Significance
Increasing evidence suggests that extracellular vesicles (EVs) can transfer genetic material to recipient cells. However, the mechanism and role of this phenomenon are largely unknown. Here we have made a remarkable discovery: EVs can harbor the full mitochondrial genome. These extracellular vesicles can in turn transfer their mtDNA to cells with impaired metabolism, leading to restoration of metabolic activity. We determined that hormonal therapy induces oxidative phosphorylation-deficient breast cancer cells, which can be rescued via the transfer of mtDNA-laden extracellular vesicles. Horizontal transfer of mtDNA occurred in cancer stem-like cells and was associated with increased self-renewal potential of these cells, leading to resistance to hormonal therapy. We propose that mtDNA transfer occurs in human cancer via EVs.
Keywords: exosomes, mitochondrial DNA, cancer stem cells, hormonal therapy, metastasis
Abstract
The horizontal transfer of mtDNA and its role in mediating resistance to therapy and an exit from dormancy have never been investigated. Here we identified the full mitochondrial genome in circulating extracellular vesicles (EVs) from patients with hormonal therapy-resistant (HTR) metastatic breast cancer. We generated xenograft models of HTR metastatic disease characterized by EVs in the peripheral circulation containing mtDNA. Moreover, these human HTR cells had acquired host-derived (murine) mtDNA promoting estrogen receptor-independent oxidative phosphorylation (OXPHOS). Functional studies identified cancer-associated fibroblast (CAF)-derived EVs (from patients and xenograft models) laden with whole genomic mtDNA as a mediator of this phenotype. Specifically, the treatment of hormone therapy (HT)-naive cells or HT-treated metabolically dormant populations with CAF-derived mtDNAhi EVs promoted an escape from metabolic quiescence and HTR disease both in vitro and in vivo. Moreover, this phenotype was associated with the acquisition of EV mtDNA, especially in cancer stem-like cells, expression of EV mtRNA, and restoration of OXPHOS. In summary, we have demonstrated that the horizontal transfer of mtDNA from EVs acts as an oncogenic signal promoting an exit from dormancy of therapy-induced cancer stem-like cells and leading to endocrine therapy resistance in OXPHOS-dependent breast cancer.
Extracellular vesicles (EVs) are mediators of juxtacrine and paracrine signaling required for metastatic progression (1, 2). Specifically, tumor- and stromal cell-derived EVs have been shown to be potent regulators of tumor progression and resistance to therapy by transferring their cargo (proteins, lipids, mRNA, miRNA) into recipient cells, promoting signaling cascades and epigenetic changes. Although, cancer cell-derived EVs have been shown to regulate premetastatic niche formation, organotropism, migration, invasion, stemness, and survival, little is known about their role in regulating the metabolism of cancers (3).
Even though the estrogen receptor (ER)-positive variant of breast cancer is touted as the most indolent and favorable, the majority of breast cancer deaths are in fact from this subtype (4). There are several features of this category of breast cancers that likely account for this outcome. The first is that metastatic relapse can occur many years after initial diagnosis of primary disease. The second is that, when metastases do occur, they are invariably in many locations. This observation suggests that these dormant/sleeping metastatic cells are globally awakened, as if by a systemic infection activating the metabolism of these formerly quiescent cells. The third is that, once the cancer cells awaken into full-blown metastatic disease, they are largely resistant to ER-directed therapies (i.e., hormonal therapy, HT) (5). We suggest that these processes are not only linked but underlie the lethal features of metastatic disease.
The metabolic features of ER+ breast cancer and its evolution from HT-sensitive (HTS)/responsive to dormant (HTD) and the eventual development of HT-resistant (HTR) disease is poorly studied, in part due to a paucity of preclinical models. We suggest that mitochondrial activity and the regulation of mitochondrial biogenesis is central to this process. MtDNA levels and mutational status within cancers have recently become of interest, as these are associated with the development of a wide variety of cancers and resistance to therapies (6–8). For example, nontumorigenic cancer cells lacking mtDNA could reacquire tumorigenic potential in vivo following the acquisition of host mtDNA (9). Complementation of deleterious mtDNA mutations increases tumorigenicity, while expression of such genetic lesions impedes cancer cell growth (10). We hypothesized that EVs could regulate the metabolic features of HTR disease. We determined that as ER+ cancers transition from an HTS/dormant state to a resistant one, they acquire host mtDNA, which promotes oxidative phosphorylation (OXPHOS), an exit from dormancy, and the development of HTR disease. We discovered a mechanism of mtDNA transfer. Specifically, we identified the full mitochondrial genome packaged in cancer-associated fibroblast (CAF)-derived EVs and in EVs from patients with HTR metastatic disease. These EVs could transfer their mitochondrial genome in HTS/HTD cancer cells, which promoted the growth of these cells. These findings suggest that as cancers evolve they may regulate their metabolism/OXPHOS capacity through the elimination and acquisition of mtDNA from “healthy” cell-derived EVs.
Results
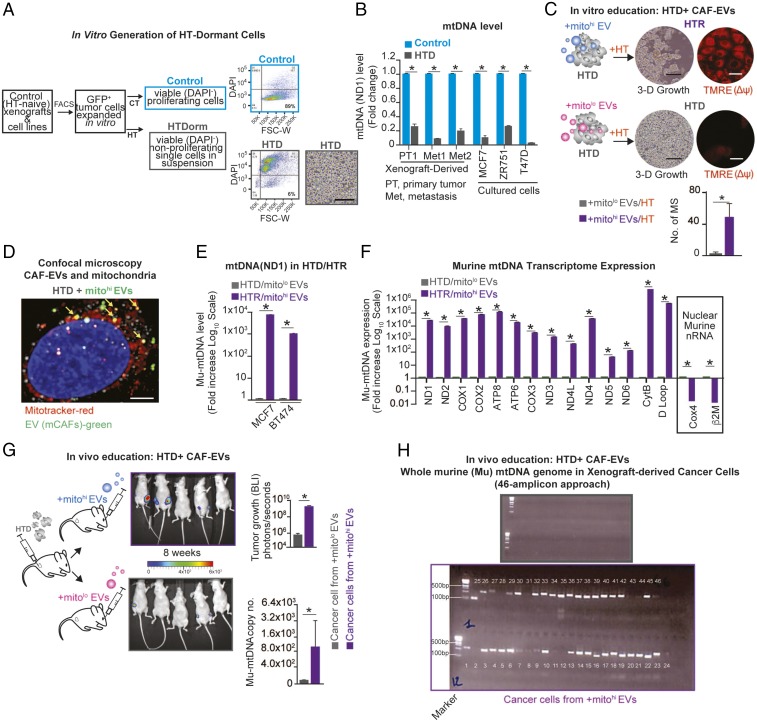
Mitochondrial Genome Identified in EVs from Patients with HTR Disease.
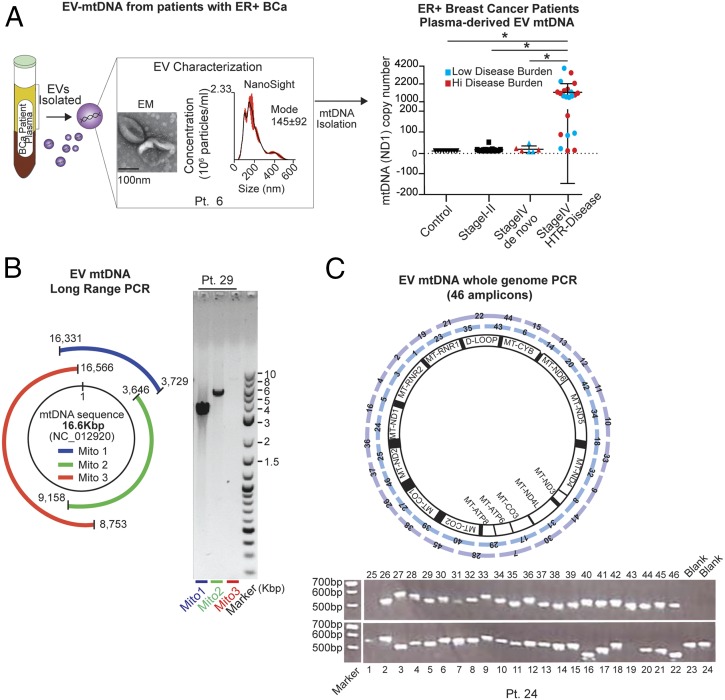
Given the growing recognition of the importance of mtDNA copy number in the evolution of cancers, and particularly in resistance to therapy (6–8), we hypothesized that these features might be captured in circulating EVs. Although the mitochondrial ND1 gene was previously identified in the EVs of glioblastoma cell lines and astrocytes (11), the presence of mitochondrial DNA has not been examined in EVs from patients with breast cancer. To address this hypothesis, we isolated and characterized EVs by electron microscopy and NanoSight analysis, which demonstrated cup-shaped membrane vesicles ∼140 nm in diameter from the plasma of patients with metastatic ER+ breast cancer who at the time of blood draw had HTR disease (Fig. 1A and SI Appendix, Table S1). Genomic DNA is commonly found in EVs. However, in agreement with the current literature, DNase treatment of EVs remarkably lowered overall DNA content, which indicates that the majority of DNA resides on the outer surface of EVs (12, 13). To determine the content of DNA inside the vesicles, EVs were treated with DNase to digest cell-free DNA collected during EV isolation. Our findings revealed that 19/22 (∼86%) of these patients had EVs containing high levels of mtDNA, irrespective of tumor burden (Fig. 1A, mitochondrial ND1 gene copy number). Conversely, EVs isolated from the plasma of (i) individuals who were never diagnosed with cancer (n = 9), (ii) patients who were newly diagnosed with early-stage (I–II) ER+ breast cancer (following tumor removal) and thus had no evidence of cancer (n = 12), and (iii) patients with de novo untreated stage IV cancer (n = 6) had either undetectable or very low levels of ND1 copies (Fig. 1A). These data suggest that the presence of the ND1 mtDNA in circulating EVs is selective for patients with HTR disease and is not simply a reflection of metastatic disease burden. We and others have previously described the presence of genomic DNA in EVs from cancer-derived cell lines and in patients with pancreatic cancer (13). However, in our cohort of patients with metastatic HTR disease, only 7/22 (32%) expressed the nuclear gene encoding GAPDH (SI Appendix, Fig. S1A and Table S1). Not only was the ND1 mitochondrial gene expressed in circulating EVs from HTR patients, but also the complete mitochondrial genome, as determined by long-range PCR (three PCRs amplifying 3.9-kb, 5.5-kb, and 7.8-kb amplicons encompassing the complete 16.6-kb circular mitochondrial genome) and by whole mtDNA genome PCR amplification of 46 amplicons (Fig. 1 B and C and SI Appendix, Fig. S1B).
