Loss of iron-nicotianamine transporters causes mislocalization of iron in Arabidopsis leaves that leads to profound defects in the long-distance signaling of iron status to roots.
Abstract
The mechanisms of root iron uptake and the transcriptional networks that control root-level regulation of iron uptake have been well studied, but the mechanisms by which shoots signal iron status to the roots remain opaque. Here, we characterize an Arabidopsis (Arabidopsis thaliana) double mutant, yellow stripe1-like yellow stripe3-like (ysl1ysl3), which has lost the ability to properly regulate iron deficiency-influenced gene expression in both roots and shoots. In spite of markedly low tissue levels of iron, the double mutant does not up- and down-regulate iron deficiency-induced and -repressed genes. We have used grafting experiments to show that wild-type roots grafted to ysl1ysl3 shoots do not initiate iron deficiency-induced gene expression, indicating that the ysl1ysl3 shoots fail to send an appropriate long-distance signal of shoot iron status to the roots. We present a model to explain how impaired iron localization in leaf veins results in incorrect signals of iron sufficiency being sent to roots and affecting gene expression there. Improved understanding of the mechanism of long-distance iron signaling will allow improved strategies for the engineering of staple crops to accumulate additional bioavailable iron in edible parts, thus improving the iron nutrition of the billions of people worldwide whose inadequate diet causes iron deficiency anemia.
Iron is an essential cofactor for many cellular redox reactions and is required for life. Plants tightly regulate iron uptake to maintain a balance between the demand for iron in photosynthetic tissues (Briat et al., 2015) and the risk of overaccumulation that could lead to cellular damage (Kampfenkel et al., 1995). Many plants, including Arabidopsis (Arabidopsis thaliana), use a combination of rhizosphere acidification and reduction to solubilize and obtain iron from the soil (Walker and Connolly, 2008; Jeong and Guerinot, 2009; Morrissey and Guerinot, 2009). In Arabidopsis, the genes most directly responsible for iron uptake are the Fe(II) transporter IRT1 (Eide et al., 1996; Henriques et al., 2002; Varotto et al., 2002; Vert et al., 2002) and the ferric chelate reductase FRO2 (Robinson et al., 1999; Connolly et al., 2003). Both these genes are strongly up-regulated by iron deficiency, and regulatory proteins governing the expression of these genes have been the subject of intense investigation (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005, 2008; Bauer et al., 2007; Wang et al., 2007; Long et al., 2010; Selote et al., 2015; Li et al., 2016; Mai et al., 2016). An additional component of iron uptake in nongraminaceous plants is the discovery that iron deficiency induces the secretion of phenolics and flavins (Rodríguez-Celma et al., 2013), later better defined as coumarins (Schmid et al., 2014) that are secreted by the ABCG37/PDR9 transporter (Fourcroy et al., 2014).
We understand less about the mechanisms by which plants signal their iron status internally. Both local and long-distance signals are clearly present. Several lines of evidence point to the idea that shoots can signal iron status to roots and that uptake processes at the root are strongly controlled by such shoot signaling. The pea (Pisum sativum) mutant dgl accumulates high levels of iron in leaves yet constitutively expresses the root iron uptake response. When dgl shoots were grafted to wild-type roots, ferric reductase activity was strongly increased, suggesting that a signal transmitted from the mutant shoots stimulated the activity of the iron uptake machinery in the roots (Grusak and Pezeshgi, 1996). In cucumber (Cucumis sativus) and sunflower (Helianthus annuus; Romera et al., 1992), tobacco (Nicotiana tabacum; Enomoto et al., 2007), and Arabidopsis (García et al., 2013), treatment of the leaves of iron-deficient plants with iron caused the down-regulation of iron uptake activities in roots growing in iron-deficient medium, again suggesting that adequate iron in the shoots affects iron deficiency responses in the roots. Moreover, in the Arabidopsis experiment, the expression of root iron deficiency-associated genes was decreased by foliar iron application (García et al., 2013). On the other hand, potato (Solanum tuberosum) roots growing from tubers without leaves, and even growing in culture in the absence of tubers and leaves, developed iron deficiency responses (acidification of the rhizosphere, increased ferric reductase activity, and morphological changes at the root tip) when growing on iron-free medium, showing that the presence of shoots is not required for the iron deficiency response (Bienfait et al., 1987).
Split-root experiments, in which one half of the root system is grown with iron while the other is grown in nutrient solution lacking iron, have been very useful for uncovering internal signals. In split-root experiments using sunflower and cucumber, differences were observed in the magnitude of the iron deficiency response, as measured by ferric reductase activity, on both the –Fe and +Fe sides of the root system. Ferric reductase activity was higher on both sides of the split +Fe/–Fe root system than it was for split roots with both sides receiving adequate iron (+Fe/+Fe), clearly indicating that long-distance signals occur (Romera et al., 1992). Interestingly, ferric reductase activity was consistently even higher on the –Fe side than on the +Fe side. Morphologically, only roots on the –Fe side developed swollen tips. Likewise, only the roots on the –Fe side acidified the growth medium (Romera et al., 1992). Thus, in these experiments, iron deficiency responses at the level of the root itself were revealed in addition to the long-distance signals from shoots. Studies using Arabidopsis also have indicated that a local signal is present at the roots (Vert et al., 2003). In these experiments, a bipyridyl wash was used to completely remove all traces of iron from the –Fe side of the split roots. This allowed a clear demonstration that some iron must be present locally in order to have detectable FRO2 and IRT1 expression, and this unequivocally demonstrates that the local environment plays a role. This was particularly true at the level of protein for IRT1, which was undetectable on the –Fe side of the root system. In the split-root experiment performed by Vert et al. (2003), the plants were never subjected to a period of iron deficiency, since iron was always available in the solution, at least on the +Fe side of the experiment. Thus, because of the experimental setup, the presence or absence of long-distance signals of iron deficiency emanating from the shoots was not tested.
Members of the well-conserved Yellow Stripe1-Like (YSL) family of proteins function as transporters of metals that are complexed with nicotianamine (NA; DiDonato et al., 2004; Koike et al., 2004; Roberts et al., 2004; Schaaf et al., 2004; Murata et al., 2006; Gendre et al., 2007). The nonproteinogenic amino acid NA is a strong complexor of various transition metals (Anderegg and Ripperger, 1989; von Wiren et al., 1999). We have shown previously that Arabidopsis plants with lesions in both YSL1 (At4g24120) and YSL3 (At5g53550) display strong interveinal chlorosis, have low Fe, Cu, Mn, and Zn levels in leaves, and exhibit strong reproductive defects. All of the defects displayed by the ysl1ysl3 double mutant (DM) can be alleviated if excess iron is applied to the soil, demonstrating that these growth defects are caused primarily by a lack of iron (Waters et al., 2006). YSL3 and YSL1 are localized to the plasma membrane and function as iron transporters in yeast functional complementation assays (Chu et al., 2010). Both YSL1 and YSL3 are expressed strongly in the vasculature of leaves. We have hypothesized that the function of these YSL transporters in vegetative tissues is to take up iron that arrives in leaves via the xylem (Waters et al., 2006). The iron deficiency response of ysl1ysl3 DM plants also is abnormal, in that it appears to be weaker than that of wild-type plants, in spite of the low tissue levels of iron (Waters et al., 2006).
Here, we expand and refine work on the iron deficiency responses of the ysl1ysl3 DM to show that they have an impaired ability to mount a robust iron deficiency response. We used split-root experiments in a new way to show that long-distance signals predominate in damping down iron deficiency responses after iron resupply to iron-deficient plants. Using grafting experiments, we demonstrate that ysl1ysl3 mutant shoots are incapable of sending long-distance signals related to iron deficiency to the roots. Our data are consistent with a model in which the amount of iron in the leaf veins determines the signaling of iron status that is transmitted to the roots.
RESULTS
Iron-Regulated Gene Expression in ysl1ysl3 DM Plants
We have shown previously that DM plants responding to iron deficiency do not up-regulate ferric chelate reductase activity as strongly as wild-type plants (Waters et al., 2006). Furthermore, the DM has low iron in leaves and is chlorotic and, thus, might be expected to up-regulate iron deficiency responses even when grown on normal medium. In early experiments (Waters et al., 2006), gene expression was monitored using nonquantitative reverse transcriptase-PCR, and the expression of IRT1 appeared to be somewhat lower in the DM than in the wild type.
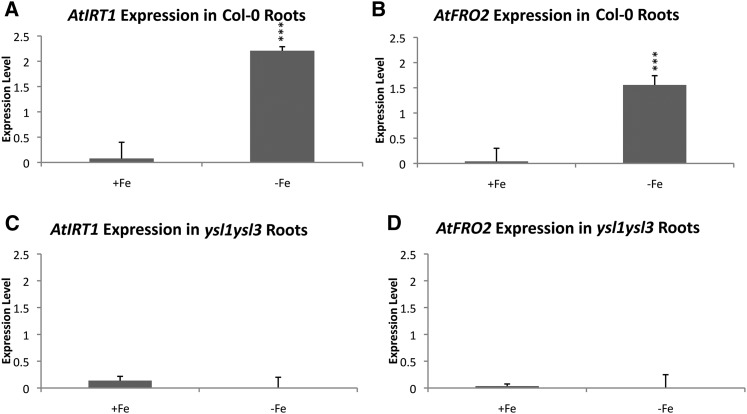
To better address iron deficiency-regulated gene expression (hereafter IDRGE) in ysl1ysl3 plants, we used quantitative reverse transcriptase (qRT)-PCR. Wild-type plants (positive control) and ysl1ysl3 mutant plants were grown for 14 d and then transferred to Murashige and Skoog medium (MS) plates prepared without iron (MS−Fe) or maintained on MS+Fe for 4 d. After this 4-d period, three biological replicates of root tissue were collected, RNA was isolated from each, and IRT1 and FRO2 transcript levels were measured (Fig. 1). In wild-type plants, as expected, IRT1 expression levels were significantly higher in the −Fe plants (Fig. 1; 28-fold change, P < 0.001 by Student’s t test). FRO2 expression levels also were significantly higher in plants grown on MS−Fe for 4 d (Fig. 1; 22-fold change, P < 0.001). However, ysl1ysl3 plants showed no change in the expression of IRT1 and FRO2 in response to the 4-d period of iron deficiency (Fig. 1), suggesting that DM plants are incapable of robustly responding to iron deficiency.
