Significance
Understanding the genetic basis of human disease can reveal mechanisms of disease pathology and guide the design of novel treatment strategies. Here, we leverage insights from genetic studies to create a blueprint for treatment of inflammatory bowel disease (IBD). We demonstrate the feasibility of using small-molecule inhibitors to recapitulate the antiinflammatory function of CARD9 mutations associated with protection from IBD.
Keywords: CARD9, inflammatory bowel disease, small molecules, therapeutics
Abstract
Advances in human genetics have dramatically expanded our understanding of complex heritable diseases. Genome-wide association studies have identified an allelic series of CARD9 variants associated with increased risk of or protection from inflammatory bowel disease (IBD). The predisposing variant of CARD9 is associated with increased NF-κB–mediated cytokine production. Conversely, the protective variant lacks a functional C-terminal domain and is unable to recruit the E3 ubiquitin ligase TRIM62. Here, we used biochemical insights into CARD9 variant proteins to create a blueprint for IBD therapeutics and recapitulated the mechanism of the CARD9 protective variant using small molecules. We developed a multiplexed bead-based technology to screen compounds for disruption of the CARD9–TRIM62 interaction. We identified compounds that directly and selectively bind CARD9, disrupt TRIM62 recruitment, inhibit TRIM62-mediated ubiquitinylation of CARD9, and demonstrate cellular activity and selectivity in CARD9-dependent pathways. Taken together, small molecules targeting CARD9 illustrate a path toward improved IBD therapeutics.
Genome-wide association studies (GWAS) have provided information on thousands of common single nucleotide polymorphisms (SNPs) associated with multifactorial diseases such as inflammatory bowel disease (IBD) (1–5), rheumatoid arthritis (6), type-2 diabetes, schizophrenia, and many other heritable traits associated with disease (7). A major challenge remains to translate genetic associations into a deeper understanding of disease pathogenesis and to guide drug discovery (7). Recent approaches combining GWAS data with genomic and biological datasets point to disease genes, pathways, or protein networks (6), but this wealth of human genetic information has yet to be fully exploited for widespread clinical and therapeutic use. However, a few seminal examples have demonstrated success. Discovery of the human CCR5-d32 variant, which conveys resistance to HIV infection, led to development of HIV-entry inhibitors targeting CCR5 (maraviroc) (8). In cardiovascular disease, gain- (9) and loss-of-function PCSK9 variants were shown to dramatically impact low-density lipoprotein cholesterol (LDLc) levels (10). These findings guided the discovery of targeted PCSK9 therapeutics (alirocumab and evolocumab) that were approved by the Food and Drug Administration in 2015 for reducing LDLc and risk for myocardial infarction.
The genetic architecture of IBD risk has been mapped in detail and suggests potential therapeutic targets. In particular, the field has benefitted from GWAS (2, 4, 11, 12) followed by deep exome resequencing analyses (3, 5). However, the translation of genetics to therapeutics has not yet been achieved, and IBD patients (2 million in the United States alone) (7) have limited treatment options. In the present work, we aim to bridge the gap between genetic knowledge in IBD and its therapeutic potential by focusing on protective variants that both reveal the mechanisms of disease pathogenesis and suggest safe and effective therapeutic strategies.
In exome-sequencing studies, CARD9 variants were shown to have significant risk as well as protective associations with IBD (3, 5). CARD9 plays a key role in mediating innate immune signaling from C-type lectin receptors, such as Dectin-1 and Mincle, which are responsible for recognition of fungi and mycobacteria (13–17) in myeloid cells (18, 19). Ligand engagement and activation of these receptors induces recruitment of Syk kinase (20), leading to activation of PKCδ (21) and assembly of the CARD9–BCL10–MALT1 ternary complex (CBM signalosome). In the CBM signalosome, the CARD9 N-terminal CARD domain interacts with the CARD domain of BCL10 and triggers downstream IKK phosphorylation, subsequent IKα and IKβ degradation, followed by NF-κB translocation and transcriptional activation (15). NF-κB–dependent cytokine production, including such proinflammatory cytokines as TNFα and IL-6, culminates in the regulation of adaptive immune responses by promoting Th1 and Th17 polarization (22–27).
The protective variant CARD9Δ11, despite having an intact N-terminal sequence, lacks activity and exerts a dominant negative affect on CBM signaling. We previously reported the protective mechanism of CARD9Δ11 and discovered that the E3 ubiquitin ligase TRIM62 specifically interacts with WT CARD9 C-terminal domain (CTD) and activates CARD9 via K27 ubiquitinylation. In stark contrast, CARD9Δ11 is incapable of recruiting TRIM62 and exhibits impaired NF-κB activation (28). These findings suggest that small molecules causing CARD9 to lose its interaction with TRIM62 may mimic the protective actions of CARD9Δ11 in IBD. Here we use a bead-based system for reliable high-throughput detection of the CARD9–TRIM62 interaction in vitro, perform a high-throughput small-molecule screen for selective disruptors of the CARD9–TRIM62 protein–protein interaction (PPI) and identify small-molecule compounds that mimic the behavior of the protective variant CARD9Δ11 by inhibiting the TRIM62-mediated activation step in CBM assembly.
Results
A Highly Sensitive Bead-Based ELISA Reveals the Key Determinants of CARD9–TRIM62 Protein–Protein Interaction.
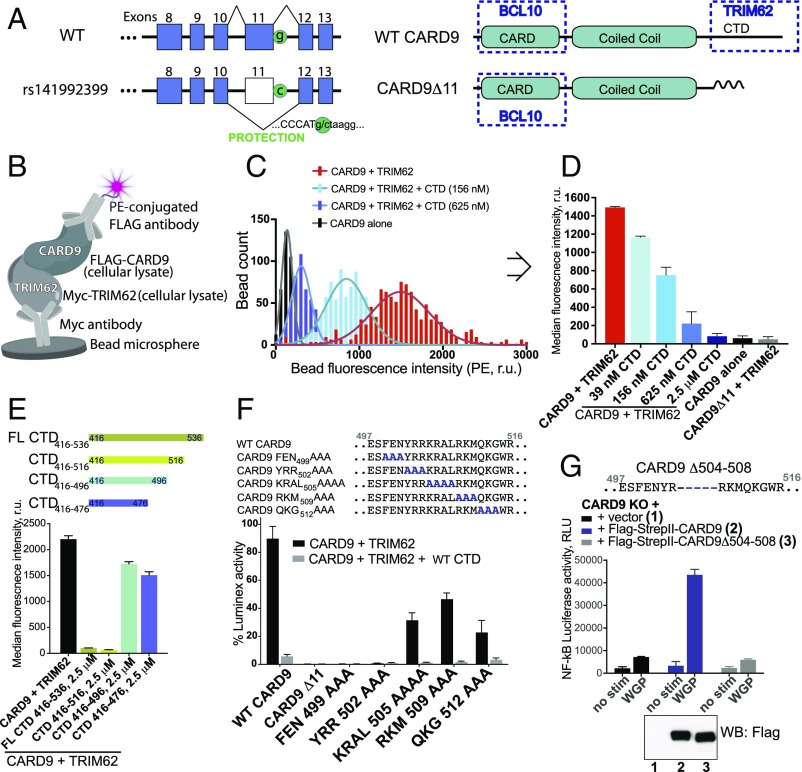
The rare protective variant CARD9Δ11 is a splice-site mutation and is predicted to result in a frameshift, premature termination, and truncation of the protein (3, 5) (Fig. 1A and Fig. S1A), which ablates TRIM62 binding and inhibits cytokine production in dendritic cells (28). Thus, we sought to define the CARD9 site of interaction required for TRIM62 binding. First, we developed a bead-based modified ELISA that uses color-coded magnetic beads (up to 500 different spectral sets of beads; Luminex technology). The beads are conjugated to anti-Myc antibodies for surface capture of Myc-tagged TRIM62 (Fig. 1B). Flag-tagged CARD9 is then added, allowing coassociation of both proteins, which is then measured by the intensity of the fluorescence signal of phycoerythrin (PE)-conjugated anti-Flag antibody associated with the bead. This bead-based format increased the signal window dramatically relative to traditional plate-based ELISA (Fig. S1B) and enabled sensitive detection of CARD9–TRIM62 PPI. Every bead in the experimental well is independently quantified by a flow-based technology, enabling a robust measure of median fluorescence intensity from multiple, aggregated measurements (Fig. 1 C and D).
