A soybean serine hydroxymethyltransferase has a unique and essential role in soybean cyst nematode resistance.
Abstract
Rhg4 is a major genetic locus that contributes to soybean cyst nematode (SCN) resistance in the Peking-type resistance of soybean (Glycine max), which also requires the rhg1 gene. By map-based cloning and functional genomic approaches, we previously showed that the Rhg4 gene encodes a predicted cytosolic serine hydroxymethyltransferase (GmSHMT08); however, the novel gain of function of GmSHMT08 in SCN resistance remains to be characterized. Using a forward genetic screen, we identified an allelic series of GmSHMT08 mutants that shed new light on the mechanistic aspects of GmSHMT08-mediated resistance. The new mutants provide compelling genetic evidence that Peking-type rhg1 resistance in cv Forrest is fully dependent on the GmSHMT08 gene and demonstrates that this resistance is mechanistically different from the PI 88788-type of resistance that only requires rhg1. We also demonstrated that rhg1-a from cv Forrest, although required, does not exert selection pressure on the nematode to shift from HG type 7, which further validates the bigenic nature of this resistance. Mapping of the identified mutations onto the SHMT structural model uncovered key residues for structural stability, ligand binding, enzyme activity, and protein interactions, suggesting that GmSHMT08 has additional functions aside from its main enzymatic role in SCN resistance. Lastly, we demonstrate the functionality of the GmSHMT08 SCN resistance gene in a transgenic soybean plant.
Soybean cyst (Heterodera glycines ‘Ichinohe’) nematode (SCN) is an obligate sedentary endoparasite on soybean (Glycine max) roots. The nematode penetrates into the root and migrates toward the vasculature, where it establishes a permanent feeding site (syncytium) by successive cell wall dissolution and the fusion of hundreds of adjacent cells (Endo, 1965). The syncytium serves as a nutrient sink on which the nematode feeds. SCN continues to be the number one pathogen affecting soybean productivity in the United States (Koenning and Wrather, 2010) and is an increasing problem in other soybean-producing countries (Mendes and Dickson, 1993; Peng et al., 2016). Management relies heavily on the use of resistant soybean cultivars. In these cultivars, necrosis of the feeding site prevents the development of the sedentary nematode (Endo, 1965; Acedo et al., 1984; Mahalingam and Skorupska, 1996; Kandoth et al., 2011).
Two major quantitative trait loci that contribute to SCN resistance are rhg1 on chromosome 18 and Rhg4 on chromosome 8 (for review, see Concibido et al., 2004). The rhg1 locus is found in most known resistance sources of soybean and acts alone, such as in plant introduction (PI) 88788, or in combination with Rhg4 in sources such as PI 548402 (Peking) and PI 437654 (Meksem et al., 2001; Brucker et al., 2005). PI 88788 is the most widely deployed source of resistance, present in more than 95% of resistant cultivars used in the north-central United States (Mitchum, 2016). Currently, there are nematode populations in the United States that can grow on these resistant cultivars, and the problem is increasing (Mitchum et al., 2007; Colgrove and Niblack, 2008; Mitchum, 2016). It is necessary, therefore, not only to identify new sources of resistance but also to understand the mechanism of resistance to design a more durable resistance in soybean.
The rhg1-b locus responsible for the PI 88788-type of resistance is encoded by three unrelated genes present in tandem in a 31-kb segment repeated nine to 10 times in the resistant cultivar from which it was cloned, whereas a single copy of this 31-kb segment is present in susceptible cultivars (Cook et al., 2012). The genes encode an amino acid transporter, an α-SNAP (soluble NSF [N-ethylmaleimide-sensitive factor] attachment protein), and a protein with a wound-inducible (WI12) domain. Thus, the high copy number of the repeat and the concerted action of these genes are the basis of resistance to SCN in PI 88788-type resistance. In sources with Peking-type resistance, only two to four copies of this repeat region are present (Cook et al., 2014; Lee et al., 2015). Furthermore, the α-SNAP gene (GmSNAP18) in the rhg1-a repeat of soybean cv Forrest (resistance from Peking) is sufficient for SCN resistance (Liu et al., 2017). Expression of GmSNAP18 (rhg1-a) from cv Forrest in an SCN-susceptible recombinant inbred line carrying the susceptible cv Essex allele at the rhg1 (rhg1-s) locus and the resistant cv Forrest allele at the Rhg4 locus resulted in a significant reduction in SCN development compared with the control (Liu et al., 2017). In contrast, expression of the GmSNAP18 gene from either cv Essex (rhg1-s) or PI 88788 (rhg1-b) did not result in a significant reduction in SCN development compared with the control, demonstrating the specificity of cv Forrest GmSNAP18 in conferring Peking-type soybean resistance to SCN. These findings suggest that, even though rhg1 is required, it functions differently in these two types of resistance.
The Rhg4 gene encodes a predicted cytosolic serine hydroxymethyltransferase (GmSHMT08; Liu et al., 2012). The Rhg4 allele found in Peking is not present in wild soybean accessions, suggesting that this allele emerged during the domestication of soybean (Wu et al., 2016). The resistant allele differs from the susceptible allele by two amino acid changes, P130R and N358Y (Liu et al., 2012). SHMT is a pyridoxal-5′-phosphate (PLP)-dependent enzyme present ubiquitously in all living organisms and is important for the synthesis of thymidylate, purines, and the amino acid Met (Blakely, 1969). The enzyme catalyzes the simultaneous interconversion of Ser and tetrahydrofolate (THF) to Gly and 5,10-methylene THF (MTHF). The reaction is central to the one-carbon metabolism of plants and animals (Christensen and MacKenzie, 2006). This reaction is a major source of methyl groups for the synthesis of S-adenosyl-Met (SAM), a methyl group donor in the methylation reactions of a number of biologically important molecules, both in plants and animals. SAM also is a precursor for the synthesis of polyamines and the plant hormone ethylene. SHMTs are encoded by a multigene family in plants. Multiple isoforms are present within a single cell and function in different cellular compartments, including the cytoplasm, mitochondria, and plastids in plants (Christensen and MacKenzie, 2006). SHMT mutations are associated with various disease states in humans, including cancer (Heil et al., 2001; Skibola et al., 2002; Kim, 2003; Lim et al., 2005). In plants, mitochondrial SHMT is required for the proper functioning of photorespiration (Somerville and Ogren, 1981). Arabidopsis (Arabidopsis thaliana) with a mutation in AtSHM1 (shmt1-1) hyperaccumulates reactive oxygen species and forms spontaneous necrotic lesions in the leaves (Moreno et al., 2005).
The identification of a SHMT as a plant resistance gene is unprecedented. The two amino acid changes in GmSHMT08 alter its enzymatic properties (Liu et al., 2012), suggesting that, in a metabolically active syncytium, this may have catastrophic effects, ultimately leading to a hypersensitive response resulting in cell death. In vitro kinetic studies suggest that the enzymatic properties of GmSHMT08 are different from those of the allele present in susceptible plants (Liu et al., 2012). A more detailed analysis, however, is required to elucidate the functional implications of these two amino acid changes and to better understand how this enzyme contributes to SCN resistance in soybean. It also is important to understand how GmSHMT08 function relates to rhg1-a, which is required for the resistance function of Rhg4.
Here, we show that Rhg4 complements resistance in whole-plant transformation experiments, corroborating our previous transgenic hairy root complementation study (Liu et al., 2012). We also report the discovery of a comprehensive allelic series of mutants in GmSHMT08 that show a loss of resistance to SCN. Molecular mapping of the mutations on the predicted structural model reveal that they disrupt oligomeric structure as well as the cofactor-binding sites of the enzyme, providing additional functional insights. Importantly, we identified two GmSHMT08 null mutants in resistant cv Forrest that show a level of susceptibility to SCN comparable to that of a susceptible variety. From these results, we conclude that Peking-type rhg1-a resistance in cv Forrest is fully dependent on Rhg4 and further demonstrate that this resistance is mechanistically different from the PI 88788-type of resistance.
