A long noncoding RNA enhances Arabidopsis tolerance to drought and salt stress by modulating the expression of stress-related genes.
Abstract
Long noncoding RNAs (lncRNAs) affect gene expression through a wide range of mechanisms and are considered as important regulators in many essential biological processes. A large number of lncRNA transcripts have been predicted or identified in plants in recent years. However, the biological functions for most of them are still unknown. In this study, we identified an Arabidopsis (Arabidopsis thaliana) lncRNA, DROUGHT INDUCED lncRNA (DRIR), as a novel positive regulator of the plant response to drought and salt stress. DRIR was expressed at a low level under nonstress conditions but can be significantly activated by drought and salt stress as well as by abscisic acid (ABA) treatment. We identified a T-DNA insertion mutant, drirD, which had higher expression of the DRIR gene than the wild-type plants. The drirD mutant exhibits increased tolerance to drought and salt stress. Overexpressing DRIR in Arabidopsis also increased tolerance to drought and salt stress of the transgenic plants. The drirD mutant and the overexpressing seedlings are more sensitive to ABA than the wild type in stomata closure and seedling growth. Genome-wide transcriptome analysis demonstrated that the expression of a large number of genes was altered in drirD and the overexpressing plants. These include genes involved in ABA signaling, water transport, and other stress-relief processes. Our study reveals a mechanism whereby DRIR regulates the plant response to abiotic stress by modulating the expression of a series of genes involved in the stress response.
Drought and high soil salinity are major abiotic stresses that can significantly limit plant productivity. Plants respond to these stresses by changing their metabolism, physiology, and development, which, to a certain extent, could mitigate the negative impact of the stresses on their growth and reproduction. Underlying many of these changes is the dramatic reprograming of gene expression in response to these stresses. Since both drought stress and salt stress cause cellular dehydration or osmotic stress, gene regulation under drought and salt stress shares some overlapping mechanisms. This is partly attributed to the activation of the biosynthesis and accumulation of the stress hormone abscisic acid (ABA) under both drought and salt stresses (Xiong and Zhu, 2003; Nambara and Marion-Poll, 2005; Barrero et al., 2006). ABA and dehydration-derived primary and secondary signals activate many stress-responsive genes through an interconnected ABA signaling and ABA-independent signaling network that involves sensing, signal transduction, and gene activation (Yamaguchi-Shinozaki and Shinozaki, 2006; Munemasa et al., 2015; Zhu, 2016)
In the core ABA signaling pathway, ABA is perceived by the PYRABACTIN RESISTANCE1/PYR1-LIKE/REGULATORY COMPONENTS OF ABA RECEPTORS (PYR/PYL/RCAR) group of receptors that, upon binding with ABA, sequester type 2C protein phosphatases and release their inhibition on downstream components, including the SUCROSE NONFERMENTING1-RELATED PROTEIN KINASES SUBFAMILY2 (SnRK2s; Ma et al., 2009; Park et al., 2009; Cutler et al., 2010). These protein kinases subsequently phosphorylate and activate various downstream targets including, among others, transcriptional factors (Cutler et al., 2010; Munemasa et al., 2015; Zhu, 2016). Among these transcription factors, the ABA-responsive element-binding factor class of bZIP transcription factors is well studied. These transcription factors bind the conserved ABA-responsive element cis-element in the promoters of many ABA- and stress-responsive genes and regulate their expression to enhance plant stress tolerance (Furihata et al., 2006; Yamaguchi-Shinozaki and Shinozaki, 2006; Cutler et al., 2010). SnRK2s also could activate other targets such as NADPH oxidases, SLOW ANION CHANNEL1, and aquaporins (Sirichandra et al., 2009; Grondin et al., 2015; Munemasa et al., 2015). These signaling molecules and channels are involved directly in stomatal closure to conserve water under drought stress (Kwak et al., 2003; Furihata et al., 2006; Geiger et al., 2009; Brandt et al., 2015; Grondin et al., 2015).
Many stress-responsive genes contain another conserved cis-element, the C-REPEAT/DEHYDRATION RESPONSIVE ELEMENT, which can be recognized by the DEHYDRATION-RESPONSIVE ELEMENT-BINDING/C-REPEAT-BINDING FACTOR class of transcription factors in response to cold, drought, or salt stress (Shinozaki and Yamaguchi-Shinozaki, 2000; Agarwal et al., 2006; Thomashow, 2010). These genes include, for example, RESPONSIVE TO DEHYDRATION29A (RD29A), RD17, KIN1, and EARLY RESPONSE TO DEHYDRATION10. The proteins encoded by these stress-responsive genes may help the plants to reduce stress-caused cellular damage and enhance the ability of the plants to withstand the stress.
Prior research on stress gene regulation focused mainly on protein-encoding genes. In recent years, non-protein-coding transcripts are emerging as important regulators of gene expression. Among them, long noncoding RNAs (lncRNAs) represent diverse classes of transcripts longer than 200 nucleotides without or with little protein-coding potential. The lncRNAs have been considered important regulators of many essential biological processes by functioning as precursors of microRNAs (miRNAs) and other small RNAs or as miRNA target mimics. They also may affect alternative splicing of pre-mRNA and regulate chromatin state and chromatin loop dynamics (Ariel et al., 2015; Liu et al., 2015a). In plants, lncRNAs are transcribed by the plant-specific RNA polymerases Pol IV and Pol V as well as by Pol II and Pol III (Wierzbicki et al., 2008; Dinger et al., 2009) and are considered potential regulators of the plant response to the environment. For example, overexpression of the lncRNA INDUCED BY PHOSPHATE STARVATION1 results in reduced shoot inorganic phosphate content (Franco-Zorrilla et al., 2007). Down-regulation of CIS-NATURAL ANTISENSE RNA impairs the transfer of phosphate from roots to shoots and decreases seed yield in rice (Oryza sativa; Jabnoune et al., 2013). Transgenic overexpression of asHSFB2a, an antisense RNA of HSFB2a, affects the plant response to heat stress (Wunderlich et al., 2014). Several reports also show that drought or salt stress could alter lncRNA expression in plants. For example, 19, 664, and 98 drought-responsive lncRNAs were identified in foxtail millet (Setaria italica), maize (Zea mays), and rice, respectively (Qi et al., 2013; Zhang et al., 2014a; Chung et al., 2016). Salt stress and dehydration stress also alter the accumulation of eight and six lncRNAs, respectively, in Arabidopsis (Arabidopsis thaliana; Ben Amor et al., 2009). Nonetheless, no lncRNA has been characterized to regulate the plant response or tolerance to drought or salt stress.
In this study, we identified and characterized the function of a novel lncRNA, DRIR (for DROUGHT INDUCED lncRNA). The expression of DRIR was induced by drought and salt stress. Increasing DRIR expression enhanced plant sensitivity to ABA and increased tolerance to drought and salt stress. In situ hybridization analysis revealed that DRIR is localized mainly in the nucleus. Transcriptome sequencing and real-time PCR analysis revealed that DRIR modulates the expression of genes involved in the stress response, including ABA response, water transport, and transcription, that may collectively contribute to an enhanced abiotic stress tolerance.
RESULTS
The Expression of DRIR Is Induced by Drought and Salt Stress
We are interested in the potential role of lncRNA in plant stress responses. We hypothesized that if lncRNAs of this nature are present, they may be induced by abiotic stress. To identify these lncRNAs, we conducted RNA sequencing (RNA-seq) of salt-treated (300 mm NaCl, 3 h) Arabidopsis seedlings (Ding et al., 2014) and searched for transcripts longer than 200 nucleotides but with no recognized open reading frames that were induced significantly by the stress treatment. Among dozens of putative lncRNAs identified, a transcript of 755 nucleotides, which we referred to as DRIR, was chosen for further study. DRIR does not seem to encode a protein, although a short reading frame encoding 41 amino acids that does not show homology to any known peptide could be predicted. The middle part of the sequence has limited similarity to transposon elements, but overall, the sequence does not show clear homology to other genes. Like many other lncRNAs, DRIR does not have homologs in other plants.
Initially identified as salt stress inducible in our RNA-seq studies, the regulation of DRIR by stress was investigated further. Total RNA was extracted from the wild-type seedlings that were treated with dehydration or 150 mm NaCl. The expression level of DRIR was examined using real-time PCR. Consistent with our RNA-seq data, higher levels of DRIR transcripts were found to accumulate in salt-treated seedlings relative to untreated seedlings. The expression level of DRIR was 168-fold that in untreated seedlings (Supplemental Fig. S1). In addition, the expression level of DRIR in dehydrated seedlings was 59-fold that in untreated seedlings (Supplemental Fig. S1). To confirm these data, a construct was made in which the GUS reporter gene was placed under the control of a 2-kb DRIR promoter. This construct (PDRIR:GUS) was introduced into Columbia-0 plants using Agrobacterium tumefaciens-mediated transformation. Transgenic lines were treated with dehydration or 150 mm NaCl, and the PDRIR:GUS activities were analyzed. As shown in Supplemental Figure S1, the PDRIR:GUS activities were increased significantly under dehydration and salt stress conditions relative to the control conditions, confirming that drought and salt stress strongly up-regulate the expression of DRIR.
