Down-regulation of CAD1 in poplar leads to different metabolic routes for coniferaldehyde and sinapaldehyde and alters lignin amount and structure, improving the physicochemical properties of wood for saccharification.
Abstract
In the search for renewable energy sources, genetic engineering is a promising strategy to improve plant cell wall composition for biofuel and bioproducts generation. Lignin is a major factor determining saccharification efficiency and, therefore, is a prime target to engineer. Here, lignin content and composition were modified in poplar (Populus tremula × Populus alba) by specifically down-regulating CINNAMYL ALCOHOL DEHYDROGENASE1 (CAD1) by a hairpin-RNA-mediated silencing approach, which resulted in only 5% residual CAD1 transcript abundance. These transgenic lines showed no biomass penalty despite a 10% reduction in Klason lignin content and severe shifts in lignin composition. Nuclear magnetic resonance spectroscopy and thioacidolysis revealed a strong increase (up to 20-fold) in sinapaldehyde incorporation into lignin, whereas coniferaldehyde was not increased markedly. Accordingly, ultra-high-performance liquid chromatography-mass spectrometry-based phenolic profiling revealed a more than 24,000-fold accumulation of a newly identified compound made from 8-8 coupling of two sinapaldehyde radicals. However, no additional cinnamaldehyde coupling products could be detected in the CAD1-deficient poplars. Instead, the transgenic lines accumulated a range of hydroxycinnamate-derived metabolites, of which the most prominent accumulation (over 8,500-fold) was observed for a compound that was identified by purification and nuclear magnetic resonance as syringyl lactic acid hexoside. Our data suggest that, upon down-regulation of CAD1, coniferaldehyde is converted into ferulic acid and derivatives, whereas sinapaldehyde is either oxidatively coupled into S′(8-8)S′ and lignin or converted to sinapic acid and derivatives. The most prominent sink of the increased flux to hydroxycinnamates is syringyl lactic acid hexoside. Furthermore, low-extent saccharification assays, under different pretreatment conditions, showed strongly increased glucose (up to +81%) and xylose (up to +153%) release, suggesting that down-regulating CAD1 is a promising strategy for improving lignocellulosic biomass for the sugar platform industry.
The depletion of fossil feedstock and the urgent need to decrease greenhouse gas emissions demand the use of renewable and sustainable energy sources (USEIA, 2013; Vanholme et al., 2013a). Second-generation biofuels are produced from nonedible biomass, such as lignocellulosic material, and are favored over first-generation biofuels that are made from feedstock that can also serve as food and feed, such as maize (Zea mays) grain (Yuan et al., 2008; Solomon, 2010). Lignocellulosic biomass consists mainly of cellulose, hemicelluloses, and lignins. Lignin accounts for 20% to 30% of the dry weight of biomass and for a considerably higher proportion of the carbon and energy, depending on the plant species. Its biosynthetic pathway is rather well described in model species, starting from the amino acid Phe and finally leading to the canonical monolignols p-coumaryl, coniferyl, and sinapyl alcohols. After their biosynthesis, these monolignols are transported to the cell wall (CW), where they are oxidized by peroxidases and laccases to monolignol radicals that combinatorially couple and cross-couple with the growing oligomer to form the lignin polymer, resulting in the formation of p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units in the lignin polymer (Freudenberg, 1959; Boerjan et al., 2003; Vanholme et al., 2010a, 2013b). The generation of biofuels or other bio-based products from lignocellulosic biomass typically requires three steps: a pretreatment to increase the accessibility to the plant CW polysaccharides, a saccharification step in which the polysaccharides are hydrolyzed by enzymes into primary sugars, and a fermentation step that converts the monosaccharides to, for example, ethanol. Lignin interferes with the saccharification process by limiting the access of the CW polysaccharides to enzymatic degradation (Chen and Dixon, 2007; Van Acker et al., 2013) and by binding with the enzymes themselves (Gao et al., 2014). Decreasing the amount of lignin is often seen as a promising strategy to improve saccharification in bioenergy crops, but it is also often accompanied by an undesired biomass yield penalty (Bonawitz and Chapple, 2013; Van Acker et al., 2013). As an alternative, lignin composition can be modified in order to make lignin easier to extract or degrade during pretreatments (Vanholme et al., 2008; Simmons et al., 2010; Mottiar et al., 2016).
Changing the ratios of the traditional monolignols responsible for producing the H, G, and S units in lignin has already been shown to affect saccharification efficiency. Based on a set of Arabidopsis (Arabidopsis thaliana) mutants with a range of S/G ratios, a high S/G ratio was found to have a negative impact on saccharification without pretreatment, whereas the S/G ratio had no impact on saccharification efficiency when an acid pretreatment preceded the saccharification (Van Acker et al., 2013). The frequency of H units also is an important parameter determining saccharification efficiency. H units are preferentially present as phenolic end groups in the lignin polymer (Huis et al., 2012). By increasing their frequency, the lignin polymers become shorter and, therefore, are easier to extract from the plant CW. This has been demonstrated in alfalfa (Medicago sativa) down-regulated in either p-COUMARATE 3-HYDROXYLASE or HYDROXYCINNAMOYL-COENZYME A:SHIKIMATE HYDROXYCINNAMOYLTRANSFERASE (Ziebell et al., 2010). In addition to the traditional monolignols, alternative monomers can be incorporated into the lignin polymer to render it easier to degrade or extract. For example, the incorporation of ferulic acid leads to the formation of acetal bonds that can be easily cleaved under mildly acidic conditions (Leplé et al., 2007; Ralph et al., 2008). The abundance of ferulic acid-derived units was positively correlated with saccharification efficiency in poplar down-regulated in CINNAMOYL-COENZYME A REDUCTASE (CCR), although a causal relationship has not yet been demonstrated (Van Acker et al., 2014). On the other hand, down-regulation of CAFFEIC ACID O-METHYLTRANSFERASE (COMT) leads to the incorporation of 5-hydroxyconiferyl alcohol (i.e. the reduction product of the enzyme’s direct substrate, 5-hydroxyconiferaldehyde), resulting in lignin-containing benzodioxane structures and generally more digestible CWs (Van Doorsselaere et al., 1995; Lapierre et al., 1999; Ralph et al., 2001; Chen and Dixon, 2007; Vanholme et al., 2010b; Weng et al., 2010; Van Acker et al., 2013). In addition to a modification of the lignin amount or the ratio of the traditional lignin monomers in the polymer, another strategy to improve biomass processing is to introduce chemically labile bonds into the lignin polymer by expressing genes leading to the biosynthesis of monolignol conjugates (Vanholme et al., 2012a; Tsuji et al., 2015; Mottiar et al., 2016). For example, engineering the biosynthesis of monolignol ferulates by expressing a FERULOYL-COENZYME A MONOLIGNOL TRANSFERASE from Angelica sinensis in poplar (Populus spp.; Wilkerson et al., 2014) or increasing the p-coumaroylation of monolignols by expressing a p-COUMAROYL-COENZYME A:MONOLIGNOL TRANSFERASE from brachypodium (Brachypodium distachyon) in Arabidopsis (Petrik et al., 2014; Smith et al., 2015; Sibout et al., 2016) make the lignin polymer more susceptible to alkaline pretreatment.
Here, lignin composition was modified by down-regulating the gene encoding a CINNAMYL ALCOHOL DEHYDROGENASE (CAD), the enzyme that catalyzes the last step of the monolignol biosynthetic pathway in which hydroxycinnamaldehydes are reduced to their corresponding hydroxycinnamyl alcohols, the monolignols. The down-regulation of CAD has been studied previously in many species: in Arabidopsis (Sibout et al., 2003, 2005; Van Acker et al., 2013; Anderson et al., 2015), poplar (Baucher et al., 1996; Lapierre et al., 1999, 2004; Pilate et al., 2002), Nicotiana spp. (tobacco; Halpin et al., 1994; Hibino et al., 1995; Chabannes et al., 2001; Kaur et al., 2012), Pinus spp. (pine; MacKay et al., 1997; Ralph et al., 1997; Wu et al., 1999), maize (Halpin et al., 1998; Vermerris et al., 2010; Fornalé et al., 2012), Panicum virgatum (switchgrass; Fu et al., 2011; Saathoff et al., 2011), Eucalyptus camaldulensis (Valério et al., 2003), Oryza sativa (rice; Zhang et al., 2006; Li et al., 2009), alfalfa (Baucher et al., 1999; Jackson et al., 2008), and brachypodium (Bouvier d’Yvoire et al., 2013). Most of these studies focused on the consequences of down-regulating CAD expression on lignin content and composition, and several studies also investigated the biomass properties of these CAD-deficient plants for industrial applications like pulping or biofuel production (Baucher et al., 1996; Lapierre et al., 1999; Pilate et al., 2002; Sibout et al., 2005; Jackson et al., 2008; Fu et al., 2011; Saathoff et al., 2011; Fornalé et al., 2012; Bouvier d’Yvoire et al., 2013; Anderson et al., 2015). Depending on the studied species, the lignin content and composition (S/G ratio) either remained equal or were reduced, independently of each other, compared with the corresponding control line. However, all studied species had the common feature that down-regulating CAD caused the incorporation of hydroxycinnamaldehydes into the lignin polymer. The presence of hydroxycinnamaldehydes during lignin polymerization typically results in a reddish-brown coloration of the xylem (Halpin et al., 1994; Baucher et al., 1996, 1999; Ralph et al., 1997; Lapierre et al., 1999, 2004; Chabannes et al., 2001; Pilate et al., 2002; Sibout et al., 2005; Zhang et al., 2006; Jackson et al., 2008; Vermerris et al., 2010; Bouvier d’Yvoire et al., 2013), a coloration that results not from the lignin itself but from side reactions of the hydroxycinnamaldehydes (Kim et al., 2002; Fournand et al., 2003; Ralph et al., 2008). In poplar, CAD has been down-regulated previously by sense and antisense constructs. In these transgenic lines, whether grown in the greenhouse or in the field, the lignin composition (S/G ratio) was unaffected but the typical incorporation of hydroxycinnamaldehydes, mainly sinapaldehyde, was seen (Baucher et al., 1996; Lapierre et al., 1999, 2004; Pilate et al., 2002). Lignin content was equal to that in the wild type in young (3-month-old) greenhouse-grown poplar trees (Baucher et al., 1996) but was reduced, by up to 11%, in older (7- and 12-month-old) greenhouse-grown CAD-deficient poplars (Lapierre et al., 2004). No biomass penalty was observed for the antisense CAD-deficient trees, whether grown in the greenhouse or in the field (Baucher et al., 1996; Lapierre et al., 1999; Pilate et al., 2002).
Kraft pulping was the first industrially relevant process that was applied to wood derived from CAD-deficient poplars. Pulp from the transgenic poplars had a lower Kappa number than that from their wild-type controls (Baucher et al., 1996; Pilate et al., 2002), indicating that less alkali or lower cooking time was required to extract lignin from the wood to reach the same degree of delignification. The reason for the increased susceptibility of lignin to alkaline degradation has been attributed to the increased presence of conjugated carbonyl functionalities (situated at the para-position relative to the 4-O-aryl-ether linkages) in the lignin that originate from the incorporation of hydroxycinnamaldehydes, the substrates of CAD (Vanholme et al., 2012a), as well as to the higher abundance of free-phenolic end groups (Lapierre et al., 2004). These data suggested that lignin also might be easier to remove during the alkaline biomass pretreatments used in the production of fermentable sugars. Indeed, in CAD-deficient alfalfa (Jackson et al., 2008), maize (Fornalé et al., 2012), switchgrass (Fu et al., 2011; Saathoff et al., 2011), and brachypodium (Bouvier d’Yvoire et al., 2013), the saccharification yield was improved when using different types of pretreatment, and in maize, the increased saccharification yield also translated into an increased bioethanol yield of 8% when expressed on a dry biomass basis (Fornalé et al., 2012). However, although poplar has economic potential to be commercialized as a second-generation bioenergy crop when grown under a short-rotation coppice culture (Littlewood et al., 2014), no data are yet available on saccharification yields in CAD-deficient poplars.
The phenotypes of the poplars that were down-regulated for CAD using sense and antisense constructs often were not uniform over the xylem, as visualized by the nonhomogenous red xylem phenotype (Baucher et al., 1996). Given the prognosis that CAD deficiency in poplar may improve saccharification efficiency to fermentable sugars on the one hand, and given the efficiency of the RNA interference (RNAi) strategy to stably and strongly down-regulate target genes on the other hand, new transgenic poplar (Populus tremula × Populus alba) lines specifically down-regulated in CAD1 expression and having only 15% residual CAD activity (hpCAD) were developed. In addition to confirming the commonly known features of CAD down-regulation, as reported before, thioacidolysis and NMR revealed that only sinapaldehyde, but not coniferaldehyde, incorporated at increased levels into the lignin polymer of hpCAD poplars. To study this in more detail, the metabolic consequences of CAD1 down-regulation were mapped using ultra-high-performance liquid chromatography-mass spectrometry (UHPLC-MS)-based phenolic profiling and structural characterization of differentially accumulating compounds by multistage mass spectrometry (MSn), NMR, and oligolignol sequencing (Morreel et al., 2010a, 2010b). Our data show that sinapaldehyde is either oxidatively cross-coupled into its homodimer S′(8-8)S′ and into the lignin polymer or, together with coniferaldehyde, converted to a plethora of cinnamic acids and derivatives, among which the accumulation of syringyl lactic acid hexoside is the most prominent. Finally, biomass processing of the CAD1 down-regulated poplar wood showed large improvements in saccharification efficiency upon alkaline pretreatments.
RESULTS
Generation of CAD1-Deficient Transgenic Poplar
The CAD gene family in poplar (Populus trichocarpa) consists of 16 members, of which two, PtrCAD1 (Potri.009G095800) and PtrCAD2 (Potri.016G078300), encode CAD enzymes able to mediate the reduction of hydroxycinnamaldehydes into monolignols (Barakat et al., 2009; Shi et al., 2010; Wang et al., 2014). PtrCAD1 has been reported to be the only CAD gene that is highly expressed in xylem (Shi et al., 2010), although both CAD genes are highly expressed in xylem according to www.popgenie.org and Sundell et al. (2017). PtrCAD1 has a higher catalytic efficiency toward the hydroxycinnamaldehydes than PtrCAD2 and, hence, is thought to play the most prominent role in lignin biosynthesis in this tissue (Shi et al., 2010; Wang et al., 2014). To down-regulate CAD1 in P. tremula × P. alba, the P. tremula × P. alba CAD1 coding sequence (PtaCAD1; Supplemental Fig. S1A), corresponding to PtrCAD1, was cloned in the pHellsgate8 gene-silencing vector to produce 18 independent cauliflower mosaic virus 35S-driven hpCAD transgenic poplar lines. Seventeen of the transgenic lines exhibited the typical red xylem phenotype, suggesting that the expression of CAD was effectively reduced in these lines (Supplemental Fig. S2A). CAD activity was determined in eight lines (1, 2, 4, 11, 14, 19, 20, and 24) showing different levels of xylem coloration. Line 20, which did not show the red wood phenotype, did not show any reduction in CAD activity, whereas CAD activity was reduced similarly in the other seven lines to approximately 15% residual alcohol dehydrogenase activity (Supplemental Fig. S2B). Of those seven remaining lines, based on their stable phenotype over multiple independent experiments, the hpCAD lines 4, 19, and 24 were chosen for all further analyses. The transcript levels of PtaCAD1 and PtaCAD2 in developing xylem were determined using quantitative reverse transcription (qRT)-PCR in the wild type and the selected hpCAD lines (Supplemental Fig. S1B). As in P. trichocarpa, both PtaCAD1 and PtaCAD2 are expressed at similar levels in the xylem of wild-type poplars. For PtaCAD1, on average, a 5% residual transcript level was observed for the hpCAD lines compared with the wild type. Transcript levels of PtaCAD2 were not affected, illustrating the specific down-regulation of CAD1.
To further identify potential off-target effects of the used silencing approach, a BLAST analysis revealed a gene that had a high sequence similarity with PtaCAD1 (E-value = 2e-153). This gene, referred to as Pta_tCAD1, appeared as a truncated version of PtaCAD1, missing the first three exons of the PtaCAD1 sequence (Supplemental Fig. S3A). The Pta_tCAD1 transcript level remained under the detection limit in developing xylem of both wild-type and hpCAD poplars, which is consistent with the low expression of the corresponding gene (Potri.001G30000) in P. trichocarpa according to www.popgenie.org. Pta_tCAD1 lacks the majority (seven out of eight conserved amino acids) of the N-terminal GroES-like structure known to contain the zinc-binding domains that act in the catalytic site of alcohol dehydrogenases (Youn et al., 2006a). In addition, seven out of 12 conserved residues, hypothesized to be involved in substrate binding, are absent from the Pta_tCAD1 protein as well as a Gly-rich (GxGxxG) motif, necessary for binding the NADP+ cofactor (Supplemental Table S1). The nucleotide-binding domain of PtaCAD1 is composed of six parallel β-sheets and five helices, but the Pta_tCAD1 protein lacks one β-sheet and two helices. Altogether, it is highly unlikely that Pta_tCAD1 is able to perform the conversion of hydroxycinnamaldehydes to the corresponding alcohols.