RESULTS
Functional Complementation in Whole-Plant Transgenic Soybean
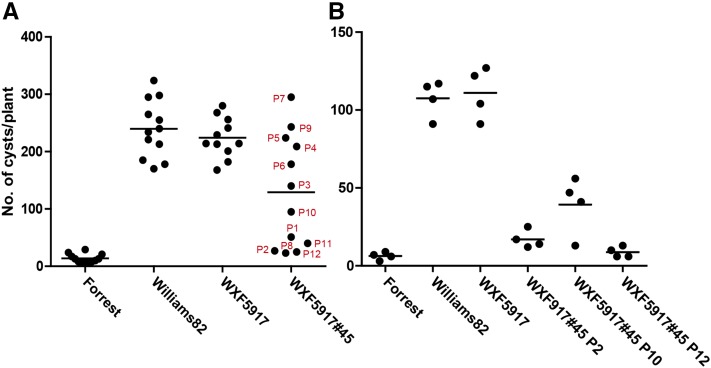
We demonstrated previously that the GmSHMT08 gene at the Rhg4 locus confers resistance to SCN using loss-of-function approaches and genetic complementation experiments in transgenic soybean hairy roots (Liu et al., 2012). Here, we extended this work to show complementation by whole-plant transformation of the susceptible soybean recombinant inbred line (RIL) WXF5917 using the same 5.1-kb GmSHMT08 gene used in our hairy root experiments. WXF5917 carries rhg1-a from the cv Forrest parent (resistant allele) and rhg4 (GmSHMT08) from the cv Williams 82 parent (susceptible allele) and is susceptible to SCN. Unlike cv Williams 82, cv Forrest, cv Essex, the EXF63 RIL used in prior hairy root complementation experiments, and the cv Forrest mutant F6266 all are found recalcitrant to transformation. Although WXF5917 is amenable to transformation with marker genes, few transformants with Rhg4 were recovered despite multiple attempts. A single transgenic line survived and produced seeds. Twelve seeds from this line (WXF5917 Rhg4 #45) were germinated and phenotyped for resistance to SCN. For comparison, we included cv Williams 82 (susceptible), untransformed WXF5917, and cv Forrest (resistant). The results of this experiment are shown in Figure 1A. The SCN-resistant cv Forrest plants had an average of 13.7 cysts. The SCN-susceptible cv Williams 82 and untransformed WXF5917 control plants had an average of 240 and 224 cysts, respectively. Genomic PCR on DNA from leaf tissues of transgenic plants showed segregation for the transgene in the T1 generation. Of the 12 plants analyzed, 10 plants were positive and two plants (P7 and P9) were negative for the transgene (Supplemental Fig. S1A). The two plants with no transgene (P7 and P9) were susceptible to SCN. Five of the WXF5917 transgenic plants showed increased resistance, with cyst numbers comparable to those of resistant cv Forrest (P1, P2, P8, P11, and P12). Three transgenic plants showed intermediate resistance (P3, P6, and P10). Despite testing positive for the transgene, two plants had cyst numbers similar to the average cyst number on the susceptible plants (P4 and P5). We collected seeds from several plants that showed increased resistance or intermediate resistance (P2, P10, and P12) and tested for SCN resistance in the subsequent generation. All resistant plants remained resistant in the T2 generation, whereas the plant (P10) that had intermediate resistance showed segregation (Fig. 1B). Reverse transcription (RT)-quantitative PCR (qPCR) analysis of transgene expression in the plants from the T2 generation showed a strong correlation between resistance and the expression of the transgene (Supplemental Fig. S1B). Lines showing an intermediate resistance showed transgenic expression but, analogous to the segregating parent plant (P10), might be heterozygous for the transgene. From these results, we conclude that the cv Forrest SHMT gene complements resistance in the SCN-susceptible soybean RIL WXF5917.
Figure 1.
Characterization of GmSHMT08 complemented transgenic soybean plants. A, SCN phenotype of T1 plants. Each dot corresponds to the number of cysts on an individual plant. B, SCN phenotype of plants in the T2 generation. The seeds from T1 plants P2, P10, and P12 were used in the greenhouse assay. Each dot corresponds to the number of cysts on an individual plant.
Forward Genetic Screen for GmSHMT08 Mutants
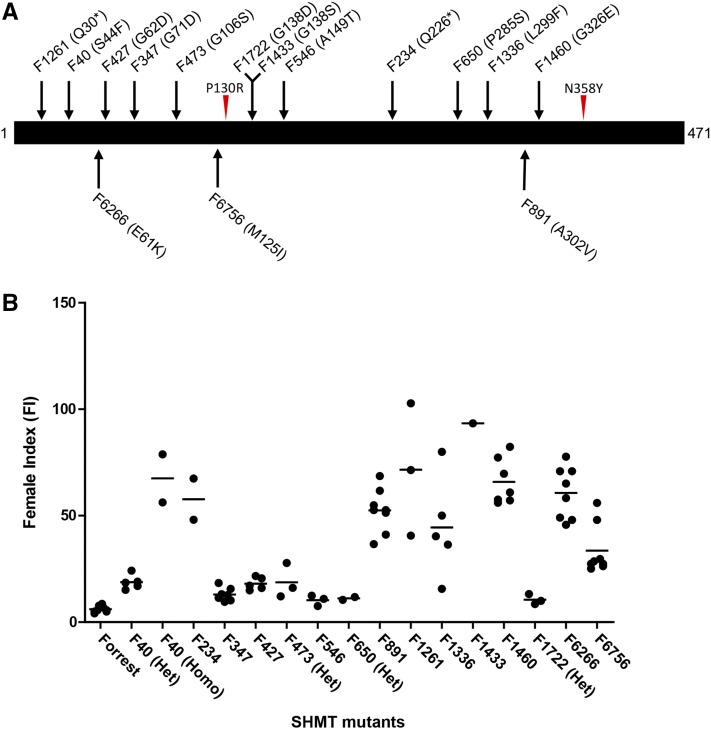
We screened two ethyl methanesulfonate (EMS)-mutagenized populations of SCN-resistant soybean cv Forrest to identify mutations that cause a loss of resistance to SCN. A single seed from each of 1,225 lines of the FM2011 population were phenotyped for altered SCN resistance. A single seed from 698 lines, two seeds from 132 lines, and three seeds from 279 lines were phenotyped from the FM2013 population, for a total of 1,799 plants. GmSHMT08 was PCR amplified and sequenced from genomic DNA of all mutants that showed a loss of resistance to SCN. In total, we identified 13 new GmSHMT08 mutants (Table I; Fig. 2A). Wherever possible, we tested additional seeds from that mutant line and/or advanced the plants to the next generation and repeated the SCN phenotyping. A summary of the female index (FI) values as a measure of nematode development on each of the mutants from data accumulated across different experiments is depicted in Figure 2B. All mutants exhibited an average FI greater than that of cv Forrest (6.1%) when challenged with the SCN PA3 population used in this study. Two new mutants, F234 (226Q*) and F1261 (F30*), carried nonsense mutations that resulted in a truncated protein. Some of the plants were heterozygous for the mutation at GmSHMT08 and exhibited an intermediate SCN resistance phenotype when compared with plants homozygous for the mutation (F40 [Table I; Fig. 2B] and F6756 [Liu et al., 2012]). These results indicate that Rhg4 has a dosage effect on SCN resistance.
