Significance
Several chromatin-modifying enzymes are mutated in human craniofacial disorders. These factors function genome-wide to regulate accessibility and expression of extensive gene sets. Therefore, understanding chromatin-modifier function requires identification of responsible cellular origins and genomic characterization of phenotypes in primary cells. We now combine reporter flow cytometry with low cell number genomics to identify neural crest stem-cell factors as molecular targets of UTX in Kabuki syndrome. UTX demethylates histones to regulate some gene expression, but many UTX-bound regions are subject to novel mechanisms of transcriptional regulation. This study identifies UTX cellular and molecular targets in craniofacial development and this methodology is broadly adaptable to study genome distribution of other chromatin factors in neural crest disorders.
Keywords: UTX, KDM6A, Kabuki syndrome, histone demethylation, neural crest
Abstract
Kabuki syndrome, a congenital craniofacial disorder, manifests from mutations in an X-linked histone H3 lysine 27 demethylase (UTX/KDM6A) or a H3 lysine 4 methylase (KMT2D). However, the cellular and molecular etiology of histone-modifying enzymes in craniofacial disorders is unknown. We now establish Kabuki syndrome as a neurocristopathy, whereby the majority of clinical features are modeled in mice carrying neural crest (NC) deletion of UTX, including craniofacial dysmorphism, cardiac defects, and postnatal growth retardation. Female UTX NC knockout (FKO) demonstrates enhanced phenotypic severity over males (MKOs), due to partial redundancy with UTY, a Y-chromosome demethylase-dead homolog. Thus, NC cells may require demethylase-independent UTX activity. Consistently, Kabuki causative point mutations upstream of the JmjC domain do not disrupt UTX demethylation. We have isolated primary NC cells at a phenocritical postmigratory timepoint in both FKO and MKO mice, and genome-wide expression and histone profiling have revealed UTX molecular function in establishing appropriate chromatin structure to regulate crucial NC stem-cell signaling pathways. However, the majority of UTX regulated genes do not experience aberrations in H3K27me3 or H3K4me3, implicating alternative roles for UTX in transcriptional control. These findings are substantiated through demethylase-dead knockin mutation of UTX, which supports appropriate facial development.
Neurocristopathies are defined as disorders originating from deficiencies in any aspect of neural crest (NC) biology including specification, migration, and differentiation (1). NC cells comprise a multipotent stem-cell lineage that is specified along the dorsal axis of the vertebrate neural tube. Upon specification, NC cells migrate ventrally and are responsible for diverse differentiation events, depending on rostral–caudal positioning upon reaching their embryonic destination (2–4). Cranial NC cells can differentiate into sensory ganglia, preosteoblasts, and chondrocytes to pattern and form all anterior facial bone and cartilage. Cardiac NC cells differentiate into smooth muscle and are required for outflow tract septation and patterning of the aortic arch arteries. Trunk NC cells develop into melanocytes and dorsal root ganglia, while vagal and sacral NC cells form the enteric nervous system. Thus, neurocristopathies will compromise common sets of organ systems featuring craniofacial, cardiac, and neurological dysfunction. Classical neurocristopathies such as Treacher Collins syndrome result from transcriptional deficiencies, while more recently, neurocristopathies have been described from mutations in chromatin remodeling machinery (5–7).
Kabuki syndrome is a human disorder defined largely by facial characteristics. Affected individuals present midfacial hypoplasia with a broad depressed nasal tip, elongated palpebral fissures with partial eyelid eversion, and large prominent abnormal earlobes (8, 9). Accompanying partially penetrant features include cleft or high arched palate, postnatal growth retardation, and cognitive disability. The disorder can also include several forms of congenital cardiac abnormalities, including ventricular septal closure defects, patent ductus arteriosus, and aortic coarctation. Kabuki syndrome results in the majority of cases (60–70%) from mutations in KMT2D (MLL2, also referred to as MLL4), which encodes a histone H3 lysine 4 (H3K4) methylase that creates an active chromatin environment for enhancing genic transcription (10–12). A smaller percentage of Kabuki individuals (6–14%) carry mutations in UTX (KDM6A), a histone H3 lysine 27 (H3K27) demethylase that removes repressive chromatin modifications (10, 11, 13). Active H3K4 trimethylation (H3K4me3) can co-occur in promoter regions together with repressive H3K27 trimethylation (H3K27me3) in the form of bivalent chromatin (14). Genes regulated by bivalent chromatin have been identified in developmental stem cells and are hypothesized to be poised for rapid gene induction in cellular differentiation events. Similarly, UTX and KMT2D coexist in a protein complex (15) that may be designed to resolve bivalent chromatin through coordinated removal of H3K27me3 and enhancement of H3K4me3. Given the spectrum of phenotypes in Kabuki syndrome resembling a neurocristopathy and the phenocopy of Kabuki causative UTX and KMT2D mutations, NC cells represent a relevant model system to study the coordinated functions of these histone-modifying enzymes.
Kabuki causative mutations in UTX have been identified in both female and male patients (13, 16). This is surprising, given UTX’s location on the X chromosome. Kabuki syndrome results from hypomorphic female heterozygous mutation and null male hemizygous mutation of UTX. Although male patients lack UTX activity, there may be compensation by UTY, a Y-chromosome homolog of UTX. Given that UTY has lost enzymatic activity (17–19), UTX-mediated H3K27me3 demethylation may not be essential for function in craniofacial development.
We have developed a NC mouse model to examine UTX molecular function in craniofacial pathogenesis. Our genetic studies indicate that UTX has both demethylase-dependent and -independent roles in NC development. Epigenomic analysis of primary NC cells supports this observation as UTX regulates H3K27me3 demethylation at a subset of gene loci, while other transcriptional targets experience unaltered histone methylation. We find that the majority of Kabuki mutations disrupt UTX demethylation; however, two point mutations that reside outside of the JmjC demethylase domain inhibit alternative UTX functions. Because demethylase-dead knockin (KI) mutation of UTX facilitates normal craniofacial development, we conclude that UTX functions mainly through demethylase-independent transcriptional control of stem-cell signaling pathways to influence NC cell viability.
Results
Mouse UTX NC Knockout Models Several Features of Human Kabuki Syndrome.
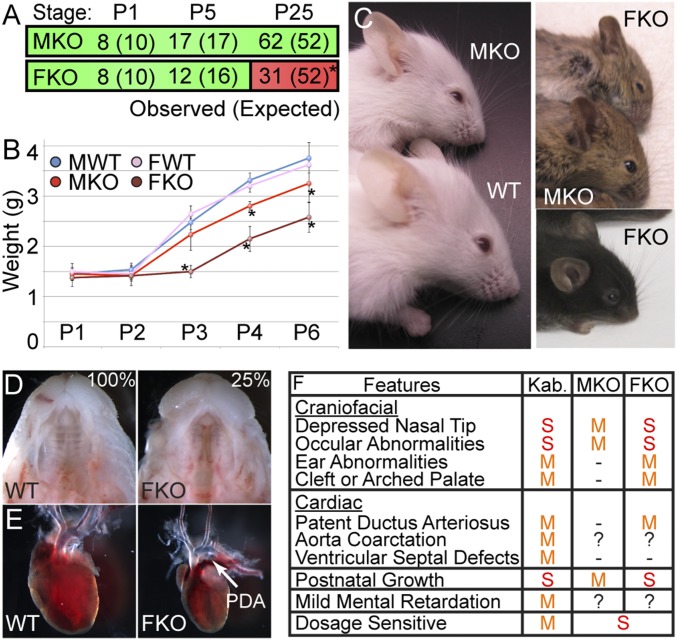
We have previously contrasted male and female mouse Utx mutant phenotypes to elucidate molecular function based on the presence of the demethylase-dead Y-chromosome homolog, UTY (17). If male Utx mutation elicits a phenotype, then UTY cannot fully compensate for UTX loss, signifying demethylase dependency. In contrast, if Utx homozygous females demonstrate enhanced phenotypic severity over Utx hemizygous males, then UTY is providing demethylase-independent compensation for loss of UTX. To model Kabuki syndrome in the mouse, we drove NC-specific deletion of a conditional Utx allele with a Wnt1-Cre transgene (20). This Utx allele has a floxed (fl) third exon, and Cre-mediated deletion produces a nonsense frameshift null for UTX protein (17). While Utxfl/y Wnt1-Cre males (hereafter abbreviated as MKO for male NC knockout) are viable based on expected Mendelian genotype frequencies, Utxfl/fl Wnt1-Cre females (abbreviated FKO for female NC knockout) demonstrate significant postnatal lethality as ∼40% die before weaning (Fig. 1A). Both MKO and FKO mice experience significant reductions in weight starting at postnatal day 3 (P3) that persist throughout adulthood with FKO mice more severely afflicted (Fig. 1B).
Fig. 1.
UTX neural crest knockout models clinical features of Kabuki syndrome. (A) Observed and expected (parentheses) genotype frequencies of MKO and FKO mice at indicated postnatal timepoints (χ2 *P = 0.02). (B) MKO and FKO mice experience growth retardation relative to WT male (MWT) and female (FWT) controls (t test *P < 0.03). (C) MKO and FKO craniofacial phenotypes. FKO mice have partially penetrant cleft palate (D, 25%, n = 5/20 E18.5 embryos) and patent ductus arteriosus (PDA) (E, n = 3/3). (F) Summary and comparison of human Kabuki clinical features (Kab.) to MKO and FKO phenotypes. M, mild; S, severe.
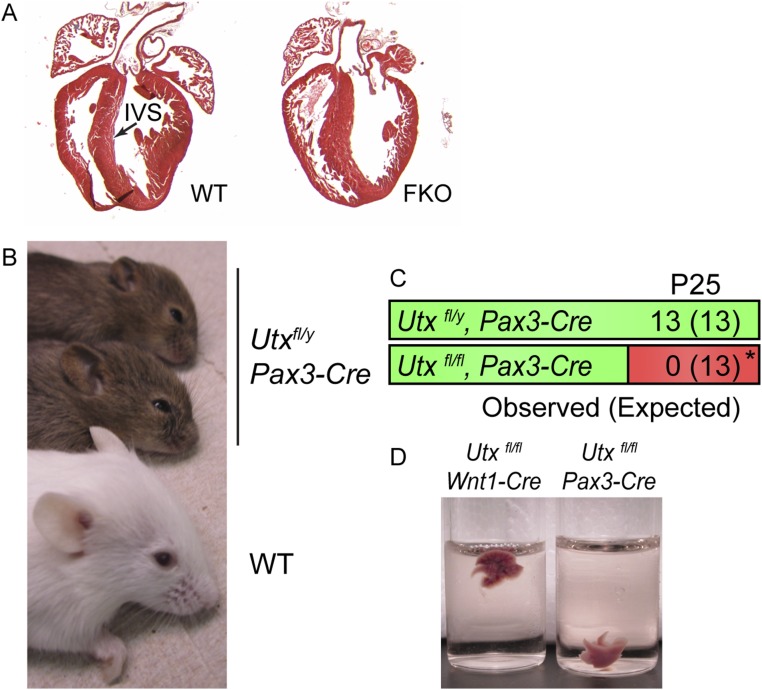
Anterior facial lineages are derived from cranial NC, and MKO mice exhibited frontonasal hypoplasia with a depressed snout, an increased facial angle producing a more prominent forehead, and reduced palpebral fissures (Fig. 1C). FKO mice displayed enhanced MKO characteristics, including greater facial depression, a broader dome-shaped forehead, severe palpebral fissure reduction, and rounded low-set earlobes (Fig. 1C). Partially penetrant cleft palate was specific to FKO embryos (Fig. 1D), which is a likely cause for lower than expected FKO recovery between P1 and P5 (Fig. 1A). Furthermore, FKO mice also exhibited patent ductus arteriosus congenital heart defects (Fig. 1E) without abnormalities in ventricular septal closure (Fig. S1A).
