The albino pap7-1 mutant of Arabidopsis reveals the relative impact of light and plastid developmental stage on the expression of nuclear genes involved in metabolism and photosynthesis.
Abstract
Plants possessing dysfunctional plastids due to defects in pigment biosynthesis or translation are known to repress photosynthesis-associated nuclear genes via retrograde signals from the disturbed organelles toward the nucleus. These signals are thought to be essential for proper biogenesis and function of the plastid. Mutants lacking plastid-encoded RNA polymerase-associated proteins (PAPs) display a genetic arrest in eoplast-chloroplast transition leading to an albino phenotype in the light. Retrograde signaling in these mutants, therefore, could be expected to be similar as under conditions inducing plastid dysfunction. To answer this question, we performed plastome- and genomewide array analyses in the pap7-1 mutant of Arabidopsis (Arabidopsis thaliana). In parallel, we determined the potential overlap with light-regulated expression networks. To this end, we performed a comparative expression profiling approach using light- and dark-grown wild-type plants as relative control for the expression profiles obtained from light-grown pap7-1 mutants. Our data indicate a specific impact of retrograde signals on metabolism-related genes in pap7-1 mutants reflecting the starvation situation of the albino seedlings. In contrast, light regulation of PhANGs and other nuclear gene groups appears to be fully functional in this mutant, indicating that a block in chloroplast biogenesis per se does not repress expression of them as suggested by earlier studies. Only genes for light harvesting complex proteins displayed a significant repression indicating an exclusive retrograde impact on this gene family. Our results indicate that chloroplasts and arrested plastids each emit specific signals that control different target gene modules both in positive and negative manner.
The buildup of the photosynthetic machinery during photomorphogenesis of angiosperms requires a tight coordination of nuclear and plastid gene expression as the photosynthesis genes are distributed over both genetic compartments (Waters and Langdale, 2009; Arsovski et al., 2012; Pogson et al., 2015). This coordination is achieved by a mutual information Exchange between nucleus and plastids that is called “anterograde” (from nucleus toward plastids) and “retrograde” (from plastids toward nucleus) signaling. This mutual signaling has been studied extensively, but is still far from being understood (Chi et al., 2013; Chan et al., 2016; de Souza et al., 2016; Kleine and Leister, 2016). The retrograde plastidial signals that are identified so far are numerous and of very diverse nature (see below). A recent proposal categorizes them according to their respective developmental context into 1) biogenic signals that act during early chloroplast biogenesis (e.g. during germination and seedling development) controlling proper organelle establishment and 2) operational signals that are send from well-developed chloroplast in later plant stages to mediate acclimation responses to environmental changes (Pogson et al., 2008). This concept has been expanded by the proposal of a third category, degradational signals sent from chloroplasts during senescence and that mediate nutrient allocation when plastids are finally degraded (Pfannschmidt and Munne-Bosch, 2013).
Current models suggest the action of various plastid metabolites, protein factors, reactive oxygen species, and redox signals from photosynthesis as mediators of retrograde signaling. The list of identified metabolites and oxidation products includes haem, singlet oxygen or hydrogen peroxide, carotenoid oxidation products, 3′-phosphoadenosine-5′-P, methylerythritol cyclodiphosphate, and oxo-phytodienoic acid (Lee et al., 2007; Galvez-Valdivieso et al., 2009; Estavillo et al., 2011; Woodson et al., 2011; Ramel et al., 2012; Xiao et al., 2012; Park et al., 2013). Proteins proposed to act as plastid signals include envelope-tethered eukaryotic transcription factors TFIIB-like and PTM, both being released from the outer plastid membrane by targeted proteolysis (Lagrange et al., 2003; Sun et al., 2011) and a plastid localized Whirly1 protein that is released from plastids upon stress (Isemer et al., 2012). A very recent study, however, puts this particular function of PTM in retrograde signaling into question (Page et al., 2017a, 2017b). Most of these retrograde signaling molecules are discussed as stress signals operating from fully developed chloroplasts. However, their mode of action and their potential interactions are largely not understood and many questions concerning their function and interaction remain unanswered.
Biogenic signals that are proposed to be active only during proplastid-to-chloroplast conversion are even less understood and the retrograde signals that contribute to chloroplast biogenesis remain to be identified. One major difficulty in studying biogenic plastid signal(s) is the large functional and temporal overlap with the photoreceptor (PR)-controlled light signaling network. Already early studies in this research field revealed that it is difficult to separate the influences of plastid signals on nuclear gene expression from those initiated by light as both occur at the same time range and on the same target genes. Transgenic reporter gene approaches could demonstrate that plastid and light signals do even use the same promoter elements in front of their target genes (Kusnetsov et al., 1996; Sullivan and Gray, 2002; Brown et al., 2005). Other studies suggest a close functional relationship between both control modes and it has been proposed that plastid signals can even remodel light signaling pathways from positive into negative signals and vice versa (Ruckle et al., 2007, 2012). Another recent study, however, suggests that light and plastid signaling routes act antagonistically in nuclear gene expression (Martín et al., 2016). The determined relative impact of light and retrograde signals on nuclear gene expression remains unclear.
Coordination of nuclear and plastid gene expression is also important for the establishment of the gene expression machinery in plastids and, in particular, for plastid localized RNA polymerases. A nuclear-encoded single-subunit phage-type RNA polymerase (NEP) and a plastid-encoded prokaryotic-type RNA polymerase (PEP) exist in plastids of green vascular plants. PEP is composed of four plastid-encoded subunits and 12 nuclear-encoded polymerase-associated proteins (PAPs). Furthermore, in Arabidopsis (Arabidopsis thaliana), PEP requires interaction with six nucleus-coded σ-factors for promoter recognition (Lerbs-Mache, 2011; Börner et al., 2015; Pfannschmidt et al., 2015).
These RNA polymerases are key players in the coordination of the gene expression between plastids and the nucleus as they transcribe the genetic information within the plastids in a developmentally well-coordinated manner that is especially important during the early stages of seedling development (Liebers et al., 2017). Although not all details are known yet, it is largely accepted that NEP represents the dominant plastid RNA polymerase activity in plastids of nongreen embryonic and meristematic cells. Its activity is essential for the expression of the plastid rpo genes and for the establishment of the core enzyme of PEP (Liere et al., 2011). During the course of chloroplast biogenesis, this basal PEP core enzyme then becomes decorated with PAPs. As far as known, PAPs are induced in their expression by light, and in silico analyses strongly suggest that they represent a tight regulon (Steiner et al., 2011; Pfannschmidt et al., 2015). The precise structural and functional roles of PAPs within the PEP complex and their regulatory relationship to plastid (and potentially nuclear) transcription are largely unknown and subject to current research.
Interestingly, all PAPs cause the same phenotypic consequences when their corresponding genes are inactivated. In Arabidopsis but also in maize (Zea mays) or rice (Oryza sativa), inactivation of pap genes results in albino, ivory, or pale-green phenotypes with arrested plastid development. Plastids of such mutants do not develop a thylakoid membrane system and display enhanced transcript accumulation of NEP-dependent genes whereas transcript accumulation of PEP-dependent genes (including those for photosynthesis) is typically diminished (Pfalz and Pfannschmidt, 2013). This expression profile of plastid-encoded genes is reminiscent of those found in plastid rpo deletion mutants of tobacco (Nicotiana tabacum; Hajdukiewicz et al., 1997; De Santis-MacIossek et al., 1999; Legen et al., 2002). The best possible explanation for this effect to date is that the lack of any of the PAPs either prevents the formation or compromises the stability of the PEP complex in developing chloroplasts. This, subsequently, leads to a lack of PEP-dependent processes and a concomitant arrest in chloroplast biogenesis because PEP is responsible for the expression of photosynthesis and tRNA genes (Williams-Carrier et al., 2014). It is important to note that the PAP assembly around the PEP core does not occur in the dark (Pfannschmidt and Link, 1994) and, consequently, pap mutants perform a normal skotomorphogenesis remaining undistinguishable from wild type (Gilkerson et al., 2012). Expression and assembly of PAPs around the PEP core complex therefore appear to represent a key initiation step in the formation of chloroplasts. The corresponding mutants represent, therefore, a useful tool to study the impact of a blocked transition from proplastids (or eoplasts) toward chloroplasts on the photomorphogenic program during seedling development.
Here, we present a study using the Arabidopsis pap7-1 mutant to elucidate the relative impact of arrested plastid development and light on nuclear gene expression. Like other PAPs, the PAP7/pTAC14 protein has been identified as a subunit of the plastid-encoded RNA polymerase (PEP; Pfalz et al., 2006; Steiner et al., 2011). In Arabidopsis, the corresponding gene (At4g20130) codes for a protein of 55 kDa that contains a chloroplast transit peptide, a predicted SET domain characteristic of protein Lys methyltransferases and a putative Rubisco LSMT substrate-binding domain. The precise function of the protein within the PEP complex as well as any evidence for methylation activity is still elusive, but an inactivation of the pap7-1/ptac14 gene in Arabidopsis results in an albino phenotype that is viable only on Suc-supplemented medium (Gao et al., 2011; Steiner et al., 2011). The mutant displays all the molecular and structural features described for other pap mutants (Gao et al., 2011) that we proposed to name as the “PAP syndrome”. Our study provides a detailed catalog of target gene modules at plastome and genomewide levels and gives unexpected and novel clues into the involvement of biogenic retrograde signaling in the regulation of nuclear genes for photosynthesis and metabolism.
