Wounding triggers callus formation in Arabidopsis through the activation of cytokinin biosynthesis and AP2/ERF-mediated developmental pathway.
Abstract
Wounding is a primary trigger of organ regeneration, but how wound stress reactivates cell proliferation and promotes cellular reprogramming remains elusive. In this study, we combined transcriptome analysis with quantitative hormonal analysis to investigate how wounding induces callus formation in Arabidopsis (Arabidopsis thaliana). Our time course RNA-seq analysis revealed that wounding induces dynamic transcriptional changes, starting from rapid stress responses followed by the activation of metabolic processes and protein synthesis and subsequent activation of cell cycle regulators. Gene ontology analyses further uncovered that wounding modifies the expression of hormone biosynthesis and response genes, and quantitative analysis of endogenous plant hormones revealed accumulation of cytokinin prior to callus formation. Mutants defective in cytokinin synthesis and signaling display reduced efficiency in callus formation, indicating that de novo synthesis of cytokinin is critical for wound-induced callus formation. We further demonstrate that type-B ARABIDOPSIS RESPONSE REGULATOR-mediated cytokinin signaling regulates the expression of CYCLIN D3;1 (CYCD3;1) and that mutations in CYCD3;1 and its homologs CYCD3;2 and 3 cause defects in callus formation. In addition to these hormone-mediated changes, our transcriptome data uncovered that wounding activates multiple developmental regulators, and we found novel roles of ETHYLENE RESPONSE FACTOR 115 and PLETHORA3 (PLT3), PLT5, and PLT7 in callus generation. All together, these results provide novel mechanistic insights into how wounding reactivates cell proliferation during callus formation.
Plant life is full of damaging stresses, and plants rapidly activate defense responses upon wounding to protect themselves from pathogenic infections. Plants also repair wound sites through the formation of unorganized cell mass, called callus, and regenerate new organs. Wound-induced calli are thought to seal wound sites, prevent water loss, and serve as cellular sources for vasculature differentiation and/or de novo organogenesis (Ikeuchi et al., 2013; Ikeuchi et al., 2016). A series of previous studies have illustrated how plants sense wound signals and activate downstream stress responses. Wound stress that causes a defense response is perceived through so-called damage-associated molecular patterns. Damage-associated molecular patterns include the cell wall derivative oligogalacturonic acid (Bishop et al., 1981) and extracellular ATP (Choi et al., 2014; Tanaka et al., 2014), the latter of which, upon perception, triggers cytoplasmic calcium signaling and a burst of reactive oxygen species. These local wound signals are then translated into electrical and chemical signals and often transmitted to other parts of plants. Wound response invokes depolarization of plasma membrane through the activity of cation channel GLU RECEPTOR-LIKEs, and this electric signal travels over a long distance (Mousavi et al., 2013). Subsequently, jasmonate (JA) synthesis is up-regulated and a JA-induced pathway mediates acquisition of herbivores resistance (Koo et al., 2009). While JA mediates substantial portions of the wound response, other aspects are independent of JA (Titarenko et al., 1997), suggesting that wounding also activates other signaling pathways in parallel.
Callus formation and organ regeneration often entail cell cycle reentry of quiescent cells, which is achieved through the reactivation of core cell cycle regulators CYCLIN (CYC) and CYCLIN-DEPENDENT KINASES (CDK; Inzé and De Veylder, 2006). Recent studies have started to reveal how cells transduce wound signals to activate cell proliferation and callus formation. A set of APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factors called WOUND-INDUCED DEDIFFERENTIATION1 (WIND1), WIND2, WIND3, and WIND4 plays key roles in wound-induced callus formation (Iwase et al., 2011). Expression of WIND genes is rapidly induced by wounding, and their expression remains high in calli formed at wound sites. Ectopic overexpression of WIND1, WIND2, WIND3, or WIND4 substitutes wounding to induce callus formation, and conversely chimeric repression of WIND in WIND1-SRDX plants results in reduced efficiency in wound-induced callus formation. WIND1 likely mediates wound-induced up-regulation of cytokinin signaling, because wound-induced cytokinin signaling is suppressed in WIND1-SRDX plants. A recent study showed that WIND1 directly up-regulates the expression of ENHANCER OF SHOOT REGENERATION1 (ESR1), encoding another AP2/ERF transcription factor in Arabidopsis, to promote callus formation and shoot regeneration (Iwase et al., 2017). The loss-of-function mutants of WINDs or ESR1 do not completely lack the ability to form calli, suggesting that there are other parallel regulatory pathways of callus formation.
The plant hormones auxin and cytokinin are well established as efficient inducers of callus in tissue culture. Recent studies have revealed that callus formation on auxin-rich callus-inducing media (CIM) follows a developmental program of root formation. Indeed, histology and gene expression profiles of CIM-induced calli resemble those of root primordia with organized expression of root meristem regulators such as WUSCHEL-RELATED HOMEOBOX5 (WOX5; Sugimoto et al., 2010; Liu et al., 2014). Accordingly, key regulators of root formation, such as LATERAL ORGAN BOUNDARY DOMAIN16 (LBD16), LBD17, LBD18, and LBD29 are required for callus formation on CIM (Fan et al., 2012). Recent studies have shown that several AP2/ERF transcription factors also play pivotal roles in organ regeneration. For example, PLETHORA3 (PLT3), PLT5, and PLT7, key regulators of lateral root development (Hofhuis et al., 2013), confer calli with pluripotency in tissue culture (Kareem et al., 2015). Another AP2/ERF transcription factor, ERF115, has been shown to regulate replacement of root stem cells after injury (Heyman et al., 2013, 2016) while its close homolog ERF113/RAP2.6L promotes shoot regeneration from calli (Che et al., 2006) as well as tissue repair in floral stems (Asahina et al., 2011). Accumulating evidence suggests that calli formed at wound sites are distinct from those formed on CIM (Iwase et al., 2011; Ikeuchi et al., 2013), but the commonalities and differences of their regulatory mechanisms still remain obscure.
Genes that encode transcription factors with important developmental roles are often under epigenetic repression, such as histone modification and DNA methylation, to prevent their ectopic expression (Ikeuchi et al., 2015b). In the absence of POLYCOMB REPRESSIVE COMPLEX2 (PRC2)-mediated repression, for example, WIND1–3, WOX5, and several other regulators of cellular reprogramming are ectopically expressed, and fully differentiated cells undergo spontaneous callus formation (Ikeuchi et al., 2015a). Mutants defective in other epigenetic regulation have higher or lower efficiency of callus formation and/or shoot regeneration in tissue culture, and these phenotypes are associated with differential expression of reprogramming regulators (Li et al., 2011; He et al., 2012; Shemer et al., 2015; Lee et al., 2016). When plants sense wound stress and generate calli at wound sites, the chromatin status of genes encoding cell fate regulators likely changes to allow their transcription. Molecular mechanisms underlying this control, however, remain largely unknown.
Time-course transcriptome analysis is a powerful approach to infer global physiological responses and identify novel regulators of these responses. Several previous studies have reported large-scale transcriptional changes during wound-induced defense response (Reymond et al., 2000; Devoto et al., 2005; Kilian et al., 2007) and regeneration (Chen et al., 2016a; Efroni et al., 2016). Lack of transcriptome analyses with sufficient time resolution, however, hinders us from understanding how wounding stimuli induce regenerative responses. In this study, we performed time course RNA-seq analyses within 24 h after wounding and investigated how plants respond to wound stress and change their transcriptional program to undergo callus formation. By combining our transcriptome datasets with hormonal measurements and mutant analyses, we identified several positive and negative pathways that regulate callus formation. Our results thus provide an important molecular framework of how plants sense wound stress and reactivate cell proliferation at wound sites.
RESULTS
Expression of Cell Cycle Genes Is Activated within 24 h after Wounding
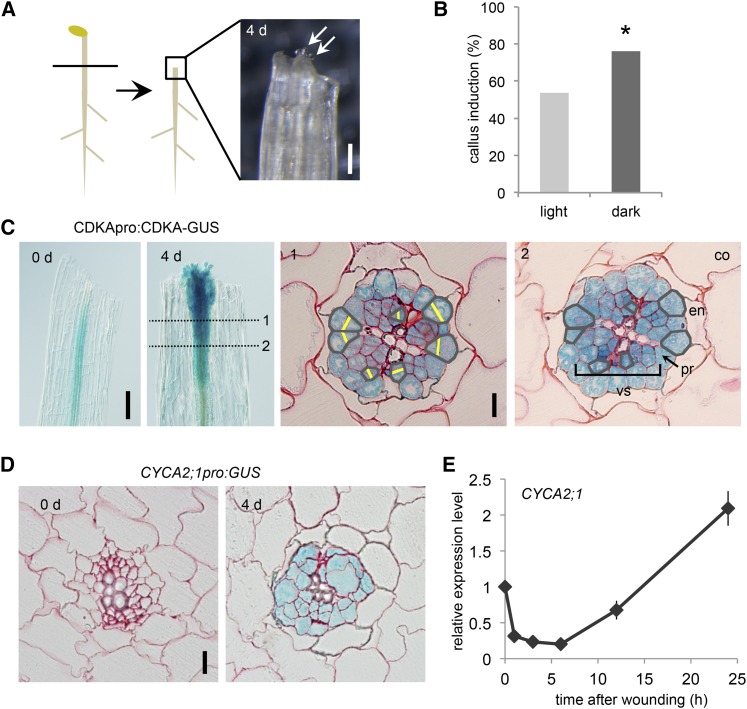
We had previously established an experimental system where we can induce callus formation in wounded hypocotyls without exogenous application of plant hormones (Fig. 1A; Iwase et al., 2011). In this study, we improved the efficiency of callus induction from ∼50% to ∼70% by incubating hypocotyl explants in the dark after wounding (Fig. 1B). To explore the cellular basis of callus formation, we induced calli in CDKApro:CDKA-GUS plants (Adachi et al., 2009) and examined the site of cell cycle activation during callus formation. As shown in Figure 1C, we detected strong activation of CDKA-GUS expression near wound sites at 4 d after cutting. Histological analysis of developing calli revealed that CDKA promoter activity is present in the vasculature and pericycle cells but is not detectable in surrounding cells, such as endodermis, cortex, and epidermis (Fig. 1C). By comparing serial cross sections, we confirmed that new cell plates are formed only in the vasculature and pericycle cells. Histological analysis of CYCA2;1pro:GUS plants, another reporter line for cell proliferation (Burssens et al., 2000), also showed that the CYCA2;1 promoter activity is restricted to the vasculature and pericycle cells (Fig. 1D), indicating that reactivation of cell cycle in these cell types gives rise to calli in wounded hypocotyls. To examine when CYCA2;1 is transcriptionally induced, we isolated RNA at 0, 1, 3, 6, 12, and 24 h after wounding and performed quantitative reverse transcription coupled with PCR (qRT-PCR) analysis. We found that the expression level of CYCA2;1 initially declines and subsequently increases within 24 h after wounding (Fig. 1E). These results thus establish that initial transcriptional modification leading to cell cycle activation occurs within 24 h after wounding.
Figure 1.