The Expression Pattern and Subcellular Localization of DRIRR
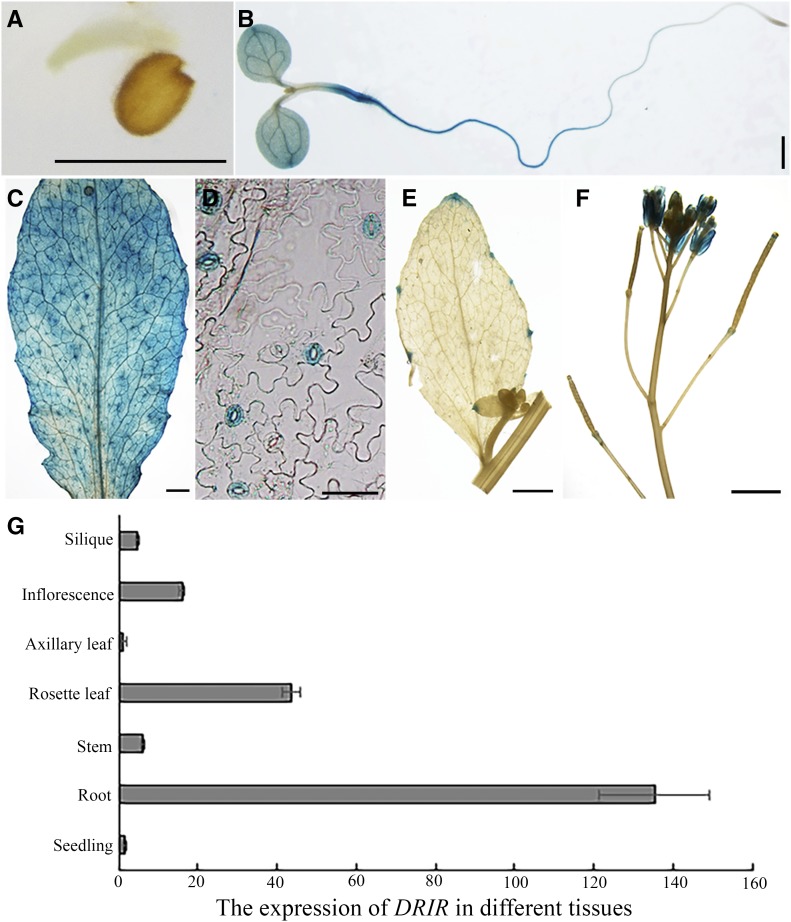
Data from the Affymetrix AG and ATH1 GeneChip arrays in the Genevestigator database (https://genevestigator.com/gv/) indicate that DRIR is expressed at a low level in plants and is expressed mainly in the root, inflorescence, embryo, shoot, and leaf. To further examine the expression pattern of DRIR, different parts of PDRIR:GUS transgenic lines were stained for GUS activity, and the results revealed that GUS signal was detected predominantly in roots, cotyledons, and stems in 6-d-old seedlings (Fig. 1B). In adult plants, strong GUS signals were detected in rosette leaves, guard cells, and flower petals (Fig. 1, C, D, and F). None or only weak GUS signal was detected in the embryo, axillary leaf, and silique (Fig. 1, A, E, and F). To verify these results, total RNA was extracted from different tissues and real-time PCR was performed. Consistent with the GUS staining data, real-time PCR showed that DRIR was highly expressed in roots and rosette leaves and to a lesser extent in stems, axillary leaves, inflorescences, and siliques (Fig. 1G).
Figure 1.
Expression pattern of DRIR in seedlings. Shown are DRIR promoter-GUS activities in germinating embryo (A), 6-d-old seedling (B), rosette leaf (C), guard cells in rosette leaf epidermis (D), axillary leaf (E), and inflorescence (F). Bars = 1 mm (A, B, C, E and F) and 50 µm (D). G shows transcript abundance of DRIR in different tissues as determined by quantitative RT-PCR. Values shown are means ± sd from three biological replicates.
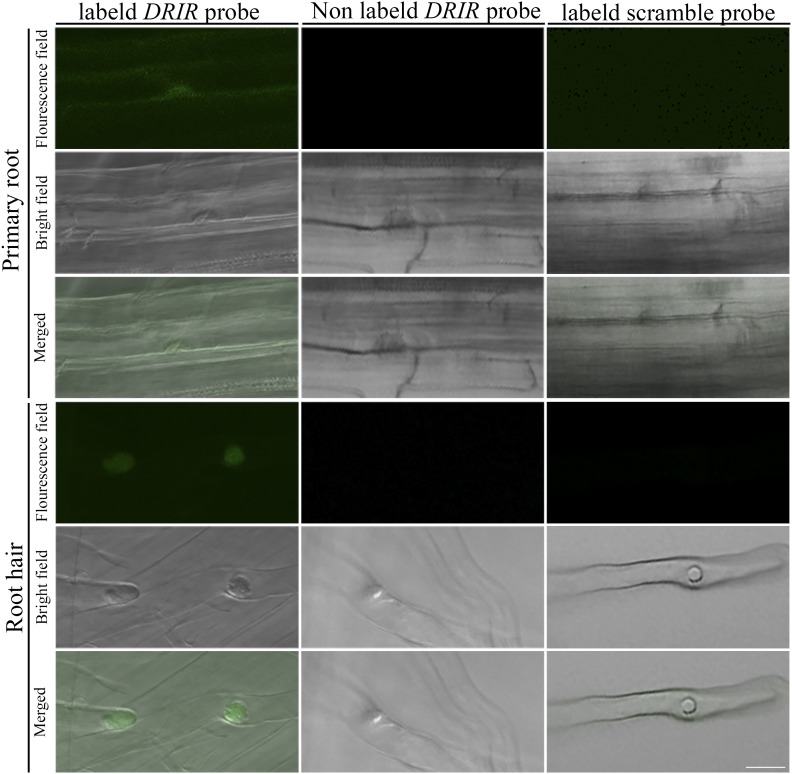
To investigate the subcellular localization of DRIR, fluorescence in situ hybridization in roots was performed with Alexa Fluor 488-labeled probe specific to DRIR. Fluorescence signal could be seen in nuclei of root cells and root hair cells that were hybridized with the Alexa Fluor 488-labeled DRIR probe but not in cells hybridized with the unlabeled DRIR probe or a labeled control probe (Fig. 2). To confirm this, nucleus was stained with 4′,6-diamino-phenylindole, and the signal of Alexa Fluor 488-labeled DRIR probe and the signal of 4′,6-diamino-phenylindole were found to colocalize in nuclei of root hairs (Supplemental Fig. S2). In addition, we isolated nuclei from a DRIR-overexpressing line (A12; see below) and extracted RNA from nuclei. One microgram of cDNA from nuclear RNA or total RNA was used to perform reverse transcription (RT)-PCR. A random mRNA (CAR4) was used as a control. The results show that a DRIR fragment could be amplified from nuclear RNA and that the gel signal strength was similar to that amplified from total RNA. In contrast, the CAR4 fragment could only be amplified from total RNA but not from nuclear RNA. These data suggest that DRIR transcripts are localized mainly in nuclei (Supplemental Fig. S3).
Figure 2.
Subcellular localization of the DRIR transcripts. Fluorescence in situ hybridization with Alexa Fluor 488-labeled antisense DRIR probe was performed in 7-d-old Columbia-0 root. Hybridization with unlabeled probe and Alexa Fluor 488-labeled scramble probe was used as a negative control. Bar = 20 µm.
DRIR Has a Function in Response to Drought and Salt Stress
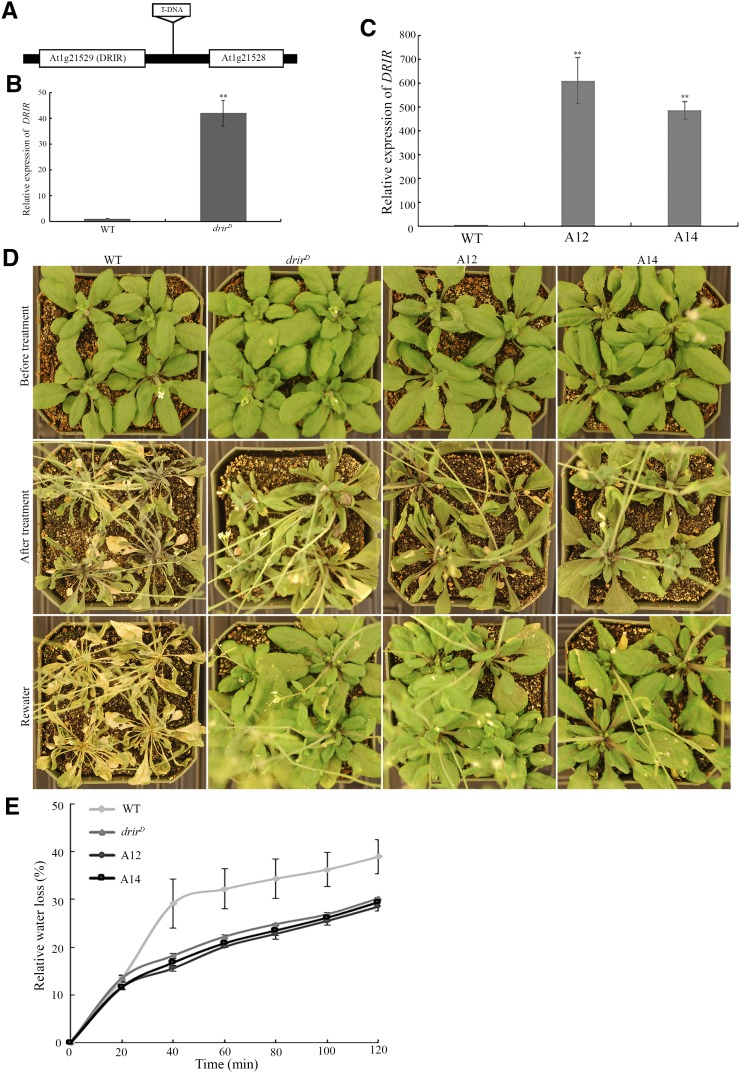
Since the expression of DRIR was up-regulated by drought and salt stress, the functions of DRIR in plant responses to drought and salt stress were analyzed. A T-DNA insertion mutant of DRIR, SAIL_813_G12, was obtained. The mutant contained a T-DNA insertion at 417 bp downstream of the gene At1g21529 (i.e. DRIR) and 467 bp upstream of the gene At1g21528 that encodes a hypothetical protein (Fig. 3A). The expression of DRIR in the mutant was significantly higher than that in the wild type (Fig. 3B). However, there was no significant difference in the expression of At1g21528 between the mutant and the wild type (Supplemental Fig. S4). Due to the activation nature of the mutation, we referred to this mutant as drirD. To test the drought tolerance of the mutant, 3-week-old seedlings of the wild type and drirD in soil were treated with drought stress by stopping watering for 20 d. All leaves of wild-type seedlings became totally withered and dry, but drirD leaves withered to a lesser extent and none was totally dry (Fig. 3D). Two days after rewatering, the wild-type seedlings were still withered; in contrast, drirD leaves became green and turgid again (Fig. 3D), indicating that the drirD mutant is more tolerant to drought stress. We examined transpirational water loss rates of fifth and sixth leaves of 4-week-old soil-grown plants. The water loss rate of drirD detached leaves was much slower than that of the wild-type leaves. drirD leaves lost 18%, 22%, 25%, 27%, and 30% of fresh weight at 40, 60, 80, 100, and 120 min after detachment, respectively, whereas wild-type leaves lost 29%, 32%, 34%, 36%, and 39% of fresh weight at the same times (Fig. 3E). These data demonstrate that DRIR is important for drought stress tolerance.
Figure 3.
The drirD mutant and overexpressing lines are more tolerant to drought stress. A, The Arabidopsis DRIR locus. The position of a T-DNA insertion in the drirD mutant (SAIL_813_G12) is shown. B, Relative transcript levels of DRIR in the wild type (WT) and drirD. **, P < 0.01 by Student’s t test. Data represent means ± sd. C, Relative transcript levels of DRIR in the wild type and two overexpressing lines, A12 and A14. **, P < 0.01 by Student’s t test. Data represent means ± sd. D, Morphology of seedlings before and after drought stress treatment. Three-week-old seedlings were drought stressed by stopping watering for 20 d before rewatering. Photographs were taken before (top row) and after (middle row) the drought treatment and 2 d after rewatering (bottom row). E, Transpirational water loss rates of detached leaves at the indicated times after detachment. Values shown are means ± sd from three replicates.
