Significance
Glioblastoma multiforme (GBM) is uniformly lethal and shows resistance to all forms of therapy. Glioma stem cells (GSCs) have been shown to support GBM maintenance and exhibit enhanced resistance to ionizing radiation, a cornerstone of GBM therapy. This study establishes that proliferating cell nuclear antigen-associated factor (PAF) depletion profoundly reduces GSC frequency and tumorigenicity, in part, by down-regulating DNA replication and pyrimidine metabolism. Moreover, PAF depletion impairs error-prone DNA translesion synthesis (TLS) and enhances sensitivity of GSCs to radiation treatment. Pharmacological impairment of DNA replication and TLS diminished GSC maintenance and radioresistance, illuminating a potential GBM treatment strategy of combined TLS inhibition and radiation therapy.
Keywords: glioma stem cells, self-renewal, DNA translesion synthesis
Abstract
An integrated genomic and functional analysis to elucidate DNA damage signaling factors promoting self-renewal of glioma stem cells (GSCs) identified proliferating cell nuclear antigen (PCNA)-associated factor (PAF) up-regulation in glioblastoma. PAF is preferentially overexpressed in GSCs. Its depletion impairs maintenance of self-renewal without promoting differentiation and reduces tumor-initiating cell frequency. Combined transcriptomic and metabolomic analyses revealed that PAF supports GSC maintenance, in part, by influencing DNA replication and pyrimidine metabolism pathways. PAF interacts with PCNA and regulates PCNA-associated DNA translesion synthesis (TLS); consequently, PAF depletion in combination with radiation generated fewer tumorspheres compared with radiation alone. Correspondingly, pharmacological impairment of DNA replication and TLS phenocopied the effect of PAF depletion in compromising GSC self-renewal and radioresistance, providing preclinical proof of principle that combined TLS inhibition and radiation therapy may be a viable therapeutic option in the treatment of glioblastoma multiforme (GBM).
Glioblastoma multiforme (GBM) is an aggressive primary brain tumor with a poor prognosis owing, in part, to resistance to both chemotherapy and radiation therapy (1). GBM possesses a subpopulation of cells exhibiting stem cell-like properties, including self-renewal activity, multilineage differentiation potential, and stem cell marker expression (2–4), as well as reliance on stem cell pathways, such as the Notch and Hedgehog/GLI1 pathways (5, 6). Numerous structural proteins, transcription factors, and cell surface proteins have been proposed to define glioma stem cells (GSCs), including OLIG2 (7), SOX2 (8), Nestin (9), CD133 (10, 11), and SSEA1 (12), although no definitive GSC marker has been identified to date (13). Thus, the assessment of stemness of glioma cells is typically complemented by functional assays, such as in vitro neurosphere formation (a readout for proliferation and self-renewal), evaluation of the cellular hierarchy within the tumor (an assessment of cellular multipotency potential), and limiting dilution measurement (the gold standard for determining stem cell frequency). Because these GSCs show robust tumorigenic potential and therapeutic resistance (4, 10, 14), significant effort has been devoted toward understanding the molecular mechanisms underlying GSC biology, with the goal of illuminating pathways and targets that may improve the treatment of GBM.
GBM is characterized by a high proliferative index (15) and aberrant constitutive DNA damage signaling (16). In response to DNA damage, normal cells activate the DNA damage response (DDR), utilizing a variety of DNA damage sensing and repair pathways (e.g., base excision repair, nucleotide excision repair, homologous recombination, nonhomologous end-joining, mismatch repair, direct reversal) to maintain genomic integrity, whereas the inability to repair DNA damage leads to apoptosis (17). In contrast, glioma cells harboring significant ongoing genome instability alter DDR processes, including the preferential activation of DNA damage sensing (e.g., ATR/CHK1, ATM/CHK2) and repair (e.g., PARP1) pathways (10, 18–20), to better cope with DNA double-stranded breaks (DSBs) when exposed to ionizing radiation. As ionizing radiation also results in replication fork stalling, one would anticipate that GSCs may also up-regulate pathways that enable the bypass of unrepaired DNA lesions during DNA replication, a process known as DNA translesion synthesis (TLS), to minimize additional DSBs that would arise from the stalled replication forks. A key step in the regulation of TLS includes the monoubiquitination of proliferating cell nuclear antigen (PCNA) by the Rad6/Rad18 enzymes to facilitate the switch from error-free DNA polymerases to low-fidelity TLS polymerases (including DNA polymerase η) to bypass the DNA lesion (21). Thus, the levels of monoubiquitinated PCNA and the frequency of cells with DNA polymerase η foci provide a reliable, albeit indirect, method by which to evaluate TLS. Whether TLS influences GSC self-renewal, tumorigenicity, and radioresistance has not been explored previously.
To identify new molecular determinants governing GSC self-renewal independent of any conventional GSC markers, and thereby enable exploration in a wider spectrum of glioma cells with GSC-like functional properties, we undertook an integrated genomic and functional approach by overlapping a list of DDR genes that were overexpressed in GBM relative to the normal brain (which included putative radioresistance genes in GBM), with a second list of genes that were derived from the transcriptomic comparison of noncancerous neural stem cells (NSCs) with or without telomere dysfunction (which included genes that influence self-renewal and radioresistance) (22–24), hypothesizing that these filters would enhance the identification of stem cell-enriched genes functioning in GBM. This approach uncovered the PCNA-associated factor (PAF) as a GSC-enriched DDR gene with a role in GSC self-renewal, tumorigenicity, and radioresistance. Our work identified an association between high-PAF expression and TLS activity in GSCs and established that inhibition of TLS enhances the radiosensitivity of GBM.
Results
An Integrated Genomic and Functional Approach Identifies PAF as a GSC-Enriched Gene with Potential Clinical Relevance.
To reveal DDR genes that have an impact on GSC maintenance, we first identified DDR genes that showed overexpression in The Cancer Genome Atlas (TCGA) dataset for GBM relative to normal brain (Fig. 1A and Datasets S1 and S2), and then integrated these with DDR genes that were differentially expressed in normal murine neural stem/progenitor cells (NSPCs) with impaired self-renewal potential due to “endogenous” DNA damage from shortened telomeres (22–24) (Fig. 1A). We reasoned that the integration of these two datasets would enrich for stem cell-relevant DDR genes that might impact GSC self-renewal and radioresistance, without overreliance on imprecise GSC markers.
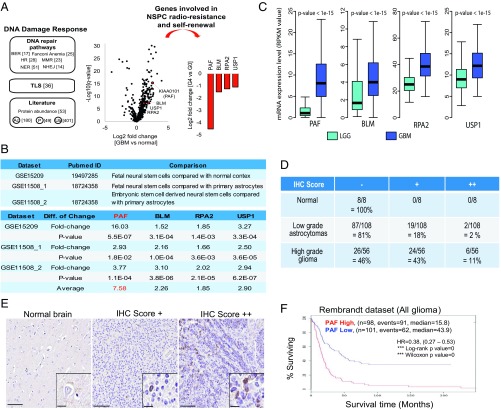
Fig. 1.