Figure 1.
qRT-PCR analysis of wild-type and ysl1ysl3 IRT1 and FRO2 expression levels after 4 d of iron deficiency. A and B, IRT1 and FRO2 expression levels in wild-type roots. C and D, IRT1 and FRO2 expression levels in ysl1ysl3 mutant roots.
To expand on this result, we used microarray analysis to determine the extent to which iron-regulated genes are misregulated in the ysl1ysl3 DM. In this experiment, we grew wild-type Columbia-0 (Col-0) plants for 10 d with iron (MS+Fe) and then shifted the plants to fresh medium containing iron (control) or to fresh medium that lacked iron (MS−Fe) for a period of 3 d. We also grew ysl1ysl3 DM plants for 10 d on MS+Fe and shifted the plants to fresh MS+Fe for a period of 3 d, the same growth regimen as for the wild-type +Fe plants. Under these growth conditions, the wild-type plants grown on MS−Fe and the DM plants have similar levels of chlorosis and similar iron levels in shoots (Waters et al., 2006). Shoots and roots were harvested separately from three biological replicates of each of the three samples, and RNA was prepared and hybridized to the Ath1 GeneChip. Comparisons were made between wild-type +Fe and wild-type –Fe samples to define the iron-regulated genes. Only these genes were used in this analysis. We used a significance cutoffs of P < 0.05 and fold difference in expression of 2 or greater. The microarray data comparisons are shown in Supplemental Data S1 and have been deposited at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) under accession number GSE92716.
In the DM plants, which are chlorotic and have low iron accumulation in their tissues, the expression level of most iron-regulated genes in roots was not significantly different from that observed in control wild-type plants grown in +Fe conditions (Tables I–III). This is the opposite of our expectation that chlorotic DM plants with low tissue levels of iron would have a gene expression pattern characteristic of iron starvation. Among several well-characterized up- and down-regulated genes in roots (Table I), all of the normally up-regulated genes were indistinguishable between the ysl1ysl3 DM and wild-type +Fe plants. Similarly, the level of expression of all of these genes was significantly different from that of wild-type –Fe plants. This is true both for up-regulated and down-regulated genes. In shoots (Table II), several of the well-characterized iron deficiency-regulated genes listed are significantly up-regulated (BHLH101, BHLH39, and FRO3) or down-regulated (FERRITIN3) in the ysl1ysl3 DM plants. However, all of the genes shown in Table II also have significantly lower expression than what was observed in wild-type –Fe plants. Based on the entire suite of iron-regulated genes in our experiments, we defined three categories of expression (Table III). Category 1, similar to wild-type +Fe: expression of these genes is statistically indistinguishable in ysl1ysl3 DM and wild-type +Fe plants. Category 2, intermediate: expression of these genes in ysl1ysl3 DM plants is statistically different from both the wild type +Fe and wild-type –Fe plants. Category 3, similar to wild type –Fe: expression of these genes is statistically indistinguishable in ysl1ysl3 DM and wild-type –Fe plants. For example (Table III), among the 359 genes that are up-regulated by –Fe in roots, 240 (67%) were statistically indistinguishable from the wild-type +Fe sample and were statistically different from the wild-type –Fe sample. Of the 119 genes remaining, only 34 genes (9%) were statistically different from wild-type +Fe and statistically similar to the wild-type –Fe level. The rest of the genes (24%) had the intermediate response, in which the level of expression was higher in the DM, but not as high as the level in the –Fe wild-type samples. The pattern observed among genes that were down-regulated by –Fe in wild-type roots was similar (Table III).
Table I. Comparison of the expression of select iron deficiency-regulated genes (Petit et al., 2001; Stacey et al., 2002, 2008; Mukherjee et al., 2006; Schaaf et al., 2006; Wang et al., 2007; Klatte et al., 2009; Morrissey et al., 2009; Haydon et al., 2012; Palmer et al., 2013; Fourcroy et al., 2016; Mai et al., 2016; Sisó-Terraza et al., 2016; Yan et al., 2016; Ziegler et al., 2017) in ysl1ysl3 DM and wild-type roots.
*, P < 0.05; **, P < 0.001; N.S. indicates that the difference was not significant (P > 0.05).
| Gene Name | Common Name | Fold Change in −Fe Wild-Type Versus +Fe Wild-Type Roots | Fold Change in +Fe Wild-Type Versus DM Roots | Fold Change in –Fe Wild-Type Versus DM Roots |
|---|---|---|---|---|
| AT3G12900 | Proposed F6′H1 coumarin synthesis | 86.3** | N.S. | 115.5** |
| AT1G56430 | NAS4 | 77.1** | N.S. | 35.6** |
| AT4G16370 | OPT3 | 38.6** | N.S. | 21.1** |
| AT3G56980 | BHLH39 | 18.0** | N.S. | 8.9** |
| AT5G04150 | BHLH101 | 16.7** | N.S. | 13.1** |
| AT5G13740 | ZIF1 | 10.2** | N.S. | 7.2** |
| AT3G12820 | MYB10 | 9.9** | N.S. | 9.2** |
| AT1G23020 | FRO3 | 9.7** | N.S. | 5.7** |
| AT5G03570 | IREG2/FPN2 | 7.9** | 0.58* | 4.6** |
| AT4G19690 | IRT1 | 5.9** | N.S. | 7.0** |
| AT3G13610 | F6′H1 coumarin synthesis | 2.6* | N.S. | 2.5* |
| AT3G53480 | PDR9 | 2.1** | N.S. | 1.9** |
| AT5G56080 | NAS2 | 0.19** | N.S. | 0.13** |
| AT3G56090 | FERRITIN3 | 0.21** | N.S. | 0.34** |
| AT2G40300 | FERRITIN4 | 0.09** | N.S. | 0.11** |
| AT5G01600 | FERRITIN1 | 0.07** | N.S. | 0.05** |
| AT1G21140 | VITL1 | 0.04** | N.S. | 0.04** |
Table III. Levels of iron-regulated gene expression in ysl1ysl3 DM plants.
Two comparisons were made: ysl1ysl3 DM compared with wild-type +Fe and ysl1ysl3 DM compared with wild-type –Fe. The expression of each gene was evaluated as statistically indistinguishable from wild-type +Fe (similar to wild-type +Fe), statistically indistinguishable from wild-type –Fe (similar to wild-type –Fe), or statistically different from both the wild-type +Fe and the wild-type –Fe samples (intermediate).
| Expression | Root |
Shoot |
||
|---|---|---|---|---|
| Up-Regulated | Down-Regulated | Up-Regulated | Down-Regulated | |
| Similar to wild-type +Fe | 67% (240) | 60% (235) | 43% (392) | 23% (225) |
| Intermediate | 24% (85) | 25% (99) | 29% (267) | 33% (314) |
| Similar to wild-type –Fe | 9% (34) | 15% (57) | 28% (255) | 44% (427) |
| Total genes | 359 | 391 | 914 | 966 |
Table II. Comparison of the expression of select iron deficiency-regulated genes (Petit et al., 2001; Stacey et al., 2002, 2008; Mukherjee et al., 2006; Cohu and Pilon, 2007; Wang et al., 2007; Klatte et al., 2009) in ysl1ysl3 DM and wild-type shoots.
*, P < 0.05; **, P < 0.001; N.S. indicates that the difference was not significant (P > 0.05).
| Gene Name | Common Name | Fold Change in –Fe Wild-Type Versus +Fe Wild-Type Shoots | Fold Change in +Fe Wild-Type Versus DM Shoots | Fold Change in –Fe Wild-Type Versus DM Shoots |
|---|---|---|---|---|
| AT4G19690 | IRT1 | 17.9** | N.S. | 22.7** |
| AT3G56980 | BHLH39 | 15.1** | 3.3* | 4.6* |
| AT5G04150 | BHLH101 | 10.5** | 3.3** | 3.2** |
| AT1G23020 | FRO3 | 6.8** | 2.5** | 2.8** |
| AT1G56430 | NAS4 | 5.4** | N.S. | 6.4** |
| AT4G16370 | OPT3 | 3.8** | N.S. | 2.9** |
| AT3G56090 | FERRITIN3 | 0.2** | 0.53* | 0.39** |
| AT4G25100 | Fe-SOD | 0.06** | N.S. | 0.10** |
| AT5G01600 | FERRITIN1 | 0.03** | N.S. | 0.02** |
| AT2G40300 | FERRITIN4 | 0.02** | N.S. | 0.06** |
The pattern in leaves was distinct from that in roots: more genes that were up- or down-regulated by iron deficiency in wild-type plants were similarly up- or down-regulated in the ysl1ysl3 DM grown on MS+Fe (Table III). For example, among the genes that are up-regulated by –Fe in leaves, 392 of 914 (42%) were statistically indistinguishable from the wild-type +Fe sample and were statistically different from the wild-type –Fe sample. Of the remaining 522 genes, 255 genes (28%) were statistically different from the wild-type +Fe and statistically similar to the wild-type –Fe level, indicating that they had a level of expression characteristic of iron deficiency. The rest of the genes (29%) had an intermediate response in which the level of expression was higher in the DM but not as high as the level in the –Fe wild-type samples. The pattern observed among genes that were down-regulated by –Fe in wild-type shoots was similar (Table III). Notably, even though the proportion of genes with appropriate expression is higher in leaves, a large fraction (32.8%) of the iron-regulated genes in leaves of DM plants had expression levels characteristic of +Fe plants in spite of their state of iron deficiency. Together, the data from this experiment demonstrate a pervasive defect in the ability of the ysl1ysl3 DM to respond appropriately to iron deficiency, particularly in roots.