Phenotypic Analysis of Greenhouse-Grown CAD1-Deficient Poplar
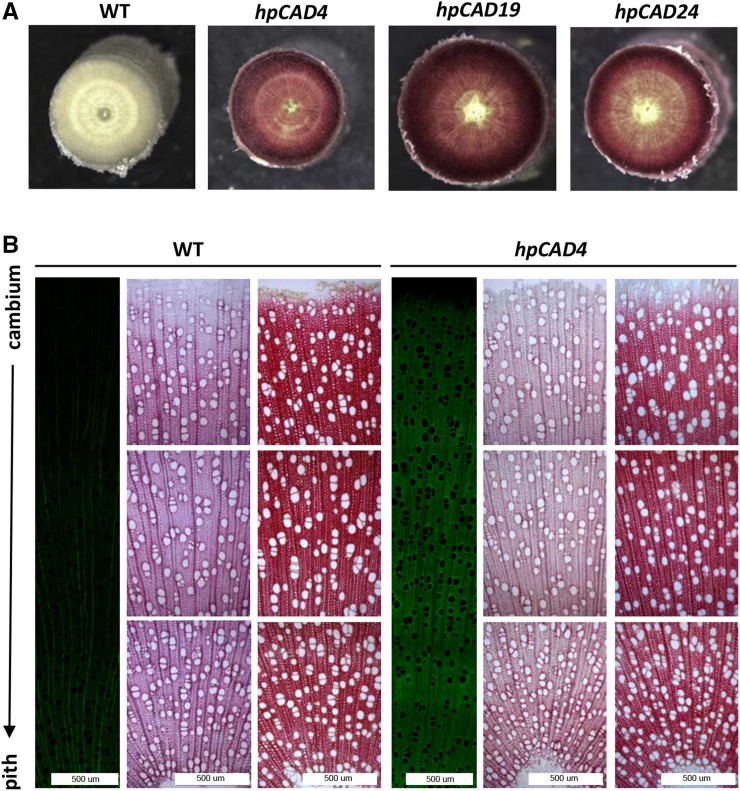
Five biological replicates of all three hpCAD lines and the wild type were obtained by in vitro micropropagation, transferred to soil, and cut back after 15 weeks of growth to ensure simultaneous and uniform regrowth. Trees originating from the regrowth were harvested after 3.5 months, when they were approximately 1.5 m tall. Stem height and weight were similar for all lines when compared with wild-type poplars (Table I). Upon debarking, the wood showed a typical red coloration distributed uniformly over the entire stem and not in patches, as was observed previously in particular poplar lines down-regulated in CAD (Baucher et al., 1996), CCR (Leplé et al., 2007; Van Acker et al., 2014), or COMT (Tsai et al., 1998) by sense or antisense methods. This uniform red coloration was not limited only to young xylem but was also observed in the entire cross section of hpCAD poplars (Fig. 1A) and is indicative of a stable down-regulation of CAD in these newly generated transgenic lines.
Table I. Biomass of CAD1-deficient poplar compared with the wild type.
Values shown are average ± sd height and fresh weight of 3.5-month-old greenhouse-grown poplar stems from wild-type and hpCAD lines (n = 5). No significant differences were found (Dunnett’s adjusted Student’s t test, P < 0.05).
| Parameter | Wild Type | hpCAD4 | hpCAD19 | hpCAD24 |
|---|---|---|---|---|
| Height (cm) | 148.3 ± 17.3 | 131.4 ± 12.9 | 150.1 ± 16.6 | 149.0 ± 10.2 |
| Fresh weight (g) | 53.4 ± 14.4 | 49.9 ± 12.5 | 60.8 ± 18.7 | 58.5 ± 10.7 |
Figure 1.
Microscopy. A, The typical red coloration associated with CAD1 down-regulation is visible across the entire stem section. B, Wood anatomy, ranging from cambium to pith, of the wild type (WT) and hpCAD4 (for hpCAD19 and hpCAD24, see Supplemental Fig. S4). Left columns show blue light autofluorescence (induced by long-wavelength excitation) of a stem section, middle columns show the results of the cross sections after Wiesner staining, and right columns are the results after treatment with Mäule reagent. For the Wiesner and Mäule staining, the three images are observations at three different spots of one single section. For all three columns, representative images are shown.
Histochemical analyses were performed to evaluate the effect of CAD1 down-regulation on wood anatomy (Fig. 1B; Supplemental Fig. S4). Inspection of the sections did not reveal any obvious differences between hpCAD lines and the wild type; in particular, no collapsed vessel (irregular xylem) phenotype could be detected. However, blue light autofluorescence (induced by a 488-nm excitation wavelength; Fig. 1B, left columns) was stronger in the sections of the hpCAD poplars compared with those of the wild type, especially in fibers and vessels, suggesting compositional shifts in the CW components. The phloroglucinol-HCl (Wiesner) reagent reacts with the aldehyde end groups in lignin (Kim et al., 2003) and generally is used to visualize the presence of lignin in a concentration-dependent manner (Fig. 1B, middle columns). On the other hand, the Mäule reagent specifically stains S lignin red (Fig. 1B, right columns). Wild-type and hpCAD poplars did not stain differently with either of these two lignin-specific stains.
CAD1-Deficiency Effects on Lignin Content and Structure
The CW composition was determined for 3.5-month-old hpCAD and wild-type poplar stems. CWs were purified from air-dried wood samples by removing extractives using water and ethanol under sonication. The recovered extract-free sample corresponds to CWs. When expressed per dry weight, the CW percentage was similar for all transgenic poplar and wild-type lines (Table II). Lignin content was determined by the Klason procedure. This method allowed a determination of the acid-insoluble lignin fraction (referred to as Klason lignin [KL]) and the acid-soluble lignin fraction (ASL). Both KL and ASL contents decreased on average by 10% and 13%, respectively, for hpCAD poplar lines compared with the wild type (Table II). These moderate reductions were not compensated for by significantly different levels of cellulose or hemicelluloses, except that hpCAD19 displayed a 9% increased hemicellulose level (Table II). The relative distribution of the main neutral sugars from the amorphous polysaccharides revealed some minor changes in hpCAD lines, mainly a moderate Xyl enrichment (Table II).
Table II. CW composition.
CW analyses of the stems from wild-type and 3.5-month-old hpCAD poplars: (A) CW content (% CW) and composition and (B) distribution of the main CW neutral sugars. Boldface or underlined values indicate significantly increased or decreased values, respectively, as compared with those of the wild type (Dunnett’s adjusted Student’s t test, P < 0.05).
| A) CW composition (in mg/g CW)a | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Line | % CW | KL | ASL | Cellulose | Hemicellulose | ||||||
| Wild type |
87.3 ± 0.8 |
194.7 ± 7.9 |
16.8 ± 0.3 |
482 ± 24 |
268 ± 12 |
||||||
|
hpCAD4 |
87.6 ± 0.0 |
175.0 ± 3.3 |
14.5 ± 0.1 |
495 ± 16 |
279 ± 17 |
||||||
|
hpCAD19 |
87.8 ± 0.7 |
183.6 ± 1.2 |
14.5 ± 0.5 |
492 ± 15 |
290 ± 10 |
||||||
|
hpCAD24 |
88.6 ± 0.5 |
177.5 ± 3.8 |
14.2 ± 0.1 |
487 ± 11 |
278 ± 16 |
||||||
| B) Relative distribution of main neutral sugars from amorphous polysaccharides (% by weight)b | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Line |
Rha |
Ara |
Xyl |
Man |
Gal |
Glc |
|||||
| Wild type |
2.1 |
1.6 |
77.0 |
3.9 |
3.0 |
12.5 |
|||||
|
hpCAD4 |
2.0 |
1.6 |
79.2 |
3.1 |
3.2 |
11.0 |
|||||
|
hpCAD19 |
2.1 |
1.5 |
79.7 |
3.4 |
2.9 |
10.4 |
|||||
| hpCAD24 | 2.2 | 1.6 | 79.5 | 3.4 | 3.0 | 10.4 | |||||
The data represent means ± sd from individually analyzed plants (n = 5).
The sd between biological replicate analyses is in the 5% to 10% range.
Lignin structure was studied by thioacidolysis, an analytical degradation method that yields lignin-derived monomers from lignin units involved only in labile 8-O-4 (β-O-4) bonds. When expressed in µmol g−1 lignin, thioacidolysis yield is reduced when the proportion of resistant interunit bonds increases, but it also may be reduced when lignins are derived from monomers other than the prototypical monolignols. The thioacidolysis yield (i.e. the sum of all released monomers, including aldehyde-derived monomers) expressed on a KL content basis was reduced by approximately 20% for the three hpCAD poplar lines compared with the wild type (Table III). The relative frequency of S thioacidolysis monomers derived from S units involved only in arylglycerol-β-aryl ether structures was found to be reduced in the hpCAD poplars, whereas that of G monomers was increased (Table III; Supplemental Table S2). This result reveals that CAD1 deficiency more specifically affects the formation of conventional syringylglycerol-β-aryl ether units. Concomitant with this reduction, S indenes released from sinapaldehyde units ether linked at their 8-positions (Kim et al., 2002) were found to increase substantially, up to 20-fold, as well as the S dithioketal compound derived from syringaldehyde end groups (Table III; for the structures of these compounds, see Supplemental Fig. S5). By contrast, CAD1 deficiency induced little change in the frequency of vanillin-derived monomers (data not shown). Coniferaldehyde end groups that are mainly responsible for the Wiesner lignin staining were found to be less abundant in the lignins from hpCAD poplars than in the wild type (Table III).
Table III. Lignin composition.
The determination of thioacidolysis monomers released from extract-free stems of wild-type and CAD1-deficient poplar lines is shown. The data represent means ± sd from individually analyzed plants (n = 5). Thioacidolysis yields are expressed in µmol g−1 KL. Boldface or underlined values indicate significantly increased or decreased values, respectively, as compared with those of the wild type (Dunnett’s adjusted Student’s t test, P < 0.05).
| Line | Main H, G, and S Monomers: Total Yield and Relative Molar % |
Monomers from Aldehyde Units |
Total Lignin-Derived Monomers (Relative % of the Wild Type) | |||||
|---|---|---|---|---|---|---|---|---|
| H + G + S | H | G | S | Coniferaldehyde End Groups | Sinapaldehyde-Derived Indene | Syringaldehyde End Groups | ||
| µmol g−1 KL | % | µmol g−1 KL | µmol g−1 KL | |||||
| Wild type | 2,535 ± 78 | 0.6 ± 0.2 | 35.0 ± 1.1 | 64.4 ± 1.0 | 5.2 ± 0.3 | 4.2 ± 0.3 | 6.1 ± 0.2 | 2,572 ± 66 (100) |
| hpCAD4 | 1,821 ± 31 | 0.5 ± 0.0 | 40.0 ± 0.4 | 59.6 ± 0.4 | 3.3 ± 0.0 | 154.0 ± 10.7 | 100.5 ± 2.5 | 2,093 ± 39 (81) |
| hpCAD19 | 1,805 ± 13 | 0.5 ± 0.1 | 40.1 ± 0.3 | 59.3 ± 0.3 | 3.4 ± 0.5 | 139.8 ± 12.6 | 80.1 ± 17.1 | 2,039 ± 42 (79) |
| hpCAD24 | 1,876 ± 111 | 0.5 ± 0.1 | 39.4 ± 0.3 | 60.1 ± 0.2 | 3.6 ± 0.2 | 141.8 ± 5.4 | 70.3 ± 3.7 | 2,105 ± 106 (81) |
The frequency of lignin units with free phenolic groups is a major structural trait that modulates important properties, such as lignin susceptibility to alkaline or oxidative treatments. This parameter was examined by thioacidolysis performed on permethylated samples (Lapierre et al., 1988, 1989; Lapierre and Rolando, 1988). In agreement with previous data (Lapierre et al., 1988; Lapierre and Rolando, 1988), and whatever the line (Table IV), about 90% of lignin-derived H monomers were found to be methylated (i.e. derived from terminal units with free, and therefore methylatable, phenolic groups). The frequency of G units with free phenolic groups also was increased substantially, from 24% in the wild type to 33% in hpCAD poplars. For S units, which are mostly internal units (Lapierre and Rolando, 1988), the frequency of free phenolic groups also was increased slightly in CAD1-deficient poplars (from 2.4% in the wild type to 3.1% in hpCAD poplars). Thus, concomitant with the lower release of the normal monomers and the increased release of sinapaldehyde and syringaldehyde units, CAD1 deficiency induced a higher frequency of terminal units with free phenolic groups in poplar lignins. This alteration is diagnostic for lignins displaying more branching structures (i.e. biphenyl and/or biphenyl ether bonds) and/or, as is most likely here, lignins with lower average molecular weight (Mr).
Table IV. Free-phenolic group analysis and low-Mr phenolics released upon alkaline treatment.
Relative percentages of free-phenolic groups in H, G, or S lignin units involved only in 8-O-4 bonds, as revealed by thioacidolysis of permethylated extract-free stems from wild-type and hpCAD poplar lines, are shown. Amounts are given for p-hydroxybenzoic acid, vanillin, and syringaldehyde, expressed in mg g−1 KL, released by mild alkaline hydrolysis of extract-free stems of wild-type and hpCAD poplar lines. Calculations are as described previously (Lapierre, 2010). The data represent means ± sd from individually analyzed plants (n = 5). Boldface values are increased significantly compared with the wild type (Dunnett’s adjusted Student’s t test, P < 0.05).
| Line | Free-Phenolic Groups of H, G, or S Units Only Involved in β/8-O-4 Bonds |
Alkali-Released Phenolic Compound |
||||
|---|---|---|---|---|---|---|
| H | G | S | p-Hydroxybenzoic Acid | Vanillin | Syringaldehyde | |
| % | mg g−1 KL | |||||
| Wild type | 90.1 ± 1.1 | 24.2 ± 0.4 | 2.4 ± 0.1 | 20.6 ± 1.3 | 1.7 ± 0.1 | 1.2 ± 0.1 |
| hpCAD4 | 87.8 ± 1.4 | 33.1 ± 0.4 | 3.2 ± 0.1 | 23.4 ± 0.7 | 6.9 ± 0.3 | 24.0 ± 0.8 |
| hpCAD19 | 89.3 ± 1.6 | 32.4 ± 1.0 | 3.0 ± 0.3 | 18.8 ± 2.7 | 6.3 ± 0.2 | 22.0 ± 1.1 |
| hpCAD24 | 89.2 ± 2.5 | 32.6 ± 0.8 | 3.1 ± 0.3 | 19.0 ± 2.3 | 6.4 ± 0.8 | 23.6 ± 2.6 |
Poplar lignins are acylated specifically by p-hydroxybenzoate, and this acylation is performed on monolignols before their polymerization (Morreel et al., 2004; Ralph, 2010; Lu et al., 2015). HPLC analysis of the low-molecular-weight phenolics released upon mild alkaline treatment of the extract-free samples revealed that CAD1 deficiency did not affect the level of lignin-linked p-hydroxybenzoate esters (Table IV). In contrast, the amounts of alkali-released vanillin and syringaldehyde were found to be increased considerably in the hpCAD poplar lines. These aldehydes originate from the oxidative degradation of labile lignin units, and their recovery is increased by the severity of the alkaline treatment (Bouvier d’Yvoire et al., 2013). CAD1 deficiency, therefore, markedly affected the structure of poplar lignins, noticeably by increasing the frequency of labile lignin units that are oxidatively degraded to C6C1 aldehydes during mild alkaline treatment (i.e. free-phenolic terminal units including sinapaldehyde moieties linked via their C8 position).
