Significance
Phylogenetic diversity is a measure of biodiversity that incorporates both the number of species and their evolutionary histories. Comparisons of biodiversity between regions that are currently ecologically similar but support different numbers of species are informed by considering differences in their geographic and evolutionary histories. Here we analyze diversity in numerous floras distributed across eastern Asia and eastern North America, and find that phylogenetic diversity in eastern Asia substantially exceeds that in eastern North America when controlling statistically for climate and species richness. This difference parallels the more complex history and geography of the Asian continent.
Keywords: flowering plants, growth forms, historical contingency, phylogenetic diversity, region effect
Abstract
Although eastern Asia (EAS) and eastern North America (ENA) have similar climates, plant species richness in EAS greatly exceeds that in ENA. The degree to which this diversity difference reflects the ages of the floras or their rates of evolutionary diversification has not been quantified. Measures of species diversity that do not incorporate the ages of lineages disregard the evolutionary distinctiveness of species. In contrast, phylogenetic diversity integrates both the number of species and their history of evolutionary diversification. Here we compared species diversity and phylogenetic diversity in a large number of flowering plant (angiosperm) floras distributed across EAS and ENA, two regions with similar contemporary environments and broadly shared floristic history. After accounting for climate and sample area, we found both species diversity and phylogenetic diversity to be significantly higher in EAS than in ENA. When we controlled the number of species statistically, we found that phylogenetic diversity remained substantially higher in EAS than in ENA, although it tended to converge at high latitude. This pattern held independently for herbs, shrubs, and trees. The anomaly in species and phylogenetic diversity likely resulted from differences in regional processes, related in part to high climatic and topographic heterogeneity, and a strong monsoon climate, in EAS. The broad connection between tropical and temperate floras in southern Asia also might have played a role in creating the phylogenetic diversity anomaly.
Species diversity is often correlated with environmental variables, particularly climate (1–5); however, regions with similar environments sometimes support substantially different numbers of species, producing species diversity anomalies (4–11). Eastern Asia (EAS) and eastern North America (ENA) are continental regions at low to mid north-temperate latitudes with similar climates. The contemporary floras of the two regions are largely derived from a single paleoflora, the “Boreotropical flora,” that was broadly distributed across the Northern Hemisphere during the early Tertiary (12). The contemporary regional floras of EAS and ENA have retained many floristic similarities that are absent from other regions within the same latitudes, even within the same continent (e.g., ENA vs. western North America). For example, EAS and ENA share approximately 60 plant genera that do not occur anywhere else in the world, an observation that drew the attention of 19th-century naturalists, including Charles Darwin and Asa Gray (13). The similarities in climate, vegetation types, and floristic composition between EAS and ENA, combined with a marked difference in species diversity favoring EAS, have inspired ecologists to seek the mechanisms that have generated this prominent species diversity anomaly (5, 6, 10, 14, 15).
Previous studies have used taxonomic diversity (e.g., number of species [species richness] or a diversity index based on species relative abundances) to compare biodiversity in each assemblage. Although species diversity is a cornerstone of biodiversity research, it considers all species equivalent and does not incorporate differences that accumulate between species over evolutionary history (16). Phylogenetic diversity is a metric that also accounts for evolved differences among species by considering the time represented in the branches of the evolutionary tree of a region’s species (17). Accordingly, the contrast between species richness and phylogenetic diversity provides insight into the origins of diversity anomalies, such as that between the EAS and ENA floras, when evolutionary history has had a role.
Here we compare species diversity and phylogenetic diversity of flowering plants (angiosperms) in assemblages occupying similar environments in EAS and ENA. We then relate phylogenetic diversity to spatial environmental variation in the two regions. The southern part of EAS and tropical Asia are considered by many as the center of origin and diversification of angiosperms (18–20), according to which the EAS angiosperm flora would have a longer evolutionary history than the ENA flora. Moreover, old tropical clades might have penetrated more readily into temperate areas in EAS, which supports a broad geographic connection between temperate and tropical floras, than in ENA, where the Gulf of Mexico and arid environments in northern Mexico largely separate temperate from tropical floras. Accordingly, phylogenetic diversity might be higher in EAS than in ENA in temperate areas with similar climates. We test this hypothesis statistically. Finally, previous studies (e.g., ref. 21) have shown that distributions of plant species with large body size (trees) respond to climate variation differently than those with small body size (herbs). Thus, we also partition analyses based on plant growth forms (i.e., herb, shrub, and tree).
Results
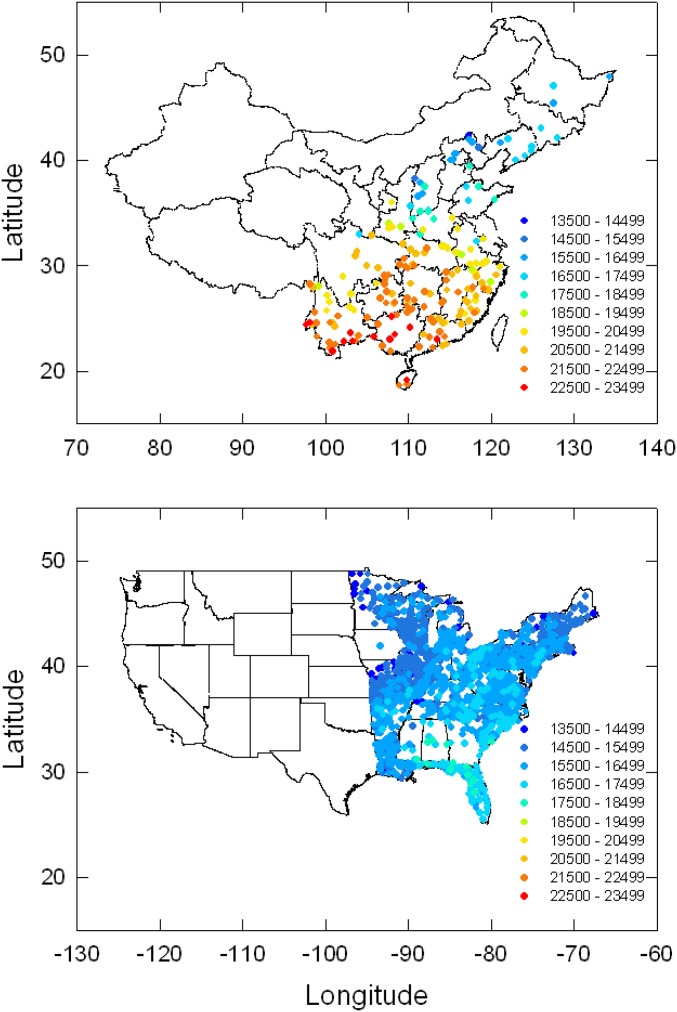
We compared species richness and phylogenetic diversity between EAS, encompassing 21 provinces or equivalent administrative regions (hereinafter provinces) in China, and ENA, based on 31 states in the US (Fig. 1). At this regional scale, the number of species of angiosperms varied among provinces from 1,820 to 14,286 in EAS and among states from 1,370 to 2,897 in ENA. At the local scale (nature reserves and parks in EAS, counties in ENA), the number of species of angiosperms varied from 506 to 5,092 in EAS and from 501 to 1,325 in ENA. Species richness was strongly correlated with phylogenetic diversity in both EAS and ENA at both spatial scales (r = 0.996 and 0.974 at regional and local scales, respectively, for EAS; r = 0.987 and 0.946 at regional and local scales, respectively, for ENA; P < 0.001 for all).