To confirm that the drought-tolerant phenotype of the T-DNA insertion mutant is attributable to increased expression of DRIR, a construct consisting of the DRIR gene driven by the cauliflower mosaic virus 35S promoter was transferred to Columbia-0, and more than 20 independent transgenic lines were obtained. Two overexpressing lines (A12 and A14) were randomly selected for detailed phenotype analysis. The expression level of DRIR in both transgenic lines was higher than in the wild type (Fig. 3C). These lines were tested for their drought tolerance along with the drirD mutant. It was found that the two overexpressing lines, similar to drirD, were more tolerant to drought stress (Fig. 3D). The water loss speed of detached leaves of the transgenic plants also was slower than that of the wild-type leaves but was similar to that of drirD (Fig. 3E). These data indicate that the drought tolerance seen in the drirD mutant is caused by the increased expression of DRIR and that DRIR plays a positive role in limiting transpirational water loss and increasing drought tolerance.
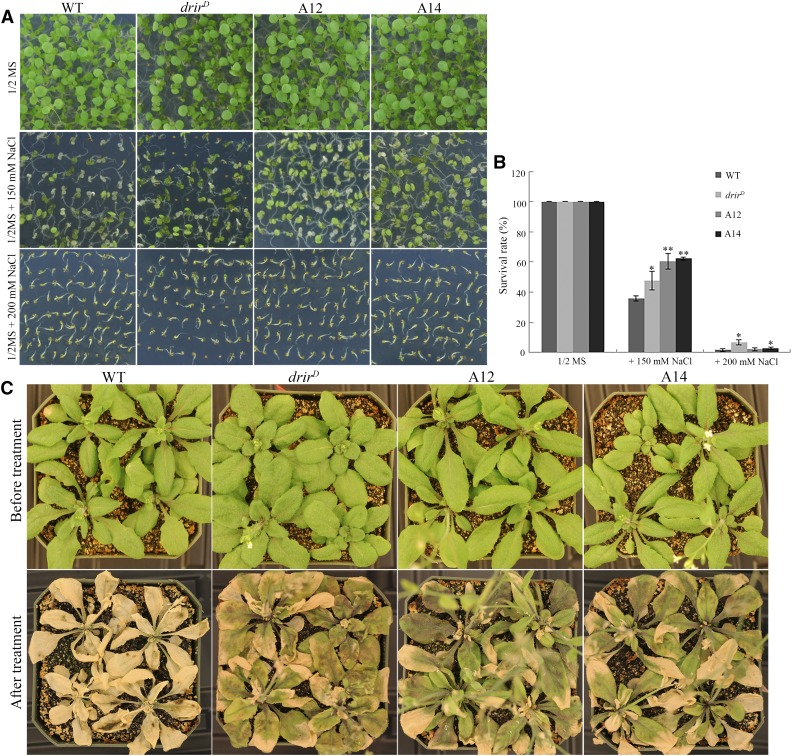
To further elucidate the role of DRIR in the Arabidopsis response to abiotic stress, seeds of the wild type, drirD, A12, and A14 were sown onto one-half-strength Murashige and Skoog (1/2 MS) medium plates containing 150 or 200 mm NaCl. As shown in Figure 4, A and B, in contrast to seedlings on 1/2 MS medium without NaCl, many seedlings were bleached and dead at 8 d after germination and growth on 1/2 MS medium containing different concentrations of NaCl. Compared with the 36% survival rate of the wild-type seedlings, the survival rates of drirD, A12, and A14 seedlings were 48%, 60%, and 62% on medium containing 150 mm NaCl, respectively (Fig. 4, A and B). To distinguish the effect of salt stress on seed germination and on seedling growth, 4-d-old seedlings of the wild type, drirD, A12, and A14 grown on 1/2 MS were transferred to 1/2 MS medium containing different concentrations of NaCl and allowed to grow for an additional 4 d. Similar to plants with seeds sown directly onto medium supplemented with salt, the transferred seedlings of drirD and overexpressing lines also were more tolerant to the salt stress than the wild type. The survival rates of drirD, A12, and A14 were 38%, 48%, and 42% under 150 mm NaCl treatment and 17%, 4%, and 8% under 200 mm NaCl treatment, respectively. In contrast, only 27% and 2% of the wild-type seedlings survived under 150 or 200 mm NaCl, respectively (Supplemental Fig. S5). To evaluate the role of DRIR in plant salt tolerance in soil, 4-week-old seedlings in soil were irrigated with 200 mm NaCl. Several days later, damage to leaves was observed. Two weeks later, leaves of drirD, A12, and A14 showed chlorosis but none died. However, nearly all wild-type seedlings were dead (Fig. 4C). These data indicate that drirD and the overexpressing lines are more tolerant to salt stress.
Figure 4.
The drirD mutant and DRIR-overexpressing lines are more tolerant to salt stress. A, Seeds of the wild type (WT), drirD, A12, and A14 were sown onto 1/2 MS agar medium plates supplemented with 0, 150, or 200 mm NaCl. Photographs were taken 8 d after the plates were incubated in the growth chamber for germination and growth. B, Seedling survival rate. At least 300 seedlings for each treatment per genotype were scored. *, P < 0.05 and **, P < 0.01 by Student’s t test compared with the wild type. Data represent means ± sd. C, Morphology of 4-week-old seedlings irrigated with 200 mm NaCl. Photographs were taken before and 2 weeks after the salt stress treatment.
drirD and Overexpressing Lines Are More Sensitive to ABA
Since phytohormones, in particular ABA, play critical roles in the plant response to abiotic stress (Yamaguchi-Shinozaki and Shinozaki, 2006; Waadt et al., 2014; Munemasa et al., 2015), the possible regulation of DRIR by phytohormones was analyzed. We found that the expression level of DRIR was increased significantly by ABA treatment. However, the effects of indole acetic acid, zeatin, 24-epibrassinolide, and GA on the expression of DRIR were not as profound as that of ABA (Supplemental Fig. S6). We also analyzed GUS activities in PDRIR:GUS transgenic plants under ABA treatment. As shown in Supplemental Figure S6, stronger PDRIR:GUS activities could be observed in ABA-treated seedlings compared with mock-treated seedlings, indicating that the expression of DRIR is induced by ABA.
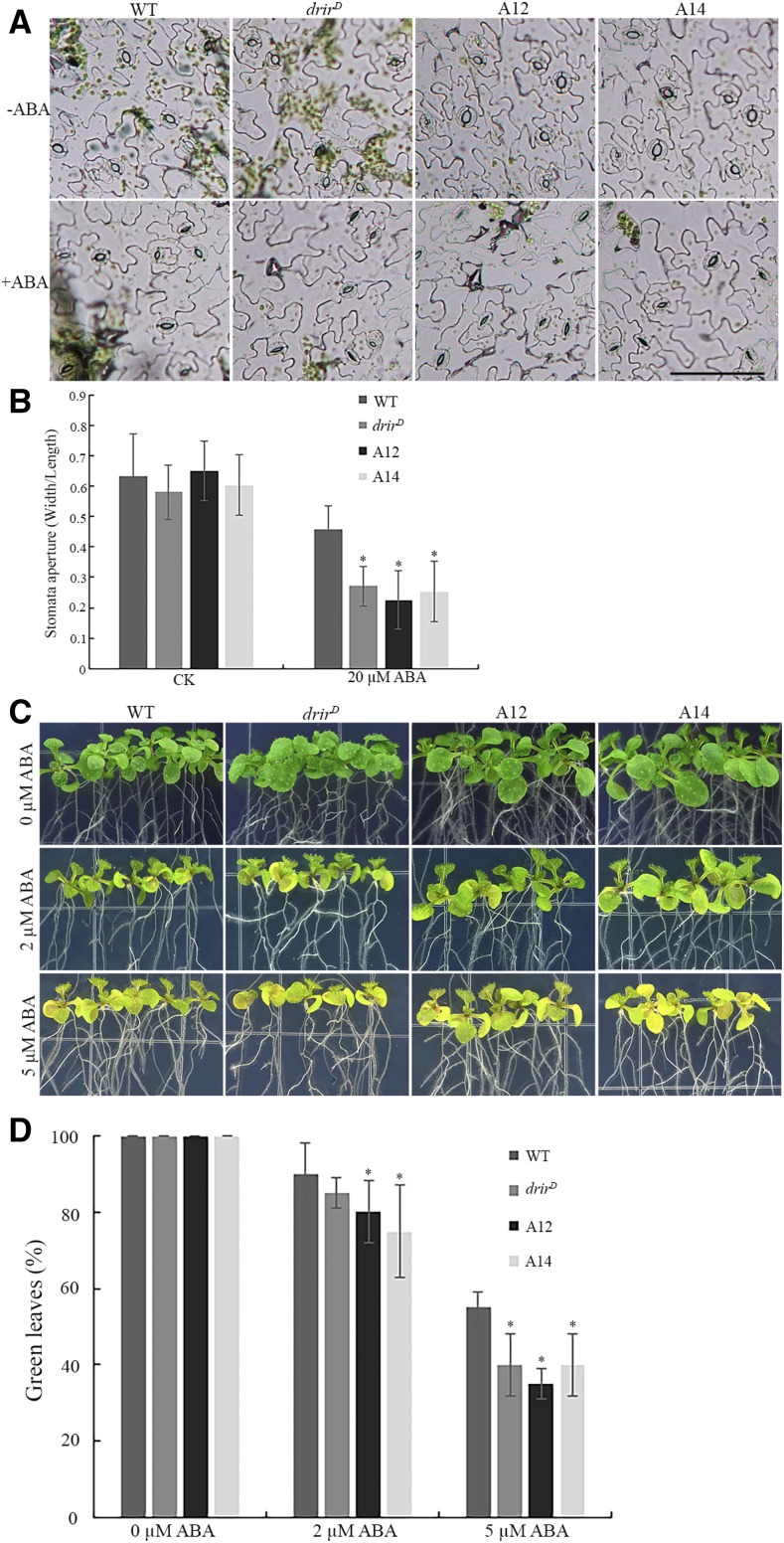
Since ABA increases the expression of DRIR, we investigated the responses of drirD and overexpressing lines to ABA. We first examined the sensitivity of stomata movement to exogenous ABA. The fifth and sixth leaves of 4-week-old plants were detached, submerged in stomata open solution, and incubated in a growth chamber for 2 h. The leaf samples were then transferred to a similar open solution with 20 µm ABA to induce stomata closure as described (Brandt et al., 2015). After incubation in the open solution for 2 h, stomata were widely open, and there was no significant difference in stomata aperture between the wild type and drirD or overexpressing lines. The width-to-length ratio for all these genotypes was about 0.6 (Fig. 5, A and B). However, the stomata aperture of drirD or overexpressing lines was much smaller than that of the wild type with the ABA treatment. The stomata width-to-length ratio of drirD and the two overexpressing lines decreased from about 0.6 to 0.27, 0.22, and 0.25, respectively, whereas that of the wild type merely decreased to 0.46 (Fig. 5, A and B). These data indicate that stomata of drirD and the overexpressing lines were more responsive to ABA-induced closure than those of the wild type.
