Determining exact epitopes for cell wall-directed monoclonal antibodies provides the basis for a detailed elucidation of polysaccharide structures at the cellular level.
Abstract
In the last three decades, more than 200 monoclonal antibodies have been raised against most classes of plant cell wall polysaccharides by different laboratories worldwide. These antibodies are widely used to identify differences in plant cell wall components in mutants, organ and tissue types, and developmental stages. Despite their importance and broad use, the precise binding epitope has been determined for only a few of these antibodies. Here, we use a plant glycan microarray equipped with 88 synthetic oligosaccharides to comprehensively map the epitopes of plant cell wall glycan-directed antibodies. Our results reveal the binding epitopes for 78 arabinogalactan-, rhamnogalacturonan-, xylan-, and xyloglucan-directed antibodies. We demonstrate that, with knowledge of the exact epitopes recognized by individual antibodies, specific glycosyl hydrolases can be implemented into immunological cell wall analyses, providing a framework to obtain structural information on plant cell wall glycans with unprecedented molecular precision.
Plant cell walls are highly complex sophisticated composites largely composed of polysaccharide networks with essential functions in the life cycle of plants. Plant physiology, growth, and development all depend on the structure and dynamics of the cell wall (Keegstra, 2010). Moreover, cell wall polysaccharides receive enormous interest as sources of renewable materials and for the production of biofuels (Pauly and Keegstra, 2010). To enhance the economic viability of biomass as a renewable resource, an increasing number of genetically engineered plants with modified cell wall compositions have been generated (Loqué et al., 2015). However, a prerequisite to performing targeted genetic modifications is a detailed knowledge of plant cell wall structure and its biosynthesis.
To characterize these very diverse plant cell wall components and the genes responsible for their synthesis, biochemical tools are required that can identify molecular structures in the cell wall with high precision. Glycosidic linkage analyses of cell wall extracts can provide quantitative information on the abundance of monosaccharides and their linkage types (Pettolino et al., 2012). This information can then be used to derive the occurrence and structures of different polysaccharide classes. However, this method can only be applied to whole organs; thus, it remains unclear which tissue and cell types contain the identified polysaccharides. To obtain high spatial resolution, a large number of monoclonal antibodies (mAbs) that bind distinct classes of cell wall polysaccharides have been developed (Meikle et al., 1994; Willats et al., 1998; Classen et al., 2004; McCartney et al., 2005; Pattathil et al., 2010; Ralet et al., 2010). These mAbs are widely used to localize polysaccharides in cells and tissues of various plant species (Classen et al., 2004; Guillon et al., 2004; da Costa et al., 2017) and to characterize mutant plants with alterations in cell wall composition (Gendre et al., 2013; Pacheco-Villalobos et al., 2016). Yet, the limited information on the precise molecular structures bound by the mAbs has hindered a comprehensive interpretation of immunological cell wall analyses.
Due to the heterogeneity and diversity of glycoforms within polysaccharide classes, the small number of well-characterized mAbs have the precision with which these molecular probes can be used to infer polysaccharide occurrence or abundance. The only viable option to precisely determine the binding epitopes of large numbers of the existing mAbs is to screen their binding capabilities with structurally well-defined oligosaccharides. Previous studies with cell wall-related oligosaccharides have demonstrated that glycan microarrays (Rillahan and Paulson, 2011) can be used to determine the binding epitopes of cell wall glycan-directed mAbs (Pedersen et al., 2012; Schmidt et al., 2015). Synthetic chemistry is ideally suited to procure these well-defined oligosaccharides, as it provides de novo-designed oligosaccharides of exceptional purity. Herein, we report the production of a microarray equipped with 88 synthetic oligosaccharides that enabled us to determine the binding epitopes of 78 plant cell wall glycan-directed mAbs. We further show that glycosyl hydrolases can be used on plant sections to specifically modify cell wall polysaccharides, generating new glycan epitopes that are detectable by specific mAbs. Integrating glycosyl hydrolases in immunological cell wall analyses thus provides additional structural information on plant cell wall polysaccharides.
RESULTS AND DISCUSSION
Generation of a Synthetic Plant Glycan Microarray and Determination of mAb Epitopes
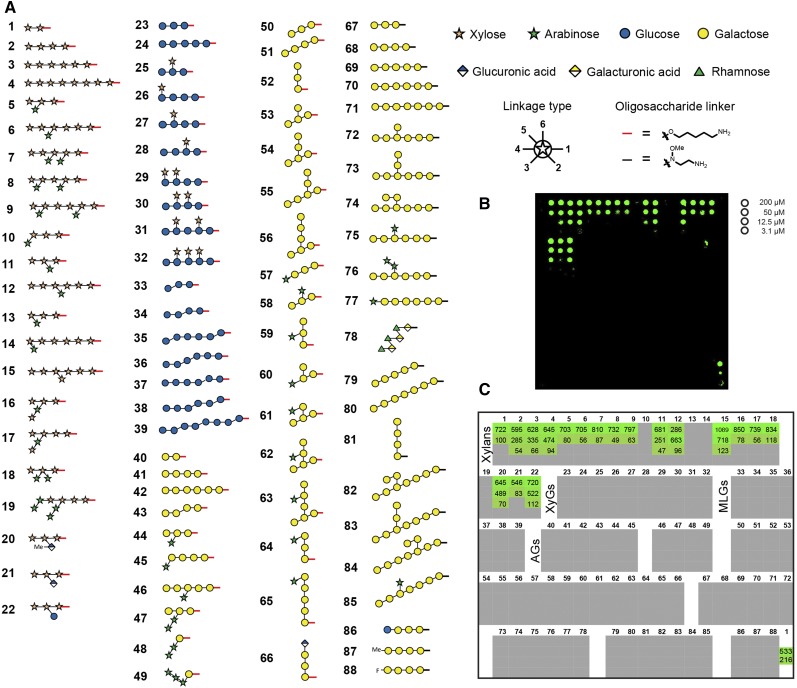
We have chemically synthesized a library of plant cell wall-derived oligosaccharides either using automated glycan assembly (compounds 1–66; Fig. 1A; Bartetzko et al., 2015, 2017; Schmidt et al., 2015; Dallabernardina et al., 2016, 2017; Senf et al., 2017) or by conventional solution-phase chemistry (compounds 67–88; Fig. 1A; Zakharova et al., 2013; Andersen and Clausen, 2014; Andersen et al., 2016a, 2016b). These include the following oligosaccharides: xylan oligosaccharides (compounds 1–22) composed of a β-1,4-linked xylan backbone with arabinofuranose substitutions in the 2- and/or 3-position of the Xyl residues or with GlcA substitutions in the 2-position; glucan oligosaccharides (compounds 23–39) with a β-1,4-linked glucan backbone that can be substituted with α-1,6-linked Xyl residues (xyloglucan; compounds 25–32) or interspersed with β-1,3-linkages (mixed-linkage glucans; compounds 33–39); pectin-derived oligosaccharides including β-1,4-linked type I (arabino)galactans (compounds 40–49, 67–77, and 86–88) and an RG-I backbone oligosaccharide (compound 78); and type II (arabino)galactan oligosaccharides composed of a β-1,3-linked backbone branched with β-1,6-linked galactan side chains that can be further substituted with α-1,3-linked arabinofuranoses or a terminal β-1,6-linked GlcA (compounds 50–66 and 79–85).
Figure 1.