Fig. 1.
The distributions and standardized phylogenetic diversities of 1,309 angiosperm floras in EAS and ENA. Phylogenetic diversity represents the sum of branch lengths (in million years) for 500 species selected at random from each flora.
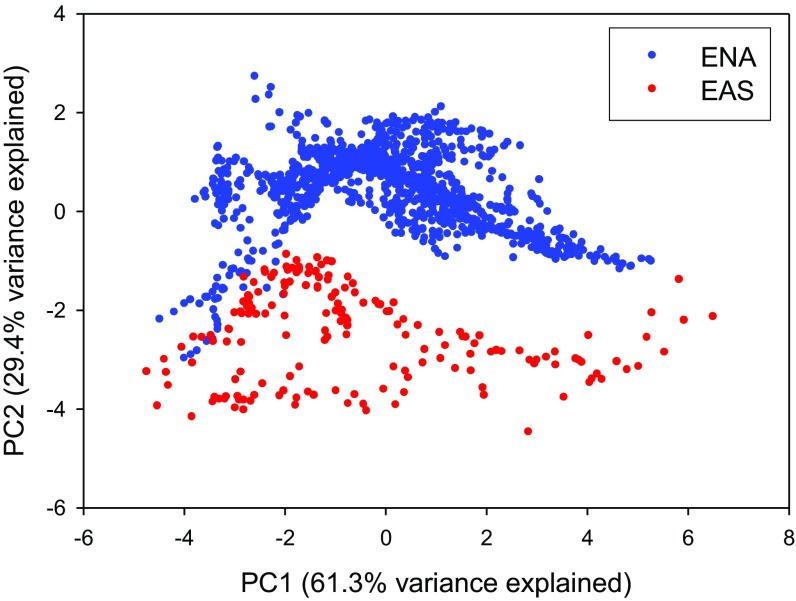
We used principal components (PC) analysis to extract axes of correlated climate variation. Values on the first principal component (PC1: primarily temperature, temperature seasonality, and precipitation) for the ENA localities completely overlapped those for the EAS localities (Fig. S1); however, PC2 values for the ENA localities uniformly exceeded those for the EAS localities (Fig. S1), reflecting the lower precipitation seasonality in ENA (Tables S1 and S2). We used ANCOVA to compare species diversity or phylogenetic diversity between EAS and ENA. Both species diversity and phylogenetic diversity were significantly higher in EAS than in ENA, under matched climate conditions and corrected for sample area, for both regional and local floras (Table 1). Climate PC1 was a significant effect in all four ANCOVAs (regional and local vs. species diversity and phylogenetic diversity; P < 0.001 for all), whereas climate PC2 was a significant effect only for species diversity at the local scale (P = 0.005; P > 0.09 in the other three cases) (Table 1).
Fig. S1.
Positions of the 1,309 floras in EAS and ENA with respect to PC1 and PC2 of the six climate variables (Tables S1 and S2).
Table S1.
The first two axes (PC1 and PC2) produced by PC analysis based on the correlation matrix between climate variables (n = 1,309)
| Variable | PC1 | PC2 |
| Eigenvalue | 3.677 | 1.765 |
| Percentage of variance | 61.288 | 29.420 |
| Cumulative % of variance | 61.288 | 90.708 |
| Eigenvectors | ||
| Tmean | −0.482 | −0.178 |
| Pmean | −0.461 | 0.049 |
| Tmin | −0.500 | −0.169 |
| Pmin | −0.265 | 0.628 |
| Tseas | 0.480 | 0.150 |
| Pseas | 0.074 | −0.722 |
Tmean, mean annual temperature; Pmean, mean annual precipitation; Tmin, minimum temperature of the coldest month; Pmin, precipitation of the driest month; Tseas, temperature seasonality; Pseas, precipitation seasonality.
Values in the lower part of the table are the loadings of each variable on the two PCs. Bold type indicates the strongest contributions to each PC axis, not statistical significance, which is indicated in Table S2.
Table S2.
Pearson’s correlation coefficients between climate variables and derived PC variables PC1 and PC2
| Variable | Tmean | Pmean | Tmin | Pmin | Tseas | Pseas | PC1 |
| Pmean | 0.732 | ||||||
| Tmin | 0.968 | 0.770 | |||||
| Pmin | 0.265 | 0.501 | 0.288 | ||||
| Tseas | −0.843 | −0.736 | −0.926 | −0.301 | |||
| Pseas | 0.067 | −0.129 | 0.050 | −0.827 | −0.047 | ||
| PC1 | −0.924 | −0.883 | −0.959 | −0.507 | 0.920 | 0.142 | |
| PC2 | −0.236 | 0.065 | −0.224 | 0.834 | 0.199 | −0.959 | 0.000 |
P < 0.05 in all cases except the three shown in italic type.
Table 1.
ANCOVA of species diversity and phylogenetic diversity of angiosperms in EAS and ENA with region (EAS vs. ENA) as the main effect and climate variables (PC1 and PC2) and area as covariates
| Source | Regional scale | Local scale | ||||||
| SS | df | F | P | SS | df | F | P | |
| Species diversity (EAS > ENA) | ||||||||
| Region | 0.083 | 1 | 6.8 | 0.012 | 3.853 | 1 | 352.1 | <0.001 |
| PC1 | 0.595 | 1 | 48.9 | <0.001 | 0.875 | 1 | 79.9 | <0.001 |
| PC2 | 0.003 | 1 | 0.3 | 0.600 | 0.088 | 1 | 8.1 | 0.005 |
| Area | 0.174 | 1 | 14.3 | <0.001 | 1.583 | 1 | 144.6 | <0.001 |
| Error | 0.572 | 47 | 13.702 | 1,252 | ||||
| Phylogenetic diversity (EAS > ENA) | ||||||||
| Region | 0.165 | 1 | 14.0 | <0.001 | 4.298 | 1 | 744.4 | <0.001 |
| PC1 | 0.664 | 1 | 56.2 | <0.001 | 1.676 | 1 | 290.2 | <0.001 |
| PC2 | 0.005 | 1 | 0.4 | 0.519 | 0.016 | 1 | 2.8 | 0.092 |
| Area | 0.117 | 1 | 9.9 | 0.003 | 0.973 | 1 | 168.5 | <0.001 |
| Error | 0.555 | 47 | 7.229 | 1,252 | ||||
SS, sums of squares.