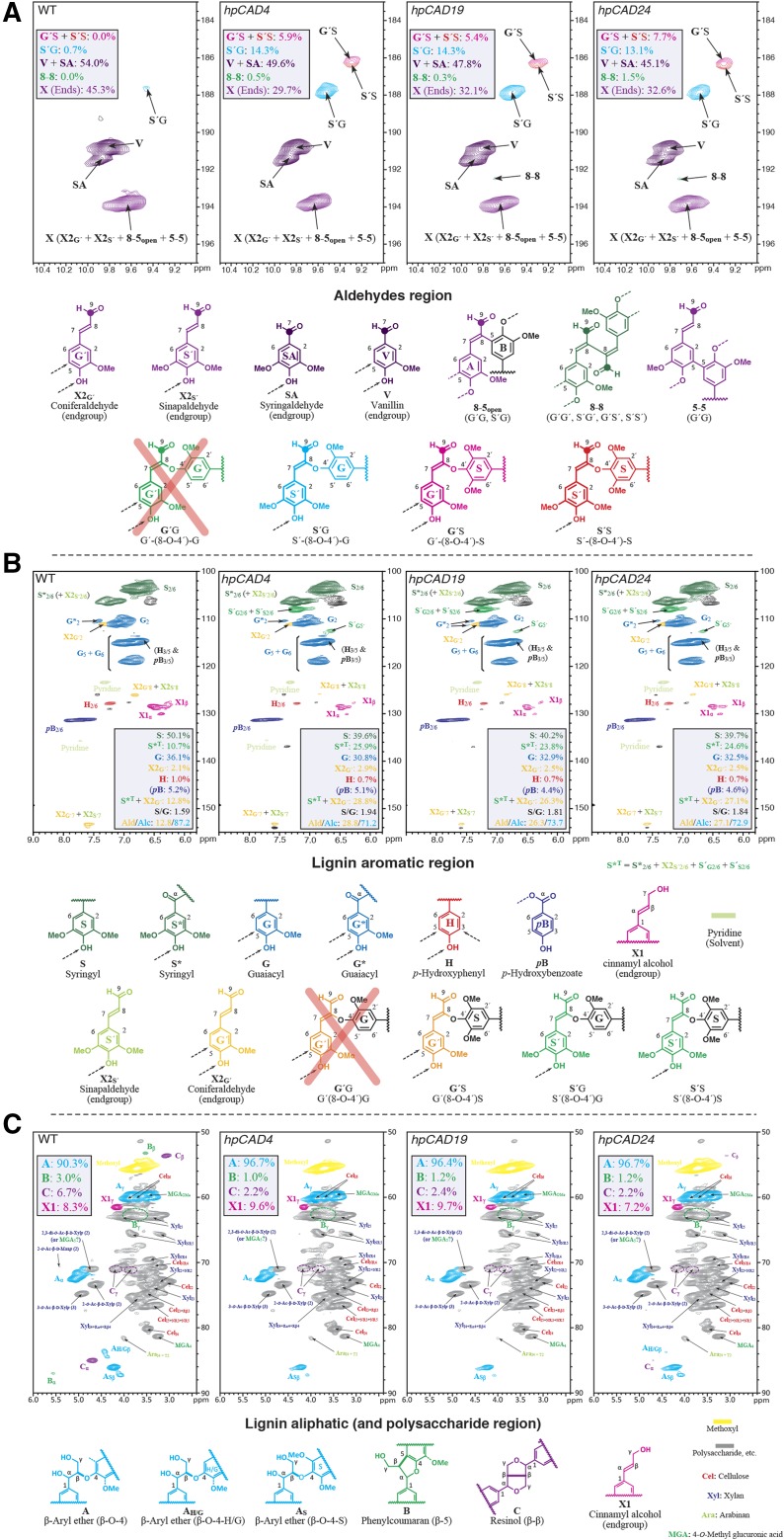
The lignin modifications observed using thioacidolysis, mainly the increase in aldehyde components, were supported by the results from 2D 1H-13C correlation heteronuclear single-quantum coherence (HSQC) NMR spectrometry, which, to our knowledge, was applied here for the first time on CAD1-deficient poplars (Fig. 2). Products of p-hydroxycinnamaldehyde's endwise coupling into the lignin polymer were seen readily from examination of both the aldehyde (Fig. 2A) and aromatic (Fig. 2B) regions of the NMR spectra, confirming that certainly sinapaldehyde and, less clearly evidently, coniferaldehyde are functioning as lignin monomers in chain extension reactions (Kim et al., 2002, 2003; Lapierre et al., 2004; Anderson et al., 2015). All three hpCAD lines showed the appearance of cross-coupled hydroxycinnamaldehyde 8-O-4-linked structures at similar levels (about 20% compared with less than 1% for the wild type) along with 8-8-linked homodimeric structural units at trace levels (Fig. 2A; Supplemental Table S3). Two points regarding these correlations and their interpretations are of note. First, the internal hydroxycinnamaldehyde’s 8-O-4-linked structures only appear less prevalent than the various V, SA, and X end groups because of the nature of these HSQC spectra (Mansfield et al., 2012), as end units are significantly overrepresented due to their higher mobility and longer relaxation. Thus, the internal units should be seen as more significant than they appear from the relative correlation peak volumes. Second, as we have proven previously (Kim et al., 2000, 2003), coniferaldehyde will not 8-O-4 cross-couple with G units, either in planta or in vitro, but only with S units. For this reason, the G′-(8-O-4)-G structure is shown with a cross through it; the structure resulting from coniferaldehyde’s 8-O-4 homocoupling, however, is possible, but it cannot be identified unambiguously from these spectra. The H/G/S distribution differences in lignin can be deduced from the aromatic region of the NMR spectra (Fig. 2B). Aromatic moieties from hydroxycinnamaldehydes (mainly from 8-O-4-linked cross-coupled structures, and mainly from sinapaldehyde) also were revealed as new products from this region; see the S′G2/6 and S′S2/6 correlations that are not evident in the wild type's spectrum. About 27% of the total lignin aromatics detected by NMR were aldehydes in the transgenic lines, as compared with only 13% in the wild type (although these figures are again distorted by the significant overestimation of end group units versus those of the internal units in the chain in the NMR of these differentially and fast-relaxing systems). The S/G ratios of units derived from the canonical monolignols only (without the hydroxycinnamaldehydes) were lower in hpCAD poplars (as also observed with thioacidolysis; Supplemental Table S2). In contrast, the total S/G ratios (with hydroxycinnamaldehydes) were higher in the transgenic lines (Fig. 2B; Supplemental Table S3).
Figure 2.
NMR. Partial short-range 1H-13C HSQC NMR spectra from the whole CWs of 3.5-month-old wild-type (WT) and CAD1-deficient poplar lines, as indicated in the top left corner of each plot. A, Aldehyde region. B, Lignin aromatic and double bond region. C, Lignin oxygenated aliphatic and polysaccharide region. The colors of the contours correspond with the structures drawn at the bottom of each plot. In B, the signals of the S, S*, G, X2G′, and H units sum to 100%. The signal of pB is given in parentheses, because it is left out of this sum. pB is not an actual lignin unit but a decoration found mainly on S units derived from lignification using monolignol p-hydroxybenzoates (Lu et al., 2015). Similarly, in C, X1 is not always a pure peak and, as an end group, is overrepresented in these spectra; it is reported on the A + B + C = 100 basis. n = 4 for the wild type and n = 3 for each CAD1-deficient line, and representative spectra are shown. Level data are from uncorrected integrals only.
The aliphatic region (Fig. 2C) validates the H/G/S distribution changes in the aromatic region and provides structural details regarding the bonding types and distribution of interunit linkage patterns present in the lignin fraction of whole CWs. However, with the unsaturation of the side chain and the aldehydic γ-position, no information regarding the abundance of aldehydes can be deduced from this region. Additional lignin compositional changes were found for the CAD1-deficient poplars compared with the wild type. The relative abundance of monolignol-derived β-aryl ether (β-O-4 [A]) structures was higher as a consequence of the relatively lower abundance of both phenylcoumaran B (8-5) and resinol C (8-8) structures in the hpCAD poplars. Cinnamyl alcohol end groups X1 also appears to be increased slightly, from integrals of 6% in the wild type to 7% to 10% in CAD1-deficient poplars, but this is due in part to the distortion caused by the lower level of monolignol-derived structures and the major monomer substitution (by sinapaldehyde; Fig. 2C; Supplemental Table S3).
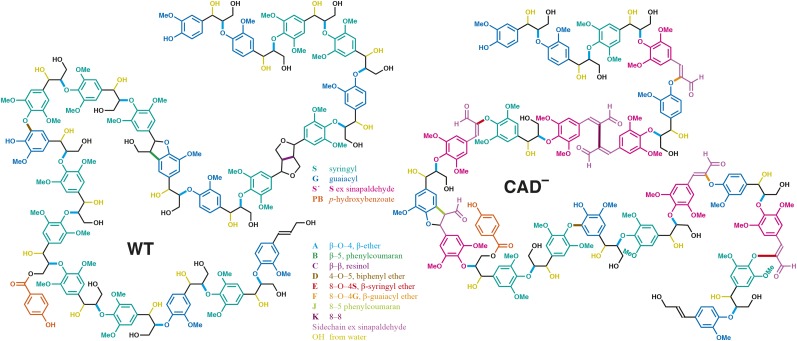
Based on these NMR data, models were generated of the lignin polymers in wild-type and hpCAD poplars (Fig. 3; Supplemental Information S1). However, some caution is necessary with these models. The models were built to confirm the determined distribution of G and S units as well as of the interunit linkage types and contain only 20 units. Hence, it was impossible to accurately represent minor units (i.e. units that were less than 5% present). Nevertheless, the structures are fully chemically legal, and various features attempt to follow the best available current information (e.g. 4-O-5-linked units D are free-phenolic and not represented as branching units; Li et al., 2016; Yue et al., 2016). Despite the clear difference in compositional unit frequencies between the wild type and the CAD-lignin model, both are primarily linear chains, akin to those drawn here.
Figure 3.
Models based on NMR. Models conforming with the available data for polymers of 20 units for the wild type (WT) and the CAD1-deficient poplar lignins are shown. For details on the features of these two models, see Supplemental Information S1.
Profiling of Methanol-Soluble Phenolics
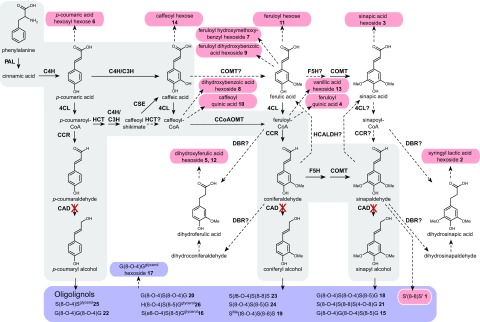
It is remarkable that CAD1 down-regulation increases sinapaldehyde incorporation into lignin whereas coniferaldehyde incorporation was not evidently increased. This suggests that coniferaldehyde is metabolized in another way. To gain insight into the effect of CAD1 down-regulation on small-Mr phenolics, xylem extracts of hpCAD and wild-type poplars were profiled via UHPLC-MS (Fig. 4; Table V; Supplemental Table S4). From an estimated 2,154 profiled compounds (see “Materials and Methods”), 64 had a lower abundance (average fold change < 0.5) in the hpCAD lines, whereas 348 showed a higher abundance (average fold change > 2), as compared with the wild type. In agreement with the lower lignin content, down-regulation of CAD1 resulted in the decrease of oligolignols formed by the combinatorial coupling of the three canonical monolignols, as identified by lignin sequencing (compounds 15–26 in Table V). Among the structurally characterized mass-to-charge ratio (m/z) features with an increased abundance upon CAD1 down-regulation, phenylpropanoids derived from sinapic and ferulic acids (3–5, 7, 9, 11, and 12 in Table V) were prevalent. In addition, p-coumaroyl hexosyl hexose (6), two caffeic acid derivatives (10 and 14), and two benzoic acid derivatives (8 and 13) were characterized. In addition, the m/z features of compounds 1 and 2 were below the detection limit in the phenolic profiles of the wild type but increased to levels of 24,156 and 8,650 times, respectively, above the detection limit in hpCAD poplars. Thereby, these m/z features were among the most prominent m/z features in the xylem extracts of hpCAD poplars. Clearly, the flux through phenylpropanoid and monolignol biosynthesis upon the down-regulation of CAD1 was massively redirected to the biosynthesis of compounds 1 and 2.
Figure 4.
Metabolic pathway based on phenolic profiling. The main pathway involved in monolignol biosynthesis is shown with a gray background. The relative increase and decrease in metabolite abundances in the three hpCAD poplar lines compared with the wild type are mapped on the pathway. Red round-cornered boxes represent significant increases and blue round-cornered boxes represent significant decreases in abundance. Dashed arrows indicate one or more proposed conversions. PAL, Phe ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumaric acid:CoA ligase; HCT, hydroxycinnamoyl-CoA shikimate hydroxycinnamoyl transferase; C3H, coumarate 3-hydroxylase; CSE, caffeoyl shikimate esterase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; F5H, ferulate 5-hydroxylase; COMT, caffeate O-methyltransferase; CAD, cinnamyl alcohol dehydrogenase; HCALDH, hydroxycinnamaldehyde dehydrogenase; DBR, double-bond reductase. The activities of PAL, C4H, 4CL, HCT, CCoAOMT, CCR, F5H, COMT, and CAD poplar enzymes have been described by Wang et al. (2014); conversions with Kcat/KM > 0.1 were considered as proven and represented with solid arrows, whereas conversions with Kcat/KM < 0.1 were considered as proposed and thus represented with dashed arrows. The β-oxidative side chain shortening of cinnamoyl-CoAs is a potential pathway toward the different benzoic acids (Widhalm and Dudareva, 2015). DBR has been found to reduce the C7-C8 double bond of p-coumaraldehyde and coniferaldehyde in Arabidopsis (Youn et al., 2006b), but it also might reduce the corresponding hydroxycinnamoyl-CoAs (Ibdah et al., 2014). The functions of a heterotrimeric C4H/C3H protein complex in the hydroxylation of p-coumaryl shikimate toward caffeoyl shikimate and a similar heterodimeric C4H/C3H protein complex that is able to convert p-coumaric acid directly into caffeic acid in poplar have been described by Chen et al. (2011). The role of HCALDH in catalyzing the oxidation of coniferaldehyde and sinapaldehyde to their corresponding carboxylic acids has been shown in Arabidopsis (Nair et al., 2004). The role of CSE in catalyzing the hydrolysis of caffeoyl shikimate to caffeic acid has been shown in Arabidopsis and Medicago truncatula, and evidence exists for a similar role of CSE in poplar (Vanholme et al., 2013b; Ha et al., 2016). The biosynthesis of hydroxycinnamate shikimate and quinate esters has been established in Nicotiana tabacum and Nicotiana benthamiana starting from their respective CoA thioesters via an HCT (Hoffmann et al., 2004). Glycosylation by UDP-glycosyltransferases of hydroxycinnamic acids has been described previously (Meissner et al., 2008).
Table V. Methanol-soluble phenolics.
UHPLC-MS-based phenolic profiling of hpCAD4, hpCAD19, hpCAD24, and wild-type stems (n = 10, 10, 10, and 30, respectively). A list of 412 compounds (348 up and 64 down) with significantly different signal intensities between the wild type and the three hpCAD lines was obtained, of which 26 compounds (14 up and 12 down) were structurally characterized. The structurally characterized compounds are shown in order of decreasing fold change in abundance. The fold change was calculated as the average peak intensity in the three hpCAD lines divided by the average peak intensity in the wild type. The tandem mass spectrometry (MS/MS) spectra of the individual metabolites can be found in Supplemental Table S4. The abundances of compounds 1 to 14 were higher, whereas those of compounds 15 to 26 were lower, in the hpCAD poplars. Whenever the compound was not detected, an arbitrary peak intensity of 50 was taken into account to compute the fold change. In the column Elucidation Level, I indicates identified, a structure that was elucidated via NMR or by spiking the synthesized compound; A indicates annotated, a structure with a rather firm structural elucidation based on comparison of the MS/MS spectrum with those of identified structural analogs and on the accurate m/z value; and C indicates structurally characterized, a fairly high degree of certainty in the structural elucidation that is based on the MS/MS spectral interpretation, accurate m/z value, and data available from the literature and public databases (Morreel et al., 2014). SMe, 7-O-Methylsyringylglycerol moiety.