A glycan microarray equipped with synthetic cell wall oligosaccharides. A, The printed oligosaccharides comprise fragments of four major polysaccharide classes: xylans (compounds 1–22), glucans (23–39), galactans (40–77 and 79–88), and rhamnogalacturonan (RG)-I (78). Red and black bars at the reducing end of the oligosaccharides indicate the different linkers of the respective compounds produced either by automated glycan assembly (1–66) or conventional solution-phase chemistry (67–88). The legend for linkage types denotes at which position the next monosaccharide is attached. B, Fluorescence signal for the binding of LM10 to xylan oligosaccharides. Each compound was printed at four different concentrations as indicated on the right. The printing pattern of the glycan microarray is depicted in C. C, Quantification of the fluorescence signal for LM10. The values denote fold change over background. Only signals of more than 4-fold above background and above 4% of the maximal value are shown. Note that the 200 and 50 µm concentrations of oligosaccharide 1 were reprinted on the array as the last spots (bottom right corner) to confirm constant printing efficiency.
The synthetic oligosaccharides were printed at four different concentrations on chemically activated glass slides using a noncontact piezoelectric microarray printer (Fig. 1). This glycan array platform was used to systematically map the epitopes of 209 plant cell wall glycan-directed mAbs that were obtained from different laboratories. The binding strength of the mAbs to the printed oligosaccharides correlates with the fluorescence intensity observed for an individual interaction (Fig. 1B; Supplemental Data File S1). To define a positive signal, we required at least a 4-fold increase of signal intensity over background and a value of at least 4% of the maximal value for a particular antibody. The quantification of the binding strength of the xylan-directed mAb LM10 for the four different glycan printing concentrations is depicted in Figure 1C.
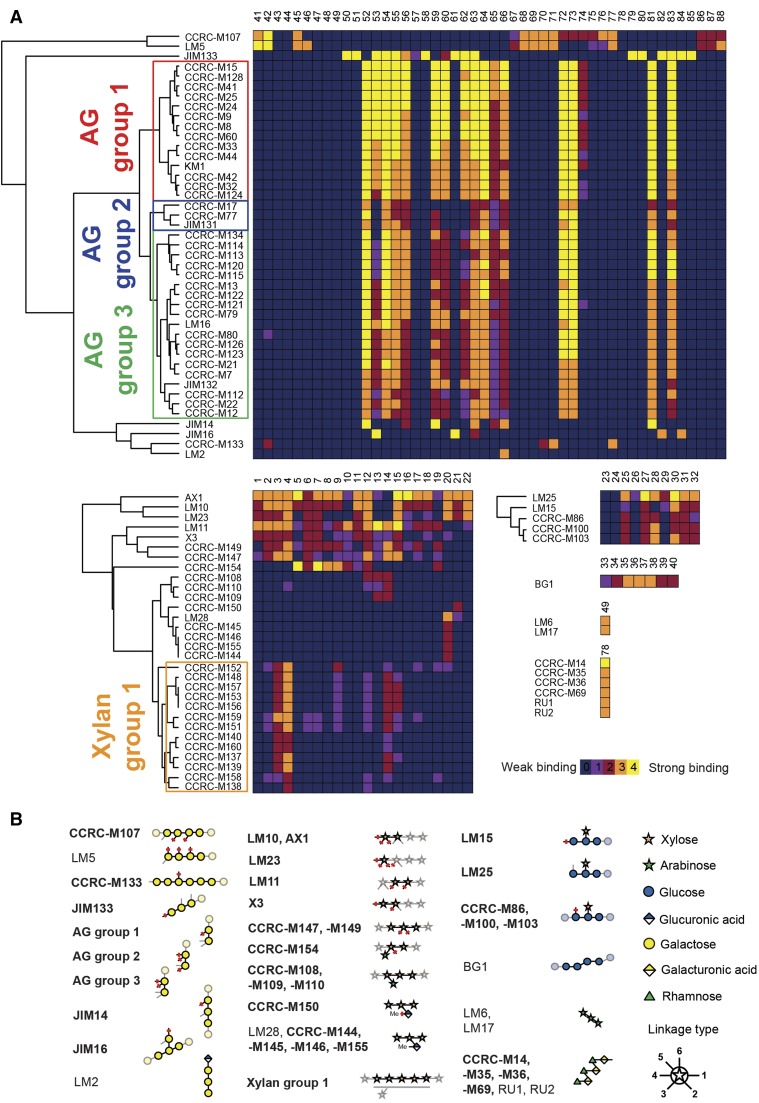
We created a summarizing heat map of the binding capabilities of individual mAbs and the 88 different synthetic glycans (Fig. 2A). Based on the binding patterns of the mAbs, we aimed to identify a common motif among the recognized oligosaccharides that represents the actual epitope. Several antibodies whose epitopes had been characterized in detail previously using defined oligosaccharides (Puhlmann et al., 1994; Steffan et al., 1995; Willats et al., 1998; Andersen et al., 2016a) were included in our analysis. We were able to confirm the specificity of these mAbs: for example, the binding of LM2 to GlcA that is terminally attached to 1,6-linked galactan (Smallwood et al., 1996) and of LM6 and LM17 to 1,5-linked arabinan (Verhertbruggen et al., 2009; Fig. 2B). Using the binding patterns of previously uncharacterized antibodies, we identified the binding epitopes for an additional 78 mAbs (Fig. 2B). To resolve similarities in the binding specificities of the mAbs, we used hierarchical clustering on the data obtained with arabinogalactan (AG)-, xylan-, and xyloglucan-binding antibodies.
Figure 2.
Identification of plant cell wall mAb epitopes. A, Heat maps of the binding interactions between individual mAbs and respective synthetic glycans. The binding strength of an antibody to a compound is visualized by a color code (0–4), which denotes how many of the four printed glycan concentrations displayed a positive fluorescence signal. Antibodies were grouped based on hierarchical clustering. A representative result of three replicates is shown. The full heat map is shown in Supplemental Figure S1. B, Identified epitopes of cell wall-directed antibodies. Linkages that are marked with red bars indicate positions that must not be occupied. Light linkages and light monosaccharide symbols indicate positions for substitutions that are allowed but not required for binding. For antibodies depicted in boldface, no or very limited epitope information was available previously. Note that mAbs of xylan group 1 tolerate different degrees of low-level substitution of the xylan backbone. The mAb from Classen et al. (2004) was named KM1.
AG-Directed Antibodies
The hierarchical clustering identified three large groups of AG-binding antibodies among the antibodies tested. Based on the oligosaccharides bound, we deduced that these three groups of antibodies bind to β-1,6-linked galactans that can be present in type I or type II (arabino)galactan. None of these antibodies bind exclusively to β-1,3-linked or β-1,4-linked galactans (Fig. 2; Supplemental Fig. S2, A–C). This observation is consistent with previous ELISA experiments that revealed binding of these mAbs to diverse classes of polysaccharides, indicating the existence of these β-1,6-linked galactan epitopes in RG-I, type II AG, and other pectic polysaccharide preparations from various plant sources (Pattathil et al., 2010).