RESULTS
Arabidopsis Pap7-1 Mutants Exhibit Normal Photomorphogenic Development but Never Develop Chloroplasts
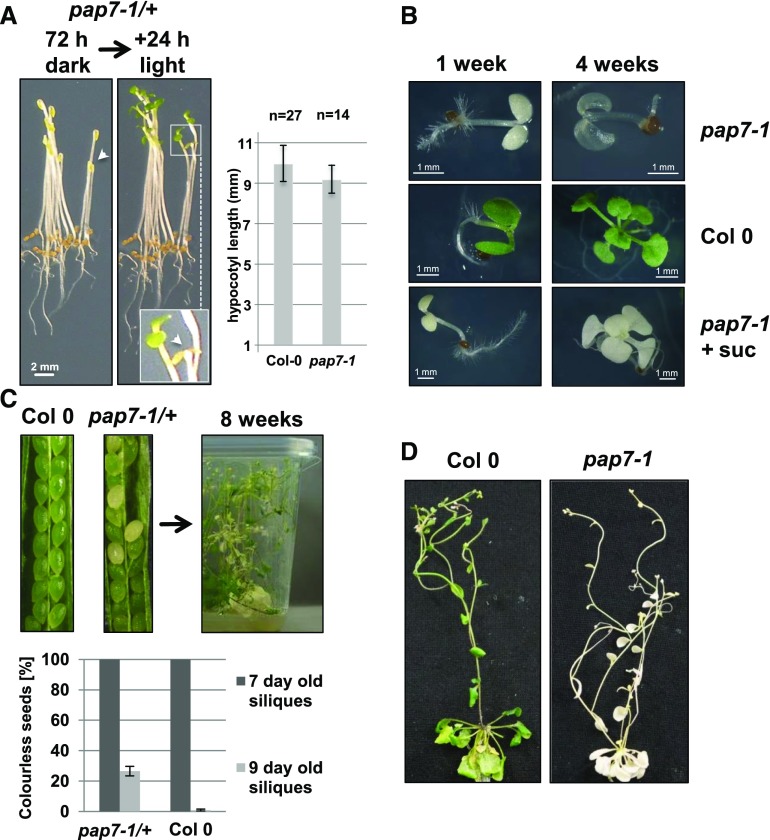
Homozygous pap7-1 mutant seedlings are known to develop an albino phenotype when grown in the light (Gao et al., 2011; Steiner et al., 2011). However, when grown in the dark, homozygous mutants develop a fully normal etiolated phenotype that remains macroscopically undistinguishable from heterozygous mutant or wild-type seedlings. They can be identified only by the missing greening process upon subsequent illumination (Fig. 1A), whereas they otherwise demonstrate a wild-type-like morphology. Grown directly in light without preceding dark phase, homozygous mutants show light-induced repression of hypocotyl elongation, opening of the apical hook, and separation and expansion of the cotyledons (Fig. 1B). When kept on standard growth medium without an additional carbon source, pap7-1 mutant seedlings could not develop further than the cotyledon stage, started to bleach, became transparent, and finally died (Fig. 1B). On Suc-supplemented medium, however, pap7-1 mutants were able to generate a rosette of almost wild-type-like appearance. The leaf blades were slightly smaller and the petioles shorter than in wild type (Fig. 1B). It has been observed that a transitory green stage with photosynthetic activities occurs in Arabidopsis embryos starting from the early torpedo stage [7 d to 9 d after fertilization (daf)] until the end of the maturation (mature green stage at 23 daf to 27 daf; Allorent et al., 2013). Checking this phase, we found in siliques of heterozygous pap7-1 mutants both green and white seeds with a segregation ratio of 3:1, indicating that already in homozygous embryos the chloroplast formation is prevented (Fig. 1C). Because these seeds were able to germinate with no bias in allelic transmission (3:1 ratio of homozygote mutants), this lack of chloroplast biogenesis appears nonessential for proper seed development and maturation. When growing the progeny of such heterozygous pap7-1 mutants on sugar-supplemented medium in short-day conditions under very dim light (approximately 8 to 12 μE WL), homozygous mutant plants developed a reasonable rosette that was even able to initiate the flowering program after shifting the plant container to long-day-conditions (Fig. 1, C and D). Architecture and size of the resulting inflorescence did not exhibit major differences in comparison to green plants, as indicated by control plants grown in the same containers (Fig. 1D). In sum, these observations indicate that the major photomorphogenic programs (and hence the action of the corresponding PRs) are functional in the mutant.
Figure 1.
Developmental characteristics of the pap7-1 mutant. A, 72-h dark-grown progeny of a pap7-1/+ heterozygote subjected to 24 h of 20 µE white light to trigger photomorphogenesis. White arrowheads indicate a pap7-1 homozygote mutant before and after subjection to light. Hypocotyl length ± sd measured after 72 h of dark growth. Genotypes were assigned after 24 h of light exposure (n = number of measurements). B, Growth of homozygous pap7-1 seedlings and wild type on 1/2 strength MS-medium in petri dishes without or with (+ suc) Suc supplementation. C, Impact of pap7-1 inactivation on chloroplast development during transient embryo greening and seed segregation in siliques of heterozygous pap7-1 mutants (pap7-1/+) in comparison to wild type (two left panels). Long-term growth (8 weeks) of the progeny of heterozygous mutants was performed on 1/2 strength MS-medium supplemented with 3% Suc in transparent plastic containers in a short-day light period (right panel). Seed segregation into green and colorless seeds was counted in 7 to 10 siliques per measurement day (7 d or 9 d after fertilization) in wild type and pap7-1/+ mutants (bottom panel). D, Long-term-grown plants in rosette stage were put into long-day conditions to induce flowering and photographed at the flowering stage.
Transcript Accumulation in Albino Pap7-1 Plastids
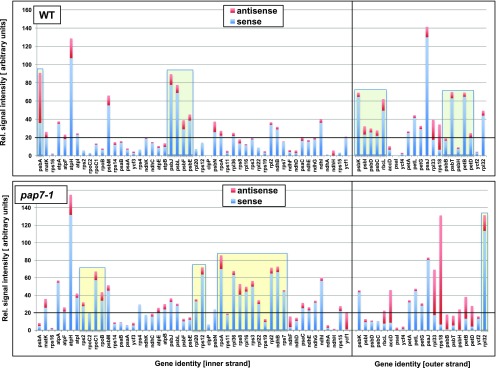
All pap mutants analyzed so far display largely reduced accumulation of PEP-dependent transcripts, whereas NEP-dependent gene transcripts do overaccumulate (Börner et al., 2015; Pfannschmidt et al., 2015). In all published reports, however, only a few representative genes per gene class were investigated. Here, we performed a comparative plastomewide analysis in which we determined the accumulation of all plastidial mRNAs in light-grown pap7-1 mutant plants and in wild-type plants grown in parallel (Fig. 2). To this end we used a custom-made macroarray that, in addition to sense RNAs, also allowed the detection of all corresponding antisense RNAs (Demarsy et al., 2012). Plastidial antisense RNAs were found, in Arabidopsis, to accumulate specifically in early phases of germination and radicle out-growth—two developmental steps preceding chloroplast biogenesis (Demarsy et al., 2012).
Figure 2.
Macroarray analysis comparing plastid transcript accumulation in light-grown wild-type and pap7-1 Arabidopsis seedlings. Given is the transcript accumulation in wild-type (top panel) and mutant (bottom panel) seedlings both for sense (blue bars) and antisense (red bars) transcripts. Hybridization signals were normalized to the total signal intensity of the membrane and are given as arbitrary units in the left margin. Genes are labeled at the bottom of each panel according to accepted nomenclatures. Sequence of genes corresponds to their organization on the plastome separated between inner strand (left parts) and outer strand of the plastome (right parts). High transcript accumulation of PEP-dependent transcripts in wild type is highlighted by green boxes. High transcript accumulation of NEP-dependent transcripts in the pap7-1 mutant is highlighted by yellow boxes. WT, wild type.
We observed reduced transcript accumulation for many PEP-dependent class-1 genes. However, mainly psb genes (encoding PSII components) were affected in the mutant (Fig. 2, light-green boxes), whereas, for psa genes (encoding PSI components), the reduction was found to be less pronounced or even nonexistent. Genes for the cytochrome b6f complex (pet genes) displayed mixed responses ranging from almost none to strong reduction of transcript accumulation. In contrast, all class-2 genes (genes with PEP and NEP promoters) displayed a stable or even enhanced transcript accumulation in the mutant. This class includes the ndh genes (encoding components of the NADH dehydrogenase complex), the genes encoding the proteins of the small (rps) and large (rpl) ribosomal subunits, and genes for components of the ATP synthase (atp). Enhanced accumulation could be also observed for class-3 genes (genes with NEP promoters only) rpoA and rpoB/C1/C2 (encoding the PEP core subunits; Fig. 2, yellow boxes), but not for the class-3 genes ycf1 and accD. A general down-regulation of PEP-dependent genes and a corresponding up-regulation of NEP-dependent genes, as suggested, thus cannot be confirmed by our macroarray analysis.
These observations were independently supported by the microarray analysis that includes the full set of plastome-located genes. In addition, the microarray also includes all tRNA genes that are not covered by the macroarray (Supplemental Table S1). These displayed all strongly reduced accumulation in the pap7-1 mutant line, suggesting that they are transcribed by PEP. This is in agreement with results obtained in a recent study on pap mutants of maize (Williams-Carrier et al., 2014).
Antisense RNA accumulation for most genes displayed no major differences between wild-type and pap7-1 mutant plants. However, we observed specific overaccumulation of antisense transcripts for the psbB/psbT/psbH/petB/petD operon, the rpl33 and rps18 genes, and the two genes ycf1 and accD (encoding import machinery subunit TIC214 and the β-subunit of the acetyl-CoA carboxylase complex, respectively; Fig. 2). This differential accumulation suggests that antisense production is not just a concomitant by-product of read-through transcription, but that a distinct unknown mechanism is at its origin.
In summary, our macroarray experiment uncovered that the disturbance in PEP activity does not cause gene-class-specific transcription changes but rather many gene-specific effects, suggesting a much more complex transcription regulation in arrested albino plastids than current models anticipate.