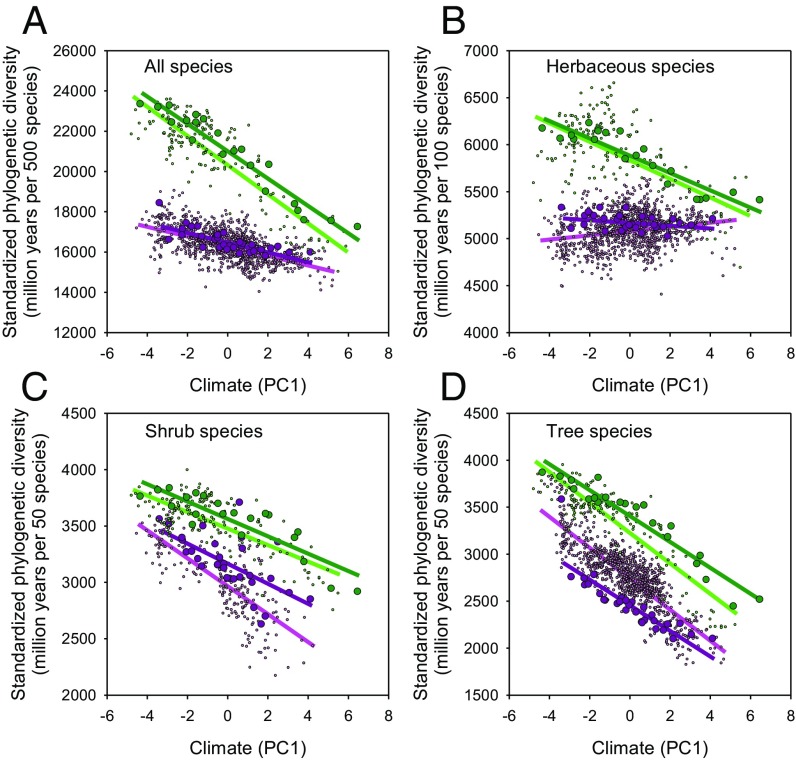
We conducted similar analyses for “standardized” phylogenetic diversity of angiosperms constructed by drawing 500 species at random from the provincial and state regional floras, as well as from the local floras, to control for the regional difference in species richness. Standardized phylogenetic diversity remained significantly higher in EAS than in ENA at both regional and local scales (P < 0.001 for both), after accounting for climate and sample area (Table 2). In both analyses, PC2 (precipitation seasonality) was not a significant effect (P > 0.8 at both regional and local scales) (Table 2). For a given value of PC1 (average temperature, temperature seasonality, and precipitation), standardized phylogenetic diversity tended to be higher in EAS than in ENA for all angiosperms (Fig. 2A), as well as for each growth form (Fig. 2 B–D).
Table 2.
ANCOVA of standardized phylogenetic diversity (based on 500 randomly selected angiosperm species) in EAS and ENA, with region (EAS vs. ENA) as the main effect and climate variables (PC1 and PC2) and area as covariates
| Source | Regional scale | Local scale | ||||||
| SS | df | F | P | SS | df | F | P | |
| Region | 0.022 | 1 | 118.0 | <0.001 | 0.484 | 1 | 1,411.5 | <0.001 |
| PC1 | 0.037 | 1 | 196.5 | <0.001 | 0.328 | 1 | 957.2 | <0.001 |
| PC2 | <0.001 | 1 | 0.1 | 0.823 | <0.001 | 1 | <0.1 | 0.854 |
| Area | <0.001 | 1 | 2.2 | 0.145 | 0.005 | 1 | 14.7 | <0.001 |
| Error | 0.009 | 47 | 0.429 | 1,252 | ||||
Model for floras at the regional scale: R2 = 0.947, adjusted least squares means, 4.314 and 4.217, respectively, for EAS and ENA. Model for floras at the local scale: R2 = 0.821; adjusted least squares means, 4.313 and 4.211, respectively, for EAS and ENA.
Fig. 2.
Comparison of standardized phylogenetic diversity of angiosperms between EAS (green) and ENA (pink) at two spatial scales (local floras with smaller pink and green dots; regional floras with larger dark-pink and dark-green dots) along a climatic gradient represented by the first principal component (PC1) of six climatic variables. Lines are linear least squares fits to the data. The four panels represent (A) all species, (B) herbs, (C) shrubs, and (D) trees.
When testing the simultaneous statistical effects of PC1, PC2, sample area, and region on either the phylogenetic diversity of a complete flora or standardized phylogenetic diversity, region was significant (P < 0.001) for floras at both regional and local scales. Similarly, when phylogenetic diversity in both regions was simultaneously regressed on PC1, PC2, and sample area, residuals of EAS floras were significantly (P < 0.05) larger than those of ENA for floras at both regional and local scales (Table 3). These results are consistent with those of the ANCOVA.
Table 3.
Average of residuals from multiple regressions with floras of both ENA and ENA included in each model
| Comparison | EAS | ENA | P* |
| Regional scale | |||
| Overall PD | 0.137 | −0.093 | <0.001 |
| Standardized PD | 0.011 | −0.007 | 0.020 |
| Local scale | |||
| Overall PD | 0.185 | −0.030 | <0.001 |
| Standardized PD | 0.028 | −0.004 | 0.023 |
From t tests.
Multiple regressions: PD ∼ PC1 + PC2 + area, where PD is phylogenetic diversity, PC1 and PC2 are the first two principal components of climate variables, and area is flora area.
Standardized phylogenetic diversity of angiosperms decreased with increasing values of PC1 (colder, more seasonal climates) in all of the regressions for EAS and ENA at both the regional and local scales (P < 0.05) (Table S3 and Fig. 2), except for herbaceous species in ENA at the regional scale (P = 0.75) and at the local scale (a positive relationship; P < 0.001) (Table S3). Of the three growth forms (i.e., herb, shrub, and tree), standardized phylogenetic diversity decreased most rapidly for trees and most slowly for herbs along the same climate gradient toward lower mean annual temperature, lower mean annual precipitation, lower minimum temperature, and higher temperature seasonality. Comparing the values of β (slopes) among the three growth forms in Table S3; Table S2 shows the relationships between PC1 and climate variables.
Table S3.