Fig. 1.
A highly sensitive bead-based ELISA reveals the key determinants of CARD9–TRIM62 interaction. (A) The CARD9Δ11 protective variant encodes a G-to-C substitution resulting in disruption of a splice site and exon 11 skipping. WT CARD9 consists of CARD domain (interacts with BCL10), coiled coil domain, and C-terminal domain (CTD) (binds TRIM62 during CARD9 activation, disrupted in the protective variant). (B) Bead-based assay designed to detect CARD9–TRIM62 PPI in vitro. (C) Histogram of fluorescence intensity of each bead for CARD9 and TRIM62 (red), CARD9, and TRIM62 in the presence of CTD (156 nM, light blue; 625 nM, dark blue), or CARD9 alone in the absence of TRIM62 (black). (D) Median fluorescence intensity is taken from integrated signals of all individual beads in a well and provides a robust measure of CARD9–TRIM62 PPI disruption, as evidenced by a dose-dependent CTD competition. CARD9Δ11 is unable to bind TRIM62. (E) CARD9–TRIM62 disruption by four truncated CTD constructs (CTD416–536, CTD416–516, CTD416–496, or CTD416–476). (F) Mutational mapping of key CARD9 residues responsible for TRIM62 binding. (G) Human variant CARD9Δ504–508 precisely corresponds to the key TRIM62-interacting residues and results in disrupted CARD9 signaling, as measured by Dectin-1–triggered NF-κB–driven luciferase activity in THP-1 cells. Western blot confirms equivalent protein expression. RLU, relative light units; r.u., relative units. Data in Fig. 1 are mean ± SD for at least triplicates.
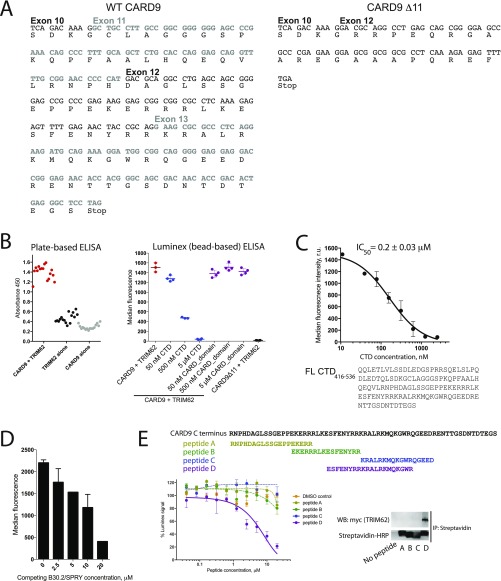
Fig. S1.
Development of a bead-based ELISA for quantifying CARD9–TRIM62 interaction. (A) DNA and protein sequences of WT CARD9 and CARD9Δ11. (B) Signal-to-noise comparison of a plate-based ELISA with bead-based readout. CARD9–TRIM62 interaction (red) can be inhibited by competition with purified CTD (5 μM, blue), but not by purified CARD domain (amino acids 2–98; 5 μM, purple). Data are expressed as median PE fluorescence per bead ± SD. (C) C-terminal B30.2/SPRY domain of TRIM62 competitively inhibits interaction between CARD9 and TRIM62, as measured by the bead-based ELISA. Data are expressed as median PE fluorescence per bead ± SD. (D) Short peptides corresponding to key TRIM62-interacting CARD9 residues, but not to other regions of CARD9 CTD, can competitively inhibit CARD9–TRIM62 interaction. Data are expressed as median PE fluorescence per bead ± SD. (E) Peptides inhibit CARD9–TRIM62 interaction by directly binding TRIM62, as evidenced by immunoprecipitation using biotinylated peptides and streptavidin pulldown from cellular lysate of HEK293T cells overexpressing myc-TRIM62. Functional inhibition data are expressed as the mean of percent Luminex signal (100% = median PE fluorescence per bead when no peptide inhibitor is present) ± SD.
To validate the specificity of the observed signal, we used recombinant purified CTD of CARD9 (amino acids 416–536), which successfully competed with formation of the TRIM62–CARD9 complex (Fig. 1 C and D), with an IC50 of 0.2 μM (Fig. S1C). As expected, the recombinant purified CARD domain of CARD9 did not affect CARD9–TRIM62 interaction (Fig. S1B). Importantly, the behavior of CARD9Δ11, incapable of engaging TRIM62 in PPI, is faithfully recapitulated by the complete abrogation of PE signal (Fig. 1D and Fig. S1B).
We used this assay system to interrogate the molecular determinants of the CARD9–TRIM62 interaction. TRIM62 contains an N-terminal B-box/RING domain, an intermediate coiled coil domain, and a C-terminal B30.2/SPRY domain. B30.2/SPRY domains are common protein–protein interaction domains in immune signaling pathways and are the substrate specificity determinants for a significant subset of TRIM E3 ligases (29). We therefore tested the effects of recombinant purified B30.2/SPRY domain of TRIM62 in a competition assay with full-length CARD9–TRIM62 binding. Indeed, purified recombinant B30.2/SPRY domain of TRIM62 competed with full-length TRIM62 for CARD9 binding (Fig. S1D), documenting the key involvement of B30.2/SPRY in CARD9 recognition.
CARD9 CTD does not possess conserved homology with known protein domain sequences and is predicted to possess a relatively high degree of disorder and a partially folded structure by modeling (PsiPred) (30). To identify the TRIM62-interacting region of CTD, we used recombinant purified CTD constructs with sequential C-terminal truncations [CTD416–536 (full length), CTD416–516, CTD416–496, or CTD416–476] and evaluated their ability to compete with full-length CARD9 for TRIM62 binding. Full-length CTD416–536 and CTD416–516 were potent, however further truncation (CTD416–496) dramatically decreased the potency of CTD as a competitor (Fig. 1E), suggesting that key binding interface residues are situated between amino acids 496 and 516 of CARD9. Among a panel of peptides representing different portions of the CTD, the 20-mer peptide corresponding to this sequence (497–516, amino acid sequence ESFENYRRKRALRKMQKGWR) was the only one capable of dose-dependent CARD9–TRIM62 PPI inhibition (Fig. S1E). This peptide directly binds TRIM62, as supported by the streptavidin bead affinity precipitation of biotinylated peptide and myc-TRIM62 in cellular lysates (Fig. S1E).
A corresponding alanine scan of this region in full-length CARD9 revealed the requirement of key residues (499–504) for mediating CARD9–TRIM62 interaction, as their alanine substitutions abrogated binding (Fig. 1F). We recently identified a naturally occurring human CARD9 variant in a pooled human monocyte cDNA library, CARD9Δ504–508, that has a deletion in precisely this region (28) (Fig. 1G). Indeed, when reconstituted in CARD9 KO THP-1 monocytes, CARD9Δ504–508 was unable to mediate signal transduction from Dectin-1 activation by whole glucan particles (WGPs). In these experiments, CARD9Δ504–508 failed to activate a reporter gene comprising the NF-κB response element driving luciferase expression (Fig. 1G). Taken together, we identified critical amino acids lacking in CARD9Δ11 that are required for TRIM62 binding. Having a greater understanding of the CARD9–TRIM62 complex, we next pursued identifying inhibitors of this interaction.
A Multiplexed High-Throughput Screen Identifies Inhibitors of CARD9–TRIM62 Protein–Protein Interaction.