Separation of Light- and Plastid-Dependent Gene Regulation during Photomorphogenesis by Trilateral Differential Gene Expression Profiling
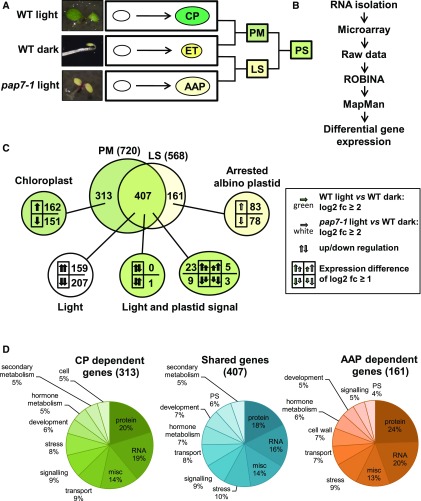
Chloroplast biogenesis is embedded into the general photomorphogenic program of seedling development that strongly impedes a clear distinction of light-, development-, and plastid-dependent signaling (López-Juez et al., 1998, 2007). However, because illuminated pap7-1 mutants develop normally on Suc-supplemented media (Fig. 1; Gao et al., 2011; Steiner et al., 2011) indicating that chloroplast biogenesis can be separated from photomorphogenesis, we used it as a tool to separate the gene groups regulated by either light or plastid developmental stage. To this end, we performed genomewide differential gene expression profiling in Arabidopsis by microarray hybridization. To unravel truly light- and plastid-dependent gene expression changes, we did a trilateral comparative profiling including light-grown pap7-1 mutant seedlings (pap7-1 light, containing white plastids), light-grown wild-type seedlings (wild-type light, containing chloroplasts), and dark-grown wild-type seedlings (wild-type dark, containing etioplasts, ETs), all at the two-cotyledon-stage (Fig. 3A). We performed a supervised analysis of the expression data using the MapMan tool (Usadel et al., 2005) and an unsupervised analysis by performing a weighted gene expression network analysis (WGCNA) using gene ontology (GO) groups to combine the advantages of both gene categorization tools (Klie and Nikoloski, 2012).
Figure 3.
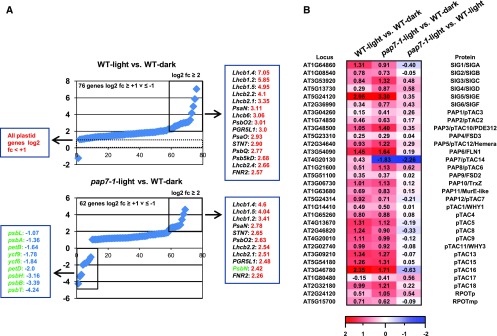
Identification of gene modules responsive to light and/or biogenic plastid signals. A, Strategy for differential expression profiling of 4-d-old to 5-d-old Arabidopsis seedlings in wild type and pap7-1 mutants (pap7-1). Left panel displays photographs of seedlings with representative phenotypic appearance. The right panel indicates the corresponding developmental transition of the plastids in each of these seedlings. Small white ovals represent undifferentiated pro/eoplasts from seeds. Upon illumination, they develop into green CPs in wild type (wild-type light) or arrested albino plastids (AAP) in pap7 mutants (pap7-1 light). Growth in the dark leads to development of yellow ETs in wild type (wild-type dark). Analysis of differences between expression profiles in these plant samples (indicated by brackets) identify genes for PM, for plastid-independent LS, and for PS (for details, see text). B, Flow diagram of bioinformatic analysis done with primary expression data from the microarray analysis. C, Detailed comparison of differentially expressed genes (indicated by numbers) within the PM and LS expression modules according to their direction of expression change (indicated by arrows). Only genes exceeding a threshold of log2 fc ≥ 2 were taken into analysis. Genes found in both modules were further separated according to their direction and degree of gene expression change. Arrows of different size but same direction indicate genes that display the same direction of expression change but with a difference of at least log2 fc ≥ 1. Green arrows are expression change in wild type. White arrows are expression change in pap7-1 mutants. For more details, see legend box. D, Proportional distribution of genes within the different gene groups defined in Fig. 3C according to their functional association within MapMan bins. WT, wild type.
In our supervised strategy, we assumed that a wild-type-light to wild-type-dark comparison reveals expression changes controlled by the photomorphogenic program (PM; Fig. 3A). This should identify all genes activated or inactivated by light and plastid signals. A parallel comparison of pap7-1 light- and wild-type-dark should identify genes being regulated only by light, whereas the developmental status of the plastid is negligible (light signaling, LS; Fig. 3A) because both samples do not develop chloroplasts. Subsequent comparison of the significantly regulated gene groups identified in PM and LS then should identify genes specifically regulated by light, by plastid stage (PS), or by both (Fig. 3A).
Significantly regulated genes in the data sets were identified (Supplemental Table S1) and were imported into the MapMan visualization tool (Fig. 3B; Supplemental Fig. S1). As expected, the PM data set revealed large genomewide changes known to be characteristic for the photomorphogenic program (Ma et al., 2001). This included a massive up-regulation of genes for photosynthesis, energy metabolism, and tetrapyrrole biosynthesis as well as sulfate reduction and a great number of other biosynthetic pathways (Supplemental Fig. S1A). In the LS data set, we observed an impact on mostly the same gene groups and with similar strength, indicating that the light-regulation in the pap7-1 mutant works in a comparable manner as in wild type (Supplemental Fig. S1B). Direct comparison of pap7-1 light versus wild-type-light gene expression profiles demonstrated only limited differences (Supplemental Fig. S1C), indicating that the gene regulation in the light-grown pap7-1 mutant resembles much more that of light-grown than dark-grown wild type. This indicates that the lack of functional chloroplasts in the pap7-1 mutant exerts only a minor impact on light-regulated gene expression networks.
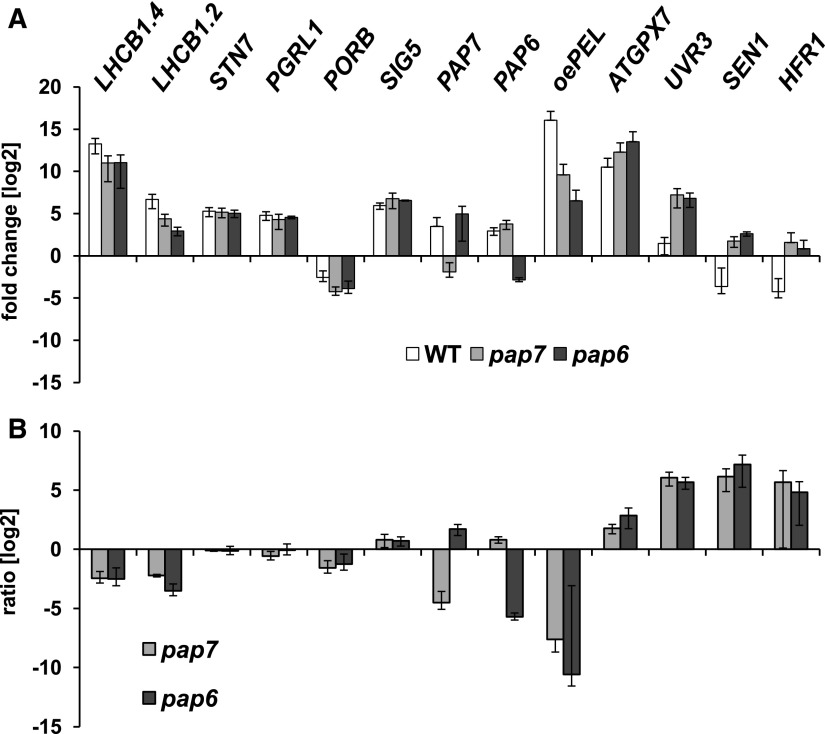
The quality of the expression profiles obtained in the microarray approaches was tested by quantitative RT-PCR of selected genes being representative for different expression classes found. To this end, RNA was isolated from identically, but independently grown plant samples from wild type and pap7-1 mutants. As a further independent control for the arrested plastid development another pap mutant, pap6-1, was used (Gilkerson et al., 2012). All genes tested in the quantitative reverse transcription PCR (qRT-PCR) displayed highly reproducible expression data (correlation factors above 0.9) when compared to the corresponding data obtained from the microarrays (Fig. 4; Supplemental Fig. S2). Furthermore, the expression data in the two different pap mutants displayed a remarkable low variation (Fig. 4), indicating that the observed expression profiles in the mutant background are robust and likely representative not only for pap7-1 but also for other pap mutants.
Figure 4.
Expression profiles of selected genes from the microarray analysis tested by qRT-PCR. A, Expression changes of genes representative for distinct expression classes in wild-type light, pap7-1, and pap6-1 mutants compared to wild-type dark. Genes for light-harvesting complex II protein1.4, light harvesting complex II protein1.2, and “overexpression leads to pseudo-etiolation in light” represent genes displaying a reduced light induction in pap7-1 when compared to wild type. Genes encoding protein state transition kinase7, PGR5-like protein 1a, and σ-factor5 represent genes displaying no effect of pap7-1 on light induction, whereas the gene for protochlorophyllide oxidoreductase B represents an example for strong light repression both in wild type and the pap7-1 mutant. The genes for glutathione peroxidase 7 and (6 to 4) DNA photolyase were used as examples displaying light induction in wild type and further promotion in pap7-1. Genes for SEN1 and HFR1 senescence-associated protein DIN1 and the transcription factor Long Hypocotyl after far-red1 represent genes displaying repression in wild type but promotion in the mutant. As additional genetic control, a defect in the gene for a phospho-fructokinase like1 protein (pap6-1/fln1) was used to detect potential mutation-specific responses. Expression of both pap genes was tested in all RNA samples as further control. B, Difference in expression change between light-grown wild-type and the pap7-1 and pap6-1 mutants. Negative values indicate lower expression, positive values higher expression than in wild type. Data given represent means of three independent experiments. Primers used are given in Supplemental Table S4. LHCB1.2, light-harvesting complex II protein 1.2; LHCB1.4, light-harvesting complex II protein 1.4; STN7, state transition kinase7; PGRL1a, PGR5-like protein1a; PORB, protochlorophyllide oxidoreductase B; SIG5, σ-factor 5; ATGPX7, glutathione peroxidase7; oePEL, “overexpression leads to pseudo-etiolation in light”; UVR3, (6 to 4) DNA photolyase; SEN1, senescence-associated protein DIN1; HFR1, transcription factor Long Hypocotyl after far-red1; WT, wild type.