Elevated PAF expression indicated poor prognosis. (A) Unbiased genomic approach to identify radioresistance-conferring DDR genes that are enriched in GSCs (FDR < 0.01). The numbers in the brackets represent the number of genes in each category. (B) mRNA expression analysis of the four stem cell-enriched DDR candidate genes in human NSCs versus differentiated brain cells. (C) Comparison of PAF, BLM, RPA2, and USP1 mRNA expression levels in low-grade glioma (LGG) and GBM in the TCGA dataset. RPKM, reads per kilobase of transcript per million mapped reads. Immunohistochemistry (IHC) scores of PAF in different grades of glioma using glioma patient tissue microarray (D) and representative images of the respective IHC scores (E) are shown. (Scale bars: 100 μm; Inset, 20 μm.) (F) Analysis of correlation between PAF mRNA expression levels and glioma patient overall survival (P < 0.01 by log-rank analysis). HR, hazard ratio.
With this approach, we identified four genes [false discovery rate (FDR) < 0.01], namely, PAF (also known as KIAA0101), BLM (Bloom Syndrome Protein), RPA2 (Replication Protein A 2), and USP1 (Ubiquitin Specific Peptidase 1) (Fig. 1A). Strikingly, all of these genes are implicated in TLS (Fig. S1A). With respect to TLS, PAF regulates the sliding of PCNA along the DNA and facilitates the switch from error-free to error-prone DNA synthesis (25). BLM recognizes DNA adducts that are associated with stalled replication forks in addition to G4 DNA junctions (26). RPA2 coats single-stranded DNA at sites of DNA damage and recruits E1 (Rad6) and E3 (Rad18) to monoubiquitinate PCNA, which leads to the switch to error-prone DNA synthesis (27), while USP1 deubiquitinates monoubiquitinated PCNA (28). In this study, PAF was chosen for further in-depth study as it was the only gene with significant enrichment in human NSPCs relative to differentiated brain cells, including primary astrocytes (approximately eightfold) (Fig. 1B), suggestive of its potential role in stem cell self-renewal/proliferation.
Fig. S1.
Related to Figs. 1 and 2. Elevated PAF expression indicated poor prognosis and identified glioma cells with GSC marker expression. (A) Current model of the TLS pathway that may be mediated by PCNA or Fanconi anemia core complex. The four identified genes are shown in red. MonoUb, monoubiquitinated. (B) Representative Western blot analysis of PAF and PCNA in transformed neurospheres (Neu) compared with isogenic tumorspheres (TS); vinculin serves as the loading control. Quantification of PAF and PCNA intensities was normalized to the respective vinculin controls. (C) Representative Western blot analysis of PAF and PCNA in CD133− and CD133+ GSCs; vinculin serves as the loading control. (D) qRT-PCR analysis of CD133 and PAF mRNA levels of GSCs after CD133 sorting (n = 3) (mean ± SD). The ribosomal protein gene, RPLP2, serves as the housekeeping gene.
To further assess the relevance of PAF in GBM, we compared its mRNA levels in low-grade gliomas versus GBM in the TCGA database, revealing PAF versus BLM, RPA2, or USP1 mRNA expression was dramatically higher in GBM relative to low-grade gliomas (approximately eightfold for PAF versus approximately twofold for BLM, RPA2, and USP1) (Fig. 1C). Tissue microarray analysis of PAF protein levels in 172 patient samples showed negligible expression in the normal brain, low expression in low-grade astrocytomas, and robust expression in high-grade gliomas (Fig. 1 D and E). The Repository for Molecular Brain Neoplasia Database datasets revealed a negative correlation between PAF and overall survival (Fig. 1F).
Next, we asked whether PAF is preferentially expressed in GSCs and noncancerous murine NSPCs. We found that PAF expression colocalized with cancer stem cell markers, including CD133, SOX2, and OLIG2 (Fig. 2A), but not with differentiated cell markers, including GFAP and TUJ1 (Fig. 2B). Western blot analysis confirmed elevated PAF expression in most GBM patient-derived GSCs compared with noncancerous human NSPCs (Fig. 2C) and in oncogenic-transduced human NSPCs and isogenic GSCs derived from experimentally generated GBM relative to the parental NSC as described previously (29) (Fig. S1B). Notably, PAF protein and mRNA levels were also higher in radioresistant CD133+ GSCs (10) (Fig. S1 C and D). Furthermore, in the normal mouse brain, PAF expression was localized to proliferating (Ki67+) NSPCs (Nestin+ and Sox2+) (Fig. 2 D and E) but not to more differentiated (GFAP+) cells from G0 mice (Fig. 2F). Importantly, PAF expression was specific to NSPCs in the mouse subventricular zone (SVZ), and, correspondingly, there was markedly decreased PAF expression with near-complete eradication of NSPCs (Nestin+ and Sox2+) in the telomere dysfunctional/G5 mouse model (Fig. 2D). Together, these clinical and experimental data raise the possibility of a role of PAF in GSC biology.
Fig. 2.
PAF is preferentially expressed in GSCs and noncancerous murine NSPCs. The immunofluorescence of PAF, cancer stem cell markers (CD133, SOX2, and OLIG2) (A) or differentiated cell markers (GFAP and Tuj1) (B), and DAPI in GBM xenograft tissue is shown. (Scale bar: 50 μm.) (C) Representative Western blot analysis of PAF protein in multiple GSC lines compared with normal human NSPCs; β-actin serves as the loading control. Immunofluorescence of Paf (red); NSPC (Nestin and Sox2, green) (D), Ki67 (green) (E) or astrocytic (Gfap, green) (F) markers; and DAPI (blue) in the SVZ of representative wild-type (WT)/G0 and telomere-dysfunctional/G5 mice are shown. (Scale bar: 50 μm.)
PAF Supports GSC Self-Renewal, Tumorigenicity, and Tumor Heterogeneity.
We next assessed whether PAF levels influenced GSC self-renewal and clonogenicity. In multiple GSC lines, PAF depletion robustly diminished the number of tumorspheres (Fig. 3A and Fig. S2A), indicating impaired GSC self-renewal with lower PAF levels. PAF depletion also significantly decreased GSC colony formation in soft agar assays (Fig. S2B). Conversely, gain-of-function analysis showed that enforced expression of PAF in the TS586 cell line with relatively low-baseline PAF expression (Fig. 2C) resulted in a dramatic increase in colony formation (Fig. S2C). Moreover, in murine intracranial injection tumor studies, firefly luciferase-labeled, PAF-depleted GSCs generated smaller volume tumors and fewer mice with tumors, whereas control GSCs produced large tumors with high penetrance (Fig. 3 B and C). Upon pathological analysis, PAF-depleted GSCs formed either small or no tumors, while PAF-expressing GSCs generated infiltrative, aggressive tumors (Fig. 3D). Thus, we conclude that PAF inhibition compromises the tumorigenic potential of GSCs.
Fig. 3.
PAF is required for GSC self-renewal and tumorigenicity. (A) Tumorsphere formation of GSCs following PAF knockdown (n = 4) (mean ± SD). (B and C) In vivo bioluminescence-based images 29 d postorthotopic injection of GSC TS543 (1 × 105 cells) transduced with NT/control or PAF shRNAs. Representative mice (B) and quantification of tumor volume based on bioluminescence (C) are shown. There were no tumors detected in three of the five mice bearing PAF-depleted GSCs for either of the shRNAs against PAF. Max, maximum; Min, minimum. (D) H&E coronal sections of mouse tumors formed after injection of GSC TS543 (1 × 105 cells) with or without PAF knockdown. Brains were harvested at the same time as mice with NT/control shRNA-treated GSCs had to be killed. (Scale bar: 1 mm.) (E) Ratio of GFP+/GFP− cells in an in vivo GFP competition assay. GFP− GSCs were transduced with NT/control or PAF shRNA (n > 5) (mean). (F) Frequency of NT/control or PAF shRNA-transduced GFP− cells, which were marked by various stem cell markers, in an in vivo GFP competition assay (n > 5) (mean). Tumor incidence (G) and survival curves (H) of mice implanted with GSCs transduced with NT or PAF shRNA in in vivo limiting dilution assays are shown.
