DCL2 is required for efficient RDR6-dependent systemic posttranslational gene silencing, recruiting RNA-DEPENDENT RNA POLYMERASE6 and promoting the production of dsRNA that is mainly processed into 21-nucleotide siRNAs by DCL4 in wild-type plants.
Abstract
Posttranscriptional gene silencing (PTGS) of transgenes involves abundant 21-nucleotide small interfering RNAs (siRNAs) and low-abundance 22-nucleotide siRNAs produced from double-stranded RNA (dsRNA) by DCL4 and DCL2, respectively. However, DCL2 facilitates the recruitment of RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) to ARGONAUTE 1-derived cleavage products, resulting in more efficient amplification of secondary and transitive dsRNA and siRNAs. Here, we describe a reporter system where RDR6-dependent PTGS is initiated by restricted expression of an inverted-repeat dsRNA specifically in the Arabidopsis (Arabidopsis thaliana) root tip, allowing a genetic screen to identify mutants impaired in RDR6-dependent systemic PTGS. Our screen identified dcl2 but not dcl4 mutants. Moreover, grafting experiments showed that DCL2, but not DCL4, is required in both the source rootstock and the recipient shoot tissue for efficient RDR6-dependent systemic PTGS. Furthermore, dcl4 rootstocks produced more DCL2-dependent 22-nucleotide siRNAs than the wild type and showed enhanced systemic movement of PTGS to grafted shoots. Thus, along with its role in recruiting RDR6 for further amplification of PTGS, DCL2 is crucial for RDR6-dependent systemic PTGS.
Plants use transcriptional gene silencing (TGS) and posttranscriptional gene silencing (PTGS) to help combat virus infection, suppress transposon activity, silence transgenes, and regulate endogenous gene expression during development (Henderson et al., 2006; Chitwood et al., 2009; Mirouze et al., 2009; Nogueira et al., 2009; Carlsbecker et al., 2010; Borges and Martienssen, 2015). TGS prevents loci from being transcribed, whereas PTGS targets mRNA and viral RNA. Both forms of gene silencing are induced by double-stranded RNA (dsRNA), which can be formed by the transcription of perfect or imperfect inverted repeats, bidirectional transcription of a locus, or the action of RNA-dependent RNA polymerases (RDRs) that convert single-stranded RNA to dsRNA (Dalmay et al., 2000; Smith et al., 2000; Borsani et al., 2005; Vaucheret, 2006; Curaba and Chen, 2008; Xie and Qi, 2008). Partially double-stranded regions of precursor microRNA transcripts are processed into microRNAs (miRNAs) by DCL1, whereas completely complementary dsRNA is processed into small interfering RNAs (siRNAs) by one of the four DICER-LIKE (DCL) RNase III enzymes (DCL1–DCL4; Park et al., 2002; Reinhart et al., 2002; Kurihara and Watanabe, 2004; Xie et al., 2004; Dunoyer et al., 2005; Henderson et al., 2006). Mature, single-stranded miRNAs or siRNAs that are bound to an ARGONAUTE (AGO) silencing effector protein guide the silencing of complementary RNA and/or DNA targets.
As key components in small RNA biogenesis and silencing, the role of DCLs has been intensively studied. In Arabidopsis (Arabidopsis thaliana), DCL1 produces miRNAs (Park et al., 2002; Reinhart et al., 2002; Kurihara and Watanabe, 2004); DCL3 is responsible for the biogenesis of 24-nucleotide siRNAs that guide chromatin modification and TGS (Xie et al., 2004; Wierzbicki et al., 2009; Johnson et al., 2014); and DCL4 is responsible for the synthesis of trans-acting siRNAs (Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005). Of particular relevance to our study, DCL4 also can act together with DCL2 at the posttranscriptional level to produce virus- and transgene-derived siRNAs that are 21 and 22 nucleotides in length, respectively (Blevins et al., 2006; Bouché et al., 2006; Fusaro et al., 2006; Henderson et al., 2006; Mallory and Vaucheret, 2009; Dadami et al., 2013; Parent et al., 2015a).
It has often been considered that DCL2 plays a subordinate and redundant role to DCL4 in siRNA biogenesis and PTGS (Borges and Martienssen, 2015). However, consistent with 22-nucleotide miRNAs initiating the RDR6-dependent, transitive production of trans-acting siRNAs from TAS transcripts (Chen et al., 2010; Cuperus et al., 2010; Borges and Martienssen, 2015), DCL2 enhances the RDR6-dependent transitivity and biogenesis of secondary siRNAs from transgenes (Mlotshwa et al., 2008; Parent et al., 2015a). Once dsRNA is produced by RDR6, DCL4 outcompetes DCL2, leading to a much greater abundance of 21-nucleotide siRNAs than 22-nucleotide siRNAs. However, the role of DCL2 in stimulating RDR6-dependent secondary siRNA production is crucial, as the level of total siRNAs, along with transgene silencing, decreased in a dcl2 mutant compared with the wild type (Parent et al., 2015a).
Remarkably, when TGS or PTGS is triggered in a cell, it can spread throughout the plant (Palauqui et al., 1997; Voinnet and Baulcombe, 1997; Brosnan et al., 2007; Molnar et al., 2010; Gursanscky and Carroll, 2012). Our previous research identified several genes that are required for the reception of graft-transmissible PTGS in Arabidopsis shoots, including RDR6 and, surprisingly, several components of the TGS pathway (Brosnan et al., 2007). In that study, scions expressing GFP were grafted onto rootstocks expressing a constitutive, GFP-specific inverted repeat (Brosnan et al., 2007). Consistent with previous reports showing that RDR6 is required for the PTGS triggered by sense transgenes (Dalmay et al., 2000; Mourrain et al., 2000; Béclin et al., 2002; Schwach et al., 2005), the reception of graft-transmissible PTGS of a sense transgene in the shoot required RDR6 (Brosnan et al., 2007). By contrast, rdr6 rootstocks expressing a constitutive GFP-specific inverted repeat were uncompromised in transmitting PTGS to scions, indicating that RDR6 was dispensable in the presence of a constitutively expressed inverted repeat (Brosnan et al., 2007). We also showed that a dcl2 dcl3 dcl4 triple mutant rootstock expressing a constitutive GFP-specific inverted repeat was able to transmit PTGS from rootstocks to scions, even though it was severely compromised in the biogenesis of 21-, 22-, and 24-nucleotide siRNAs (Brosnan et al., 2007). These results suggested that a larger RNA derived from the inverted repeat and/or low levels of DCL1-generated 21-nucleotide siRNAs acted as systemic PTGS signals (Brosnan et al., 2007).
In contrast to our earlier study based on the constitutive expression of a GFP-specific inverted repeat in rootstocks (Brosnan et al., 2007), and to identify genetic determinants required specifically for the transmission of RDR6-dependent PTGS from rootstocks, we have developed a new GFP reporter system. The reporter line constitutively expresses a GFP target mRNA throughout the plant as well as restricted expression of a GFP-specific inverted repeat specifically in the root tip. The root tip-specific expression of the GFP-specific inverted repeat was designed to ensure that RDR6-dependent PTGS of GFP was initiated in the root tip and then spread systemically throughout the root and into the shoot.
We carried out a forward genetic screen using this reporter system in Arabidopsis and recovered independent root-to-shoot transmission of PTGS (rtp) mutants. We amplified and sequenced genes known to be involved in PTGS from these mutants and showed that two, rtp5-1 and rtp5-2, carried mutations in DCL2 that were responsible for the defect in systemic PTGS. Importantly, to our knowledge, these are the first dcl2 mutants that have been recovered in a forward genetic screen for defects in gene silencing in plants. Grafting dcl2 and wild-type plants showed that DCL2 is required in both the source rootstock and the recipient shoot tissue for efficient systemic PTGS. Furthermore, dcl4 roots produce more DCL2-dependent 22-nucleotide siRNAs than the wild type and show enhanced systemic PTGS. Our results indicate that DCL2 promotes systemic PTGS by facilitating the recruitment of RDR6 to produce dsRNA from target mRNA. While the predominant activity of DCL4 in processing dsRNA into 21-nucleotide siRNAs has obscured the important role of DCL2 in systemic PTGS, combining dcl2 and dcl4 mutations completely abolished graft-transmissible PTGS, suggesting that DCL4 can contribute inefficiently to systemic PTGS. However, DCL2 is more effective than DCL4 at inducing systemic PTGS, and in wild-type plants, DCL4 limits the capacity of DCL2 to promote systemic PTGS.
RESULTS
A GFP Reporter System for Studying Systemic PTGS
The GFP reporter line 10027-3 carries a single copy of the pUQC10027 T-DNA, integrated in an intergenic region of chromosome 1, about 700 bp upstream of At1g09840 (Supplemental Fig. S1). The T-DNA is composed of a two-component GFP reporter system: a constitutively expressed p35S:GFP transgene linked to an intron-splicible GF inverted repeat (nucleotides 9–400 of GFP) driven by the root tip-specific RCH1 promoter (pRCH1; Casamitjana-Martínez et al., 2003; Brosnan et al., 2007; Fig. 1A; Supplemental Figs. S1–S3). In the wild-type background, PTGS of GFP is initiated in the root tip and spreads into the developing hypocotyl 1 to 2 d after germination (Supplemental Fig. S2B) and subsequently into leaves as they form at the shoot apex (Fig. 1A). The reporter phenotype closely resembles root-to-shoot graft-transmissible PTGS (Fig. 1B), except that spreading of PTGS to the shoot apex is initiated earlier in plant development (Fig. 1A). In the wild type, cotyledon tissue formed during embryogenesis and before the onset of systemic PTGS maintained GFP fluorescence in germinated seedlings (Supplemental Fig. S2B), but with time, silencing slowly advanced into the cotyledons (Fig. 1A). Grafting experiments showed that PTGS was transmitted from 10027-3 rootstocks to leaf tissue of GFP-expressing scions (Fig. 1B) but not to GFP and YFP reporters expressed in pollen (Eady et al., 1994) or in female gamete precursor cells in developing ovules (Tucker et al., 2012; Supplemental Fig. S4; Supplemental Table S1). These results are consistent with earlier reports showing that PTGS initiated in the root is transmitted to newly formed leaf tissue but not through to the next generation (Brosnan et al., 2007; Liang et al., 2012; Supplemental Fig. S4C).