Cell cycle activation during callus formation at wound sites. A, A schematic diagram describing how callus is induced at wound sites. Hypocotyls of 7-d-old etiolated seedlings are cut and incubated on hormone-free Murashige and Skoog medium. White arrows mark callus cells at 4 d after wounding. Scale bar = 0.1 mm. B, Incubation of hypocotyl explants in the dark promotes callus induction efficiency. An asterisk indicates statistical significance determined by proptest (n > 600; P < 2.2E-16). C, CDKApro:CDKA-GUS plants show stronger CDKA expression near wound sites at 4 d after wounding (left). Serial transverse sections of CDKApro:CDKA-GUS plants show formation of new cell plates during callus formation (right). Numbers (1, 2) indicate the position of transverse sections. Gray lines mark cells in same cell files, and yellow lines highlight newly formed cell plates. vs, Vasculature; pr, pericycle; en, endodermis; co, cortex. Scale bar = 0.1 mm (left), 10 μm (right). D, Transverse sections of CYCA2;1pro:GUS plants reveal stronger CYCA2;1 expression in the vasculature and pericycle at 4 d after wounding. Scale bar = 10 μm. Transverse sections in C and D are counterstained by safranin-O. E, Real-time qRT-PCR analysis showing relative CYCA2;1 expression level after wounding. Data are mean ± se (n = 3, biological replicates).
Wounding Induces Multiple Cellular Events with Specific Temporal Patterns
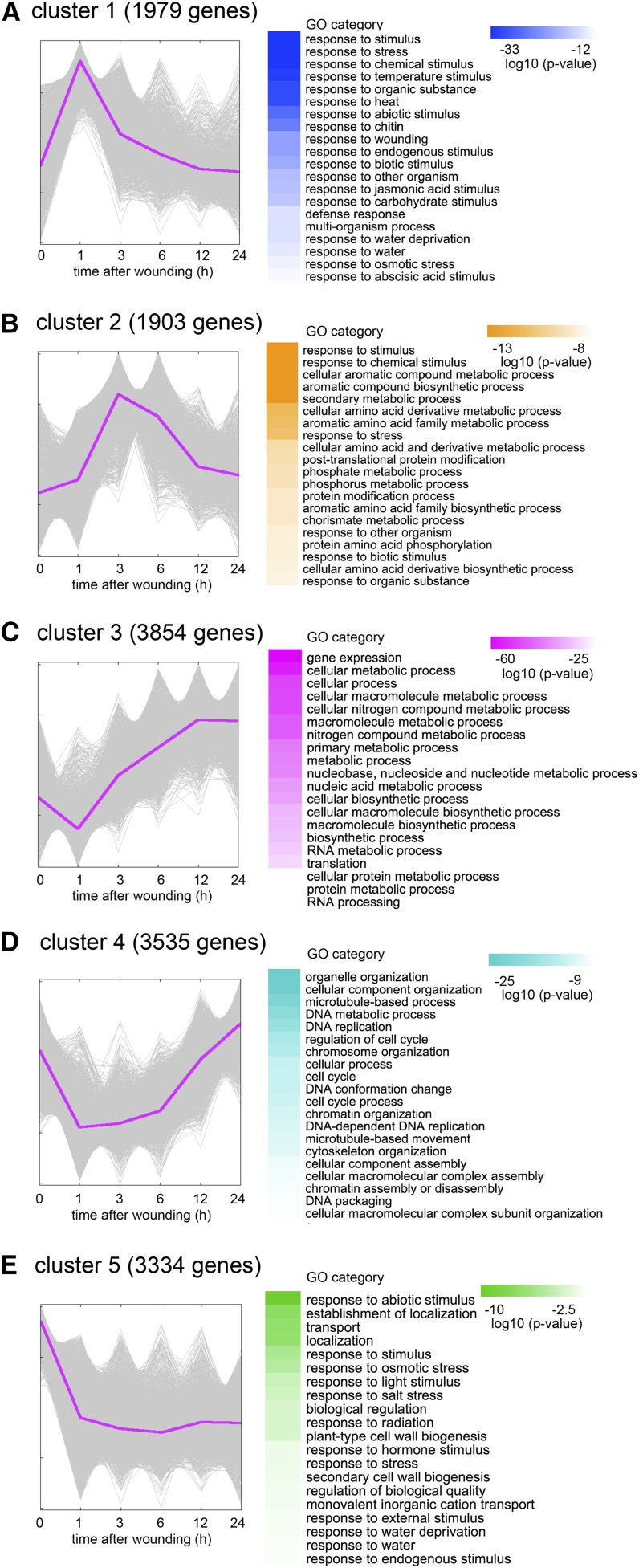
In order to unravel global transcriptional dynamics leading to cell cycle activation and callus initiation, we performed time course RNA-seq analysis for hypocotyl explants harvested at 0, 1, 3, 6, 12, and 24 h after wounding. Taking genes with reads per kilobase million mapped reads (RPKM) higher than 0.1, we identified 18,332 genes that are expressed at least at one time point within 24 h after wounding, and among them 14,605 genes display significant transcriptional changes based on Bayesian estimation of temporal regulation (false discovery rate [FDR] = 0.05; Aryee et al., 2009). We verified the expression profile of two selected genes by qRT-PCR (Supplemental Fig. S1A), supporting the reliability of our RNA-seq data. To decipher the general trend of gene expression profiles, we subjected the 14,605 differentially expressed genes to k-means clustering analysis. We first grouped these genes into different numbers of clusters and finally found that grouping them into five clusters conveys the major characteristic temporal expression patterns induced by wounding (Fig. 2; Supplemental Fig. S1B).
Figure 2.
Clustering analysis of time-course RNA-seq revealed five characteristic transcriptional changes induced by wounding. A, The 1,979 genes in cluster 1 are transiently induced within 1 h after wounding. GO categories such as response to stimuli and response to wounding are overrepresented. B, Cluster 2 contains 1,903 genes induced between 3 and 6 h after wounding. Genes involved in various metabolic processes are enriched in this cluster. C, Cluster 3 includes 3,854 genes induced after 3 h. GO categories such as gene expression, translation, and cellular biosynthetic processes are overrepresented. D, The 3,535 genes in cluster 4 are initially down-regulated but subsequently up-regulated after 12 h. Genes involved in the regulation of cell cycle are enriched in this cluster. E, Cluster 5 contains 3,334 genes that are down-regulated after wounding. Genes involved in stress response and cell wall biogenesis are overrepresented in this cluster. Magenta lines highlight the centroid of each cluster. Top 20 GO categories are shown with color codes representing the log10 (P value).
This clustering analysis revealed that key transcriptional activation or repression occurs in several transient waves after wounding. The first set of 1,979 genes in cluster 1, for instance, show rapid induction after wounding to reach maximum expression levels within 1 h (Fig. 2A). Gene ontology (GO) analysis using BiNGO revealed that GO categories such as response to stimuli (FDR corrected P value = 3.02E-53) and response to wounding (3.91E-22) are highly represented in this cluster (Fig. 2A). As previously shown by Reymond et al. (2000), response to biotic stresses (3.92E-21), defense responses (9.03E-16), and response to water deprivation (1.07E-15) are also overrepresented (Fig. 2A). Interestingly, we found that response to other abiotic stimuli such as heat (3.14E-31) is also highly enriched (Fig. 2A), implying that part of wound-induced transcriptomes might overlap with other stress-induced transcriptomes. As expected, genes involved in JA response (5.37E-19) and another stress-associated plant hormone abscisic acid (ABA; 2.15E-13) are also overrepresented in this cluster (Fig. 2A).
In cluster 2, where transcript levels of 1,903 genes are highest between 3 and 6 h after wounding, various metabolic categories are overrepresented (Fig. 2B). Genes that encode enzymes for aromatic compound synthesis (1.00E-15) and amino acid derivative metabolic processes (3.63E-12) are highly represented, and in particular those in the chorismate pathway (8.87E-10) are enriched. Chorismate is a precursor of Trp, which can then be turned into defensive substances such as camalexin (Ren et al., 2008). Genes encoding enzymes that convert Trp into camalexin, such as CYP79B2 and PAD3, are also included in cluster 2, suggesting that camalexin synthesis pathway is activated by wounding. The GO categories such as posttranslational protein modification (2.61E-10) and phosphate metabolic process (3.93E-10) are represented in cluster 2 by genes encoding calcium-dependent kinases, calmodulin-binding protein kinases, and other mediators of calcium signaling (Supplemental Table S1). Moreover, other kinases such as those involved in defense response are also included in cluster 2 (Supplemental Table S1), implying that wounding activates calcium-mediated phosphorylation signaling cascades as reported previously (Hettenhausen et al., 2016).
In cluster 3, where transcript abundance of 3,854 genes starts to increase at 3 h after wounding, GO categories such as gene expression (1.00E-62), translation (3.24E-31), and RNA processing (8.41E-22) are overrepresented (Fig. 2C). This cluster also contains genes involved in cellular macromolecule metabolic processes (2.08E-48) and cellular biosynthetic processes (1.43E-38), suggesting that cells start to generate a new proteome.
Transcript abundance of 3,535 genes in cluster 4 is reduced within 1 h after wounding, but later increased at around 12 h. GO categories represented in this cluster include microtubule-based process (1.74E-22), organelle organization (8.72E-31), and regulation of cell cycle (1.08E-16; Fig. 2D). GO categories such as chromatin assembly (2.13E-10) and DNA replication (8.08E-13) are also enriched (Fig. 2D), suggesting that basic machineries needed for cell division are down-regulated in the initial stress response but are reactivated within 24 h to prepare for cell cycle reactivation.
In cluster 5, where transcript levels of 3,334 genes rapidly decline within 1 h after wounding and remain down-regulated up to 24 h, response to abiotic stimulus (5.80E-11) is most highly enriched (Fig. 2E). Cell wall biogenesis (1.08E-04) is also highly represented, and 8 out of 10 CELLULOSE SYNTHASE genes are included in this cluster (Supplemental Table S1). Transcriptional down-regulation of these genes might reflect the physiological behavior of wounded explants to shut down unnecessary cellular events and save metabolic energy.
These data suggest that wounding triggers a series of cellular processes, ranging from early stress response and production of defense molecules to later reactivation of protein synthesis and cell proliferation. To examine the generality of these observations, we compared our dataset with another wound-induced transcriptome dataset reported by Kilian et al. (2007). Interestingly, genes responding rapidly to wound stress, i.e. those in clusters 1, 2, and 5, overlap significantly with genes induced at early time points in Kilian et al. (2007) (Supplemental Fig. S2), suggesting that they are involved in general wound response. In contrast, genes that are induced later, i.e. those in clusters 3 and 4, do not have significant overlaps with the dataset in Kilian et al. (2007) (Supplemental Fig. S2), implying that they are associated with callus formation occurring only in our experimental condition.