We were interested to know what functional categories of genes in the ysl1ysl3 DM either responded as if in iron deficiency (responded appropriately) or responded as if iron was sufficient (responded inappropriately). For roots, the genes responding appropriately (34 up-regulated genes and 54 down-regulated genes) could not be classified into specific groups using the DAVID Bioinformatics Resources 6.7 Gene Functional Classification Tool (Supplemental Data S2). In contrast, for shoots, the genes responding appropriately could be classified into several specific categories (Supplemental Data S2). Overrepresented up-regulated genes included five groups that could be characterized generally as regulatory: response to auxin, ATP-binding (mostly kinases), zinc-binding (mostly zinc-finger containing), and transcription factors (two groups). Overrepresented down-regulated genes in shoots included groups that could be characterized as related to plastid functions: chloroplast metal binding, thylakoid, pentatricopeptide repeat-containing. This is not surprising, since the leaves of the plants are chlorotic. Iron-containing oxidoreductases also were identified as an overrepresented group of genes, possibly reflecting the unavailability of iron for the production of these enzymes. Also overrepresented were genes related to apoplasmic functions (glycosylphosphatidylinositol-anchored proteins, secreted glycoproteins, glucosyltransferases), genes related to cellular regulation (kinases, zinc finger-containing, and transcription factors), and glutathione-S-transferases.
Split-Root Experiment to Reveal Local and Long-Distance Signaling
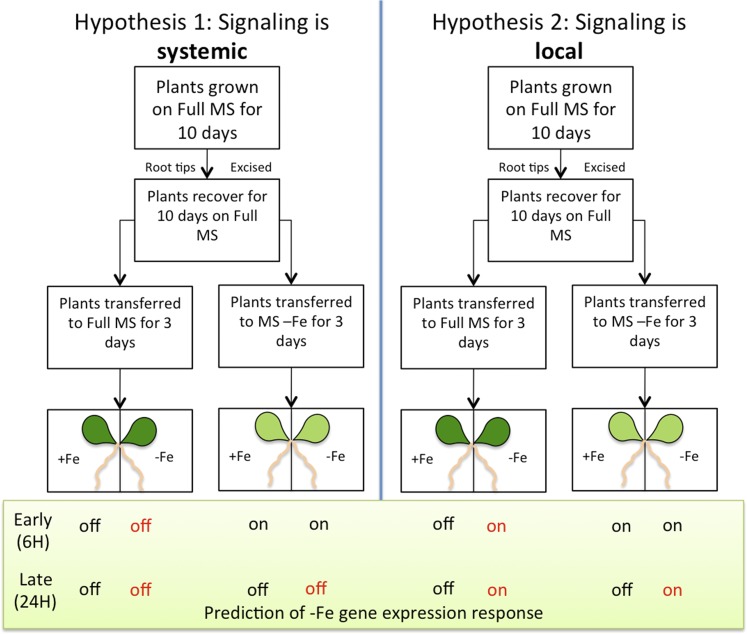
Given that YSL1 and YSL3 are expressed predominantly in leaves and yet have a strong effect on IDRGE throughout the plant, we became curious to understand whether DM plants might have defects in long-distance signaling of iron status. In several published split-root experiments pertaining to iron signaling (Vert et al., 2003), the plants were never subjected to a period of iron deficiency during the experiment. Instead, the root systems of iron-replete plants were placed into –Fe and +Fe medium, and because the plant always had at least part of its root system in iron-containing medium, the levels of iron in shoots in these experiments were similar to those of plants grown entirely on +Fe medium. We used a protocol in which plants were subjected to a period of iron starvation and then moved to the split plates. In this experiment, the plants will recover from the previously induced iron deficiency, because iron is taken up through the roots placed on +Fe medium. We then measured whether gene expression changes during the recovery from iron deficiency, namely the down-regulation of IRT1 and FRO2 expression, occurs equally on the +Fe and –Fe sides of the plate. The split-root experimental design used is shown in Figure 2. Twenty-one-day-old plants were either transferred to regular MS (+Fe; control for movement of the plants) or to MS with no added iron (MS−Fe) for 3 d to induce iron deficiency. After 3 d, plants were transferred to split plates on which one-half of the root system was exposed to MS+Fe and the other half was exposed to MS−Fe. Samples are described based on the initial 3-d treatment (first symbol) and the side of the split plate the roots were placed on (second symbol). For example, the roots of plants that were plated on MS−Fe for 3 d and then transferred to the MS+Fe side of the split plate are indicated as (−/+).
Figure 2.
Split-root experimental design. Hypothesis 1 (left) shows the gene expression changes of IRT1 and FRO2 at the 6- and 24-h time points, reflecting a systemic (long-distance) signal. Hypothesis 2 (right) shows the gene expression changes of IRT1 and FRO2 at the 6- and 24-h time points, reflecting a local signal.
Tissue was collected at 6 and 24 h after placement on the split plate. These time points were chosen to provide an early point when the shoots are still iron deficient and a late time point when the shoots would have recovered, at least partially, from iron deficiency. The 24-h time period also would potentially allow root systems to acclimate to their local iron environment, if local signaling occurs.
To measure whether the plants had time to perceive iron in the shoots after being moved from –Fe to a split plate, we monitored the level of ferritin mRNA in leaves at the 6- and 24-h time points (Fig. 3). Arabidopsis FER1 levels respond closely to the iron status (Gaymard et al., 1996) of the shoots. Plants that entered the split plate after a period of iron starvation had an 11-fold increase in FER1 expression between the 6- and 24-h time points (P ≤ 0.05 by Student’s t test), indicating that, during this time, iron reached the shoots from the +Fe side and gene expression responded to this new iron-sufficient status (Fig. 3). This result confirms that the times for sampling were appropriate. Interestingly, control plants that entered the split plates without iron starvation, which should not have experienced iron deficiency, also had a significant (P ≤ 0.05 by Student’s t test) rise in FER1 expression between the 6- and 24-h time points, although the magnitude (∼4-fold increase) was smaller. This likely indicates that simply replating on fresh medium results in additional iron uptake, even when plants are not overtly iron deficient at the time of transfer.
Figure 3.
qRT-PCR analysis of the split-root experiment. A, FER1 expression in shoots. Leaf samples were taken from plants at 6 and 24 h after placement on the split-root plates. In this context, +Fe indicates plants that had been grown on normal iron-containing medium prior to being moved to the split plate. –Fe indicates plants that had been grown on –Fe medium for 3 d prior to being moved to the split plate. B, IRT1 root expression after 6 and 24 h on a split plate. C, FRO2 root expression after 6 and 24 h on a split plate. D, FER1 shoot expression. E, IRT1 root expression after 6 and 24 h on a split plate. F, FRO2 root expression after 6 and 24 h on a split plate. For A and D, 6 and 24 h denote the time points at which the shoot was harvested after transfer to the split plate. +Fe indicates that plants experienced iron-replete conditions prior to transfer. –Fe indicates that plants were started on iron-deficient conditions prior to transfer. For B, C, E, and F, +/+ denotes a plant that was started on iron-replete medium and placement of roots on the replete side of the split plate. +/− denotes a plant that was started on iron-replete medium and placement of roots on the iron-deficient side of the split plate. −/+ denotes a plant that was started on iron-deficient medium and placement of roots on the iron-replete side of the split plate. −/− denotes a plant that was started on iron-deficient medium and placement of roots on the iron-deficient side of the split plate. The results reflect three biological replicates. In cases where multiple significantly different levels of expression were observed, these are indicated using lower case letters.
For the control Col-0 plants that were grown with sufficient iron prior to being moved onto the split-root plate, the expression of both IRT1 and FRO2 in the roots was relatively low compared with the levels observed for plants that entered the split plate after a period of iron deficiency (Fig. 3). After 24 h of growth on the split plate, no significant differences between the +Fe and –Fe sides of the plate were observed for these control plants. This indicates that, after this brief, 24-h period on the split plate, local signaling was not observed. In contrast, the expression levels of both IRT1 and FRO2 in plants that had been grown without iron before placement into the split-root system were high at the 6-h time point (∼75× and ∼120× higher than levels in the control, respectively; P ≤ 0.01; Fig. 3). After 24 h, the expression levels of both genes went down on both sides of the plate, as the shoots began to recover from iron deficiency. The extent of this decreased expression was not significantly different on the two sides of the plate. This result indicates that long-distance signals affect gene expression during the recovery from iron deficiency. Overall, these results suggest that long-distance signals originating from the shoots are the predominant factor controlling the expression of iron deficiency-related genes during iron resupply. Our data do not rule out the existence of local signals, which could occur over a longer period.
