In Phaedactylum, NO produced from nitrite by the nitrate reductase up-regulates the expression of genes involved in nitrite assimilation into amino acids and triggers a remodeling of lipids.
Abstract
Nitric oxide (NO) is an intermediate of the nitrogen cycle, an industrial pollutant, and a marker of climate change. NO also acts as a gaseous transmitter in a variety of biological processes. The impact of environmental NO needs to be addressed. In diatoms, a dominant phylum in phytoplankton, NO was reported to mediate programmed cell death in response to diatom-derived polyunsaturated aldehydes. Here, using the Phaeodactylum Pt1 strain, 2E,4E-decadienal supplied in the micromolar concentration range led to a nonspecific cell toxicity. We reexamined NO biosynthesis and response in Phaeodactylum. NO inhibits cell growth and triggers triacylglycerol (TAG) accumulation. Feeding experiments indicate that NO is not produced from Arg but via conversion of nitrite by the nitrate reductase. Genome-wide transcriptional analysis shows that NO up-regulates the expression of the plastid nitrite reductase and genes involved in the subsequent incorporation of ammonium into amino acids, via both Gln synthesis and Orn-urea pathway. The phosphoenolpyruvate dehydrogenase complex is also up-regulated, leading to the production of acetyl-CoA, which can feed TAG accumulation upon exposure to NO. Transcriptional reprogramming leading to higher TAG content is balanced with a decrease of monogalactosyldiacylglycerol (MGDG) in the plastid via posttranslational inhibition of MGDG synthase enzymatic activity by NO. Intracellular and transient NO emission acts therefore at the basis of a nitrite-sensing and acclimating system, whereas a long exposure to NO can additionally induce a redirection of carbon to neutral lipids and a stress response.
Nitric oxide (NO•) is an intermediate of the nitrogen (N) cycle (Fowler et al., 2013). This cycle is a complex network of reactions interconverting dinitrogen, the most abundant gas in the atmosphere, into gaseous nitrogen oxides, or NOx (NO•, nitrogen dioxide, NO2, and nitrous gas), water soluble ions (ammonium, NH4+; nitrite, NO2−; and nitrate, NO3−), organic molecules (from small soluble compounds like urea to proteins, lipids, and nucleic acids), and mineral forms (Holloway and Dahlgren, 2002; Fowler et al., 2013). NO• is an important pollutant in industrial flue gases (Vunjak-Novakovic et al., 2005; Zhu et al., 2015) and wastewaters (Kampschreur et al., 2009; Pan et al., 2015). All gases in the N cycle, including NO•, are present in oceans (Zehr and Ward, 2002; Nicholls et al., 2007), either because of gas exchanges at the air-water interface (Nicholls et al., 2007) or because they are produced within oceans themselves. NO• is generated in seawater by nonbiological photochemical reactions (Olasehinde et al., 2010), large-scale electrical discharges (Gallardo and Rhodes, 1997), and enzymatic activities in organisms living in the aerobic photic zone (Zhang et al., 2006; Olasehinde et al., 2010; Kumar et al., 2015; Eroglu et al., 2016) or in oxygen minimum zones (Naqvi et al., 1998; Nicholls et al., 2007; Martens-Habbena et al., 2015). Among the key biogeochemical cycles on which ecosystems depend for their sustainability, the N cycle is clearly the most perturbed by human activities (Fowler et al., 2013), marked by massive anthropogenic leakage of nitrate and ammonia from fertilized soils (Nicholls et al., 2007; Fowler et al., 2013) and by emissions of NOx (Nicholls et al., 2007; IPCC, 2014; Michalski et al., 2014).
There are no reliable reports on NO• concentrations in aquatic ecosystems, because this reactive molecule has a lifetime of only a few seconds (Naqvi et al., 1998; Zehr and Ward, 2002; Zhang et al., 2006; Olasehinde et al., 2010). In natural seawater, NO• concentration has been estimated between 0.01 and 10 nm (Zhang et al., 2006). In industrial microalgae cultivation systems supplied with industrial flue gas (Vunjak-Novakovic et al., 2005), we estimate that NOx (including NO•) concentrations could reach the micro- to millimolar range, that is, one thousand- to one million-fold above the natural level. NO• diffuses freely, and even minor and transient variations could have high impacts on living organisms, in which NO• is also known to act as a signaling molecule.
NO• has been reported to act as a gasotransmitter in a plethora of biological functions in prokaryotes and eukaryotes, in nonphotosynthetic and photosynthetic cells, and in terrestrial or aquatic ecosystems (Wendehenne et al., 2001; Moreau et al., 2010; Kumar et al., 2015; Eroglu et al., 2016). Two major enzymatic pathways can produce NO• in aerobic conditions, via either a nitric oxide synthase (NOS) using Arg as a substrate (Wilson et al., 2008), or a nitrate reductase (NR) using nitrite as a substrate (Yamasaki and Sakihama, 2000; Stöhr et al., 2001; Rockel et al., 2002). The role of NO• in phytoplankton has been recently reviewed based on the available published data, showing that it could be synthesized in photosynthetic eukaryotes containing primary plastids (chlorophyta, rhodophyta) or secondary plastids (e.g. diatoms, haptophytes, etc.; Kumar et al., 2015).
In the green alga Chlamydomonas, nitrite is assumed to be the only NO• source, since the addition of Arg or the analog Nω-Nitro-l-Arg had no effect on its production (Sakihama et al., 2002). NO• was shown to act on nitrogen assimilation by repressing nitrate assimilation at two levels. Firstly, NO• represses the expression of the NR and the transporters of nitrate and ammonium (de Montaigu et al., 2010). Secondly, it directly regulates the activities of NR and nitrate and ammonium transporters in Chlamydomonas (Sanz-Luque et al., 2013). Following nitrogen depletion, NO• is produced from intracellular nitrite and operates in the specific cytochrome b6f degradation pathway (Wei et al., 2014). In Chlamydomonas, NO• appears therefore to have a dual role, repressing nitrogen assimilation and acting in response to nitrogen starvation. In other photosynthetic eukaryotes, NO• may have opposite effects depending on the nitrogen status (Jin et al., 2009). To date, no clear scenario has arisen for a general model linking NO• signaling with the nitrogen status.
In the marine diatom Phaeodactylum tricornutum, NO• has been proposed to act in population size control, being involved in a “stress surveillance system” upon exposure to high concentrations of the diatom-derived aldehyde 2E,4E/Z-decadienal (DD; Vardi et al., 2006; Vardi et al., 2008). The treatment of Phaeodactylum with micromolar concentrations of DD was reported to (1) induce a calcium spike followed by the synthesis of NO•, (2) trigger the expression of the NO associated protein (NOA), coding for a homolog of a plant chloroplast protein involved in NO• production; and eventually (3) promote the entry into caspase-dependent programmed cell death (PCD; Vardi et al., 2006, 2008). The decrease in NO• produced by Phaeodactylum in the presence of an NOS-inhibitor led to the conclusion that NO• was produced by an NOS-like enzyme (Vardi et al., 2006), which seems contradictory with the absence of any NOS gene in the Phaeodactylum genome (Di Dato et al., 2015). NO• was therefore proposed to be derived from Arg and to act on the diatom itself, triggering PCD, and to diffuse outside the plasma membrane, spread rapidly through diatom population, triggering death in surrounding cells, eventually acting in the control of the population size (Vardi et al., 2008; Bidle, 2015).
Because climate change is marked by an increase in NO• emissions, the biological perturbations it could create on phytoplankton must be evaluated. Phaeodactylum appears as an appropriate model for this evaluation, firstly because the biosynthesis and physiological roles of NO• have been previously investigated (Vardi et al., 2006; Vardi et al., 2008) and secondly because some diatom strains have been considered for biotechnological applications, based on their neutral lipid content (Levitan et al., 2014; Abida et al., 2015). In this article, we analyzed the response of Phaeodactylum to various doses of NO• and found unanticipated results that led us to reexamine the pathway of NO• production by this diatom and the physiological responses this gasotransmitter could trigger.
RESULTS AND DISCUSSION
Treatment of P. tricornutum with DD Leads to Caspase/Metacaspase-Independent Cell Death and Does Not Support a Role of NO in PCD
We started our study by setting up an experimental protocol previously reported to trigger PCD in P. tricornutum, in response to potent infochemicals deriving from disrupted diatoms cells (Vardi et al., 2006, 2008). Following wounding or nutrient stresses, diatom-derived polyunsaturated fatty acids are oxidized enzymatically into volatile polyunsaturated aldehydes (PUAs), including DD (Miralto et al., 1999). PUAs have been considered as potent interspecific and intraspecific signaling compounds (Casotti et al., 2005; Vardi et al., 2006). By treating P. tricornutum and Thalassiosira weissflogii with increasing doses of DD from 33 to 66 µm, a burst of NO• was measured using fluorophore 4-amino-5-methylamino-2',7'-difluorescein diacetate (DAF-FM) as a NO•-reporter, appearing within 5 min, with an intensity proportional to DD concentration (Vardi et al., 2006). Both DD and externally provided NO• were then reported to trigger PCD.
The concentration required to induce PCD was previously reported to be >20 µm, with DD solubilized in methanol at a final 1% concentration (Vardi et al., 2006). The role of DD as a cell-to-cell infochemical led us to consider that DD effects should not rely on any solvent, especially methanol that could promote loss of the volatile DD molecule at the air-water interface. We sought therefore to detect the effect of DD in the absence of methanol and after 3-h treatments to detect rapid responses. Previous studies have been performed using the P. tricornutum CCMP 632 strain (Vardi et al., 2006, 2008). Here, we used Pt1 8.6 (accessions CCAP1055/1 or CCMP 2561), isolated from the CCMP 632 strain, and now used as a main reference (De Martino et al., 2007; Bowler et al., 2008). We detected a far more sensitive effect after the addition of DD without methanol compared to the same treatments in presence of methanol. Growth was inhibited when DD was supplied at 3.3 µm without methanol, whereas growth was apparently unaffected when DD was solubilized in methanol (Supplemental Fig. S1A). The chlorophyll a fluorescence normalized to cell concentration decreased to one-half the value of untreated cells, 3 h after addition of 33 µm DD without methanol and after addition of >100 µm DD in presence of methanol (Supplemental Fig. S1B). The Fv/Fm measure of PSII efficiency was one-half of that of untreated cells after addition of 3.3 µm DD in the absence of methanol, whereas a similar effect required 66 µm DD in presence of methanol (Supplemental Fig. S1C). Eventually, photochemical quenching capacity Y(II) was completely abolished after addition of 3.3 µm DD without methanol and after addition of 100 µm DD in presence of methanol (Supplemental Fig. S1D), indicating an interruption of electron transport to PSI. This functional disruption of photosynthesis could be due to a defect of components of the photosynthetic electron transport chain, like cytf or PSI, or of the Calvin cycle, or more simply to a nonspecific effect of DD triggering membrane disruptions and leakage. Together, these results show that methanol decreases the impact of DD on Phaeodactylum (Supplemental Fig. S1; Supplemental Table S1), very likely by promoting its volatility. The sensitivity of Phaeodactylum to DD was therefore higher than previously observed (Vardi et al., 2006, 2008), and was consistent with susceptibilities reported for other phytoplankton including other diatoms, that is, T. weissflogii and Skeletonema marinoi, with half-lethal doses between 0.1 and 2.2 µm DD (Ribalet et al., 2007). In addition, whereas the caspase/metacaspase inhibitor carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone had been reported to partly rescue Phaeodactylum treated with DD (Vardi et al., 2008), we could not detect any effect of carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone supplied at 20 µm on the impairment of Fv/Fm or Y(II) (Supplemental Fig. S1, E and F). These results are therefore consistent with the known nonspecific membrane-disrupting properties of PUAs (Casotti et al., 2005; Ribalet et al., 2007).
In Phaeodactylum, the cellular level of polyunsaturated fatty acids (mainly eicosapentaenoic acid) is about 2 nmol/106 cells in nutrient replete or nitrogen- or phosphorus-deprived media (Abida et al., 2015). Supposing that all eicosapentaenoic acid could be converted into PUAs, one should suppose that 1.65.107 to 3.3.107 Phaeodactylum cells per mL should be lysed to reach 33 and 66 µm PUAs (Vardi et al., 2006). The density of diatom cells required to reach micromolar concentrations of DD is therefore higher than that found in natural conditions, even after the formation of a bloom, with <105 cells/mL (Tiselius and Kuylenstierna, 1996). Consistently, the most recent survey of PUA levels in oceans shows that the highest concentration is in the nanomolar range (Ribalet et al., 2014). Thus, in our attempts to use DD as a physiological trigger of NO•, we could not confirm that it could act as a cell-to-cell specific signal in a submicromolar concentration range. Future works should therefore help clarify whether nanomolar concentrations of PUAs, without methanol, could effectively trigger any physiological response in Phaeodactylum.