Fig. 1.
Mitochondrial genome identified in EVs from the plasma of patients with HTR disease. (A) Circulating EVs were isolated from the plasma (5–10 mL) of 22 patients with HTR metastatic disease [patients with high-volume disease (>10% of organ involvement) are denoted in red, and those with low-volume disease (<1% of organ involvement) are denoted in blue], 9 healthy controls, 12 patients with early-stage breast cancer following removal of their cancer, and 6 patients with de novo metastatic breast cancer who had not yet received treatment. MtDNA copy number quantification was determined after DNase treating EVs by qPCR for the ND1 gene (10 ng of DNA was used). A representative NanoSight plot (mode and size) and electron micrographs are also shown (patient 6). Data are reported as the mean ± SD for experiments performed in triplicate; each point represents a patient value. *P < 0.005 (Student’s t test). (B) Schematic and representative gel electrophoresis image of long-range PCR (three contiguous amplicons: Mito1, 3.9 kbp; Mito2, 5.5 kbp; Mito3, 7.8 kbp) encoding the complete 16.6-kbp circular mitochondrial genome purified from EVs in the plasma of patient 29 (2 ng of total DNA). (C) Schematic and a representative gel electrophoresis image of whole-genome amplification (using 46 overlapping PCR amplicons covering the complete mtDNA genome) from patient-derived EV-DNA (1 ng for each PCR) (EVs were derived from patient 24, see uncropped gel SI Appendix, Fig. S1B). For EV analysis of all patients, see SI Appendix, Table S1.
Development of Xenograft Models of HTS to HTR Disease.
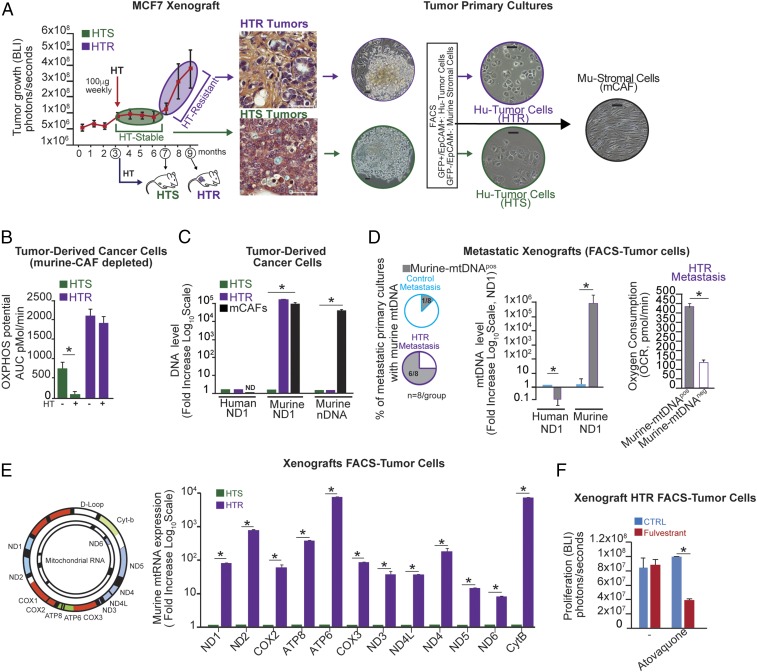
To study these phenomena in vivo, we developed experimental models that capture some of the essential features of the clinical scenario described above (the development of HTR disease) (14). ER+ (MCF7-Luciferase/GFP+) mammary fat pad (MFP) xenografts were established in the absence of estradiol supplementation (14). To identify the distinguishing characteristics of HTR disease, tumor-bearing mice were treated with HT (fulvestrant). Interestingly, we observed a bimodal growth pattern with no growth for 4 mo (HTS) followed by exponential growth (HTR) (Fig. 2A).
Fig. 2.
The horizontal transfer of host (murine) mtDNA associates with HTR disease. (A) Representative tumor-growth kinetics (determined by bioluminescence imaging, BLI) of GFP+/Luciferase+MCF7 xenografts (n = 10) treated weekly with fulvestrant once MFP tumors were established (3 mo after inoculation), which led to a 4-mo period of disease stability on HT (HTS) followed by exponential growth on HT (HTR). Representative H&E images of tumors and primary cultures from unsorted as well as FACS-purified cancer cells (GFP+/Epcam+) and mCAFs (GFP−/Epcam) are shown. (Scale bars, 100 μm.) (B) OXPHOS potential in tumor-derived cancer cells ± HT (fulvestrant, 10 μM) in A was measured by Seahorse technology. (C) DNA level expressed as fold change (human and murine nuclear and mitochondrial) (log10 scale) in cancer cells and mCAFs isolated from HTS and HTR disease in A. (D) Percentage of metastatic lesions isolated by FACS from tumor-bearing mice (n = 8 per group) expressing mu-mtDNA as determined by ND1 qPCR (2 ng of total DNA). OCRs in mu-mtDNA+ and control HTR cells were determined by Seahorse technology. (E) Mu-mtRNA expression of 12 genes (ND1, ND2, COX2, ATP8, ATP6, COX3, ND3, ND4L, ND4, ND5, ND6, CytB) is shown as fold change (log10 scale) determined by qRT-PCR in the HTR and HTS cells in A. (F) Proliferation potential in the presence of the mitochondrial complex III inhibitor atovaquone (1 μM) in HTR cells with/without HT (fulvestrant, 10 μM). The bar graph shows BLI values at the end point of the experiment (7 d). Data in B–F are reported as the mean of three independent experiments; error bars indicate SD; *P < 0.05 (student’s t test).
Metabolic Features of HTS Versus HTR Disease.
We, and others, have demonstrated that luminal (ER+) breast cancers are metabolically dependent on OXPHOS rather than on aerobic glycolysis (SI Appendix, Fig. S2 A and B) (14, 15). We hypothesized that the metabolic profile of HTR tumor cells might differ from that of HTS tumor cells.
To test this, we established primary cultures from two different tumor phases (HTS and HTR) and noted an enrichment of murine cancer-associated fibroblasts (mCAFs) in the HTR primary cultures compared with HTS ones (Fig. 2A).
These data were also corroborated in our recent paper in which increased CAF proliferation in HTR disease was quantified by different techniques (16). Additionally, desmin immunohistochemical analysis of these HTR tissues confirmed the presence of CAFs, suggesting that the communication between cancer and stroma cells could instigate resistance to HT (SI Appendix, Fig. S2C). By FACS analysis we separated cancer cells (EpCAM+/GFP+) from mCAFs (EpCAM−/GFP−) in these tumor specimens (Fig. 2A) and examined OXPHOS capacity in these FACS-sorted tumor-derived cells in the presence or absence of HT (fulvestrant). The OXPHOS potential of HTS cells was inhibited with HT, while HTR tumor-derived cells, which were unaffected by HT, had a threefold greater OXPHOS capacity than HTS cells (Fig. 2B).
Host mtDNA Transfer in Xenograft Models of HTR Disease.