| No. | Trivial Name | Elucidation Level | Retention Time | m/z | Average Peak Intensity |
Fold Change | |||
|---|---|---|---|---|---|---|---|---|---|
| Wild Type | hpCAD4 | hpCAD24 | hpCAD19 | ||||||
| min | |||||||||
| 1 | S′(8-8)S′ | I | 12.39 | 413.1244 | 50 | 1,278,235 | 1,155,155 | 1,190,069 | 24,156 |
| 2 | Syringyl lactic acid hexoside | I | 3.21 | 403.1248 | 50 | 528,780 | 403,322 | 365,399 | 8,650 |
| 3 | Sinapic acid hexoside | A | 4.47 | 385.1091 | 50 | 39,568 | 26,677 | 7,034 | 488 |
| 4 | Feruloyl quinic acid | A | 7.97 | 367.1036 | 71 | 20,980 | 12,293 | 5,362 | 181 |
| 5 | Dihydroferulic acid hexoside | A | 3.86 | 357.1198 | 50 | 8,089 | 7,847 | 3,480 | 129 |
| 6 | p-Coumaric acid hexosyl hexose | A | 3.74 | 487.1564 | 78 | 12,240 | 7,252 | 6,467 | 111 |
| 7 | Feruloyl hydroxyl-methoxybenzyl hexoside | C | 15.83 | 461.1452 | 417 | 52,927 | 37,003 | 34,703 | 100 |
| 8 | Dihydroxybenzoic acid hexoside | A | 1.67 | 153.0170 | 105 | 11,307 | 9,008 | 9,266 | 94 |
| 9 | Feruloyl dihydroxybenzoic acid hexoside | A | 12.88 | 491.1199 | 50 | 1,736 | 1,462 | 2,137 | 36 |
| 10 | Caffeoyl quinic acid | A | 4.26 | 353.0882 | 3,499 | 50,745 | 47,716 | 41,313 | 13 |
| 11 | Feruloyl hexose | A | 5.4 | 355.1038 | 13,876 | 129,240 | 92,359 | 83,730 | 7 |
| 12 | Dihydroferulic acid hexoside | A | 4.74 | 357.1195 | 10,772 | 48,579 | 33,555 | 34,210 | 4 |
| 13 | Vanillic acid hexoside | A | 2.28 | 329.0898 | 90,929 | 236,429 | 176,961 | 211,144 | 2 |
| 14 | Caffeoyl hexose | A | 4.09 | 341.0887 | 126,360 | 305,682 | 261,863 | 297,608 | 2 |
| 15 | G(8-O-4)G(8-O-4)S(8-5)G | A | 15.13 | 779.2904 | 684 | 50 | 50 | 50 | 0.0731 |
| 16 | S(e8-O-4)S(8-5)Gglycerol | A | 9.47 | 647.2289 | 12,863 | 387 | 709 | 35 | 0.0293 |
| 17 | G(8-O-4)Gglycerol hexoside | A | 3.65 | 571.2014 | 7,851 | 429 | 61 | 59 | 0.0233 |
| 18 | G(8-O-4)S(8-O-4)S(8-5)G | A | 16.29 | 809.3010 | 2,163 | 50 | 50 | 50 | 0.0231 |
| 19 | SMe(t8-O-4)S(8-8)S | A | 19.88 | 657.2535 | 2,288 | 50 | 50 | 50 | 0.0218 |
| 20 | G(8-O-4)S(8-O-4)G | A | 10.84 | 601.2276 | 10,845 | 57 | 311 | 188 | 0.0171 |
| 21 | S(8-O-4)S(8-8)S(4-O-8)G | A | 17.91 | 839.3119 | 3,246 | 50 | 50 | 50 | 0.0154 |
| 22 | G(8-O-4)G(8-O-4)G | A | 9.89 | 571.2171 | 5,416 | 104 | 50 | 50 | 0.0126 |
| 23 | S(t8-O-4)S(8-8)S | A | 16.57 | 643.2380 | 13,169 | 180 | 122 | 69 | 0.0094 |
| 24 | S(8-O-4)S(8-5)G | A | 14.64 | 613.2274 | 15,593 | 71 | 129 | 232 | 0.0092 |
| 25 | S(8-O-4)Sglycerol | A | 4.64 | 469.1718 | 6,738 | 50 | 50 | 50 | 0.0074 |
| 26 | H(8-O-4)S(8-5)Gglycerol | A | 9.31 | 587.2120 | 9,804 | 50 | 50 | 50 | 0.0051 |
Structural elucidation of compounds 1 and 2 involved both mass spectrometry (MS) and NMR analyses. The MS data of compound 1 (m/z 413.1244, C22H21O8, Δppm = 0.51) indicated a highly conjugated structure that preferentially fragmented via the loss of one or two methyl radicals (the most abundant MS/MS product ions were at m/z 398.10 and 383.07), losses that are typical for methyl aryl ethers (Supplemental Table S4; Bowie, 1990). Furthermore, the third and fourth most abundant MS/MS product ions were observed at m/z 369.09 and 354.07 due to the combined loss of one or two methyl radicals (−15.02 or −30.05 Da, respectively) and a formaldehyde radical (−29 Da). Based on the chemical formula and the main MS/MS fragmentations, compound 1 was tentatively characterized as a sinapaldehyde dimer. Examination via NMR of the purified compound 1 and spectral comparison with that of a previously synthesized model (Kim et al., 2003) identified this compound as S′(8-8)S′ 1, a dimer consisting of two units derived from sinapaldehyde (Supplemental Fig. S6).
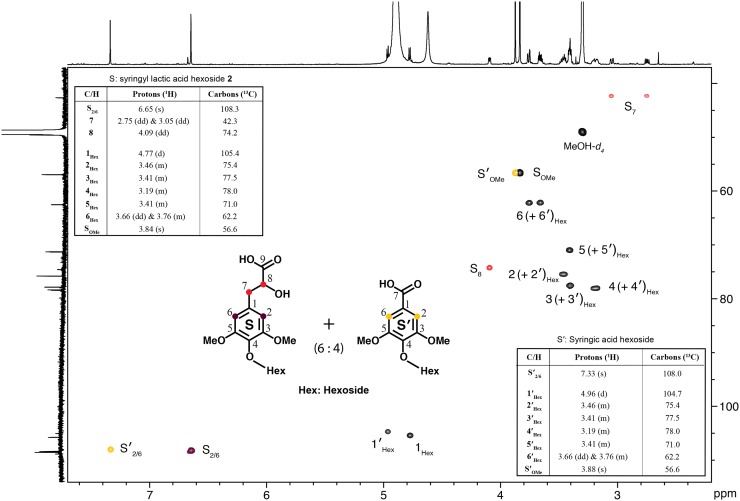
The MS/MS spectrum of compound 2 (precursor ion at m/z 403.1248, C17H23O11, Δppm = 0.53) showed a base peak at m/z 241.07 due to the loss of anhydrohexose (−162.05 Da; Supplemental Fig. S7). Other product ions at m/z 223.06, 208.04, 179.07, 164.05, and 149.02 were reminiscent of a sinapic acid-derived moiety (Morreel et al., 2014). The sinapic acid product ion at m/z 223.06 results from the m/z 241.07 product ion due to water loss. Therefore, the latter product ion represents a hydrated sinapic acid moiety. Because the m/z 241.07 product ion also fragmented to m/z 195.06 via formic acid loss, a gas-phase reaction typical for 2-hydroxycarboxylic acids (Bandu et al., 2006; Greene et al., 2013), this indicates that the hydroxyl function is attached to the 2-position relative to the carboxylic acid. The latter was confirmed by the presence of a product ion at m/z 72.99 derived from the lactic acid moiety of the compound 2 ion (for fragmentation pathways, see Supplemental Fig. S7). Therefore, this compound was characterized as syringyl lactic acid hexoside 2 (Table V; Fig. 5). To verify the identity of compound 2, it was purified from xylem extracts from hpCAD poplar for structural elucidation by NMR; the HSQC spectrum along with high-resolution proton and carbon data (plotted on the projection axes) are shown in Figure 5. The syringyl lactic acid hexoside structure was elucidated via 1D proton and carbon and 2D correlation spectroscopy (COSY), HSQC, and heteronuclear multiple-bond correlation (HMBC) NMR experiments. The chemical shifts were logically matched with the structure, and the proton NMR coupling constants and patterns showed the two protons at the C/H 7-position and one proton at an obviously oxygenated 8-position (based on its proton and carbon chemical shifts). A phenolic glycoside was identified by the 2D HMBC NMR data that showed a connection between the phenolic carbon (C4) and the anomeric proton (H1) of the glycoside. The glycoside peaks were assigned based on a COSY experiment, but we were not able to identify the exact sugar. We also recognized later that the isolated fraction of syringyl lactic acid hexoside was contaminated with syringic acid glycoside and assigned the peaks. The two structurally characterized compounds, S′(8-8)S′ 1 and syringyl lactic acid hexoside 2, were identified here, to our knowledge for the first time in plants, as deduced from searching the CAS database (https://scifinder.cas.org/scifinder/login).
Figure 5.
Identification of syringyl lactic acid hexoside by MS/MS and NMR. 2D 1H-13C correlation HSQC NMR spectra of a chromatic fraction of hpCAD xylem extracts containing syringyl lactic acid hexoside (2) are shown. Syringic acid hexoside appeared to be present in the same chromatic fraction. The structural formulas of both compounds are shown.
It was surprising that, among the 348 profiled compounds with increased abundance, only one feature, S′(8-8)S′ 1, was composed of a hydroxycinnamaldehyde, whereas we expected many others to be derived from either coniferaldehyde or sinapaldehyde, the substrates of CAD, in analogy with the accumulation of numerous 5-hydroxyconiferyl alcohol derivatives in COMT-deficient plants (Vanholme et al., 2010b; Weng et al., 2010). This suggested that many of the m/z features that we could not structurally resolve by MS fragmentation might be more complex oxidative coupling products from the coupling of hydroxycinnamaldehydes with coniferyl and/or sinapyl alcohol. To investigate whether additional oligolignol-containing units derived from hydroxycinnamaldehydes were among the compounds that accumulated in the hpCAD plants, various combinations of horseradish peroxidase/hydrogen peroxide-based dehydrogenation polymers (DHPs) were made with coniferyl alcohol, sinapyl alcohol, coniferaldehyde, and sinapaldehyde. UHPLC-MS analysis of the low-Mr fraction resulting from these oxidative coupling assays revealed a multitude of dimers and trimers (Supplemental Table S5), resulting in a database of approximately 550 unique DHP dimers, trimers, and tetramers. Using R (www.r-project.org), we automatically searched for the presence of any of these oligomers, or derivatives thereof (e.g. glucosides), in the phenolic profiles of the CAD1-deficient poplar samples. However, except for S′(8-8)S′, none of the coupling products detected in DHPs could be detected in the hpCAD extracts (Table VI). In the oxidative coupling assays, S′(8-8)S′ also coupled to coniferyl alcohol or sinapaldehyde via an 8-O-4 linkage. However, these trimers were below the detection limit in vivo (Table VI). None of the many coupling products of coniferaldehyde that were found in the DHPs were detected in the poplar extracts of either hpCAD or the wild type.
Table VI. Oxidative coupling assay.
S′(8-8)S′-derived oligolignols in the oxidative coupling assays and in the metabolite profiles of hpCAD are shown. The shorthand name is the unique identifier of the peak, resulting from merging the retention time and m/z value. ND, Not detected.
| Oligolignols | Theoretical Mass | Shorthand Name |
|
|---|---|---|---|
| Oxidative Coupling Assays | hpCAD | ||
| m/z | |||
| S′(8-8)S′ | 413.12 | 12.21_413.1246 m/z | 12.39_413.1244 m/z |
| S′(8-8)S′(4-O-8)G | 609.19 | 14.10_609.1971 m/z | ND |
| S′(8-8)S′(4-O-8)S′ | 619.18 | 19.94_619.1811 m/z | ND |
| S′(8-8)S′(4-O-8)S | 639.21 | ND | ND |
| S′(8-8)S′(4-O-8)G(4-O-8)G | 805.27 | ND | ND |
| S′(8-8)S′(4-O-8)G(5-8)G | 787.26 | ND | ND |
| S′(8-8)S′(4-O-8)G(4-O-8)S′ | 815.25 | ND | ND |
| S′(8-8)S′(4-O-8)S′(4-O-8)S′ | 825.24 | ND | ND |
We also investigated whether p-coumaraldehyde, coniferaldehyde, and sinapaldehyde, the substrates of CAD, were present in the hpCAD lines, but all three compounds remained below the detection limit in the plant extracts.
Saccharification Efficiency
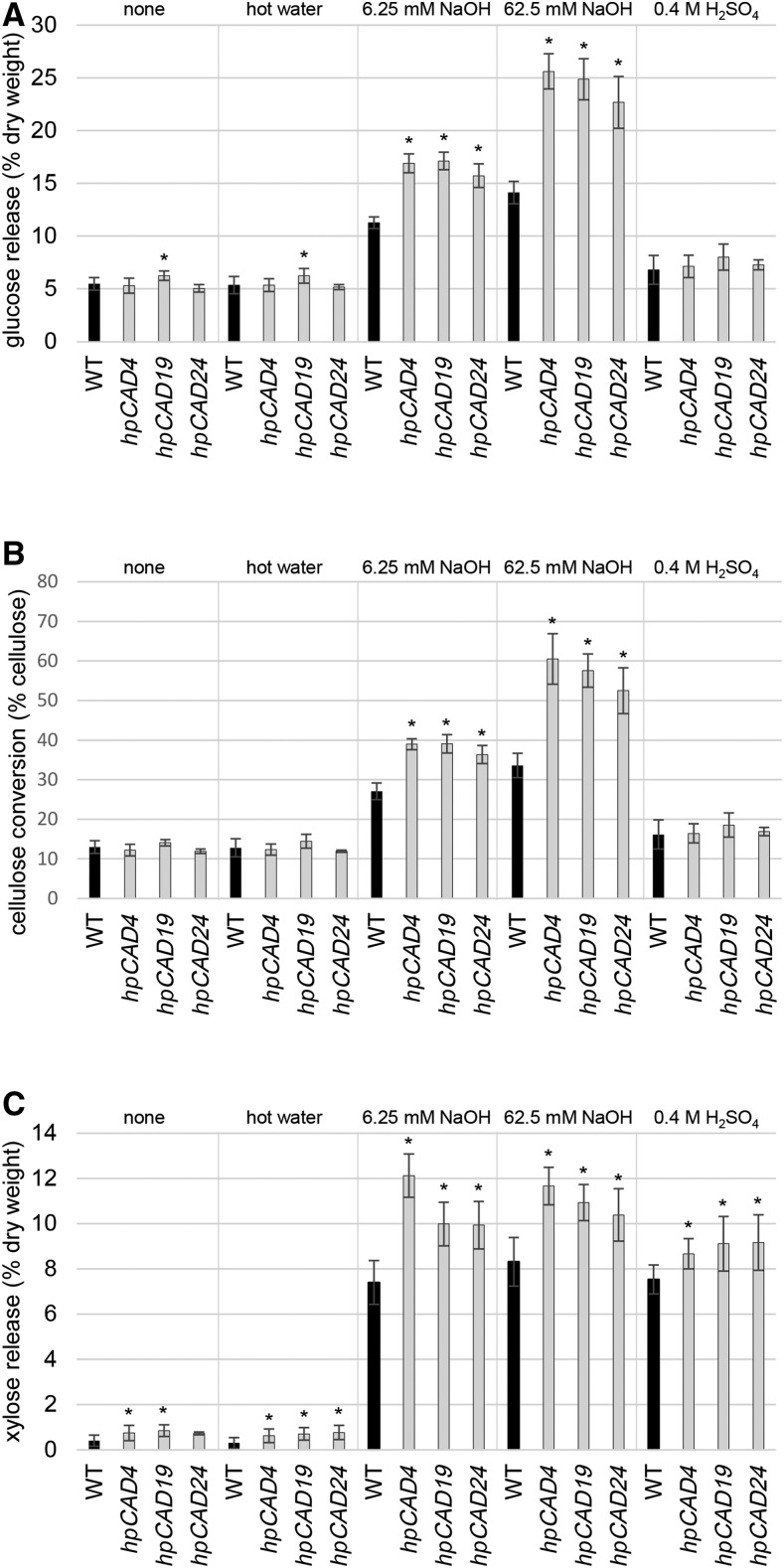
The hydrolysis of CW polysaccharides into primary sugars, a process called saccharification, was determined for the three hpCAD lines and compared with that of the wild type. Saccharifications were preceded by a pretreatment to loosen the plant CW and, hence, make the CW polysaccharides more accessible to enzymatic hydrolysis. Five different pretreatment conditions were applied: no pretreatment, hot water, acid, and two alkaline pretreatments (with different concentrations of sodium hydroxide). As has been noted previously, lignins from CAD-deficient plants, even in hardwoods, are more readily solubilized and extracted in alkali, in part due to their higher phenolic contents (Lapierre et al., 1989). For saccharification, an enzyme mixture containing multiple hydrolyzing activities, including exo-1,4-β-glucanase, endo-1,4-β-glucanase, hemicellulases, and β-glucosidase, was used. Hence, both cellulose and hemicellulose polymers were hydrolyzed, and the released Glc and Xyl were measured and expressed as percentages of dry weight. The released Glc was higher in all three hpCAD lines compared with the wild type by up to 81% when alkaline pretreatments were applied (Fig. 6A; Supplemental Table S6), but the Glc release remained comparable with that of the wild type for other pretreatment conditions. Cellulose conversions were calculated based on the amount of cellulose (Table II) and the amount of Glc released upon saccharification (Supplemental Table S6). Because there was no significant difference in cellulose content observed for the hpCAD lines compared with the wild type, the conclusions regarding cellulose conversion were similar: only with alkaline pretreatments, cellulose conversions were higher, from 27% in the wild type up to 39% in hpCAD with a 6.25 mm NaOH pretreatment and from 34% in the wild type up to 61% in hpCAD with a 62.5 mm NaOH pretreatment, all under the same partial saccharification conditions (Fig. 6B; Supplemental Table S6). The enzyme cocktail used in the saccharification experiments contained both cellulases and hemicellulases. Hemicelluloses account for 27% to 29% of the CW (Table II) and contain on average 10% to 13% Glc (Table II). Therefore, a part of the improved saccharification yields for hpCAD poplars has to be attributed to the hemicelluloses. The amount of Xyl released from hemicellulosic polymers was higher for almost every transgenic line and treatment condition (Fig. 6C; Supplemental Table S6). The highest increase, up to 153%, was observed with hot water pretreatment, although the total release of Xyl under these conditions remained low. Under alkaline conditions, the wild type released on average 44% of the Xyl present in the hemicellulose fraction, whereas the hpCAD lines released up to 61% (Supplemental Table S6).