Identification of Gene Groups Responding to Either Light or Biogenic Plastid Signals
Next, we performed a more detailed analysis of the various regulated gene groups. To restrict the analysis to strongly regulated genes, we introduced a threshold of log2-fold change (fc) ≥ 2 for definition of significantly regulated genes (881 genes in total). Identities and expression changes of these genes are given in Supplemental Table S2. Seven-hundred-and-twenty genes in the PM data set and 568 genes in the LS data set met this criterion (Fig. 3C). Four-hundred-and-seven genes were identified in both data sets with a majority being regulated in the same direction in both conditions (159 up-regulated and 207 down-regulated genes; Fig. 3C). These 366 equally regulated genes were not influenced by the developmental state of the plastid, but by illumination only. Thus, they represent plastid-independent, light-regulated genes. Only 41 of the 407 genes displayed either opposite (one gene) or different strength in their regulation, where we regarded only differences of at least log2 fc ≥ 1 as significant. These genes appear to be partly affected by both plastid stage and light. The largest group (23 genes) displayed up-regulation in wild-type light versus wild-type dark, but significantly less up-regulation in pap7-1- light versus wild-type dark. This expression pattern corresponds to the regulation mode attributed to the classical definition of a plastid signal that causes a lower accumulation of light-induced nuclear gene transcripts when chloroplast development is inhibited by chemical or genetic means (Pfannschmidt 2010).
In addition, we identified 313 genes that were exclusively up- (162) or down- (151) regulated only when chloroplast biogenesis (CP) occurred (CP-dependent genes, Fig. 3). These genes may either cause chloroplast biogenesis or are related to specific functions exerted by this fully developed plastid type such as retrograde redox regulation from photosynthesis. Interestingly, we identified 161 genes that displayed exclusive up- (83) or down- (78) regulation only in the presence of the arrested albino plastid (AAP-dependent genes, Fig. 3). These genes are likely regulated because of the missing chloroplast biogenesis or function (i.e. photosynthesis or connected biosynthesis pathways). Both developmental plastid stages apparently send distinct, light-independent signals to the nucleus that either repress or enhance separate sets of genes. Because of their light-independency, these signals are not identical with the classical plastid signal and imply the existence of plastid-type specific positive and negative signals not yet defined by current models of retrograde signaling.
In total, 41.5% of the 881 strongly regulated genes appear to be light-regulated, 4.6% are regulated by combined light and plastid signals, and 53.8% (corresponding to 474 genes) are regulated by plastid signals only (CP and AAP). Decreasing the threshold for significant regulation to log2 fc ≥ 1 identified 3658 regulated genes, which is roughly four times more than with log2 fc ≥ 2 as threshold (Supplemental Table S3). In this larger group, 46.4% of the genes appear to be light-regulated, 3.9% are regulated by light and plastid signals, and 49.5% are regulated by plastid signals only (Supplemental Fig. S3). Thus, enlargement of the number of investigated genes did not result in major relative changes between the three regulation modes, indicating that the arbitrarily chosen thresholds did not produce a bias for a specific regulation pattern. The inclusion of dark-grown plants as an additional reference point instead of doing a direct comparison of wild-type light and pap7-1 light provides a much more precise separation of plastid- and light-regulated genes. Both factors control very distinct sets of genes and the overlap between the two signaling pathways/expression networks appears to be very limited.
Among Photosynthesis-Associated Nuclear Genes, Only LHCB Genes Are Affected by Plastid Signals in Pap7-1
The CP-, light-, and AAP-dependent genes were finally sorted according to their functional categories (MapMan bin; Fig. 3D). The majority of functional categories were identical for all three groups comprising the MapMan bins “protein”, “RNA”, “stress”, “transport”, “signaling”, “development”, and “hormone metabolism”. Interestingly, the group of CP-dependent genes did not include “photosynthesis genes”, whereas the group of AAP-dependent genes did not include the bin “secondary metabolism”, but instead the bin “cell wall”. The large overlap in functional categories, however, is not reflected at the level of individual gene identities (Supplemental Table S1), and our analysis clearly demonstrates that each gene group represents a distinct set of regulated genes (see also results from WGCNA).
The lack of photosynthesis genes in the CP-dependent group was surprising with respect to the notion that photosynthesis-associated nuclear genes (PhANGs) are considered to be a prime target for plastid signals during biogenic control. We, therefore, analyzed these genes separately and compared their relative expression changes in the PM and LS data sets (Fig. 5A). In wild-type light versus wild-type dark, 76 genes exhibited a light-induced expression increase of log2 fc ≥ 1. Ninety-three photosynthesis genes (including all plastome-localized genes) displayed expression variations that remained below this threshold, suggesting that they are not or just mildly affected by light. Only 15 genes exhibited a strong light induction of log2 fc ≥ 2. The top five of these genes all encode proteins of the light-harvesting complex of PSII (LHCB; framed in Fig. 5A). In the pap7-1 light versus wild-type dark comparison, we observed a similar expression profile for photosynthesis genes. Here, 62 genes exhibited light-induced expression of log2 fc ≥ 1 and 11 genes displayed expression changes of log2 fc ≥ 2. Ten of these genes were also found as top-regulated genes in wild type. However, we observed that, specifically, LHCB genes displayed significant less accumulation in pap7-1 compared to wild type, whereas all other genes remained fairly constant. Comparable results were obtained in the independent qRT-PCR controls (Fig. 4). This reduced expression can be attributed to the impact of biogenic signals from the arrested albino plastid. In addition, we also observed nine significantly down-regulated genes in this comparison, all of them being plastome-localized (Fig. 5A). This confirms the observation using the MapMan visualization (Supplemental Fig. S1) in which inhibition of chloroplast development had only limited impact on the overall nuclear gene expression profile. We conclude that inhibition of chloroplast development in the pap7-1 mutant represses specifically plastome-localized photosynthesis genes and a small set of nuclear LHCB genes, whereas all other PhANGs are either not affected or are just mildly attenuated.
Figure 5.
Light-induced expression changes of genes for photosynthesis and plastid transcription. A, Graphs display the expression values of significantly light-affected photosynthesis genes (log2 ≥ 1) detected by comparison of expression profiles for wild-type light versus wild-type dark (top graph) and pap7-1 light versus wild-type dark (bottom graph). From the 169 photosynthesis genes present in the corresponding MapMan bin, 72 and 62 genes, respectively, exhibited an expression change of at least log2 ≥ 1. In the wild-type light versus wild-type dark comparison, all plastid genes remained below this threshold. Genes exceeding an expression change of log2 ≥ 2 were boxed in the graph and listed in a panel aside. Nuclear-encoded genes are given in black, plastid-encoded genes are given in green. The difference in the expression change of a particular gene that occurs between the top and the bottom panel reflects the plastid influence on its expression. Strongly down-regulated genes in pap7-1 light versus wild-type dark (log2 ≥ −1) are given in a left box. B, Expression changes for all 31 nuclear genes encoding components of the plastid gene transcription machinery in wild type and pap7-1. The respective comparisons are indicated in the top panel, gene identities in the left, and protein identities in the right panels. Given values represent log2-fold changes; a corresponding color-code is depicted at the bottom level. WT, wild type.
Another interesting question was whether other nuclear genes for components of the plastid transcription machinery were affected in their expression by the repressed PEP activity in pap7-1 plastids. We, therefore, analyzed the expression data of genes for all σ-factors, PAPs, PTAC components, and for NEP. The majority of these components displayed light induction in wild type when compared to the dark control (Fig. 5B). This is in accordance with earlier bioinformatic analyses. The observed regulation patterns were largely maintained in the mutant with only minor deviations from the wild type, indicating that the developmental state of the plastid has also no significant impact on the expression of these genes whereas light appears to be a dominant regulator even in the mutant. The functional PEP deficiency does not exert a retrograde repressive control of other nuclear-encoded PEP or NEP components.
WGCNA
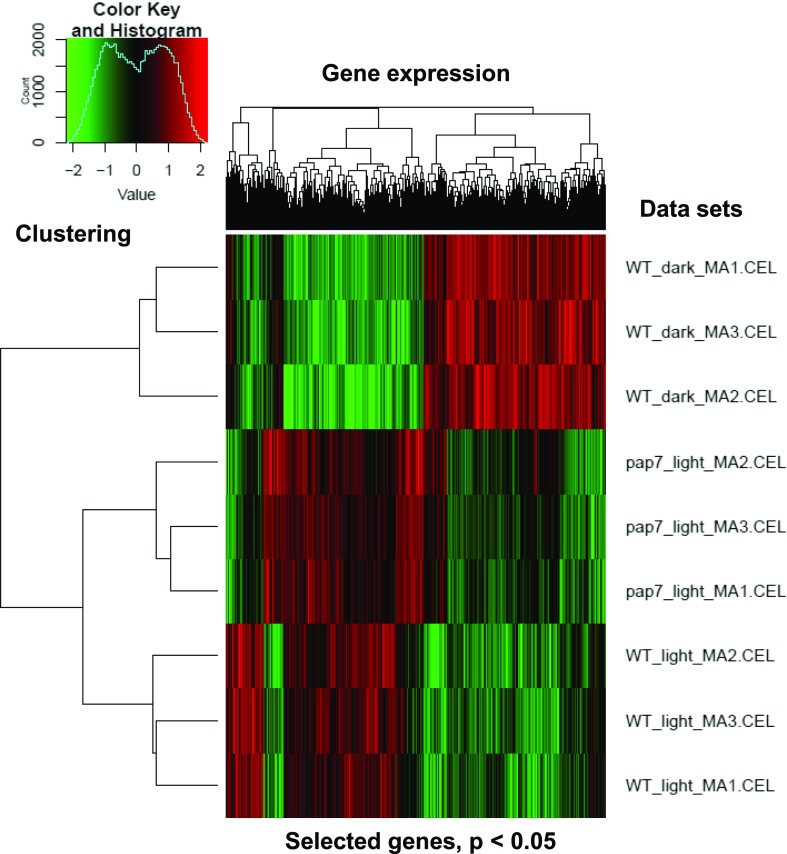
The supervised analysis (Fig. 3) used presettings based on assumptions drawn from literature and our own experiences. To avoid any unwanted bias, we performed an additional unsupervised analysis of the gene expression data sets employing a WGCNA (Supplemental Fig. S4). As in our supervised analysis, the cluster analysis of the expression data revealed a much closer correlation between pap7-1 light and wild-type light samples than between pap7-1 light and wild-type dark samples (Fig. 6). Nevertheless, the gene expression profiles of pap7-1 light and wild-type light samples displayed a number of specific differences. In total, six significant gene expression modules could be identified by WGCNA within the data set (Supplemental Fig. S4). These modules describe characteristic similarities and differences between the three plant samples and largely correspond to the groups defined in the supervised differential analysis. Because the unsupervised analysis did not include expression thresholds, it covered much larger gene numbers. The identified expression modules divide into two groups (Supplemental Fig. S4B). Group 1 comprises three modules (modules blue, yellow, and green) in which two samples are highly similar, whereas the third one is opposing. Group 2 comprises three modules (modules turquoise, brown, and red) in which two samples are opposing, whereas the third one is in an intermediate state between the two others.