Because depletion of mtDNA, as well as mtDNA mutations, abrogates OXPHOS function along with tumorigenic potential, we hypothesized that HTR lesions would carry wild-type mtDNA and higher mtDNA copy number, explaining their increased OXPHOS capacity. We performed detailed genetic characterization of mtDNA for the presence of mutations. MtDNA mutations were found in HTS cells, suggesting that reduced OXPHOS potential in HTS disease may be due to the presence of such mtDNA genotypes (SI Appendix, Table S2). In contrast, HTR cells harbored no such mutations and expressed murine mtDNA (mu-mtDNA) sequences (but not nuclear DNA, mu-nDNA) at levels equal to those found in mCAFs, whereas there was no evidence of mu-mtDNA in HTS tumor-derived cells (Fig. 2C). Additionally, HTS-derived xenograft cultures did not show outgrowth of mCAFs in vitro (Fig. 2A). As in the MCF7 xenograft model, we found the presence of mu-mtDNA in one of three (33%) patient-derived tumor graft (PDX) cancer cell lines that we recently established from HRT bone metastases (SI Appendix, Fig. S2D) (16).
To study the relevance of OXPHOS in the context of HTR metastatic disease, tumor-bearing mice (MCF7 model) underwent mastectomies followed by adjuvant HT (tamoxifen) or vehicle control and were followed for ∼9 mo. One hundred percent of the control mice had evidence of one or two metastases. Although 90% of the HT-treated mice had no evidence of disease, 10% had widespread (>20) HTR metastases involving multiple organs (SI Appendix, Fig. S2E) (14). Cancer cells were from HTR metastatic lesions isolated by FACS (for GFP), and the presence of mtDNA was determined. Six of eight HTR metastases but only one of eight treatment-naive metastases expressed mu-mtDNA (Fig. 2D). Interestingly, mu-mtDNA–harboring (mtDNAhi) HTR cells had a higher oxygen consumption rate (OCR) than HTR cells lacking mu-mtDNA (Fig. 2D). Additionally, in agreement with a functional role of mu-mtDNA transfer, these mu-mtDNAhi HTR cells expressed the complete mu-mtDNA–encoded transcriptome (Fig. 2E).
To determine the role of mitochondrial bioenergetics in HTR disease, mu-mtDNAhi HTR cells and HTS cells (with no mu-mtDNA) were treated with the OXPHOS inhibitor atovaquone with and without HT (fulvestrant). We demonstrated that atovaquone resensitized HTR cells to HT (Fig. 2F). On the contrary, HTS cells displayed the same sensitivity to fulvestrant and mitochondrial respiratory complex inhibitors, suggesting that OXPHOS inhibitors and HT did not have a synergistic effect on cell proliferation and/or cell death (SI Appendix, Fig. S2F). Taken together, our data suggest that host (murine)-derived mtDNA is acquired as cancer cells or metastases transition from an HTS state to one characterized by HTR.
CAF EVs Contain the Full Mitochondrial Genome.
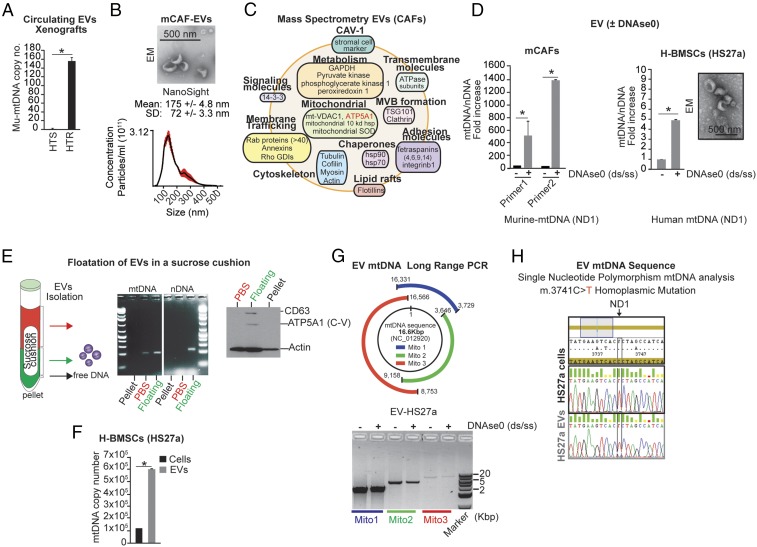
We have described and quantified the enrichment of CAFs in HTR disease from two different models, MCF7 and ZR751 (16). It is well accepted that CAFs and recruited bone marrow-derived stromal cells play a critical role in cancer initiation, growth, invasion, metastasis, and therapeutic resistance through the production of growth factors, cytokines, chemokines, catabolites, extracellular matrix proteins, and EVs, which modulate the behavior of cancers, including their metabolism (15–18). Given that EVs have been shown to express and horizontally transfer genetic material (e.g., DNA, miRNA) to recipient cells (19), we hypothesized that CAF-derived EVs could (i) harbor the mitochondrial genome and (ii) transfer the mtDNA into the mitochondria of HT-treated tumor cells. We demonstrated that mice harboring HTR xenografts had circulating EVs with a 150-fold higher mu-mtDNA ND1 level than mice HTS xenografts (Fig. 3A). Moreover, the ratio of mtDNA/genomic DNA was markedly elevated in these EVs (SI Appendix, Fig. S2G).
Fig. 3.
Stromal-derived EVs harbor the mitochondrial genome. (A) qPCR of mu-mtDNA copy number (ND1 gene) in circulating EV DNA isolated from HTR and HTS tumor-bearing mice (n = 3 per group). (B) Electron microscopy (scale bar, 500 nm) and NanoSight analyses of EVs isolated from mCAFs. The concentration of particles is reported as mean ± SD. (C) Representative exosomal proteins identified by quantitative mass spectrometry of mCAF-derived EVs (SI Appendix, Table S3). (D) Ratio of mtDNA/nDNA level by qPCR (two unique primer sets were used for murine ND1, and one set each was used for human ND1, murine GAPDH, and human GAPDH) in EVs isolated from mCAFs and human bone marrow stromal cells (H-BMSCs, HS27a) ± DNase0 treatment to eliminate exogenous DNA contamination (Materials and Methods). A representative electron microscopy image of HS27a EVs is shown. (Scale bar, 500 nm.) (E) Schematic and representative PCR gel electrophoresis of mtDNA (ND1) and nDNA (GAPDH) from fractions following sucrose cushion purification (SI Appendix, Materials and Methods) of mCAF-derived EVs. Free DNA was eliminated by DNase0 digestion before extraction of EV-DNA. Western blot analysis of Actin, the EV marker CD63, and the mitochondria marker ATP5A1 is also reported in each EV isolated component. (F) mtDNA level as absolute copy number (qPCR, ND1) from HS27a EVs and cells. (G) Schematic and representative gel electrophoresis image of long-range PCR (three contiguous amplicons; SI Appendix, Table S4) encoding the 16-kbp mtDNA genome from purified EVs ± DNase0. (H) Electropherogram of the DNA sequence of the ND1 gene showing a mutation conserved between HS27a cells and EVs (SI Appendix, Table S2). Data are presented as the mean of three independent experiments; error bars indicate SD; *P < 0.05 (student’s t test).
We isolated EVs from the conditioned media of cultured mCAFs using a sucrose gradient approach. Electron microscopy and NanoSight analysis of these EVs demonstrated cup-shaped membrane vesicles ∼150 nm in diameter (Fig. 3B). Although these vesicles are larger than standard exosomes (∼100 nm), quantitative mass spectrometry of these CAF-derived EVs revealed the presence of some mitochondrial proteins, such as ATP5A and VDAC1, together with numerous canonical proteins identified in exosomes (i.e., vesicles of endocytic origin) (Fig. 3C and SI Appendix, Table S3) (20). By qPCR, using specific primers for mtDNA and nDNA, we determined that mtDNA was markedly enriched in EVs compared with nDNA, as DNase0 treatment of EVs eliminated most free-DNA contamination but had no effect on mtDNA levels (Fig. 3D). Flotation of the centrifuged pellets on a sucrose cushion followed by EV isolation and DNA and Western blot analyses further demonstrated that mtDNA is enriched in the CD63hi EV component (Fig. 3E). In agreement with the mass spectrometry data (Fig. 3B), we confirmed the up-regulation of mitochondrial complex V subunit F0 (ATP5A1) in the mtDNAhi/CD63hi EV component (Fig. 3E). These data suggest that components of mtDNAhi EVs may originate from mitochondria.
