Transcriptional regulation of TAA1 by various developmental, hormonal and environmental signals is directly regulated by Arabidopsis type B ARRs, which serve as central integrators.
Abstract
During embryogenesis and organ formation, establishing proper gradient is critical for auxin function, which is achieved through coordinated regulation of both auxin metabolism and transport. Expression of auxin biosynthetic genes is often tissue specific and is regulated by environmental signals. However, the underlying regulatory mechanisms remain elusive. Here, we investigated the transcriptional regulation of a key auxin biosynthetic gene, l-Tryptophan aminotransferase of Arabidopsis1 (TAA1). A canonical and a novel Arabidopsis (Arabidopsis thaliana) response regulator (ARR) binding site were identified in the promoter and the second intron of TAA1, which were required for its tissue-specific expression. C-termini of a subset of the type B ARRs selectively bind to one or both cis elements and activate the expression of TAA1. We further demonstrated that the ARRs not only mediate the transcriptional regulation of TAA1 by cytokinins, but also mediate its regulation by ethylene, light, and developmental signals. Through direct protein-protein interactions, the transcriptional activity of ARR1 is enhanced by ARR12, DELLAs, and ethylene-insenstive3 (EIN3). Our study thus revealed the ARR proteins act as key node that mediate the regulation of auxin biosynthesis by various hormonal, environmental, and developmental signals through transcriptional regulation of the key auxin biosynthesis gene TAA1.
Auxin is a key plant hormone that regulates cell growth and division, organ formation, and responses of plants to various external stimuli (Swarup et al., 2002; Benjamins and Scheres, 2008; Kasahara, 2015). As a plant morphogen, formation of proper auxin gradient is critical for functions of auxin. It is widely accepted that the coordinated actions of various auxin transporters play an essential role in establishing auxin gradient (Petrásek and Friml, 2009; Vandenbussche et al., 2010; Sawchuk et al., 2013). However, with the discovery of genes encoding auxin biosynthetic enzymes, it is noted that these genes exhibit tissue-specific expression pattern. So far, it is unclear how the expression of these genes are regulated and how their specific expression contributes to the establishment of the auxin gradient.
TAA1 catalyzes the formation of indole-3-pyruvic acid (IPA) from l-Trp, which is the first step in the IPA-dependent IAA biosynthesis pathway (Stepanova et al., 2008; Tao et al., 2008; Zhao, 2010). taa1/sav3 mutant displays defects in responses to vegetative shade, gravity, and hormones including cytokinin and ethylene, suggesting that TAA1-mediated auxin biosynthesis is an important component in these responses (Stepanova et al., 2008; Tao et al., 2008; Zhou et al., 2011). TAA1 exhibits tissue-specific and dynamic expression patterns during plant development. Expression of TAA1 is also regulated by hormones including ethylene and cytokinin and by environmental stimuli such as aluminum treatment (Stepanova et al., 2008; Zhou et al., 2011; Yang et al., 2014). REVOLUTA, a CLASS III HOMEODOMAIN LEU ZIPPER transcription factor, was reported to regulate auxin biosynthesis through direct transcriptional regulation of TAA1 and YUCCA5 (YUC5), another gene required for IPA-dependent IAA biosynthesis (Brandt et al., 2012). However, details of the underlying regulatory mechanism remain elusive.
Cytokinin is known to act together with auxin in regulating many aspects of plant growth and development, including embryogenesis, meristem development and maintenance, lateral root initiation, and development (Moubayidin et al., 2009; Chandler and Werr, 2015). Cytokinin is perceived by membrane-localized cytokinin receptors, which are autophosphorylated after binding cytokinins. Through a multistep phosphorelay, the signal was transferred to the Arabidopsis (Arabidopsis thaliana) His phosphotransfer proteins and then to the receiver domain of Arabidopsis response regulators (ARRs), which regulate the transcription of downstream target genes in the nucleus (To and Kieber, 2008; Werner and Schmülling, 2009). The ARRs fall into four categories: type A, B, C, and the Arabidopsis pseudoresponse regulators (Schaller et al., 2007; To et al., 2007). The 10 type A ARRs are transcriptionally activated by cytokinin signal, and most of them act as negative regulators of the cytokinin signaling (To et al., 2004; To et al., 2007). The type-B ARRs are positive regulators containing an N-terminal receiver domain and a C-terminal DNA-binding and transactivation domain. Of the 11 type-B ARRs, ARR1, 10, and 12 are considered to be essential, as their elimination severely reduced the cytokinin response (Mason et al., 2005).
The relationship between auxin and cytokinin is rather complicated. For example, they act antagonistically in controlling root meristem size but act synergistically during the response to Aluminum (Dello Ioio et al., 2008; Ruzicka et al., 2009; Yang et al., 2017). Recent studies begin to shed light on the molecular mechanisms underlying the cross talk between auxin and cytokinin at the levels of hormone metabolism, transport, and signaling (Coenen and Lomax, 1997; Moubayidin et al., 2009; Jones et al., 2010; Schaller et al., 2015; Šimášková et al., 2015). A homeostatic feedback loop was proposed that regulates IAA and cytokinin concentration in Arabidopsis root tissue (Dello Ioio et al., 2008; Ruzicka et al., 2009). At the metabolic level, it was shown that auxin down-regulates cytokinin biosynthesis in Arabidopsis, while application of cytokinin increases the biosynthesis rate of auxin in young, developing shoot and root tissue. Such regulation is believed to be achieved at least partially through a direct transcriptional regulation of key genes involved in cytokinin and auxin biosynthesis (Miyawaki et al., 2004; Nordström et al., 2004; Tanaka et al., 2006; Jones et al., 2010). However, the underlying regulatory mechanism is still unclear.
In this study, we performed TAA1 promoter analysis and identified two key regulatory cis elements. We further demonstrated that the C terminus of ARR1 (ARR1C) binds directly to these cis elements and mediates the transcriptional regulation of TAA1 by cytokinins and developmental and environmental cues. As TAA1 catalyzes the first step of the IPA-mediated auxin biosynthesis, transcriptional regulation of TAA1 may act as prerequisite for further regulation on auxin biosynthesis.
RESULTS
Identification of Cis Elements Required for TAA1 Expression
Using 5-d old TAA1pro:TAA1g-GUS transgenic lines (Tao et al., 2008), we observed that TAA1 is expressed in the young leaves, the vascular tissues, and the quiescent center (QC; Fig. 1A). Similar expression pattern was observed by Stepanova et al. (2008) using the TAA1pro:GFP-TAA1 transgenic line. We constructed TAA1pro:GUS transgenic lines for TAA1 promoter analysis. Surprisingly, in these lines, GUS signal was only detected in young leaves, but not in the roots (Fig. 1A). TAA1 contains a long second intron (918 bp). We performed Blast analysis using a 3.5 kb fragment that contains the TAA1 promoter and the first and the second exons and introns. A Brassica rapa ssp. Pekinensis clone KBrB084M08 was identified. As shown in Supplemental Figure S1, five regions showing high homology between the two sequences were identified. We thus divided the promoter region of TAA1 into six segments: P0–P4 and a core promoter region (Fig. 1B). The core promoter contains a putative TATA-box. A fragment in the second intron, P5, was also used in our study (Fig. 1B). We fused P0–P5 with the core promoter to drive GUS expression and constructed P0/1/2/3/4/5-COREpro:GUS transgenic lines. Through GUS expression pattern analysis (more than six independent lines for each construct), we detected GUS signal only in P1- and P5-COREpro:GUS lines. As shown in Figure 1C, in 5-d-old light-grown seedlings, GUS signal was only detected in the above-ground tissues of the P1-COREpro:GUS line and in the roots of the P5-COREpro:GUS line. Their expression patterns were similar to those in the corresponding tissues of the TAA1pro:TAA1g-GUS lines. We thus conclude that in light grown seedlings, the cis elements in P1 and P5 control the expression of TAA1 in the above ground leaf tissues and roots, respectively.
Figure 1.