Figure 6.
Cluster analysis of genes differentially regulated in the three growth setups as defined by ANOVA. Genes displaying an FDR of < 0.05 were included. The right margin identified the data sets from the microarray analysis; the left margin indicates a cladogram defining the correlation in gene expression profiles between them. On top, clustering of gene groups according to their expression is indicated. Diagram in top-left corner gives the color key and numbers of genes with corresponding gene expression values. Red indicates up-regulation; green indicates down-regulation. WT, wild type.
Genes in module “blue” (in total 1838 genes) display highly similar expression in pap7-1 light and wild-type light samples and an opposing expression in wild-type dark. The dominant regulating factor in this module is the light, whereas the plastid state appears to play no role or only a very minor one. Module “blue”, therefore, covers the light-regulated genes. Genes in module “yellow” (in total 769 genes) display highly similar expression in pap7-1 light and wild-type-dark samples and an opposing expression in wild-type-light. The dominant regulating factor here is the plastid state, whereas light appears to have no major impact. Module “yellow”, therefore, covers genes under a plastidial regulation that is exerted from a normally developed chloroplast. Genes in module “green” (in total 671 genes) display a highly similar expression in wild-type-light and wild-type-dark samples and an opposing expression in pap7-1-light. Light appears to be of no importance for the regulation of these genes, but the arrested plastid development and the genetic background of the mutant. Module “green”, therefore, covers genes that are misregulated because of the disturbance in plastid development and the pap7-1 protein deficiency. The expression profiles of the genes covered by these three modules correspond to those in the three gene groups defined in our supervised analysis (Fig. 3C), and provide independent analytical confirmation for the correctness of our selection criteria. Understanding and interpretation of the modules in group 2 appeared to be much more difficult as the intermediate expression profiles in each module prevent an identification of the dominant regulating factor. Gene groups defined by these modules are likely under multifactorial control and were, therefore, excluded from further analysis.
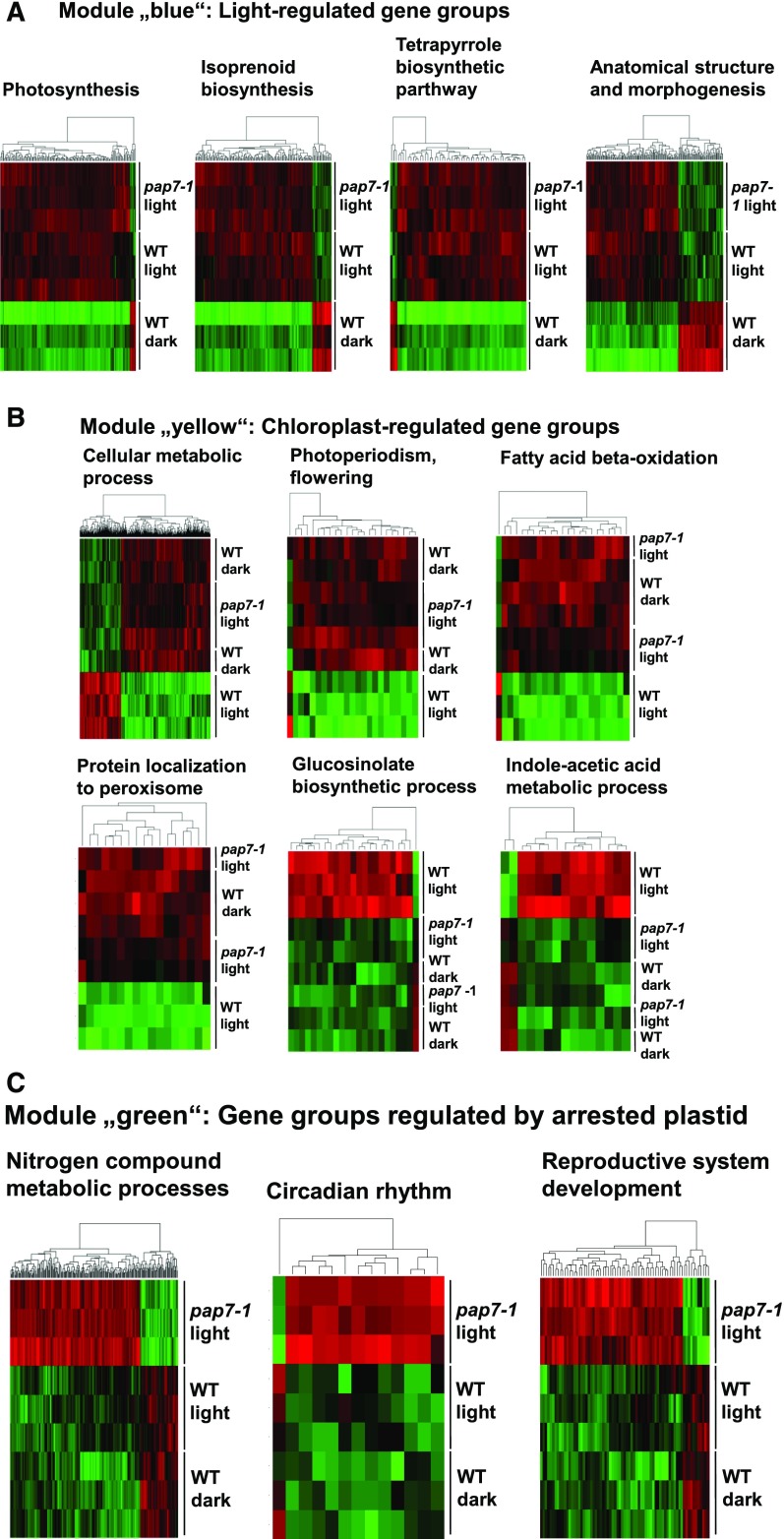
We analyzed the three gene modules for enriched GO groups (Supplemental Figs. S5 and S7) and indicated most important GO groups with respect to the mutant phenotype below. Light-regulated genes in module “blue” included the GO groups for “Photosynthesis”, “Isoprenoid biosynthesis”, “Tetrapyrrole biosynthesis”, and for “Anatomical structure and morphogenesis” (Fig. 7A). All these gene groups are apparently activated by light without displaying a major impact of the developmental state of the plastid. Like in the supervised analysis, PhANGs occur under the light-regulated gene groups supporting the notion that the impact of the plastid stage on the expression of this gene group is very limited. Chloroplast-regulated genes in module “yellow” (Fig. 7B) contained GO groups for “Cellular metabolic processes”, “Photoperiodism and flowering”, “Fatty acid beta oxidation”, “Protein localization to peroxisome”, “Glucosinolate biosynthetic process”, and “Indoleacetic acid metabolic processes”. Although the first four groups appear to be repressed, the latter two are mainly activated by the presence of functional chloroplasts. Finally, in the pap7-1-regulated module “green” (Fig. 7C), we found GO groups for “Nitrogen compound metabolism processes”, “Circadian rhythm”, and “Reproductive system development”. These gene groups were either activated or stayed active upon the genetic arrest of chloroplast biogenesis. Our triplicate analyses describe distinct gene sets that are specifically targeted by the three factors of light, chloroplasts, and arrested albino plastids, in both a positive and a negative manner.
Figure 7.
GO groups of differentially expressed gene sets within the gene modules identified by WGCNA. A, Gene module “blue” with four major gene groups regulated by light. B, Gene module “yellow” with six major gene groups regulated by the chloroplast. C, Gene module “green” with three major gene groups specifically regulated by the arrested plastid. Genes displaying an FDR < 0.05 were included. On top of each heat map, the selected GO group is given; underneath, a clustering of the genes in this group, according to their expression, is indicated. The right margin identifies the data sets from the microarray analysis. Red indicates up-regulation; green indicates down-regulation. For further gene groups, see Supplemental Figs. S8 to S10. WT, wild type.
DISCUSSION
Our plastome- and genomewide gene expression analysis in the pap7-1 mutant provides the first detailed view of the disturbances occurring at the transcript level of this mutant. This revealed interesting and unexpected facts that help us to better understand the causes for the albinism in this mutant. In addition, the obtained data provide important novel clues for our understanding of other pap mutants as well as retrograde biogenic signals and their target genes.
Impact of Pap7-1 Deficiency on Plastid Gene Expression
Elucidation of the plastid transcriptome in the pap7-1 mutant uncovered that many PEP-dependent class I transcripts displayed only low accumulation whereas NEP-dependent rpo transcripts exhibited enhanced accumulation (Fig. 2). This is in coincidence with earlier reports. However, we observed significant differences between psa and psb gene groups, suggesting differential effects on class-I transcription. In addition, NEP-dependent ycf1 and accD gene transcripts did not overaccumulate as rpo transcripts, but exhibited clearly reduced transcript accumulation (Fig. 2). Furthermore, class-II transcripts revealed largely wild-type-like accumulation in the mutant (Fig. 2). These effects cannot be reconciled with this model of plastid transcription in albino plastids of pap mutants, which assumes a general inactivation of PEP activity, whereas the NEP activities are up-regulated. Instead, these data imply differential and gene-specific effects on transcription activities in the albino plastids. Because all class-I transcripts did accumulate to a certain degree in pap7-1 plastids, it is likely that these arise from a basal activity of the PEP core-complex. Such a basal activity was identified in ETs of mustard, and was shown to be able in faithful promoter recognition using σ-factors, but in a different manner than the corresponding activity from chloroplasts (Eisermann et al., 1990; Tiller and Link, 1993; Pfannschmidt and Link, 1994). This scenario could provide a realistic explanation for the observed transcriptome in the pap7-1 mutant plastids, and suggest that the gene expression mechanisms in the albino plastids are likely arrested in a stage similar to ETs. Alternatively, the residual class-I transcripts may arise from read-through transcription performed by NEP enzyme activities; however, such a model would lack an explanation for differential gene group transcription. Regardless of their origin, the residual class-I transcripts apparently are not sufficient to elicit the formation of a photosynthetic apparatus, suggesting the involvement of additional constraints for the chloroplast formation.