JA and ABA Are Not Required for Callus Formation at Wound Sites
Since many plant hormone-associated genes are rapidly induced in our RNA-seq datasets, we sought to investigate functional roles of plant hormones in wound-induced callus formation. Among well-described plant hormones, JA is best characterized as a wound-inducible hormone (Creelman et al., 1992). Accordingly, we found that among 51 JA biosynthesis and response genes we tested (Nemhauser et al., 2006; Supplemental Table S3), the transcript level of 19 JA biosynthesis genes, such as ALLENE OXIDE SYNTHASE (AOS) and LIPOXYGENASE3, 4, 6, and 20 JA-responsive genes, such as TYR AMINOTRANSFERASE3, are strongly up-regulated within 1 h after wounding (Fig. 3A). Consistently, our quantitative hormone measurements revealed that the endogenous level of JA starts to increase within 1 h and peaks at 6 h after wounding before it declines back to its initial level (Fig. 3B). To test whether this increase in JA level is required for callus formation, we performed a wound-induced callus-induction assay using aos (Park et al., 2002) and coronatine insensitive1-16B (coi1-16B: Noir et al., 2013), mutants defective in JA biosynthesis or signaling, respectively. As shown in Figure 3C, both mutants show slightly higher efficiency of callus formation compared to wild type, suggesting that JA synthesis and signaling are slightly inhibitory to callus formation at wound sites.
Figure 3.
Wound-induced JA synthesis and response are inhibitory for callus formation. A, Expression profiles retrieved from RNA-seq show that many genes involved in JA biosynthesis and response are induced within 1 h after wounding. Note that only genes with RPKM > 1 that change significantly along the time course are shown (20/27 genes for JA synthesis; 22/24 genes for JA responses). The color code indicates the log2 value of relative fold change compared to 0 h. B, Overall levels of endogenous JA are increased transiently after wounding. Alphabetical letters indicate statistical significance determined by ANOVA and Tukey’s multicomparison test (n = 3; P < 0.01). C, Callus induction is enhanced in aos and coi1-16B compared to wild type (Col-gl). Asterisks indicate statistical significance determined by proptest (n > 200 for each genotype; P < 0.05).
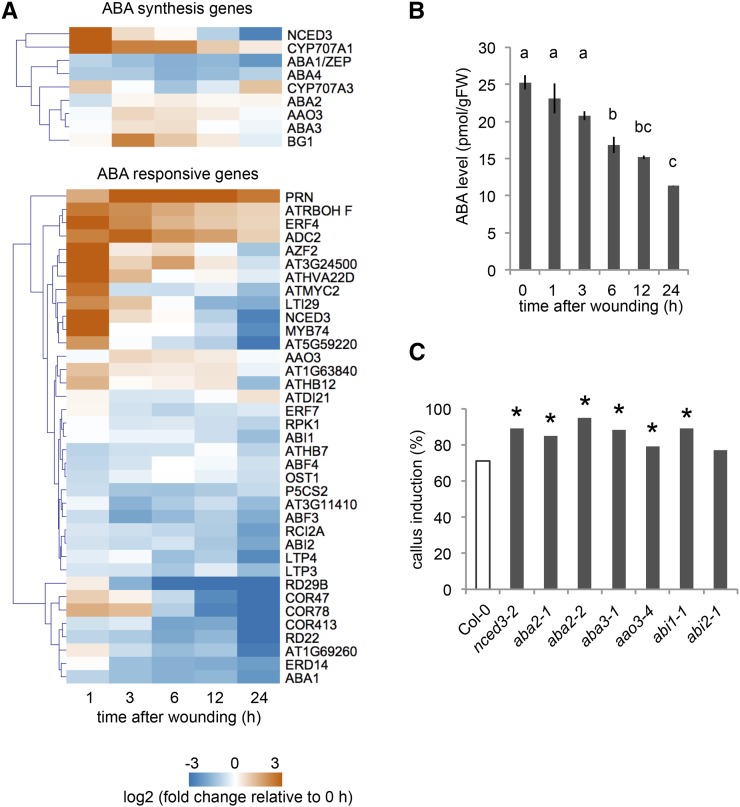
ABA transduces multiple stress responses, such as drought and heat stress, but only a few studies have reported its involvement in wound-induced signaling (Pēna-Cortés et al., 1989; Dammann et al., 1997). Our transcriptome data show that NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3 (NCED3), encoding a key enzyme for ABA synthesis, is strongly up-regulated by wounding, while ABA-catabolizing enzyme genes CYP707A1 and CYP707A3 are also induced (Fig. 4A). In addition, we found that among 41 ABA response genes we tested (Supplemental Table S3), 21 genes, such as ZINC-FINGER PROTEIN2, are induced at least at one time point within 24 h after wounding, but others are downregulated within the same time frame (Fig. 4A). Our hormonal analysis did not detect an increase in endogenous ABA levels, and instead, overall ABA levels appear to decrease within 24 h after wounding (Fig. 4B). Interestingly, mutants with reduced ABA synthesis or signaling are consistently more efficient in callus formation than wild type (Fig. 4C). These results, therefore, suggest that ABA is not required but rather slightly inhibitory for callus formation after wounding.
Figure 4.
ABA-mediated signaling is inhibitory for callus formation at wound sites. A, Expression profiles retrieved from RNA-seq datasets show that a subset of ABA biosynthesis and signaling genes are induced within 24 h after wounding. Note that only genes with RPKM > 1 that change significantly along the time course are shown (9/15 genes for ABA synthesis; 37/41 genes for ABA responses). B, Overall endogenous ABA content gradually decreases after wounding. Alphabetical letters indicate statistical significance determined by ANOVA and Tukey’s multicomparison test (n = 3; P < 0.01). C, Callus formation tends to be enhanced in both ABA synthesis mutants, i.e. nced3-2, aba2-1, aba2-2, aba3-1, and aao3-4, and signaling mutants, i.e. abi1-1. Asterisks indicate statistical significance determined by proptest (n > 100 for each genotype; P < 0.05).
Wound-Induced Cytokinin Synthesis Is Required for Callus Formation
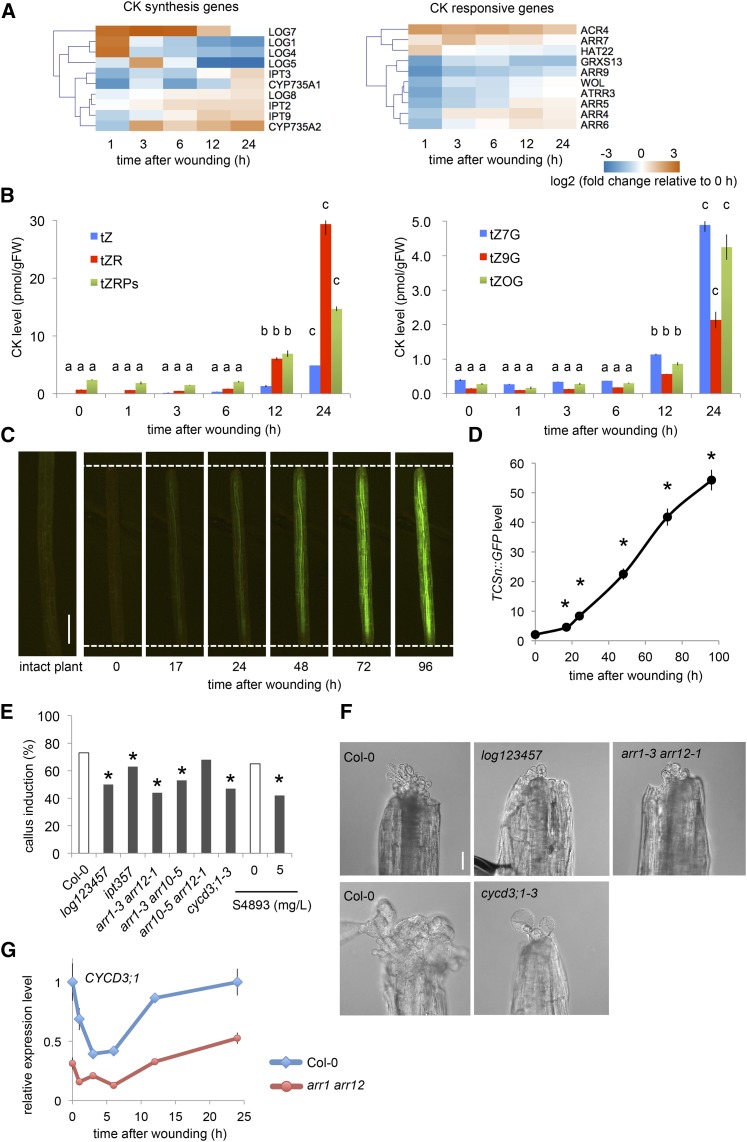
We have previously reported that cytokinin response is elevated in wounded explants (Iwase et al., 2011), but how wounding up-regulates cytokinin response remained unclear. Our RNA-seq analysis revealed that among 19 cytokinin biosynthesis genes we examined (Supplemental Table S3), 10 genes, including ISOPENTENYL TRANSFERASEs (IPTs; Kakimoto, 2001; Takei et al., 2001), CYP735As (Kiba et al., 2013), and LONELY GUYs (LOGs; Tokunaga et al., 2012), are up-regulated within 24 h after wounding (Fig. 5A). Quantitative measurement indeed revealed that the endogenous levels of an active cytokinin called trans-Zeatin (tZ) and its precursor tZR start to accumulate within 12 h after wounding (Fig. 5B). We also found that their glucosidated derivatives, tZ7G, tZ9G, and tZOG, accumulate similarly after 12 h (Fig. 5B), further corroborating the activation of tZ synthesis pathway after wounding. In contrast, another active form of cytokinin cis-Zeatin (cZ) and its precursor cZR appear to accumulate earlier after wounding, but their levels decline to the basal level by 24 h (Supplemental Fig. S3). We did not detect significant increase in the level of isopentenyladenine (iP) and its precursor iPR after wounding, although some of their derivatives, such as iPR and iP7G, are mildly increased (Supplemental Fig. S3A). To test whether wound-induced accumulation of cytokinin is mediated by WIND1, we next quantified tZ and tZR levels in WIND1-SRDX plants (Iwase et al., 2011). As shown in Supplemental Figure S3B, tZ and tZR levels tend to be higher in WIND1-SRDX, implying that WIND1 is not required for the accumulation of cytokinin. Our transcriptome analysis further showed that some of cytokinin-responsive genes, such as type-A ARABIDOPSIS RESPONSE REGULATORs (ARRs; D’Agostino et al., 2000), are upregulated by wounding (Fig. 5A). We also visualized the cytokinin response using the Two Component signaling Sensor new (TCSn)::GFP reporter line (Zürcher et al., 2013) and detected a clear activation of GFP expression from 17 h onward after wounding, indicative of activated cytokinin signaling (Fig. 5, C and D). Some species of cytokinin, like tZ, are normally produced in roots and transported to shoots through the vasculature (Kiba et al., 2013). Importantly, TCSn::GFP expression is activated in hypocotyl explants where roots are cut off (Fig. 5, C and D), supporting that de novo biosynthesis within explants, rather than reallocation of cytokinin from roots, is the primary cause of increased cytokinin response.
Figure 5.