Figure 5.
drirD and overexpressing lines are more sensitive to ABA. A and B, Stomata of drirD and the overexpressing lines are more sensitive to ABA-induced stomata closure. A, Representative images of stomata on leaf epidermis in response to ABA treatment. The fifth and sixth rosette leaves of 4-week-old plants were incubated in a stomata open solution for 2 h and then transferred to the open solution supplemented with 20 µm ABA. Images were taken before and 2 h after ABA treatment. Bar = 100 µm. B, Stomata aperture (measured by width over length) in response to the ABA treatment in different genotypes. At least 500 stomata from 30 leaves of each genotype were measured. CK, control with no ABA treatment. *, P < 0.05 by Student’s t test compared with the wild type (WT). Error bars represent sd. C, Seedling sensitivity to ABA. Four-day-old seedlings of the wild type, drirD, A12, and A14 on 1/2 MS plates were transferred to 1/2 MS plates supplemented with 0, 2, or 5 µm ABA. Representative images show the morphology of seedlings 8 d after growth on ABA medium plates. D, Green leaf percentage of each genotype. At least 100 seedlings for each treatment were scored. *, Significant between the sample and the wild type at P < 0.05 by Student’s t test. Data represent means ± sd.
To exclude the possibility that DRIR affects the plant response to drought stress by regulating stomatal density, stomatal densities of the fifth and sixth leaves of 4-week-old plants were examined, but no significant difference was found among these genotypes (Supplemental Fig. S7), suggesting that DRIR does not regulate the morphogenesis and differentiation of stomatal cells.
We further examined the sensitivity of drirD and overexpressing seedlings to ABA. Four-day-old seedlings of the wild type, drirD, A12, and A14 grown on 1/2 MS agar plates were transferred to 1/2 MS agar plates supplemented with different concentrations of ABA. Eight days later, more seedlings of drirD and overexpressing lines were found to have etiolated leaves compared with the wild type (Fig. 5C). While almost 90% of leaves of wild-type seedlings were green, less than 85% of leaves of drirD and overexpressing seedlings were green under 2 µm ABA treatment. Although the percentage of green leaves of wild-type seedlings decreased to 55% under 5 µm ABA treatment, less than 40% of green leaves could be found in drirD and overexpressing seedlings (Fig. 5D), indicating that drirD and overexpressing seedlings are more sensitive to ABA than the wild type.
DRIR Regulates the Expression of Genes Involved in the Stress Response
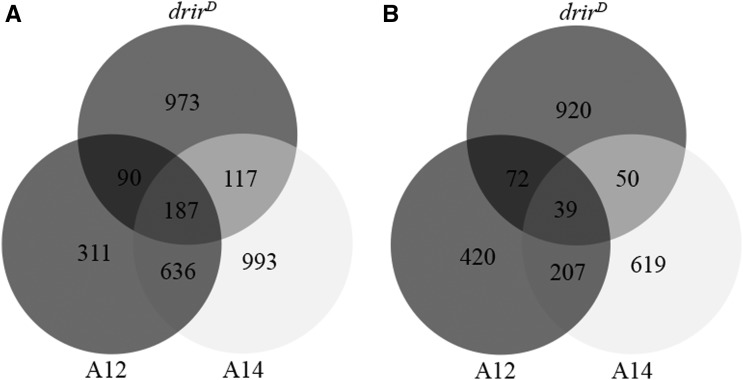
To elucidate the molecular mechanism of DRIR function in the plant response to drought stress, 10-d-old seedlings treated with dehydration were used for transcriptome sequencing analysis. Compared with the wild type, there were 1,367, 1,224, and 1,933 genes up-regulated and 1,081, 738, and 915 genes down-regulated more than 2-fold in drirD, A12, and A14, respectively (Fig. 6; Supplemental Table S1). Among them, 187 genes were up-regulated and 39 genes were down-regulated both in drirD and the two overexpressing lines (Fig. 6; Supplemental Table S2). Among those genes up-regulated both in drirD and the overexpressing lines, 28 genes have been suggested to be involved in the plant response to drought stress, salt stress, or ABA (Table I).
Figure 6.
Number of up- and down-regulated genes in dehydration-treated drirD and overexpressing lines in transcriptome sequencing analysis compared with dehydration-treated wild-type seedlings. A, Number of genes up-regulated more than 2-fold in dehydration-treated drirD and overexpressing seedlings. B, Number of genes down-regulated more than 2-fold in dehydration-treated drirD and overexpressing seedlings.
Table I. Genes reported to be involved in the plant response to drought stress, salt stress, or ABA that are up-regulated both in drirD and in the overexpressing lines.
| Gene Identifier | Gene Name | Description | Reference |
|---|---|---|---|
| AT1G02920 | ATGST11 | Glutathione transferase | Yang et al. (1998) |
| AT1G09090 | AtrbohB | NADPH oxidase AtrbohB | Kwak et al. (2003) |
| AT1G65970 | TPX2 | Thioredoxin-dependent peroxidase2 | Kumar et al. (2015) |
| AT1G67760 | TCP-1/cpn60 chaperonin family protein | Gong et al. (2001) | |
| AT1G73260 | ATKTI1 | Trypsin inhibitor | Chini et al. (2004) |
| AT1G74080 | ATMYB122 | Putative transcription factor | Ding et al. (2013) |
| AT1G79840 | GL2 | Homeodomain protein | Wang and Li (2008) |
| AT2G15390 | FUT4 | α-(1,2)-Fucosyltransferase | Tryfona et al. (2014) |
| AT2G18550 | ATHB21 | Homeodomain Leu zipper class I (HD-Zip I) protein | Ding et al. (2013) |
| AT2G25810 | TIP4 | Tonoplast intrinsic protein4;1 | Alexandersson et al. (2005); Regon et al. (2014) |
| AT2G26290 | ARSK1 | Root-specific kinase1 (ARSK1) | Hwang and Goodman (1995) |
| AT2G29090 | CYP707A2 | Protein with ABA 8′-hydroxylase activity | Arc et al. (2013); Sasaki et al. (2015) |
| AT2G38905 | Low temperature- and salt-responsive protein family | Medina et al. (2007) | |
| AT3G09940 | ATMDAR3 | Member of the monodehydroascorbate reductase gene family | Brini et al. (2011) |
| AT3G29035 | NAC3 | Protein with transcription factor activity | Nakashima et al. (2012) |
| AT3G45700 | NPF2.4 | Member of the NAXT NPF subfamily | Li et al. (2016a) |
| AT3G54770 | ARP1 | Putative RNA-binding protein | Jung et al. (2013) |
| AT4G19030 | NIP1 | Aquaporin | Alexandersson et al. (2005) |
| AT4G25820 | XTR9 | Xyloglucan endotransglycosylase | Li et al. (2008) |
| AT4G28520 | CRU3 | 12S seed storage protein | Nambara et al. (1995) |
| AT4G40090 | AGP3 | Arabinogalactan protein3 (AGP3) | Ma and Bohnert (2007) |
| AT5G01550 | LECRKA4.2 | Member of the lectin receptor kinase subfamily A4 | Xin et al. (2009) |
| AT5G06630 | Pro-rich extensin-like family protein | Dinneny et al. (2008) | |
| AT5G10230 | ANNAT7 | Calcium-binding protein annexin (ANNAT7) | Cantero et al. (2006) |
| AT5G13080 | WRKY75 | Transcription factor induced during inorganic phosphate deprivation | Ding et al. (2013) |
| AT5G44610 | MAP18 | Protein with seven repeated VEEKK motifs | Kato et al. (2010) |
| AT5G46350 | WRKY8 | Member of the WRKY transcription factors | Hu et al. (2013) |
| AT5G48010 | THAS | Oxidosqualene cyclase | de Silva et al. (2011) |
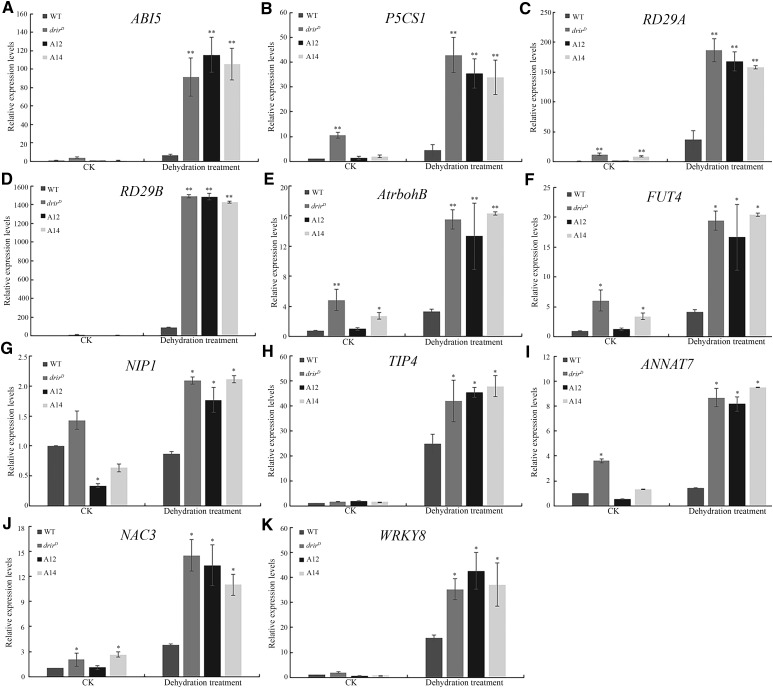
To further validate the expression of those genes that are known to be critical in the plant response to stress, total RNA was extracted from 10-d-old drirD and overexpression seedlings treated with dehydration stress, and real-time PCR was performed to analyze their expression. As shown in Figure 7, the expression of Arabidopsis thaliana respiratory burst oxidase B (AtrbohB), an NADPH oxidase gene, increased more than 15-fold in dehydration-treated drirD and the overexpressing lines, whereas less than 2-fold increase was found in dehydration-treated wild-type seedlings (Fig. 7E). The expression of FUCOSYLTRANSFERASE4 (FUT4) increased 19-, 16-, and 20-fold in dehydration-treated drirD and the A12 and A14 overexpressing lines, respectively, but only 4-fold increase was observed in wild-type seedlings (Fig. 7F). Whereas not significantly up-regulated by dehydration stress in the wild type, NOD26-LIKE INTRINSIC PROTEIN1 (NIP1), a member of the NIP aquaporin subfamily, was expressed about 2-fold in dehydrated drirD and the overexpressing lines compared with the untreated wild type (Fig. 7G). The expression level of another aquaporin protein gene, TONOPLAST INTRINSIC PROTEIN4 (TIP4), increased 24-fold in dehydration-treated wild-type seedlings, but it increased more than 40-fold in drirD and the overexpressing lines (Fig. 7H). The expression of an Arabidopsis annexin gene, ANNAT7, increased more than 8-fold in dehydration-treated drirD and the overexpressing lines, while it increased only 2-fold in wild-type seedlings (Fig. 7I). Two transcription factor genes, NAM, ATAF, AND CUC PROTEIN3 (NAC3) and WRKY PROTEIN8 (WRKY8), also were expressed significantly higher in dehydrated drirD and the overexpressing lines than in wild-type seedlings (Fig. 7, J and K).