One such constraint could be the enhanced antisense transcript accumulation in the mutant that was observed notably for the psbB/T/H/petB/D operon, the rpl33/rps18 operon, and the ycf1 and accD genes (Fig. 2). As this effect appears to be rather gene-specific, it likely does not represent just an arbitrary accumulation of read-through transcription of the respective opposite strand of the plastome. Mechanistically, one could expect that enhanced antisense transcript accumulation interferes with translation efficiency of the corresponding sense transcript due to duplex formation, as it was suggested for the plastid psbT gene (Zghidi-Abouzid et al., 2011). This would provide a gene-group-specific mechanism that could prevent the formation of functional photosystem II and Cytb6f complexes. Whether this antisense production is based on a specific transcription event will be an interesting field of future research.
Our microarray analysis of pap7-1 RNA samples indicated a strongly reduced accumulation of most plastidial tRNAs (Supplemental Fig. S1). This is in agreement with a recent study performed in several maize pap mutants, proposing that PEP activity has a major role in the expression of plastidial tRNAs (Williams-Carrier et al., 2014). Reduction in plastid tRNA accumulation restricts plastid translation, as the tRNA molecules transfer the amino acids. Furthermore, because the tRNAE is the precursor of amino-levulinic acid, tetrapyrrole biosynthesis and the generation of chlorophylls might be severely affected.
In sum, the differential accumulation of gene-specific transcripts of all classes implies the existence of specific, yet unknown, transcription events in the arrested albino plastids, suggesting a more defined and diversified division of labor between the PEP and NEP enzymes during early steps of chloroplast biogenesis than the current models propose.
Retrograde Control of Nuclear Gene Expression by Biogenic Plastidial Signals
A major improvement of our study when compared to earlier work in this field arose from our experimental setup, which included dark-grown wild-type plants as an additional reference point. Inclusion of homozygous pap7-1 mutants grown in the dark as further control was not feasible, as homozygous mutant seedlings in this stage remain macroscopically indistinguishable from wild-type or heterozygous mutants (Fig. 1A). Discrimination between wild-type and mutant genotypes at this stage would be technically possible only at the molecular level of individual seedlings, exacerbating largely the harvest of sufficient material for RNA preparation. The indistinguishability of dark-grown homozygous mutants is in accordance with our working hypothesis that the mutation becomes effective only under illumination, although it does not affect the skotomorphogenic program. Consequently, segregation of the progeny of heterozygous pap7-1 mutant plants becomes macroscopically visible only in the light when the transition toward chloroplasts is arrested. Nevertheless, the trilateral expression profiling allowed us to define unambiguously distinct gene sets responding specifically to 1) light, 2) chloroplast signals, and 3) signals from arrested albino plastids. This distinction is impossible by a simple bilateral mutant-wild-type comparison in the light as this provides only the relative differences between the two conditions. The impact of light in each of them requires the additional comparison with the dark control that then reveals whether light has a promoting, inhibiting, or no effect and allows finally for the distinction between retrograde and light control.
The three gene modules identified by our approach contain both activated and repressed genes, implying the existence of retrograde biogenic signals that are of either a positive or a negative nature. Because the gene sets regulated by chloroplasts and arrested plastids are different, it is likely that the controlling signals are transmitted via separate pathways. Whether these are fully independent from each other or mutually exclude each other remains to be investigated. The question of whether a weaker nuclear gene expression indicates the action of a negative signal or the missing action of a positive signal has been debated in the past (Pfannschmidt, 2010; Terry and Smith, 2013; Hills et al., 2015). A recent study (Page et al., 2017a, 2017b) and our data presented here indicate that the action of both positive and negative signals needs to be considered in current models.
The phenotypic analysis (Fig. 1) revealed that despite chloroplast deficiency, the general photomorphogenic program in the pap7-1 mutant appears to be operational as long as an external carbon source is available. Older mutant plants generate a complete albino rosette that is comparable to those of dark-grown mutants with constitutively active phytochromes (Su and Lagarias, 2007), confirming that general plant development and chloroplast biogenesis represent largely separate processes. Even the initiation of the flowering transition was found to be functional, which corresponds to observations reported for rpo deletion mutants of tobacco (De Santis-MacIossek et al., 1999). We assume that illumination activates the PR systems in both wild-type and pap7-1 mutant seedlings in the same manner, suggesting that synthesis and action of the PRs (including their chromophores) are fully functional in the albino mutant.
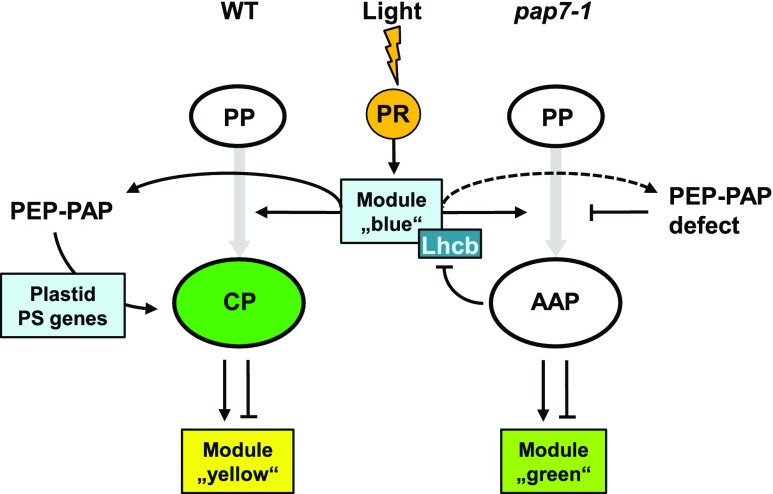
This explains the similarity of the expression profiles of light-grown wild-type and pap7-1 mutants (Supplemental Fig. S1), including the expression of the corresponding light-regulated gene module (module “blue”; Fig. 8). The module contains the GO groups for isoprenoid and tetrapyrrole biosynthesis (Fig. 7) that produce important precursors for primary products such as carotenoids, chlorophylls, haem, phytochromobilin, plastoquinones, or different plant hormones (Pulido et al., 2012). Both biosynthesis pathways provide essential metabolites for plant metabolism and development and, therefore, are likely active in the albino plastids with the apparent exception of Chl biosynthesis. For the same reasons, the GO group for morphogenesis and anatomical structure (Fig. 7) may appear in this module because it is part of the light-initiation of photomorphogenesis. Unexpectedly, however, was the identification of PhANGs that, in contrast to general assumptions, did not exhibit any major repression by retrograde signals from the arrested plastid development. Only a few Lhcb genes appeared to be selectively targeted by plastid signals (Fig. 5) and these, in addition, seem to interact with light (Figs. 3 and 5). This may explain the manifold connections identified between light and plastid signaling using promoters of these genes in genetic screens and corresponding mutants (Larkin, 2014). Interestingly, the structural and functional defect in the PEP-PAP complex in pap7-1 mutants does not affect the transcript accumulation of other nuclear-encoded components of the plastid transcription machinery (Fig. 5B), implying that these genes are not under retrograde control.
Figure 8.
Model of anterograde and retrograde signaling during early steps of chloroplast biogenesis in wild-type and pap7-1 mutants. Light signals (flash) are perceived by PRs (orange circle). Small white ovals represent undifferentiated proteoplasts present in seeds. Large ovals in green and white represent the different plastid types in wild-type (CP) and pap7-1 (AAP) seedlings. Gray-shaded arrows indicate the developmental process leading to these plastid types. Boxes indicate and name the genes or gene modules regulated during these processes. Thin black arrows represent positive regulation; lines with a blocking bar represent negative regulation. WT, wild type.
It remains for us to understand why chloroplast biogenesis does not work in the mutant. A major determinant of this developmental block certainly is the reduced expression of plastid photosystem II genes, but the finding of residual transcripts suggests that other molecular reasons also likely play a role, such as the obvious inhibition of Chl biosynthesis (Gao et al., 2012) and further defects that are unknown to date. We conclude that in the pap7-1 mutant, plastidial photosystem II genes are especially strongly repressed, whereas retrograde signals from the arrested albino plastid neither modulate nor antagonistically counteract light regulation of PhANGs. This is different from conclusions obtained in recent studies on retrograde signaling using lincomycin, an inhibitor of plastid translation (Ruckle et al., 2007; Martín et al., 2016). Because pap7-1 mutants developed rather normally in the first few days even without sugar, we regard it as likely that either the genetic block in pap7-1 mutants results in milder effects than a lincomycin treatment, or the observed differences are due to technical differences in the respective setups, e.g. light intensity. Both scenarios suggest that there exists a threshold for the effectiveness of retrograde signals. Elucidating the molecular nature for this will be an interesting topic for future research. The retrograde-controlled gene groups identified in this study provide a useful first tool for such investigations.
An interesting result of our expression profiling was the identification of separate biogenic signals from chloroplasts and albino plastids. Chloroplast signal-dependent GO groups were mostly related to metabolism, likely because chloroplast biogenesis initiates the conversion from a heterotrophic to autotrophic life style. This includes the down-regulation of β-oxidation of fatty acids that takes place in glyoxysomes. Oppositely, GO groups for glucosinolate biosynthesis and indoleacetic acid biosynthesis were activated, both being highly important for pathogen defense and growth of green plants (Fig. 7B). GO groups regulated by signals from the arrested albino plastid relate mostly to starvation processes and the mobilization of storage energies (nitrogen compound metabolic processes, reproductive system development) being indicative of the nonautotrophic metabolism in the albino plantlets that requires the mobilization of all internal resources. Surprisingly, a significant impact on the GO group of circadian rhythms was observed that suggests an influence of the plastid developmental stage on circadian clock genes. A circadian control from the nucleus influencing chloroplast transcription has been recently reported (Noordally et al., 2013). Furthermore, iron metabolism and plastid developmental stage were demonstrated to have a significant influence on the period of the circadian clock (Salomé et al., 2013). These observations suggest that our results reflect a mutual influence between plastids and the circadian clock of larger significance, providing an interesting target for future research (Fig. 7C).