Table I. Summary of SHMT mutants.
| Line | Nucleotide | Amino Acid | Zygosity | Average FI |
|---|---|---|---|---|
| cv Forrest | – | – | – | 6.1 |
| F40 | C131T | S44F | Heterozygous | 18.8 |
| F40 | C131T | S44F | Homozygous | 67.5 |
| F234 | C676T | Q226* | Homozygous | 57.8 |
| F347 | G184A | G62S | Homozygous | 13 |
| F427 | G212A | G71D | Homozygous | 18.1 |
| F473 | G316A | G106S | Heterozygous | 18.7 |
| F546 | G445A | A149T | Homozygous | 10.3 |
| F650 | C853T | P285S | Heterozygous | 11.2 |
| F891 | C905T | A302V | Homozygous | 52.4 |
| F1261 | C88T | Q30* | Homozygous | 72 |
| F1336 | C895T | L299F | Homozygous | 44.5 |
| F1433 | G413A | G138S | Homozygous | 93.4 |
| F1460 | G977A | G326E | Homozygous | 65.9 |
| F1722 | G412A | G138D | Heterozygous | 10.6 |
| F6266a | G181A | E61K | Homozygous | 60.4 |
| F6756a | G375A | M125I | Homozygous | 33.6 |
Identified by TILLING (Liu et al., 2012).
Figure 2.
GmSHMT08 mutants identified by forward genetic screening. SCN phenotyping of EMS-mutagenized soybean cv Forrest identified GmSHMT08 mutants exhibiting a loss of resistance. A, Schematic depicting the positions of GmSHMT08 mutations within the GmSHMT08 protein sequence. The mutant line name and the amino acid change (parentheses) are shown. Mutants F6266 and F6756 were discovered previously by Liu et al. (2012). The red arrowheads correspond to amino acid polymorphisms in GmSHMT08 between resistant cv Forrest and susceptible cv Essex. B, The SCN FI of newly identified GmSHMT08 mutants in comparison with resistant cv Forrest. Each dot corresponds to the FI of an individual plant.
Testing of Advanced SHMT Mutant Lines
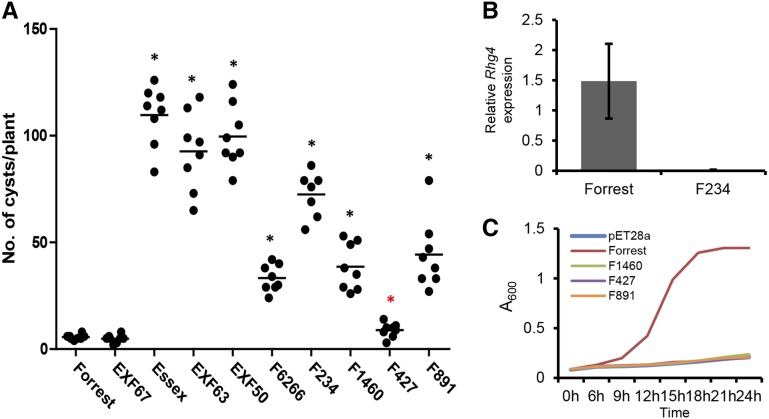
A subset of cv Forrest mutants, including F234, F427, F891, and F1460, for which we obtained sufficient seeds for further characterization, were advanced to homozygosity and phenotyped for SCN resistance in a side-by-side comparison alongside cv Forrest, cv Essex, the previously identified cv Forrest GmSHMT08 mutant F6266 (Liu et al., 2012), and RILs EXF50, EXF63, and EXF67 (Fig. 3A). All mutants showed a statistically significant loss of resistance compared with the cv Forrest and EXF67 resistant controls. The truncation mutant F234 (Q226*) was highly susceptible to SCN, with cyst numbers comparable to those of the EXF63 and EXF50 RILs that carry susceptible alleles at either rhg4 (GmSHMT08) or rhg1-a, respectively (average FI, 65.8%). All other mutants displayed a partial loss of resistance, with average FI values ranging from 8.1% (F427) to 40.2% (F891).
Figure 3.
Characterization of GmSHMT08 mutants. A, SCN phenotype of GmSHMT08 mutants in comparison with resistant (cv Forrest and EXF67) and susceptible (cv Essex, EXF63, and EXF50) soybean lines. Each dot corresponds to the number of cysts on an individual plant. Asterisks denote statistically significant differences in resistance in comparison with cv Forrest (paired Student’s t test, P < 0.01, red; P < 0.0001, black). B, Rhg4 expression in cv Forrest and the F234 mutant. Root cDNA was used as a template for qPCR. C, E. coli complementation assay using mutant GmSHMT08 proteins. The proteins were expressed under the control of the IPTG-inducible T7 promoter in E. coli shmt mutant GS245 pLysS strain. Proteins were induced with 0.25 mm IPTG at 37°C. The absorbance of bacterial cultures at 600 nm was measured at the time intervals shown and plotted.
We attempted to clone the GmSHMT08 cDNAs from all four mutants advanced to M5. Despite multiple attempts, we failed to clone the GmSHMT08 cDNA from the F234 mutant. A quantitative RT-PCR assay using primers that specifically amplify GmSHMT08 at the Rhg4 locus showed that GmSHMT08 mRNA levels are undetectable in this mutant (Fig. 3B), further confirming that this is a GmSHMT08 null allele. The GmSHMT08 cDNAs for F427, F891, and F1460 were cloned into protein expression vector pET28a. The expressed proteins were tested for enzyme activity using an Escherichia coli shmt mutant complementation assay (Liu et al., 2012). At 0.25 mm isopropyl β-D-1-thiogalactopyranoside (IPTG) induction, cv Forrest GmSHMT08 used as a positive control supported the growth of the mutant bacteria. In contrast, the GmSHMT08 mutations found in F427, F891, and F1460 resulted in proteins that are enzymatically inactive, as they were unable to support the growth of the bacteria (Fig. 3C). Our results demonstrate that mutations that affect the activity of the enzyme lead to partial loss of resistance in plants. A complete loss of the GmSHMT08 protein reverts plants to a susceptible phenotype. These results indicate that GmSHMT08 at the Rhg4 locus is essential for Peking-type resistance to SCN even when an intact resistant rhg1-a allele is present.
SCN Virulence Experiments
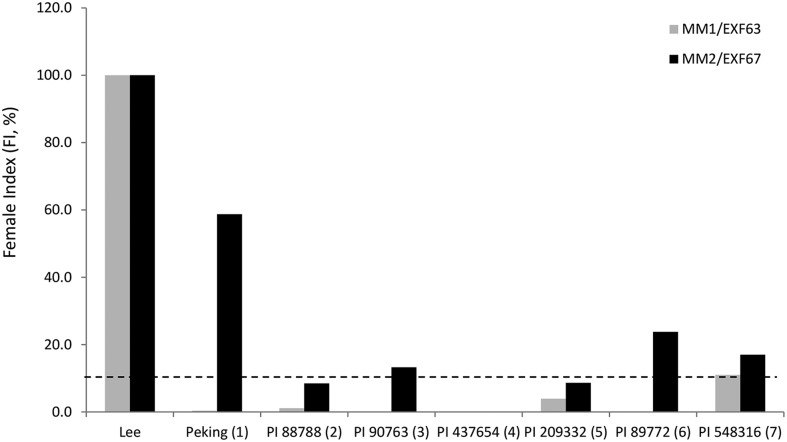
To further examine the bigenic nature of resistance and the essentiality of Rhg4 in mediating Peking-type resistance, we carried out a nematode virulence study. We inoculated EXF63, which has the rhg1-a allele from cv Forrest (resistant) and the rhg4 allele of cv Essex (susceptible), and EXF67, which inherited rhg1-a and Rhg4 alleles from cv Forrest, with SCN PA3 (race 3; HG type 7). These nematode populations were inbred by mass selection for more than 5 years, which represented at least 60 generations. HG type tests were then conducted on each population, and the results are shown in Figure 4. The SCN population MM2 (for Melissa Mitchum 2) selected on EXF67 had an FI of nearly 60% on Peking and slightly elevated FI values on PI 90763 (13%) and PI 89772 (24%), indicating a shift in the population from HG type 7 (race 3) to HG type 1.3.6.7 (race 14). In contrast, the SCN population (MM1) selected on EXF63 remained HG type 7. This result indicates that the rhg1-a allele from cv Forrest alone does not pose any selection pressure to the nematode to shift from HG type 7 and further validates the bigenic nature of this resistance.