Identification of the regulatory elements that control tissue-specific expression of TAA1. A, GUS expression patterns of 5-d-old TAA1 pro:TAA1g-GUS and TAA1pro:GUS seedlings. B, Schematic diagram showing DNA fragments used for cis-element analysis. C, GUS expression patterns of 5-d-old P1-COREpro:GUS and P5-COREpro:GUS seedlings.
To fine map the cis elements, we generated a series of deletion mutants using the P1/P5-COREpro:GUS constructs. As shown in Supplemental Figure S2A, in the P1 deletion series (P1-del-a/b/c/d-COREpro:GUS), deletion of b segment (126 bp) resulted in loss of GUS signal (Supplemental Fig. S2C, more than 12 independent lines for each construct). While in the P5 deletion series (P5-del-a/b/c -COREpro:GUS; Supplemental Fig. S2B), deletion of segment b (228 bp) resulted in a complete loss of GUS signal (Supplemental Fig. S2D). Using similar approach, we further analyzed P1b and P5b. As shown in Supplemental Figure S3, A and B, the cis element in P1 was narrowed down to a 15 bp region, including both P1b2-1 and -2, while the cis element in P5 was narrowed down to P5b1 and P5b3, including 47 and 45bp of DNAs, respectively (Supplemental Fig. S3C).
A Subset of ARRs Bind Specifically to P1 and/or P5
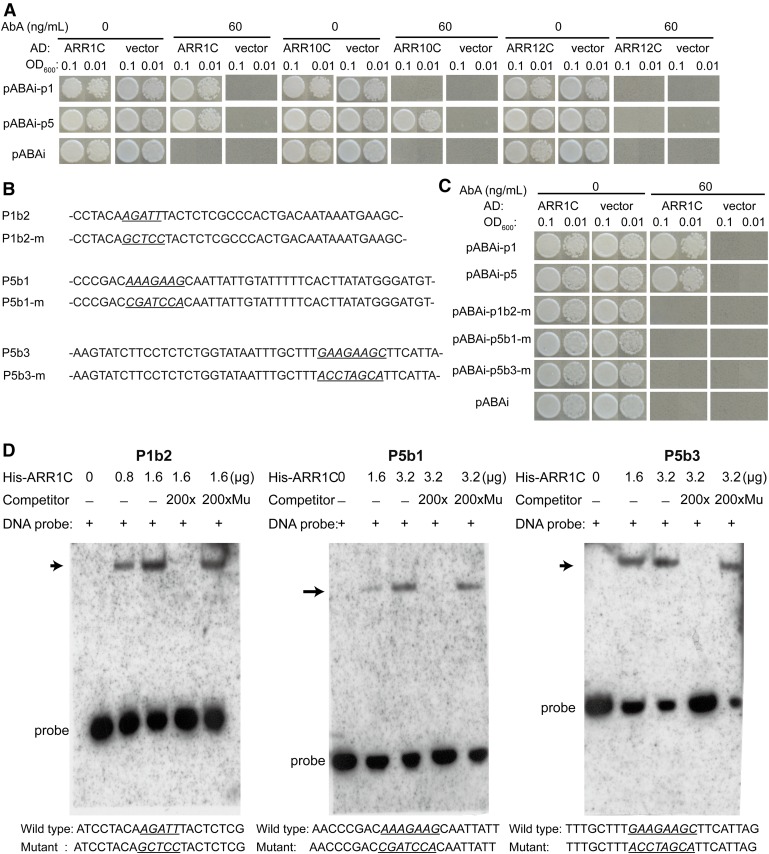
Through the Genevestigator website (www.genevestigator.com), we found the expression of TAA1 increased in the ARR21C (ARR21 C-terminal) overexpression lines (Tajima et al., 2004). Through yeast 1-hybrid (Y1H) assay, we found that ARR21C interacted with both P1 and P5 (Supplemental Fig. S4). As the expression of ARR21 was mainly detected in the reproductive organs but was hardly detected in seedlings (Tajima et al., 2004), we turned to type B ARRs expressed in seedlings, including ARR1, 10, and 12, which also play key roles in the cytokinin signaling pathway (Hwang et al., 2012). As shown in Figure 2A, ARR1C interacted with both P1 and P5, ARR10C only interacted with P5, while ARR12C did not interact with either P1 or P5. These results suggest that only a subset of ARRs can directly bind to P1/P5.
Figure 2.
Interactions between ARRs and P1/P5 in vitro. A, Interactions among ARR1C, ARR10C, ARR12C, and P1/P5 in yeast. pABAi, empty vector control. ABA: Aureobasidin A; AD:pGADT7. B, Partial sequences of P1b2, P5b1, P5b3, and their corresponding mutant forms. Predicted ARR binding sites and their corresponding mutant forms were italicized and underlined. C, Y1H results showing interactions between ARR1C and P1/P5 were abolished by mutations in the putative ARR binding sites. D, EMSA assay showing ARR1C binds specifically to P1b2, P5b1, and P5b3, His-ARR1C:His-tagged ARR1C. Probe and the mutant competitor (Mu) sequences are shown at the bottom. The putative ARR binding sites and their corresponding mutant forms were italicized and underlined. Arrowheads mark the shifted bands.
To further investigate how ARRs regulate the expression of TAA1, we focused on the role of ARR1 (Sakai et al., 2001). A putative ARR binding site (AAGATTT) was identified at the junction of P1b2-1 and -2 (Fig. 2B; Sakai et al., 2001; Taniguchi et al., 2007). Mutation in this binding site (Fig. 2C) abolished the interaction between P1 and ARR1C in yeast. The above result is consistent with our promoter analysis result (Supplemental Fig. S3B) showing both P1b2-1 and P1b2-2 are required for TAA1 expression. P5, on the other hand, contains no canonical ARR binding site within the P5b1 or P5b3 region. As two cis elements were required for P5 function (Supplemental Fig. S3C), we searched for homologous sequences between P5b1 and P5b3 and identified an AAGAAG element in both regions (Fig. 2B). Mutation in either one of the putative ARR1 binding site abolished its interaction with ARR1 (Fig. 2C), suggesting that they are both required for the interaction between P5 and ARR1.
To confirm the interaction between ARR1C and P1/P5, we performed an electrophoretic mobility shift assay (EMSA). Three biotin-labeled probes corresponding to the ARR binding-site-containing region of P1b2, P5b1, and P5b3 were used. Bacterial expressed ARR1C binds to all three probes (Fig. 2D). An excess amount of unlabeled cold probes, but not those with mutations in the ARR binding sites, could effectively compete with the labeled probes, indicating that ARR1C binds to these probes in a sequence-specific manner (Fig. 2D).
ARR1C Activates Downstream Gene Expression through P1 and P5 In Vivo
To demonstrate the interaction between ARR1C and P1/P5 in vivo, we performed Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) assay. HSPpro:ARR1C-3FL lines expressing C-terminal FLAG-tagged ARR1C under the control of a heat-inducible promoter (HSP; Severin and Schöffl, 1990) were generated. Primers covering P1, P5, Exon3 and 3′ UTR were designed (Fig. 3A). Five-day-old light-grown seedlings were heat shocked at 37°C for 1 h, followed by 2 h of recovery at 22°C before sample collection. The expression of ARR6, a well-known target of ARR1, was first examined to verify ARR1C function using quantitative reverse-transcriptase real-time PCR (qRT-PCR; Supplemental Fig. S5). Expression of endogenous TAA1 was also significantly induced in the transgenic seedlings (Fig. 3B), indicating that overexpression of ARR1C indeed induces the expression of TAA1. ChIP experiments were then performed. As shown in Figure 3C, the P1 and P5 regions were specifically enriched, indicating that ARR1C binds to P1 and P5 in vivo.
Figure 3.
ARR1C binds to both P1 and P5 and activates TAA1 expression in vivo. A, A cartoon showing regions amplified in the ChIP-qPCR experiment. B, Expression of endogenous TAA1 is induced in heat-treated HSPpro:ARR1C-3FL seedlings (FL, FLAG tag). C, ChIP-qPCR assays showing that P1 and P5 regions were specifically enriched as the ARR1C binding regions. Representative data from three biological replicates were shown. D and E, Protoplast transient expression assay showing ARR1C activates the expression of the LUC through its specific interaction with the ARR binding sites on P1/P5. Error bars represent se of mean (sem; n = 3). **Significant difference to corresponding control with P < 0.01.