In addition to mCAFs, we also examined EVs from distinct human cell lines including cancer cell lines (HeLa, Caski, MCF7), bone marrow-derived stromal cell lines (HS27a, HS5), normal fibroblasts (HMF, MRC5), and patient-derived CAFs (primary cultures from bone metastases of breast cancers). The levels of mtDNA in EVs were proportional to cellular mtDNA expression (SI Appendix, Fig. S3A). Using multiple approaches, including absolute DNA copy number, long-range PCR, 46-amplicon–based PCR, and rolling circle PCR coupled with the selective amplification of mtDNA sequences, we identified the complete circular mitochondrial genome packaged within EVs with the exception of those from HS5 cells (SI Appendix, Fig. S3 B–E). Interestingly, we also determined the presence of high levels of mtDNA-laden EVs in CAF primary cultures derived from breast cancer metastases (SI Appendix, Fig. S3B). Among all the examined models, HS27a-derived EVs expressed the highest levels of mtDNA relative to intracellular mtDNA (Fig. 3 F and G). Consistent with their cell of origin, mtDNA SNPs and mutations were also conserved between cells and their EVs, suggesting that this process does not necessarily serve simply to clear damaged mtDNA genomes, e.g., after a mitophagic trigger (Fig. 3H and SI Appendix, Fig. S3F) (21). Taken together these data demonstrate the presence and packaging of the entire mitochondrial genome within many, but not all, cell-derived EVs.
Functional Consequences of EV mtDNA Transfer.
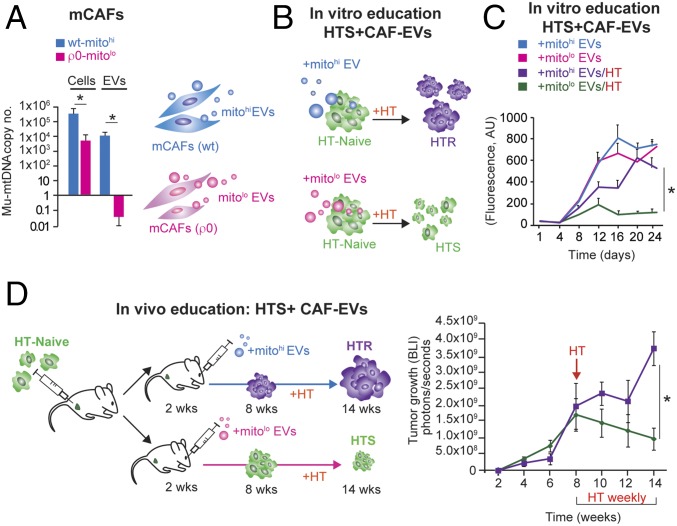
Given our observation that mu-mtDNA was acquired in the transition from HTS to HTR tumors (Fig. 2) and the presence of the mitochondrial genome within CAF EVs, we hypothesized that EVs could transfer mtDNA to cancer cells, promoting HTR disease. To test this hypothesis, we isolated EVs from wild-type and mtDNA-deficient CAFs using the ρ0 protocol (Materials and Methods). Notably, the depletion of mtDNA in CAFs did not affect their proliferation potential but resulted in a profound decrease in EV mtDNA copy number as well as mitochondrial transcripts (Fig. 4A and SI Appendix, Fig. S4 A and B). To test the role of mtDNA transfer in mediating the transition from HTS to HTR disease, we isolated EVs from murine CAFs (wild type or ρ0) and administered them to HT-naive cells once weekly for 4 wk in the presence or absence of HT (Fig. 4B). The number of EVs administered (3 × 109) was determined experimentally (108–1011 EVs were tested) and represents the amount produced by ∼1 × 109 cells in 48 h. No effect of EVs was observed in the absence of HT, while CAF-derived mitohi-EVs conferred HTR growth (Fig. 4 B and C).
Fig. 4.
mCAF-derived EVs educate tumor cells, mediating HTR disease. (A) Mu-mtDNA copy number by qPCR in mCAFs and their EVs; Wt, wild type; ρ0, cells depleted for mtDNA (Materials and Methods). The bar graph reports the mean copy number (log10 scale) of three independent experiments; error bars indicate SD. *P < 0.05 (Student's t test). (B) Schematic showing HTS cells (green) treated with 3 × 109 mCAF-derived EVs (wild-type mtDNAhi or ρ0-mtDNAlo) once weekly for 4 wk + HT (fulvestrant, 10 μM/wk), leading to the growth of wild-type mtDNAhi-EV–educated cells. (C) Proliferation (shown by Calcein AM staining) of BT474 (HTS cells) cultured for 24 d ± HT (fulvestrant, 10 μM/wk) and treated weekly with 3 × 109 mCAF-derived EVs (wild-type mtDNAhi or ρ0 mtDNAlo). The mean ± SD for each time point of the growth curve is reported; *P < 0.05 (post hoc t test corrected for multiple comparisons after GLM for repeated measures). (D) Schematic and tumor growth curve of HTS cells (MCF7) injected into the MFP and subsequently educated weekly via retroorbital injection of 3 × 109 with wild-type mitohi or ρ0-mtDNAlo EVs. After 8 wk, HT was administered (fulvestrant, 100 μg/wk) for 6 wk. The mean ± SEM for each time point of the growth curve is reported; *P < 0.05 (post hoc t test corrected for multiple comparisons after GLM for repeated measures).
Further characterization of the OXPHOS capacity of HTR cells demonstrated an increase in OXPHOS in those cells educated with mitohi-EVs, but no effect was observed in the HT-treated cells educated with mitolo-EVs (SI Appendix, Fig. S4C). In addition, the HTR mitohi-EV–educated cells contained EV-derived mu-mtDNA (SI Appendix, Fig. S4D). We next asked if the transfer of mtDNA in vivo could promote HTR disease. To test this hypothesis, we injected HT-naive cells into the MFP of mice followed by weekly injections of either CAF-derived mitohi-EVs or EVs lacking mtDNA (mitolo-EVs). After 2 mo, no difference in tumor growth was observed in the two cohorts. However, 6 wk following the administration of HT (fulvestrant), the mice educated with mitohi-EVs had developed HTR tumors showing increased mu-mtDNA copy number, while those educated with mitolo-EVs had evidence of HTS disease, as their tumor burden decreased (Fig. 4D and SI Appendix, Fig. S4E). Thus, we demonstrated that the presence of mtDNA is required for the EV-mediated HTR phenotype both in vitro and in vivo.
EV-mtDNA Transfer Mediates an Exit from Metabolic Dormancy.
Since human mtDNA was reduced but still present following HT, we asked whether mu-mtDNA alone could rescue the loss of OXPHOS potential in cancer cells depleted of endogenous mtDNA. To do so we generated luminal breast cancer cells depleted of mtDNA (ρo) and educated these cells with mu-mtDNAhi CAF-EVs for 2 mo. Not surprisingly, these ρo cells did not proliferate in vitro or in vivo, and EV-mediated mtDNA transfer did not rescue their proliferation potential (SI Appendix, Fig. S5A). These latter observations suggested that the residual presence of endogenous mtDNA is necessary for the exogenous mtDNA-mediated proliferation potential of recipient cells.
We next hypothesized that EV-mediated mtDNA transfer may also occur in dormant or metabolically quiescent tumor cells, leading to an exit from dormancy. To address this hypothesis, we first considered the recipient cells. We previously showed that HT (fulvestrant or tamoxifen) treatment of HT-naive tumor cells led to the generation of cells that were metabolically OXPHOS low and expressed low levels of mtDNA (14).
Similarly, here we treated ER+ tumor-derived cells and cell lines with vehicle or HT (fulvestrant) and isolated viable (DAPI−) cells by FACS (Fig. 5A). These HTD cells (i) did not proliferate in culture but remained viable as single cells and (ii) displayed decreased expression of genes related to protein synthesis and OXPHOS, as determined by microarray analysis (SI Appendix, Fig. S5B). In several models of HT-induced tumor dormancy, we demonstrated that mtDNA level (copy number) was reduced by 70–90% in the HT-treated cells compared with controls (Fig. 5B). Additionally, OXPHOS potential, mitochondrial complex protein expression, and mitochondrial complex I and IV activity were reduced in HT-treated cells (SI Appendix, Fig. S5 C–E). Overall, these data indicate that HT can induce an OXPHOS-dormant phenotype characterized by the loss of mtDNA and mitochondrial complex activity (14).