Iron Localization in ysl1ysl3 Mutants
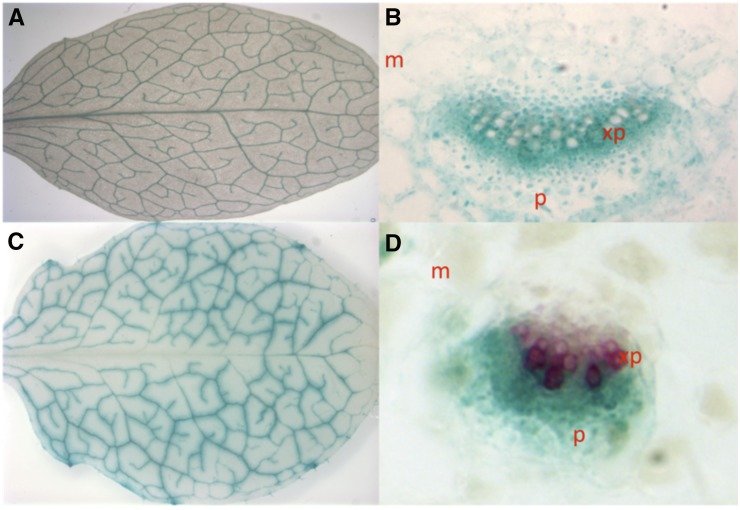
It is difficult to understand why mutations in Fe-NA transporters would result in a defect in iron signaling. We have hypothesized that the function of YSL1 and YSL3 in vegetative tissues is to take up iron into vascular parenchyma cells after it arrives in leaves via the xylem (Waters et al., 2006). This hypothesis was based initially on the vascular parenchyma localization of YSL1 and YSL3 transcription determined using promoter:GUS constructs and on the iron regulation of YSL1 and YSL3, in which iron deficiency represses expression. We have hypothesized that down-regulation of these transporters during iron deficiency would prevent the removal of iron from the vasculature in mature leaves. This could allow the iron to be further translocated to younger, still developing tissues, possibly via xylem-to-phloem exchange. Translatome profiling (Mustroph et al., 2009) indicates that both YSL1 and YSL3 are expressed strongly in leaf phloem companion cells. We analyzed cross sections of the leaves of soil-grown YSL1p::GUS and YSL3p::GUS plants (Waters et al., 2006) and observed that the expression of both genes was very tightly associated with the vasculature in 32-d-old rosette leaves (Fig. 4). YSL1 is expressed in many cell types in and around the vasculature (Fig. 4). YSL1p::GUS expression is strongest in the xylem parenchyma, although expression in the mesophyll and phloem is visible, consistent with the hypothesis that this transporter functions in the unloading of Fe-NA from the xylem into surrounding tissues. YSL3p::GUS expression was seen in the phloem and in parts of the xylem parenchyma (Fig. 4). YSL3p::GUS cross sections were counterstained with phloroglucinol, which stains lignin, to better differentiate between xylem and phloem cells.
Figure 4.
GUS staining of plants with YSL1 (A and B) and YSL3 (C and D) promoters fused to the GUS reporter gene. Mesophyll is indicated as m, and phloem is indicated as p. A, YSL1p::GUS rosette leaf shows YSL1 expression specific to vasculature tissues. B, Cross section of the midrib of YSL1p::GUS rosette leaf reveals darkest staining in the xylem parenchyma (xp). C, YSL3p::GUS rosette leaf shows YSL3 expression specific to the vasculature tissues. D, Cross section of the midrib of YSL3p::GUS rosette leaf reveals staining largely in the phloem. Phloroglucinol is used as a counterstain for lignified tissue.
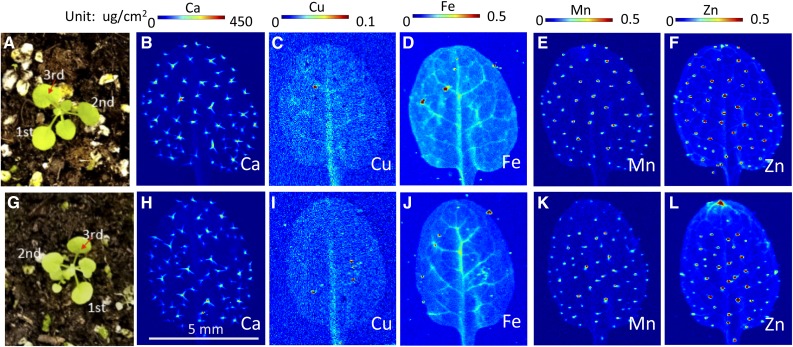
One hypothesis consistent with the localization of YSL1 and YSL3 expression patterns is that ysl1ysl3 DM plants remove iron inefficiently from the veins, causing iron levels in veins to remain relatively high at all times. Many authors have hypothesized that that iron sensing might occur in the vasculature (Maas et al., 1988; García et al., 2013; Zhai et al., 2014), and so, in DM plants, this sensing mechanism would detect normal or even high iron levels. Therefore, the expression of the iron acquisition machinery in roots would be suppressed, owing to the signal of adequate iron that would emanate from shoots. We used synchrotron x-ray fluorescence spectroscopy (SXRF) of leaves to observe patterns of metal accumulation in the leaf veins of wild-type and DM plants (Fig. 5). Plants were grown in potting soil for 18 d, and the third leaf was used for metal analysis (Fig. 5). Chlorosis is not strongly evident in the ysl1ysl3 DM at this stage. Although iron is low in the DM, as expected from bulk measurements (Waters et al., 2006; Chu et al., 2010), iron is concentrated in the veins of the DM plants. The level of iron in the ysl1ysl3 DM veins is at least as high as that of matched wild-type samples, but levels of iron outside of the veins are low compared with the wild type (Fig. 5). The patterns of Ca, Mn, and Zn are similar in wild-type and DM plants; thus, iron seems to be affected differently from the other transition metals, Zn and Mn. The low level of Cu in the DM makes interpretation of the pattern of accumulation for Cu difficult to judge. Similar results were observed in older (4 weeks) plants, where the DM showed more obvious interveinal chlorosis (Supplemental Fig. S1). These results are consistent with the idea that the iron-sensing defect of the ysl1ysl3 DM may be caused by a relative excess of iron in the vasculature.
Figure 5.
Synchrotron x-ray fluorescence microscopy map of elemental distribution in leaves of the wild type and the ysl1ysl3 DM of Arabidopsis. A and G, The third leaf of 18-d-old soil-grown wild-type (A) and ysl1ysl3 (G) plants was used. B to F, Spatial distribution of Ca, Cu, Fe, Mn, and Zn in wild-type leaves. H to L, Spatial distribution of Ca, Cu, Fe, Mn, and Zn in ysl1ysl3 leaves.
We measured the ionome (Salt et al., 2008) using inductively coupled plasma mass spectrometry (ICP-MS) of the phloem of both wild-type and DM plants to make a quantitative assessment of the metals that are contained in phloem sap. One caveat to any experiment in which phloem contents are measured is that the collected phloem samples can be contaminated with contents of cut cells and/or with contents of the cut xylem. To collect phloem exudates, the sample is cut under EDTA to prevent sealing of the sieve plates and consequent blockage of the sieve tubes. Cut samples are moved to fresh EDTA and then recut, and the contents are allowed to bleed into the EDTA for 30 min at 100% humidity in the dark to prevent xylem uptake of the EDTA solution. Most of the contents of the cut cells should be released during this period. The samples are then moved into fresh distilled water to collect phloem exudate for a period of 2 h. The collection is performed in 100% humidity in the dark, again to prevent transpiration as much as possible. As a control to reveal the extent of contamination of the resulting phloem exudate samples, a set of control samples is treated in the same way, except that both cutting steps are performed in water rather than EDTA. In these samples, the phloem will seal, and substances released into solution during the 2-h exudation period will come primarily from cut cells and xylem and, thus, can be taken as the extent of this contamination of the phloem sample. For the majority of measured elements, the total exuded amount of the element in the water-cut control samples was markedly less than that in the phloem exudate samples (Supplemental Fig. S2), indicating that the extent of ion leakage during the exudation period was generally low relative to the extent of bona fide phloem exudation. However, it is important to realize that the phloem samples are not absolutely pure and that, because it is not possible to compare the control and phloem exudate samples after normalization, we can only estimate, in nonnormalized samples, the extent to which contamination from cut cells and/or xylem leaching occurs. For example, the majority (73%) of the boron in the phloem samples is estimated to have derived from the cut cells, indicating that very little boron is in phloem, so even small amounts of boron leaked from other sources containing abundant soluble boron will make a large contribution to the total. In contrast, only about 5.1% of the manganese in the phloem samples came from the cut cells, likely reflecting that only a small amount of soluble manganese is present in the control samples. For the transition metals iron, zinc, and copper, which are most central to this analysis, we estimate, based on the wild-type samples, that 17%, 14%, and 17%, respectively, of the metal measured in phloem will have resulted from contamination from other tissues.
In this analysis, the amount of material collected during the exudation period varies from one plant to another. Because the volume of the exuded fluid is extremely small, even small sample losses due to evaporation during weighing become very significant. Thus, we were not able to reliably measure the volume of exuded liquid in our samples. Instead, we normalized the samples to elements present in large quantities and that are not thought to vary between mutant and wild-type plants. To test which elements should be used for normalization, we calculated the correlation coefficients of all measured elements with all other elements in every sample. When taken across all samples (water-cut control and EDTA cut) or in phloem samples (EDTA cut), the concentrations of K, Ca, and P were highly correlated (R2 > 0.90), suggesting that the concentrations of these elements are strongly related to the amount of exudate and not to differences between genotypes. However, when the correlation coefficients were calculated for the control (water-cut) sample, only a correlation between manganese and calcium (R2 > 0.7) was observed. This likely highlights the very small amounts of material that were present in these control exudations. Since calcium was the best-correlated element for all the samples, we used this element for normalization in the data presented here and note that normalization to potassium gave very similar results.
The amounts of iron, zinc, copper, and manganese in the phloem of ysl1ysl3 plants are not significantly different from the amounts of these elements in wild-type plants (Fig. 6). Thus, it is possible that these normal iron levels in phloem give rise to a signal of iron sufficiency in the shoots.
Figure 6.
Metal analysis of phloem exudates. Normalization for exudate amount is relative to calcium. Error bars indicate se. A minimum of 10 replicates per sample type were performed. WT, Wild type.