Figure 4.
HG type tests of selected nematode populations MM1 and MM2. The nematode populations MM1 and MM2 were derived by mass selection of PA3 for more than 60 generations on RILs EXF63 (MM1) and EXF67 (MM2). The FI values on each resistance indicator line with respect to susceptible soybean cv Lee74 were plotted. The dotted line denotes the 10% FI used as the cutoff value in determining resistance to SCN.
Computational Modeling of GmSHMT08 Mutations
A computational homology modeling approach was used previously to gain insights into the impact of mutations to GmSHMT08 (Liu et al., 2012). The new mutations identified here were mapped onto the homology model and grouped according to their impact on the GmSHMT08 oligomeric structure and cofactor-binding sites.
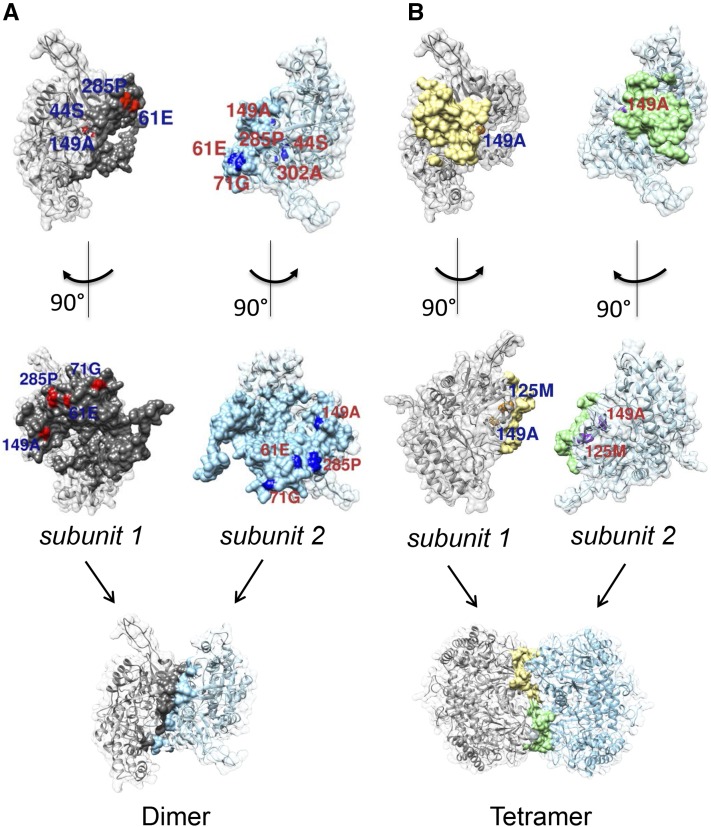
Mutations Annotated with the Protein-Binding Sites
The protein-binding site of GmSHMT08 involved in dimerization included 141 residues (Fig. 5A). From our set of mutations, six were located on the protein-binding site (residues 61, 71, 149, 285, and 302) and one in close proximity (residue 44; Fig. 5A). The protein-binding site involved in tetramerization was smaller and included only 31 residues. The mutated residues associated with the tetramerization binding site included three residues in close proximity (residues 125, 138, and 149) but none residing on the binding site itself (Fig. 5B).
Figure 5.
Computation modeling and mapping of mutants on the GmSHMT08 structure. A, Mutation positions on or in close proximity to the dimer binding sites (solid gray and solid light blue) for both interacting subunits. Shown in red (with blue labels) are mutation positions for the first subunit and in navy (with red labels) for the second subunit. B, Mutation positions on or in close proximity to the tetramer binding sites (solid yellow and solid green) for both interacting subunits. Shown in orange (with blue labels) are mutation positions for the first subunit and in purple (with red labels) for the second subunit.
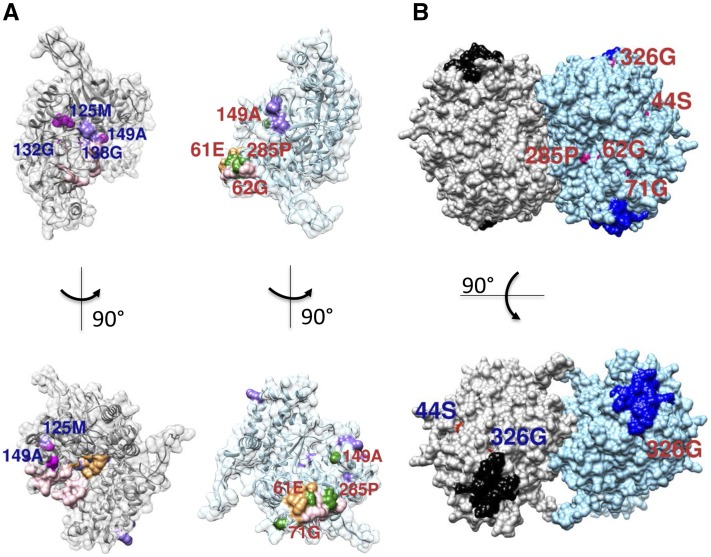
Mutations Annotated with the Functional Sites
The identified PLP/PLP-glycine site included 16 residues, the THF/MTHF/5-formyl THF (FTHF) site included 10 residues, and the Gly-binding sites included four residues. In total, four GmSHMT08 mutations are in close proximity to functional sites (residues 62, 125, 138, and 149; Fig. 6A). Not shown are 299L and 106G, which are internal residues and not annotated with any of the functional sites. However, 106G is in close proximity with the protein-binding site (residues 105 and 107 belong to the binding sites and are surface residues) and may affect dimerization. 299L is an internal residue and could impact the structure of the monomer, thus affecting GmSHMT08 function.
Figure 6.
Computation modeling and mapping of mutants on the GmSHMT08 structure. A, Mutation positions on or in close proximity to the functional sites (THF, MTHF, and FTHF in light pink, PLP-serine/PLP-glycine (PLS/PLG) in light orange, and Gly in light purple) for both interacting subunits. Shown in magenta are mutation positions for the first subunit and in green for the second subunit. B, GmSNAP18 protein-binding sites mapped on the GmSHMT08 tetramer together with its surface mutations. Shown are the surface mutations mapped on only one of the dimer subunits. The two GmSHMT08 dimers are colored gray and light blue. The two putative GmSNAP18 binding sites for each dimer are shown in black and navy. Shown in red (with blue labels) are mutation positions for one monomer of the first dimer and in magenta (with red labels) for a monomer of the second dimer.
Mutations Annotated with Interaction Sites
Using the advanced remote homology modeling protocol described above, we predicted a possible interaction between GmSHMT08 and GmSNAP18 and mapped the putative SNAP protein-binding site on the surface of the GmSHMT08 tetramer. The putative protein-binding site in the interaction with GmSNAP included 21 residues that were located closer to the C-terminal part of GmSHMT08. Interestingly, one of the GmSHMT08 mutations, 326G, was found adjacent to the protein-binding site (Fig. 6B).
DISCUSSION
The Rhg4 gene, cloned in 2012, encodes a cytosolic SHMT (Liu et al., 2012). The GmSHMT08 protein conferring resistance in cv Forrest has two amino acid polymorphisms compared with the susceptible version in cv Essex, resulting in a gain of function in SCN resistance. In this study, we demonstrated the genetic complementation of GmSHMT08 by a whole-plant transformation of soybean. The cv Forrest mutant F6266 and susceptible RIL EXF63, which we used for prior hairy root complementation experiments, were recalcitrant to transformation. Therefore, WXF5917, a RIL that retained the resistant rhg1-a allele of cv Forrest and the susceptible rhg4 allele from cv Williams 82, was used for the transgenic experiment. Resistant T1 plants remained resistant in T2, and plants with intermediate resistance were shown to be segregating in T2. The analysis of transgene expression showed that the transgene was expressed in all the plants analyzed, but some of them remained partially susceptible. This may be explained by the presence of two copies of the wild-type susceptible allele of the gene along with a single copy of the transgene. The expression ratio between the resistant and susceptible versions of SHMT may be critical for conferring full resistance in transgenic plants. To our knowledge, this is the first time the functionality of any SCN resistance gene has been demonstrated in a transgenic soybean plant.