To examine if ARR1C can activate the expression of TAA1 via the P1/P5 region, we performed transient expression analysis using the Arabidopsis protoplast system. P1 and P5, together with the core promoter, were cloned to the upstream of a luciferase (LUC) reporter gene (P1pro:LUC and P5pro:LUC). Constitutively expressed Renilla luciferase (R-LUC) was used as a transformation efficiency control (Takai et al., 2007). As shown in Figure 3, D and E, the expression of luciferase was strongly up-regulated in the presence of ARR1C. Such activation was completely abolished when the corresponding ARR binding site in P1 or P5 was mutated (Fig. 3, D and E), indicating ARR1C can activate downstream gene expression through its specific interaction with the ARR binding sites.
ARR12 Interacts with ARR1C and Enhances Its Transcriptional Activity
As shown below, ARR12 is involved in the transcriptional regulation of TAA1 even though it does not bind to either P1 or P5. We investigated if ARR1C interacts with ARR12C using bimolecular fluorescence complementation (BiFC). As shown in Figure 4A, ARR1C interacts specifically with ARR12C, but not with the control protein PKS1 in the protoplast. Using the protoplast transient expression system, we demonstrated that ARR12C enhances the transcriptional activity of ARR1C on P1/P5. As shown in Figure 4B, ARR12C by itself did not activate the LUC reporter gene expression, confirming that ARR12C does not bind to P1/P5. In the presence of ARR1C, ARR12C significantly enhances the reporter gene expression. Together, the above data indicate that ARR12C may regulate TAA1 expression through interacting with ARR1C and enhancing its transcriptional activity.
Figure 4.
ARR12C directly interacts with ARR1C to promote its transcriptional activity on TAA1. A, BiFC showing interaction between ARR12C and ARR1C. Scale bar, 25μm. B and C, Protoplast transient expression assay showing ARR12C promotes ARR1C-activated expression of LUC reporter gene driven by P1 (B) or P5 (C). Error bars represent se of mean (sem; n = 3). **Significant difference to corresponding control with P < 0.01.
Exogenous Cytokinin Regulates TAA1 Expression through ARRs
Transcriptional activity of the type B ARRs is activated by cytokinins (Kim et al., 2006; Veerabagu et al., 2012). Using TAA1pro:TAA1g-GUS line, we showed that cytokinin 6-BA (6-Benzylaminopurine, 6-BA) enhanced TAA1 expression in cotyledons and hypocotyls, and such enhancement was compromised in the arr1,12 mutant (Fig. 5A). We further confirmed the induction of TAA1 by BA treatment in cotyledons and true leaves using qRT-PCR. Such induction was reduced in arr1 and arr12 mutant and was almost completely abolished in the arr1,12 double mutant, indicating that both ARR1 and ARR12 are required for TAA1 expression even though ARR12 does not bind either P1 or P5 (Fig. 5B). Furthermore, the expression of TAA1 in arr1 or arr1,12 mutant was slightly lower than that in the wild type, suggesting the ARRs are required for TAA1 basal expression in light-grown seedlings. To investigate if induction of TAA1 is a primary response to cytokinins, we performed a time course assay. Five-day-old light-grown seedlings were treated with 6-BA for 3, 6, 9, and 12 h. As shown in Supplemental Figure S6A, induction of TAA1 can be observed after 3 h of BA treatment. Blocking protein synthesis with Cycloheximide significantly reduced TAA1 expression (as indicated by the difference in scales). However, the induction of TAA1 by 6-BA remains normal (Supplemental Fig. S6B), indicating that cytokinin-activated TAA1 expression is a primary response that occurs independent of newly synthesized proteins.
Figure 5.
The relationship between cytokinin and TAA1. A, 6-BA alters TAA1 expression pattern in an ARR-dependent manner. B, 6-BA enhances TAA1 expression in cotyledons and true leaves in an ARR-dependent manner. Error bars represent sem (n = 3). C, Response to 6-BA in roots requires functional TAA1. Regions between arrowheads are primary roots. Quantitative measurement of root length is shown in the right. Error bars represent sem (n ≥ 15). * and **Significant difference to Col-0 with P < 0.01 and 0.001, respectively (Student’s t test).
In roots, BA treatment inhibited root growth and altered the TAA1 expression pattern: Strong GUS signal was detected in tissues surrounding the vasculature, presumably in the ground tissues, while the expression at root tips (around QC) became less prominent or disappeared (Fig. 5A; Supplemental Fig. S6C). The above BA-induced expression changes were also compromised in the arr1.12 mutant. Further examination revealed that root growth inhibition exerted by BA was reduced in both sav3-1 and arr1,12, suggesting TAA1 is required for the responses to BA in roots (Fig. 5C).
To examine if the ARR binding sites are required for the transcriptional regulation of TAA1 by BA, we examined the GUS expression patterns of the P1/P5 deletion serious. As shown in Supplemental Figure S6D, P1-del-b2-3,b2-4,b3-5 all maintained BA responsiveness, while P1-del-b2-1 and b2-2, each containing only a 8-bp deletion in the P1 region, exhibited no GUS signal. The ARR binding site locates at the junction of b1 and b2. We thus concluded that the ARR binding site does confer the cytokinin responsiveness. Similarly, GUS expression in P5-del-b2,4,5 exhibited BA-induced expression pattern change, while P5-del b1 and b3, each containing an ARR binding site, exhibited no GUS signal (Supplemental Figure S6E). Thus, for both P1 and P5, the ARR binding sites are required not only for the reporter expression, but also for the cytokinin responsiveness.
Exogenous Cytokinin Affects Auxin Signaling Partially through TAA1
Using an auxin reporter line, DR5:GUS, whose expression often correlates with the levels of auxin (Mallory et al., 2005), we found exogenous BA significantly increased GUS expression in cotyledons and upper roots but reduced its expression at root tips (Fig. 6; Supplemental Fig. S7). In sav3-1/taa1 background, BA-induced GUS expression was less prominent, suggesting BA affects the auxin pathway partially through TAA1 (Fig. 6; Supplemental Fig. S7).
Figure 6.
Exogenous cytokinin enhances auxin signaling in a TAA1-dependent manner. Expression pattern of DR5:GUS in the wild type or sav3-1/taa1 background.
arr Mutants Have Reduced Auxin Levels
As the above data indicate, ARRs are involved in the transcriptional regulation of TAA1, we speculate that arr mutants may exhibit phenotypes consistent with having reduced auxin levels. It was previously reported that auxin is required for seed dormancy regulation (Liu et al., 2013). We measured the germination rate of freshly harvested Col-0, taa1/sav3-1, and arr1,12 seeds with or without imbibition. As shown in Figure 7A, germination rates of all three genotypes were similar when seeds were imbibed at 4°C for 4 d before germination. Without imbibition, the germination rates of both taa1/sav3-1 and arr1,12 were much higher than that of the wild type, indicating these mutants are defective in dormancy regulation.
Figure 7.
arr mutants display phenotypes consistent with having reduced auxin level. A, Germination rates of freshly harvested seeds with (right) or without (left) imbibition (n = 3, ≥70 seeds/sample). B, Hypocotyl length of Col-0, sav3-1, and arr1,10,12 seedlings grown in Wc or shade (n ≥ 15). C, arr1,12 is hypersensitive to NPA in shade (n ≥ 15). D, Quantification of the free IAA levels in 5-d-old light-grown (Wc) Col-0 and arr1,10,12 seedlings treated with Wc or shade for 2 h. E, Quantification of the free IAA levels in 2-d-old dark-grown Col-0 and arr1,10,12 seedlings. Error bars represent sem. * and **Significant difference to controls with P < 0.05 and 0.01, respectively (Student’s t test).
TAA1 is required for shade-induced hypocotyl elongation (Tao et al., 2008). Although arr1, 10, and 12 single or double mutants did not display defects in shade-induced hypocotyl elongation, arr1,10,12 triple mutant was significantly shorter than the wild type in shade (Fig. 7B). Furthermore, arr1,12 double mutant were hypersensitive to auxin transport inhibitor naphthylphthalamic acid (NPA) in shade (Fig. 7C), suggesting the auxin level may be reduced in the double mutant.