Fig. S1.
FKO and Pax3-Cre driven Utx phenotypes. (A) WT or FKO hearts were dissected at P2 and sectioned. Interventricular septum (IVS) appeared normal in FKO mice (n = 3/3). (B) Male Utxfl/y, Pax3-Cre mice demonstrate shortened snouts, narrow palpebral fissures, and a dome-shaped forehead. (C) Utxfl/y, Pax3-Cre mice are viable; however, female Utxfl/fl, Pax3-Cre mice die at birth and are not recovered at weaning (χ2 *P < 0.01). (D) Utxfl/fl, Pax3-Cre lungs fail to inflate at birth (n = 3) and float in PBS.
We observed a similar spectrum of phenotypes when UTX knockout is driven by another NC Cre line derived by insertion into the Pax3 locus (21). The dysmorphic facial phenotypes were also observed when NC knockout was driven by Pax3-Cre (Fig. S1B). Utxfl/y, Pax3-Cre males had enhanced frontonasal depression, and Utxfl/fl, Pax3-Cre females could not be recovered at weaning, due to deficiencies in lung inflation at birth (Fig. S1 C and D). These contrasts to Wnt1-Cre–driven phenotypes may be due to leakiness of Pax3-Cre in intercostal skeletal muscle and/or hypomorphic loss of Pax3 in combination with Utx knockout (21, 22).
In summary, NC-specific knockout of Utx establishes a mouse model for Kabuki syndrome that exhibits many features of the human disorder (Fig. 1F). MKO mice have a Kabuki-like manifestation of mild craniofacial features and postnatal growth retardation, indicative of demethylase-dependent UTX function. FKO mice have more severe growth, craniofacial, and cardiac abnormalities. Thus, UTY performs some demethylase-independent UTX compensation in NC development. UTY only partially compensates for loss of UTX in mouse NC, creating an allelic phenotypic gradient. Heterozygous Utx females having one copy of Utx are asymptomatic for facial phenotypes, MKO having one copy of Uty manifest mild phenotypes, and FKO completely lacking both homologs are more severely affected. Similar dosage sensitivity exists in Kabuki patients. UTX−/y males have more severe developmental delay and learning disabilities compared with UTX+/− females (11, 16, 23). Therefore, UTY has some function, but a single copy of UTY (UTX−/y) is not as functional as a single copy of UTX (UTX+/−).
NC Cells Require UTX for Appropriate Anterior Cranial Bone Structure.
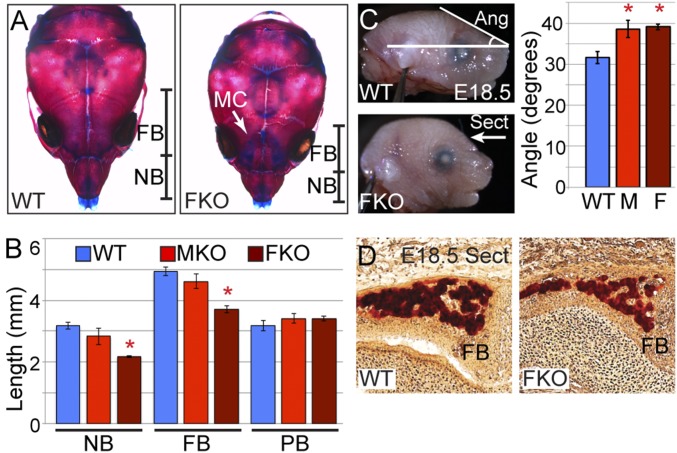
To detail the skeletal structure that may underlie MKO and FKO facial dysmorphism, we performed alizarin red (bone) and alcian blue (cartilage) staining on P10 skull preparations. Gross bone composition was normal; however, FKO mice exhibited dramatic shortening of nasal and frontal bones (Fig. 2A). Measurement of bone lengths verified significant shortening of FKO frontal and nasal structures, while MKO mice experienced mild reductions that failed to reach statistical significance (Fig. 2B). Posterior cranial structures such as parietal bones that are derived from non-NC origins were unaffected in MKO and FKO mice (Fig. 2B). Mutant mice displayed no deficiencies in cartilage distribution, but exhibited a persistence of mandibular cartilage (Fig. 2A). Skeletal abnormalities manifested in embryonic development as similar craniofacial phenotypes were present at late embryonic stages (Fig. 2C). Quantification of E18.5 facial angle revealed significant alterations in both MKO and FKO embryos (Fig. 2C). Alizarin red stained transverse sections of E18.5 cranial regions revealed decreased frontal bone thickness in FKO embryos (Fig. 2D). Therefore, the primary defects in MKO and FKO anterior cranial structure occur earlier in embryonic NC development.
Fig. 2.
NC cells require UTX for appropriate anterior cranial bone structure. (A) Whole mount alizarin red and alcian blue staining of P10 WT and FKO mice. Nasal bone (NB) and frontal bone (FB) lengths are illustrated. MC, Meckel’s cartilage. (B) Nasal (NB), frontal (FB), and parietal bone (PB) lengths from P10 skeletal preparations (n = 3 for each genotype, t test *P < 0.003). (C) FKO (F) and MKO (M) increases in forehead slope are present at E18.5 (t test *P < 0.02, n ≥ 3). (D) At E18.5 FKO mice have decreased FB thickness (section depicted in C: Sect).
UTX NC Loss of Function Results in Postmigratory Embryonic Neural Crest Deficiencies.
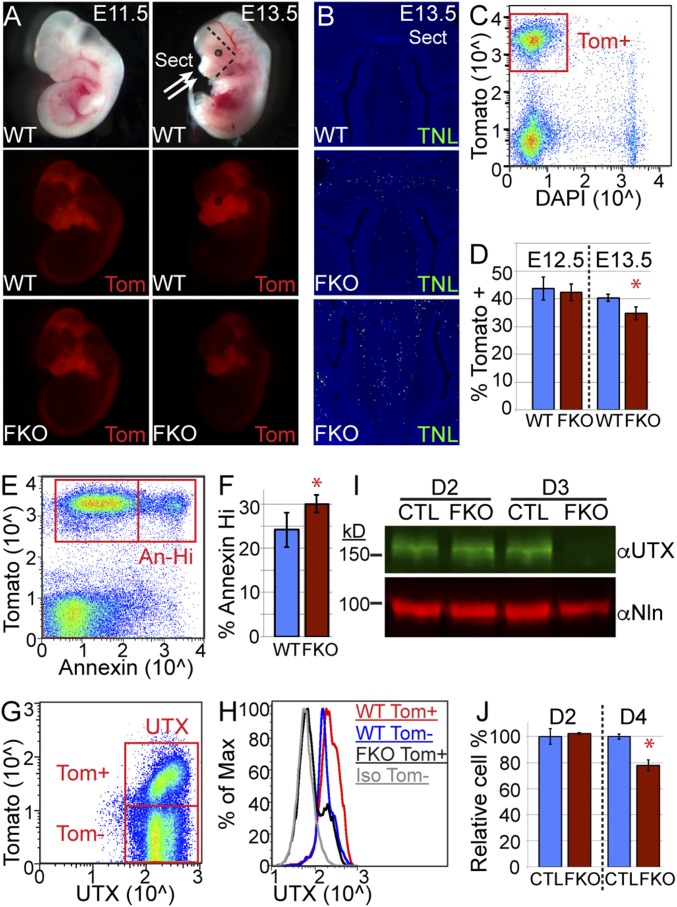
We traced embryonic NC cells utilizing a Cre-activated Tomato reporter inserted into the Rosa locus (24). Cranial NC cells complete migration to ventral facial regions by E9 (25). They continue to proliferate and expand before differentiation to facial lineages at E12 (26). Compared with Tomato reporter fluorescence driven by Wnt1-Cre in WT embryos, FKO NC fluorescence was normal subsequent to migration completion and across proliferative phases (E11.5), but was diminished at differentiation onset (E13.5, Fig. 3A). E13.5 sectioning and Tunel assay of anterior facial regions indicated elevated FKO NC cell death, particularly in the future frontonasal suture and nasal septum regions (Fig. 3B).
Fig. 3.
UTX regulates postmigratory embryonic neural crest viability. (A) Tomato (Tom), activated in NC by Wnt1-Cre, traced NC embryonic development at E11.5 and E13.5. (B) Tunel assay (TNL) of E13.5 WT and FKO anterior facial regions (depicted in Fig. 4A: Sect). (C) Anterior facial regions were dissected (dashed lines in Fig. 4A), dissociated, and flow sorted to isolate live (DAPI excluded) cranial NC cells (Tom+). (D) FKO experience loss of relative NC cell percentages at E13.5 (t test *P = 0.01). (E and F) Tom+ E13.5 cranial NC were stained for annexin V and gated on annexin high (An-Hi) populations to determine relative percentage (t test *P < 0.05). (G and H) Tom+ or Tom− cranial NC were stained for UTX and plotted as a histogram (Iso, isotype antibody). (I) O9-1 NC cells were transduced with CRISPR lentivirus for UTX (FKO) or a control guide RNA (CTL), selected for the given number of days, and Western blotted for UTX or nucleolin (Nln). (J) Cell numbers were quantified with CellTiter-Glo at given timepoints. FKO was graphed as a relative percentage of CTL (t test *P < 0.02).