We used a forward genetic screening approach coupled with sequencing of the GmSHMT08 gene to identify 13 new mutants covering the entire length of the GmSHMT08 protein. The average FI values of these mutants ranged from 13% to 93.4%. Although all mutants exhibited some loss of resistance relative to the resistant wild-type cv Forrest, the calculated FI values are from data accumulated over different experiments and, therefore, are not directly comparable. Some mutants were segregating for the GmSHMT08 mutation, and we observed an intermediate SCN phenotype for these plants compared with plants homozygous for the mutation. Although Rhg4 showed dominant action in mapping studies, these results demonstrate that the gene has a dosage effect on SCN resistance and further support the results of our transgenic experiment. More significantly, we identified two truncation mutants, one of which is a confirmed GmSHMT08 null allele. Altogether, we now have 15 mutants in this gene, including two mutants identified previously by TILLING (F6266 and F6756; Liu et al., 2012). Our results illustrate the power of mutagenesis to characterize gene function and represent the most extensive allelic series of mutations in any soybean gene to date.
Although SHMTs are present ubiquitously in all organisms, very little is known about the structure and function of these enzymes in plants. The Arabidopsis mitochondrial SHMT is encoded by AtSHM1 and is required for the proper functioning of photorespiration (Somerville and Ogren, 1981; Voll et al., 2006). Its ortholog in rice (Oryza sativa) is OsSHM1 (Wang et al., 2015; Wu et al., 2015). AtSHM3 (Zhang et al., 2010) encodes an SHMT protein that is targeted to plastids, and AtSHM7 encodes an SHMT that is a nonfunctional enzyme and is found to be required for SAM biosynthesis in the nucleus and that epigenetically affects sulfur homeostasis (Huang et al., 2016). None of the cytosolic SHMTs have been characterized in detail. The soybean genome contains at least 14 SHMT genes. The Rhg4 GmSHMT08 belongs to subgroup 1a. Subgroup 1a underwent a recent duplication in soybean, and Rhg4 GmSHMT08 on chromosome 8 accumulated more nonsynonymous mutations than its homolog on chromosome 5 and, therefore, evolved a function in resistance to SCN during the domestication of soybean (Wu et al., 2016). The mutants we identified will aid in a thorough biochemical characterization and a better understanding of the functional roles of this SHMT in resistance to SCN.
We used homology modeling and computational mapping to group the identified mutations into those that affect oligomeric structure, functional sites, and protein interaction sites. Mutations that affect oligomeric structure or functional sites have been shown to alter the enzymatic activity of this protein by several site-directed mutagenesis studies (for review, see Appaji Rao et al., 2003). Our analysis of a subset of mutants showed that the mutant enzymes are incapable of supporting the growth of an E. coli shmt mutant (Fig. 3C), demonstrating that the mutations affect catalytic activity. The effect of these mutations on SCN susceptibility ranged from an average FI of 8.1% to 65.8% (Fig. 3A), with the truncation mutant F234 showing the greatest impact on SCN resistance in a simultaneous comparison with other mutants. The F6266 mutation that we identified previously is a dead enzyme (Liu et al., 2012). Nevertheless, this mutant is still partially resistant to SCN in a direct comparison with the truncation mutant F234. The F234 mutant theoretically encodes for a protein of only 226 amino acids. Moreover, we were unable to detect the corresponding cDNA in this mutant, suggesting that it is a null mutation. Taken together, these findings clearly demonstrate that the GmSHMT08 protein has additional functions aside from its main enzymatic role in SCN resistance.
Peking-type resistance has been shown to be bigenic, requiring two genetic loci, Rhg4 and rhg1-a (Meksem et al., 2001). The rhg1-a in the Peking-type encodes an α-SNAP protein (GmSNAP18; Liu et al., 2017). Our bioinformatic analysis predicted an interaction between GmSHMT08 and the GmSNAP18 protein, and we mapped the interaction site on the surface of the GmSHMT08 protein (Fig. 6B). Although this prediction needs to be verified experimentally, it can be hypothesized that this interaction may be important for resistance and could be an explanation for the partial resistance observed for F6266 and other enzymatically inactive mutants. Our in vitro assays suggest a regulatory function for GmSHMT08 (Liu et al., 2012); therefore, the regulation of metabolite flux through sequestration or altered channeling is still a possibility with an inactive protein and cannot be ruled out.
The epistatic relationship between rhg1 and Rhg4 is not fully understood. The two SCN-susceptible RILS, EXF50, which has the Rhg4 resistant allele and the rhg1-a susceptible allele, and EXF63, which has the susceptible rhg4 allele but the resistant rhg1-a allele, suggest that neither of the genes confers any resistance individually. However, the residual resistance observed in our null mutant argues against this. We show here that the GmSHMT08 null mutant F234 has an average FI of 65.8%. Although rhg1-a seems to confer some amount of resistance in the cv Forrest GmSHMT08 null mutant, the contribution from other minor quantitative trait loci cannot be ignored (Lakhssassi et al., 2017). Yu et al. (2016) showed that rhg1-a alone may confer some level of resistance in crosses using PI 437654, a PI with a similar copy number (three copies) to that in cv Forrest. PI 437654 has a similar epistatic relationship between rhg1 and Rhg4 in conferring resistance to SCN (Wu et al., 2009). Unlike cv Forrest, PI 437654 is resistant to multiple SCN HG types, and although its resistance is poorly understood, it likely involves other unknown loci in addition to rhg1 and Rhg4. In support of the bigenic nature of resistance, our SCN virulence experiments demonstrate that rhg1-a alone does not provide any selection pressure to SCN. Despite years of selection on EXF63 (rhg1F rhg4E), the PA3 population remained HG type 7, whereas PA3 selected on EXF67 (rhg1F Rhg4F) shifted to HG type 1.3.6.7, indicative of a shift in virulence of the SCN population on Peking-type resistance (Fig. 4).
We are only beginning to understand the mechanism of resistance to SCN conferred by rhg1-a and Rhg4. A recent report suggests that the rhg1 α-SNAP confers resistance by cytotoxic effects of the large-scale accumulation of the aberrant α-SNAP protein at the nematode feeding sites, which disrupts SNARE complexes and vesicle trafficking (Bayless et al., 2016). How GmSHMT08 conditions this process is not clear. The allelic series of mutants identified here will play a key role in helping to understand how GmSHMT08 contributes to this process. SCN resistance in soybean uses a mechanism that is unique so far; nevertheless, it is one of the best-characterized resistances in an agricultural context. Understanding this resistance will help us design and develop broad-spectrum resistance not only to SCN but also to other economically important pathogens.
MATERIALS AND METHODS
Plant and Nematode Material
Inbred SCN population PA3 (Prakash Arelli Race 3; HG type 7) was maintained on susceptible soybean (Glycine max) ‘Williams 82’ according to standard procedures (Niblack et al., 2002). The soybean RILs used in this study included WXF5917, EXF50, EXF63, and EXF67 derived from crosses between the SCN-resistant cv Forrest and the SCN-susceptible cv Williams 82 or cv Essex, confirmed to have the genetic background (genotype) of rhg1Frhg1Frhg4W82rhg4W82, rhg1Erhg1ERhg4FRhg4F, rhg1Frhg1Frhg4Erhg4E, and rhg1Frhg1FRhg4FRhg4F, respectively, using simple sequence repeat and other markers, or by sequencing (Liu et al., 2012, 2017). MM1 and MM2 were generated by mass inbreeding of PA3 on RILs EXF63 and EXF67, respectively. HG type tests were conducted according to standard protocols (Niblack et al., 2002). The seven PIs used in the HG type test are PI 548402 or Peking (1), PI 88788 (2), PI 90763 (3), PI 437654 (4), PI 209332 (5), PI 89772 (6), and PI 548316 or Cloud (7) and are numbered 1 to 7. The FI, where FI = (mean number of females on a test soybean line)/(mean number of females on the susceptible standard) × 100, is used to describe the percentage of an SCN population that can reproduce on a resistant line. Populations with an FI of 10% or greater on a PI are given the number corresponding to the PI to assign an HG type. For example, the HG type 1.3.6.7 designation means that the SCN population has an FI of 10% or greater on Peking (1), PI 90763 (3), PI 89772 (6), and PI 548316 (7).