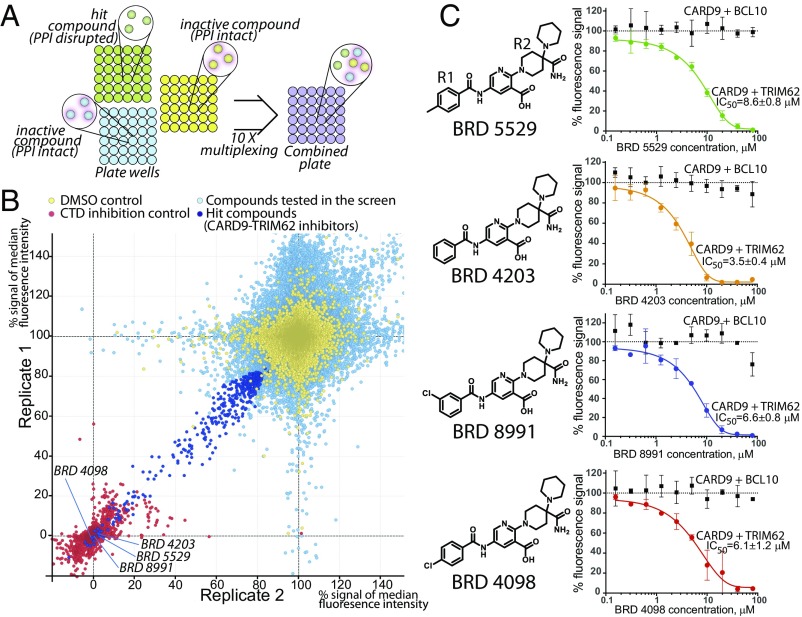
To increase the throughput of the bead-based ELISA, we introduced a multiplexing step, taking advantage of the Luminex technology that includes multiple color-coded beads that distinguishes the color-coded beads by their spectral signature during the flow-based readout. Each PPI reaction is arrayed in a 384-well format such that each of 10 plates contains a distinct color-coded bead set. Compound treatments are coded by well position, and plate identity is coded by bead fluorescence. Each compound treatment can be individually quantified after pooling multiple plates for detection and readout steps (Fig. 2A). Importantly, each bead behaves individually, and PPIs do not reequilibrate between individual beads when mixed. Titration of CARD9 C-terminal domain (positive control) matched precisely when measured in independent bead sets that were pooled after CTD exposure (Fig. S2A). This unique advantage enables measurement of multiple small-molecules’ effects independently in one well while avoiding the pitfalls of pooled small-molecule screens such as high total small-molecule concentration and need for pool deconvolution, while also shortening the readout times by a factor of >10.
Fig. 2.
A high-throughput screen identifies selective CARD9–TRIM62 PPI inhibitors. (A) Color-coded beads are plated and incubated with compounds in individual wells in the presence of CARD9 and TRIM62. After a wash step, beads are then combined for antibody staining and detection steps. (B) Replicate 1 vs. replicate 2 plot for all of the compounds (132,813) screened in duplicate in the CARD9–TRIM62 disruption assay. Yellow, neutral controls (DMSO); red, positive control (purified CTD, 5 μM); light blue, screened compounds (20 μM); dark blue, hits selected for followup investigation. Axis units are median PE fluorescence per bead within sample, normalized with respect to CTD positive control (0%) and DMSO negative control (100%). (C) Structural family of hit compounds are potent dose-dependent inhibitors in the CARD9–TRIM62 PPI assay (colored lines and data points) and inactive in the CARD9–BCL10 PPI assay (black lines and data points). Data are mean of percent Luminex signal (100% = median PE fluorescence per bead with DMSO control) ± SD of at least triplicates.
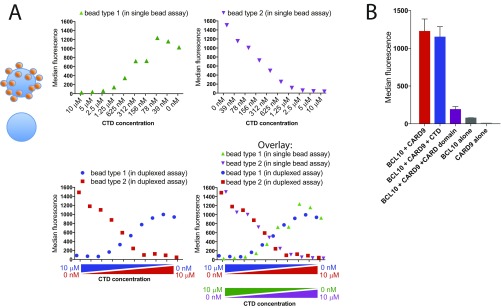
Fig. S2.
Multiplexing of bead-based ELISA for high-throughput screening. (A) To confirm that there is no reequilibration of the TRIM62-bound CARD9 between the different beads derived from different plates, single bead assays (Top two panels) were compared with the duplexed assay composed of two bead types/color codes (Bottom Left). The ability of CTD to disrupt CARD9–TRIM62 fully overlapped in duplexed versus single bead assays, confirming the independence of CARD9–TRIM62 interactions on each bead during simultaneous readout. Data are expressed as median PE fluorescence per bead. (B) CARD9–BCL10 assay was developed in which Myc-BCL10 produced in HEK293T lysates was immobilized on anti–myc-conjugated beads for binding to Flag-CARD9. The BCL10–CARD9 signal was present only when both lysates were present (red bar), and not in the case of either protein alone (dark gray and light gray bard). BCL10–CARD9 complexes were quantified with phycoerythrin (PE)-labeled anti-Flag antibodies and expressed as median PE fluorescence per bead (y axis). BCL10–CARD9 interaction was inhibited by the presence of purified CARD domain (amino acids 2–98; 5 μM, purple), but not by purified CTD (amino acids 416–536; 5 μM, blue). Data are expressed as median PE fluorescence per bead ± SD.
We collected a set of 132,813 small molecules from commercial vendors and diversity-oriented synthesis (DOS) (31) performed at the Broad Institute. Compounds were screened in duplicate at 20-μM concentration (Fig. 2B, light blue data points), using DMSO as a neutral control (yellow data points) and purified recombinant CTD of CARD9 as a positive control (red data points). From the top-ranked hits of the primary screen (dark blue data points, 449 hits, 0.3% hit rate), a subset of 228 was available for resupply. We verified the purity of all obtained compounds by liquid chromatography (LC)-MS analysis. Upon retest, a family of four structurally related compounds emerged as effective dose-dependent CARD9–TRIM62 PPI inhibitors: BRD5529, 8.6 μM; BRD4203, 3.5 μM; BRD8991, 6.6 μM; and BRD4098, 6.1 μM (Fig. 2C), all of which compare favorably with CTD competitive inhibition. The molecular structures of this series of compounds comprised a 2,5-diaminonicotinic acid core with various benzoyl (R1) and aliphatic quaternary carbon-containing (R2) groups (Fig. 2C).
To demonstrate compound specificity, we developed a CARD9–BCL10 bead-based ELISA, analogous to that used for CARD9–TRIM62 detection, by using Myc-BCL10 and Flag-CARD9. BCL10 interaction with CARD9 occurs via binding of their respective N-terminal CARD domains, and as such should not be influenced by inhibitors of the C-terminal interactions between CARD9 and TRIM62. As expected, purified CARD domain of CARD9 competitively disrupted the signal in the BCL10–CARD9 bead-based ELISA, while CARD9 CTD had no effect (Fig. S2B). Small-molecule CARD9–TRIM62 inhibitors did not interfere with CARD9–BCL10 interaction at up to 80 μM of the tested dose (Fig. 2C, black lines).
Small-Molecule Screening Hits Directly Bind CARD9.