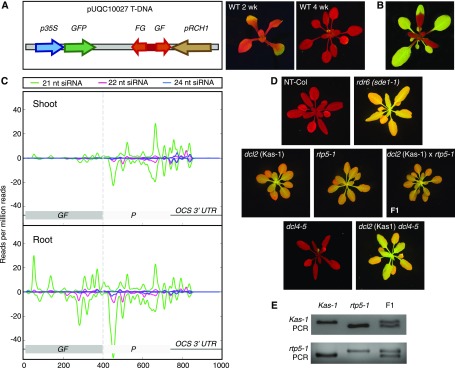
Figure 1.
DCL2 is required for RDR6-dependent root-to-shoot transmission of systemic PTGS. A, Left, pUQC10027 T-DNA showing the cauliflower mosaic virus p35S promoter driving the expression of the GFP coding sequence (720 nucleotides) and the root tip-specific RCH1 promoter driving the expression of a GF-specific dsRNA (nucleotides 9–400 of the GFP coding sequence). Right, A 10027-3 wild-type (WT) plant at 2 and 4 weeks old. Under blue light, chlorophyll and GFP fluorescence appear red and green, respectively. B, Graft-transmissible silencing of GFP in a wild-type scion of transgenic line 214 (Brosnan et al., 2007; Supplemental Fig. S2A) 4 weeks after grafting onto a 10027-3 rootstock. C, GFP-specific siRNA profile in shoots and roots of 10027-3 wild-type seedlings grown in vitro on Murashige and Skoog medium for 1 week. Positive and negative scales on the y axis represent sense and antisense siRNA profiles, respectively. The x axis shows nucleotide positions in the GFP coding sequence and 3′ UTR. Sequences homologous to the GF-specific dsRNA expressed specifically in the root tip to initiate systemic PTGS are indicated in darker gray. The data presented here are for one biological replicate; two additional biological replicates are shown in Supplemental Figure S5. D, Typical GFP phenotypes of 10027-3 wild-type and mutant plants. All 10027-3 lines were homozygous for the 10027-3 T-DNA locus. A nontransgenic wild-type (NT-Col) plant is shown as a negative control. rdr6 and dcl2 mutants (Kas-1 and rtp5-1) and the dcl2 (Kas-1) dcl4-5 double mutant are defective in systemic PTGS. By contrast, dcl4-5 shows complete PTGS of GFP in leaf tissue. E, PCR zygosity assays for dcl2 mutations in the dcl2 (Kas-1) and rtp5-1 mutants and their F1 progeny. The dcl2 (Kas-1) and rtp5-1 mutations failed to complement in the F1 (D), indicating that the dcl2 mutations were responsible for defective systemic PTGS. Further complementation tests between dcl2 (Kas-1), rtp5-1, and a third dcl2 mutant, rtp5-2, confirmed that DCL2 is required for systemic PTGS (Supplemental Fig. S7). In A, B, and D, plants were grown in soil under long days.
To further characterize the nature of GFP silencing in roots and leaves of 10027-3 wild-type plants, we performed small RNA sequencing (RNA-Seq). As mentioned above, GFP silencing was initiated by restricted expression of a GF-specific inverted repeat specifically in the root tip. Importantly, the 3′ P portion of the GFP coding sequence (nucleotides 401–720) and the OCS 3′ untranslated region (UTR) are missing from the inverted repeat. Following the initiation of GFP silencing by GF-specific dsRNAs and primary siRNAs in the root tip, secondary siRNAs were produced from the 3′ P portion of GFP and the OCS 3′ UTR in both root and shoot tissue (Fig. 1C; Supplemental Fig. S5). These secondary siRNAs were collectively referred to as P-specific siRNAs. In 10027-3 wild-type shoot tissue, almost all of the siRNAs were P specific (Fig. 1C), indicating that 3′ fragments of cleaved GFP mRNA are a more efficient template than 5′ fragments for RDR6-dependent biogenesis of secondary siRNAs (Yoshikawa et al., 2005; Brosnan et al., 2007).
When we introduced the 10027-3 reporter into an rdr6 mutant (sde1-1; Dalmay et al., 2000), no PTGS was observed (Fig. 1D). Only GF-specific primary siRNAs were detected in 10027-3 rdr6 roots (Supplemental Fig. S6), and these primary siRNAs expressed in the root tip were incapable of inducing systemic PTGS in the rdr6 mutant (Fig. 1D). Thus, while GF-specific siRNAs detected in 10027-3 wild-type plants could be either primary siRNAs produced from the GF inverted repeat expressed in the root tip or RDR6-dependent secondary siRNAs produced throughout the plant, all P-specific siRNAs in wild-type plants were RDR6-dependent, transitive secondary siRNAs.
A Genetic Screen for Mutants Defective in Root-to-Shoot Transmission of PTGS Identifies ago1, dcl2, rdr6, and sgs3 Mutants
In order to identify the genes required for systemic spreading of PTGS, we carried out a mutagenesis screen using the 10027-3 GFP reporter line. After ethyl methanesulfonate mutagenesis, we recovered 44 independent rtp mutants exhibiting defects in root-to-shoot transmission of PTGS. We carried out complementation tests involving crosses to mutants known to be defective in PTGS spontaneously triggered by sense transgenes (Dalmay et al., 2000; Fagard et al., 2000; Mourrain et al., 2000) and identified six new mutant alleles of rdr6 (rtp2-1 to rtp2-6), two new alleles of sgs3 (rtp3-1 and rtp3-2), and a new hypomorphic allele of ago1 (rtp4-1; Supplemental Table S2). All three of these genes are known to be required for the production of transitive, secondary siRNAs (Mlotshwa et al., 2008; Parent et al., 2015a), indicating that this pathway was required for systemic PTGS in 10027-3 wild-type plants.
To further characterize our collection of rtp mutants, we amplified and sequenced other genes known to be involved in PTGS from each mutant, including DCL4 and DCL2. This approach identified two candidate dcl2 mutants, rtp5-1 (Fig. 1D) and rtp5-2 (Supplemental Fig. S7), that carried nonsense (W-796-*) and missense (A-1098-V) mutations in DCL2, respectively. We then combined the 10027-3 reporter with a natural dcl2 mutant derived from ecotype Kas-1 (Parent et al., 2015a), which also was defective in root-to-shoot transmission of PTGS (Fig. 1D). This natural dcl2 allele exhibits a three-nucleotide deletion within the DCL2 coding sequence, which makes it nonfunctional (Y-564 del; Parent et al., 2015a). Complementation tests involving crossing dcl2 (Kas-1), rtp5-1, and rtp5-2 confirmed that the dcl2 mutations were indeed the causative mutations in each mutant (Fig. 1, D and E; Supplemental Fig. S7). Some rtp5-2 plants, and F1 plants from crosses of rtp5-2 to dcl2 (Kas-1) and rtp5-1, showed delayed onset of PTGS (Supplemental Fig. S7), indicating that the rtp5-2 missense mutation is a leaky dcl2 allele.
No dcl4 mutants were recovered in our screen, which raised a question about the role of DCL4 in systemic PTGS. To address this question, the 10027-3 reporter was introduced into dcl4-5 (Dunoyer et al., 2005). In contrast to dcl2 mutants, the GFP reporter was completely silenced in leaves of 10027-3 dcl4-5 plants (Fig. 1D).
We also produced a 10027-3 dcl2 (Kas-1) dcl4-5 double mutant, which showed full expression of GFP in leaves that was comparable to 10027-3 rdr6 (Fig. 1D), further indicating that both DCL2 and DCL4, and not DCL1 or DCL3, contribute to the RDR6-dependent PTGS of transgenes in Arabidopsis (Parent et al., 2015a).
DCL2 Stimulates the Production of RDR6-Dependent, P-Specific siRNAs
In 1- and 2-week-old 10027-3 wild-type plants, GFP expression was barely detectable in roots, hypocotyls, and shoot apices (Fig. 2; Supplemental Fig. S8). By contrast, strong GFP fluorescence was observed throughout 10027-3 rdr6 plants (Fig. 2). There was also a complete absence of PTGS in all tissues of 10027-3 dcl2 (Kas-1) dcl4-5 double mutant seedlings (Fig. 2A), consistent with both DCL4 and DCL2, but not DCL1 or DCL3, contributing to RDR6-dependent PTGS in Arabidopsis (Parent et al., 2015a). Indeed, GFP-specific siRNAs were not detected in the roots of the dcl2 (Kas-1) dcl4-5 or dcl2 (Kas-1) dcl4-2 double mutants (Fig. 2B; Supplemental Fig. S8). The dcl2 single mutants Kas-1 and rtp5-1 showed minimal GFP expression in root tissue (Fig. 2A; Supplemental Fig. S8). However, in comparison with wild-type roots, dcl2 roots had substantially lower amounts of RDR6-dependent P-specific siRNAs in roots (Figs. 2B and 3; Supplemental Figs. S8 and S9; Supplemental Table S3). Based on small RNA-Seq, the levels of P-specific siRNAs in 1- and 2-week-old dcl2 (Kas-1) roots were only 17% and 64% of the wild-type control level, respectively (Supplemental Table S3). dcl2 (Kas-1) and rtp5-1 showed silencing of GFP in the lower part of the hypocotyl, but PTGS failed to spread completely throughout the dcl2 hypocotyl and leaf tissue (Figs. 1D and 2A).