Regression coefficients (β) and coefficients of determination (R2) for the regression of standardized phylogenetic diversity on PC1 of six climatic variables
| EAS | ENA | |||||
| Sample | β | R2 | P | β | R2 | P |
| Regional scale | ||||||
| All angiosperms | −0.008 | 0.872 | <0.001 | −0.002 | 0.232 | <0.001 |
| Herbs | −0.005 | 0.750 | <0.001 | <0.001 | 0.004 | 0.747 |
| Shrubs | −0.010 | 0.757 | <0.001 | −0.013 | 0.400 | <0.001 |
| Trees | −0.019 | 0.897 | <0.001 | −0.023 | 0.797 | <0.001 |
| Local scale | ||||||
| All angiosperms | −0.009 | 0.635 | <0.001 | 0.001 | 0.009 | 0.002 |
| Herbs | −0.005 | 0.239 | <0.001 | 0.004 | 0.151 | <0.001 |
| Shrubs | −0.009 | 0.557 | <0.001 | −0.018 | 0.583 | <0.001 |
| Trees | −0.023 | 0.780 | <0.001 | −0.026 | 0.698 | <0.001 |
For the purpose of comparison among four different groups of species, the same threshold for selecting species at random (n = 50) was used to calculate standardized phylogenetic diversity for all four groups of species in this analysis.
Discussion
Plant species richness in EAS and ENA has been compared in several studies (e.g., refs. 5, 10, and 15). Phylogenetic diversity, which also incorporates the history of evolutionary diversification, is directly related to species richness (17), but whether the EAS−ENA anomaly extends to phylogenetic diversity has not been investigated until now. A comparison of the two measures of diversity might distinguish between contemporary ecology on one hand and regional history and geography on the other hand as causes of the diversity anomaly. The fact that phylogenetic diversity is strongly correlated with species diversity (r >0.97) suggests that the contrast in species richness between the regions reflects deep historical differences. Previous discussions of plant species diversity in EAS and ENA (5, 10, 13, 22) have suggested several factors that might be associated with the higher diversity in EAS: (i) greater climatic and topographic heterogeneity, (ii) more complex geological history, (iii) greater floristic antiquity, (iv) closer geographic connection of temperate forest floras to tropical forest floras in southern Asia, and (v) absence of extensive Quaternary glaciation (9, 10, 23). Phylogeographic studies (23–26) also point to the collision of the Indian plate with Eurasia, which began 55–40 Ma (27), as contributing to phylogenetic diversity in EAS by adding phylogenetic lineages to the flora and increasing the early topographic complexity of the region.
The India-Eurasia collision also dramatically modified climates in EAS (28, 29), which might have accelerated species formation and, over the long term, increased the phylogenetic diversity in EAS (24, 27). Analyses of several clades of vascular plants [e.g., those including Taiwania cryptomerioides (30), Cercidiphyllum japonicum (31), Tetracentron sinense (25), and Cyclocarya paliurus (24)] have revealed diversification coinciding with intensifcation of the Asian monsoon (24), suggesting that the Asian monsoon climate, with abundant summer precipitation, might have promoted species formation in EAS. Furthermore, the mild monsoon climate, in conjunction with the complex topography of EAS, provided refugia for many ancient lineages, including so-called “Tertiary relics” currently endemic to EAS (7, 32, 33).
Phylogenetic diversity in angiosperm floras (whether trees, shrubs, or herbs) is substantially higher in EAS than in ENA in areas with similar climates, independent of the number of species (Fig. 2). Two factors might account for this. First, North America was separated from tropical South America until the late Tertiary rise of the Isthmus of Panama (34–36), and the eastern part of the continent has remained isolated from American tropical floras by the Gulf of Mexico. In contrast, temperate floras in EAS have been directly connected across a broad geographic expanse with Asian tropical floras for tens of millions of years (37). Second, unlike ENA, in which many Tertiary relics (families and genera of both gymnosperms and angiosperms) became extinct with climate cooling and extensive glaciation in the late Tertiary, EAS suffered the loss of few Tertiary relics. Eucommia, Cercidiphylum, Sargentodoxa, Cyclocarya, Engelhardia, Platycarya, and Pterocarya are among the angiosperm genera characteristic of the Tertiary paleoflora of ENA (7, 38, 39) that became extinct there, while persisting in the modern flora of EAS. Most of these Tertiary relict genera are represented by single species with deep evolutionary relationships—stem lineages mostly reaching back 50−100 Ma.
Species in regional floras tend to be more closely related to each other across the phylogeny in ENA than in EAS. For example, the tree family Juglandaceae (walnuts and hickories) has 26 species in EAS and 22 species in ENA. The 26 species in EAS are spread across seven genera, four of which (Cyclocarya, Engelhardia, Platycarya, and Pterocarya) are ancient (early Tertiary) lineages that disappeared from North America, as noted above. In contrast, 20 of the 22 species (including nine hybrids) in ENA belong to a single genus, Carya, and are closely related, while the other two are species of Juglans.
Phylogenetic diversity decreases among regional and local floras with increasing values of the first climate PC axis, and hence with decreasing temperature and precipitation. This is consistent with the tropical niche conservatism hypothesis (6, 40), which suggests that stressful environments exclude lineages lacking cold tolerance mechanisms. However, phylogenetic diversity of angiosperms decreases with increasing PC1 much more rapidly in EAS than in ENA (Fig. 2A), and, at least among herbaceous species, phylogenetic diversity in the two continental regions tends to converge toward the higher end of the PC1 axis (Fig. 2A), i.e., at higher latitudes, confirming several previous studies (8, 15). The EAS–ENA anomaly in species and phylogenetic diversity of angiosperms emerges at mid-latitudes (45−55° N) and increases toward the subtropics.
Our finding that the region effect on species richness and phylogenetic diversity diminishes northward might reflect greater sharing by the northern parts of the two continents of the regional and historical processes that have generated contemporary biodiversity patterns in northern latitudes. The northernmost parts of Asia and North America were connected by the Bering Land Bridge during the mid-Cretaceous (41) and throughout most of the Tertiary, including the Pleistocene glacial periods (42). Thus, the paleofloras of the northern parts of Asia and North America were connected for a long period and share the same recent history of floristic development. In contrast, the southern portions of the two continents have differed substantially, owing to mountain building and the direct connection of temperate and tropical floras in Asia.
The strength of the relationship between phylogenetic diversity and climate increases from herbs through shrubs to trees. Specifically, phylogenetic diversity decreases more quickly with increasing PC1 (corresponding to decreasing temperature and precipitation) for trees than for herbs and shrubs (Table S3). This suggests that plants with larger body sizes are more sensitive to stressful climatic conditions than those with smaller body sizes. In a related study, Ricklefs and Latham (21) found that geographic ranges of plant genera distributed disjunctly between EAS and ENA were more strongly correlated among herbs than among trees. Qian et al. (43) showed that compared with herbs, woody angiosperms exhibited more phylogenetic conservatism in colder environments over an elevation gradient in the Changbai Mountains of northeastern China.
The weaker relationship between phylogenetic diversity and climate shown by herbs, compared with trees, might reflect the greater ability of smaller plants to take advantage of protected microhabitats. Herbs can avoid extreme cold by being annual, by producing underground buds and stems, or by remaining under snow cover during winter. Ricklefs and Latham (21) suggested that large woody plants have climate-dominated niches, whereas herbaceous plants have edaphic- and microhabitat-dominated niches. Interestingly, although standardized phylogenetic diversity in a given climate is higher in EAS than in ENA for both herbs (Fig. 2B) and trees (Fig. 2D), standardized phylogenetic diversity for herbs varies little with respect to climate PC1 in ENA (Fig. 2B), whereas the relationship for trees tends to converge between the two continental regions (Fig. 2D). This suggests that, regardless of the different evolutionary histories of the two continental regions, the relationship of phylogenetic diversity to climate is more similar for trees, the distributions of which are more sensitive to climate.