Figure 7.
Relative expression levels of selected genes in dehydration-treated seedlings. RNA was extracted from 10-d-old seedlings that were dehydrated to lose about 40% fresh weight, and gene expression level was measured by RT-quantitative PCR normalized against the 18S rRNA gene. Relative expression levels are shown for ABI5 (A), P5CS1 (B), RD29A (C), RD29B (D), AtrbohB (E), FUT4 (F), NIP1 (G), TIP4 (H), ANNAT7 (I), NAC3 (J), and WRKY8 (K). CK, controls without dehydration treatment. *, P < 0.05 and **, P < 0.01 by Student’s t test compared with untreated (CK) or dehydration-treated wild-type (WT) seedlings. Data represent means ± sd.
In our RNA-seq analysis, we also found that several other ABA-signaling or ABA-inducible genes were expressed at a higher level in dehydration-treated drirD and the overexpressing seedlings than in wild-type seedlings, although the increasing levels were less than 2-fold (Supplemental Table S1). These genes include, for example, ABSCISIC ACID-INSENSITIVE5 (ABI5), Δ1-PYRROLINE-5-CARBOXYLATE SYNTHETASE1 (P5CS1), RD29A, and RD29B. Real-time PCR analysis was performed to validate the expression levels of these genes. Compared with untreated seedlings, the expression of these genes was increased significantly in dehydration-treated wild-type seedlings, but the extent of increase was much smaller than in drirD or overexpressing seedlings. The expression of ABI5 increased more than 90-fold in dehydration-treated drirD and overexpressing seedlings, while it increased only 6-fold in dehydrated wild-type seedlings (Fig. 7A). The expression of P5CS1 increased 4-, 42-, 35-, and 33-fold in dehydration-treated wild-type, drirD, A12, and A14 seedlings, respectively (Fig. 7B). The expression of RD29A and RD29B also increased more than 160- and 1,400-fold in dehydration-treated drirD and overexpressing seedlings, respectively, while it increased only 37-and 86-fold in dehydration-treated wild-type seedlings (Fig. 7, C and D). These data indicate that DRIR could regulate plant tolerance to drought stress by modulating the expression of genes involved in ABA signaling or stress responses.
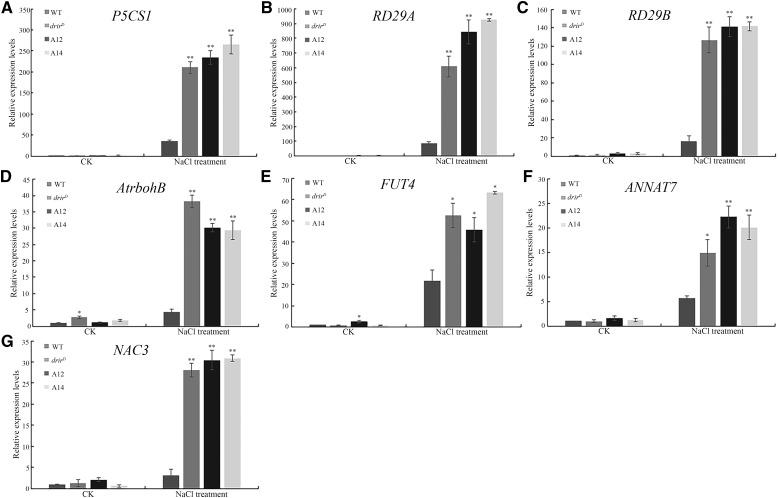
We further investigated the regulation of these stress-related genes by DRIR in response to salt stress. Ten-day-old seedlings were treated with 200 mm NaCl for 3 h, and real-time PCR was performed to analyze the expression of the above-mentioned genes. The results showed that the expression of P5CS1, RD29A, RD29B, AtrbohB, FUT4, ANNAT7, and NAC3 increased dramatically in salt-treated drirD and overexpressing seedlings, whereas their expression increased to a much lesser extent in salt-treated wild-type seedlings (Fig. 8), indicating that DRIR also regulates the expression of these genes under salt stress. Nonetheless, no significant difference was found in the expression of ABI5 and NIP1 between salt-treated wild-type and drirD or overexpressing seedlings (Supplemental Fig. S8, A and B). Furthermore, the expression of TIP4 and WRKY8 in salt-treated wild-type seedlings was even higher than in drirD and overexpressing seedlings (Supplemental Fig. S8, C and D). These results suggest that DRIR may differentially regulate stress-related genes through complex mechanisms.
Figure 8.
Relative expression levels of selected genes in salt stress-treated seedlings. RNA was extracted from 10-d-old seedlings that were treated with 200 mm NaCl for 3 h, and gene expression level was measured by RT-quantitative PCR normalized against the 18S rRNA gene. Relative expression levels are shown for P5CS1 (A), RD29A (B), RD29B (C), AtrbohB (D), FUT4 (E), ANNAT7 (F), and NAC3 (G). CK, controls without NaCl treatment. *, P < 0.05 and **, P < 0.01 by Student’s t test compared with untreated (CK) or salt-treated wild-type (WT) seedlings. Data represent means ± sd.
DRIR Affects the Accumulation of Pro and Reactive Oxygen Species
Pro is an important osmolyte with cellular protection functions in plants. Abiotic stress conditions as well as ABA treatment could promote the accumulation of Pro by inducing P5CS1 expression (Abrahám et al., 2003; Sharma et al., 2011). Since drirD and the overexpressing lines are more sensitive to ABA and the expression of P5CS1 in drirD and the overexpressing lines is increased dramatically when treated with dehydration and salt stress, we analyzed the contents of Pro in 10-d-old seedlings treated with dehydration or salt stress. As shown in Supplemental Figure S9, the content of Pro increased from 10 to 30 µg g−1 fresh weight in wild-type seedlings when treated with dehydration stress, while it increased to more than 60 µg g−1 fresh weight in drirD and the overexpressing lines. When treated with salt stress, the content of Pro increased to 30 µg g−1 fresh weight in wild-type seedlings, yet it increased to more than 42 µg g−1 fresh weight in drirD and the overexpressing lines. These results suggest that the synthesis of Pro was likely enhanced in drirD and the overexpressing lines under drought and salt stress.
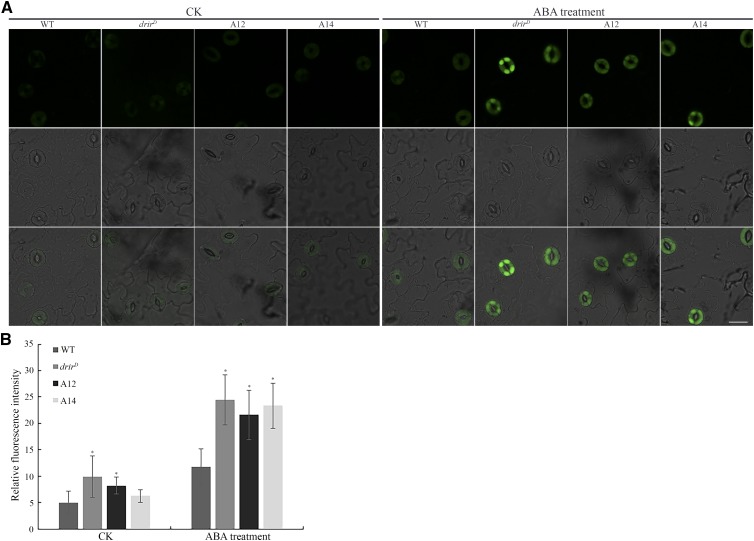
Reactive oxygen species (ROS) are critical second messengers in the ABA regulation of stomata closure, and their synthesis is catalyzed by NADPH oxidases (Kwak et al., 2003; Watkins et al., 2014). Since stomata closure of drirD and the overexpressing lines was more sensitive to ABA and the expression of the NADPH oxidase catalytic subunit gene AtrbohB was dramatically up-regulated in dehydration- and salt-treated drirD and overexpressing seedlings, we examined whether DRIR affects ROS accumulation in guard cells during ABA regulation of stomata closure. Peeled epidermises from leaves treated with or without ABA were stained by 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA) and examined with a confocal microscope. As shown in Figure 9, the intensity of the fluorescence signal of H2DCF-DA in guard cells of leaves without ABA treatment was very low but increased dramatically after ABA treatment, indicating that ROS accumulation was increased significantly in ABA-treated leaves. Interestingly, the fluorescence signal of H2DCF-DA in ABA-treated drirD and overexpressing guard cells was much higher than that in ABA-treated wild-type guard cells (Fig. 9A). The relative fluorescence intensity in guard cells of ABA-treated wild-type leaves was 11.7, while those of drirD and overexpressing leaves were 24.5, 21.6, and 23.3, respectively (Fig. 9B). These data demonstrate that more ROS accumulated in guard cells of drirD and overexpressing leaves during ABA induction of stomata closure.
Figure 9.
More ROS accumulated in guard cells of drirD and overexpressing leaves in response to ABA treatment. A, Representative images of ROS production stained by the fluorescent dye H2DCF-DA in guard cells without (left) or with (right) ABA treatment. Top row, Fluorescence images; middle row, bright-field images; bottom row, merged images. Bar = 20 µm. B, Relative fluorescence intensity of H2DCF-DA. More than 100 guard cells from 10 leaves of each genotype were measured. CK, controls without ABA treatment. *, P < 0.05 by Student’s t test compared with untreated or ABA-treated wild type (WT) seedlings. Error bars represent sd.