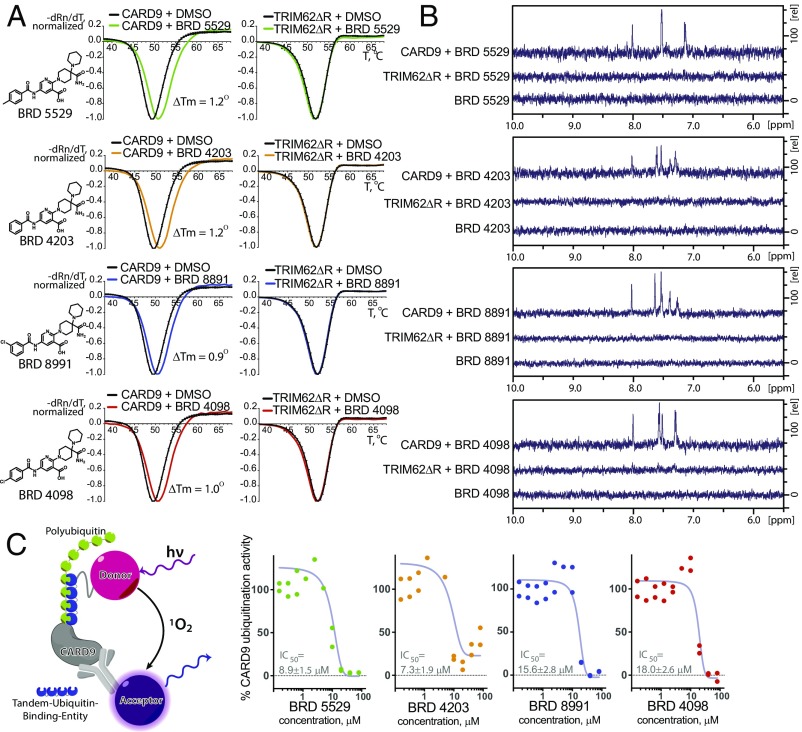
We next investigated whether the hit compounds directly bind CARD9 or TRIM62, or score as positives by indirect means, by biophysical binding experiments. Differential scanning fluorimetry (DSF) revealed a right shift of the purified CARD9 protein melting temperature (Tm) in the presence of all four small-molecule inhibitors identified above (Fig. 3A), suggesting the compounds directly bind and stabilize the protein. Moreover, BRD5529 did not produce a detectable Tm shift when incubated with CARD9Δ11 in DSF (Fig. S3A), suggesting that C terminus of CARD9, responsible for TRIM62 interaction, also engages in the compound binding. No detectable Tm shift was observed for compounds incubated with purified TRIM62ΔRING (full-length TRIM62 protein was refractory to high-yield expression required for biophysical assays). We verified these findings in an orthogonal nonfluorimetric and label-free assay by performing saturation transfer difference NMR (STD NMR) experiments (32–35). This technique requires direct association of a protein and small molecule to observe the small molecule’s NMRs. For all four compounds in the series, addition of CARD9–TRIM62 inhibitors (200 μM) to purified CARD9 protein (5 μM) induced significant STD signals (Fig. 3B), whereas the STD signals were not detectable in the presence of TRIM62ΔRING (5 μM) under identical conditions. The compounds did not exhibit “self” STD signals in the absence of protein, which suggests against the possibility of false positives due to nonspecific aggregation. Collectively, these data are consistent with a direct, TRIM62-mutually exclusive interaction between compounds in the series and CARD9.
Fig. 3.
CARD9–TRIM62 PPI inhibitors directly bind CARD9, but not TRIM62, and disrupt its ubiquitinylation in vitro. (A) Differential scanning fluorimetry (DSF) profiles of CARD9 and TRIM62ΔRING (labeled as TRIM62ΔR) in the presence or absence of the hit compounds. DMSO was used as neutral control. x axis, temperature, degrees Celsius; y axis, −dRn/dT (negative first derivative of fluorescence intensity, normalized). Data represent the mean ± SD of triplicates. (B) STD NMR spectra for mixtures of CARD9 with compound, TRIM62ΔRING with compound, and compound alone. x axis, chemical shift change [Δ] ppm (parts per million); y axis, relative signal intensity. (C) An alphaLISA assay was designed to detect polyubiquitinylation of CARD9 in the presence of reconstituted E1–E2–E3 TRIM62 complex. Data represent percent AlphaLISA signal (100% = mean AlphaLISA signal with DMSO control).
Fig. S3.
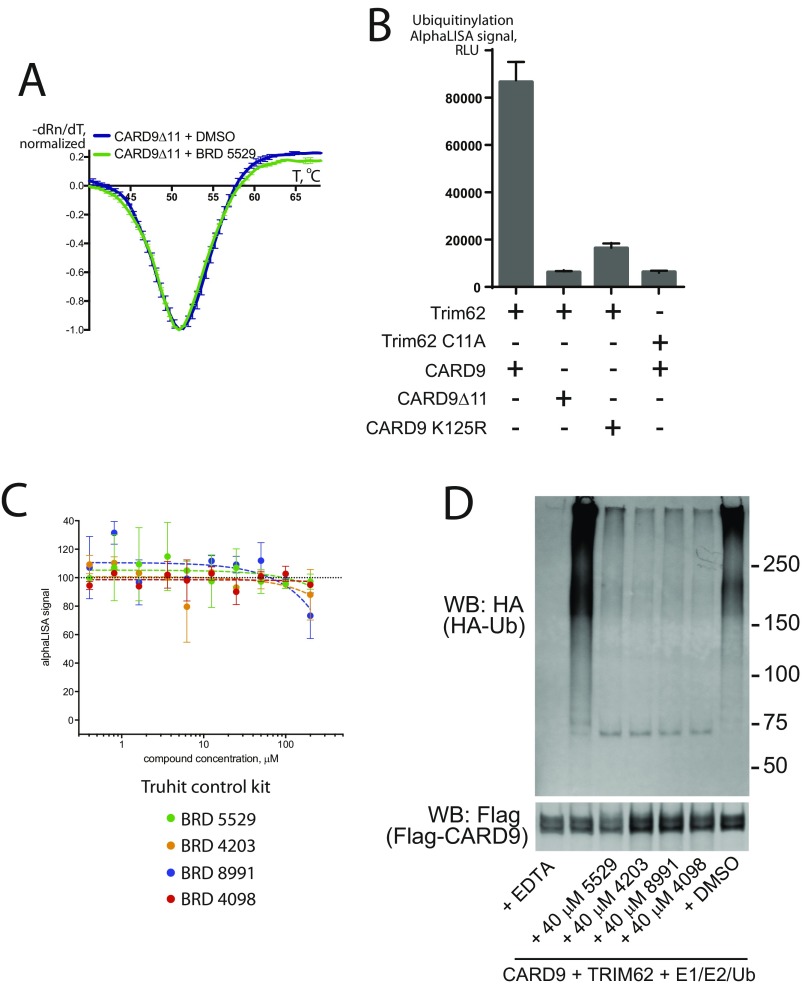
Development of a functional assay for CARD9 ubiquitinylation by TRIM62. (A and B) AlphaLISA assay detects TRIM62-mediated CARD9 ubiquitinylation in the presence of WT TRIM62 and WT CARD9. Ubiquitinylation of CARD9 was significantly decreased with TRIM62 C11A (ligase-dead mutant), CARD9Δ11, or CARD9 K125R lacking a key lysine for TRIM62-mediated ubiquitinylation. Data are expressed as mean AlphaLISA signal ± SD. (C) The family of hit compounds tested in the Truhit AlphaLISA assay designed for detection of nonspecific AlphaLISA inhibitors. Data are expressed as mean of percent AlphaLISA signal (100% = mean AlphaLISA signal with DMSO control) ± SD. (D) Western blotting detects the inhibition of in vitro CARD9 ubiquitinylation reaction by the family of the hit compounds. Results are representative of three independent experiments.
CARD9-Targeting Molecules Inhibit Functional TRIM62-Mediated CARD9 Ubiquitinylation in Vitro.
The functional consequence of TRIM62 association with CARD9, upon ligand recognition by C-type lectin receptors, is the engagement of the E1 and E2 ubiquitinylation machinery and subsequent CARD9 ubiquitinylation (28). To determine whether disruption of the CARD9–TRIM62 interaction via direct binding to CARD9 protein results in a functional response, we reconstituted the E1, E2, and E3 (UBE1–UBE2D2–TRIM62) (36, 37) complex in vitro. To detect and quantify CARD9 ubiquitinylation in plate-based format, we used AlphaLISA donor and acceptor beads. Successful polyubiquitinylation allows affinity reagents to bring into proximity donor beads (affinity for polyubiquitin) and acceptor beads (affinity to Flag-tagged CARD9). This proximity produces luminescence proportional to the extent of CARD9 ubiquitinylation (Fig. 3C). We confirmed that the signal is specific to the conditions when WT CARD9 and WT TRIM62 are both present in the reaction and is not observed when catalytically dead TRIM62 C11A was substituted for WT TRIM62 (Fig. S3B). Accordingly, CARD9Δ11 or CARD9K125R (with TRIM62-targeted lysine 125 mutated to arginine) were inactive as well (Fig. S3B). Using this system, we observed that compounds in the series produce dose-dependent inhibition of TRIM62-mediated CARD9 ubiquitinylation in vitro (Fig. 3C, BRD5529, 8.9 μM; BRD4203, 7.3 μM; BRD8991, 15.6 μM; and BRD4098, 18.0 μM), consistent with the effects on CARD9–TRIM62 PPI disruption. Importantly, nonspecific inhibition of AlphaLISA signal was not observed, as assessed by the AlphaLISA Truhit control kit (Fig. S3C). To additionally confirm the activity of compounds, we performed TRIM62-mediated ubiquitinylation assay coupled with Western blot detection (Fig. S3D), which corroborated our alphaLISA findings.