We then directly measure the free IAA levels in arr1,10,12 mutants under white light (Wc) and shade. Compared to the wild type, the free IAA level is significantly lower in arr1,10,12 mutants under both Wc and shade conditions (Fig. 7D). We also measured the free IAA levels in dark-grown Col-0 and arr1,10,12 mutant seedlings and found that the IAA level in the mutant is also significantly lower than that in the wild type, suggesting that the ARRs are required for IAA biosynthesis in dark-grown seedlings as well (Fig. 7E).
In summary, the above data demonstrated that the arr mutants exhibit phenotypes related to auxin deficiency.
ARRs Are Required for the Regulation of TAA1 Expression In Planta
Expression of TAA1 is regulated by both developmental and environmental signals. To investigate if alteration in endogenous cytokinin level would affect TAA1 expression, we took advantage of the recent discovery that SCARECROW (SCR) represses the expression of ARR1 in root tip, which regulates auxin production and cell differentiation (Moubayidin et al., 2013). ARR1 is normally expressed in all tissues of the root transition zone. In the scr-1 mutant, ARR1 is ecotopically expressed in the proximal meristem at high level (Moubayidin et al., 2013). We found the expression of TAA1 was also elevated in the scr-1 mutant in an ARR1-dependent manner (Fig. 8A). Furthermore, mutation in TAA1 partially suppressed the short root phenotype of scr-1 (Fig. 8B), suggesting elevated TAA1 expression may contribute to the short root phenotype of scr-1.
Figure 8.
ARRs are required for transcriptional regulation of TAA1 in planta. A, Expression of TAA1 in Col-0, scr-1, and arr1-3 scr-1 roots (n = 3). B, Root phenotypes of 7-d-old light-grown seedlings. Regions between arrowheads are primary roots. Quantitative measurements of the root length are shown at the bottom (n ≥ 15). Error bars represent sem. * and **Significant difference between samples with P < 0.05 and 0.01, respectively (Student’s t test).
ARRs Are Required for the Regulation of TAA1 Expression in Response to Light
Expression of TAA1 increases during germination (Supplemental Fig. S8A; www.genevestigator.com). We found that compared to the wild type, only a slight reduction in TAA1 level was observed in light-grown arr1,12 seedlings, which is consistent with our previous observation (Supplemental Fig. S8, B and C). Interestingly, the expression of TAA1 is transiently induced in seedlings transferred from dark to light, and such induction is ARR-dependent (Fig. 9A; Supplemental Fig. S8C). Using TAA1pro:TAA1g-GUS seedlings, we observed the GUS reporter was highly expressed in hypocotyls of dark-grown seedlings, while the expression in cotyledons was weak and was limited to the cotyledon tips (Fig. 9B). Upon light exposure, GUS signal increased dramatically in cotyledons but was gradually reduced in hypocotyls (Fig. 9B). In the arr1,12 mutant, GUS signal in hypocotyls reduced normally upon light exposure, while light-induced increase of GUS signal in cotyledons was compromised (Fig. 9B).
Figure 9.
Transcriptional regulation of TAA1 by light requires ARRs. A, Expression of TAA1 is transiently induced by light. Two-day-old dark-grown (D) seedlings were either kept in dark for 1 day or exposed to light (L) for 12 or 24 h. B, Light-induced changes in GUS expression pattern of TAA1pro:TAA1g-GUS in wild type or arr1,12 mutant background. Scale bar, 1 mm. C, Cotyledon opening rate of 2-d-old dark-grown seedlings transferred to light for the indicated amount of time (n ≥ 60). D, Cotyledon areas of 2-d-old dark-grown seedlings transferred to light for the indicated amount of time (n = 30). E, Expression of TAA1, SAUR14, SAUR50, and SAUR65 in 2-d-old dark (D)-grown seedlings exposed to light (L) for 0, 2, and 4 h. Error bars represent sem. A, C, D, and E, * and **Significant difference to the corresponding controls with P < 0.05 and 0.01 (Student’s t test), respectively.
To investigate the biological consequence of this transient induction of TAA1 in cotyledons, we examined phenotypes of the wild type, sav3-1, and arr1,12 during dark-to-light transition. As shown in Figure 9C, there was a significant delay in cotyledon opening in both sav3-1 and arr1,12. After light treatment, expression of TAA1 is expanded to the leaf margin (Fig. 9B). As light promotes cotyledon growth, we measured cotyledon area in response to light. As shown in Figure 9D, light-induced cotyledon expansion was also significantly reduced in both sav3-1 and arr1,12 mutants. The above results indicate that the transient induction of TAA1 by light may be required for light-induced cotyledon opening and expansion.
It was recently reported that the expression of several small auxin up RNAs (SAURs) were altered in cotyledons and hypocotyls during the dark-to-light transition (Sun et al., 2016). We examined the expression of TAA1, SAUR14, SAUR50, and SAUR65 in cotyledons and hypocotyls during dark-to-light transition. As shown in Figure 9E, the expression of TAA1 was increased in cotyledons but was reduced in hypocotyls upon light exposure. In dark-grown arr1,12 mutants, the expression of TAA1 was slightly lower than that in the wild type in hypocotyls. Light-induced expression of TAA1 in cotyledons was abolished in the mutants, but light-induced reduction of TAA1 in hypocotyls occurred normally (Fig. 9E). In response to light, the expression of SAUR14 and 50, but not SAUR65, increased in cotyledons, while the expression of SAUR50 and 65, but not SAUR14, decreased in hypocotyls (Sun et al., 2016). Our results confirmed the previous observation and showed that light-induced expression of SAUR14 and 50 in cotyledons was compromised in both sav3-1 and arr1,12, while light-induced reduction of SAUR50 and 65 in hypocotyls remains normal in these mutants, despite their lower expression in hypocotyls of the dark-grown seedlings. The expression pattern of the SAURs thus correlates well with that of the TAA1.
DELLA Protein Partially Mediated the Transcriptional Regulation of TAA1 by Light
DELLA proteins are key signaling components of the GA pathway. It was previously reported that the stability of DELLA proteins is regulated by light, and DELLA proteins interact directly with ARR1C to promote its transcriptional activity (Achard et al., 2007; Marín-de la Rosa et al., 2015). We found that in the presence of GA, light-induced TAA1 expression is partially suppressed (Supplemental Fig. S8D), suggesting that DELLA proteins are also involved in this process. Through a yeast two-hybrid assay, we confirmed that DELLA protein GAI does interact directly with ARR1C (Supplemental Fig. S8E). We then used the protoplast system to investigate if GAI participates in the transcriptional regulation of TAA1 through P1 and P5 cis elements. As shown in Figure 10, A and B, GAI by itself did not significantly change the expression of the reporter gene driven by P1 or P5. However, in the presence of ARR1C, it significantly promoted the activation of the reporter gene by ARR1C. As light activates TAA1 expression in cotyledons through ARR1 and 12, we further examined if stabilizing DELLA proteins in dark-grown seedlings by GA biosynthesis inhibitor Paclobutrazol (PAC) would enhance TAA1 expression in cotyledons. Using P1-COREpro:GUS, we found that PAC treatment significantly increased the expression of GUS in cotyledons, indicating that stabilizing DELLA proteins does increase P1-mediated transcription of the reporter gene (Fig. 10C) and mimics the effect of light. To further examine if the influence of PAC on TAA1 expression requires functional ARR1 and 12, we examined the GUS expression of TAA1pro:TAA1g-GUS in wild type and arr1,12 mutant background. As shown in Figure 10D, PAC treatment did enhance GUS expression in cotyledons of the wild-type seedlings, and such enhancement was abolished in the arr1,12 mutant. Similar results were obtained using qRT-PCR to quantify TAA1 expression (Fig. 10E). We thus conclude that PAC treatment enhances TAA1 expression in an ARR-dependent manner.
Figure 10.