DISCUSSION
In this study, we identified and functionally characterized a novel abiotic stress-related lncRNA from Arabidopsis, DRIR. Our experimental data demonstrate that DRIR positively regulates plant tolerance to drought and salt stress by modulating the expression of genes critical to the stress response.
DRIR Is a Novel lncRNA Regulating Plant Tolerance to Drought and Salt Stress
Plant lncRNAs are expected to play important roles in plant development and the response to environment conditions, and thousands of lncRNAs have been identified in plants (Liu et al., 2012; Li et al., 2014; Shuai et al., 2014; Wang et al., 2014; Zhang et al., 2014b; Chen et al., 2015). However, revealing the functions of plant lncRNA is still challenging. This may not be due only to the fact that most plant lncRNAs are expressed at low levels and their expression may be confined to specific cell types or specific conditions, but also to the fact that plant lncRNAs are not evolutionarily conserved in general (Liu et al., 2012, 2015a, 2015b).
Among the lncRNAs identified so far, some of them are probably responsive to drought and salt stress. By using a deep transcriptome sequencing approach, Qi et al. (2013) identified 584 lncRNAs in foxtail millet. Among them, 17 lincRNAs and two NATs are drought responsive. Chung et al. (2016) also found 98 drought-responsive lncRNAs in rice by using RNA-seq. By genome-wide analysis of full-length cDNA databases, Ben Amor et al. (2009) identified 76 Arabidopsis lncRNAs and found that the accumulation of 22 lncRNAs was altered by abiotic stress. These lncRNAs include three up-regulated and five down-regulated under salt treatment and four up-regulated and two down-regulated under dehydration treatment. Transgenic analyses showed that overexpressing npc536 increases lateral root number under salt treatment. Using RNA-seq, 664 drought-responsive lncRNAs were identified in maize. Among them, 567 lncRNAs were up-regulated and 97 lncRNAs were down-regulated in drought-stressed leaves of maize (Zhang et al., 2014a). Nonetheless, no lncRNA that functions in the plant response to drought or salt stress tolerance has been characterized in detail.
In this study, we identified a lncRNA whose length is 755 nucleotides and does not seem to encode a protein. The transcriptional locus of DRIR is initiated and terminated between two genes, and DRIR should be an intergenic lincRNA. We showed that the drirD activation mutant and DRIR-overexpressing lines are more tolerant to drought and salt stress. Three-week-old drirD and overexpressing seedlings were able to survive 20 d of drought stress treatment, whereas wild-type seedlings were killed by the stress. Leaves of the drirD mutant and overexpressing lines also had slower transpirational water loss rate than the wild type. This is likely due to DRIR’s role in regulating the transpiration rate, since leaf stomata closure of drirD and overexpressing lines was more sensitive to ABA. Furthermore, growth assays either on culture medium or in soil showed that the drirD mutant and overexpressing seedlings are more tolerant to salt stress than the wild type. These results indicate that DRIR regulates the plant response to both drought and salt stress.
DRIR Regulates ABA-Mediated Responses to Drought and Salt Stress
It has been reported that some plant lncRNAs may participate in a phytohormone-mediated response to the environment. The Arabidopsis lincRNA APOLO is transcribed by RNA polymerases Pol II and Pol V in response to auxin from a locus located about 5 kb upstream of PINOID (PID), which is a key regulator of polar auxin transport. The dual transcription of APOLO regulates the formation of a chromatin loop encompassing the promoter of PID and, thus, affects auxin transport (Ariel et al., 2014). Liu et al. (2015b) suggested that lncRNAs may participate in ABA-induced complex assembly and the relocalization of RNA-BINDING PROTEIN. However, lncRNAs involved in the ABA-mediated stress response are still unknown.
In this study, DRIR may regulate ABA-mediated drought and salt stress responses. There appears to exist a positive feed-forward mechanism for DRIR regulation of plant tolerance to drought and salt stress. The expression of DRIR is induced by ABA, and the resulting increased DRIR level further enhances the sensitivity of the plants to ABA. This was shown by increased etiolated leaves in drirD and overexpressing seedlings under ABA treatment as well as by increased sensitivity to ABA in stomata closure in these plants relative to the wild-type plants. At the molecular level, ABA-signaling or ABA-responsive genes were expressed at higher levels in drirD and overexpressing plants. For example, ABI5 is a transcription factor in ABA signaling and has a function in the plant response to abiotic stress (Tanaka et al., 2012; Hopper et al., 2016). P5CS1 is involved in ABA modulation of the stress response in Arabidopsis by controlling Pro accumulation (Abrahám et al., 2003; Sharma et al., 2011). RD29A and RD29B play roles in the plant response to drought or salt stress, and their expression is controlled by ABA (Nakashima et al., 2006). AtrbohB is a member of the NADPH oxidase catalytic subunit genes, and the enzyme catalyzes the synthesis of the second messengers ROS in ABA signaling (Kwak et al., 2003). The expression of these genes in drirD and overexpressing lines is much higher than in the wild type when treated with dehydration. Consistent with the increased gene expression, increased accumulation of Pro and ROS was observed in drirD and overexpressing lines under dehydration stress or ABA treatment. Since ROS are critical second messengers in the ABA regulation of stomata closure (Kwak et al., 2003; Watkins et al., 2014), the increased sensitivity of stomata closure to ABA likely resulted from enhancing ABA-induced ROS accumulation in guard cells. This increased guard cell responsiveness to ABA, rather than alteration in stomatal density, should be responsible for the reduced transpirational water loss seen in the mutant and overexpressing lines (Fig. 3E). Thus, together with increased Pro accumulation and the enhanced expression of other ABA- and stress-responsive genes, DRIR significantly improves plant drought tolerance.
Under salt stress conditions, the expression levels of P5CS1, RD29A, RD29B, and AtrbohB are notably increased in drirD and overexpressing lines as well. These data suggest that DRIR regulates plant tolerance to drought and salt stress at least partly by mediating the expression of those genes involved in ABA signaling and ABA-regulated stress tolerance. The expression levels of other stress-related genes, including FUT4, NIP1, TIP4, ANNAT7, NAC3, and WRKY8, also are increased significantly in drirD and overexpression lines under dehydration stress. FUT4 has been reported to affect plant sensitivity to salt stress by coding a fucosyltransferase that catalyzes the synthesis of fucosylated AGPs in leaves and roots (Tryfona et al., 2014). TIP4 and NIP1, coding for two aquaporin proteins, play important roles in water uptake (Alexandersson et al., 2005; Regon et al., 2014). ANNAT7 encodes a calcium-binding protein annexin and likely has a function in salt and dehydration stress response (Cantero et al., 2006). The NAC3 and WRKY8 transcription factors also function in drought or salt stress response (Nakashima et al., 2012; Hu et al., 2013). The regulation of these many stress-related proteins by DRIR suggests that its mechanism in regulating plant tolerance to drought or salt stress is complex. For example, DRIR could regulate plant tolerance to drought stress by enhancing water transport, increasing the accumulation of stress-relief proteins, and sensitizing the stomata response to ABA. DRIR also could regulate plant tolerance to salt stress by affecting the activity of fucosyltransferase or NAC3 transcription factor or by regulating redox status. The difference between gene expression in dehydration-treated and salt-treated seedlings is likely due to different mechanisms of DRIR in regulating plant tolerance to drought and salt stress. For example, the expression levels of NIP1 and TIP4 were increased to much higher levels in drirD and overexpressing seedlings than in wild-type seedlings when treated with dehydration, but the difference was not as significant when treated with salt stress. This could suggest that aquaporins participate in the DRIR regulation of drought stress tolerance but not in salt stress tolerance.
The lncRNAs may execute their functions through a multitude of mechanisms. Cytoplasm-localized lncRNAs could function as target mimics of miRNAs and, thus, promote its target mRNA translation (Ariel et al., 2015). The lncNATs usually trigger their complementary mRNA degradation by binding with them or promote their translation through recruitment to polysomes (Zubko and Meyer, 2007; Jabnoune et al., 2013). lncRNAs commonly function to regulate target RNA alternative splicing or affect chromatin topology and regulate neighboring gene transcription (Ariel et al., 2014; Bardou et al., 2014). In addition, lncRNAs could function as precursors of miRNAs and other small RNAs (Pant et al., 2008). In this study, in situ hybridization showed that DRIR is localized mainly in the nucleus, suggesting that DRIR may function mainly in nuclear processes such as transcription but may not function as a target mimic of miRNAs, as do other cytoplasm-localized lncRNAs. Furthermore, no known miRNA in Arabidopsis could be found either at the DRIR locus or to match well with the DRIR sequence. Thus, DRIR may not be a precursor of miRNAs. The observation that the expression of many stress-responsive genes was altered in drirD and the overexpressing lines suggests that DRIR may function at or upstream of the stage of gene transcription in the stress or ABA signal transduction pathways. One intriguing observation is that constitutive expression of DRIR did not significantly increase the expression levels of most stress-responsive genes under normal conditions. Rather, the expression of these genes was potentiated significantly by the overexpression of DRIR only under stress conditions (Figs. 7 and 8). As DRIR is transcribed at the chromatin locus between two genes, it is unlikely that DRIR functions by binding directly with cognate RNA and modulating their degradation or promoting their translation, as lncNATs do. However, the molecular mechanisms of how DRIR exerts its functions are still an open question.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants and materials used in this study were in the Columbia-0 ecotype background. Unless stated otherwise, seeds were sterilized and planted on 1/2 MS medium (Sigma-Aldrich) plates containing 0.8% (w/v) agar and 1% (w/v) Suc. Plates were moved to 22°C chambers with a 16-h-light/8-h-dark cycle for germination and growth after stratification at 4°C for 3 d. Ten days later, seedlings were transferred to soil and placed in a growth room at 22°C with a 16-h-light/8-h-dark cycle.
Analysis of DRIR Promoter:GUS Activity
The DRIR promoter fragment (2 kb) was amplified and inserted into the pMDC162 vector using the Gateway cloning technology. After sequence confirmation, the construct was transformed into Arabidopsis using Agrobacterium tumefaciens (strain GV3101). The GUS staining procedure was performed as described previously (Chen et al., 2013). Samples were stained in GUS staining buffer at 37°C overnight followed by decoloring with 70% ethanol and 30% acetic acid. Samples were observed and photographed using a microscope.