Methods
Study Area and Floristic Data.
We assembled checklists of angiosperm species at two spatial scales (regional and local) from published sources. For ENA, each regional species checklist included all of the species in a particular state, compiled based on the work of Kartesz (44); a local checklist included all species in a particular county. Species lists for counties in ENA were compiled according the US Department of Agriculture’s (USDA) Plants database (https://plants.usda.gov/java/), supplemented by local floras listed in appendix A in ref. 15. For EAS, a regional species checklist included all of the species in a province and was compiled based on the work of Wu et al. (45). Because few counties in mainland China have been botanized with the aim of generating complete species checklists (46, 47), most Chinese counties do not have reliable species checklists; however, more than 200 nature reserves or national parks in China have been well botanized for the purpose of compiling complete species checklists. In the present study, a local species checklist in China included all species in a nature reserve, national park, or similar local area. We compiled species lists for local floras in EAS based on the summaries provided by several authors (32, 48–50). For both EAS and ENA, to reduce the possibility of including incomplete checklists, we included only those species checklists that contained 500 or more angiosperm species. Our final dataset included 21 regional and 173 local species checklists for EAS and 31 regional and 1,084 local species checklists for ENA (Fig. 1 and Table S3). On average, each regional flora covered an area of 208,519 km2 in EAS and 96,115 km2 in ENA, and each local flora covered 1,104.9 km2 in EAS and 656.4 km2 in ENA.
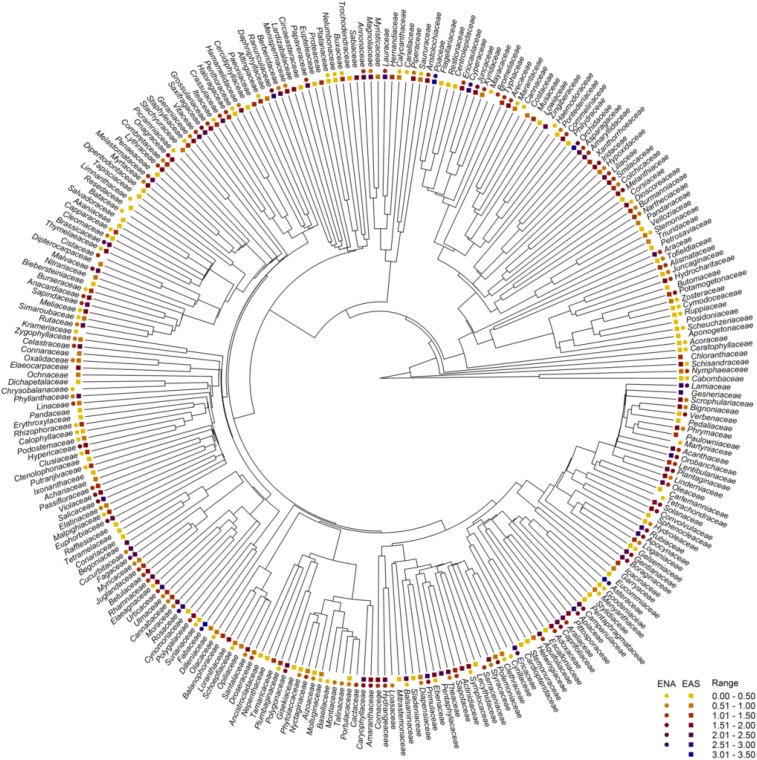
The botanical nomenclature of the species in the floras was standardized according to The Plant List (version 1.1; www.theplantlist.org). Infraspecific taxa were combined at the species level. Nonnative species in each flora were excluded. As a result, the 1,309 floras included 29,727 species of angiosperms, belonging to 3,312 genera and 267 families (51) (Fig. S2). The regional and local angiosperm floras in EAS included 23,753 species in 2,615 genera and 247 families, and those in ENA included 6,352 species in 1,303 genera and 186 families. Approximately 89% of the families and 47% of the genera in ENA also occurred in EAS. Each species was assigned a growth form (i.e., herb, shrub, tree, liana) according to Kartesz (44), Wu et al. (45), and the USDA Plants database (https://plants.usda.gov/java/).
Fig. S2.
Phylogeny showing the 267 families of angiosperms included in the study floras from EAS and ENA. Species diversity in each family in each of the two continental regions is shown by color on a log10 scale (indicated in the “Range” column). The phylogeny was extracted from that of Qian and Zhang (72) for all families of seed plants worldwide.
Phylogeny Reconstruction and Phylogenetic Diversity.
We used the megaphylogeny “Phytophylo” (52) (available at https://github.com/jinyizju/) as a backbone to generate a phylogenetic tree for the species included in this study. Phytophylo is an updated version of the megaphylogeny published by Zanne et al. (53), which was generated based on seven gene regions and 39 fossil calibrations. All angiosperm families worldwide have been completely resolved in Phytophylo. Of the 3,306 genera in EAS and ENA, 2,484 (75.1%) are included in Phytophylo. Among the woody plants, a higher proportion of the genera (87.0%) are included in Phytophylo. Thus, the phylogenetic tree used in this study was completely resolved at the family level and well resolved at the genus level. For those genera and species in our dataset that were absent from Phytophylo, we used the software S.PhyloMaker (52) (available at https://github.com/jinyizju/) to add them to their respective families (in the case of genera) and genera (in the case of species) in the megaphylogeny using Scenario 3, which is analogous to using Phylomatic with Bladj to generate a phylogeny (54), an approach commonly used in phylogenetic community studies (55, 56). We pruned the megaphylogeny to include only the 29,727 species present in our study floras. The resulting phylogenetic tree was much better resolved than those used in previous large-scale phylogenetic studies on plants, which were generated on the basis of megaphylogenies with Phylomatic (e.g., megaphylogenies R20031202, R20080417, R20091110, and R20120829; ref. 57) as backbones for generating phylogenetic trees; the majority of genera in a phylogenetic tree generated using these megaphylogenies are unresolved (52).