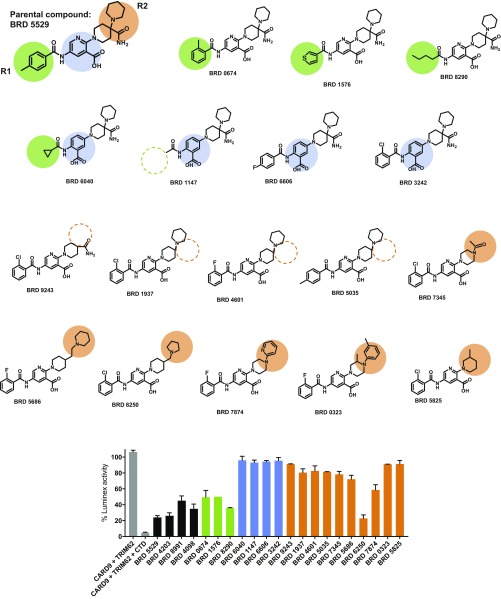
To understand the activity determinants of the small-molecule hits, we performed a round of structure–activity relationship (SAR) studies (Fig. S4). In the primary series, we observed that CARD9-targeting compounds tolerate substitutions in the aromatic ring at −R1 (phenyl in BRD4203; p-toluyl in BRD5529; m-chloro-phenyl in BRD8991; and p-chloro-phenyl in BRD4098). Consistent with these findings, examination of an expanded series of R1 group analogs (green circles) revealed that changes in the R1 group mostly maintained activity (green bars in Fig. S4 compared with black bars for the parental series). We also determined that modification of the 2,5-aminonicotinic acid core was not tolerated and analogs with an altered core lost activity completely (blue circles and blue bars, Fig. S4). Substitutions in the aliphatic part (R2, orange circles and orange bars, Fig. S4), including the quaternary carbon center, differentially affected competitive CARD9 binding and may be amenable for future optimizations.
Fig. S4.
Structure–activity relationship in the family of hit compounds as measured by CARD9–TRIM62 interaction assay. Tested structures are depicted, along with color codes highlighting the differences between the parental and the SAR compounds. Gray, CARD9–TRIM62 signal and the CTD inhibitor control. Black, parental compounds BRD5529, BRD4203, BRD8991m and BRD4098. Green, substitutions in the (−R1) portion of the structure, which appear to be well tolerated. Blue, substitutions in the pyridinecarboxylic acid core of the structure, which are detrimental to the compounds’ activity. Orange, substitutions in the aliphatic region of the structure, which result in variable activity, and are likely possible to develop further. Functional inhibition data are expressed as the mean of percent Luminex signal (100% = median PE fluorescence with DMSO control) ± SD.
BRD5529 Inhibits CARD9-Dependent Signaling in Innate Immune Cells.
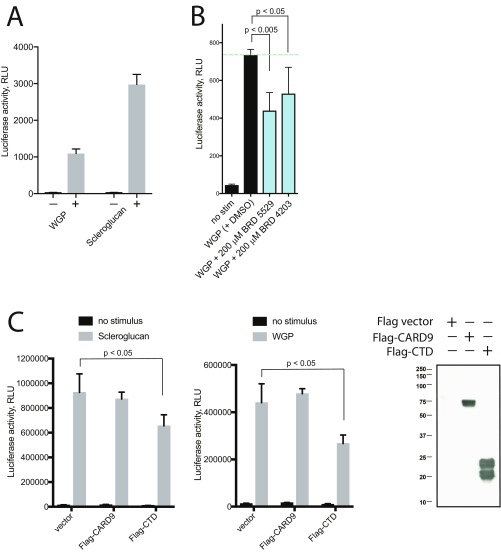
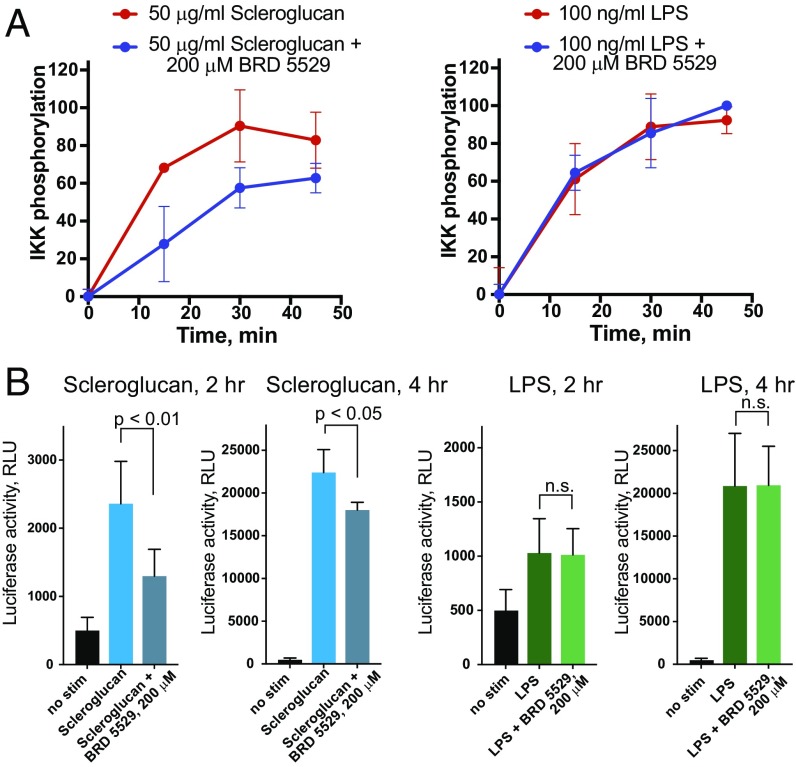
We next investigated whether the CARD9-binding compounds could attenuate CARD9-dependent signaling in a cellular context. We selected BRD5529 based on its potency and complete inhibition of CARD9 ubiquitinylation in vitro, as well as for its favorable solubility. We used primary bone marrow-derived dendritic cells (BMDCs) and assessed phosphorylation of IKK, the immediate downstream event of CARD9 signalosome formation (15). We employed stimulation of Dectin-1 by scleroglucan, a highly specific ligand, more potent than other Dectin-1 ligands, such as depleted zymosan whole glucan particles (WGPs) (Fig. S5A), curdlan, and schyzophyllan, which enabled a sufficient dynamic range to detect the IKK phosphorylation. Upon stimulation of BMDCs with scleroglucan (Dectin-1 ligand), we observed a time-dependent increase in IKK phosphorylation. When treated with BRD5529, our selected lead compound, IKK phosphorylation was significantly reduced in the context of Dectin-1 activation (scleroglucan treatment), but was not affected upon LPS-triggered CARD9-independent TLR4 activation (Fig. 4A). These findings suggest that BRD5529 functions in a CARD9-specific manner. We further conducted a series of assays that probe transcriptional effects of CARD9 signaling downstream of IKK activation by using THP-1 monocytes transduced with an NF-κB–driven luciferase reporter. Notably, BRD5529 attenuated activation of the NF-κB reporter in THP-1 monocytes treated with scleroglucan (Fig. 4B) or WGPs (Fig. S5B) but had no effect on LPS-driven CARD9-independent NF-κB activation (Fig. 4B).
Fig. S5.
WGP (whole glucan particle, dispersible) Dectin-1–mediated signaling to NF-κB is inhibited by the CARD9-targeting compounds. (A) Scleroglucan is a more potent ligand than WGP that activates Dectin-1–mediated signaling. NF-κB activation in THP-1 cells reconstituted with Dectin-1 was measured by NF-κB–driven luciferase reporter in the presence or absence of 50 μg/mL WGP or scleroglucan. (B) Cells were stimulated with WGP (50 μg/mL) in the presence or absence of 200 μM BRD5529 or BRD4203. At the 4-h time point, luciferase activity was quantified. RLU, relative light units. For A and B, data represent the mean ± SD of at least six replicates and are representative of two independent experiments. P values were calculated by Student’s t test. (C) Inhibition of Dectin-1–mediated (scleroglucan or WGP stimulated) NF-κB luciferase reporter inhibition by heterologous expression of CTD of CARD9 in wild-type THP-1 cells. Flag-vector plasmid or Flag-CARD9 were used as negative controls, and CTD expression was confirmed by anti-Flag Western blot. Data represent mean ± SD of three replicates and are representative of two independent experiments.