In sum, the arrest of chloroplast development in the pap7-1 mutant can be best explained by a specific disturbance in the light-induced buildup of the PEP complex during the pro/eoplast-to-chloroplast transition. This leads to concomitant defects in the expression of PSII components and tRNAs, which in turn limits Chl biosynthesis and (likely) translation. The albinism of the mutant likely is the result of a multifactorial syndrome that prevents chloroplast biogenesis without destroying the plastid. Therefore, the functional and metabolic state of the arrested albino plastid resembles that of an ET despite it perceiving light. This likely allows the mutant to develop normally when the lack of photosynthesis is complemented by an external carbon source. Correspondingly, the overall gene expression profiles of pap7-1 mutants do exhibit a chimerical character, with metabolic genes regulated like in dark-grown seedlings and photoregulated genes like in light-grown plants. In sum, the pap7-1 mutant, but likely also other pap mutants, provide an interesting tool for dissecting further the molecular processes during the early steps in chloroplast biogenesis.
MATERIALS AND METHODS
Plant Material
We used Arabidopsis (Arabidopsis thaliana) Columbia (Col) 0 as wild type throughout the study. As null alleles for the genes pap7-1/ptac14 and pap6/fln1 we used the Arabidopsis T-DNA inactivation lines SAIL_566_F06 and GK-443A08, respectively. These lines (named pap7-1 and pap6-1 in this study) were characterized earlier in detail for singularity of T-DNA insertion and for causal connection between the albino phenotype and the corresponding gene defect (Arsova et al., 2010; Gao et al., 2011; Steiner et al., 2011; Gilkerson et al., 2012). Seeds of wild-type and heterozygous mutants were surface-sterilized and spread on half-strength standard Murashige & Skoog (MS) medium supplemented with indicated amounts of Suc in petri dishes, stratified for 3 d, and grown to the two-cotyledon stage at 21°C for further analyses. Light-grown plants used for array analyses were grown under permanent white light of 120 μE to 150 µE photon flux density. In long-term growth experiments, the illumination intensity of the white light source was reduced to 8 μE to 12 μE.
Plastid Macroarray Analysis
Plant material was grown 6 d on MS medium supplemented with 0.5% Suc. Approximately 500 mg each of green wild-type and albino mutant cotyledons were harvested separately and shock-frozen in liquid nitrogen. Total RNA was isolated following published procedures (Demarsy et al., 2006, 2012). Potential DNA contaminations were removed by DNase treatment and its absence was proven by PCR. For preparation of the hybridization probe, 4 μg of each RNA preparation was reverse-transcribed using a gene-specific primer mix annealing to 80 protein-coding genes and their corresponding antisense sequences. Subsequently, a reverse transcription reaction was performed using Superscript II Reverse Transcriptase (Invitrogen) in the presence of all four nucleotides and 100 µCi [α-32P] dATP (Demarsy et al., 2012). Unincorporated nucleotides were finally removed from the labeled cDNAs by gel-filtration through a Sephadex G50 column. For evaluation of relative differences in plastid transcription between the biological backgrounds of the two samples, we measured the total radioactivity of each cDNA sample after synthesis and verified the cDNA profile on a sequencing gel. In the experiment described here, radioactively labeled cDNAs of wild type and pap7-1 differed by just 3% (wild type > pap7-1) in incorporation, indicating that total plastid gene expression is not significantly different in wild-type and pap7-1 plantlets. Subsequent hybridization of the radiolabeled probes with the macroarray, washing conditions, subsequent signal detection in a phosphorimager (FLA-8000; Fujifilm), and final data analysis were done essentially as described in Demarsy et al. (2012). Construction of the plastome macroarray, including description of the gene-specific sense and antisense probes and their spotting pattern, was published elsewhere (Lerbs-Mache, 2011).
Genomewide Microarray Analysis
Plant material was grown on medium supplemented with 1% (w/v) sugar to harmonize the germination. Illuminated wild-type and homozygous pap7-1 mutants were grown for 5 d for full expansion of the cotyledons, whereas dark-grown wild-type seedlings were grown for 4 d to avoid mechanical stress imposed by physical contact with the lid of the petri dishes. Plant materials were harvested at 10:00 a.m. and shock-frozen in liquid N. Dark-grown material was harvested and shock-frozen at the same time point of the day, but under a green safe-light to exclude any light effects in the profiles. Total RNA from these materials was then basically prepared as described in Logemann et al. (1987). In brief, 250 mg of frozen plant material was ground in a mortar and purified with the RNeasy purification kit (Qiagen). Concentration and purity of RNA samples were determined spectroscopically and intactness was proven by ethidium bromide staining after separation on 1.2% (w/v) agarose gels. Purified samples from three biological replicates each were sent on dry ice to a commercial service (Kompetenzzentrum für Fluoreszente Bioanalytik) where a second quality check, cDNA synthesis, and labeling was performed according to the GeneChip 3′ IVT Express Kit protocol (Affymetrix). Hybridization and reading of signals was performed using the Arabidopsis Genome Array ATH1 (Affymetrix) and according to standard protocols of the service.
qRT-PCR
Reverse transcription was performed with 1 μg total RNA isolated from three independent biological replicates of wild-type, pap7-1, and pap6 mutants grown identically to those for the microarray analyses. cDNA was synthesized using oligo(dT) primers and the SuperScript II Reverse Transcriptase (Invitrogen) following the manufacturers’ recommendations. The qRT-PCR analysis was performed using the GoTaq qPCR Master Mix (Promega) and the Rotor-Gene 3000 equipment (Qiagen). Primer sequences for genes of interest were designed using the software ApE-A Plasmid Editor (v2.0.47) and NCBI/Primer-BLAST with preference to intron spanning amplicons. Each primer pair was tested for amplification efficiency using the synthesized cDNA. Only primer pairs with amplification efficiencies between 90% and 110% were used for further analysis. For used primer sequences and gene identities, see Supplemental Table S4. The relative quantification was calculated according to described methods (Pfaffl, 2001). The Arabidopsis genes for actin 7 and ubiquitin 5 were used as internal control and reference genes for quantification.
Bioinformatics
In the supervised analysis, the hybridization signal data from the microarray analysis performed by a commercial service (KFB Regensburg) were analyzed with the ROBINA (http://www.mapman.gabipd.org/web/guest) and MapMan (http://www.mapman.gabipd.org/web/guest) programs (Usadel et al., 2005, 2009). Statistical analysis by t test and subsequent calculation of the false-discovery rate (FDR) were performed according to the ROBINA program. Gene expression changes with an FDR of P ≤ 0.05 were regarded as statistically significant. The microarray data given in the Supplemental File S1 are based on three biological replicates each. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002; Barrett et al., 2013) and are accessible through GEO Series accession no. GSE88988 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE88988). Visualization of the cellular pathways and functional categories of the expression data were carried out using the MapMan and PegMan packages according to Ath_AFFY_ATH1_TAIR8_Jan 2010 (http://mapman.gabipd.org; Usadel et al., 2005). The visualization tool of MapMan was used to identify similarities and differences in the different metabolic pathways. A Wilcoxon rank sum test was used to visualize significantly expressed genes in PegMan (Usadel et al., 2009). Venn diagrams were calculated using the expression log values of the MapMan package. In the unsupervised WGCNA, the original CEL files supplied by the commercial service were imported into R with Bioconductor (package oligo; Carvalho and Irizarry, 2010) followed by normalization and background correction of the raw data using RMA. After selecting 27,296 annotated genes, a scaling and centering followed by a cluster analysis was performed. Differentially expressed genes were selected with ANOVA and an FDR threshold of 0.05, followed by a WGCNA (Langfelder and Horvath, 2008) and a GO enrichment analysis using the R package topGO (http://www.mpi-sb.mpg.de/∼alexa).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Relative gene expression profiles of wild-type and pap7-1 mutants visualized using the MapMan tool.
Supplemental Figure S2. Correlation of expression data from selected genes obtained from microarrays and qRT-PCR.
Supplemental Figure S3. Identification of gene modules responsive to light and/or biogenic plastid signals with a significance threshold of log2 fc ≥ 1.
Supplemental Figure S4. Weighted gene coexpression network cluster analysis.
Supplemental Figure S5. Gene ontology groups within module “blue”.
Supplemental Figure S6. Gene ontology groups within module “yellow”.
Supplemental Figure S7. Gene ontology groups within module “green”.
Supplemental Table S1. Gene expression changes of genes sorted according to the encoding genomic compartment.
Supplemental Table S2. Gene sets with an expression change larger than the threshold 2 [log2].
Supplemental Table S3. Gene sets with an expression change larger than the threshold 1 [log2].
Supplemental Table S4. Nucleotide sequences of primers used for qRT-PCR.
Footnotes
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) to T.P. (PF323-5-2) and the Deutsche Forschungsgemeinschaft (DFG) research group FOR 804. The study also received institutional support from the French National Research Agency (ANR-10-LABEX-04 GRAL Labex, Grenoble Alliance for Integrated Structural Cell Biology).
Articles can be viewed without a subscription.