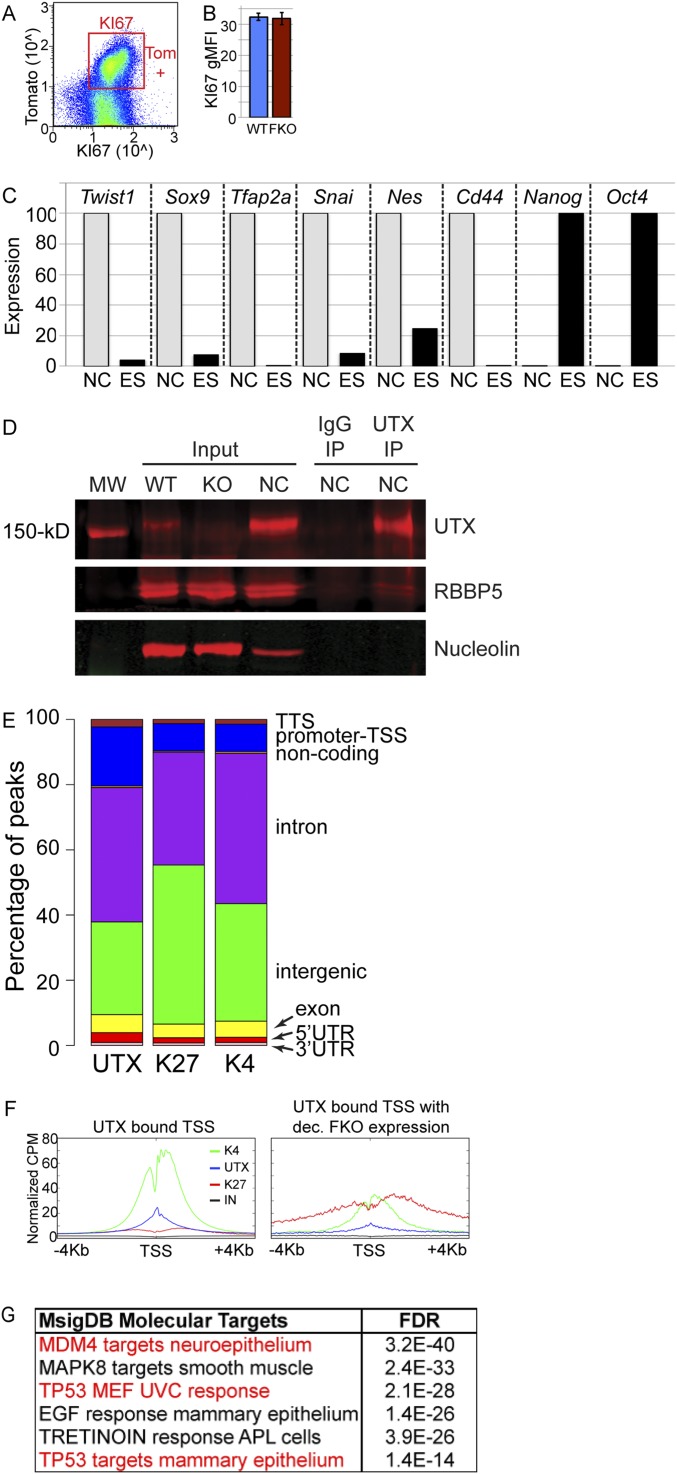
Cranial NC cells were isolated by dissection of frontal facial regions rostral from the maxilla and anterior to the otic vesicle (dashed lines in Fig. 3A). This tissue was dissociated and flow sorted for live (based on DAPI exclusion) cranial NC (based on Tomato fluorescence, Fig. 3C). Based on relative sorted cell percentages, FKO NC cell numbers were normal at E12.5, but were significantly reduced by E13.5 (Fig. 3D). Nuclear staining of isolated cranial NC for KI67 and flow cytometry revealed no differences in cellular proliferation (Fig. S2 A and B); however, surface staining for annexin V (recognizing plasma membrane phosphatidylserine and early apoptosis) demonstrated a greater percentage of FKO Tomato-positive cells exhibiting high annexin staining (Fig. 3 E and F). Flow cytometry revealed UTX protein levels were elevated in Tomato-positive WT NC relative to Tomato-negative non-NC cells, and verified that UTX was lost in FKO NC (Fig. 3 G and H).
Fig. S2.
Overview of H3K4me3, H3K27me3, and UTX genomics and NC properties. (A and B) Tomato+ (Tom+) E13.5 cranial NCs were stained for KI67 flow cytometry (gMFI, geometric mean fluorescence intensity). (C) RT-PCR of the O9-1 NC cell line relative to ES cells confirmed the presence of several NC markers and absence of Nanog and Oct4 ES markers. (D) UTX antibody Western blot of input lysates from WT or UTX KO ES cell lines (WT and KO) and a cranial NC cell line (NC). The UTX antibody is specific and immunoprecipitates (IP) RBBP5 of the KMT2D protein complex. Nucleolin is blotted as a loading control and a 150-kDa molecular weight (MW) ladder band is indicated. (E) Homer analysis identified relative regional locations of UTX, H3K27me3 (K27), and H3K4me3 (K4) peaks at indicated genic and intergenic locations. (F) Profiles across all UTX-bound transcription start sites (TSSs) or UTX TSSs where FKO RNA expression is decreased (dec.). (G) MsigDB identified FKO-dependent targets as being enriched for P53-regulated datasets.
We examined UTX function in the O9-1 self-renewing female primary cranial NC cell line that was obtained by similar sorting methodology at E8.5 (27). The O9-1 cranial NC line identity was verified by RT-PCR for NC stem-cell markers (Fig. S2C). UTX was knocked out by lentiviral infection of the NC line with Cas9 and Utx guide RNAs followed by puromycin selection for transduction. UTX protein was lost after 3 d of selection for UTX knockout (FKO) but was unaffected by a nontargeting control guide RNA (CTL) (Fig. 3I). Knockout of UTX in this cell line resulted in a significant reduction in cranial NC cells across the onset of UTX protein depletion (Fig. 3J). In summary, UTX is required for postmigratory cranial NC viability.
UTX Molecular Analysis of Cranial NC Cells.
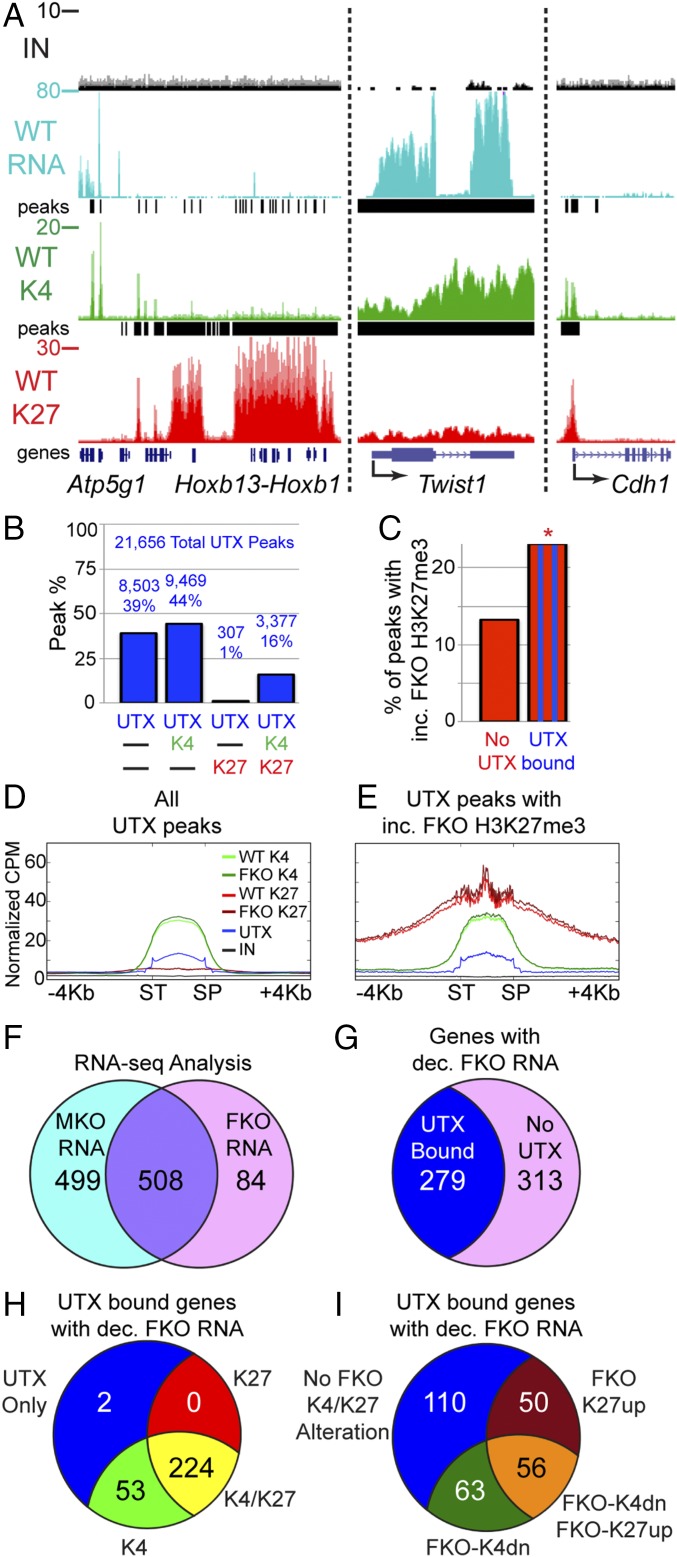
Isolated primary cranial NC cells were subjected to molecular analysis at E12.5, before onset of FKO NC loss. We performed mRNA-sequencing (RNA-seq) on biological replicates of flow-sorted WT, MKO, and FKO NC cells. These expression data were correlated with histone profiles of both H3K27me3 and H3K4me3 ChIP-seq on pooled biological replicates of WT and FKO. The UTX antibody specifically immunoprecipitated UTX (Fig. S2D), but was not as efficient for ChIP analysis and required greater quantities of cells. Therefore, UTX ChIP-seq was performed on the O9-1 self-renewing primary cranial NC cell line (27). To confirm the validity of expression and ChIP-seq data, we examined WT enrichment at the Hoxb genomic region. As Hox gene expression is repressed in cranial NC cells, the Hoxb cluster has high enrichment of H3K27me3 without RNA expression (Fig. 4A). This bipartite pattern of H3K27me3 enrichment is similar to what has been observed at Hoxb in undifferentiated mouse ES cells (28). Proximal to Hoxb is a set of genes, including Atp5g1, that is highly expressed by RNA-seq, have strong peaks of H3K4me3 at genic transcription start sites (TSSs), and lack H3K27me3 enrichment (Fig. 4A). Similarly, Twist1, a highly expressed gene essential for NC viability and differentiation (29), has high RNA-seq signal and H3K4me3 accumulation, but low H3K27me3. Cdh1, a facial ectodermal marker (30), was repressed with H3K27me3 and H3K4me3 promoter peaks.
Fig. 4.
Demethylase-dependent and -independent UTX targets. (A) RNA-seq (RNA), H3K4me3 ChIP-seq (K4), and H3K27me3 ChIP-seq (K27) of isolated E12.5 WT cranial NC at indicated genes (IN, input; peaks, Chip-seq enrichment peaks). (B) Percentage of UTX peaks that overlap with H3K4me3 (K4) and/or H3K27me3 (K27). Total peak numbers are indicated. (C) Percentage of peaks with significantly increased (inc.) FKO H3K27me3 were significantly greater at sites overlapping with UTX binding compared with unbound H3K27me3 sites (23% vs. 13%, Fisher’s exact test *P < 0.001). (D and E) Profiles comparing WT to FKO across all scaled UTX peaks (D, SP, stop; ST, start) or UTX peaks that overlap with sites of increased (inc.) FKO H3K27me3 (E, same cpm scale). (F) RNA-seq identified 508 genes significantly reduced in both MKO and FKO expression relative to WT or 84 FKO specific genes (592 total). (G) The total set of 592 genes with decreased (dec.) FKO expression were overlaid with UTX-bound genes (UTX ChIP-seq peak within 10 kb of gene body) to identify direct UTX gene targets. (H) A total of 279 direct UTX targets were classified as having peaks of H3K27me3 (K27), H3K4me3 (K4), K4/K27, or UTX only within 10 kb of the gene body. (I) A total of 279 direct UTX targets were classified as experiencing enhanced FKO H3K27me3 (FKO-K27up), reduced FKO H3K4me3 (FKO-K4dn), both alterations, or bound by UTX with no alterations within 10 kb of the gene body.
Demethylase-Dependent and -Independent Roles for UTX in Cranial NC Gene Regulation.