The carboxy-2-phenyl-4,4,5,5-tetramethyl-imidazoline-1-oxyl-3-oxide (cPTIO) reagent donates an oxygen atom to generate NO2 and is often used to scavenge NO• and act as an antidote (Keszler et al., 2010). We sought whether DD-treated cells could be rescued by cPTIO, as previously reported (Vardi et al., 2008). Unexpectedly, Phaeodactylum cells proved to be sensitive to cPTIO, with more than one-third decrease of chlorophyll and a 20% decline in Fv/Fm after incubation with 100 μm cPTIO (dissolved in Enriched Seawater, Artificial Water [ESAW]; Supplemental Fig. S2; Supplemental Table S2). Therefore, cPTIO could be used neither to detect a possible involvement of NO• in DD response, nor to reverse any effect observed after NO• treatments. In addition, none of the NO• treatments reported in this study could suggest a role of NO in cell death.
A comparison of our study with previous works is summarized in Supplemental Tables S1 and S2. Taken together, we could not obtain results consistent with a specific DD → NO• → PCD cascade, but rather a caspase/metacaspase-independent nonspecific cell death triggered by high concentrations of DD. We reconsidered the question of the physiological role of NO•.
External Supply of NO• Impairs the Growth of P. tricornutum
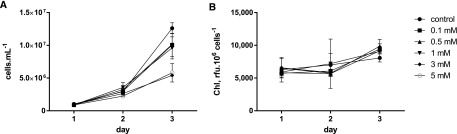
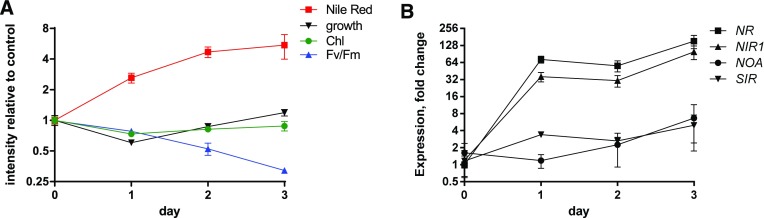
We used two different methods for supplying NO, either by providing NO• directly to the cells through the addition of an NO•-saturated solution or by adding a NO•-donor agent, S-Nitroso-N-acetylpenicillamine (SNAP). Nitroso-acetylpenicillamine (NAP), a structural analog of SNAP, which lacks the NO-moiety, was used as a control for SNAP experiments. We verified the production of NO• following SNAP dissolution by two methods. Firstly, using Membrane Inlet Mass Spectrometry (NRC Halifax Laboratories), we observed an increase of the NO• signal (m/z = 30) with increasing doses of SNAP, up to a concentration of 600 µm (Supplemental Fig. S3A). Secondly, using the DAF-FM probe, we detected a signal in <30 min following dilution (Supplemental Fig. S3B), showing the efficiency of this NO• donor in our experimental design. Both NO• donated by SNAP (Fig. 1A) or supplied directly as a gas (not shown) slowed down the growth of Phaeodactylum, as previously reported (Vardi et al., 2006, 2008). A concentration of 1 mm SNAP reduced growth by only 20% and was selected for further experiments. In contrast to DD, SNAP treatment did not affect chlorophyll levels (Fig. 1B).
Figure 1.
Effect of NO• supplied by SNAP on the growth of Phaeodactylum. The incubation was performed in a volume of 500 µL, inoculated at 106 cells · mL−1, with immediate addition of SNAP. A, Cell concentrations. Cell concentrations were estimated using a Malassez counting chamber. B, Chlorophyll content. Spectrophotometric measurement of fluorescence was performed at 680 nm at room temperature using 106 cells.
External Supply of NO• Triggers the Accumulation of TAG within P. tricornutum Cells
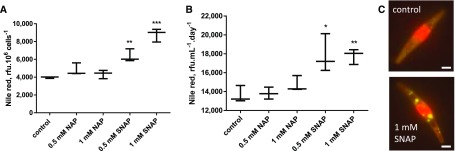
Cell division arrest following general stress responses is known to coincide with the accumulation of TAG (Kohlwein, 2010; Meï et al., 2017). We analyzed the accumulation of TAG droplets in Phaeodactylum by Nile Red staining. Following exposure to NO•, we did not see any evidence of chloroplast disintegration or of cell death. We observed that a 2-d incubation with 1 mm SNAP in a 500-µL culture volume triggered a 2.2-fold increase of TAG per cell (Fig. 2A) and a >2-fold increase of TAG productivity, corresponding to the level of TAG per volume of culture and per day (Fig. 2B). Interestingly, upon nitrogen starvation, two large lipid droplets become visible in Phaeodactylum, flanking the chloroplast (Abida et al., 2015); in SNAP-treated cells, numerous lipid droplets of variable sizes can be observed (Fig. 2C).
Figure 2.
Effect of increasing concentrations of SNAP on the production of TAG in P. tricornutum. The incubation was performed in a volume of 500 µL, inoculated at 106 cells · mL−1, with immediate addition of chemicals. Measurements were performed after 2 d of incubation. A, Effect of increasing concentrations of SNAP on TAG level per cell. TAG level per cell were measured using Nile Red staining and is given in relative fluorescence units per 106 cells. Stars indicate statistical relevant treated compared to untreated samples, with P values of 5.10−3 and 4.10−4 for 0.5 mm SNAP and 1 mm SNAP, respectively. B, Effect of increasing concentrations of SNAP on TAG productivity. TAG productivity is given in Rfu corresponding to the fluorescence of Nile Red per milliliter and per day. Statistically relevant responses are indicated by stars after treatments with 0.5 mm SNAP (P value 3.10−2) and 1 mm SNAP (P value 4.10−4). C, Epifluorescence images of Nile red stained treated and untreated cells. Bar, 5 µm.
We sought to determine whether this effect observed following an external supply of NO• could be also attributed to NO• as an internal signal, enzymatically produced by the diatom itself. To that purpose, we reexamined the question of the endogenous production of NO•.
Biosynthesis of NO• in Phaeodactylum Is Arg Independent and Nitrite Dependent
Data from previous studies on NO• production in Phaeodactylum are confusing. On the one hand, it was shown that NO• production was dependent on Arg (Vardi et al., 2006) and authors concluded on the action of a NOS-like enzyme. Intriguingly, it was not possible to detect any NOS-like sequence in the genome of P. tricornutum (Di Dato et al., 2015).
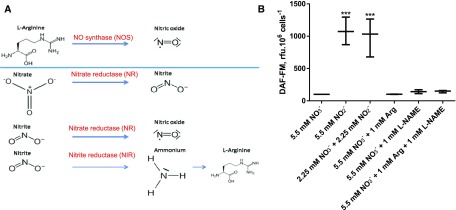
On the other hand, an ortholog of the plant chloroplast NOA protein was identified (Vardi et al., 2008). In Arabidopsis (Arabidopsis thaliana), AtNOA acts in NO• production, but the protein does not possess any NOS function (Moreau et al., 2008). AtNOA1 and NR are responsible for the majority of NO• release (Gas et al., 2009; Moreau et al., 2010). The mode of action of AtNOA1 remains elusive, although it might be involved in the regulation of NR activity (Jin et al., 2009; Mandal et al., 2012). In this NR-dependent pathway, NO• is produced by reduction of nitrite (NO2−) by NR (Fig. 3A). Nitrite is reduced to ammonia (NH3) by the NIR, and ammonium is a substrate for the biosynthesis of Arg. Therefore, addition of Arg could have a negative feedback regulation on NIR, thereby providing high nitrite levels for NR-dependent NO• production (Fig. 3A). This could explain a positive action of an Arg supply on NO• production via NR and in the absence of any NOS. One way to make the distinction between NO• produced from Arg via a NOS or via a NR is to detect an inhibition of the NOS pathway by a blocking agent, for example, L-NAME (Nω-nitro-L-Arg methyl ester) in cofeeding experiments with Arg.
Figure 3.
Biosynthesis of NO• in Phaeodactylum is Arg-independent and nitrite dependent. A, Different pathways for NO• production in microorganisms. Two general pathways exist in aerobic conditions, either by a NOS pathway using Arg as substrate (top) or by a side activity of NR using nitrite as substrate (bottom). Nitrite reduction generates ammonium, which can be incorporated into amino acids, including Arg (lowest reactions). It is thus possible that a supply of Arg exerts a negative feedback, leading to nitrite accumulation and a NR-dependent production of NO•. B, Detection of NO• in Phaedocatylum based on nitrogen sources. Cells were harvested from complete media and inoculated in nitrogen-free media at 2.106 cells ·mL−1. DAF-FM was added at a concentration of 10 µm. After dark incubation and washing, cells were collected into 500-µL culture aliquots and supplemented with the indicated nitrogen sources: nitrate (NO3−, standard medium), nitrite (NO2−), 1 mm Arg, and the NOS-blocking agent l-NAME. DAF-FM fluorescence was recorded after 3 h of incubation. ***P < 10−4.
We therefore analyzed the endogenous level of NO• in Phaeodactylum culture in presence of the two potential substrates, Arg and NO2−. The detection of NO• in Phaeodactylum was much higher when the diatoms were cultured in presence of NO2−, either in the presence or absence of NO3− (Fig. 3B), supporting a production via NR. By contrast, addition of Arg did not impact on the production of NO•. Similarly, addition of L-NAME did not lead to any significant change, when cells were cofed with Arg (Fig. 3B; Supplemental Table S2).
Taken together, our results are consistent with the absence of any NOS activity and with the presence of a unique NR-dependent production of NO• in Phaeodactylum. This NR-dependent pathway is conserved in photosynthetic eukaryotes from distant clades, such as Arabidopsis (Moreau et al., 2010; Sanz-Luque et al., 2015), the red alga Gracilaria chilensis (Chow et al., 2013), and the green alga Chlamydomonas (Wei et al., 2014).
NOA Overexpression Correlates with an Increased Endogenous Production of NO• by Phaeodactylum
We reexamined the role of the NOA protein in the endogenous production of NO• and designed genetic constructs for NOA overexpression. Genomic DNA extracted from P. tricornutum Pt1 strain was used as substrate, and a 2,352-bp sequence was amplified by PCR using oligonucleotides designed from Phatr2_56150 (Vardi et al., 2008), now referenced as Phatr3_J40200. The NOA PCR product was cloned into the pH4 vector (De Riso et al., 2009), transformed into P. tricornutum wild-type cells, and we obtained overexpressing lines following resistance to Zeocin selection, including NOAOE4 and NOAOEf generated by two independent series of transformation experiments. The RNA level of the NOA gene was more than 30-fold induced in the two independent overexpressing lines and led to an elevated NO• production (Supplemental Fig. S4), which was consistent with previous reports (Vardi et al., 2008). We thus confirmed that NOA played a role in NO• production and used these overexpressing lines as models for the analysis of the physiological role of NO• in Phaeodactylum. We obtained similar results with both independent overexpressing lines and present below the results obtained with NOAOE4.
NOA Overexpression Activates the Transcription of the Plastid Ferredoxin-Nitrite Reductase
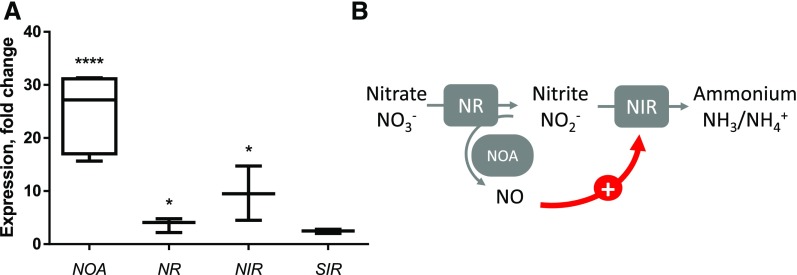
In plants, the precise molecular function of NOA is unresolved, but some reports suggest an interplay with nitrogen assimilation (Yamasaki and Sakihama, 2000; Moreau et al., 2010). NO• production was also correlated with a transcriptional control of nitrogen assimilation genes, as described for Chlamydomonas (de Montaigu et al., 2010). We sought whether such transcriptional control could exist in Phaeodactylum. We analyzed by quantitative reverse transcription PCR (RT-qPCR) the expression level of the genes coding for the cytosolic NR (Phatr3_J54983), the plastid ferredoxin-dependent nitrite reductase, NIR (Phatr3_J12902), and the sulfite reductase SIR (Phatr3_J9538), used as a nitrogen-independent negative control. In nutrient-replete conditions (10× ESAW), the NIR gene was highly induced in NOAOE4, whereas NR expression was slightly increased and that of SIR did not change significantly, compared to the respective wild type and cells transformed with the empty vector (Fig. 4A). A nitrite-sensing module can therefore be proposed based on a positive regulatory loop triggering NIR transcription downstream of NR-dependent transient NO• emission (Fig. 4B).
Figure 4.