Fig. 4.
BRD5529 attenuates CARD9 signaling in a stimulus-dependent manner. (A) Phosphorylation of IKK in primary BMDCs was assessed by flow cytometry. Cells were treated with the indicated stimuli (Dectin-1, scleroglucan; TLR4 control, LPS) in the presence or absence of 200 μM BRD5529. y axis, percent IKK phosphorylation compared with maximum phospho-IKK within experimental series. Data are represented as mean of at least triplicate ± SD. (B) NF-κB activation in THP-1 cells reconstituted with Dectin-1 and NF-κB–driven luciferase reporter. Cells were stimulated with scleroglucan (Left, blue bars) or LPS (Right, green bars) ± 200 μM BRD5529 for 2 h or 4 h. RLU, relative light units. Data are mean ± SD of at least quadruplicates. P values were calculated by Student’s t test. n.s., nonsignificant.
Taken together, we propose that the discovered “BRD” small molecules directly bind CARD9 in vitro and inhibit its association with TRIM62, while also inhibiting functional ubiquitinylation and activation of CARD9. When applied to innate immune cells, CARD9-targeting molecule BRD5529 selectively inhibits Dectin-1–mediated CARD9 activation, while leaving LPS-activated signaling intact, supporting the concept of targeting CARD9 with small molecules to attenuate its activity in proinflammatory settings.
Discussion
IBD includes Crohn’s disease (CD) and ulcerative colitis (UC), two chronic inflammatory conditions associated with overly aggressive immune responses to otherwise commensal bacteria in the gut. IBD affects ∼2 million people in the United States (38), and there is urgent need for effective and targeted treatments. Immunosuppressants and biological agents are often the best options; however, therapy is limited by loss of efficacy, underscoring the unmet clinical need for further therapeutic development. In the present work, human genetic data generated by exome sequencing, biochemical analyses of variant proteins, and cellular functional genomics inspired a precision approach to therapeutic intervention.
IBD is a classic example of a multifactorial genetic disease, which is provoked by a combination of disparate factors (e.g., environmental exposure and diet) including a significant genetic component (38). In such cases, genome-wide association studies are among the most powerful tools for deciphering the genetic basis of disease risk and related pathological mechanisms. Among IBD risk genes, CARD9 is unique in that both risk and protective variants have been identified. A common risk allele (40% of the population) encodes an amino acid substitution S12N in the CARD9 protein that is associated with heightened CARD9 activation, although the mechanistic basis for this phenotype remains to be defined conclusively (1, 4). The rare protective variant CARD9Δ11 is a splice site mutation that results in premature CARD9 truncation (Fig. 1 A and B) and loss of key C-terminal functions, despite having an intact N terminus involved in the canonical CARD9–BCL10–MALT1 signalosome interactions. Even with the risk allele also present, this truncation of CARD9 strongly protects against IBD [odds ratio (OR) = 0.29, P < 10−16]. These findings suggest that decreased CARD9 signaling may be beneficial in IBD treatment; however, persons completely deficient in CARD9 function via missense or early stop codon mutations are immunocompromised and susceptible to life-threatening fungal infections (22, 39–43). Thus, we hypothesized that the safe and effective targeting approach may be to mimic precisely the protective CARD9Δ11 mechanism rather than completely ablating CARD9 activity.
We have previously characterized the protective CARD9Δ11 variant and its interaction with TRIM62, and in this current work, aimed to develop small molecules that mimic the phenotype of CARD9Δ11. Such CARD9 domain-specific small-molecule inhibitors must target and disrupt CARD9–TRIM62 interaction, which occurs in neutral and/or risk alleles of CARD9, but is disrupted in CARD9Δ11. Protein–protein interactions, despite the fact that they are frequently impacted by genetic variants associated with disease (44), are challenging therapeutic targets and have been classified as “undruggable” (45). Moreover, the CARD9 C-terminal domain likely possesses an only partially folded structure, as predicted by structural modeling, increasing the challenge of finding molecules that directly bind this domain of CARD9.
To address the challenge of measuring PPI disruption by compounds in a high-throughput format, we developed a multiplexed method with a bead-based readout and simple relative quantification. We applied this technology to enable a high-throughput PPI disruption screen that yielded small molecules that disrupt CARD9–TRIM62 interactions in a manner that mimics the CARD9 protective variant. We report the discovery of structurally related compounds that specifically and directly bind CARD9, potently inhibit its association with TRIM62, but not with the N-terminal binding partner BCL10, and effectively inhibit functional TRIM62-mediated CARD9 ubiquitination in cells. We further show that the lead compound in this series selectively modulates CARD9-dependent signaling in the context of innate immune activation both in a monocytic cell line THP-1 and primary immune cells (BMDCs), thus recapitulating the behavior of CARD9Δ11 and providing an example of an approach of precision immunomodulatory intervention rather than broad immunosuppression. As the selective effect appeared moderate in magnitude, we employed expression of CTD of CARD9 as an estimate of the maximum possible effect of CARD9–TRIM62 inhibition and obtained a similar decrease in NF-κB activity (Fig. S5C), with the caveat that the CTD (which binds TRIM62) slightly differs in its mechanism of action from CARD9-targeting compounds.
The present work continues a thread from genetics (5), through functional biology (28) to therapeutic discovery. By using a human genetics-informed strategy and combining it with a state-of-the-art screening technology, we develop small-molecule probes that directly engage a target lacking enzymatic activity—a common goal of many therapeutic projects guided by human genetics.
Materials and Methods
Flag-StrepII-CARD9 and TRIM62 were purified as described (28). TRIM62ΔRING and B30.2/SPRY were expressed in Sf9 cells in pFastBac as His8 fusion. For Luminex assay, HEK293F cells were transfected with 3XMyc-TRIM62 in pCMV and separately with Flag-CARD9 in pcDNA4/TO. Anti-Myc tag antibody (9E10) was conjugated to the beads and used for TRIM62 affinity capture from cleared lysates. Following the incubation with compounds, Flag-CARD9 lysate was added; after a series of washes and secondary PE-conjugated antibody incubation, readout was performed on MagPlex instruments. Extended experimental details on Luminex bead-based ELISA, screening libraries, DSF and STD NMR assays, ubiquitinylation assays, cell culture, phospho-IKK FACS, CARD9 rescue in THP-1 cells, and NF-κB assay are provided in Supporting Information.
SI Materials and Methods
Protein Constructs and Purification.
Full-length Flag-StrepII-CARD9 for AlphaLISA, DSF, and NMR assay was expressed as previously described (28). Full-length TRIM62 for AlphaLISA assay was expressed as previously described (28). TRIM62ΔRING was expressed in Sf9 cells in pFastBac construct as His8 fusion, and purified on Ni affinity column followed by size exclusion chromatography. B30.2/SPRY domain of TRIM62 was expressed as His8–MBP fusion in Sf9 cells and purified on Ni affinity column followed by size exclusion chromatography. Full-length CTD416–536 and truncated constructs CTD416–516, CTD416–496, and CTD416–476 were expressed as previously described (28). The CARD domain was expressed as His8 fusion in Escherichia coli [BL21(DE3)] and purified on Ni affinity column followed by size exclusion chromatography.
Luminex Bead-Based ELISA.