Fig. 5.
mCAF-derived EVs promote the exit from HT-induced tumor dormancy. (A) Schematic of the experimental design and representative flow plots: HT-naive cells were FACS isolated from xenografts (GFP+) or luminal breast cancer cell lines and were treated with HT (fulvestrant, 10 μM/wk) for 2 mo; HTD cancer cells (6% of the viable population, HTDorm) were FACS purified (Dapi−) and displayed a single-cell (nonproliferating) morphology in 3D. (Scale bar,100 μm.) (B) mtDNA levels (determined by qPCR and expressed as fold change; naive cells were used as reference) in luminal breast cancer cells and xenograft-derived cells (multiple models) ± HT (fulvestrant, 10 μM) for 2 mo. (C) Dormant MCF7 cells (HTD) were isolated (SI Appendix, Fig. S5) and treated weekly for 4 wk with 3 × 109 mCAF-derived EVs (wt-mtDNAhi or ρ0-mtDNAlo) + HT (fulvestrant, 10 μM/wk). After 40 d, mammosphere (MS) number and mitochondrial membrane potential (Δψ) were determined by TMRE staining (red) in HTR/mitohi-EV– and HTD/mitolo-EV–educated cells. (Scale bars, 15 μm.) (D) Confocal microscopy of HTD cells incubated for 48 h with PKH67 Green-labeled mCAF EVs (green), MitoTracker-labeled mitochondria (red), and EVs colocalized with mitochondria (yellow). (Scale bar, 5 μm.) (E) Mu-mtDNA level (qPCR, ND1) shown as the fold increase (log10 scale) of the reference HTD from the two HTD–HTR models (MCF7 and BT474) described in C. (F) Mu-mtRNA expression of 14 genes (shown as fold change, −log10 scale) determined by qRT-PCR in HTR and HTS cells in E (reference HTD cells; MCF7); the expression of nuclear-encoded (nonmitochondrial) murine RNA transcripts was also determined (Cox4, β2M). (G) Schematic, representative photographs, and bar graph showing BLI of tumor growth derived from HTD cells (ZR751 GFP+/Luciferase+ cells) injected in the MFP of mice (n = 5 per group) that were treated weekly for 8 wk via retroorbital injection with 3 × 109 wt-mitohi or ρ0-mitolo mCAF-derived EVs. Data are shown as mean ± SD at the end point (4 mo); *P < 0.05 (student’s t test). The mu-mtDNA level is reported as the copy number (qPCR, ND1) in FACS-purified ZR751 EV-educated cells at the end point of the experiment. (H) Representative gel electrophoresis from whole mu-mtDNA PCR amplification using a set of nuclear mitochondrial sequences (NumtS) excluding overlapping primers in cancer cells from G (grey, mitolow EV-derived tumor cells; purple mitohi EV-derived tumor cells) (Materials and Methods). Data in B, C, and E–G are reported as the mean of three independent experiments; error bars indicate SD; In B, C, E, and F, *P < 0.05 (student’s t test).
Given the observation that HTR disease harbors mu-mtDNA (Fig. 2), we asked if EVs could transfer their mtDNA to HTD cells, resulting in an exit from metabolic quiescence. To test this, we isolated EVs from murine CAFs (wild type or ρ0) and administered them to HTD cells once weekly for 4 wk in the presence of HT (Fig. 5C, schematic). After 6 wk we observed the proliferation of HTD cells as mammospheres with self-renewing capacity (HTR-EV cells) (Fig. 5C). The functional consequences of the mtDNA transfer led to the activation of mitochondrial membrane potential [determined by tetramethylrhodamine, ethyl ester (TMRE) staining] observed in (MCF7, BT474) HTR-EV cell models (Fig. 5C and SI Appendix, Fig. S6A). By confocal microscopy and electron microscopy we observed the colocalization of labeled (PKH67 and TSG101) EVs with the mitochondria 48 h after their administration to breast cancer cells, including HTD cells, suggesting that EVs can fuse with recipient cells’ mitochondria (Fig. 5D and SI Appendix, Fig. S6B). Additionally, in agreement with the molecular studies, the majority of CAF-EVs harbor DNA, which could be transferred to recipient cells as determined by confocal microscopy analysis (SI Appendix, Fig. S6C). In contrast to HTD cells, OXPHOS-proficient cancer cells (control cells never treated with HT) did not contain mu-mtDNA following mtDNAhi-EV administration either in vitro or in vivo (copy number = 0), suggesting that OXPHOS-deficient cancer cells were more permissive for the uptake and retention of exogenous mtDNA (up to 10,000 copy numbers) (SI Appendix, Fig. S6D). In agreement with this observation, HTR mitohi-EV–educated cells (MCF7 and BT474) demonstrated the presence of mu-mtDNA and the expression of the complete murine mitochondrial transcriptome (Fig. 5 E and F and SI Appendix, Fig. S6E). In multiple luminal breast cancer models of HTD to HTR (mitohi-EV–educated) cells, we observed increased mitochondrial complex I and IV activity and restoration of normal-appearing mitochondria (SI Appendix, Fig. S6 F and G). Additionally these HTR mitohi cells displayed sensitivity to OXPHOS inhibition in vitro (Fig. 2F).
We next asked if the transfer of mtDNA in vivo could promote exit from metabolic dormancy. We injected HTD cells into the MFP of mice followed by weekly injections of either CAF-derived mitohi-EVs or EVs lacking mtDNA (mitolo-EVs). After 2 mo, three of five mice from the EV-injected cohort had developed large tumors, while two of five control mice had very small tumors (Fig. 5G). Analysis of tumors from mitohi-EV–educated mice demonstrated the presence of the whole mu-mtDNA genome by PCR and sequencing analyses of the mu-mtDNA only in FACS-purified tumor cells from mitohi-EV–educated xenografts (GFP+) (Fig. 5 G and H and SI Appendix, Fig. S7A). We then asked whether a higher mu-mtDNA level would persist following ex vivo passages. Cancer cells isolated from mitohi-EV xenografts were cultured for up to 120 d, and the mtDNA level was determined. We found that mu-mtDNA was lost after 30 d of ex vivo culture, suggesting that continuous EV education is needed for exogenous mtDNA transfer and activity (SI Appendix, Fig. S7B).
Finally, we asked whether the loss or decreased production of mtDNAhi-EVs (ρ0-CAFs) could interfere with the CAF-induced exit from dormancy of HTD cells. We coinjected wild-type or ρ0 mCAFs with HTD cells in the MFP (MCF7-Luciferase/GFP+). After 6 mo, 90% (9/10) of mice bearing mCAFs/HTD cells had tumors and metastases, whereas only 17% (2/12) of mice harboring mtDNAloCAF/HTD cells generated tumors, and these mice developed no metastases (SI Appendix, Fig. S7C). Molecular analysis showed that cancer cells isolated from HTD cells that had exited dormancy expressed mu-mtDNA, determined by ND1 mtDNA copy number and the presence of murine COX3 and ND5 mtDNA genes (SI Appendix, Fig. S7D). Taken together, these data demonstrated that mtDNAhi-EVs promoted the exit from HT-induced metabolic dormancy in luminal breast cancer.
MtDNA Horizontal Transfer Preferentially Occurs in Cancer Stem Cell-Like Cells in Vivo, Leading to Increased Self-Renewal with HT.
Many mechanisms of HT resistance have been postulated, including Her2 overexpression, ERα mutations, and cancer stem cell-like cell (CSC) propagation (14, 16, 22, 23). Notably, we have determined that these CSCs are localized in stromal niches (16) and require OXPHOS potential to self-renew (14, 24).
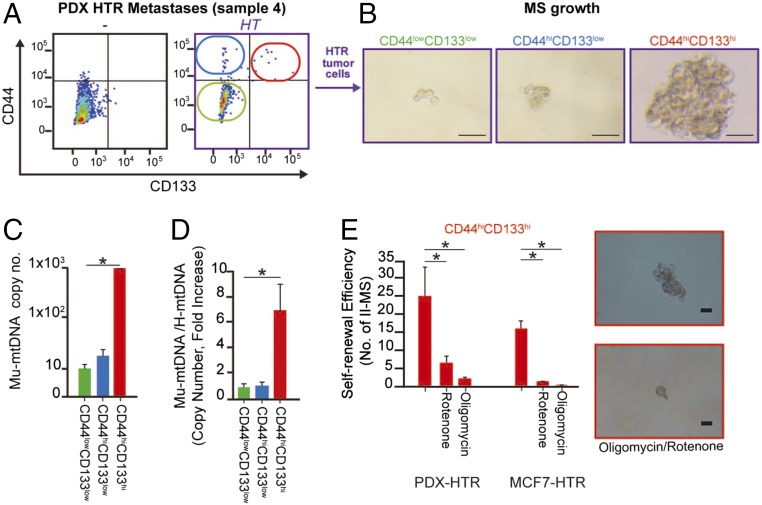
Here, we asked whether the horizontal transfer of mtDNA could occur in CSCs. To do so, we used recently generated primary cultures from HTR PDXs (16). In agreement with previous studies (14, 16, 22, 23), using CD133 and CD44 as markers, we found that HT promoted the expansion of self-renewing CD133hi/CD44hi CSCs from these PDX cultures (Fig. 6 A and B). Further analysis of FACS-purified populations demonstrated the selective enrichment of mu-mtDNA in CD44hiCD133hi CSCs compared with noncancer stem cell-like populations (nCSCs) (Fig. 6C).
Fig. 6.
mtDNA horizontal transfer in vivo preferentially occurs in CSCs, leading to increased self-renewal with hormonal therapy. (A) Representative CD133/CD44 expression by flow analysis of HRT PDX cancer cells and controls (no HT). Briefly, cancer cells were isolated from patient-derived bone metastases (sample 4), grown in culture dishes for 2 wk, and then injected into the MFP of NOD/SCID mice treated with HT (fulvestrant, 1 mg/wk). After 5 mo, tumor tissues were digested, and cancer cells were cultured in vitro and FACS sorted. (B) Representative images of 3D cell cultures (low-attachment plates at day 7) of the three cell populations derived from A. (Scale bars, 100 μm.) (C) The mu-mtDNA level is reported as copy number (qPCR, ND1) in FACS-purified cancer cell populations from A. (D) Bar graph showing mtDNA copy number as fold increase of the ratio of murine versus human mtDNA in cell populations from A. (E) Self-renewal potential as secondary mammosphere formation (II-MS) in the presence of mitochondrial poisons administered every 48 h in CD44hi/CD133hi CSCs at day 7 of 3D culture (rotenone 100 nM, oligomycin 200 nM). Data in C–E are reported as the mean of three independent experiments; error bars indicate SD; *P < 0.05 (student’s t test).