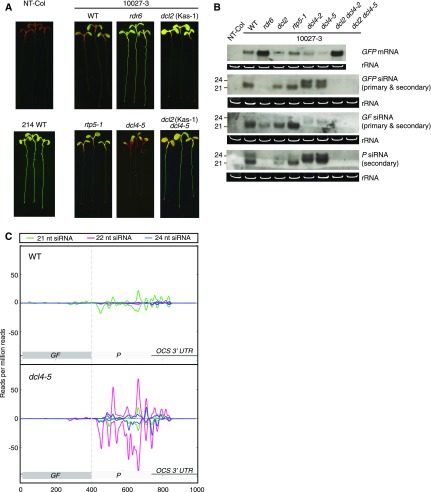
Figure 2.
dcl2 and dcl4 mutants display contrasting systemic PTGS phenotypes and levels of RDR6-dependent secondary siRNAs. Seedlings were grown in vitro under long days for 1 week. A, Typical GFP phenotypes of 10027-3 wild-type (WT) and mutant seedlings. All 10027-3 lines were homozygous for the 10027-3 T-DNA locus. Nontransgenic wild-type (NT-Col) and WT 214 (the wild-type genotype expressing a p35S:GFP transgene) seedlings are shown as GFP-negative and -positive controls, respectively. rdr6 and dcl2 (Kas-1) mutant and dcl2 (Kas-1) dcl4-5 double mutant seedlings are defective in systemic PTGS; by contrast, dcl4-5 seedlings showed enhanced systemic PTGS compared with wild-type seedlings. B, Northern blot of root GFP mRNA and GFP-, GF-, or P-specific siRNAs in nontransgenic wild-type (NT-Col) and 10027-3 genotypes. Both dcl4-5 and dcl4-2 mutant roots showed enhanced biogenesis of P-specific siRNAs, and both dcl2 dcl4-5 and dcl2 dcl4-2 double mutant roots were devoid of GFP-specific siRNAs. The GFP siRNA probe was the full-length 720-nucleotide GFP coding sequence. The GF and P siRNA probes were nucleotides 9 to 400 and 401 to 720 of the coding sequence, respectively. In A and B, dcl2 refers to the dcl2 (Kas-1) mutant and rdr6 refers to the sde1-1 allele. C, Enhanced accumulation of GFP-specific siRNAs in 10027-3 dcl4-5 shoots. The positive and negative scales on the y axis represents sense and antisense siRNA alignments, respectively. The x axis shows nucleotide positions in the GFP coding sequence and 3′ UTR. The data presented are averages of three independent biological replicates. The individual replicates are shown in Supplemental Figure S10.
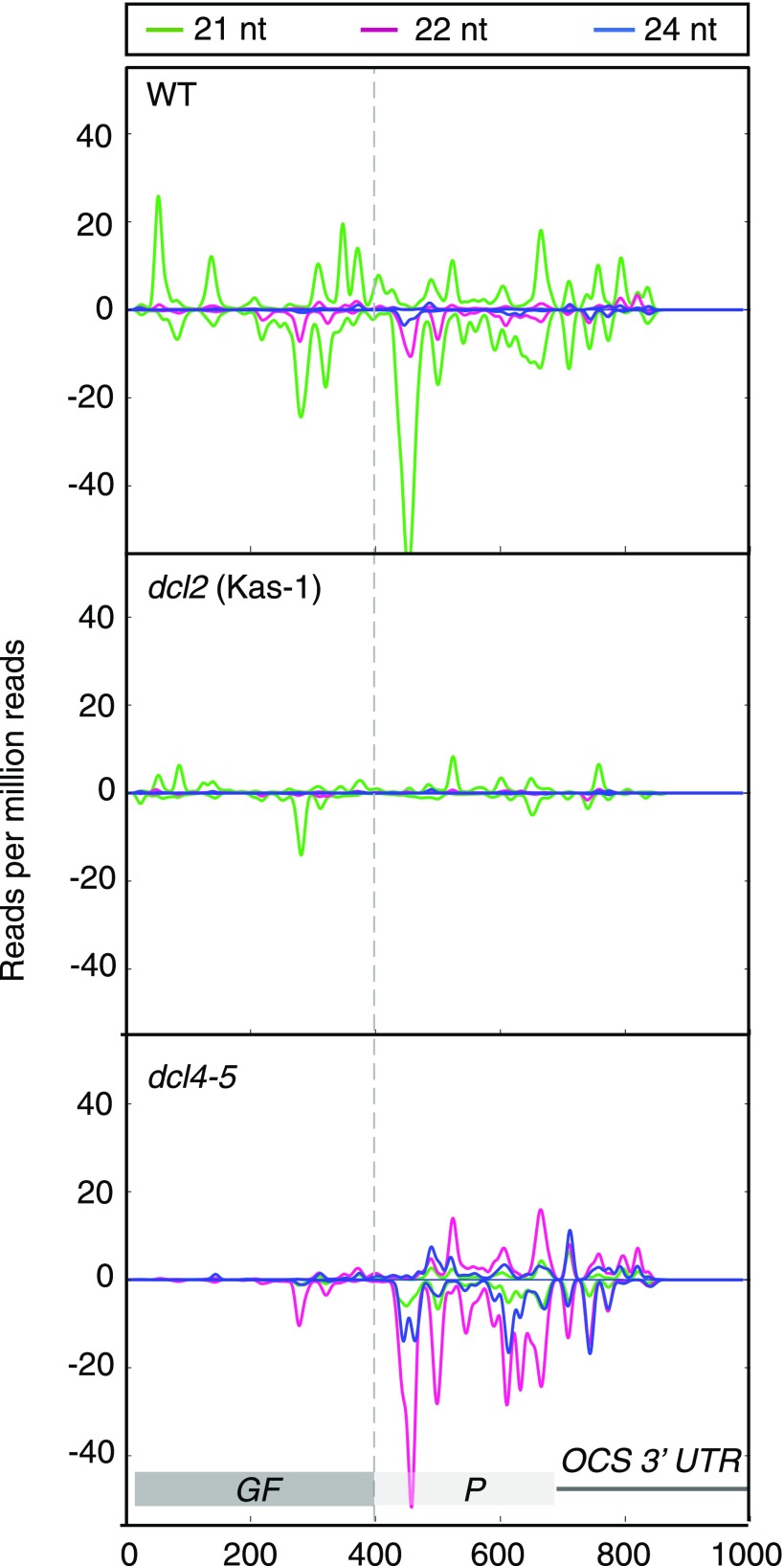
Figure 3.
DCL2 enhances the biogenesis of RDR6-dependent secondary siRNAs. GFP-specific siRNA profiles are shown in roots of 10027-3 wild-type (WT), dcl2 (Kas-1), and dcl4-5 seedlings. All 10027-3 lines were homozygous for the 10027-3 T-DNA locus. Plants were grown in vitro under long days for 1 week. Positive and negative scales on the y axis represent sense and antisense siRNA profiles, respectively. The x axis shows nucleotide positions in the GFP coding sequence and 3′ UTR. Sequences homologous to the GF-specific dsRNA expressed specifically in the root tip to initiate systemic PTGS are indicated in darker gray. The data presented are averages of two or three biological replicates that are shown individually in Supplemental Figure S9.
Consistent with an earlier report for the GUS reporter in Arabidopsis (Parent et al., 2015a), the extent of GFP silencing was more pronounced in dcl4-5 seedlings than in wild-type seedlings, with GFP silencing extending farther into the cotyledons of dcl4 seedlings (Fig. 2A). There were very few GF-specific siRNAs in dcl4-5 or dcl4-2 roots, but there were up to 1 order of magnitude more 22- and 24-nucleotide P-specific siRNAs in the roots of dcl4-5 mutants compared with the wild type (Figs. 2B and 3; Supplemental Figs. S8 and S9; Supplemental Table S3). There was about 4 times more total GFP-specific siRNAs in 1-week-old shoots of dcl4-5 compared with the wild type, and almost all of these were transitive, P-specific siRNAs (Fig. 2C; Supplemental Fig. S10; Supplemental Table S3). To further investigate the relationship between the loss of DCL4 and the enhanced biogenesis of 22-nucleotide and total GFP-specific siRNAs in 1-week-old dcl4-5 shoots, we measured DCL2 mRNA levels in the shoots of 1-week-old wild-type versus dcl4-5 seedlings. We found that DCL2 mRNA levels were not significantly different in the dcl4 shoots compared with the wild type (Supplemental Fig. S11), indicating that the higher amount of 22-nucleotide and total GFP-specific siRNAs observed in dcl4-5 was not due to an increased expression of DCL2 mRNA in this mutant. Nevertheless, these data further confirmed the findings of Parent et al. (2015a) that, in the absence of DCL4, both DCL2 activity and RDR6-dependent secondary siRNA biogenesis are enhanced.