DELLA protein GAI promotes the transcriptional activation of TAA1 by ARR1C in cotyledons. A and B, Protoplast transient expression assay showing GAI promotes ARR1C-activated expression of LUC reporter gene driven by P1 (A) or P5 (B). C, PAC treatment enhanced GUS expression in cotyledons of dark-grown P1-COREpro:GUS seedlings. Scale bar, 1 mm. D, GUS expression pattern of TAA1pro:TAA1g-GUS in wild type or arr1,12 mutant background in response to PAC treatment. Scale bar, 1 mm. E, qRT-PCR results showing TAA1 expression in cotyledons in response to PAC. For experiments in C to E, seedlings were grown in dark on 1/2 MS medium supplemented with or without 0.5 μm of PAC for 2 d (2D). Error bars represent se of mean (sem; n = 3). **Significant difference between the two samples with P < 0.01 (Student’s t test).
EIN3 Directly Interacts with ARR1 to Promote Its Transcriptional Activity on TAA1
Ethylene inhibits root elongation in etiolated seedlings in a TAA1-dependent manner (Stepanova et al., 2008). We found that similar to the sav3/taa1 mutant, both arr1 and arr1,12 mutant exhibited reduced root response to the ethylene precursor 1-aminocyclopropane-1- carboxylic acid (ACC; Fig. 11A), suggesting the type B ARRs are also required for the ethylene response. It was previously demonstrated that ethylene induces TAA1 expression in roots of dark-grown seedlings (Stepanova et al., 2008). We found ACC-induced TAA1 expression in roots, especially in root cortex, also depends on ARR1, ARR12, and the ARR binding sites on P5 (Supplemental Fig. S10). EIN3 is a key transcription factor mediating the ethylene response. We found that EIN3 interacts with ARR1C (Fig. 11B) in the BiFC test. Using the protoplast system and P1pro:LUC/P5pro:LUC reporters, we showed that EIN3 by itself does not activate the reporter expression (Fig. 11, C and D). However, in the presence of ARR1C, EIN3 significantly enhanced the transcriptional activity of ARR1C, suggesting that EIN3 may enhance TAA1 expression through interacting with ARR1. Thus, the transcriptional regulation of TAA1 by ethylene also requires the type B ARRs, and such regulation is achieved through direct interaction between EIN3 and ARR1, which subsequently affects ethylene-induced root growth inhibition.
Figure 11.
EIN3 directly interacts with ARR1 to promote its transcriptional activity on TAA1. A, Ethylene response of Col-0, sav3-1,arr1, and arr1,12 roots. B, BiFC showing interactions between ARR1C and EIN3. Scale bar, 25 μm. C and D, Protoplast transient expression assay showing EIN3 promotes ARR1C-activated expression of LUC reporter gene driven by P1 (C) or P5 (D). E, A proposed model showing that the type B ARRs function as central transcriptional regulators of TAA1. The type B ARRs can directly bind to the cis elements on P1 and/or P5 to promote TAA1 expression. N terminus of the type B ARRs inhibits the transcriptional activity of the C terminus, which can be relieved by the activation of the cytokinin pathway. Developmental and environmental signals may regulate TAA1 expression through modulating cytokinin metabolism or signaling. Alternatively, they may regulate transcription factors such as DELLAs and EIN3 that directly interact with the type B ARRs and modulate their transcriptional activities. Arrows with dashed lines indicate proposed interactions. Error bars represent se of mean (sem; n = 3). **Significant difference to the corresponding controls with P < 0.01 (Student’s t test).
We propose that transcriptional regulation of TAA1 by developmental or environmental signals may be achieved through two ways: one is to regulate the activities of the type B ARRs through modulating cytokinin levels, signaling, or dimerization of the ARRs, and the other is to regulate transcription factors such as the DELLAs and EIN3 that can interact with the type B ARRs to modulate their transcriptional activities (Fig. 11E). In this model, the ARRs that can directly bind to the corresponding ARR binding sites in the TAA1 genome play a key role in integrating various internal and external signals.
DISCUSSION
It has long been recognized that the ratio between auxin and cytokinin determines cell fate. The levels of these two hormones are both regulated by developmental and environmental cues, which then direct plant growth and development. How the levels of these two hormones are regulated and coordinated is a key question to understand the mechanisms underlying the plant growth regulation. In this study, we investigate the transcriptional regulation of TAA1, a key gene in IAA biosynthesis. Expression of TAA1 exhibits tissue specificity and correlates well with where IAA is actively synthesized (Stepanova et al., 2008; Tao et al., 2008; Zhao, 2010). Furthermore, the transcript level of TAA1 is regulated by various environmental cues (Stepanova et al., 2008; Tao et al., 2008; Zhou et al., 2011; Yang et al., 2014). We thus hypothesize that the transcriptional regulation of TAA1 may contribute to the establishment of proper auxin gradient in response to both developmental and environmental cues.
Through promoter analysis, we identified two regions (P1 and P5) that are both required and sufficient for the tissue-specific expression of TAA1 in cotyledons and roots of light-grown seedlings, respectively. In dark-grown seedlings, TAA1 is strongly expressed in hypocotyls, which is regulated by P5 (Supplemental Fig. S9) instead of P1. Furthermore, light-induced change of TAA1 expression pattern in cotyledons and hypocotyls are mediated by P1 and P5, respectively (Supplemental Fig. S9), suggesting both cis elements are responsive to light.
Through Y1H, we discovered that both ARR1C and ARR21C bind to P1 and P5, ARR10C only binds to P5, while ARR12C does not bind to either P1 or P5 (Supplemental Fig. S4; Fig. 2A). Although ARR12 does not interact with either cis element, mutation in ARR12 does seem to enhance phenotypes of arr1 (Fig. 5B). We found that ARR12C interacts with ARR1C and enhances the transcriptional activity of ARR1C, indicating that ARR12 may influence TAA1 expression through forming a heterodimer with ARR1. As only one canonical ARR binding site (AAGATTT) was identified in P1, the above results suggest only one subunit of the homo- or heterodimer of ARR proteins is required to bind to P1.
Two copies of a novel ARR binding site (AAGAAG) were discovered in P5. These binding sites are required for the specific binding of ARR1C to P1 or P5 (Fig. 2, C and D). The interaction between ARR1C and P1/P5 in vivo was further demonstrated through ChIP-qPCR (Fig. 3C). Using the protoplast transient expression system, we also showed that the specific binding of ARR1C to P1/P5 can induce the expression of a downstream reporter gene (Fig. 3, D and E).
To investigate if the expression patterns of the ARRs and TAA1 overlaps, we generated ARR1pro:eGFP-GUS and ARR12pro:eGFP-GUS lines. As shown in Supplemental Figure S11, ARR1 is strongly expressed at the tips of the cotyledons, leaf and root vasculature, and emerging new leaves, which are similar to the expression pattern of TAA1. ARR12 also highly expressed in cotyledons and the vascular tissues. In samples stained for a short period of time, GUS signals were observed in cells around stomata. Interestingly, neither ARR1 nor ARR12 is expressed in root meristematic region, suggesting that either other ARRs such as ARR10 or other transcription factors are required for TAA1 expression in that region.
Previously, Jones et al. (2010) showed that application of exogenous cytokinins led to a rapid increase in auxin biosynthesis in young, developing root and shoot tissues, while reducing endogenous cytokinin level caused a reduction in auxin biosynthesis. We found that cytokinin enhanced the expression of TAA1 in the above-ground tissues and alters its expression pattern in the root system. Both changes occur in an ARR-dependent manner (Fig. 5, A and B; Supplemental Fig. S6C). After BA treatment, roots of majority of the seedlings displayed the pattern shown in Figure 5A, while small percentage of seedlings displayed a pattern shown in Supplemental Fig. S6C. In both patterns, GUS signal increased in cortex but decreased around QC. Cytokinin treatment alters DR5:GUS expression in a way similar to its effect on TAA1, and these changes are TAA1 dependent (Fig. 6). We thus propose that cytokinin influences auxin pathway partially through ARR-mediated transcriptional regulation of TAA1. The above results would also suggest cytokinin may exert its effect partially through regulating auxin biosynthesis and that TAA1 would be required for responses to cytokinin. Indeed, like the arr1,12 mutant, taa1/sav3-1 mutant was hyposensitive to 6-BA (Fig. 5C), which was consistent with the previous report (Zhou et al., 2011). In summary, our results revealed that cytokinin enhances auxin response in the aboveground tissues in young seedlings in a TAA1-dependent manner (Fig. 6; Supplemental Fig. S7), indicating cytokinin acts synergistically with auxin. Interestingly, in the root tissue, although cytokinin also enhances auxin response in regions close to the hypocotyl-root junction, it reduces auxin response at root tips (Fig. 6). This result suggests that cytokinin can act either synergistically or antagonistically with auxin, depending on in which tissues they interact. Recently, Meng et al. (2017) reported that the type B ARRs can directly bind to the promoters of YUC1 and 4 and repress their expression during shoot stem cell specification. As most of these ARRs are transcription activators, they may interact with other transcription regulators to repress YUC expression. Identifying ARR interactors and unraveling their expression patterns thus would further advance our knowledge on the interactions between the cytokinin and the auxin pathway.