While the binding pattern of these mAbs for the tested AG oligosaccharides is largely similar, a major difference between these groups is their binding properties to oligosaccharides 53, 60, and 62. AG group 1 antibodies bind strongly, AG group 3 antibodies bind weakly, and AG group 2 antibodies do not bind to these oligosaccharides at all (Fig. 2A; Supplemental Fig. S2, A–C). In these three oligosaccharides, the lower Gal of a β-1,6-linked galactan disaccharide epitope is substituted in the 3-position either with Gal or with arabinose. The antibody groups may be distinguished further based on their binding to oligosaccharide 74. The AG group 1 antibodies bind to oligosaccharide 74, while AG groups 2 and 3 do not. In oligosaccharide 74, the upper Gal residue of a β-1,6-linked galactan disaccharide epitope is substituted at the C4-position. Based on the oligosaccharides tested, we conclude that AG group 2 antibodies bind to a minimal epitope of a β-1,6-linked disaccharide and the lower Gal may be part of a β-1,4-linked galactan backbone. The minimal epitope for the AG group 1 and 3 antibodies cannot be delineated completely with the oligosaccharides tested, although it appears that AG group 1 and 3 antibodies bind to a β-1,6-linked disaccharide of which the lower Gal can be part of both β-1,3-linked and β-1,4-linked galactan backbones (as in oligosaccharides 53 and 72). Unlike AG group 3 antibodies, AG group 1 antibodies tolerate a substitution at the 4-position on the upper Gal.
More differentiated binding patterns to AGs were observed for antibodies JIM14 and JIM16. The binding pattern JIM14 revealed that at least three consecutive β-1,6-linked Gal units are required for binding (Fig. 2A; Supplemental Fig. S2D). Arabinose substitution at the central Gal unit did not affect binding, indicating that also arabinosylated galactan polysaccharides can be recognized by JIM14. However, lack of binding to compound 64 showed that the third Gal toward the nonreducing terminus must not be arabinosylated. To investigate if JIM14 binds an internal epitope or the nonreducing end of β-1,6-galactans, we synthesized two glycans that do not permit binding to the terminal Gal unit (compounds 65 and 66). Based on JIM14 binding to compounds 65 and 66, we conclude that JIM14 binds an internal epitope on β-1,6-linked galactans. JIM16, on the other hand, recognizes a β-1,3-linked galactan backbone when substituted with a single β-1,6-linked Gal residue (Fig. 2B), as this antibody bound strongly only to compounds 53 and 61 (Fig. 2A).
We identified an antibody that specifically recognizes the β-1,3-linked galactan backbone in AGs. JIM133 selectively binds all tested β-1,3-linked galactooligosaccharides, with tolerance for various β-1,6-linked arabinose and Gal substitutions. The strongly reduced binding of JIM133 to compound 57 compared with compound 50 indicates that this antibody requires the free nonreducing end of the galactan for binding. Thus, JIM133 can be used to detect the nonreducing ends of the β-1,3-linked galactan backbone in AG structures of arabinogalactan proteins (AGPs).
We identified two antibodies in addition to the previously characterized LM5 antibody (Jones et al., 1997; Andersen et al., 2016a) that recognize β-1,4-linked type I galactan, a prominent side chain in RG-I. CCRC-M107 requires a minimum degree of polymerization (DP) = 4 (Supplemental Fig. S2E; compounds 41, 42, and 68–71). In contrast, CCRC-M133 only binds to β-1,4-linked galactan with DP = 6 or greater (Supplemental Fig. S2F; compounds 42, 70, and 71). Their strong binding to oligosaccharide 77 indicates that CCRC-M107 and CCRC-M133 bind to internal parts of type I galactan. Our results further show that CCRC-M107 tolerates limited arabinose substitution, whereas CCRC-M133 does not bind to any substituted β-1,4-linked galactooligosaccharides tested here.
Xylan-Directed Antibodies
We identified xylan-directed mAbs that cover most epitopes found in natural xylans. Antibodies in xylan group 1 recognize low-substituted xylans, tolerating to varying degrees different patterns of limited arabinose substitution (Fig. 2). These mAbs bind strongly to oligosaccharides 3 and 4, indicating that longer stretches of β-1,4 xylan backbone represent the preferred epitope for these antibodies. Weak binding to substituted xylan oligosaccharides also was observed (e.g. oligosaccharides 9, 12, and 15). However, the available oligosaccharides do not permit an exhaustive determination of the tolerated substitution patterns. The xylan group 1 antibodies with the least tolerance for arabinose substitution of the backbone are CCRC-M140, CCRC-M160, CCRC-M137, and CCRC-M139. CCRC-M152 stands out from the xylan 1 group by the observation that this antibody binds more strongly to oligosaccharide 9 than to oligosaccharide 14, which is different from the binding pattern of the remaining xylan group 1 antibodies. Contrary to these xylan group 1 mAbs, we found LM11, CCRC-M147, and CCRC-M149 to bind the β-1,4-linked xylan backbone with a high tolerance for backbone substitution. With slight differences in the binding patterns, most arabinose-substituted xylan oligosaccharides were recognized.
In addition to these mAbs that recognize the xylan backbone, we also identified several mAbs that specifically detect distinct substituents present on xylan polymers and do not bind to unsubstituted xylan oligosaccharides (compounds 1–4). For example, we found that CCRC-M154 selectively binds to xylan oligosaccharides with an arabinose substitution in the 3-position. However, compound 18 with two arabinose substituents linked to the same Xyl residue was not recognized. On the other hand, we found that CCRC-M108, CCRC-M109, and CCRC-M110 bind specifically to xylan oligosaccharides with arabinose in the 2-position (Fig. 2). Interestingly, CCRC-M108, CCRC-M109, and CCRC-M110, specific for 2-substituted arabinoxylans, all were raised against Phormium tenax (New Zealand flax) and had been shown to bind exclusively to isolated P. tenax xylan and not to xylans from other plants in ELISA experiments carried out with polysaccharide ligands (Pattathil et al., 2010). Thus, 2-substituted arabinoxylans might be specific to certain plants, although monosaccharide linkage analyses previously suggested a broader occurrence of single arabinose substitution in the 2-position of xylans (Izydorczyk and Biliaderis, 1995). Furthermore, we confirm the binding of LM28 to xylan oligosaccharides substituted with GlcA in the 2-position (Cornuault et al., 2015). While LM28 binds both methylated and unmethylated GlcA side chains, we here identify mAbs that bind exclusively to either unmethylated GlcA (CCRC-M150, a correction of previously published data [Schmidt et al., 2015]) or 4-O-methyl GlcA (CCRC-M144, CCRC-M145, CCRC-M146, and CCRC-M155). None of these antibodies bind to the Glc-substituted compound 22, demonstrating that the carboxyl group is essential for the binding of these mAbs. Since our collection did not contain esterified xylan oligosaccharides, we were not able to detect antibodies that recognize these epitopes. While LM12 binds to ferulated xylans (Pedersen et al., 2012), antibodies specifically recognizing acetylated xylan have yet to be developed.
The most unexpected result was obtained for LM10, as this frequently used antibody was previously thought to bind low or unsubstituted xylan (McCartney et al., 2005). The glycan microarray experiments show that LM10 binds to all xylan oligosaccharides tested except compounds 10, 13, 14, and 19 (Fig. 1, B and C). These compounds are all substituted with arabinose either at the 2- or 3-position of the terminal Xyl residue. While a lack of binding to compounds 10, 13, and 19 could still be explained by their relatively high arabinose content, the absence of binding to compound 14, which was designed specifically to characterize this antibody, unequivocally established that LM10 binds to the nonreducing end of xylans. Similar results were obtained for AX1, X3, and LM23, which also bind to terminal Xyl residues but tolerate slightly different substitution patterns.