HEK293F (R790-07, Life Technologies) cells were grown in Freestyle media (cat. no. 12338) according to manufacturer’s instructions. Cells were transiently transfected with 3XMyc-TRIM62 in pCMV and separately with Flag-CARD9 in pcDNA4/TO. Anti-Myc tag antibody (9E10, Sigma) was conjugated to the Luminex beads (10 spectral range beads in 10 separate reactions) using Pierce reagents/Luminex coupling kit. Flag-CARD9 and Myc-TRIM62 transfected cells were separately lysed in the lysis buffer [50 mM Tris pH 7.4, 150 mM NaCl, 0.5% Nonidet P-40, protease inhibitors (Complete, 11873580001, Roche), phosphatase inhibitors (PhosStop, 4906845001, Roche)] and cleared by centrifugation at 10,000 × g. Cleared TRIM62 lysates were added to the beads for Myc-mediated capture, incubated at room temperature for 1 h, then washed once and reconstituted in 1% BSA in PBS.
Screening compounds were predispensed into assay-ready plates (Twin.tec, Eppendorf; 50 nL of 10 mM compound stock in DMSO per well). Cleared CARD9 lysate (20 μL per well) was added to the assay-ready plate and preincubated with the screening compounds for 30 min before addition of TRIM62-bound beads (300 beads per well, in 5 μL of 1% BSA in PBS). Plates were mixed on high-speed microplate thermomixer Bioshake IQ (Quantifoil Instruments) at 2,400 rpm for 2 min and incubated for 40 min at room temperature. Plates were then washed in a Biomek NXP instrument with magnetic plate holders once with 1% BSA in PBS. Each of the 10 plates with nonidentical spectral regions of beads were combined into one deep-well plate, and incubated with Flag-PE antibody conjugate (ab72469, Abcam) at a final dilution of 1:400 for 1 h. After one wash with 1% BSA in PBS, the readout was performed on Luminex MagPlex instruments with standard operating protocol. Bead quantity per well was employed as quality-control measure; wells with bead count less than 20 were rejected from the analysis.
Screening Libraries and Compounds.
The screening library was assembled from compounds derived from diversity-oriented synthesis and purchased from vendors (autophagy-focused library from ChemBridge; 14,000 compound informer set from Maybridge; autophagy-focused library from ChemDiv; 10,000 compound informer set from WuXi; compound collection from Molecular Libraried Program).
Differential Scanning Fluorimetry.
DSF assay was performed on a Roche LightCycler 480 II with ramp speed of 4.8 °C per min and 12 acquisitions per °C. Experimental buffer was 50 mM Tris, pH 7.5, 150 mM NaCl, 0.5 mM Tris(2-carboxyethyl)phosphine hydrochloride (reducing agent). Freshly purified or flash frozen in 10% glycerol, protein was dialyzed overnight into experimental buffer. Purified Flag-StrepII-CARD9 (2.5 μM) or TRIM62ΔRING (2.5 μM) were mixed with the corresponding small molecule (100 μM) and SYPRO dye (1:1,000 final dilution; Life Technologies) in 384-well polypropylene PCR plates (10 μL per well). As part of the assay optimization, lower concentrations of protein (0.8–2.5 μM) and higher concentrations of the compounds (100–400 μM) were all successfully tested. The plates were sealed and heated in the instrument across a temperature range of 25 °C–95 °C. The minimum of the negative first derivative curve was used to estimate the protein melting temperature (Tm).
STD NMR.
STD experiments were performed using CARD9 and TRIM62ΔRING purified by size exclusion on a S200 column in a completely deuterated buffer, since CARD9 precipitation occurs during dialysis or concentration. Proteins were diluted to 5 μM and compounds were at 200 μM with 2% d6-DMSO. Experiments were performed on a Bruker spectrometer equipped with a cryoprobe and operating at a proton frequency of 600 MHz. Data were collected at 280 K in interleaved mode as described (35) with 16 scans, 14 ppm sweep width, 2.2-s saturation at a power level of 6.78 W, and offset of −0.25 ppm.
AlphaLISA and Western Blot Ubiquitinylation Assay.
FlagStrep-CARD9 and TRIM62 were expressed and purified as previously described (28). Assay-ready plates containing 80 nL of 10 mM compound in DMSO were used. Ubiquitinylation reactions contained 12.5 nM Flag-Strep-CARD9, 1.25 nM TRIM62, 500 nM ubiquitin, 10 nM UBE1 (LifeSensors), 200 nM UBE2D2 (LifeSensors), 50 mM Tris pH = 7.5, 5 mM MgCl2, 2.5 mM ATP, 1 mM DTT, 0.005% Tween 20 in a final volume of 10 µL. To assemble reactions, all reaction components except ubiquitin and TRIM62 were combined and dispensed at a 2× concentration in a volume of 5 µL using a Multi-Drop Combi Reagent Dispenser. Reactions were initiated by adding ubiquitin and TRIM62 at a 2× concentration in a volume of 5 µL, in the same manner. The reactions were incubated at ambient temperature for 90 min then stopped by the addition of 10 µL of 2× quench/detect solution using a 16-ViaFlow dispenser. The 2× quench/detect solution contained 100 mM Hepes pH = 7.5, 300 mM NaCl, 0.01% Tween 20, 30 mM ETDA, 40 µg/mL streptavidin-donor beads, 10 µg/mL α-Flag-acceptor beads (both from PerkinElmer) and 100 nM biotin-TUBE1 (LifeSensors). Plates were incubated at room temperature for an additional 1 h, then read on a PerkinElmer Envision.
For Western blot detection, ubiquitinylation reactions contained 250 nM Flag-Strep-CARD9, 5 nM TRIM62, 5 µM of HA-tagged ubiquitin (Boston Biochem), 25 nM UBE1 (LifeSensors), 300 nM UBE2D2 (LifeSensors), 50 mM Tris pH = 7.5, 5 mM MgCl2, 2.5 mM ATP, 1 mM DTT in a final volume of 20 µL. Reactions were quenched by the addition of the LDS sample buffer with 50 mM DTT. HA antibody F-7 (Santa Cruz) was used for the detection of polyubiquitin chains, and Flag M2 antibody from Sigma was used to detect Flag-StrepII-CARD9 as a loading control.
Cell Culture and Primary Cell Isolation.
THP-1 and HEK293T cells were obtained from ATCC and cultured according to ATCC instructions. Cell cultures were regularly checked for mycoplasma contamination using mycoplasma PCR test. BMDCs were isolated from femur and tibia of C57B/6 mice by harvesting bone marrow, followed by hypotonic red blood cell lysis. Obtained bone marrow cells were cultured in DMEM (10% FBS, 1% penicillin/streptomycin, 1% glutamine) with 25 ng/mL GM-CSF.
Phospho-IKK FACS.
For phospho-IKK FACS analysis, the BMDCs were plated on day 7 in U-bottom 96-well plates at 150,000 cells per well in DMEM, 10% FBS, 1% penicillin/streptomycin, glutamine, and GM-CSF the day before the assay. Growth media was replaced with reduced serum media (2% FBS), BRD5529 compound was added to the appropriate wells (at 200 μM), and cells were incubated for 2 h. Cells were stimulated with 50 μg/mL scleroglucan (Invivogen) or 100 ng/mL LPS (Sigma) for the indicated times at 37 °C. At the end of the incubation, media were aspirated and cells were fixed in 4% PFA for 10 min at 37 °C, then resuspended in blocking buffer (3% BSA in permeabilization buffer; eBioscience). Cells were harvested by scraping with pipette tips, spun at 350 × g for 3 min, resuspended in staining mixture [blocking buffer + rat anti-IA/IE APC 1:1,000 (BD Biosciences), rabbit anti phospho-IKK PE 1:50 (Cell Signaling Technology)] and stained at room temperature in the dark for 60 min. Cells were washed once in blocking buffer and resuspended in FACS buffer (PBS, 2% FBS). FACS experiments were run on a Cytoflex instrument (Beckman Coulter) and analyzed using FlowJo software by sequentially gating on live/single cells/MHCII high-cell population.
CARD9 Rescue in THP-1 Cell Lines and NF-κB Reporter Assay.