Wound-induced cytokinin synthesis and signaling are required for callus formation. A, Expression profiles retrieved from RNA-seq datasets show that a subset of cytokinin biosynthesis and signaling genes are induced within 24 h after wounding. Note that only genes with RPKM > 1 that change significantly along the time course are shown (10/19 genes for cytokinin synthesis; 10/17 genes for cytokinin responses). B, Endogenous content of several tZ-type cytokinins starts to increase at 12 h and reaches more than 10-fold at 24 h compared to intact plants. Alphabetical letters indicate statistical significance determined by ANOVA and Tukey’s multicomparison test (n = 3; P < 0.01). C, Time-course imaging of TCSn::GFP plants reveals up-regulation of cytokinin signaling in wounded hypocotyls. Dashed lines indicate wound sites. Scale bar = 0.5 mm. D, Quantitative analysis of TCSn::GFP signals in wounded hypocotyls. Data are mean ± se (n = 11), and asterisks indicate statistical significance compared to 0 h determined by t test (P < 0.05). E, Mutants defective in cytokinin biosynthesis, i.e. log123457 and ipt357, and signaling, i.e. arr1-3 arr12-1, arr1-3 arr10-5, display reduced callus initiation phenotypes at 4 d after wounding. The cycd3;1-3 triple mutants have similar reduction in callus initiation. S-4893 is a cytokinin signaling inhibitor which blocks callus formation in wounded hypocotyls. Asterisks indicate statistical significance determined by proptest (n > 180 for each genotype, P < 0.05). F, Representative images of calli generated 14 d after wounding in wild type (Col-0), log123457, arr1-3 arr12-1, and cycd3;1-3 hypocotyls. Scale bar = 0.1 mm. G, The expression of CYCD3;1 is reduced in arr1-3 arr12-1. Data are mean ± se (n = 3, biological replicates).
To test whether cytokinin plays a role in callus induction at wound sites, we examined callus formation efficiency in cytokinin biosynthesis mutants log123457 and ipt357 (Tokunaga et al., 2012; Miyawaki et al., 2006) and found that they are less efficient in callus formation (Fig. 5, E and F). We further corroborated the importance of cytokinin by showing that double mutants of type B ARRs, namely arr1-3 arr12-1 and arr1-3 arr10-5 (Argyros et al., 2008), are defective in callus formation. We also found that arr10-5 arr12-1 do not exhibit reduced callus formation phenotypes (Fig. 5E), suggesting that ARR1 likely plays more profound roles than ARR10 and ARR12 in wound-induced callus formation. Moreover, application of anticytokinin S4893 (Arata et al., 2010) at the time of wounding hampers callus formation (Fig. 5E), further supporting that cytokinin response induced after wounding regulates callus formation.
A previous study by Dewitte et al. (2007) demonstrated that CYCD3s function downstream of cytokinin signaling. Consistently, our qRT-PCR analysis showed that the expression of CYCD3;1 is reduced in arr1-3 arr12-1 (Fig. 5G), supporting an involvement of CYCD3s in the cytokinin-signaling pathway. Furthermore, we found that the cycd3;1-3 triple mutant is less efficient in wound-induced callus formation compared to wild type (Fig. 5, E and F). Taken together, these results show that wounding induces cytokinin synthesis and subsequent activation of its signaling pathway promotes cell cycle activation at wound sites.
Wound Stress Does Not Modify Overall Auxin Biosynthesis or Response
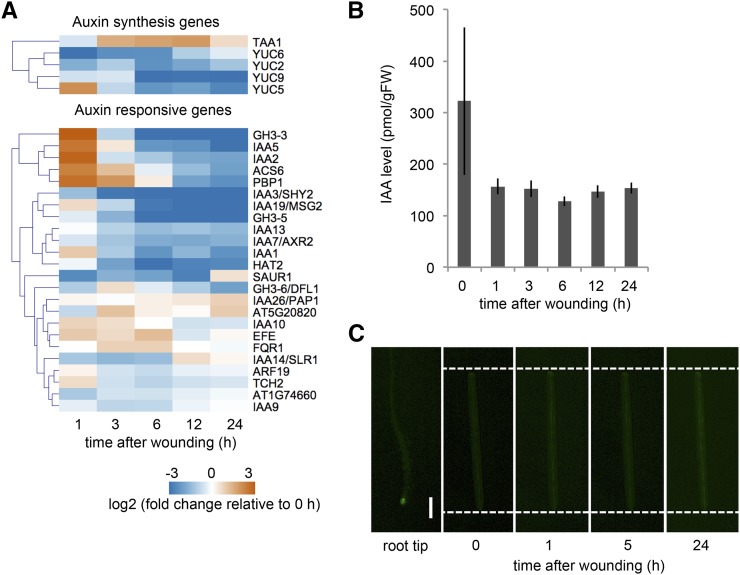
Auxin plays pivotal roles in the control of wound-induced root regeneration (Chen et al., 2016b), and it also promotes callus formation in tissue culture (Skoog and Miller, 1957). We examined transcriptional profiles of 13 auxin biosynthesis genes including YUCCA1–10 (YUC1–10) and TRP AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1) and found activation of only YUC5 and TAA1 after wounding (Fig. 6A). Our transcriptome data also showed transient up-regulation of a small subset of auxin response genes, while majority of previously described auxin response genes remain unchanged or down-regulated after wounding (Fig. 6A; Supplemental Table S3). Moreover, the endogenous level of indole acetic acid (IAA) does not change significantly after wounding (Fig. 6B) and auxin response, visualized by an auxin reporter line DR5rev:GFP, is also unaffected up to 24 h after wounding (Fig. 6C). These observations suggest that the endogenous auxin level or response is not significantly changed in wounded hypocotyls.
Figure 6.
Wounding does not modify overall auxin biosynthesis and signaling. A, Expression profiles retrieved from RNA-seq datasets show that the expression of auxin biosynthesis genes YUC5 and TAA1 are induced after wounding while other biosynthesis genes, YUC2, YUC6, and YUC9, are repressed. A subset of auxin response genes is transiently up-regulated at 1 h after wounding, but others remain unchanged or downregulated. Note that only genes with RPKM > 1 that change significantly along the time course are shown (5/13 genes for IAA synthesis; 24/50 genes for IAA responses). B, An overall content of endogenous IAA level does not change significantly after wounding (n = 3; P = 0.273; ANOVA). C, Time-course imaging of DR5rev:GFP plants shows that auxin response is not elevated at wound sites of hypocotyl explants. Dashed lines indicate wound sites. The strong DR5rev:GFP expression in root tips confirms the reporter activity. Scale bar = 0.5 mm.
A Set of Wound-Induced AP2/ERF Transcriptional Regulators Contribute to Callus Formation
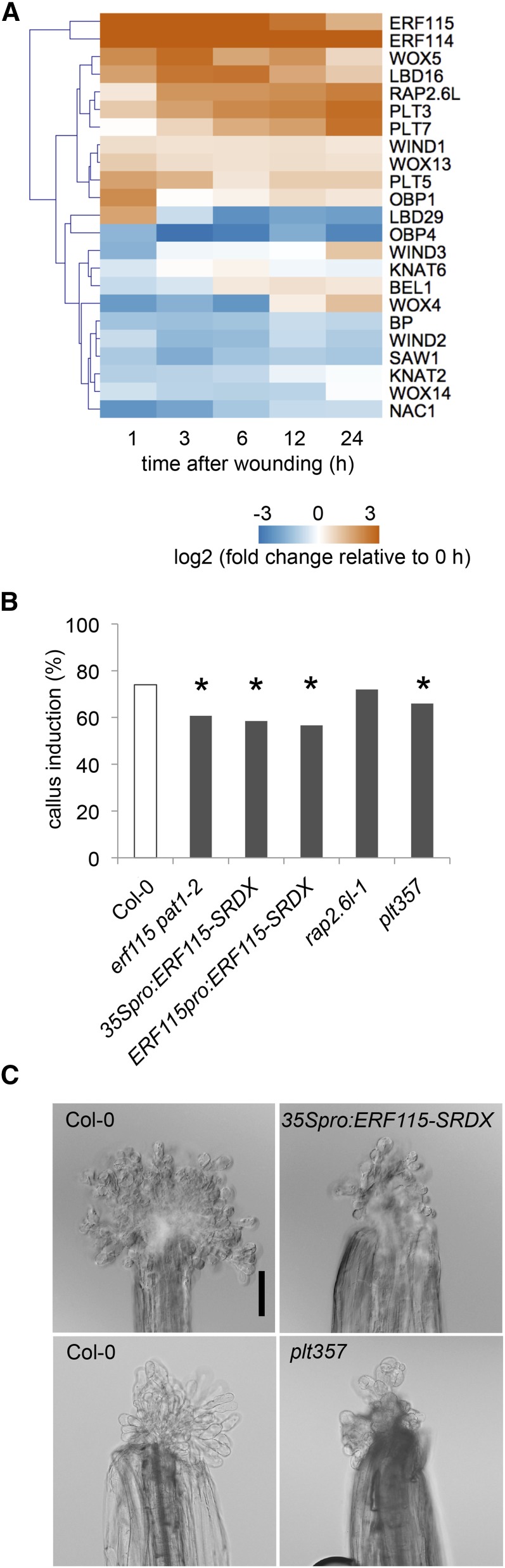
In addition to the hormonal regulators described above, recent studies have identified a set of developmental regulators that control cell proliferation and/or cell fate change in plants (Ikeuchi et al., 2013, 2016). Using our RNA-seq datasets, we investigated whether wounding modifies the expression of 49 transcriptional regulators that are implicated in callus formation or regeneration, which we collectively refer to reprogramming regulators (Supplemental Table S3). As expected, WIND1 is induced within 1 h after wounding, and its close homolog WIND3 is up-regulated at around 12 h (Fig. 7A). A recent study has shown that another member of AP2/ERF family transcription factors, ERF115, is upregulated by DNA damage and wounding in Arabidopsis roots (Heyman et al., 2016). Interestingly, we found that the expression of ERF115 and two of its close homologs ERF113/RAP2.6L and ERF114 is strongly up-regulated within 1 h in wounded hypocotyls (Fig. 7A). Our callus induction assay using wounded hypocotyls, in addition, revealed that a loss-of-function mutant of ERF115 and its interaction partner PHYTOCHROME A SIGNAL TRANSDUCTION1 (PAT1) is less effective in callus formation at wound sites (Fig. 7B), indicating that the ERF115-PAT1 complex plays key roles in wound-induced callus formation. We confirmed the callus formation defects using two chimeric repressor lines of ERF115, 35Spro:ERF115-SRDX, and ERF115pro:ERF115-SRDX, where we drove the expression of ERF115-SRDX chimeric protein by the cauliflower mosaic virus 35S (35S) promoter and ERF115 promoter, respectively (Fig. 7, B and C). On the contrary, callus induction efficiency in erf113/rap2.6l single mutants is comparable to that in wild type (Fig. 7B), suggesting that ERF113/RAP2.6L alone is dispensable for wound-induced callus formation.
Figure 7.
A set of wound-induced transcriptional regulators participate in callus formation. A, Expression profiles retrieved from RNA-seq datasets show that reprogramming regulator genes, such as WIND1, WIND3, ERF113/RAP2.6L, ERF114, ERF115, PLT3, PLT5, and PLT7 is up-regulated after wounding. Note that only genes with RPKM > 1 that change significantly along the time course are shown (23/49 genes). B, A subset of wound-induced AP2/ERF transcriptional regulators, ERF115, PLT3, PLT5, and PLT7, is required for callus formation in wounded hypocotyls. C, Representative images of calli generated 14 d after wounding in Col-0, 35Spro::ERF115-SRDX, and plt357 hypocotyls. Scale bar = 0.1 mm.