Similar to the PDX model, MCF7-HTR showed increased mu-mtDNA copy number in CSCs (CD44hiCD133hi) (SI Appendix, Fig. S8A). Interestingly, a higher mu-mtDNA copy number was associated with a slight decrease in human mtDNA levels in CSCs (SI Appendix, Fig. S8B). Thus, the ratio of murine/human mtDNA was much higher in CD44hiCD133hi cells (Fig. 6D). Notably, these observations were in agreement with our previous findings of increased expansion of dormant CD133hi CSCs with decreased mtDNA copy number (14).
To investigate the functional consequences of OXPHOS activity in the expansion of HTR-derived mu-mtDNAhi CSCs, we treated these cells with selective inhibitors of mitochondrial respiratory chain complexes (RCC), oligomycin and rotenone. Consistently, self-renewal potential, quantified as secondary mammosphere potential, was dramatically reduced following RCC inhibition (Fig. 6E).
Overall, these observations in multiple models of ER+ breast cancer demonstrate that CAFs can package relatively large amounts of DNA—the full mitochondrial genome—into EVs, which is released and taken up by dormant CSCs followed by the transcription of the donor mtDNA, resulting in higher OXPHOS potential and exit from metabolic quiescence or dormancy of CSCs and the development of HT resistance. Our data highlight how the horizontal transfer of mtDNA via EVs from the tumor microenvironment promotes OXPHOS-proficient CSCs, leading to therapy-resistant metastases (25, 26). Thus, we propose that EVs play a pivotal role in cancer metabolism, with potentially broad implications for their control (27).
Discussion
The role of EV DNA transfer in human disease remains poorly investigated (3). Furthermore, the composition of EV DNA and whether this molecule is inside the vesicles is still a conundrum (28).
Since the discovery of the Warburg effect, many efforts have been made to study metabolic reprogramming in cancer cells from the gain of oncogenes, metastatic progression, and in response to therapy. In agreement with other investigators (26, 29, 30), we demonstrated that luminal breast cancer cells (comprising ≥70% of human breast cancer) require efficient mitochondrial respiration to maintain their tumorigenic potential (30). Both triple-negative and Her2+ breast cancer cell lines are more dependent on anaerobic glycolysis, as depletion of mtDNA decreases their metastatic capacity, suggesting that functional respiration is required for metastatic dissemination (9). These observations are supported by the association between the occurrence of deleterious mtDNA mutations and a low tumorigenic phenotype (31). Conversely, genetic reconstitution of mitochondrial function (using cybrids) of mtDNA-depleted cancer cells promoted reactive oxygen species production, leading to cell proliferation (32).
Cross-species exchange of mtDNA did not restore OXPHOS potential in xenocybrids (33). However, whether the transfer of mtDNA from a murine cell to a human cancer cell via EVs could rescue mitochondrial bioenergetics was never postulated before. Therefore, our model differs from xenocybrids in several respects. (i) We do not use enucleated donor cells, which function as “mitochondrial” donors containing full mitochondria, cytoplasm, and organelles that fuse with recipient cells. (ii) Our recipient cells are not mtDNA depleted but have reduced mtDNA from hormonal therapy. (iii) We perform experiments with EVs, which are vehicles capable of transferring genomic DNA from one cell type to another, conferring profound phenotypes, including transformation (19). (iv) EVs fuse with resident mitochondria (this paper) and are known to traffic to mitochondria via Rabs (34). Additionally, work by Kukat, et al. (35) has determined that the 16-kb mtDNA genome can fit in a nucleoid which is ∼100 nm in diameter, similar to the size of our vesicles. Our experimental model demonstrates the transfer of exogenous mtDNA in vivo and in vivo via EVs and provides its functional consequence in OXPHOS-dependent breast cancers.
Mutations, deletions, and changes in mtDNA copy number have been observed in cancers, particularly in response to therapy (36–38). However, the mechanisms and the clinical relevance of these phenomena remain unclear. These mutations may represent passenger events during therapy-driven cancer cell selection (39); alternatively, when accumulating toward homoplasmy, they may be drivers of disease (40), playing a prominent role in chemoresistance. While decreased mtDNA copy number reduces replication, conferring resistance to antineoplastic drugs such as anthracyclines and taxanes (41, 42), efficient mitochondria biogenesis would make cells more resistant to antimitochondrial agents, as was demonstrated in melanomas treated with BRAF inhibitors (43).
In contrast to the dogma that mtDNA and mitochondria are retained and propagated within somatic cells of higher organisms, recent studies have pointed out that mitochondria and mtDNA can move between mammalian cells via dynamic intercellular organelle highways or nanotubes (9, 44, 45). Seminal work from Giuseppe Attardi’s laboratory demonstrated the complementation of respiratory function via exogenous mitochondria in the ρo model (46). Additionally, it was recently shown that astrocytes and mesenchymal stem cells could release mitochondrial particles in microvesicles, which were taken up by damaged neurons or macrophages, enhancing their survival or OXPHOS activity (47, 48). In agreement with these observations, our data provide evidence that mtDNA can be transferred via EVs from stromal cells to cancer cells to sustain OXPHOS potential and mediate an exit from therapy-induced metabolic dormancy.
The mechanisms by which the complete mitochondrial genome is selectively packaged within EVs are not known. We suggest that the cells most capable of producing mtDNAhi-EVs are those that can readily reprogram their metabolism in response to oxidative stress, resulting in reduced mitochondrial activity (OXPHOS) and increased mtDNA replication. Most normal cells respond to oxidative stress, hypoxia, DNA-damaging agents, and/or nutrient deprivation by undergoing autophagy, apoptosis, and senescence (48). The cells that survive or even continue to proliferate in these conditions are those we hypothesize to be the optimal producers of functional mtDNA+-EVs. It was recently shown by Phinney, et al. (48) that in response to oxidative stress mesenchymal stem cells package depolarized mitochondria in very large microvesicles. Although we did find entire mitochondria in our mass spectrometry and electron microscopy experiments, it is possible that mitochondria generate EVs that contain some mitochondria components, including mtDNA. These mitochondria-derived EVs are released and transfer their cargo in recipient cells. Indeed, by semiquantitative mass spectrometry, we found some mitochondrial proteins (e.g., ATP5A, VDAC1, mitochondrial phosphate carrier protein and mitochondrial peroxiredoxin-5) (Fig. 3B and SI Appendix, Table S3) in addition to an abundance of proteins commonly found in vesicles of endocytic origin. Importantly, once EVs are taken up by cells, it is not known how the mtDNA is incorporated within the mitochondria, which would require traversing multiple EV and mitochondrial membranes, possibly through the translocase of the outer/inner membrane (TOM/TIM) complexes. Our study provides an important platform for further studies to elucidate how the full mtDNA genome is packaged, released, taken up, and incorporated into recipient cells.
ER+ disease can recur years to decades after the primary tumor diagnosis, suggesting that tumor cells have the capacity to remain dormant for prolonged periods of time (49, 50). Determining how tumor cells are induced into a dormant state and the systemic cues that eventually cause dormant CSCs to exit dormancy (increased CSC self-renewal), leading to hormone-refractory metastatic disease, are poorly understood, in large part due to the paucity of clinical material and relevant animal tumor models. In our current study, we have generated such models and identified the presence of mtDNA-EVs in the circulation of patients with HTR ER+ metastatic disease and of HTR tumor-bearing mice. We demonstrated in preclinical models that (i) although HT eradicates many cancer cells, some undergo a state of viable dormancy, characterized by the loss of mtDNA; (ii) the development of HTR full-blown metastatic disease occurs through the restoration of mitochondrial function; and (iii) this may occur through the transfer of mtDNA to CSCs via CAF-EVs.
Additionally, we demonstrate a specific genome packaged in EVs (mtDNA) and the phenotype derived from its horizontal transfer in breast cancer. In summary, our data support the hypothesis that in cancer mtDNA+-EVs/exosomes are found and act as “infectious” mediators of therapy resistance in OXPHOS-dependent human cancers, leading to metastatic progression (SI Appendix, Fig. S9).
Materials and Methods
Isolation and Characterization of EVs.
EV isolation from the plasma of patients (SI Appendix, Table S1) and the conditioned media of cancer and stromal cell cultures was performed using sequential centrifugation. The EVs were treated or not, depending on the experiment, with 1 U of Baseline-ZERO DNase0 solution (Epicentre) for 1 h at 37 °C to digest DNA adherent to the surface of EVs or present in solution and were subsequently inactivated for 10 min at 65 °C. Vesicle preparations were verified by electron microscopy. Exosome size and particle number were analyzed using the LM10 or DS500 nanoparticle characterization system (NanoSight; Malvern Instruments) equipped with a blue laser (405 nm).