Intriguingly, all antibodies that bind the nonreducing end were raised against short oligosaccharides as immunogens, except for JIM133. All mAbs that were raised against polysaccharide antigens (72 of 78 mAbs) recognized an internal epitope within the polymer. These observations have important implications for the choice of oligosaccharide antigens to be used for the generation of new cell wall-directed antibodies. Instead of internal sequences of complex polymers such as RG-II, terminal oligosaccharides should be used as immunogens.
Xyloglucan-Directed Antibodies
Many mAbs have been raised against xyloglucan using either oligosaccharides (e.g. LM15 and LM25; Marcus et al., 2008; Pedersen et al., 2012) or polysaccharides (e.g. CCRC-M86, CCRC-M100, and CCRC-M103) as antigens (Pattathil et al., 2010). The majority of these mAbs did not bind to any of our compounds (Supplemental Data File S1), probably because these antibodies require Gal or Fuc substitutions for binding, which are not included in our xyloglucan oligosaccharide library. Here, we could determine the epitopes for five xyloglucan antibodies. These xyloglucan-directed antibodies recognize a xyloglucan epitope with at least one α-1,6-linked Xyl residue linked to a β-1,4-linked glucan backbone (no binding to unsubstituted β-1,4-linked glucan oligosaccharides 23 and 24 was observed; Fig. 2). While we could not derive whether LM25 binds to terminal or internal parts of xyloglucan, this distinction was possible for LM15 and CCRC-M103 (similarly, CCRC-M86 and CCRC-M100). We observed similar binding patterns for these antibodies except for the fact that LM15 does not bind to compound 28, whereas CCRC-M103 does not bind to compound 29 (Fig. 2A; Supplemental Fig. S2, G and H). Compound 28 is an elongated version of compound 25 with one additional Glc unit at the nonreducing end. This additional Glc abolished the binding of LM15, suggesting that LM15 only tolerates a single unsubstituted backbone glucosyl residue at the nonreducing end of the oligosaccharide. Based on the importance of the nonreducing end of the oligosaccharides (25 versus 28) for binding, we conclude that LM15 is directed toward the nonreducing end, with the requirement for the second to last Glc to be substituted with an α-1,6-linked Xyl. Contrary to LM15, CCRC-M103 binds more strongly to compound 28 compared with compound 25, suggesting that CCRC-M103 probably binds to internal parts of xyloglucan. The binding of CCRC-M103 to compounds 25, 27, 28, and 30 to 32 and the lack of binding to compounds 26 and 29 further suggest that CCRC-M103 requires a free Glc at the position next to the Xyl substituent toward the nonreducing end.
RG-I-Directed Antibodies
Six antibodies directed at the backbone of RG-I were detected (Fig. 2). For antibodies CCRC-M14, CCRC-M35, CCRC-M36, and CCRC-M69, indirect evidence suggested that they bind the backbone of RG-I (Pattathil et al., 2010), but definitive proof was missing. In addition, we could confirm the previously published binding epitope for INRA-RU1 and INRA-RU2, which was determined using purified RG oligosaccharides in competitive ELISA experiments (Ralet et al., 2010).
Glycosyl Hydrolase Digests on the Glycan Microarray
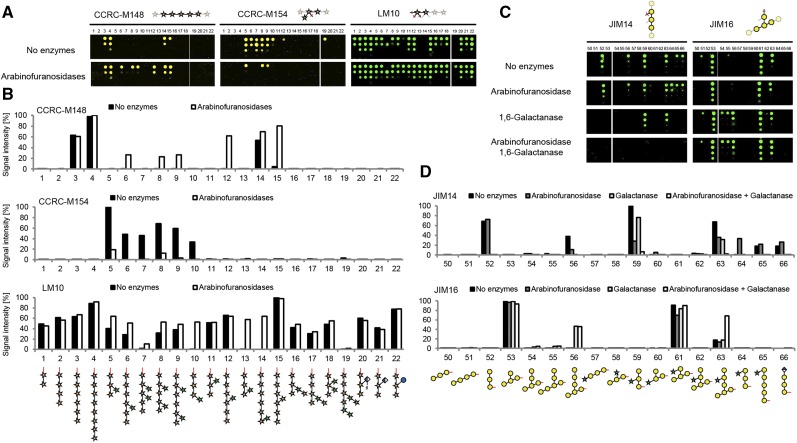
Next, we investigated if the immobilized glycans can be modified on the array using carbohydrate-active enzymes. Incubation of the microarray slides with arabinofuranosidases acting on arabinoxylan results in dearabinosylation of the xylan oligosaccharides, as detected with the xylan-directed mAbs LM10, CCRC-M154, and the xylan group 1 antibody CCRC-M148 (Fig. 3, A and B). As expected, CCRC-M148 now recognized oligosaccharides that were previously too heavily arabinosylated to permit binding of this antibody. For example, oligosaccharide 8 was bound after dearabinosylation (Fig. 3, A and B), suggesting that CCRC-M148 binds to unsubstituted xylan backbones of DP = 5 or greater, a refinement of previous characterization data for this antibody that indicated CCRC-M148 binding to xylan oligosaccharides of DP = 6 or greater (Schmidt et al., 2015). The binding of CCRC-M154, which is selective for 3-substituted arabinofuranoses, was strongly reduced after arabinofuranosidase treatment. Restoration of LM10 binding to oligosaccharides with an arabinose attached to the nonreducing Xyl unit (compounds 10, 13, and 14) upon dearabinosylation further confirmed the proposed binding mode of LM10 directed at the nonreducing end of xylans.
Figure 3.
The glycan microarray as a platform to characterize glycosyl hydrolases. A, On-array treatment of xylan oligosaccharides with arabinofuranosidases and assessment of arabinose cleavage with CCRC-M148, CCRC-M154, and LM10. Note that the 200 µm concentration for oligosaccharide 7 was misprinted, resulting in a low signal intensity after quantification (see LM10). B, Quantification of the fluorescence signal with and without arabinofuranosidase treatment as shown in A. C, AG digest on the array with 1,6-galactanase and arabinofuranosidase detected with JIM14 and JIM16. D, Quantification of the fluorescence signals in C. Representative results of three independent experiments are shown.