RNA in Situ Hybridization
RNA in situ hybridization was performed as described (Gong et al., 2005) with minor modifications. Roots were taken from 7-d-old Columbia-0 seedlings and fixed in a glass vial by adding 10 mL of fixation buffer (120 mm NaCl, 7 mm Na2HPO4, 3 mm NaH2PO4, 2.7 mm KCl, 0.1% [v/v] Tween 20, 80 mm EGTA, 5% [v/v] paraformaldehyde, and 10% [v/v] dimethyl sulfoxide). The samples were shaken gently for 2 h at room temperature. After dehydration twice for 5 min each in absolute methanol and three times for 5 min each in absolute ethanol, the samples were incubated for 30 min in 1:1 ethanol:xylene and then washed twice for 5 min each with absolute ethanol, twice for 5 min each with absolute methanol, and once for 5 min with 1:1 methanol:fixation buffer without 5% (v/v) paraformaldehyde. The samples were postfixed in the fixation buffer for 30 min at room temperature and rinsed twice with fixation buffer without 5% (v/v) paraformaldehyde and once with 1 mL of perfect HybPlus hybridization buffer (Sigma-Aldrich; H-7033). Each glass vial was then added with 1 mL of hybridization buffer and prehybridized in an incubator for 1 h at 50°C. After prehybridization, 5 pmol of probe specific to DRIR or scramble probe was added into the vial and hybridized at 50°C in darkness for more than 8 h. The sequence of probe specific to DRIR is 5′-CTCCAAACTCCTTTATTTCTTAACCAAAAGTTACAATTCATGAGAAGATGATCTAGAACATCATTTCTAGACTCATCTTCTAAATCTCACACACGAGATTGTTTACACAAATTGCATAAAGCTCTCTAAACAATGAGAGTACCTATTTATAACCAAAAAGCAGTAAAAGATAGATGCGGATATTACCTCAGAATATCTTC-3′, and the sequence of scramble probe is 5′-GTGTAACACGTCTATACGCCCAGTGTAACACGTCTATACGCCCAGTGTAACACGTCTATACGCCCAGTGTAACACGTCTATACGCCCAGTGTAACACGTCTATACGCCCAGTGTAACACGTCTATACGCCCAGTGTAACACGTCTATACGCCCAGTGTAACACGTCTATACGCCCAGTGTAACACGTCTATACGCCCAGT-3′. Probes were labeled with or without Alexa Fluor 488 by using the ULYSiS Nucleic Acid Labeling kit (Molecular Probes; U21650). After hybridization, the samples were washed once for 60 min in 2× SSC (1× SSC is 0.15 m NaCl and 0.015 m sodium citrate) and 0.1% SDS at 50°C and once for 20 min in 0.2× SSC and 0.1% (w/v) SDS at 50°C in darkness. The samples were observed immediately using a Zeiss LSM 710 confocal microscope with a 488-nm excitation laser.
Nuclear RNA Extraction
Nuclei of the DRIR-overexpressing line A12 were isolated as described (Zhang and Jiang, 2015) with minor modifications. One gram of 1-week-old A12 seedlings was ground to fine powder in liquid nitrogen. The ground powder was transferred into a 2-mL ice-cold centrifuge tube, and 1 mL of ice-cold NIB buffer (10 mm Tris-HCl, 80 mm KCl, 10 mm EDTA, 1 mm spermidine, 1 mm spermine, 0.15% [v/v] mercaptoethanol, 0.5 m Suc, and 10 µL of RNase inhibitor, pH 9.5) was added to suspend the powder. After being agitated gently on ice for 6 min, the mixture was filtered through a folded four- layer Miracloth into a new 2-mL tube and centrifuged at 1,100g for 10 min at 4°C. After the supernatant was decanted as much as possible, 1 mL of NIB buffer with the addition of 0.5% (v/v) Triton X-100 was added to resuspend the pellet, and the tube was centrifuged subsequently at 1,100g for 10 min at 4°C. The supernatant was discarded, and the above step was repeated three times. The final pellet was used to extract nuclear RNA by using the Plant RNeasy Kit (Qiagen).
Stress Treatment
For the drought tolerance test, 3-week-old well-watered seedlings had watering withheld for 20 d and then were rewatered to allow recovery for 2 d. The plants before and after the treatment were photographed and surveyed as described previously (Li et al., 2016b). To detect the rate of water loss, detached leaves from 4-week-old plants were exposed to air at room temperature and weighed at the indicated times as described previously (Xiong et al., 2001).
For NaCl treatment, seeds were sown onto 1/2 MS medium plates containing different concentrations of NaCl and were stratified at 4°C for 3 d. The plates were then incubated in a growth chamber for germination and grown for 8 d before scoring the phenotypes. Alternatively, 4-d-old seedlings on 1/2 MS plates were transferred to 1/2 MS plates containing different concentrations of NaCl and grown for an additional 4 d before scoring the phenotypes. Seedling survival rate referred to the percentage of seedlings with at least one green leaf among total seedlings treated. For NaCl treatment of soil-grown seedlings, 4-week-old seedlings were watered with 200 mm NaCl and photographs were taken 2 weeks later.
ABA Treatment
For ABA sensitivity analysis, 4-d-old seedlings on 1/2 MS plates were transferred to 1/2 MS plates supplemented with different concentrations of ABA. Eight days later, photographs were taken and green leaves of each seedling were counted. ABA-induced stomata closure assays were performed as described previously (Brandt et al., 2015) with minor modifications. The fifth and sixth rosette leaves of 4-week-old plants were detached and immersed in a stomata open solution (5 mm KCl, 50 µm CaCl2, and 10 mm MES-Tris, pH 5.6) and incubated under cool-white light for 2 h. To induce stomata closure, leaves were transferred to the stomata open solution with 20 µm ABA added. After incubation under cool-white light for 2 h, leaf epidermis peels were prepared and imaged with a BX52M microscope. Apertures were measured using ImageJ software.
Transcriptome Sequencing Analysis
Ten-day-old seedlings were dehydrated on dry filter paper in petri dishes until loss of 40% fresh weight and then incubated for 2 h in sealed plastic bags to prevent further water loss. Total RNA was extracted using the RNeasy Mini Kit (Invitrogen), and DNA was cleaned by DNase I (New England Biolabs). About 2 to 4 µg of cleaned total RNA was used to construct RNA-seq libraries by using the Illumina Whole Transcriptome Analysis Kit following the standard protocol (Illumina HiSeq system) and sequenced on the HiSeq 2000 platform. The gene expression levels (fragments per kilobase of transcript per million mapped reads, FPKM) were calculated with Cufflinks (2.0.2) as described previously (Trapnell et al., 2010; Chen et al., 2013). Two biological replicates were performed.
Real-Time Quantitative PCR
Total RNA was extracted from seedlings by using the Plant RNeasy Kit with DNase I treatment (Qiagen). cDNAs were synthesized from total RNA by using SuperScript III reverse transcriptase (Invitrogen). Real-time quantitative PCR was performed on the ABI 7900HT Fast Real-Time PCR System using 18S rRNA as a control. Primers used in this study are presented in Supplemental Table S3.
Pro and ROS Content
For free Pro measurement, 10-d-old seedlings of the wild type, drirD, A12, and A14 grown on 1/2 MS plates were transferred to empty plates to lose 40% fresh weight or transferred to 1/2 MS solution with 200 mm NaCl for 3 h. Free Pro was assayed using the ninhydrin assay as described (Bates et al., 1973).
ROS content in guard cells was measured as described (Watkins et al., 2014) with minor modifications. Detached leaves from 4-week-old seedlings were immersed in water with or without 20 µm ABA for 1 h. Epidermises were then peeled and stained with 2.5 µm H2DCF-DA (Sigma-Aldrich) for 30 min. After being washed three times with water, guard cells were examined using a Zeiss LSM 710 confocal microscope with excitation at 488 nm and emission at 525 nm.
Accession Numbers
Sequence data described in this article can be found in the Arabidopsis Genome Initiative under accession number At1g21529 (DRIR). RNA-seq data from this article can be found in the Short Read Archive database (National Center for Biotechnology Information) under accession number SRP113651.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression of DRIR was induced by dehydration and salt stress.
Supplemental Figure S2. Localization of DRIR RNA in the nucleus.
Supplemental Figure S3. The DRIR fragment amplified from total RNA or nuclear RNA by RT-PCR.
Supplemental Figure S4. Transcript levels of the DRIR neighboring gene At1g21528 in the wild type and the drirD mutant.
Supplemental Figure S5. drirD and overexpressing lines are more tolerant to salt stress.
Supplemental Figure S6. Expression of DRIR was induced by ABA.
Supplemental Figure S7. Stomata density of rosette leaves of the wild type, drirD, and overexpressing lines.
Supplemental Figure S8. Relative expression levels of selected genes in response to salt stress treatment.
Supplemental Figure S9. Free Pro in dehydration- and salt-treated seedlings.
Supplemental Table S1. Results of transcriptome sequencing analysis.
Supplemental Table S2. List of genes up-regulated or down-regulated more than 2-fold in drirD or overexpressing lines compared with the wild type in transcriptome sequencing analysis.
Supplemental Table S3. Primers used in real-time PCR analysis.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing the T-DNA insertion lines.
Footnotes
Articles can be viewed without a subscription.