Figure 6.
Saccharification yields. A, Average Glc release, expressed as percentages of dry weight, for the different CAD1-deficient lines and the wild type (WT; n = 5, each biological repeat measured in triplicate). B, Cellulose conversion, expressed as percentages of cellulose, calculated based on the amounts of released Glc and the quantified amount of cellulose. C, Average Xyl release, expressed as percentages of dry weight. The results are grouped per treatment condition, which is indicated at the top of each graph. Error bars represent sd. Asterisks indicate significant differences from the wild type within the same treatment condition (Dunnett’s adjusted Student’s t test, P < 0.05).
DISCUSSION
CAD converts hydroxycinnamaldehydes into hydroxycinnamyl alcohols in the last step of the monolignol biosynthetic pathway. Previously, CAD was down-regulated in poplar using sense and antisense strategies. The newly generated CAD1-deficient poplar lines described here were obtained by a 35S-driven hairpin RNAi strategy and had residual CAD1 transcript levels of about 5% (Supplemental Fig. S1B) and CAD activity of about 15% (Supplemental Fig. S2B). This effective down-regulation is in contrast with previous studies claiming that the 35S promoter is not the most effective promoter for RNAi silencing of secondary CW-related genes compared with xylem-specific promoters such as the GT43B promoter (Ratke et al., 2015). However, this apparent contradiction might be explained by the specific nature of lignification, which is under both cell-autonomous and non-cell-autonomous control (Pesquet et al., 2013; Smith et al., 2013, 2017). Indeed, the CAD gene is expressed in poplar rays (Regan et al., 1999), and CAD protein has been localized in ray cells (Goffner et al., 1998; Šamaj et al., 1998). It has been hypothesized that monolignols synthesized in the ray cells diffuse toward the neighboring xylem cells (Hawkins et al., 1997). Because the 35S promoter confers strong expression in rays in secondary xylem (Nilsson et al., 1996; Chen et al., 2000; Ratke et al., 2015), it is expected that the expression of PtaCAD1 is silenced in these cells, reducing a major source of monolignols destined for lignification of the neighboring xylem cells.
The uniform and intense red coloration (Fig. 1A), which was maintained over successive cycles of vegetative propagation, indicates that the selected transgenic lines are stably down-regulated in CAD1. Nowadays, even better techniques, such as genome editing using CRISPR/Cas9, allow the generation of complete, stable, and heritable knockouts (Zhou et al., 2015), but such methods were not available at the outset of this study.
CAD1 Down-Regulation Results in the Incorporation of Sinapaldehyde into the Lignin Polymer, But Coniferaldehyde Has Another Fate
The hpCAD lines had a 10% lower lignin content (Table II), and their lignin structures were dramatically affected. The hpCAD samples displayed the hallmarks of CAD deficiency (i.e. the increased frequency of free-phenolic units and of hydroxycinnamaldehyde and benzaldehyde units in lignins). In the case of poplars and in agreement with previous results (Kim et al., 2002; Lapierre et al., 2004), these additional aldehyde units are mostly composed of sinapaldehyde and syringaldehyde units, with little change noted in coniferaldehyde and vanillin units. In conjunction, the level of sinapyl alcohol-derived S units was relatively more reduced than the level of coniferyl alcohol-derived G units (Table III; Supplemental Table S2). Consistent with thioacidolysis data revealing the preferential accumulation of sinapaldehyde units in the lignins of hpCAD poplar lines, the phenolic profiling results showed a more than 24,000-fold accumulation of a sinapaldehyde homodimer [S′(8-8)S′; compound 1 in Table V and Supplemental Table S4]. Taken together, these data suggest that sinapaldehyde in hpCAD lines was at least partially oxidatively coupled into a dimer or polymerized into the lignin, whereas coniferaldehyde (which also was expected to be produced in higher amounts, given the reduction of coniferyl alcohol-derived G units in the lignin; Supplemental Table S2) did not end up in the lignin and, thus, must have been metabolized in a different way.
One possible explanation for why only sinapaldehyde, but not coniferaldehyde, is incorporated in the lignin is the involvement of PtaCAD2 or other proteins with CAD activity. PtaCAD2 is expressed at similar levels to PtaCAD1 in the xylem of the P. tremula × P. alba hybrid used here, as quantified using qRT-PCR (Supplemental Fig. S1B). According to Sundell et al. (2017), other genes sharing sequence similarity with CAD1 and CAD2 also are expressed in poplar xylem, albeit not as high as CAD1 and CAD2. Therefore, we cannot exclude the possibility that PtaCAD2 or any other CAD-like protein is responsible for the residual 15% CAD activity and that down-regulation of PtaCAD2 would result in a stronger accumulation of coniferaldehyde. However, aside from the fact that only 15% of CAD activity remained, the catalytic efficiency for coniferaldehyde of the corresponding P. trichocarpa PtrCAD2 is much lower than that of PtrCAD1 (Shi et al., 2010; Wang et al., 2014), making this scenario questionable.
A more likely explanation is that coniferaldehyde is metabolized in a different way. Insight into how coniferaldehyde is metabolized in the hpCAD lines was provided by both lignin analysis and phenolic profiling. The CAD1 down-regulation resulted in the accumulation of dihydroferulic acid hexoside 5 and 12, feruloyl dihydroxybenzoic acid hexoside 9, feruloyl quinic acid 4, feruloyl hydroxybenzyl hexoside 7, and feruloyl hexose 11 in the xylem (Fig. 4; Table V; Supplemental Table S4). The excess of coniferaldehyde, therefore, was likely metabolized into ferulic acid rather than being exported to the CW (Sibout et al., 2005). An oxidation route for cinnamaldehydes to their corresponding carboxylic acids has been established in Arabidopsis and is catalyzed by HYDROXYCINNAMALDEHYDE DEHYDROGENASE/REDUCED EPIDERMAL FLUORESCENCE1 (Goujon et al., 2003; Nair et al., 2004). Transcriptomics in an Arabidopsis cad-c cad-d double mutant showed higher expression of the corresponding gene encoding this aldehyde dehydrogenase (Sibout et al., 2005). The accumulation of dihydroferulic acid hexosides 5 and 12 can possibly be explained by the activity of an alkenal DBR. The activities of an Arabidopsis DBR reducing the C7-C8 double bond of p-coumaraldehyde and coniferaldehyde, and an apple (Malus × domestica) DBR reducing the double bond in p-coumaroyl-CoA and feruloyl-CoA, have been shown by enzymatic assays (Youn et al., 2006b; Ibdah et al., 2014). Vanillic acid hexoside 13 also accumulated in the CAD1-deficient poplars (Table V). This benzenoid is derived via an as yet largely unexplored chain-shortening pathway from feruloyl-CoA, the latter likely being derived from ferulic acid via 4-COUMARATE COENZYME A LIGASE activity (Widhalm and Dudareva, 2015).
As compared with the ferulic acid derivative pools, sinapic acid derivative pools were much more elevated in the xylem of hpCAD lines (Table V): syringyl lactic acid hexoside 2 and sinapic acid hexoside 3 accumulated 8,650- and 488-fold, respectively. Syringyl lactic acid and its aglycone have not yet been described as plant metabolites and, consequently, nothing is known about their biosynthesis. The structural similarity to phenyl lactic acid and 4-hydroxyphenyl lactic acid suggests that syringyl lactic acid could be formed from these compounds: starting from 4-hydroxyphenyl lactic acid, subsequent hydroxylations and methylations would introduce the necessary methoxyl substituents on the benzene ring to produce syringyl lactic acid. The biosynthetic route from Tyr to 4-hydroxyphenyl lactic acid has been described for plants and holds a transamination followed by a reduction (Petersen et al., 1993).
Alternatively, syringyl lactic acid might be synthesized from sinapic acid; the high sinapic acid hexoside 3 levels support such a route. Mechanistically, the reaction might be similar to the hydroxylation step during α-oxidation of fatty acids (Hitchcock and Rose, 1971). Opposite to the hydroxylation step in fatty acid β-oxidation, α-oxidation-associated hydroxylation occurs on a single bond rather than a double bond. Thus, for this mechanism to operate, a prior double bond reduction of sinapic acid to dihydrosinapic acid (or from sinapaldehyde to dihydrosinapaldehyde followed by its oxidation to dihydrosinapic acid) is necessary, after which dihydrosinapic acid is hydroxylated to syringyl lactic acid. As stated above, DBR enzymes from Arabidopsis and apple reducing the C7-C8 double bond of phenylpropanoids have been described (Youn et al., 2006b; Ibdah et al., 2014).
The observations that syringyl lactic acid hexoside 2 and sinapic acid hexoside 3 accumulated in the phenolic pool, and that syringaldehyde appeared to accumulate in the lignin (Table IV), could indicate that only part of the sinapaldehyde ended up in the lignin, with a part metabolized into sinapic acid and syringaldehyde, via metabolic routes similar to those for coniferaldehyde. Alternatively, sinapic acid and derivatives were made via parallel hydroxylation of their G-type homologs via 5-hydroxylation and 5-O-methyltransferase reactions on the free acid at the cinnamoid/benzenoid levels (e.g. from ferulic acid via 5-hydroxyferulic acid to sinapic acid and from vanillin via 5-hydroxyvanillin to syringaldehyde). The more than 100-fold increase in p-coumaric acid hexosyl hexose 6 suggests a similar conversion of p-coumaraldehyde to p-coumaric acid. Caffeoyl quinic acid 10, caffeoyl hexose 14, and dihydroxybenzoic acid (protocatechuic acid) 8 are then potentially made via p-coumaroyl-CoA (Fig. 4).
The suggested efficient conversion of cinnamaldehydes to cinnamic acids by a putative poplar HCALDH homolog, followed by hexosylation to detoxify the accumulating cinnamic acids, is in agreement with the absence of any noticeable accumulation of glycosylated oligolignols derived from the cinnamaldehyde's coupling with a normal monolignol. Indeed, we have shown previously that, in Arabidopsis leaf protoplasts, monolignols are coupled not only in the CW but also in the cytoplasm, upon which the coupling products are glycosylated for sequestration in the vacuole (Dima et al., 2015). Also in Arabidopsis stems, glycosylation of oligolignols is a commonly observed phenomenon, especially in plants perturbed in the lignin biosynthetic pathway (Vanholme et al., 2010b, 2012b). In poplar xylem, this process appears to be less common, although a glycosylated oligolignol 20 was detected with low abundance among the metabolites in the hpCAD lines (Table V).
Based on a predictive kinetic metabolic flux (PKMF) model of the phenylpropanoid pathway, it has been predicted that a reduction in CAD activity in poplar would result in the accumulation of coniferaldehyde and sinapaldehyde (Wang et al., 2014). However, the PKMF model does not predict the increased fluxes observed in hpCAD poplars toward other pathway intermediates, such as ferulic acid and sinapic acid, that we propose to be likely derived from the excess of free cinnamaldehydes, nor toward metabolites outside the core phenylpropanoid pathway, such as syringaldehyde, vanillic acid, dihydroferulic acid hexoside, or the newly identified syringyl lactic acid hexoside (Tables III and V). Our data show the relevance of broad phenolic profiling (lignomics; Morreel et al., 2010b) in suggesting metabolites and enzymes (e.g. HCALDH and DBR) as input parameters into the current PKMF model of the phenylpropanoid pathway to allow for a more accurate prediction of the fluxes through this pathway.
Sinapaldehyde 8-8 Homodimerization Can Start the Lignin Chain
The observation that the S′(8-8)S′ dimer 1 was the most accumulating compound suggests that S′(8-8)S′ less readily allows further coupling with monolignols. This was also observed in the oxidative coupling assays, where only two trimers were detected in which S′(8-8)S′ was 4-O-8 coupled with a coniferyl alcohol or a sinapaldehyde. Part of the S′(8-8)S′, however, was detected as a new structure in the lignin by NMR (Fig. 2A), suggesting that sinapaldehyde dimerization is a way of starting lignin chains in the hpCAD lines. Lignin chains in hardwoods start by monolignol dimerization, producing either syringaresinol (that results in resinol units C in the lignin) from 8-8-coupling of sinapyl alcohol (primarily) or a hydroxycinnamyl alcohol end group X1 from coniferyl or sinapyl alcohol coupling at its 8-position with coniferyl alcohol at its 5- or 4-O-position (to result in phenylcoumaran B or β-ether A structures, respectively; Fig. 2B). As noted in the aliphatic regions of the NMR spectra (Fig. 2C), the normal resinol units C are sharply reduced in the hpCAD lines, whereas the level of hydroxycinnamyl alcohol end groups X1 is increased mildly. Again, as observed in the aldehyde region of the NMR spectra (Fig. 2A), lignin chains also are apparently started from sinapaldehyde 8-8 homodimerization, as evidenced by the appearance of this new structure in the lignin.
Repercussions of Syringaldehyde and Sinapaldehyde Incorporation on Lignin Structure
The increased incorporation of syringaldehydes into lignin has further repercussions on the structure of lignin and its physicochemical properties (Table III). Benzenoids lack an 8- (or β)-carbon; therefore, they can only start a lignin polymer chain (Vanholme et al., 2012a). Generally speaking, benzenoids could enter into 5-5- or 4-O-5-coupling with a lignin oligomer/polymer, in which the units still occupy terminal positions on the polymer (5-5-coupling is not possible for syringaldehyde, as the C5 is methoxylated). In other words, benzenoids cannot incorporate into the backbone of growing chains. The increased abundance of syringaldehyde in hpCAD lines (Table III), together with the reduced total lignin amount, implies that the average lignin polymer chain in hpCAD lines is shorter than that in the wild type. This hypothesis is supported by the observation that the relative fraction of phenolic end groups (detected as methylated thioacidolysis products) was increased in hpCAD lines (Table IV).
The incorporation of sinapaldehyde into the lignin (Fig. 2; Table III) resulted in structures and properties that are predicted to be substantially different from those derived from the canonical monolignols. When monolignols incorporate via their β- or C8-position into β-aryl ether structures, the quinone methide intermediate gets rearomatized via a nucleophilic attack. Generally, the nucleophile is water and results in the formation of an α-hydroxy functionality (Vanholme et al., 2012a). However, when hydroxycinnamaldehydes incorporate, rearomatization by elimination of the acidic proton on the C8-position (which is also termed the α-position relative to the aldehyde functionality) outcompetes nucleophilic addition. This mechanism consequently leads to (1) lignin with fewer hydroxyl functions (i.e. one aldehydic carbonyl function for each hydroxycinnamaldehyde coupled into a β-ether structure, as opposed to two hydroxyl functions for canonical monolignols coupled into a β-ether structure), and consequently lignin that is more hydrophobic, which may reduce its noncovalent associations with hemicelluloses (Carmona et al., 2015). In addition, the mechanism leads to (2) β-ether moieties with conjugated γ-carbonyl functionalities. Such structures are more prone to alkaline and/or oxidative degradation as compared with the typical β-ether moieties derived from canonical monomers (Tsuji et al., 2015). Moreover, this mechanism (3) also prevents the nucleophilic attack by nucleophiles other than water (e.g. alcohol and carboxylic acid groups from hemicelluloses), thereby avoiding the creation of benzyl ether or benzyl ester bonds that putatively contribute to recalcitrant lignin-carbohydrate complexes (Mottiar et al., 2016).
In summary, the lignin in hpCAD lines differs from that in wild-type lignin by the substantial incorporation of sinapaldehyde into the polymer. Lignification involving copolymerization of monolignols with sinapaldehyde results in seven characteristics that potentially influence the physicochemical properties of the wood and that could be of relevance for industrial processing of the wood: (1) 10% less lignin; (2) shorter lignin polymer chains with (3) a consequently higher proportion of free-phenolic end groups, resulting in lignin that is more alkali-soluble; (4) conjugated carbonyl functions that facilitate lignin cleavage under alkaline conditions; (5) a more hydrophobic lignin (because the β-ether units, for example, have two fewer hydroxyl groups per unit); (6) the possibility of a weaker noncovalent association with hemicelluloses; and possibly (7) lower actual lignin-hemicellulose bonding, making the CW more readily accessible.
CAD1 Down-Regulation Results in Improved Saccharification after Alkaline Pretreatment
Although the hpCAD lines had ∼10% less KL, the saccharification efficiencies for hpCAD lines were increased compared with the wild type only when an alkaline pretreatment preceded the saccharification protocol. We observed increased saccharification efficiencies ranging from 40% (hpCAD24, 6.25 mm NaOH) up to 81% (hpCAD4, 62.5 mm NaOH; Fig. 6A; Supplemental Table S6) in incomplete digestion experiments aimed at determining saccharification ease. Together with a similar cellulose content for the hpCAD lines and the wild type, the cellulose conversion increased from 27% in the wild type up to 39% in hpCAD19 (6.25 mm NaOH) and from 34% in the wild type up to 61% in hpCAD4 (62.5 mm NaOH; Fig. 6B; Supplemental Table S6). When an acid pretreatment (with sulfuric acid), hot water, or no pretreatment preceded the saccharification step, the saccharification efficiencies (and cellulose conversions) were similar for the hpCAD poplar lines and the wild type. The same was observed in alfalfa, where no increase in saccharification yield could be detected in both untreated and sulfuric acid-pretreated CAD-deficient plants (Jackson et al., 2008). In contrast, and presumably largely because of the substantial CW compositional differences between monocots and dicots (Popper et al., 2011), CAD-deficient switchgrass showed an increased saccharification efficiency (the ratio of sugars released by enzymatic hydrolysis to the total sugars available in the CW) of 19% to 89% without pretreatment and of 19% to 44% with a similar sulfuric acid pretreatment to that used in alfalfa (Fu et al., 2011). Also, CAD-deficient maize stems showed a 25% increased saccharification yield when no pretreatment was applied (Fornalé et al., 2012).