Effect of NOA overexpression on the expression level of nitrogen assimilation genes in nutrient-replete condition. A, NOA overexpression activates the transcription of the plastid ferredoxin-nitrite reductase. RNA was extracted from a 108-cell pellet of NOAOE4, wild type, and pH4 and reversely transcribed. Quantitative real time PCR was conducted on 20 ng cDNA using oligonucleotides binding TUB as internal control and NOA, NR, NIR, and SIR as genes of interest. Quantification cycles of NOAOE4-derived cDNA were normalized to wild-type and pH4 values. Significant changes were observed on the level of NOA (P < 10−4), NR (P = 1.25.10−2), and NIR (P = 2.10−2). B, Nitrite-sensing module. SIR, sulfite reductase.
NOA Is Not Involved in the Onset of the Response of Phaeodactylum to Nitrogen Starvation
Since Phaeodactylum increases NR expression levels when nitrogen is limited (Levitan et al., 2015a; Yang et al., 2016), and since NR expression was slightly increased in the NOAOE background, we sought to determine whether NOA was also responsive to nitrogen availability. In a 3-d time course of cells shifted to nitrogen-depleted conditions, the typical nitrogen starvation responses occurred, that is, reduction of Fv/Fm, chlorophyll fluorescence, and cell growth and induction of neutral lipids as measured by Nile Red staining (Fig. 5A). We confirmed a rapid, strong, and parallel induction of NR and NIR in response to nitrogen starvation (Fig. 5B). SIR and NOA responded to general nutrient deprivation with a late induction pattern, reaching a maximal 4- to 6-fold induction after 3 d of cultivation (Fig. 5B). Thus, although in nutrient-replete culture we had observed the overexpression of NOA correlated with higher NR and NIR levels, under nitrogen starvation the regulation of these genes is uncoupled. This is in contrast with Chlamydomonas, in which NO• was shown to be produced from cytosolic nitrite and act in the orchestrated response upon nitrogen shortage (Miller et al., 2010; Wei et al., 2014). In Phaeodactylum, NO• does not appear as a mediator in the onset of the nitrate deprivation response.
Figure 5.
Expression of NOA, NR, NIR, and SIR in nitrogen-depleted wild-type Phaeodactylum cells. Cells were harvested and resuspended in nitrogen-free media in 100-mL cultures a concentration of 106 cells · mL−1. Each day, a 20-mL culture aliquot was harvested for RNA extraction and a 300-µL culture aliquot was used for physiological measurements. A, Physiological parameters. Chl was measured by the absorption at 680 nm at room temperature, Nile red fluorescence was quantified to indicate neutral lipid contents, Fv/Fm was measured using fast chlorophyll fluorescence kinetics, and cell concentrations were estimated via the absorption at 730 nm. B, NOA, NR, NIR, and SIR gene expression. RNA was extracted from a cell pellet and reversely transcribed. Quantitative real-time PCR was conducted on 20 ng cDNA using oligonucleotides binding TUB as internal control and NOA, NR, and NIR as genes of interest. The SIR gene was used as a nitrogen-unrelated control. Data are normalized with value measured with cells harvested before the shift. Data are the results of three independent biological replicates.
Altogether, these results support a role of NOA and NO• in a fine tuning of the regulation of the nitrogen assimilation system more selective for nitrite over nitrate (uncoupled and higher up-regulation of NIR over NR), but this NO•-mediated pathway does not operate during nitrogen starvation, where a strong and parallel up-regulation of NR and NIR is triggered. We sought therefore to characterize the extent of the specific transcriptional changes induced by NO• at the whole genome scale.
Whole Genome Transcriptome Analysis in Response to Increasing Doses of NO• Highlights a Role in Nitrite Sensing, High-Nitrite Adaptation, and Reprogramming of Lipid Synthesis
We analyzed the effects of increasing doses of NO• on the whole transcriptome of Phaeodactylum. To that purpose, we supplied NO• from a saturated solution at concentrations of 3 and 10 µm. Of the 12,393 referenced genes, reads were obtained for 11,464. We found 804 genes differentially expressed with a |Log2(fold change)| > 1 in at least one of the contrasts (i.e. comparing NO• supplies at 0 µm vs. 3 µm, 0 µm vs. 10 µm or 3 µm vs. 10 µm), and with P value < 0.05. A partition of differentially expressed genes was performed using a K-mean method, with the number of partitions set to 6 (Liu et al., 2014). Each group or cluster consisted of genes with similar expression profiles following treatments with NO• (Supplemental Fig. S5; Supplemental Table S3). Two clusters were comprised of genes down-regulated following NO• treatments, that is, group 1 (DR1; 55 genes) containing genes with the strongest magnitude in decreased expression (in the -2 to -4 Log2FC range) and group 2 (DR2; 282 genes) with moderate but significant decreased expression (Log2FC approximately −1). Likewise, two clusters were comprised of genes up-regulated following NO• treatments, that is, group 6 (UR1; 54 genes) containing genes with the strongest magnitude in expression increase (in the +2 to +4 Log2FC range) and group 3 (UR2; 291 genes) with moderate but significant increased expression (Log2FC approximately +1; Supplemental Fig. S5; Supplemental Table S3). For each group, we sought whether gene ontology (GO) terms could be enriched, either by the DAVID method (http://david.abcc.ncifcrf.gov; Huang et al., 2007; Supplemental Table S4) or using the GOseq R package (Young et al., 2010; Supplemental Table S5). We then mined information in the groups corresponding to a differential expression that could be correlated with the dose, that is, DR1, DR2, UR1, and UR2.
Focusing on genes up-regulated by NO•, we examined the possible metabolic pathways or cellular processes responsible for this reprogramming. Clearly, GO terms and metabolic pathways corresponding to nitrogen assimilation, glycolysis, phosphoenolpyruvate (PEP) production, pyruvate (Pyr) production, acetyl-CoA production, and glycerolipid production are enriched in UR1 and UR2 (in bold characters and underlined in Supplemental Tables S4 and S5). More specific GO terms retrieved concerning nitrogen metabolism include the carbamoyl synthase activity, arginosuccinate synthetase activity, Gln synthesis, and Arg synthesis. These terms highlight a possible activation of two pathways generating amino acids, that is, the Gln synthase/Gln oxoglutarate aminotransferase (GS/GOGAT) route producing Gln and Glu and the Orn-urea pathway. More specific GO terms retrieved concerning carbon metabolism include glycolysis, phosphopyruvate hydratase activity, phosphoglycerate kinase activity, and lipid metabolism.
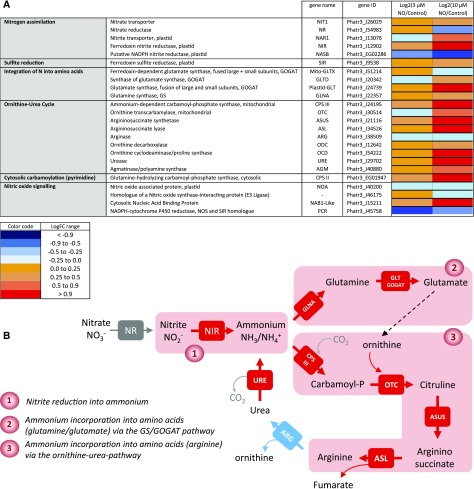
We thus examined the corresponding metabolic pathways in detail. Figure 6 shows the dose-dependent response of genes involved in nitrogen (nitrate, nitrite, ammonium) assimilation, incorporation into amino acids, and entry into the Orn-urea pathway, which was shown to be a specific metabolic feature of diatoms (Allen et al., 2011). The most evident response to low-dose treatments with NO• among the set of genes known to be involved in nitrogen assimilation is the specific expression of the gene coding for the plastid NIR protein, reducing NO2− into NH3/NH4+ (Fig. 6A). We included in this analysis the gene putatively coding for the large subunit of a prokaryotic-type NADPH-dependent nitrite reductase (NASB; Allen et al., 2011; Levitan et al., 2015b), which is not transcriptionally modified in the high-nitrite adaptive response shown here (Fig. 6A). Genes coding for enzymes involved in nitrogen incorporation into amino acids via the GS/GOGAT route (Guerra et al., 2013) are up-regulated starting with the incorporation of NH3 into Gln by GLNA, followed by the production of Glu by the Glu synthase systems, Mito-GLTX, Plastid-GLT, and GLTD. This up-regulated pathway is partly localized in the plastid, where NIR also resides (Fig. 6).
Figure 6.
Dose-response expression of nitrogen assimilation genes following treatment with increasing concentrations of NO•. A, Gene differential expression after treatments with NO•. Phaeodactylum cells were treated with 0 (control), 3, and 10 µm NO• using a calibrated NO•-saturated aqueous solution and after 96 h of NO• treatment, RNA was extracted and subjected to a RNAseq analysis. Libraries for each sample were prepared for stranded 100-bp paired end sequencing and samples were combined and analyzed in a single Illumina HisEquation 2000 lane. Reads were mapped on the most recent genome version of Phaeodactylum tricornutum (http://protists.ensembl.org/Phaeodactylum_tricornutum). Annotated genes coding for nitrogen assimilation were then used to probe changes in three metabolic modules, that is, nitrogen assimilation, integration of nitrogen into amino acids, and integration of nitrogen via the Orn-urea pathway (or cycle). NOA expression was also examined. Eventually, genes with some sequence similarity with genes acting in NO• signaling in other organisms were also included, although this function seems unlikely based on our study. B, Pathways of nitrogen assimilation and incorporation into amino acids up-regulated by NO•.
Genes encoding the Orn-urea pathway are clearly and coordinately activated, to the notable exception of one of its components, the arginase gene, ARG (Fig. 6A). Firstly, the mitochondrial carbamoyl phosphate synthase, CPS III, combining CO2 and NH4+ into carbamoyl phosphate, is up-regulated. Following this capture of ammonium, carbamoyl phosphate is combined with Orn by the OTC, forming citrulline, then combined with Asp by the argininosuccinate synthase, ASUS. Argininosuccinate is broken down into Arg and fumarate by the argininosuccinate lyase, ASL. Whereas CPS III, OTC, ASUS, and ASL are all activated, ARG encoding the enzyme regenerating Orn and producing urea is unchanged, even slightly down-regulated. The expression level of ARG in control conditions, evaluated by DESeq2-normalized counts (in a total of 11,465 genes; Supplemental Table S3), is not low (approximately 1,800). Nevertheless, it is lower than that of CPS III (approximately 6,443) that reaches very high expression levels following NO• treatments (>18,000; Supplemental Table S3). A similar specific decrease of ARG expression compared to the other genes of the Orn-urea cycle has been reported previously in Phaeodactylum cells, following a shortage in phosphate (Yang et al., 2014). The so-called Orn-urea cycle is therefore likely not cyclic, interrupted, or highly slowed down at the level of Arg (Fig. 6B) and it appears therefore as a system mainly tuned to capture NH4+, without driving the production of urea. Consistently, the gene encoding the urease, URE, is also activated in parallel with the others, breaking down urea back into NH4+ and CO2. Again, a similar activation of URE has been previously observed upon phosphate shortage (Yang et al., 2014). Orn, which needs to be recycled to allow this pathway to operate, can alternatively derive from Glu and acetyl-CoA, produced by an acetyl-Orn deacetylase. Two proteins containing zinc-peptidase-like domains in Phaeodactylum (Phatr3_J15083 and Phatr3_J18404) might possibly produce Orn by this pathway, but this function needs to be demonstrated. Taken together, all genes of the Orn-urea pathway seem to be mobilized to trap ammonium produced by NIR and use it to produce Arg.
Interestingly, exogenous NO• does not trigger the up-regulation of NOA (Fig. 6A). We sought sequences in the genome of Phaeodactylum, which might be homologous to genes involved in NO• signaling in other eukaryotes. As mentioned above, no NOS gene could be identified (Di Dato et al., 2015). Genes coding for a putative NOS-interacting protein and an NADPH-cytochrome P450 NOS homolog have been found, but their annotation is based on partial sequence similarities and they do not respond to the NO• treatment (Fig. 6A). Thus, our analysis could not lead us to identify any component acting in the synthesis of NO• besides NOA.
Recently, NO• has been shown to nitrosylate an RNA-binding protein in the cytosol of Chlamydomonas, NAB1, which represses the translation of some mRNAs of photosynthetic components such as LHC II isoforms (Berger et al., 2016). A NAB1-like sequence was identified, lacking the nitrosylated domain identified in the Chlamydomonas sequence; nevertheless, the NAB1-like gene appeared as up-regulated by the treatment.