Fig. S2.
Related to Fig. 3. PAF is required for GSC self-renewal and tumorigenicity. (A) Knockdown efficiency of PAF by the shRNAs used in this study. (B) Soft agar colony formation of PAF-depleted GSCs (n = 4) (mean ± SD). (C) Soft agar colony formation assays after PAF overexpression in GSCs having low-baseline PAF expression (n = 4) (mean ± SD). Gating strategies for the quantification of TRA-1-85+/GFP− (D) and TRA-1-85+/594+/GFP− (E) GSCs in the GFP competition experiment are shown. FSC, forward-scattered light. The AF594+ cells represent either stem or progenitor cell markers. (F) Frequency of GFP− GSCs transduced with NT/control or PAF shRNA in various progenitor lineages in an in vivo GFP competition assay (n > 5) (mean). (G) Immunohistochemistry staining of stem cell and proliferation markers in xenograft tumors generated from PAF-expressing or -depleted GSC TS543 (n = 3). (H) Cell viability of GSCs and their differentiated counterparts with or without PAF knockdown (n = 4) (mean ± SD). (I) In vitro limiting dilution assays of GSC TS603 transduced with NT/control or PAF shRNA calculated with extreme limiting dilution analysis.
We next performed systematic analyses to substantiate the role of PAF in enabling GSCs to generate the cellular hierarchy within the tumor using a GFP competition experiment involving orthotopic implantation of GFP-expressing control (GFP+) GSCs and (GFP−) GSCs with or without PAF depletion. PAF-depleted GFP− GSCs showed a significant reduction in relative fitness, as evidenced by their diminished representation in the tumor mass (Fig. 3E and Fig. S2D). In the competitive setting, PAF-depleted GFP− GSCs also generated fewer stem-like cells (marked by CD133+, SOX2+, and OLIG2+) compared with PAF-expressing GFP− GSCs (Fig. 3F and Fig. S2E). Analysis of more differentiated cells in the tumor showed that PAF depletion decreased the frequency of GFAP+ and MAP2+ cells compared with PAF-expressing controls (Fig. S2F). Consistent with the above multilineage analyses illuminating the importance of PAF on cancer cells with stem cell-like properties, PAF-depleted GSCs generated tumors that had reduced Nestin and OLIG2 staining compared with PAF-expressing GSCs, even in a noncompetitive setting (Fig. S2G). Given that the silencing of PAF only affected the proliferation of GSCs but not their differentiated counterpart in vitro (Fig. S2H), these collective data support the interpretation that PAF contributes to the cellular hierarchy within the tumor by regulating GSC self-renewal, rather than serving as a deciding factor between GSC self-renewal and differentiation.
To further validate the role of PAF in supporting GSC stemness, we evaluated the impact of PAF depletion on GSC tumor initiating potential in well-established limiting dilution tumor assays. In in vivo xenografting studies, we demonstrated that mice implanted with PAF-expressing cells developed tumors with a higher incidence rate compared with mice implanted with PAF-depleted cells (Fig. 3G), consistent with the possibility that PAF supports the frequency of tumor initiating cells. Mice with PAF-expressing GSCs also developed tumors with a shorter latency and decreased survival compared with mice implanted with PAF-depleted cells (Fig. 3H). The loss of GSC stemness upon PAF depletion was further confirmed by performing the in vitro limiting dilution assays with another GSC line (Fig. S2I). Together, these results underscore the importance of maintenance of adequate PAF levels in supporting GSC self-renewal and tumorigenic potential.
PAF Influences DNA Replication and Pyrimidine Metabolism in GSCs.
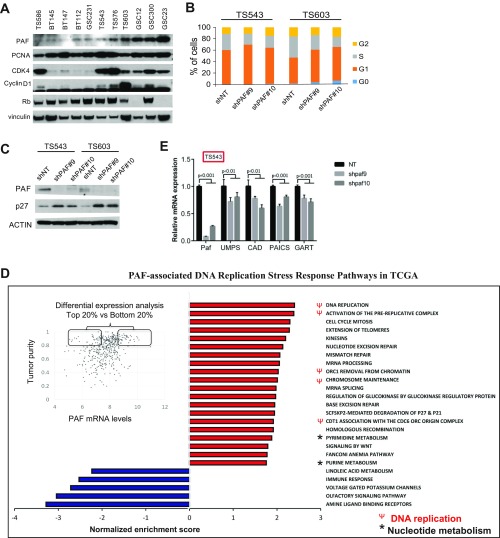
Next, we explored the potential mechanistic bases by which PAF supports GSC maintenance. PAF can influence DNA replication, cell cycle, and TLS pathways (30–32); recently, PAF has been shown to promote cancer through PCNA-independent mechanisms, including the activation of the Wnt/β-catenin pathway in colon cancer cells (33, 34) and MAPK signaling via LAMTOR3 in pancreatic cancer cells (35). Therefore, we employed immunoprecipitation coupled with mass spectrometry analysis to identify interactors of PAF that may illuminate its important role in GSC maintenance. We found that FLAG-tagged PAF immunoprecipitated only PCNA in GSC lysates (Fig. 4A), and flow cytometry analysis confirmed that PAF was predominantly coexpressed with PCNA in GSCs (Fig. 4B), suggesting that PAF may regulate DNA replication and cell cycle progression through its interaction with PCNA in GSC. Western blot analysis of multiple GSC lysates also showed that PAF expression correlated with the expression of CDK4 and Cyclin D1, (Fig. S3A), supporting the idea that PAF expression may be coordinated with entry into the DNA synthesis phase of the cell cycle. To validate the potential role of PAF in DNA replication, we first performed transient BrdU labeling studies to audit cells that were progressing through DNA replication (S phase of the cell cycle). As expected, PAF depletion resulted in a significant reduction of BrdU+ cells relative to the short hairpin nontargeting (shNT)/control shRNA (Fig. 4C). This S-phase entry blockade was further confirmed by cell cycle propidium iodide (PI) staining (Fig. S3B) and increased p27 protein levels in PAF-depleted GSCs by Western blot analysis (Fig. S3C).
Fig. 4.
PAF influences DNA replication and pyrimidine metabolism in GSCs. (A) Silver-stained gel showing protein interactors of FLAG-PAF in GSC TS543. (B) Frequency of GSCs expressing PAF+/PCNA+ or PAF+/PCNA− (n = 5). The gating strategy is shown below. (C) Frequency of BrdU+ GSCs after PAF depletion (n = 4) (mean ± SD). 7-AAD, 7-aminoactinomycin D. (D) Significantly down-regulated (blue bars) and up-regulated (red bars) pathways identified by GSEA in multiple GSC lines after PAF knockdown (FDR < 0.05). (E) Metabolomic analysis of GSC TS543, with or without PAF knockdown, revealing metabolites that are significantly down-regulated (blue font) or up-regulated (red font) (n = 3). SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine. (F) Representative images of soft agar colony formation of CAD-depleted GSCs (n = 4). (G) Comparison of CAD mRNA expression levels in normal brain and GBM in the TCGA dataset. (H) Analysis of correlation between CAD mRNA expression levels and IDH1 wild-type GBM patient overall survival (P < 0.05 by log-rank analysis). HR, hazard ratio.
Fig. S3.