The enzyme mix that was used for saccharification also contained hemicellulases, resulting in the release of Xyl. Remarkably, and in contrast to the Glc release, the amount of released Xyl during saccharification was increased for the hpCAD poplar lines compared with the wild type, not only upon alkaline pretreatment but upon every pretreatment that was applied (Fig. 6C; Supplemental Table S6). The highest increase was observed with a hot water pretreatment (up to +153%); however, the absolute amount of Xyl released with no or hot water pretreatment remained low. The Xyl released upon saccharification with an acid pretreatment resulted in equivalent amounts as upon alkaline pretreatment (6.25 mm NaOH). The higher Xyl releases during saccharification of hpCAD-derived wood samples could be explained in part by the hemicellulose composition of the hpCAD poplars, which is enriched in Xyl by ∼3% (Table II). However, this increase in Xyl content cannot explain the total increase. Based on the shifts in lignin composition, a reduced interaction between lignin and hemicelluloses was predicted, both noncovalently and covalently, as compared with the wild type, potentially explaining the increased hemicellulose degradation. For industrial bioethanol production, the Xyl sugars also can be fermented into bioethanol (e.g. when using yeast genetically engineered to ferment pentoses; Demeke et al., 2013).
CAD1 Down-Regulation Does Not Affect Plant Yield under Greenhouse Conditions
Although the lignin amount of hpCAD poplars was reduced, and its structure heavily altered, the trees had no biomass yield penalty when grown for 3.5 months in the greenhouse (Table I). In contrast, yield penalties have been observed previously in other plants in which CAD activity was severely reduced. For example, Arabidopsis cad-c cad-d double mutants were reported to have shorter and weaker stems (Sibout et al., 2005). Nevertheless, the hpCAD poplar lines will ultimately need to be evaluated in a field trial to investigate whether they can grow outdoors without a yield penalty while maintaining the beneficial effects on saccharification efficiency. Environmental factors have been shown previously to interact with CAD deficiency in CAD-deficient M. truncatula: the phenotype of these plants was equal to that of the wild type when grown at ambient temperatures, but the plants were dwarfed when grown at elevated temperatures (30°C; Zhao et al., 2013). In contrast, down-regulating CAD in Nicotiana attenuata produced plants with thin and structurally unstable stems when grown in the greenhouse, but the normal phenotype was restored when plants were grown outdoors (Kaur et al., 2012). Hence, it is important to investigate how the hpCAD poplars with this level of lignin alteration will respond when grown in the field, especially regarding biomass and bioethanol yield. A field trial with the three lines described here was established under a short-rotation culture (VIB, 2013) to answer these questions and to provide sufficient biomass for semi-industrial processing in a pilot biorefinery.
MATERIALS AND METHODS
Generation of hpCAD Poplar Transgenic Lines
To prepare the binary vector for poplar transformation, the full-length PtaCAD1 coding sequence from Populus tremula × Populus alba (INRA 717-1B4), corresponding to PtrCAD1 (Potri.009G095800.1) from Populus trichocarpa (99% identity; Supplemental Fig. S1A), was first PCR amplified from CF233412 plasmid clone (Déjardin et al., 2004) with Pfx polymerase and the following forward and reverse primers (5′-CACCCTGGTGCCGCGCGGCAGCATGGGTAGCCTTGAAACA-3′ and 5′-TCAGGGAATAAGCTTGCTAC-3′). The PCR product was then cloned into the pENTR vector using the pENTR/D-TOPO cloning kit (Invitrogen, Life Technologies) according to the manufacturer’s instructions to generate pENTR_CAD, an entry clone for the Gateway cloning system (Invitrogen). The pENTR_CAD construct was verified by sequencing. The CAD1 coding sequence was then shuttled from pENTR_CAD to pHellsgate8 (Helliwell and Waterhouse, 2003) using Gateway LR Clonase. The generated pHG8_CAD binary vector was suited for intron-spliced hairpin RNA-mediated gene silencing. It was transferred into Agrobacterium tumefaciens strain C58 (pMP90) using triparental mating. Eighteen independent transgenic lines were obtained from A. tumefaciens-mediated transformation of the INRA clone 717-1B4, according to Leplé et al. (1992).
Sequence Analysis and Protein Modeling
Homologs of PtaCAD1 were identified by BLAST from the P. tremula × P. alba 717-1B4 genome (http://aspendb.uga.edu/s717) using the PtaCAD1 coding sequence. Three-dimensional protein structures of both PtaCAD1 and Pta_tCAD1 were generated using the crystal structure of AtCAD5 (Protein Data Bank no. 2cf6) as a modeling template in the fully automated protein structure homology-modeling server SWISS-MODEL (Youn et al., 2006a; Biasini et al., 2014).
Plant Material
The 18 transgenic lines and the wild type were micropropagated, acclimatized in vitro, and grown in three replicates for 3 months in the greenhouse. A 5-cm stem fragment was collected at the base of the plant for first phenotypic screenings (xylem color and anatomy). These stem sections were photographed with a Zeiss Stemi 2000-C stereomicroscope. An additional 10-cm stem fragment was debarked, and the xylem was scraped and ground in liquid nitrogen for CAD activity assays.
Three CAD1-deficient lines (hpCAD4, hpCAD19, and hpCAD24) were chosen and, together with the wild-type poplar, simultaneously micropropagated in vitro to obtain 40 ramets of each line that were grown randomly in the greenhouse. For metabolite profiling, stems of 10 ramets of the three hpCAD lines and 30 ramets of the wild type were cut 10 cm above soil level after 3 months of growth. A basal 10-cm stem fragment was sampled and debarked. Subsequently, the debarked stem samples were frozen immediately in liquid nitrogen and stored at −70°C. Two weeks later (after 3.5 months of growth), another five ramets per line were cut 10 cm above soil level. Height and fresh weight were determined immediately after harvesting. The stem was debarked, air dried, ground, sieved to pass a mesh of 0.5 mm, and used for CW analyses, NMR structural profiling, and saccharification assays. For microscopy purposes, cross sections were made from three independent 4-month-old hpCAD and wild-type poplars.
Assay for CAD Activity
Proteins were extracted from 200 mg of scraped developing xylem by grinding for 1 min in 3 mL of ice-cold 100 mm Tris-HCl, pH 7.5, 2% (w/v) polyethylene glycol 6000, 2% (w/v) polyvinylpolypyrrolidone, 200 mm sodium ascorbate, and 5 mm DTT. After two centrifugations at 13,000 rpm at 4°C for 10 min, the proteins in the final supernatant were quantified using the Bradford test (Bradford, 1976), and a calibration curve was prepared from BSA. CAD activity was assayed at 30°C on 2 µg of extracted proteins in 350 µL of 100 mm Tris-HCl, pH 8.8, 0.1 mm coniferyl alcohol, and 0.2 mm β-NADP. The reduction of NADP in NADPH was followed at 405 nm on a Multiskan Spectrum.
qRT-PCR
Transcript levels of PtaCAD1 (corresponding to PtrCAD1 [Potri.009G095800.1]), PtaCAD2 (corresponding to PtrCAD2 [Potri.016G078300.1]), and Pta_tCAD1 (corresponding to Potri.001G30000.1) were determined in stems of 10-week-old hpCAD and wild-type poplars using qRT-PCR. The top 50 cm of the stem was removed, and the 10-cm stem piece below the cut was immediately debarked and frozen in liquid nitrogen. Xylem was scraped, and total RNA was isolated with the RNeasy Plant Mini Kit (Qiagen) and treated with Ambion DNA-free (Life Technologies) to remove contamination with genomic DNA. A total of 1 μg was used as a template for the synthesis of cDNA using the iScript cDNA Synthesis Kit (Bio-Rad). Samples were run in triplicate on a LightCycler 480 Real-Time SYBR Green PCR System (Roche) according to the manufacturer’s instructions. Fluorescence values were exported from the LightCycler program, whereupon Ct values, normalization factors, and primer efficiencies were calculated according to Ramakers et al. (2003) using two reference genes: 18S (AF206999) and UBQ (BU879229; Brunner et al., 2004). qRT-PCR primers are listed in Supplemental Figure S1C (for PtaCAD1 and PtaCAD2) and Supplemental Figure S3B (for Pta_tCAD1).
Microscopy
Fresh 30-µm-thick transverse stem sections were prepared with a rotary microtome (Leica RM 2155; Leica Microsystems) equipped with disposable blades (Low Profile Sec35p; Microm). For laser scanning confocal microscopy, the sections were mounted in fluoromount G medium between a slide and coverslip and examined with a Zeiss LSM 700 microscope with 2% laser energy. The excitation wavelength of 488 nm and emission wavelength of 495 to 700 nm were used for imaging autofluorescence. Lignins were detected in stem cross sections using Wiesner and Mäule stains. For Wiesner staining, which detects cinnamaldehyde end groups on lignins, giving a pink-violet color, fresh sections were mounted directly on slides in phloroglucinol-HCl solution and observed immediately with a light microscope (Leica DMR; Leica Microsystems) with a Leica DFC 320 digital camera. For Mäule staining, which visualizes S lignins, fresh cross sections were immersed in 1% (w/v) potassium permanganate aqueous solution for 15 min, followed by 10 s of water washing and then 30 s of treatment with 10% HCl. After a subsequent treatment with 5% (w/v) sodium bicarbonate, a red-purple color developed. Sections were mounted on slides in Aqueous Mount-quick Medium and observed using a light microscope, as described above.
CW Analysis
KL was measured according to the standard procedure, starting from 350 mg of extractive-free CW residue (Méchin et al., 2014). The ASL was measured spectrophotometrically as described previously (Dence, 1992). Lignin composition was determined by thioacidolysis (Lapierre et al., 1999), and the frequency of free-phenolic end groups in lignin was determined by thioacidolysis after exhaustive phenol methylation of the samples with diazomethane (Pitre et al., 2007). The determination of p-hydroxybenzoate esters and other phenolics linked to poplar lignins was by mild alkaline hydrolysis according to a previously published procedure (Lapierre et al., 1999). Crystalline cellulose content was determined by the Updegraff method, and compositional analyses of hemicelluloses was by the alditol-acetate assay (Foster et al., 2010).
NMR
The whole-plant CW gel-state NMR samples in dimethyl sulfoxide-d6/pyridine-d5 (4:1) were prepared as described previously (Kim et al., 2008; Kim and Ralph, 2010; Mansfield et al., 2012). NMR experiments were performed on a Bruker Biospin AVANCE 700-MHz spectrometer fitted with a cryogenically cooled 5-mm quadruple-resonance 1H/31P/13C/15N QCI gradient probe with inverse geometry (proton coils closest to the sample) as described previously (Kim et al., 2008; Kim and Ralph, 2010; Mansfield et al., 2012). Volume integration of contours in HSQC plots was performed on data processed without linear prediction and used Bruker’s TopSpin 3.2 (Mac version) software. Note that integrals are to be used for relative comparisons: they do not represent true quantification and, in particular, end groups are substantially overestimated (Mansfield et al., 2012). The peaks characteristic of the lignin structures and their compositions were selected and estimated after the contour-level adjustment to achieve optimal peak separation. Quantification was performed for wild-type (four replicates) and CAD1-deficient (three replicates of each line) poplars.
To confirm the identity of the S′(8-8)S′ dimer, this compound was chemically synthesized by radical reactions in two different ways. (1) Sinapaldehyde (1 g, 4.81 mmol), acetone:water (20 mL:980 mL), horseradish peroxidase (5 mg), and hydrogen peroxide (1%, 20 mL) were combined. (2) Sinapaldehyde (530 mg, 2.59 mmol) was dissolved in ethyl acetate (30 mL). Silver(I) oxide (720.7 mg, 3.11 mmol) was added, and the reaction mixture was stirred overnight at room temperature. The mixture was filtered through a fine-sintered glass filter to remove Ag and evaporated to give a red solid. Separation of the crude products was performed on preparative thin-layer chromatography plates (CHCl3:ethyl acetate, 1:1, v/v). The S′(8-8)S′ dehydrodimer was obtained as the major product from both methods. The structure of S′(8-8)S′ was confirmed by NMR: 1H NMR (acetone-d6) δ 9.67 (s, 2H, 9), 7.99 (br s, 2H, ArOH), 7.80 (s, 2H, 7), 7.03 (s, 4H, 2/6), 3.71 (s, 12H, OMe); 13C NMR (acetone-d6) δ 192.7 (9), 153.0 (7), 148.6 (3/5), 140.0 (4), 134.7 (8), 126.0 (1), 109.0 (2/6), 56.5 (OMe).
Metabolomics
Debarked 10-cm basal stem fragments from 3-month-old, greenhouse-grown poplar were scraped (30–200 mg dry weight) with a scalpel, and the resulting isolated xylem was ground using mortar and pestle. Liquid-liquid extraction of the homogenized plant material was performed with 5 mL of methanol. Of the supernatants, 1 mL was lyophilized and redissolved in 0.8 mL of milliQ water:cyclohexane (1:1, v/v). The tubes were vortexed and centrifuged at 14,000 rpm (20,000g) for 10 min. A 200-µL aliquot of the lower water phase was transferred to an ultra-HPLC vial. A 15-µL aliquot of the water phase was injected on an ultra-HPLC system (Waters Acquity UPLC) equipped with a BEH C18 column (2.1 × 150 mm, 1.7 µm; Waters) and hyphenated to a time-of-flight mass spectrometer (Synapt Q-Tof; Waters) using gradient elution. Buffer A was composed of water containing 1% (v/v) acetonitrile and 0.1% (v/v) formic acid (pH 3). Buffer B was composed of acetonitrile containing 1% (v/v) water and 0.1% (v/v) formic acid (pH 3). The following gradient was applied: 95% A for 0.1 min, decreased to 50% A in 30 min at a flow of 350 µL min−1 and a column temperature of 40°C. The autosampler temperature was maintained at 10°C. For MS analysis, the flow was diverted to the mass spectrometer equipped with an electrospray ionization source and lockspray interface for accurate mass measurements. The MS source parameters were as follows: capillary voltage, 2.6 kV; sampling cone, 37 V; extraction cone, 3.5 V; source temperature, 120°C; desolvation temperature, 400°C; cone gas flow, 50 L h−1; and desolvation gas flow, 550 L h−1. The collision energy for the trap and transfer cells were 4 and 3 V, respectively. For data acquisition, the dynamic range enhancement mode was activated. Full-scan data were recorded in negative centroid V-mode; the mass range between m/z 100 and 1,000, with a scan speed of 0.2 s scan−1, was recorded with Masslynx software (Waters).
For structural elucidation, MS/MS was used. For MS/MS, all settings were the same as in full MS, except the collision energy was ramped from 15 to 45 eV in the trap and the scan time was set at 0.5 s. Leucin-enkephalin (250 pg μL−1 solubilized in water:acetonitrile 1:1 [v/v] with 0.1% formic acid) was used for the lock mass calibration, with scanning every 10 s with a scan time of 0.5 s. Chromatograms were processed via Progenesis QI version 2.1 (Nonlinear Dynamics), yielding 7,521 m/z features. The m/z features that were associated with the same compound were grouped based on similar retention time and a high correlation in abundances between the biological replicates (Morreel et al., 2014), yielding 2,154 m/z feature groups. Normalization of the m/z feature abundances was performed to the dry weight of the samples. The m/z features that had average abundance lower than 500 in each of the four lines were filtered away. All statistics were performed in R version 3.1.2 on arcsinh transformed ion intensities. One-way ANOVA followed by Bonferroni posthoc tests (P < 0.05 for each of the three transgenic lines) were performed with the lm function and the pairwise.t.test function, respectively. An experiment-wise significance threshold (P < 0.05) was computed using the qvalue package in R. The m/z features for which the abundances were significantly different, and for which the averaged fold change of the three transgenic lines was greater than 2 or less than 0.5, were retained. This resulted in a list of 589 m/z features (corresponding to 410 m/z feature groups), of which 508 (corresponding to 348 m/z feature groups) were higher and 81 (corresponding to 64 m/z feature groups) were lower in abundance in the xylem of hpCAD lines than in the wild type.