In this reprogramming, the NO2− → NH3/NH4+ → amino acid route is uncoupled from nitrate assimilation and activates both the GS/GOGAT and the Orn-urea pathway as the two gates to incorporate NH3/NH4+ (Fig. 6B). The GS/GOGAT route alone was reported previously to be a key regulatory site for the nitrate-dependent control of carbon partitioning between protein and lipid biosynthesis (Guerra et al., 2013). Lipid should therefore not accumulate. By contrast, because the ARG gene is uncoupled from the general up-regulation of the Orn-urea pathway, fumarate is expected to accumulate and feed carbon metabolism via malate. Based on the GO enrichment analysis, the PEP/Pyr hub and acyl-lipid metabolism were highlighted in differentially expressed genes. We thus examined the acyl-lipid anabolism and catabolism in more details, using a table of genes annotated for this purpose (Supplemental Table S6).
The most striking feature in carbon metabolism reprogramming is the coordinated up-regulation of the pathway synthesizing PEP, Pyr, and acetyl-CoA in the plastid (Supplemental Table S6). These include the gene coding for the plastid glyceraldehyde-3-phosphate dehydrogenase, Plastid-GAP (Phatr3_J22122), the phosphoglycerate kinase, Plastid-PGK (Phatr3_J29157), and the enolase, Plastid-ENOL (Phatr3_J41515), leading to the synthesis of PEP. The Pyr kinase, Plastid-PKP (Phatr3_J22404), leading to the production of Pyr, and two genes coding for dihydrolipoamide acetyltransferase E2 components of the Pyr dehydrogenase complex, Plastid-DHLATa and Plastid-DHLATb (Phatr3_J23850 and Phatr3_EG02309), are up-regulated, acting in the direction of a production of acetyl-CoA (Supplemental Table S6). Based on this transcriptome reprogramming, carbon metabolism, including the excess of fumarate generated by the activated Orn-urea cycle, can converge toward a production of acetyl-CoA.
Downstream of acetyl-CoA, the activation of fatty acid and TAG biosynthesis is not obvious based on the transcriptomic response to NO• (Supplemental Table S6). The production of a diacyl moiety in the plastid appears slightly stimulated via a coordinated up-regulation of a glycerol-3-phosphate acyltransferase, ATS1 (Phatr3_J3262), and two 1-acylglycerol-3-phosphate acyltransferases, ATS2a and ATS2b (Phatr3_J11916 and Phatr3_J43099). The production of TAG could be sustained by a moderate up-regulation of an ER localized acyl-CoA:diacylglycerol acyltransferase, DGAT2B (Phatr3_J49544). Genes coding for enzymes involved in TAG and fatty acid degradation do not respond significantly to the NO• treatment (Supplemental Table S6).
The gene coding for the lipid droplet protein, LDP (Phatr3_J48859; Yoneda et al., 2016) is up-regulated as an unambiguous marker of TAG accumulation. In addition, two lipases that could possibly degrade TAG are down-regulated, that is, a TAG lipase gene, TAGL-Like-B (Phatr3_J48799 and a phospholipase A, PLA-A (Phatr3_J44005; Supplemental Table S6). An accumulation of TAG can therefore occur via a push/protect activated process, pushing at the level of acetyl-CoA production and to some extent fatty acid synthesis and protecting by producing LDP and preventing TAG hydrolysis.
An intriguing coordinated response was the clear up-regulation of fatty acid (FA) desaturase genes (Dolch and Marechal, 2015): the plastid ω6-acyl-lipid desaturase, FAD6 (Phatr3_J48423); the plastid Δ6-FA desaturase, Plastid-Delta6FAD (Phatr3_EG02619); the ER localized ω6-acyl-lipid desaturase, FAD2 (Phatr3_J25769); an ER localized Δ5-FA front-end desaturase, ER-delta5-FAD-A (Phatr3_J46830); and Δ6-FA front-end desaturase, ER-delta6-FAD (Phatr3_J29488; Supplemental Table S6). Following NO• treatment, it is likely that polyunsaturated FAs (PUFAs) become nitrated via NO•-derived radicals, forming NO2-FAs (Baker et al., 2009; Sánchez-Calvo et al., 2013; Fazzari et al., 2014). One should therefore expect that the treatment leads to a degradation of PUFAs, which might be compensated by the up-regulation of FA desaturase genes.
Taken together, the remodeling of lipid upon exposure to NO• seems not solely based on gene expression reprogramming, since only TAG accumulation could possibly be activated transcriptionally, whereas a compensating mechanism for a loss of very-long chain PUFAs (VLC-PUFAs) seems to be activated. We sought therefore to refine our analysis of lipid changes and advance in the possible mechanism of action of NO• in this remodeling.
NOA Induces a Specific Glycerolipid Remodeling in Phaeodactylum, Marked by a Change in the MGDG/TAG Balance
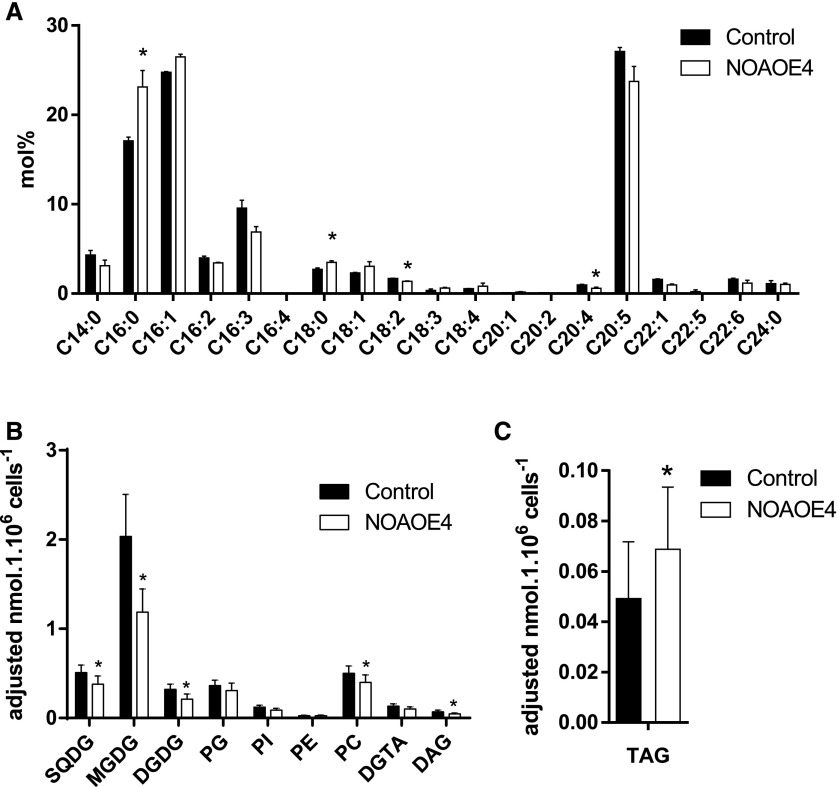
Comprehensive lipid analyses need cell amounts that could not be obtained with the treatment of wild-type cells with NO•. Using NOAOE lines as a model of a nontransient exposure to NO•, we could observe that in nutrient-replete conditions, the level of TAG per cell estimated by Nile Red staining was higher, consistent with a larger number of intracellular lipid droplets, as observed for SNAP-treated cells (Supplemental Fig. S6). We extracted lipids and performed fatty acid analyses and whole glycerolipidomic profiling. Consistent with the Nile Red measurements, the total fatty acid profile of NOAOE lines compared to the wild type indicated significantly higher 16:0 and 18:0 levels and an approximately 1.5-fold increase in TAG productivity (Fig. 7, A and C). By contrast, VLC-PUFAs displayed trends of reductions and accordingly the greatest sink for VLC-PUFAs, that is, chloroplast monogalactosyldiacylglycerol (MGDG), was significantly reduced in the overexpression lines (Fig. 7B). The distribution of fatty acids in the different lipid classes was reported previously (Abida et al., 2015) and was similar in this study. The fatty acid profiles in TAG and MGDG lipid classes were unaltered (Supplemental Fig. S7).
Figure 7.
Effect of NOA overexpression on total fatty acid and glycerolipid profiles in Phaeodactylum cells grown in nutrient-replete conditions. Three-day-old 50-mL cultures inoculated at 106 cells · mL−1 were harvested and lipids extracted. A, Total FA distributions. The FA distribution in total lipid extracts was based on gas chromatography coupled with flame ionization detection analyses of FAME. B, Membrane glycerolipid profiles. C, TAG content. Glycerolipid class abundances per million cells were assessed based on high pressure liquid chromatography-tandem mass spectrometry analyses. Data are the result of triplicate analyses. DGDG, digalactosyldiacylglycerol; DGTA, diacylglyceryl hydroxymethyltrimethyl-β-Ala, PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositide; SQDG, sulfoquinovosyldiacylglycerol. Stars indicate significant changes with P < 0.05.
According to the lipid profiles, the most affected lipids in NOAOE lines were MGDG and TAG. The increase in TAG is consistent with the transcriptomic analyses reported above, as evidenced by the coordinated up-regulation of genes involved in acetyl-CoA production, TAG biosynthesis, and the gene coding for the lipid droplet protein, LDP. Among the putative genes possibly involved in MGDG biosynthesis, MGD1, MGD2, and MGD3 (Phatr3_J54168, Phatr3_J9619, Phatr3_J14125), no read could be counted for MGD3 and the expression of MGD1 and MGD2 was not significantly altered upon NO• treatment (Supplemental Table S6). We sought to determine whether the decrease in galactolipids could occur by a direct effect of NO• on MGD activity.
Direct Inhibitory Effect of NO• on the Enzymatic Activity of Phaeodactylum MGDG Synthase
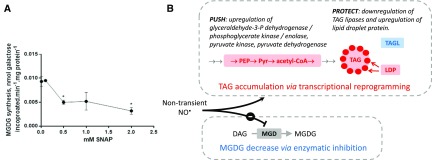
We used the enzymatic assay developed for plant MGDG synthase, providing radioactive uridine di-phosphogalactose (UDP-Gal) to Phaeodactylum protein extracts, and measured the incorporation of the radioactivity in galactolipids extracted by solvents, following incubation with increasing concentrations of SNAP (Fig. 8A). MGDG synthase activity was altered, with a 50% decrease following incubation with 0.5 mm SNAP. This result showed that NO• can affect MGDG synthases, likely by S-nitrosylation of the thiol groups known to be essential for the activity of this enzyme (Maréchal et al., 1994b, 1995). This enzymatic inhibition could be responsible for a decrease of plastid glycerolipid biosynthesis and thus contribute to the partial redirection of glycerolipid flux toward TAG (Fig. 8B).
Figure 8.
A, Effect of NO• on the MGDG synthase activity in Phaeodactylum membrane extracts. Activity was assayed based on the incorporation of Gal in MGDG, following incubation with radiolabeled UDP-[14C]Gal. Data are the results of three technical replicates. Stars indicate significant changes with P < 0.05. B, Lipid remodeling via transcriptional reprogramming and enzymatic regulation.
CONCLUSION
In summary, this study was motivated by evaluating the possible impact of environmental variations of the NO• level in oceans on phytoplankton, and more particularly diatoms. NO• was previously reported to be emitted by diatoms in response to high concentrations of PUAs such as DD and then to act in cell-to-cell communication, triggering PCD and allowing diatom population size control (Vardi et al., 2006, 2008; Bidle, 2015). We could not confirm this physiological response, in line with previous reports suggesting that DD triggered membrane leakage and nonspecific toxicity in Phaeodactylum (Ribalet et al., 2007). We do not exclude that a physiological response to DD might occur in the pico- to nanomolar range, which should be evaluated in the future, in the absence of methanol. Likewise, we could not confirm the previously reported production of NO• by a NOS-like activity in Phaeodactylum using Arg as a substrate (Vardi et al., 2006, 2008). Rather, our work shows that in Phaeodactylum, NO• is produced via a nitrite-dependent pathway, by a side activity of the NR, acting therefore in a nitrite-sensing system. The nitrite-sensing role of NO• is likely transient, since the assimilation of nitrite and its reduced form, ammonium, is coordinately up-regulated by NO• at the transcriptional level, therefore consuming nitrite and activating both amino acid biosynthesis and the Orn-urea pathway. One should keep in mind that not all diatoms lack an NOS gene (Di Dato et al., 2015) and that the nitrite-sensing system might be different in NOS-containing diatoms. The NO•-dependent remodeling of carbon metabolism seems to depend, at least partly, on the presence of the Orn-urea pathway, producing fumarate as a side-product. Our analyses are consistent with a role of NO• on the remodeling of glycerolipids redirecting carbon flux toward the production of TAG, at least partly via transcriptomic reprogramming, and the specific decrease of MGDG, by enzymatic inhibition of MGD activity. This specific feature of diatoms is consistent with previous studies showing that lipid production in P. tricornutum could be optimized when nitrate assimilation was inhibited and ammonium was the sole nitrogen source (Frada et al., 2013). We propose a revision of the physiological and ecophysiological role of NO• in diatoms, which appears related to the environmental nitrogen status and more specifically the level of nitrite. An anthropogenic increase of NO• in the environment could therefore alter nitrogen assimilation systems and act as an important stressor at the ecosystem level. Future works include the characterization of the interplay between nitrate, nitrite, and NO in diatoms involving NR, NIR, amino acid biosynthesis, Orn-urea pathway, and NOA; the functional characterization of these genes; and eventually a reevaluation of possible roles of NO• in the response to abiotic or biotic stresses.