CARD9 KO THP-1 cells were generated by a CRIPSR-based approach as previously described (28). For the NF-κB luciferase assay, CARD9 KO or WT THP-1 cell lines were transduced with Dectin-1 (NM_022570) and a lentivirus-based NF-κB luciferase reporter, containing NF-κB responsive element upstream of minimal promoter and Luc2P (46). For experiments described in Fig. 1G, CARD9 KO cells overexpressing Dectin-1 and the NF-κB luciferase reporter were rescued with the indicated CARD9 constructs (CARD9 WT or CARD9Δ504–508). For compound treatment (Fig. 4B and Fig. S3), BRD5529 (at 200 μM) was preincubated with Dectin-1, NF-κB reporter-expressing THP-1 cells for 2 h, followed by stimulation with scleroglucan (50 μg/mL), whole glucan particles (WGP Dispersible, 50 μg/mL; Invivogen) or LPS (100 ng/mL). Reporter luciferase activity was measured using Steadylite Plus reagent (PerkinElmer).
Supplementary Material
Acknowledgments
We thank John Davis and David Peck for their help in establishing Luminex high-throughput assay and readout, Wenting Xu for help with THP-1 reconstitution experiments, and the Broad Institute Connectivity Map platform. This research was funded by the Leona M. and Harry B. Helmsley Charitable Trust and NIH Grants GM-038627 (to S.L.S.) and DK043351 (to R.J.X.). S.L.S. is an Investigator at the Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: S.L.S. and B.F.C. were coauthors on a 2015 review article. S.L.S. and K.M.S. were coauthors on a 2015 research paper. B.F.C. and K.M.S. independently supplied reagents to the same laboratory. There was no active collaboration.
Data deposition: Compound high-throughput data and results are deposited in PubChem, https://pubchem.ncbi.nlm.nih.gov/ (assay IDs 1259337–1259342).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705748114/-/DCSupplemental.
References
- 1.Jostins L, et al. International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaudoin M, et al. Quebec IBD Genetics Consortium; NIDDK IBD Genetics Consortium; International IBD Genetics Consortium Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 2013;9:e1003723. doi: 10.1371/journal.pgen.1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivas MA, et al. National Institute of Diabetes and Digestive Kidney Diseases Inflammatory Bowel Disease Genetics Consortium (NIDDK IBDGC); United Kingdom Inflammatory Bowel Disease Genetics Consortium; International Inflammatory Bowel Disease Genetics Consortium Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okada Y, et al. RACI consortium; GARNET consortium Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilliam BL, Riedel DJ, Redfield RR. Clinical use of CCR5 inhibitors in HIV and beyond. J Transl Med. 2011;9(Suppl 1):S9. doi: 10.1186/1479-5876-9-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abifadel M, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 10.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 11.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkes M, et al. Wellcome Trust Case Control Consortium Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross O, et al. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature. 2006;442:651–656. doi: 10.1038/nature04926. [DOI] [PubMed] [Google Scholar]
- 14.Ishikawa E, et al. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med. 2009;206:2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth S, Ruland J. Caspase recruitment domain-containing protein 9 signaling in innate immunity and inflammation. Trends Immunol. 2013;34:243–250. doi: 10.1016/j.it.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Schoenen H, et al. Cutting edge: Mincle is essential for recognition and adjuvanticity of the mycobacterial cord factor and its synthetic analog trehalose-dibehenate. J Immunol. 2010;184:2756–2760. doi: 10.4049/jimmunol.0904013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Werninghaus K, et al. Adjuvanticity of a synthetic cord factor analogue for subunit Mycobacterium tuberculosis vaccination requires FcRgamma-Syk-Card9-dependent innate immune activation. J Exp Med. 2009;206:89–97. doi: 10.1084/jem.20081445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hara H, et al. The adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and Toll-like receptors. Nat Immunol. 2007;8:619–629. doi: 10.1038/ni1466. [DOI] [PubMed] [Google Scholar]
- 19.Goodridge HS, et al. Differential use of CARD9 by dectin-1 in macrophages and dendritic cells. J Immunol. 2009;182:1146–1154. doi: 10.4049/jimmunol.182.2.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abram CL, Lowell CA. The expanding role for ITAM-based signaling pathways in immune cells. Sci STKE. 2007;2007:re2. doi: 10.1126/stke.3772007re2. [DOI] [PubMed] [Google Scholar]
- 21.Strasser D, et al. Syk kinase-coupled C-type lectin receptors engage protein kinase C-σ to elicit Card9 adaptor-mediated innate immunity. Immunity. 2012;36:32–42. doi: 10.1016/j.immuni.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, et al. CARD9 mutations linked to subcutaneous phaeohyphomycosis and TH17 cell deficiencies. J Allergy Clin Immunol. 2014;133:905–908.e3. doi: 10.1016/j.jaci.2013.09.033. [DOI] [PubMed] [Google Scholar]
- 23.Drummond RA, Saijo S, Iwakura Y, Brown GD. The role of Syk/CARD9 coupled C-type lectins in antifungal immunity. Eur J Immunol. 2011;41:276–281. doi: 10.1002/eji.201041252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LeibundGut-Landmann S, et al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8:630–638. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- 25.Robinson MJ, et al. Dectin-2 is a Syk-coupled pattern recognition receptor crucial for Th17 responses to fungal infection. J Exp Med. 2009;206:2037–2051. doi: 10.1084/jem.20082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saijo S, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32:681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Sokol H, et al. Card9 mediates intestinal epithelial cell restitution, T-helper 17 responses, and control of bacterial infection in mice. Gastroenterology. 2013;145:591–601.e3. doi: 10.1053/j.gastro.2013.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Z, et al. Ubiquitin ligase TRIM62 regulates CARD9-mediated anti-fungal immunity and intestinal inflammation. Immunity. 2015;43:715–726. doi: 10.1016/j.immuni.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozato K, Shin DM, Chang TH, Morse HC., 3rd TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burke MD, Schreiber SL. A planning strategy for diversity-oriented synthesis. Angew Chem Int Ed Engl. 2004;43:46–58. doi: 10.1002/anie.200300626. [DOI] [PubMed] [Google Scholar]
- 32.Begley DW, Moen SO, Pierce PG, Zartler ER. Saturation transfer difference NMR for fragment screening. Curr Protoc Chem Biol. 2013;5:251–268. doi: 10.1002/9780470559277.ch130118. [DOI] [PubMed] [Google Scholar]
- 33.Klein J, Meinecke R, Mayer M, Meyer B. Detecting binding affinity to immobilized receptor proteins in compound libraries by HR-MAS STD NMR. J Am Chem Soc. 1999;121:5336–5337. [Google Scholar]
- 34.Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 35.Mayer MMB. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew Chem. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 36.Huang F, Xiao H, Sun BL, Yang RG. Characterization of TRIM62 as a RING finger E3 ubiquitin ligase and its subcellular localization. Biochem Biophys Res Commun. 2013;432:208–213. doi: 10.1016/j.bbrc.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 37.Streich FC, Jr, Ronchi VP, Connick JP, Haas AL. Tripartite motif ligases catalyze polyubiquitin chain formation through a cooperative allosteric mechanism. J Biol Chem. 2013;288:8209–8221. doi: 10.1074/jbc.M113.451567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanternier F, et al. Deep dermatophytosis and inherited CARD9 deficiency. N Engl J Med. 2013;369:1704–1714. doi: 10.1056/NEJMoa1208487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lanternier F, et al. Inherited CARD9 deficiency in otherwise healthy children and adults with Candida species-induced meningoencephalitis, colitis, or both. J Allergy Clin Immunol. 2015;135:1558–1568.e2. doi: 10.1016/j.jaci.2014.12.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gavino C, et al. CARD9 deficiency and spontaneous central nervous system candidiasis: Complete clinical remission with GM-CSF therapy. Clin Infect Dis. 2014;59:81–84. doi: 10.1093/cid/ciu215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glocker EO, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009;361:1727–1735. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rieber N, et al. Extrapulmonary Aspergillus infection in patients with CARD9 deficiency. JCI Insight. 2016;1:e89890. doi: 10.1172/jci.insight.89890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sahni N, et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell. 2015;161:647–660. doi: 10.1016/j.cell.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Makley LN, Gestwicki JE. Expanding the number of ‘druggable’ targets: Non-enzymes and protein-protein interactions. Chem Biol Drug Des. 2013;81:22–32. doi: 10.1111/cbdd.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Connell DJ, et al. Simultaneous pathway activity inference and gene expression analysis using RNA sequencing. Cell Syst. 2016;2:323–334. doi: 10.1016/j.cels.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]