References
- Abrahám E, Rigó G, Székely G, Nagy R, Koncz C, Szabados L (2003) Light-dependent induction of proline biosynthesis by abscisic acid and salt stress is inhibited by brassinosteroid in Arabidopsis. Plant Mol Biol 51: 363–372 [DOI] [PubMed] [Google Scholar]
- Agarwal PK, Agarwal P, Reddy MK, Sopory SK (2006) Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep 25: 1263–1274 [DOI] [PubMed] [Google Scholar]
- Alexandersson E, Fraysse L, Sjövall-Larsen S, Gustavsson S, Fellert M, Karlsson M, Johanson U, Kjellbom P (2005) Whole gene family expression and drought stress regulation of aquaporins. Plant Mol Biol 59: 469–484 [DOI] [PubMed] [Google Scholar]
- Arc E, Sechet J, Corbineau F, Rajjou L, Marion-Poll A (2013) ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front Plant Sci 4: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F, Jegu T, Latrasse D, Romero-Barrios N, Christ A, Benhamed M, Crespi M (2014) Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol Cell 55: 383–396 [DOI] [PubMed] [Google Scholar]
- Ariel F, Romero-Barrios N, Jégu T, Benhamed M, Crespi M (2015) Battles and hijacks: noncoding transcription in plants. Trends Plant Sci 20: 362–371 [DOI] [PubMed] [Google Scholar]
- Bardou F, Ariel F, Simpson CG, Romero-Barrios N, Laporte P, Balzergue S, Brown JW, Crespi M (2014) Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev Cell 30: 166–176 [DOI] [PubMed] [Google Scholar]
- Barrero JM, Rodríguez PL, Quesada V, Piqueras P, Ponce MR, Micol JL (2006) Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ 29: 2000–2008 [DOI] [PubMed] [Google Scholar]
- Bates L, Waldren R, Teare I (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39: 205–207 [Google Scholar]
- Ben Amor B, Wirth S, Merchan F, Laporte P, d’Aubenton-Carafa Y, Hirsch J, Maizel A, Mallory A, Lucas A, Deragon JM, et al. (2009) Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res 19: 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt B, Munemasa S, Wang C, Nguyen D, Yong T, Yang P, Poretsky E, Belknap TF, Waadt R, Aleman F, et al. (2015) Calcium specificity signaling mechanisms in abscisic acid signal transduction in Arabidopsis guard cells. eLife 4: e03599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brini F, Yamamoto A, Jlaiel L, Takeda S, Hobo T, Dinh HQ, Hattori T, Masmoudi K, Hanin M (2011) Pleiotropic effects of the wheat dehydrin DHN-5 on stress responses in Arabidopsis. Plant Cell Physiol 52: 676–688 [DOI] [PubMed] [Google Scholar]
- Cantero A, Barthakur S, Bushart TJ, Chou S, Morgan RO, Fernandez MP, Clark GB, Roux SJ (2006) Expression profiling of the Arabidopsis annexin gene family during germination, de-etiolation and abiotic stress. Plant Physiol Biochem 44: 13–24 [DOI] [PubMed] [Google Scholar]
- Chen J, Quan M, Zhang D (2015) Genome-wide identification of novel long non-coding RNAs in Populus tomentosa tension wood, opposite wood and normal wood xylem by RNA-seq. Planta 241: 125–143 [DOI] [PubMed] [Google Scholar]
- Chen T, Cui P, Chen H, Ali S, Zhang S, Xiong L (2013) A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet 9: e1003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Grant JJ, Seki M, Shinozaki K, Loake GJ (2004) Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J 38: 810–822 [DOI] [PubMed] [Google Scholar]
- Chung PJ, Jung H, Jeong DH, Ha SH, Choi YD, Kim JK (2016) Transcriptome profiling of drought responsive noncoding RNAs and their target genes in rice. BMC Genomics 17: 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- de Silva K, Laska B, Brown C, Sederoff HW, Khodakovskaya M (2011) Arabidopsis thaliana calcium-dependent lipid-binding protein (AtCLB): a novel repressor of abiotic stress response. J Exp Bot 62: 2679–2689 [DOI] [PubMed] [Google Scholar]
- Ding F, Cui P, Wang Z, Zhang S, Ali S, Xiong L (2014) Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genomics 15: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Liu N, Virlouvet L, Riethoven JJ, Fromm M, Avramova Z (2013) Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol 13: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS (2009) NRED: a database of long noncoding RNA expression. Nucleic Acids Res 37: D122–D126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2006) Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 103: 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al. (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Dong CH, Lee H, Zhu J, Xiong L, Gong D, Stevenson B, Zhu JK (2005) A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17: 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Koiwa H, Cushman MA, Ray A, Bufford D, Kore-eda S, Matsumoto TK, Zhu J, Cushman JC, Bressan RA, et al. (2001) Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol 126: 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin A, Rodrigues O, Verdoucq L, Merlot S, Leonhardt N, Maurel C (2015) Aquaporins contribute to ABA-triggered stomatal closure through OST1-mediated phosphorylation. Plant Cell 27: 1945–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper DW, Ghan R, Schlauch KA, Cramer GR (2016) Transcriptomic network analyses of leaf dehydration responses identify highly connected ABA and ethylene signaling hubs in three grapevine species differing in drought tolerance. BMC Plant Biol 16: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Chen L, Wang H, Zhang L, Wang F, Yu D (2013) Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J 74: 730–745 [DOI] [PubMed] [Google Scholar]
- Hwang I, Goodman HM (1995) An Arabidopsis thaliana root-specific kinase homolog is induced by dehydration, ABA, and NaCl. Plant J 8: 37–43 [DOI] [PubMed] [Google Scholar]
- Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu Q, Poirier Y (2013) A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 25: 4166–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HJ, Kim MK, Kang H (2013) An ABA-regulated putative RNA-binding protein affects seed germination of Arabidopsis under ABA or abiotic stress conditions. J Plant Physiol 170: 179–184 [DOI] [PubMed] [Google Scholar]
- Kato M, Nagasaki-Takeuchi N, Ide Y, Maeshima M (2010) An Arabidopsis hydrophilic Ca2+-binding protein with a PEVK-rich domain, PCaP2, is associated with the plasma membrane and interacts with calmodulin and phosphatidylinositol phosphates. Plant Cell Physiol 51: 366–379 [DOI] [PubMed] [Google Scholar]
- Kumar D, Datta R, Hazra S, Sultana A, Mukhopadhyay R, Chattopadhyay S (2015) Transcriptomic profiling of Arabidopsis thaliana mutant pad2.1 in response to combined cold and osmotic stress. PLoS ONE 10: e0122690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JDG, Schroeder JI (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Byrt C, Qiu J, Baumann U, Hrmova M, Evrard A, Johnson AA, Birnbaum KD, Mayo GM, Jha D, et al. (2016a) Identification of a stelar-localised transport protein that facilitates root-to-shoot transfer of chloride in Arabidopsis. Plant Physiol 170: 1014–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Eichten SR, Shimizu R, Petsch K, Yeh CT, Wu W, Chettoor AM, Givan SA, Cole RA, Fowler JE, et al. (2014) Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol 15: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Wu XY, Li H, Song JH, Liu JY (2016b) A dual-function transcription factor, AtYY1, is a novel negative regulator of the Arabidopsis ABA response network. Mol Plant 9: 650–661 [DOI] [PubMed] [Google Scholar]
- Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20: 2238–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH (2012) Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24: 4333–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang H, Chua NH (2015b) Long noncoding RNA transcriptome of plants. Plant Biotechnol J 13: 319–328 [DOI] [PubMed] [Google Scholar]
- Liu X, Hao L, Li D, Zhu L, Hu S (2015a) Long non-coding RNAs and their biological roles in plants. Genomics Proteomics Bioinformatics 13: 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Bohnert HJ (2007) Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol 8: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Medina J, Ballesteros ML, Salinas J (2007) Phylogenetic and functional analysis of Arabidopsis RCI2 genes. J Exp Bot 58: 4333–4346 [DOI] [PubMed] [Google Scholar]
- Munemasa S, Hauser F, Park J, Waadt R, Brandt B, Schroeder JI (2015) Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr Opin Plant Biol 28: 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol Biol 60: 51–68 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819: 97–103 [DOI] [PubMed] [Google Scholar]
- Nambara E, Keith K, McCourt P, Naito S (1995) A regulatory role for the Abi3 gene in the establishment of embryo maturation in Arabidopsis-thaliana. Development 121: 629–636 [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Xie S, Liu Y, Yi F, Yu J (2013) Genome-wide annotation of genes and noncoding RNAs of foxtail millet in response to simulated drought stress by deep sequencing. Plant Mol Biol 83: 459–473 [DOI] [PubMed] [Google Scholar]
- Regon P, Panda P, Kshetrimayum E, Panda SK (2014) Genome-wide comparative analysis of tonoplast intrinsic protein (TIP) genes in plants. Funct Integr Genomics 14: 617–629 [DOI] [PubMed] [Google Scholar]
- Sasaki K, Kim MH, Kanno Y, Seo M, Kamiya Y, Imai R (2015) Arabidopsis cold shock domain protein 2 influences ABA accumulation in seed and negatively regulates germination. Biochem Biophys Res Commun 456: 380–384 [DOI] [PubMed] [Google Scholar]
- Sharma S, Villamor JG, Verslues PE (2011) Essential role of tissue-specific proline synthesis and catabolism in growth and redox balance at low water potential. Plant Physiol 157: 292–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3: 217–223 [PubMed] [Google Scholar]
- Shuai P, Liang D, Tang S, Zhang Z, Ye CY, Su Y, Xia X, Yin W (2014) Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J Exp Bot 65: 4975–4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C, Gu D, Hu HC, Davanture M, Lee S, Djaoui M, Valot B, Zivy M, Leung J, Merlot S, et al. (2009) Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett 583: 2982–2986 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Osakabe Y, Katsura S, Mizuno S, Maruyama K, Kusakabe K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J 70: 599–613 [DOI] [PubMed] [Google Scholar]
- Thomashow MF. (2010) Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol 154: 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryfona T, Theys TE, Wagner T, Stott K, Keegstra K, Dupree P (2014) Characterisation of FUT4 and FUT6 α-(1 → 2)-fucosyltransferases reveals that absence of root arabinogalactan fucosylation increases Arabidopsis root growth salt sensitivity. PLoS ONE 9: e93291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Hitomi K, Nishimura N, Hitomi C, Adams SR, Getzoff ED, Schroeder JI (2014) FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. eLife 3: e01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chung PJ, Liu J, Jang IC, Kean MJ, Xu J, Chua NH (2014) Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res 24: 444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li X (2008) Salt stress-induced cell reprogramming, cell fate switch and adaptive plasticity during root hair development in Arabidopsis. Plant Signal Behav 3: 436–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins JM, Hechler PJ, Muday GK (2014) Ethylene-induced flavonol accumulation in guard cells suppresses reactive oxygen species and moderates stomatal aperture. Plant Physiol 164: 1707–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich M, Gross-Hardt R, Schoffl F (2014) Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol Biol 85: 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Wang A, Yang G, Gao P, Zheng ZL (2009) The Arabidopsis A4 subfamily of lectin receptor kinases negatively regulates abscisic acid response in seed germination. Plant Physiol 149: 434–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu JK (2001) The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13: 2063–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhu JK (2003) Regulation of abscisic acid biosynthesis. Plant Physiol 133: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yang KY, Kim EY, Kim CS, Guh JO, Kim KC, Cho BH (1998) Characterization of a glutathione S-transferase gene ATGST 1 in Arabidopsis thaliana. Plant Cell Rep 17: 700–704 [DOI] [PubMed] [Google Scholar]
- Zhang W, Han Z, Guo Q, Liu Y, Zheng Y, Wu F, Jin W (2014a) Identification of maize long non-coding RNAs responsive to drought stress. PLoS ONE 9: e98958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Jiang J (2015) Genome-wide mapping of DNase I hypersensitive sites in plants. Methods Mol Biol 1284: 71–89 [DOI] [PubMed] [Google Scholar]
- Zhang YC, Liao JY, Li ZY, Yu Y, Zhang JP, Li QF, Qu LH, Shu WS, Chen YQ (2014b) Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol 15: 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2016) Abiotic stress signaling and responses in plants. Cell 167: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko E, Meyer P (2007) A natural antisense transcript of the Petunia hybrida Sho gene suggests a role for an antisense mechanism in cytokinin regulation. Plant J 52: 1131–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]