MATERIALS AND METHODS
Chemicals
The chemicals used in the composition of growth media, the solvents as well as CHAPS, 1,2-dioleoyl-sn-glycerol (DAG), MOPS, NAP, SNAP, phosphatidylglycerol (PG), and unlabeled UDP-Gal were obtained from Sigma-Aldrich. The DAF-FM was purchased from Thermofisher Scientific. The diatom-derived aldehyde DD was obtained from Acros organics. [14C]labeled (11.0 GBq.mmol-1) UDP-Gal was obtained from New England Nuclear. The caspase/metacaspase inhibitor Z-VAD-FMK was purchased from Promega.
Cultivation of Phaeodactylum tricornutum
P. tricornutum (Pt1) Bohlin Strain 8.6 CCMP2561 (Culture Collection of Marine Phytoplankton, now known as NCMA: National Center for Marine Algae and Microbiota) was used in all experiments. Pt1 cells were maintained and grown in 20 mL or 50 mL at 20°C, in 250-mL flasks, in a modified ESAW medium (NaCl 362.7 mm; Na2SO4 25 mm; KCl 8.03 mm; NaHCO3 2.067 mm; KBr 0.725 mm; H3BO3 0.372 mm; NaF 0.0657 mm; MgCl2 47.18 mm; CaCl2 9.134 mm; SrCl2 0.082 mm; Na2-glycerophosphate 21.8 µm; Na2SiO3 105.6 µm; Na2EDTA 14.86 µm; Fe(NH4)2(SO4)2 5.97 µm; FeCl3 0.592 µm; MnSO4 2.42 µm; ZnSO4 0.254 µm; CoSO4 0.0569 µm; Na2MoO4 0.52 µm; H3BO3 61.46 µm; Na2SeO3 10 nm; biotin [vitamin H] 8.18 nm; cobalamin [vitamin B12] 2.94 nm; thiamine [vitamin B1] 0.594 µm; Falciatore et al., 2000), using either 10 times enriched nitrogen and phosphate sources (10× ESAW, containing 5.49 mm NaNO3 and 0.22 mm NaH3PO4; Abida et al., 2015) or the same medium without nitrogen. Cells were grown on a 12:12 light (30 μE m−2 · sec−1) / dark cycle. Cells were subcultured twice a week by inoculating 1 · 106 cells · mL−1 with fresh media. Growth was evaluated by cell counting using a Malassez counting chamber or by the absorption at 730 nm using a TECAN plate reader.
Incubation of Phaeodactylum with NO•-Saturated Solutions or NO• Donors
Incubation of P. tricornutum with gaseous NO• was performed by using NO•-saturated aqueous solutions. In this approach, a tank of pure NO• was bubbled into a small quantity of distilled water until the saturation point was reached, confirmed by mass spectrometric analysis of the solution. The concentration of NO• in a saturated solution was taken to be 1.9 mm (Gerrard, 1980). NO• saturates were then typically diluted 20- to 200-fold for use with live Phaeodactylum suspensions. Alternatively, an NO•-donor agent, SNAP, was used. This compound releases NO• when dissolved (Miller and Megson, 2007) and was therefore prepared freshly immediately before use. NAP was used as a nonactive compound for control experiments.
Incubation of Phaeodactylum with DD
Since DD is a highly toxic and volatile compound, all experiments were performed under a fume hood safety cabinet, and only freshly prepared DD solutions were used, either solubilized in pure methanol or in the growth medium. A 1-mL volume of a Phaeodactylum culture was inoculated at a cell density of 1.2 · 106 cell · mL−1 in 24-well plates, with different concentrations of DD in the absence or presence of methanol (final concentration 0.5%) during 3 h. For experiments with the caspase/metacaspase inhibitor, Z-VAD-FMK was used at a final concentration of 20 µm. Growth was quantified based on the absorption at 730 nm using a TECAN infinite M1000Pro plate reader.
Measure of NO Using DAF-FM, a Fluorescent Reporter Molecule
The DAF-FM allows the sensitive detection of low levels of nitric peroxide (ONOO−), which is in equilibrium with NO• and thus indicates NO• levels (St Laurent et al., 2015). DAF-FM was previously used to detect NO• levels in P. tricornutum cells (Vardi et al., 2008). Cultures were diluted to obtain 106 cells mL−1 in 10 mL, and cells were incubated under gentle shaking with 20 µL of 5 mm DAF-FM (in DMSO) for 1.5 h at room temperature and in the dark. Cells were washed and resuspended in 10 mL of fresh 10× ESAW medium. Aliquot fractions (500 µL) were transferred to a 48-well culture plate, to which SNAP was added as indicated. For the examination of DAF-FM-dependent detection of nitric peroxide, 150-µL samples were transferred into a 96-well plate and fluorescence was measured with a TECAN infinite M1000Pro plate reader (excitation wavelength at 488 nm, emission at 529 nm).
Chlorophyll Fluorescence Measurements
To determine photosynthesis parameters in cell cultures, room temperature fast chlorophyll fluorescence kinetics were measured using a Speedzen MX fluorescence imaging system (JBeamBio) with settings previously described (Johnson et al., 2009; Allorent et al., 2013). To this end, a 150-µL volume of P. tricornutum culture was transferred to a transparent 96 well-plate and dark-incubated for 15 to 30 min before measurements. Excitation was performed in the blue range (λ = 450 nm, F0). F0 is the steady-state fluorescence in dark-adapted cultures, Fm is the maximal fluorescence after a saturating light pulse with green light (520 nm) of dark-adapted cultures, Fm’ the same in light adapted cultures, and Fv is the difference between Fm and F0. With these parameters, the maximum efficiency of energy conversion of PSII can be calculated as Fv/Fm and photochemical quenching capacity indicated by the quantum yield of PSII as Y(II) = (Fm’ − F)/Fm’ (Butler and Kitajima, 1975; Misra et al., 2012).
Extraction and Determination of Chlorophyll a Level
A 700-µL culture of Phaeodactylum incubated in presence of various concentrations of DD was centrifuged for 3 min at 13,000g. The supernatant was removed and the pellet was suspended in 1 mL pure methanol. Samples were then centrifuged for 3 min at 13,000g, and the supernatant was used to measure A652 (A652), 665 nm (A665), and 750 nm (A750) using a Jasco V-650 spectrophotometer. Chlorophyll a level (Chl a) was determined following the equation Chl a = 16.29 (A665 − A750) − 8.54 (A665 − A750) (Porra et al., 1989).
Measure of TAG Accumulation by Nile Red Staining
Accumulation of TAG droplets was monitored by Nile Red (Sigma-Aldrich) fluorescent staining (excitation wavelength at 485 nm; emission at 525 nm) as previously described (Cooksey et al., 1987; Abida et al., 2015). In brief, cells were diluted and adjusted to a cell density that was linearly correlated with Nile Red fluorescence. Nile Red solution (40 μL of a 2.5 μg · mL−1 stock solution in DMSO) was added to 160-μL cell suspensions. Oil bodies stained with Nile Red were then visualized using a Zeiss AxioScope.A1 microscope (FITC filter; excitation wavelength at 488 nm; emission at 519 nm). The productivity, corresponding to the accumulation of TAG per volume and per time unit, was calculated based on the staining by Nile Red and expressed in relative fluorescence unit of Nile Red per milliliter and per day of incubation. Alternatively, Nile red fluorescence values were normalized to the cell concentration.
Genetic Construction for NOA Overexpression
Genomic DNA was extracted from P. tricornutum Pt1 cells using the following procedure. Firstly, 108 cells were harvested and frozen in liquid nitrogen. A volume of 20 µL of Edward-buffer (Tris-HCl 200 mm, pH 7.5; NaCl 250 mm; EDTA 25 mm; SDS 0.5%, w/v) was added, then samples were homogenized and debris removed by centrifugation. The supernatant was transferred to the same volume of isopropanol to precipitate DNA. After an additional 15-min centrifugation at 10,000g, the pellet was washed with ethanol 70%, dried, and solubilized in TE buffer (10 mm Tris-HCL, pH7, 1 mm EDTA). DNA concentration was measured using a Nanodrop 2000 spectrophotometer (Thermo Scientific). Using genomic DNA as matrix, a 2,352-bp sequence was amplified by PCR with the following oligonucleotides designed from Phatr2_56150 (Vardi et al., 2008) and carrying respectively XbaI and EcoRI restriction sites (underlined sequence): NOA-Fw XbaI 5′-TTTATCTAGAATGGTCCCCACTGGTTGTATG-3′, NOA-Rev EcoRI 5′-TTTAGAATTCCTAATTACGCCCTACACCTTTTCTTC-3′. The gene ID of NOA in the third version of the genome (http://protists.ensembl.org/Phaeodactylum_tricornutum) is Phatr3_J40200. PCR was performed using Phusion High Fidelity polymerase (Thermo Scientific) according to the manufacturer’s instructions. The PCR product was digested by EcoRI and XbaI, purified, and cloned in the linearized expression vector. The expression vector used for overexpression corresponds to the pH4-GUS vector (De Riso et al., 2009). The vector contains a gene coding for resistance to Zeocin (Shble), allowing selection of transformed cells. Expression of the NOA gene is controlled by the constitutive histone 4 promoter.
Transformation of Phaeodactylum and Selection of Strain Overexpressing NOA
Wild-type P. tricornutum cells were transformed via particle-bombardment under aseptic conditions (Kikkert, 1993) using a protocol described previously (Falciatore et al., 1999) modified as follows. A 3- to 4-d-old Pt1 culture was concentrated to 4.107 cells in 500 µL and spread onto a 1% agar-plate containing 1× ESAW medium. While shaking vigorously, 2 to 3 µg of nonlinearized plasmid was added to 25 µL ethanol-sterilized tungsten particles (Sigma), together with 25 µL of 2.5 m CaCl2 and 10 µL of 0.1 m spermidine. After mixing for 3 min using a vortex, the pellet was washed two times (1,500 g; 5 s; room temperature) with 700 µL precooled 100% ethanol. DNA-coated tungsten particles were then resuspended in 25 µL 100% ethanol. A 12-µL fraction of the mix was transferred onto a microcarrier, and bombardment was carried out using 1,550-psi rupture disks (BioRad). After 2 to 3 d of incubation under continuous illumination, cells were transferred to similar agar-plates containing 100 µg ∙ mL−1 Zeocin (Promega) for the selection of resistant transformed cells. Colonies appearing after 4 to 6 weeks were transferred to a new plate for 1 week, prior to inoculation of 20-mL liquid cultures.
Analysis of NOA Gene Expression in P. tricornutum Cells
To quantify the NOA mRNA level in overexpressing lines, quantitative PCR (qPCR) was performed after reverse transcription (RT) of extracted RNA (RT-qPCR). RNA was extracted from 107 cells that were previously pelleted, frozen in liquid nitrogen, and stored at −80°C until processing. A volume of 1 mL TriReagent (Sigma) was added to the frozen pellet and transferred to a new Eppendorf tube. After mixing for 30 s using a vortex, samples were incubated for 5 min at room temperature. Chloroform (200 µL) was added and tubes inverted and incubated for 15 min at room temperature. Phase separation was achieved by centrifugation (1,500g; 30 min; 4°C). The upper phase was transferred to a new tube, and RNA was precipitated using 1 volume isopropanol (1,500g; 30 min; 4°C), washed with 75% ice cold ethanol (1,500g; 5 min; 4°C), and the pellet was dried in a Speed Vac system (Eppendorf Concentrator 5301) prior to suspension in 30 µL diethyl dicarbonate-treated water (Sigma) at 65°C for 10 min. RNA was purified following a second ethanol precipitation using 1 volume of 5 m ammonium, acetate (2.5 m final concentration), and 1 volume isopropanol. Samples were incubated for 10 min on ice and centrifuged, washed, dried, and resuspended as above. Concentration was determined using a NanoDrop device (Thermo Scientific). The RNA obtained (1 µg) was used for RT after DNAse treatment (Qiagen) following manufacturer’s instructions to yield 1 µg cDNA, which was diluted to 10 ng · µL−1. For quantitative real time PCR, housekeeping gene oligonucleotides previously described (Siaut et al., 2007), namely RPS (5′-CGAAGTCAACCAGGAAACCAA-3′ and 5′-GTGCAAGAGACCGGACATACC-3′) and TUBA (5′-CTGGGAGCTTTACTGCTTGGA-3′ and 5′-ATGGCTCGAGATCGACGTAAA-3′), were used as internal controls. NOA-specific oligonucleotides were 5′-CCTGAAAAGTTCGCTACGCA-3′ and 5′-CGGATCCTTTTTGCCCTGAG-3′. The total qPCR reaction volume was 10 µL (120 nm oligonucleotide, 20 ng cDNA, 5 µL 2X SYBR Green Sso Advanced [Bio-Rad]). A two-step thermo-profile in 40 cycles was applied after 3 min at 95°C initial denaturation (95°C for 10 s; 58°C for 30 s) and a melt curve was detected (from 65°C to 95°C with a 0.5°C increment) (Bio-Rad CFX Connect Real-Time System). Evaluation of gene expression was carried out in three biological replicates, each one with technical triplicates, using the CFX Connect Real-Time System software, with TUBA and RPS as internal controls.