As the expression of TAA1 is reduced in the arr mutants without BA treatment (Fig. 5B; Supplemental Fig. S8B), we speculate that the auxin level may be reduced in the arr mutants. Indeed, arr1,10,12 triple mutants were defective in shade-induced hypocotyl elongation, which requires functional TAA1 (Fig. 7B). Although arr1,12 mutants were not defective in shade-induced hypocotyl elongation, they were hypersensitive to NPA in shade (Fig. 7C) and were defective in dormancy regulation, a phenotype that is also exhibited by auxin-deficient mutant (Fig. 7A; Liu et al., 2013). Through quantification of the free IAA levels, we found that indeed the auxin level is reduced in the arr1,10,12 mutants under Wc, shade, and dark conditions.
Expression of TAA1 is regulated by both developmental and environmental stimuli. It was recently reported that SCR represses ARR1 expression at QC, which controls auxin production via transcriptional regulation of ANTHRANILATE SYNTHASE BETA SUBUNIT1 (Moubayidin et al., 2013). We observed that in the scr-1 mutant, ectopically expressed ARR1 also activated TAA1 transcription and the taa1/sav3-1 mutant partially suppressed the short root phenotype of scr-1, indicating ARR1 also regulates TAA1 expression in planta (Fig. 8).
As an important environmental signal, light plays a critical role in plant development. We found that light induces TAA1 expression in cotyledons but represses its expression in hypocotyls (Fig. 9, B and E). Mutation in ARR1 and 12 reduced TAA1 expression in hypocotyls of dark-grown seedlings, abolished light-induced TAA1 expression in cotyledons, but did not affect the repression of TAA1 in hypocotyls by light (Fig. 9, B and E). These results indicate that transcriptional up-regulation of TAA1 in cotyledons by light requires ARR1 and 12, while other ARRs or transcriptional regulator may be responsible for the down-regulation of TAA1 in hypocotyls by light. Furthermore, the light-induced auxin responsive SAUR14 and SAUR50 expression in cotyledons depends on both TAA1 and ARR1 and 12; while light-repressed SAUR50 and SAUR65 expression in hypocotyls remained normal, though SAUR50 and SAUR65 exhibited lower expression in hypocotyls of dark grown arr1,12 seedlings (Fig. 9E). SAURs participate in light-regulated cotyledon opening and leaf expansion (Sun et al., 2016). Both taa1/sav3-1 and arr1,12 mutants exhibited defects in light regulated cotyledon opening and leaf expansion (Fig. 9, C and D), indicating ARRs mediate the transcriptional regulation of TAA1 by light, which plays an important role in the light response of seedlings.
DELLA proteins are not only key players in the GA signaling pathway but also regulate plant growth through interacting with various transcription factors and subsequently repress or enhance their transcriptional activity (Schwechheimer, 2012; Davière and Achard, 2013; Locascio et al., 2013). Rosa et al. demonstrated that DELLAs interact directly with the C terminus of the ARR1 and function as a transcription coactivator of the type B ARRs in regulating the expression of cytokinin target genes (Marín-de la Rosa et al., 2015). We confirmed the interaction between GAI and ARR1C and showed that GAI can also promote the transcriptional activity of ARR1C on reporter genes driven by P1/P5 (Fig. 10, A and B). It is well known that light treatment increases the protein level of DELLAs (Achard et al., 2007; Alabadí et al., 2008). We found that stabilizing DELLA proteins using PAC was sufficient to enhance TAA1 expression in cotyledons of dark-grown seedlings in an ARR-dependent manner (Fig. 10, C –E), suggesting that light treatment may enhance TAA1 expression through stabilizing DELLAs. Interestingly, same as the light treatment, PAC treatment inhibited TAA1 expression in hypocotyls (Fig. 10D). We propose that in hypocotyls, stabilized DELLAs may interact with other transcriptional coactivators of ARR1 to repress their functions. Alternatively, DELLAs may interact with other type B ARRs to inhibit their transcriptional activity. Further studies are required to test these hypotheses.
In summary, our study established a direct link between cytokinin and auxin biosynthesis. We revealed a direct transcriptional regulation of TAA1 by a subgroup of the type B ARRs through two types of ARR binding sites. As mutations in these ARR binding sites abolished TAA1 expression in specific tissues, we propose the type B ARRs serve as central regulators that integrate various signals and regulate the expression of TAA1. Developmental or environmental signals may alter TAA1 expression through regulating either the cytokinin pathway or transcription factors that can directly interact with the type B ARRs to affect their transcriptional activities (Fig. 11E). Furthermore, there are many type B ARRs and each exhibits a characteristic tissue-specific expression pattern. These ARRs may act together to regulate TAA1 expression, which remains to be unraveled.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
arr1-3 (Mason et al., 2005; Argyros et al., 2008), arr12-1 (Mason et al., 2005; Yokoyama et al., 2007; Argyros et al., 2008), arr1-3 arr12-1 (Mason et al., 2005; Argyros et al., 2008), arr1-3 arr10-5 arr12-1 (Mason et al., 2005; Argyros et al., 2008), scr-1 (Moubayidin et al., 2013), sav3-1 (Tao et al., 2008), DR5:GUS (Mallory et al., 2005; Cheng et al., 2006) were used in this study. For all experiments, seeds were surface-sterilized, planted on 1/2 Murashige and Skoog (MS) medium (Sigma-Aldrich) with 0.8% agar, and then imbibed at 4°C for 2 to 4 d. Seedlings were grown under Wc or darkness at 22°C.
Transgenic plants were generated by the floral-dipping method (Clough and Bent, 1998) and screened with 50 μg/mL kanamycin on 1/2 MS medium (except for ARR1/12 pro: eGFP-GUS, which were screened using Basta).
Quantitative measurements of hypocotyl, root length, and cotyledon area were performed on scanned images of seedlings using Scion Image software (http://www.scioncorp.com). For cotyledon opening, cotyledons open more than 0 degrees were counted as “opened cotyledon.” More than 15 seedlings were used for each treatment or genotype. In all figures, error bars represent se of mean (sem).
GUS staining was performed using 5-Bromo-4-chloro-3-indoxyl-beta-d-glucuronide cyclohexylammonium salt (Gold Biotechnology) as described by Richard et al. (Jefferson et al., 1987).
To assay responses to cytokinin, PAC, and ACC, seedlings were grown on 1/2 MS medium supplemented with 1 μm of 6-BA, 0.5 μm of PAC, and 0.1/1 μm of ACC.
Constructs
The first A in the ATG initiation codon was defined as nucleotide 1. Positional information was shown in Supplemental Table S1. Primers for all constructs are given in Supplemental Dataset 1. Most constructs were generated using ligation independent cloning method (Aslanidis et al., 1994). For constructs used in TAA1 expression pattern analysis, the uidA gene was first cloned into pJHA212K (Yoo et al., 2005) using SalI/BamHI; the 2-kb promoter of TAA1 was PCR amplified and cloned in pJHA212K-GUS using KpnI/SacI; finally the 800-bp DNA downstream of TAA1 stop codon was PCR-amplified and inserted using the SalI site to generate TAA1 pro:GUS.