Vector Construction
The 5,103-bp cv Forrest GmSHMT08 genomic sequence (GenBank accession no. JQ714083) containing the coding sequence and the 5′ regulatory region was digested with restriction enzyme SbfI from binary vector pAKK:GmSHMT08 (Liu et al., 2012) and subcloned into the SpeI site of the pSPH2 transformation vector (Jacobs et al., 2015) by blunt-end cloning to create pSPH2:GmSHMT08. The orientation of GmSHMT08 in the clone was determined by sequencing, and a clone in the reverse orientation carrying the selectable marker for hygromycin resistance was used for transformation.
Biolistic Transformation of Somatic Embryos
Biolistic transformation was performed using WXF5917 somatic embryos. Transformation, embryo formation, and plant recovery were performed as described previously (Hancock et al., 2011). DNA was isolated from embryogenic cultures with a cetyl-trimethyl-ammonium bromide extraction procedure (Murray and Thompson, 1980). PCR was performed to confirm the presence of the GmSHMT08 transgene with primers WGCT-F and pSMART 1715F (Supplemental Table S1). Thermal cycling conditions consisted of a first step of denaturation at 94°C for 4 min, followed by 35 cycles of denaturation for 20 s at 94°C, annealing for 20 s at 58°C, and extension for 30 s at 72°C, and a final extension for 5 min at 72°C.
Transgenic Plant Characterization
Transgenic WXF5917 plants (carrying the cv Forrest GmSHMT08 transgene) were inoculated with SCN eggs as described by Brown et al. (2010). WXF5917 untransformed plants were used as a control. At 30 d postinoculation, cysts on the roots were counted. Each plant in the experiment was analyzed by genomic PCR using primers specific to the transgene as described above. To check DNA quality, two primers that amplify the endogenous GmSHMT08 gene were used (Supplemental Table S1). The seeds of transgenic plants showing intermediate or full resistance were used to test SCN resistance in the subsequent generation to ascertain if the resistance was inherited in a Mendelian fashion. The expression of the transgene in these plants was further confirmed and quantitated using the KAPA SYBR FAST One-Step qRT-PCR kit (Kapa Biosystems) and primers described in Supplemental Table S1 with RT-qPCR conditions as follows: RT at 42°C for 5 min followed by 95°C for 3 min, and the following PCR cycling parameters: 95°C for 10 s, 65°C to 58°C at 1°C per cycle gradient, followed by 42 cycles at 58°C and 72°C for 1 s (total of 50 cycles). The qPCR data were analyzed according to Pfaffl (2001).
Mutant Screening and Phenotyping
Two EMS mutant M2 populations (FM2011 and FM2013) were developed from the SCN-resistant soybean cv Forrest at Southern Illinois University in Carbondale according to the methods described by Cooper et al. (2008) and Meksem et al. (2008). The FM2011 and FM2013 populations contained 1,588 and 2,827 lines, respectively.
A forward genetic screen of these mutants for a loss of resistance to the SCN was conducted. A single seed for each mutant line (M2) was germinated, and the seedlings were inoculated with 1,200 SCN PA3 eggs in a greenhouse environment (Brown et al., 2010). The wild-type resistant cv Forrest, susceptible cv Essex, and the previously identified GmSHMT08 mutant F6266 (Liu et al., 2012) were used as controls. At 30 d postinoculation, roots were rinsed free of soil and the cysts were dislodged using a high-powered hand sprayer and counted. Each cyst represents one female nematode. The FI, where FI = (mean number of females on a mutant soybean line)/(mean number of females on the susceptible standard) × 100, was used as a measure of nematode reproduction. Plants with FI > 10% were transplanted to increase seeds, and leaf tissues from these plants were harvested. DNA was isolated from leaves of mutants showing increased susceptibility, and the GmSHMT08 gene at the Rhg4 locus was amplified by PCR using the primers listed in Supplemental Table S1. The thermal cycling conditions consisted of a first step of denaturation at 98°C for 2 min, followed by 35 cycles of denaturation for 30 s at 98°C, annealing for 30 s at 59°C, and extension for 90 s at 72°C, and a final extension for 10 min at 72°C. The sequence data were analyzed using Sequencher software 5.2.4 (Gene Codes). The seeds of plants with GmSHMT08 gene mutations were collected and further tested in subsequent generations to confirm the loss of resistance. A subset of the mutants was advanced to homozygosity and simultaneously tested in a direct phenotypic comparison. The data were analyzed using GraphPad Prism software, and means were compared for significant differences using an unpaired Student’s t test.
Cloning of GmSHMT08 Mutant cDNAs
The cDNAs of newly identified GmSHMT08 mutants used in the direct phenotypic comparison experiment, F891-1, F1460-2, and F427-2, were cloned by RT-PCR from leaves of respective mutant plants into the pGEM-T Easy (Promega) vector using the primers listed in Supplemental Table S1. It was not possible to clone the truncated GmSHMT08 mutant F234 cDNA from mutant plants despite repeated attempts. The clones were sequence verified and used for expression cloning.
Escherichia coli Complementation Assay
The pGEM-T Easy clones of F891-1, F1460-2, and F427-2 were digested with NdeI and HindIII restriction enzymes and subcloned into the pET28a expression vector at the NdeI-HindIII sites to express the GmSHMT08 protein as an N-terminal (His)6 tag fusion. PCR-positive clones were sequence verified to ensure that they were in frame with the N-terminal (His)6 tag. These clones were then used to transform GS245(DE3)pLysS (glyA strain) for protein expression. The expression of mutant proteins was verified in this strain after IPTG induction. The bacterial cultures expressing mutant proteins were used for complementation assays as described by Liu et al. (2012). Strains transformed with cv Forrest GmSHMT08 and vector only were used as controls. The complementation experiments were repeated at least three times.
RT Reaction and qPCR Assays
Total RNA was isolated from root tissues using the Miniprep Kit (Qiagen). RT was done using the Primescript first-strand cDNA synthesis kit (Takara, Clontech). Real-time quantitative RT-PCR was conducted as described by Kandoth et al. (2011). Samples were normalized relative to the soybean ubiquitin gene (accession no. D28123) as an endogenous control.
Computational Models of GmSHMT08 Dimer and Tetramer
A computational template-based approach was performed to structurally characterize the GmSHMT08 protein in its dimeric and tetrameric forms. First, we obtained a homology model of the GmSHMT08 homodimer using cv Essex sequence as a target. The search for structural templates of GmSHMT08 in the Protein Data Bank (PDB; Berman et al., 2000) resulted in 43 structurally resolved SHMT homologs from a diverse set of bacterial and mammalian species; no structurally resolved plant SHMTs were found. We selected the structure with the largest coverage of the GmSHMT08 sequence and highest sequence identity. The resulting structure, mouse SHMT homodimer (PDB identifier 1EJI; target-template sequence identity of 57%, template coverage of 100%), was selected as a template for homology modeling of GmSHMT08. Homology modeling was done using MODELER-9 (Sali and Blundell, 1993), and the top-ranked model was selected from the set of candidate models using the building MODELER scoring function. The resulting dimer model of GmSHMT08 was used to obtain protein-binding sites involved in dimerization.