Cell Lines and Primary Cultures.
Human cancer cell lines are described in SI Appendix, Materials and Methods. Mitochondria-depleted cells (ρ0 cells) (51) were generated by administering CAFs/cancer cultures with ρ0 medium (2 mM glucose, 1% FCS, 0.4 μg/mL ethidium bromide, 50 μg/mL uridine, 100 μg/mL pyruvate) for 2 mo. Primary cultures of CAFs and cancer cells from breast cancer xenografts (Memorial Sloan Kettering Cancer Center Institutional Review Board approval no. 97-094) and PDX were established from primary and metastatic tissues of xenografts treated with and without HT (16). Cancer cells were obtained following tissue enzymatic digestion (collagenase/hyaluronidase) and cell-sorting purification (GFP+/DAPI−) and were passaged in vitro. Fulvestrant (HT) was purchased from Sigma-Aldrich. For all EV studies, cells were grown in exosome-depleted FBS (Gibco, Thermo Fisher Scientific).
EV Education of HTR Experimental Breast Cancer.
The in vivo role of CAF EVs in the promotion of HTR luminal breast cancer was determined by injecting CAF EV (mCAFs, isolated from HTR xenografts and cultured in vitro; SI Appendix, Materials and Methods) and mtDNA-depleted EVs (from mtDNA-depleted CAFs) into the venous circulation of tumor-bearing mice (weekly retroorbital injection of 3 × 109 particles per mouse). MtDNA-EV education of HTD cells was performed as described in SI Appendix, Materials and Methods.
Mitochondrial Activity Assay.
Mitochondrial respiration and OXPHOS potential in cells was assessed using Seahorse technology (www.agilent.com/en-us/promotions/xftechnologyoverview) (51). The OCR, extracellular acidification rate, and glycolytic potential were determined using the Seahorse extracellular flux analyzer (XF-96, XF-24; Seahorse Bioscience). To allow comparisons between experiments, data are presented as OCR (pmol/min). Graphs showing the kinetics of OCR under the basal condition followed by the sequential addition of oligomycin (1 μM), 2-deoxy-d-glucose (2DG) (50 mM), rotenone (300 nM), and glucose (20 mM) are in Fig. 2B (Cell Mito Stress Kit; Seahorse Bioscience). The progress curve is annotated to show the relative contribution of basal, ATP-linked (oligomycin) oxygen consumption and the reserve capacity of the cells (after the addition of 2DG + rotenone). Furthermore, OXPHOS potential was determined by measuring the area of the progress curve after the addition of glucose and rotenone/oligomycin.
MtDNA/nDNA Copy Number Quantification.
Human and murine mtDNA and housekeeping DNA were amplified by standard PCR (mitochondrial: ND1 and ND5; nuclear: GAPDH and ACTIN) (SI Appendix, Table S4) and subsequently were extracted from agarose gels using the Nucleospin Gel and PCR Clean-Up Kit (Macherey Nagel) and quantified using an Agilent 2100 Bioanalyzer. DNA copy number was calculated using the following formula: Copy number = (Ng × 6.022 × 1,023)/(length × 1 × 109 × 650) where Ng represents the concentration of the eluted sample, 6.022 × 1,023 represents Avogadro’s number, length represents the length of the amplicon in base pairs, and 1 × 109 × 650 represents the average weight of a base pair in nanograms. Standard curves were created by qPCR amplifying serial dilutions of the amplicon of interest and were used to interpolate the cycle threshold (CT) data for quantification. For our experiments, we calculated the absolute copy number of mtDNA in 10 ng of total EV-DNA (from 1013 particles). The total amount of EV DNA ranged from 500 ng to 2.5 μg, depending on the model.
Whole-mtDNA Amplification and Sequencing Assays.
Total DNA (1–5 ng) was used for mtDNA amplification with the MitoALL Resequencing kit (Applera). PCR amplification and Sanger sequencing of 46 amplicons were performed as previously described (52). Electropherograms were analyzed by SeqScape software (Applied Biosystems). Functional annotation was performed by applying previously described methods (21) and consulting MitoMap (53) and HmtDB (54). Mu-mtDNA was also sequenced using a specific 46-amplicon PCR technique (this method is under patent approval, and its details cannot be included). Long-range PCR and qPCR (SI Appendix, Materials and Methods) were used to amplify the whole mu-mtDNA using three pairs of overlapping primers (55). The 5′ extremity of the primers was modified with an amino C6 sequence so that the annealing temperature could reach 68 °C and lead to a more specific amplification. Each amplification reaction was performed on a total of 20 ng of DNA with a GeneAmp PCR System 9700 version 2.5 model (Applied Biosystems). Amplicons were visualized on a 0.8% agarose gel using the ChemiDoc XRS+ System (Bio-Rad).
Statistical Analysis.
Statistical analysis was performed by SPSS (SPSS Incorporation). Continuous variables were analyzed by unequal variance t test, paired t test (for samples, n = 2), general linear model (GLM) ANOVA, or GLM for repeated measures (samples, n > 2). Mann–Whitney, Wilcoxon, and Friedman tests were used to analyze ordinal variables. P values were adjusted for multiple comparisons according to Bonferroni correction. All tests were two-sided. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Mesruh Turkekul, Afsar Barlas, Sho Fujisawa, Romin Yevgeniy of the Memoria Sloan Kettering Cancer Center (MSKCC) Molecular Cytology Core, and Leah Blitstein of Brandeis University for technical assistance. This work was supported by US Department of Defense Grant W81XWH-10-1-1013 (to P.S.), MSKCC Support Grant/Core Grant P30 CA008748 (to J.B.), Charles and Marjorie Holloway Foundation (J.B.), Sussman Family Fund (J.B.), Lerner Foundation (J.B.), Beth C. Tortolani Foundation (J.B. and D.L.), US Department of Defense W81XWH-13-1-0425 (to D.L. and J.B.), National Cancer Institute Grant CA169538 (to D.L.), US Department of Defense W81XWH-13-1-0427 (to D.L.), Manning Foundation (D.L.), Paduano Foundation (D.L.), Champalimaud Foundation (D.L.), Mary Kay Foundation (D.L.), Malcolm Hewitt Weiner Foundation (D.L.), Rapp Foundation (D.L.), American Hellenic Educational Progressive Association 5th District Cancer Research Foundation (D.L. and A.H), Marco Polo Fellowship Università di Bologna (C.C.), Italian Association for Cancer Research IG14242_JANEUTICS (to G.G), Italian Ministry of Health Grant GR-2013-02356666 (to G.G.), EUFP7 People Marie Curie Action, Mitochondrial European Educational Training Grant 317433 (to G.G.), Medical Research Council UK MR/L007339/1 (to A.G.), and Palloti Legacy (M.B.).
Footnotes
Conflict of interest statement: P.S. and J.B. hold a patent on this discovery (US 62/451,453).
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE84104).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704862114/-/DCSupplemental.