We used Faith’s (17) phylogenetic diversity (PD; i.e., the length of all of the phylogenetic branches required to span a given set of species) as a metric of phylogenetic diversity (e.g., refs. 58 and 59) in each flora. Faith’s PD consistently increases with species richness in an assemblage. To account for this effect of species richness, we took a rarefaction approach to calculate a standardized phylogenetic diversity. Specifically, for each angiosperm flora, we calculated PD for a randomly selected set of 500 species, and repeated this simulation 1,000 times to estimate a mean of randomized PD values. The approach that we used to standardize phylogenetic diversity based on a fixed number of species is commonly used in the current literature (60–62). In addition to calculating a standardized PD based on 500 species randomly selected from all growth forms in a flora, we calculated a standardized PD for each of the three growth forms: herb, shrub, and tree. We did not calculate standardized PD values for lianas, which were represented by few or no species in most of the floras. Because the number of species in each growth form was smaller than that for all angiosperm species, and also varied greatly among the three growth forms, we included floras in the analysis only if the number of species was ≥100 for herbs, ≥50 for shrubs, and ≥50 for trees. We then calculated a standardized PD for each growth form in each flora based on 1,000 randomizations.
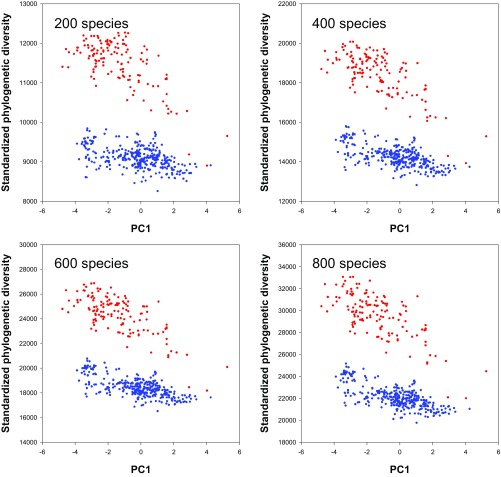
We used PhyloMeasures version 2.1 to calculate PD (63). To determine whether the use of different thresholds to select species might have influenced the result of an analysis, we conducted the following test. For those local floras in both continental regions with ≥800 angiosperm species (n = 468), we used four different thresholds to set species numbers: 200, 400, 600, and 800. We calculated standardized phylogenetic diversity for each threshold. As shown in Fig. S3, patterns of standardized phylogenetic diversity are nearly identical among the four thresholds, indicating that using different thresholds has no effect on the conclusions of our study.
Fig. S3.
The effect of using different numeric thresholds for selecting species on the relationship between standardized phylogenetic diversity and the score of each flora on PC1. As shown below, patterns of standardized phylogenetic diversity are nearly identical among the four thresholds, indicating that using different thresholds has no effect on conclusions drawn from the analysis. Red symbols pertain to EAS; blue symbols, to ENA.
Climate Data.
Mean annual temperature and mean annual precipitation are two key climate variables that determine plant distributions at a broad spatial extent (64). Stressful climates (including extreme cold and drought) and seasonal variation in climate (temperature and precipitation seasonality) also constrain the distributions of species (65–67). Accordingly, we used six variables to characterize the climate of each flora: mean annual temperature, mean annual precipitation, minimum temperature of the coldest month, precipitation during the driest month, temperature seasonality, and precipitation seasonality. We obtained values for these variables from the WorldClim database (68) (www.worldclim.org: variables bio1, bio12, bio6, bio14, bio4, and bio15, respectively). The mean value of each of the six climate variables was calculated for each flora using 30-arc-s resolution data. For local floras in China that lack electronic maps, we extracted climate data based on the location and area of each flora.
To remove redundancy among the climate variables and provide uncorrelated synthetic climate axes, we subjected the six climate variables to a principal components analysis (PCA) based on their correlation matrix. The first two climatic axes (PC1 and PC2) accounted for 90.7% of the variance in the six raw climate variables; PC1 accounted for more than twice as much variance as PC2 (Table S1). The other four axes each explained <5% of the variance in the climate variables. Thus, we used PC1 and PC2 as synthetic climate variables in this study. PC1 was strongly correlated with four of the six climate variables (mean annual temperature [r = −0.92], mean annual precipitation [−0.88], minimum temperature of the coldest month [−0.96], and temperature seasonality [0.92]; Table S2), whereas PC2 was strongly correlated with the other two climatic variables (precipitation of the driest month [0.83] and precipitation seasonality [−0.96]; Table S2). This suggests that PC1 represents a gradient of temperature seasonality and cold stress, while PC2 represents a gradient of precipitation seasonality and water stress.
Statistical Analysis.
We used Pearson’s correlation analysis and regression analysis to explore the relationships between species and phylogenetic diversity, and between diversity and climatic variables, within EAS and ENA. We used ANCOVA to explore differences in species richness and phylogenetic diversity between EAS and ENA, with continent as a main effect and the first two PC axes as covariates. This approach has been used to determine differences in species richness between regions (e.g., refs. 5 and 19). Because species diversity in an assemblage increases with sample area (69), as does the number of tip branches across a phylogenetic tree for the assemblage, and because sample area varied among floras both between and within the two continents, we included sample area as a covariate in analyses of covariance.
We also conducted multiple regression analyses to explore region effects on phylogenetic diversity after accounting for climate and sample area. Specifically, we used two multiple regression models. First, we regressed standardized phylogenetic diversity simultaneously on PC1, PC2, sample area, and region (coded as 1 for EAS and 2 for ENA). Second, we regressed standardized phylogenetic diversity simultaneously on PC1, PC2, and sample area, and used t tests to determine whether residuals from a regression differed between the two continental regions. To compare the relationship (regression slope) of standardized phylogenetic diversity to climate among different plant growth forms within regions, we selected 50 species of each growth form at random from each flora, and repeated this 1,000 times. Species richness, phylogenetic diversity, and sample area were log10-transformed in ANCOVA and regression analyses. SYSTAT version 7 (70) was used to conduct statistical analyses, and PC-ORD version 4 (71) was used for PCA. Species diversity, phylogenetic diversity, and sample area of each flora were log10-transformed in all statistical analyses to normalize the distributions of the data.
Acknowledgments
This study was partly supported by the Major Program of the National Natural Science Foundation of China (Grant 31590823) and the National Key R&D Program of China (Grant 2017YFC0505200).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703985114/-/DCSupplemental.