To develop an overview of the relationship between UTX binding and histone methylation, we examined the overlap of UTX, H3K27me3, and H3K4me3 in cranial NC cells on a genomic level. UTX, H3K27me3, and H3K4me3 peaks of enrichment were identified by the MACS peak calling algorithm (version 2) (31). Due to the use of male and female samples in our analysis, all genes and peaks on the X and Y chromosomes were excluded from analysis. MACS identified 21,656 UTX peaks, 38,134 H3K4me3 peaks, and 17,867 H3K27me3 peaks of enrichment. UTX peaks overlapped more frequently with H3K4me3 peaks than H3K27me3 peaks, possibly due to the sheer number of H3K4me3 peaks (Fig. 4B). UTX did frequently overlap with H3K27me3 (17% of UTX peaks), and most of these sites had a corresponding peak of H3K4me3 (92%, 3,377 total peaks, Fig. 4B).
UTX is an H3K27me3 demethylase utilized to remove repressive H3K27me3 in gene activation events. Therefore, we expected direct UTX targets to experience elevated H3K27me3 in FKO NC cells relative to WT at sites of UTX binding. EdgeR compared read counts at prominent peaks (with cpm >1) to identify those with significantly [false discovery rate (FDR) < 0.05] elevated FKO H3K27me3. Overall, 15% of all H3K27me3 peaks were significantly increased in FKO NC (2,705 peaks). We assessed the frequency with which elevated FKO H3K27me3 co-occurred with WT UTX binding as evidence of direct UTX-mediated demethylation. A total of 23% of all UTX-occupied H3K27me3 peaks were elevated in FKO NC cells, compared with only 13% of all UTX-unoccupied H3K27me3 peaks (Fig. 4C); thus UTX binding significantly correlates with H3K27me3 demethylation. A profile across all UTX peaks demonstrated overlap with H3K4me3 enrichment and H3K27me3 depletion (Fig. 4D). Only a subset of these UTX peaks fit criteria for demethylation (UTX peaks that overlap with elevated FKO K3K27me3) and display particularly high H3K27me3 (even in WT samples) with an enrichment spike at the UTX peak center (Fig. 4E).
Based on gene annotation data, UTX and H3K4me3 peaks were found at more genic regions than H3K27me3 (Fig. S2E). UTX was bound more frequently to promoters and TSSs, while H3K27me3 had more intergenic localization. To characterize NC UTX transcriptional targets, EdgeR identified RNA-seq transcripts that were significantly (FDR < 0.05) reduced in FKO cells relative to WT. A profile of all UTX-bound TSSs reveals extensive H3K4me3 enrichment (Fig. S2F, Left). UTX binding spans these TSSs with little H3K27me3 accumulation. However, UTX-bound transcriptional targets with expression significantly reduced in FKO NC cells demonstrate elevated H3K27me3 as a potential regulatory feature (Fig. S2F, Right).
RNA-seq analysis identified a high confidence set of 508 potential demethylase-dependent genes (expression reduced in both MKO and FKO NC relative to WT, FDR < 0.05, Fig. 4F). An additional set of 84 FKO-specific reductions (relative to WT levels) comprise potential demethylase-independent genes (Fig. 4F). We used this total set of 592 FKO-affected genes to identify direct UTX-bound targets (Fig. 4G). Notably, of 592 genes with altered NC expression, only 279 were in proximity (±10 kb from the gene body) to direct UTX binding. UTX-bound transcriptional targets frequently (80%) had peaks of H3K27me3 that coincided with genic H3K4me3 enrichment (Fig. 4H). However, only 40% of UTX direct transcriptional targets experienced a demethylation phenotype (FKO-K27up, 106 total genes, Fig. 4I). An equally large set of UTX direct targets of activation (110 genes) had no significant alterations of H3K27me3 or H3K4me3 distribution upon UTX loss of function (Fig. 4I); therefore, UTX has both histone demethylase-dependent and -independent roles in transcriptional regulation of NC target genes.
UTX Regulates Transcription of Craniofacial NC Stem-Cell Signaling Pathways.
We prioritized UTX targets as having a strong effect on NC transcription/methylation based on genes/peaks with higher read counts [reads per kilobase of region per million mapped reads (RPKM) ≥ 4] and more dramatic changes (absolute log fold changes ≥0.5) in FKO NC cells. UTX bound only five genes to strongly regulate both H3K27me3 demethylation and H3K4me3 methylation (Dataset S1). Whereas few UTX-regulated genes exhibited strong H3K27me3-specific demethylation (15 genes) or H3K4me3-specific methylation (10 genes), a larger set of UTX-regulated transcripts displayed weak or no effect on histone methylation levels (174 genes). Several UTX targets have previously characterized function in NC stem-cell biology, osteogenesis/chondrogenesis, or craniofacial biology including strong H3K27me3 demethylase targets: Ddr1, Gadd45g, Nes, Dpf1; weak demethylase: Arid3b, Otx1, Otx2, Gbx2, Sox2; and demethylase independent: Chd7 (32–40) (Dataset S1). Comparison of the FKO misexpression dataset to the Molecular Signatures Database (MsigDB) identified many UTX targets as P53-regulated genes (Fig. S2G).
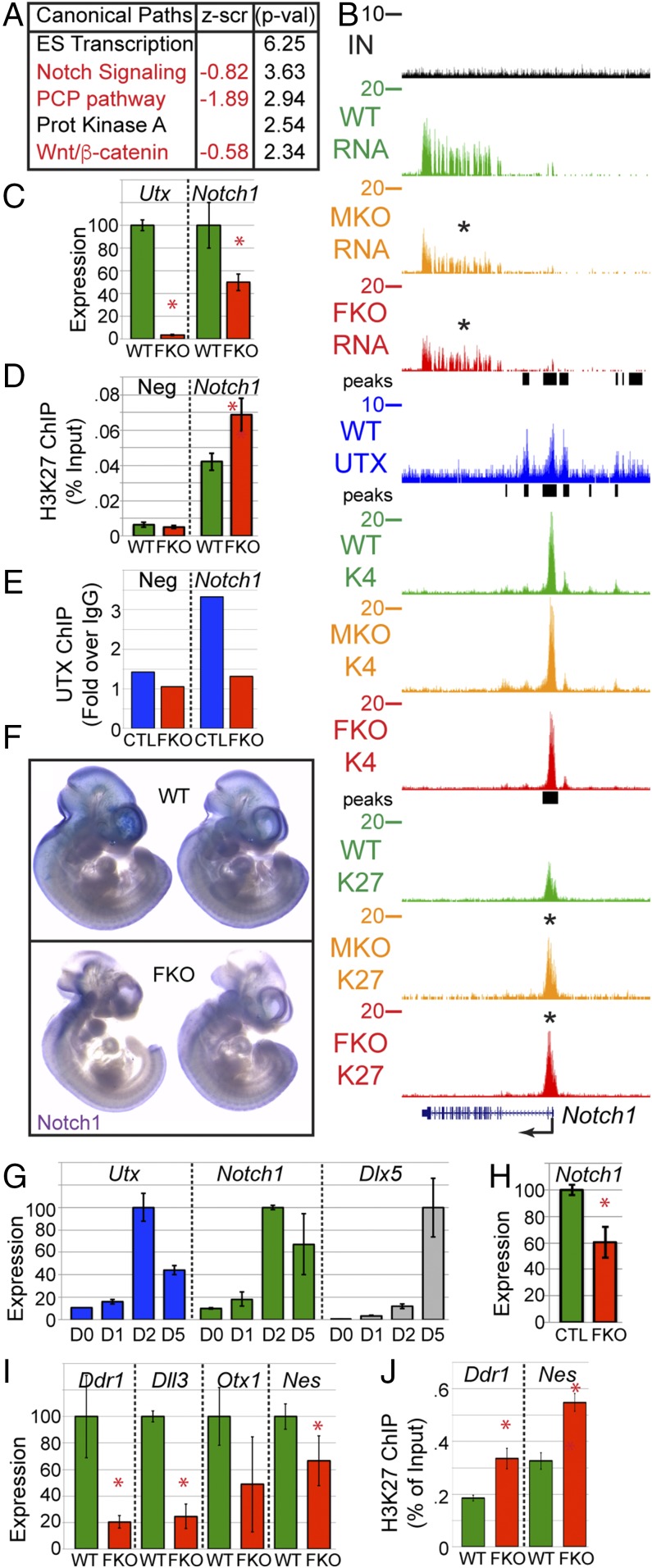
FKO altered transcripts were imported into Ingenuity Pathway Analysis (IPA) to identify affected UTX pathways based on the transcriptional profile. The most significantly down-regulated pathways included Notch signaling and PCP/Wnt/β-catenin signaling with both pathways playing significant roles in neural crest biology (Fig. 5A). UTX binding had strong effects on expression, but weak effects on demethylation of several Wnt pathway members, including Wnt1, Wnt7b, and Tcf7l2 (Dataset S1). UTX strongly demethylated H3K27me3 from Notch1 and Lfng within the Notch pathway with weaker effect on Dll1 (Dataset S1). Both MKO and FKO cranial NC experience significant reduction in Notch1 expression. While there was no effect on H3K4me3, Notch1 H3K27me3 was elevated in both MKO and FKO NC (Fig. 5B). WT UTX peaks were located at the transcription start site where demethylation is occurring as well as several sites upstream and downstream, which may consist of other regulatory elements. We sought to verify UTX activity through other nonsequencing methods. Notch1 RNA-seq and H3K27me3 ChIP-seq results were verified by qRT-PCR and ChIP-qPCR (Fig. 5 C and D). UTX ChIP-qPCR verified UTX Notch1 binding in the O9-1 cranial NC cell line and was lost with UTX CRISPR knockout (Fig. 5E). In situ hybridization demonstrated reduced Notch1 expression in E10.5 FKO embryos, particularly in cranial regions (Fig. 5F). Notch1 expression increased at the onset of osteoprogenitor differentiation of the O9-1 NC line at a time that correlated with elevated Utx expression (Fig. 5G), and CRISPR knockout of UTX reduced Notch1 expression (Fig. 5H). We validated additional UTX-regulated factors by qRT-PCR and H3K27me3 ChIP-qPCR (Fig. 5 I and J).
Fig. 5.
UTX regulates cranial NC targets. (A) Ingenuity Pathway Analysis of FKO-affected gene. Z score represents the overall alteration in FKO pathway expression. P = −log. (B) UCSC tracks of RNA-seq (RNA), H3K27me3 ChIP-seq (K27), H3K4me3 ChIP-seq (K4), and UTX ChIP-seq (UTX) at the Notch1 locus. (C) Quantitative RT-PCR of isolated primary NC (t test *P < 0.05, n ≥ 3). (D) H3K27me3 ChIP-qPCR. Neg is a control locus (Npm1) lacking H3K27me3 (t test *P < 0.03, n ≥ 3). (E) UTX ChIP-qPCR in O9-1 NC line treated with control (CTL) or Utx guide RNAs (FKO). Neg is a control locus (Slamf6). ChIP was normalized as percentage input and plotted as fold increase over IgG ChIP control. (F) Whole mount in situ hybridization for Notch1 in E10.5 WT or FKO embryos. (G) O9-1 maintained as NC stem cells (D0) or osteoprogenitor differentiated for indicated days. (H) Notch1 qRT-PCR at 2D of osteogenic differentiation with CRISPR for CTL or UTX FKO (t test *P < 0.03, n = 3). (I) Verification of RNA-seq targets by qRT-PCR (t test *P < 0.05, n ≥ 3). (J) Verification of H3K27me3 ChIP-seq targets by ChIP qPCR (t test *P < 0.02, n ≥ 3).
Altogether, genomic analysis has revealed UTX regulation of several factors and pathways that regulate NC cell biology and craniofacial development, but only a small subset of these targets experience UTX-mediated histone demethylation.