DCL2 Is Required in Both Source and Recipient Tissue for Efficient Root-to-Shoot Transmission of PTGS
To further investigate the role of DCL2 in systemic silencing, we conducted grafting experiments using various combinations of scions and rootstocks. First, we grafted 10027-3 wild-type roots onto wild-type scions constitutively expressing a p35S:GFP transgene (transgenic line 214; Fig. 2A; Supplemental Fig. S2A; Brosnan et al., 2007). Under long-day conditions, the grafted plants started to flower around 5 weeks, and by this time, about 65% of line 214 wild-type scions grafted onto 10027-3 wild-type rootstocks showed GFP silencing (Fig. 4A). By contrast, line 214 wild-type scions grafted onto nontransgenic Columbia-0 (Col-0) rootstocks remained GFP positive (Supplemental Table S1), confirming that grafting per se does not induce spontaneous silencing of GFP.
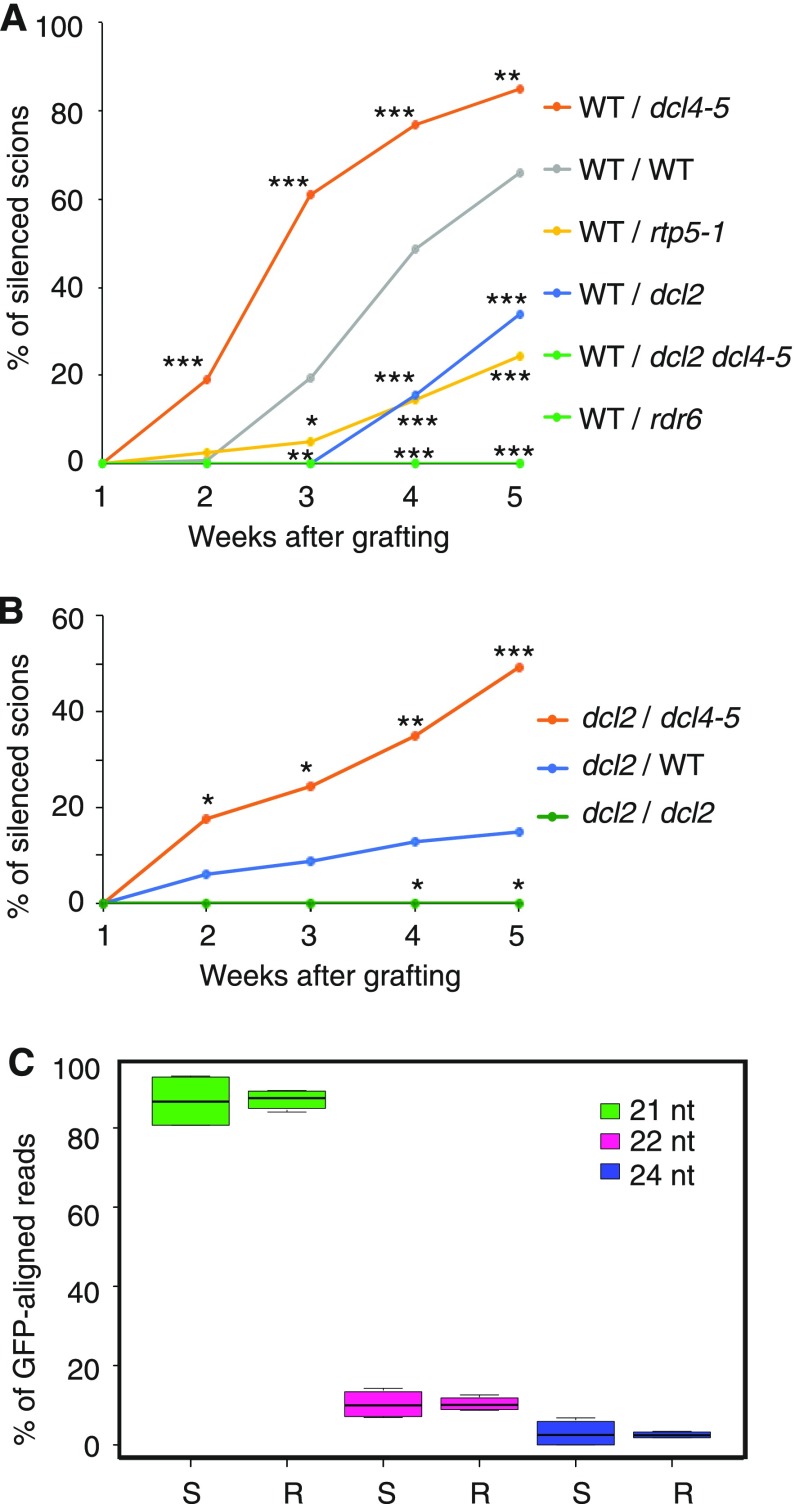
Figure 4.
DCL requirements in rootstocks and scions for the transmission and reception of systemic, RDR6-dependent PTGS. A, The transmission of PTGS is compromised from dcl2 rootstocks but enhanced from dcl4 rootstocks. Wild-type (WT) scions expressing GFP (transgenic line 214) were grafted onto 10027-3 wild-type and mutant rootstocks. All 10027-3 lines were homozygous for the 10027-3 T-DNA locus. In total, 39 to 129 grafted plants were assessed over at least three independent experiments for each combination of genotypes. P values for pairwise comparison of each treatment with wild-type/wild-type grafts are indicated for 2 to 5 weeks postgrafting. B, Reception of PTGS is compromised in dcl2 scions but enhanced by grafting dcl2 scions onto dcl4 rootstocks. 10027-3 dcl2 (Kas-1) scions were grafted onto 10027-3 wild-type, dcl4-5, or dcl2 (Kas-1) rootstocks. Scions of self-grafted 10027-3 dcl2 (Kas-1) control plants showed no silencing. In total, 38 to 101 grafted plants were assessed over at least three independent experiments for each combination of genotypes. P values for pairwise comparison between dcl2/wild type and dcl2/dcl4-5 or dcl2/dcl2 are indicated for 2 to 5 weeks postgrafting. In A and B, plants were transferred to soil 1 week after grafting; GFP silencing in shoots was monitored every week and is expressed as a percentage of the total number of scions. Statistical analysis was performed using Fisher’s exact tests followed by Benjamini and Hochberg multiple correction (*, P < 0.05; **, P < 0.01; and ***, P < 0.001). dcl2 refers to the dcl2 (Kas-1) allele, and rdr6 refers to the sde1-1 allele. C, Size distributions of GFP-specific siRNAs in floral buds of nontransgenic scions grafted onto 10027-3 rootstocks (S) and in 2-week-old 10027-3 roots used for grafting (R). The data are based on eight small RNA libraries (nscion = 4, nrootstock = 4). Nontransgenic scions were grafted onto either 10027-3 wild-type (n = 2) or rdr6 (n = 2) rootstocks. The GFP-specific siRNA profiles for these roots are shown in Supplemental Figure S5. Box plots were generated based on the percentage of each GFP-aligned siRNA size class relative to all GFP-aligned sRNA size classes (21, 22, and 24 nucleotides) for each sample. Combined analysis of the eight small RNA libraries showed that there was no difference in the relative abundance of each siRNA size class in the scion or rootstock tissues analyzed.
We then grafted line 214 wild-type scions onto various 10027-3 mutant rootstocks to test for defects in the production of the mobile PTGS signal and root-to-shoot transmission of PTGS. Transmission of PTGS from 10027-3 dcl2 (Kas-1 or rtp5-1) rootstocks compared with 10027-3 wild-type rootstocks was significantly impaired (Fig. 4A; P < 0.001 at 4 and 5 weeks postgrafting). This poor transmission of PTGS was associated with lower amounts of total GFP-specific siRNAs but, particularly, with the low amount of 22-nucleotide P-specific siRNAs in roots of 10027-3 dcl2 mutants (Figs. 2B and 3; Supplemental Table S3).
By contrast, 10027-3 dcl4-5 rootstocks transmitted GFP silencing to line 214 wild-type scions more effectively than 10027-3 wild-type rootstocks (Fig. 4A; P < 0.001 or 0.01, depending on the time after grafting). For example, at 3 weeks after grafting, about 20% of line 214 scions grafted onto wild-type rootstocks showed silencing, whereas 60% of the same scions grafted onto dcl4-5 rootstocks showed silencing (P < 0.001). Similarly, at 5 weeks after grafting, about 65% of line 214 scions grafted onto wild-type rootstocks showed silencing, whereas over 80% of the same scions grafted onto dcl4-5 rootstocks showed silencing (P < 0.01). This enhanced transmission of PTGS from dcl4 roots was associated with higher levels of DCL2-dependent, 22-nucleotide P-specific siRNAs in the dcl4 roots (Figs. 2B and 3; Supplemental Figs. S8 and S9; Supplemental Table S3) and provided further evidence of the importance of DCL2 in promoting systemic PTGS.
As expected, 10027-3 rdr6 rootstocks and 10027-3 dcl2 dcl4 double mutant rootstocks failed to transmit PTGS to 214 wild-type scions (Fig. 4A).
Next, we tested whether DCL2 is required for the efficient reception of PTGS in scions by grafting 10027-3 dcl2 (Kas-1) scions onto 10027-3 wild-type rootstocks. Only 15% of 10027-3 dcl2 (Kas-1) scions grafted onto 10027-3 wild-type rootstocks showed silencing of GFP 5 weeks after grafting (Fig. 4B). We also grafted 10027-3 dcl2 (Kas-1) scions onto 10027-3 dcl4-5 rootstocks, and remarkably, reception of systemic PTGS was observed in about 50% of 10027-3 dcl2 (Kas-1) scions at 5 weeks after grafting (Fig. 4B). These data indicate that the low efficiency of reception of PTGS observed in 10027-3 dcl2 scions could be partly alleviated by grafting onto dcl4 rootstocks.