As a second example, we used a β-1,6-endogalactanase (Kotake et al., 2004; Kitazawa et al., 2013) to trim the side chains of synthetic AG oligosaccharides. This endogalactanase was reported to hydrolyze β-1,6-linked galactan side chains of AGP polysaccharides until one or two Gal residues remain on the β-1,3-linked galactan backbone (Kitazawa et al., 2013). Binding of JIM14, which recognizes glycans with at least three consecutive β-1,6-linked Gal residues (Fig. 2B), was abolished upon treatment with β-1,6-endogalactanase for all synthetic galactan oligosaccharides that were not substituted with arabinose at the central Gal unit (oligosaccharides 52, 56, 65, and 66) but not for the oligosaccharides that were arabinosylated (oligosaccharides 59 and 63; Fig. 3, C and D). To remove the arabinoses from oligosaccharides 59 and 63, we used an arabinofuranosidase and unexpectedly found slightly reduced binding of JIM14 to oligosaccharides 56, 59, and 63, probably due to traces of galactanase activity in the enzyme preparation. However, when both arabinofuranosidase and galactanase were applied together on the glycan microarray, the recognition of oligosaccharides 59 and 63 was completely abolished (Fig. 3, C and D). This indicated that the galactanase only tolerates arabinose substitution in certain positions of the galactan to be hydrolyzed. JIM16 recognizes the 1,3-linked galactan backbone when substituted with a single 1,6-linked Gal residue (Fig. 2B). Binding of JIM16 to oligosaccharide 56 after galactanase digestion and oligosaccharide 63 after arabinofuranosidase and galactanase digestion confirmed that a single 1,6-linked Gal unit remained on the 1,3-linked backbone of these oligosaccharides (Fig. 3D). Although exact substrate specificities of glycosyl hydrolases acting on cell wall glycans can only be determined by structural characterization of the reaction products after incubation of oligosaccharides or polysaccharides, these data demonstrate that the synthetic plant glycan microarray platform provides a useful tool, in combination with well-characterized mAbs, to collect valuable information on the substrate specificities of glycosyl hydrolases in a high-throughput fashion. As our platform provides specific information on the enzyme’s substrate specificities, it represents a powerful extension of previous polysaccharide-based array platforms for the identification of new glycosyl hydrolases (Vidal-Melgosa et al., 2015; Walker et al., 2017).
Implementation of Glycosyl Hydrolases into Cell Wall Labeling Studies
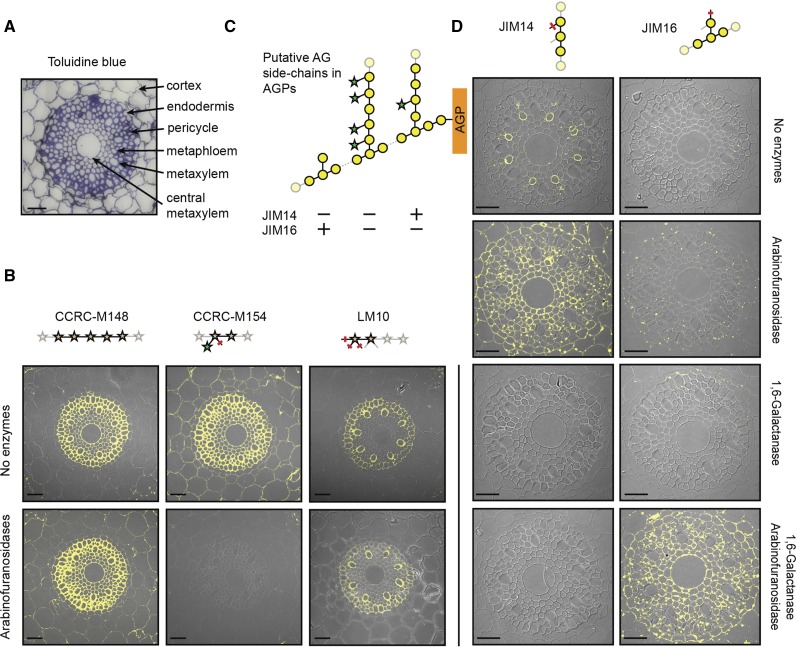
Specific knowledge of the molecular structure bound by a particular mAb and the substrate specificities of glycosyl hydrolases acting on the very same polysaccharide allows for a detailed immunological elucidation of glycan structures in the cell wall. We performed antibody staining of xylans and AGs in roots of Brachypodium distachyon (Brachypodium), which is a model system in which to study grass roots (Hardtke and Pacheco-Villalobos, 2015). We used CCRC-M148 to detect unsubstituted xylan stretches, CCRC-M154 to identify arabinosylated xylan, and LM10 to specifically track the nonreducing ends of xylan polymers (Fig. 4). Interestingly, these three mAbs showed distinct binding patterns on sections of the root elongation zone. CCRC-M148 bound to walls of all cells of the stele (central metaxylem to pericycle cells), CCRC-M154 bound to walls of cells in the stele and in the surrounding cortex, and LM10 selectively bound to walls of pericycle and metaxylem cells. Thus, the secondary cell walls in the stele contain xylans with a low degree of arabinosylation, whereas the primary walls in cortex cells contain only highly arabinosylated xylans (Christensen et al., 2010). To confirm that the binding of CCRC-M154 to the cortex cells results from their high arabinoxylan content, we applied arabinoxylan-specific arabinofuranosidases (McCleary et al., 2015) to the sections prior to antibody staining. Indeed, CCRC-M154 binding was abolished completely in all cell walls, while CCRC-M148 now also bound to the cortex cell walls in addition to the stele walls. Apparently, dearabinosylating the xylan exposed unsubstituted xylan stretches for CCRC-M148 binding in cortex cell walls. LM10 binding was not affected by arabinofuranosidase treatment, suggesting that arabinosylation of the terminal Xyl unit of the xylan chain was not causative for the observed differential binding of LM10 to the walls of different cell types. Instead, we hypothesize that the nonreducing ends of the xylan chains are covalently modified or covered by interactions with other components of the cell wall matrix.
Figure 4.
Immunological analyses reveal detailed molecular glycan structures in Brachypodium root sections. A, Toluidine Blue was used to stain sections and identify cell types (Hardtke and Pacheco-Villalobos 2015). B, Immunolabeling of (arabino)xylan using CCRC-M148, CCRC-M154, and LM10 in root tips of Brachypodium. C, Putative AG side chains with expected binding of JIM14 and JIM16. D, Immunolabeling of AG structures using JIM14 and JIM16. Arabinofuranosidases and galactanases were incubated on the sections prior to the antibody staining to reveal xylan and galactan epitopes. Representative results of three independent biological replicates are shown. Bars = 50 µm.
JIM14 and JIM16 represent an interesting pair of antibodies with which to visualize differentially branched galactan in type II AG polysaccharides that are present on AGPs (Fig. 4). Yariv stain-based localization of AGPs showed that all cells in the root sections contained AGPs in their cell wall (Supplemental Fig. S3). Lack of JIM16 binding to these root sections suggests that the AG structures contain no single β-1,6-linked Gal substitutions of the backbone but rather longer galactan side branches. However, only metaphloem cells were stained by JIM14. We hypothesized that all other cells might display higher degrees of arabinosylation than are recognizable by JIM14. To test this hypothesis, we incubated the sections with arabinofuranosidase prior to antibody staining. Indeed, dearabinosylation resulted in JIM14 binding to all cells. As we were able to convert the JIM14 epitope into the JIM16 epitope by enzymatically trimming the β-1,6-linked galactan side chains on the microarray (Fig. 3, C and D), we tested if the JIM16 epitope could be revealed in the root sections as well. We incubated the sections with the β-1,6-galactanase that was used on the microarray but did not observe any JIM16 binding upon subsequent antibody staining. Only when both arabinofuranosidase and β-1,6-galactanase were applied together prior to antibody staining was the JIM16 epitope revealed in all root cells. Apparently, the metaphloem cells contain at least some arabinose substitution, which inhibited complete enzymatic cleavage of the β-1,6-linked galactan branches. Thus, we conclude that the AG structures in metaphloem cells contain a low degree of arabinosylation, whereas all other root cells display highly arabinosylated AGPs. The complexity of the AG structure in AGPs was associated previously with growth phenotypes in Brachypodium mutants and an Arabidopsis (Arabidopsis thaliana) mutant (Knoch et al., 2013; Pacheco-Villalobos et al., 2016). Both studies used glycosidic linkage analyses on whole roots or root segments to show differences in type II AG-related Gal linkages (Knoch et al., 2013; Pacheco-Villalobos et al., 2016). To resolve which cell types display the differences in AGP complexity, more detailed analyses are now feasible based on the newly derived epitope information for type II AG-specific antibodies and the possibility for integrating specific glycosyl hydrolases into cell wall analyses.