Location of the Iron-Sensing Defect
There are two ways that YSL1 and YSL3 could be involved in iron sensing or signaling. YSL1 and YSL3 could be involved in the transmission of signals from the leaves that move to the roots to trigger changes in iron uptake. Alternatively, the signal from the leaves could be generated independently of YSL1 and YSL3, but these proteins could be required in roots to receive the signal and set iron uptake in motion. Thus, we used seedling grafting to address whether YSL1 and YSL3 are required in the roots, in the shoots, or in both for appropriate IDRGE.
To facilitate the scoring of iron-regulated gene expression, we made use of a wild-type Col-0 line that contains a FRO3p-GUS transgene (Mukherjee et al., 2006; kindly provided by Erin Connolly, Pennsylvania State University). The FRO3p-GUS gene is up-regulated by –Fe in both shoots and roots (Jeong and Connolly, 2009), giving a convenient read out of IDRGE in the wild-type tissue used in the grafting experiments. Four types of grafts were performed: FRO3p-GUS shoot::wild-type root (F3G/WT), FRO3p-GUS shoot::ysl1ysl3 root (F3G/ysl1ysl3), FRO3p-GUS shoot::FRO3p-GUS root (F3G/F3G), and ysl1ysl3 shoot::FRO3p-GUS root (ysl1ysl3/F3G). Self-grafts of ysl1ysl3 were not made, since no reporter gene would be present in either scion or stock. Moreover, self grafts of ysl1ysl3 seedlings were very difficult to obtain because graft establishment was very low. After the graft junction had formed, the plants were moved either to normal MS (containing iron; +Fe) or to MS prepared without iron (–Fe) for a period of 72 h. The plants were visually inspected for the presence of adventitious roots during the graft-healing process (1 week), and any adventitious roots that began to form were immediately excised. The plants were then stained for GUS activity and again visually inspected for the presence of adventitious roots. Samples with adventitious roots were not used in the analysis. The results are given in Table IV, and the typical appearance of the GUS-stained plants is shown in Supplemental Figure S3. No blue color was observed in any FRO3p-GUS wild-type tissue in any graft growing on +Fe, indicating that the promoter, as expected, is not active in +Fe conditions. When grown on –Fe, FRO3p-GUS-containing tissues (root, shoot, or both) of self-grafted wild-type plants stained blue, as expected, in the majority of the grafted plants (Table IV). When FRO3p-GUS wild-type shoots were grafted to ysl1ysl3 roots, blue color was observed in the shoots of seven of the nine successfully grafted plants, similar to the results from grafting of FRO3p-GUS onto wild-type roots. This suggests that YSL1 and YSL3 are not required in the root for correct iron deficiency-induced gene expression in the shoot. In contrast, FRO3p-GUS wild-type roots that were grafted to ysl1ysl3 shoots were never blue (11 out of 11 successful grafts), while eight of nine FRO3p-GUS self-grafted plants had blue roots. This indicates that ysl1ysl3 shoots are incapable of sending the signal necessary for IDRGE in roots.
Table IV. Results from seedling grafting experiments.
| Graft Shoot/Root | +Fe Shoots |
+Fe Roots |
–Fe Shoots |
–Fe Roots |
||||
|---|---|---|---|---|---|---|---|---|
| No. GUS Positive | No. GUS Negative | No. GUS Positive | No. GUS Negative | No. GUS Positive | No. GUS Negative | No. GUS Positive | No. GUS Negative | |
| F3G/WT | 0 | 5 | 0 | 5 | 4 | 2 | 0 | 4 |
| F3G/ysl1ysl3 | 0 | 4 | 0 | 4 | 7 | 2 | 0 | 9 |
| F3G/F3G | 0 | 3 | 0 | 3 | 8 | 1 | 8 | 1 |
| ysl1ysl3/F3G | 0 | 8 | 0 | 8 | 0 | 11 | 0 | 11 |
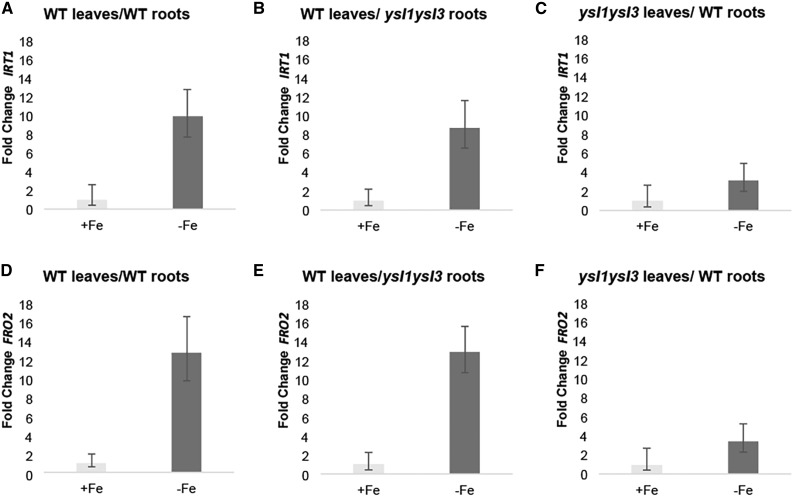
We were unable to generate ysl1ysl3 plants containing the FRO3p-GUS transgene; thus, this experiment could not conclusively tell us whether ysl1ysl3 roots are capable of perceiving a signal of iron deficiency from shoots. To test this, qRT-PCR was performed to test the expression of IRT1 and FRO2 in the roots from wild-type/wild-type, wild-type/ysl1ysl3, and ysl1ysl3/wild-type grafts (Fig. 7). As in the previous grafting experiment, plants were grown for 3 d on either +Fe or –Fe medium, then RNA was prepared from roots, and qRT-PCR was performed. For the wild-type/wild-type grafts, mean IRT1 and FRO2 expression were 9.9- and 12.7-fold higher, respectively, on –Fe medium (RQmin-RQmax = 7.7–12.8 for IRT1 and 9.7–16.6 for FRO2, P = 0.003 and P = 0.001. RQmin and RQmax define the minimum and maximum change in expression level based on 3 standard deviations.). For ysl1ysl3/wild-type grafts, the up-regulation of IRT1 and FRO2 was muted, with mean IRT1 and FRO2 expression 3.1- and 3.4-fold higher, respectively, on –Fe medium (RQmin-RQmax = 2–5 for IRT1 and 2.2–5.3 for FRO2). The difference between expression on +Fe and –Fe was not statistically significant by Welch’s t test (P = 0.07 and P = 0.08 for IRT1 and FRO2). This is consistent with the idea that IDRGE in roots depends on YSL1 and YSL3 in shoots. In the grafts of wild-type shoots onto ysl1ysl3 roots, the expression of IRT1 and FRO2 was up-regulated normally, 8.7- and 12.9-fold higher, respectively, on –Fe medium (RQmin-RQmax = 6.5–11.7 for IRT1 and 10.7–15.6 for FRO2, P = 0.003 and P = 0.004). This demonstrates that roots lacking YSL1 and YSL3 are fully capable of perceiving the shoot signals that control IDRGE.
Figure 7.
qRT-PCR analysis of grafted plants. Samples were prepared from roots of the indicated graft combinations that were transferred to +Fe (control) and –Fe conditions for 72 h. Fold change in expression relative to the +Fe control for each graft type is shown. Error bars indicate 95% confidence intervals. Graft combinations are expressed as shoot/root. A, IRT1 expression in roots of wild-type (WT)/wild-type grafts. B, IRT1 expression in roots of wild-type/ysl1ysl3 grafts. C, IRT1 expression in roots of ysl1ysl3/wild-type grafts. D, FRO2 expression in roots of wild-type/wild-type grafts. E, FRO2 expression in roots of wild-type/ysl1ysl3 grafts. F, FRO2 expression in roots of ysl1ysl3/wild-type grafts.
DISCUSSION
Reinterpretation of the Phenotype of ysl1ysl3 DM Plants
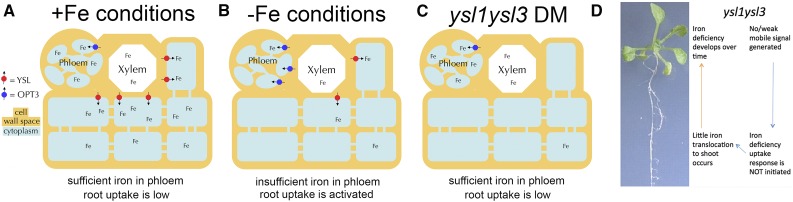
YSL1 and YSL3 are Fe(II)-NA transporters (Chu et al., 2010), and ysl1ysl3 DM plants have low levels of foliar iron relative to wild-type plants (Waters et al., 2006). However, the iron in the leaves of the DM is located near or in the veins (Fig. 4), indicating that loss of Fe-NA transport causes iron to be retained in vascular tissues, with nonvascular tissues having lower iron. Using ICP-MS, the level of iron in the phloem sap was found to be indistinguishable from the level of phloem iron in wild-type plants. Here, we showed that ysl1ysl3 DM plants exhibit a pervasive inability to regulate gene expression in response to iron deficiency (Fig. 3; Table I), and the loss of YSL1 and YSL3 function in shoots prevents normal iron deficiency responses from occurring in roots (Table II). This new feature of the ysl1ysl3 phenotype suggests that the decreased iron levels and leaf chlorosis of the DM may not be entirely due to the loss of iron-NA transport but, instead, may be the result of inadequate root iron uptake resulting from failed long-distance iron signaling (Fig. 8). In wild-type plants, a rheostatic control presumably exists so that iron deficiency signals are sent and stopped repeatedly throughout development as the iron status of the leaf undergoes mild oscillations. In the ysl1ysl3 mutant, permanent basal-level iron uptake that cannot respond to changing conditions in the leaves leads gradually to iron deficiency (Fig. 8). Consistent with a gradual worsening of iron deficiency in ysl1ysl3 plants, chlorosis develops gradually over the first 3 weeks of growth, becoming severe at the vegetative-to-reproductive transition when the demand for iron in aboveground parts peaks owing to rapid increases in tissue mass.