Lipidomic Profiling by Liquid Chromatography–Tandem Mass Spectrometry
Glycerolipids were extracted from freeze-dried P. tricornutum cells grown in 50 mL of medium. About 50 · 106 to 100 ·106 cells are required for a triplicate analysis. First, cells were harvested by centrifugation and immediately frozen in liquid nitrogen. Once freeze-dried, the pellet was suspended in 4 mL of boiling ethanol for 5 min to prevent lipid degradation, and lipids were extracted as described previously (Simionato et al., 2013) by addition of 2 mL methanol and 8 mL chloroform at room temperature. The mixture was then saturated with argon and stirred for 1 h at room temperature. After filtration through glass wool, cell debris was rinsed with 3 mL chloroform/methanol 2:1 (v/v), and 5 mL of 1% NaCl was then added to the filtrate to initiate phase separation. The chloroform phase was dried under argon before solubilizing the lipid extract in 1 mL of chloroform. Total glycerolipids were quantified from their FAs: in a 10-µL aliquot fraction, a known quantity of saturated 15-carbon FA (15:0) was added and all FAs were methanolyzed into methyl esters (FAME) by a 1-h incubation in 3 mL 2.5% H2SO4 in pure methanol at 100°C (Jouhet et al., 2003). The reaction was stopped by addition of 3 mL water, and 3 mL hexane was added for phase separation. After 20 min of incubation, the hexane phase was transferred to a new tube. FAMEs were extracted a second time via the addition, incubation, and extraction of another 3 mL hexane. The combined collected hexane fractions (6 mL) were argon-dried and FAMEs were suspended in 30 µL hexane for analysis by gas chromatography coupled with flame ionization detection (Perkin Elmer) using a BPX70 (SGE) column. FAMEs were identified by comparison of their retention times with those of standards (Sigma) and quantified by the surface peak method using 15:0 for calibration. Extraction and quantification were performed with at least three biological replicates. Glycerolipids were then analyzed and quantified by high pressure liquid chromatography-tandem mass spectrometry with appropriate standard lipids. The lipid extracts corresponding to 25 nmol of total FAs were dissolved in 100 µL of chloroform/methanol 2/1 (v/v) containing 125 pmol of each internal standard. Internal standards used were PE 18:0-18:0 and DAG 18:0-22:6 from Avanti Polar Lipid and sulfoquinovosyldiacylglycerol (SQDG) 16:0-18:0 extracted from spinach (Spinacia oleracea) thylakoid (Déme et al., 2014) and hydrogenated (Buseman et al., 2006). Lipids were then separated by HPLC and quantified by MS/MS. The HPLC separation method was adapted from a previously described procedure (Rainteau et al., 2012). Lipid classes were separated using an Agilent 1200 HPLC system using a 150-mm × 3-mm (length × internal diameter) 5-µm diol column (Macherey-Nagel) at 40°C. The mobile phases consisted of hexane/isopropanol/water/1 m ammonium acetate, pH 5.3 [625/350/24/1 (v/v/v/v)] (A) and isopropanol/water/1 m ammonium acetate, pH 5.3 [850/149/1 (v/v/v)] (B). The injection volume was 20 µL. After 5 min, the percentage of B was increased linearly from 0% to 100% in 30 min and kept at 100% for 15 min. This elution sequence was followed by a return to 100% A in 5 min and an equilibration for 20 min with 100% A before the next injection, leading to a total runtime of 70 min. The flow rate of the mobile phase was 200 µL/min. The distinct glycerophospholipid classes were eluted successively as a function of the polar head group. Mass spectrometric analysis was performed on a 6460 triple quadrupole mass spectrometer (Agilent) equipped with a Jet stream electrospray ion source under following settings: drying gas heater at 260°C, drying gas flow at 13 L · min−1, sheath gas heater at 300°C, sheath gas flow at 11 L · min−1, nebulizer pressure at 25 psi, capillary voltage at ±5000 V, and nozzle voltage at ±1000 V. Nitrogen was used as collision gas. The quadrupoles Q1 and Q3 were operated at widest and unit resolution respectively. Phosphatidylcholine and diacylglyceryl hydroxymethyltrimethyl-β-Ala analyses were carried out in positive ion mode by scanning for precursors of m/z 184 and 236 respectively at a collision energy (CE) of 34 and 52 eV. SQDG analysis was carried out in negative ion mode by scanning for precursors of m/z −225 at a CE of −56eV. PE, phosphatidylinositol (PI), phosphatidylglycerol, MGDG, and digalactosyldiacylglycerol measurements were performed in positive ion mode by scanning for neutral losses of 141 D, 277 D, 189 D, 179 D, and 341 D at CEs of 20 eV, 12 eV, 16 eV, 8 eV, and 8 eV, respectively. DAG and TAG species were identified and quantified by multiple reaction monitoring as singly charged ions [M+NH4]+ at a CE of 16 and 22 eV, respectively. Quantification was done for each lipid species by multiple reaction monitoring with 50 ms dwell time with the various transitions previously recorded (Abida et al., 2015). Mass spectra were processed using the MassHunter Workstation software (Agilent) for identification and quantification of lipids. Lipid amounts (pmol) were corrected for response differences between internal standards and endogenous lipids.
MGDG Synthase Enzymatic Assay
A culture of P. tricornutum (50 mL in 10× ESAW medium) was arrested in exponential phase and protein concentration determined using the Lowry method (Lowry et al., 1951). Cells were harvested by a centrifugation for 10 min at 1,500g at 4°C. The pellet was resuspended in 1 volume of 12 mm CHAPS and 20 mm MOPS-KOH, pH 7.8, and incubated for 20 min at 4°C to solubilize membrane proteins. Detergent solubilized proteins (10 µg) were then incubated for 1 h with variable concentrations of SNAP (from 0 to 2 mm, as indicated) at room temperature in the dark in a final volume of 66.5 µL. After incubation with SNAP, galactosyltransferase enzyme activity was assayed in mixed micelles at 25°C, as described previously (Maréchal et al., 1994a, 1994b). Phosphatidylglycerol (1.3 mm) and 1,2-dioleoyl-sn-glycerol (160 µm) dissolved in chloroform were first introduced into clean glass tubes. After evaporation of chloroform under a stream of argon, 300 µL of incubation medium adjusted to contain 10 µg proteins, 6 mm CHAPS, 250 mm KCl, 250 mm KH2PO4/K2HPO4, and 10 mm MOPS-KOH, pH 7.8, were added. The mixture was mixed vigorously and kept 1 h at 25°C for equilibration of mixed micelles. The reaction was then started by addition of 1 mm UDP-[14C]Gal (37 Bq · µmol−1) and stopped after 20 min by addition of chloroform/methanol (1:2, v/v). The lipids were subsequently extracted (Bligh and Dyer, 1959) and the radioactivity of the [14C]labeled MGDG produced, determined by liquid scintillation counting. Activity is expressed in nmol incorporated Gal · h−1 · mg protein−1.
RNAseq Analysis
An actively growing stock culture of P. tricornutum was used to inoculate duplicate 100-mL experimental cultures in 250-mL Erlenmeyer flasks at an initial cell density of 2.106 cells · mL−1. Using a calibrated saturated NO• solution, single additions at the start of the experiment were made to treated cultures to produce initial concentrations of either 3 or 10 µm NO•. Untreated cultures received no NO•. Cultures were allowed to grow for 4 d before harvesting cells by centrifugation at 5,000 rpm for 15 min at room temperature. After discarding the supernatant, cell pellets were flash frozen in liquid nitrogen and stored at −80°C before processing. In brief, cell aliquots were processed individually by grinding in liquid nitrogen in a mortar and pestle followed by extraction in RNA Pro lysis solution (MP Biomedicals) using a FastPrep homogenizer (two 40-s cycles, with power set at level 6, following manufacturer’s instructions). After centrifugation, the supernatant was recovered and extracted with chloroform; RNA was precipitated by addition of an equal volume of cold absolute ethanol and incubation at −20°C overnight. Precipitated RNA was recovered by centrifugation, washed in 75% ethanol, and air dried. The pellet was dissolved in RNase-free water and further purified using the clean-up protocol for the Qiagen RNeasy Mini Kit. RNA samples were quantified with a NanoDrop spectrophotometer and analyzed on an Agilent BioAnalyzer using the Plant RNA Nano program. Then 2.5 μg of each RNA was sent for RNA-Seq analysis at the McGill University and Genome Quebec Innovation Centre. Libraries for each sample were prepared for stranded 100-bp paired end sequencing, and samples were combined and analyzed in a single Illumina HisEquation 2000 lane. Reads were mapped on the most recent genome version of P. tricornutum (http://protists.ensembl.org/Phaeodactylum_tricornutum) using the Star (Spliced Transcripts Alignment to a Reference) method (Dobin et al., 2013; Engström et al., 2013). Data were filtered based on the detection of one read in at least one sample per treatment or genomic mutation and then normalized using the DESseq2 method (Varet et al., 2016). Only genes being differentially expressed with a |Log2(fold change)| >1 in at least one of the contrasts (i.e. comparing NO• supplies at 0 µm vs. 3 µm, 0 µm vs. 10 µm, or 3 µm vs. 10 µm), and with P < 0.05, were considered for further analyses. A partition of differentially expressed genes was performed using a K-mean method, with a number of partitions set to six and a clustering based on a Euclidian distance (Liu et al., 2014). For each group, we sought whether GO terms could be enriched, either by the DAVID method (http://david.abcc.ncifcrf.gov) (Huang et al., 2007), using the corresponding Refseq gene IDs and with a P value threshold set at 0.1, or using the GOseq R package (Young et al., 2010) with an identical P value threshold. Based on GO-enriched terms, a focused analysis of acyl-lipid pathways and of nitrogen assimilation was performed using a list of gene sequences with curated annotations.
Accession Numbers
Sequences from the Phaedoactylum genome analyzed more carefully in this study can be found via Ensembl! under the following accession numbers: Phatr3_EG01524; Phatr3_EG01947; Phatr3_EG01955; Phatr3_EG02208; Phatr3_EG02286; Phatr3_EG02309; Phatr3_EG02454; Phatr3_EG02496; Phatr3_EG02521; Phatr3_EG02619; Phatr3_J10068; Phatr3_J10497; Phatr3_J11319; Phatr3_J11390; Phatr3_J11811; Phatr3_J11916; Phatr3_J12420; Phatr3_J12431; Phatr3_J12642; Phatr3_J12740; Phatr3_J12884; Phatr3_J12902; Phatr3_J13073; Phatr3_J13076; Phatr3_J14202; Phatr3_J15031; Phatr3_J15211; Phatr3_J15730; Phatr3_J16364; Phatr3_J16376; Phatr3_J17086; Phatr3_J17720; Phatr3_J18029; Phatr3_J18064; Phatr3_J18940; Phatr3_J18940; Phatr3_J1971; Phatr3_J19979; Phatr3_J20143; Phatr3_J20310; Phatr3_J20342; Phatr3_J20460; Phatr3_J20508; Phatr3_J21116; Phatr3_J21201; Phatr3_J21988; Phatr3_J22122; Phatr3_J2215; Phatr3_J22274; Phatr3_J22357; Phatr3_J22404; Phatr3_J22459; Phatr3_J22510; Phatr3_J22554; Phatr3_J22677; Phatr3_J22803; Phatr3_J23639; Phatr3_J23850; Phatr3_J23913; Phatr3_J24195; Phatr3_J24610; Phatr3_J24739; Phatr3_J25769; Phatr3_J25932; Phatr3_J26029; Phatr3_J26714; Phatr3_J28009; Phatr3_J28068; Phatr3_J28652; Phatr3_J28797; Phatr3_J29157; Phatr3_J29488; Phatr3_J29702; Phatr3_J30113; Phatr3_J30145; Phatr3_J30282; Phatr3_J30514; Phatr3_J31367; Phatr3_J31440; Phatr3_J31492; Phatr3_J31662; Phatr3_J31994; Phatr3_J31994; Phatr3_J32083; Phatr3_J3262; Phatr3_J32902; Phatr3_J33198; Phatr3_J33435; Phatr3_J33720; Phatr3_J33864; Phatr3_J34485; Phatr3_J34526; Phatr3_J35240; Phatr3_J36821; Phatr3_J37086; Phatr3_J37367; Phatr3_J37372; Phatr3_J37652; Phatr3_J38509; Phatr3_J39710; Phatr3_J39949; Phatr3_J40163; Phatr3_J40200; Phatr3_J40261; Phatr3_J40880; Phatr3_J40988; Phatr3_J41423; Phatr3_J41515; Phatr3_J41570; Phatr3_J41886; Phatr3_J41969; Phatr3_J42398; Phatr3_J42683; Phatr3_J42872; Phatr3_J43010; Phatr3_J43099; Phatr3_J43116; Phatr3_J43320; Phatr3_J43352; Phatr3_J43463; Phatr3_J43469; Phatr3_J43678; Phatr3_J43773; Phatr3_J44005; Phatr3_J44028; Phatr3_J44066; Phatr3_J44231; Phatr3_J44401; Phatr3_J44584; Phatr3_J44645; Phatr3_J44806; Phatr3_J44831; Phatr3_J45223; Phatr3_J45510; Phatr3_J45518; Phatr3_J45551; Phatr3_J45758; Phatr3_J45947; Phatr3_J46175; Phatr3_J46453; Phatr3_J46570; Phatr3_J46595; Phatr3_J46830; Phatr3_J47389; Phatr3_J48423; Phatr3_J48664; Phatr3_J48778; Phatr3_J48799; Phatr3_J48859; Phatr3_J48977; Phatr3_J49163; Phatr3_J4918; Phatr3_J49339; Phatr3_J49462; Phatr3_J49524; Phatr3_J49544; Phatr3_J49771; Phatr3_J49867; Phatr3_J50356; Phatr3_J50742; Phatr3_J50770; Phatr3_J51214; Phatr3_J51454; Phatr3_J51519; Phatr3_J52268; Phatr3_J52368; Phatr3_J52648; Phatr3_J5271; Phatr3_J54017; Phatr3_J54068; Phatr3_J54151; Phatr3_J54168; Phatr3_J54222; Phatr3_J54494; Phatr3_J54528; Phatr3_J54709; Phatr3_J54974; Phatr3_J54983; Phatr3_J55069; Phatr3_J55111; Phatr3_J55153; Phatr3_J55157; Phatr3_J55192; Phatr3_J55209; Phatr3_J5527; Phatr3_J7164; Phatr3_J7678; Phatr3_J8663; Phatr3_J8860; Phatr3_J8975; Phatr3_J9255; Phatr3_J9316; Phatr3_J9538; Phatr3_J9619; Phatr3_J9709; Phatr3_J9794; Phatr3_Jdraft559.