Full Dosage of Demethylase-Dead UTX Can Support Normal Craniofacial Development.
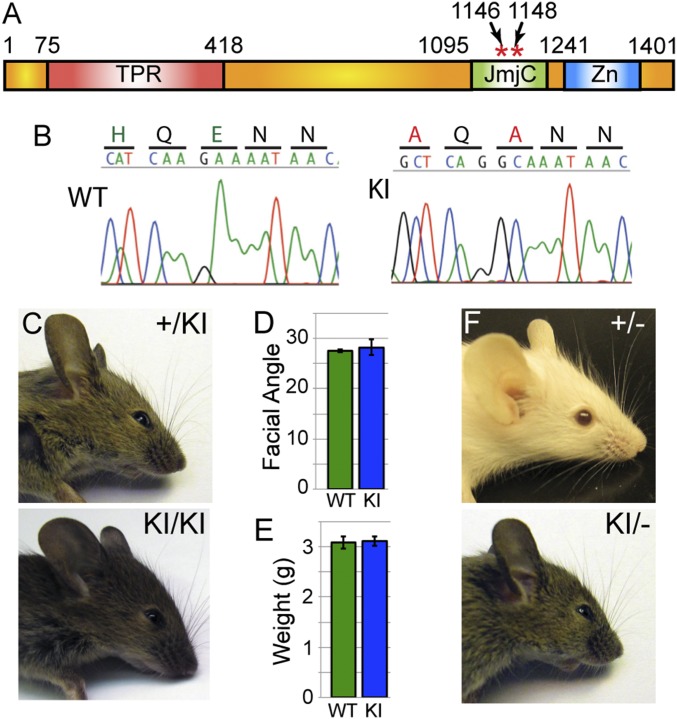
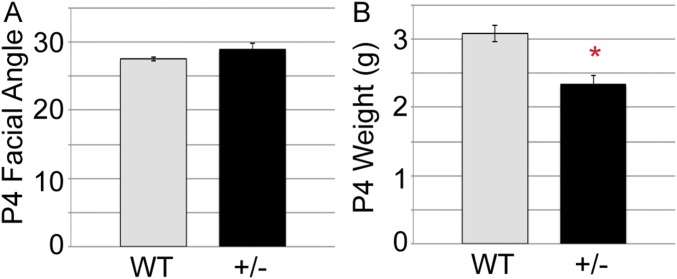
Genetic analysis of NC UTX versus UTY function has revealed that UTY can partially compensate for loss of UTX. The reason for only partial function relative to UTX may occur because NC requires some form of demethylation for establishing the appropriate cellular transcriptome. Alternatively, UTY may have additional deficiencies in other functions such as altered protein associations or chromatin binding properties that render the protein hypomorphic compared with UTX. To specifically assay NC demethylase requirements, we utilized a demethylase-dead knockin point mutation of Utx (KI allele) (allele schematic in Fig. 6A) (41, 42). Mutation of UTX H1146 and E1148 to alanine will disrupt JmjC-mediated iron cofactor binding and histone demethylation. These point mutations have been validated to eliminate histone demethylation through both in vitro assays with purified KI complexes (41) and in vivo overexpression assays (17). The KI mutant line has deficiencies in muscle regeneration and fails to demethylate several UTX targets of muscle differentiation from primary myoblasts (42). KI mutations were verified by sequencing of PCR and RT-PCR products (Fig. 6B). Both heterozygous +/KI and homozygous KI/KI females had normal facial characteristics lacking any previously observed MKO and FKO phenotypic appearance (Fig. 6C). Compared with WT mice, KI/KI homozygotes had normal body weight and facial angular measurements at early postnatal timepoints (Fig. 6 D and E).
Fig. 6.
Facial development does not require JmjC-mediated demethylation. (A) Schematic of UTX protein with location of H1146/E1148 knockin demethylase-dead point mutation (KI). (B) Sequencing of RT-PCR products from WT and KI mice verified H1146A/E1148A mutation. (C) Female heterozygous (+/KI) or homozygous (KI/KI) mice exhibit normal facial development. Compared with WT mice, KI/KI mice exhibit normal postnatal day 4 (P4) weight (D) and facial angle (E, measured as depicted in Fig. 2C). However, hypomorphic KI (monoallelic over a null allele, KI/−) exhibit some dysmorphic phenotypes such as snout hypoplasia, whereas heterozygous null (+/−) do not (F, n ≥ 5 analyzed for each genotype).
One caveat in contrasting KI/KI phenotypes to NC-specific Utx deletion is that KI/KI homozygotes have biallelic demethylase-dead UTX, while MKO hemizygotes have monoallelic demethylase-dead WT UTY. For better comparison, the KI allele was placed over a Utx null allele (−, gene trap, UtxGT1) (17). The monoallelic KI (KI/−) female mice displayed mild snout depression phenotypes similar to MKO (Fig. 6F). In contrast, Utx+/− heterozygous female mice lacked facial phenotypes but did exhibit mild postnatal growth retardation (Fig. 6F and Fig. S3). Therefore, UTX can support craniofacial development through demethylase-independent mechanisms; however, under hypomorphic conditions, UTX-mediated demethylation may be required for appropriate NC cellular gene activation.
Fig. S3.
Quantification of Utx heterozygous female phenotypes. (A) Utx+/− mice (carrying one copy of a null Utx gene trap) have normal facial angle at postnatal day 4 (P4, measured as depicted in Fig. 2C). (B) Utx+/− mice exhibit significant weight reductions at P4 (t test *P < 0.002).
Kabuki Syndrome Can Manifest Through Demethylase-Independent UTX Mutation.
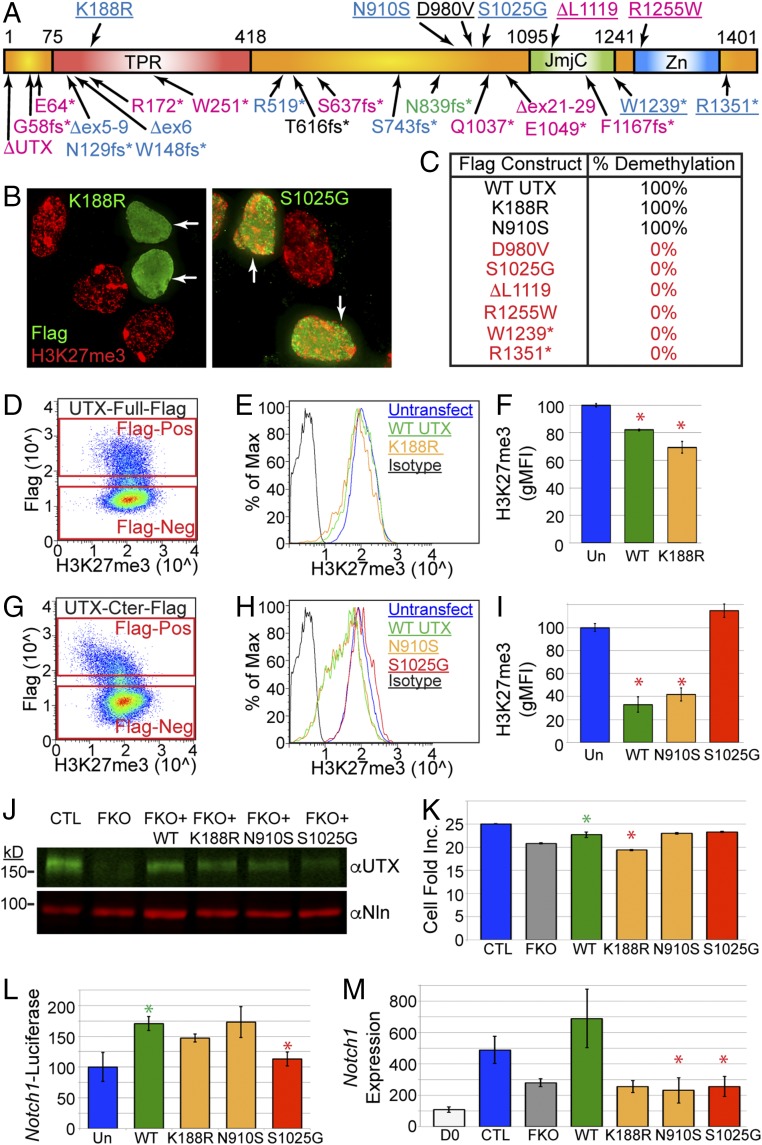
To investigate the molecular requirements of UTX that give rise to human Kabuki syndrome, we assessed the spectrum of mutations identified in Kabuki patients (10, 11, 13, 16, 23, 43–46). Kabuki causative UTX protein coding mutations vary in extent from complete UTX deletion to single amino acid point substitutions (Fig. 7A). Despite no obvious correlations in patient sex or domain-specific mutation, one commonality is the prevalence of nonsense mutations through frameshift or the introduction of stop codons. As the JmjC demethylation domain is downstream in a carboxyl-terminal (C terminal) region, the majority of these mutations will eliminate UTX-dependent demethylation activity (15 of 23 mutations). A smaller set of point mutations is predominantly located in proximity to the JmjC domain (Fig. 7A, Top); however, the molecular consequence of these mutations is unknown.
Fig. 7.
Demethylase-independent Kabuki causative UTX mutations. (A) Schematic of UTX protein with Kabuki causative mutations. Pink, female mutations; blue, male; black, undescribed sex; and green, familial male and female. Point mutations are listed Above the protein schematic and nonsense mutations are listed Below. Underlining means tested for demethylation. (B) Demethylation assay whereby HEK293 cells overexpressing Flag-UTX K188R lose H3K27me3 relative to untransfected cells. Flag-UTX S1025G fails to demethylate H3K27me3. (C) Summary of demethylation assay as percentage of scored cells with observable H3K27me3 demethylation (n = 50). (D) Full-length UTX-Flag transfected cells were subject to flow cytometry for Flag and H3K27me3. Cells were gated as transfected (Flag-Pos, positive) or untransfected (Flag-Neg, negative). (E) Histogram of H3K27me3 levels for untransfected or Flag-Pos WT UTX or K188R. (F) Full-length WT UTX and K188R transfected cells had significant reduction in H3K27me3 (Un, untransfected, t test *P < 0.05, 50,000 cells scored, n = 2). (G) C-terminal WT UTX-Flag (amino acids 880–1,401) transfected cells were subject to flow cytometry for Flag and H3K27me3. (H) Histogram of H3K27me3 levels for C-terminal UTX transfections. (I) C-terminal WT UTX and N910S transfected cells had significant reduction in H3K27me3 (t test *P < 0.02, 50,000 cells scored, n = 2). (J) O9-1 CRISPR UTX knockout (FKO) rescued with exogenous lentivirus for WT UTX or indicated point mutations. Nln, nucleolin control. (K) CellTiter-Glo numbers for rescue treatments (J), plotted as fold increase across D2–D5 of selection. (t test green *P < 0.03 for WT over FKO, t test red *P < 0.01 for K188R relative to WT, n = 3). (L) Notch1-Luciferase was transfected alone into HEK293 (Un) or cotransfected with indicated full-length UTX constructs (t test green *P < 0.02 for WT over Un, t test red *P < 0.01 for S1025G vs. WT, n = 3). (M) O9-1 NC were undifferentiated (D0) or differentiated to osteoprogenitors for 2 d with UTX FKO CRISPR or rescued with indicated WT or UTX mutant lentivirus (t test *P < 0.05 relative to WT, n = 3).