Collectively, these data clearly demonstrate that DCL2 plays a crucial role in systemic PTGS, not only in source root tissue but also in recipient shoot tissue.
To test if 22-nucleotide GFP-specific siRNAs produced by DCL2 are more efficiently transmitted from roots to shoots than 21- or 24-nucleotide siRNAs, we grafted 10027-3 wild-type and rdr6 rootstocks onto nontransgenic Col-0 scions and used small RNA-Seq to compare the relative abundance of 21-, 22-, and 24-nucleotide GFP-specific siRNAs in the source root tissue and recipient nontransgenic floral bud tissue (Fig. 4C). No obvious differences were observed in the relative abundance of 21-, 22-, and 24-nucleotide GFP-specific siRNAs between the source and recipient tissues (Fig. 4C). Thus, 22-nucleotide GFP-specific siRNAs produced in the rootstock do not appear to be transmitted more efficiently from roots to shoots than 21- or 24-nucleotide GFP-specific siRNAs.
A GUS Reporter System Confirms the Important Role of DCL2 in Systemic PTGS
We conducted additional experiments using the well-characterized GUS PTGS reporter system (Elmayan et al., 1998), which provided further evidence of the crucial role of DCL2 in RDR6-dependent systemic PTGS. We used two p35S:GUS lines: 6b4 stably expresses a p35S:GUS transgene, whereas L1 carries a p35S:GUS transgene located elsewhere in the genome that spontaneously undergoes PTGS early in development (Elmayan et al., 1998). Initially, siRNAs are produced only from the 3′ end of GUS in L1 plants, but as the plants develop, siRNAs are produced from the central portion and, eventually, also from the 5′ portion of the GUS transgene (Parent et al., 2015b).
As observed in the GFP reporter system, L1 PTGS of GUS also was impaired in rdr6 (sgs2-1), dcl2 (Kas-1), and a dcl2 (Kas-1) dcl4-5 double mutant, but not in dcl4-5 (Fig. 5A; Parent et al., 2015a). However, also in line with the GFP system (Fig. 2; Supplemental Fig. S5), GUS activity in L1 dcl2 (Kas-1) was lower than in L1 rdr6 or L1 dcl2 (Kas-1) dcl4-5 (Fig. 5A), indicating partial but not complete PTGS of both p35S:GFP and p35:GUS in dcl2 mutants.
Figure 5.
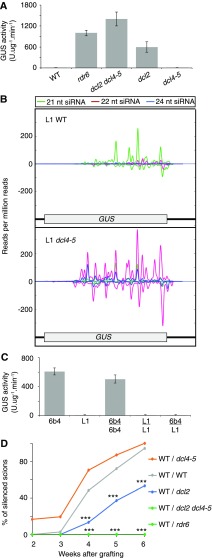
DCL2 is required for RDR6-dependent, systemic PTGS of GUS. A, GUS activity in shoots of L1 genotypes, showing defective PTGS of p35:GUS in rdr6 and dcl2 mutants but not in dcl4-5. B, Contrasting GUS-specific siRNA profiles in 1-week-old roots of L1 wild-type (WT) and L1 dcl4-5 plants. Positive and negative scales on the y axis represent sense and antisense siRNA profiles, respectively. The x axis shows nucleotide positions in the GUS coding sequence and 3′ UTR. C, GUS activity in shoots of grafted and ungrafted 6b4 and L1 wild-type plants. 6b4 and L1 both carry a single p35S:GUS transgene but at different locations in the genome (Elmayan et al., 1998). For A and C, ungrafted and grafted plants were grown for up to 6 weeks in soil under long days. Ten to 20 plants were analyzed in each of two independent experiments (n = 2). Error bars represent se. D, Transmission of RDR6-dependent PTGS of GUS is compromised from dcl2 rootstocks but enhanced from dcl4 rootstocks. 6b4 wild-type scions were grafted onto L1 wild-type or mutant rootstocks. A piece of leaf was harvested each week after grafting for the measurement of GUS activity. The results are expressed as percentages of silenced scions. P values for pairwise comparison of each treatment with wild type/wild type are indicated (Fisher’s exact tests followed by Benjamini and Hochberg multiple correction: *, P < 0.05; **, P < 0.01; and ***, P < 0.001). For individual week 2 to 4 data, P values for dcl4-5 rootstocks compared with wild-type rootstocks were less than 0.1 but not significant (i.e. P > 0.05). However, pairwise comparison between wild-type and each mutant rootstock for combined week 2 to week 6 data generated highly significant P values of 3.67 × 10−10 (dcl2), 0.003 (dcl4-5), and 4.67 × 10−10 (rdr6 and dcl2 dcl4-5). In total, 100 to 243 grafted plants were assessed for each combination of genotypes. In A, C, and D, GUS activity was determined by 4-methylumbelliferyl-β-d-glucuronide assays (fluorescence units per minute per microgram of total proteins quantified by the Bradford protocol). dcl2 refers to the dcl2 (Kas-1) allele, and rdr6 refers to the sgs2-1 allele. All L1 and 6b4 lines were homozygous for the respective GUS reporter T-DNA locus.
We also performed small RNA-Seq on L1 wild-type and dcl4-5 roots, and much higher levels of GUS-specific siRNAs, particularly 22-nucleotide siRNAs, were detected in L1 dcl4-5 roots compared with L1 wild-type roots (Fig. 5B). These results further indicated that, in the absence of DCL4, increased DCL2 activity resulted in an increase in RDR6-dependent secondary siRNAs along the GUS transgene.
We also conducted grafting experiments with the GUS reporter lines. Grafting 6b4 wild-type scions onto L1 wild-type rootstocks resulted in graft-transmissible, systemic silencing of the 6b4 p35S:GUS transgene (Fig. 5C). We then grafted 6b4 wild-type scions onto various L1 mutant rootstocks to determine the impact of rdr6, dcl2, and dcl4 mutations on root-to-shoot transmission of PTGS. As expected, none of the 6b4 scions grafted onto L1 rdr6 or L1 dcl2 (Kas-1) dcl4-5 rootstocks became silenced (Fig. 5D), confirming that an rdr6 single mutation or a dcl2 dcl4 double mutation completely prevented PTGS and the production of a graft-transmissible PTGS signal. Also consistent with the data obtained for our GFP lines, the L1 dcl2 (Kas-1) and L1 dcl4-5 rootstocks displayed defective and enhanced root-to-shoot transmission of PTGS to 6b4 wild-type scions, respectively (Fig. 5D).
Thus, use of the GUS reporter system confirmed that DCL2 is clearly required for efficient root-to-shoot transmission of RDR6-dependent PTGS and that, in wild-type roots, DCL4 outcompetes DCL2, thereby limiting the extent of systemic spreading of RDR6-dependent PTGS.
DISCUSSION
Our work has provided new insights into the contribution of DCLs to root-to-shoot transmission of RDR6-dependent systemic PTGS. DCL2 has often been considered to be a redundant backup for DCL4 in executing PTGS against viruses and transgenes (Bouché et al., 2006; Fusaro et al., 2006; Borges and Martienssen, 2015). However, DCL2 is more efficient than DCL4 at inducing the RDR6-dependent transitivity and biogenesis of secondary siRNAs from transgenes (Mlotshwa et al., 2008; Parent et al., 2015a). Furthermore, our genetic screen described here has uncovered a crucial role for DCL2 in root-to-shoot transmission of RDR6-dependent systemic PTGS.
Our previous work using a constitutively expressed GFP-specific inverted repeat to induce PTGS in rootstocks indicated that RDR6, DCL2, DCL3, and DCL4 were all dispensable for the production of graft-transmissible PTGS signals from roots to shoots (Brosnan et al., 2007). By contrast, the root tip-restricted expression of the GFP-specific inverted repeat in this study necessitated a requirement for RDR6 in the transmission of systemic PTGS signals from roots to shoots. Thus, DCL2 is required for efficient RDR6-dependent systemic PTGS (Figs. 4A and 5D) but not for RDR6-independent transmission of systemic PTGS from rootstocks expressing a constitutive inverted-repeat transgene (Brosnan et al., 2007). By contrast, DCL4 activity is dispensable for the transmission of both RDR6-dependent (Figs. 4A and 5D) and RDR6-independent PTGS from rootstocks to scions (Brosnan et al., 2007). Indeed, the absence of DCL4 enhanced the efficiency of root-to-shoot transmission of RDR6-dependent PTGS (Figs. 4A and 5D), indicating that, in wild-type plants, DCL4 outcompetes DCL2, thereby limiting the extent of systemic spreading of RDR6-dependent PTGS.
A recent report suggested that the greater affinity of DCL4 for RDR6-dependent dsRNA products has obscured the important role of DCL2 in recruiting RDR6 (Parent et al., 2015a). Our data indicate that this role of DCL2 is crucial in both the source root tissue and the recipient shoot for RDR6-dependent systemic PTGS. The absence of DCL2 in the root greatly compromised the transmission of RDR6-dependent systemic PTGS to the shoot, whereas the loss of DCL4 increased DCL2 activity and, concomitantly, enhanced the transmission of RDR6-dependent PTGS to the shoot (Figs. 4, A and B, and 5). Furthermore, loss of DCL2 in the shoot also compromised the reception of PTGS from wild-type rootstocks (Fig. 4, B and C), but this defect could be suppressed by grafting dcl2 scions onto dcl4 rootstocks (Fig. 4B).