Our RNA-seq data showed that the expression of another set of AP2/ERF family transcription factors, PLT3, PLT5, and PLT7, is also activated after wounding (Fig. 7A). We further found that the plt357 triple mutant displays significant defects in callus induction (Fig. 7, B and C), demonstrating that these PLTs participate in callus formation at wound sites. Other developmental regulators that show strong up-regulation after wounding include LBD16, WOX5, WOX13, and a Dof family transcription factor OBF BINDING PROTEIN1 known to be associated with cell proliferation (Skirycz et al., 2008; Fig. 7A). Their functional link to callus induction, however, remains unclear since some of their close homologs such as LBD29, WOX4, WOX14, and OBF BINDING PROTEIN4 are downregulated after wounding (Fig. 7A). It is interesting to note that regulators of shoot meristem formation such as BREVIPEDICELUS and KNOTTED-LIKE FROM ARABIDOPSIS THALIANA2 from class I KNOTTED1-like HOMEOBOX family, are rapidly down-regulated after wounding (Fig. 7A). A dimerization partner of class I KNOTTED1-like HOMEOBOX, SAWTOOTH1 (Kumar et al., 2007), is also abruptly down-regulated (Fig. 7A), together implying that hypocotyls cells start to lose the shoot identity soon after wounding.
Wounding Induces Dynamic Transcriptional Changes of Chromatin Regulators
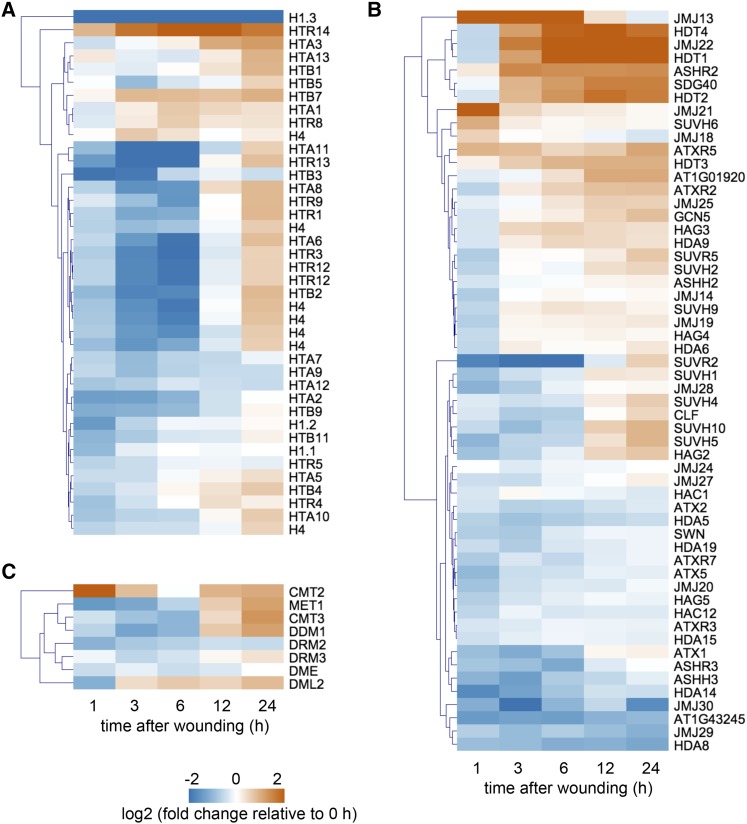
Under unstressed conditions, epigenetic mechanisms such as histone modification prevent callus formation by maintaining the repressive status of callus-inducing genes (Ikeuchi et al., 2015b). Indeed, the loci of many callus regulators including WIND3, ERF115, RAP2.6L, PLT3, PLT5, PLT7, and LBD16 have repressive H3K27me3 marks in unstressed plants (Supplemental Fig. S4; Lu et al., 2011). Given that all of these genes are transcriptionally activated by wounding before the onset of cell proliferation (Fig. 7A), marked histones cannot be diluted through nucleosome replication during cell cycle progression. Accordingly, induction of these genes likely entails active modification of the chromatin environment, for instance, by erasure of repressive histone marks, addition of active histone marks and replacement of histones within nucleosomes. Therefore, we examined whether the expression profiles of histones and histone modifiers change upon wounding in our RNA-seq datasets. As shown in Figure 8A, most genes encoding core histones such as H2A, H2B, H3, and H4, display transcript profiles associated with cell cycle reactivation; thus, the majority of these histone genes are initially down-regulated and subsequently up-regulated after wounding. What is very distinct from these general trends is the steep decline in the expression of a linker histone H1.3 within 1 h after wounding (Fig. 8A), implying that the loss of H1.3 shortly after wounding might have some impact on the chromatin status and associated transcriptional changes.
Figure 8.
Wounding induces dynamic transcriptional changes of chromatin regulators. A, Expression profiles retrieved from RNA-seq datasets show that most of histone genes are initially down-regulated but subsequently up-regulated by 24 h after wounding. B, A subset of genes encoding histone-modifying enzymes undergoes significant transcriptional changes after wounding. C, Genes that encode DNA methyltransferases and demethylases seem to be also transcriptionally affected by wounding. Note that only genes with RPKM > 1 that change significantly along the time course are shown (40/45 genes for histones; 56/92 genes for histone modifier; 8/11 DNA methyltransferases and demethylases).
Among putative histone methyltransferases harboring a SET domain, CURLY LEAF (CLF) and SWINGER (SWN), encoding a methyltransferase subunit of PRC2 that deposits H3K27me3 (Chanvivattana et al., 2004), display transient down-regulation within 6 h and subsequent mild up-regulation after 12 h (Fig. 8B). Other histone methyltransferases, such as ARABIDOPSIS TRITHORAX-LIKE PROTEIN2 that deposit an active H3K4me2 mark (Saleh et al., 2008), are rapidly down-regulated, while SUVH4/KRYPTONITE, SUVH5, and SUVH6 mediating repressive H3K9 methylation (Ebbs and Bender, 2006) are up-regulated after wounding (Fig. 8B). Our RNA-seq data also showed that JUMONJI30 (JMJ30), encoding a demethylase acting on H3K27me3 (Gan et al., 2014), is rapidly down-regulated after wounding (Fig. 8B). Among other histone demethylase genes, JMJ22, a demethylase targeting H4R3 (Cho et al., 2012), JMJ13 and JMJ21 are strongly induced within 24 h after wounding (Fig. 8B).
Histone acetylation and deacetylation are generally associated with gene activation and inactivation, respectively (Struhl, 1998). In our dataset, most of the acetyl transferase genes remain unchanged upon wounding, except GCN5-RELATED N-ACETYLTRANSFERASE (GNAT) family members GENERAL CONTROL NONDEREPRESSIBLE5 (GCN5; Benhamed et al., 2006), HISTONE ACETYLTRANSFERASE OF THE GNAT FAMILY2 (HAG2), and HAG3 (Earley et al., 2007), which are induced around 3 to 6 h after wounding (Fig. 8B). Among 18 histone deacetylases (Liu et al., 2014), a plant-specific HD2 family of histone deacetylases, HISTONE DEACETYLASE1 (HDT1), HDT2, HDT3, and HDT4, are up-regulated between 3 and 12 h after wounding, while two of RPD3/HDA1 family genes, HDA5 and HDA8, are down-regulated within 1 h (Fig. 8B).
DNA methylation is another important component of epigenetic regulation, and indeed DNA methyltransferase genes show dynamic expression changes after wounding (Fig. 8C). The expression of CHROMOMETHYLASE2 (CMT2) is most rapidly induced, i.e. within 1 h, after wounding, followed by the activation of METHYLTRANSFERASE 1 (MET1), CMT3, DECREASED DNA METHYLATION1, and DEMETER LIKE2 at later time points (Fig. 8C). We also noticed that the expression of DOMAINS REARRANGED METHYLTRANSFERASE2 and DEMETER is reduced after wounding (Fig. 8C).
Overall, these dynamic transcriptional changes in histone-modifying enzymes and DNA methyltransferases suggest that wound-induced modification of transcriptomes likely involves active modulation of the chromatin landscape.
DISCUSSION
In this study, we employed time course transcriptome analysis and quantitative hormonal analysis to study how wound stress triggers callus formation in Arabidopsis. Our data showed that wounded hypocotyls initially activate stress responses but subsequently commit cells for reentry into the cell cycle through up-regulation of the cytokinin-mediated developmental pathway (Fig. 9). We also demonstrated that wound stress activates a set of AP2-ERF transcriptional regulators, and they contribute to callus formation at wound sites (Fig. 9). This study thus illustrates how plants convey a diverse range of signals in response to wounding and yet selectively induce cell cycle reactivation through the combined activation of multiple signaling cascades. Previous studies have shown that other hormones such as auxin, ethylene, and GA3 play important roles in different forms of tissue regeneration (Asahina et al., 2011; Melnyk et al., 2015; Efroni et al., 2016; Chen et al., 2016b; Matsuoka et al., 2016). Identification of cytokinin as a key player of callus formation therefore highlights diverse and specific roles of endogenous hormones in regenerating wounded tissues.
Figure 9.
A schematic diagram describing how callus formation is regulated after wounding. Wounding transiently increases the endogenous level of JA in Arabidopsis hypocotyls, but this is inhibitory for callus induction. Overall ABA level is decreased after wounding, which allows efficient callus formation at wound sites. Wounding up-regulates cytokinin biosynthesis and signaling, leading to the activation of cell proliferation and callus formation. Several AP2-ERF transcriptional regulators, such as ERF115, WIND1, ESR1, PLT3, PLT5, and PLT7, are also up-regulated by wounding and contribute to callus formation.
Global Transcriptional Changes during Wound-Induced Callus Formation
Our RNA-seq data showed that wounding causes dramatic changes in gene expression profiles, and indeed ∼80% of genes expressed at least once in our time course dataset display differential expression within 24 h after wounding. Many genes that show transcriptional changes relatively early, i.e. those in clusters 1, 2, and 5, are known to be responsive to wounding (Fig. 2; Supplemental Fig. S2). Genes encoding biosynthesis enzymes for defense substances, for example, are wound inducible (Reymond et al., 2000; Devoto et al., 2005) and are up-regulated within 3 h after wounding in our dataset. Similarly, wound-inducible protein kinases that constitute pathogen recognition or calcium signaling are activated within 3 h. It is also interesting to note that genes annotated as responsive to other stresses such as heat, chitin, or biotic stimulus are strongly activated at 1 h after wounding. Previous studies have reported that heat stress induces somatic embryogenesis in some plant species (Fehér 2015), and biotic interactions can trigger cellular proliferation and reprogramming during symbiosis and parasitism (Suzaki et al., 2012; Ishida et al., 2016). It will be thus interesting to test if these stress-induced developmental changes share regulatory mechanisms with wound-induced callus formation.