Figure 8.
Model to explain the role of YSL1 and YSL3 in iron signaling. A, In wild-type plants grown in iron-sufficient conditions, YSLs function to move Fe from veins into surrounding parenchyma cells. The iron can then move in the symplasm to supply cells in the leaf lamina. A sufficient level of iron in the phloem causes Fe sufficiency to be sensed. B, In wild-type plants under Fe deficiency, OPT3 expression is activated and YSL1 and YSL3 expression is repressed. Low levels of Fe in phloem cause Fe insufficiency to be sensed. C, In ysl1ysl3 DM plants, iron arrives via xylem but is not taken up into surrounding tissues. Xylem-to-phloem transfer of Fe, mediated by OPT3, causes the phloem levels of Fe to remain relatively high. The resulting sufficient level of Fe in the phloem causes Fe sufficiency to be sensed. D. A model to explain the ysl1ysl3 phenotype. Because iron deficiency is never signaled to the roots, iron deficiency uptake response is always muted. This results in little iron accumulation in the shoots. Over time, iron deficiency results.
Local and Long-Distance Signaling of Iron Status
The existence of a long-distance signal of iron status that is sent from shoot to root is well supported by reports that foliar application of iron to the shoots of iron-deficient plants causes a decrease in the iron deficiency responses of roots (Maas et al., 1988; Enomoto et al., 2007; García et al., 2013), by the observation of mutants in which the genotype of the shoot controls iron uptake at the root (Grusak and Pezeshgi, 1996), and by split-root experiments (Romera et al., 1992).The split-root system also has been used previously to understand the subtle dynamics of IRT1 and FRO2 expression that depend on the local root environment (Vert et al., 2003). Vert et al. (2003) showed convincingly that, in roots treated with bipyridyl so as to contain virtually no residual (apoplast) iron, the expression of IRT1 and FRO2 was low, indicating that the local root environment affects the expression of these genes. In our study, plants entered the split-root system after a period of iron deficiency, so that we could specifically test whether the roots on the –Fe side of the plate would respond to a long-distance signal from shoots. In this experiment, iron-deficient plants become iron sufficient over a 24-h period, and during that time, the shoot shifts from communicating iron deficiency to communicating iron sufficiency. After only 24 h of iron resupply through the +Fe side of the split plate, previously iron-starved plants decreased steady-state mRNA levels for IRT1 and FRO2 by ∼10× to 20×. This decrease occurred on both the –Fe and +Fe sides of the plate.
Mutants Impaired in Long-Distance Signaling of Iron Status
The grafting experiments presented here (Fig. 7; Table IV) clearly indicate that ysl1ysl3 DM plants are impaired in long-distance signaling of iron status. Since YSL1 and YSL3 are metal-NA transporters, potential roles for NA in this process were considered. It is notable that mutants lacking NA due to the loss of nicotianamine synthase (NAS) are able to signal iron deficiency; in fact, they constitutively signal iron deficiency and accumulate excess iron in aboveground organs (Stephan and Scholz, 1993; Schuler et al., 2012). Plants lacking NA include the tomato (Solanum lycopersicum) chloronerva mutant (Herbik et al., 1999; Higuchi et al., 1999; Ling et al., 1999) and the Arabidopsis quadruple NAS nas4X mutant (Klatte et al., 2009). Both of these mutants can accumulate iron in phloem, but there are problems in unloading it from the phloem (Stephan and Scholz, 1993; Schuler et al., 2012). IDRGE in these mutants can be repressed by external iron application to leaves (García et al., 2013). Thus, NA is not likely to be involved directly in shoot-to-root signaling.
Other mutants have been implicated in long-distance iron signaling, including opt3-2 mutants, which constitutively activate IDRGE and have high iron levels in shoot tissue (Stacey et al., 2008), thus exhibiting a phenotype that is the exact opposite of the ysl1ysl3 DM. OPT3 expression in leaves is induced strongly by iron deficiency, while the expression of YSL1 and YSL3 is repressed (Waters et al., 2006; Stacey et al., 2008). Both localized expression experiments (Mendoza-Cózatl et al., 2014) and grafting experiments (Zhai et al., 2014) indicate that OPT3 activity in shoots is needed for correct long-distance iron signaling to roots, again paralleling the effect in the ysl1ysl3 DM, which also is impaired, albeit in an opposite way, in long-distance iron signaling. OPT3 has iron transport activity, and its likely role is to import iron into phloem companion cells (Zhai et al., 2014). YSL1 and YSL3 likely help to move iron away from the veins for distribution to the leaf lamina, thus explaining why the ysl1ysl3 DM plants accumulate iron in veins while having overall low levels of iron in leaves (Fig. 8). We speculate that iron in the veins of ysl1ysl3 plants could be redistributed into the phloem, likely by OPT3, and the resulting normal levels of iron in the phloem trigger the signal that leaves contain adequate iron in ysl1ysl3 DM plants.
MATERIALS AND METHODS
Growth Conditions
For soil growth, Arabidopsis (Arabidopsis thaliana) seeds were placed in sterile Eppendorf tubes (1.5 mL) containing 1 to 1.5 mL of distilled water and allowed to imbibe for 3 to 5 d at 4°C. Seeds were planted directly onto Promix potting soil (Fafard Canadian Growing Mix 2) pretreated with Gnatrol (Valent Bioscience). Growth chamber conditions for soil-grown plants were 16 h of light and 8 h of dark at 24°C.
For growth in sterile culture, seeds were sterilized and plated directly on sterile petri plates containing 1× MS+Fe (prepared with 10× Macronutrient Solution and 10× Micronutrient Solution from Phytotechnologies) and placed at 4°C for 3 to 5 d. MS−Fe was prepared using Phytotechnologies 10× Macronutrients and then supplemented with 6.2 mg L−1 boric acid, 0.025 mg L−1 cobalt chloride∙6H2O, 0.025 mg L−1 cupric sulfate∙5H2O, 16.9 mg L−1 manganese sulfate∙H2O, 0.25 mg L−1 molybdic acid (sodium salt)∙2H2O, 0.83 mg L−1 potassium iodide, and 8.6 mg L−1 zinc sulfate∙7H2O. Growth chamber conditions for plate-grown plants were 16 h of light and 8 h of dark at 24°C.
Split-Root Experiments
Plants were grown for 10 d on sterile petri plates containing 1× MS. On day 10, the root tips were excised to promote lateral root formation and plants were allowed to grow for an additional 10 d. Plants were then transferred to either 1× MS−Fe or to normally prepared MS+Fe (containing 100 μm FeSO4 EDTA) for 3 d. Sterile split petri plates were prepared so that one-half of the plate contained MS−Fe and the other half contained MS+Fe. Plants were transferred to split plates so that each half of the root system was exposed to different conditions. Root and shoot tissues were harvested separately at 6 and 24 h after placement on the split plate and stored at −80°C until subsequent analyses could be performed.
Grafting
For grafting, plants were grown on one-half-strength (1/2×) MS with 12 h of light at 22°C and 12 h of dark at 15°C. Plates were placed in a vertical position to encourage straight root growth. Seedlings were grown under these conditions for 5 to 7 d before grafting. Using a small, angled scalpel (Micro Knife-Angled 150/13 cm; Fine Science Tools), scion and rootstock were severed by making a straight horizontal cut across the hypocotyl. The grafts were formed by inserting scion and stock into a small piece of silicone tube (0.012 inch [0.3 mm] i.d. × 0.025 inch [0.64 mm]) that was rinsed in ethanol and then sterile distilled water. To reduce adventitious root formation, plants were moved to 27°C for 2 to 3 d (16 h of light and 8 h of dark) after grafting, before being returned to normal temperatures and 12 h of light. Each grafted plant was inspected for adventitious root formation 3 to 4 d after grafting. An example of a successful graft can be seen in Supplemental Figure S3. Any adventitious roots were removed with a sharp scalpel and fine forceps. Visual inspection for adventitious roots was continued every 24 to 48 h until grafts were successfully formed, typically within 5 to 7 d. Successfully grafted plants were transferred to sterile petri plates containing 1/2× MS+Fe or 1/2× MS−Fe. Plants were maintained in growth chambers at 24°C (16 h of light and 8 h of dark) for 72 h and then stained for GUS activity or harvested for RNA isolation.
GUS Histochemical Staining
Tissues were prefixed in ice-cold 10% (v/v) acetone for 10 min prior to placement on either six- or 24-well polystyrene tissue culture plates. Samples were covered with GUS buffer [0.1 m NaPO4, pH 7.2, 0.5 mm K3Fe(CN6), 10 mm EDTA, 0.01% (v/v) Triton X-100, and 0.5 mg mL−1 5-bromo-4-chloro-3-indolyl-β-glucuronic acid] and placed at 37°C for 2 to 24 h. If tissues contained chlorophyll, they were soaked in 70% (v/v) ethanol and then washed repeatedly in 70% (v/v) to 95% (v/v) ethanol until samples were clear.
RNA Isolation
Root and shoot tissues were ground in 1.5-mL Eppendorf tubes using a tissue lyser (Retsch) and 3.2-mm Chrome Steel Beads (454 g; BioSpec Products). Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen), followed by DNase I treatment (Ambion). Total RNA concentrations were quantified using a NanoDrop (Thermo Scientific). RNA integrity was evaluated using a 1× TAE (40 mm Tris-acetate, 1 mm EDTA) gel stained with ethidium bromide.
cDNA Synthesis and Reverse Transcriptase-PCR
First-strand cDNA was synthesized from 480 ng of total RNA with SuperScript III reverse transcriptase (Invitrogen) using Oligo-dT primers. Equal amounts of the reverse transcriptase reaction were used as templates in 25-μL PCRs. Primers used to test cDNA quality were AtAct2-forward (5′-GCTGAGAGATTCAGATGCCCA-3′) and AtAct2-reverse (5′-GTGGATTCCAGCAGCTTCCAT-3′). PCRs were performed using Ex-Taq polymerase (Takara) under these conditions: initial denaturing step at 94°C for 1 min, 30 cycles of 94°C for 30 s, 57°C for 45 s, and 72°C for 1 min, an elongation step at 72°C for 10 min, and 16°C for 5 min.