Related to Fig. 4. PAF influences DNA replication and pyrimidine metabolism in GSCs. (A) Representative Western blot analysis of PAF, PCNA, CDK4, Cyclin D1, and Rb in multiple GSCs. Vinculin serves as the loading control. (B) Cell cycle analysis of GSCs upon PAF knockdown using PI staining (n = 3). (C) Representative Western blot analysis of p27 in PAF-depleted GSCs. PAF and vinculin serve as the positive and loading controls, respectively. (D) Pathways that are significantly associated with PAF expression in TCGA GBM tumors: positive correlation is shown in red, and negative correlation is shown in blue (FDR < 0.05). (E) qRT-PCR analysis of PAF, UMPS, CAD, PAICS, and GART mRNA levels of PAF-depleted GSCs (n = 3) (mean ± SD). The ribosomal protein gene, RPLP2, serves as the housekeeping gene.
In addition to the protein interaction analysis, we performed gene expression profiling of multiple human GSC lines with or without PAF knockdown to identify genes and pathways that correlated with loss of GSC maintenance upon PAF depletion. Gene set enrichment analysis (GSEA) of the differentially expressed genes revealed an enrichment of DNA replication stress response pathways, including cell cycle, DNA replication, DDR, and nucleotide metabolism in PAF-depleted GSCs (Fig. 4D). These cell cycle, DNA replication, DDR, and nucleotide metabolism pathways also correlated significantly with PAF expression in TCGA GBM tumors, supporting the clinical relevance of PAF in GBM treatment (FDR < 0.05; Fig. S3D). Of potential significance was the down-regulation of nucleotide metabolism pathways in PAF-depleted GSCs (Fig. 4D and Fig. S3E) as purine synthesis was recently shown to be important for GSC maintenance (36).
To determine if altered nucleotide metabolism could also contribute to the loss of GSC maintenance, we complemented our transcriptomic analysis with metabolomic analysis of PAF-expressing versus -depleted GSCs, revealing a significant down-regulation of the pyrimidine and arginine metabolism pathways that would result in a reduction of orotate, the precursor of uridine monophosphate for subsequent cytidine triphosphate synthesis (Fig. 4E). This tracked well with the down-regulation of carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase (CAD) mRNA expression in PAF-depleted GSCs (Fig. S3E). Consistent with the potential role of pyrimidine metabolism in GSC maintenance, CAD knockdown resulted in a profound decrease in GSC colony formation in soft agar assays (Fig. 4F). Reinforcing the clinical relevance of CAD and pyrimidine synthesis in GBM, we also observed that CAD mRNA expression was significantly increased in GBM tumors compared with the normal brain (Fig. 4G), and that higher CAD mRNA expression correlated with shorter survival of patients with IDH1 wild-type GBM (Fig. 4H). Therefore, we conclude that the loss of GSC maintenance upon PAF depletion can be attributed, in part, to decreased DNA replication and pyrimidine synthesis.
PAF-Associated TLS Activity Influences GSC Radioresistance.
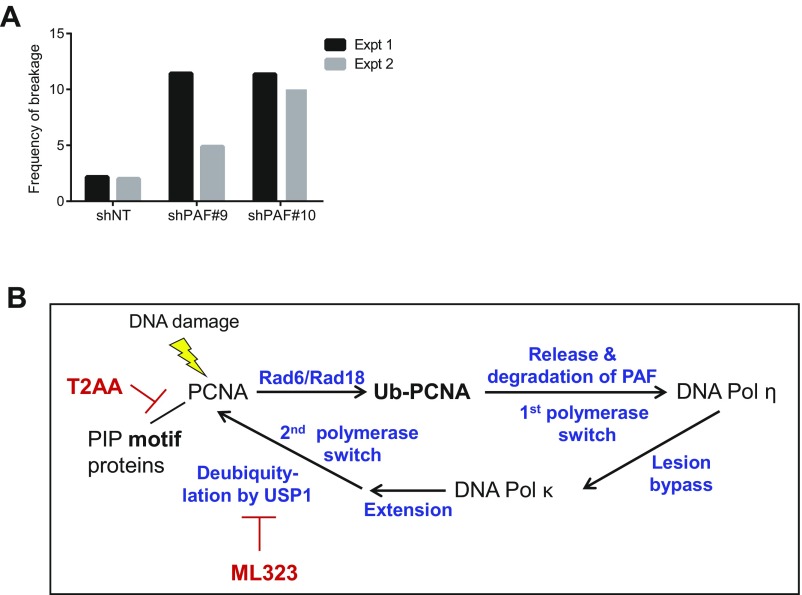
Due to its interaction with PCNA, we next evaluated the potential role of PAF in TLS by monitoring the levels of monoubiquitinated PCNA and frequency of cells with DNA Pol η foci in PAF-depleted GSCs. PAF knockdown led to an increase in monoubiquitinated PCNA in the Triton-insoluble fraction of GSCs compared with cells transduced with control shRNA (Fig. 5A), a level comparable to that seen when DNA damage is induced by UV light (37, 38). Furthermore, immunofluorescence microscopy of DNA Pol η revealed an approximately twofold increase in GSCs with DNA Pol η foci upon PAF depletion relative to cells transduced to control shRNA, consistent with possible slowed kinetics of switching to DNA Pol κ in PAF-depleted cells (Fig. 5 B and C). The increase in monoubiquitinated PCNA and GSCs with DNA Pol η foci upon PAF knockdown is consistent with the model that loss of PAF impedes the ability of GSCs to bypass DNA lesions by switching from error-free DNA synthesis to low-fidelity DNA synthesis, which is particularly important given the high level of endogenous DNA damage signaling in GSCs. In accordance with impaired TLS, there was a slight increase in baseline genomic instability of PAF-depleted GSCs (Fig. S4A). We also examined the connection between PAF and TLS in GBM tumors and found that PAF mRNA levels correlated positively with known components of the PCNA-mediated TLS pathway, including PCNA, Rad6, DNA Pol η, DNA Pol κ, and USP1 in IDH1 wild-type GBM tumors (Fig. 5D), reinforcing the link between PAF and TLS in this disease.
Fig. 5.
PAF levels influence GSC radioresistance. (A) Representative Western blot analysis of PCNA in the Triton-insoluble and Triton-soluble fractions of GSC TS543 lysates. Monoubiquitinated PCNA is marked by a red asterisk. exp, exposure. Quantification of GSC TS543 with DNA Pol η foci (red) with or without PAF knockdown, followed by UV light treatment (30 J/m2) (B), and representative immunofluorescence microscopy images (C) (n = 5) (mean ± SD) are shown. (Scale bar: 20 μm.) (D) Correlative analysis of PAF mRNA levels and the TLS signature score in IDH1 wild-type GBM tumors. RMA, robust multi-array average. (E) Frequency of tumorsphere formation of PAF-depleted GSC TS543, with or without radiation treatment, in a single cell per well assay (n = 3) (mean ± SD). (F) Tumorsphere formation of PAF-depleted GSC GSC23, with or without radiation treatment (n = 6) (mean ± SD).
Fig. S4.
Related to Figs. 5 and 6. PAF regulates PCNA-mediated TLS and radioresistance in GSCs. (A) Frequency of chromosomal breakages in PAF-depleted GSC TS603 in two independent experiments (Expt). (B) Known targets of T2AA and ML323 in the PCNA-mediated TLS pathway. Ub, ubiquitin.
