidstra R, Scheres B (2003) Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr Biol 13: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Chang F, Wang Y, Wang S, Ma H (2011) Molecular control of microsporogenesis in Arabidopsis. Curr Opin Plant Biol 14: 66–73 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cock JM, McCormick S (2001) A large family of genes that share homology with CLAVATA3. Plant Physiol 126: 939–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI (2005) Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17: 3350–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, You C, Zhu E, Huang Q, Ma H, Chang F (2016) Feedback regulation of DYT1 by interactions with downstream bHLH factors promotes DYT1 nuclear localization and anther development. Plant Cell 28: 1078–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, Clark SE (2006) The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J 45: 1–16 [DOI] [PubMed] [Google Scholar]
- Edlund AF, Swanson R, Preuss D (2004) Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell (Suppl) 16: S84–S97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Lu D, Ma X, Peng Y, Sun Y, Ning G, Ma H (2012) Regulation of the Arabidopsis anther transcriptome by DYT1 for pollen development. Plant J 72: 612–624 [DOI] [PubMed] [Google Scholar]
- Ferguson AC, Pearce S, Band LR, Yang C, Ferjentsikova I, King J, Yuan Z, Zhang D, Wilson ZA (2017) Biphasic regulation of the transcription factor ABORTED MICROSPORES (AMS) is essential for tapetum and pollen development in Arabidopsis. New Phytol 213: 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu CM (2005) The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17: 2542–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Hause G, Boutilier K, Casamitjana-Martinez E, Weijers D, Offringa R, van der Geest L, van Lookeren Campagne M, Liu CM (2004) Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene 327: 37–49 [DOI] [PubMed] [Google Scholar]
- Fiume E, Fletcher JC (2012) Regulation of Arabidopsis embryo and endosperm development by the polypeptide signaling molecule CLE8. Plant Cell 24: 1000–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Ge X, Chang F, Ma H (2010) Signaling and transcriptional control of reproductive development in Arabidopsis. Curr Biol 20: R988–R997 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Beals TP, Sanders PM (1993) Anther development: basic principles and practical applications. Plant Cell 5: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu JN, Zhu J, Yu Y, Teng XD, Lou Y, Xu XF, Liu JL, Yang ZN (2014) DYT1 directly regulates the expression of TDF1 for tapetum development and pollen wall formation in Arabidopsis. Plant J 80: 1005–1013 [DOI] [PubMed] [Google Scholar]
- Guilford WJ, Schneider DM, Labovitz J, Opella SJ (1988) High resolution solid state C NMR spectroscopy of sporopollenins from different plant taxa. Plant Physiol 86: 134–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hord CL, Chen C, Deyoung BJ, Clark SE, Ma H (2006) The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 18: 1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hord CL, Sun YJ, Pillitteri LJ, Torii KU, Wang H, Zhang S, Ma H (2008) Regulation of Arabidopsis early anther development by the mitogen-activated protein kinases, MPK3 and MPK6, and the ERECTA and related receptor-like kinases. Mol Plant 1: 645–658 [DOI] [PubMed] [Google Scholar]
- Huang J, Zhang T, Linstroth L, Tillman Z, Otegui MS, Owen HA, Zhao D (2016) Control of anther cell differentiation by the small protein ligand TPD1 and its receptor EMS1 in Arabidopsis. PLoS Genet 12: e1006147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Nagata N, Yoshiba Y, Ohme-Takagi M, Ma H, Shinozaki K (2007) Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19: 3549–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Liu X, Owen HA, Zhao D (2008) Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proc Natl Acad Sci USA 105: 2220–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Yang H, Wei Z, Ma H, Ge X (2013) Rice male development under drought stress: phenotypic changes and stage-dependent transcriptomic reprogramming. Mol Plant 6: 1630–1645 [DOI] [PubMed] [Google Scholar]
- Jun J, Fiume E, Roeder AH, Meng L, Sharma VK, Osmont KS, Baker C, Ha CM, Meyerowitz EM, Feldman LJ, et al. (2010) Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol 154: 1721–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kim SS, Douglas CJ (2013) Sporopollenin monomer biosynthesis in Arabidopsis. J Plant Biol 56: 1–6 [Google Scholar]
- Laux T, Mayer KF, Berger J, Jürgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Li H, Zhang D (2010) Biosynthesis of anther cuticle and pollen exine in rice. Plant Signal Behav 5: 1121–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang Y, Huang J, Ahsan N, Biener G, Paprocki J, Thelen JJ, Raicu V, Zhao D (2017) Two SERK receptor-like kinases interact with EMS1 to control anther cell fate determination. Plant Physiol 173: 326–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Ma X, Feng B, Ma H (2012) AMS-dependent and independent regulation of anther transcriptome and comparison with those affected by other Arabidopsis anther genes. BMC Plant Biol 12: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani C, Beuckeleer MD, Truettner J, Leemans J, Goldberg R (1990) Induction of male sterility in plants by a chimaeric ribonuclease gene. Nature 347: 737–741 [Google Scholar]
- Mascarenhas JP. (1975) The biochemistry of angiosperm pollen development. Bot Rev 41: 259–314 [Google Scholar]
- Mayfield JA, Preuss D (2000) Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nat Cell Biol 2: 128–130 [DOI] [PubMed] [Google Scholar]