While a degree of RDR6-dependent PTGS occurred in the roots of dcl2 mutants, the levels of siRNA were lower than in the wild type (Figs. 2B and 3; Supplemental Figs. S8 and S9; Supplemental Table S3). Decreased transgene expression also was observed in dcl2 compared with rdr6 or dcl2 dcl4 double mutant leaf tissue (Fig. 1D; Parent et al., 2015a). This suggests that a degree of RDR6-dependent PTGS also occurs in dcl2 leaves, but in the absence of DCL2, RDR6 is not sufficiently engaged to bring about complete and systemic PTGS. These dcl2 phenotypes suggest that DCL2 is not only required for efficient RDR6-dependent graft-transmissible PTGS but also for the efficient cell-to-cell movement of RDR6-dependent PTGS throughout both root and shoot tissue. Consistent with this proposition, a mosaic pattern of GUS expression was observed in L1 dcl2 (Kas-1) leaves (Parent et al., 2015a).
Virus-induced RNA silencing (VIGS) also can spread from cell to cell (Bouché et al., 2006). A recent study on turnip crinkle virus-induced VIGS in Nicotiana benthamiana showed that RNA interference (RNAi) against DCL2 and DCL4 significantly decreased and enhanced the intercellular spreading of VIGS in leaf tissue, respectively (Qin et al., 2017). Therefore, this report is consistent with our demonstrated important role of DCL2 in systemic and cell-to-cell movement of PTGS of transgenes.
Another recent report, also in N. benthamiana, showed that RNAi against DCL4 enhanced RNAi-based resistance against potato spindle tuber viroid infection, but in this case, RNAi against DCL2 alone did enhance susceptibility (Katsarou et al., 2016). However, RNAi against both DCL2 and DCL3 simultaneously resulted in enhanced susceptibility to the viroid. Therefore, this report is consistent with DCL4 limiting an important role for not only DCL2, but also DCL3, in RNAi-based defense against viroids (Katsarou et al., 2016). The possibility clearly exists for a mechanistic overlap between the RNAi-based viroid defense mechanism and the systemic PTGS of transgenes. While DCL3 cannot substitute for DCL2 and DCL4 in the production of an RDR6-dependent, graft-transmissible PTGS signal in rootstocks (Figs. 4A and 5D), DCL3 and other components of the TGS pathway are required for the reception of RDR6-dependent PTGS in scions (Brosnan et al., 2007).
It is unclear how DCL2 activity enhances the recruitment of RDR6 for transitive siRNA biogenesis associated with PTGS of transgenes. It is well documented that the cleavage of TAS transcripts and some other mRNAs by AGO1 in complex with DCL1-dependent 22-nucleotide miRNAs results in the recruitment of RDR6 and the biogenesis of trans-acting siRNAs and phased siRNAs (phasiRNAs), respectively (Chen et al., 2010; Cuperus et al., 2010; Borges and Martienssen, 2015). One possibility is that the cleavage of transgene transcripts by AGO1 in complex with DCL2-dependent 22-nucleotide siRNAs results in an enhanced recruitment of RDR6 and biogenesis of transitive siRNAs. However, there also is evidence that miRNA-induced transitivity is triggered when the passenger strand, and not the mature miRNA strand of the miRNA duplex, is 22 nucleotides in length (Manavella et al., 2012). Thus, enhanced recruitment of RDR6 may occur upstream of the cleavage of transgene transcripts by AGO1 in complex with DCL2-dependent 22-nucleotide siRNAs.
The molecular nature of the systemic PTGS signal(s) also needs to be elucidated. In our earlier work using a constitutively expressed GFP-specific inverted repeat to induce RDR6-independent PTGS in rootstocks, we showed that the level of GFP-specific siRNAs was severely compromised in dcl2 dcl3 dcl4 triple mutant rootstocks, and yet, these rootstocks could still transmit PTGS to scions (Brosnan et al., 2007). These dcl2 dcl3 dcl4 rootstocks generated very low levels of 21-nucleotide siRNAs, presumably produced by DCL1, but also considerable amounts of larger RNAs derived from the inverted repeat. Thus, either or both of these RNA species derived from a constitutively expressed inverted repeat could serve as systemic PTGS signals (Brosnan et al., 2007). Similarly, RDR6-derived large RNAs and/or siRNAs also could serve as mobile signals in RDR6-dependent systemic PTGS of transgenes.
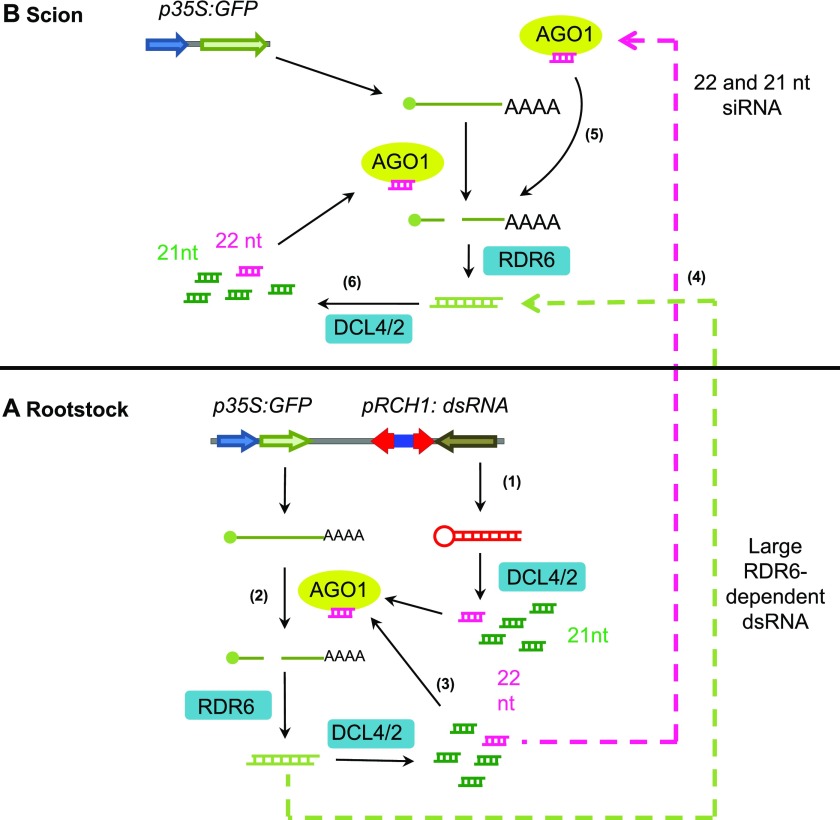
To summarize, we present a model for root-to-shoot transmission of RDR6-dependent systemic PTGS of transgenes (Fig. 6). RDR6-dependent dsRNA produced in the root is processed into abundant 21-nucleotide siRNAs and low-abundance 22-nucleotide primary siRNAs by DCL4 and DCL2, respectively. DCL2 activity efficiently recruits RDR6, forming an amplification feedback loop to enhance the production of additional, secondary siRNAs. The extent of engagement of the RDR6-dependent amplification loop, and the extent of PTGS, depend on the activity of DCL2 and its capacity to recruit RDR6, possibly via the biogenesis of 22-nucleotide siRNAs. RDR6-dependent large RNAs and/or 21- and 22-nucleotide siRNAs could act as mobile PTGS signals and move from the rootstock to the scion to induce systemic PTGS. In the scion, mobile 21- and 22-nucleotide siRNAs coming from the rootstock could be loaded into AGO1 to direct the cleavage of GFP mRNA and the induction of PTGS. In this scenario, cleavage of GFP mRNA by AGO1 in complex with 22-nucleotide siRNAs could recruit RDR6 and induce PTGS more efficiently than cleavage guided by 21-nucleotide siRNAs. Alternatively, a large RDR6-dependent systemic RNA signal derived from the root could engage the RDR6-dependent amplification loop directly in the shoot to induce PTGS.
Figure 6.
Model for the production and reception of systemic PTGS signals in wild-type plants. A, GF-specific dsRNA produced in the root tip is processed into abundant 21-nucleotide (green) and low-abundance 22-nucleotide (pink) primary siRNAs by DCL4 and DCL2, respectively (step 1). These siRNAs are loaded into AGO1 and guide the cleavage of GFP mRNA. DCL2 activity more efficiently recruits RDR6, which produces dsRNA from the cleaved mRNA (step 2). The RDR6-dependent dsRNA is processed by DCL4 and DCL2 into secondary siRNAs, forming an amplification feedback loop to direct the cleavage of GFP mRNAs and the production of additional secondary siRNAs (step 3). The extent of engagement of the RDR6-dependent amplification loop, and the extent of PTGS, depend on the activity of DCL2 and its capacity to recruit RDR6, perhaps directly or via the biogenesis of 22-nucleotide siRNAs. RDR6-dependent 21- to 22-nucleotide siRNAs and/or large RNA act as mobile PTGS signals and move from the rootstock to the scion to induce systemic PTGS (step 4). It is unknown whether (1) siRNA or large RNA systemic PTGS signals move as double-stranded or single-stranded molecules, (2) the movement of systemic PTGS signals is by diffusion or is facilitated by an active process, or (3) further amplification of systemic PTGS signals occur en route to the scion (Gursanscky and Carroll, 2012). B, In the scion, mobile 21- and 22-nucleotide siRNAs coming from the rootstock could be loaded into AGO1 to direct the cleavage of GFP mRNA and the induction of PTGS. In this scenario, cleavage of GFP mRNA by AGO1 in complex with 22-nucleotide siRNAs would recruit RDR6 and induce PTGS more efficiently than cleavage guided by 21-nucleotide siRNAs (step 5). Alternatively, large RDR6-dependent systemic RNA signals could engage the RDR6-dependent amplification loop directly to induce PTGS (step 6).