Expression levels were measured in reference to the housekeeping gene AtAct2. Primer efficiencies were calculated empirically by amplifying from serial dilutions of a sample cDNA with each primer set. The primers used in this study were, for AtAct2, AtAct2-forward (5′-GCTGAGAGATTCAGATGCCCA-3′) and AtAct2-reverse (5′-GTGGATTCCAGCAGCTTCCAT-3′), for AtFRO2, AtFRO2-forward (5′-GCTTCCGCCGATTTCTTAAGGC-3′) and AtFRO2-reverse (5′-AACGGAGTTATCCCGCTTCCTC-3′), for AtIRT1, AtIRT1-forward (5′-ACTTCAAACTGCGCCGGAAGAATG-3′) and AtIRT1-reverse (5′-AGCTTTGTTGACGCACGGTTC-3′), and for AtFER1, AtFER1-forward (5′-CAACGTTGCTATGAAGGGACTAGC-3′) and AtFER1-reverse (5′-ACTCTTCCTCCTCTTTGGTTCTGG-3′).
Brilliant 2× SYBR Green Master Mix (Stratagene) was used to carry out qRT-PCR. A two-step thermal cycling profile was used with the following parameters: initial denaturing for 15 min at 95°C, and 40 cycles of 10 s at 95°C and 30 s at 60°C. After amplification with the two-step profile, a melting curve was used to ensure that a single amplification product had been formed. All qRT-PCRs were carried out with three replicates. The final threshold values (Ct) were calculated as the average of these three replicates. The comparative ΔCt method was used to analyze the quantities of each amplified product by comparing the Ct values of the sample of interest with the Ct value of a housekeeping gene under the same condition. Primer efficiencies were taken into account using the geNorm algorithm (Vandesompele et al., 2002). A negative control using water instead of cDNA was used for each quantitative PCR set.
Microarray Analysis
Plants were grown on sterile petri plates containing 1× MS. Plates were placed in an upright position so that the roots grew along the surface rather than inside the agar, which allowed for easy transfer. Incubator conditions for plate-grown plants were 16 h of light and 8 h of dark at 22°C. After 10 d, plants were moved either to fresh iron-containing plates (+Fe samples) or to plates prepared without iron (–Fe samples). Growth was continued for 3 d before samples were harvested. Separate shoot and root samples were collected from three independent biological replicates of wild-type plants grown with normal iron (wild-type +Fe), wild-type plants grown without iron for 3 d (wild-type –Fe), and ysl1ysl3 DM plants grown with iron. Chlorophyll measurements were made at the time of harvest to ensure that the levels of iron deficiency chlorosis were equal in the leaves of the ysl1ysl3 DM samples and the leaves of the wild-type –Fe samples. RNA was prepared using the Qiagen RNeasy kit according to the manufacturer’s instructions. Biotinylated cRNA was prepared according to the Affymetrix 3′ IVT Express Kit Manual from 500 ng of total RNA. Following fragmentation, 10 μg of cRNA was hybridized for 16 h at 45°C on a GeneChip Arabidopsis Genome Array. GeneChips were washed and stained in the Affymetrix Fluidics Station 450. GeneChips were scanned using the Affymetrix GeneChip 3000 7G Scanner. The data were analyzed with Microarray Suite version 5.0 (MAS 5.0) using Affymetrix default analysis settings and global scaling as the normalization method. The trimmed mean target intensity of each array was arbitrarily set to 250. Differential expression between pairs of samples was determined using Bioconductor Limma (Gentleman et al., 2004). All microarray data have been deposited at NCBI GEO (accession no. GSE92716).
Phloem Exudate Collection
Sterile seeds were germinated on 1/2× MS. Plates were positioned vertically so that the roots grew along the surface rather than inside the agar, which allowed for easy transfer. Growth chamber conditions for plates were 16 h of light and 8 h of dark at 22°C. After the plants made the second set of true leaves, they were transferred to sterile petri plates containing 1/2 MS+Fe or 1/2 MS−Fe for 3 d. Whole rosettes were harvested after 3 d from plants on 1/2 MS+Fe and those on 1/2 MS−Fe by making a cut below the hypocotyls in the uppermost part of the root. Keeping the stem submerged in 5 mm EDTA, pH 6, a second cut was made in the hypocotyl region, approximately 5 mm above the first cut. Each rosette (after the second cut) was arranged on a 96-well plate so that the stem was submerged in 100 µL of 5 mm EDTA solution, and the plate was immediately placed into a humidity chamber to minimize the amount of EDTA drawn into the leaves by transpiration and xylem transport. After 20 min of incubation in dark, the cut stems were submerged in 100 µL of sterile distilled, deionized water on a fresh 96-well plate and incubated in a dark humid chamber for 2 h. The exudate solution was immediately transferred into labeled microcentrifuge tubes, frozen in liquid nitrogen, and stored at −80C until use.
Exudate Treatment of Roots
FRO3p-GUS plants (FRO3p-GUS reporter construct in the Col-0 wild-type background) were grown for 2 weeks on 1/2 MS+Fe, and then roots were excised using a scalpel. About five roots were placed per well of a 12-well microtiter plate. The wells were filled with 1 mL of either distilled, deionized water (negative control) or 1 mL of phloem exudate collected as described. The plates were incubated in the dark for 48 h, and then GUS staining was performed on the roots.
SXRF
Seeds of wild-type and ysl1ysl3 mutant plants were stratified overnight at 4°C in water prior to sowing in soil, type Lambert 111. After germination, plants were fertilized every week with the standard NPK fertilizer. The first true leaf and the second leaf from the top were detached immediately prior to analysis, placed in the wet chamber made between two layers of metal-free Kapton film, and mounted onto 35-mm slide mounts. The spatial distribution of iron in hydrated leaf tissues was imaged via SXRF at the F3 station, a bending magnet beamline with multilayer monochromator, at the Cornell High Energy Synchrotron Source. The 2D iron raster maps were acquired at 40-µm resolution, with a 0.3 s pixel−1 dwell time, using a focused, monochromatic incident x-ray beam at 13.2 keV and photon flux of approximately 3 × 1010 photons s−1. These settings did not cause damage to plant tissues within the 4- to 6-h scans required for the analysis of the full set of genotypes. Element-specific x-ray fluorescence was detected using a Vortex ME-4 Silicon Drift Detector positioned 90° to the incident beam. Samples were positioned 45° to both the incident beam and the Silicon Drift Detector. Calibration of x-ray fluorescence spectroscopy equipment was done using a uniform thin metal film standard during each experiment. Absolute concentration maps of trace elements were processed with PyMca [1] through a GUI designated Praxes, the software wrapper developed at the Cornell High Energy Synchrotron Source.
Accession Numbers
Microarray data have been deposited at NCBI GEO under accession number GSE92716.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Synchrotron x-ray fluorescence microscopy-based imaging of the spatial distribution of indicated elements in leaves of the wild type and the ysl1ysl3 DM of Arabidopsis.
Supplemental Figure S2. Comparison of total metals in control and phloem exudations.
Supplemental Figure S3. Typical grafting results.
Supplemental Data S1. Microarray data.
Supplemental Data S2. Gene functional classification tool analysis of iron-regulated genes.
Footnotes
This work was supported by grants to E.L.W. (NSF IOS-1441450 and NSF IOS-153980). Work in the O.K.V. lab is supported by NIFA/USDA Hatch under 2014-15-151. CHESS is supported by the NSF and NIH/NIGMS via NSF award DMR-1332208.
Articles can be viewed without a subscription.