Compound Purification
Debarked stems of poplars grown for 3 months in the greenhouse were homogenized using GRINDOMIX GM 200 (Retch). One liter of methanol was added to 100 g of fresh plant material. A total of 500 mL of the supernatant was lyophilized and redissolved in 11 mL of milliQ water:cyclohexane (1:1, v/v). Subsequently, 10 mL of the aqueous phase was injected onto a Waters Autopurification System (Waters) hyphenated to an Acquity QDa detector (Waters). Reverse-phase separation was performed on a Sunfire C18 prep column (19 × 150 mm, 5 μm). Except for the flow (10 mL min−1), solvent compositions, gradient conditions, and column temperature were in agreement with those used for metabolomics.
Oxidative Coupling Assays
The oxidative coupling assays were performed as described previously (Niculaes et al., 2014). Coniferyl alcohol, sinapyl alcohol, coniferaldehyde, and sinapaldehyde were incubated with horseradish peroxidase in all possible combinations. The assays were initiated by adding hydrogen peroxide. After incubation, the resulting combinatorial coupling products were subjected to UHPLC-MS analysis as described above.
Saccharification Assay
Debarked poplar stems were air dried, ground extensively using a Retsch MM300 mixer mill (Retsch), and sieved to pass a mesh of 0.5 mm. Material was weighed, and saccharification assays were performed using the iWALL custom-designed robot (Labman Automation) as has been described in detail (Santoro et al., 2010). The dilute base pretreatment solutions consisted of 6.25 and 62.5 mm NaOH. The dilute acid pretreatment solution was 0.4 m H2SO4, as described (Santoro et al., 2010). The conditions for the hot water pretreatment were as similar as possible to the other pretreatments (i.e. 90°C for 3 h).
Statistics
All statistics, unless specified, were performed using SAS Enterprise Guide 6 (SAS Institute). One-way ANOVA determinations followed by posthoc two-sided Dunnett’s tests were applied to test for significant differences between the transgenic lines and the wild type. Differences with a Dunnett adjusted P < 0.05 were considered significant.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers Potri.009G095800.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence and qRT-PCR of P. tremula × P. alba CAD1.
Supplemental Figure S2. Phenotype and residual CAD activity.
Supplemental Figure S3. Gene structure of Pta_tCAD1.
Supplemental Figure S4. Microscopy for hpCAD19 and hpCAD24.
Supplemental Figure S5. Structures of thioacidolysis-derived compounds.
Supplemental Figure S6. Identification of S′(8-8)S′ by NMR.
Supplemental Figure S7. Gas-phase fragmentation of syringyl lactic acid hexoside.
Supplemental Table S1. Protein sequence analysis of PtaCAD1 and Pta_tCAD1.
Supplemental Table S2. Yields of conventional lignin thioacidolysis monomers.
Supplemental Table S3. Lignin NMR data.
Supplemental Table S4. UHPLC-MS-based phenolic profiling.
Supplemental Table S5. Dimers and trimers made by in vitro oxidative coupling assays.
Supplemental Table S6. Saccharification results.
Supplemental Information S1. Features of the lignin models.
Acknowledgments
We thank Nadège Millet for the production of the hpCAD lines and Nathalie Boizot, Camille Grand-Perret, Véronique Lainé-Prade, and Marie-Claude Lesage-Descauses for technical assistance. AGPF experiments at Orléans relied on the greenhouse facilities of GBFOR Experimental Unit and the XYLOBIOTECH technical facility belonging to the XYLOFOREST platform (ANR-10-EQPX-16).
Glossary
- CW
cell wall
- H
p-hydroxyphenyl
- G
guaiacyl
- S
syringyl
- RNAi
RNA interference
- UHPLC-MS
ultra-high-performance liquid chromatography-mass spectrometry
- qRT
quantitative reverse transcription
- KL
Klason lignin
- ASL
acid-soluble lignin
- HSQC
heteronuclear single-quantum coherence
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- DHP
dehydrogenation polymer
- PKMF
predictive kinetic metabolic flux
Footnotes
This work was supported by grants from the Multidisciplinary Research Partnership Biotechnology for a Sustainable Economy of Ghent University, the Agency for Innovation by Science and Technology (IWT) through the SBO project BIOLEUM and the SBO-FISH project ARBOREF, the European Union’s Seventh Framework Programme for research, technological development, and demonstration (ENERGYPOPLAR FP7-211917 and MultiBioPro grant agreement 311804), and the Hercules Foundation (Grant AUGE/014). H.K. and J.R. were funded by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-FC02-07ER64494).
Articles can be viewed without a subscription.
References
- Anderson NA, Tobimatsu Y, Ciesielski PN, Ximenes E, Ralph J, Donohoe BS, Ladisch M, Chapple C (2015) Manipulation of guaiacyl and syringyl monomer biosynthesis in an Arabidopsis cinnamyl alcohol dehydrogenase mutant results in atypical lignin biosynthesis and modified cell wall structure. Plant Cell 27: 2195–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandu ML, Grubbs T, Kater M, Desaire H (2006) Collision induced dissociation of alpha hydroxy acids: evidence of an ion-neutral complex intermediate. Int J Mass Spectrom 251: 40–46 [Google Scholar]
- Barakat A, Bagniewska-Zadworna A, Choi A, Plakkat U, DiLoreto DS, Yellanki P, Carlson JE (2009) The cinnamyl alcohol dehydrogenase gene family in Populus: phylogeny, organization, and expression. BMC Plant Biol 9: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher M, Bernard-Vailhé MA, Chabbert B, Besle JM, Opsomer C, Van Montagu M, Botterman J (1999) Down-regulation of cinnamyl alcohol dehydrogenase in transgenic alfalfa (Medicago sativa L.) and the effect on lignin composition and digestibility. Plant Mol Biol 39: 437–447 [DOI] [PubMed] [Google Scholar]
- Baucher M, Chabbert B, Pilate G, Van Doorsselaere J, Tollier MT, Petit-Conil M, Cornu D, Monties B, Van Montagu M, Inzé D, et al. (1996) Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in poplar. Plant Physiol 112: 1479–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, et al. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42: W252–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C (2013) Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr Opin Biotechnol 24: 336–343 [DOI] [PubMed] [Google Scholar]
- Bouvier d’Yvoire M, Bouchabke-Coussa O, Voorend W, Antelme S, Cézard L, Legée F, Lebris P, Legay S, Whitehead C, McQueen-Mason SJ, et al. (2013) Disrupting the cinnamyl alcohol dehydrogenase 1 gene (BdCAD1) leads to altered lignification and improved saccharification in Brachypodium distachyon. Plant J 73: 496–508 [DOI] [PubMed] [Google Scholar]
- Bowie JH. (1990) The fragmentations of even-electron organic negative ions. Mass Spectrom Rev 9: 349–379 [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brunner AM, Yakovlev IA, Strauss SH (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona C, Langan P, Smith JC, Petridis L (2015) Why genetic modification of lignin leads to low-recalcitrance biomass. Phys Chem Chem Phys 17: 358–364 [DOI] [PubMed] [Google Scholar]
- Chabannes M, Barakate A, Lapierre C, Marita JM, Ralph J, Pean M, Danoun S, Halpin C, Grima-Pettenati J, Boudet AM (2001) Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J 28: 257–270 [DOI] [PubMed] [Google Scholar]
- Chen C, Meyermans H, Burggraeve B, De Rycke RM, Inoue K, De Vleesschauwer V, Steenackers M, Van Montagu MC, Engler GJ, Boerjan WA (2000) Cell-specific and conditional expression of caffeoyl-coenzyme A-3-O-methyltransferase in poplar. Plant Physiol 123: 853–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Chen HC, Li Q, Shuford CM, Liu J, Muddiman DC, Sederoff RR, Chiang VL (2011) Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc Natl Acad Sci USA 108: 21253–21258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déjardin A, Leplé JC, Lesage-Descauses MC, Costa G, Pilate G (2004) Expressed sequence tags from poplar wood tissues: a comparative analysis from multiple libraries. Plant Biol (Stuttg) 6: 55–64 [DOI] [PubMed] [Google Scholar]
- Demeke MM, Dumortier F, Li Y, Broeckx T, Foulquié-Moreno MR, Thevelein JM (2013) Combining inhibitor tolerance and D-xylose fermentation in industrial Saccharomyces cerevisiae for efficient lignocellulose-based bioethanol production. Biotechnol Biofuels 6: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dence CW. (1992) The determination of lignin. In Lin SY, Dence CW, eds, Methods in Lignin Chemistry. Springer-Verlag, Berlin, pp 33–61 [Google Scholar]
- Dima O, Morreel K, Vanholme B, Kim H, Ralph J, Boerjan W (2015) Small glycosylated lignin oligomers are stored in Arabidopsis leaf vacuoles. Plant Cell 27: 695–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornalé S, Capellades M, Encina A, Wang K, Irar S, Lapierre C, Ruel K, Joseleau JP, Berenguer J, Puigdomènech P, et al. (2012) Altered lignin biosynthesis improves cellulosic bioethanol production in transgenic maize plants down-regulated for cinnamyl alcohol dehydrogenase. Mol Plant 5: 817–830 [DOI] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M (2010) Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass). Part II. Carbohydrates. J Vis Exp 1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournand D, Cathala B, Lapierre C (2003) Initial steps of the peroxidase-catalyzed polymerization of coniferyl alcohol and/or sinapyl aldehyde: capillary zone electrophoresis study of pH effect. Phytochemistry 62: 139–146 [DOI] [PubMed] [Google Scholar]
- Freudenberg K. (1959) Biosynthesis and constitution of lignin. Nature 183: 1152–1155 [DOI] [PubMed] [Google Scholar]
- Fu C, Xiao X, Xi Y, Ge Y, Chen F, Bouton J, Dixon RA, Wang ZY (2011) Downregulation of cinnamyl alcohol dehydrogenase (CAD) leads to improved saccharification efficiency in switchgrass. BioEnergy Res 4: 153–164 [Google Scholar]
- Gao D, Haarmeyer C, Balan V, Whitehead TA, Dale BE, Chundawat SP (2014) Lignin triggers irreversible cellulase loss during pretreated lignocellulosic biomass saccharification. Biotechnol Biofuels 7: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffner D, Van Doorsselaere J, Yahiaoui N, Samaj J, Grima-Pettenati J, Boudet AM (1998) A novel aromatic alcohol dehydrogenase in higher plants: molecular cloning and expression. Plant Mol Biol 36: 755–765 [DOI] [PubMed] [Google Scholar]
- Goujon T, Sibout R, Pollet B, Maba B, Nussaume L, Bechtold N, Lu F, Ralph J, Mila I, Barrière Y, et al. (2003) A new Arabidopsis thaliana mutant deficient in the expression of O-methyltransferase impacts lignins and sinapoyl esters. Plant Mol Biol 51: 973–989 [DOI] [PubMed] [Google Scholar]
- Greene LE, Grossert JS, White RL (2013) Correlations of ion structure with multiple fragmentation pathways arising from collision-induced dissociations of selected α-hydroxycarboxylic acid anions. J Mass Spectrom 48: 312–320 [DOI] [PubMed] [Google Scholar]
- Ha CM, Escamilla-Trevino L, Yarce JCS, Kim H, Ralph J, Chen F, Dixon RA (2016) An essential role of caffeoyl shikimate esterase in monolignol biosynthesis in Medicago truncatula. Plant J 86: 363–375 [DOI] [PubMed] [Google Scholar]
- Halpin C, Holt K, Chojecki J, Oliver D, Chabbert B, Monties B, Edwards K, Barakate A, Foxon GA (1998) Brown-midrib maize (bm1): a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J 14: 545–553 [DOI] [PubMed] [Google Scholar]
- Halpin C, Knight ME, Foxon GA, Campbell MM, Boudet AM, Boon JJ, Chabbert B, Tollier MT, Schuch W (1994) Manipulation of lignin quality by down-regulation of cinnamyl alcohol-dehydrogenase. Plant J 6: 339–350 [Google Scholar]
- Hawkins S, Samaj J, Lauvergeat V, Boudet A, Grima-Pettenati J (1997) Cinnamyl alcohol dehydrogenase: identification of new sites of promoter activity in transgenic poplar. Plant Physiol 113: 321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C, Waterhouse P (2003) Constructs and methods for high-throughput gene silencing in plants. Methods 30: 289–295 [DOI] [PubMed] [Google Scholar]
- Hibino T, Takabe K, Kawazu T, Shibata D, Higuchi T (1995) Increase of cinnamaldehyde groups in lignin of transgenic tobacco plants carrying an antisense gene for cinnamyl alcohol-dehydrogenase. Biosci Biotechnol Biochem 59: 929–931 [Google Scholar]
- Hitchcock C, Rose A (1971) The stereochemistry of alpha-oxidation of fatty acids in plants: the configuration of biosynthetic long-chain 2-hydroxy acids. Biochem J 125: 1155–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M (2004) Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16: 1446–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huis R, Morreel K, Fliniaux O, Lucau-Danila A, Fénart S, Grec S, Neutelings G, Chabbert B, Mesnard F, Boerjan W, et al. (2012) Natural hypolignification is associated with extensive oligolignol accumulation in flax stems. Plant Physiol 158: 1893–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibdah M, Berim A, Martens S, Valderrama ALH, Palmieri L, Lewinsohn E, Gang DR (2014) Identification and cloning of an NADPH-dependent hydroxycinnamoyl-CoA double bond reductase involved in dihydrochalcone formation in Malus×domestica Borkh. Phytochemistry 107: 24–31 [DOI] [PubMed] [Google Scholar]
- Jackson LA, Shadle GL, Zhou R, Nakashima J, Chen F, Dixon RA (2008) Improving saccharification efficiency of alfalfa stems through modification of the terminal stages of monolignol biosynthesis. BioEnergy Res 1: 180–192 [Google Scholar]
- Kaur H, Shaker K, Heinzel N, Ralph J, Gális I, Baldwin IT (2012) Environmental stresses of field growth allow cinnamyl alcohol dehydrogenase-deficient Nicotiana attenuata plants to compensate for their structural deficiencies. Plant Physiol 159: 1545–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ralph J (2010) Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6/pyridine-d5. Org Biomol Chem 8: 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ralph J, Akiyama T (2008) Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6. BioEnergy Res 1: 56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ralph J, Lu F, Pilate G, Leplé JC, Pollet B, Lapierre C (2002) Identification of the structure and origin of thioacidolysis marker compounds for cinnamyl alcohol dehydrogenase deficiency in angiosperms. J Biol Chem 277: 47412–47419 [DOI] [PubMed] [Google Scholar]
- Kim H, Ralph J, Lu F, Ralph SA, Boudet AM, MacKay JJ, Sederoff RR, Ito T, Kawai S, Ohashi H, et al. (2003) NMR analysis of lignins in CAD-deficient plants. Part 1. Incorporation of hydroxycinnamaldehydes and hydroxybenzaldehydes into lignins. Org Biomol Chem 1: 268–281 [DOI] [PubMed] [Google Scholar]
- Kim H, Ralph J, Yahiaoui N, Pean M, Boudet AM (2000) Cross-coupling of hydroxycinnamyl aldehydes into lignins. Org Lett 2: 2197–2200 [DOI] [PubMed] [Google Scholar]
- Lapierre C. (2010) Determining lignin structure by chemical degradations. In Heitner C, Dimmel DR, Schmidt JA, eds, Lignin and Lignans. CRC Press, Boca Raton, FL, pp 11–48 [Google Scholar]
- Lapierre C, Jouin D, Monties B (1989) On the molecular-origin of the alkali solubility of Gramineae lignins. Phytochemistry 28: 1401–1403 [Google Scholar]
- Lapierre C, Monties B, Rolando C (1988) Thioacidolyses of diazomethane-methylated pine compression wood and wheat straw in situ lignins. Holzforschung 42: 409–411 [Google Scholar]
- Lapierre C, Pilate G, Pollet B, Mila I, Leplé JC, Jouanin L, Kim H, Ralph J (2004) Signatures of cinnamyl alcohol dehydrogenase deficiency in poplar lignins. Phytochemistry 65: 313–321 [DOI] [PubMed] [Google Scholar]
- Lapierre C, Pollet B, Petit-Conil M, Toval G, Romero J, Pilate G, Leplé JC, Boerjan W, Ferret V, De Nadai V, et al. (1999) Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have an opposite impact on the efficiency of industrial kraft pulping. Plant Physiol 119: 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre C, Rolando C (1988) Thioacidolyses of pre-methylated lignin samples from pine compression and poplar woods. Holzforschung 42: 1–4 [Google Scholar]
- Leplé JC, Brasileiro ACM, Michel MF, Delmotte F, Jouanin L (1992) Transgenic poplars: expression of chimeric genes using four different constructs. Plant Cell Rep 11: 137–141 [DOI] [PubMed] [Google Scholar]