SUPPLEMENTAL DATA
The following supplemental materials are available.
Supplemental Figure S1. Effect of DD on P. tricornutum cells after a 3-h treatment in presence or absence of methanol.
Supplemental Figure S2. Impairment of Phaeodactylum photosynthesis following treatment with cPTIO.
Supplemental Figure S3. Verification of NO• production by SNAP. A, Membrane inlet mass spectrometer detection of NO• (m/z = 30 amu).
Supplemental Figure S4. P. tricornutum NOA overexpressing lines produce more NO•. A, NOA mRNA level in cells transformed with the empty vector (pH4 (EV)) and the NOA overexpression construct.
Supplemental Figure S5. K-mean clustering of differentially expressed genes in response to increasing doses of NO•.
Supplemental Figure S6. Effect of NOA overexpression on the level of neutral lipids in Phaeodactylum cells.
Supplemental Figure S7. Fatty acid distribution in MGDG and TAG in NOAOE4 and wild type.
Supplemental Table S1. Comparison of the effects of DD on P. tricornutum obtained in this work with previous studies.
Supplemental Table S2. Comparison of the effects of various compounds on P. tricornutum obtained in this work with previous studies.
Supplemental Table S3. Phaeodactylum differentially expressed genes following treatments with increasing doses of NO.
Supplemental Table S4. Functional annotation enrichment in Phaeodactylum gene clusters differentially expressed following NO treatments using the DAVID method.
Supplemental Table S5. GOseq analysis of GO enrichment in Phaeodactylum gene clusters differentially expressed following NO• treatments.
Supplemental Table S6. Lipid gene expression following NO treatment.
Acknowledgments
The authors are grateful to Giovanni Finazzi and Dimitris Petroutsos for fruitful discussions and advices. The authors thank Colleen Murphy for conducting the RNA extractions used for the RNASeq experiments.
Footnotes
This work was supported by grants from Agence Nationale de la Recherche (ANR DiaDomOil), CEA (Irtelis PhD grant program), the CEA-Total partnership, Programme Investissement d’Avenir (Océanomics), and the National Research Council of Canada’s Algal Carbon Conversion Program.
References
- Abida H, Dolch LJ, Mei C, Villanova V, Conte M, Block MA, Finazzi G, Bastien O, Tirichine L, Bowler C, et al. (2015) Membrane glycerolipid remodeling triggered by nitrogen and phosphorus starvation in Phaeodactylum tricornutum. Plant Physiol 167: 118–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen AE, Dupont CL, Obornik M, Horak A, Nunes-Nesi A, McCrow JP, Zheng H, Johnson DA, Hu H, Fernie AR, et al. (2011) Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473: 203–207 [DOI] [PubMed] [Google Scholar]
- Allorent G, Courtois F, Chevalier F, Lerbs-Mache S (2013) Plastid gene expression during chloroplast differentiation and dedifferentiation into non-photosynthetic plastids during seed formation. Plant Mol Biol 82: 59–70 [DOI] [PubMed] [Google Scholar]
- Baker PR, Schopfer FJ, O’Donnell VB, Freeman BA (2009) Convergence of nitric oxide and lipid signaling: anti-inflammatory nitro-fatty acids. Free Radic Biol Med 46: 989–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger H, De Mia M, Morisse S, Marchand CH, Lemaire SD, Wobbe L, Kruse O (2016) A light switch based on protein S-nitrosylation fine-tunes photosynthetic light harvesting in chlamydomonas. Plant Physiol 171: 821–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidle KD. (2015) The molecular ecophysiology of programmed cell death in marine phytoplankton. Ann Rev Mar Sci 7: 341–375 [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, et al. (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456: 239–244 [DOI] [PubMed] [Google Scholar]
- Buseman CM, Tamura P, Sparks AA, Baughman EJ, Maatta S, Zhao J, Roth MR, Esch SW, Shah J, Williams TD, et al. (2006) Wounding stimulates the accumulation of glycerolipids containing oxophytodienoic acid and dinor-oxophytodienoic acid in Arabidopsis leaves. Plant Physiol 142: 28–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler WL, Kitajima M (1975) Fluorescence quenching in photosystem II of chloroplasts. Biochim Biophys Acta 376: 116–125 [DOI] [PubMed] [Google Scholar]
- Casotti R, Mazza S, Brunet C, Vantrepotte V, Ianora A, Miralto A (2005) Growth inhibition and toxicity of the diatom aldehyde 2-trans, 4-trans-decadienal on Thalassiosira weissflogii (Bacillariophyceae). J Phycol 41: 7–20 [Google Scholar]
- Chow F, Pedersen M, Oliveira MC (2013) Modulation of nitrate reductase activity by photosynthetic electron transport chain and nitric oxide balance in the red macroalga Gracilaria chilensis (Gracilariales, Rhodophyta). J Appl Phycol 25: 1847–1853 [Google Scholar]
- Cooksey KE, Guckert B, Williams SA, Callis PR (1987) Fluorometric determination of the neutral lipid content of microalgal cells using Nile Red. J Microbiol Methods 6: 333–345 [Google Scholar]
- De Martino A, Meichenin A, Shi J, Pan K, Bowler C (2007) Genetic and phenotypic characterization of Phaeodactylum tricornutum (Bacillariophyceae) accessions. J Phycol 43: 992–1009 [Google Scholar]
- Demé B, Cataye C, Block MA, Marechal E, Jouhet J (2014) Contribution of galactoglycerolipids to the 3-dimensional architecture of thylakoids. FASEB J 28: 3373–3383 [DOI] [PubMed] [Google Scholar]
- de Montaigu A, Sanz-Luque E, Galvan A, Fernandez E (2010) A soluble guanylate cyclase mediates negative signaling by ammonium on expression of nitrate reductase in Chlamydomonas. Plant Cell 22: 1532–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Riso V, Raniello R, Maumus F, Rogato A, Bowler C, Falciatore A (2009) Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res 37: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Dato V, Musacchia F, Petrosino G, Patil S, Montresor M, Sanges R, Ferrante MI (2015) Transcriptome sequencing of three Pseudo-nitzschia species reveals comparable gene sets and the presence of Nitric Oxide Synthase genes in diatoms. Sci Rep 5: 12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolch LJ, Marechal E (2015) Inventory of fatty acid desaturases in the pennate diatom Phaeodactylum tricornutum. Mar Drugs 13: 1317–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström PG, Steijger T, Sipos B, Grant GR, Kahles A, Ratsch G, Goldman N, Hubbard TJ, Harrow J, Guigo R, et al. (2013) Systematic evaluation of spliced alignment programs for RNA-seq data. Nat Methods 10: 1185–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu E, Gottschalk B, Charoensin S, Blass S, Bischof H, Rost R, Madreiter-Sokolowski CT, Pelzmann B, Bernhart E, Sattler W, et al. (2016) Development of novel FP-based probes for live-cell imaging of nitric oxide dynamics. Nat Commun 7: 10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falciatore A, Casotti R, Leblanc C, Abrescia C, Bowler C (1999) Transformation of nonselectable reporter genes in marine diatoms. Mar Biotechnol (NY) 1: 239–251 [DOI] [PubMed] [Google Scholar]
- Falciatore A, d’Alcala MR, Croot P, Bowler C (2000) Perception of environmental signals by a marine diatom. Science 288: 2363–2366 [DOI] [PubMed] [Google Scholar]
- Fazzari M, Trostchansky A, Schopfer FJ, Salvatore SR, Sanchez-Calvo B, Vitturi D, Valderrama R, Barroso JB, Radi R, Freeman BA, et al. (2014) Olives and olive oil are sources of electrophilic fatty acid nitroalkenes. PLoS One 9: e84884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler D, Coyle M, Skiba U, Sutton MA, Cape JN, Reis S, Sheppard LJ, Jenkins A, Grizzetti B, Galloway JN, et al. (2013) The global nitrogen cycle in the twenty-first century. Philos Trans R Soc Lond B Biol Sci 368: 20130164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frada MJ, Burrows EH, Wyman KD, Falkowski PG (2013) Quantum requirements for growth and fatty acid biosynthesis in the marine diatom Phaeodactylum tricornutum (Bacillariophyceae) in nitrogen replete and limited conditions. J Phycol 49: 381–388 [DOI] [PubMed] [Google Scholar]
- Gallardo L, Rhodes H (1997) Oxidized nitrogen in the remote pacific: the role of electrical discharges over the oceans. J Atmos Chem 26: 147–168 [Google Scholar]
- Gas E, Flores-Perez U, Sauret-Gueto S, Rodriguez-Concepcion M (2009) Hunting for plant nitric oxide synthase provides new evidence of a central role for plastids in nitric oxide metabolism. Plant Cell 21: 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard W. (1980) Gas solubilities widespread applications. Pergamon Press, Oxford, p 497
- Guerra LT, Levitan O, Frada MJ, Sun JS, Falkowski PG, Dismukes GC (2013) Regulatory branch points affecting protein and lipid biosynthesis in the diatom Phaeodactylum tricornutum. Biomass Bioenergy 59: 306–315 [Google Scholar]
- Holloway JM, Dahlgren RA (2002) Nitrogen in rock: occurrences and biogeochemical implications. Global Biogechem Cyc 16: 65-61–65-17. [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, et al. (2007) DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 35: W169–W175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC (2014) Clim Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. IPCC, Geneva, Switzerland [Google Scholar]
- Jin CW, Du ST, Zhang YS, Lin XY, Tang CX (2009) Differential regulatory role of nitric oxide in mediating nitrate reductase activity in roots of tomato (Solanum lycocarpum). Ann Bot 104: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Vandystadt G, Bujaldon S, Wollman FA, Dubois R, Roussel P, Alric J, Beal D (2009) A new setup for in vivo fluorescence imaging of photosynthetic activity. Photosynth Res 102: 85–93 [DOI] [PubMed] [Google Scholar]
- Jouhet J, Marechal E, Bligny R, Joyard J, Block MA (2003) Transient increase of phosphatidylcholine in plant cells in response to phosphate deprivation. FEBS Lett 544: 63–68 [DOI] [PubMed] [Google Scholar]
- Kampschreur MJ, Temmink H, Kleerebezem R, Jetten MSM, van Loosdrecht MCM (2009) Nitrous oxide emission during wastewater treatment. Water Res 43: 4093–4103 [DOI] [PubMed] [Google Scholar]
- Keszler A, Zhang Y, Hogg N (2010) Reaction between nitric oxide, glutathione, and oxygen in the presence and absence of protein: How are S-nitrosothiols formed? Free Radic Biol Med 48: 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkert JR. (1993) The Biolistic(R) Pds-1000 He Device. Plant Cell Tissue Organ Cult 33: 221–226 [Google Scholar]
- Kohlwein SD. (2010) Triacylglycerol homeostasis: insights from yeast. J Biol Chem 285: 15663–15667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Castellano I, Patti FP, Palumbo A, Buia MC (2015) Nitric oxide in marine photosynthetic organisms. Nitric Oxide 47: 34–39 [DOI] [PubMed] [Google Scholar]
- Levitan O, Dinamarca J, Hochman G, Falkowski PG (2014) Diatoms: a fossil fuel of the future. Trends Biotechnol 32: 117–124 [DOI] [PubMed] [Google Scholar]
- Levitan O, Dinamarca J, Zelzion E, Gorbunov MY, Falkowski PG (2015a) An RNA interference knock-down of nitrate reductase enhances lipid biosynthesis in the diatom Phaeodactylum tricornutum. Plant J 84: 963–973 [DOI] [PubMed] [Google Scholar]