References
- 1.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527:100–104. doi: 10.1038/nature15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desrochers LM, Antonyak MA, Cerione RA. Extracellular vesicles: Satellites of information transfer in cancer and stem cell biology. Dev Cell. 2016;37:301–309. doi: 10.1016/j.devcel.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fadoukhair Z, et al. Evaluation of targeted therapies in advanced breast cancer: The need for large-scale molecular screening and transformative clinical trial designs. Oncogene. 2016;35:1743–1749. doi: 10.1038/onc.2015.249. [DOI] [PubMed] [Google Scholar]
- 5.Ignatiadis M, Sotiriou C. Luminal breast cancer: From biology to treatment. Nat Rev Clin Oncol. 2013;10:494–506. doi: 10.1038/nrclinonc.2013.124. [DOI] [PubMed] [Google Scholar]
- 6.Zong WX, Rabinowitz JD, White E. Mitochondria and cancer. Mol Cell. 2016;61:667–676. doi: 10.1016/j.molcel.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weerts MJ, et al. Mitochondrial DNA content in breast cancer: Impact on in vitro and in vivo phenotype and patient prognosis. Oncotarget. 2016;7:29166–29176. doi: 10.18632/oncotarget.8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh JC, et al. Adaptive mitochondrial reprogramming and resistance to PI3K therapy. J Natl Cancer Inst. 2015;107:dju502. doi: 10.1093/jnci/dju502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan AS, et al. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab. 2015;21:81–94. doi: 10.1016/j.cmet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Calabrese C, et al. Respiratory complex I is essential to induce a Warburg profile in mitochondria-defective tumor cells. Cancer Metab. 2013;1:11. doi: 10.1186/2049-3002-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guescini M, et al. C2C12 myoblasts release micro-vesicles containing mtDNA and proteins involved in signal transduction. Exp Cell Res. 2010;316:1977–1984. doi: 10.1016/j.yexcr.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Fischer S, et al. Indication of horizontal DNA gene transfer by extracellular vesicles. PLoS One. 2016;11:e0163665. doi: 10.1371/journal.pone.0163665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thakur BK, et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014;24:766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sansone P, et al. Self-renewal of CD133(hi) cells by IL6/Notch3 signalling regulates endocrine resistance in metastatic breast cancer. Nat Commun. 2016;7:10442. doi: 10.1038/ncomms10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Outschoorn UE, Peiris-Pagés M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: A therapeutic perspective. Nat Rev Clin Oncol. 2017;14:11–31. doi: 10.1038/nrclinonc.2016.60. [DOI] [PubMed] [Google Scholar]
- 16.Sansone P, et al. Evolution of cancer stem-like cells in endocrine-resistant metastatic breast cancers is mediated by stromal microvesicles. Cancer Res. 2017;77:1927–1941. doi: 10.1158/0008-5472.CAN-16-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Outschoorn UE, Lisanti MP, Sotgia F. Catabolic cancer-associated fibroblasts transfer energy and biomass to anabolic cancer cells, fueling tumor growth. Semin Cancer Biol. 2014;25:47–60. doi: 10.1016/j.semcancer.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Gascard P, Tlsty TD. Carcinoma-associated fibroblasts: Orchestrating the composition of malignancy. Genes Dev. 2016;30:1002–1019. doi: 10.1101/gad.279737.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai J, et al. Extracellular vesicle-mediated transfer of donor genomic DNA to recipient cells is a novel mechanism for genetic influence between cells. J Mol Cell Biol. 2013;5:227–238. doi: 10.1093/jmcb/mjt011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 21.Santorsola M, et al. A multi-parametric workflow for the prioritization of mitochondrial DNA variants of clinical interest. Hum Genet. 2016;135:121–136. doi: 10.1007/s00439-015-1615-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simões BM, et al. Anti-estrogen resistance in human breast tumors is driven by JAG1-NOTCH4-dependent cancer stem cell activity. Cell Reports. 2015;12:1968–1977. doi: 10.1016/j.celrep.2015.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raffo D, et al. Tamoxifen selects for breast cancer cells with mammosphere forming capacity and increased growth rate. Breast Cancer Res Treat. 2013;142:537–548. doi: 10.1007/s10549-013-2760-2. [DOI] [PubMed] [Google Scholar]
- 24.Kitajima S, et al. The RB-IL-6 axis controls self-renewal and endocrine therapy resistance by fine-tuning mitochondrial activity. Oncogene. 2017;36:5145–5157. doi: 10.1038/onc.2017.124. [DOI] [PubMed] [Google Scholar]
- 25.Simões RV, et al. Metabolic plasticity of metastatic breast cancer cells: Adaptation to changes in the microenvironment. Neoplasia. 2015;17:671–684. doi: 10.1016/j.neo.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Outschoorn UE, et al. Anti-estrogen resistance in breast cancer is induced by the tumor microenvironment and can be overcome by inhibiting mitochondrial function in epithelial cancer cells. Cancer Biol Ther. 2011;12:924–938. doi: 10.4161/cbt.12.10.17780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao H, et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. eLife. 2016;5:e10250. doi: 10.7554/eLife.10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahlert C, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289:3869–3875. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sotgia F, Martinez-Outschoorn UE, Lisanti MP. Cancer metabolism: New validated targets for drug discovery. Oncotarget. 2013;4:1309–1316. doi: 10.18632/oncotarget.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Outschoorn UE, Pavlides S, Sotgia F, Lisanti MP. Mitochondrial biogenesis drives tumor cell proliferation. Am J Pathol. 2011;178:1949–1952. doi: 10.1016/j.ajpath.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gasparre G, et al. A mutation threshold distinguishes the antitumorigenic effects of the mitochondrial gene MTND1, an oncojanus function. Cancer Res. 2011;71:6220–6229. doi: 10.1158/0008-5472.CAN-11-1042. [DOI] [PubMed] [Google Scholar]
- 32.Martínez-Reyes I, et al. TCA Cycle and Mitochondrial Membrane Potential Are Necessary for Diverse Biological Functions. Mol Cell. 2016;61:199–209. doi: 10.1016/j.molcel.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenzie M, Chiotis M, Pinkert CA, Trounce IA. Functional respiratory chain analyses in murid xenomitochondrial cybrids expose coevolutionary constraints of cytochrome b and nuclear subunits of complex III. Mol Biol Evol. 2003;20:1117–1124. doi: 10.1093/molbev/msg132. [DOI] [PubMed] [Google Scholar]
- 34.Jin RU, Mills JC. RAB26 coordinates lysosome traffic and mitochondrial localization. J Cell Sci. 2014;127:1018–1032. doi: 10.1242/jcs.138776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kukat C, et al. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci USA. 2011;108:13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reznik E, et al. Mitochondrial DNA copy number variation across human cancers. eLife. 2016;5:5. doi: 10.7554/eLife.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong B, et al. Mitochondrial dynamics regulating chemoresistance in gynecological cancers. Ann N Y Acad Sci. 2015;1350:1–16. doi: 10.1111/nyas.12883. [DOI] [PubMed] [Google Scholar]
- 38.Cormio A, et al. Mitochondrial changes in endometrial carcinoma: Possible role in tumor diagnosis and prognosis (review) Oncol Rep. 2015;33:1011–1018. doi: 10.3892/or.2014.3690. [DOI] [PubMed] [Google Scholar]
- 39.Ju YS, et al. ICGC Breast Cancer Group ICGC Chronic Myeloid Disorders Group ICGC Prostate Cancer Group Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. eLife. 2014;3:3. doi: 10.7554/eLife.02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurelac I, Romeo G, Gasparre G. Mitochondrial metabolism and cancer. Mitochondrion. 2011;11:635–637. doi: 10.1016/j.mito.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 41.Guerra F, et al. Mitochondrial DNA mutation in serous ovarian cancer: Implications for mitochondria-coded genes in chemoresistance. J Clin Oncol. 2012;30:e373–e378. doi: 10.1200/JCO.2012.43.5933. [DOI] [PubMed] [Google Scholar]
- 42.Hsu CW, Yin PH, Lee HC, Chi CW, Tseng LM. Mitochondrial DNA content as a potential marker to predict response to anthracycline in breast cancer patients. Breast J. 2010;16:264–270. doi: 10.1111/j.1524-4741.2010.00908.x. [DOI] [PubMed] [Google Scholar]
- 43.Vazquez F, et al. PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pasquier J, et al. Preferential transfer of mitochondria from endothelial to cancer cells through tunneling nanotubes modulates chemoresistance. J Transl Med. 2013;11:94. doi: 10.1186/1479-5876-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackson MV, et al. Mitochondrial transfer via tunneling nanotubes is an important mechanism by which mesenchymal stem cells enhance macrophage phagocytosis in the in vitro and in vivo models of ARDS. Stem Cells. 2016;34:2210–2223. doi: 10.1002/stem.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King MP, Attardi G. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 47.Hayakawa K, et al. Transfer of mitochondria from astrocytes to neurons after stroke. Nature. 2016;535:551–555. doi: 10.1038/nature18928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phinney DG, et al. Mesenchymal stem cells use extracellular vesicles to outsource mitophagy and shuttle microRNAs. Nat Commun. 2015;6:8472. doi: 10.1038/ncomms9472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hart CD, et al. Challenges in the management of advanced, ER-positive, HER2-negative breast cancer. Nat Rev Clin Oncol. 2015;12:541–552. doi: 10.1038/nrclinonc.2015.99. [DOI] [PubMed] [Google Scholar]
- 50.Arnedos M, et al. Precision medicine for metastatic breast cancer–limitations and solutions. Nat Rev Clin Oncol. 2015;12:693–704. doi: 10.1038/nrclinonc.2015.123. [DOI] [PubMed] [Google Scholar]
- 51.Qian W, Van Houten B. Alterations in bioenergetics due to changes in mitochondrial DNA copy number. Methods. 2010;51:452–457. doi: 10.1016/j.ymeth.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 52.Kurelac I, et al. Somatic complex I disruptive mitochondrial DNA mutations are modifiers of tumorigenesis that correlate with low genomic instability in pituitary adenomas. Hum Mol Genet. 2013;22:226–238. doi: 10.1093/hmg/dds422. [DOI] [PubMed] [Google Scholar]
- 53.Lott MT, et al. mtDNA variation and analysis using MitoMap and Mitomaster. Curr Bioinformatics. 2013;44:1.23.21–1.23.26. doi: 10.1002/0471250953.bi0123s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rubino F, et al. HmtDB, a genomic resource for mitochondrion-based human variability studies. Nucleic Acids Res. 2012;40:D1150–D1159. doi: 10.1093/nar/gkr1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dames S, Eilbeck K, Mao R. A high-throughput next-generation sequencing assay for the mitochondrial genome. Methods Mol Biol. 2015;1264:77–88. doi: 10.1007/978-1-4939-2257-4_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.