In contrast, genes that respond transcriptionally later, i.e. those in clusters 3 and 4, represent protein synthesis, organelle organization, and cell proliferation (Fig. 2; Supplemental Fig. S2). Interestingly, genes in clusters 3 and 4 significantly overlap with genes that are highly expressed in calli on CIM (Supplemental Fig. S5; Sugimoto et al., 2010), suggesting that these genes are generally associated with callus formation. Cellular capacity of protein synthesis is tightly controlled according to the developmental and environmental conditions. Transcript level of ribosomal genes, for example, is higher in developing organs where cells actively proliferate than in maturing organs (Noir et al., 2013). Therefore, activation of genes encoding ribosomal proteins in cluster 3 may be associated with preparation for cell proliferation during callus formation, which is also observed before the onset of protoplast cell proliferation (Chupeau et al., 2013). Components of protein synthesis machineries enriched in cluster 3 might be required for the generation of a new proteome during cellular reprogramming and callus formation, even though dynamic changes of transcript profiling take place as soon as 1 h after wounding. Cell cycle regulators found in cluster 4 are transiently down-regulated and later activated at 12 h. A similar trend is observed during protoplast culture, where the expression of cell cycle regulators declines transiently upon protoplast isolation and is subsequently reactivated (Chupeau et al., 2013). Such transcriptional dynamics of cell cycle regulators may be common in stress-triggered cellular reprogramming and proliferation.
Endogenous Hormonal Changes That Impact Callus Formation at Wound Sites
Our hormonal analysis revealed substantial accumulation of tZ- and cZ-type cytokinin in wounded hypocotyls, which results in the activation of cytokinin response prior to callus formation. Our phenotypic analyses demonstrated that de novo synthesis of cytokinin is a key driver of callus formation at wound sites (Fig. 5). Cytokinin is known to promote cell proliferation through transcriptional up-regulation of CYCD3s (Riou-Khamlichi et al., 1999). Our data show that CYCD3;1 expression is compromised in the arr1-3 arr12-1 mutant and, in addition, the cycd3;1-3 mutant displays reduced callus formation at wound sites (Fig. 5), further supporting an involvement of CYCD3s in cytokinin-dependent regulation of cell proliferation. Interestingly, the cytokinin biosynthesis genes LOG1, LOG4, LOG7, and CYP735A2, which are strongly induced after wounding in our RNA-seq datasets, do not show similar transcriptional changes in other wound-induced transcriptome data like those reported by Kilian et al. (2007). This is likely because these transcriptional responses are not part of the general wound response and probably occur only under the physiological condition where wounding triggers callus formation. It will be interesting to explore how wound-induced transcriptional regulatory pathways incorporate different physiological cues to optimize downstream gene expression.
We should note that cytokinin biosynthesis and signaling mutants are not completely defective in callus formation (Fig. 5), suggesting that there are other signaling pathways promoting callus formation at wound sites. Intriguingly, contribution of auxin in wound-induced callus formation appears to be marginal, since we do not detect accumulation of endogenous auxin or activation of auxin response after wounding (Fig. 6). These observations are consistent with our previous report that solitary root mutants, defective in auxin signaling, have no obvious defects in wound-induced callus formation (Iwase et al., 2011). Our data also show that wounding rapidly activates JA synthesis and signaling, yet JA is not required for callus formation and is rather inhibitory to callus induction (Fig. 3). JA is known to block plant growth (Yan et al., 2007), and JA represses both cell proliferation and cell expansion to limit organ growth (Noir et al., 2013). Lack of JA’s contribution in callus formation is therefore in line with previous studies, strongly suggesting that JA primarily mediates defense-related responses after wounding. Another stress hormone, ABA, negatively regulates plant growth by repressing cell proliferation under stress conditions (Wang et al., 1998). Our phenotypic analyses also show that ABA is repressive for callus formation (Fig. 4), confirming the negative impact of ABA on cell proliferation. In contrast to JA, for which the endogenous level increased by more than 2-fold within 6 h after wounding, we detected a significant decrease in the overall ABA level within 24 h after wounding (Fig. 4), although biological significance of this reduction is not clear. Given that the transcript levels of ABA biosynthesis and response genes are strongly increased within 1 h after wounding, it is also possible that the ABA level is transiently and/or locally increased earlier than 1 h and we could not detect these differences at 1 h.
ERF115 and PLT3, 5, 7 as Transcriptional Regulators of Wound-Induced Callus Formation
Recent studies have identified key regulators that control cell proliferation and/or differentiation in plant meristem development and/or regeneration, and only a subset of them have been linked to hormonal regulation (Ikeuchi et al., 2013). Given that some of these genes are rapidly induced after wounding, they are good candidates that might also function in wound-induced callus formation, and our genetic analysis indeed uncovered that ERF115 is required for callus induction at wound sites (Fig. 7). Given that ERF115 acts upstream of WIND1 during root meristem repair (Heyman et al., 2016), one plausible hypothesis is that ERF115-WIND1 promotes callus formation in parallel to the pathway driven by increased cytokinin accumulation (Fig. 9). We have recently shown that both ARR1-, ARR12-mediated cytokinin signaling and WIND1-mediated wound signaling activate ESR1 expression to promote callus formation at wound sites (Iwase et al., 2017). Thus, these hormonal and transcriptional pathways likely converge and/or cross talk to promote wound-induced callus formation, and future studies should reveal the complex functional relationships between these pathways.
In addition, we show that another set of AP2/ERF transcription factors, PLT3, PLT5, and PLT7, also participate in callus formation at wound sites (Fig. 7). Importantly, ESR1 does not regulate the expression of PLT3, PLT5, and PLT7 (Iwase et al., 2017); thus, they may function in a pathway independent from the WIND1-ESR1-mediated pathway (Fig. 9). These members of the PLT family are known to act downstream of the cytokinin pathway (Kareem et al., 2015). However, given that they are all up-regulated within 1 h, which is well before we detect increased cytokinin response after wounding, we suspect that their up-regulation is controlled by a cytokinin-independent wound signaling (Fig. 9). Interestingly, plt357 mutants as well as WIND1-SRDX and esr1 are not defective in developing calli on CIM, although their calli fail to regenerate shoots on shoot-inducing media (Kareem et al., 2015; Iwase et al., 2017). These observations suggest that these regulators play distinct roles in different contexts, i.e. promoting cell proliferation at wound sites and conferring callus with pluripotency on CIM.
Epigenetic Modification Induced by Wounding
A previous transcriptome study by Chupeau et al. (2013) reported dynamic transcriptional modification of various epigenetic regulators during cell cycle reentry of Arabidopsis protoplast cells. Similarly, our RNA-seq data highlighted that many genes encoding histones, histone modification enzymes and DNA methyltransferases undergo transcriptional changes within 24 h after wounding (Fig. 8), suggesting that global modification of gene expression patterns after wounding likely involves regulation at the chromatin level. One key question in the context of callus formation and organ regeneration is how wound stress modulates the epigenetic landscape and allows rapid activation of PRC2-targeted reprogramming regulator genes. Our data show that among all the histone genes we examined, only a linker histone H1.3 displays very specific transcriptional down-regulation within 1 h after wounding (Fig. 8). Linker histone H1s are associated with chromatin compaction and histone H1.3, in particular, is localized at genomic loci distinct from those occupied by H1.1 and H1.2 (Rutowicz et al., 2015). It is thus possible that the reduction of H1.3 in response to wounding may cause the release of H1.3-occupied heterochromatic region, leading to activation of gene expression. Future studies should investigate the physiological roles of H1.3 in wound response and callus formation using Arabidopsis mutants. Interestingly, a previous study showed that histone H1.3 is up-regulated under drought or low-light conditions in an ABA-dependent manner (Rutowicz et al., 2015); thus, the reduction of H1.3 after wounding might also be associated with down-regulation of endogenous ABA.
Another mechanism that may permit activation of PRC2-repressed reprogramming regulator genes is to reduce the level of H3K27me3. Our RNA-seq data show that the transcript levels of PRC2 subunits, CURLY LEAF and SWINGER, are transiently down-regulated after wounding (Fig. 8), but it is unlikely that this transient reduction contributes to the loss of H3K27me3 at already PRC2-marked loci. We also detected several putative histone demethylase JMJ genes up-regulated within 24 h after wounding (Fig. 8), raising the possibility that active removal of H3K27me3 by these demethylases may help to activate gene expression. Alternatively, reprogramming regulator genes may be transcriptionally activated through the deposition of active histone marks. An increase in the level of acetylation, for instance, may contribute to the wound-induced gene activation, since genes encoding histone acetyl transferases, such as GCN5 and HAG2, are induced after wounding (Fig. 8).
Our RNA-seq data also showed that DNA methylation enzymes display dynamic transcriptional changes after wounding. Among them, CMT2 encoding a CHH methyltransferase is most rapidly induced after wounding (Fig. 8). Functional roles of DNA methylation in plant stress response are not fully established (Kim et al., 2015), but a previous study by Shen et al. (2014) reported that the expression of CMT2 is down-regulated upon heat stress. Loss-of-function mutations of CMT2 are found in natural Arabidopsis populations that display high-temperature habitats, and these mutations are shown to contribute to heat stress tolerance in plants (Shen et al., 2014). It would be interesting to test whether CMT2 impacts wound-induced defense response and/or regeneration.
Given that some of the PRC2-targetted reprogramming genes, such as ERF115 and PLT5, are transcriptionally activated within 1 h after wounding (Fig. 7; Supplemental Fig. S4), chromatin status of these loci has to change even earlier than 1 h. It is worth noting that stress-induced modification of chromatin status does not necessarily involve transcriptional changes of epigenetic regulators, and alternatively it might be mediated by the recruitment of existing regulators to specific loci. Future studies should, therefore, explore how the status of histone variants, histone modification, and DNA methylation changes at each reprogramming gene locus upon wounding and how these epigenetic changes affect its transcription.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
For phenotypic analyses, we used the following mutants: aos (Park et al., 2002); jar1-1 (Staswick et al., 2002); coi1-16B (Noir et al., 2013); nced3-2 (Gálvez-Valdivieso et al., 2009); aba2-1 and aba3-1 (Léon-Kloosterziel et al., 1996); aba2-2 (Nambara et al., 1998); aao3-4 (Seo et al., 2004); abi1-1 and abi2-1 (Koornneef et al., 1984); log123457 (Tokunaga et al., 2012); ipt357 (Miyawaki et al., 2006); arr1-3 arr12-1, arr1-3 arr10-5, and arr10-5 arr12-1 (Argyros et al., 2008); cycd3;1-3 (Dewitte et al., 2007); erf115 pat1-2 and 35Spro:ERF115-SRDX (Heyman et al., 2013); rap2.6l-1 (Che et al., 2006); plt357 (Prasad et al., 2011); and WIND1-SRDX (Iwase et al., 2011). For the expression analysis of cell cycle genes, we used CDKApro:CDKA-GUS (Adachi et al., 2009) and CYCA2;1pro:GUS (Burssens et al., 2000). As reporters of hormonal responses, we used TCSn::GFP (Zürcher et al., 2013) and DR5rev:GFP (Benková et al., 2003). All plants were in Columbia-0 (Col-0) background except aos and coi1-16B, which are in the gl1 background and CYCA2;1pro:GUS in C24 background. Plants were grown on 0.6% (w/v) gelzan plates containing Murashige and Skoog (MS) salts and 1% Suc on nylon mesh at 22°C. For callus induction, 7-d-old etiolated hypocotyls were cut with disposable knives and incubated in the dark. Phenotypic analyses were performed on hypocotyls that were cut once at 7 mm from the hypocotyl-root junction, and callus formation at the apical end of cut hypocotyls was observed. Callus induction was quantified as a percentage of explants with more than one callus cell developing from wound sites.