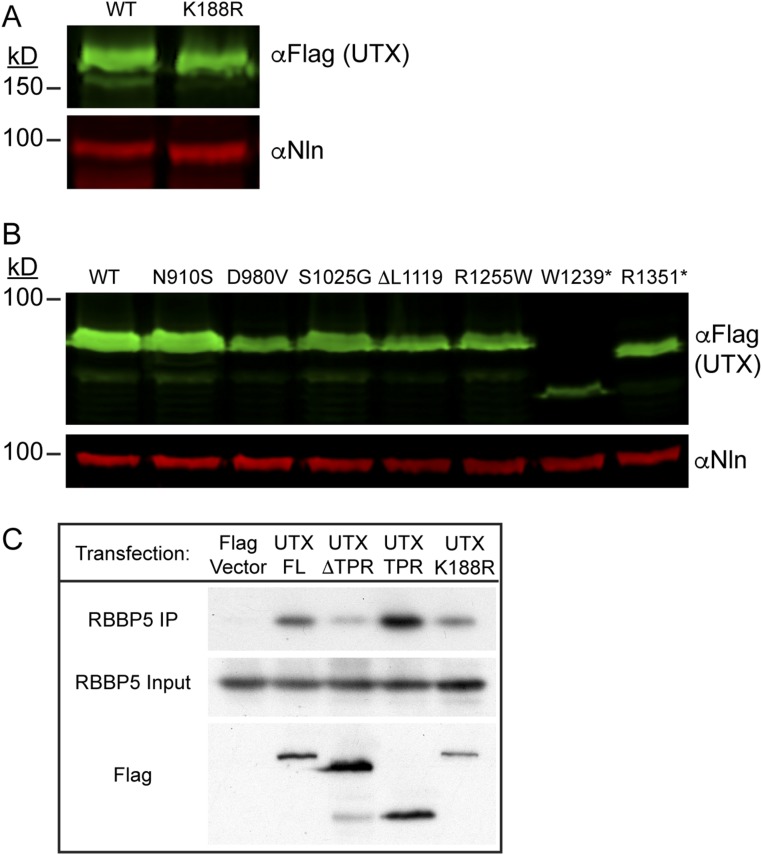
Demethylation activity can be assayed by transiently transfecting HEK293T cells to overexpress WT Flag-UTX and dramatically reduce H3K27me3 levels by immunofluorescence relative to untransfected cells (17, 19, 47). By introducing Kabuki causative point mutations into Flag-UTX, we assayed the effect that these mutations have on demethylase activity (Fig. 7B). We scored cells expressing moderate to high levels of Flag-UTX protein for any observable reduction in H3K27me3 immunofluorescence over untransfected cells in the same field. These assays were performed with the UTX C terminus (C-ter), encoding the JmjC and surrounding domains identified as essential for proper structure and function (48). Most point mutations or small deletions (D980V, S1025G, ΔL1119, R1255W, W1239*, and R1351*) eliminated UTX demethylation activity as no cells experienced H3K27me3 reduction (Fig. 7 B and C). Only two Kabuki causative point mutations, K188R (assayed in full-length UTX) and N910S, maintained enzymatic activity (Fig. 7 B and C) with 100% of medium-high cells exhibiting loss of H3K27me3. Most of these constructs exhibited overall expression levels similar to WT (Fig. S4 A and B).
Fig. S4.
UTX association with KMT2D complex is not disrupted by K188R. (A and B) Western blots of transfected HEK293 cells used in the demethylation assays from Fig. 7 B and C using full-length Flag-tagged UTX constructs (A) or C-terminal constructs (B). (C) IP of indicated Flag-tagged UTX constructs. WT Flag-UTX immunoprecipitated RBBP5 of the KMT2D protein complex. This association occurs through the TPR domain and is not disrupted by K188R mutation.
To quantify demethylation activity, these transfected cells were subjected to flow cytometry. Cells were gated based on untransfected (Flag negative) or transfected (Flag positive) signal (Fig. 7D). Cells expressing WT full-length UTX and K188R mutation exhibited a significant, mild reduction in H3K27me3 levels compared with untransfected cells (Fig. 7 E and F). Expression of the WT C-ter UTX construct was much more efficient in demethylating H3K27me3 (Fig. 7 G–I). The demethylation-inactivating S1025G mutation exhibited H3K27me3 levels similar and even higher than untransfected cells (Fig. 7 H and I). Both WT C-ter UTX and N910S mutation dramatically reduced the profile of H3K27me3 (Fig. 7H) resulting in highly significant demethylation (Fig. 7I). We conclude that K188R and N910S mutations have demethylation activities similar to WT and disrupt alternative UTX functions in Kabuki patients.
As the K188R mutation resides in the tetratricopeptide (TPR) protein association domain, we reasoned that K188R might disrupt UTX protein association with the KMT2D H3K4 methylation complex. The TPR domain is required for this association, as its removal in Flag-UTX (ΔTPR) reduces immunoprecipitation of RBBP5, a KMT2D complex protein (Fig. S4C). The TPR domain alone is sufficient for RBBP5 association; however, K188R does not appreciably affect KMT2D complex interaction.
To test the function of demethylase-independent Kabuki causative point mutations, we attempted to rescue NC cellular deficits caused by UTX knockout (Fig. 3J). CRISPR lentiviral UTX knockout of the O9-1 NC cell line was performed in combination with additional exogenous UTX lentivirus carrying a synonymous substitution in the guide targeting sequence. Expression of WT UTX from this exogenous lentivirus partially restored knocked out protein levels (Fig. 7J) and deficits in cellular numbers (Fig. 7K). Expression of exogenous UTX K188R failed to rescue UTX knockout cellular phenotypes; however, N910S and S1025G appeared similar to WT. We also examined regulated expression of one UTX target, Notch1. A Notch1-Luciferase reporter construct was utilized containing a 6-kb promoter region upstream of the Notch1 TSS. Upon transfection into HEK293 cells, this reporter was activated by cotransfection with WT UTX (Fig. 7L). Only S1025G mutation reduced Notch1 promoter activation relative to WT UTX (Fig. 7L). Notch1 activation was also examined through O9-1 osteoprogenitor differentiation following UTX knockout/lentiviral rescue (Fig. 7M). UTX N910S and S1025G mutation failed to restore Notch1 activation to WT levels (Fig. 7M). Overall, elimination of UTX demethylase activity is a common feature of Kabuki syndrome, but the disorder can manifest through disruption of alternative UTX functions, which have varying effects on NC cellular culture properties.
Discussion
Chromatin-modifying enzymes are increasingly prevalent in biological research pertaining to neural crest cellular function. These enzymes that catalyze histone posttranslational modifications or nucleosomal remodeling have been mutated in several distinct craniofacial human syndromes with overlapping clinical features, including histone methylases (KMT2D: Kabuki syndrome), histone demethylases (UTX: Kabuki syndrome), chromatin remodelers (CHD7: CHARGE syndrome and SWI/SNF: Coffin-Siris syndrome), and histone acetylases (CBP: Rubinstein-Taybi syndrome) (12, 13, 40, 49–51). Many of these craniofacial factors form protein associations (17, 52, 53) and may function not only within the confines of given enzymatic activity, but may play enzymatic-independent roles in the recruitment of alternative chromatin machinery within a common neural crest cellular precursor. BRG1, ARID1A, and CHD7 chromatin remodeling enzymes all produce NC cell intrinsic functions in migration and differentiation based on tissue-specific knockout studies (6, 54, 55). In fact BRG1 and CHD7 cooperate in activation of NC transcription factors (7). In contrast to chromatin remodelers, histone posttranslational enzymatic function has remained largely unstudied with regards to modeling human disorders by neural crest function. Utx morphant zebrafish demonstrate cardiac, brain, and cranial defects (46); however, the cellular and molecular contribution of UTX function is not known.
We have now characterized the genetic, molecular, and genomic function of UTX in NC-based craniofacial development. Female FKO UTX NC-specific mouse knockout has faithfully modeled many clinical features of Kabuki syndrome, including craniofacial, cardiac, and growth abnormalities. Male MKO UTX NC knockout has more moderate phenotypes due to partial demethylase-independent compensation by UTY. Notably, Utx+/− heterozygous female mice exhibit postnatal growth deficits, but fail to model craniofacial phenotypes present in heterozygous human patients. This may occur because mice often fail to reproduce human haploinsufficient phenotypes; UTX human point mutations and partial deletions may be dominant negative; or human phenotypes may be influenced by other modifier loci. The combination of MKO and FKO demethylase dependency and independency is also observed at the genomic level as UTX binding has variable effects on the chromatin landscape of regulated transcripts ranging from strong, weak, or no effect on H3K27me3 demethylation. It is possible that weaker demethylation observations may arise due to cellular heterogeneity of pooled primary cells rather than a lack of enzymatic activity. However, UTX demethylation is not crucial for NC cellular development as KI/KI catalytic point mutation lacks any discernible craniofacial phenotype. UTX-mediated demethylation is required in vivo for muscle regeneration and antiviral immunity; however, many developmental decisions do not require catalytic activity including mesoderm induction, mammary luminal differentiation, T-cell differentiation, and gross embryonic development (17, 28, 41, 42, 52, 56–58). Furthermore, two Kabuki causative human point mutations, K188R and N910S, exhibit normal H3K27me3 demethylation and produce the disorder through disruption of alternative, demethylase-independent UTX function. These mutations have varying effects on NC cell culture, and knockin mouse models will be required to identify more precise cellular and molecular causative mechanisms.
Several crucial NC cellular pathways and genes related to craniofacial development are altered with UTX loss of function. We observed a UTX-dependent effect on P53-based targets. UTX associates with P53 and may play a direct role in P53-based gene activation (59). P53 alleles have been shown to modify the phenotypic severity of NC mutations in chromatin remodeling machinery (60). UTX mutant NC also misexpressed CHD7, which is frequently mutated in CHARGE syndrome and can have overlapping clinical features to Kabuki syndrome (40, 61).
Pathway analysis identified the strongest transcriptional deficiencies in Notch and Wnt signaling pathways. These pathways experience moderate reductions in gene expression of several pathway members. Embryos with complete knockout of NC Wnt signaling fail to form any anterior facial structures; however, genetic modulation by hypomorphic mutations in multiple components (Wnt1 and Wnt3a) can produce similar facial hypoplasial phenotypes to UTX NC mutation (62, 63). Similar facial characteristics can also be derived from combinatorial hypomorphic mutation of Notch signaling (Notch1/Dll3) (64). The FKO and MKO phenotypes may be due to a combination of genes and pathways identified in our study, and future research will uncover the contribution of various pathways to MKO and FKO pathogenesis.
MKO mice with a single copy of demethylase-dead UTY or KI/− mice with a single copy of demethylase-dead UTX both develop mild craniofacial phenotypes. However, KI/KI catalytic UTX mutation has normal facial development. Therefore, UTX demethylation events (such as Notch1) can be overcome under normal UTX dosage (KI/KI) by alternative UTX functions. UTX physically associates with KMT2D and CBP, which function in enhancer activation through H3K27 acetylation (53, 65–68). UTX has previously been localized to both promoters and enhancers in mouse mammary tissue (57) and has been correlated with ES cell enhancer activation (69). We found UTX at a large percentage of distal elements (Fig. 5B and Fig. S2E) and UTX may regulate NC enhancer activation through demethylase-independent acetylase recruitment. Future research will be required to determine the extent of UTX and KMT2D overlap at NC enhancers and identify roles in NC enhancer activation. UTX and UTY can also physically associate with SWI/SNF chromatin remodeling machinery as well as lineage-specific transcription factors (17, 52, 53, 70). Now that we have established a model system capable of assessing NC genomics, we can begin to examine genomic overlap and coregulation potential of alternative demethylase-independent UTX regulatory factors.