References
- 1.Currie DJ, Paquin V. Large-scale biogeographical patterns of species richness of trees. Nature. 1987;329:326–327. [Google Scholar]
- 2.Kleidon A, Mooney HA. A global distribution of biodiversity inferred from climatic constraints: Results from a process-based modelling study. Glob Change Biol. 2000;6:507–523. [Google Scholar]
- 3.Kreft H, Jetz W. Global patterns and determinants of vascular plant diversity. Proc Natl Acad Sci USA. 2007;104:5925–5930. doi: 10.1073/pnas.0608361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qian H, Ricklefs RE. A latitudinal gradient in large-scale beta diversity for vascular plants in North America. Ecol Lett. 2007;10:737–744. doi: 10.1111/j.1461-0248.2007.01066.x. [DOI] [PubMed] [Google Scholar]
- 5.Ricklefs RE, Qian H, White PS. The region effect on mesoscale plant species richness between eastern Asia and eastern North America. Ecography. 2004;27:129–136. [Google Scholar]
- 6.Latham RE, Ricklefs RE. Global patterns of tree species richness in moist forests: Energy-diversity theory does not account for variation in species richness. Oikos. 1993;67:325–333. [Google Scholar]
- 7.Latham RE, Ricklefs RE. Continental comparisons of temperate-zone tree species diversity. In: Ricklefs RE, Schluter D, editors. Species Diversity: Historical and Geographical Perspectives. Univ of Chicago Press; Chicago: 1993. pp. 294–314. [Google Scholar]
- 8.Qian H. A comparison of the taxonomic richness of temperate plants in East Asia and North America. Am J Bot. 2002;89:1818–1825. doi: 10.3732/ajb.89.11.1818. [DOI] [PubMed] [Google Scholar]
- 9.Qian H, Ricklefs RE. A comparison of the taxonomic richness of vascular plants in China and the United States. Am Nat. 1999;154:160–181. doi: 10.1086/303230. [DOI] [PubMed] [Google Scholar]
- 10.Qian H, Ricklefs RE. Large-scale processes and the Asian bias in species diversity of temperate plants. Nature. 2000;407:180–182. doi: 10.1038/35025052. [DOI] [PubMed] [Google Scholar]
- 11.Schluter D, Ricklefs RE. Convergence and the regional component of species diversity. In: Ricklefs RE, Schluter D, editors. Species Diversity in Ecological Communities. Historical and Geographical Perspectives. Univ of Chicago Press; Chicago: 1993. pp. 230–242. [Google Scholar]
- 12.Wolfe JA. Some aspects of plant geography of the Northern Hemisphere during the Late Cretaceous and Tertiary. Ann Mo Bot Gard. 1975;62:264–279. [Google Scholar]
- 13.Qian H. Floristic relationships between Eastern Asia and North America: Test of Gray’s hypothesis. Am Nat. 2002;160:317–332. doi: 10.1086/341523. [DOI] [PubMed] [Google Scholar]
- 14.Ricklefs RE. Cladogenesis and morphological diversification in passerine birds. Nature. 2004;430:338–341. doi: 10.1038/nature02700. [DOI] [PubMed] [Google Scholar]
- 15.Qian H, Fridley JD, Palmer MW. The latitudinal gradient of species-area relationships for vascular plants of North America. Am Nat. 2007;170:690–701. doi: 10.1086/521960. [DOI] [PubMed] [Google Scholar]
- 16.Cisneros LM, et al. Multiple dimensions of bat biodiversity along an extensive tropical elevational gradient. J Anim Ecol. 2014;83:1124–1136. doi: 10.1111/1365-2656.12201. [DOI] [PubMed] [Google Scholar]
- 17.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- 18.Takhtajan AL. Flowering Plants: Origin and Dispersal. Oliver & Boyd; Edinburgh: 1969. [Google Scholar]
- 19.Smith AC. The Pacific as a Key to Flowering Plant History. Univ of Hawaii Press; Honolulu: 1970. pp. 1–26. [Google Scholar]
- 20.Wu C-Y, editor. The Vegetation of China. Science Press; Beijing: 1980. [Google Scholar]
- 21.Ricklefs RE, Latham RE. Intercontinental correlation of geographical ranges suggests stasis in ecological traits of relict genera of temperate perennial herbs. Am Nat. 1992;139:1305–1321. [Google Scholar]
- 22.Qian H, White PS, Song J-S. Effects of regional vs. ecological factors on plant species richness: An intercontinental analysis. Ecology. 2007;88:1440–1453. doi: 10.1890/06-0916. [DOI] [PubMed] [Google Scholar]
- 23.Qiu Y-X, Fu C-X, Comes HP. Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol Phylogenet Evol. 2011;59:225–244. doi: 10.1016/j.ympev.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 24.Kou Y, et al. The antiquity of Cyclocarya paliurus (Juglandaceae) provides new insights into the evolution of relict plants in subtropical China since the late Early Miocene. J Biogeogr. 2016;43:351–360. [Google Scholar]
- 25.Sun Y-X, et al. Chloroplast phylogeography of the East Asian Arcto-Tertiary relict Tetracentron sinense (Trochodendraceae) J Biogeogr. 2014;41:1721–1732. [Google Scholar]
- 26.Renner SS. Available data point to a 4-km-high Tibetan Plateau by 40 Ma, but 100 molecular-clock papers have linked supposed recent uplift to young node ages. J Biogeogr. 2016;43:1479–1487. [Google Scholar]
- 27.Favre A, et al. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol Rev Camb Philos Soc. 2015;90:236–253. doi: 10.1111/brv.12107. [DOI] [PubMed] [Google Scholar]
- 28.Guo ZT, et al. Onset of Asian desertification by 22 Myr ago inferred from loess deposits in China. Nature. 2002;416:159–163. doi: 10.1038/416159a. [DOI] [PubMed] [Google Scholar]
- 29.Harris N. The elevation history of the Tibetan Plateau and its implications for the Asian monsoon. Palaeogeogr Palaeoclimatol Palaeoecol. 2006;241:4–15. [Google Scholar]
- 30.Chou Y-W, Thomas PI, Ge X-J, LePage BA, Wang C-N. Refugia and phylogeography of Taiwania in East Asia. J Biogeogr. 2011;38:1992–2005. [Google Scholar]
- 31.Qi X-S, et al. Molecular data and ecological niche modelling reveal a highly dynamic evolutionary history of the East Asian Tertiary relict Cercidiphyllum (Cercidiphyllaceae) New Phytol. 2012;196:617–630. doi: 10.1111/j.1469-8137.2012.04242.x. [DOI] [PubMed] [Google Scholar]
- 32.Qian H, et al. Phytogeographical analysis of seed plant genera in China. Ann Bot (Lond) 2006;98:1073–1084. doi: 10.1093/aob/mcl192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ying T-S, Zhang Y-L, Boufford DE. The Endemic Genera of Seed Plants of China. Science Press; Beijing: 1993. [Google Scholar]
- 34.Coates AG. The geologic evolution of the Central American isthmus. In: Jackson JBC, Budd AF, Coates AG, editors. Evolution and Environment in Tropical America. Univ of Chicago Press; Chicago: 1996. pp. 21–56. [Google Scholar]
- 35.Hallam A. An Outline of Phanerozoic Biogeography. Oxford Univ Press; Oxford, UK: 1994. [Google Scholar]
- 36.Stehli FG, Webb SD, editors. The Great American Biotic Interchange. Plenum; New York: 1985. [Google Scholar]