Cloning and Generation of Transgenic Plants
To generate ERF115pro:ERF115-SRDX lines, the ERF115-SRDX dominant-negative allele was generated by fusing the SRDX-repressor domain behind the ERF115 ORF using adaptor ligation PCR (as described by Heyman et al., 2013). The 2000-bp upstream promoter region of ERF115 was recombined in front of the ERF115-SRDX allele into the pK7m24GW3 vector. Transgenic plants were selected using 45mg/L kanamycin.
Transcriptome Analysis
We isolated total RNA from 3- to 5-mm hypocotyl explants at 0, 1, 3, 6, 12, and 24 h after wounding using the RNeasy plant mini kit (Qiagen). We had biological triplicates for each time point with the exception of 24 h where we had biological duplicates. Isolated RNA was subject to library preparation with Truseq stranded mRNA sequencing kit (Illumina) and then subjected to single-end sequencing with the Illumina HisEquation 2000 platform. Read mapping was performed on CLC Genomics Workbench (CLC Bio), and over 90% of reads were uniquely mapped on TAIR10 Arabidopsis genome, resulting in 8 to 20 million mapped reads per sample. Among 18,332 expressed genes (>0.1 RPKM), expression of 14,605 genes was significantly changed during the time course according to the Bayesian estimation of temporal regulation (Aryee et al., 2009; FDR = 0.05). For genes with significantly changed expression, we performed k-means clustering and figures of merit analysis using Multiple Experiment Viewer (Saeed et al., 2003). GO analyses were performed with BiNGO (Maere et al., 2005) in Cytoscape. For the heat maps, genes with >1 RPKM were selected and log2-fold changes against 0 h were shown for each gene. Gene clustering analysis was performed based on Eucledian distance, an average linking method in Multiple Experiment Viewer software.
Quantitative Real-Time RT-PCR
Total RNA of 400 ng, isolated from hypocotyl explants as described for transcriptome analysis, were subjected to the first-strand cDNA synthesis with the primescript RT reagent kit (Takara). Quantitative real-time PCR was performed with Thunderbird SYBR qPCR mix (Toyobo) and a set of primers listed in Supplemental Table S4.
Histological Analysis
GUS staining was performed as previously described (Kertbundit et al., 1991), and stained samples were observed using a Leica M165 C stereomicroscope. For Technovit sectioning, GUS-stained hypocotyls were fixed in a formaldehyde-acetic acid solution (formalin: acetic acid: 70% ethanol, 1:1:18), dehydrated through a graded ethanol series and embedded in Technovit 7100 (Heraeus Kulzer). Sections of 4-μm thickness were prepared with RM2135 (Leica), counterstained by safranin O, and observed under a Olympus BX51 microscope.
Quantitative Analysis of Plant Hormones
Hypocotyls were cut into 3- to 5-mm explants and incubated for 0, 1, 3, 6, 12, and 24 h after wounding in the dark. Three biological replicates were prepared for each time point and 30 to 50 mg (fresh weight) of hypocotyl explants were harvested for each replicate. Contents of cytokinins were quantified with ultra-performance liquid chromatography-electrospray interface and tandem quadrupole mass spectrometer (AQUITY UPLC System/Xevo-TQS; Waters) as described previously (Kojima et al., 2009). Auxin, JA, and ABA were quantified with ultra-high-performance liquid chromatography (HPLC)-electrospray interface and quadrupole-orbitrap mass spectrometer (UHPLC/Q-Exactive; Thermo Scientific) as described previously (Kojima and Sakakibara 2012; Shinozaki et al., 2015).
Accession Numbers
RNA sequencing data have been deposited to Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101422) under accession number GSE101422. Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CDKA (AT3G48750), CYCA2;1 (AT5G25380), AOS (AT5G42650), COI1 (AT2G39940), NCED3 (AT3G14440), ABA2 (AT1G52340), ABA3 (AT1G16540), AAO3 (AT2G27150), ABI1 (AT5G67030), ABI2 (AT1G52340), LOG1 (AT2G28305), LOG2 (AT2G35990), LOG3 (AT2G37210), LOG4 (AT3G53450), LOG5 (AT4G35190), LOG7 (AT5G06300), IPT3 (AT3G63110), IPT5 (AT5G19040), IPT7 (AT3G23630), ARR1 (AT3G16857), ARR10 (AT4G31920), ARR12 (AT2G25180), CYCD3;1 (AT4G34160), CYCD3;2 (AT5G67260), CYCD3;3 (AT3G50070), ERF115 (AT5G07310), PAT1 (AT5G48150), RAP2.6L (AT5G13330), PLT3 (AT5G10510), PLT5 (AT5G57390), and PLT7 (AT5G65510). For additional accession numbers, see Supplemental Table S3.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Real-time qRT-PCR analysis validates expression profiles deduced from RNA-seq.
Supplemental Figure S2. Comparison of the gene expression profile with another wound-induced
transcriptome.
Supplemental Figure S3. Quantitative analysis of endogenous cytokinin levels.
Supplemental Figure S4. A subset of wound-induced genes is marked by H3K27me3 in intact plants.
Supplemental Figure S5. Comparison of the gene expression profile with calli on CIM.
Supplemental Table S1. A list of genes included in each of five clusters.
Supplemental Table S2. A list of gene ontology categories overrepresented in five clusters.
Supplemental Table S3. A list of genes selected for detailed transcriptional analyses.
Supplemental Table S4. A list of primers used in this study.
Acknowledgments
The authors are grateful to the members of Sugimoto’s lab for discussions and comments on the manuscript. RNA-seq was performed at the University of Utah sequencing facility. The authors thank Mariko Ohnuma, Mariko Mouri, Chika Ikeda, Yasuko Yatomi, Mieko Ito, and Noriko Doi for technical assistance.
Footnotes
This work was supported by the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry, a grant from the Ministry of Education, Culture, Sports, Science and Technology to K.S. (15H05961), and grants from the Japan Society for the Promotion of Science (JSPS) to M.I. (15K18564, 17K15146, 17J40121), A.I. (15K18565, 15KK0265, 17K07461), and K.S. (17H03704). M.I. is supported by a RIKEN Special Postdoctoral Researcher Programme, JSPS fellowship and the Naito Foundation. J.H. thanks the Research Foundation-Flanders for a postdoctoral fellowship.
Articles can be viewed without a subscription.
References
- Adachi S, Nobusawa T, Umeda M (2009) Quantitative and cell type-specific transcriptional regulation of A-type cyclin-dependent kinase in Arabidopsis thaliana. Dev Biol 329: 306–314 [DOI] [PubMed] [Google Scholar]
- Arata Y, Nagasawa-Iida A, Uneme H, Nakajima H, Kakimoto T, Sato R (2010) The phenylquinazoline compound S-4893 is a non-competitive cytokinin antagonist that targets Arabidopsis cytokinin receptor CRE1 and promotes root growth in Arabidopsis and rice. Plant Cell Physiol 51: 2047–2059 [DOI] [PubMed] [Google Scholar]
- Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee MJ, Gutiérrez-Pabello JA, Kramnik I, Maiti T, Quackenbush J (2009) An improved empirical bayes approach to estimating differential gene expression in microarray time-course data: BETR (Bayesian Estimation of Temporal Regulation). BMC Bioinformatics 10: 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahina M, Azuma K, Pitaksaringkarn W, Yamazaki T, Mitsuda N, Ohme-Takagi M, Yamaguchi S, Kamiya Y, Okada K, Nishimura T, et al. (2011) Spatially selective hormonal control of RAP2.6L and ANAC071 transcription factors involved in tissue reunion in Arabidopsis. Proc Natl Acad Sci USA 108: 16128–16132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M, Bertrand C, Servet C, Zhou D-X (2006) Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18: 2893–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bishop PD, Makus DJ, Pearce G, Ryan CA (1981) Proteinase inhibitor-inducing factor activity in tomato leaves resides in oligosaccharides enzymically released from cell walls. Proc Natl Acad Sci USA 78: 3536–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burssens S, de Almeida Engler J, Beeckman T, Richard C, Shaul O, Ferreira P, Van Montagu M, Inzé D (2000) Developmental expression of the Arabidopsis thaliana CycA2;1 gene. Planta 211: 623–631 [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon Y-H, Sung ZR, Goodrich J (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276 [DOI] [PubMed] [Google Scholar]
- Che P, Lall S, Nettleton D, Howell SH (2006) Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol 141: 620–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Cheng J, Chen L, Zhang G, Huang H, Zhang Y, Xu L (2016a) Auxin-independent NAC pathway acts in response to explant-specific wounding and promotes root tip emergence during de novo root organogenesis in Arabidopsis. Plant Physiol 170: 2136–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Tong J, Xiao L, Ruan Y, Liu J, Zeng M, Huang H, Wang J-W, Xu L (2016b) YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J Exp Bot 67: 4273–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JN, Ryu JY, Jeong YM, Park J, Song JJ, Amasino RM, Noh B, Noh YS (2012) Control of seed germination by light-induced histone arginine demethylation activity. Dev Cell 22: 736–748 [DOI] [PubMed] [Google Scholar]
- Choi J, Tanaka K, Cao Y, Qi Y, Qiu J, Liang Y, Lee SY, Stacey G (2014) Identification of a plant receptor for extracellular ATP. Science 343: 290–294 [DOI] [PubMed] [Google Scholar]
- Chupeau M-C, Granier F, Pichon O, Renou J-P, Gaudin V, Chupeau Y (2013) Characterization of the early events leading to totipotency in an Arabidopsis protoplast liquid culture by temporal transcript profiling. Plant Cell 25: 2444–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman RA, Tierney ML, Mullet JE (1992) Jasmonic acid/methyl jasmonate accumulate in wounded soybean hypocotyls and modulate wound gene expression. Proc Natl Acad Sci USA 89: 4938–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino IB, Deruère J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann C, Rojo E, Sánchez-Serrano JJ (1997) Abscisic acid and jasmonic acid activate wound-inducible genes in potato through separate, organ-specific signal transduction pathways. Plant J 11: 773–782 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V, et al. (2007) Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci USA 104: 14537–14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A, Ellis C, Magusin A, Chang H-S, Chilcott C, Zhu T, Turner JG (2005) Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol Biol 58: 497–513 [DOI] [PubMed] [Google Scholar]
- Earley KW, Shook MS, Brower-Toland B, Hicks L, Pikaard CS (2007) In vitro specificities of Arabidopsis co-activator histone acetyltransferases: Implications for histone hyperacetylation in gene activation. Plant J 52: 615–626 [DOI] [PubMed] [Google Scholar]
- Ebbs ML, Bender J (2006) Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18: 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Mello A, Nawy T, Ip P-L, Rahni R, DelRose N, Powers A, Satija R, Birnbaum KD (2016) Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 165: 1721–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Xu C, Xu K, Hu Y (2012) LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res 22: 1169–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehér A. (2015) Somatic embryogenesis - Stress-induced remodeling of plant cell fate. Biochim Biophys Acta 1849: 385–402 [DOI] [PubMed] [Google Scholar]
- Gálvez-Valdivieso G, Fryer MJ, Lawson T, Slattery K, Truman W, Smirnoff N, Asami T, Davies WJ, Jones AM, Baker NR, et al. (2009) The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 21: 2143–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan E-S, Xu Y, Wong J-Y, Goh JG, Sun B, Wee W-Y, Huang J, Ito T (2014) Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat Commun 5: 5098. [DOI] [PubMed] [Google Scholar]