The association of PAF and TLS, coupled with the known radioresistance of GSCs and the use of radiation therapy as a standard of care for GBM, prompted us to assess whether manipulation of PAF levels or TLS activity would impact GSC maintenance in the context of ionizing radiation (10). To that end, we observed that PAF-depleted GSCs generated fewer tumorspheres than cells transduced with the control nontargeting shRNA following radiation treatment (2 Gy versus no radiation: shNT = 66%, shPAF#9 = 48%, shPAF#10 = 29%); this effect was not observed at higher radiation dose as this GSC line was highly sensitive to 4 Gy alone (Fig. 5E). Similar results were obtained with a second GSC line (4 Gy versus no radiation: shNT = 60%, shPAF#9 = 29%, shPAF#10 = 44%) (Fig. 5F). We conclude that PAF promotes GSC radioresistance.
Pharmacological Inhibition of PCNA-Associated DNA Replication and TLS Impairs GSC Maintenance and Radioresistance.
As there are no inhibitors of PAF available, we determined the impact of pharmacological inhibition of PCNA-associated DNA replication and TLS using compounds T2AA and ML323 (Fig. S4B): T2AA binds to PCNA at a PIP-box cavity and blocks the interaction between PCNA and PIP box-containing proteins, including PAF and DNA Pol η (39), and ML323 is a potent USP1-UAF1 inhibitor that inhibits the deubiquitination of PCNA and association of PCNA with error-free DNA polymerase (40). GSC treatment with these PCNA inhibitors (10 μM, 24 h) resulted in significant down-regulation of the mRNA levels of known GSC markers, suggesting that DNA replication and TLS function support the maintenance of the GSC subpopulation. Specifically, upon PCNA inhibitor treatment, we observed down-regulation of SOX2 and OLIG2 (and a weak trend with Nestin) in TS543, CD133 and SOX2 down-regulation in GSC2, and SOX2 down-regulation in TS603 (Fig. 6A). Finally, we determined the effect of PCNA inhibition on GSC self-renewal and radioresistance. Both PCNA inhibitors significantly reduced GSC tumorsphere formation in a dose-dependent manner, indicating compromised self-renewal (Fig. 6B). Moreover, both compounds further impaired GSC tumorsphere formation when combined with radiation treatment (4 Gy versus no radiation: DMSO = 32%, 2 μM T2AA = 12%, 2 μM ML323 = 11%) (Fig. 6B). Collectively, these results support the theory that inhibitors of PCNA-associated DNA replication and TLS may enhance radiosensitivity of PAF-expressing GBM tumors.
Fig. 6.
Pharmacological impairment of TLS compromises hallmark features of GSC. (A) qRT-PCR analysis of CD133, SOX2, Nestin, and OLIG2 mRNA levels of GSCs after 24 h treatment with TLS inhibitors (n = 3) (mean ± SD). The ribosomal protein gene, RPLP2, serves as the housekeeping gene. (B) Frequency of tumorsphere formation of GSC TS543, with or without radiation treatment, in the presence of TLS inhibitors in a single cell per well assay (n = 3) (mean ± SD).
Discussion
In this study, an integrated approach identified four TLS-relevant genes, including PAF, BLM, RPA2, and USP1, as stem cell-enriched DDR genes in GBM. We observed that PAF expression was enriched in normal NPSCs and in glioma cells expressing “stem cell” markers and contributed to GSC self-renewal, tumorigenicity, and multipotency in vivo. These findings qualify PAF as a GSC maintenance factor, and the connection of PAF and TLS illuminates a potential strategy to overcome the high radioresistance of GSCs.
During self-renewal and proliferation, GSCs encounter various challenges, including replication stress, DNA damage insults, and checkpoint induction, which have a direct impact on genome stability and stem cell maintenance. While DNA damage sensing and repair pathways (18–20), as well as chromosomal instability (41), have been shown to influence GSC function, this study provides experimental evidence that the capacity of GSCs to tolerate DNA damage is crucial for their maintenance and tumorigenic properties. Our study also links PAF and PCNA-mediated DNA replication and TLS in GSC self-renewal and radioresistance. PAF is a known DNA damage-regulated factor that regulates both DNA replication and accessibility of TLS enzymes to PCNA (32); the former would explain why PAF depletion alone could profoundly reduce the GSC pool in the absence of radiation treatment. PAF influences TLS by facilitating polymerase switching at PCNA by promoting its proteasome-dependent removal from PCNA in the presence of DNA damage (32). Following lesion bypass, the reassociation of PAF with PCNA enables the release of TLS Pol η and restoration of error-free DNA synthesis (Fig. S4B). As both of these polymerase exchanges are critical for efficient lesion bypass, this would explain why PAF depletion could sensitize GSCs to radiation treatment. Furthermore, we demonstrate that the pharmacological perturbation of PCNA-mediated DNA replication and TLS with T2AA or ML323 phenocopies the effect of PAF knockdown in compromising GSC proliferation and radioresistance. Although TLS enzymes are expressed in both normal and cancer cells, the higher expression of the TLS enzymes in cancer cells would suggest that there could be a therapeutic window for the use of TLS inhibitors in cancer. Moreover, we would expect that the use of targeted radiation therapy (as opposed to systemic chemotherapeutic agents) with TLS inhibitors would serve to minimize systemic toxicities on highly proliferative tissues. Finally, the PAF knockout mouse is viable (42), suggesting that the selective depletion of PAF may be a viable approach as opposed to perturbing TLS through PCNA inhibitors. Together, our findings support the development of selective, potent TLS inhibitors for use in radiation therapy to improve the outcome of GBM treatment.
Methods and Materials
Isolation of Murine NSPCs.
All experiments were performed under preapproved institutional animal care and use committee (IACUC) protocols at MD Anderson Cancer Center. All mice were housed in specific pathogen-free conditions in animal research facilities at MD Anderson Cancer Center, which are fully Association for the Assessment and Accreditation of Laboratory Animal Care-accredited and meet NIH standards as set forth in the Guide for Care and Use of Laboratory Animals (43). The heterozygous (G0 TERTER) and late-generation homozygous (G4/G5 TERTER) mice were generated based on the standard breeding protocol of successive generations of telomerase-deficient mice (23). All studies were performed on age- and gender-matched adult (3–6 wk old) G0 TERTER and telomere dysfunctional G4/G5 TERTER mice. The SVZ of brain from 3- to 6-wk-old TERTER mice was dissected, and NSPCs were isolated using a Neural Tissue Dissociation Kit (P) (Miltenyi) and plated in NeuroCult Proliferation medium (Stem Cell Technologies) supplemented with EGF and β-FGF (20 ng⋅mL−1 each). Primary neurospheres were dissociated and expanded to form secondary neurospheres before RNA extraction.
Gene Expression Profiling.
Gene expression profiling was performed at the Sequencing and Noncoding RNA Program at MD Anderson Cancer Center. RNA was isolated and hybridized on the GeneChip Mouse Genome 430 2.0 Array or GeneChip Human Genome U133 plus 2.0 Array (Affymetrix) according to the manufacturer’s instructions. Raw data (CEL files) were preprocessed using robust multiarray analysis. To identify pathways that were enriched in the G4/G5 TERTER NSPCs or PAF-depleted GSCs, GSEA (software.broadinstitute.org/gsea/login.jsp;jsessionid=97D630B8CAD363F0BD517A72CE8F74CA) was performed on genes that were consistently up-regulated or down-regulated compared with the respective controls.
Retrospective Analysis of PAF Gene Expression in Human Gliomas.
Correlations between glioma grade and gene expression were determined through analysis of TCGA (https://tcga-data.nci.nih.gov/docs/publications/tcga/) and Oncomine.