Materials and Methods
Mice.
All mouse experimental procedures were approved by the University of North Carolina Institutional Animal Care and Use Committee. All mice were maintained on outbred genetic backgrounds. The Utxfl and UtxGT1 alleles are described (17, 28). The Wnt1-Cre transgene, Pax3-Cre knockin, and Tomato reporter were imported from The Jackson Laboratory (20, 21, 24). Utx demethylase-dead knockin mice were developed as described (41, 42).
Constructs, Transfections, Immunofluorescence, Western Blotting, and Antibodies.
Transfection of HEK293T was accomplished with Lipofectamine 3000 as directed (Invitrogen). Fixation, extraction, and immunofluorescence were performed as described (17). Immunofluorescence antibodies include anti-Flag (Sigma, F3165; 1:500) and anti-H3K27me3 (Cell Signaling, 9733S; 1:500). Tunel was performed with the Roche cell death detection kit as directed (11684795910). Cells were imaged with Zeiss axiovision software. Image stacks were deconvolved and Z projected. For Western blotting, nuclear lysates were prepared according to Invitrogen’s nuclear extraction protocol. Immunoprecipitations were carried out with 10 μL antibody and 50 µL protein A/G beads (Santa Cruz, sc-2003) in buffer A as described, using 500 µg of lysate (15). Immunoprecipitation reactions were boiled off beads and run with 10% input on a 6% SDS/PAGE gel. Western blotting was performed as described (17) with anti-UTX (Cell Signaling, 33510S; 1:4,000), anti-RBBP5 (Bethyl Labs, A300-109A; 1:5,000), and anti-nucleolin (Bethyl Labs, A300-711A; 1:5,000).
Cell Culture and Constructs.
HEK293T were maintained in DMEM supplemented with glutamine, penicillin–streptomycin, and 10% FBS. Flag-human UTX is described (17). The Flag-human UTX C-terminal construct containing residues 880–1,401 (GenBank: NP_066963.2) was described (17). Site-directed mutagenesis was performed via QuikChange Lightning (Agilent) as directed to produce point mutations or small deletions. All mutations were introduced into the UTX C-terminal construct for demethylase assay except for UTX K188R, which was mutated in the full-length construct. To assay demethylase activity, constructs were transfected into HEK293T and processed for immunofluorescence or flow cytometry 2 d later. The O9-1 cranial NC cell line was generated, cultured, and differentiated as described (27). Luciferase assays were performed following transfection of a 6-kb Notch1 promoter-Luciferase construct (71) as described (17).
Histology.
Embryonic fixation and sectioning was performed as described (17). Alizarin red and alcian blue staining was performed as described (72).
Primary NC Isolation.
Anterior facial regions from E12.5 WT (male), MKO, or FKO embryos carrying the Tomato reporter were isolated by forceps dissection of facial regions rostral from the maxilla and anterior to the otic vesicle. This tissue was dissociated in HBSS containing 0.25% trypsin and 0.7 mg/mL DNase I for 8 min at 37 °C followed by pipetting and neutralization with 12% FBS as described (73, 74). Cells were washed on ice and resuspended in RPMI with 10% FBS, 2% BSA, and 100 ng/mL DAPI. Cells were filtered through a 70-μm cell strainer and sorted on a Beckman Coulter MoFlo XDP [University of North Carolina (UNC) Flow Cytometry Core] based on Tomato fluorescence and DAPI exclusion into either TRIzol-LS (Thermo Fisher) for RNA preparation or RPMI 10% FBS for ChIP. Cells for ChIP were washed in PBS and the cell pellet was frozen. Cellular staining and flow cytometry were performed as described (56) with anti-UTX (Cell Signaling, 33510S; 1:300), anti-KI67 (BioLegend, 350507; 1:30), and annexin V (BioLegend, 640919).
RNA-Seq and ChIP-Seq Library Preparation.
RNA was isolated with TRIzol-LS as directed (Thermo Fisher) from two biological replicates of 2 × 105 WT, MKO, or FKO E12.5 sorted NC cells and mRNA was isolated for cDNA synthesis, ligation of Truseq adapters, and library amplification (Kappa KK8420 stranded RNA-seq kit). For native ChIP-seq, frozen E12.5 sorted cranial NC cell pellets were subjected to MNase digestions followed by overnight ChIP with preconjugated antibody-Dynabead complexes (50% each protein A/G; Thermo Fisher 100-01D and 10003D) as described (75). Anti-H3K27me3 (Abcam, ab6002; 1 µL) or anti-H3K4me3 (Abcam, ab8580; 1 µL) antibodies were used for ChIP on 3 WT, 3 FKO, and 1 MKO biological replicates of pooled samples totaling 8 × 105 or 2 × 105 cells, respectively, for H3K27me3 and H3K4me3. Anti-UTX (Cell Signaling, 33510S; 10 μL) was used for ChIP on 20 × 106 O9-1 cranial NC cells that were fixed with 0.3% formaldehyde for 30 min at 4 °C and processed for MNase and ChIP as described (76). ChIP DNA was purified with ChIP DNA Clean and Concentrator Kit (D5205; Zymo Research) and Truseq adapter ligation and library amplification was performed with Hyper Prep Kit (Kappa KK8502).
RNA-Seq and ChIP-Seq Analysis.
Library samples were multiplexed and sequenced with the HiSEq. 2500 Analyzer (UNC High Throughput Sequencing Facility). The quality of the sequence reads was evaluated with FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were then mapped to the MM9 B6 genome using Tophat2 (RNA-seq, ref. 77) or Bowtie2 (ChIP-seq, ref. 78). We removed any sequencing data on the X and Y chromosomes due to sex differences between WT male embryos and FKO females. Genic RNA-seq reads were counted with htseq-count (79) and significant (FDR < 0.05) differential expression was assessed at expressed genes (with cpm >1) with EdgeR (80). For ChIP-seq, significant enrichment (peaks) were called using MACS version 2 (ref. 31; liulab.dfci.harvard.edu/MACS/index.html) with the input ChIP-seq datasets for the background model, on pooled replicates. Multicov within the Bedtools suite was utilized to count reads for individual biological replicates across MACS peaks (81) and EdgeR assessed significant (FDR < 0.05) differential read enrichment at identified peaks (with cpm >1). Bedtools identified closest genes in proximity to ChIP-seq peaks (within 10 kb of gene body), and Bedtools intersect identified direct overlap of different ChIP-seq datasets. UTX targets were prioritized as having a strong effect on transcription/methylation based on genes/peaks with higher read counts (RPKM ≥ 4) and more dramatic changes (absolute log fold changes ≥0.5). Direct UTX H3K27me3 demethylation was characterized as a gene bound by UTX (within 10 kb of gene body) with significantly elevated FKO H3K27me3 (within 10 kb of gene body). Biological replicates were merged and Bedtools genomecov created bedgraph files that were uploaded to the University of California Santa Cruz (UCSC) genome browser for visualization (82). Deeptools was utilized to view profiles and heatmaps of sequencing data (83). Ingenuity Pathway Analysis and the MsigDB were utilized to analyze common signaling components based on the RNA-seq up-regulated and down-regulated genic expression data (https://www.qiagenbioinformatics.com/, ref. 84). Peak annotations were performed using the HOMER annotatePeaks.pl program (85).
UTX Knockdown, ChIP-PCR, and UTX Rescue.
A Utx guide sequence (ACCTCACGAACCCGAAAAGG) or control nontargeting guide sequence (ACGGAGGCTAAGCGTCGCAA) were cloned into lentiCRISPR v2 (Addgene, 52961; ref. 86) and transfected into HEK293T cells with psPAX2 (Addgene, 12260) and pMD2.G (Addgene, 12259) packaging plasmids provided by Didier Trono, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland. Cell media were collected after 2 d and incubated with the O9-1 cranial NC cell line (1:4 dilution). The next day, transduced cells were placed under 2 μg/mL puromycin selection for 3 d. Cells were either processed for protein Western blotting or fixed for ChIP. UTX ChIP was performed followed by quantitative PCR for control or UTX targeted peak locations (CFX96 real-time PCR system; BIO-RAD). Quantitative PCR for either IgG ChIP or UTX ChIP in control or Utx-targeted knockdown were normalized relative to input DNA and graphed as fold increase over IgG. A full-length lentiviral UTX construct was utilized to rescue NC cellular phenotypes (87). Site-directed mutagenesis altered the PAM sequence adjacent to the Utx guide target sequence so that expression from this construct would not be affected by CRISPR knockout constructs. Site-directed mutagenesis also introduced K188R, N910S, and S1025G mutations.
In Situ Hybridization.
A Notch1 in situ hybridization probe was amplified, corresponding to a previously utilized region of Notch1 (88). Whole mount in situ hybridization was performed as described (6, 89).
Primers.
All genotyping, mutagenesis, and qPCR primers are available on request.
Supplementary Material
Acknowledgments
We thank Robert E. Maxson for generous contribution of the O9-1 cranial neural crest cell line, Sebastian Brenner for generous contribution of the UTX lentiviral construct, and the members of the T.M. laboratory for review of this manuscript. This work was financially supported by a University of North Carolina Junior Faculty Development Award (to K.B.S.) and NIH Award R01GM101974 (to T.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession GSE103849).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705011114/-/DCSupplemental.