- 37.Hall R, Holloway JD, editors. Biogeography and Geological Evolution of SE Asia. Backhuys Publishers; Leiden, The Netherlands: 1998. [Google Scholar]
- 38.Call V, Dilcher D. The fossil record of Eucommia (Eucommiaceae) in North America. Am J Bot. 1997;84:798–814. [PubMed] [Google Scholar]
- 39.Tiffney BH. Fruits and seeds of the Tertiary Brandon lignite, VII: Sargentodoxa (Sargentodoxaceae) Am J Bot. 1993;80:517–523. doi: 10.1002/j.1537-2197.1993.tb13834.x. [DOI] [PubMed] [Google Scholar]
- 40.Wiens JJ, Donoghue MJ. Historical biogeography, ecology and species richness. Trends Ecol Evol. 2004;19:639–644. doi: 10.1016/j.tree.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 41.Sanmartín I, Enghoff H, Ronquist F. Patterns of animal dispersal, vicariance and diversification in the Holarctic. Biol J Linn Soc Lond. 2001;73:345–390. [Google Scholar]
- 42.McKenna MC. Holarctic landmass rearrangements, cosmic events, and Cenozoic terrestrial organisms. Ann Mo Bot Gard. 1983;70:459–489. [Google Scholar]
- 43.Qian H, Hao Z, Zhang J. Phylogenetic structure and phylogenetic diversity of angiosperm assemblages in forests along an elevational gradient in Changbaishan, China. J Plant Ecol. 2014;7:154–165. [Google Scholar]
- 44.Kartesz JT. A synonymized checklist and atlas with biological attributes for the vascular flora of the United States, Canada, and Greenland. In: Kartesz JT, Meacham CA, editors. Synthesis of the North American Flora. Version 1.0. North Carolina Botanical Garden; Chapel Hill, NC: 1999. [Google Scholar]
- 45.Wu C-Y, Raven PH, Hong D-Y, editors. Flora of China. Vol 2–25 Science Press, Beijing; Missouri Botanical Garden; St. Louis: 1994–2013. [Google Scholar]
- 46.Qian H. Environmental determinants of woody plant diversity at a regional scale in China. PLoS One. 2013;8:e75832. doi: 10.1371/journal.pone.0075832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qian H, Ricklefs RE. Latitude, tree species diversity and the metabolic theory of ecology. Glob Ecol Biogeogr. 2011;20:362–365. [Google Scholar]
- 48.Qian H, Chen S. Reinvestigation on species richness and environmental correlates of bryophytes at a regional scale in China. J Plant Ecol. 2016;9:734–741. [Google Scholar]
- 49.Wang K. National Nature Reserves of China. Anhui Scientific and Technology; Hefei, China: 2003. [Google Scholar]
- 50.Zhao SQ, Fang JY, Peng CH, Tang ZY. Relationships between species richness of vascular plants and terrestrial vertebrates in China: Analyses based on data of nature reserves. Divers Distrib. 2006;12:189–194. [Google Scholar]
- 51.APG An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 2016;181:1–20. [Google Scholar]
- 52.Qian H, Jin Y. An updated megaphylogeny of plants, a tool for generating plant phylogenies and an analysis of phylogenetic community structure. J Plant Ecol. 2016;9:233–239. [Google Scholar]
- 53.Zanne AE, et al. Three keys to the radiation of angiosperms into freezing environments. Nature. 2014;506:89–92. doi: 10.1038/nature12872. [DOI] [PubMed] [Google Scholar]
- 54.Webb CO, Ackerly DD, Kembel SW. Phylocom: Software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics. 2008;24:2098–2100. doi: 10.1093/bioinformatics/btn358. [DOI] [PubMed] [Google Scholar]
- 55.Kooyman R, Rossetto M, Cornwell W, Westoby M. Phylogenetic tests of community assembly across regional to continental scales in tropical and subtropical rain forests. Glob Ecol Biogeogr. 2011;20:707–716. [Google Scholar]
- 56.Moro MF, et al. The role of edaphic environment and climate in structuring phylogenetic pattern in seasonally dry tropical plant communities. PLoS One. 2015;10:e0119166. doi: 10.1371/journal.pone.0119166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Webb CO, Ackerly DD, Kembel SW. 2011 Phylocom: Software for the analysis of phylogenetic community structure and character evolution, with phylomatic. User’s Manual Version 4.2. Available at phylodiversity.net/phylocom/. Accessed January 10, 2017.
- 58.Chao A, Chiu C-H, Jost L. Unifying species diversity, phylogenetic diversity, functional diversity, and related similarity and differentiation measures through Hill numbers. Annu Rev Ecol Evol Syst. 2014;45:297–324. [Google Scholar]
- 59.Li R, Kraft NJ, Yu H, Li H. Seed plant phylogenetic diversity and species richness in conservation planning within a global biodiversity hotspot in eastern Asia. Conserv Biol. 2015;29:1552–1562. doi: 10.1111/cobi.12586. [DOI] [PubMed] [Google Scholar]
- 60.Slik JWF, et al. Environmental correlates for tropical tree diversity and distribution patterns in Borneo. Divers Distrib. 2009;15:523–532. [Google Scholar]
- 61.Tang G, et al. Phylogenetic support for the Tropical Niche Conservatism Hypothesis despite the absence of a clear latitudinal species richness gradient in Yunnan’s woody flora. Biogeosci Discuss. 2014;11:7055–7077. [Google Scholar]
- 62.Qian H, Field R, Zhang J, Zhang J, Chen S. Phylogenetic structure and ecological and evolutionary determinants of species richness for angiosperm trees in forest communities in China. J Biogeogr. 2016;43:603–615. [Google Scholar]
- 63.Tsirogiannis C, Sandel B. PhyloMeasures: A package for computing phylogenetic biodiversity measures and their statistical moments. Ecography. 2016;39:709–714. [Google Scholar]
- 64.Whittaker RH. Communities and Ecosystems. Macmillan; London: 1975. [Google Scholar]
- 65.Kamilar JM, Beaudrot L, Reed KE. Climate and species richness predict the phylogenetic structure of African mammal communities. PLoS One. 2015;10:e0121808. doi: 10.1371/journal.pone.0121808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patrick LE, Stevens RD. Phylogenetic community structure of North American desert bats: Influence of environment at multiple spatial and taxonomic scales. J Anim Ecol. 2016;85:1118–1130. doi: 10.1111/1365-2656.12529. [DOI] [PubMed] [Google Scholar]
- 67.Weigelt P, et al. Global patterns and drivers of phylogenetic structure in island floras. Sci Rep. 2015;5:12213. doi: 10.1038/srep12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. [Google Scholar]
- 69.Rosenzweig ML. Species Diversity in Space and Time. Cambridge Univ Press; Cambridge, UK: 1995. [Google Scholar]
- 70.Wilkinson L, Hill M, Welna JP, Birkenbeuel GK. 1992. SYSTAT for Windows: Statistics (SYSTAT, Evanston, IL)
- 71.McCune B, Mefford MJ. 1999. PC-ORD: Multivariate Analysis of Ecological Data, version 4.0 (MjM Software Design, Gleneden Beach, OR)
- 72.Qian H, Zhang J. Using an updated time-calibrated family-level phylogeny of seed plants to test for non-random patterns of life forms across the phylogeny. J Syst Evol. 2014;52:423–430. [Google Scholar]