It is important to note that integrating glycosyl hydrolases to specifically reveal antibody epitopes is different from previous studies, in which plant sections were pretreated with a pectate lyase (Marcus et al., 2008, 2010; Hervé et al., 2009) to unmask xylan, xyloglucan, and mannan epitopes (Marcus et al., 2008, 2010; Hervé et al., 2009). The rational use of glycosyl hydrolases to alter the molecular structures of specific cell wall glycans provides additional structural information on the respective glycan. As this approach is limited by the availability of glycosyl hydrolases and antibodies with precisely known specificities, further expansion of the repertoire of suitable enzymes and antibodies will be crucial. This is particularly important as we cannot fully rule out the possibility that, in cases where glycosyl hydrolases with broader substrate specificities are used, multiple cell wall glycans may be affected, and this may lead to masking or demasking of epitopes.
CONCLUSION
Using a synthetic plant carbohydrate microarray, we determined the binding epitopes for a large number of plant cell wall glycan-directed mAbs, providing the required information for the specific detection of many complex molecular structures in plant cell wall polysaccharides. We further demonstrate that integrating specific glycosyl hydrolases with immunolabeling studies of cell walls enables an even more comprehensive analysis of the distribution patterns of cell wall glycans in plant cells. Our integrated mAb-based analyses provided detailed information on glycan structures in roots of the important model grass species Brachypodium, highlighting the importance of the structural understanding of epitopes for the comprehensive interpretation of in situ cell wall labeling studies.
MATERIALS AND METHODS
Chemical Synthesis of Oligosaccharides
Compounds 1 to 66 were prepared by automated glycan assembly following previously published protocols (Bartetzko et al., 2015, 2017; Schmidt et al., 2015; Dallabernardina et al., 2016, 2017; Senf et al., 2017). Briefly, using an automated synthesizer, suitably protected monosaccharide building blocks were added in iterative glycosylation and deprotection cycles to a linker-functionalized Merrifield resin. After assembly was complete, the linker was cleaved and the oligosaccharides were globally deprotected to yield the desired glycans after HPLC purification. Compounds 67 to 88 were prepared by conventional solution-phase synthesis (Zakharova et al., 2013; Andersen and Clausen, 2014; Andersen et al., 2016a, 2016b).
Glycan Microarray Printing
The oligosaccharides were printed on CodeLink N-hydroxyl succinimide ester-activated glass slides (SurModics) using a noncontact piezoelectric spotting device (S3; Scienion). The printing was performed at room temperature and 40% humidity. To ensure constant printing efficiency, the first compound (oligosaccharide 1) was reprinted at the end of the printing run. To assess the binding strength of the antibodies, we diluted the oligosaccharides to four different printing concentrations (200, 50, 12.5, and 3.1 µm) in the coupling buffer (80% [v/v] 50 mm sodium phosphate, pH 8.5, 0.005% [v/v] CHAPS, and 20% [w/v] PEG400 [Roth]). After printing, the microarray slides were quenched for 1 h at room temperature in 100 mm ethanolamine and 50 mm sodium phosphate, pH 9, and washed three times with deionized water.
Determining mAb Epitopes
We obtained mAbs from different sources as indicated in Supplemental Data File S1 (i.e. the Complex Carbohydrate Research Center, the University of Leeds, the Institut National de la Recherche Agronomique in Nantes, the Classen laboratory at the University of Kiel, and the University of Melbourne). To incubate 16 different antibodies per microarray slide, we applied a FlexWell 16 grid (Grace Bio-Labs) to the slide. The slides were blocked with 1% (w/v) BSA in phosphate-buffered saline (PBS) for 1 h at room temperature. Then, hybridoma supernatants containing the antibodies were diluted 1:10 in PBS containing 1% (w/v) BSA and incubated for 1 h on the slides. After three washes with PBS, the slides were incubated with the respective secondary antibodies for 1 h (goat anti-rat IgG AF555 and goat anti-mouse IgM/IgG AF488; Invitrogen). Unbound secondary antibodies were removed using consecutive washes with 0.1% (v/v) Tween 20 in PBS, PBS, and deionized water. After drying the slides by centrifugation (300g, 2 min), the fluorescent signal on the slides was scanned with a GenePix 4300A microarray scanner (Molecular Devices). Image analysis and quantification of the fluorescent signal were carried out with the GenePix Pro 7 software (Molecular Devices) using the same settings for each antibody. Based on the raw data (Supplemental Data File S1), we defined positive signals for each of the four printed glycan concentrations based on two thresholds that we cross-validated using antibody binding to known epitopes: (1) a 4-fold increase of fluorescent signal intensity over background and (2) a fold change value of at least 4% of the maximal value of the respective antibody. The resulting numbers 0 to 4 indicate the minimum printing concentrations (0 = no binding, 1 = 200 µm, 2 = 50 µm, 3 = 12.5 µm, and 4 = 3.1 µm) required for the binding of an antibody. To group similar antibodies, we separately performed hierarchical clustering on AG-, xylan-, and xyloglucan-binding antibodies. To this end, we applied the hclust function in R using manhattan distance as a distance measure.
Glycosyl Hydrolase Digests on Glycan Microarrays
For the arabinoxylan digests, we used two GH43 arabinofuranosidases from Bacteroides ovatus (ABFBO17 and ABFBO25) that were purchased from Megazyme. ABFBO17 removes arabinose substituents linked to the 3-position of double-substituted Xyl residues, whereas ABFBO25 cleaves single arabinoses in the C2- and C3-positions of Xyl (Senf et al., 2017). The enzymes were used at a concentration of 10 units mL−1 in 100 mm sodium phosphate buffer, pH 6.5. For the AG digests, we used a GH51 arabinofuranosidase from Aspergillus niger purchased from Megazyme at a concentration of 0.1 units mL−1 and an endo-1,6-galactanase from Trichoderma viride (Tv6GAL) at a concentration of 5 units mL−1, both in sodium citrate buffer, pH 4.5. Tv6GAL was expressed recombinantly in Pichia pastoris as described previously (Kotake et al., 2004). In both arabinoxylan and AG digests, the enzymes were applied directly in the 16-well grid of a microarray slide and incubated overnight at 37°C. The corresponding buffers without enzymes were used as controls. After washing the slides twice with PBS, blocking and antibody staining were performed as described above. Secondary antibodies used were goat anti-rat IgG AF555 for LM10, JIM14, and JIM16 and goat anti-mouse IgM AF594 for CCRC-M148 and CCRC-M154. The fluorescent signal was recorded with a GenePix 4300A microarray scanner and quantified with the GenePix Pro 7 software.