Human-Derived GSCs.
All human research using GBM tissues and GSCs was conducted under MD Anderson IRB approved protocols. Tissues were collected from patients after obtaining written informed consent (LAB04-0001) and were subsequently analyzed (PA16-0408). Patient-derived GSCs were provided by Cameron Brennan (Memorial Sloan–Kettering Cancer Center, New York, NY) and Keith Ligon (Dana–Farber Cancer Institute, Boston, MA), as well as Frederick F. Lang and Erik P. Sulman from the Brain Tumor Center (MD Anderson Cancer Center). The GSCs were cultured in human NSC Maintenance Media (Millipore), supplemented with EGF and β-FGF (20 ng⋅mL−1 each).
Tissue Microarray Immunohistochemistry.
The glioma tissue microarray was purchased from US Biomax, and tumors were semiquantified on a relative scale of 0 to 2 (0 = negative, 2 = strongest) in a blind manner.
Vectors and Lentiviral Constructs.
FLAG-PAF constructs were generated from pENTR221 vector (clone IOH5885) using QuikChange II site-directed mutagenesis (Stratagene) as specified by the manufacturer, using the following primers: 5′-CTTTGTACAAAAAAGCAGGCACCATGGATTACAAGGATGACGATGACAAGGCGGTGCGGACTAAAGCAGACAG-3′ and 5′-CTGTCTGCTTTAGTCCGCAC CGCCTTGTCATCGTCATCCTTGTAATCCATGGTGCCTGCTTTTTTGTACA AAG-3′. These were then cloned into the pHAGE-EF1a-IRES-GFP lentiviral vector by Gateway cloning. The proper construction of all plasmids was confirmed by DNA sequencing. The shRNAs against human PAF (shPAF#9, TRCN0000278497; shPAF#10, TRCN0000278496; and shPAF#11, TRCN0000278552) were purchased from Sigma. GSCs were transduced with viral particles. Down-regulation of the target was evaluated by real-time PCR or Western blot at 72 h postinfection.
Antibodies and Chemical Reagents.
Primary antibodies used for flow cytometry, immunofluorescence, immunohistochemistry, and immunoblotting were as follows: phycoerythrin (PE)-CD133 (AC133; Miltenyi), BrdU (clone no. Bu20a; Dako), Sox2 (catalog no. ab97959; Abcam), Nestin (catalog no. ab105389; Abcam), Olig2 (catalog no. ab9610; Millipore), GFAP (catalog no. Z0334; Dako), TUJ1 (catalog no. MMS-435P; Covance), MAP2 (catalog no. ab5622; Millipore), Ki67 (clone no. SP6; Vector), TRA-1-85 (MAB3195; R&D Systems), PAF (clone no. 3C11-1F11; Abnova), Paf [clone no. Jyld12 (42), mouse-specific], PCNA (clone no. PC10; Santa Cruz Biotechnology), PCNA (clone no. EPR3821; Abcam), p27 (clone no. C-19; Santa Cruz Biotechnology), CDK4 (clone no. DCS156; Cell Signaling), Cyclin D1 (clone no. DCS6; Cell Signaling), vinculin (clone no. hVIN-1; Sigma), β-actin (clone no. AC-74; Sigma), DNA Pol η (ab186677; Abcam), and Rb (catalog no. sc-50; Santa Cruz Biotechnology). Chemical reagents were T2AA (Tocris Bioscience) and ML323 (Millipore).
Anchorage-Independent Growth Assay.
Anchorage-independent growth assays were performed in replicates of four in six-well plates. Indicated cells were seeded (1 × 104 cells per well) in NSC proliferation media with EGF and β-FGF containing 0.4% low-melting agarose on the top of bottom agar containing 1% low-melting agarose NSC proliferation media with EGF and FGF. After 14–21 d, colonies were stained with iodonitrotetrazoliumchloride (Sigma) and counted.
Intracranial Injection.
Female SCID mice aged 6–8 wk were bolted at MD Anderson’s Brain Tumor Center Animal Core, as previously described (44). GSCs were injected in 5 μL of Hank’s buffered salt solution. Animals were followed daily for the development of tumors and were killed once they showed neurological deficits or appeared moribund. The brains were removed for tumor cell isolation, paraffin embedding, or optimal cutting temperature (OCT) frozen tissue blocks. All mice manipulations were performed with the approval of the MD Anderson Cancer Center IACUC.
Cell Sorting and Cell Cycle Analysis.
The sorting and analysis of CD133+ and CD133− GSCs were performed with PE-conjugated CD133 (clone no. AC133; Miltenyi). To study the cell cycle of GSCs, BrdU was added to cells, and cells were collected after 2 h for staining using a BrdU Flow Kit (BD Pharmingen). Alternatively, cell cycle analysis was performed by standard PI staining. Briefly, cells were fixed, stained with PI solution (0.1 mg/mL PI, 2 mg/mL ribonuclease A, 0.6% Triton X-100 in PBS) for 30 min at 37 °C, spun down, and resuspended in PBS. Samples were acquired with a BCI Gallios Analyzer (Beckman Coulter). Gating strategies to exclude doublets and dead cells (DAPI) were always employed. After staining, samples were acquired using an LSR Fortessa flow cytometer or sorted using a BD Influx cell sorter. Data were analyzed by BD FACS Diva.
Western Blotting, Immunohistochemistry, and Immunofluorescence.
For Western blotting, protein lysates were resolved on 4–12% gradient polyacrylamide SDS gels and transferred onto nitrocellulose membranes according to standard procedures. Membranes were incubated with indicated primary antibodies, washed, and probed with HRP-conjugated secondary antibodies. The detection of bands was carried out upon chemiluminescence reaction followed by film exposure. To evaluate PCNA ubiquitination, Triton-soluble and -insoluble fractions (DNase-treated) of GSC lysates were extracted as previously described (45). For immunohistochemical staining, brain sections were incubated with indicated primary antibodies for 1 h at room temperature or overnight at 4 °C after deparaffinization, rehydration, antigen retrieval, quenching of endogenous peroxidase, and blocking. The sections were incubated with HRP-conjugated polymer (DAKO) for 40 min and then with diaminobenzidine using an Ultravision DAB Plus Substrate Detection System (Thermo Fisher Scientific) for 1–10 min at room temperature, followed by hematoxylin staining. Images were captured with a Nikon DS-Fi1 digital camera using a wide-field Nikon EclipseCi microscope. For immunofluorescence, secondary antibodies conjugated with Alexa 488 and 594 (Molecular Probes) were used. Images were captured with a Hamamatsu C11440 digital camera, using a wide-field Nikon EclipseNi microscope.
In Vitro Limiting Dilution and Tumorsphere Formation Assays.
GSCs were stained with PI, and PI-negative cells (n > 6) were flow-sorted with a decreasing number of cells per well (100, 10, and 1 cells) plated in 96-well plates. The percentage of wells with tumorspheres was quantified after 10 d. Extreme limiting dilution analysis was performed using software available at bioinf.wehi.edu.au/software/elda/. The tumorsphere formation assay involved seeding GSCs at a density of one cell per microliter, and the number of tumorspheres in each well was quantified after 7 d. In single-cell tumorsphere formation assay, GSCs were stained with Sytox Green and live cells were sorted using FACS into 96-well plates, followed by radiation treatment. Tumorspheres were scanned using a SCREEN Cell3iMager plate reader (InSphero) and quantified.
GFP Competition Experiment.