A number of other questions remain regarding the mechanism of systemic spreading of PTGS. It is unclear whether systemic PTGS signals move as dsRNA or single-stranded RNA molecules, whether systemic spreading of PTGS occurs cell to cell or via the phloem, and if movement occurs by diffusion or is facilitated by an active transport process (Gursanscky and Carroll, 2012). Whether further amplification of the systemic PTGS signals occurs en route to the shoot apex also is unknown (Gursanscky and Carroll, 2012). As mentioned above, we have shown that components of the TGS pathway, including DCL3, are required for the reception of systemic PTGS in the shoot (Brosnan et al., 2007). Clearly, the interaction of DCL2 and components of the TGS pathway in the reception of systemic PTGS in recipient cells also needs to be investigated further.
Finally, our forward genetic screen described here, to our knowledge the first to recover dcl2 mutants, could be implemented further to identify additional genes required for transitive siRNA biogenesis and RDR6-dependent systemic PTGS in plants. The identification and characterization of such genes could further elucidate the mechanism by which DCL2 enhances the recruitment of RDR6 and, perhaps, also the mechanism of 22-nucleotide miRNA-dependent, transitive siRNA biogenesis in plants.
CONCLUSION
DCL2 plays a crucial role in both transmission and the reception of root-to-shoot transmission of RDR6-dependent systemic PTGS. In wild-type plants, DCL4 outcompetes DCL2 for the dsRNA template and thereby limits the extent of systemic PTGS. Our findings provide further evidence that DCL2 enhances the recruitment of RDR6 and the biogenesis of secondary siRNAs and show that this level of engagement of RDR6 is essential for efficient systemic PTGS. While the causal link between DCL2 and the efficiency of transmission of RDR6-dependent PTGS from rootstocks to scions is now clearly established, the nature of systemic PTGS signals and additional details of the PTGS reception mechanism in recipient tissue require further investigation.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All plants used in this study were in the genetic background of Arabidopsis (Arabidopsis thaliana) ecotype Col-0. The rdr6 (sde1-1; Dalmay et al., 2000), rdr6 (sgs2-1; Mourrain et al., 2000), dcl2 (Kas-1; Parent et al., 2015a), dcl4-5 (Dunoyer et al., 2005), and dcl4-2 (Xie et al., 2005) mutant lines have been described previously, as well as the p35S:GUS L1 and 6b4 lines (Elmayan et al., 1998). The dcl2 (Kas-1) mutation had been introgressed into ecotype Col-0 by 10 generations of backcrosses (Parent et al., 2015a). All 10027-3, L1, and 6b4 reporter lines were homozygous for the respective T-DNA reporter locus. The collection of rtp mutants, including dcl2 ethyl methanesulfonate mutants (rtp5-1 and rtp5-2), was obtained as follows. 10027-3 wild-type seeds were mutagenized with ethyl methanesulfonate as described by Weigel and Glazebrook (2002). Approximately 30,000 M1 seeds were then sown in soil. M2 seeds were collected as bulks from 100 M1 plants, and about 100 M2 seeds from each bulk were sown on soil for screening for defects in systemic PTGS. Two weeks after germination of M2 plants, GFP-positive M2 plants were transferred to pots for further growth and analysis.
Arabidopsis plants were grown in soil or in vitro under long-day conditions (16 h of light, 8 h of dark), with fluorescent lighting (140 μmol m−2 s−1) at a constant 22°C.
For grafting experiments and siRNA analysis, plants were grown in axenic conditions in upright petri dishes on Murashige and Skoog medium, supplemented with 4 g L−1 Suc and 0.33% Phytagel (Sigma-Aldrich).
Crosses were carried out as described by Weigel and Glazebrook (2002).
Molecular Cloning and Plant Transformation
Agrobacterium tumefaciens-mediated transformation of Arabidopsis via the floral dip immersion protocol was as described by Clough and Bent (1998).
To confirm the expression pattern of the 2.6-kb RCH1 promoter (AT5G48940), the promoter was cloned upstream of the coding sequences for the reporter GFP (pRCH1:GFP) and GUS (pRCH1:GUS) coding sequences, and the transgenes were then transformed into Arabidopsis and characterized in T1 plants. Subsequently, a pRCH1:GF hairpin linked to p35S:GFP and p35S:BAR was cloned into a derivative of the pUQC214 binary vector (Brosnan et al., 2007). GF-specific sequences refer to nucleotides 9 to 400 of the GFP coding sequence, whereas downstream P-specific sequences refer to the remaining 317 nucleotides of the GFP coding sequence.
Genome sequencing and Southern-blot analysis were used to demonstrate that the 10027-3 line contained a single-copy insertion of the pUQC10027 T-DNA. Southern-blot analysis was performed using a probe and restriction enzymes that detected unique DNA fragments for each T-DNA insertion. The detection of single EcoRI and HindIII DNA fragments extending from the T-DNA right border into the flanking genomic sequence confirmed that the 10027-3 line carried a single T-DNA insertion. To identify the genomic DNA sequence flanking the right border of the 10027-3 T-DNA, we used GenomeWalker adapter-mediated PCR (Clontech), followed by sequencing of the PCR product and authentication by PCR zygosity assays.
Grafting Techniques
Grafting of Arabidopsis seedlings was done as described previously (Turnbull et al., 2002), with modifications as described by Brosnan et al. (2007). Wild-type and mutant 10027-3 or L1 rootstocks were grafted onto either line 214 or 6b4 wild-type and mutant scions expressing GFP (Brosnan et al., 2007) and GUS (Elmayan et al., 1998), respectively. PTGS of GFP and GUS was assessed in the rosette leaves.
DNA/RNA Extraction and Analysis
Rapid DNA extraction for PCR-based genotyping assays was conducted according to Edwards et al. (1991). Tissue was ground to a fine powder in liquid nitrogen for RNA extraction with TRIzol (Invitrogen), as per the manufacturer’s instructions. For mRNA and siRNA analysis, total RNA was extracted from roots or shoots of 1-week-old seedlings or from roots plus hypocotyls of 2-week-old seedlings.
RNA-gel (northern) blotting was carried out as described by Mitter et al. (2003), except that total RNAs rather than small RNA-enriched samples, and chemiluminescence rather than radioactive detection, were used. Digoxigenin-labeled probes were generated by using the PCR DIG Probe Synthesis Kit (Roche Diagnostics). Oligonucleotide primers for producing GFP-, GF-, and P-specific probes are listed in Supplemental Table S4.
After hybridization (PerfectHyb Plus Hybridization Buffer; Sigma-Aldrich) at 45°C or 60°C overnight, detection was performed following the instructions of the supplier (DIG Application Manual for Filter Hybridization; Roche Diagnostics). Membranes were exposed to chemiluminescent film (Carestream BioMax; Sigma-Aldrich) in a cassette at room temperature before developing.
Deep Sequencing Analysis of siRNAs
Small RNA libraries were generated from total RNA and sequenced by the Beijing Genomics Institute and the Australian Genome Research Facility. All raw small RNA sequence data are publicly available online at the National Center for Biotechnology Information Sequence Read Archive (BioProject PRJNA347864). Files containing single-end 50-bp reads were processed to collapsed reads using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Read files were filtered to remove any sequences that did not align to the Arabidopsis genome or the particular transgene present. Subsequently, collapsed reads of discrete sizes (21, 22, and 24 nucleotides) were aligned to reference sequences using the SCRAM small RNA aligner (https://carroll-lab.github.io/scram/). Only reads that were an exact match to the Arabidopsis ecotype Col-0 reference sequence, and the GFP and GUS transgenes, were accepted. SCRAM normalized aligned read counts at each reference position based on reads per million reads between 18 and 32 nucleotides in the processed library. Line plots (siRNA density profiles) for the GFP and GUS coding sequences were smoothed using a Blackman algorithm with a window size of 30 nucleotides for plots against the GFP reference sequence and 60 nucleotides for plots against the GUS reference sequence.
Imaging and Image Analysis
Images of micrografts and plants were taken using a Canon EOS 50D digital camera mounted on an Optiphot upright microscope (Nikon Instruments) with a Nikon 100-W mercury arc lamp attached to provide UV/blue light. Whole-plant photographs were taken with a Canon EOS 600D digital camera with an orange filter, and blue light illumination was provided by six Dark Reader Hand Lamps (Clare Chemical Research). Images were uniformly adjusted for white balance and contrast with Adobe Camera Raw.
GUS Activity Analysis
GUS activity was quantified from plant leaves by monitoring the quantity of 4-methylumbelliferone products generated from the substrate 4-methylumbelliferyl-β-d-glucuronide on a fluorometer (Thermo Scientific Fluoroskan Ascent), as described previously (Elmayan and Vaucheret, 1996).
Oligonucleotides
The sequences of oligonucleotides used in this study are listed in Supplemental Table S4.
Statistical Analysis of Silencing
All statistical tests were implemented in RStudio using custom R scripts. For grafting experiments, Fisher’s exact tests were performed on silenced and unsilenced count data for pairs of treatments arranged in 2 × 2 contingency tables. Multiple correction of the generated P values for each figure was carried out using the Benjamini and Hochberg method.