P0/1/2/3/4/5-CORE pro:GUS, a series of P1/P5-CORE pro:GUS deletion constructs were generated based on TAA1 pro:GUS. The CORE promoter of TAA1 was first PCR amplified and cloned in TAA1 pro:GUS by replacing the 2-kb promoter of TAA1 using KpnI/SacI to generate CORE pro:GUS. P0/1/2/3/4/5 and P1/P5 deletion series were then amplified from the promoter region and cloned into the CORE pro:GUS using EcoRI/SacI. Deletions were generated through soeing PCR.
For Y1H, P1, P5, P1b2-m, P5b1-m, and P5b3-m regions were amplified from the above constructs and cloned into pABAi, The coding region of ARR1C (179–690 amino acids), ARR10C (146–552 amino acids), ARR12C (181–596 amino acids), and ARR21C (147–621 amino acids) were cloned into pGADT7 (Clontech).
For Y2H, The coding region of GAI,M5-GAI (Willige et al., 2007) and ARR1C (179–690 amino acids) were cloned into pGBKT7 and pGADT7 (Clontech), respectively.
For EMSA construct, the coding fragment of ARR1C was cloned into pET28a to produce His-ARR1C.
To generate HSPpro:ARR1C-3FL transgenic plants, the coding fragment of ARR1C was cloned into vector pCHF3-HSPpro-3FL, which was derived from pCHF3-3FL (Wang et al., 2005) by replacing the 35S promoter with the promoter of soybean GmHSP 17.6L (Severin and Schöffl, 1990).
For the protoplast transient expression assay, P1/P5/P1mu/P5b1mu/P5b3mu with the CORE promoter were amplified from the above constructs and cloned into pKEX4tr-LUC (Tao et al., 2000), replacing the 35S promoter. 35S:R-LUC (from pRL-TK plasmids [Promega]), 35S:GFP, 35S:ARR1C, 35S:ARR12C, 35S:GAI, and 35S:EIN3 were generated by inserting the corresponding coding sequences into pKEX4tr.
To construct ARR1/12pro:eGFP-GUS lines, ARR1/12 promoter (from −2,000 bp to the base before the translation start codon) was cloned into pB7WG2 using HindIII/SpeI. pDONR201-eGFP-GUS (a gift from Dr. Tao Huang) was used to perform the Gateway® LR reaction (Invitrogen).
ChIP-qPCR Assay
ChIP was performed as previously described (Saleh et al., 2008) using 4 g of 5-d-old HSPpro:ARR1C-3FL seedlings and 40 μL of anti-FLAG M2 beads (Sigma) per sample. The enrichment of DNA fragments was detected by qRT-PCR. Three independent experiments were performed, and one representative result was shown.
Y1H and Y2H Assay
Y1H and Y2H assay were carried out using the Matchmaker Gold Yeast One-Hybrid and Two-Hybrid System according to the manufacturer’s protocol (Clontech). The interactions were determined based on the growth ability of the cotransformants on medium supplemented with Aureobasidin A (AbA).
qRT-PCR and Expression Analysis
Total RNAs were extracted following Roche TriPure RNA isolation protocol (www.roche-applied-science.com). One microgram of total RNAs were reverse transcribed using the First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). qRT-PCR was performed using SYBR green method with a Stratagene Mx3000P real-time system cycler (Agilent). REF3 (At1g13320) was used as a reference gene (Hong et al., 2010). At least three replicates were included in each experiment. The primers are listed in Supplemental Dataset 1.
Protoplast Transient Expression Assay
Protoplast transient expression assays were carried out according to a protocol described by Yoo et al. (2007). 35S:R-LUC was used as a transformation efficiency control. LUC and R-LUC activities were assayed using Trans Detect Double-Luciferase Reporter Assay kit (TransGen Biotech). The analysis was carried out using VARIOSKAN FLASH Multiwavelength Microplate Reader (Thermo Fisher Scientific) according to the manufacturer’s instruction. Three independent experiments (biological replicates) were performed.
EMSA
The recombinant His-ARR1C was purified using Ni Sepharose High Performance (GE Healthcare) according to the manufacturer’s instructions. EMSA was performed using Chemiluminescent EMSA Kit (Beyotime, http://www.beyotime.com). Sequences of the biotin-labeled probes, wild type (wt, with same sequence as the probe) and mutant (mut) competitors used for EMSA are listed in Supplemental Dataset 1. Unlabeled competitors were added in 200-fold excess.
Quantification of Free IAA
Two-day-old dark-grown whole seedlings or the aerial parts of 5-d-old light-grown Col-0, arr1,10,12 seedlings treated with or without simulated shade for 2 h were collected (around 15 mg tissues/sample, 3 replicates). IAA quantification were performed as previously described (Andersen et al., 2008)
BiFC
BiFC was performed using the pSATN vector series (Citovsky et al., 2006). Coding sequence of ARR1C and PKS1 were cloned into pSAT6-neYFP-C1, and those of EIN3, ARR12C, and PKS1 were cloned into pSAT6-ceYFP-C1. Fluorescence analysis was performed on an LSM 780 confocal laser-scanning system (Zeiss).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: TAA1 (At1g70560), ARR21 (At5g07210), ARR1 (At3g16857), ARR10 (At4g31920), ARR12 (At2g25180), ARR6 (At5g62920), SCR (At3g54220), SAUR14 (At4g38840), SAUR50 (At4g34760), SAUR65 (At1g29460), GAI (At1g14920),REF3 (At1g13320), EIN3 (At3g20770), PKS1 (At2g02590).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Genomic sequence of TAA1 exhibits homology with the Brassica rapa ssp. Pekinensis clone KBrB084M08.
Supplemental Figure S2. Fine mapping of the cis elements in P1 and P5.
Supplemental Figure S3. Expression patterns of GUS in the P1/P5-COREpro:GUS deletion series.
Supplemental Figure S4. ARR21C interacts with both P1 and P5 in yeast.
Supplemental Figure S5. The expression of ARR6 was up-regulated in heat-treated HSPpro:ARR1C-3FL transgenic line, but not in the Col-0 seedlings.
Supplemental Figure S6. Exogenous cytokinin alters TAA1 expression.
Supplemental Figure S7. Cytokinin-induced DR5:GUS expression requires functional TAA1.
Supplemental Figure S8. Expression of TAA1 is altered in the arr mutants, and GAI interacts with ARR1C.
Supplemental Figure S9. Light-induced change of TAA1 expression in cotyledons and hypocotyls is mediated by P1 and P5, respectively.
Supplemental Figure S10. ACC-induced TAA1 expression in root cortex depends on ARR1, ARR12, and the ARR binding sites on P5.
Supplemental Figure S11. Expression patterns of ARR1 and ARR12.
Supplemental Table S1. Positional nucleotide sequence information for TAA1 cis elements analysis.
Supplemental Dataset 1. Primers used in this study.
Acknowledgments
The authors thank Dr. Guang-Qin Guo and Dr. Xiao-Rong Li for providing arr mutants and Dr. Chuan-You Li for providing scr-1 mutants.
Footnotes
This work was supported by the National Natural Science Foundation of China, 90917013, 30870210 and 31271298 to Y.T; Fundamental Research Funds for the Central Universities 2010121090, 2012121041 to Y.T; National Key Research and Developmental Program of China 2016YFD0100604 to Y.T. This work was also supported by 111 Project B12001.