We next modeled GmSHMT08 in its homotetrameric form using as a template a homotetramer of rabbit cytosolic SHMT (PDB identifier 1RVU). Specifically, the two copies of GmSHMT08 modeled were structurally aligned against the rabbit SHMT tetramer structure (the structural alignment root-mean-square deviation for each dimer is 1.64 Å). The resulting tetramer model of GmSHMT08 was used to obtain the protein-binding sites involved in tetramerization.
Annotation of GmSHMT08 Single-Nucleotide Variants with Protein-Binding and Functional Sites
We first extracted dimerization and tetramerization protein-binding sites. To do so, a set of contact residue pairs was defined between each pair of interacting proteins. A pair of residues was defined as a residue-residue contact pair if the shortest distance between their heavy atoms was no greater than 6 Å. Once all contact pairs for a protein-protein interaction are defined, the protein-binding site of one of the two proteins will include all residues from the contact pairs that belong to that protein. Each mutation that was either part of the protein-binding site or in close proximity (6 Å or less) was identified.
We next annotated the single-nucleotide variants within functional sites onto the surface of GmSHMT08 using the structural information of ligand binding by SHMT homologs. To determine the ligand-binding sites for the PLP-Ser complex, the PLP-Gly complex, and THF/MTHF/FTHF, the obtained model of GmSHMT08 was structurally aligned with each of the orthologous SHMTs known to interact with the small ligands, and the ligand-binding site from each homolog was mapped onto the surface of the GmSHMT08 model through the structural alignment. The residues constituting the Gly-binding sites GBS1 and GBS2 in the SHMT homologs were identified in the literature and then mapped onto the structure of GmSHMT08 in a similar way, using the structural alignment of GmSHMT08 with its homolog.
Predicting a Novel GmSHMT08-GmSNAP18 Interaction and Extracting a Putative GmSNAP18 Protein-Binding Site
Using an advanced remote homology modeling protocol, we predicted a possible interaction between GmSHMT08 and GmSNAP and mapped the putative SNAP protein-binding site on the surface of the GmSHMT08 tetramer. First, we determined a structural template for a remote homologous complex. A search protocol that involves an advanced, profile-based sequence search using GmSHMT08 and GmSNAP18 sequences as seeds did not return any structural templates. However, the fact that GmSNAP18 belongs to a structurally conserved family of TPR repeat proteins (Lakhssassi et al., 2017) suggested that a structure-based remote homology approach to find an interaction could be used (Corominas et al., 2014). For this protocol, we first determined SCOP superfamilies of each protein using SCOP annotation (Lo Conte et al., 2000) and the SUPERFAMILY tool (Gough and Chothia, 2002). The resulting superfamilies for GmSHMT08 and GmSNAP18 are PLP-dependent transferases (SCOP identifier 53383) and TPR-like proteins (SCOP identifier 48452), respectively. Next, we searched DOMMINO, our comprehensive database of macromolecular interactions (Kuang et al., 2012, 2016), for a protein complex that involves the interaction between two proteins, one from the PLP-dependent transferases superfamily and another from the TPR-like superfamily. As a result, we found one complex, involving the interaction between human Ala-glyoxylate aminotransferase and the TPR domain of human PEX5P (PDB identifier 3R9A). Importantly, the high structural similarity between the GmSHMT08 modeled monomer and aminotransferase (the root-mean-square deviation between the two structures with the trimmed N and C termini is 6.44 Å) and the fact that both GmSNAP18 and the TPR domain belong to the TPR family ensure an accurate mapping of a putative GmSNAP18 binding site from the structural interaction template. The mapping was done through (1) extracting the protein-binding site of aminotransferase from the protein complex using the same protocol as above, (2) structurally aligning the aminotransferase with GmSHMT08, and (3) mapping the protein-binding residue onto the surface of GmSHMT08 using the obtained structural alignment.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. PCR analysis of the Rhg4 transgene in the transgenic plants.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank the many technicians and student workers for their assistance with greenhouse work, field planting, and harvesting.
Footnotes
This work was supported by United Soybean Board projects 1520-532-5607 (to M.G.M. and K.M.) and 1720-172-0132 (to M.G.M., K.M., and W.A.P.), Missouri Soybean Merchandising Council project 258 (to M.G.M.), and National Science Foundation grant DBI-1458267 (to D.K.). We are grateful for support from a Monsanto Undergraduate Research Fellowship to E.P. and MU Discovery Fellows and Life Sciences Undergraduate Research Opportunity Fellowships to A.L.
Articles can be viewed without a subscription.
References
- Acedo JR, Dropkin VH, Luedders VD (1984) Nematode population attrition and histopathology of Heterodera glycines-soybean associations. J Nematol 16: 48–56 [PMC free article] [PubMed] [Google Scholar]
- Appaji Rao N, Ambili M, Jala VR, Subramanya HS, Savithri HS (2003) Structure-function relationship in serine hydroxymethyltransferase. Biochim Biophys Acta 1647: 24–29 [DOI] [PubMed] [Google Scholar]
- Bayless AM, Smith JM, Song J, McMinn PH, Teillet A, August BK, Bent AF (2016) Disease resistance through impairment of α-SNAP-NSF interaction and vesicular trafficking by soybean Rhg1. Proc Natl Acad Sci USA 113: E7375–E7382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely RL. (1969) The biochemistry of folic acid and related pteridines. Frontiers in Biology 13: 189–218 [Google Scholar]
- Brown S, Yeckel G, Heinz R, Clark K, Sleper D, Mitchum MG (2010) A high-throughput automated technique for counting females of Heterodera glycines using a fluorescence-based imaging system. J Nematol 42: 201–206 [PMC free article] [PubMed] [Google Scholar]
- Brucker E, Carlson S, Wright E, Niblack T, Diers B (2005) Rhg1 alleles from soybean PI 437654 and PI 88788 respond differentially to isolates of Heterodera glycines in the greenhouse. Theor Appl Genet 111: 44–49 [DOI] [PubMed] [Google Scholar]
- Christensen KE, MacKenzie RE (2006) Mitochondrial one-carbon metabolism is adapted to the specific needs of yeast, plants and mammals. BioEssays 28: 595–605 [DOI] [PubMed] [Google Scholar]
- Colgrove AL, Niblack TL (2008) Correlation of female indices from virulence assays on inbred lines and field populations of Heterodera glycines. J Nematol 40: 39–45 [PMC free article] [PubMed] [Google Scholar]
- Concibido VC, Diers BW, Arelli PR (2004) A decade of QTL mapping for cyst nematode resistance in soybean. Crop Sci 44: 1121–1131 [Google Scholar]
- Cook DE, Bayless AM, Wang K, Guo X, Song Q, Jiang J, Bent AF (2014) Distinct copy number, coding sequence, and locus methylation patterns underlie Rhg1-mediated soybean resistance to soybean cyst nematode. Plant Physiol 165: 630–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Lee TG, Guo X, Melito S, Wang K, Bayless AM, Wang J, Hughes TJ, Willis DK, Clemente TE, et al. (2012) Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338: 1206–1209 [DOI] [PubMed] [Google Scholar]
- Cooper JL, Till BJ, Laport RG, Darlow MC, Kleffner JM, Jamai A, El-Mellouki T, Liu S, Ritchie R, Nielsen N, et al. (2008) TILLING to detect induced mutations in soybean. BMC Plant Biol 8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corominas R, Yang X, Lin GN, Kang S, Shen Y, Ghamsari L, Broly M, Rodriguez M, Tam S, Trigg SA, et al. (2014) Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism. Nat Commun 5: 3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo BY. (1965) Histological responses of resistant and susceptible soybean varieties, and backcross progeny to entry and development of Heterodera glycines. Phytopathology 55: 375–381 [Google Scholar]