Accession Numbers
Sequence data from this article can be found in the NCBI Sequence Read Archive BioProject PRJNA347864.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Arabidopsis transgenic line 10027-3 carries a single copy of the pUQC10027 T-DNA.
Supplemental Figure S2. Onset of systemic PTGS in 10027-3 wild-type seedlings.
Supplemental Figure S3. The pRCH1 promoter drives root tip-specific expression of GFP and GUS.
Supplemental Figure S4. Root-to-shoot transmission of PTGS from rootstocks to newly formed leaves, stems, and siliques, but not to pollen or female reproductive cells, in immature ovules.
Supplemental Figure S5. GFP-specific siRNA profile in shoots and roots of the 10027-3 wild type.
Supplemental Figure S6. Systemic PTGS in 10027-3 is RDR6 dependent.
Supplemental Figure S7. rtp5-2 is a new dcl2 allele.
Supplemental Figure S8. dcl2 and dcl4 mutants display contrasting levels of RDR6-dependent secondary siRNAs in roots.
Supplemental Figure S9. DCL2 enhances the biogenesis of RDR6-dependent secondary siRNAs in roots.
Supplemental Figure S10. Enhanced accumulation of GFP-specific siRNAs in 10027-3 dcl4-5 shoots.
Supplemental Figure S11. DCL2 mRNA expression is not significantly different in shoot tissue of 1-week-old wild-type and dcl4-5 seedlings.
Supplemental Table S1. Root-to-shoot transmission of PTGS from rootstocks to newly formed leaves, stems, and siliques, but not to pollen or female reproductive cells, in immature ovules.
Supplemental Table S2. New mutant alleles of rdr6, sgs3, and ago1.
Supplemental Table S3. Quantification of GFP-specific siRNAs in wild-type, dcl2, and dcl4 roots.
Supplemental Table S4. Oligonucleotides used in this study.
Acknowledgments
We thank Maud Rivard, Nathalie Bouteiller, and Nurafiqahtul Hikmah Seribini for assistance in genotyping and grafting, Chris Brosnan and Taline Elmayan for fruitful discussions, and Marjori Matzke, David Baulcombe, Steve Jacobsen, and Patrice Dunoyer for providing mutant seeds.
Footnotes
This work was supported by the Australian Research Council (ARC; B.J.C. and J.L.B.), ANR and Fondation Louis D. (H.V.), and CJS-INRA (French National Fellowship grant to C.T.). C.T. was supported by a post-doctoral fellowship from INRA and from N.M. and B.J.C. grants. Research in the Vaucheret laboratory is supported by grants from the French Agence Nationale pour la Recherche (ANR-10-LABX-40 and ANR-11-BSV6-007) and the Fondation Louis D. de l’Institut de France. N.R.G. was supported by a GRDC postgraduate research scholarship (GRS114), and J.C. by the China Scholarship Council. We are grateful for support from ARC grants DP0988294, DP120103966, DP150104048 (B.J.C.) and DP160100892 (J.L.B.).
Articles can be viewed without a subscription.
References
- Béclin C, Boutet S, Waterhouse P, Vaucheret H (2002) A branched pathway for transgene-induced RNA silencing in plants. Curr Biol 12: 684–688 [DOI] [PubMed] [Google Scholar]
- Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F Jr, Hohn T, et al. (2006) Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res 34: 6233–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Martienssen RA (2015) The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16: 727–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Lauressergues D, Gasciolli V, Vaucheret H (2006) An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J 25: 3347–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan CA, Mitter N, Christie M, Smith NA, Waterhouse PM, Carroll BJ (2007) Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc Natl Acad Sci USA 104: 14741–14746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al. (2010) Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamitjana-Martínez E, Hofhuis HF, Xu J, Liu CM, Heidstra R, Scheres B (2003) Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr Biol 13: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH (2010) 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA 107: 15269–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Nogueira FT, Howell MD, Montgomery TA, Carrington JC, Timmermans MC (2009) Pattern formation via small RNA mobility. Genes Dev 23: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC (2010) Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol 17: 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curaba J, Chen X (2008) Biochemical activities of Arabidopsis RNA-dependent RNA polymerase 6. J Biol Chem 283: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadami E, Boutla A, Vrettos N, Tzortzakaki S, Karakasilioti I, Kalantidis K (2013) DICER-LIKE 4 but not DICER-LIKE 2 may have a positive effect on potato spindle tuber viroid accumulation in Nicotiana benthamiana. Mol Plant 6: 232–234 [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553 [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O (2005) DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet 37: 1356–1360 [DOI] [PubMed] [Google Scholar]
- Eady C, Lindsey K, Twell D (1994) Differential activation and conserved vegetative cell-specific activity of a late pollen promoter in species with bicellular and tricellular pollen. Plant J 5: 543–550 [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan T, Balzergue S, Béon F, Bourdon V, Daubremet J, Guénet Y, Mourrain P, Palauqui JC, Vernhettes S, Vialle T, et al. (1998) Arabidopsis mutants impaired in cosuppression. Plant Cell 10: 1747–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan T, Vaucheret H (1996) Expression of single copies of a strongly expressed 355 transgene can be silenced post-transcriptionally. Plant J 9: 787–797 [Google Scholar]
- Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H (2000) AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci USA 97: 11650–11654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro AF, Matthew L, Smith NA, Curtin SJ, Dedic-Hagan J, Ellacott GA, Watson JM, Wang MB, Brosnan C, Carroll BJ, et al. (2006) RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep 7: 1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H (2005) Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 15: 1494–1500 [DOI] [PubMed] [Google Scholar]
- Gursanscky NR, Carroll BJ (2012) Mechanism of small RNA movement. In Kragler F, Hulskamp M, eds, Short and Long Distance Signaling. Springer, New York, pp 99–130 [Google Scholar]
- Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE (2006) Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 38: 721–725 [DOI] [PubMed] [Google Scholar]
- Katsarou K, Mavrothalassiti E, Dermauw W, Van Leeuwen T, Kalantidis K (2016) Combined activity of DCL2 and DCL3 is crucial in the defense against potato spindle tuber viroid. PLoS Pathog 12: e1005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Du J, Hale CJ, Bischof S, Feng S, Chodavarapu RK, Zhong X, Marson G, Pellegrini M, Segal DJ, et al. (2014) SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 507: 124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, White RG, Waterhouse PM (2012) Gene silencing in Arabidopsis spreads from the root to the shoot, through a gating barrier, by template-dependent, nonvascular, cell-to-cell movement. Plant Physiol 159: 984–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Vaucheret H (2009) ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep 10: 521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella PA, Koenig D, Weigel D (2012) Plant secondary siRNA production determined by microRNA-duplex structure. Proc Natl Acad Sci USA 109: 2461–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze M, Reinders J, Bucher E, Nishimura T, Schneeberger K, Ossowski S, Cao J, Weigel D, Paszkowski J, Mathieu O (2009) Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461: 427–430 [DOI] [PubMed] [Google Scholar]
- Mitter N, Sulistyowati E, Dietzgen RG (2003) Cucumber mosaic virus infection transiently breaks dsRNA-induced transgenic immunity to Potato virus Y in tobacco. Mol Plant Microbe Interact 16: 936–944 [DOI] [PubMed] [Google Scholar]
- Mlotshwa S, Pruss GJ, Peragine A, Endres MW, Li J, Chen X, Poethig RS, Bowman LH, Vance V (2008) DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS ONE 3: e1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC (2010) Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328: 872–875 [DOI] [PubMed] [Google Scholar]
- Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542 [DOI] [PubMed] [Google Scholar]
- Nogueira FT, Chitwood DH, Madi S, Ohtsu K, Schnable PS, Scanlon MJ, Timmermans MC (2009) Regulation of small RNA accumulation in the maize shoot apex. PLoS Genet 5: e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui JC, Elmayan T, Pollien JM, Vaucheret H (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J 16: 4738–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent JS, Bouteiller N, Elmayan T, Vaucheret H (2015a) Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J 81: 223–232 [DOI] [PubMed] [Google Scholar]
- Parent JS, Jauvion V, Bouché N, Béclin C, Hachet M, Zytnicki M, Vaucheret H (2015b) Post-transcriptional gene silencing triggered by sense transgenes involves uncapped antisense RNA and differs from silencing intentionally triggered by antisense transgenes. Nucleic Acids Res 43: 8464–8475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, Li B, Fan Y, Zhang X, Yu Z, Ryabov E, Zhao M, Wang H, Shi N, Zhang P, et al. (2017) Roles of Dicer-Like Proteins 2 and 4 in intra- and intercellular antiviral silencing. Plant Physiol 174: 1067–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach F, Vaistij FE, Jones L, Baulcombe DC (2005) An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol 138: 1842–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM (2000) Total silencing by intron-spliced hairpin RNAs. Nature 407: 319–320 [DOI] [PubMed] [Google Scholar]
- Tucker MR, Okada T, Hu Y, Scholefield A, Taylor JM, Koltunow AMG (2012) Somatic small RNA pathways promote the mitotic events of megagametogenesis during female reproductive development in Arabidopsis. Development 139: 1399–1404 [DOI] [PubMed] [Google Scholar]
- Turnbull CGN, Booker JP, Leyser HMO (2002) Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J 32: 255–262 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20: 759–771 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Baulcombe DC (1997) Systemic signalling in gene silencing. Nature 389: 553. [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Wierzbicki AT, Ream TS, Haag JR, Pikaard CS (2009) RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet 41: 630–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Qi X (2008) Diverse small RNA-directed silencing pathways in plants. Biochim Biophys Acta 1779: 720–724 [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]