Immunofluorescence Analysis of Brachypodium distachyon Root Sections
To analyze arabinoxylan and AG structures in Brachypodium roots, we grew Brachypodium (accession Bd21) on vertical Murashige and Skoog agar plates under a 16-h-light/8-h-dark cycle. Root tips were harvested 4 d after germination and fixed for 1 h in a 2.5% (v/v) glutaraldehyde solution. Dehydration and embedding into LR White were performed as described previously (Lee et al., 2012). The root tips were cut into 1-µm sections using a Leica Ultracut UCT ultramicrotome. Antibody staining was performed as described for the glycan microarray experiments using goat anti-rat IgG AF555 for LM10, JIM14, and JIM16 and goat anti-mouse IgM AF594 for CCRC-M148 and CCRC-M154. AGP localization in the sections was carried out as described previously (Göllner et al., 2013). In brief, after blocking the sections with BSA buffer, (β-d-Glc)3 Yariv phenylglycoside (βGlcY; 400 µg mL−1 in 0.15 m NaCl) was applied for 90 min. After three washes with PBS, first a βGlcY-antibody and then the corresponding fluorescein isothiocyanate-labeled anti-rabbit secondary antibody were incubated on the sections. Imaging was carried out with an LSM700 confocal microscope (Zeiss). Glycosyl hydrolase digests of arabinoxylan and AG in the sections were performed using the same enzymes and conditions as for the digests on the glycan microarray.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Summarizing heat map of binding interactions between individual mAbs and synthetic glycans.
Supplemental Figure S2. Selected glycan microarray scans.
Supplemental Figure S3. Yariv-based staining of AGPs in Brachypodium root sections and antibody staining negative controls.
Supplemental Data File S1. Quantification of the fluorescence signals (raw data) for all tested antibodies.
Acknowledgments
We thank Andreas Geissner for help with printing the microarrays, Rona Pitschke for technical support, and Dr. Dirk Walther for advice with the hierarchical clustering. We thank Dr. Alexandra Zakharova for the synthesis of oligosaccharide 78. Provision of anti-βGlcY antibody and mAbs KM1 by Dr. Birgit Classen, BG1 by Dr. Wei Zeng, AX1 and X3 by Dr. Fabienne Guillon, and RU1 and RU2 by Dr. Marie Christine Ralet is gratefully acknowledged.
Footnotes
We gratefully acknowledge support from the Max Planck Society, the Fonds der Chemischen Industrie (Liebig Fellowship to F.P.), and the German Research Foundation (DFG, Emmy Noether program PF850/1-1 to F.P.). M.H.C. acknowledges support from the Carlsberg Foundation, the Danish Strategic Research Council (GlycAct and SET4Future projects), and the Villum Foundation (PLANET project). Generation of the CCRC series of monoclonal antibodies used in this work was supported by grants from the U.S. Natural Science Foundation Plant Genome Program (DBI-0421683 and IOS-0923992 to M.G.H.).
References
- Andersen MC, Boos I, Marcus SE, Kračun SK, Rydahl MG, Willats WG, Knox JP, Clausen MH (2016a) Characterization of the LM5 pectic galactan epitope with synthetic analogues of β-1,4-d-galactotetraose. Carbohydr Res 436: 36–40 [DOI] [PubMed] [Google Scholar]
- Andersen MC, Kračun SK, Rydahl MG, Willats WG, Clausen MH (2016b) Synthesis of β-1,4-linked galactan side-chains of rhamnogalacturonan I. Chemistry 22: 11543–11548 [DOI] [PubMed] [Google Scholar]
- Andersen MCF, Clausen MH (2014) Synthesis and Application of Plant Cell Wall Oligogalactans. PhD thesis. Technical University of Denmark, Kemitorvet, Denmark [Google Scholar]
- Bartetzko MP, Schuhmacher F, Hahm HS, Seeberger PH, Pfrengle F (2015) Automated glycan assembly of oligosaccharides related to arabinogalactan proteins. Org Lett 17: 4344–4347 [DOI] [PubMed] [Google Scholar]
- Bartetzko MP, Schuhmacher F, Seeberger PH, Pfrengle F (2017) Determining substrate specificities of β1,4-endogalactanases using plant arabinogalactan oligosaccharides synthesized by automated glycan assembly. J Org Chem 82: 1842–1850 [DOI] [PubMed] [Google Scholar]
- Christensen U, Alonso-Simon A, Scheller HV, Willats WG, Harholt J (2010) Characterization of the primary cell walls of seedlings of Brachypodium distachyon: a potential model plant for temperate grasses. Phytochemistry 71: 62–69 [DOI] [PubMed] [Google Scholar]
- Classen B, Csávás M, Borbás A, Dingermann T, Zündorf I (2004) Monoclonal antibodies against an arabinogalactan-protein from pressed juice of Echinacea purpurea. Planta Med 70: 861–865 [DOI] [PubMed] [Google Scholar]
- Cornuault V, Buffetto F, Rydahl MG, Marcus SE, Torode TA, Xue J, Crépeau MJ, Faria-Blanc N, Willats WG, Dupree P, et al. (2015) Monoclonal antibodies indicate low-abundance links between heteroxylan and other glycans of plant cell walls. Planta 242: 1321–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa RM, Pattathil S, Avci U, Lee SJ, Hazen SP, Winters A, Hahn MG, Bosch M (2017) A cell wall reference profile for Miscanthus bioenergy crops highlights compositional and structural variations associated with development and organ origin. New Phytol 213: 1710–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallabernardina P, Schuhmacher F, Seeberger PH, Pfrengle F (2016) Automated glycan assembly of xyloglucan oligosaccharides. Org Biomol Chem 14: 309–313 [DOI] [PubMed] [Google Scholar]
- Dallabernardina P, Schuhmacher F, Seeberger PH, Pfrengle F (2017) Mixed-linkage glucan oligosaccharides produced by automated glycan assembly serve as tools to determine the substrate specificity of lichenase. Chemistry 23: 3191–3196 [DOI] [PubMed] [Google Scholar]
- Gendre D, McFarlane HE, Johnson E, Mouille G, Sjödin A, Oh J, Levesque-Tremblay G, Watanabe Y, Samuels L, Bhalerao RP (2013) Trans-Golgi network localized ECHIDNA/Ypt interacting protein complex is required for the secretion of cell wall polysaccharides in Arabidopsis. Plant Cell 25: 2633–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göllner EM, Gramann JC, Classen B (2013) Antibodies against Yariv’s reagent for immunolocalization of arabinogalactan-proteins in aerial parts of Echinacea purpurea. Planta Med 79: 175–180 [DOI] [PubMed] [Google Scholar]
- Guillon F, Tranquet O, Quillien L, Utille JP, José JOO, Saulnier L (2004) Generation of polyclonal and monoclonal antibodies against arabinoxylans and their use for immunocytochemical location of arabinoxylans in cell walls of endosperm of wheat. J Cereal Sci 40: 167–182 [Google Scholar]
- Hardtke CS, Pacheco-Villalobos D (2015) The Brachypodium distachyon root system: a tractable model to investigate grass roots. Plant Genet Genomics 18: 245–258 [Google Scholar]
- Hervé C, Rogowski A, Gilbert HJ, Knox JP (2009) Enzymatic treatments reveal differential capacities for xylan recognition and degradation in primary and secondary plant cell walls. Plant J 58: 413–422 [DOI] [PubMed] [Google Scholar]