References
- Allorent G, Courtois F, Chevalier F, Lerbs-Mache S (2013) Plastid gene expression during chloroplast differentiation and dedifferentiation into non-photosynthetic plastids during seed formation. Plant Mol Biol 82: 59–70 [DOI] [PubMed] [Google Scholar]
- Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustün S, Melzer M, Petersen K, Lein W, Börnke F (2010) Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22: 1498–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski AA, Galstyan A, Guseman JM, Nemhauser JL (2012) Photomorphogenesis. Arabidopsis Book 10: e0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, et al. (2013) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41: D991–D995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner T, Aleynikova AY, Zubo YO, Kusnetsov VV (2015) Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim Biophys Acta 1847: 761–769 [DOI] [PubMed] [Google Scholar]
- Brown NJ, Sullivan JA, Gray JC (2005) Light and plastid signals regulate the expression of the pea plastocyanin gene through a common region at the 5′ end of the coding region. Plant J 43: 541–552 [DOI] [PubMed] [Google Scholar]
- Carvalho BS, Irizarry RA (2010) A framework for oligonucleotide microarray preprocessing. Bioinformatics 26: 2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016) Learning the languages of the chloroplast: retrograde signaling and beyond. Annu Rev Plant Biol 67: 25–53 [DOI] [PubMed] [Google Scholar]
- Chi W, Sun X, Zhang L (2013) Intracellular signaling from plastid to nucleus. Annu Rev Plant Biol 64: 559–582 [DOI] [PubMed] [Google Scholar]
- De Santis-MacIossek G, Kofer W, Bock A, Schoch S, Maier RM, Wanner G, Rüdiger W, Koop HU, Herrmann RG (1999) Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: molecular biology, biochemistry and ultrastructure. Plant J 18: 477–489 [DOI] [PubMed] [Google Scholar]
- de Souza A, Wang JZ, Dehesh K (2016) Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu Rev Plant Biol 68: 85–108 [DOI] [PubMed] [Google Scholar]
- Demarsy E, Buhr F, Lambert E, Lerbs-Mache S (2012) Characterization of the plastid-specific germination and seedling establishment transcriptional programme. J Exp Bot 63: 925–939 [DOI] [PubMed] [Google Scholar]
- Demarsy E, Courtois F, Azevedo J, Buhot L, Lerbs-Mache S (2006) Building up of the plastid transcriptional machinery during germination and early plant development. Plant Physiol 142: 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisermann A, Tiller K, Link G (1990) In vitro transcription and DNA binding characteristics of chloroplast and etioplast extracts from mustard (Sinapis alba) indicate differential usage of the psbA promoter. EMBO J 9: 3981–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, Brearley C, Hell R, et al. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez-Valdivieso G, Fryer MJ, Lawson T, Slattery K, Truman W, Smirnoff N, Asami T, Davies WJ, Jones AM, Baker NR, Mullineaux PM (2009) The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 21: 2143–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao ZP, Chen GX, Yang ZN (2012) Regulatory role of Arabidopsis pTAC14 in chloroplast development and plastid gene expression. Plant Signal Behav 7: 1354–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao ZP, Yu QB, Zhao TT, Ma Q, Chen GX, Yang ZN (2011) A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol 157: 1733–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson J, Perez-Ruiz JM, Chory J, Callis J (2012) The plastid-localized pfkB-type carbohydrate kinases FRUCTOKINASE-LIKE 1 and 2 are essential for growth and development of Arabidopsis thaliana. BMC Plant Biol 12: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz PT, Allison LA, Maliga P (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16: 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills AC, Khan S, López-Juez E (2015) Chloroplast biogenesis-associated nuclear genes: control by plastid signals evolved prior to their regulation as part of photomorphogenesis. Front Plant Sci 6: 1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isemer R, Mulisch M, Schäfer A, Kirchner S, Koop HU, Krupinska K (2012) Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Lett 586: 85–88 [DOI] [PubMed] [Google Scholar]
- Kleine T, Leister D (2016) Retrograde signaling: organelles go networking. Biochim Biophys Acta 1857: 1313–1325 [DOI] [PubMed] [Google Scholar]
- Klie S, Nikoloski Z (2012) The choice between MapMan and gene ontology for automated gene function prediction in plant science. Front Genet 3: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnetsov V, Bolle C, Lübberstedt T, Sopory S, Herrmann RG, Oelmüller R (1996) Evidence that the plastid signal and light operate via the same cis-acting elements in the promoters of nuclear genes for plastid proteins. Mol Gen Genet 252: 631–639 [DOI] [PubMed] [Google Scholar]
- Lagrange T, Hakimi MA, Pontier D, Courtois F, Alcaraz JP, Grunwald D, Lam E, Lerbs-Mache S (2003) Transcription factor IIB (TFIIB)-related protein (pBrp), a plant-specific member of the TFIIB-related protein family. Mol Cell Biol 23: 3274–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin RM. (2014) Influence of plastids on light signalling and development. Philos Trans R Soc Lond B Biol Sci 369: 20130232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Kim C, Landgraf F, Apel K (2007) EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 10270–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM (2002) Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J 31: 171–188 [DOI] [PubMed] [Google Scholar]
- Lerbs-Mache S. (2011) Function of plastid sigma factors in higher plants: regulation of gene expression or just preservation of constitutive transcription? Plant Mol Biol 76: 235–249 [DOI] [PubMed] [Google Scholar]
- Liebers M, Grübler B, Chevalier F, Lerbs-Mache S, Merendino L, Blanvillain R, Pfannschmidt T (2017) Regulatory shifts in plastid transcription play a key role in morphological conversions of plastids during plant development. Front Plant Sci 8: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere K, Weihe A, Börner T (2011) The transcription machineries of plant mitochondria and chloroplasts: composition, function, and regulation. J Plant Physiol 168: 1345–1360 [DOI] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L (1987) Improved method for the isolation of RNA from plant tissues. Anal Biochem 163: 16–20 [DOI] [PubMed] [Google Scholar]
- López-Juez E. (2007) Plastid biogenesis, between light and shadows. J Exp Bot 58: 11–26 [DOI] [PubMed] [Google Scholar]
- López-Juez E, Jarvis RP, Takeuchi A, Page AM, Chory J (1998) New Arabidopsis cue mutants suggest a close connection between plastid- and phytochrome regulation of nuclear gene expression. Plant Physiol 118: 803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E (2016) Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun 7: 11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordally ZB, Ishii K, Atkins KA, Wetherill SJ, Kusakina J, Walton EJ, Kato M, Azuma M, Tanaka K, Hanaoka M, Dodd AN (2013) Circadian control of chloroplast transcription by a nuclear-encoded timing signal. Science 339: 1316–1319 [DOI] [PubMed] [Google Scholar]
- Page MT, Kacprzak SM, Mochizuki N, Okamoto H, Smith AG, Terry MJ (2017a) Seedlings lacking the PTM protein do not show a genomes uncoupled (gun) mutant phenotype. Plant Physiol 174: 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page MT, McCormac AC, Smith AG, Terry MJ (2017b) Singlet oxygen initiates a plastid signal controlling photosynthetic gene expression. New Phytol 213: 1168–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Li W, Viehhauser A, He B, Kim S, Nilsson AK, Andersson MX, Kittle JD, Ambavaram MM, Luan S, Esker AR, Tholl D, et al. (2013) Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc Natl Acad Sci USA 110: 9559–9564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R (2006) pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Pfannschmidt T (2013) Essential nucleoid proteins in early chloroplast development. Trends Plant Sci 18: 186–194 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T. (2010) Plastidial retrograde signalling—a true “plastid factor” or just metabolite signatures? Trends Plant Sci 15: 427–435 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Blanvillain R, Merendino L, Courtois F, Chevalier F, Liebers M, Grübler B, Hommel E, Lerbs-Mache S (2015) Plastid RNA polymerases: orchestration of enzymes with different evolutionary origins controls chloroplast biogenesis during the plant life cycle. J Exp Bot 66: 6957–6973 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Link G (1994) Separation of two classes of plastid DNA-dependent RNA polymerases that are differentially expressed in mustard (Sinapis alba L.) seedlings. Plant Mol Biol 25: 69–81 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Munne-Bosch S (2013) Plastidial signaling during the plant life cycle. In Biswal B, Krupinska K, eds, Plastid Development in Leaves during Growth and Senescence. Springer, Dordrecht, the Netherlands [Google Scholar]
- Pogson BJ, Ganguly D, Albrecht-Borth V (2015) Insights into chloroplast biogenesis and development. Biochim Biophys Acta 1847: 1017–1024 [DOI] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Förster B, Small ID (2008) Plastid signalling to the nucleus and beyond. Trends Plant Sci 13: 602–609 [DOI] [PubMed] [Google Scholar]
- Pulido P, Perello C, Rodriguez-Concepcion M (2012) New insights into plant isoprenoid metabolism. Mol Plant 5: 964–967 [DOI] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylidès C, Havaux M (2012) Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA 109: 5535–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle ME, Burgoon LD, Lawrence LA, Sinkler CA, Larkin RM (2012) Plastids are major regulators of light signaling in Arabidopsis. Plant Physiol 159: 366–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM (2007) Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, Oliva M, Weigel D, Krämer U (2013) Circadian clock adjustment to plant iron status depends on chloroplast and phytochrome function. EMBO J 32: 511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner S, Schröter Y, Pfalz J, Pfannschmidt T (2011) Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol 157: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YS, Lagarias JC (2007) Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell 19: 2124–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Gray JC (2002) Multiple plastid signals regulate the expression of the pea plastocyanin gene in pea and transgenic tobacco plants. Plant J 32: 763–774 [DOI] [PubMed] [Google Scholar]
- Sun X, Feng P, Xu X, Guo H, Ma J, Chi W, Lin R, Lu C, Zhang L (2011) A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat Commun 2: 477. [DOI] [PubMed] [Google Scholar]
- Terry MJ, Smith AG (2013) A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front Plant Sci 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller K, Link G (1993) Sigma-like transcription factors from mustard (Sinapis alba L.) etioplast are similar in size to, but functionally distinct from, their chloroplast counterparts. Plant Mol Biol 21: 503–513 [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, Scheible WR, Gibon Y, et al. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M (2009) A guide to using MapMan to visualize and compare omics data in plants: a case study in the crop species, maize. Plant Cell Environ 32: 1211–1229 [DOI] [PubMed] [Google Scholar]
- Waters MT, Langdale JA (2009) The making of a chloroplast. EMBO J 28: 2861–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Carrier R, Zoschke R, Belcher S, Pfalz J, Barkan A (2014) A major role for the plastid-encoded RNA polymerase complex in the expression of plastid transfer RNAs. Plant Physiol 164: 239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Chory J (2011) Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol 21: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Savchenko T, Baidoo EE, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K (2012) Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149: 1525–1535 [DOI] [PubMed] [Google Scholar]
- Zghidi-Abouzid O, Merendino L, Buhr F, Malik Ghulam M, Lerbs-Mache S (2011) Characterization of plastid PsbT sense and antisense RNAs. Nucleic Acids Res 39: 5379–5387 [DOI] [PMC free article] [PubMed] [Google Scholar]