GFP-labeled GSCs (n = 50,000) were combined with 50,000 GFP nonlabeled GSCs, which were transduced with nontargeting (NT)/control or PAF shRNAs, and intracranially injected into mice under anesthesia. After 4–5 wk, mice were killed and tumors were harvested for isolation of GSCs. GSCs were transiently (2–3 d) cultured for tumorsphere formation before flow cytometry analysis. Cells were fixed and permeabilized for immunostaining with indicated primary antibodies using a BD Cytofix/Cytoperm Kit (BD Biosciences).
In Vivo Tumor Initiation Assay.
For in vivo limiting dilution studies, viable GSCs (3 d postinfection) at dilutions of 1 × 105, 1 × 104, and 1 × 103 cells (n = 5–15 mice per group) were counted and intracranially injected into mice. Tumor incidence was determined at indicated time points by luciferase imaging of mice using a Xenogen IVIS instrument (PerkinElmer) according to the manufacturer’s instructions. All mouse manipulations were performed with the approval of the MD Anderson Cancer Center IACUC.
Metabolomic Analysis.
Metabolomic profiling was performed at the Beth Israel Deaconess Medical Center Mass Spectrometry Core. Polar metabolites were extracted with 80% (vol/vol) methanol (cooled at −80 °C) according to established protocols and quantified using a 5500 QTRAP hybrid quadrupole mass spectrometer (46). Metabolic pathways were identified based on metabolites that were altered and close to each other in the reaction network.
Analysis of Chromosomal Aberrations.
Exponentially growing cells were treated with colcemid (0.04 μg/mL) for 2 h at 37 °C. The cells were then trypsinized, transferred to 15-mL conical tubes, and centrifuged at 350 × g for 10 min. Cells were resuspended in hypotonic solution (0.075 M KCl) for 15 min at room temperature, fixed in methanol and acetic acid (3:1 vol/vol), and washed three times in the fixative. Air-dried preparations were made, and the slides were stained with 4% Giemsa. The slides were analyzed for chromosomal aberrations, including chromosome and chromatid breaks, fragments, tetraploidy, and fusions. At least 35 metaphases were analyzed from each sample. Images were captured using a Nikon 80i microscope equipped with karyotyping software from Applied Spectral Imaging, Inc.
Immunoprecipitation and Mass Spectrometry.
FLAG-PAF was overexpressed in GSCs, and cells were lysed with lysis buffer [50 mM Tris (pH 7.5), 150 mM NaCl, 1% (vol/vol) Triton X-100] supplemented with complete protease inhibitor mixture (Roche). Mouse monoclonal anti-FLAG (Sigma) was added to the cell lysate and incubated overnight at 4 °C. After the addition of 30 μL of GammaBind G Sepharose bead slurry (∼50% vol/vol in lysis buffer; GE Healthcare), the resulting mixture was placed on a rocker for 2 h at 4 °C. Nonspecifically bound proteins were removed from the beads by washing three times with lysis buffer. Bound proteins were eluted by boiling the beads in SDS loading buffer in the presence of DTT, resolved in SDS/PAGE gel, and detected using silver staining (Pierce). Silver-stained gel pieces were washed, destained, and digested in-gel with 200 ng of trypsin or chymotrypsin (sequencing grade; Promega) for 18 h at 37 °C. Resulting peptides were extracted and analyzed by high-sensitivity liquid chromatography/tandem MS on an Orbitrap Elite or Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) using collision-induced dissociation, higher energy collision dissociation, or electron transfer dissociation. Proteins were identified by database searching of the fragment spectra against the SwissProt (European Bioinformatics Institute) protein database using Mascot (v 2.3; Matrix Science). Typical search settings were as follows: mass tolerances, 10-ppm precursor, 0.8-d fragments; variable modifications, methionine sulphoxide, pyroglutamate formation; up to two missed cleavages for trypsin and up to six missed cleavages for chymotrypsin.
RNA Isolation and qRT-PCR.
RNA was isolated with an RNeasy Mini or Micro Kit (Qiagen), and then used for first-strand cDNA synthesis using random primers and SuperScriptIII Reverse Transcriptase (Invitrogen). qRT-PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems). Primers are listed in Table S1. The relative expression of genes was normalized using ribosomal protein LP2 (RPLP2) as a housekeeping gene.
Table S1.
| Gene | HGNC gene symbol | GenBank accession code | Forward primer | Reverse primer |
| RiboP | RPLP2 | NM_001004 | 5′-CTC CAT CCG CAC CTA CAC C-3′ | 5′-ATC AGG GGT ATC TTG AGC TTG G-3′ |
| CD133 | PROM1 | NM_001145848 | 5′-GGA CCC ATT GGC ATT CTC-3′ | 5′-CAG GAC ACA GCA TAG AAT AAT C-3′ |
| Sox2 | SOX2-OT | NM_003106 | 5′-CAC ACT GCC CCT CTC AC-3′ | 5′-TCC ATG CTG TTT CTT ACT CTC C-3′ |
| Nestin | NES | NM_006617.1 | 5′-AGA GGG GAA TTC CTG GAG-3′ | 5′-GAC TCC TGG TCC TGA GAG AT-3′ |
| OLIG2 | OLIG2 | NM_005806.3 | 5′-GGT AAG TGC GCA ATG CTA AGC TGT-3′ | 5′-TAC AAA GCC CAG TTT GCA ACG CAG-3′ |
| PAF | KIAA0101 | NM_014736 | 5′-ATG GTG CGG ACT AAA GCA GAC-3′ | 5′-CCT CGA TGA AAC TGA TGT CGA AT-3′ |
| CAD | CAD | NM_004341 | 5′-AGT GGT GTT TCA AAC CGG CAT-3′ | CAGAGGATAGGTGAGCACTAAGA |
| UMPS | UMPS | NM_000373.3 | 5′-TTG GTG ACG GGT CTG TAC GA-3′ | 5′-GAA GAC GCG GTC GAG ACA C-3′ |
| PAICS | PAICS | NM_001079525.1 | 5′-TTG CAG AAG AAT AGC AAC TGG TT-3′ | 5′-CAC TGT GGG TCA TTA TTG GCA T-3′ |
| GART | GART | NM_000819.4 | 5′-CTG GAA AAG GGG TGA TTG TTG C-3′ | 5′-TCA GTG AAA CAC AGA CAC GAC-3′ |
HGNC, HUGO Gene Nomenclature Committee.
Intracranial Tumor Formation in Vivo.
SCID mice (Charles River Laboratories) aged 6–8 wk were bolted as previously described (44) at MD Anderson’s Brain Tumor Center Animal Core. GSCs were transduced with lentiviral vectors expressing PAF shRNA or NT shRNA control for the knockdown experiments. Viable cells (1 × 104 and 1 × 105) were counted 72 h postinfection and grafted intracranially into SCID mice. Animals were maintained until neurological signs were apparent, at which point they were killed. In parallel survival experiments, animals were monitored until they developed neurological signs. Mouse brains were removed by transcardial perfusion with 4% paraformaldehyde (PFA) and were fixed in formalin or postfixed in 4% PFA for processing paraffin-embedded or OCT frozen tissue blocks. All mouse manipulations were performed with the approval of the MD Anderson Cancer Center IACUC.
Statistical Analysis.
Data acquisition and analysis were not blind. Tumor-free survivals were analyzed based on the log-rank test using GraphPad Prism 6. All data were analyzed by an unpaired, two-tailed Student’s t test (P < 0.05 is considered to be statistically significant). For all experiments with error bars, SD was calculated to indicate the variation within each experiment and data, and values represent mean ± SD.