- Izydorczyk MS, Biliaderis CG (1995) Cereal arabinoxylans: advances in structure and physicochemical properties. Carbohydr Polym 28: 33–48 [Google Scholar]
- Jones L, Seymour GB, Knox JP (1997) Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1→4)-β-d-galactan. Plant Physiol 113: 1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K. (2010) Plant cell walls. Plant Physiol 154: 483–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa K, Tryfona T, Yoshimi Y, Hayashi Y, Kawauchi S, Antonov L, Tanaka H, Takahashi T, Kaneko S, Dupree P, et al. (2013) β-Galactosyl Yariv reagent binds to the β-1,3-galactan of arabinogalactan proteins. Plant Physiol 161: 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoch E, Dilokpimol A, Tryfona T, Poulsen CP, Xiong G, Harholt J, Petersen BL, Ulvskov P, Hadi MZ, Kotake T, et al. (2013) A β-glucuronosyltransferase from Arabidopsis thaliana involved in biosynthesis of type II arabinogalactan has a role in cell elongation during seedling growth. Plant J 76: 1016–1029 [DOI] [PubMed] [Google Scholar]
- Kotake T, Kaneko S, Kubomoto A, Haque MA, Kobayashi H, Tsumuraya Y (2004) Molecular cloning and expression in Escherichia coli of a Trichoderma viride endo-beta-(1→6)-galactanase gene. Biochem J 377: 749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KJ, Dekkers BJ, Steinbrecher T, Walsh CT, Bacic A, Bentsink L, Leubner-Metzger G, Knox JP (2012) Distinct cell wall architectures in seed endosperms in representatives of the Brassicaceae and Solanaceae. Plant Physiol 160: 1551–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqué D, Scheller HV, Pauly M (2015) Engineering of plant cell walls for enhanced biofuel production. Curr Opin Plant Biol 25: 151–161 [DOI] [PubMed] [Google Scholar]
- Marcus SE, Blake AW, Benians TAS, Lee KJD, Poyser C, Donaldson L, Leroux O, Rogowski A, Petersen HL, Boraston A, et al. (2010) Restricted access of proteins to mannan polysaccharides in intact plant cell walls. Plant J 64: 191–203 [DOI] [PubMed] [Google Scholar]
- Marcus SE, Verhertbruggen Y, Hervé C, Ordaz-Ortiz JJ, Farkas V, Pedersen HL, Willats WGT, Knox JP (2008) Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney L, Marcus SE, Knox JP (2005) Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J Histochem Cytochem 53: 543–546 [DOI] [PubMed] [Google Scholar]
- McCleary BV, McKie VA, Draga A, Rooney E, Mangan D, Larkin J (2015) Hydrolysis of wheat flour arabinoxylan, acid-debranched wheat flour arabinoxylan and arabino-xylo-oligosaccharides by β-xylanase, α-L-arabinofuranosidase and β-xylosidase. Carbohydr Res 407: 79–96 [DOI] [PubMed] [Google Scholar]
- Meikle PJ, Hoogenraad NJ, Bonig I, Clarke AE, Stone BA (1994) A (1→3,1→4)-beta-glucan-specific monoclonal antibody and its use in the quantitation and immunocytochemical location of (1→3,1→4)-beta-glucans. Plant J 5: 1–9 [DOI] [PubMed] [Google Scholar]
- Pacheco-Villalobos D, Díaz-Moreno SM, van der Schuren A, Tamaki T, Kang YH, Gujas B, Novak O, Jaspert N, Li Z, Wolf S, et al. (2016) The effects of high steady state auxin levels on root cell elongation in Brachypodium. Plant Cell 28: 1009–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Baldwin D, Swennes AG, McGill JA, Popper Z, Bootten T, Albert A, Davis RH, Chennareddy C, et al. (2010) A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol 153: 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M, Keegstra K (2010) Plant cell wall polymers as precursors for biofuels. Curr Opin Plant Biol 13: 305–312 [DOI] [PubMed] [Google Scholar]
- Pedersen HL, Fangel JU, McCleary B, Ruzanski C, Rydahl MG, Ralet MC, Farkas V, von Schantz L, Marcus SE, Andersen MC, et al. (2012) Versatile high resolution oligosaccharide microarrays for plant glycobiology and cell wall research. J Biol Chem 287: 39429–39438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettolino FA, Walsh C, Fincher GB, Bacic A (2012) Determining the polysaccharide composition of plant cell walls. Nat Protoc 7: 1590–1607 [DOI] [PubMed] [Google Scholar]
- Puhlmann J, Bucheli E, Swain MJ, Dunning N, Albersheim P, Darvill AG, Hahn MG (1994) Generation of monoclonal antibodies against plant cell-wall polysaccharides. I. Characterization of a monoclonal antibody to a terminal α-(1→2)-linked fucosyl-containing epitope. Plant Physiol 104: 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralet MC, Tranquet O, Poulain D, Moïse A, Guillon F (2010) Monoclonal antibodies to rhamnogalacturonan I backbone. Planta 231: 1373–1383 [DOI] [PubMed] [Google Scholar]
- Rillahan CD, Paulson JC (2011) Glycan microarrays for decoding the glycome. Annu Rev Biochem 80: 797–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schuhmacher F, Geissner A, Seeberger PH, Pfrengle F (2015) Automated synthesis of arabinoxylan-oligosaccharides enables characterization of antibodies that recognize plant cell wall glycans. Chemistry 21: 5709–5713 [DOI] [PubMed] [Google Scholar]
- Senf D, Ruprecht C, de Kruijff GH, Simonetti SO, Schuhmacher F, Seeberger PH, Pfrengle F (2017) Active site mapping of xylan-deconstructing enzymes with arabinoxylan oligosaccharides produced by automated glycan assembly. Chemistry 23: 3197–3205 [DOI] [PubMed] [Google Scholar]
- Smallwood M, Yates EA, Willats WGT, Martin H, Knox JP (1996) Immunochemical comparison of membrane-associated and secreted arabinogalactan-proteins in rice and carrot. Planta 198: 452–459 [Google Scholar]
- Steffan W, Kovác P, Albersheim P, Darvill AG, Hahn MG (1995) Characterization of a monoclonal antibody that recognizes an arabinosylated (1→6)-beta-D-galactan epitope in plant complex carbohydrates. Carbohydr Res 275: 295–307 [DOI] [PubMed] [Google Scholar]
- Verhertbruggen Y, Marcus SE, Haeger A, Verhoef R, Schols HA, McCleary BV, McKee L, Gilbert HJ, Knox JP (2009) Developmental complexity of arabinan polysaccharides and their processing in plant cell walls. Plant J 59: 413–425 [DOI] [PubMed] [Google Scholar]
- Vidal-Melgosa S, Pedersen HL, Schückel J, Arnal G, Dumon C, Amby DB, Monrad RN, Westereng B, Willats WG (2015) A new versatile microarray-based method for high throughput screening of carbohydrate-active enzymes. J Biol Chem 290: 9020–9036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JA, Pattathil S, Bergeman LF, Beebe ET, Deng K, Mirzai M, Northen TR, Hahn MG, Fox BG (2017) Determination of glycoside hydrolase specificities during hydrolysis of plant cell walls using glycome profiling. Biotechnol Biofuels 10: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WG, Marcus SE, Knox JP (1998) Generation of monoclonal antibody specific to (1→5)-alpha-L-arabinan. Carbohydr Res 308: 149–152 [DOI] [PubMed] [Google Scholar]
- Zakharova AN, Madsen R, Clausen MH (2013) Synthesis of a backbone hexasaccharide fragment of the pectic polysaccharide rhamnogalacturonan I. Org Lett 15: 1826–1829 [DOI] [PubMed] [Google Scholar]