References
- 1.Bolande RP. Neurocristopathies–Unifying concept of disease arising in neural crest maldevelopment. Hum Pathol. 1974;5:409–429. doi: 10.1016/s0046-8177(75)80115-8. [DOI] [PubMed] [Google Scholar]
- 2.Dupin E, Creuzet S, Le Douarin NM. The contribution of the neural crest to the vertebrate body. Adv Exp Med Biol. 2006;589:96–119. doi: 10.1007/978-0-387-46954-6_6. [DOI] [PubMed] [Google Scholar]
- 3.Trainor PA. Specification of neural crest cell formation and migration in mouse embryos. Semin Cell Dev Biol. 2005;16:683–693. doi: 10.1016/j.semcdb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Santagati F, Rijli FM. Cranial neural crest and the building of the vertebrate head. Nat Rev Neurosci. 2003;4:806–818. doi: 10.1038/nrn1221. [DOI] [PubMed] [Google Scholar]
- 5.Dixon J, et al. The Treacher Collins Syndrome Collaborative Group Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. Nat Genet. 1996;12:130–136. doi: 10.1038/ng0296-130. [DOI] [PubMed] [Google Scholar]
- 6.Chandler RL, Magnuson T. The SWI/SNF BAF-A complex is essential for neural crest development. Dev Biol. 2016;411:15–24. doi: 10.1016/j.ydbio.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bajpai R, et al. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niikawa N, Matsuura N, Fukushima Y, Ohsawa T, Kajii T. Kabuki make-up syndrome: A syndrome of mental retardation, unusual facies, large and protruding ears, and postnatal growth deficiency. J Pediatr. 1981;99:565–569. doi: 10.1016/s0022-3476(81)80255-7. [DOI] [PubMed] [Google Scholar]
- 9.Adam MP, Hudgins L, Hannibal M. Kabuki syndrome. In: Pagon RA, et al., editors. GeneReviews(R) University of Washington; Seattle: 1993. [Google Scholar]
- 10.Miyake N, et al. MLL2 and KDM6A mutations in patients with Kabuki syndrome. Am J Med Genet A. 2013;161A:2234–2243. doi: 10.1002/ajmg.a.36072. [DOI] [PubMed] [Google Scholar]
- 11.Bögershausen N, et al. Mutation update for Kabuki syndrome genes KMT2D and KDM6A and further delineation of X-linked Kabuki syndrome subtype 2. Hum Mutat. 2016;37:847–864. doi: 10.1002/humu.23026. [DOI] [PubMed] [Google Scholar]
- 12.Ng SB, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lederer D, et al. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am J Hum Genet. 2012;90:119–124. doi: 10.1016/j.ajhg.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 15.Cho YW, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banka S, et al. Novel KDM6A (UTX) mutations and a clinical and molecular review of the X-linked Kabuki syndrome (KS2) Clin Genet. 2015;87:252–258. doi: 10.1111/cge.12363. [DOI] [PubMed] [Google Scholar]
- 17.Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 2012;8:e1002964. doi: 10.1371/journal.pgen.1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 19.Hong S, et al. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 21.Lang D, et al. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005;433:884–887. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- 22.Jarad G, Miner JH. The Pax3-Cre transgene exhibits a rostrocaudal gradient of expression in the skeletal muscle lineage. Genesis. 2009;47:1–6. doi: 10.1002/dvg.20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lederer D, Shears D, Benoit V, Verellen-Dumoulin C, Maystadt I. A three generation X-linked family with Kabuki syndrome phenotype and a frameshift mutation in KDM6A. Am J Med Genet A. 2014;164A:1289–1292. doi: 10.1002/ajmg.a.36442. [DOI] [PubMed] [Google Scholar]
- 24.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serbedzija GN, Bronner-Fraser M, Fraser SE. Vital dye analysis of cranial neural crest cell migration in the mouse embryo. Development. 1992;116:297–307. doi: 10.1242/dev.116.2.297. [DOI] [PubMed] [Google Scholar]
- 26.Goodnough LH, et al. Distinct requirements for cranial ectoderm and mesenchyme-derived wnts in specification and differentiation of osteoblast and dermal progenitors. PLoS Genet. 2014;10:e1004152. doi: 10.1371/journal.pgen.1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishii M, et al. A stable cranial neural crest cell line from mouse. Stem Cells Dev. 2012;21:3069–3080. doi: 10.1089/scd.2012.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shpargel KB, Starmer J, Yee D, Pohlers M, Magnuson T. KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development. PLoS Genet. 2014;10:e1004507. doi: 10.1371/journal.pgen.1004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bildsoe H, et al. Requirement for Twist1 in frontonasal and skull vault development in the mouse embryo. Dev Biol. 2009;331:176–188. doi: 10.1016/j.ydbio.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 30.Li H, Williams T. Separation of mouse embryonic facial ectoderm and mesenchyme. J Vis Exp. 2013 doi: 10.3791/50248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dullin C, Missbach-Guentner J, Vogel WF, Grabbe E, Alves F. Semi-automatic classification of skeletal morphology in genetically altered mice using flat-panel volume computed tomography. PLoS Genet. 2007;3:e118. doi: 10.1371/journal.pgen.0030118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaufmann LT, Niehrs C. Gadd45a and Gadd45g regulate neural development and exit from pluripotency in Xenopus. Mech Dev. 2011;128:401–411. doi: 10.1016/j.mod.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Park D, et al. Nestin is required for the proper self-renewal of neural stem cells. Stem Cells. 2010;28:2162–2171. doi: 10.1002/stem.541. [DOI] [PubMed] [Google Scholar]
- 35.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takebe A, et al. Microarray analysis of PDGFR alpha+ populations in ES cell differentiation culture identifies genes involved in differentiation of mesoderm and mesenchyme including ARID3b that is essential for development of embryonic mesenchymal cells. Dev Biol. 2006;293:25–37. doi: 10.1016/j.ydbio.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 37.Suda Y, Nakabayashi J, Matsuo I, Aizawa S. Functional equivalency between Otx2 and Otx1 in development of the rostral head. Development. 1999;126:743–757. doi: 10.1242/dev.126.4.743. [DOI] [PubMed] [Google Scholar]
- 38.Byrd NA, Meyers EN. Loss of Gbx2 results in neural crest cell patterning and pharyngeal arch artery defects in the mouse embryo. Dev Biol. 2005;284:233–245. doi: 10.1016/j.ydbio.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 39.Ellis P, et al. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- 40.Vissers LE, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 41.Wang C, et al. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci USA. 2012;109:15324–15329. doi: 10.1073/pnas.1204166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faralli H, et al. UTX demethylase activity is required for satellite cell-mediated muscle regeneration. J Clin Invest. 2016;126:1555–1565. doi: 10.1172/JCI83239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheon CK, et al. Identification of KMT2D and KDM6A mutations by exome sequencing in Korean patients with Kabuki syndrome. J Hum Genet. 2014;59:321–325. doi: 10.1038/jhg.2014.25. [DOI] [PubMed] [Google Scholar]
- 44.Micale L, et al. Molecular analysis, pathogenic mechanisms, and readthrough therapy on a large cohort of Kabuki syndrome patients. Hum Mutat. 2014;35:841–850. doi: 10.1002/humu.22547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyake N, et al. KDM6A point mutations cause Kabuki syndrome. Hum Mutat. 2013;34:108–110. doi: 10.1002/humu.22229. [DOI] [PubMed] [Google Scholar]
- 46.Van Laarhoven PM, et al. Kabuki syndrome genes KMT2D and KDM6A: Functional analyses demonstrate critical roles in craniofacial, heart and brain development. Hum Mol Genet. 2015;24:4443–4453. doi: 10.1093/hmg/ddv180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 48.Sengoku T, Yokoyama S. Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A. Genes Dev. 2011;25:2266–2277. doi: 10.1101/gad.172296.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsurusaki Y, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012;44:376–378. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- 50.Petrij F, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 51.Sanka M, Tangsinmankong N, Loscalzo M, Sleasman JW, Dorsey MJ. Complete DiGeorge syndrome associated with CHD7 mutation. J Allergy Clin Immunol. 2007;120:952–954. doi: 10.1016/j.jaci.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 52.Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tie F, Banerjee R, Conrad PA, Scacheri PC, Harte PJ. Histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine 27. Mol Cell Biol. 2012;32:2323–2334. doi: 10.1128/MCB.06392-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li W, et al. Brg1 governs distinct pathways to direct multiple aspects of mammalian neural crest cell development. Proc Natl Acad Sci USA. 2013;110:1738–1743. doi: 10.1073/pnas.1218072110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sperry ED, et al. The chromatin remodeling protein CHD7, mutated in CHARGE syndrome, is necessary for proper craniofacial and tracheal development. Dev Dyn. 2014;243:1055–1066. doi: 10.1002/dvdy.24156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cook KD, et al. T follicular helper cell-dependent clearance of a persistent virus infection requires T cell expression of the histone demethylase UTX. Immunity. 2015;43:703–714. doi: 10.1016/j.immuni.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoo KH, et al. Histone demethylase KDM6A controls the mammary luminal lineage through enzyme-independent mechanisms. Mol Cell Biol. 2016;36:2108–2120. doi: 10.1128/MCB.00089-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Welstead GG, et al. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc Natl Acad Sci USA. 2012;109:13004–13009. doi: 10.1073/pnas.1210787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akdemir KC, et al. Genome-wide profiling reveals stimulus-specific functions of p53 during differentiation and DNA damage of human embryonic stem cells. Nucleic Acids Res. 2014;42:205–223. doi: 10.1093/nar/gkt866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Nostrand JL, et al. Inappropriate p53 activation during development induces features of CHARGE syndrome. Nature. 2014;514:228–232. doi: 10.1038/nature13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verhagen JM, Oostdijk W, Terwisscha van Scheltinga CE, Schalij-Delfos NE, van Bever Y. An unusual presentation of Kabuki syndrome: Clinical overlap with CHARGE syndrome. Eur J Med Genet. 2014;57:510–512. doi: 10.1016/j.ejmg.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 62.Brault V, et al. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- 63.Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 64.Loomes KM, et al. Dll3 and Notch1 genetic interactions model axial segmental and craniofacial malformations of human birth defects. Dev Dyn. 2007;236:2943–2951. doi: 10.1002/dvdy.21296. [DOI] [PubMed] [Google Scholar]
- 65.Jin Q, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee JE, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife. 2013;2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ang SY, et al. KMT2D regulates specific programs in heart development via histone H3 lysine 4 di-methylation. Development. 2016;143:810–821. doi: 10.1242/dev.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang C, et al. Enhancer priming by H3K4 methyltransferase MLL4 controls cell fate transition. Proc Natl Acad Sci USA. 2016;113:11871–11876. doi: 10.1073/pnas.1606857113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang SP, et al. A UTX-MLL4-p300 transcriptional regulatory network coordinately shapes active enhancer landscapes for eliciting transcription. Mol Cell. 2017;67:308–321.e6. doi: 10.1016/j.molcel.2017.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee S, Lee JW, Lee SK. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell. 2012;22:25–37. doi: 10.1016/j.devcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yashiro-Ohtani Y, et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23:1665–1676. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lufkin T, et al. Homeotic transformation of the occipital bones of the skull by ectopic expression of a homeobox gene. Nature. 1992;359:835–841. doi: 10.1038/359835a0. [DOI] [PubMed] [Google Scholar]
- 73.Kubota H, Brinster RL. Culture of rodent spermatogonial stem cells, male germline stem cells of the postnatal animal. Methods Cell Biol. 2008;86:59–84. doi: 10.1016/S0091-679X(08)00004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Singh S, et al. Fluorescence-activated cell sorting of EGFP-labeled neural crest cells from murine embryonic craniofacial tissue. J Biomed Biotechnol. 2005;2005:232–237. doi: 10.1155/JBB.2005.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brind’Amour J, et al. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Commun. 2015;6:6033. doi: 10.1038/ncomms7033. [DOI] [PubMed] [Google Scholar]
- 76.Raab JR, Resnick S, Magnuson T. Genome-wide transcriptional regulation mediated by biochemically distinct SWI/SNF complexes. PLoS Genet. 2015;11:e1005748. doi: 10.1371/journal.pgen.1005748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anders S, Pyl PT, Huber W. HTSeq–A python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramírez F, et al. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thieme S, et al. The histone demethylase UTX regulates stem cell migration and hematopoiesis. Blood. 2013;121:2462–2473. doi: 10.1182/blood-2012-08-452003. [DOI] [PubMed] [Google Scholar]
- 88.Reaume AG, Conlon RA, Zirngibl R, Yamaguchi TP, Rossant J. Expression analysis of a Notch homologue in the mouse embryo. Dev Biol. 1992;154:377–387. doi: 10.1016/0012-1606(92)90076-s. [DOI] [PubMed] [Google Scholar]
- 89.Jacques-Fricke BT, Roffers-Agarwal J, Gammill LS. DNA methyltransferase 3b is dispensable for mouse neural crest development. PLoS One. 2012;7:e47794. doi: 10.1371/journal.pone.0047794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.