Supplementary Material
Acknowledgments
We thank L. K. Denzin (Rutgers University) for the PAF antibody (clone Jyld12); C. G. Liu, X. Liu, J. Zhang, and the MD Anderson Sequencing and Noncoding RNA Program for gene expression profiling; the MD Anderson Flow Cytometry and Cellular Imaging Core Facility supported by Grant NCI P30CA16672 for flow cytometers and FACS; and Drs. J. Chen, H. Piwnica-Worms, G. Peng, and R. Woods (MD Anderson Cancer Center) for critical reading and comments. We also appreciate all members of the R.A.D., L.C., and G. F. Draetta laboratories for fruitful suggestions and discussions. This research was supported by NIH Grant R01 CA084628 (to R.A.D.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708122114/-/DCSupplemental.
References
- 1.Huse JT, Holland EC. Targeting brain cancer: Advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10:319–331. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- 2.Lathia JD, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Z, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 5.Gaiano N, Fishell G. The role of notch in promoting glial and neural stem cell fates. Annu Rev Neurosci. 2002;25:471–490. doi: 10.1146/annurev.neuro.25.030702.130823. [DOI] [PubMed] [Google Scholar]
- 6.Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ligon KL, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemmati HD, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tunici P, et al. Genetic alterations and in vivo tumorigenicity of neurospheres derived from an adult glioblastoma. Mol Cancer. 2004;3:25. doi: 10.1186/1476-4598-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 11.Singh SK, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 12.Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CLL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kayaselçuk F, Zorludemir S, Gümürdühü D, Zeren H, Erman T. PCNA and Ki-67 in central nervous system tumors: Correlation with the histological type and grade. J Neurooncol. 2002;57:115–121. doi: 10.1023/a:1015739130208. [DOI] [PubMed] [Google Scholar]
- 16.Bartkova J, et al. Replication stress and oxidative damage contribute to aberrant constitutive activation of DNA damage signalling in human gliomas. Oncogene. 2010;29:5095–5102. doi: 10.1038/onc.2010.249. [DOI] [PubMed] [Google Scholar]
- 17.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 18.Carruthers R, et al. Abrogation of radioresistance in glioblastoma stem-like cells by inhibition of ATM kinase. Mol Oncol. 2015;9:192–203. doi: 10.1016/j.molonc.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmed SU, et al. Selective inhibition of parallel DNA damage response pathways optimizes radiosensitization of glioblastoma stem-like cells. Cancer Res. 2015;75:4416–4428. doi: 10.1158/0008-5472.CAN-14-3790. [DOI] [PubMed] [Google Scholar]
- 20.Venere M, et al. Therapeutic targeting of constitutive PARP activation compromises stem cell phenotype and survival of glioblastoma-initiating cells. Cell Death Differ. 2014;21:258–269. doi: 10.1038/cdd.2013.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosal G, Chen J. DNA damage tolerance: A double-edged sword guarding the genome. Transl Cancer Res. 2013;2:107–129. doi: 10.3978/j.issn.2218-676X.2013.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrón S, et al. Telomere shortening and chromosomal instability abrogates proliferation of adult but not embryonic neural stem cells. Development. 2004;131:4059–4070. doi: 10.1242/dev.01215. [DOI] [PubMed] [Google Scholar]
- 23.Jaskelioff M, et al. Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature. 2011;469:102–106. doi: 10.1038/nature09603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong KK, et al. Telomere dysfunction impairs DNA repair and enhances sensitivity to ionizing radiation. Nat Genet. 2000;26:85–88. doi: 10.1038/79232. [DOI] [PubMed] [Google Scholar]
- 25.De Biasio A, et al. Structure of p15(PAF)-PCNA complex and implications for clamp sliding during DNA replication and repair. Nat Commun. 2015;6:6439. doi: 10.1038/ncomms7439. [DOI] [PubMed] [Google Scholar]
- 26.Wu L. Role of the BLM helicase in replication fork management. DNA Repair (Amst) 2007;6:936–944. doi: 10.1016/j.dnarep.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol Cell. 2008;29:625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang TT, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 29.Hu B, et al. Epigenetic activation of WNT5A drives glioblastoma stem cell differentiation and invasive growth. Cell. 2016;167:1281–1295.e18. doi: 10.1016/j.cell.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang CN, Feng MJ, Chen YL, Yuan RH, Jeng YM. p15(PAF) is an Rb/E2F-regulated S-phase protein essential for DNA synthesis and cell cycle progression. PLoS One. 2013;8:e61196. doi: 10.1371/journal.pone.0061196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emanuele MJ, Ciccia A, Elia AEH, Elledge SJ. Proliferating cell nuclear antigen (PCNA)-associated KIAA0101/PAF15 protein is a cell cycle-regulated anaphase-promoting complex/cyclosome substrate. Proc Natl Acad Sci USA. 2011;108:9845–9850. doi: 10.1073/pnas.1106136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Povlsen LK, et al. Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nat Cell Biol. 2012;14:1089–1098. doi: 10.1038/ncb2579. [DOI] [PubMed] [Google Scholar]
- 33.Jung HY, et al. PAF and EZH2 induce Wnt/β-catenin signaling hyperactivation. Mol Cell. 2013;52:193–205. doi: 10.1016/j.molcel.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, et al. PAF-Wnt signaling-induced cell plasticity is required for maintenance of breast cancer cell stemness. Nat Commun. 2016;7:10633. doi: 10.1038/ncomms10633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jun S, et al. PAF-mediated MAPK signaling hyperactivation via LAMTOR3 induces pancreatic tumorigenesis. Cell Rep. 2013;5:314–322. doi: 10.1016/j.celrep.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, et al. Purine synthesis promotes maintenance of brain tumor initiating cells in glioma. Nat Neurosci. 2017;20:661–673. doi: 10.1038/nn.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diamant N, et al. DNA damage bypass operates in the S and G2 phases of the cell cycle and exhibits differential mutagenicity. Nucleic Acids Res. 2012;40:170–180. doi: 10.1093/nar/gkr596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cotto-Rios XM, Jones MJK, Busino L, Pagano M, Huang TT. APC/CCdh1-dependent proteolysis of USP1 regulates the response to UV-mediated DNA damage. J Cell Biol. 2011;194:177–186. doi: 10.1083/jcb.201101062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inoue A, et al. A small molecule inhibitor of monoubiquitinated proliferating cell nuclear antigen (PCNA) inhibits repair of interstrand DNA cross-link, enhances DNA double strand break, and sensitizes cancer cells to cisplatin. J Biol Chem. 2014;289:7109–7120. doi: 10.1074/jbc.M113.520429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang Q, et al. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat Chem Biol. 2014;10:298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Godek KM, et al. Chromosomal instability affects the tumorigenicity of glioblastoma tumor-initiating cells. Cancer Discov. 2016;6:532–545. doi: 10.1158/2159-8290.CD-15-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amrani YM, et al. The Paf oncogene is essential for hematopoietic stem cell function and development. J Exp Med. 2011;208:1757–1765. doi: 10.1084/jem.20102170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
- 44.Lal S, et al. An implantable guide-screw system for brain tumor studies in small animals. J Neurosurg. 2000;92:326–333. doi: 10.3171/jns.2000.92.2.0326. [DOI] [PubMed] [Google Scholar]
- 45.Ziv O, et al. Identification of novel DNA-damage tolerance genes reveals regulation of translesion DNA synthesis by nucleophosmin. Nat Commun. 2014;5:5437. doi: 10.1038/ncomms6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7:872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.