- He C, Chen X, Huang H, Xu L (2012) Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet 8: e1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettenhausen C, Sun G, He Y, Zhuang H, Sun T, Qi J, Wu J (2016) Genome-wide identification of calcium-dependent protein kinases in soybean and analyses of their transcriptional responses to insect herbivory and drought stress. Sci Rep 6: 18973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman J, Cools T, Canher B, Shavialenka S, Traas J, Vercauteren I, Van den Daele H, Persiau G, De Jaeger G, Sugimoto K, et al. (2016) The heterodimeric transcription factor complex ERF115-PAT1 grants regeneration competence. Nat Plants 2: 16165. [DOI] [PubMed] [Google Scholar]
- Heyman J, Cools T, Vandenbussche F, Heyndrickx KS, Van Leene J, Vercauteren I, Vanderauwera S, Vandepoele K, De Jaeger G, Van Der Straeten D, et al. (2013) ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342: 860–863 [DOI] [PubMed] [Google Scholar]
- Hofhuis H, Laskowski M, Du Y, Prasad K, Grigg S, Pinon V, Scheres B (2013) Phyllotaxis and rhizotaxis in Arabidopsis are modified by three PLETHORA transcription factors. Curr Biol 23: 956–962 [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Iwase A, Rymen B, Harashima H, Shibata M, Ohnuma M, Breuer C, Morao AK, de Lucas M, De Veylder L, et al. (2015a) PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis. Nat Plants 1: 15089. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Iwase A, Sugimoto K (2015b) Control of plant cell differentiation by histone modification and DNA methylation. Curr Opin Plant Biol 28: 60–67 [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Ogawa Y, Iwase A, Sugimoto K (2016) Plant regeneration: Cellular origins and molecular mechanisms. Development 143: 1442–1451 [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Sugimoto K, Iwase A (2013) Plant callus: Mechanisms of induction and repression. Plant Cell 25: 3159–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D, De Veylder L (2006) Cell cycle regulation in plant development. Annu Rev Genet 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Ishida JK, Wakatake T, Yoshida S, Takebayashi Y, Kasahara H, Wafula E, dePamphilis CW, Namba S, Shirasu K (2016) Local auxin biosynthesis mediated by a YUCCA flavin monooxygenase regulates haustorium development in the parasitic plant Phtheirospermum japonicum. Plant Cell 28: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase A, Harashima H, Ikeuchi M, Rymen B, Ohnuma M, Komaki S, Morohashi K, Kurata T, Nakata M, Ohme-Takagi M, et al. (2017) WIND1 promotes shoot regeneration through transcriptional activation of ENHANCER OF SHOOT REGENERATION1 in Arabidopsis. Plant Cell 29: 54–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K, et al. (2011) The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol 21: 508–514 [DOI] [PubMed] [Google Scholar]
- Kakimoto T. (2001) Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltransferases. Plant Cell Physiol 42: 677–685 [DOI] [PubMed] [Google Scholar]
- Kareem A, Durgaprasad K, Sugimoto K, Du Y, Pulianmackal AJ, Trivedi ZB, Abhayadev PV, Pinon V, Meyerowitz EM, Scheres B, et al. (2015) PLETHORA genes control regeneration by a two-step mechanism. Curr Biol 25: 1017–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertbundit S, De Greve H, Deboeck F, Van Montagu M, Hernalsteens JP (1991) In vivo random beta-glucuronidase gene fusions in Arabidopsis thaliana. Proc Natl Acad Sci USA 88: 5212–5216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Takei K, Kojima M, Sakakibara H (2013) Side-chain modification of cytokinins controls shoot growth in Arabidopsis. Dev Cell 27: 452–461 [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horák J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kim J-M, Sasaki T, Ueda M, Sako K, Seki M (2015) Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front Plant Sci 6: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M, Kamada-Nobusada T, Komatsu H, Takei K, Kuroha T, Mizutani M, Ashikari M, Ueguchi-Tanaka M, Matsuoka M, Suzuki K, et al. (2009) Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: An application for hormone profiling in Oryza sativa. Plant Cell Physiol 50: 1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M, Sakakibara H (2012) Highly sensitive high-throughput profiling of six phytohormones using MS-probe modification and liquid chromatography–tandem mass spectrometry. In Normanly J, ed, High-Throughput Phenotyping in Plants. Humana Press, New York, pp 151–164. [DOI] [PubMed] [Google Scholar]
- Koo AJK, Gao X, Jones AD, Howe GA (2009) A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J 59: 974–986 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant 61: 377–383 [Google Scholar]
- Kumar R, Kushalappa K, Godt D, Pidkowich MS, Pastorelli S, Hepworth SR, Haughn GW (2007) The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell 19: 2719–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Park O-S, Jung S-J, Seo PJ (2016) Histone deacetylation-mediated cellular dedifferentiation in Arabidopsis. J Plant Physiol 191: 95–100 [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10: 655–661 [DOI] [PubMed] [Google Scholar]
- Li W, Liu H, Cheng ZJ, Su YH, Han HN, Zhang Y, Zhang XS (2011) DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet 7: e1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang S, Zhao M, Luo M, Yu C-W, Chen C-Y, Tai R, Wu K (2014) Transcriptional repression by histone deacetylases in plants. Mol Plant 7: 764–772 [DOI] [PubMed] [Google Scholar]
- Lu F, Cui X, Zhang S, Jenuwein T, Cao X (2011) Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 43: 715–719 [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Sugawara E, Aoki R, Takuma K, Terao-Morita M, Satoh S, Asahina M (2016) Differential cellular control by cotyledon-derived phytohormones involved in graft reunion of Arabidopsis hypocotyls. Plant Cell Physiol 57: 2620–2631 [DOI] [PubMed] [Google Scholar]
- Melnyk CW, Schuster C, Leyser O, Meyerowitz EM (2015) A developmental framework for graft formation and vascular reconnection in Arabidopsis thaliana. Curr Biol 25: 1306–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K, Tarkowski P, Matsumoto-Kitano M, Kato T, Sato S, Tarkowska D, Tabata S, Sandberg G, Kakimoto T (2006) Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Natl Acad Sci USA 103: 16598–16603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi SAR, Chauvin A, Pascaud F, Kellenberger S, Farmer EE (2013) GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500: 422–426 [DOI] [PubMed] [Google Scholar]
- Nambara E, Kawaide H, Kamiya Y, Naito S (1998) Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant Cell Physiol 39: 853–858 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Noir S, Bömer M, Takahashi N, Ishida T, Tsui TL, Balbi V, Shanahan H, Sugimoto K, Devoto A (2013) Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol 161: 1930–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-H, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R (2002) A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Pẽna-Cortés H, Sánchez-Serrano JJ, Mertens R, Willmitzer L, Prat S (1989) Abscisic acid is involved in the wound-induced expression of the proteinase inhibitor II gene in potato and tomato. Proc Natl Acad Sci USA 86: 9851–9855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K, Grigg SP, Barkoulas M, Yadav RK, Sanchez-Perez GF, Pinon V, Blilou I, Hofhuis H, Dhonukshe P, Galinha C, et al. (2011) Arabidopsis PLETHORA transcription factors control phyllotaxis. Curr Biol 21: 1123–1128 [DOI] [PubMed] [Google Scholar]
- Ren D, Liu Y, Yang K-Y, Han L, Mao G, Glazebrook J, Zhang S (2008) A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 105: 5638–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JA (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Rutowicz K, Puzio M, Halibart-Puzio J, Lirski M, Kotliński M, Kroteń MA, Knizewski L, Lange B, Muszewska A, Śniegowska-Świerk K, et al. (2015) A specialized histone H1 variant is required for adaptive responses to complex abiotic stress and related DNA methylation in Arabidopsis. Plant Physiol 169: 2080–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Yilmaz M, Le O, Hou G, Sadder M, Al-Abdallat A, Xia Y, Lu G, Ladunga I, et al. (2008) The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell 20: 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Aoki H, Koiwai H, Kamiya Y, Nambara E, Koshiba T (2004) Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol 45: 1694–1703 [DOI] [PubMed] [Google Scholar]
- Shemer O, Landau U, Candela H, Zemach A, Eshed Williams L (2015) Competency for shoot regeneration from Arabidopsis root explants is regulated by DNA methylation. Plant Sci 238: 251–261 [DOI] [PubMed] [Google Scholar]
- Shinozaki Y, Hao S, Kojima M, Sakakibara H, Ozeki-Iida Y, Zheng Y, Fei Z, Zhong S, Giovannoni JJ, Rose JK, et al. (2015) Ethylene suppresses tomato (Solanum lycopersicum) fruit set through modification of gibberellin metabolism. Plant J 83: 237–251 [DOI] [PubMed] [Google Scholar]
- Skirycz A, Radziejwoski A, Busch W, Hannah MA, Czeszejko J, Kwaśniewski M, Zanor M-I, Lohmann JU, De Veylder L, Witt I, et al. (2008) The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J 56: 779–792 [DOI] [PubMed] [Google Scholar]
- Skoog F, Miller CO (1957) Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol 11: 118–130 [PubMed] [Google Scholar]
- Shen X, De Jonge J, Forsberg SKG, Pettersson ME, Sheng Z, Hennig L, Carlborg Ö (2014) Natural CMT2 variation is associated with genome-wide methylation changes and temperature seasonality. PLoS Genet 10: e1004842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe ML (2002) Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14: 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev 12: 599–606 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Jiao Y, Meyerowitz EM (2010) Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell 18: 463–471 [DOI] [PubMed] [Google Scholar]
- Suzaki T, Yano K, Ito M, Umehara Y, Suganuma N, Kawaguchi M (2012) Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 139: 3997–4006 [DOI] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Sugiyama T (2001) Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J Biol Chem 276: 26405–26410 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Choi J, Cao Y, Stacey G (2014) Extracellular ATP acts as a damage-associated molecular pattern (DAMP) signal in plants. Front Plant Sci 5: 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titarenko E, Rojo E, León J, Sánchez-Serrano JJ (1997) Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol 115: 817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga H, Kojima M, Kuroha T, Ishida T, Sugimoto K, Kiba T, Sakakibara H (2012) Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J 69: 355–365 [DOI] [PubMed] [Google Scholar]
- Wang H, Qi Q, Schorr P, Cutler AJ, Crosby WL, Fowke LC (1998) ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J 15: 501–510 [DOI] [PubMed]
- Yan Y, Stolz S, Chételat A, Reymond P, Pagni M, Dubugnon L, Farmer EE (2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed]
- Zürcher E, Tavor-Deslex D, Lituiev D, Enkerli K, Tarr PT, Müller B (2013) A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol 161: 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]