References
- Achard P, Liao L, Jiang C, Desnos T, Bartlett J, Fu X, Harberd NP (2007) DELLAs contribute to plant photomorphogenesis. Plant Physiol 143: 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D, Gallego-Bartolomé J, Orlando L, García-Cárcel L, Rubio V, Martínez C, Frigerio M, Iglesias-Pedraz JM, Espinosa A, Deng XW, et al. (2008) Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J 53: 324–335 [DOI] [PubMed] [Google Scholar]
- Andersen SU, Buechel S, Zhao Z, Ljung K, Novák O, Busch W, Schuster C, Lohmann JU (2008) Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell 20: 88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslanidis C, de Jong PJ, Schmitz G (1994) Minimal length requirement of the single-stranded tails for ligation-independent cloning (LIC) of PCR products. PCR Methods Appl 4: 172–177 [DOI] [PubMed] [Google Scholar]
- Benjamins R, Scheres B (2008) Auxin: The looping star in plant development. Annu Rev Plant Biol 59: 443–465 [DOI] [PubMed] [Google Scholar]
- Brandt R, Salla-Martret M, Bou-Torrent J, Musielak T, Stahl M, Lanz C, Ott F, Schmid M, Greb T, Schwarz M, et al. (2012) Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J 72: 31–42 [DOI] [PubMed] [Google Scholar]
- Chandler JW, Werr W (2015) Cytokinin-auxin crosstalk in cell type specification. Trends Plant Sci 20: 291–300 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V, Lee LY, Vyas S, Glick E, Chen MH, Vainstein A, Gafni Y, Gelvin SB, Tzfira T (2006) Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol 362: 1120–1131 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coenen C, Lomax TL (1997) Auxin-cytokinin interactions in higher plants: Old problems and new tools. Trends Plant Sci 2: 351–356 [DOI] [PubMed] [Google Scholar]
- Davière JM, Achard P (2013) Gibberellin signaling in plants. Development 140: 1147–1151 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S (2008) A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384 [DOI] [PubMed] [Google Scholar]
- Hong SM, Bahn SC, Lyu A, Jung HS, Ahn JH (2010) Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant Cell Physiol 51: 1694–1706 [DOI] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B (2012) Cytokinin signaling networks. Annu Rev Plant Biol 63: 353–380 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Gunnerås SA, Petersson SV, Tarkowski P, Graham N, May S, Dolezal K, Sandberg G, Ljung K (2010) Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell 22: 2956–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H. (2015) Current aspects of auxin biosynthesis in plants. Biosci Biotechnol Biochem 80: 34–42 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 103: 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang H, Zhao Y, Feng Z, Li Q, Yang HQ, Luan S, Li J, He ZH (2013) Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc Natl Acad Sci USA 110: 15485–15490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A, Blázquez MA, Alabadí D (2013) Genomic analysis of DELLA protein activity. Plant Cell Physiol 54: 1229–1237 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B (2005) MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17: 1360–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-de la Rosa N, Pfeiffer A, Hill K, Locascio A, Bhalerao RP, Miskolczi P, Grønlund AL, Wanchoo-Kohli A, Thomas SG, Bennett MJ, et al. (2015) Genome wide binding site analysis reveals transcriptional coactivation of cytokinin-responsive genes by DELLA proteins. PLoS Genet 11: e1005337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng WJ, Cheng ZJ, Sang YL, Zhang MM, Rong XF, Wang ZW, Tang YY, Zhang XS (2017) Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 29: 1357–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: Tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37: 128–138 [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Di Mambro R, Sabatini S (2009) Cytokinin-auxin crosstalk. Trends Plant Sci 14: 557–562 [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Di Mambro R, Sozzani R, Pacifici E, Salvi E, Terpstra I, Bao D, van Dijken A, Dello Ioio R, Perilli S, et al. (2013) Spatial coordination between stem cell activity and cell differentiation in the root meristem. Dev Cell 26: 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström A, Tarkowski P, Tarkowska D, Norbaek R, Astot C, Dolezal K, Sandberg G (2004) Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: A factor of potential importance for auxin-cytokinin-regulated development. Proc Natl Acad Sci USA 101: 8039–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J, Friml J (2009) Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Ruzicka K, Simásková M, Duclercq J, Petrásek J, Zazímalová E, Simon S, Friml J, Van Montagu MC, Benková E (2009) Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc Natl Acad Sci USA 106: 4284–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Sawchuk MG, Edgar A, Scarpella E (2013) Patterning of leaf vein networks by convergent auxin transport pathways. PLoS Genet 9: e1003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Bishopp A, Kieber JJ (2015) The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 27: 44–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Doi K, Hwang I, Kieber JJ, Khurana JP, Kurata N, Mizuno T, Pareek A, Shiu SH, Wu P, et al. (2007) Nomenclature for two-component signaling elements of rice. Plant Physiol 143: 555–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C. (2012) Gibberellin signaling in plants—the extended version. Front Plant Sci 2: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin K, Schöffl F (1990) Heat-inducible hygromycin resistance in transgenic tobacco. Plant Mol Biol 15: 827–833 [DOI] [PubMed] [Google Scholar]
- Šimášková M, O’Brien JA, Khan M, Van Noorden G, Ötvös K, Vieten A, De Clercq I, Van Haperen JM, Cuesta C, Hoyerová K, et al. (2015) Cytokinin response factors regulate PIN-FORMED auxin transporters. Nat Commun 6: 8717. [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Sun N, Wang J, Gao Z, Dong J, He H, Terzaghi W, Wei N, Deng XW, Chen H (2016) Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc Natl Acad Sci USA 113: 6071–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Parry G, Graham N, Allen T, Bennett M (2002) Auxin cross-talk: Integration of signalling pathways to control plant development. Plant Mol Biol 49: 411–426 [DOI] [PubMed] [Google Scholar]
- Tajima Y, Imamura A, Kiba T, Amano Y, Yamashino T, Mizuno T (2004) Comparative studies on the type-B response regulators revealing their distinctive properties in the His-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol 45: 28–39 [DOI] [PubMed] [Google Scholar]
- Takai R, Kaneda T, Isogai A, Takayama S, Che FS (2007) A new method of defense response analysis using a transient expression system in rice protoplasts. Biosci Biotechnol Biochem 71: 590–593 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H (2006) Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J 45: 1028–1036 [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A (2007) ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol 48: 263–277 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Yuan F, Leister RT, Ausubel FM, Katagiri F (2000) Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell 12: 2541–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Deruère J, Maxwell BB, Morris VF, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ (2007) Cytokinin regulates type-A Arabidopsis Response Regulator activity and protein stability via two-component phosphorelay. Plant Cell 19: 3901–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Kieber JJ (2008) Cytokinin signaling: Two-components and more. Trends Plant Sci 13: 85–92 [DOI] [PubMed] [Google Scholar]
- Vandenbussche F, Petrásek J, Zádníková P, Hoyerová K, Pesek B, Raz V, Swarup R, Bennett M, Zazímalová E, Benková E, et al. (2010) The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 137: 597–606 [DOI] [PubMed] [Google Scholar]
- Veerabagu M, Elgass K, Kirchler T, Huppenberger P, Harter K, Chaban C, Mira-Rodado V (2012) The Arabidopsis B-type response regulator 18 homomerizes and positively regulates cytokinin responses. Plant J 72: 721–731 [DOI] [PubMed] [Google Scholar]
- Wang X, Li X, Meisenhelder J, Hunter T, Yoshida S, Asami T, Chory J (2005) Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell 8: 855–865 [DOI] [PubMed] [Google Scholar]
- Werner T, Schmülling T (2009) Cytokinin action in plant development. Curr Opin Plant Biol 12: 527–538 [DOI] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, Schwechheimer C (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZB, Geng X, He C, Zhang F, Wang R, Horst WJ, Ding Z (2014) TAA1-regulated local auxin biosynthesis in the root-apex transition zone mediates the aluminum-induced inhibition of root growth in Arabidopsis. Plant Cell 26: 2889–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZB, Liu G, Liu J, Zhang B, Meng W, Müller B, Hayashi KI, Zhang X, Zhao Z, De Smet I, et al. (2017) Synergistic action of auxin and cytokinin mediates aluminum-induced root growth inhibition in Arabidopsis. EMBO Rep 18: 1213–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Yamashino T, Amano Y, Tajima Y, Imamura A, Sakakibara H, Mizuno T (2007) Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol 48: 84–96 [DOI] [PubMed] [Google Scholar]
- Yoo SY, Bomblies K, Yoo SK, Yang JW, Choi MS, Lee JS, Weigel D, Ahn JH (2005) The 35S promoter used in a selectable marker gene of a plant transformation vector affects the expression of the transgene. Planta 221: 523–530 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhao Y. (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZY, Zhang CG, Wu L, Zhang CG, Chai J, Wang M, Jha A, Jia PF, Cui SJ, Yang M, et al. (2011) Functional characterization of the CKRC1/TAA1 gene and dissection of hormonal actions in the Arabidopsis root. Plant J 66: 516–527 [DOI] [PubMed] [Google Scholar]