- Gough J, Chothia C (2002) SUPERFAMILY: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucleic Acids Res 30: 268–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock CN, Zhang F, Floyd K, Richardson AO, Lafayette P, Tucker D, Wessler SR, Parrott WA (2011) The rice miniature inverted repeat transposable element mPing is an effective insertional mutagen in soybean. Plant Physiol 157: 552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil SG, Van der Put NM, Waas ET, den Heijer M, Trijbels FJ, Blom HJ (2001) Is mutated serine hydroxymethyltransferase (SHMT) involved in the etiology of neural tube defects? Mol Genet Metab 73: 164–172 [DOI] [PubMed] [Google Scholar]
- Huang XY, Chao DY, Koprivova A, Danku J, Wirtz M, Müller S, Sandoval FJ, Bauwe H, Roje S, Dilkes B, et al. (2016) Nuclear localised MORE SULPHUR ACCUMULATION1 epigenetically regulates sulphur homeostasis in Arabidopsis thaliana. PLoS Genet 12: e1006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA (2015) Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotech 15: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth PK, Ithal N, Recknor J, Maier T, Nettleton D, Baum TJ, Mitchum MG (2011) The soybean Rhg1 locus for resistance to the soybean cyst nematode Heterodera glycines regulates the expression of a large number of stress- and defense-related genes in degenerating feeding cells. Plant Physiol 155: 1960–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI. (2003) Role of folate in colon cancer development and progression. J Nutr (Suppl 1) 133: 3731S–3739S [DOI] [PubMed] [Google Scholar]
- Koenning SR, Wrather JA (2010) Suppression of soybean yield potential in the continental United States from plant diseases estimated from 2006 to 2009. Plant Health Prog
- Kuang X, Dhroso A, Han JG, Shyu CR, Korkin D (2016) DOMMINO 2.0: integrating structurally resolved protein-, RNA-, and DNA-mediated macromolecular interactions. Database (Oxford) 2016: bav114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang X, Han JG, Zhao N, Pang B, Shyu CR, Korkin D (2012) DOMMINO: a database of macromolecular interactions. Nucleic Acids Res 40: D501–D506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhssassi N, Liu S, Bekal S, Zhou Z, Colantonio V, Lambert K, Barakat A, Meksem K (2017) Characterization of the Soluble NSF Attachment Protein gene family identifies two members involved in additive resistance to a plant pathogen. Sci Rep 7: 45226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TG, Kumar I, Diers BW, Hudson ME (2015) Evolution and selection of Rhg1, a copy-number variant nematode-resistance locus. Mol Ecol 24: 1774–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim U, Peng K, Shane B, Stover PJ, Litonjua AA, Weiss ST, Gaziano JM, Strawderman RL, Raiszadeh F, Selhub J, et al. (2005) Polymorphisms in cytoplasmic serine hydroxymethyltransferase and methylenetetrahydrofolate reductase affect the risk of cardiovascular disease in men. J Nutr 135: 1989–1994 [DOI] [PubMed] [Google Scholar]
- Liu S, Kandoth PK, Lakhssassi N, Kang J, Colantonio V, Heinz R, Yeckel G, Zhou Z, Bekal S, Dapprich J, et al. (2017) The soybean GmSNAP18 gene underlies two types of resistance to soybean cyst nematode. Nat Commun 8: 14822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Kandoth PK, Warren SD, Yeckel G, Heinz R, Alden J, Yang C, Jamai A, El-Mellouki T, Juvale PS, et al. (2012) A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature 492: 256–260 [DOI] [PubMed] [Google Scholar]
- Lo Conte L, Ailey B, Hubbard TJ, Brenner SE, Murzin AG, Chothia C (2000) SCOP: a structural classification of proteins database. Nucleic Acids Res 28: 257–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam R, Skorupska HT (1996) Cytological expression of early response to infection by Heterodera glycines Ichinohe in resistant PI 437654 soybean. Genome 39: 986–998 [DOI] [PubMed] [Google Scholar]
- Meksem K, Liu S, Liu X, Jamai A, Mitchum GM, Bendahmane A, Ei-mellouki T (2008) TILLING: a reverse genetics and a functional genomics tool in soybean. In Kahl G, Meksem K, eds, The Handbook of Plant Functional Genomics: Concepts and Protocols. Wiley, Hoboken, NJ, pp 251–265 [Google Scholar]
- Meksem K, Pantazopoulos P, Njiti VN, Hyten LD, Arelli PR, Lightfoot DA (2001) ‘Forrest’ resistance to the soybean cyst nematode is bigenic: saturation mapping of the Rhg1 and Rhg4 loci. Theor Appl Genet 103: 710–717 [Google Scholar]
- Mendes ML, Dickson DW (1993) Detection of Heterodera glycines on soybean in Brazil. Plant Dis 77: 499–500 [Google Scholar]
- Mitchum MG. (2016) Soybean resistance to the soybean cyst nematode Heterodera glycines: an update. Phytopathology 106: 1444–1450 [DOI] [PubMed] [Google Scholar]
- Mitchum MG, Wrather JA, Heinz RD, Shannon JG, Danekas G (2007) Variability in distribution and virulence phenotypes of Heterodera glycines in Missouri during 2005. Plant Dis 91: 1473–1476 [DOI] [PubMed] [Google Scholar]
- Moreno JI, Martín R, Castresana C (2005) Arabidopsis SHMT1, a serine hydroxymethyltransferase that functions in the photorespiratory pathway influences resistance to biotic and abiotic stress. Plant J 41: 451–463 [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niblack TL, Arelli PR, Noel GR, Opperman CH, Orf JH, Schmitt DP, Shannon JG, Tylka GL (2002) A revised classification scheme for genetically diverse populations of Heterodera glycines. J Nematol 34: 279–288 [PMC free article] [PubMed] [Google Scholar]
- Peng DL, Pend H, Wu DQ, Huang WK, Ye WX, Cui JK (2016) First report of soybean cyst nematode (Heterodera glycines) on soybean from Gansu and Ningxia China. Plant Dis 100: 229 [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 243: 779–815 [DOI] [PubMed] [Google Scholar]
- Skibola CF, Smith MT, Hubbard A, Shane B, Roberts AC, Law GR, Rollinson S, Roman E, Cartwright RA, Morgan GJ (2002) Polymorphisms in the thymidylate synthase and serine hydroxymethyltransferase genes and risk of adult acute lymphocytic leukemia. Blood 99: 3786–3791 [DOI] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL (1981) Photorespiration-deficient mutants of Arabidopsis thaliana lacking mitochondrial serine transhydroxymethylase activity. Plant Physiol 67: 666–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voll LM, Jamai A, Renné P, Voll H, McClung CR, Weber APM (2006) The photorespiratory Arabidopsis shm1 mutant is deficient in SHM1. Plant Physiol 140: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liu H, Li S, Zhai G, Shao J, Tao Y (2015) Characterization and molecular cloning of a serine hydroxymethyltransferase 1 (OsSHM1) in rice. J Integr Plant Biol 57: 745–756 [DOI] [PubMed] [Google Scholar]
- Wu J, Zhang Z, Zhang Q, Han X, Gu X, Lu T (2015) The molecular cloning and clarification of a photorespiratory mutant, oscdm1, using enhancer trapping. Front Genet 6: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Blake S, Sleper DA, Shannon JG, Cregan P, Nguyen HT (2009) QTL, additive and epistatic effects for SCN resistance in PI 437654. Theor Appl Genet 118: 1093–1105 [DOI] [PubMed] [Google Scholar]
- Wu XY, Zhou GC, Chen YX, Wu P, Liu LW, Ma FF, Wu M, Liu CC, Zeng YJ, Chu AE, et al. (2016) Soybean cyst nematode resistance emerged via artificial selection of duplicated serine hydroxymethyltransferase genes. Front Plant Sci 7: 998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Lee TG, Rosa DP, Hudson M, Diers BW (2016) Impact of Rhg1 copy number, type, and interaction with Rhg4 on resistance to Heterodera glycines in soybean. Theor Appl Genet 129: 2403–2412 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sun K, Sandoval FJ, Santiago K, Roje S (2010) One-carbon metabolism in plants: characterization of a plastid serine hydroxymethyltransferase. Biochem J 430: 97–105 [DOI] [PubMed] [Google Scholar]