References
- Anderegg G, Ripperger H (1989) Correlation between metal complex formation and biological activity of nicotianamine analogues. J Chem Soc Chem Commun 10: 647–650 [Google Scholar]
- Bauer P, Ling HQ, Guerinot ML (2007) FIT, the FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR in Arabidopsis. Plant Physiol Biochem 45: 260–261 [DOI] [PubMed] [Google Scholar]
- Bienfait HF, de Weger LA, Kramer D (1987) Control of the development of iron-efficiency reactions in potato as a response to iron deficiency is located in the roots. Plant Physiol 83: 244–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat JF, Dubos C, Gaymard F (2015) Iron nutrition, biomass production, and plant product quality. Trends Plant Sci 20: 33–40 [DOI] [PubMed] [Google Scholar]
- Chu HH, Chiecko J, Punshon T, Lanzirotti A, Lahner B, Salt DE, Walker EL (2010) Successful reproduction requires the function of Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 metal-nicotianamine transporters in both vegetative and reproductive structures. Plant Physiol 154: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohu CM, Pilon M (2007) Regulation of superoxide dismutase expression by copper availability. Physiol Plant 129: 747–755 [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Campbell NH, Grotz N, Prichard CL, Guerinot ML (2003) Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol 133: 1102–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato RJ Jr, Roberts LA, Sanderson T, Eisley RB, Walker EL (2004) Arabidopsis Yellow Stripe-Like2 (YSL2): a metal-regulated gene encoding a plasma membrane transporter of nicotianamine-metal complexes. Plant J 39: 403–414 [DOI] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto Y, Hodoshima H, Shimada H, Shoji K, Yoshihara T, Goto F (2007) Long-distance signals positively regulate the expression of iron uptake genes in tobacco roots. Planta 227: 81–89 [DOI] [PubMed] [Google Scholar]
- Fourcroy P, Sisó-Terraza P, Sudre D, Savirón M, Reyt G, Gaymard F, Abadía A, Abadia J, Alvarez-Fernández A, Briat JF (2014) Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol 201: 155–167 [DOI] [PubMed] [Google Scholar]
- Fourcroy P, Tissot N, Gaymard F, Briat JF, Dubos C (2016) Facilitated Fe nutrition by phenolic compounds excreted by the Arabidopsis ABCG37/PDR9 transporter requires the IRT1/FRO2 high-affinity root Fe2+ transport system. Mol Plant 9: 485–488 [DOI] [PubMed] [Google Scholar]
- García MJ, Romera FJ, Stacey MG, Stacey G, Villar E, Alcántara E, Pérez-Vicente R (2013) Shoot to root communication is necessary to control the expression of iron-acquisition genes in strategy I plants. Planta 237: 65–75 [DOI] [PubMed] [Google Scholar]
- Gaymard F, Boucherez J, Briat JF (1996) Characterization of a ferritin mRNA from Arabidopsis thaliana accumulated in response to iron through an oxidative pathway independent of abscisic acid. Biochem J 318: 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendre D, Czernic P, Conéjéro G, Pianelli K, Briat JF, Lebrun M, Mari S (2007) TcYSL3, a member of the YSL gene family from the hyper-accumulator Thlaspi caerulescens, encodes a nicotianamine-Ni/Fe transporter. Plant J 49: 1–15 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusak MAP, Pezeshgi S (1996) Shoot-to-root signal transmission regulates root Fe(III) reductase activity in the dgl mutant of pea. Plant Physiol 110: 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon MJ, Kawachi M, Wirtz M, Hillmer S, Hell R, Krämer U (2012) Vacuolar nicotianamine has critical and distinct roles under iron deficiency and for zinc sequestration in Arabidopsis. Plant Cell 24: 724–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Jásik J, Klein M, Martinoia E, Feller U, Schell J, Pais MS, Koncz C (2002) Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Mol Biol 50: 587–597 [DOI] [PubMed] [Google Scholar]
- Herbik A, Koch G, Mock HP, Dushkov D, Czihal A, Thielmann J, Stephan UW, Bäumlein H (1999) Isolation, characterization and cDNA cloning of nicotianamine synthase from barley: a key enzyme for iron homeostasis in plants. Eur J Biochem 265: 231–239 [DOI] [PubMed] [Google Scholar]
- Higuchi K, Suzuki K, Nakanishi H, Yamaguchi H, Nishizawa NK, Mori S (1999) Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol 119: 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P (2004) FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett 577: 528–534 [DOI] [PubMed] [Google Scholar]
- Jeong J, Connolly EL (2009) Iron uptake mechanisms in plants: functions of the FRO family of ferric reductases. Plant Sci 176: 709–714 [Google Scholar]
- Jeong J, Guerinot ML (2009) Homing in on iron homeostasis in plants. Trends Plant Sci 14: 280–285 [DOI] [PubMed] [Google Scholar]
- Kampfenkel K, Van Montagu M, Inze D (1995) Effects of iron excess on Nicotiana plumbaginifolia plants: implications to oxidative stress. Plant Physiol 107: 725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatte M, Schuler M, Wirtz M, Fink-Straube C, Hell R, Bauer P (2009) The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiol 150: 257–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike S, Inoue H, Mizuno D, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2004) OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J 39: 415–424 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang H, Ai Q, Liang G, Yu D (2016) Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiol 170: 2478–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Koch G, Bäumlein H, Ganal MW (1999) Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proc Natl Acad Sci USA 96: 7098–7103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas FM, van de Wetering DAM, van Beusichem ML, Bienfait HF (1988) Characterization of phloem iron and its possible role in the regulation of Fe-efficiency reactions. Plant Physiol 87: 167–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai HJ, Pateyron S, Bauer P (2016) Iron homeostasis in Arabidopsis thaliana: transcriptomic analyses reveal novel FIT-regulated genes, iron deficiency marker genes and functional gene networks. BMC Plant Biol 16: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Cózatl DG, Xie Q, Akmakjian GZ, Jobe TO, Patel A, Stacey MG, Song L, Demoin DW, Jurisson SS, Stacey G, et al. (2014) OPT3 is a component of the iron-signaling network between leaves and roots and misregulation of OPT3 leads to an over-accumulation of cadmium in seeds. Mol Plant 7: 1455–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J, Baxter IR, Lee J, Li L, Lahner B, Grotz N, Kaplan J, Salt DE, Guerinot ML (2009) The ferroportin metal efflux proteins function in iron and cobalt homeostasis in Arabidopsis. Plant Cell 21: 3326–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J, Guerinot ML (2009) Iron uptake and transport in plants: the good, the bad, and the ionome. Chem Rev 109: 4553–4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee I, Campbell NH, Ash JS, Connolly EL (2006) Expression profiling of the Arabidopsis ferric chelate reductase (FRO) gene family reveals differential regulation by iron and copper. Planta 223: 1178–1190 [DOI] [PubMed] [Google Scholar]
- Murata Y, Ma JF, Yamaji N, Ueno D, Nomoto K, Iwashita T (2006) A specific transporter for iron(III)-phytosiderophore in barley roots. Plant J 46: 563–572 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CM, Hindt MN, Schmidt H, Clemens S, Guerinot ML (2013) MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet 9: e1003953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit JM, Briat JF, Lobréaux S (2001) Structure and differential expression of the four members of the Arabidopsis thaliana ferritin gene family. Biochem J 359: 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts LA, Pierson AJ, Panaviene Z, Walker EL (2004) Yellow stripe1: expanded roles for the maize iron-phytosiderophore transporter. Plant Physiol 135: 112–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Lin WD, Fu GM, Abadía J, López-Millán AF, Schmidt W (2013) Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by Arabidopsis and Medicago truncatula. Plant Physiol 162: 1473–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romera FJ, Alcantara E, Delaguardia MD (1992) Role of roots and shoots in the regulation of the Fe efficiency responses in sunflower and cucumber. Physiol Plant 85: 141–146 [Google Scholar]
- Salt DE, Baxter I, Lahner B (2008) Ionomics and the study of the plant ionome. Annu Rev Plant Biol 59: 709–733 [DOI] [PubMed] [Google Scholar]
- Schaaf G, Honsbein A, Meda AR, Kirchner S, Wipf D, von Wirén N (2006) AtIREG2 encodes a tonoplast transport protein involved in iron-dependent nickel detoxification in Arabidopsis thaliana roots. J Biol Chem 281: 25532–25540 [DOI] [PubMed] [Google Scholar]
- Schaaf G, Ludewig U, Erenoglu BE, Mori S, Kitahara T, von Wirén N (2004) ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. J Biol Chem 279: 9091–9096 [DOI] [PubMed] [Google Scholar]
- Schmid NB, Giehl RFH, Döll S, Mock HP, Strehmel N, Scheel D, Kong X, Hider RC, von Wirén N (2014) Feruloyl-CoA 6′-Hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiol 164: 160–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M, Rellán-Álvarez R, Fink-Straube C, Abadía J, Bauer P (2012) Nicotianamine functions in the phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. Plant Cell 24: 2380–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selote D, Samira R, Matthiadis A, Gillikin JW, Long TA (2015) Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol 167: 273–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisó-Terraza P, Luis-Villarroya A, Fourcroy P, Briat JF, Abadía A, Gaymard F, Abadía J, Álvarez-Fernández A (2016) Accumulation and secretion of coumarinolignans and other coumarins in Arabidopsis thaliana roots in response to iron deficiency at high pH. Front Plant Sci 7: 1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, Koh S, Becker J, Stacey G (2002) AtOPT3, a member of the oligopeptide transporter family, is essential for embryo development in Arabidopsis. Plant Cell 14: 2799–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, Patel A, McClain WE, Mathieu M, Remley M, Rogers EE, Gassmann W, Blevins DG, Stacey G (2008) The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol 146: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan UW, Scholz G (1993) Nicotianamine: mediator of transport of iron and heavy-metals in the phloem. Physiol Plant 88: 522–529 [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D (2002) The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J 31: 589–599 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert GA, Briat JF, Curie C (2003) Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiol 132: 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wiren N, Klair S, Bansal S, Briat JF, Khodr H, Shioiri T, Leigh RA, Hider RC (1999) Nicotianamine chelates both FeIII and FeII: implications for metal transport in plants. Plant Physiol 119: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EL, Connolly EL (2008) Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Curr Opin Plant Biol 11: 530–535 [DOI] [PubMed] [Google Scholar]
- Wang HY, Klatte M, Jakoby M, Bäumlein H, Weisshaar B, Bauer P (2007) Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226: 897–908 [DOI] [PubMed] [Google Scholar]
- Waters BM, Chu HH, Didonato RJ, Roberts LA, Eisley RB, Lahner B, Salt DE, Walker EL (2006) Mutations in Arabidopsis yellow stripe-like1 and yellow stripe-like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol 141: 1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan JY, Li CX, Sun L, Ren JY, Li GX, Ding ZJ, Zheng SJ (2016) A WRKY transcription factor regulates Fe translocation under Fe deficiency. Plant Physiol 171: 2017–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ (2005) AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res 15: 613–621 [DOI] [PubMed] [Google Scholar]
- Zhai Z, Gayomba SR, Jung HI, Vimalakumari NK, Piñeros M, Craft E, Rutzke MA, Danku J, Lahner B, Punshon T, et al. (2014) OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and cadmium in Arabidopsis. Plant Cell 26: 2249–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J, Schmidt S, Strehmel N, Scheel D, Abel S (2017) Arabidopsis transporter ABCG37/PDR9 contributes primarily highly oxygenated coumarins to root exudation. Sci Rep 7: 3704. [DOI] [PMC free article] [PubMed] [Google Scholar]