- Mizuno S, Osakabe Y, Maruyama K, Ito T, Osakabe K, Sato T, Shinozaki K, Yamaguchi-Shinozaki K (2007) Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J 50: 751–766 [DOI] [PubMed] [Google Scholar]
- Ni J, Clark SE (2006) Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol 140: 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacini E, Juniper BE (1979) The ultrastructure of pollen-grain development in the olive (Olea europaea). 2. Secretion by the tapetal cells. New Phytol 83: 165–174 [Google Scholar]
- Phan HA, Iacuone S, Li SF, Parish RW (2011) The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23: 2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D, Lemieux B, Yen G, Davis RW (1993) A conditional sterile mutation eliminates surface components from Arabidopsis pollen and disrupts cell signaling during fertilization. Genes Dev 7: 974–985 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, Mcintire KN, Hsu YC, Pei YL, Mai TT, Beals TP, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Plant Reprod 11: 297–322 [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG (2004) Stamen structure and function. Plant Cell (Suppl) 16: S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, Lakeman MB, Torii KU (2003) Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell 15: 1095–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XF, Guo P, Ren SC, Xu TT, Liu CM (2013) Antagonistic peptide technology for functional dissection of CLV3/ESR genes in Arabidopsis. Plant Physiol 161: 1076–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XF, Yu DL, Xu TT, Ren SC, Guo P, Liu CM (2012) Contributions of individual amino acid residues to the endogenous CLV3 function in shoot apical meristem maintenance in Arabidopsis. Mol Plant 5: 515–523 [DOI] [PubMed] [Google Scholar]
- Sorensen AM, Kröber S, Unte US, Huijser P, Dekker K, Saedler H (2003) The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J 33: 413–423 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Simon R (2009) Is the Arabidopsis root niche protected by sequestration of the CLE40 signal by its putative receptor ACR4? Plant Signal Behav 4: 634–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglitz H. (1977) Role of β-1,3-glucanase in postmeiotic microspore release. Dev Biol 57: 87–97 [DOI] [PubMed] [Google Scholar]
- Strabala TJ, O’Donnell PJ, Smit AM, Ampomah-Dwamena C, Martin EJ, Netzler N, Nieuwenhuizen NJ, Quinn BD, Foote HC, Hudson KR (2006) Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol 140: 1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom I, Grote M, Abraham-Peskir J, Wiermann R (1998) Electron and x-ray microscopic analyses of reaggregated materials obtained after fractionation of dissolved sporopollenin. Protoplasma 204: 13–21 [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ (2001) The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J 28: 27–39 [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Zhang DB (2009) From Arabidopsis to rice: pathways in pollen development. J Exp Bot 60: 1479–1492 [DOI] [PubMed] [Google Scholar]
- Xiong SX, Lu JY, Lou Y, Teng XD, Gu JN, Zhang C, Shi QS, Yang ZN, Zhu J (2016) The transcription factors MS188 and AMS form a complex to activate the expression of CYP703A2 for sporopollenin biosynthesis in Arabidopsis thaliana. Plant J 88: 936–946 [DOI] [PubMed] [Google Scholar]
- Xu J, Ding Z, Vizcay-Barrena G, Shi J, Liang W, Yuan Z, Werck-Reichhart D, Schreiber L, Wilson ZA, Zhang D (2014) ABORTED MICROSPORES acts as a master regulator of pollen wall formation in Arabidopsis. Plant Cell 26: 1544–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang C, Yuan Z, Zhang D, Gondwe MY, Ding Z, Liang W, Zhang D, Wilson ZA (2010) The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22: 91–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu TT, Ren SC, Song XF, Liu CM (2015) CLE19 expressed in the embryo regulates both cotyledon establishment and endosperm development in Arabidopsis. J Exp Bot 66: 5217–5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Qian X, Chen M, Fei Q, Meyers B, Liang W, Zhang D (2016) Regulatory role of a receptor-like kinase in specifying anther cell identity. Plant Physiol 171: 2085–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SL, Jiang L, Puah CS, Xie LF, Zhang XQ, Chen LQ, Yang WC, Ye D (2005) Overexpression of TAPETUM DETERMINANT1 alters the cell fates in the Arabidopsis carpel and tapetum via genetic interaction with EXCESS MICROSPOROCYTES1/EXTRA SPOROGENOUS CELLS. Plant Physiol 139: 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SL, Xie LF, Mao HZ, Puah CS, Yang WC, Jiang L, Sundaresan V, Ye D (2003) Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15: 2792–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Zhu W, Li L, Zhang S, Yin Y, Ma H, Wang X (2010) Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc Natl Acad Sci USA 107: 6100–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H (2006) Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133: 3085–3095 [DOI] [PubMed] [Google Scholar]
- Zhao DZ, Wang GF, Speal B, Ma H (2002) The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev 16: 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu E, You C, Wang S, Cui J, Niu B, Wang Y, Qi J, Ma H, Chang F (2015) The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J 83: 976–990 [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN (2008) Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J 55: 266–277 [DOI] [PubMed] [Google Scholar]
- Zhu J, Lou Y, Xu X, Yang ZN (2011) A genetic pathway for tapetum development and function in Arabidopsis. J Integr Plant Biol 53: 892–900 [DOI] [PubMed] [Google Scholar]
- Zinkl GM, Zwiebel BI, Grier DG, Preuss D (1999) Pollen-stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development 126: 5431–5440 [DOI] [PubMed] [Google Scholar]