- Leplé JC, Dauwe R, Morreel K, Storme V, Lapierre C, Pollet B, Naumann A, Kang KY, Kim H, Ruel K, et al. (2007) Downregulation of cinnamoyl-coenzyme A reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19: 3669–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang Y, Yao J, Chen G, Li X, Zhang Q, Wu C (2009) FLEXIBLE CULM 1 encoding a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol Biol 69: 685–697 [DOI] [PubMed] [Google Scholar]
- Li Y, Akiyama T, Yokoyama T, Matsumoto Y (2016) NMR assignment for diaryl ether structures (4-O-5 structures) in pine wood lignin. Biomacromolecules 17: 1921–1929 [DOI] [PubMed] [Google Scholar]
- Littlewood J, Guo M, Boerjan W, Murphy RJ (2014) Bioethanol from poplar: a commercially viable alternative to fossil fuel in the European Union. Biotechnol Biofuels 7: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Karlen SD, Regner M, Kim H, Ralph SA, Sun RC, Kuroda K, Augustin MA, Mawson R, Sabarez H, et al. (2015) Naturally p-hydroxybenzoylated lignins in palms. BioEnergy Res 8: 934–952 [Google Scholar]
- MacKay JJ, O’Malley DM, Presnell T, Booker FL, Campbell MM, Whetten RW, Sederoff RR (1997) Inheritance, gene expression, and lignin characterization in a mutant pine deficient in cinnamyl alcohol dehydrogenase. Proc Natl Acad Sci USA 94: 8255–8260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SD, Kim H, Lu F, Ralph J (2012) Whole plant cell wall characterization using solution-state 2D NMR. Nat Protoc 7: 1579–1589 [DOI] [PubMed] [Google Scholar]
- Méchin V, Laluc A, Legée F, Cézard L, Denoue D, Barrière Y, Lapierre C (2014) Impact of the brown-midrib bm5 mutation on maize lignins. J Agric Food Chem 62: 5102–5107 [DOI] [PubMed] [Google Scholar]
- Meissner D, Albert A, Böttcher C, Strack D, Milkowski C (2008) The role of UDP-glucose:hydroxycinnamate glucosyltransferases in phenylpropanoid metabolism and the response to UV-B radiation in Arabidopsis thaliana. Planta 228: 663–674 [DOI] [PubMed] [Google Scholar]
- Morreel K, Dima O, Kim H, Lu F, Niculaes C, Vanholme R, Dauwe R, Goeminne G, Inzé D, Messens E, et al. (2010a) Mass spectrometry-based sequencing of lignin oligomers. Plant Physiol 153: 1464–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K, Kim H, Lu F, Dima O, Akiyama T, Vanholme R, Niculaes C, Goeminne G, Inzé D, Messens E, et al. (2010b) Mass spectrometry-based fragmentation as an identification tool in lignomics. Anal Chem 82: 8095–8105 [DOI] [PubMed] [Google Scholar]
- Morreel K, Ralph J, Kim H, Lu F, Goeminne G, Ralph S, Messens E, Boerjan W (2004) Profiling of oligolignols reveals monolignol coupling conditions in lignifying poplar xylem. Plant Physiol 136: 3537–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K, Saeys Y, Dima O, Lu F, Van de Peer Y, Vanholme R, Ralph J, Vanholme B, Boerjan W (2014) Systematic structural characterization of metabolites in Arabidopsis via candidate substrate-product pair networks. Plant Cell 26: 929–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottiar Y, Vanholme R, Boerjan W, Ralph J, Mansfield SD (2016) Designer lignins: harnessing the plasticity of lignification. Curr Opin Biotechnol 37: 190–200 [DOI] [PubMed] [Google Scholar]
- Nair RB, Bastress KL, Ruegger MO, Denault JW, Chapple C (2004) The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 16: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculaes C, Morreel K, Kim H, Lu F, McKee LS, Ivens B, Haustraete J, Vanholme B, Rycke RD, Hertzberg M, et al. (2014) Phenylcoumaran benzylic ether reductase prevents accumulation of compounds formed under oxidative conditions in poplar xylem. Plant Cell 26: 3775–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson O, Little CHA, Sandberg G, Olsson O (1996) Expression of two heterologous promoters, Agrobacterium rhizogenes rolC and cauliflower mosaic virus 35S, in the stem of transgenic hybrid aspen plants during the annual cycle of growth and dormancy. Plant Mol Biol 31: 887–895 [DOI] [PubMed] [Google Scholar]
- Pesquet E, Zhang B, Gorzsás A, Puhakainen T, Serk H, Escamez S, Barbier O, Gerber L, Courtois-Moreau C, Alatalo E, et al. (2013) Non-cell-autonomous postmortem lignification of tracheary elements in Zinnia elegans. Plant Cell 25: 1314–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Häusler E, Karwatzki B, Meinhard J (1993) Proposed biosynthetic pathway for rosmarinic acid in cell cultures of Coleus blumei Benth. Planta 189: 10–14 [Google Scholar]
- Petrik DL, Karlen SD, Cass CL, Padmakshan D, Lu F, Liu S, Le Bris P, Antelme S, Santoro N, Wilkerson CG, et al. (2014) p-Coumaroyl-CoA:monolignol transferase (PMT) acts specifically in the lignin biosynthetic pathway in Brachypodium distachyon. Plant J 77: 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilate G, Guiney E, Holt K, Petit-Conil M, Lapierre C, Leplé JC, Pollet B, Mila I, Webster EA, Marstorp HG, et al. (2002) Field and pulping performances of transgenic trees with altered lignification. Nat Biotechnol 20: 607–612 [DOI] [PubMed] [Google Scholar]
- Pitre FE, Pollet B, Lafarguette F, Cooke JEK, MacKay JJ, Lapierre C (2007) Effects of increased nitrogen supply on the lignification of poplar wood. J Agric Food Chem 55: 10306–10314 [DOI] [PubMed] [Google Scholar]
- Popper ZA, Michel G, Hervé C, Domozych DS, Willats WGT, Tuohy MG, Kloareg B, Stengel DB (2011) Evolution and diversity of plant cell walls: from algae to flowering plants. Annu Rev Plant Biol 62: 567–590 [DOI] [PubMed] [Google Scholar]
- Ralph J. (2010) Hydroxycinnamates in lignification. Phytochem Rev 9: 65–83 [Google Scholar]
- Ralph J, Kim H, Lu F, Grabber JH, Leplé JC, Berrio-Sierra J, Derikvand MM, Jouanin L, Boerjan W, Lapierre C (2008) Identification of the structure and origin of a thioacidolysis marker compound for ferulic acid incorporation into angiosperm lignins (and an indicator for cinnamoyl CoA reductase deficiency). Plant J 53: 368–379 [DOI] [PubMed] [Google Scholar]
- Ralph J, Lapierre C, Marita JM, Kim H, Lu F, Hatfield RD, Ralph S, Chapple C, Franke R, Hemm MR, et al. (2001) Elucidation of new structures in lignins of CAD- and COMT-deficient plants by NMR. Phytochemistry 57: 993–1003 [DOI] [PubMed] [Google Scholar]
- Ralph J, MacKay JJ, Hatfield RD, O’Malley DM, Whetten RW, Sederoff RR (1997) Abnormal lignin in a loblolly pine mutant. Science 277: 235–239 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Ratke C, Pawar PMA, Balasubramanian VK, Naumann M, Duncranz ML, Derba-Maceluch M, Gorzsás A, Endo S, Ezcurra I, Mellerowicz EJ (2015) Populus GT43 family members group into distinct sets required for primary and secondary wall xylan biosynthesis and include useful promoters for wood modification. Plant Biotechnol J 13: 26–37 [DOI] [PubMed] [Google Scholar]
- Regan S, Bourquin V, Tuominen H, Sundberg B (1999) Accurate and high resolution in situ hybridization analysis of gene expression in secondary stem tissues. Plant J 19: 363–369 [DOI] [PubMed] [Google Scholar]
- Saathoff AJ, Sarath G, Chow EK, Dien BS, Tobias CM (2011) Downregulation of cinnamyl-alcohol dehydrogenase in switchgrass by RNA silencing results in enhanced glucose release after cellulase treatment. PLoS ONE 6: e16416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šamaj J, Hawkins S, Lauvergeat V, Grima-Pettenati J, Boudet A (1998) Immunolocalization of cinnamyl alcohol dehydrogenase 2 (CAD 2) indicates a good correlation with cell-specific activity of CAD 2 promoter in transgenic poplar shoots. Planta 204: 437–443 [DOI] [PubMed] [Google Scholar]
- Santoro N, Cantu SL, Tornqvist CE, Falbel TG, Bolivar JL, Patterson SE, Pauly M, Walton JD (2010) A high-throughput platform for screening milligram quantities of plant biomass for lignocellulose digestibility. BioEnergy Res 3: 93–102 [Google Scholar]
- Shi R, Sun YH, Li Q, Heber S, Sederoff R, Chiang VL (2010) Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol 51: 144–163 [DOI] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Séguin A (2005) CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17: 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Pollet B, Goujon T, Mila I, Granier F, Séguin A, Lapierre C, Jouanin L (2003) Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis: isolation and characterization of the corresponding mutants. Plant Physiol 132: 848–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Le Bris P, Legée F, Cézard L, Renault H, Lapierre C (2016) Structural redesigning Arabidopsis lignins into alkali-soluble lignins through the expression of p-coumaroyl-CoA:monolignol transferase PMT. Plant Physiol 170: 1358–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons BA, Loqué D, Ralph J (2010) Advances in modifying lignin for enhanced biofuel production. Curr Opin Plant Biol 13: 313–320 [DOI] [PubMed] [Google Scholar]
- Smith RA, Gonzales-Vigil E, Karlen SD, Park JY, Lu F, Wilkerson CG, Samuels L, Ralph J, Mansfield SD (2015) Engineering monolignol p-coumarate conjugates into poplar and Arabidopsis lignins. Plant Physiol 169: 2992–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RA, Schuetz M, Karlen SD, Bird D, Tokunaga N, Sato Y, Mansfield SD, Ralph J, Samuels AL (2017) Defining the diverse cell populations contributing to lignification in Arabidopsis stems. Plant Physiol 174: 1028–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RA, Schuetz M, Roach M, Mansfield SD, Ellis B, Samuels L (2013) Neighboring parenchyma cells contribute to Arabidopsis xylem lignification, while lignification of interfascicular fibers is cell autonomous. Plant Cell 25: 3988–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon BD. (2010) Biofuels and sustainability. Ann N Y Acad Sci 1185: 119–134 [DOI] [PubMed] [Google Scholar]
- Sundell D, Street NR, Kumar M, Mellerowicz EJ, Kucukoglu M, Johnsson C, Kumar V, Mannapperuma C, Delhomme N, Nilsson O, et al. (2017) AspWood: high-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 29: 1585–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Popko JL, Mielke MR, Hu WJ, Podila GK, Chiang VL (1998) Suppression of O-methyltransferase gene by homologous sense transgene in quaking aspen causes red-brown wood phenotypes. Plant Physiol 117: 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji Y, Vanholme R, Tobimatsu Y, Ishikawa Y, Foster CE, Kamimura N, Hishiyama S, Hashimoto S, Shino A, Hara H, et al. (2015) Introduction of chemically labile substructures into Arabidopsis lignin through the use of LigD, the Cα-dehydrogenase from Sphingobium sp. strain SYK-6. Plant Biotechnol J 13: 821–832 [DOI] [PubMed] [Google Scholar]
- USEIA (2013) http://www.eia.gov/forecasts/ieo/. Accessed May 2017
- Valério L, Carter D, Rodrigues JC, Tournier V, Gominho J, Marque C, Boudet AM, Maunders M, Pereira H, Teulières C (2003) Down regulation of cinnamyl alcohol dehydrogenase, a lignification enzyme, in Eucalyptus camaldulensis. Mol Breed 12: 157–167 [Google Scholar]
- Van Acker R, Leplé JC, Aerts D, Storme V, Goeminne G, Ivens B, Légée F, Lapierre C, Piens K, Van Montagu MCE, et al. (2014) Improved saccharification and ethanol yield from field-grown transgenic poplar deficient in cinnamoyl-CoA reductase. Proc Natl Acad Sci USA 111: 845–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker R, Vanholme R, Storme V, Mortimer JC, Dupree P, Boerjan W (2013) Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels 6: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorsselaere J, Baucher M, Chognot E, Chabbert B, Tollier MT, Petit-Conil M, Leplé JC, Pilate G, Cornu D, Monties B, et al. (1995) A novel lignin in poplar trees with a reduced caffeic acid 5-hydroxyferulic acid O-methyltransferase activity. Plant J 8: 855–864 [Google Scholar]
- Vanholme B, Desmet T, Ronsse F, Rabaey K, Van Breusegem F, De Mey M, Soetaert W, Boerjan W (2013a) Towards a carbon-negative sustainable bio-based economy. Front Plant Sci 4: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, et al. (2013b) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010a) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Morreel K, Darrah C, Oyarce P, Grabber JH, Ralph J, Boerjan W (2012a) Metabolic engineering of novel lignin in biomass crops. New Phytol 196: 978–1000 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Morreel K, Ralph J, Boerjan W (2008) Lignin engineering. Curr Opin Plant Biol 11: 278–285 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Ralph J, Akiyama T, Lu F, Pazo JR, Kim H, Christensen JH, Van Reusel B, Storme V, De Rycke R, et al. (2010b) Engineering traditional monolignols out of lignin by concomitant up-regulation of F5H1 and down-regulation of COMT in Arabidopsis. Plant J 64: 885–897 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Storme V, Vanholme B, Sundin L, Christensen JH, Goeminne G, Halpin C, Rohde A, Morreel K, Boerjan W (2012b) A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24: 3506–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermerris W, Sherman DM, McIntyre LM (2010) Phenotypic plasticity in cell walls of maize brown midrib mutants is limited by lignin composition. J Exp Bot 61: 2479–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VIB (2013) http://www.vib.be/en/news/Pages/Field-trial-with-lignin-modified-poplars-shows-potential-for-bio-based-economy,-but-also-that-work-still-needs-to-be-done.aspx. Accessed August 2017
- Wang JP, Naik PP, Chen HC, Shi R, Lin CY, Liu J, Shuford CM, Li Q, Sun YH, Tunlaya-Anukit S, et al. (2014) Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin in Populus trichocarpa. Plant Cell 26: 894–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng JK, Mo H, Chapple C (2010) Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. Plant J 64: 898–911 [DOI] [PubMed] [Google Scholar]
- Widhalm JR, Dudareva N (2015) A familiar ring to it: biosynthesis of plant benzoic acids. Mol Plant 8: 83–97 [DOI] [PubMed] [Google Scholar]
- Wilkerson CG, Mansfield SD, Lu F, Withers S, Park JY, Karlen SD, Gonzales-Vigil E, Padmakshan D, Unda F, Rencoret J, et al. (2014) Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science 344: 90–93 [DOI] [PubMed] [Google Scholar]
- Wu RL, Remington DL, Mackay JJ, McKeand SE, O’Malley DM (1999) Average effect of a mutation in lignin biosynthesis in loblolly pine. Theor Appl Genet 99: 705–710 [DOI] [PubMed] [Google Scholar]
- Youn B, Camacho R, Moinuddin SGA, Lee C, Davin LB, Lewis NG, Kang C (2006a) Crystal structures and catalytic mechanism of the Arabidopsis cinnamyl alcohol dehydrogenases AtCAD5 and AtCAD4. Org Biomol Chem 4: 1687–1697 [DOI] [PubMed] [Google Scholar]
- Youn B, Kim SJ, Moinuddin SGA, Lee C, Bedgar DL, Harper AR, Davin LB, Lewis NG, Kang C (2006b) Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970. J Biol Chem 281: 40076–40088 [DOI] [PubMed] [Google Scholar]
- Yuan JS, Tiller KH, Al-Ahmad H, Stewart NR, Stewart CN Jr (2008) Plants to power: bioenergy to fuel the future. Trends Plant Sci 13: 421–429 [DOI] [PubMed] [Google Scholar]
- Yue F, Lu F, Ralph S, Ralph J (2016) Identification of 4-O-5-units in softwood lignins via definitive lignin models and NMR. Biomacromolecules 17: 1909–1920 [DOI] [PubMed] [Google Scholar]
- Zhang K, Qian Q, Huang Z, Wang Y, Li M, Hong L, Zeng D, Gu M, Chu C, Cheng Z (2006) GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol 140: 972–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Tobimatsu Y, Zhou R, Pattathil S, Gallego-Giraldo L, Fu C, Jackson LA, Hahn MG, Kim H, Chen F, et al. (2013) Loss of function of cinnamyl alcohol dehydrogenase 1 leads to unconventional lignin and a temperature-sensitive growth defect in Medicago truncatula. Proc Natl Acad Sci USA 110: 13660–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Jacobs TB, Xue LJ, Harding SA, Tsai CJ (2015) Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4-coumarate:CoA ligase specificity and redundancy. New Phytol 208: 298–301 [DOI] [PubMed] [Google Scholar]
- Ziebell A, Gracom K, Katahira R, Chen F, Pu Y, Ragauskas A, Dixon RA, Davis M (2010) Increase in 4-coumaryl alcohol units during lignification in alfalfa (Medicago sativa) alters the extractability and molecular weight of lignin. J Biol Chem 285: 38961–38968 [DOI] [PMC free article] [PubMed] [Google Scholar]