- Levitan O, Dinamarca J, Zelzion E, Lun DS, Guerra LT, Kim MK, Kim J, Van Mooy BA, Bhattacharya D, Falkowski PG (2015b) Remodeling of intermediate metabolism in the diatom Phaeodactylum tricornutum under nitrogen stress. Proc Natl Acad Sci USA 112: 412–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Zhang S, Chen J, Jiang K, Zhang Q, Guo K, Liu Y (2014) The transcriptional profiling of glycogenes associated with hepatocellular carcinoma metastasis. PLoS One 9: e107941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the folin phenol reagent. J Biol Chem 193: 265–275 [PubMed] [Google Scholar]
- Mandal MK, Chandra-Shekara AC, Jeong RD, Yu K, Zhu S, Chanda B, Navarre D, Kachroo A, Kachroo P (2012) Oleic acid-dependent modulation of NITRIC OXIDE ASSOCIATED1 protein levels regulates nitric oxide-mediated defense signaling in Arabidopsis. Plant Cell 24: 1654–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal E, Block MA, Joyard J, Douce R (1994a) Comparison of the kinetic properties of MGDG synthase in mixed micelles and in envelope membranes from spinach chloroplast. FEBS Lett 352: 307–310 [DOI] [PubMed] [Google Scholar]
- Maréchal E, Block MA, Joyard J, Douce R (1994b) Kinetic properties of monogalactosyldiacylglycerol synthase from spinach chloroplast envelope membranes. J Biol Chem 269: 5788–5798 [PubMed] [Google Scholar]
- Maréchal E, Miege C, Block MA, Douce R, Joyard J (1995) The catalytic site of monogalactosyldiacylglycerol synthase from spinach chloroplast envelope membranes. Biochemical analysis of the structure and of the metal content. J Biol Chem 270: 5714–5722 [DOI] [PubMed] [Google Scholar]
- Martens-Habbena W, Qin W, Horak RE, Urakawa H, Schauer AJ, Moffett JW, Armbrust EV, Ingalls AE, Devol AH, Stahl DA (2015) The production of nitric oxide by marine ammonia-oxidizing archaea and inhibition of archaeal ammonia oxidation by a nitric oxide scavenger. Environ Microbiol 17: 2261–2274 [DOI] [PubMed] [Google Scholar]
- Meï CE, Cussac M, Haslam RP, Beaudoin F, Wong YS, Marechal E, Rébeillé F (2017) C1 metabolism inhibition and nitrogen deprivation trigger triacylglycerol accumulation in Arabidopsis thaliana cell cultures and highlithgt a role of NPC in phosphatidylcholine-to-triacylglycerol pathway. Front Plant Sci doi: 10.3389/fpls.2016.02014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski G, Bhattachary SK, Girsh G (2014) NOx cycle and the tropospheric ozone isotope anomaly: an experimental examination. Atmos Chem Phys 14: 4935–4953 [Google Scholar]
- Miller MR, Megson IL (2007) Recent developments in nitric oxide donor drugs. Br J Pharmacol 151: 305–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R, Wu G, Deshpande RR, Vieler A, Gartner K, Li X, Moellering ER, Zauner S, Cornish AJ, Liu B, et al. (2010) Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol 154: 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralto A, Barone G, Romano G, Poulet SA, Ianora A, Russo GL, Buttino I, Mazzarella G, Laabir M, Cabrini M, et al. (1999) The insidious effect of diatoms on copepod reproduction. Nature 402: 173–176 [Google Scholar]
- Misra AN, Misra M, Singh R (2012) Chlorophyll fluorescence in plant biology. In Misra AN, ed, Biophysics. InTech, Rijeka, Croatia, pp 171–192 [Google Scholar]
- Moreau M, Lee GI, Wang Y, Crane BR, Klessig DF (2008) AtNOS/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. J Biol Chem 283: 32957–32967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M, Lindermayr C, Durner J, Klessig DF (2010) NO synthesis and signaling in plants–where do we stand? Physiol Plant 138: 372–383 [DOI] [PubMed] [Google Scholar]
- Naqvi SWA, Yoshinari T, Jayakumar DA, Altabet MA, Narvekar PV, Devol AH, Brandes JA, Codispoti LA (1998) Budgetary and biogeochemical implications of N2O isotope signatures in the Arabian Sea. Nature 394: 462–464 [Google Scholar]
- Nicholls JC, Davies CA, Trimmer M (2007) High-resolution profiles and nitrogen isotope tracing reveal a dominant source of nitrous oxide and multiple pathways of nitrogen gas formation in the central Arabian sea. Limnol Oceanogr 52: 156–168 [Google Scholar]
- Olasehinde EF, Takeda K, Sakugawa H (2010) Photochemical production and consumption mechanisms of nitric oxide in seawater. Environ Sci Technol 44: 8403–8408. [DOI] [PubMed] [Google Scholar]
- Pan Y, Ni BJ, Lu H, Chandran K, Richardson D, Yuan Z (2015) Evaluating two concepts for the modelling of intermediates accumulation during biological denitrification in wastewater treatment. Water Res 71: 21–31 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous-equations for assaying chlorophyll-a and chlorophyll-B extracted with 4 different solvents: verification of the concentration of chlorophyll standards by atomic-absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Rainteau D, Humbert L, Delage E, Vergnolle C, Cantrel C, Maubert MA, Lanfranchi S, Maldiney R, Collin S, Wolf C, et al. (2012) Acyl chains of phospholipase D transphosphatidylation products in Arabidopsis cells: a study using multiple reaction monitoring mass spectrometry. PLoS One 7: e41985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribalet F, Bastianini M, Vidoudez C, Acri F, Berges J, Ianora A, Miralto A, Pohnert G, Romano G, Wichard T, et al. (2014) Phytoplankton cell lysis associated with polyunsaturated aldehyde release in the Northern Adriatic Sea. PLoS One 9: e85947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribalet F, Berges JA, Ianora A, Casotti R (2007) Growth inhibition of cultured marine phytoplankton by toxic algal-derived polyunsaturated aldehydes. Aquat Toxicol 85: 219–227 [DOI] [PubMed] [Google Scholar]
- Rockel P, Strube F, Rockel A, Wildt J, Kaiser WM (2002) Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J Exp Bot 53: 103–110 [PubMed] [Google Scholar]
- Sakihama Y, Nakamura S, Yamasaki H (2002) Nitric oxide production mediated by nitrate reductase in the green alga Chlamydomonas reinhardtii: an alternative NO production pathway in photosynthetic organisms. Plant Cell Physiol 43: 290–297 [DOI] [PubMed] [Google Scholar]
- Sánchez-Calvo B, Barroso JB, Corpas FJ (2013) Hypothesis: nitro-fatty acids play a role in plant metabolism. Plant Sci 199-200: 1–6 [DOI] [PubMed] [Google Scholar]
- Sanz-Luque E, Chamizo-Ampudia A, Llamas A, Galvan A, Fernandez E (2015) Understanding nitrate assimilation and its regulation in microalgae. Front Plant Sci 6: 899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Luque E, Ocana-Calahorro F, Llamas A, Galvan A, Fernandez E (2013) Nitric oxide controls nitrate and ammonium assimilation in Chlamydomonas reinhardtii. J Exp Bot 64: 3373–3383 [DOI] [PubMed] [Google Scholar]
- Siaut M, Heijde M, Mangogna M, Montsant A, Coesel S, Allen A, Manfredonia A, Falciatore A, Bowler C (2007) Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 406: 23–35 [DOI] [PubMed] [Google Scholar]
- Simionato D, Block MA, La Rocca N, Jouhet J, Marechal E, Finazzi G, Morosinotto T (2013) The response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganization of the photosynthetic apparatus. Eukaryot Cell 12: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent CD, Moon TC, Befus AD (2015) Measurement of nitric oxide in mast cells with the fluorescent indicator DAF-FM diacetate. Methods Mol Biol 1220: 339–345 [DOI] [PubMed] [Google Scholar]
- Stöhr C, Strube F, Marx G, Ullrich WR, Rockel P (2001) A plasma membrane-bound enzyme of tobacco roots catalyses the formation of nitric oxide from nitrite. Planta 212: 835–841 [DOI] [PubMed] [Google Scholar]
- Tiselius P, Kuylenstierna M (1996) Growth and decline of a diatom spring bloom: phytoplankton species composition, formation of marine snow and the role of heterotrophic dinoflagellates. J Plankton Res 18: 133–155 [Google Scholar]
- Vardi A. (2008) Cell signaling in marine diatoms. Commun Integr Biol 1: 134–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardi A, Bidle KD, Kwityn C, Hirsh DJ, Thompson SM, Callow JA, Falkowski P, Bowler C (2008) A diatom gene regulating nitric-oxide signaling and susceptibility to diatom-derived aldehydes. Curr Biol 18: 895–899 [DOI] [PubMed] [Google Scholar]
- Vardi A, Formiggini F, Casotti R, De Martino A, Ribalet F, Miralto A, Bowler C (2006) A stress surveillance system based on calcium and nitric oxide in marine diatoms. PLoS Biol 4: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varet H, Brillet-Gueguen L, Coppee JY, Dillies MA (2016) SARTools: a DESeq2- and EdgeR-Based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS One 11: e0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vunjak-Novakovic G, Kim Y, Wu XX, Berzin I, Merchuk JC (2005) Air-lift bioreactors for algal growth on flue gas: mathematical modeling and pilot-plant studies. Ind Eng Chem Res 44: 6154–6163 [Google Scholar]
- Wei L, Derrien B, Gautier A, Houille-Vernes L, Boulouis A, Saint-Marcoux D, Malnoe A, Rappaport F, de Vitry C, Vallon O, et al. (2014) Nitric oxide-triggered remodeling of chloroplast bioenergetics and thylakoid proteins upon nitrogen starvation in Chlamydomonas reinhardtii. Plant Cell 26: 353–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendehenne D, Pugin A, Klessig DF, Durner J (2001) Nitric oxide: comparative synthesis and signaling in animal and plant cells. Trends Plant Sci 6: 177–183 [DOI] [PubMed] [Google Scholar]
- Wilson ID, Neill SJ, Hancock JT (2008) Nitric oxide synthesis and signalling in plants. Plant Cell Environ 31: 622–631 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Sakihama Y (2000) Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: in vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett 468: 89–92 [DOI] [PubMed] [Google Scholar]
- Yang J, Pan YF, Bowler C, Zhang LX, Hu HH (2016) Knockdown of phosphoenolpyruvate carboxykinase increases carbon flux to lipid synthesis in Phaeodactylum tricornutum. Algal Res 15: 50–58 [Google Scholar]
- Yang ZK, Zheng JW, Niu YF, Yang WD, Liu JS, Li HY (2014) Systems-level analysis of the metabolic responses of the diatom Phaeodactylum tricornutum to phosphorus stress. Environ Microbiol 16: 1793–1807 [DOI] [PubMed] [Google Scholar]
- Yoneda K, Yoshida M, Suzuki I, Watanabe MM (2016) Identification of a major lipid droplet protein in a marine diatom Phaeodactylum tricornutum. Plant Cell Physiol 57: 397–406 [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr JP, Ward BB (2002) Nitrogen cycling in the ocean: new perspectives on processes and paradigms. Appl Environ Microbiol 68: 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xing L, Wu Z, Liu C, Lin C, Liu L (2006) Discovery of nitric oxide in marine ecological system and the chemical characteristics of nitric oxide. Sci. Cina Ser B Chem. 49: 475–481 [Google Scholar]
- Zhu W, Xiao S, Zhang D, Liu P, Zhou H, Dai W, Liu F, Li H (2015) Highly efficient and stable Au/CeO2-TiO2 photocatalyst for nitric oxide abatement: potential application in flue gas treatment. Langmuir 31: 10822–10830 [DOI] [PubMed] [Google Scholar]